Abstract
Oxysterols are oxidized derivatives of cholesterol that play regulatory roles in lipid biosynthesis and homeostasis. How oxysterol signaling coordinates different lipid classes such as sterols and triglycerides remains incompletely understood. Here, we show that 4β-hydroxycholesterol (HC) (4β-HC), a liver and serum abundant oxysterol of poorly defined functions, is a potent and selective inducer of the master lipogenic transcription factor, SREBP1c, but not the related steroidogenic transcription factor SREBP2. By correlating tracing of lipid synthesis with lipogenic gene expression profiling, we found that 4β-HC acts as a putative agonist for the liver X receptor (LXR), a sterol sensor and transcriptional regulator previously linked to SREBP1c activation. Unique among the oxysterol agonists of the LXR, 4β-HC induced expression of the lipogenic program downstream of SREBP1c and triggered de novo lipogenesis both in primary hepatocytes and in the mouse liver. In addition, 4β-HC acted in parallel to insulin-PI3K–dependent signaling to stimulate triglyceride synthesis and lipid-droplet accumulation. Thus, 4β-HC is an endogenous regulator of de novo lipogenesis through the LXR-SREBP1c axis.
Supplementary key words: oxysterol, SREBP1c, liver-X-Receptor, de-novo-lipogenesis, lipid droplets, insulin
Abbreviations: 4β-HC, 4β-hydroxycholesterol; DNL, de novo lipogenesis; DPBS, dulbecco’s phosphate buffered saline; ent-4HC, enantiomer of 4β-HC; EtOAc, ethyl acetate; HC, hydroxycholesterol; LD, lipid droplet; LDS, lipid-depleted serum; mTOR, mechanistic Target of Rapamycin; NAFLD, nonalcoholic fatty liver disease; PI3K, phosphatidylinositol 3-kinase; THF, tetrahydrofuran
All cells must achieve and maintain a balanced composition of their internal membranes to grow, proliferate, or adapt to sudden changes in external conditions and nutrient availability (1). Dedicated biosynthetic pathways mediate the synthesis of fatty acids, sterols, phospholipids, and sphingolipids, but how these pathways communicate with each other to coordinate their respective activities and respond to changing metabolic needs is poorly understood (2, 3).
The liver X receptor (LXR) α and β are transcription factors belonging to the nuclear receptor superfamily that play key roles in maintaining lipid homeostasis in multiple cells and organs (4, 5, 6, 7). The LXRα and LXRβ dimerize with the retinoid X receptor (RXR) and activate target genes that mediate cholesterol efflux from cells, including ABC-family transporters, as well as genes that mediate conversion of cholesterol into bile acids in the liver to facilitate cholesterol elimination from the body, such as cytochrome p450 7a-hydroxylase (8, 9, 10). Accordingly, mice lacking the LXRα exhibit impaired bile acid metabolism and defective cholesterol elimination (9), along with enhanced inflammation and formation of atherosclerotic plaques (11). Conversely, synthetic LXRα agonists have shown promise in reducing atherosclerosis and preventing cardiovascular disease in animal models (12, 13, 14).
Another key mediator of lipid homeostasis is the helix-loop-helix-leucine zipper transcription factor, SREBP1c. SREBP1c is a master regulator of biosynthesis of fatty acids and triglycerides [collectively referred to as de novo lipogenesis (DNL)] that is subject to tight transcriptional and posttranslational regulation. Along with its paralogue, the master steroidogenic transcription factor SREBP2, SREBP1c resides at the endoplasmic reticulum (ER) membrane, to which it is anchored via a single transmembrane helix. When cholesterol concentration in the ER membrane is low, SREBP1c and SREBP2 are transported to the Golgi apparatus via interaction with SREBP cleavage-activating protein, a cholesterol-sensing chaperone that favors their loading into COPII vesicles. At the Golgi membrane, resident proteases cleave the DNA-binding portion of SREBP1c and SREBP2 from the transmembrane portion, enabling their translocation to the nucleus and activation of downstream programs for DNL and de novo steroidogenesis, respectively.
In addition to their homeostatic regulation by cholesterol levels, the SREBPs lie downstream of metabolic hormone signaling. For example, in the liver, both the expression and proteolytic activation of SREBP1c are stimulated by the insulin-phosphatidylinositol 3-kinase (PI3K)-mechanistic Target of Rapamycin (mTOR) pathway, as part of a mechanism that converts excess of glucose into lipids, which are required for energy storage (15, 16, 17). However, the range of regulatory inputs to SREBP1c and their respective interplay remain to be fully elucidated.
The LXRα and LXRβ were shown to directly bind to the promoter of the SREBP1c gene and trigger activation of its downstream lipogenic genes (6). Accordingly, synthetic LXR ligands strongly promote DNL and increased plasma triglyceride levels (13, 18, 19), providing evidence for cross-talk between LXR- and SREBP1c-dependent programs.
Although the physiological significance of LXR-dependent regulation of DNL through SREBP1c remains unclear, this cross-talk has important clinical implications. In particular, LXR-dependent upregulation of SREBP1c potentially limits the usefulness of LXR agonists to improve cholesterol metabolism, as the resulting induction of lipogenic programs could lead to undesirable effects, such as nonalcoholic fatty liver disease (NAFLD), a condition that has risen to epidemic proportions in recent years (20). Thus, understanding how LXR-dependent activation of SREBP1c occurs and its functional interaction with other pathways controlling lipid homeostasis such as PI3K-mTOR signaling are key open questions.
Oxysterols are a family of metabolites that originate from an oxygenation reaction of cholesterol. Some oxysterols are signaling molecules involved in a wide range of physiological processes controlling cholesterol, glucose, and lipid metabolisms (21). Levels of oxysterols are known to change in pathological situations such as obesity, atherosclerosis, and Alzheimer's disease (22, 23). A subset of oxysterols function as endogenous LXR ligands and were shown to activate LXRα-dependent gene expression in vitro, including those bearing hydroxyl groups in positions 4, 7, 20, 22, 24, 25, and 27 on the cholesterol backbone (4, 24, 25). Interestingly, although these oxysterols are considered bona fide LXR activators, none is known to activate SREBP1c and its downstream lipogenic programs, whereas several oxysterols have been shown to promote LXR-dependent cholesterol efflux. In contrast, synthetic LXR ligands including T0901317 and GW3965 can induce both cholesterol efflux and SREBP1c-dependent DNL (18, 19). This leads to the question of whether DNL is a physiologically relevant LXR-dependent response, and if so, the identity of the endogenous ligand that triggers LXR-dependent SREBP1c expression.
Here we identify 4β-hydroxycholesterol (HC) (4β-HC) as an LXR activator that selectively triggers SREBP1c activation and de novo fatty acid and triglyceride synthesis. 4β-HC promoted the expression and proteolytic processing of SREBP1c but not of the related steroidogenic factor SREBP2, thus triggering de novo synthesis of fatty acids but not cholesterol. In primary mouse hepatocytes, 4β-HC additively enhance insulin action in promoting SREBP1c expression and activation, leading to increased triglyceride synthesis and storage. Thus, 4β-HC may be a novel lipogenic factor that can shift lipid homeostasis toward triglyceride accumulation via regulation on SREBP1c.
Results
4β-HC is a unique oxysterol that drives SREBP1c gene expression
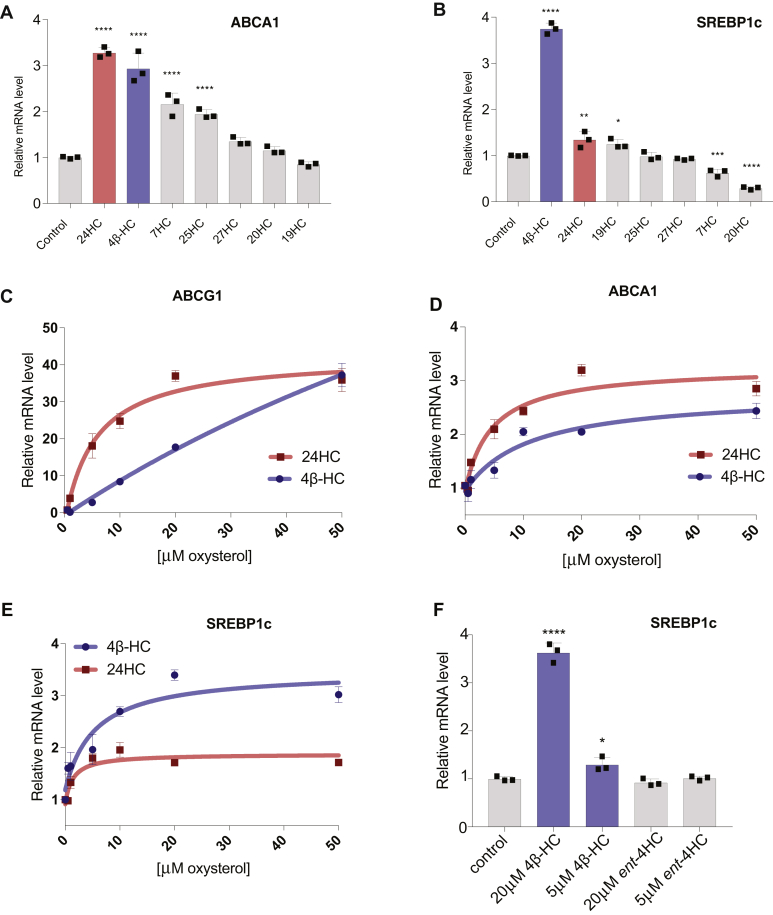
To identify oxysterol ligands that could promote SREBP1c expression, we treated liver carcinoma–derived Huh7 cells with a panel of oxysterols selected among the most abundant in the bloodstream, including 4β-, 7β-, 19-, 20-, 24(S)-, 25-, and 27-HC. By quantitative PCR, several oxysterols previously identified as LXR activators, including 4β-HC, 7β-HC, 24(S)-HC and 25-HC, induced the expression of a canonical LXR target gene, ABCA1, with variable potency (Fig. 1A). In contrast, 4β-HC was the only oxysterol to induce significant upregulation of the SREBP1c transcript (Fig. 1B). A dose-response comparison between 4β-HC and 24(S)-HC showed that 24(S)-HC is a more potent activator than 4β-HC toward ABCA1 (Fig. 1C) and another canonical LXR gene target, ABCG1 (Fig. 1D). Conversely, 4β-HC activated SREBP1c, more potently than 24(S)-HC (Fig. 1E). 4β-HC–mediated induction of the SREBP1c gene was enantioselective, as the nonnatural enantiomer of 4β-HC (ent-4HC) was unable to induce SREBP1c mRNA expression even at the highest concentration used (20 μM) (Fig. 1F). These data suggest that SREBP1c induction depends on unique structural features of 4β-HC.
Fig. 1.
