Abstract
Current methods for determining “LDL-C” in clinical practice measure the cholesterol content of both LDL and lipoprotein(a) [Lp(a)-C]. We developed a high-throughput, sensitive, and rapid method to quantitate Lp(a)-C and improve the accuracy of LDL-C by subtracting for Lp(a)-C (LDL-Ccorr). Lp(a)-C is determined following isolation of the Lp(a) on magnetic beads linked to monoclonal antibody LPA4 recognizing apolipoprotein(a). This Lp(a)-C assay does not detect cholesterol in plasma samples lacking Lp(a) and is linear up to 747 nM Lp(a). To validate this method clinically over a wide range of Lp(a) (9.0–822.8 nM), Lp(a)-C and LDL-Ccorr were determined in 21 participants receiving an Lp(a)-specific lowering antisense oligonucleotide and in eight participants receiving placebo at baseline, at 13 weeks during peak drug effect, and off drug. In the groups combined, Lp(a)-C ranged from 0.6 to 35.0 mg/dl and correlated with Lp(a) molar concentration (r = 0.76; P < 0.001). However, the percent Lp(a)-C relative to Lp(a) mass varied from 5.8% to 57.3%. Baseline LDL-Ccorr was lower than LDL-C [mean (SD), 102.2 (31.8) vs. 119.2 (32.4) mg/dl; P < 0.001] and did not correlate with Lp(a)-C. It was demonstrated that three commercially available “direct LDL-C” assays also include measures of Lp(a)-C. In conclusion, we have developed a novel and sensitive method to quantitate Lp(a)-C that provides insights into the Lp(a) mass/cholesterol relationship and may be used to more accurately report LDL-C and reassess its role in clinical medicine.
Supplementary key words: lipoprotein(a), low density lipoprotein, cholesterol, biomarker, cardiovascular disease risk, therapy
Abbreviations: [apo(a)], apolipoprotein(a); apoB, apolipoprotein B-100; ASO, antisense oligonucleotide; CV, coefficient of variation; EACA, epsilon aminocaproic acid; hApoB, human apolipoprotein B-100; [Lp(a)], lipoprotein(a); TC, total cholesterol; TGs, triglycerides; UCSD, University of California San Diego
“LDL-C” is routinely used to assess LDL-mediated CVD risk and response to therapy. All LDL-C assays used in clinical practice, including the reference method “beta-quantification,” measure the cholesterol content of LDL, IDL-C, and lipoprotein(a) [Lp(a)-C], of which the latter contains cholesterol in its LDL moiety. Lp(a)-C can constitute a significant portion of measured LDL-C, especially if Lp(a) is elevated. Without this methodological confounder, the correct LDL-C in such patients can be significantly lower than the laboratory measurement of LDL-C (1).
Lp(a) is composed of apolipoprotein(a) [apo(a)] covalently bound to the apolipoprotein B-100 (apoB) moiety of LDL (2). Like LDL, Lp(a) contains cholesterol esters, free cholesterol, phospholipids, triglycerides (TGs), and carbohydrates on its apolipoprotein components. While LDL-C, measured by beta-quantification, Friedewald or Martin-Hopkins calculations, or by direct LDL-C assays, has been generally accepted as an accurate biomarker for LDL-mediated CVD risk, LDL-C is actually a composite measurement of the cholesterol content on LDL, Lp(a), and IDL particles (1). Almost all individuals have circulating Lp(a), and approximately 20% of the population have highly elevated levels >50 mg/dl (3). Lp(a) and LDL have distinct biological activities, each mediating CVD risk independently (4). Moreover, LDL-C and Lp(a) respond differently to lipid-lowering therapies, with statins causing an increase or no change in Lp(a), compared with a decrease in LDL-C (5).
In order to more accurately understand an individual's LDL-C attributable risk and to more accurately monitor treatment effects on LDL and Lp(a) individually, correct LDL-C without its Lp(a)-C component needs to be quantified. We describe a sensitive, high-throughput, and rapid assay to measure Lp(a)-C, which can complement the traditional lipid profile for determination of LDL-C.
Methods
Study population
The specificity of the Lp(a)-C assay was determined in the following populations: 1) 28 individuals with low Lp(a), less than 6 mg/dl and 2) a subset of subjects with elevated baseline Lp(a) (>125 nM) from a phase II and placebo-controlled randomized trial of antisense oligonucleotide (ASO) mediated Lp(a) lowering (6). Plasma samples from eight subjects who received placebo and 21 subjects who received Lp(a) lowering (IONIS-APO(a)Rx) ASO at baseline, peak treatment effect (day 85/99), and at the end of the study (day 190) when Lp(a) levels have recovered following ASO washout were included in this analysis. This cohort was intentionally chosen to evaluate the specificity of the Lp(a)-C assay over a wide range of Lp(a), as Lp(a) molar concentrations decreased by an average of 63.8% ± 19.5% (mean ± SD), whereas total cholesterol (TC), LDL-C, and HDL-C were not significantly changed with IONIS-APO(a)Rx ASO. This study was approved by the University of California San Diego (UCSD) Human Research Protections Program.
Lp(a) purification
Purified Lp(a) used in spike-in experiments was isolated from the Liposorber postapheresis eluent from a single donor undergoing lipid apheresis for the treatment of homozygous familial hypercholesterolemia. This subject's preapheresis plasma Lp(a) level was 85 nM with a predominant (>95%) apo(a) isoform size of 24 kringle IV repeats. To prevent oxidation in storage, final concentrations of EDTA (2 mM) and beta-hydroxybutyrate (20 μM), respectively, were added to the eluent. To prevent nonspecific association between Lp(a) and other lipoproteins, proline and epsilon aminocaproic acid (EACA) were added at a final concentration of 200 mM each. The density of the eluent was adjusted by addition of NaBr for sequential ultracentrifugation in a Type-50 Ti rotor (Beckman) for 16–24 h at 10°C at 50,000 rpm, and the 1.063 g/ml < density < 1.090 g/ml fractions were harvested. This fraction was applied to an SW-400 gel filtration column (General Electric) and eluted into 0.5 ml fractions. Each fraction was assayed for the presence of apo(a), apoB, and apolipoprotein A1 by ELISA (see supplemental methods). The fractions containing apo(a) and apoB in proportion to apo(a), but not those containing apolipoprotein A1, were pooled (supplemental Fig. S1), concentrated, and buffer exchanged into PBS with 0.5 mM EDTA using Amicon centrifugal filter units (Millipore). Lp(a) purity was assessed by lipoprotein agarose gel electrophoresis and SDS-PAGE (supplemental Fig. S2).
