Abstract
SLCO1B1 (solute carrier organic anion transporter family member 1B1) is an important transmembrane hepatic uptake transporter. Genetic variants in the SLCO1B1 gene have been associated with altered protein folding, resulting in protein degradation and decreased transporter activity. Next-generation sequencing (NGS) of pharmacogenes is being applied increasingly to associate variation in drug response with genetic sequence variants. However, it is difficult to link variants of unknown significance with functional phenotypes using “one-at-a-time” functional systems. Deep mutational scanning (DMS) using a “landing pad cell–based system” is a high-throughput technique designed to analyze hundreds of gene open reading frame (ORF) missense variants in a parallel and scalable fashion. We have applied DMS to analyze 137 missense variants in the SLCO1B1 ORF obtained from the Exome Aggregation Consortium project. ORFs containing these variants were fused to green fluorescent protein and were integrated into “landing pad” cells. Florescence-activated cell sorting was performed to separate the cells into four groups based on fluorescence readout indicating protein expression at the single cell level. NGS was then performed and SLCO1B1 variant frequencies were used to determine protein abundance. We found that six variants not previously characterized functionally displayed less than 25% and another 12 displayed approximately 50% of wild-type protein expression. These results were then functionally validated by transporter studies. Severely damaging variants identified by DMS may have clinical relevance for SLCO1B1-dependent drug transport, but we need to exercise caution since the relatively small number of severely damaging variants identified raise questions with regard to the application of DMS to intrinsic membrane proteins such as organic anion transporter protein 1B1.
Significance Statement
The functional implications of a large numbers of open reading frame (ORF) “variants of unknown significance” (VUS) in transporter genes have not been characterized. This study applied deep mutational scanning to determine the functional effects of VUS that have been observed in the ORF of SLCO1B1(solute carrier organic anion transporter family member 1B1). Several severely damaging variants were identified, studied, and validated. These observations have implications for both the application of deep mutational scanning to intrinsic membrane proteins and for the clinical effect of drugs and endogenous compounds transported by SLCO1B1.
Introduction
The SLCO1B1 (solute carrier organic anion transporter family member 1B1) gene encodes a transmembrane organic anion transporter protein 1B1 (OATP1B1) that transports endogenous compounds such as 17-β-glucuronosyl estradiol and bilirubin as well as drugs such as statins and certain oral antidiabetic agents (Kitamura et al., 2008; van de Steeg et al., 2013). Genetic polymorphisms in or near a transporter gene can result in large individual variation in transporter-facilitated drug uptake (Niemi, 2010; Oshiro et al., 2010). For example, the SLCO1B1*5 missense variant (rs4149056) is associated with decreased plasma clearance of statins such as simvastatin, which can result in statin-induced myopathy (Giacomini et al., 2013). This same variant has been associated with increased plasma concentrations of estrone conjugates (Dudenkov et al., 2017; Moyer et al., 2018). The mechanism for decreased function associated with SLCO1B1*5 may be related to alternation in its translocation to the cell membrane, as reported by previous studies (Kameyama et al., 2005; Voora et al., 2009). The Mayo Clinic recently completed the RIGHT 10K pharmacogenomic study during which next-generation sequencing (NGS) was performed using DNA from more than 10,000 Mayo Clinic Biobank participants to identify variants in 77 pharmacogenes, including SLCO1B1, to make it possible to study the clinical implications of pharmacogenomic variants in these genes (Bielinski et al., 2014, 2020). The Exome Aggregation Consortium based at the Broad Institute has aggregated exome sequencing data for 60,706 individuals of diverse ancestries (Lek et al., 2016). Most of the variants observed in these subjects were variants of unknown significance (VUS). Most VUS—unlike common pharmacogenomics variants—are less frequent or rare, so they will be observed only occasionally in clinical practice, but when they do occur, their consequences can be highly clinically relevant. Therefore, the application of high-throughput assays to begin the process of determining which variants might have functional implications represents a significant step forward in terms of practical clinical utility.
Deep mutational scanning (DMS) is a technique that provides a platform with which a large number of missense variants can be interrogated in parallel, making it much more efficient than conventional “one variant at a time” methods (Matreyek et al., 2017). We recently functionally characterized 230 CYP2C9 and CYP2C19 missense variants using a DMS landing pad system. During those studies we identified and functionally validated a series of severely damaging variants (Zhang et al., 2020). Fowler's group, pioneers in this field, and Yang's group have used this landing pad system to study the function of a series of important proteins such as TPMT (thiopurine S-methyltransferase), PTEN (phosphatase and tensin homolog), and NUDT15 (nudix hydrolase 15), all of which are primarily located in the cytosol (Matreyek et al., 2018; Suiter et al., 2020). Although the OATP1B1 transporter is an intrinsic membrane protein, one of the mechanisms that regulates transporter activity involves variation in protein expression as a result of lysosome-mediated or other mechanisms for protein degradation (Alam et al., 2016). We should also note the limited applicability of DMS for the study of missense variants leading to loss of function via other mechanisms such as variants that result in changes in subcellular localization or post-translational regulation.
In the present study, we set out to analyze the functional implications of missense variants that have been observed in the SLCO1B1 open reading frame (ORF). We analyzed 137 missense variants that have been observed in the ORF of this gene (Lek et al., 2016). Specifically, we included genetic variants with minor allele frequencies (MAFs) > 0.00001 as reported by the Exome Aggregation Consortium as well as novel ORF VUS observed by the Mayo RIGHT 10K project.
We found that 6 of the 137 SLCO1B1 missense variants that we studied displayed less than approximately 25% of wild-type (WT) protein expression, a level that might significantly decrease transporter activity. We also compared variant functional information determined by DMS with the predictions of computational algorithms, and, finally, we experimentally validated variants found to be severely damaging by the use of Western blot analysis and transport studies. Our findings indicate that DMS can be an efficient high-throughput method for the identification of low protein abundance ORF VUS that might have potential clinical implications for drug transport. However, they also suggest that caution will have to be exercised in the interpretation of this type of data for intrinsic membrane proteins like OATP1B1.
Materials and Methods
Generation of DMS Variant Library.
The landing pad cell line clone#20 with a single landing pad was previously generated to integrate SLCO1B1 expression cassettes, and SLCO1B1 promotorless cassettes were created by Gibson Assembly as previously described (Zhang et al., 2020). The attachment site on promoterless cassettes and the plasmid attachment site on landing pad clone#20 were integrated by using Bxb1 recombinase. Human SLCO1B1 ORF cDNA plasmids were obtained from Genscript (Piscataway, NJ). Nicking mutagenesis methods were modified from Wrenbeck et al. (2016) to construct variant libraries for ORFs containing SLCO1B1 missense variants. Phosphorylated oligonucleotides for SLCO1B1 variants were purchased from IDT (Coralville, IW). Sanger sequencing was used to validate sequences of the variant clones.
Cell Culture and Plasmid Transfection.
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, 100 μg/ml penicillin, and 0.1 mg/ml streptomycin. Long-term passage of the landing pad cell line used the medium described above with 2 µg/ml doxycycline (Sigma-Aldrich, St. Louis, MO). Doxycycline medium was removed 1 day before adding Bxb1 recombinase by transfection. The expression vector pCAG-NLS-HA-Bxb1 (#51271; Addgene) was used to express Bxb1 recombinase–mediated integration of variant libraries performed with plasmid DNA using 5 × 105 cells transfected with 3 µg of plasmid DNA using 6 µl of Fugene6 (Promega, Fitchburg, WI) in a six-well plate.
Fluorescence-Activated Cell Sorting.
