Abstract
Background:
MicroRNAs (miRNAs) are “master post-transcriptional regulators” of gene expression. Here we investigate miRNAs involved in the incentive motivation for cocaine elicited by exposure to cocaine-associated cues.
Methods:
We conducted NanoString nCounter analyses of microRNA expression in the nucleus accumbens shell of male rats that had been tested for cue reactivity in a previous study. These rats had been trained to self-administer cocaine while living in isolate housing, then during a subsequent 21-day forced abstinence period they either stayed under isolate housing or switched to environmental enrichment (EE), as this EE intervention is known to decrease cocaine seeking. This allowed us to create groups of “high” and “low” cocaine seekers using a median split of cocaine-seeking behavior.
Results:
Differential expression analysis across high- and low-seekers identified 33 microRNAs that were differentially expressed in the nucleus accumbens shell. Predicted mRNA targets of these microRNAs are implicated in synaptic plasticity, neuronal signaling, and neuroinflammation signaling, and many are known addiction-related genes. Of the 33 differentially-expressed microRNAs, 8 were specifically downregulated in the low-seeking group and another set of 8 had expression levels that were significantly correlated with cocaine-seeking behavior.
Conclusion:
These findings not only confirm the involvement of previously identified microRNAs (e.g., miR-212, miR-495) but also reveal novel microRNAs (e.g., miR-3557, miR-377) that alter, or are altered by, processes associated with cocaine-seeking behavior. Further research examining the mechanisms involved in these microRNA changes and their effects on signaling may reveal novel therapeutic targets for attenuating drug craving.
Keywords: Drug abuse, Substance use disorders, Gene expression, miRNA, Isolation stress, Motivation
1. Introduction
Psychostimulant abuse is a significant, ongoing problem in the U.S with devastating economic and social costs to drug abusers and their communities (National Drug Intelligence Center, 2011; National Institute on Drug Abuse, 2015; Pomara et al., 2012). Cocaine use disorders (CUDs) in particular are a serious issue, as cocaine-related deaths have increased substantially over the past few decades, even as general use has declined (Center for Behavioral Health Statistics and Quality, 2015; McCall Jones et al., 2017; National Institute on Drug Abuse, 2020). Unfortunately, there are few treatment options that are effective in promoting long-term abstinence from drug use, especially psychostimulant use. Consequently, 40–60 % of drug users relapse within the first year of abstinence (National Institute on Drug Abuse, 2018). This is in part because drug-associated cues elicit drug craving that strengthens over prolonged abstinence, leaving those with CUDs vulnerable to relapse despite efforts to cease drug use (Dackis and O’Brien, 2001; Gawin and Kleber, 1986; Neisewander et al., 2000). For example, cues such as a crack pipe or crack house acquire conditioned stimulus effects which can trigger craving and relapse (Ciccocioppo and Martin-Fardon, 2004; Conklin and Tiffany, 2002; Ehrman et al., 1992; Weiss et al., 2001). Motivational effects of cocaine-conditioned cues persist even after months without drug use in animal models of drug-seeking, a phenomenon referred to as incubation of craving (Grimm et al., 2001; Tran-Nguyen et al., 1998). Thus, treatments that reduce cue-elicited craving are needed to promote long-term abstinence.
In both animals and humans, various forms of enrichment are effective in attenuating cocaine-related behaviors throughout the abstinence-relapse cycle (Lynch et al., 2013; Vannan et al., 2018). Typically, environmental enrichment (EE) in animal models consists of social housing in small groups that are given access to novel toys and/or exercise equipment. Importantly, EE is effective in reducing drug-seeking behavior when given as an intervention during abstinence, as measured in cocaine conditioned place preference and operant behavior animal models (Chauvet et al., 2012; Ma et al., 2016a; Solinas et al., 2008; Thiel et al., 2011, 2010, 2009). Thus, EE can be used as a tool experimentally to create groups of animals with differing levels of cocaine-seeking behavior.
There is growing interest in the epigenetics of drug abuse, including the role of microRNAs (miRNAs). In general, mammalian miRNAs post-transcriptionally silence gene expression by the imperfect base-pairing of nucleotides at positions 2–8 in the 5′ end of the miRNA (widely referred to as the “seed sequence”) and other miRNA sequences to the 3′ untranslated regions (UTRs) of target mRNAs (Bartel, 2009; Schirle et al., 2014). Because hundreds to thousands of genes have miRNA target sequences in the 3′UTRs, miRNAs function as “master regulators” of gene expression (Plotnikova et al., 2019). The capacity of miRNAs to manipulate and alter gene expression has made this class of RNAs an exciting avenue for finding new therapeutic targets for CUDs treatment development. So far, several miRNAs have been shown to play a role in the motivational processes underlying CUDs, including the let-7 family (Chandrasekar and Dreyer, 2011, 2009), miR-212 (Hollander et al., 2010; Im et al., 2010), miR-495 (Bastle et al., 2018), and others (Kenny, 2014). It is likely that many miRNAs that contribute to resilience or susceptibility to CUDs have not yet been identified. Furthermore, many previous studies have examined the NAc as a whole, including both the core and shell, yet these subregions interface differently with corticolimbic inputs and play different roles in cocaine-seeking behavior. For instance, the NAc shell receives input from the basolateral amygdala, and this pathway is involved in processing incentive salience of cocaine-associated cues (Ma et al., 2016b; Millan and McNally, 2011).
The present study employed NanoString nCounter analysis to identify miRNAs that are differentially-expressed in the NAc shell in rats with “high” vs. “low” levels of cocaine-seeking behavior. Tissue was harvested from a subset of rats utilized in a previous experiment (Powell et al., 2020) that had confirmed that rats with a history of cocaine self-administration exhibit less operant responding reinforced by cocaine-associated light/tone cues when they were housed in EE for 21 days of abstinence than when housed in isolation (ISO) [90.25 ± 20.96 and 207.4 ± 33.55 mean responses ± SEM, respectively (n = 15–16/group)]. Here, we took advantage of the varying degrees of cue-elicited motivation for cocaine across a subset of the animals from this study to explore miRNAs as possible mediators of cue-elicited cocaine-seeking behavior.
2. Methods
2.1. Animals and tissue collection
All experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal care and Use Committee at Arizona State University. Male Sprague-Dawley rats (N = 12) used in a previous study (Powell et al., 2020) were sacrificed by isoflurane overdose immediately after a 1-h test for cocaine cue reactivity. In the previous study, single-housed rats had been trained to self-administer cocaine (0.75 mg/kg, IV) delivered response-contingently with light and tone cues. After ≥15, 2-h sessions of training, rats underwent 21 days of forced abstinence, either remaining in single-housing or switching to an enriched environment with 3–5 cage mates, a running wheel, tubes, toys to enhance novelty, and extra nesting materials. Upon completion of forced abstinence, animals were placed back into the self-administration chamber with the cues, but not cocaine, available response-contingently (i.e., cue reactivity test) (Acosta et al., 2008; Kufahl et al., 2009). The number of times that a rat pressed the lever resulting in cue presentations without cocaine delivery was used as a measure of cocaine-seeking behavior and is thought to reflect the degree of incentive motivation for cocaine elicited by the cues. Within 5 min of completing the test, brains were harvested and rapidly frozen in 2-methylbutane that was placed on dry ice to achieve a temperature of approximately −20 °C. Later, 2 mm coronal sections containing the NAc shell were excised using a brain matrix to place razor blades at the appropriate location on the ventral surface of the brain for capturing the NAc in the tissue section. Tissue punches were then taken containing the NAc core and anterior commissure (1 mm diameter). Secondary punches (2 mm diameter) were taken containing the NAc shell using the previously punched location of the core as a landmark. RNA was isolated from the NAc shell samples using the standard Trizol method as performed previously (Bastle et al., 2018). Samples (100–150 ng of RNA) were then analyzed for miRNA expression using the Nanostring® nCounter Rat miRNA Expression Assay Kit v1.5 at the University of Arizona Genetics Core. The panel quantifies expression of 423 rat miRNAs in version v1.5, slightly fewer than the 496 listed in the miRBase Rattus norvegicus miRNA database.
2.2. Bioinformatics analyses
Nanostring nCounter analysis provided expression levels of miRNAs as raw counts for each miRNA in the sample. 100 ng total RNA was used in a multiplexed reaction to anneal specific miRNA tags followed by ligation and enzymatic purification to remove excess unincorporated tags in the assay, using the manufacturer protocol. Sequence specific fluorescent reporter probes and biotinylated capture probes were hybridized to ligated target nucleic acid complexes overnight at 65 °C for >12 h, followed by a series of automated washes and immobilization onto a streptavidin lined cartridge for data collection. Digital images from the cartridges were obtained over 4 h with the nCounter Digital Analyzer (CCD camera and microscope objective lens) using 555 FOV data resolution. Digital counts were tabulated and exported as comma separated values.