Lp(a) ELISA, LPA isoforms, and oxidized phospholipids on apoB measurements
Plasma Lp(a) molar concentration (nanomolars) and mass (milligrams per deciliter) in the antisense trial were measured by the Northwest Lipid Research Laboratories and UCSD assays, respectively, as previously reported (6). Otherwise, Lp(a) mass (milligrams per deciliter) was measured with the Roche assay, which has a lower limit of detection of 6 mg/dl. Measurement of oxidized phospholipids on apoB was described (6). TC, LDL-C determined from fasting plasma using the Friedewald method, HDL-C, TGs, and apoB were measured using commercial assays (6). For the UCSD Lp(a) assay, a previously validated sandwich ELISA using an anti-apoB-100 capture antibody and the monoclonal anti-apo(a) detection antibody, LPA4, was performed as previously described (7). LPA isoforms were measured as previously described (6).
Generation of murine monoclonal antibody LPA4-coated magnetic beads
LPA4 was harvested from hybridomas expanded in mouse ascites and purified using a protein G affinity column (Abcore, Poway, CA). Conjugation of LPA4 to dynabeads MyOne Epoxy magnetic beads (Life Technologies) was performed according to the manufacturer's protocol. Briefly, the coupling reaction involved 30 μg of LPA4 per milligram of dynabeads. Once coupled, unbound antibody was washed off according to the manufacturer's protocol, and the LPA4-coupled dynabeads were stored in PBS.
Measurement of Lp(a)-C
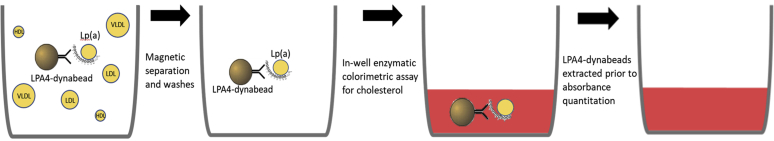
Fifteen microliters of each plasma sample were added to 15 μl of PBS containing 1% BSA, 200 mM proline, and 200 mM EACA. The presence of proline and EACA prevents the association between Lp(a) and TG-rich lipoproteins (8). Each diluted plasma sample was assayed in duplicate and added to 1 mg of LPA4-dynabeads in U-bottom 96-well plate. The beads were resuspended on a plate shaker at 900 rpm for 10 s, followed by a 45-min room temperature incubation with gentle shaking at 500 rpm to prevent the beads from precipitating. Lp(a) bound onto LPA4-dynabeads were then extracted from each well using a magnetic bead extraction replicator (V&P Scientific, Inc.; catalog no. 407AM-N1) and released into a parallel 96-well plate containing 200 μl of PBS, 1% BSA, 200 mM proline, and 200 mM EACA in each well to wash off any nonspecifically bound lipoproteins. This process was repeated for a total of three washes. Then LPA4-dynabeads containing Lp(a) were transferred to a parallel, clear, flat-bottom, 96-well plate containing 200 μl of enzymatic cholesterol reagent (Pointe Scientific), resuspended with shaking at 500 rpm, then incubated at 37°C for 5 min. Each flat-bottom plate also contains a dedicated row that had been prepopulated with 2-fold serial dilutions of cholesterol standard (Pointe Scientific), ranging from 0.0375 to 1.5 μg cholesterol for generation of a standard curve. The plates were analyzed for absorbance at 500 (primary) and 700 nm (background). The amount (micrograms) of Lp(a)-C in each sample was determined based on the absorbance at 500 to 700 nm value calibrated against the standard curve. Then, the concentration of Lp(a)-C was determined based on the input volume of plasma. In the case of a 15 μl input, the milligrams per deciliter of Lp(a)-C = μg Lp(a) × 100/15. A simplified schematic of the Lp(a)-C assay is depicted in Fig. 1.
Fig. 1.
Schematic of the lipoprotein(a)-cholesterol [Lp(a)-C] assay. Lp(a) in plasma is affinity captured by LPA4-dynabeads and separated from other cholesterol carrying lipoproteins (depicted as yellow circles in the left panel) in each well by magnetic extraction and washes. Then, an enzymatic colorimetric cholesterol reagent is added to each well, generating a red color with intensity proportional to the amount of cholesterol present. Following a 5-min incubation period to ensure all cholesterol on Lp(a) has been processed, LPA4-dynabeads are extracted by magnet, and the absorbance at 500 (primary) and 700 nm (background) quantified.
Determination of LDL-Ccorr
LDL-Ccorr was determined by subtracting the directly measured Lp(a)-C from the reported LDL-C.
Mouse model
Transgenic mice expressing human apolipoprotein B-100 (hApoB) only and human-like LDL, because of a mutation in codon 2153 preventing apoB-48 synthesis, which were previously generated (9).
TC and direct LDL-C assays
TC measurements were performed using an enzymatic and colorimetric assay (Pointe Scientific) according to the manufacturer's protocol. Direct LDL-C assays are assumed to only measure cholesterol on LDL particles; however, these assays are referenced to beta-quantification, a method that cannot distinguish Lp(a)-C from LDL-C because of overlapping densities of LDL and Lp(a) particles. To assess whether these direct LDL-C assays also inadvertently quantitate the Lp(a)-C content of the samples, colorimetric direct LDL-C reagents were purchased from Roche (LDLC3; catalog no. 07005717), Sekisui (direct LDL-C; catalog no. 7120), and Wako (L-Type LDL-C; catalog no. 993-00404). Cholesterol assays were performed in clear flat-bottom 96-well plates. Two microliters of human plasma, purified Lp(a) as noted above, and assay calibrator from Wako (catalog no. 990-28011) containing 150 mg/dl LDL-C were added to 2 μl of PBS for a total of 4 μl of input for each assay. For spike-in experiments, 2 μl of purified Lp(a) was added to 2 μl of plasma as input. Incubation times, temperature, and absorbance wavelengths were performed according to the manufacturer's protocol. Absorbance was quantified on a BioTek Synergy HTX plate reader.
Statistics
Descriptive statistical analysis, correlation analysis using Spearman's rho test, analysis between parametric data sets using ANOVA and nonparametric data sets using Kruskal-Wallis testing were performed with SPSS, version 26 (IBM).
Results
Immunoprecipitation of Lp(a) and direct quantification of its cholesterol content
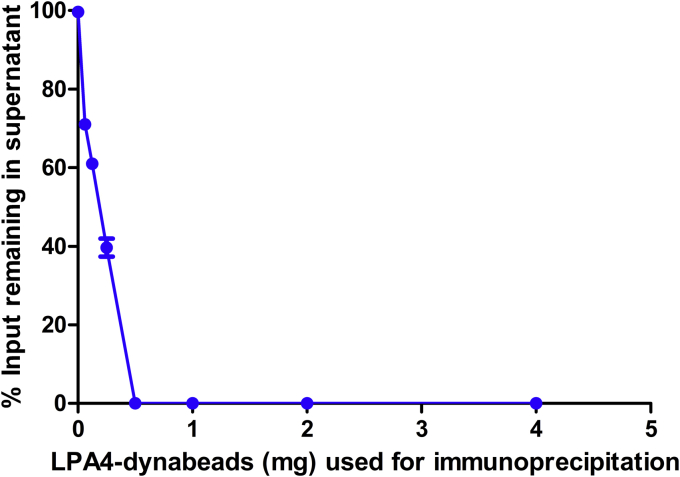
Lp(a) was immunoprecipitated from plasma from a patient with an elevated Lp(a) particle concentration of 573 nM (value at the 99th percentile in the population) using a monoclonal antibody against apo(a), LPA4, directly coupled to nonporous magnetic beads (LPA4-dynabeads). One-half a milligram (0.5 mg) of LPA4-dynabeads was able to deplete all the Lp(a) from 15 μl of this plasma (Fig. 2). To accommodate for even higher Lp(a) levels in the population, and to accommodate a margin of error, all subsequent reactions described in this article utilized 1 mg of LPA4-dynabeads.
