ABSTRACT
Xanthomonas is a well-studied genus of bacterial plant pathogens whose members cause a variety of diseases in economically important crops worldwide. Genomic and functional studies of these phytopathogens have provided significant understanding of microbial-host interactions, bacterial virulence and host adaptation mechanisms including microbial ecology and epidemiology. In addition, several strains of Xanthomonas are important as producers of the extracellular polysaccharide, xanthan, used in the food and pharmaceutical industries. This polymer has also been implicated in several phases of the bacterial disease cycle. In this review, we summarise the current knowledge on the infection strategies and regulatory networks controlling virulence and adaptation mechanisms from Xanthomonas species and discuss the novel opportunities that this body of work has provided for disease control and plant health.
Keywords: plant disease, adaptation, extracellular polysaccharides, biofilm, type III effectors, regulatory circuits
Here, we discuss the current knowledge surrounding regulatory networks and systems that control virulence and adaption mechanisms in Xanthomonas species. Additionally, we detail how study of these pathogens has provided novel opportunities for disease control and plant health.
INTRODUCTION
Xanthomonas (two Greek words; xanthos, meaning ‘yellow’, and monas, meaning ‘entity’) is a large genus of plant-associated Gram-negative bacteria. These yellow-pigmented bacteria are generally rod shaped with a single polar flagellum, are obligate aerobes and have an optimal growth temperature of between 25 and 30°C. (Bradbury 1984) The genus, which resides at the base of the gamma subdivision of the proteobacteria (Jun et al. 2010), comprises 27 species that cause serious diseases in almost 400 plants (124 monocots and 268 dicots) including a wide variety of important crops such as rice, citrus, cabbage and pepper (Ryan et al. 2011). Pathogenic species of Xanthomonas show a high degree of host plant specificity and species can be further differentiated into pathovars that are defined by characteristic host range and/or tissue specificity, invading either the xylem elements of the vascular system or the intercellular spaces of the mesophyll parenchyma tissue of the host (Ryan et al. 2011; Jacques et al. 2016) (Fig. 1, Table 1). Along with host range being defined at the plant species or genus level, several Xanthomonas intrapathovar groups of strains interact with intraspecies variants of hosts. Races have been described within several pathovars, including Xanthomonascampestris pv. campestris and Xanthomonas oryzae pv. oryzae to group strains that interact specifically with some host cultivars near isogenic lines carrying specific resistance genes or varieties (Vicente and Holub 2013; Jacques et al. 2016).
Figure 1.
Life cycle and disease symptoms of Xanthomonas. (A), Model illustrating the life cycle of the black rot pathogen Xanthomonas campestris pv. campestris (Xcc). Like most Xanthomonads, Xcc can survive in plant debris in soil for up to two years, but not more than six weeks in free soil. Xcc also has the ability to colonise plant seeds which represents a major route of disease transmission. Xcc can also be spread from infected plants to healthy plants by various environmental and mechanical means. After germination of colonised seeds, the seedling becomes infected. This may manifest as shrivelling and the blackening of the margins of the seedling. Xcc may also invade mature plants via the hydathodes, although leaf damage caused by insects and the root system also serve as portals of entry. These entry points usually provide a direct path to the plant vascular system leading to systemic host infection. V-shaped necrotic lesions extending from the leaf margins manifest as the infection develops. The disease draws its name from the blackened veins within the necrotic lesions. (B), Examples of disease symptoms caused by various Xanthomonas species. (i, ii) Black rot of cabbage caused by Xanthomonas campestris pv. campestris. (iii, iv) Citrus canker of citrus caused by Xanthomonas citri pv. citri. (v, vi) Bacterial leaf streak of rice caused by Xanthomonas oryzae pv. oryzicola. (vii, viii) Bacterial blight of rice caused by Xanthomonas oryzae pv. oryzae.
Table 1.