4β-HC is a unique LXR ligand that drives SREBP1c expression. A: Oxysterol screen for LXR target gene expression. Huh7 cells were treated with indicated oxysterols (20 μM) in 24-h time course. ABCA1 mRNA levels or (B) SREBP1c mRNA level were measured by RT-PCR (N = 3). C: Dose-response curves of 4β-HC and 24-HC in Huh7 cells were treated for 24 h. mRNA levels of ABCA1, (D) ABCG1, and (E) SREBP1c were measured by RT-PCR. Line plotted by nonlinear fit (N = 3). E: SREBP1c induction by 4β-HC is stereospecific. Huh7 cells were treated with 4β-HC or an enantiomer of 4β-HC (ent-4HC) for 24 h in the indicated concentration (N = 3). Bars are the mean + SD. Statistical significance calculated by one-way ANOVA. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. NS, not significant; 4β-HC, 4β-hydroxycholesterol.
4β-HC induces expression and activation of SREBP1 but not SREBP2
Oxysterols such as 25- and 27-HC suppress SREBP1 and SREBP2 activation by blocking their trafficking to the Golgi, where proteolytic processing of the SREBPs to the mature nuclear form occurs (26, 27).
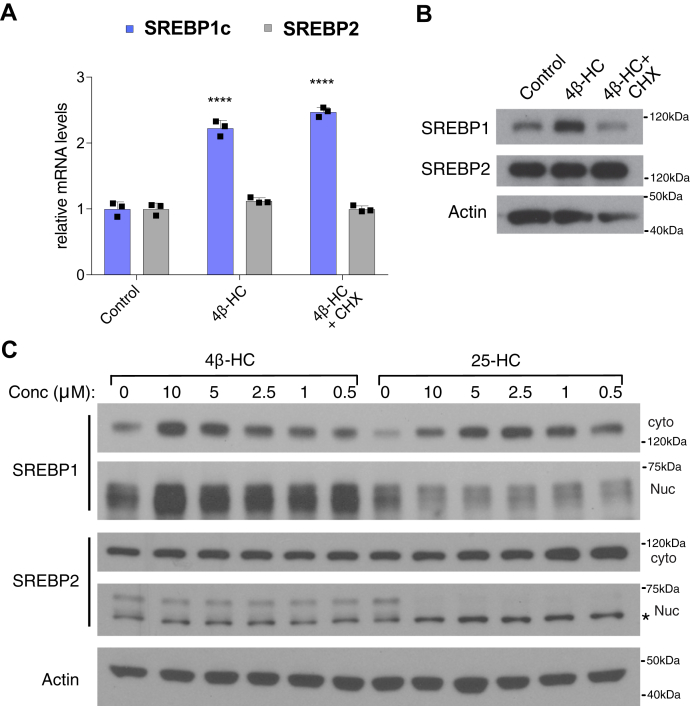
In contrast to these oxysterols, 4β-HC significantly increased SREBP1c mRNA levels (Fig. 2A) and protein levels in a cycloheximide-sensitive manner (Fig. 2B). However, 4β-HC did not increase either mRNA or protein levels of SREBP2 (Fig. 2A, B). In keeping with the increased total levels of SREBP1c, 4β-HC increases both cytosolic and nuclear forms of SREBP1 in a dose-dependent manner, whereas levels of cytoplasmic or nuclear SREBP2 protein levels did not change (Fig. 2C). Consistent with previous reports (26, 27) and in contrast to 4β-HC, 25-HC reduced the nuclear forms of both SREBP1 and SREBP2, thereby causing the accumulation of the unprocessed cytoplasmic form of both proteins but without transcriptional upregulation (Fig. 1B).
Fig. 2.
4β-HC induces expression and activation of SREBP1 but not SREBP2. A: 4β-HC increases SREBP1 protein expression. Huh7 cells were treated with 20 μM 4β-HC and a translation inhibitor, cycloheximide (CHX), for 4 h followed by measurement of SREBP1 and SREBP2 mRNA (N = 3) and (B) protein level (N = 1). C: 4β-HC increases SREBP1 cytosolic and nuclear levels while not affecting SREBP2. Huh7 cells were treated with 4β-HC or 25-HC for 24 h followed cytosolic-nuclear fractionation to measure protein level of SREBP1 and SREBP2 cytoplasmic and nuclear levels (N = 1). Asterisk denotes unspecific band in SREBP2 nuclear blot. Bars are the mean + SD. Statistical significance calculated by one-way ANOVA. ∗∗∗∗P < 0.0001. Cyto, cytosolic; Nuc, nuclear; 4β-HC, 4β-hydroxycholesterol.
These data suggest that, unlike other oxysterols that function as inhibitors of both SREBP1c and SREBP2, 4β-HC is a specific inducer of SREBP1c expression and activation.
4β-HC induce lipogenic programs through the LXRs
Along with other oxysterols, 4β-HC was previously shown to activate LXRα-dependent transcription in luciferase assays in vitro, supporting its role as a putative LXR ligand (4, 28). In turn, the LXR transcriptionally activates SREBP1c by directly binding to its promoter region (6). Combining these observations, we thus hypothesized that 4β-HC may transcriptionally activate SREBP1c and its downstream lipogenic programs via the LXR. Consistent with this possibility, cotreating cells with 4β-HC together with an LXR antagonist (GSK-2033) abolished 4β-HC–dependent induction of SREBP1c gene expression (Fig. 3A).
Fig. 3.
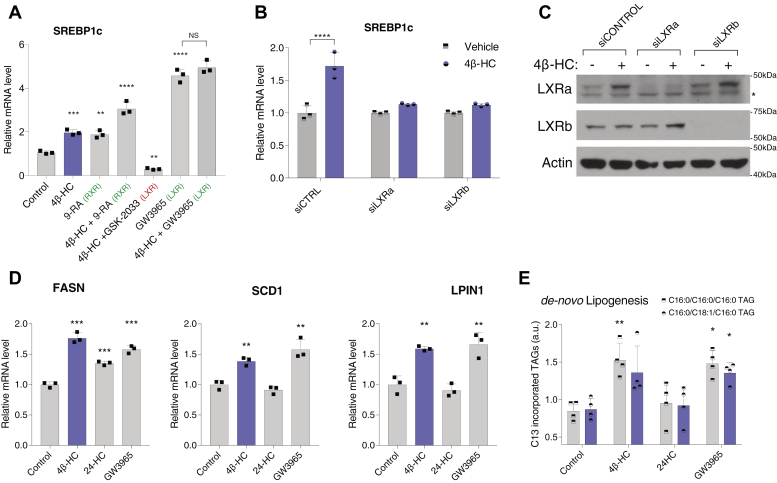
4β-HC induces lipogenic programs through the LXRs. A: 4β-HC interacts with LXR and RXR agonists and antagonists like an LXR ligand. Huh7 cells were treated with 20 μM 4β-HC, RXR agonist, 9-cis-retinoic acid (9-RA), LXR antagonist (GSK-2033), and LXR agonist (GW3965). For convenience, agonists are marked in green and antagonists are marked in red (N = 3). B: LXRα and LXRβ are required for SREBP1c induction by 4β-HC in Huh7 cells. Knockdown of LXRα or LXRβ by siRNA for 72 h followed by treatment with 5 μM 4β-HC for 24 h followed by RT-PCR of SREBP1c. (N = 3). C: Knockdown efficiency was evaluated by measurement of LXRα and LXRβ protein levels (N = 3). D: 4β-HC induction of lipogenic genes. Huh7 cells were treated for 24 h with 4β-HC, 24-HC, or LXR agonist (GW3965) followed by mRNA measurement of fatty acid synthase (FASN), stearoyl-CoA desaturase 1 (SCD1), and lipin1 (LPIN1) (N = 3). E: 4β-HC increases de novo lipogenesis. Huh7 cells were treated for 24 h with 5 μM 4β-HC, 24-HC, or LXR agonist (GW3965) with media containing C13 glucose followed by lipid extraction. C13 incorporation into TAGs was measured via LC/MS (N = 5). The asterisk denotes an unspecific band in the LXRα blot. Bars are the mean + SD. Statistical significance was calculated by one-way ANOVA. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. 4β-HC, 4β-hydroxycholesterol; 24-HC, 24-hydroxycholesterol.
The effect of 4β-HC on SREBP1c induction was additive with an RXR ligand, 9-cis-retinoic acid. Moreover, coincubation of 4β-HC with the LXR agonist, GW3965, used at concentrations that activate the LXR maximally, caused no additional increase in SREBP1c expression over GW3965 alone (Fig. 3A). siRNA-mediated knockdown of either the LXRα or LXRβ (both of which are expressed in Huh7 cells) largely abolished 4β-HC–dependent SREBP1c mRNA expression (Fig. 3B). Interestingly, we noticed that 4β-HC treatment increased LXRα protein levels, a stabilizing effect observed for other established LXR ligands (29) (Fig. 3C). Together, and combined with previous reports these data support the hypothesis that 4β-HC induces SREBP1c gene expression by acting as an LXR agonist.
We next compared the ability of 4β-HC to induce SREBP1c-dependent lipogenic programs with that of the LXR agonist, GW3965. Fatty acid synthase (FASN), Stearoyl-CoA desaturase (SCD1) and Lipin1 (LPIN1) are validated SREBP1c downstream targets in Huh7 cells (30, 31). Treatment with either 4β-HC or GW3965 significantly increases the expression of these genes (Fig. 3D). In contrast, 24-HC, another putative LXR ligand that failed to induce SREBP1c in our hands (Fig. 1B, E), had minimal or no effect on these SREBP1c target genes (Fig. 3D).
Previous work had shown that GW3965 induces FASN to a greater extent than the 1.6-fold we observed in Huh7 (32). Huh7, a hepatocellular carcinoma line, is known to hyperactivated DNL to supply membranal lipids required for rapid division and growth (33, 34). We speculate that the modest increase in FASN by GW3965 or 4β-HC is due to already elevated baseline expression that cannot be increased much further. To further substantiate the prolipogenic effect of 4β-HC, we directly measured DNL by C13 incorporation into triglycerides using LC/MS. Similarly to lipogenic gene induction, both GW3965 and 4β-HC had a modest but statistically significant 1.5-fold increase in C13-labeled C16:C16:C16 TAG, or trending toward significance for C16:C18:C16 TAG, whereas 24-HC caused no significant change (Fig. 3E). Combined, these data suggest that the prolipogenic action of 4β-HC is comparable, in mechanism and potency, to known LXR agonists.