Fig. 2.
Efficacy of lipoprotein(a) [Lp(a)] affinity capture using LPA4-dynabeads. The amount of Lp(a) remaining in each 15 μl aliquot of plasma following incubation with increasing amounts of LPA4-dynabeads. Lp(a) was quantified by ELISA and expressed as a percentage of Lp(a) in plasma not exposed to LPA4-dynabeads. Each data point represents the mean ± SD of three independent experiments.
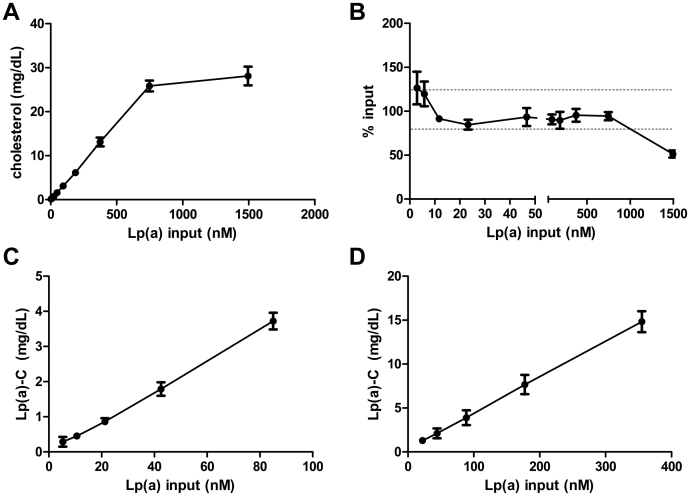
To test the specificity and linearity of cholesterol measured on LPA4-dynabead immunoprecipitated Lp(a) [Lp(a)-C], plasma from mice that do not express apo(a) or Lp(a) but express hApoB (and therefore human-like LDL) with or without spiked-in purified Lp(a) was assayed for Lp(a)-C. Mice expressing hApoB have a TC of 150 mg/dl, non-HDL-C of 120 mg/dl, and no circulating Lp(a) had an Lp(a)-C of 0 mg/dl. Lp(a)-C was measured in hApoB mouse plasma with serial 2-fold increments of purified Lp(a) spiked in, ranging from 2.9 to 1494.0 nM (Fig. 3A, B). There was a linear relationship between Lp(a)-C and the amount of spiked-in Lp(a) particle number up to 747.0 nM Lp(a), beyond which the assay is saturated (Fig. 3A, B). Based on the TC of the spiked-in purified Lp(a) measured in parallel, the percent recovery (SD) was 126.3 (18.5), 119.7 (14.0), 91.3 (0.5), 84.7 (5.5), 93.3 (10.2), 90.7 (5.5), 89.7 (9.5), 95.3 (7.2), and 94.3 (4.5)% with 2.9, 5.8, 11.7, 23.3, 46.7, 93.4, 186.8, 373.5, and 747.0 nM spiked-in purified Lp(a), respectively (Fig. 3B). The average percent recovery of purified Lp(a)-C was 91.3% after exclusion of the values >100% associated with an Lp(a) input of 5.8 nM or less.
Fig. 3.
Lipoprotein(a)-cholesterol [Lp(a)-C] assay sensitivity and linearity across a range of Lp(a) molar concentrations. Lp(a)-C measured in human apolipoprotein B-100 mouse plasma spiked in with purified Lp(a) (A) and expressed as a percentage of total cholesterol directly measured on purified Lp(a) (B). The dotted lines in panel B delineate 120% and 80% recovery rates. Lp(a)-C measured in serial dilutions of plasma with Lp(a) particle number of 85.0 nM (C) and 355.0 nM (D). Each data point represents the mean ± SD of three independent experiments.
To further evaluate linearity of the assay, Lp(a)-C was determined in serial 2-fold dilutions of plasma from a patient with Lp(a) level of 85.0 nM (normal <75 nM) (Fig. 3C) and another patient with elevated Lp(a) of 355.0 nM (Fig. 3D). The R2 correlation coefficient between Lp(a)-C and Lp(a) particle concentration was 0.998 and 0.999, respectively.
Intra-assay coefficient of variation (CV) determined by five replicate Lp(a)-C measurements from plasma with Lp(a) particle concentrations of 2.7, 12.2, 26.2, 51.9, 54.4, 165.8, 181.9, and 522.2 nM was 2.2, 7.7, 9.5, 4.6, 5.2, 1.0, 5.6, and 4.7%, respectively. Interassay CVs determined by Lp(a)-C measurements performed on 4 consecutive days from plasma with Lp(a) particle concentrations of 5.2, 15.5, 54.0, 68.1, 85.0, 100.9, and 390.0 were 0.8, 4.1, 10.0, 5.0, 12.0, 6.0, and 10.0%, respectively.
Lp(a)-C measurements in individuals with low Lp(a) mass
The Lp(a)-C assay specificity was further evaluated in 28 individuals with Lp(a) mass reported to be less than 6 mg/dl, the Roche assay's lower limit of detection (Table 1). In this group, the median (range) of TC, LDL-C, HDL-C, and TG was 131.0 (36.0–421.0), 71.5 (7.0–342.0), 38.0 (24.0–64.0), and 103.0 (23.0–417.0) mg/dl, respectively. The median (range) of Lp(a)-C for this group was 0.8 (0.0–3.0) mg/dl. Lp(a)-C measured 0.0 mg/dl in five samples, which presumably have negligible Lp(a) mass. Lp(a)-C did not significantly correlate with TC (Spearman's r = 0.1; P = 0.5), LDL-C (r = 0.3; P = 0.1), HDL-C (r = −0.3; P = 0.1), or TG (r = 0.2; P = 0.3) in this group of individuals with very low Lp(a) mass, suggesting negligible interference from non-Lp(a) particles.
Table 1.