List of names and acronyms ofXanthomonas strains discussed in this review.
| Xanthomonas spp. | Pathovar | Acronym | Host | Disease | Taxonomy ID |
|---|---|---|---|---|---|
| X. albilineans | Sugarcane | Leaf scald | NCBI:txid29447 | ||
| X. alfalfae | Alfalfa | Bacterial leaf spot | NCBI:txid366650 | ||
| X. arboricola | X. arboricola pv. pruni | Xap | Prunus | Bacterial spot | NCBI:txid69929 |
| X. arboricola pv. punicae | Xcp | Pomegranate (Punica granatum) | Leaf blight | NCBI:txid487838 | |
| X. arboricola pv. juglandis | Xaj | Persian (English) walnut (Juglans regia) | Walnut blight | NCBI:txid195709 | |
| X. axonopodis | X. axonopodis pv. manihotis | Xam | Cassava | Bacterial blight | NCBI:txid43353 |
| X. campestris | X. campestris pv. armoraciae | Xca | Horseradish | Bacterial leaf spot | NCBI:txid329463 |
| X. campestris pv. campestris | Xcc | Brassicaceae | Black rot | NCBI:txid340 | |
| X. campestris pv. leersiae | Xcl | Perennial grass | Bacterial streak | NCBI:txid487875 | |
| X. campestris pv. musacearum | Xvm | Banana | Enset wilt | NCBI:txid454958 | |
| X. campestris pv. raphani | Xcr | Brassica oleracea | Bacterial leaf spot | NCBI:txid359385 | |
| X. campestris pv. vitians | Xcv | Lettuce | Bacterial leaf spot | NCBI:txid83224 | |
| X. cannabis | Cannabis (Cannabis sativa L.) | Bacterial leaf spot | NCBI:txid1885674 | ||
| X. citri | X. citri pv. citri | Xcci | Citrus | Citrus canker | NCBI:txid611301 |
| X. citri pv. fuscans | Xcf | Bean (Phaseolus vulgaris L.) | Bacterial blight | NCBI:txid366649 | |
| X. citri pv. glycines | Xcg | Soybean (Glycine max) | Bacterial pustule | NCBI:txid473421 | |
| X. citri pv. malvacearum | Xcm | Cotton (Gossypium spec.) | Bacterial blight | NCBI:txid86040 | |
| X. citri pv. mangiferaeindicae | Xmi | Mango (Mangifera indica) | Bacterial black spot | NCBI:txid454594 | |
| X. citri pv. punicae | Xcp | Pomegranate (Punica granatum) | Leaf blight | NCBI:txid487838 | |
| X.cucurbitae | Cucurbits | Bacterial spot | NCBI:txid56453 | ||
| X. cynarae | Artichoke (Cynara scolymus L.) | Bacterial bract spot | NCBI:txid10214 | ||
| X. euvesicatoria | X. campetris pv. vesicatoria | Xav | Pepper and tomato | Bacterial leaf spot | NCBI:txid456327 |
| X. floridensis | Watercress | – | NCBI:txid1843580 | ||
| X. fragariae | Strawberry | Bacterial angular leaf spot | NCBI:txid48664 | ||
| X. gardneri | Pepper and tomato | Bacterial spot | NCBI:txid90270 | ||
| X. maliensis | Rice | – | NCBI:txid1321368 | ||
| X. nasturtii | Watercress | – | NCBI:txid1843581 | ||
| X. oryzae | X. oryzae pv. oryzae | Xoo | Rice | Bacterial blight | NCBI:txid64187 |
| X. oryzae pv. oryzicola | Xoc | Rice | Bacterial streak | NCBI:txid129394 | |
| X. perforans | Tomato | Bacterial spot | NCBI:txid442694 | ||
| X. phaseoli | X. phaseoli pv. phaseoli | Xcp | Bean (Phaseolus vulgaris L) | Bacterial blight | NCBI:txid1985254 |
| X. prunicola | nectarine (Prunus persica var. nectarina) trees | – | NCBI:txid2053930 | ||
| X. pseudoalbilineans | Not in NCBI list yet | ||||
| X. sacchari | Sugarcane | Chlorotic streak disease | NCBI:txid56458 | ||
| X. translucens | X. translucens pv. translucens | Xtt | Wheat | Black chaff | NCBI:txid134875 |
| X. translucens pv. undulosa | Xtu | Wheat | Black chaff | NCBI:txid487909 | |
| X. vasicola | X. vasicola pv. vasculorum | Xvv | Sugarcane | Gumming disease | NCBI:txid325776 |
Note: Xanthomonasarboricola pv. punicae is currently listed as X. citri pv. punicae in NCBI taxonomy.