The promoterless SLCO1B1 plasmids with attachment sites, as shown graphically in Fig. 1A, were transfected 24 hours after recombinase Bxb1 transfection into landing pad clone#20. The expression of blue fluorescent protein (BFP) in landing pad cells was inducted by doxycycline. After 5 days, candidate clones were trypsinized, washed with PBS, and fixed in 4% formaldehyde at 4°C for 10 minutes. The cells were analyzed by flow cytometer FACS CantoX (BD Biosciences, San Jose, CA) and by the use of FACSDiva version 8.0 software and FlowJo software version 10 (BD Biosciences). The FACS CantoX instrument utilizes colinear 405, 488, and 561 nm lasers plus forward and side angle light scatter. Library cells were washed, trypsinized, and resuspended in PBS containing 5% FBS. Cells were then sorted into four bins using a FACSAria with 407, 488, and 532 nm lasers (BD Biosciences), and the cells were collected in culture medium. BFP−/mCherry+ cells containing SLCO1B1 variants were flow sorted and grown for 5 days. BFP−/mCherry+ cells were sorted again to determine the protein expression of SLCO1B1 variants based on their GFP/mCherry ratios. Gates were set based on GFP/mCherry ratios for cells integrating known SLCO1B1 variants and WT proteins as gating references. Four gates were set to dissect the pooled libraries into four different bins based on GFP/mCherry ratios. The data were analyzed by FACSDiva version 8.0.1 software.
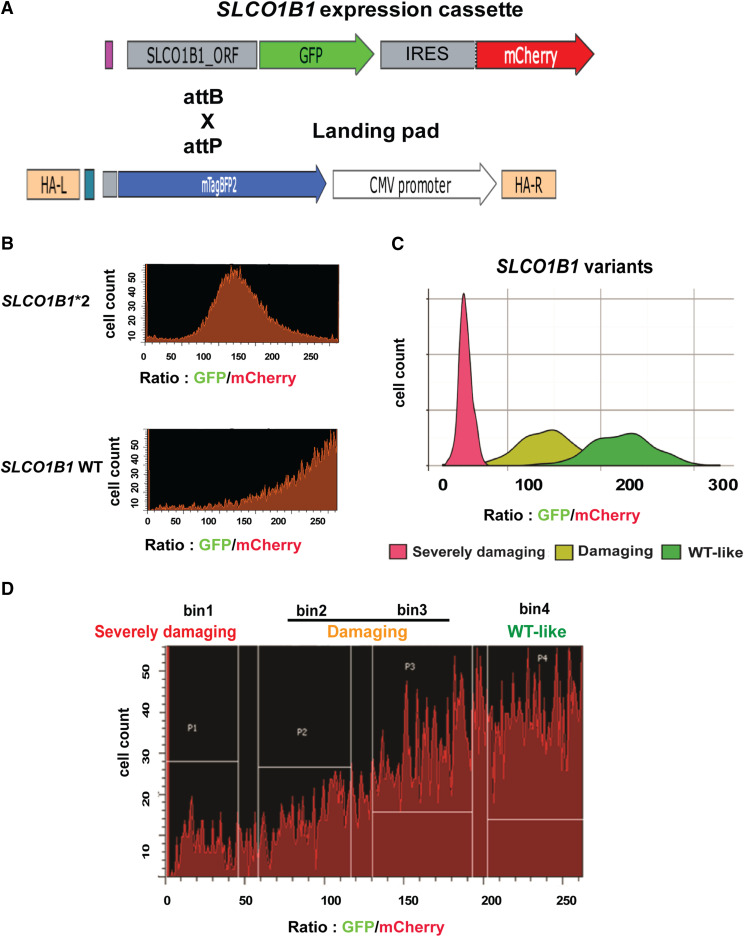
Fig. 1.
Flow cytometry of SLCO1B1 constructs with known variants and FACS of pooled SLCO1B1 variant libraries. (A) The SLCO1B1 expression cassette is depicted diagrammatically. When this vector is integrated into a “landing pad” in HEK293 cells, it results in the expression of recombinant protein that is labeled with GFP-labeled SLCO1B1, whereas the cell itself will express mCherry, so the ratio of GFP to mCherry serves as an indication of the stability of the expressed protein, i.e., the higher that ratio, the more stable the protein encoded by the expressed variants. The SLCO1B1 expression cassette was integrated into landing pad through attB and attP recombination. (B and C) Flow cytometry analysis of BFP−/mCherrry+ cells that had integrated wild-type or known damaging variant such as SLCO1B1*2. Note that for the WT protein, most of the cells eluted toward higher GFP/mCherry ratios, whereas cells containing damaging variants eluted at significantly lower GFP/mCherry ratios than did cells expressing the WT. Mean GFP/mCherry ratios for those variants were consistent with Western blot results obtained during our previous study. (D) Cells integrating SLCO1B1 pooled variant libraries were sorted into four bins based on their GFP/mCherry ratios. The variants were categorized into three groups: severely damaging variants fell into bin 1, damaging variants fell into bin 2 and bin 3, and tolerated variants fell into bin 4. Gates were set based on WT SLCO1B1 and SLCO1B1*2. Pools of sorted cells in each bin were collected and used as input material for subsequent amplicon DNA sequencing. HA-L, left homologous arm; HA-R, right homologous arm; IRES, internal ribosome entry site.
Sequencing Library Preparation and Sequencing.
Amplicons for SLCO1B1 were amplified from 250 ng genomic DNA using KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA). Primers were designed to bind to common nonmutated regions of the cassette sequences. Polymerase chain reaction products were purified by use of the QIAquick PCR Purification Kit (Qiagen, Germany) and were quantified by Qubit dsDNA HS Reagent (Fisher Scientific, Hampton, NH). The amplicon DNA (1 ng) was used as the starting material for library preparation by use of the Nextera XT DNA Preparation Kit (Illumina, San Diego, CA). Barcode adapters (Genewiz, South Plainfield, NJ) were used for library preparation, and samples were pooled after indexing and were sequenced using the Illumina HiSeq4000 Sequencing System in rapid run mode using the TruSeq Rapid SBS Kit (Illumina) with 300-cycle and 2 × 150 bp paired-end read capability. Files were aligned to the SLCO1B1 reference sequence.
Variant Calling.
The fastq files were aligned with the SLCO1B1 reference sequence using Burrows-Wheeler Aligner version 0.7.15. Samtools mpileup version 1.5 was used together with a custom Python script for single nucleotide variant calling. A base quality score cutoff of 20 and a mapping quality score cutoff of 20 were applied for single nucleotide variant calling. Custom scripts were used to summarize the data and add allele frequencies for each base at all positions in the reference sequence (Supplemental Script 1).
Western Blots.
BFP−/mCherry+ cells containing individual SLCO1B1 variants were lysed, and proteins were separated by SDS-PAGE prior to transfer to polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with rabbit polyclonal OATP1B1 antibody directed against a recombinant fragment corresponding to human OATP1B1 aa426-537. (cat. no. ab224610; Abcam) at a 1:1000 dilution. mCherry protein was measured using mouse monoclonal mCherry antibody at a 1:2000 dilution (cat. no. SAB2702291; Sigma), and its expression was used as a loading control. Proteins were detected using the SuperSignal West Dura Extended Duration Substrate (Thermo Scientific, Waltham, MA), and Western blot images were captured by use of the ChemiDoc Touch Image System (Bio-Rad, Hercules, CA).
Transporter Assay.
Radioactively labeled estradiol 17-β-d-glucuronide [Estradiol-6,7-3H(N)] 51.5 Ci/mmol (PerkinElmer, Boston, MA) was used to measure the uptake of this compound by SLCO1B1 transporter variants. Specifically, BFP−/mCherry+ cells were seeded at a density of 4 × 105 cells per well on 24-well plates and were grown to confluence for 24 hours (van de Steeg et al., 2013). Prior to the start of the experiment, cells were washed twice with prewarmed Hanks’ balanced salt solution (HBSS)/HEPES (pH 7.4) and were incubated with increasing concentrations of [3H]-labeled estradiol 17-β-d-glucuronide ranging from 1.5 to 48 nM for 1 minute. The highest concentration that we used was higher than the physiologic range, but this concentration range was used for the in vitro uptake study (Parvez et al., 2016). Uptake was terminated by washing the cells with 0.4 ml ice-cold HBSS/HEPES plus 0.5% bovine serum albumin and twice with 0.4 ml ice-cold HBSS/HEPES, followed by the addition of 200 µl M-PER buffer per well (Thermo Scientific). The cell lysate (150 μl) was transferred to a 5-ml plastic scintillation vial for the measurement of radioactivity by liquid scintillation counting (Beckman Coulter, Indianapolis, IN). Protein concentrations for each sample were measured using the Bradford method (BioRad). The amount of radioactively labeled estradiol 17-β-d-glucuronide that accumulated within the cells was determined by using liquid scintillation counting. The data were expressed in counts per minute (CPM) normalized by the protein content in milligrams.