Plasma lipid parameters in 28 individuals with Lp(a) mass <6 mg/dL
| Lp(a) mass (mg/dl) | TC (mg/dl) | LDL-C (mg/dl) | HDL-C (mg/dl) | TG (mg/dl) | Lp(a)-C (mg/dl) |
|---|---|---|---|---|---|
| <6 | 421 | 342 | 42 | 147 | 0 |
| <6 | 421 | 72 | 42 | 51 | 0 |
| <6 | 161 | 71 | 41 | 95 | 0 |
| <6 | 123 | 54 | 47 | 81 | 0 |
| <6 | 84 | 25 | 54 | 30 | 0 |
| <6 | 36 | 7 | 24 | NA | 0.1 |
| <6 | 129 | 99 | 27 | 101 | 0.1 |
| <6 | 55 | 12 | 29 | NA | 0.2 |
| <6 | 203 | 128 | 36 | 193 | 0.2 |
| <6 | 56 | 17 | 44 | NA | 0.2 |
| <6 | 91 | 36 | 38 | 99 | 0.3 |
| <6 | 331 | 168 | 45 | 388 | 0.4 |
| <6 | 82 | 35 | 34 | 136 | 0.5 |
| <6 | 133 | 72 | 41 | NA | 0.8 |
| <6 | 59 | 30 | 28 | 41 | 0.8 |
| <6 | 38 | 7 | 26 | NA | 0.8 |
| <6 | 77 | 30 | 38 | 23 | 1 |
| <6 | 185 | 128 | 31 | NA | 1.3 |
| <6 | 358 | 262 | 64 | 242 | 1.3 |
| <6 | 255 | 146 | 43 | 338 | 1.9 |
| <6 | 103 | 54 | 36 | 101 | 2 |
| <6 | 221 | 152 | 57 | 103 | 2.1 |
| <6 | 67 | 31 | 31 | 78 | 2.4 |
| <6 | 191 | 78 | 38 | 417 | 2.7 |
| <6 | 223 | 153 | 38 | 113 | 2.8 |
| <6 | 354 | 291 | 28 | 252 | 2.8 |
| <6 | 147 | 85 | 28 | 227 | 2.9 |
| <6 | 114 | 77 | 24 | 54 | 3 |
NA, data not available.
Lp(a)-C in a cohort of individuals with elevated plasma Lp(a) molar concentrations and following Lp(a) reduction with an ASO
In this cohort, 21 participants received Lp(a)-lowering therapy [IONIS-APO(a)Rx], with median (range) of Lp(a) molar concentrations, as measured by the Northwest Lipid Research Laboratories assay (6), of 344.6 (149.3–822.8), 113.7 (9.0–505.8), and 244.5 (67.2–697.0) nM at baseline, trough, and recovery time points, respectively (Kruskal-Wallis test; P < 0.001) (Table 2 and supplemental Table S1). The mean (SD) of Lp(a) molar concentrations was 36.1 (19.5)% and 82.5 (29.5)% of baseline levels at trough and recovery time points, respectively (ANOVA; P < 0.001). In addition, for the current study, Lp(a) mass levels were also measured at each time point using the UCSD assay reporting data as total Lp(a) mass in milligrams per deciliter (6). At baseline, trough, and recovery, Lp(a) mass levels were 111.4 (55.3–157.8), 54.6 (2.8–103.2), and 87.0 (27.3–132.9) mg/dl, respectively (Kruskal-Wallis test; P < 0.001). Plasma lipid parameters at each time point, including LDL-C determined by Friedewald calculation, are described in Table 2 and supplemental Table S1. The median (range) of Lp(a)-C levels measured in baseline, trough, and recovery time points was 14.2 (5.6–35.0), 7.4 (0.6–19.7), and 12.9 (5.5–25.7) mg/dl, respectively (Kruskal-Wallis test; P < 0.001). For completeness, eight individuals receiving placebo ASO were also evaluated. In the treatment group, none of the plasma lipid or Lp(a) parameters differed significantly between the three time points. The median (range) of Lp(a) molar concentration, Lp(a) mass, and Lp(a)-C was 209.0 (131.9–542.4) nM, 73.9 (52.1–166.8) mg/dl, and 15.5 (8.4–30.8) mg/dl, respectively (Table 2).
Table 2.
Plasma lipoprotein parameters in 29 individuals with elevated baseline Lp(a) enrolled in a Lp(a)-lowering ASO clinical trial
| Placebo ASO (n = 8) |
IONIS-APO(a)Rx ASO (n = 21) |
|||
|---|---|---|---|---|
| All time points | Baseline | Trough | Recovery | |
| TC; mean (SD) (mg/dl) | 208 (47.7) | 197.0 (39.7) | 181.3 (45.7) | 199.2 (33.4) |
| LDL-C; mean (SD) (mg/dl) | 122.1 (28.0) | 116.3 (36.8) | 102.2 (41.7) | 118.1 (34.6) |
| HDL-C; mean (SD) (mg/dl) | 58.0 (28.6) | 54.4 (12.8) | 52.0 (13.9) | 54.2 (15.4) |
| TG; median (range) (mg/dl) | 95.0 (46.0–436.0) | 114.0 (64.0–319.0) | 131.0 (65.0–305.0) | 131.5 (61.0–271.0) |
| apoB-100; mean (SD) (mg/dl) | 98.0 (23.6) | 95.2 (25.5) | 85.5 (29.0) | 93.5 (22.1) |
| Lp(a) molar concentration; median (range) (nM) | 209.0 (131.9–542.4) | 344.6 (149.3–822.8)a | 113.7 (9.0–505.8)a | 244.5 (67.2–697.0)a |
| Lp(a) mass; median (range) (mg/dl) | 73.9 (52.1–166.8) | 111.4 (55.3–157.8)a | 54.6 (2.8–103.2)a | 87.0 (27.3–132.9)a |
| apo(a) major isoform; median (range) | 16.0 (14.0–20.0) | 17.0 (13.0–19.0) | 17.0 (13.0–19.0) | 17.0 (13.0–19.0) |
| apo(a) minor isoform; median (range) | 20.0 (null–30.0) | 20.0 (null–29.0) | 20.0 (null–29.0) | 20.0 (null–29.0) |
| OxPL-apoB; median (range) (nM) | 21.3 (13.6–37.2) | 25.2 (16.8–41.2)a | 16.6 (7.5–25.2)a | 22.0 (12.3–37.3)a |
| Lp(a)-C (mg/dl); median (range) | 15.5 (8.4–30.8) | 14.2 (5.6–35.0)a | 7.4 (0.6–19.7)a | 12.9 (5.5–25.7)a |
| % Lp(a)-C; median (range) | 17.9 (9.6–57.3) | 15.6 (6.9–41.5) | 18.7 (5.8–45.4) | 16.0 (8.4–31.4) |
| LDL-Ccorr; median (range) (mg/dl) | 98.5 (65.1–165.4) | 95.2 (38.0–162.6) | 90.3 (33.9–214.9) | 110.5 (47.5–169.0) |
| % Lp(a)-C/LDL-C; median (range) | 11.3 (7.2–25.0) | 15.0 (5.4–43.4)b | 7.3 (0.7–25.1)b | 11.8 (5.2–28.1)b |
% Lp(a)-C, percent of Lp(a)-C/Lp(a) mass; LDL-Ccorr, LDL-C – Lp(a)-C; % Lp(a)-C/LDL-C, percent of Lp(a)-C/LDL-C; OxPL-apoB, oxidized phospholipid on apolipoprotein B-100.