Differential expression analysis was performed using the R package “limma” (Ritchie et al., 2015) to identify differences in miRNA expression between the high- and low-seeking groups. Briefly, normalization factors were calculated to scale the raw library sizes. Raw counts were converted to counts per million reads (CPM) for each miRNA, and miRNAs with very low expression (CPM < 30) were filtered out, which removed 68 of 420 miRNAs before the analysis. Weighted least squares were calculated for each miRNA and sample, then a linear model was fit. Contrasts were performed on the fitted linear model to compare expression of each miRNA between high- and low-seeking groups based on log2 fold change values.
TargetScan 7.2, a miRNA target predictor, was used to determine possible mRNA targets of the differentially-expressed miRNAs (http://www.targetscan.org/vert_72) (Agarwal et al., 2015). TargetScan predicts targets based on the miRNA’s seed sequence, as well as conserved sites on mRNAs that fully or partially match this sequence. For some miRNAs, the available data on the predicted mRNA targets applied not to a single miRNA, but to a miRNA family. For example, mir-3573-5p is part of the miR-423-5p/3573-5p family and predicted targets for all miRNAs in this family are shared. To obtain accurate TargetScan predictions, MIMAT accession numbers provided by Nanostring were used to determine whether each mature miRNA originated from the 5′ or 3′ arm of its precursor. Prior literature does not always follow this convention; thus, in this paper, the 3p/5p label is only included when discussing results of the current study exclusive of comparisons to other work, or when prior authors have made the miRNA designations clear.
Because some miRNAs were upregulated, while others were downregulated, predicted mRNA targets were given “impact scores” to signify the levels of up- and downregulation that might result from the miRNA changes. The predicted targets and impact scores were input into IPA (version 51963813; QIAGEN Inc.) to identify significantly regulated pathways (Krämer et al., 2014). We then compared the TargetScan predicted mRNA targets of the differentially-expressed miRNAs to the known addiction-related genes in the Knowledgebase of Addiction-Related Genes (KARG) database (http://karg.cbi.pku.edu.cn) (Li et al., 2008). At the time of analysis, the rat KARG contained 1135 genes, of which 347 had an evidence score of 2 or more (were supported by 2 or more lines of evidence), and these were the only genes included in the analysis.
2.3. miRNA validation with RT-qPCR
Leftover RNA from the same samples used for the Nanostring analyses was used to validate select miRNAs. For each sample, approximately 45 ng of purified RNA were used to prepare cDNA using the Taqman® Advanced MicroRNA cDNA synthesis kit (Applied Biosystems, Foster City, CA, USA, # A28007), Taqman® Advanced MicroRNA Assay primers (Life Technologies, Grand Island, NY, USA) for miR-376c-3p, miR-107-3p, and miR-212-3p, and Taqman® MicroRNA Assay primer for the control transcript U6. cDNA for each sample was diluted 1:100 with nuclease-free water, then run in triplicate for each miRNA and U6. Relative expression was determined using the comparative 2−ΔCt method (Livak and Schmittgen, 2001).
2.4. Statistical analysis
Rats were divided into groups based on median split of cocaine-seeking behavior. Statistical calculations were performed in GraphPad Prism 8, or R 3.6 (R Core Team, 2019). Linear regressions were used to analyze the correlation between miRNA levels (CPM) and cocaine-seeking behavior. For differential expression analysis, p-values and false discovery rates (FDR) using the Benjamini-Hochberg method were calculated using the R package “limma” (Ritchie et al., 2015). The statistical threshold for all tests was p < 0.05.
2.5. Data availability
Nanostring data are deposited in the Gene Expression Omnibus (GSE153524). R code used for data analyses are available at https://gitlab.com/neisewander_asu/vannan-powell-2020.
3. Results
3.1. Differentially-expressed miRNAs in the NAc shell correlate with cocaine-seeking behavior
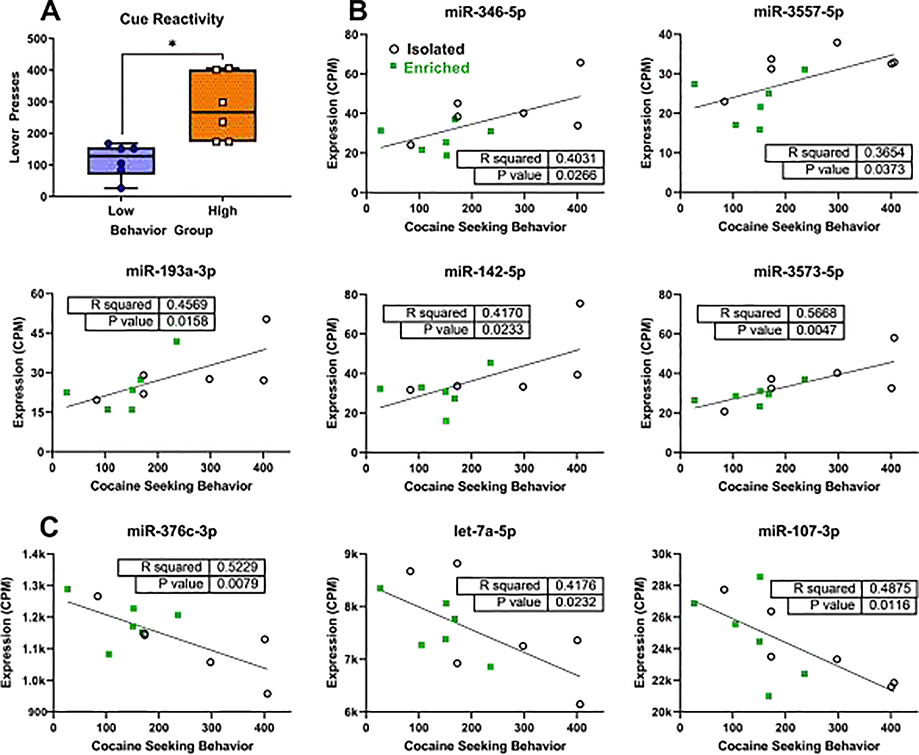
The high and low cocaine-seeking groups (n = 6/group) derived from the median split of cocaine seeking values were significantly different in their cocaine-seeking behavior [t(10) = 3.452, p = 0.0062, Fig. 1A]. Cocaine-seeking values aligned well with housing condition: the “high” cocaine-seeking group consisted of 83.3 % (5) ISO and 16.7 % (1) EE rats and vice versa for the “low” cocaine-seeking group. In total, expression levels of 75 miRNAs were significantly correlated with cocaine-seeking behavior (Supplementary Table 1), although not all of these miRNAs were differentially expressed in the low vs. high seeking groups. Analysis of Nanostring counts using limma identified 33 miRNAs that were differentially expressed in the NAc shell in animals that displayed high versus low cocaine-seeking behavior (Table 1). Of these, 8 were downregulated and 25 were upregulated (Fold Change >1 and <1 on Table 1, respectively) in the low-seeking group relative to the high-seeking group. For Table 1, log2 fold change values have been converted to linear values where Fold Change = 2^(log2 values). In addition, 8 of the 33 miRNAs had expression levels that correlated with cocaine-seeking behavior, 5 positively and 3 negatively (Fig. 1C).
Fig. 1.