The main route of Xanthomonas species (spp.) transmission is via contaminated seeds, although weeds and infected plant debris are also potential sources of inoculum (Gitaitis and Walcott 2007). Initially, bacteria grow epiphytically (on leaf surfaces), and then enter into the host through either hydathodes or wounds to spread systemically through the vascular system or through stomata to colonise the mesophyll parenchyma (Ryan et al. 2011; Fig. 1). For example, X. campestris pv. campestris (Xcc) causes a systemic vascular disease of brassicas known as black rot, characterised by V-shaped chlorotic lesions spreading from the leaf margins, whereas X. campestris pv. armoraciae (Xca) causes a leaf spotting disease of the brassica mesophyllic tissue but does not colonise the vascular system (Fargier, Fischer-Le Saux and Manceau 2011).
Here, we review the lifestyle and properties of Xanthomonas spp. with an emphasis on adaptability, virulence and epidemiology. We describe the mechanisms that contribute to the ability of Xanthomonas spp. to survive during epiphytic and endophytic growth and to cause disease, considering the role of diverse regulatory and sensing systems, secreted effectors and the biosynthesis of extracellular polysaccharide (EPS) and lipopolysaccharides (LPS). We discuss the insight that genome sequencing of Xanthomonas spp., has had on taxonomical classifications of the bacteria and understanding pathogen evolution and host adaptation. Finally, we consider how recent work understanding Xanthomonas spp. pathophysiology is being exploited for disease suppression and biocontrol.
XANTHOMONAS LIFESTYLE
Xanthomonas in the plant microbiome
Xanthomonas is known to live part of its life cycle outside of the plant host such as in the lesion of fallen leaves or freely in the soil, which might serve as an inoculum for further infection of plant hosts (Zhao, Damicone and Bender 2002). Multiple studies, covering numerous hosts and geographic locations, have identified the Xanthomonadales order as an important component in both the rhizosphere and soil microbiome environments, where they compose between 2% and 7% of the bacteria in the microbial community (Bulgarelli et al. 2015; Bhattacharyya et al. 2018). However, metagenomic analysis at higher resolution indicated that the abundance of the Xanthomonas genus ranges from below detection levels to 0.7% in the rhizosphere and soil microbiomes (Souza et al. 2016; Xu et al. 2018).
The abundance of Xanthomonas within the plant phyllosphere microbial community varies greatly between plant species, tissues, sampling season and geographic locations. Xanthomonas has been identified in the aerial parts of tomato, lettuce, rapeseed, clover, soybean, arabidopsis and rice (Delmotte et al. 2009; Knief et al. 2012; Rastogi et al. 2012; Ottesen et al. 2019). Several metagenomics analyses from the United States recognise Xanthomonas as a component of the tomato phyllosphere (Ottesen et al. 2015, 2019). Anatomical based analysis in tomato plants also identified Xanthomonas as an important component of the tomato microbiome (Ottesen et al. 2013). Notably, Xanthomonas represented 10%–40% of the whole bacterial communities of fruits, leaves and flowers, while, similarly to other studies, was not found in a significant abundance in the rhizosphere (Ottesen et al. 2013). Xanthomonas was also found as a dominant member in the microbiome of field grown Romain lettuce phyllosphere (Rastogi et al. 2012; Burch et al. 2016). In particular, one large scale microbiome study with samples collected from 88 different lettuce growing areas has identified Xanthomonas to be present in about a third of all samples, where it comprises, on average, 4% of the lettuce microbiome (Rastogi et al. 2012). It should be noted that in many cases Xanthomonas was not found as a major component of the phyllosphere, indicating that Xanthomonas is not an integral component of the core plant microbial community (Liu et al. 2017; Suhaimi et al. 2017).