Values are reported as median (range). Baseline, trough, and recovery refers to time points associated with respective Lp(a) molar concentrations in IONIS-APO(a)Rx ASO treated subjects.
Kruskal-Wallis P values comparing changes in subjects receiving apo(a)rx ASO across time points.
Denotes P < 0.001.
Denotes P = 0.01.
Relationship of Lp(a)-C to Lp(a) mass and concentration
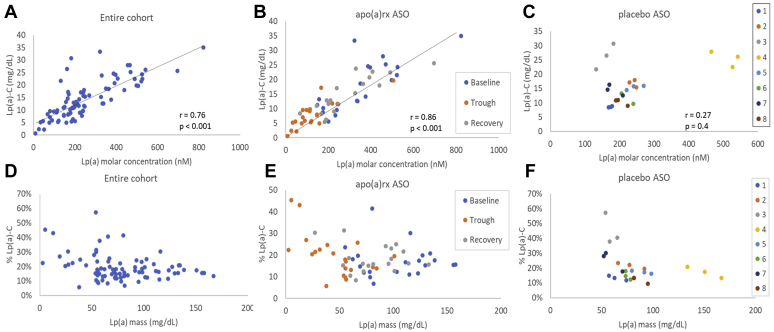
In the entire cohort of 29 individuals over three time points, Lp(a)-C levels correlated well with Lp(a) molar concentration and mass measured in nanomolar and milligrams per deciliter (Fig. 4A), with Spearman's rho (r) of 0.76 (P < 0.001) and 0.69 (P < 0.001), respectively (Table 3). Other statistically significant correlations with Lp(a)-C include oxidized phospholipids on apoB (r = 0.65; P < 0.001), TC (r = 0.39; P < 0.001), and LDL-C (r = 0.29; P = 0.008). However, Lp(a)-C did not significantly correlate with LDL-Ccorr (r = 0.10; P = 0.35). Correlations between Lp(a)-C and lipid parameters were also analyzed within the placebo and treatment groups separately (supplemental Table S2). When only the baseline ASO and placebo samples were analyzed together, the correlation between Lp(a)-C and Lp(a) molar concentration was 0.65 (P < 0.001) (supplemental Table S3). In the treatment group over three time points, Lp(a)-C correlated highly with Lp(a) molar concentrations (r = 0.86; P < 0.001) but not with TC, HDL-C, or TG (Fig. 4B and supplemental Table S2). The finding that changes in Lp(a)-C tracks with ASO-mediated specific changes in Lp(a) further demonstrates the specificity of this Lp(a)-C assay.
Fig. 4.
Relationship between lipoprotein(a)-cholesterol [Lp(a)-C] and Lp(A) molar concentrations in all individuals (A), those treated with IONIS-APO(a)Rx ASO (B), and those treated with placebo ASO (C). Relationship between % Lp(a)-C and Lp(A) mass in all individuals (D), those treated with IONIS-APO(a)Rx ASO (E), and those treated with placebo ASO (F). Data from baseline, trough, and recovery time points are represented in each panel. In panels C and F, each color in the legend references one individual. apo(a), apolipoprotein(a); ASO, antisense oligonucleotide.
Table 3.
Spearman correlation coefficients (P values) between Lp(a)-C and various Lp(a) and plasma lipoprotein parameters in 29 individuals with elevated baseline Lp(a) enrolled in a Lp(a)-lowering ASO clinical trial
| Lp(a) molar (nM) | Lp(a)-C (mg/dl) | % Lp(a)-C | |
|---|---|---|---|
| TC (mg/dl) | 0.24 (P = 0.03) | 0.39 (P < 0.001) | 0.36 (P = 0.001) |
| LDL-C (mg/dl) | 0.25 (P = 0.02) | 0.29 (P = 0.008) | 0.25 (P = 0.024) |
| LDL-Ccorr (mg/dl) | 0.1 (P = 0.4) | 0.1 (P = 0.35) | 0.19 (P = 0.09) |
| HDL-C (mg/dl) | 0.02 (P = 0.9) | 0.05 (P = 0.687) | 0.11 (P = 0.320) |
| TG (mg/dl) | −0.06 (P = 0.6) | 0.16 (P = 0.162) | 0.28 (P = 0.010) |
| Lp(a) mass (mg/dl) | 0.91 (P <0.001) | 0.69 (P < 0.001) | −0.25 (P = 0.023) |
| Lp(a) molar (nM) | 1 | 0.76 (P < 0.001) | −0.07 (P = 0.541) |
| OxPL-apoB (nM) | 0.85 (P < 0.001) | 0.65 (P < 0.001) | 0.1 (P = 0.104) |
| Lp(a)-C (mg/dl) | 0.76 (P < 0.001) | 1 | 0.42 (P < 0.001) |
| % Lp(a)-C | −0.07 (P = 0.5) | 0.42 (P < 0.001) | 1 |
%Lp(a)-C, percent Lp(a)-C/Lp(a) mass; OxPL-apoB, oxidized phospholipid on apolipoprotein B-100.
Three time points (baseline, trough, and recovery) from each of the 29 individuals were included in this correlation analysis.
Statistically significant correlations are given in bold.
In baseline samples across the entire cohort, the proportion of LDL-C that was Lp(a)-C based on direct measurement was median (range) of 13.2 (5.4–42.4)%. Baseline LDL-Ccorr was significantly lower than laboratory-measured LDL-C [mean (SD), 102.2 (31.8) vs. 119.2 (32.4) mg/dl, respectively, with P < 0.001].
To understand the interindividual heterogeneity of the relationship between Lp(a)-C and Lp(a) mass, Lp(a)-C was expressed as a percentage of Lp(a) mass measured in milligrams per deciliter using the UCSD assay (% Lp(a)-C). The median (range)% of Lp(a)-C across the entire cohort was 17.3 (5.8–57.3)% (Fig. 4D). In individuals receiving IONIS-APO(a)Rx ASO, % Lp(a)-C was similar at baseline, trough, and recovery; 15.6 (6.9–41.5), 18.7 (5.8–45.4), and 16.0 (8.4–31.4)%, respectively (Kruskal-Wallis test; P = 0.5) (Fig. 4E and Table 2). In the placebo group, % Lp(a)-C was higher in those with lower Lp(a) mass, with Spearman correlation r = −0.54, P = 0.009 (Fig. 4F). To further evaluate whether the high variation in % Lp(a)-C was due to intraindividual or interindividual differences, the mean (SD) intraindividual CV of % Lp(a)-C across the three predetermined study time points was determined to be 20.4 (10.2)% in the entire cohort. The CV of % Lp(a) was 21.6 (10.9)% in the IONIS-APO(a)Rx ASO group, 16.8 (7.5)% in the placebo group, without a statistically significant difference between the two treatment groups (P = 0.23). Moreover, there was no statistically significant correlation between neither % Lp(a)-C and Lp(a) molar concentration (r = −0.04; P = 0.7) nor Lp(a) mass (r = −0.2; P = 0.1).