Correlation of miRNA expression levels and the number of active lever presses during the cue reactivity test in each animal. (A). Separation of groups of the low- (blue) and high-seeking (orange) groups determined by median split of active lever presses. Boxes indicate median and quartiles; whiskers indicate minimum and maximum. * indicates difference from low-seeking group, p < 0.05, independent samples t-test. Positive (B) and negative (C) Pearson correlations of miRNA expression with cocaine-seeking behavior, as measured by active lever presses during the cue reactivity test. Isolated and enriched animals are depicted with open black circles and green squares, respectively. Only miRNAs that had both significantly different expression between high- and low-seeking groups and that correlated significantly with behavior (table inset on each graph) are shown here. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Table 1:
Differentially-expressed miRNAs between the high- and low-seeking groups.
| miRNA | High-Seeking (CPM)1 | Low-Seeking (CPM) | Fold Change2 | P-Value | FDR3 |
|---|---|---|---|---|---|
| miRNAs with higher expression in high cocaine-seeking animals | |||||
| miR-463-3p | 37.04 | 23.26 | 1.447 | 0.0188 | 0.3885 |
| miR-346-5p | 42.42 | 26.40 | 1.439 | 0.0114 | 0.3291 |
| miR-483-3p | 45.76 | 29.62 | 1.431 | 0.0036 | 0.3108 |
| miR-3557-5p | 33.24 | 21.68 | 1.417 | 0.0074 | 0.3108 |
| miR-193a-3p | 32.98 | 20.81 | 1.390 | 0.0275 | 0.4400 |
| miR-133a-3p | 68.68 | 46.95 | 1.378 | 0.0133 | 0.3291 |
| miR-142-5p | 43.47 | 28.52 | 1.342 | 0.0385 | 0.4906 |
| mir-3573-5p | 39.58 | 26.59 | 1.331 | 0.0204 | 0.3993 |
| miRNAs with higher expression in low cocaine-seeking animals | |||||
| miR-29a-3p | 68099.60 | 70947.29 | 0.878 | 0.0360 | 0.4868 |
| miR-16-5p | 10815.91 | 11290.58 | 0.875 | 0.0465 | 0.5104 |
| miR-93-5p | 306.12 | 324.61 | 0.864 | 0.0326 | 0.4783 |
| miR-495-3p | 6727.51 | 7266.45 | 0.850 | 0.0422 | 0.4906 |
| miR-376c-3p | 1106.98 | 1197.90 | 0.845 | 0.0289 | 0.4429 |
| miR-410-3pa | 2722.19 | 2973.03 | 0.841 | 0.0239 | 0.4205 |
| miR-329-3p | 5361.27 | 5863.05 | 0.834 | 0.0432 | 0.4906 |
| let-7a-5p | 7226.04 | 7915.63 | 0.833 | 0.0258 | 0.4320 |
| miR-652-3p | 734.49 | 800.55 | 0.832 | 0.0486 | 0.5104 |
| miR-377-3p | 290.24 | 320.87 | 0.832 | 0.0418 | 0.4906 |
| miR-107-3p | 23171.08 | 25691.54 | 0.828 | 0.0150 | 0.3291 |
| miR-138-5p | 1727.38 | 1915.73 | 0.822 | 0.0225 | 0.4175 |
| miR-487b-3p | 4030.70 | 4628.25 | 0.802 | 0.0066 | 0.3108 |
| miR-344b-1-3p/miR-344b-2-3pa | 357.45 | 411.75 | 0.793 | 0.0026 | 0.3108 |
| miR-128-3p | 1644.39 | 1883.53 | 0.791 | 0.0346 | 0.4868 |
| miR-137-3p | 2451.13 | 2888.88 | 0.790 | 0.0147 | 0.3291 |
| miR-323-3p | 1155.50 | 1336.43 | 0.790 | 0.0088 | 0.3108 |
| miR-337-3p | 317.90 | 377.92 | 0.776 | 0.0051 | 0.3108 |
| miR-125b-3p | 1341.30 | 1589.26 | 0.772 | 0.0125 | 0.3291 |
| miR-409-5p | 326.65 | 389.53 | 0.771 | 0.0049 | 0.3108 |
| miR-218a-5p | 6391.04 | 7740.13 | 0.759 | 0.0393 | 0.4906 |
| miR-212-3pb | 354.77 | 448.44 | 0.728 | 0.0078 | 0.3108 |
| miR-130b-3p | 1279.74 | 1616.11 | 0.719 | 0.0104 | 0.3291 |
| miR-221-3p | 862.09 | 1078.44 | 0.718 | 0.0087 | 0.3108 |
| miR-132-3pb | 25643.46 | 32814.25 | 0.710 | 0.0188 | 0.3885 |
Indicate miRNAs in the same family.
miRNA expression levels were calculated as counts per million (CPM) and where available, 3p or 5p designations are included.
Fold change is an estimate of the effect derived from the log2 fold changes from “limma”, which were then converted to linear values with the equation 2^(log2 value). Values <1 signifies higher expression in high-seeking animals, and values >1 signifies higher expression in low-seeking animals.
FDR = false discovery rate.
Nanostring results for miR-376c-3p, miR-107-3p, and miR-212-3p, which were all elevated in low-seeking animals, were validated using RT-qPCR (Supplementary Fig. 1). Subsequent t-tests including all rats (n = 6 of each group) did not show significant differences in expression between high- and low-seeking groups for these miRNAs. However, separating the seeking groups into quartiles (including only the 3 highest and 3 lowest cocaine-seeking animals), revealed significantly higher expression in the low-seeking group for miR-376c-3p [t(4) = 3.05, p = 0.0379, Suppl. Fig. 1A] and miR-107-3p [t(4) = 2.81, p = 0.0481, Suppl. Fig. 1B]. Although thevalues for miR-212-3p did not quite meet the threshold for significance after separating animals into quartiles [t(4) = 2.63, p = 0.0583], they did correlate significantly with behavior overall ([F(1,10) = 6.16, p = 0.0324]) (Suppl. Fig. 1C).
3.2. Predicted mRNA targets of the differentially-expressed miRNAs
TargetScan 7.2 was utilized to identify predicted mRNA targets of the differentially-expressed miRNAs for the rat, which were then utilized in subsequent analyses. The number of predicted targets varied for each miRNA and ranged from 11 (miR-487b-3p) to 3,972 (miR-3557-5p).
To create a list for input into IPA, first we determined the overall impact of our miRNAs on the list of predicted targets (mRNAs) relative to the low-seeking condition. If a miRNA was upregulated in low cocaine-seeking animals, its predicted targets were given a score of −1, as they would be downregulated by that miRNA, whereas targets of downregulated miRNAs were given a score of +1. For example, the mRNA Nuclear factor I B (Nfib) is a predicted target of 16 differentially-expressed miRNAs, of which 3 were downregulated in the low-seeking group (+3) and 13 of which were upregulated (−13), leading to a total impact score of −10 (Supplementary Table 2). We began with 9,761 predicted targets, which comprised 23.76 % of the transcribed genes in the tissue analyzed. Because more miRNAs were upregulated rather than downregulated in the low-seeking condition, predicted targets were more likely to have a negative impact score, and negative impact scores were greater in magnitude: 6,456 targets had a negative impact score (range: −1 to −14); 3,305 had a positive or 0 impact score (range: 0 to +3.) Because IPA analysis is more robust with smaller lists of genes, we prioritized candidates based on impact score. Only mRNA targets that were mapped to IPA and had an impact score of −3 or lower and +1 or higher were included, reducing the list to 3,600 predicted mRNA targets.
The majority of mRNAs were predicted targets of only 1 or 2 differentially-expressed miRNAs (6,128; 62.8 %). However, 9 mRNAs were targets of 15 or more miRNAs: Zinc finger and BTB domain containing 20 (Zbtb20), Nuclear factor of activated T-cells 5 (Nfat5), Argonaute RNA-induced silencing complex (RISC) component 1 (Ago1), Nfib, Protein quaking (Qki), Phosphodiesterase 3A (Pde3a), Retinoic acid receptor-related orphan receptor B (Rorb), Transcription factor 4 (Tcf4), and Kruppel like factor 7 (Klf7), with impact scores ranging from −8 to −14. In total, 136 mRNAs (1.39 %) were predicted targets of at least 10 differentially-expressed miRNAs.