Epiphytic lifestyle
The infection cycle of Xanthomonas can be divided into the epiphytic stage and the endophytic stage. The epiphytic stage initiates once bacteria are introduced into the aerial tissues of a new host, usually leaf or fruit tissue and continues until the entrance into the host tissue via the plant natural openings and wounds. Once inside a host plant, the bacteria enter the endophytic stage and colonise the host. Upon reaching high population bacteria re-emerge onto the leaf surface and are transmitted mostly through wind or rain to a new host starting the infection cycle again (Moreira et al. 2015). Many aspects of the Xanthomonas endophytic infection have been studied and factors that are important have been described (see below). In contrast, less attention has been given to the investigation of the epiphytic stage of the Xanthomonas lifestyle.
Traditionally, the plant surface area is considered to be a hostile environment for bacteria (Lindow and Brandl 2003). In this environment bacteria are exposed to severe radiation, unstable humidity, limited nutrient resources and competition with other bacteria occupying the same niche. In spite of these obstacles, Xanthomonas can sustain in the leaf surface areas for extended periods of time (Rigano et al. 2007b; Zarei et al. 2018). Xanthomonas strains were reported to be capable of maintaining stable bacterial populations in their host or non-host plant. Studies comparing epiphytic survival in host and non-host plant in different pathosystems reported better epiphytic fitness in the host, indicating that host specificity plays a role in epiphytic fitness (Rigano et al. 2007b; Zarei et al. 2018). The duration that Xanthomonas bacteria can sustain on the leaf surface is less clear and the survival ranges between a few weeks to several months in different reports (Rigano et al. 2007b; Zarei et al. 2018). We speculate that these variations may result from the use of different bacterial species, host plants or field environments.
Attachment and biofilm formation on the plant surface
When introduced to the plant surface Xanthomonas utilises multiple adhesion strategies to attach to the plant surface, including bacterial surface polysaccharides (Vorhölter, Niehaus and Pühler 2001; Petrocelli et al. 2012), adhesion proteins (Pradhan, Ranjan and Chatterjee 2012) and the type IV pilus (Dunger et al. 2014; Petrocelli et al. 2016). After initial attachment, Xanthomonas forms biofilm-like structures. Similar structures were found in a multitude of leaf surface dwelling bacteria and it was hypothesized that creation of a microenvironment within the leaf surface biofilm protects the bacteria from harsh abiotic stress conditions in the phyllosphere (Yu et al. 2013). Biofilm is a general term for structures created by bacterial communities on the surface areas and composed of bacterial cells connected through extracellular polymeric substance matrix composed of exopolysaccharides (EPS), extracellular DNA (eDNA), proteins and lipids (Castiblanco and Sundin 2016). Xanthomonas biofilm is a dynamic structure and its assembly and dispersal is mediated by the quorum sensing signal molecule diffusible signal factor (DSF). Through the internal second messenger cyclic di-GMP, DSF promotes biofilm formation through induction of EPS production and pilus assembly (Tang et al. 1991; Guo et al. 2011; Rai et al. 2012; Ryan 2013). In parallel, DSF negatively controls the biofilm by positively regulating β-1,4-mannanase, ManA, which disperses EPS and disassembles biofilm (Dow et al. 2003; Tao, Swarup and Zhang 2010; Chen et al. 2010b).
The biofilm matrix in Xanthomonas spp. is considered to be mainly composed of Xanthan gum (discussed in detail below) (Crossman and Dow 2004). Xanthan gum biosynthesis is a complex procedure mediated by the gum operon, a large transcriptional unit containing 12 enzyme coding genes (gumB-gumM) (Vojnov et al. 2001a). Disruption of either gumB, gumC, gumD, gumE, gumH, gumK, gumI and gumK compromises both Xanthan gum production (Vojnov et al. 1998; Kim et al. 2009) and biofilm formation (Rigano et al. 2007b; Li and Wang 2011a). In addition to the gum cluster, biofilm EPS composition was identified to be dependent on xagABC, a glycosyltransferase system associated with polysaccharide biosynthesis (Tao, Swarup and Zhang 2010; Li and Wang 2012). eDNA has long been considered to be an integral part in bacterial biofilms (Okshevsky and Meyer 2015) but only recently shown to play a role in Xanthomonas biofilm formation. Specifically, Sena-Vélez and colleagues have shown that eDNA is highly abundant in Xanthomonas citri mature biofilm and accumulates throughout biofilm development (Sena-Vélez et al. 2016). In addition, extracellular DNase treatment was able to inhibit biofilm formation but only partially dispersed mature biofilm, suggesting that eDNA plays a significant role in early stage of biofilm establishment (Sena-Vélez et al. 2016). Other factors that were reported to be critical for Xanthomonas biofilm formation include cell surface LPS and lipoproteins (Li and Wang 2011a; Li and Wang 2011b; Petrocelli et al. 2012; Yan, Hu and Wang 2012), the bacterial flagellum and type IV pili (Malamud et al. 2011; Dunger et al. 2014).