4β-HC induces lipid-droplet formation and triglyceride accumulation
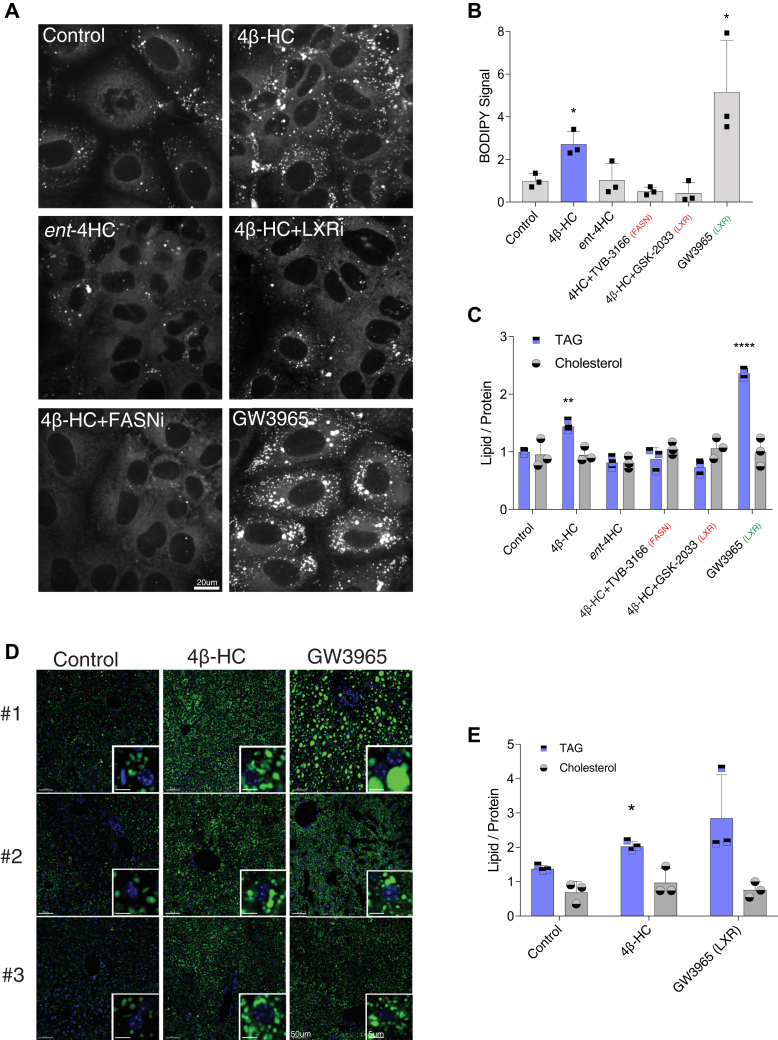
In keeping with the ability of 4β-HC to upregulate fatty acid biosynthetic genes via SREBP1c, treating Huh7 cells with 4β-HC (but not with its unnatural enantiomer, ent-4HC) for 72 h resulted in marked accumulation of lipid droplets (LDs), as revealed by staining with the lipophilic dye BODIPY 493/503 (Fig. 4A, B). LD accumulation induced by 4β-HC was suppressed by simultaneous treatment with a FASN inhibitor, TVB-3166, or with the LXR inhibitor GSK-2033. Measurement of triglyceride content in cell extracts confirmed the ability of 4β-HC to induce triglyceride accumulation, albeit with lower potency than the LXR agonist GW3965, whereas cholesterol levels remained unchanged (Fig. 4C). Consistent with the BODIPY staining, both LXR and FASN inhibitors hindered 4β-HC–induced triglyceride accumulation (Fig. 4C). Moreover, as seen with SREBP1c induction, the ent-4HC failed to induce triglyceride accumulation (Fig. 4C). Thus, 4β-HC is sufficient to induce the formation of triglyceride-containing LDs in an LXR- and FASN-dependent manner in cell culture.
Fig. 4.
4β-HC induces lipid-droplet formation and triglyceride accumulation. A: 4β-HC increases the lipid droplet size and number. Huh7 cells were treated with 5 μM 4β-HC with indicated drugs for 72 h followed staining with lipid droplet dye, BODIPY 493/503, and visualization by confocal microscopy and (B) quantified using ImageJ (N = 3). C: 4β-HC increases triglycerides (TAG) levels. Huh7 cells were treat as (C) followed by measurement of triglycerides, total cholesterol, and protein levels using commercial kits (N = 3). D: 4β-HC increases lipid droplet in the mouse liver. Mice were fed normal chew with either vehicle 50 mg/kg/day 4β-HC or 10 mg/kg/day GW3965 for 5 days. Liver samples were fixed and stained with BODIPY 493/503 and DAPI to observe lipid droplet and nuclei ultrastructure (N = 3). E: 4β-HC increases triglycerides (TAG) levels in the mouse liver treated as above, followed by measurement of triglycerides, total cholesterol, and protein levels using commercial kits (N = 3). For convenience, agonists are marked in green and antagonists are marked in red. Bars are the mean + SD. Statistical significance calculated by one-way ANOVA.∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001. ent-4HC, stereo enantiomer-4HC, FASNi, TVB-3166; LXRi, GSK-2033; 4β-HC, 4β-hydroxycholesterol.
Next, we tested the effect of 4β-HC on in vivo lipogenesis by feeding mice a normal diet supplemented with either 4β-HC or GW3965. After 7 days, the livers were harvested, LDs were assessed by BODIPY staining, and the liver lipid content (normalized to protein mass) was measured. Consistent with the results in Huh7 cells, 4β-HC significantly increased the size and number of LDs in liver sections (Fig. 4D) and liver triglyceride content (Fig. 4E), albeit with lower potency than the synthetic LXR agonist, GW3965. Collectively, these data suggest that 4β-HC is a prolipogenic factor that can increase liver lipid content in vivo.
4β-HC acts in parallel to insulin-PI3K signaling to drive SREBP1c expression
Insulin is a key hormone that drives SREBP1c transcription, proteolytic processing, and DNL in the postprandial state. Insulin regulates SREBP1c transcription via poorly understood mechanisms, which include AKT-dependent transcriptional downregulation of Insig-2a, the ER-retention factor that blocks translocation of SREBP cleavage-activating protein-SREBP1c to the Golgi (35, 36). The LXR was shown to be required for insulin-dependent activation on SREBP1c (37), but whether and how insulin activates LXR is not understood.
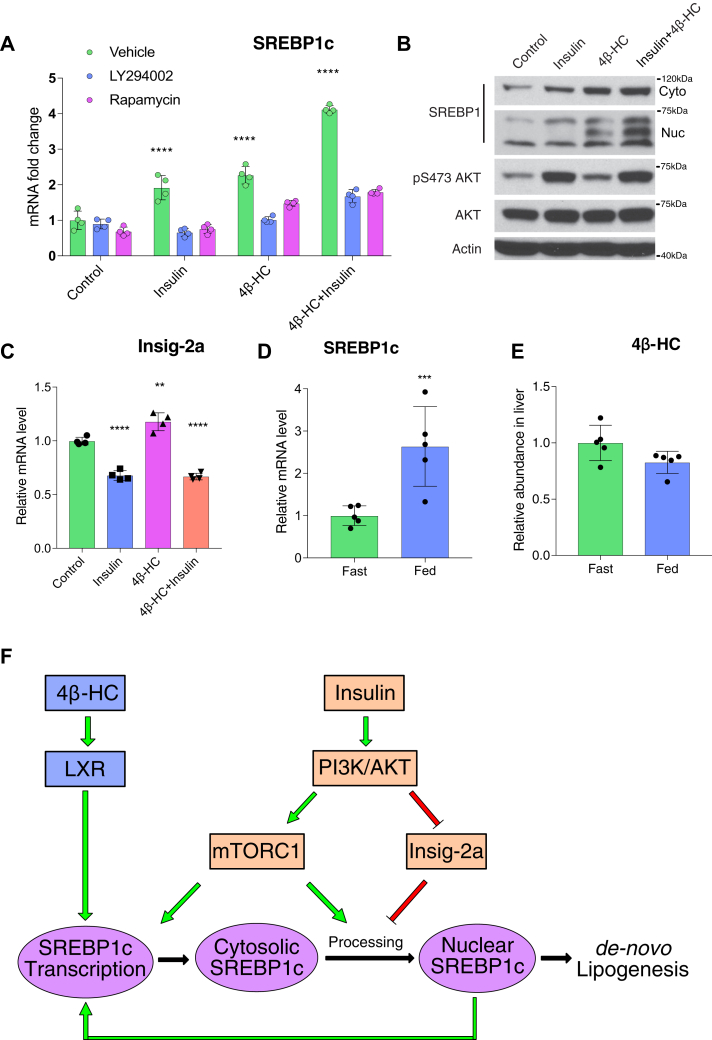
To interrogate the relationship between 4β-HC and insulin signaling in driving SREBP1c transcription and processing, we used an insulin-responsive primary mouse hepatocyte (38). In these cells, stimulation with either 4β-HC or insulin alone increased the mRNA levels of SREBP1c, while combined 4β-HC and insulin increased SREBP1c mRNA levels additively [as previously shown for LXR agonists (37)] (Fig. 5A). Interestingly, treatment with PI3K or mTORC1 inhibitors abolished SREBP1c induction by both insulin and 4β-HC (Fig. 5A), raising the possibility of a ‘coincidence detection’ model, in which a minimal amount of both insulin-PI3K-mTORC1 and 4β-HC signaling must be present for SREBP1c induction to occur.
Fig. 5.
4β-HC acts in parallel to insulin-PI3K signaling to drive SREBP1c expression. A: SREBP1c transcription is additive by 4β-HC and insulin. Primary hepatocytes were treated overnight with vehicle or 5 μM 4β-HC followed with 6 h stimulation with combinations of insulin, PI3K inhibitor (LY294002), or rapamycin. The SREBP1c mRNA level was measured by RT-PCR (N = 4). B: 4β-HC and insulin have an additive effect on SREBP1c expression and nuclear processing. Primary hepatocytes were treated overnight with vehicle or 4β-HC followed by addition of insulin for 40 min. Proteins were extracted and SREBP1 and AKT protein levels were measured (N = 2). C: Primary hepatocytes were treated with 4β-HC and insulin as described in (A) followed by RT-PCR measurement of Insig-2a mRNA level (N = 4). D: Insulin does not induce 4β-HC synthesis. Mice were fasted for 16 h and then refed for 4 h, followed by liver extraction and RT-PCR for SREBP1c mRNA level (N = 4) and (E) 4β-HC levels by MS (N = 5). F: Model; the 4β-HC-LXR pathway acts in parallel to the insulin-PI3K pathway to drive SREBP1c expression in an additive fashion. ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Similar to their effects on transcriptional induction, insulin and 4β-HC stimulated proteolytic processing of SREBP1c in an additive manner (Fig. 5B). 4β-HC did not affect AKT phosphorylation significantly, suggesting that insulin-PI3K-AKT and 4β-HC signaling act in parallel and converge at the level of the SREBP1 gene promoter (Fig. 5B and supplemental Fig. 1A).
Consistent with previous reports, we also detected a marked decrease in Insig-2a mRNA in insulin-stimulated hepatocytes (Fig. 5C). In contrast, 4β-HC caused a mild increase of Insig-2a mRNA levels, and combined insulin and 4β-HC was similar to insulin alone, suggesting Insig-2a downregulation is not required for 4β-HC–dependent SREBP1c activation (Fig. 5C).