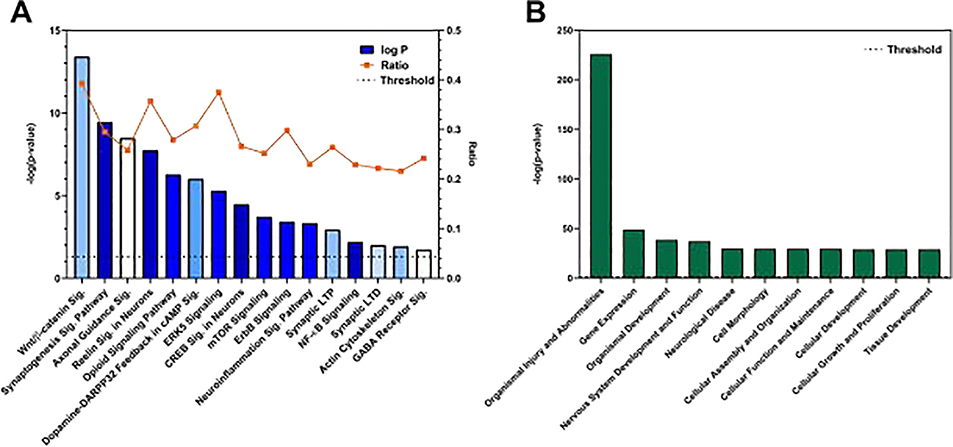
IPA revealed many significant pathways, Including Wnt/ β-catenin Signaling, Synaptogenesis Signaling Pathway, Axonal Guidance Signaling, Dopamine/DARPP34 Feedback in cAMP Signaling, CREB Signaling in Neurons, and ERK5 Signaling, among others (Fig. 2A, Supplementary Table 3). Significant Diseases and Functions were also provided by IPA (Fig. 2B, Supplementary Table 4) and included: Learning in the category Behavior; Development of Neurons and Morphogenesis of Neurons in the categories Cellular Development and Cellular Growth and Proliferation; Migration of Neurons in the category Cellular Movement; Transcription of RNA and Expression of RNA in the category Gene Expression (Supplementary Table 9); and Neurotransmission in the category Nervous System Development and Function (Supplementary Tables 5–10). Including Wnt/ β-catenin Signaling, Synaptogenesis Signaling Pathway, Axonal Guidance Signaling, Dopamine/DARPP34 Feedback in cAMP Signaling, CREB Signaling in Neurons, and ERK5 Signaling, among others (Fig. 2A, Supplementary Table 3).
Fig. 2.
Summary of pathway analysis using IPA. Panels depict several pathways (A) and diseases and functions (B) related to the predicted targets of the differentially-expressed miRNAs. Threshold levels indicate −[log (p = 0.05)]. Bar colors represent the ranges of the z-score calculated by IPA. Darker shades indicate z-scores farther from zero, and blue indicates a negative z-score. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.3. miRNAs and addiction-related genes
For each miRNA, the number of predicted targets found in TargetScan was also compiled and then cross-referenced to addiction-related genes in the KARG database (Table 2). Three miRNAs, miR-3557-5p, miR-377-3p, and miR-337-3p, targeted a large percentage of KARG (between 17.00 % and 24.21 %). Together, the putative targets of significant miRNAs covered 205 (59.1 %) of the 347 KARG genes included in our analysis. Several addiction-related mRNAs were predicted to be targets of at least 10 differentially-expressed miRNAs: Nuclear factor I A (Nfia), Nfib, Circadian locomoter output cycles protein kaput (Clock), Ataxin 1 (Atxn1), cyclic AMP (cAMP)-responsive element binding protein 1 (Creb1), Zinc finger and BTB domain containing 16 (Zbtb16), Purine rich element binding protein A (Pura), and Gamma-aminobutyric acid (GABA) type B receptor subunit 2 (Gabbr2), potentially indicative of their key role in cocaine-seeking and drug motivation (Supplementary Table 11). Notably, all these mRNAs were predicted to be downregulated in the low-seeking group compared to high-seeking, with impact scores between −4 (Zbtb16) and −11 (Atxn1) (Supplementary Table 2).
Table 2:
Overlap of the predicted mRNA targets for each miRNA in TargetScan and KARG databases.
| miRNA | TargetScan1 | KARG1 | % KARG2 |
|---|---|---|---|
| miRNAs with higher expression in high cocaine-seeking animals | |||
| miR-463-3p | 1396 | 26 | 7.49 % |
| miR-346-5p | 114 | 6 | 1.73 % |
| miR-483-3p | 79 | 4 | 1.15 % |
| miR-3557-5p | 3972 | 84 | 24.21 % |
| miR-193a-3p | 190 | 5 | 1.44 % |
| miR-133a-3p | 549 | 25 | 7.20 % |
| miR-142-5p | 704 | 21 | 6.05 % |
| mir-3573-5p | 129^ | 6 | 1.73 % |
| miRNAs with higher expression in low cocaine-seeking animals | |||
| miR-29a-3p | 1013 | 32 | 9.22 % |
| miR-16-5p | 1090^ | 30 | 8.65 % |
| miR-93-5p | 1056^ | 29 | 8.36 % |
| miR-495-3p | 616 | 22 | 6.34 % |
| miR-376c-3p | 188^ | 7 | 2.02 % |
| miR-410-3pa | 497^ | 14 | 4.03 % |
| miR-329-3p | 255^ | 7 | 2.02 % |
| let-7a-5p | 1022^ | 19 | 5.48 % |
| miR-652-3p | 14 | 1 | 0.29 % |
| miR-377-3p | 2217 | 59 | 17.00 % |
| miR-107-3p | 558^ | 22 | 6.34 % |
| miR-138-5p | 537 | 16 | 4.61 % |
| miR-487b-3p | 11 | 1 | 0.29 % |
| miR-344b-1-3p/miR-344b-2-3pa | 497^ | 14 | 4.03 % |
| miR-128-3p | 950 | 25 | 7.20 % |
| miR-137-3p | 1019 | 26 | 7.49 % |
| miR-323-3p | 357 | 10 | 2.88 % |
| miR-337-3p | 3243 | 65 | 18.73 % |
| miR-125b-3p | 443 | 9 | 2.59 % |
| miR-409-5p | 100 | 2 | 0.58 % |
| miR-218a-5p | 869 | 26 | 7.49 % |
| miR-212-3pb | 360^ | 14 | 4.03 % |
| miR-130b-3p | 748^ | 19 | 5.48 % |
| miR-221-3p | 366^ | 14 | 4.03 % |
| miR-132-3pb | 360^ | 14 | 4.03 % |
Represents miRNAs that were present on TargetScan as miRNA families, not individual miRNAs. miRNAs in the same family share their seed sequence and thus the same predicted miRNA targets.
Indicate miRNAs in the same family.
Values are the number of predicted mRNA targets found in each respective database.
Percentage of KARG that the predicted targets comprise. Colors indicate miRNAs relative to the other group.
4. Discussion
NanoString nCounter analysis of the NAc shell of male rats with a history of cocaine self-administration that were tested for cocaine-seeking behavior after 21 days of abstinence identified 33 miRNAs displaying differential expression in the “high” and “low” cocaine-seeking groups. Of these, expression of 8 miRNAs correlated significantly with cocaine-seeking behavior. Predicted mRNA targets of the 33 miRNAs were analyzed using IPA, which revealed several significant pathways including Synaptogenesis and Opioid Signaling. Cross-reference of TargetScan and the KARG database showed that many of these miRNAs were predicted to target mRNAs of addiction-related genes.
Many of the differentially-expressed miRNAs identified in this study have been previously implicated in drug abuse, including miR-29a, miR-16, miR-495, mir-376c, miR-329, miR-138, miR-137, miR-337, miR-125b, miR-212, miR-130b, miR-221, and miR-132 (Bastle et al., 2018; Dave and Khalili, 2010; Eipper-Mains et al., 2011; Hollander et al., 2010; Im and Kenny, 2012; Lippi et al., 2011; Schaefer et al., 2010; Shin et al., 2010). Interestingly, these miRNAs all had greater expression in the low-seeking group compared to the high-seeking group. Of these, miR-212, a CREB-induced activity-dependent miRNA in the same family as miR-132, is perhaps the best-studied in the addiction field. For example, Sadakierska-Chudy et al. found that 2-h daily access to cocaine increases both miR-212 and miR-132 in the dorsal striatum compared to saline-yoked controls, and this increase is persistent, lasting 10 days into subsequent extinction training (Sadakierska-Chudy et al., 2017). In addition, Hollander et al. demonstrated that miR-212 and miR-132 may be involved in the transition from casual to compulsive drug use, as both are upregulated in the dorsal striatum following extended (6-h) daily cocaine self-administration compared to cocaine-naïve rats (Hollander et al., 2010). They also found that striatal miR-212 overexpression reduces compulsive-like cocaine-taking behavior during extended access (6-h daily sessions), while knockdown produces more compulsive cocaine consumption. Due to the close relatedness of miR-132 to miR-212, the authors suggest miR-132 may play a similar role in compulsive cocaine-taking. In the present study, the increased expression of miR-212 and miR-132 in the NAc shell of rats with low cocaine-seeking behavior suggests that these miRNAs are protective against motivation for cocaine. Together, it appears that striatal miR-212 and miR-132 expression may shield against two defining phases of CUDs: transition to an addicted-like phenotype as well as craving during protracted abstinence.