Direct LDL-C assays detect Lp(a)-C
To ascertain whether commercially available direct LDL assays also detect Lp(a)-C, cholesterol was quantified on purified Lp(a) that was free of any LDL using a TC assay as reference, and by three independent direct LDL-C assays. The mean (SD) of cholesterol content of Lp(a) was 54.9 (1.3) mg/dl by the TC assay, 44.4 (0.6) mg/dl by the Roche LDLC3 assay, 57.3 (1.9) mg/dl by the Sekisui direct LDL-C assay, and 49.7 (2.3) mg/dl by the Wako L-type LDL-C assay (Table 4). Using the TC on purified Lp(a) as a reference, 87 (3.1)%, 104 (5.4)%, and 90 (6.2)% of Lp(a)-C was measured as LDL-C by the Roche, Sekisui, and Wako assays, respectively. When purified Lp(a), with a TC content of 54.9 mg/dl, was spiked-into plasma from a patient with a TC of 155.6 mg/dl and Lp(a) mass of 5 mg/dl, 84 (2.3)%, 98 (0.2)%, and 98 (0.2)% of the additional cholesterol from the exogenously added Lp(a) was measured as LDL-C by the Roche, Sekisui, and Wako assays, respectively (Table 4).
Table 4.
Performance of three direct LDL-C assays with detection of cholesterol on purified Lp(a) and Lp(a) spiked-in plasma
| Input |
% Pure Lp(a)-C detected | % Spiked-in Lp(a)-C detected | |||
|---|---|---|---|---|---|
| Plasma sample | Purified Lp(a) | Plasma + purified Lp(a) | |||
| TC | 155.6 (3.2) | 54.9 (1.3) | 200.5 (0.6) | 100 | 100 |
| Roche direct LDL-C | 97.7 (2.2) | 44.4 (0.6) | 144.0 (1.1) | 87 (3.1) | 84 (2.3) |
| Sekisui direct LDL-C | 113.1 (6.4) | 57.3 (1.9) | 166.8 (3.3) | 104 (5.4) | 98 (0.2) |
| Wako direct LDL-C | 105.1 (5.2) | 49.7 (2.3) | 158.9 (2.9) | 90 (6.2) | 98 (0.1) |
Purified Lp(a) with a cholesterol content of 54.9 mg/dl was used for these experiments. % pure Lp(a)-C detected = Lp(a)-C measured by the respective direct LDL-C assay divided by Lp(a)-C measured by the TC assay. % spiked-in Lp(a)-C = cholesterol measured in plasma spiked with pure Lp(a) − cholesterol measured in plasma without added Lp(a) divided by cholesterol measured in purified Lp(a), using each respective assay. Data described are from three separate experiments and expressed as mean (SD). Units are in milligrams per deciliter unless otherwise specified.
Discussion
In this study, we demonstrate the development and validation of a rapid, high-throughput, specific, and sensitive assay to quantify Lp(a)-C. We demonstrate several important observations from this work: 1) Lp(a)-C can be measured and are in the linear range in subjects up to 99th percentile of population levels; 2) the percent of Lp(a) cholesterol relative to its mass, which was previously reported as 30% by Dahlen (10) and other studies (11, 12, 13) and now used widely to correct the LDL-C for Lp(a) mass, is more variable than previously reported, with a range of 6–57% among individuals; 3) the contribution of Lp(a)-C to LDL-C can be substantial and clinically relevant, with an average of 17 mg/dl in subjects with elevated Lp(a), which can translate to 10% difference in relative risk based on therapeutic studies; 4) subjects with substantially elevated Lp(a) have significantly lower correct LDL-C than appreciated; and 5) direct LDL assays also measure Lp(a)-C in proportion to the amount present in the sample.
This Lp(a)-C method has several important implications for clinical care, including assessing the role of Lp(a)-C and corrected LDL-C in risk prediction, reclassifying LDL-C thresholds in clinical diagnosis, and assessing treatment effects. Importantly, conventional lipid-lowering therapies such as statins do not lower Lp(a) and may increase it (14). With the advent of highly effective Lp(a)-lowering therapies (15), understanding an individual's correct LDL-C can guide the choice of the appropriate intensity and combination of therapies required to achieve guideline-directed LDL-C goals. The importance on clinical risk prediction of a corrected LDL-C compared with the laboratory LDL-C was recently demonstrated in a large meta-analysis from the Lipoprotein(a) Studies Collaboration (18,043 patients; 5,390 events; 4.7 years of median follow-up) (16). When comparing top versus bottom quartiles, the multivariable-adjusted hazard ratio for CVD was significant for LDL-C but not for corrected LDL-C, which in this study was estimated using Lp(a)-C calculated by range of 20–45% of Lp(a) mass. Furthermore, in a routine laboratory database involving 531,144 patients, reclassification of patients across guideline-recommended LDL-C categories when using LDL-Ccorr30 reassigned ∼30–40% of subjects to lower LDL-C categories. Validation of these findings with a quantitative method, such as the one described here, would have far-reaching implications in clinical care and guideline-recommended targets for LDL-C.
As the absence of plasma Lp(a) is rare, all LDL-C assays, including Friedewald, Martin-Hopkins, beta-quantification, and direct LDL-C assays, do not accurately reflect the correct LDL-C. To date, the inclusion of Lp(a)-C in LDL-C measurements stems from the inability to separate Lp(a) from LDL because of shared composition and overlapping densities. Clinical assays are referenced to beta-quantification, which shares this limitation. Therefore, LDL-C calculated by Friedewald will also inaccurately reflect correct LDL-C. The Martin-Hopkins formula is an advance over Friedewald but suffers from the same limitations in being referenced to beta-quantitation, which also includes the Lp(a)-C content. As shown here, direct LDL-C assays have the same limitation in measuring 84–98% of the cholesterol content on Lp(a). While the inaccuracy of LDL-C may be negligible in individuals with low Lp(a) levels, it can be significant in individuals with elevated Lp(a), who are common in the population. With the technique reported here, traditional reporting of LDL-C can be complemented by directly measured Lp(a)-C, and LDL-Ccorr (LDL-C − Lp(a)-C) determined as a more accurate reflection of correct LDL-C. Alternatively, Lp(a) immunodepleted plasma, which can be accomplished using LPA4-conjugated magnetic beads without any change in plasma volume (therefore preserving its non-Lp(a) component concentrations), can be assayed for correct LDL-C using conventional assays such as a direct LDL-C assay or by Friedewald or Martin-Hopkins calculation.
The Lp(a)-C assay described here is the only one that has been validated with spike-in experiments with purified Lp(a), to the best of our knowledge. The Lp(a)-C assay is linear across a range of 2.9–747.0 nM Lp(a) input, although there was a 20–26% overestimation bias with Lp(a) levels of 5.8 nM or less, which are not clinically important. Despite the borderline performance with very low Lp(a) input, the assay is suitable for the majority of the population, even those with Lp(a) mass levels at the 99th percentile. Importantly, those with elevated Lp(a) mass would more likely have a significant component of LDL-C as Lp(a)-C. The Lp(a)-C assay is specific and does not detect cholesterol in plasma from transgenic mice expressing hApoB that have human LDL, along with endogenous lipoproteins, but not Lp(a). Lp(a)-C correlated well with Lp(a) molar concentrations, further supporting the high specificity of this assay.