Results
Generation of SLCO1B1 Variant Libraries.
We used the high-throughput DMS system to study the protein expression of 137 SLCO1B1 missense variants. The DMS system includes a landing pad cell line and promoterless SLCO1B1 cassettes. Landing pad cell line clone#20 with a single landing pad was used in these studies, as described in our previous publication (Zhang et al., 2020). Briefly, this landing pad cell line was generated using HEK293T cells, which have been reported to have hypotriploid karyotypes. Therefore, we screened different clones and found clone#20 with one copy of the landing pad, which enabled us to integrate a single SLCO1B1 variant per cell (Zhang et al., 2020). A promotorless SLCO1B1 cassette was constructed that included the SLCO1B1 ORF sequence and C terminus of the ORF was fused with GFP to indicate protein expression (Fig. 1A). mCherry was expressed after the internal ribosome entry site (IRES) component, which was used as a control for transfection. Once the SLCO1B1 ORF cassette landed on the landing pad by use of the Bxb1 recombinase, BFP in landing pad cells was disrupted and the BFP−/mCherry+ cells were collected for flow cytometry or fluorescence-activated cell sorting (FACS) analysis performed in the subsequent experiments. GFP/mCherry ratios were used as an indicator for SLCO1B1 protein expression in the DMS system. An earlier study (Tirona et al., 2001), using Western blotting alone had shown that the SLCO1B1*2 (rs56101265) variant allele affected final transporter protein quantity. For SLCO1B1*2, the mean GFP/mCherry ratio was 61.5% of WT GFP/mCherry ratio, in good agreement with the Western blot results of Tirona et al. (2001) (see Fig. 1B). These results were used as flow cytometry gating controls for subsequent experiments. Specifically, we used nicking mutagenesis to create 137 SLCO1B1 missense variants with a MAF higher than 0.001% from the Exome Aggregation Consortium and the Mayo Clinic RIGHT 10K project. The pooled SLCO1B1 variant expression cassettes were integrated into landing pad clone#20. As the next step, we used the known damaging SLCO1B1*2 variant together with the WT SLCO1B1 construct as references to establish FACS gating. Specifically, the SLCO1B1 variant libraries were sorted by FACS into four different “bins” based on the values of GFP/mCherry ratios, which was an indicator of the protein expression for each variant, i.e., the higher that ratio, the more was the protein abundance of the expressed SLCO1B1 variant (Fig. 1C). We used three categories of variant classification—“severely damaging” variants in bin 1, “damaging” variants in bins 2 and 3, or “tolerated” variants in bin 4—on the basis of flow cytometry validation (Fig. 1, C and D). The DMS system, as shown in Fig. 1, made it possible to determine the quantity of variant protein expressed for each of the variants encoded by constructs containing VUS.
Effect of SLCO1B1 Variants on Protein Levels.
Pools of BFP−/mCherry+ cells expressing SLCO1B1 missense variants were sorted by four-way FACS as shown in Fig. 1D. DNA was extracted from the cells collected in each bin and was then subjected to NGS amplicon sequencing. Variant frequencies for each variant in each bin were called by custom scripts (see Supplemental Script 1). Abundance scores for each SLCO1B1 individual variant were determined using the following equation in which Fv = variant frequency of the SLCO1B1 variant in each bin:
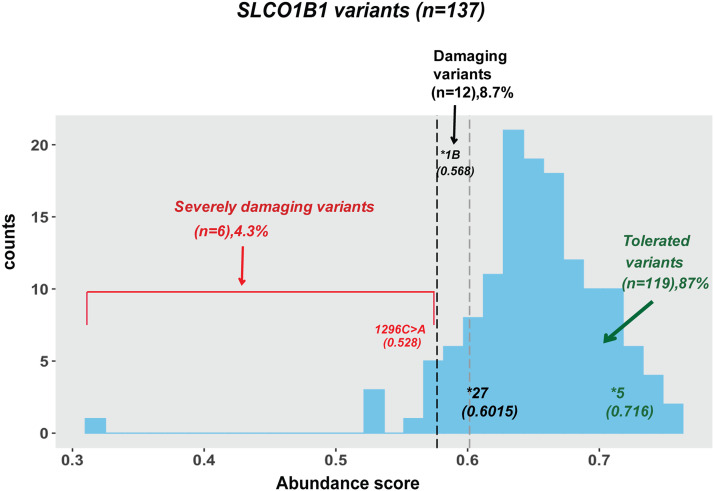
The “abundance score” for each variant was calculated by multiplying Fv with weighted values from 0.25 to 1 across the four bins, with the weighted values being assigned on the basis of the percentage of protein expression compared with WT (Fowler and Fields, 2014; Matreyek et al., 2017, 2018; Zhang et al., 2020). The mean abundance score for each individual variant was calculated based on at least three independent replicate assays. The abundance scores for SLCO1B1 variants shown graphically in Fig. 2 and in Supplemental Fig. 1 and Supplemental Table 1. “Severely damaging” variants fell into bin 1, “damaging” variants fell into bin 2 and bin 3, and “tolerated” variants fell into bin 4 on the basis of the flow cytometry results. Specifically, “severely damaging” SLCO1B1 variants had approximately 25% protein expression or less as compared with WT, with abundance scores of less than 0.5768 (SLCO1B1*1B 388A>G, rs2306283), whereas variants with abundance scores equal to or above that threshold but lower than 0.6015 (SLCO1B1*27 1200C>G, rs59113707) were considered as “damaging,” expressing approximately 50% of the OATP1B1 WT protein abundance. As a result, SLCO1B1 variants with abundance scores above 0.6015 were categorized as “tolerated” (Fig. 2). In summary, we performed FACS to separate the cells into four bins based on fluorescence readout. The amplicon sequencing of DNA in each bin, followed by computational analysis of variant frequencies in each bin, was then used to determine the level of OATP1B1 expression for constructs expression each VUS (Fig. 3). We observed six severely damaging SLCO1B1 variants (1462G>A, 1246G>A, 215G>A, 1508A>G 1828C>T, and 1296C>A) as determined by abundance scores calculated from variant frequencies.
Fig. 2.
Protein abundance scores for 137 SLCO1B1 variants. Variants having abundance scores less than or equal to 0.5728 SLCO1B1 (1296C>A) were classified as “severely damaging” variants, whereas variants having abundance scores equal to or above 0.5768 (SLCO1B1*1B, 388A>G, rs2306283) but less than 0.6015 (SLCO1B1*27, 1200C>G, rs59113707) were classified as “damaging.” Variants having abundance scores higher than 0.6015 were classified as “tolerated.” The results shown are averages abundance scores for four replicates. S.D. values are listed in Supplemental Table 1.
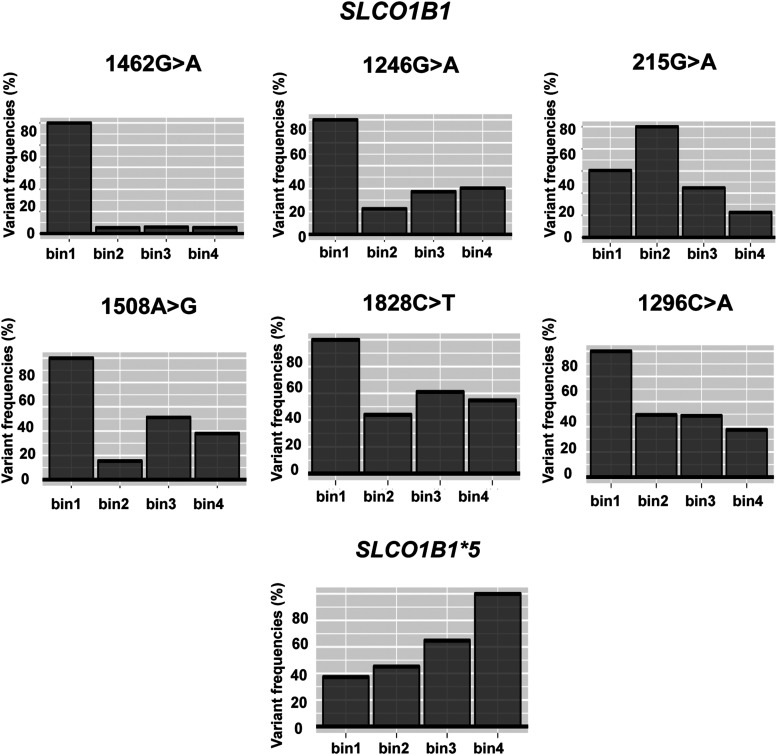
Fig. 3.