Among the miRNAs previously associated with motivation for cocaine is miR-495, a miRNA that we identified because its levels decrease during cocaine self-administration and its over-expression in the NAc shell attenuates motivation for cocaine (Bastle et al., 2018). We have shown that miR-495 regulates expression of multiple addiction-related genes including Brain derived neurotrophic factor (Bdnf), Calcium-calmodulin activated protein kinase IIα (Camk2a) and Activity-regulated cytoskeleton-associated protein (Arc) among other mRNAs such as Per2 and Gria3 (Bastle et al., 2018). Here, we found that miR-495 has higher expression in low-seeking animals, supporting our prior research that suggests upregulation of miR-495 is protective against motivation for drugs of abuse.
To identify novel candidate miRNAs that may regulate cocaine-related behavior in this study we cross-referenced TargetScan and KARG. Of the 33 differentially-expressed miRNAs, miR-3557-5p, miR-377-3p, and miR-337-3p were predicted to target particularly high percentages of the KARG database (>17.00 %, up to 24.14 %). Of these 3, miR-3557-5p and miR-377-3p have not been studied in substance abuse or psychiatric illness to our knowledge, suggesting they may be novel targets for addiction research. However, miRNAs that were predicted to target a large percentage of KARG also had many TargetScan predicted targets (>2000). Caution is needed when the number of predicted targets is so large, due to an increased likelihood of false positives and limitations of these databases. By contrast, miRNAs with fewer than 200 predicted targets (miR-346-5p, miR-483-3p, miR-193a-3p, mir-3573-5p, miR-376c-3p, miR-652-3p, miR-487b-3p, miR-409-5p) had little overlap with the KARG database (under 2.02 %).
Among the miRNAs predicted to target expression of a large number of addiction-related genes, miR-337-3p has been the subject of prior drug abuse research. Here, we found that miR-337-3p has significantly higher expression in the low-seeking group, suggesting it may be protective against cocaine-seeking behavior. However, in striatal Dopamine Receptor 2 (Drd2)-expressing neurons, miR-337-3p is upregulated after acute cocaine injection in mice (Schaefer et al., 2010), suggesting that this miRNA may be associated with the initial neurobiological changes after drug exposure. Similarly, miR-376c, miR-138, and miR-137, which all have significantly higher expression in the low-seeking group in our study, are also upregulated in striatal Drd2-expressing neurons after acute cocaine injection (Schaefer et al., 2010). Therefore, these miRNAs may either protect against or facilitate addictive behaviors depending on factors such as previous drug experience. Other factors that may contribute to these seemingly contradictory results include differences in the brain region studied and cell-type specificity.
Several addiction studies have identified the importance of let-7 miRNAs, particularly let-7d (Chandrasekar and Dreyer, 2011, 2009; He et al., 2010; Hollander et al., 2010), although these miRNAs have few addiction-related targets according to KARG (5.48 %). For example, let-7d expression is decreased in regions of the mesolimbic reward pathway including the NAc core and shell, striatum, and ventral tegmental area after 15 days of daily cocaine injections compared to saline controls (Chandrasekar and Dreyer, 2009), whereas overexpression of let-7d in the NAc attenuates cocaine conditioned place preference (Chandrasekar and Dreyer, 2011). Here, we demonstrate that another let-7 family member, let-7a, has higher expression in low-seeking animals and displays a significant negative correlation with cocaine-seeking. Together, these data suggest let-7 miRNAs may modulate different aspects of cocaine-related behavior. Although TargetScan assumes the same predicted targets for all let-7 miRNAs, differences in their biogenesis and expression patterns may contribute to distinct roles in neuronal function and thus drug abuse (Roush and Slack, 2008).
Many of the miRNAs identified here have been explored primarily in relation to other psychiatric illnesses, such as schizophrenia and depression, which have high comorbidity with substance abuse (Batel, 2000; Kessler et al., 2005; Kosten et al., 1998; Paykel et al., 2005). For example, miR-16, miR-495, miR-652, miR-107, miR-138, and miR-137 have been linked to schizophrenia (Beveridge et al., 2010; Moreau et al., 2011; Ripke et al., 2011; Santarelli et al., 2011; Wright et al., 2013) and were all upregulated in the low-seeking group compared to the high-seeking animals in the present study. Similarly, miR-16 has been implicated in depression, and is believed to underlie the therapeutic effects of the antidepressant fluoextine through targeted downregulation of the serotonin transporter (SERT) mRNA (Slc6a4) (Baudry et al., 2010). Importantly, antidepressants that blocks serotonin reuptake through SERT, encoded by the gene Slc6a4, are effective in reducing cocaine-seeking and -taking in some preclinical models (Baker et al., 2001; Burmeister et al., 2003; Harris et al., 2001; Richardson and Roberts, 1991).This suggests that miR-16-5p, which was found here to have higher expression in low-seeking rats, may be therapeutic for treating addiction by reducing depressive symptoms. The overlap of miRNAs implicated in addiction and comorbid psychiatric illnesses may help inform treatments for those suffering from addiction occurring in conjunction with, or exacerbated by, other conditions.
We utilized IPA to uncover pathways that are potentially regulated by the 33 differentially-expressed miRNAs by inputting the miRNAs’ predicted mRNA targets and the estimated impact of the miRNAs on their expression (i.e. impact score). This analysis revealed several pathways, including Axonal Guidance Signaling, Opioid Signaling, and ERK5 signaling, that are potentially regulated by the differentially-expressed miRNAs. Many of these pathways have previously been implicated in addiction, including WNT/β-catenin signaling (Cuesta and Pacchioni, 2017) and Neuroinflammation signaling pathway (Clark et al., 2013). These results also validate our recent RNA-seq analysis of the NAc shell in animals that underwent the same training and testing procedures as the present study, except that in addition to EE and ISO housing, animals also underwent different lengths of forced abstinence (1 or 21 days) (Powell et al., 2020). We found that contrasting EE and ISO animals given 21 days of abstinence, similar to the present study, implicated several of the pathways found here, including Synaptogenesis Signaling, Reelin Signaling, Neuroinflammation Signaling, Synaptic Long-Term Potentiation, and CREB Signaling in Neurons, and that Bdnf, a widely studied addiction gene, (Li and Wolf, 2015) was a top upstream regulator of this comparison. In the present study, 7 miRNAs are predicted to target Bdnf with an impact score of −7. IPA also revealed significant functions of the predicted targets including Dendritic Growth/Branching and Morphology of Dendritic Spines, which support prior research that several of our miRNAs of interest, including miR-29a, miR-329, miR-137 and miR-132, regulate dendritic spine formation and morphology (Impey et al., 2010; Lippi et al., 2011; Smrt et al., 2010). Indeed, our IPA results both validate and expand on current knowledge by implicating several miRNAs in cue-elicited cocaine-seeking behavior.
Of the pathways identified in the current study, CREB signaling is one of the most highly studied in addiction (Carlezon et al., 2005; Gomez et al., 2015; Krasnova et al., 2016; Kreibich et al., 2009; Larson et al., 2011; Mattson et al., 2005). As mentioned earlier, increased miR-212 expression is related to reduced compulsive-like cocaine-taking behavior, which is thought to involve increasing CREB signaling and decreasing MeCP2/BDNF signaling (Hollander et al., 2010; Im et al., 2010). Similarly, our prior study suggested CREB Signaling in Neurons is an important mechanism underlying cocaine-seeking behavior, and found that Creb1 was a top upstream regulator of the differentially-expressed RNAs in the contrast between EE and ISO animals with 21 days of abstinence (Powell et al., 2020). Here, we found that 11 of the 33 differentially-expressed miRNAs in high- vs. low-cocaine seeking animals are predicted to target Creb1 (impact score = −7), suggesting that these miRNAs may be important regulators of the pathways identified in our previous study.