To further probe possible connections between insulin-PI3K and 4β-HC-LXR signaling, we tested whether insulin signaling promotes 4β-HC synthesis. Previous reports had shown that in humans, 4β-HC has a very slow kinetics, with an extremely long half-life in plasma (60 h) (39). Pharmacological induction of the main 4β-HC–synthesizing enzyme, cytochrome P450 3A, doubles 4β-HC concentration in human plasma in 8 days (40), a very different pattern from insulin, which peaks within 1–2 h after a meal and drops in between. On the other hand, in vitro work in primary rat hepatocytes led to the hypothesis that insulin signaling may produce an unknown LXR ligand that, in turn, induces SREBP1c (37). To test the possibility of insulin-dependent 4β-HC production, we compared 4β-HC levels in the liver of mice that were either fasted or refed. Although mice that were refed showed significant induction of SREBP1c transcription, consistent with SREBP1c regulation by insulin (Fig. 5D), the levels of 4β-HC did not increase accordingly in the liver (Fig. 5E) or serum (Supplemental Fig. 1B). Collectively, these data suggest that insulin does not induce 4β-HC production according to fasting/feeding cycles and that 4β-HC most likely acts in parallel to insulin-PI3K signaling in driving SREBP1c transcription and SREBP1c-dependent DNL (Fig. 5F).
Discussion
Here we identify 4β-HC as a unique oxysterol that activates SREBP1c expression and promotes lipogenic gene programs, resulting in induction of fatty acid biosynthesis and cellular accumulation of triglycerides in LDs both in cell culture and in vivo. Our results are most consistent with a model in which 4β-HC acts in parallel to insulin-PI3K-mTOR signaling, and the two pathways have additive effects on SREBP1c activation. A simple mechanism that explains the additive effect is that the SREBP1c promoter contains both an LXR-binding element and an SREBP-binding element and that transcription can be stimulated by the two transcription factors independently (41). However, the observation that inhibition of PI3K-AKT signaling also blunts 4β-HC–dependent SREBP1 induction (Fig. 5A) points to a possible ‘coincidence detection’ model, where at least some signaling by one input (i.e., insulin) has to be present for the other input (4β-HC) to be effective, and vice versa. From a temporal standpoint, 4β-HC kinetics suggest that it stimulates SREBP1c expression in a chronic manner, whereas insulin acts acutely in the postprandial state.
A recent publication by Salonurmi et al. (42) showed that 4β-HC induces cholesterol efflux from peripheral mononuclear cells in vivo via transcriptional upregulation of ABCA1 and concomitant suppression of influx transporters. In our hands, 4β-HC did not induce ABCA1 expression in primary hepatocytes (supplemental Fig. 1C), whereas its induction was observed in Huh7 cells. Thus, 4β-HC–dependent regulation of cholesterol efflux versus DNL may be cell type specific and tied to different physiological settings.
Several groups using different animal models (mice, rats, rabbits, and swine) had all observed that 4β-HC levels increase when animals are fed a high cholesterol diet (43, 44, 45, 46), whereas a high-fat but with low-cholesterol diet reduces 4β-HC levels in mice (47). Dietary cholesterol was shown to increase SREBP1c expression in an LXR-dependent manner (6, 9). Furthermore, genetically disrupting hepatic cholesterol synthesis through SREBP2 KO also causes SREBP1c downregulation, which can be rescued by an LXR agonist (48). This study also determined that 4β-HC levels are decreases in young SREBP2-null mice, defining a correlation between SREBP2-dependent cholesterol synthesis, 4β-HC levels, and SREBP1c expression. Together with this published literature, our results strongly suggest that 4β-HC may be the cholesterol-derived molecule that induces SREBP1c activation via the LXR.
An important question is why 4β-HC is the sole oxysterol ligand of LXRs to activate SREBP1 expression in our hands. Several possibilities can be envisioned. The LXR-RXR heterodimer can recruit coactivators (PGC-1α, TRRAP, ACS-2, p300, SRC-1) and corepressors (NCoR, SMRT) to the promoters of target genes in a ligand-dependent manner (49, 50, 51, 52, 53), but whether all LXR ligands are equally effective in recruiting specific combinations of cofactors is unclear. Supporting this model was an observation in macrophages that the ability of the LXR to recruit RNA polymerase II to SREBP1c promoter requires a specific LXR ligand, while recruitment of RNA polymerase II to the ABCA1 promoter is more promiscuous (29). Thus, 4β-HC may be able to direct a unique set of coactivators and RNA polymerase II to the SREBP1c promoter, resulting in its activation.
Consistent with previous reports, the synthetic LXR agonist GW3965 was also able to trigger SREBP1 expression (18, 19). Synthetic LXR agonists are generally more potent than natural LXR ligands, possibly reflecting higher affinity for the ligand-binding site of the LXR. By analogy, 4β-HC may bind to the LXR with higher affinity than other oxysterol ligands. In turn, higher affinity may translate into longer residence time on the SREBP1c promoter DNA, a possible prerequisite for its efficient activation.
Our data point to the importance of the enzyme that produces 4β-HC, Cyp3A4 (Cyp3A11 in mice) (54), as a crucial regulator of lipogenesis. Consistent with that, several groups have reported that increased Cyp3A4 expression by overexpressing its activator, pregnane X receptor, correlated with increases in lipogenic gene expression and liver triglyceride levels (55, 56). Conversely, decreased Cyp3A4 expression (57) or its pharmacological inhibition (58) was associated with lower lipogenic gene expression and liver triglyceride levels. Taken together, these data suggest that Cyp3A4 and 4β-HC may regulate diet-induced lipogenic genes and liver triglyceride levels.
From a more clinical perspective, 4β-HC might have an aggravating effect on the development of NAFLD. NAFLD is characterized by elevated liver triglycerides not due to alcohol consumption or any other known causes (59). Elevated triglyceride levels are associated with LXR and SREBP1c upregulation in NAFLD (60). Patients with NAFLD show a significant increase in 4β-HC plasma levels compared with healthy patients (61). Thus, it is plausible that elevated 4β-HC levels could be an unrecognized driver of triglyceride accumulation in NAFLD. It would be interesting to determine the effect of pharmacologic Cyp3A4 inhibition on disease progression in patients with NAFLD.
In conclusion, this work highlights a role for 4β-HC, which was long viewed as an ‘orphan’ oxysterol, in regulating lipid metabolism in the liver together with insulin. Future work, dissecting the role of 4β-HC in other organs and in different pathological settings, will provide a full picture on the function and significance of this highly abundant oxysterol.
Materials and methods
Materials
Reagents were purchased from the following sources. Antibodies used are as follows: SREBP1 (2A4, Santa Cruz Biotechnology), SREBP2 (30682, Abcam), LXRα (PP-PPZ0412-00, R&D systems), LXRβ (K8917, R&D systems), phospho-T308 AKT (C31E5E), AKT (11E7) (Cell Signaling Technology).
Drugs used are as follows: Cycloheximide (Cell Signaling Technology) was used at 10 μg/ml. 9-cis-retinoic acid (Sigma) was used at 50 μM. The LXR antagonist GSK-2033 (Axon Medchem) was used at 500 nM. The LXR agonist GW3965 (Fisher Scientific) was used at 500 nM. PI3K inhibitor, LY294002 (Cell Signaling Technology), was used at 10 μM. Rapamycin was used at 100 nM and received as a gift from David Sabatini. Methyl-beta-cyclodextrin was purchased from Sigma. All sterols except for custom-synthesized ent-4β-HC (see below) were purchased from Steraloids. C13 glucose was purchased from Cambridge Isotope Laboratories.
Sterol: methyl-beta-cyclodextrin precomplexing
All sterols were made to 50 mM stocks in ethanol. To deliver the sterols to cells, 1.25 mM sterol was complexed with 25-mM methyl-beta-cyclodextrin and vortexed until the solution was clear. Sterols were added to the media in an indicated concentration and incubation time. Control samples were treated by adding the same volume of ethanol to methyl-beta-cyclodextrin, which then was delivered to cells in the same corresponding volume.
Cell culture
Huh7 cells were maintained on DMEM (5 g/l glucose + glutamine, Gibco) supplemented with 10% FBS (VMR) and p/s (Gibco). Lipid-depleted serum (LDS) was made as described (62). For assays, on day one; 105 cells were plates in 6 cm plates. On day 2, media was changed to 1% LDS and 1 g/l glucose DMEM. On day 3; plates were spiked with precomplexed sterols for indicated times, concentrations, and additional compounds.
Primary mouse hepatocytes were purchased from the UCSF liver center. The isolation protocol is based on the study by Li, Brown, and Goldstein (63) and adjusted in the following manner. Mice were fasted overnight before isolation. Hepatocytes were isolated by the perfusion protocol (64) and plated at density of 7 × 105/well on 6-well collagen-coated plates (Corning) in DMEM supplemented with 10% FBS. Once cells adhere, the media was replaced to Medium 199 (GIBCO) containing 100 nM dexamethasone (Sigma), 100 nM 3,3,5-triiodo-L-thyronine (T3, Sigma), and Insulin-Transferrin-Selenium (Gibco). Next day, the same media was used without Insulin-transferrin-Selenium to assay insulin, 4β-HC, and inhibitors at indicated times and concentrations.
Real-time PCR analysis for gene expression
RNA was extracted using the RNeasy kit (Qiagen). One microgram of RNA was reverse-transcribed using Super Script III (Invitrogen). Quantitative PCR was performed using Ssoadvanced (Bio-Rad) in StepOnePlus (ABI). The list of primers is in Table 1.
Table 1.
RT-PCR primers
| Gene | Species | Forward | Reverse |
|---|---|---|---|
| TBP | Human | TTGTACCGCAGCTGCAAAAT | TATATTCGGCGTTTCGGGCA |
| SREBP1c | Human | GCGCCTTGACAGGTGAAGTC | GCCAGGGAAGTCACTGTCTTG |
| FASN | Human | CTTCAAGGAGCAAGGCGTGA | ACTGGTACAACGAGCGGATG |
| SCD1 | Human | TCTAGCTCCTATACCACCACCA | TCGTCTCCAACTTATCTCCTCC |
| ABCA1 | Human | TGTTCGCGGCCCTCAT | CGAGATATGGTCCGGATTGC |
| ABCG1 | Human | TGCAATCTTGTGCCATATTTGA | CCAGCCGACTGTTCTGATCA |
| LPIN1 | Human | CCAGCCCAATGGAAACCTCC | AGGTGCATAGGGATAACTTCCTG |
| SREBP2 | Human | GAGCTGGGTGGTCTGGAG | TTGCAGCATCTCGTCGATGT |
| SREBP1C | Mouse | CGGAAGCTGTCGGGGTAG | GTTGTTGATGAGCTGGAGCA |
| SREBP2 | Mouse | GCGTTCTGGAGACCATGGA | ACAAAGTTGCTCTGAAAACAAATCA |
| INSIG-2A | Mouse | TGTGAGCTGGACTAGCTTGCT | CCTAAGCCGTAAAACAAAATG |
| TBP | Mouse | ACCCTTCACCAATGACTCCTATG | ATGATGACTGCAGCAAATCGC |
Protein extraction and Western blot
Cells were harvested with the RIPA buffer supplemented with Phosphatase inhibitor and protease inhibitor (10 mM Tris Cl (pH 8.0), 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl, 10 mM Na-PPi, 10 mM Na-Beta-glycerophosphate), sonicated with Bioruptor (Diagenode), and normalized using the BCA kit (Thermo Scientific).