Variant frequencies by bin for the six newly identified “severely” damaging variants (1462G>A, 1246G>A, 215G>A, 1508A>G 1828C>T, and 1296C>A) for SLCO1B1, their distribution into each of the four bins, and similar data for the common SLCO1B1*5 allele.
Using DMS, the variant calling results for 137 SLCO1B1 variants (MAF > 0.00001) from the Exome Aggregation Consortium browser (currently the gnomAD database) and SLCO1B1 variants from the Mayo RIGHT 10K study are also listed in the order of classification of variants from DMS results in Table 1. In addition, we compared the DMS results with other prediction algorithms using SIFT (sorting intolerant from tolerant), Provean, Polyphen2, and CADD (combined annotation dependent depletion) and found two severely damaging variants (215G>A and 1296C>A) and eight damaging variants (388A>G, 1200C>G, 671T>A, 1015G>C, 235C>T, 38C>A, 991A>G, and 154A>G) that were identified by the DMS method that were missed by one of the four algorithms. Those results are listed in Supplemental Table 3. We also searched PharmVar, a database that includes, among other information, the possible impact of pharmacogenetic sequence variation on drug response, but that database does not include reports of the function of these variants (Gaedigk et al., 2018). One of the six severely damaging variants shown in Fig. 3, SLCO1B1 c.1296C>A, rs534931824, which had a MAF of 0.01%, might also provide clinically useful information.
TABLE 1.
Protein abundance scores of SLCO1B1 variants from ExAC Browser and Mayo Right 10K Study
| EXACT cDNA | EXACT Amino acid | RSID | Common Allele Name | Allele Frequency | Right 10K (Variant Prevalence) | DMS | |||
|---|---|---|---|---|---|---|---|---|---|
| WT | Heterozygous | Homozygous | Functional Study | Abundance Score | |||||
| c.1296C>A | p.Asn432Lys | rs534931824 | 0.000108 | Severely Damaging | 0.5728 | ||||
| c.1828C>T | p.Arg610Cys | rs748860610 | 5.77E-05 | 10082 | 2 | 0 | Severely Damaging | 0.5679 | |
| c.1246G>A | p.Val416Met | rs77468276 | 1.66E-05 | Severely Damaging | 0.5225 | ||||
| c.1462G>A | p.Gly488Ser | rs774471564 | 1.65E-05 | Severely Damaging | 0.3205 | ||||
| c.1508A>G | p.Asn503Ser | rs368423244 | 1.65E-05 | Severely Damaging | 0.5332 | ||||
| c.215G>A | p.Ser72Asn | rs780686282 | 8.77E-06 | Severely Damaging | 0.5322 | ||||
| c.388A>G | p.Asn130Asp | rs2306283 | SLCO1B1*1B | 0.4795 | 3525 | 4920 | 1639 | Damaging | 0.5768 |
| c.1200C>G | p.Phe400Leu | rs59113707 | SLCO1B1*27 | 0.004341 | Damaging | 0.6015 | |||
| c.169C>T | p.Arg57Trp | rs139257324 | SLCO1B1*33 | 0.000108 | Damaging | 0.5848 | |||
| c.671T>A | p.Phe224Tyr | rs756431817 | 7.42E-05 | Damaging | 0.5925 | ||||
| c.1015G>C | p.Val339Leu | rs758315826 | SLCO1B1*61 | 3.42E-05 | Damaging | 0.577 | |||
| c.235C>T | p.Leu79Phe | rs370130036 | 3.40E-05 | Damaging | 0.5867 | ||||
| c.695A>C | p.Lys232Thr | rs374328647 | 3.30E-05 | Damaging | 0.594 | ||||
| c.38C>A | p.Ala13Glu | rs778214174 | 2.49E-05 | Damaging | 0.5994 | ||||
| c.593A>G | p.Asp198Gly | rs376755211 | 2.47E-05 | Damaging | 0.5944 | ||||
| c.991A>G | p.Ser331Gly | rs774845200 | 1.79E-05 | Damaging | 0.5875 | ||||
| c.1796G>A | p.Cys599Tyr | rs531488136 | 1.65E-05 | Damaging | 0.5799 | ||||
| c.154A>G | p.Ile52Val | rs762874802 | 1.65E-05 | Damaging | 0.5984 | ||||
| c.521T>C | p.Val174Ala | rs4149056 | SLCO1B1*5 | 0.1294 | 7057 | 2768 | 259 | TOLERATED | 0.7167 |
| c.463C>A | p.Pro155Thr | rs11045819 | SLCO1B1*4 | 0.1166 | 7181 | 2656 | 247 | TOLERATED | 0.6718 |
| c.1929A>C | p.Leu643Phe | rs34671512 | SLCO1B1*19 | 0.04632 | 9074 | 986 | 24 | TOLERATED | 0.7172 |
| c.733A>G | p.Ile245Val | rs11045852 | SLCO1B1*24 | 0.007622 | TOLERATED | 0.6492 | |||
| c.633A>G | p.Ile211Met | rs201722521 | 0.004007 | 10074 | 9 | 1 | TOLERATED | 0.7314 | |
| c.1463G>C | p.Gly488Ala | rs59502379 | SLCO1B1*9 | 0.003196 | TOLERATED | 0.6747 | |||
| c.1495A>G | p.Ile499Val | rs74064213 | 0.002482 | TOLERATED | 0.6654 | ||||
| c.664A>G | p.Ile222Val | rs79135870 | SLCO1B1*29 | 0.00099 | TOLERATED | 0.7112 | |||
| c.317T>C | p.Ile106Thr | rs200227560 | 0.000693 | 10076 | 8 | 0 | TOLERATED | 0.6273 | |
| c.758G>A | p.Arg253Gln | rs11045853 | SLCO1B1*25 | 0.00042 | 10083 | 1 | 0 | TOLERATED | 0.6323 |
| c.170G>A | p.Arg57Gln | rs61760182 | 0.000356 | 10083 | 1 | 0 | TOLERATED | 0.7455 | |
| c.452A>G | p.Asn151Ser | rs2306282 | SLCO1B1 *16 | 0.000347 | TOLERATED | 0.6227 | |||
| c.1034C>T | p.Thr345Met | rs61760243 | 0.000253 | 10082 | 2 | 0 | TOLERATED | 0.6214 | |
| c.2032C>T | p.His678Tyr | rs200995543 | SLCO1B1*34 | 0.000249 | TOLERATED | 0.6821 | |||
| c.1309G>A | p.Gly437Arg | rs142965323 | SLCO1B1*26 | 0.0002 | 10078 | 6 | 0 | TOLERATED | 0.6296 |
| c.1622A>T | p.Gln541Leu | rs71581988 | 0.000132 | TOLERATED | 0.6445 | ||||
| c.2045C>T | p.Ser682Phe | rs140790673 | SLCO1B1*28 | 0.000108 | TOLERATED | 0.6382 | |||
| c.1007C>G | p.Pro336Arg | rs72559747 | 0.000104 | 10083 | 1 | 0 | TOLERATED | 0.634 | |
| c.1732G>A | p.Val578Ile | rs201001269 | 9.10E-05 | 10083 | 1 | 0 | TOLERATED | 0.6291 | |
| c.1322C>A | p.Thr441Asn | rs141779296 | 8.38E-05 | TOLERATED | 0.6661 | ||||
| c.1226A>G | p.Lys409Arg | rs199859384 | 8.32E-05 | TOLERATED | 0.7054 | ||||
| c.1373A>T | p.Tyr458Phe | rs750798503 | 7.44E-05 | TOLERATED | 0.687 | ||||
| c.601A>G | p.Lys201Glu | rs556914358 | 7.42E-05 | TOLERATED | 0.6046 | ||||