A caveat of the median split used to examine the effects of housing environment during forced abstinence in the present study is that animals at the outer range of the housing groups showed nearly the same lever pressing during the cue reactivity tests, as can be seen in Fig. 1A. We considered using a quartile analysis, which includes only the 3 animals with the greatest and least cue reactivity (i.e., 6 total animals representing the first and fourth quartiles of the total dataset), however, a drawback to this approach is the loss of power due to smaller sample size, reducing the capacity to find potentially relevant changes in miRNA expression. Our follow up analyses showing significant correlations of cocaine-seeking behavior with expression of 75 individual miRNAs, including 8 of the 33 miRNAs that are differentially expressed between “low” and “high” reactivity groups, mitigated concern with our approach. Furthermore, a preliminary comparison to the quartile analysis showed 11 differentially-expressed miRNAs, 4 of which are shared with the original analysis (miR-346-5p, miR-193a-3p, miR-3573-3p, and miR-107-3p). These 4 miRNAs, along with 4 of the 7 uniquely identified miRNAs (miR-301b-3p, miR-3561-3p, miR-3558-3p, and miR-208b-3p), are significantly correlated with behavior. We also used the quartile analysis in our follow-up RT-qPCRvalidation experiments, which confirmed increased expression of miR-276c and miR-107-3p in the low-seeking group relative to high-seeking animals, and additionally showed a significant correlation of miR-212-3p expression with cocaine-seeking behavior with all 12 animals. Finally, Nanostring nCounter employs dual probes and hybridization to directly measure target molecules without the bias of amplification-dependent techniques (e.g. RT-qPCR) (Eastel et al., 2019), and though not as sensitive as small RNA-seq, Nanostring displays improved detection of lowly-expressed miRNAs that might be missed by techniques such as microarrays (Eastel et al., 2019; Kulkarni, 2011). This is especially important for detecting disease biomarkers, which may be present in low abundance (Foye et al., 2017). Indeed, for the present study, all 8 differentially-expressed miRNAs that were elevated in the high-seeking group displayed low CPM values. Still, Nanostring is a proprietary platform, which somewhat limits flexibility and future use. In addition, the specificity of the assay depends largely on the design of the probes (Eastel et al., 2019); here, however, we used a pre-designed assay from Nanostring that has been widely used by other researchers (e.g. Chaudhuri et al., 2018; Mellios et al., 2018; Murphy et al., 2014), bolstering confidence in our approach.
5. Conclusion
In this study, we identified 33 miRNAs that are differentially-expressed in rats displaying high versus low cocaine-seeking behavior. Although this study focused on motivation for cocaine during abstinence, it is possible that the miRNAs identified may be relevant to other aspects of CUDs, as others have linked miRNAs to acute cocaine exposure (Bastle et al., 2018), escalation of cocaine self-administration (Hollander et al., 2010), and cocaine CPP (Chandrasekar and Dreyer, 2011; Viola et al., 2016). Understanding the role of these miRNAs in motivation for cocaine may lead to novel treatments as currently pioneered in the cancer field, where some miRNA therapeutics have even advanced to clinical trials (Bonneau et al., 2019; Wahid et al., 2010). Further research on the role of miRNAs in CUDs will aid in understanding the underlying mechanisms involved and position the field to capitalize on the knowledge for development of treatments.
Supplementary Material
Acknowledgements
The authors thank Raul Garcia, Samantha Scott, John Paul Bonadonna, Mark Namba, Ayleen Mokbel, Aracely Esquer, Trisha Chaudhury, Thomas Benson, Daniela Alcazar, Madeleine St. Peter, and Ryan Bastle for their technical and editing assistance
Funding source
This work was supported by the National Institute on Drug Abuse [DA034097]. The sponsor had no role in the study design, data collection, data analysis and interpretation, writing of the article, or decision to submit the article for publication.
Footnotes
Declaration of Competing Interest
The authors report no declarations of interest.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.drugalcdep.2021.108585.
References
- Acosta JI, Thiel KJ, Sanabria F, Browning JR, Neisewander JL, 2008. Effect of schedule of reinforcement on cue-elicited reinstatement of cocaine-seeking behavior. Behav. Pharmacol 19, 129–136. 10.1097/FBP.0b013e3282f62c89. [DOI] [PubMed] [Google Scholar]
- Agarwal V, Bell GW, Nam J-W, Bartel DP, 2015. Predicting effective microRNA targets in mammalian mRNAs. Elife 4, e05005. 10.7554/eLife.05005.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Tran-Nguyen LTL, Fuchs RA, Neisewander JL, 2001. Influence of individual differences and chronic fluoxetine treatment on cocaine-seeking behavior in rats. Psychopharmacology (Berl.) 155, 18–26. 10.1007/s002130000676. [DOI] [PubMed] [Google Scholar]
- Bartel DP, 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastle RM, Oliver RJ, Gardiner AS, Pentkowski NS, Bolognani F, Allan AM, Chaudhury T, St. Peter M, Galles N, Smith C, Neisewander JL, Perrone-Bizzozero NI, 2018. In silico identification and in vivo validation of miR-495 as a novel regulator of motivation for cocaine that targets multiple addiction-related networks in the nucleus accumbens. Mol. Psychiatry 23, 434–443. 10.1038/mp.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batel P, 2000. Addiction and schizophrenia. Eur. Psychiatry 15, 115–122. 10.1016/S0924-9338(00)00203-0. [DOI] [PubMed] [Google Scholar]
- Baudry A, Mouillet-Richard S, Schneider B, Launay J-M, Kellermann O, 2010. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science 329, 1537–1541. 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ, 2010. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry 15, 1176–1189. 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V, 2019. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. Electron. J. Int. Fed. Clin. Chem. Lab. Med. [PMC free article] [PubMed] [Google Scholar]
- Burmeister JJ, Lungren EM, Neisewander JL, 2003. Effects of fluoxetine and d-fenfluramine on cocaine-seeking behavior in rats. Psychopharmacology (Berl.) 168, 146–154. 10.1007/s00213-002-1307-8. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Duman RS, Nestler EJ, 2005. The many faces of CREB. Trends Neurosci. 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Center for Behavioral Health Statistics and Quality, 2015. Results from the 2014 National Survey on Drug Use and Health: Detailed Tables. Substance Abuse and Mental Health Services Administration, Rockville, MD. [Google Scholar]
- Chandrasekar V, Dreyer J-LL, 2009. microRNAs miR-124, let-7d and miR-181a regulate Cocaine-induced Plasticity. Mol. Cell. Neurosci 42, 350–362. [DOI] [PubMed] [Google Scholar]
- Chandrasekar V, Dreyer J-L, 2011. Regulation of miR-124, let-7d, and miR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 36, 1149–1164. 10.1038/npp.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri AD, Dastgheyb RM, Yoo SW, Trout A, Talbot CC, Hao H, Witwer KW, Haughey NJ, 2018. TNFα and IL-1β modify the miRNA cargo of astrocyte shed extracellular vesicles to regulate neurotrophic signaling in neurons article. Cell Death Dis. 9 10.1038/s41419-018-0369-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet C, Goldberg SR, Jaber M, Solinas M, 2012. Effects of environmental enrichment on the incubation of cocaine craving. Neuropharmacology 63, 635–641. 10.1016/j.neuropharm.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccocioppo R, Martin-Fardon R, Weiss F, 2004. Stimuli associated with a single cocaine experience elicit long-lasting cocaine-seeking. Nat. Neurosci 7, 495–496. 10.1038/nn1219. [DOI] [PubMed] [Google Scholar]
- Clark KH, Wiley CA, Bradberry CW, 2013. Psychostimulant abuse and neuroinflammation: emerging evidence of their interconnection. Neurotox. Res 23, 174–188. 10.1007/s12640-012-9334-7. [DOI] [PubMed] [Google Scholar]
- Conklin CA, Tiffany ST, 2002. Applying extinction research and theory to cue-exposure addiction treatments. Addiction 97, 155–167. 10.1046/j.1360-0443.2002.00014.x. [DOI] [PubMed] [Google Scholar]
- Cuesta S, Pacchioni AM, 2017. Are changes in the Wnt/β-catenin pathway involved in cocaine and stress-induced long-term neuroadaptations? J. Addict. Prev. Med 2, 112. [Google Scholar]
- Dackis CA, O’Brien CP, 2001. Cocaine dependence: a disease of the brain’s reward centers. J. Subst. Abuse Treat 21, 111–117. 10.1016/S0740-5472(01)00192-1. [DOI] [PubMed] [Google Scholar]
- Dave RS, Khalili K, 2010. Morphine treatment of human monocyte-derived macrophages induces differential miRNA and protein expression: impact on inflammation and oxidative stress in the central nervous system. J. Cell. Biochem 110, 834–845. 10.1002/jcb.22592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastel JM, Lam KW, Lee NL, Lok WY, Tsang AHF, Pei XM, Chan AKC, Cho WCS, Wong SCC, 2019. Application of NanoString technologies in companion diagnostic development. Expert Rev. Mol. Diagn 19, 591–598. 10.1080/14737159.2019.1623672. [DOI] [PubMed] [Google Scholar]
- Ehrman RN, Robbins SJ, Childress AR, O’Brien CP, 1992. Conditioned responses to cocaine-related stimuli in cocaine abuse patients. Psychopharmacology (Berl.) 107, 523–529. 10.1007/BF02245266. [DOI] [PubMed] [Google Scholar]