Knockdown using siRNA
siRNA ON-TARGET plus smart pool against LXRα (cat# L-003413-00-0005), LXRβ (cat# L-003412-02-0005), or nontargeted siRNA ON-TARGETplus Non-targeting Pool (cat# D-001810-10-05) was purchased from Dharmacon. Five micromolar siRNA was mixed with 5 μl Lipofectamine RNAiMAX (Life Technologies) in Opti-MEM (Gibco). siRNA is added to preplated Huh7 (105 cells/6 cm plate) in regular media without penicillin streptomycin for 5 h followed by replacement to regular media for 72 h.
LD microscopy
Huh7 were plated on a coverslip coated with fibronectin (Corning) and treated as indicated with sterols and drugs. Cells were fixed with paraformaldehyde and stained with 1 μg/ml BODIPY 493/503 for 1 h. Coverslips were mounted with VECTASHIELD with DAPI (Vector Laboratories) and imaged on a spinning disk confocal system (Andor Revolution on a Nikon Eclipse Ti microscope). The BODIPY signal was measured using ImageJ and normalized by the number of nuclei.
Triglyceride and cholesterol measurements
Liver samples were powdered with a pestle and mortar and lysed in the RIPA buffer. Huh7 cells were also harvested in the RIPA buffer. Five microliters of the samples was used to measure triglyceride using Triglyceride Infinity (Thermo Fisher) or cholesterol using the Amplex red cholesterol measuring kit (Invitrogen) in a clear 96-well sample. The BCA kit (Thermo Scientific) was used for normalization of the protein level. Absorbance and fluorescence were measured by the PerkinElmer Envision Multilabel plate reader.
C13 incorporation into triglycerides
Huh7 cells were seeded at 200K per 6-cm plates. The next day, DMEM media with glutamine, containing 5 mM C13 glucose (Cambridge Isotope Laboratories) and 1% LDS including oxysterols and the LXR agonist were added for 24 h. C12 glucose–treated plates were used as reference. Cells were washed twice with ice-cold PBS and scraped, and pellets were snap-frozen and kept in −80°C for later analysis. Lipid extraction and analysis by LC/MS was performed as described (65).
Husbandry and diets
All mouse procedures were performed and approved under the University of California, Berkeley Animal Care and Use Committee. Ten-week-old C57BL/6J male mice were purchased from the Jackson Laboratory and housed for one week in our facility under standard conditions before experiments were performed. Free access to water and chow (Lab Diets, #3038) was provided throughout this acclimation period. Afterward, mice were placed on a diet with 50 mg/kg/day 4β-HC or LXR agonist GW3965 10 mg/kg/day for 7 days. Powdered 10% by kCal fat diet (Research Diets Inc. #D12450J) was used as the base of each treatment food, forming pellets that were dried overnight at room temperature in a laminar flow hood. After 7 days, mice were euthanized using CO2 and cervical dislocation.
Cryosectioning and fluorescent histochemistry
Liver samples were fixed using 4% (v/v) paraformaldehyde overnight at 4°C. The next day, samples were cryopreserved using sterile-filtered 30% sucrose (w/v) in dulbecco’s phosphate buffered saline (DPBS) (Gibco, 14190-144). After 3 days, each sample was placed in a 1:1 30% sucrose:Neg-50 (Richard-Allan Scientific) solution and incubated overnight at 4°C. The samples were frozen on dry ice using undiluted Neg-50 at −50°C and stored at −80°C until sectioning. Sequential 20 μM thick sections were obtained from each sample using a Leica CM3050S cryostat.
For nuclei and LD labeling, sectioned tissue was washed three times at room temperature in DPBS for 5 min each. Afterward, DPBS containing 10 μM BODIPY (Invitrogen, #D3922) was placed on the samples and incubated for 30 min at room temperature in the dark. Next, the slides were washed with DPBS twice before incubating in DPBS containing 5 μg/ml DAPI (Invitrogen, D1306) for 10 min at room temperature in the dark. After DAPI staining, the slides were washed three times in DPBS for 5 min each before being mounted using SlowFade Diamond antifade (Invitrogen, #S36972) and sealing with nail polish overnight. Slides were imaged immediately using a Zeiss LSM710 confocal microscope. Images were developed using the IMARIS (Bitplane) image analysis software suite.
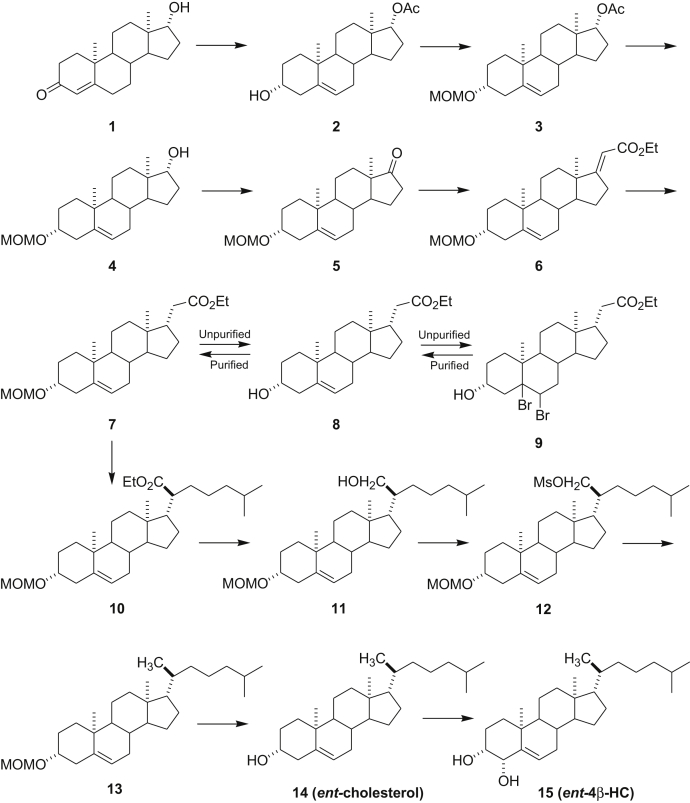
Synthesis of ent-4β-HC
ent-steroid 2
ent-Testosterone (1) was prepared as described previously [(66); see also references therein]. To a solution of ent-testosterone (1, 3.8 g, 13.2 mmol) in acetic anhydride (80 ml) was added NaI (7.92 g, 52 mmol) and trimethylsilyl chloride (5.8 ml, 52 mmol) at 0°C under N2. After addition, the reaction was allowed to warm to room temperature for 2 h. The reaction was added to Et3N (40 ml) in diethyl ether (100 ml). The ether solution was washed with brine (50 ml × 4) and aqueous NaHCO3 (50 ml × 2) and dried over Na2SO4. After filtration, the solvent was removed under reduced pressure and the residue was purified by flash column chromatography (silica gel eluted with 25% ethyl acetate (EtOAc) in hexanes) to give ent-steroid 2 (3.05 g, 70%): 1H NMR (400 MHz, CDCl3) δ 5.33–5.32 (m, 1H), 4.60 (t, J = 8.3 Hz, 1H), 3.52–3.47 (m, 1H), 2.30–0.90 (m), 2.02 (s, 3H), 1.00 (s, 3H), 0.79 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 171.2, 140.9, 121.1, 82.7, 71.5, 51.0, 50.0, 42.3, 42.2, 37.2, 36.7, 36.5, 31.6, 31.5, 31.4, 27.4, 23.5, 21.1, 20.5, 19.3, 11.8.
ent-steroid 3
ent-Steroid 2 (3.05 g, 4.04 mmol) was dissolved in CH2Cl2 (50 ml) and cooled to 0°C. (i-Pr)2EtN (3.0 ml) and ClCH2OMe (1.35 ml, 18.0 mmol) were added, and the reaction was stirred at room temperature for 16 h. The reaction was made basic by adding aqueous NaHCO3 solution, and the product was extracted into CH2Cl2. The combined extracts were washed with brine, dried over Na2SO4, filtered, and solvent removed to give a viscous liquid that was purified by flash column chromatography (silica gel eluted with 10% EtOAc in hexanes) to give ent-steroid 3 as a colorless liquid (2.65 g, 77%): 1H NMR (400 MHz, CDCl3) δ 5.33–5.32 (m, 1H), 4.65 (s, 2H), 4.59 (t, J = 8.2 Hz, 1H), 3.39–3.35 (m, 1H), 3.34 (s, 3H), 2.35–0.89 (m), 2.01 (s, 3H), 0.99 (s, 3H), 0.78 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 171.0, 140.7, 121.2, 94.6, 82.6, 76.7, 55.0, 50.9, 50.0, 42.3, 39.4, 37.1, 36.7, 31.6, 31.4, 28.8, 27.4, 23.5, 21.0, 20.4, 19.3, 11.8.
ent-steroid 4
To a solution of ent-steroid 3 (2.65 g, 7.05 mmol) in methanol (60 ml), K2CO3 (4.0 g) was added at room temperature. The mixture was refluxed for 16 h. Methanol was removed under reduced pressure, and the residue was purified by flash column chromatography (silica gel eluted with 25% EtOAc in hexanes) to give ent-steroid 4 (2.31 g, 99%): 1H NMR (400 MHz, CDCl3) δ 5.32–5.30 (m, 1H), 4.64 (s, 2H), 3.61 ( t, J = 8.6 Hz, 1H), 3.40–3.34 (m, 1H), 3.33 (s, 3H), 2.31–0.87 (m), 0.95 (s, 3H), 0.72 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 140.7, 121.3, 94.5, 81.6, 76.7, 55.0, 51.2, 50.2, 42.6, 39.4, 37.2, 36.7, 36.5, 31.8, 31.4, 30.3, 28.8, 23.3, 20.5, 19.3, 10.9.