While biofilm formation, stability and composition have been extensively examined in Xanthomonas spp., very few studies directly tested the effect of biofilm disruption on epiphytic fitness or analysed biofilm composition under environmental conditions. EPS biosynthesis deficient mutants (gumB and gumD, in particular) displayed significant reduction in leaf surface survival compared to the wild type X. citri pv. citri (Xcci) and X. axonopodis pv. manihotis (Xam) (Kemp et al. 2004; Dunger et al. 2007; Rigano et al. 2007b). Furthermore, Li and Wang have demonstrated that chemical inhibition of biofilm formation by D-leucine and 3-indolylacetonitrile disrupts leaf surface colonisation and disease development by Xcci (Li and Wang 2013).
Surface movement and chemotaxis
Only a handful of studies have focused on motility and chemotactic movement on the plant aerial surfaces by bacterial pathogens (Hockett, Burch and Lindow 2013; Moreira et al. 2015) and the importance of chemotactic motility in the epiphytic environment is not completely clear. It is hypothesized that motility is required for clustering of bacteria on the leaf surface and directing the bacteria towards potential entry points into the plant (Hockett, Burch and Lindow 2013; Moreira et al. 2015).
A few studies have monitored GFP-labelled Xanthomonas on host leaves, showing similar patterns of behaviour. A few hours after inoculation, attachment of single bacterial colonies can be observed on the leaf surface. In the following days, bacteria start clustering together in microcolonies and later aggregate in the proximity of plant natural openings such as stomata or hydathodes, then enter the host endophytic spaces (Zhang et al. 2009; Cubero et al. 2011; Cerutti et al. 2017). Different Xanthomonas spp. have preferences for particular plant openings according to their colonisation strategy (Ryan et al. 2011). Non-systemic apoplastic Xanthomonas such as X. euvesicatoria or X. citri display high preference for stomata while systemic xylem colonising species like Xcc or X. oryzae pv. oryzae (Xoo) usually cluster around the hydathodes (Shekhawat and Srivastava 1972; Zhang et al. 2009; Cubero et al. 2011; Cerutti et al. 2017).
Directional movement of the bacteria to the plant openings is potentially mediated through chemotaxis. Chemotactic movement of X. oryzae towards rice xylem sap, xylem specific sugars and certain amino acids was demonstrated previously (Kumar Verma, Samal and Chatterjee 2018). Kumar Verma et al. have linked xylem sap- and amino acid-triggered movement to the MCP2 chemoreceptor. Deletion of either mcp2 or chemosensory signalling protein coding genes cheW2, cheA2, cheY1 and cheB2 disrupts chemotactic movement towards these metabolites (Kumar Verma, Samal and Chatterjee 2018). In addition these mutants were not able to epiphytically infect rice leaves (Kumar Verma, Samal and Chatterjee 2018), indicating that X. oryzae uses xylem chemotactic signals leaked from hydathodes to direct themselves into the xylem vessels.
Most Xanthomonas spp. harbour motility-associated apparatuses like flagella and type IV pili and motility of these bacteria in vitro was reported to be mostly attributed to flagella (Yang et al. 2009; Malamud et al. 2011; Tian et al. 2015). It is yet to be shown if flagella directly play a role in endophytic fitness or plant surface motility but it was reported to be significant to infectivity (Malamud et al. 2011; Bae et al. 2018; Kumar Verma, Samal and Chatterjee 2018), suggesting that flagellar motility might play a role in the epiphytic phase.