This Lp(a)-C assay is a high-throughput one and can be performed on 96-well plates or adapted for use with existing clinical analyzers using magnetic beads. The entire assay is completed within 1 h. Other Lp(a)-C assays have been described, including those using electrophoretic (17, 18), single-density gradient ultracentrifugation (19, 20), and affinity based on porous matrices using wheat germ agglutinin (which can bind glycoproteins such as apo(a)) (21) or polyclonal anti-Lp(a) serum (22). However, neither a gold-standard assay nor reference materials to standardize or harmonize Lp(a)-C assays currently exist. Therefore, additional studies will be required to understand which assay(s) will be accurate and clinically useful.
It had been generally accepted that the cholesterol content of Lp(a) is ∼30%. However, it is important to note that this estimation was based on a small number of studies that had biochemically characterized Lp(a) purified from only three to four individuals in each study (11, 12, 13). In these studies, both unesterified and esterified cholesterol were quantified. Clinical cholesterol assays used for TC, LDL-C, HDL-C, and Lp(a)-C measurements are calibrated against unesterified cholesterol mass only, and thereby underestimate TC mass. This may be one reason that % Lp(a)-C determined by the method reported in this study is lower than what was originally described. However, Lp(a)-C determined by this method is more clinically relevant, as it can be directly compared with LDL-C and used to calculate correct LDL-C, since it is the unesterified cholesterol that is quantified in both assays.
It is also very important to note that the Lp(a) mass assays in milligrams per deciliter, used as a denominator for % Lp(a)-C, are deeply flawed. Although it is implied that the protein, lipid, and carbohydrate components of Lp(a) are measured, in reality only the apo(a) component is detected immunologically. Relative units corresponding to the amount of apo(a) detected are converted to milligrams per deciliter values based on calibrators with milligrams per deciliter values assigned to them in a nonstandardized manner. Because Lp(a) mass is highly heterogenous between individuals, not only because of differences in apo(a) isoform size, but multiple variables such as glycosylation on apo(a) and lipid content, there is no primary reference material for standardization of Lp(a) measurement in milligrams per deciliter. A recent National Heart, Lung and Blood Institute working group for Lp(a) has recommended against using milligrams per deciliter assays for Lp(a) measurement (23). Therefore, estimation of Lp(a)-C based on a fixed assumed percent cholesterol content of Lp(a) mass in milligrams per deciliter may be a currently “expedient” first-step estimate but will not be accurate for most individuals and not optimally informative of the importance of Lp(a)-C as a risk factor or its response to therapy. The wide variability in the % Lp(a)-C per Lp(a) mass suggests that the historical 30% value should be discontinued in clinical studies for estimating corrected LDL-C because of the high likelihood of error at the individual level. For more precise CVD risk assessment and management, directly measured Lp(a)-C or measurement of LDL-C in Lp(a) immunodepleted plasma will be necessary.
Limitations
While no gold-standard Lp(a)-C assay exists for comparison, we believe that this study provides sufficient internal validation of the magnetic monoclonal antibody affinity Lp(a)-C assay methodology to allow its use in further clinical studies. Further evaluation of this Lp(a)-C assay in larger and more diverse populations, such as those with dyslipidemia, and those on various lipid-lowering therapies, will be required to validate its clinical utility, and more thoroughly understand the interindividual heterogeneity of % Lp(a)-C observed in this study. Ultimately, Lp(a)-C and correct LDL-C will need to be determined in cohorts followed for CVD outcomes in order to define whether these improved quantitative and empirical measures are better predictors of the current state of the art. Finally, IDL-C will remain a component of LDL-C after subtraction of Lp(a)-C.
Supplemental data
This article contains supplemental data.
Conflict of interest
Drs Tsimikas and Witztum are coinventors and receive royalties from patents owned by UCSD on oxidation-specific antibodies and of biomarkers related to oxidized lipoproteins and are cofounders and have equity interests in Oxitope, Inc. and Kleanthi Diagnostics, LLC. Although these relationships have been identified for conflict of interest management based on the overall scope of the project and its potential benefit to Oxitope and Kleanthi, the research findings included in this particular publication may not necessarily relate to the interests of Oxitope and Kleanthi. The terms of this arrangement have been reviewed and approved by the UCSD in accordance with its conflict of interest policies. Dr Tsimikas has a dual appointment at UCSD and Ionis Pharmaceuticals, and Dr Witztum is a consultant to Ionis Pharmaceuticals. The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Ionis Pharmaceuticals for providing clinical samples with extensive plasma lipid and lipoprotein(a) characterization and Amber Sanchez, MD (UCSD Therapeutic Apheresis Program) for making apheresis eluent available.
Author contributions
C. Y. conceptualized and designed the study, conducted the experiments, analyzed the data, and wrote the original draft of the article. J. L. W. analyzed the data and provided critical review and editing of the article. S. T. conceptualized the study, analyzed the data, and provided critical review and editing of the article.
Funding and additional information
Funding was provided by the Fondation Leducq (S. T. and C. Y.), National Heart, Lung and Blood Institute [HL148188 (J. L. W.)], and C. Y. is supported by National Institutes of Health 1K08HL150271 and UL1TR001442. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Calvin Yeang, Email: cyeang@health.ucsd.edu.
Sotirios Tsimikas, Email: stsimikas@health.ucsd.edu.