ent-steroid 5
To a solution of ent-steroid 4 (1.5 g, 4.54 mmol) in CH2Cl2 (60 ml), Dess–Martin periodinane (2.5 g, 6 mmol) was added at room temperature. After 1 h, water (50 ml) was added, the product was extracted into CH2Cl2 (150 ml × 3), and the combined extracts were washed with brine (50 ml × 2). The organic layer was dried over Na2SO4 and filtered and the solvents were removed. The residue was purified by flash column chromatography (silica gel eluted with 10% EtOAc in hexanes) to give ent-steroid 5 (1.5 g, 100%): 1H NMR (400 MHz, CDCl3) δ 5.39–5.38 (m, 1H), 4.68 (s, 2H), 3.45–3.38 (m, 1H), 3.37 (s, 3H), 2.49–0.98 (m), 1.03 (s, 3H), 0.88 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 221.0, 140.9, 120.9, 94.7, 76.7, 55.1, 51.7, 50.2, 47.5, 39.5, 37.1, 36.8, 35.8, 31.4, 31.3, 30.8, 28.8, 21.8, 20.3, 19.3, 13.5.
ent-steroid 6
A solution of freshly prepared sodium ethoxide (sodium 0.4 g, 15 mmol dissolved in ethanol 15 ml) was added dropwise slowly to a solution of ent-steroid 5 (1.5 g, 4.54 mmol) and triethyl phosphonoacetate (3.44 g, 15 mmol) in anhydrous ethanol (25 ml) under N2 while stirring at 35–40°C. After addition, the reaction was refluxed for 16 h. After cooling to room temperature, the ethanol was removed and the residue was dissolved in ether, which was washed with water, dried over Na2SO4, and filtered. The solvent was removed, and the residue was purified by flash column chromatography (silica gel eluted with 10% EtOAc in hexanes) to give ent-steroid 6 (1.68 g, 87%): 1H NMR (400 MHz, CDCl3) δ 5.52 (s, 1H), 5.35–5.34 (m, 1H), 4.66 (s, 2H), 4.15–4.09 (m, 2H), 3.43–3.33 (m, 1H), 3.35 (s, 3H), 2.84–2.79 (m, 2H), 2.36–0.93 (m), 1.01 (s, 3H), 0.82 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 176.1, 167.3, 140.7, 121.3, 108.6, 94.6, 76.7, 59.4, 55.1, 53.7, 50.2, 46.0, 39.5, 37.2, 36.8, 35.1, 31.6, 31.5, 30.4, 28.8, 24.4, 20.9, 19.3, 18.2, 14.3.
The reaction sequence reported below that converts ent-steroid 6 into ent-steroid 16 (ent-VP1-001) is based on that reported previously for the preparation of the natural stereoisomer of ent-steroid 16 (67).
Unpurified ent-steroid 7
To a solution of ent-steroid 6 (1.4 g, 3.48 mmol) in EtOAc (150 ml), PtO2 (15 mg) was added at room temperature. Hydrogenation was carried out under 20 psi for 6 h. The solvent was removed, and the residue was purified by flash column chromatography (silica gel eluted with 10% EtOAc in hexanes) to give unpurified ent-steroid 7 (1.4 g, 100%): 1H NMR δ 4.63–4.60 (m, 1H), 4.08–4.03 (m, 2H), 3.48–3.32 (m, 1H), 3.31 (s, 3H), 2.34–0.57 (m), 0.76 (s, 3H), 0.54 (s, 3H); 13C NMR δ 176.1, 140.7, 121.3, 94.4, 76.2, 60.0, 55.3, 55.0, 54.5, 46.9, 44.9, 42.1, 37.4, 37.0, 35.6, 35.5, 35.3, 35.2, 32.1, 28.7, 28.1, 24.4, 20.9, 14.2, 12.5.
Unpurified ent-steroid 7 contains minor amounts of the ent-steroid in which the Δ5 double bond has been hydrogenated. This saturated ent-steroid could not be removed easily by chromatography on silica gel. To separate the two compounds chromatographically, ent-steroid 7 was converted first into ent-steroid 8 and then into ent-steroid 9, which is easily purified. ent-Steroid 9 was then converted back via ent-steroid 8 into ent-steroid 7 and then subsequently into ent-steroid 10.
Unpurified ent-steroid 8
Acetyl chloride (2 ml) was slowly added to unpurified hydrogenation product ent-steroid 7 (1.4 g, 3.48 mmol) in ethanol (30 ml) at room temperature. After 2 h, water was added and the product was extracted into CH2Cl2 (100 ml × 2). The combined extracts were dried over Na2SO4 and filtered, and the solvent was removed under reduced pressure. The residue was purified by flash column chromatography (silica gel eluted with 25% EtOAc in hexanes) to give unpurified ent-steroid 8 (1.2 g): 1H NMR (400 MHz, CDCl3) δ 5.35–5.34 (m, 1H), 4.13–4.07 (m, 2H), 3.55–3.47 (m, 1H), 2.38–0.81 (m), 1.10 (s, 3H), 0.61 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 173.9, 140.8, 121.5, 71.6, 60.1, 55.5, 50.3, 46.8, 42.2, 41.9, 37.3, 37.2, 36.5, 35.2, 31.9, 31.8, 31.6, 28.1, 24.5, 20.8, 19.4, 14.2, 12.4.
ent-steroid 9
To a solution of unpurified ent-steroid 8 (1.2 g, 3.33 mmol) in diethyl ether (100 ml) and acetic acid (5 ml), Br2 in HOAc (3 ml) was added slowly until brown color persisted. After 5 min, aqueous Na2S2O3 was added and the reaction became colorless. EtOAc (100 ml) was added, and the EtOAc solution was washed with aqueous NaHCO3 (50 ml × 2), brine (50 ml), and dried over anhydrous Na2SO4. After filtration, the solvent was removed under reduced pressure, and the residue was purified by flash column chromatography (silica gel eluted with 20% EtOAc in hexanes) to give ent-steroid 9 (1.4 g, 81%): 1H NMR (400 MHz, CDCl3) δ 4.82–4.81 (m, 1H), 4.44–4.37 (m, 1H), 4.12–4.06 (m, 2H), 2.72–1.08 (m), 1.43 (s, 3H), 0.62 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 173.8, 89.6, 68.9, 60.1, 56.0, 54.0, 47.6, 46.6, 45.6, 42.2, 42.0, 37.2, 37.0, 36.7, 35.2, 30.9, 30.1, 28.0, 24.2, 21.0, 20.3, 14.2, 12.7.
Purified ent-steroid 8
Zinc dust (6.0 g) was added to a solution of ent-steroid 9 (1.4 g, 2.7 mmol) in HOAc (20 ml) and EtOAc (30 ml) at room temperature. After 16 h, the mixture was filtered through Celite and washed with EtOAc (200 ml). The solvent was removed under reduced pressure, and the residue was purified by flash column chromatography (silica gel eluted with 25% EtOAc in hexanes) to give purified ent-steroid 8 (925 mg, 95%): 1H NMR (400 MHz, CDCl3) δ 5.26–5.25 (m, 1H), 4.06–4.01 (m, 2H), 3.85 (s, br, 1H), 3.47–3.40 (m, 1H), 2.31–0.73 (m), 0.93 (s, 3H), 0.54 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 173.8, 140.7, 121.1, 71.2, 60.0, 55.4, 50.1, 46.6, 41.9, 41.7, 37.1, 37.0, 36.3, 35.0, 31.7, 31.7, 31.2, 27.9, 24.3, 20.6, 19.2, 14.0, 12.2.
Purified ent-steroid 7
Purified ent-steroid 8 (925 mg, 2.57 mmol) was dissolved in CH2Cl2 (20 ml) and cooled to 0°C. (i-Pr)2EtN (1.3 ml, 7.5 mmol) and ClCH2OMe (0.45 ml, 6.0 mmol) were added, and the reaction was stirred at room temperature for 16 h. The reaction mixture was made basic by adding aqueous saturated NaHCO3 solution and the product extracted into CH2Cl2. The combined extracts were washed with brine, dried over anhydrous Na2SO4, and the solvent removed to give a viscous liquid that was purified by flash column chromatography (silica gel eluted with 20% EtOAc in hexanes) to give purified ent-steroid 7 as a colorless liquid (1.02 g, 98%): 1H NMR (400 MHz, CDCl3) δ 5.34–5.33 (m, 1H), 4.67 (s, 2H), 4.12 (q, J = 7.0 Hz, 2H), 3.42–3.36 (m, 1H), 3.35 (s, 3H), 2.37–0.80 (m), 1.00 (s, 3H), 0.60 (s, 3H); 13C NMR (CDCl3) δ 173.8, 140.7, 121.5, 94.6, 76.8, 60.0, 55.5, 55.1, 50.3, 46.7, 41.9, 39.5, 37.2, 37.1, 36.7, 35.2, 31.9, 31.8, 28.9, 28.1, 24.5, 20.7, 19.3, 14.2, 12.3.
ent-steroid 10
To a solution of the ent-steroid 7 (202 mg, 0.5 mmol) in tetrahydrofuran (THF) (10 ml), lithium diisopropylamide (0.75 ml, 2.0 M in THF, 1.5 mmol) and HMPA (0.29 ml, 1.65 mmol) were added at –78°C. After 1 h, 1-bromo-4-methylpentane (0.44 ml, 3 mmol) was added. After addition, the reaction was warmed to room temperature for 16 h. Aqueous NH4Cl was added and extracted with EtOAc (100 ml × 2), and the combined extracts were dried over anhydrous Na2SO4. The solvent was removed under reduced pressure, and the residue was purified by flash column chromatography (silica gel eluted with 20% EtOAc in hexanes) to give ent-steroid 10 (236 mg, 97%): 1H NMR (400 MHz, CDCl3) δ 5.34–5.33 (m, 1H), 4.67 (s, 2H), 4.13–4.08 (q, J = 7.4 Hz, 2H), 3.41–3.37 (m, 1H), 3.35 (s, 3H), 2.35–0.79 (m), 0.98 (s, 3H), 0.70 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 176.2, 140.7, 121.5, 94.6, 76.9, 59.6, 56.0, 55.1, 52.6, 50.1, 47.4, 41.9, 39.5, 38.8, 37.5, 37.2, 36.7, 32.2, 31.8, 31.7, 28.9, 27.8, 27.0, 25.0, 23.8, 22.7, 22.3, 20.8, 19.3, 14.2, 12.0.
ent-steroid 11
To a solution of ent-steroid 10 (236 mg, 0.5 mmol) in diethyl ether (20 ml), LiAlH4 (2.0 M in diethyl ether, 4.0 ml, 8.0 mmol) was added at room temperature. After 2 h, water (0.32 ml), 10% of NaOH (0.64 ml), and water (0.96 ml) were slowly added sequentially. After stirring for 30 min, the mixture was filtered through Celite and washed with CH2Cl2 (100 ml). The solvent was removed under reduced pressure and the residue was purified by flash column chromatography (silica gel eluted with 25% EtOAc in hexanes) to give ent-steroid 11 (212 mg, 98%): 1H NMR (400 MHz, CDCl3) δ 5.34–5.33 (m, 1H), 4.66 (s, 2H), 3.71–3.61 (m, 2H), 3.44–3.36 (m, 1H), 3.34 (s, 3H), 2.35–0.88 (m), 0.99 (s, 3H), 0.68 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 140.6, 121.6, 94.6, 76.7, 62.5, 56.6, 55.1, 50.3, 50.1, 42.3, 42.0, 39.5, 39.1, 37.2, 36.6, 31.8, 29.5, 28.8, 27.9, 27.5, 24.1, 24.0, 22.7, 22.5, 21.0, 19.3, 12.1.