Response to radiation and light
On the leaf surface, bacteria are exposed to solar radiation, which includes UV-A and UV-B wavelengths that cause damage to DNA, membranes and proteins. Epiphytic bacteria adopted multiple strategies to reduce radiation exposure and its damage by blocking radiation through pigments and biofilm structures, and acquiring DNA repair complexes (Jacobs, Carroll and Sundin 2005; Cazorla et al. 2008). Most Xanthomonas spp. produce yellow membrane bound pigments called xanthomonadins, whose synthesis is dependent on a gene cluster encoding seven transcriptional units (pigA-pigG) (Poplawsky, Kawalek and Schaad 1993). Deletion of pigC and pigG in X. campestris resulted in enhanced susceptibility to radiation from natural light but not to UV (Poplawsky et al. 2000). Several UV sensitive mutants were identified in different Xanthomonas strains; most are related to response to oxidative stress and DNA repair (Martínez et al. 1997; Shen et al. 2007; Tondo et al. 2016).
It was reported that response to visible light plays a significant role in modulating pathogenicity and epiphytic fitness. XccBphP of X. campestris encodes a functional bacteriophytochrome protein displaying altered response between red and far red (Otero et al. 2016). Characterisation of WT and XccBphP mutant in light and dark conditions revealed that light negatively regulates epiphytic survival associated traits such as motility, xanthan production and extracellular hydrolase activity in a XccBphP dependent manner (Bonomi et al. 2016). LOV (Light, Oxygen, Voltage), a blue light photosensory protein of X. citri, was reported to regulate surface attachment and biofilm formation in a light dependent manner (Kraiselburd et al. 2012). Interestingly, Kraiselburd et al. (2012) reported that light plays a positive role in biofilm formation in X. citri, suggesting that the response to light is not the same in different Xanthomonas spp.
Interaction with other bacteria on the aerial surfaces
Plant aerial surfaces are colonised by a multitude of microorganisms that form a complex communal structure. The interactions between different bacteria on the leaf surface have not been extensively examined, however. Similarly to other leaf surface habitating bacteria like Pseudomonas syringae and Pantoea agglomerans (Monier and Lindow 2005), aggregates of Xanthomonas are usually considered as a single species aggregates (Monier and Lindow, 2005). Xanthomonas utilizes the type IV and type VI secretion systems to interact with the microflora of the phyllosphere. Xanthomonas citri utilizes a type IV secretion system to kill other Gram-negative bacteria through delivery of antibacterial proteins (Souza et al. 2015). The group also reported that the type IV secretion system contributes to competitive fitness when inoculated with other bacteria, indicating that Xanthomonas utilises this system to establish itself on the leaf surface by actively killing bacteria that occupy the same niche (Souza et al. 2015). The same group also recently reported that the X. citri type VI secretion system is required for protection against the predatory amoeba Dictyostelium (Bayer-Santos et al. 2018). These studies indicate that Xanthomonas relies on multiple systems to respond to microorganisms in its habitat.
Cuticle interactions, resource utilisation and metabolic activity on the plant aerial surfaces
Microorganisms on plant aerial surfaces are exposed to limited amount of macro- and micro- nutrients and unstable humidity (Lindow and Brandl 2003). A few studies have examined Xanthomonas metabolism on the leaf surface. Cubero and colleagues monitored the expression of an unstable GFP variant in X. citri colonising leaf and fruit surfaces as a marker for metabolic activity (Cubero et al. 2011). The study concluded that although metabolic activity was significantly reduced during the incubation on the leaf surface, it increased once bacteria reached the stomatal area (Cubero et al. 2011). Epiphytic survival studies do occasionally show an increase in the bacterial population on the leaf surface (Zarei et al. 2018), indicating that the bacteria can utilize the scarce resources in the phyllosphere to some extent. It is unclear, however, what metabolic pathways are induced in Xanthomonas in this environment. Déjean and colleagues reported that disruption of xylanase-coding genes in X. campestris resulted in reduced epiphytic growth on the surface of host and non-host plants, suggesting that xylan is available to the bacteria and can be utilised as a carbon source on the plant aerial surfaces (Déjean et al. 2013).