| c.518A>G | p.Tyr173Cys | rs141467543 | 7.42E-05 | TOLERATED | 0.6644 | ||||
| c.1213G>A | p.Val405Ile | rs376060151 | 6.67E-05 | TOLERATED | 0.6274 | ||||
| c.1865C>T | p.Ser622Leu | rs368052440 | 6.65E-05 | TOLERATED | 0.6501 | ||||
| c.638A>G | p.Asn213Ser | rs372477451 | 6.61E-05 | 10083 | 1 | 0 | TOLERATED | 0.6644 | |
| c.455G>C | p.Arg152Thr | rs145144129 | 5.79E-05 | TOLERATED | 0.7588 | ||||
| c.639T>A | p.Asn213Lys | rs752897663 | 5.78E-05 | TOLERATED | 0.6908 | ||||
| c.542G>A | p.Arg181His | rs142101690 | 5.77E-05 | TOLERATED | 0.7027 | ||||
| c.211G>A | p.Gly71Arg | rs373327528 | 5.18E-05 | 10081 | 3 | 0 | TOLERATED | 0.7462 | |
| c.1080C>G | p.Phe360Leu | rs140674443 | 4.99E-05 | TOLERATED | 0.6719 | ||||
| c.66A>T | p.Arg22Ser | rs142087529 | 4.99E-05 | TOLERATED | 0.71 | ||||
| c.410C>T | p.Ser137Leu | rs151204465 | 4.96E-05 | 10083 | 1 | 0 | TOLERATED | 0.659 | |
| c.152C>T | p.Ser51Phe | rs769900186 | 4.96E-05 | TOLERATED | 0.7129 | ||||
| c.577C>T | p.Leu193Phe | rs376996580 | 4.95E-05 | TOLERATED | 0.6597 | ||||
| c.1978G>C | p.Glu660Gln | rs368443740 | 4.20E-05 | TOLERATED | 0.6279 | ||||
| c.1178G>A | p.Gly393Glu | rs768154342 | 4.19E-05 | TOLERATED | 0.6814 | ||||
| c.380C>G | p.Thr127Ser | rs569028384 | SLCO1B1*33 | 4.14E-05 | 10083 | 1 | 0 | TOLERATED | 0.7218 |
| c.298G>A | p.Gly100Ser | rs144508550 | 4.13E-05 | TOLERATED | 0.6233 | ||||
| c.850A>G | p.Asn284Asp | rs779059572 | 4.12E-05 | TOLERATED | 0.651 | ||||
| c.508A>T | p.Met170Leu | rs764816711 | 4.12E-05 | TOLERATED | 0.6552 | ||||
| c.238G>T | p.Val80Leu | rs781021072 | 3.39E-05 | TOLERATED | 0.6433 | ||||
| c.1739G>A | p.Arg580Gln | rs763991908 | 3.31E-05 | TOLERATED | 0.6099 | ||||
| c.385A>G | p.Ile129Val | rs759691773 | 3.31E-05 | TOLERATED | 0.6807 | ||||
| c.1573C>T | p.Pro525Ser | rs71581987 | 3.30E-05 | TOLERATED | 0.6178 | ||||
| c.766G>A | p.Gly256Arg | rs754247932 | 3.30E-05 | TOLERATED | 0.6289 | ||||
| c.728G>A | p.Ser243Asn | rs558073276 | 3.30E-05 | TOLERATED | 0.6366 | ||||
| c.485G>A | p.Cys162Tyr | rs138374684 | 0.000033 | 10083 | 1 | 0 | TOLERATED | 0.6533 | |
| c.1829G>A | p.Arg610His | rs769518588 | 0.000033 | TOLERATED | 0.6645 | ||||
| c.743C>T | p.Thr248Ile | rs774398133 | 3.30E-05 | TOLERATED | 0.6676 | ||||
| c.703G>A | p.Val235Met | rs147421160 | 0.000033 | 10082 | 2 | 0 | TOLERATED | 0.6918 | |
| c.106C>T | p.Leu36Phe | rs751767004 | 3.30E-05 | TOLERATED | 0.7133 | ||||
| c.992G>A | p.Ser331Asn | rs760313969 | 2.68E-05 | 10082 | 2 | 0 | TOLERATED | 0.6478 | |
| c.212G>A | p.Gly71Glu | rs540723056 | 2.61E-05 | TOLERATED | 0.7123 | ||||
| c.250G>T | p.Val84Leu | rs750031541 | 2.52E-05 | TOLERATED | 0.6253 | ||||
| c.1878G>C | p.Leu626Phe | rs200526972 | 2.51E-05 | 10083 | 1 | 0 | TOLERATED | 0.6499 | |
| c.1087G>A | p.Val363Ile | rs764782382 | 2.49E-05 | 10083 | 1 | 0 | TOLERATED | 0.6104 | |
| c.1742C>T | p.Ala581Val | rs751309254 | 2.49E-05 | TOLERATED | 0.6202 | ||||
| c.944G>A | p.Gly315Glu | rs373619379 | 2.49E-05 | TOLERATED | 0.6837 | ||||
| c.904A>T | p.Asn302Tyr | rs770854976 | 2.48E-05 | TOLERATED | 0.6022 | ||||
| c.314G>T | p.Gly105Val | rs773434165 | 2.48E-05 | TOLERATED | 0.6312 | ||||
| c.629G>T | p.Gly210Val | rs766417954 | 2.48E-05 | TOLERATED | 0.6446 | ||||
| c.1671G>A | p.Met557Ile | rs770420484 | 2.48E-05 | TOLERATED | 0.6606 | ||||
| c.1441T>C | p.Tyr481His | rs745708956 | 2.48E-05 | 10083 | 1 | 0 | TOLERATED | 0.6722 | |
| c.1444A>G | p.Ile482Val | rs769428117 | 2.48E-05 | TOLERATED | 0.7065 | ||||
| c.1729A>G | p.Met577Val | rs371102023 | 2.48E-05 | TOLERATED | 0.7287 | ||||
| c.778C>T | p.Leu260Phe | rs756955511 | 2.47E-05 | TOLERATED | 0.6275 | ||||
| c.1793C>T | p.Thr598Met | rs201861991 | 2.47E-05 | 10082 | 2 | 0 | TOLERATED | 0.6285 | |
| c.598G>A | p.Ala200Thr | rs540112224 | 2.47E-05 | TOLERATED | 0.6502 | ||||
| c.541C>T | p.Arg181Cys | rs138965366 | 2.47E-05 | TOLERATED | 0.6519 | ||||
| c.1616C>T | p.Ala539Val | rs558485740 | SLCO1B1*46 | 2.47E-05 | TOLERATED | 0.6642 | |||
| c.875C>T | p.Ala292Val | rs778642823 | 2.47E-05 | TOLERATED | 0.6772 | ||||
| c.1784T>C | p.Ile595Thr | rs139026094 | 2.47E-05 | TOLERATED | 0.7203 | ||||
| c.1564G>T | p.Gly522Cys | rs112909948 | 2.47E-05 | TOLERATED | 0.7355 | ||||
| c.981G>T | p.Gln327His | 1.90E-05 | TOLERATED | 0.6342 | |||||
| c.986T>G | p.Phe329Cys | rs764497327 | 1.84E-05 | TOLERATED | 0.6703 | ||||
| c.1966A>G | p.Ile656Val | rs757219127 | 1.69E-05 | TOLERATED | 0.635 | ||||
| c.1159G>A | p.Ala387Thr | rs775082787 | 1.69E-05 | TOLERATED | 0.6456 | ||||
| c.1319T>G | p.Met440Arg | rs139797371 | SLCO1B1*43 | 1.68E-05 | 10082 | 2 | 0 | TOLERATED | 0.682 |
| c.1298A>G | p.Lys433Arg | rs772057264 | 1.67E-05 | TOLERATED | 0.639 | ||||
| c.193C>G | p.Leu65Val | rs766895771 | 1.67E-05 | 10082 | 2 | 0 | TOLERATED | 0.6759 | |
| c.1214T>C | p.Val405Ala | 1.67E-05 | TOLERATED | 0.6979 | |||||
| c.1100A>G | p.Tyr367Cys | rs757036708 | 1.66E-05 | TOLERATED | 0.6216 | ||||
| c.481G>A | p.Gly161Ser | rs749356996 | 1.66E-05 | TOLERATED | 0.6364 | ||||
| c.1076T>C | p.Val359Ala | rs147750118 | 1.66E-05 | TOLERATED | 0.6436 | ||||
| c.47C>T | p.Ser16Leu | rs753618172 | 1.66E-05 | TOLERATED | 0.7199 | ||||
| c.128T>C | p.Leu43Pro | rs770472561 | 1.65E-05 | TOLERATED | 0.6088 | ||||
| c.1729A>C | p.Met577Leu | rs371102023 | 1.65E-05 | TOLERATED | 0.6142 | ||||