- Eipper-Mains JE, Kiraly DD, Palakodeti D, Mains RE, Eipper BA, Graveley BR, 2011. microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA 17, 1529–1543. 10.1261/rna.2775511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foye C, Yan IK, David W, Shukla N, Habboush Y, Chase L, Ryland K, Kesari V, Patel T, 2017. Comparison of miRNA quantitation by Nanostring in serum and plasma samples. PLoS One 12, e0189165. 10.1371/journal.pone.0189165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawin FH, Kleber HD, 1986. Abstinence symptomatology and psychiatric diagnosis in cocaine abusers. Arch. Gen. Psychiatry 43, 107. 10.1001/archpsyc.1986.01800020013003. [DOI] [PubMed] [Google Scholar]
- Gomez AM, Altomare D, Sun W-L, Midde NM, Ji H, Shtutman M, Turner JR, Creek KE, Zhu J, 2015. Prefrontal microRNA-221 mediates environmental enrichment-induced increase of locomotor sensitivity to nicotine. Int. J. Neuropsychopharmacol 1–12. 10.1093/ijnp/pyv090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JW, Hope BT, Wise RA, Shaham Y, 2001. Incubation of cocaine craving after withdrawal. Nature 412, 141–142. 10.1038/35084134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris GC, Altomare K, Aston-Jones G, 2001. Preference for a cocaine-associated environment is attenuated by augmented accumbal serotonin in cocaine withdrawn rats. Psychopharmacology (Berl.) 156, 14–22. 10.1007/s002130100693. [DOI] [PubMed] [Google Scholar]
- He Y, Yang C, Kirkmire CM, Wang ZJ, 2010. Regulation of opioid tolerance by let-7 family microRNA targeting the μ opioid receptor. J. Neurosci. 30, 10251–10258. 10.1523/JNEUROSCI.2419-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JA, Im H-II, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ, 2010. Striatal microRNA controls cocaine intake through CREB signalling. Nature 466, 197–202. 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Kenny PJ, 2012. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ, 2010. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci 13, 1120–1127. 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Davare M, Lasiek A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA, 2010. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol. Cell. Neurosci 43, 146–156. 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PJ, 2014. Epigenetics, microRNA, and addiction. Dialogues Clin. Neurosci 16, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Wai TC, Demler O, Walters EE, 2005. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch. Gen. Psychiatry 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosten TR, Markou A, Koob GF, 1998. Depression and stimulant dependence: neurobiology and pharmacotherapy. J. Nerv. Ment. Dis 186, 737–745. 10.1097/00005053-199812000-00001. [DOI] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J, Tugendreich S, 2014. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530. 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnova IN, Justinova Z, Cadet JL, 2016. Methamphetamine addiction: involvement of CREB and neuroinflammatory signaling pathways. Psychopharmacology (Berl.). 10.1007/s00213-016-4235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreibich AS, Briand L, Cleck JN, Ecke L, Rice KC, Blendy JA, 2009. Stress-induced potentiation of cocaine reward: a role for CRF R1 and CREB. Neuropsychopharmacology 34, 2609–2617. 10.1038/npp.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufahl PR, Zavala AR, Singh A, Thiel KJ, Dickey ED, Joyce JN, Neisewander JL, 2009. c-Fos expression associated with reinstatement of cocaine-seeking behavior by response-contingent conditioned cues. Synapse 63, 823–835. 10.1002/syn.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, 2011. Digital multiplexed gene expression analysis using the nanostring ncounter system. Curr. Protoc. Mol. Biol 94, 25B.10.1–25B.10.17. 10.1002/0471142727.mb25b10s94. [DOI] [PubMed] [Google Scholar]
- Larson EB, Graham DL, Arzaga RR, Buzin N, Webb J, Green TA, Bass CE, Neve RL, Terwilliger EF, Nestler EJ, Self DW, 2011. Overexpression of CREB in the nucleus accumbens shell increases cocaine reinforcement in self-administering rats. J. Neurosci 31, 16447–16457. 10.1523/JNEUROSCI.3070-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wolf ME, 2015. Multiple faces of BDNF in cocaine addiction. Behav. Brain Res. 10.1016/j.bbr.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-Y, Mao X, Wei L, 2008. Genes and (common) pathways underlying drug addiction. PLoS Comput. Biol 4, e2. 10.1371/journal.pcbi.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G, Steinert JR, Marczylo EL, D’Oro S, Fiore R, Forsythe ID, Schratt G, Zoli M, Nicotera P, Young KW, 2011. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J. Cell Biol 194, 889–904. 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lynch WJ, Peterson AB, Sanchez V, Abel J, Smith MA, 2013. Exercise as a novel treatment for drug addiction: a neurobiological and stage-dependent hypothesis. Neurosci. Biobehav. Rev 37, 1622–1644. 10.1016/j.neubiorev.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y-Y, Wang X, Huang Y, Marie H, Nestler EJ, Schlüter OM, Dong Y, 2016a. Re-silencing of silent synapses unmasks anti-relapse effects of environmental enrichment. Proc. Natl. Acad. Sci. U. S. A 113, 5089–5094. 10.1073/pnas.1524739113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y-Y, Wang X, Huang Y, Marie H, Nestler EJ, Schlüter OM, Dong Y, 2016b. Re-silencing of silent synapses unmasks anti-relapse effects of environmental enrichment. Proc. Natl. Acad. Sci. U. S. A 113, 5089–5094. 10.1073/PNAS.1524739113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson BJ, Bossert JM, Simmons DE, Nozaki N, Nagarkar D, Kreuter JD, Hope BT, 2005. Cocaine-induced CREB phosphorylation in nucleus accumbens of cocaine-sensitized rats is enabled by enhanced activation of extracellular signal-related kinase, but not protein kinase A. J. Neurochem 95, 1481–1494. 10.1111/j.1471-4159.2005.03500.x. [DOI] [PubMed] [Google Scholar]
- McCall Jones C, Baldwin GT, Compton WM, 2017. Recent increases in cocaine-related overdose deaths and the role of opioids. Am. J. Public Health 107, 430–432. 10.2105/AJPH.2016.303627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Feldman DA, Sheridan SD, Ip JPK, Kwok S, Amoah SK, Rosen B, Rodriguez BA, Crawford B, Swaminathan R, Chou S, Li Y, Ziats M, Ernst C, Jaenisch R, Haggarty SJ, Sur M, 2018. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol. Psychiatry 23, 1051–1065. 10.1038/mp.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan EZ, McNally GP, 2011. Accumbens shell AMPA receptors mediate expression of extinguished reward seeking through interactions with basolateral amygdala. Learn. Mem 18, 414–421. 10.1101/lm.2144411. [DOI] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM, 2011. Altered MicroRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol. Psychiatry 69, 188–193. 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SJ, Lusardi TA, Phillips JI, Saugstad JA, 2014. Sex differences in microRNA expression during developmentin rat cortex. Neurochem. Int. 77, 24–32. 10.1016/j.neuint.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Economic Impact of Illicit Drug Use on American Society, 2011. National Drug Intelligence Center, Washington, DC, DC. [Google Scholar]
- National Institute on Drug Abuse, 2015. Trends & Statistics [WWW Document]. https://www.drugabuse.gov/related-topics/trends-statistics.
- National Institute on Drug Abuse, 2018. The Science of Drug Use and Addiction: The Basics |. National Institute on Drug Abuse (NIDA). https://www.drugabuse.gov/publications/media-guide/science-drug-use-addiction-basics. [Google Scholar]
- National Institute on Drug Abuse, 2020. DrugFacts: Nationwide Trends [WWW Document]. https://www.drugabuse.gov/publications/drugfacts/nationwide-trends.
- Neisewander JL, Baker DA, Fuchs Ra, Tran-Nguyen LT, Palmer A, Marshall JF, 2000. Fos protein expression and cocaine-seeking behavior in rats after exposure to a cocaine self-administration environment. J. Neurosci 20, 798–805. 10.1523/JNEUROSCI.20-02-00798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paykel ES, Brugha T, Fryers T, 2005. Size and burden of depressive disorders in Europe. Eur. Neuropsychopharmacol 15, 411–423. 10.1016/j.euroneuro.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Plotnikova O, Baranova A, Skoblov M, 2019. Comprehensive analysis of human microRNA–mRNA interactome. Front. Genet 10, 933. 10.3389/fgene.2019.00933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomara C, Cassano T, D’Errico S, Bello S, Romano AD, Riezzo I, Serviddio G, Riezzo I, 2012. Data available on the extent of cocaine use and dependence: biochemistry, pharmacologic effects and global burden of disease of cocaine abusers. Curr. Med. Chem 19, 5647–5657. 10.2174/092986712803988811. [DOI] [PubMed] [Google Scholar]
- Powell GL, Vannan A, Bastle RM, Wilson MA, Dell’Orco M, Perrone-Bizzozero NI, Neisewander JL, 2020. Environmental enrichment during forced abstinence from cocaine self-administration opposes gene network expression changes associated with the incubation effect. Sci. Rep 10 10.1038/s41598-020-67966-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2019. R: a Language and Environment for Statistical Computing.