ent-steroid 12
To a solution of ent-steroid 11 (212 mg, 0.48 mmol) in CH2Cl2 (10 ml), mesyl chloride (1 mmol, 0.08 ml) and Et3N (0.28 ml, 2 mmol) were added at 0°C. After 1 h, aqueous NH4Cl was added and the product was extracted into CH2Cl2 (100 ml × 2). The combined extracts were dried over anhydrous Na2SO4 and filtered and the solvents removed. The residue was purified by flash column chromatography (silica gel eluted with 10% EtOAc in hexanes) to give ent-steroid 12 (241 mg, 97%): 1H NMR (400 MHz, CDCl3) δ 5.33–5.32 (m, 1H), 4.66 (s, 2H), 4.36–4.32 (m, 1H), 4.18–4.09 (m, 1H), 3.42–3.37 (m, 1H), 3.34 (s, 3H), 2.97 (s, 3H), 2.34–0.89 (m), 0.98 (s, 3H), 0.69 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 140.6, 121.4, 94.6, 76.8, 70.0, 56.4, 55.1, 50.0, 49.9, 42.0, 39.7, 39.4, 39.2, 39.0, 37.2, 37.1, 36.6, 31.7, 31.6, 29.4, 28.8, 27.7, 27.4, 24.0, 23.4, 22.6, 22.4, 20.9, 19.3, 12.1.
ent-steroid 13
To a solution of ent-steroid 12 (241 mg, 0.46 mmol) in diethyl ether (30 ml), LiAlH4 (2.0 M in diethyl ether, 4.0 ml, 8.0 mmol) was added at room temperature. After 2 h, water (0.32 ml), 10% of NaOH (0.64 ml), and water (0.96 ml) were slowly added sequentially. After stirring for 30 min, the mixture was filtered through Celite and washed with CH2Cl2 (100 ml). The solvent was removed under reduced pressure and the residue was purified by flash column chromatography (silica gel eluted with 10% EtOAc in hexanes) to give ent-steroid 13 (188 mg, 95%): 1H NMR (400 MHz, CDCl3) δ 5.35–5.34 (m, 1H), 4.68 (s, 2H), 3.46–3.38 (m, 1H), 3.36 (s, 3H), 2.37–0.86 (m), 1.01 (s, 3H), 0.68 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 140.7, 121.7, 94.6, 76.9, 56.7, 56.1, 55.1, 50.1, 42.3, 39.8, 39.5, 39.4, 37.2, 36.7, 36.2, 35.8, 31.9, 31.8, 28.9, 28.2, 28.0, 24.3, 23.8, 22.8, 22.5, 21.0, 19.3, 18.7, 11.8.
ent-steroid 14 (ent-cholesterol)
To a solution of ent-steroid 13 (188 mg, 0.44 mmol) in THF (20 ml), 6 N HCl (10 ml) was added at room temperature. After 4 h, the product was extracted into CH2Cl2 (100 ml × 2) and the combined extracts were washed with aqueous NaHCO3 (50 ml × 2), dried over anhydrous Na2SO4, and filtered. The solvent was removed under reduced pressure, and the residue was purified by flash column chromatography (silica gel eluted with 20% EtOAc in hexanes) to give ent-steroid 14 (165 mg, 98%); 1H NMR (400 MHz, CDCl3) δ 5.36–5.35 (m, 1H), 3.57–3.49 (m, 1H), 2.33–0.86 (m), 1.01 (s, 3H), 0.68 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 140.7, 121.7, 71.8, 56.7, 56.1, 50.1, 42.3, 42.2, 39.8, 39.5, 37.2, 36.5, 36.2, 35.8, 31.9(2C), 31.6, 28.2, 28.0, 24.3, 23.8, 22.8, 22.6, 21.1, 19.4, 18.7, 11.8.
ent-steroid 15 (ent-4β-HC)
A procedure previously reported to convert cholesterol to 4β-HC was used (68) to convert ent-cholesterol 14 into ent-4β-HC 15.
To a solution of ent-cholesterol 14 (29 mg, 0.0747 mmol) in dioxane (5 ml) and water (2 drops), SeO2 (17 mg, 0.15 mmol) was added at room temperature. The mixture was heated to 90°C for 16 h. After cooling to room temperature, the solvent was removed under reduced pressure. The residue was purified by flash column chromatography (silica gel eluted with 30% EtOAc in hexanes) to give ent-4β-HC 15 (17 mg, 58%): mp 169–171°C; [α]D20 +41.7 (c = 0.12, CHCl3); 1H NMR (400 MHz, CDCl3) δ 5.69–5.68 (m, 1H), 4.15–4.14 (m, 1H), 3.58–3.55 (m, 1H), 2.20–0.78 (m), 1.19 (s, 3H), 0.69 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 142.7, 128.8, 77.3, 72.5, 56.9, 56.1, 50.2, 42.3, 39.7, 39.5, 36.9, 36.2, 36.0, 35.8, 32.1, 31.8, 28.2, 28.0, 25.4, 24.2, 23.8, 22.8, 22.5, 21.0, 20.5, 18.7, 11.8; IR (film, cm−1) 3406, 1455, 1366, 1072.
Supplemental data
This article contains supplemental data.
Conflict of interest
R. Z. is cofounder, scientific advisor, and stockholder with Frontier Medicines Corp. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank all members of the Zoncu Lab for helpful insights. The research reported in this publication was supported in part by the National Institutes of Health S10 program under award number 1S10RR026866-01.
Author contributions
O. M. and R. Z. conceived and designed the study; O. M. performed all experiments except for the following; P. J. H. Z. harvested and stained liver sections; C. B. extracted and analyzed lipids for DNL measurements by mass spectrometry; R. V. E. carried out the oxysterol screen; X. J. extracted, analyzed, and quantified oxysterols in serum and liver; M. Q. synthesized ent-4HC; O. M. and R. Z. wrote the article. D. S. O., D. F. C., D. K. N., A. S., and E. J. W. helped with data analysis and editing of the article.
Funding and additional information
This work was supported by National Institutes of Health R01GM127763 and R01GM130995 to R. Z., a National Niemann-Pick foundation postdoctoral fellowship to O. M., a National Institutes of Health R01 HL067773 to D. S. O. and D. F. C. The Taylor Family Institute for Innovative Psychiatric Research to D. F. C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Primary mouse hepatocytes were provided by core services of the UCSF Liver Center (grant no. P30 DK026743).
Supplemental data
References
- 1.Nohturfft A., Zhang S.C. Coordination of lipid metabolism in membrane biogenesis. Annu. Rev. Cell Dev. Biol. 2009;25:539–566. doi: 10.1146/annurev.cellbio.24.110707.175344. [DOI] [PubMed] [Google Scholar]
- 2.Thelen A.M., Zoncu R. Emerging roles for the lysosome in lipid metabolism. Trends Cell Biol. 2017;27:833–850. doi: 10.1016/j.tcb.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janowski B.A., Willy P.J., Devi T.R., Falck J.R., Mangelsdorf D.J. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 5.Kalaany N.Y., Mangelsdorf D.J. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- 6.Repa J.J., Liang G., Ou J., Bashmakov Y., Lobaccaro J.M., Shimomura I. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshikawa T., Shimano H., Amemiya-Kudo M., Yahagi N., Hasty A.H., Matsuzaka T. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costet P., Luo Y., Wang N., Tall A.R. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 2000;275:28240–28245. doi: 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]
- 9.Peet D.J., Turley S.D., Ma W., Janowski B.A., Lobaccaro J.M., Hammer R.E. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 10.Svensson S., Ostberg T., Jacobsson M., Norstrom C., Stefansson K., Hallen D. Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation. EMBO J. 2003;22:4625–4633. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong C., Bradley M.N., Rong X., Wang X., Wagner A., Grijalva V. LXRalpha is uniquely required for maximal reverse cholesterol transport and atheroprotection in ApoE-deficient mice. J. Lipid Res. 2012;53:1126–1133. doi: 10.1194/jlr.M022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calkin A.C., Tontonoz P. Liver x receptor signaling pathways and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2010;30:1513–1518. doi: 10.1161/ATVBAHA.109.191197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joseph S.B., McKilligin E., Pei L., Watson M.A., Collins A.R., Laffitte B.A. Synthetic LXR ligand inhibits the development of atherosclerosis in mice. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7604–7609. doi: 10.1073/pnas.112059299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terasaka N., Hiroshima A., Koieyama T., Ubukata N., Morikawa Y., Nakai D. T-0901317, a synthetic liver X receptor ligand, inhibits development of atherosclerosis in LDL receptor-deficient mice. FEBS Lett. 2003;536:6–11. doi: 10.1016/s0014-5793(02)03578-0. [DOI] [PubMed] [Google Scholar]
- 15.Azzout-Marniche D., Becard D., Guichard C., Foretz M., Ferre P., Foufelle F. Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem. J. 2000;350 Pt 2:389–393. [PMC free article] [PubMed] [Google Scholar]
- 16.Horton J.D., Goldstein J.L., Brown M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricoult S.J., Manning B.D. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 2013;14:242–251. doi: 10.1038/embor.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grefhorst A., Elzinga B.M., Voshol P.J., Plosch T., Kok T., Bloks V.W. Stimulation of lipogenesis by pharmacological activation of the liver X receptor leads to production of large, triglyceride-rich very low density lipoprotein particles. J. Biol. Chem. 2002;277:34182–34190. doi: 10.1074/jbc.M204887200. [DOI] [PubMed] [Google Scholar]
- 19.Schultz J.R., Tu H., Luk A., Repa J.J., Medina J.C., Li L. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai J., Zhang X.J., Li H. Progress and challenges in the prevention and control of nonalcoholic fatty liver disease. Med. Res. Rev. 2018 doi: 10.1002/med.21515. [DOI] [PubMed] [Google Scholar]
- 21.Mutemberezi V., Guillemot-Legris O., Muccioli G.G. Oxysterols: From cholesterol metabolites to key mediators. Prog. Lipid Res. 2016;64:152–169. doi: 10.1016/j.plipres.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Guillemot-Legris O., Mutemberezi V., Muccioli G.G. Oxysterols in metabolic syndrome: from bystander molecules to bioactive lipids. Trends Mol. Med. 2016;22:594–614. doi: 10.1016/j.molmed.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Poli G., Biasi F., Leonarduzzi G. Oxysterols in the pathogenesis of major chronic diseases. Redox Biol. 2013;1:125–130. doi: 10.1016/j.redox.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nury T., Zarrouk A., Vejux A., Doria M., Riedinger J.M., Delage-Mourroux R. Induction of oxiapoptophagy, a mixed mode of cell death associated with oxidative stress, apoptosis and autophagy, on 7-ketocholesterol-treated 158N murine oligodendrocytes: impairment by alpha-tocopherol. Biochem. Biophys. Res. Commun. 2014;446:714–719. doi: 10.1016/j.bbrc.2013.11.081. [DOI] [PubMed] [Google Scholar]