Leaf and fruit epidermal surfaces are covered by the cuticle, a waxy layer that prohibits water evaporation and serves as a physical barrier against microbial pathogens, preventing microorganisms from both entering the plant and accessing the plant water and nutrient resources (Ziv et al. 2018). Alteration and disruption of cuticle structure was reported in several microorganisms. Pseudomonas syringae bacteria secrete multiple surfactants and toxins to increase leaf wettability while multiple fungal pathogens actively degrade host cutin polymer using cutinases, lipolytic/esterolytic enzymes (Lindow and Brandl 2003). No data are available regarding Xanthomonas-cuticle interactions and further research is required to establish the strategies utilised by Xanthomonas to alter or disrupt the plant cuticle.
XANTHOMONAS DISPERSAL AND SPREAD
Environmental factors
Dispersal of pathogens and the transfer between hosts play an extremely important role in the pathogen infection cycle. Environmental factors that facilitate pathogen dispersal can dictate why susceptible crops are routinely infected in some areas but stay pathogen free in others. Xanthomonas spp. thrive better on slightly higher temperatures (on average 25–35°C) and are capable to reach high enough inoculum in the host to allow dispersal (Christiano et al. 2009; Morales et al. 2018a; 2018b). In addition, high humidity was found to be required for epiphytic survival and transmission (Christiano et al. 2009). In this condition, the high abundance of bacteria on the surface area of a infected host will be a more significant inoculum source for dispersal, while, in parallel, the same condition will contribute to epiphytic survival of the bacteria upon arrival to a leaf of a new host.
Plant response to warm and humid environment exacerbate disease necrotic symptoms, which increases the availability of the pathogen to the outside environment (Diab et al. 1982; Kumar et al. 2006). In high humidity, plants open stomata and hydathodes to sustain water homoeostasis (Zeng, Melotto and He 2010; Singh 2013). This open state allows both Xanthomonas entry from the plant surface (Zeng, Melotto and He 2010) and exit of high inoculum of bacteria from infected plants. Guttation of xylem sap from hydathodes at night and during high humidity is considered as the route of exit and entry of xylem-inhabiting Xanthomonas and plays a significant role in acquisition and dispersal of these pathogens (Guo and Leach 1989; Cerutti et al. 2017).
Dispersal through wind and rain
Wind and rain are considered as the main carriers of most Xanthomonass spp. in short- and long-distance dispersal of most Xanthomonas spp. Tropical storms, tornadoes and heavy rainfalls are considered as significant facilitators of long distance dispersal of many Xanthomonas spp. and were found to be highly correlated to outbreaks (Gottwald and Irey 2007; Champoiseau et al. 2009; Jha and Sonti 2009). The effect of wind on pathogen dispersal has been extensively studied in regard to citrus canker, caused by X. citri. Multiple epidemiological studies have identified that physical proximity to infection areas and rainfall significantly increase disease occurrence (Gottwald et al. 2002; Gottwald et al. 2007). Several studies have focused on pathogen dispersal during simulated wind and rain in laboratory conditions. These studies demonstrated a direct correlation between wind intensity and the pathogen dispersal distance between infected and neighbouring plants in X. citri and X. alfalfae, indicating that wind is a major abiotic vector of the bacteria (Bock et al. 2012). The understanding of the importance of wind and rain to dispersal of Xanthomonas has greatly contributed to disease management of citrus canker. Application of windbreaks successfully reduced pathogen spread between groves (Behlau et al. 2008; Moschini et al. 2014). In addition, tighter surveillance for canker symptomatic plants after heavy rain and storms has enabled early detection of potential outbreaks (Gottwald and Irey 2007; Canteros, Gochez and Moschini 2017).