Supplemental data
References
- 1.Yeang C., Witztum J.L., Tsimikas S. ‘LDL-C’ = LDL-C + Lp(a)-C: implications of achieved ultra-low LDL-C levels in the proprotein convertase subtilisin/kexin type 9 era of potent LDL-C lowering. Curr. Opin. Lipidol. 2015;26:169–178. doi: 10.1097/MOL.0000000000000171. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt K., Noureen A., Kronenberg F., Utermann G. Structure, function, and genetics of lipoprotein (a) J. Lipid Res. 2016;57:1339–1359. doi: 10.1194/jlr.R067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varvel S., McConnell J.P., Tsimikas S. Prevalence of elevated Lp(a) mass levels and patient thresholds in 532 359 patients in the United States. Arterioscler. Thromb. Vasc. Biol. 2016;36:2239–2245. doi: 10.1161/ATVBAHA.116.308011. [DOI] [PubMed] [Google Scholar]
- 4.Tsimikas S. A test in context: lipoprotein(a): diagnosis, prognosis, controversies, and emerging therapies. J. Am. Coll. Cardiol. 2017;69:692–711. doi: 10.1016/j.jacc.2016.11.042. [DOI] [PubMed] [Google Scholar]
- 5.Willeit P., Ridker P.M., Nestel P.J., Simes J., Tonkin A.M., Pedersen T.R., Schwartz G.G., Olsson A.G., Colhoun H.M., Kronenberg F., Drechsler C., Wanner C., Mora S., Lesogor A., Tsimikas S. Baseline and on-statin treatment lipoprotein(a) levels for prediction of cardiovascular events: individual patient-data meta-analysis of statin outcome trials. Lancet. 2018;392:1311–1320. doi: 10.1016/S0140-6736(18)31652-0. [DOI] [PubMed] [Google Scholar]
- 6.Viney N.J., van Capelleveen J.C., Geary R.S., Xia S., Tami J.A., Yu R.Z., Marcovina S.M., Hughes S.G., Graham M.J., Crooke R.M., Crooke S.T., Witztum J.L., Stroes E.S., Tsimikas S. Antisense oligonucleotides targeting apolipoprotein(a) in people with raised lipoprotein(a): two randomised, double-blind, placebo-controlled, dose-ranging trials. Lancet. 2016;388:2239–2253. doi: 10.1016/S0140-6736(16)31009-1. [DOI] [PubMed] [Google Scholar]
- 7.Tsimikas S., Fazio S., Viney N.J., Xia S., Witztum J.L., Marcovina S.M. Relationship of lipoprotein(a) molar concentrations and mass according to lipoprotein(a) thresholds and apolipoprotein(a) isoform size. J. Clin. Lipidol. 2018;12:1313–1323. doi: 10.1016/j.jacl.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Gaubatz J.W., Hoogeveen R.C., Hoffman A.S., Ghazzaly K.G., Pownall H.J., Guevara J., Jr., Koschinsky M.L., Morrisett J.D. Isolation, quantitation, and characterization of a stable complex formed by Lp[a] binding to triglyceride-rich lipoproteins. J. Lipid Res. 2001;42:2058–2068. [PubMed] [Google Scholar]
- 9.Skalen K., Gustafsson M., Rydberg E.K., Hulten L.M., Wiklund O., Innerarity T.L., Boren J. Subendothelial retention of atherogenic lipoproteins in early atherosclerosis. Nature. 2002;417:750–754. doi: 10.1038/nature00804. [DOI] [PubMed] [Google Scholar]
- 10.Gaubatz J.W., Heideman C., Gotto A.M., Morrisett J.D., Dahlen G.H. Human plasma lipoprotein [a]. Structural properties. J. Biol. Chem. 1983;258:4582–4589. [PubMed] [Google Scholar]
- 11.Fless G.M., Rolih C.A., Scanu A.M. Heterogeneity of human plasma lipoprotein (a). Isolation and characterization of the lipoprotein subspecies and their apoproteins. J. Biol. Chem. 1984;259:11470–11478. [PubMed] [Google Scholar]
- 12.Albers J.J., Hazzard W.R. Immunochemical quantification of human plasma Lp(a) lipoprotein. Lipids. 1974;9:15–26. doi: 10.1007/BF02533209. [DOI] [PubMed] [Google Scholar]
- 13.Fless G.M., ZumMallen M.E., Scanu A.M. Physicochemical properties of apolipoprotein(a) and lipoprotein(a-) derived from the dissociation of human plasma lipoprotein (a) J. Biol. Chem. 1986;261:8712–8718. [PubMed] [Google Scholar]
- 14.Tsimikas S., Gordts P., Nora C., Yeang C., Witztum J.L. Statin therapy increases lipoprotein(a) levels. Eur. Heart J. 2020;41:2275–2284. doi: 10.1093/eurheartj/ehz310. [DOI] [PubMed] [Google Scholar]
- 15.Tsimikas S., Karwatowska-Prokopczuk E., Gouni-Berthold I., Tardif J.C., Baum S.J., Steinhagen-Thiessen E., Shapiro M.D., Stroes E.S., Moriarty P.M., Nordestgaard B.G., Xia S., Guerriero J., Viney N.J., O'Dea L., Witztum J.L. Lipoprotein(a) Reduction in Persons with Cardiovascular Disease. N. Engl. J. Med. 2020;382:244–255. doi: 10.1056/NEJMoa1905239. [DOI] [PubMed] [Google Scholar]
- 16.Willeit P., Yeang C., Moriarty P.M., Tschiderer L., Varvel S.A., McConnell J.P., Tsimikas S. Low-density lipoprotein cholesterol corrected for lipoprotein(a) cholesterol, risk thresholds, and cardiovascular events. J. Am. Heart Assoc. 2020;9 doi: 10.1161/JAHA.119.016318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baudhuin L.M., Hartman S.J., O'Brien J.F., Meissner I., Galen R.S., Ward J.N., Hogen S.M., Branum E.L., McConnell J.P. Electrophoretic measurement of lipoprotein(a) cholesterol in plasma with and without ultracentrifugation: comparison with an immunoturbidimetric lipoprotein(a) method. Clin. Biochem. 2004;37:481–488. doi: 10.1016/j.clinbiochem.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Nauck M., Winkler K., Wittmann C., Mayer H., Luley C., Marz W., Wieland H. Direct determination of lipoprotein(a) cholesterol by ultracentrifugation and agarose gel electrophoresis with enzymatic staining for cholesterol. Clin. Chem. 1995;41:731–738. [PubMed] [Google Scholar]
- 19.Yeang C., Clopton P.C., Tsimikas S. Lipoprotein(a)-cholesterol levels estimated by vertical auto profile correlate poorly with Lp(a) mass in hyperlipidemic subjects: Implications for clinical practice interpretation of Lp(a)-mediated risk. J. Clin. Lipidol. 2016;10:1389–1396. doi: 10.1016/j.jacl.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkarni K.R., Garber D.W., Marcovina S.M., Segrest J.P. Quantification of cholesterol in all lipoprotein classes by the VAP-II method. J. Lipid Res. 1994;35:159–168. [PubMed] [Google Scholar]
- 21.Seman L.J., Jenner J.L., McNamara J.R., Schaefer E.J. Quantification of lipoprotein(a) in plasma by assaying cholesterol in lectin-bound plasma fraction. Clin. Chem. 1994;40:400–403. [PubMed] [Google Scholar]
- 22.Kinpara K., Okada H., Yoneyama A., Okubo M., Murase T. Lipoprotein(a)-cholesterol: a significant component of serum cholesterol. Clin. Chim. Acta. 2011;412:1783–1787. doi: 10.1016/j.cca.2011.05.036. [DOI] [PubMed] [Google Scholar]
- 23.Tsimikas S., Fazio S., Ferdinand K.C., Ginsberg H.N., Koschinsky M.L., Marcovina S.M., Moriarty P.M., Rader D.J., Remaley A.T., Reyes-Soffer G., Santos R.D., Thanassoulis G., Witztum J.L., Danthi S., Olive M. Recommendations to reduce lipoprotein(a)-mediated risk of cardiovascular disease and aortic stenosis. J. Am. Coll. Cardiol. 2018;71:177–192. doi: 10.1016/j.jacc.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.