| c.1628T>G | p.Leu543Trp | rs72661137 | 1.65E-05 | TOLERATED | 0.6165 | ||||
| c.1765A>G | p.Ile589Val | rs779674373 | 1.65E-05 | TOLERATED | 0.6209 | ||||
| c.529G>C | p.Gly177Arg | rs750234871 | 1.65E-05 | TOLERATED | 0.6362 | ||||
| c.395C>T | p.Ser132Leu | rs763429608 | 1.65E-05 | TOLERATED | 0.6368 | ||||
| c.560C>T | p.Pro187Leu | rs779195754 | 1.65E-05 | TOLERATED | 0.641 | ||||
| c.1384G>A | p.Asp462Asn | rs778655808 | 1.65E-05 | TOLERATED | 0.6418 | ||||
| c.674C>T | p.Thr225Ile | rs370943869 | 1.65E-05 | TOLERATED | 0.6468 | ||||
| c.1784T>G | p.Ile595Ser | rs139026094 | 1.65E-05 | TOLERATED | 0.6508 | ||||
| c.808A>C | p.Ile270Leu | rs201438350 | 1.65E-05 | TOLERATED | 0.6511 | ||||
| c.331A>C | p.Thr111Pro | rs759510840 | 1.65E-05 | TOLERATED | 0.6515 | ||||
| c.1837T>C | p.Cys613Arg | rs377350683 | SLCO1B1*30 | 1.65E-05 | TOLERATED | 0.6571 | |||
| c.1778C>G | p.Ala593Gly | rs768644633 | 1.65E-05 | TOLERATED | 0.6588 | ||||
| c.763G>C | p.Val255Leu | rs766769140 | 1.65E-05 | 10083 | 1 | 0 | TOLERATED | 0.6662 | |
| c.145A>G | p.Lys49Glu | rs745339838 | 1.65E-05 | TOLERATED | 0.6715 | ||||
| c.1856C>T | p.Thr619Ile | rs760486881 | 1.65E-05 | TOLERATED | 0.6726 | ||||
| c.1430A>G | p.Asn477Ser | rs781211732 | 1.65E-05 | TOLERATED | 0.6797 | ||||
| c.1664A>G | p.His555Arg | rs781111529 | 1.65E-05 | TOLERATED | 0.6803 | ||||
| c.133G>A | p.Ala45Thr | rs555367334 | 1.65E-05 | 10083 | 1 | 0 | TOLERATED | 0.681 | |
| c.1781T>C | p.Leu594Pro | rs761720319 | 1.65E-05 | TOLERATED | 0.6918 | ||||
| c.527T>C | p.Met176Thr | rs548326440 | 1.65E-05 | TOLERATED | 0.6921 | ||||
| c.1805G>T | p.Trp602Leu | rs778178385 | 1.65E-05 | 10082 | 2 | 0 | TOLERATED | 0.6926 | |
| c.1589G>A | p.Cys530Tyr | rs184762532 | 1.65E-05 | TOLERATED | 0.6941 | ||||
| c.1451C>A | p.Pro484His | rs568944276 | 1.65E-05 | TOLERATED | 0.6949 | ||||
| c.610C>T | p.His204Tyr | rs767379248 | 1.65E-05 | TOLERATED | 0.6953 | ||||
| c.713G>A | p.Gly238Glu | rs374113543 | 1.65E-05 | 10081 | 3 | 0 | TOLERATED | 0.7046 | |
| c.1414C>T | p.Pro472Ser | rs746507861 | 1.65E-05 | TOLERATED | 0.7267 | ||||
| c.1612G>A | p.Val538Ile | rs760163504 | 1.65E-05 | TOLERATED | 0.7384 | ||||
| c.1465T>A | p.Cys489Ser | rs144733213 | 1.65E-05 | TOLERATED | 0.7535 | ||||
| c.222A>T | p.Glu74Asp | rs745392993 | 9.18E-06 | TOLERATED | 0.6631 | ||||
| c.1000A>T | p.Thr334Ser | rs77871475 | 8.75E-06 | TOLERATED | 0.6152 | ||||
Functional Validation of SLCO1B1 Severely Damaging Variants.
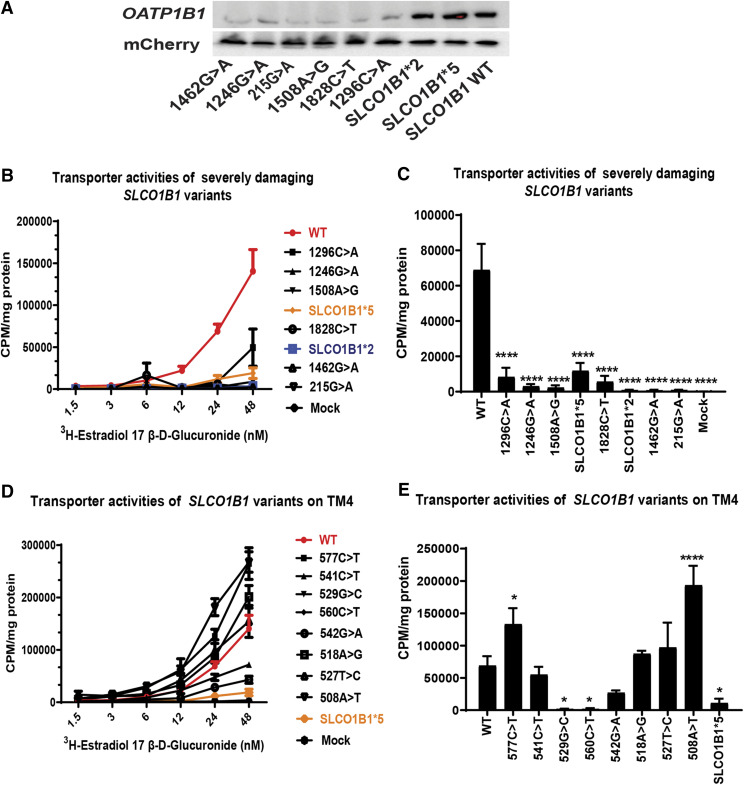
We next attempted to confirm our results for the severely damaging variants that we identified by DMS by the use of functional studies. We validated the protein expression data for these newly identified severely damaging variants (1462G>A, 1246G>A, 215G>A, 1508A>G, 1828C>T, and 1296C>A) by applying Western blot analyses. The results are shown in Fig. 4A. The six variants for SLCO1B1 predicted to be severely damaging displayed less than 25% protein expression when compared with the OATP1B1 WT protein. SLCO1B1*2 and SLCO1B1*5 were also studied as comparators. Finally, we performed transporter assays to determine the transporter activity of these newly identified severely damaging variants. Transport by the severely damaging variants was significantly decreased when compared with the WT protein as measured by the uptake of radioactive 17-estradiol β-d-glucuronide, a prototypic substrate for transport by SLCO1B1. The concentration-dependent 17-estradiol β-d-glucuronide uptake by severely damaging variants and WT OATP1B1 protein is shown graphically in Fig. 4B. All six newly identified functional SLCO1B1 variants revealed significantly lower transporter activities, as shown in Fig. 4B and by the bar graph in Fig. 4C, which depicts the level of reduction in transport at optimal concentrations of radioactive 17-estradiol β-d-glucuronide. Protein degradation of variants represents a common mechanism by which missense variants can alter protein abundances and, as a result, transport function. However, there are also examples in which alterations in transport are clearly not related to variation in transporter protein quantity. For example, SLCO1B1*5 displays WT-like protein abundance but is associated with decreased transporter activity. The mechanism for decreased function associated with SLCO1B1*5 may be related to alternation in its translocation to the cell membrane as reported previously (Kameyama et al., 2005; Voora et al., 2009). The amino acid changed by the *5 variant maps to SLCO1B1 transmembrane domain 4 (TM4), so we also studied transport of a prototypic SLCO1B1 substrate by eight additional variants that we studied that mapped to the same transmembrane domain. We found that, of the eight variants with WT-like abundance scores, six displayed normal or even elevated transport, but two (SLCO1B1 529G>C and 560C>T) displayed relatively decreased transporter capacity, as shown in Fig. 4D and by the bar graph in Fig. 4E, which depict the transporter activities at 24 nM radioactive 17-estradiol β-d-glucuronide. These observations suggest that these additional two variants in TM4 may also display impaired transport just as does SLCO1B1*5. Furthermore, two variants (SLCO1B1 508A>T and 577C>T) showed significantly increased activity as compared to WT, tested statistically by one-way ANOVA P < 0.05, as shown in Fig. 4E.