- 25.Janowski B.A., Grogan M.J., Jones S.A., Wisely G.B., Kliewer S.A., Corey E.J. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. U.S.A. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams C.M., Reitz J., De Brabander J.K., Feramisco J.D., Li L., Brown M.S. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 27.Radhakrishnan A., Ikeda Y., Kwon H.J., Brown M.S., Goldstein J.L. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nury T., Samadi M., Varin A., Lopez T., Zarrouk A., Boumhras M. Biological activities of the LXRalpha and beta agonist, 4beta-hydroxycholesterol, and of its isomer, 4alpha-hydroxycholesterol, on oligodendrocytes: effects on cell growth and viability, oxidative and inflammatory status. Biochimie. 2013;95:518–530. doi: 10.1016/j.biochi.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Ignatova I.D., Angdisen J., Moran E., Schulman I.G. Differential regulation of gene expression by LXRs in response to macrophage cholesterol loading. Mol. Endocrinol. 2013;27:1036–1047. doi: 10.1210/me.2013-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishimoto K., Nakamura H., Tachibana K., Yamasaki D., Ota A., Hirano K. Sterol-mediated regulation of human lipin 1 gene expression in hepatoblastoma cells. J. Biol. Chem. 2009;284:22195–22205. doi: 10.1074/jbc.M109.028753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimomura I., Shimano H., Korn B.S., Bashmakov Y., Horton J.D. Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J. Biol. Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]
- 32.Peng D., Hiipakka R.A., Xie J.T., Dai Q., Kokontis J.M., Reardon C.A. A novel potent synthetic steroidal liver X receptor agonist lowers plasma cholesterol and triglycerides and reduces atherosclerosis in LDLR(-/-) mice. Br. J. Pharmacol. 2011;162:1792–1804. doi: 10.1111/j.1476-5381.2011.01202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calvisi D.F., Wang C., Ho C., Ladu S., Lee S.A., Mattu S. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 2011;140:1071–1083. doi: 10.1053/j.gastro.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L., Che L., Tharp K.M., Park H.M., Pilo M.G., Cao D. Differential requirement for de novo lipogenesis in cholangiocarcinoma and hepatocellular carcinoma of mice and humans. Hepatology. 2016;63:1900–1913. doi: 10.1002/hep.28508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yabe D., Komuro R., Liang G., Goldstein J.L., Brown M.S. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yecies J.L., Zhang H.H., Menon S., Liu S., Yecies D., Lipovsky A.I. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen G., Liang G., Ou J., Goldstein J.L., Brown M.S. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foretz M., Pacot C., Dugail I., Lemarchand P., Guichard C., Le Liepvre X. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol. Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bodin K., Andersson U., Rystedt E., Ellis E., Norlin M., Pikuleva I. Metabolism of 4 beta -hydroxycholesterol in humans. J. Biol. Chem. 2002;277:31534–31540. doi: 10.1074/jbc.M201712200. [DOI] [PubMed] [Google Scholar]
- 40.Kasichayanula S., Boulton D.W., Luo W.L., Rodrigues A.D., Yang Z., Goodenough A. Validation of 4beta-hydroxycholesterol and evaluation of other endogenous biomarkers for the assessment of CYP3A activity in healthy subjects. Br. J. Clin. Pharmacol. 2014;78:1122–1134. doi: 10.1111/bcp.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herschlag D., Johnson F.B. Synergism in transcriptional activation: a kinetic view. Genes Dev. 1993;7:173–179. doi: 10.1101/gad.7.2.173. [DOI] [PubMed] [Google Scholar]
- 42.Salonurmi T., Nabil H., Ronkainen J., Hyotylainen T., Hautajarvi H., Savolainen M.J. 4beta-hydroxycholesterol signals from the liver to regulate peripheral cholesterol transporters. Front. Pharmacol. 2020;11:361. doi: 10.3389/fphar.2020.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim E.J., Kim B.H., Seo H.S., Lee Y.J., Kim H.H., Son H.H. Cholesterol-induced non-alcoholic fatty liver disease and atherosclerosis aggravated by systemic inflammation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serviddio G., Bellanti F., Villani R., Tamborra R., Zerbinati C., Blonda M. Effects of dietary fatty acids and cholesterol excess on liver injury: a lipidomic approach. Redox Biol. 2016;9:296–305. doi: 10.1016/j.redox.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimabukuro M., Okawa C., Yamada H., Yanagi S., Uematsu E., Sugasawa N. The pathophysiological role of oxidized cholesterols in epicardial fat accumulation and cardiac dysfunction: a study in swine fed a high caloric diet with an inhibitor of intestinal cholesterol absorption, ezetimibe. J. Nutr. Biochem. 2016;35:66–73. doi: 10.1016/j.jnutbio.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 46.Wooten J.S., Wu H., Raya J., Perrard X.D., Gaubatz J., Hoogeveen R.C. The influence of an obesogenic diet on oxysterol metabolism in C57BL/6J mice. Cholesterol. 2014;2014:843468. doi: 10.1155/2014/843468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guillemot-Legris O., Mutemberezi V., Cani P.D., Muccioli G.G. Obesity is associated with changes in oxysterol metabolism and levels in mice liver, hypothalamus, adipose tissue and plasma. Sci. Rep. 2016;6:19694. doi: 10.1038/srep19694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rong S., Cortes V.A., Rashid S., Anderson N.N., McDonald J.G., Liang G. Expression of SREBP-1c requires SREBP-2-mediated generation of a sterol ligand for LXR in livers of mice. Elife. 2017;6 doi: 10.7554/eLife.25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X., Li S., Wu J., Xia C., Lala D.S. Liver X receptors interact with corepressors to regulate gene expression. Mol. Endocrinol. 2003;17:1019–1026. doi: 10.1210/me.2002-0399. [DOI] [PubMed] [Google Scholar]
- 50.Huuskonen J., Fielding P.E., Fielding C.J. Role of p160 coactivator complex in the activation of liver X receptor. Arterioscler. Thromb. Vasc. Biol. 2004;24:703–708. doi: 10.1161/01.ATV.0000121202.72593.da. [DOI] [PubMed] [Google Scholar]
- 51.Oberkofler H., Schraml E., Krempler F., Patsch W. Potentiation of liver X receptor transcriptional activity by peroxisome-proliferator-activated receptor gamma co-activator 1 alpha. Biochem. J. 2003;371(Pt 1):89–96. doi: 10.1042/BJ20021665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner B.L., Valledor A.F., Shao G., Daige C.L., Bischoff E.D., Petrowski M. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol. Cell Biol. 2003;23:5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Castellani L.W., Sinal C.J., Gonzalez F.J., Edwards P.A. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18:157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bodin K., Bretillon L., Aden Y., Bertilsson L., Broome U., Einarsson C. Antiepileptic drugs increase plasma levels of 4beta-hydroxycholesterol in humans: evidence for involvement of cytochrome p450 3A4. J. Biol. Chem. 2001;276:38685–38689. doi: 10.1074/jbc.M105127200. [DOI] [PubMed] [Google Scholar]
- 55.Huang J.H., Zhang C., Zhang D.G., Li L., Chen X., Xu D.X. Rifampicin-induced hepatic lipid accumulation: association with up-regulation of peroxisome proliferator-activated receptor gamma in mouse liver. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J., Zhai Y., Mu Y., Gong H., Uppal H., Toma D. A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J. Biol. Chem. 2006;281:15013–15020. doi: 10.1074/jbc.M511116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He J., Gao J., Xu M., Ren S., Stefanovic-Racic M., O'Doherty R.M. PXR ablation alleviates diet-induced and genetic obesity and insulin resistance in mice. Diabetes. 2013;62:1876–1887. doi: 10.2337/db12-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chudnovskiy R., Thompson A., Tharp K., Hellerstein M., Napoli J.L., Stahl A. Consumption of clarified grapefruit juice ameliorates high-fat diet induced insulin resistance and weight gain in mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neuschwander-Tetri B.A., Caldwell S.H. Nonalcoholic steatohepatitis: summary of an AASLD Single Topic Conference. Hepatology. 2003;37:1202–1219. doi: 10.1053/jhep.2003.50193. [DOI] [PubMed] [Google Scholar]
- 60.Higuchi N., Kato M., Shundo Y., Tajiri H., Tanaka M., Yamashita N. Liver X receptor in cooperation with SREBP-1c is a major lipid synthesis regulator in nonalcoholic fatty liver disease. Hepatol. Res. 2008;38:1122–1129. doi: 10.1111/j.1872-034X.2008.00382.x. [DOI] [PubMed] [Google Scholar]
- 61.Ikegami T., Hyogo H., Honda A., Miyazaki T., Tokushige K., Hashimoto E. Increased serum liver X receptor ligand oxysterols in patients with non-alcoholic fatty liver disease. J. Gastroenterol. 2012;47:1257–1266. doi: 10.1007/s00535-012-0585-0. [DOI] [PubMed] [Google Scholar]
- 62.Goldstein J.L., Basu S.K., Brown M.S. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 63.Li S., Brown M.S., Goldstein J.L. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seglen P.O. Preparation of rat liver cells. I. Effect of Ca 2+ on enzymatic dispersion of isolated, perfused liver. Exp. Cell Res. 1972;74:450–454. doi: 10.1016/0014-4827(72)90400-4. [DOI] [PubMed] [Google Scholar]
- 65.Benjamin D.I., Li D.S., Lowe W., Heuer T., Kemble G., Nomura D.K. Diacylglycerol metabolism and signaling is a driving force underlying FASN inhibitor sensitivity in cancer cells. ACS Chem. Biol. 2015;10:1616–1623. doi: 10.1021/acschembio.5b00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Covey D.F. Ent-steroids chemistry and biology. Polish J. Chem. 2006;80:511–522. [Google Scholar]
- 67.Wicha J., Bal K. Synthesis of 21-hydroxycholesterol and 25-hydroxycholesterol from 3β-hydroxyandrost-5-en-17-one. A method for the stereospecific construction of sterol side-chains. J. C. S. Perkin I. 1978:1282–1288. [Google Scholar]
- 68.Nury T., Samadi M., Zarrouk A., Riedinger J.M., Lizard G. Improved synthesis and in vitro evaluation of the cytotoxic profile of oxysterols oxidized at C4 (4α- and 4β-hydroxycholesterol) and C7 (7-ketocholesterol, 7α- and 7β-hydroxycholesterol) on cells of the central nervous system. Eur. J. Med. Chem. 2013;70:558–567. doi: 10.1016/j.ejmech.2013.09.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.