- Richardson NR, Roberts DCS, 1991. Fluoxetine pretreatment reduces breaking points on a progressive ratio schedule reinforced by intravenous cocaine self-administration in the rat. Life Sci. 49, 833–840. 10.1016/0024-3205(91)90248-A. [DOI] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, Scolnick E, Cichon S, St. Clair D, Corvin A, Gurling H, Werge T, Rujescu D, Blackwood DHR, Pato CN, Malhotra AK, Purcell S, Dudbridge F, Neale BM, Rossin L, Visscher PM, Posthuma D, Ruderfer DM, Fanous A, Stefansson H, Steinberg S, Mowry BJ, Golimbet V, De Hert M, Jönsson EG, Bitter I, Pietiläinen OPH, Collier DA, Tosato S, Agartz I, Albus M, Alexander M, Amdur RL, Amin F, Bass N, Bergen SE, Black DW, Børglum AD, Brown MA, Bruggeman R, Buccola NG, Byerley WF, Cahn W, Cantor RM, Carr VJ, Catts SV, Choudhury K, Cloninger CR, Cormican P, Craddock N, Danoy PA, Datta S, De Haan L, Demontis D, Dikeos D, Djurovic S, Donnelly P, Donohoe G, Duong L, Dwyer S, Fink-Jensen A, Freedman R, Freimer NB, Friedl M, Georgieva L, Giegling I, Gill M, Glenthøj B, Godard S, Hamshere M, Hansen M, Hansen T, Hartmann AM, Henskens FA, Hougaard DM, Hultman CM, Ingason A, Jablensky AV, Jakobsen KD, Jay M, Jürgens G, Kahn RS, Keller MC, Kenis G, Kenny E, Kim Y, Kirov GK, Konnerth H, Konte B, Krabbendam L, Krasucki R, Lasseter VK, Laurent C, Lawrence J, Lencz T, Lerer FB, Liang KY, Lichtenstein P, Lieberman JA, Linszen DH, Lönnqvist J, Loughland CM, MacLean AW, Maher BS, Maier W, Mallet J, Malloy P, Mattheisen M, Mattingsdal M, McGhee KA, McGrath JJ, McIntosh A, McLean DE, McQuillin A, Melle I, Michie PT, Milanova V, Morris DW, Mors O, Mortensen PB, Moskvina V, Muglia P, Myin-Germeys I, Nertney DA, Nestadt G, Nielsen J, Nikolov I, Nordentoft M, Norton N, Nöthen MM, O’Dushlaine CT, Olincy A, Olsen L, O’Neill FA, Ørntoft TF, Owen MJ, Pantelis C, Papadimitriou G, Pato MT, Peltonen L, Petursson H, Pickard B, Pimm J, Pulver AE, Puri V, Quested D, Quinn EM, Rasmussen HB, Réthelyi JM, Ribble R, Rietschel M, Riley BP, Ruggeri M, Schall U, Schulze TG, Schwab SG, Scott RJ, Shi J, Sigurdsson E, Silverman JM, Spencer CCA, Stefansson K, Strange A, Strengman E, Stroup TS, Suvisaari J, Terenius L, Thirumalai S, Thygesen JH, Timm S, Toncheva D, Van Den Oord E, Van Os J, Van Winkel R, Veldink J, Walsh D, Wang AG, Wiersma D, Wildenauer DB, Williams HJ, Williams NM, Wormley B, Zammit S, Sullivan PF, O’Donovan MC, Daly MJ, Gejman PV, 2011. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet 43, 969–978. 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ, 2008. The let-7 family of microRNAs. Trends Cell Biol. 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Sadakierska-Chudy A, Frankowska M, Miszkiel J, Wydra K, Jastrzębska J, Filip M, 2017. Prolonged induction of miR-212/132 and REST expression in rat striatum following cocaine self-administration. Mol. Neurobiol 54, 2241–2254. 10.1007/s12035-016-9817-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ, 2011. Upregulation of dicer and MicroRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol. Psychiatry 69, 180–187. 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Im HI, Venø MT, Fowler CD, Min A, Intrator A, Kjems J, Kenny PJ, O’Carroll D, Greengard P, 2010. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J. Exp. Med 207, 1843–1851. 10.1084/jem.20100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, MacRae IJ, 2014. Structural basis for microRNA targeting. Science (80-.) 346, 608–613. 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin V, Jin H, Ng E, Ng E, Cheng A, Chong W, Wong C, Leung W, Sung J, Chu K, 2010. NF-κB targets miR-16 and miR-21 in gastric cancer: involvement of prostaglandin E receptors. Carcinogenesis 32, 240–245. 10.1093/carcin/bgq240. [DOI] [PubMed] [Google Scholar]
- Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, Jin P, Zhao X, 2010. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28, 1060–1070. 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas M, Chauvet C, Thiriet N, El Rawas R, Jaber M, 2008. Reversal of cocaine addiction by environmental enrichment. Proc. Natl. Acad. Sci. U. S. A 105, 17145–17150. 10.1073/pnas.0806889105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel KJ, Sanabria F, Pentkowski NS, Neisewander JL, 2009. Anti-craving Effects of Environmental Enrichment. 10.1017/S1461145709990472. [DOI] [PMC free article] [PubMed]
- Thiel KJ, Pentkowski NS, Peartree NA, Painter MR, Neisewander JL, 2010. Environmental living conditions introduced during forced abstinence alter cocaine-seeking behavior and Fos protein expression. Neuroscience 171, 1187–1196. 10.1016/J.NEUROSCIENCE.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel KJ, Engelhardt B, Hood LE, Peartree NA, Neisewander JL, 2011. The interactive effects of environmental enrichment and extinction interventions in attenuating cue-elicited cocaine-seeking behavior in rats. Pharmacol. Biochem. Behav 97, 595–602. 10.1016/j.pbb.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Nguyen LTL, Fuchs RA, Coffey GP, Baker DA, O’Dell LE, Neisewander JL, O’Dell LE, Neisewander JL, 1998. Time-dependent changes in cocaine-seeking behavior and extracellular dopamine levels in the amygdala during cocaine withdrawal. Neuropsychopharmacology 19, 48–59. 10.1016/S0893-133X(97)00205-4. [DOI] [PubMed] [Google Scholar]
- Vannan A, Powell GL, Scott SN, Pagni BA, Neisewander JL, 2018. Animal models of the impact of social stress on cocaine use disorders. In: Olive MF, Tomek SE (Eds.), Animal Models for Examining Social Influences on Drug Addiction. International Review of Neurobiology. Academic Press, pp. 131–169. 10.1016/BS.IRN.2018.07.005. [DOI] [PubMed] [Google Scholar]
- Viola TW, Wearick-Silva LE, De Azeredo LA, Centeno-Silva A, Murphy C, Marshall P, Li X, Singewald N, Garcia F, Bredy TW, Grassi-Oliveira R, 2016. Increased cocaine-induced conditioned place preference during periadolescence in maternally separated male BALB/c mice: the role of cortical BDNF, microRNA-212, and MeCP2. Psychopharmacology (Berl.) 233, 3279–3288. 10.1007/s00213-016-4373-z. [DOI] [PubMed] [Google Scholar]
- Wahid F, Shehzad A, Khan T, Kim YY, 2010. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta - Mol. Cell Res 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Weiss F, Martin-Fardon R, Ciccocioppo R, Kerr TM, Smith DL, Ben-Shahar O, 2001. Enduring resistance to extinction of cocaine-seeking behavior induced by drug-related cues. Neuropsychopharmacology 25, 361–372. 10.1016/S0893-133X(01)00238-X. [DOI] [PubMed] [Google Scholar]
- Wright C, Turner JA, Calhoun VD, Perrone-Bizzozero N, 2013. Potential impact of miR-137 and its targets in schizophrenia. Front. Genet 4 10.3389/fgene.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nanostring data are deposited in the Gene Expression Omnibus (GSE153524). R code used for data analyses are available at https://gitlab.com/neisewander_asu/vannan-powell-2020.