Fig. 4.
Validation of SLCO1B1 variants identified as containing severely damaging variants. (A) Western blot validation of SLCO1B1 variants identified as containing severely damaging variants. The protein expression of SLCO1B1 in BFP−/mCherry+ cells integrating severely damaging variants were validated by Western blot analysis. mCherry was used as a loading control. A control lane contained WT SLCO1B1. (B) Concentration-dependent uptake of estradiol 17-β-d-glucuronide by SLCO1B1 WT BFP−/mCherry+ cells and the six newly identified severely damaging SLCO1B1 variant BFP−/mCherry+ cells after 1-minute incubations. The quantity of radioactively labeled estradiol 17-β-d-glucuronide that accumulated within the cells was determined by liquid scintillation counting. The data are expressed in CPM normalized by the amount of protein content in milligrams. Data are presented as mean uptake for three replicate experiments. (C) The bar graph shows the uptake of estradiol 17-β-d-glucuronide (24 nM) for variants in SLCO1B1 TM4 in BFP−/mCherry+ cells after 1-minute incubations. The uptake activities of variants in severely damaging variants against WT were tested by one-way ANOVA; ****P < 0.0001. (D) Concentration-dependent uptake of estradiol 17-β-d-glucuronide for variants in SLCO1B1 TM4 in BFP−/mCherry+ cells after 1-minute incubations. Data are presented as means ± S.D. of CPM per mg protein for three replicated experiments. (E) The bar graph shows the uptake of estradiol 17- β-d-glucuronide (24 nM) for variants in SLCO1B1 TM4 in BFP−/mCherry+ cells after 1-minute incubations. The uptake activities of variants in TM4 against WT were tested by one-way ANOVA; *P < 0.05; ****P < 0.0001.
Discussion
There have been functional studies of a limited number of clinically relevant SLCO1B1 drug transporter variants which have applied “one-at-a-time” systems that are labor intensive and require time-consuming assays. In this study, we have used the DMS landing pad platform to functionally characterize naturally occurring ORF missense variants for SLCO1B1 in a high-throughput fashion (Fowler and Fields, 2014; Matreyek et al., 2017, 2018; Zhang et al., 2020). The landing pad cell line clone#20 with a single landing pad was used to screen variant protein expression in a high-throughput manner (Zhang et al., 2020). Missense variants in SLCO1B1 may result in altered protein expression as a result of proteasome- or lysosome-mediated degradation, a major mechanism responsible for decreased protein expression for pharmacogenomic variants (Wang et al., 2004; Alam et al., 2016; Matreyek et al., 2018; Suiter et al., 2020; Zhang et al., 2020). Loss of function by variants containing nonsynonymous SLCO1B1 ORF single nucleotide polymorphisms due to decreased protein expression made it possible for us to analyze that function by the use of fluorescence reporter assays. FACS was used to separate variants associated with differing protein expression levels, all of which were subsequently identified by NGS to make it possible to calculate the frequency of each of the variants. We chose to study focused variant libraries, that is, libraries that included variants above a specified level of natural occurrence rather than using saturation mutant libraries for SLCO1B1 missense variants. Specifically, we analyzed 137 nonsynonymous ORF variants for SLCO1B1 from the Exome Aggregation Consortium study that had MAF > 0.00001 (see Fig. 2) (Lek et al., 2016). We validated the transporter activities for severely damaging variants, and those results were in good agreement with protein expression levels, as shown in Fig. 4A. The crystal structure of SLCO1B1 has not yet been reported, but 12 transmembrane domains have been identified in OATP1B1 transporter sequences (Hong et al., 2010). Four of six newly identified severely damaging variants (1462G>A, 1508A>G, 1828C>T, 1296C>A) were located in extracellular domains and two variants (1246G>A, 215G>A) were located in transmembrane domains. In silico predictions with regard to how damaging individual variants might be were not always consistent with our DMS results, as shown in Supplemental Table 3, and previous publication suggested that decreased protein expression of SLCO1B1 variants is only one of the mechanisms that can result in impaired function (Kameyama et al., 2005). Obviously, proteins that include SLCO1B1 nonsynonymous variants can display WT-like protein abundance joined with decreased transporter activity. That fact is emphasized in dramatic fashion by SLCO1B1 *5, which displayed significantly reduced transporter activity, together with a protein level similar to that of WT SLCO1B1. The list of variants included in the study included eight variants that mapped to gene sequence encoding TM4, the domain that includes SLCO1B1*5. Most of those TM4 variants displayed WT-like or higher levels of transport, but two of the eight showed decreased transport (see Fig. 4C). One possible limitation of the use of DMS to study OATP1B1 and other intrinsic membrane proteins might be related to the fact that mechanisms for loss of function or decreased activity for these proteins may be missed by the type of assay which we applied—i.e., protein expression. In silico predictions have been widely applied to predict variation in protein function that has implications for pharmacogenomics and other aspects of drug effect (Flanagan et al., 2010; Kircher et al., 2014; Choi and Chan, 2015; Vaser et al., 2016). Our own previous work and that of others supports the importance of the application of a variety of functional methods to validate results obtained by using predictive algorithms. Therefore, we compared calling variant function by the use of DMS with the predictions of computational algorithms, and significant differences were found between our results and those of predictive algorithms, differences which may be due to underlying molecular mechanisms responsible for SLOC1B1 decreased function, as listed in Supplemental Table 3.
Based on our results and the experience of other groups, DMS appears to be a useful and sensitive method for the study of cytosolic proteins such as TPMT (thiopurine S-methyltransferase), PTEN (phosphatase and tensin homolog), and NUDT15 (nudix hydrolase 15) and of endoplasmic reticulum proteins such as CYP2C9 and CYP2C19, for which a major mechanism of loss of function is protein degradation in which case damaging variants would be expected to display clear fluorescence separation from WT-like variants (Wang et al., 2005; Li et al., 2008; Matreyek et al., 2018; Devarajan et al., 2019; Suiter et al., 2020). The functional implications of genetic variation that alters amino acid sequence in the SLCO1B1 gene is clearly a complex process involving multiple mechanisms, which could include changes in plasma membrane localization and integration, protein degradation, and transcriptional and post-translational variation (Alam et al., 2016, 2018). For intrinsic transmembrane proteins like OATP1B1, DMS may be one of a series of methods that will be needed to predict alterations in SLCO1B1 function.
In summary, we have identified and validated six SLCO1B1 severely damaging variants that had not previously been reported in PharmVar. Those variants are potentially actionable clinically if they can be linked to individual variation in drug response phenotypes or disease pathophysiology. Functional studies of the variants that we found to display decreased protein expression supported the functional consequences predicted by DMS.
Abbreviations
- BFP
blue fluorescent protein
- CPM
counts per minute
- DMS
deep mutational scanning
- FACS
fluorescence-activated cell sorting
- HBSS
Hanks’ balanced salt solution
- MAF
minor allele frequency
- NGS
next-generation sequencing
- OATP1B1
organic anion transporter protein 1B1
- ORF
open reading frame
- TM4
transmembrane domain 4
- VUS
variants of unknown significance
- WT
wild type
Authorship Contributions
Participated in research design: Zhang, Ho, Wang, Weinshilboum.
Conducted experiments: Zhang, Moon.
Performed data analysis: Zhang, Sarangi, Kalari.
Contributed to the writing of the manuscript: Zhang, Ho, Weinshilboum.
Note Added in Proof:Table 1 was accidentally listed as Figure 5 in the Fast Forward version that appeared online March 3, 2021. Table 1 has now been correctly listed.
Footnotes
This study was funded by National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) [Grant U19-GM61388] (The Pharmacogenomics Research Network), NIGMS [Grant R01-GM28157]; NIH National Institute on Alcohol Abuse and Alcoholism (NIAAA) [Grant R01-AA27486] and [Grant K01-AA2850], NIGMS [Grant R01-GM125633], and the Mayo Clinic Center for Individualized Medicine.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Alam K, Crowe A, Wang X, Zhang P, Ding K, Li L, Yue W (2018) Regulation of organic anion transporting polypeptides (OATP) 1B1- and OATP1B3-mediated transport: an updated review in the context of OATP-mediated drug-drug interactions. Int J Mol Sci 19:855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam K, Pahwa S, Wang X, Zhang P, Ding K, Abuznait AH, Li L, Yue W (2016) Downregulation of organic anion transporting polypeptide (OATP) 1B1 transport function by lysosomotropic drug chloroquine: implication in OATP-mediated drug-drug interactions. Mol Pharm 13:839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, Ryu E, Targonski PV, Van Norstrand MD, Hathcock MA, et al. (2014) Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 89:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinski SJ, St Sauver JL, Olson JE, Larson NB, Black JL, Scherer SE, Bernard ME, Boerwinkle E, Borah BJ, Caraballo PJ, et al. (2020) Cohort profile: the right drug, right dose, right time: using genomic data to individualize treatment protocol (RIGHT protocol). Int J Epidemiol 49:23–24k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan S, Moon I, Ho MF, Larson NB, Neavin DR, Moyer AM, Black JL, Bielinski SJ, Scherer SE, Wang L, et al. (2019) Pharmacogenomic next-generation DNA sequencing: lessons from the identification and functional characterization of variants of unknown significance in CYP2C9 and CYP2C19. Drug Metab Dispos 47:425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudenkov TM, Ingle JN, Buzdar AU, Robson ME, Kubo M, Ibrahim-Zada I, Batzler A, Jenkins GD, Pietrzak TL, Carlson EE, et al. (2017) SLCO1B1 polymorphisms and plasma estrone conjugates in postmenopausal women with ER+ breast cancer: genome-wide association studies of the estrone pathway. Breast Cancer Res Treat 164:189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SE, Patch AM, Ellard S (2010) Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers 14:533–537. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Fields S (2014) Deep mutational scanning: a new style of protein science. Nat Methods 11:801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE, PharmVar Steering Committee (2018) The Pharmacogene Variation (PharmVar) Consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin Pharmacol Ther 103:399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini KM, Balimane PV, Cho SK, Eadon M, Edeki T, Hillgren KM, Huang SM, Sugiyama Y, Weitz D, Wen Y, et al. International Transporter Consortium (2013) International Transporter Consortium commentary on clinically important transporter polymorphisms. Clin Pharmacol Ther 94:23–26. [DOI] [PubMed] [Google Scholar]
- Hong M, Li S, Zhou F, Thomas PE, You G (2010) Putative transmembrane domain 12 of the human organic anion transporter hOAT1 determines transporter stability and maturation efficiency. J Pharmacol Exp Ther 332:650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K (2005) Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics 15:513–522. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S, Maeda K, Wang Y, Sugiyama Y (2008) Involvement of multiple transporters in the hepatobiliary transport of rosuvastatin. Drug Metab Dispos 36:2014–2023. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Exome Aggregation Consortium (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang L, Burgess RJ, Weinshilboum RM (2008) Thiopurine S-methyltransferase pharmacogenetics: autophagy as a mechanism for variant allozyme degradation. Pharmacogenet Genomics 18:1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek KA, Starita LM, Stephany JJ, Martin B, Chiasson MA, Gray VE, Kircher M, Khechaduri A, Dines JN, Hause RJ, et al. (2018) Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat Genet 50:874–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek KA, Stephany JJ, Fowler DM (2017) A platform for functional assessment of large variant libraries in mammalian cells. Nucleic Acids Res 45:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer AM, de Andrade M, Faubion SS, Kapoor E, Dudenkov T, Weinshilboum RM, Miller VM (2018) SLCO1B1 genetic variation and hormone therapy in menopausal women. Menopause 25:877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi M (2010) Transporter pharmacogenetics and statin toxicity. Clin Pharmacol Ther 87:130–133. [DOI] [PubMed] [Google Scholar]
- Oshiro C, Mangravite L, Klein T, Altman R (2010) PharmGKB very important pharmacogene: SLCO1B1. Pharmacogenet Genomics 20:211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez MM, Jung JA, Shin HJ, Kim DH, Shin JG (2016) Characterization of 22 antituberculosis drugs for inhibitory interaction potential on organic anionic transporter polypeptide (OATP)-mediated uptake. Antimicrob Agents Chemother 60:3096–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suiter CC, Moriyama T, Matreyek KA, Yang W, Scaletti ER, Nishii R, Yang W, Hoshitsuki K, Singh M, Trehan A, et al. (2020) Massively parallel variant characterization identifies NUDT15 alleles associated with thiopurine toxicity. Proc Natl Acad Sci USA 117:5394–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirona RG, Leake BF, Merino G, Kim RB (2001) Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem 276:35669–35675. [DOI] [PubMed] [Google Scholar]
- van de Steeg E, Greupink R, Schreurs M, Nooijen IH, Verhoeckx KC, Hanemaaijer R, Ripken D, Monshouwer M, Vlaming ML, DeGroot J, et al. (2013) Drug-drug interactions between rosuvastatin and oral antidiabetic drugs occurring at the level of OATP1B1. Drug Metab Dispos 41:592–601. [DOI] [PubMed] [Google Scholar]
- Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC (2016) SIFT missense predictions for genomes. Nat Protoc 11:1–9. [DOI] [PubMed] [Google Scholar]
- Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, Ginsburg GS (2009) The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol 54:1609–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Nguyen TV, McLaughlin RW, Sikkink LA, Ramirez-Alvarado M, Weinshilboum RM (2005) Human thiopurine S-methyltransferase pharmacogenetics: variant allozyme misfolding and aggresome formation. Proc Natl Acad Sci USA 102:9394–9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yee VC, Weinshilboum RM (2004) Aggresome formation and pharmacogenetics: sulfotransferase 1A3 as a model system. Biochem Biophys Res Commun 325:426–433. [DOI] [PubMed] [Google Scholar]
- Wrenbeck EE, Klesmith JR, Stapleton JA, Adeniran A, Tyo KE, Whitehead TA (2016) Plasmid-based one-pot saturation mutagenesis. Nat Methods 13:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Sarangi V, Moon I, Yu J, Liu D, Devarajan S, Reid JM, Kalari KR, Wang L, Weinshilboum R (2020) CYP2C9 and CYP2C19: deep mutational scanning and functional characterization of genomic missense variants. Clin Transl Sci 13:727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]