Dispersal by people and non-specific vectors
Agricultural practices are a major means for pathogen transmission. While it has been claimed that people and agricultural machinery are one of the main sources of short distance and long-distance transmission, few studies have been dedicated to establishing the exact effect of these factors on Xanthomonas dispersal. Dispersal through people and farming equipment was reported as the major source of spread of the banana and plantain pathogen X.campestris pv. musacearum (Xvm) (Nakato, Mahuku and Coutinho 2018). The bacteria were reported to survive on farming equipment for between one and three weeks and shown to be actively transferred from one plant to another through farming tools (Addis et al. 2010; Nakato, Mahuku and Coutinho 2018). Education of local farmers about the importance of cleaning of farming tools and proper destruction of infected crops is currently considered a key element in the prevention of banana Xanthomonas wilt in east and central Africa (Nakato, Mahuku and Coutinho 2018).
The lifecycle of several vascular tissue-residing plant pathogens such as Candidatus Liberibacter, Xylella fastidiosa and Pantoea stewartii includes a phase of habitation inside or associated with a specific insect vector which allows plant-to-plant transmission (Chatterjee, Almeida and Lindow 2008; Roper 2011; Wang et al. 2017). A similar mechanism is not known for any Xanthomonas spp. although speculation of a role of non-specific biotic vector has arisen (Belasque et al. 2005; Zandjanakou-Tachin et al. 2007). Xanthomonas bacteria have been isolated from field specimens of potential biotic vectors such as insects, birds and bats; the bacteria were shown to survive on these alleged vectors for a duration of a few days (Belasque et al. 2005; Zandjanakou-Tachin et al. 2007). Studies of active plant-to-plant transmission conducted with leafminers, flea beetles and grasshoppers have shown that insect to plant transmission of Xanthomonas spp. can occur but in a very low capacity, indicating that vector transmission probably only plays a relatively small role in short distance transmission (Belasque et al. 2005; Zandjanakou-Tachin et al. 2007; van der Wolf and van der Zouwen 2010; Buregyeya et al. 2014). Van der Wolf and colleagues showed that X. campestris can be transmitted to cabbage flowers though flies and bumble bees and demonstrated that flower infection can be carried on to the seeds (van der Wolf and van der Zouwen 2010; van der Wolf et al. 2019). The prevalence of such transmission in the field is still unknown and should be subjected to further study. Transmission through pollinating vectors is speculated to be one of the main means of dispersal for Xvm (Nakato, Mahuku and Coutinho 2018). It was found that stingless bees and fruit flies carry high bacterial inoculum of Xvm when isolated from infected fields (Tinzaara et al. 2006) and that floral parts are usually the initial inoculation point of the bacteria (Ocimati et al. 2012; Rutikanga et al. 2015). Following these observations, floral debudding is now commonly practiced in banana to prevent pathogen infection through any insect vector (Blomme et al. 2017). It should be noted that while insect infection is very likely linked to Xvm dispersal, direct vector-mediated plant-to-plant transmission has yet to be experimentally verified. Future research of this unique system can provide new insights into Xanthomonas-vector interactions.
Seed dispersal
Seed transmission is reported for many Xanthomonas spp. and contaminated seeds are considered as a dominant factor in epidemics of black rot of crucifers, bacterial spot of tomato, bacterial blight of cassava and many others (Gitaitis and Walcott 2007). Although a significant amount of work has been dedicated towards standardising screening methods for identification of infected seeds and general seed treatment (Gitaitis and Walcott 2007), our understanding of the mechanisms of seed transport and of the bacterial lifestyle within the seed is fragmentary. Xanthomonas spp. can survive inside seeds for periods of several weeks to several months (He and Munkvold 2013) and studies of X. campestris and X. oryzae have also detected that duration of survival is different between seeds from different hosts (Dutta et al. 2016). Transmission efficiency from seed to seedling for several Xanthomonas spp. was found to be directly dependent on the amount of bacteria inoculum in the seed (Darrasse et al. 2007). Transmission from plant to seeds usually occurs when plants are heavily infected but the frequency appears to be pathogen and host dependent, varying from less than 1% to around 90% (Humeau et al. 2006; Giovanardi et al. 2015). No consistent data is available regarding which part of the seed is contaminated by the bacteria. Contamination of the outer seed coat was reported in most cases while contamination of the inner seed and embryo was reported in some cases but not others. For instance, only external colonisation was reported in lettuce seeds infected with X. campestris pv. vitians (Xcv) (Barak, Koike and Gilbertson 2002) while Xcc, X. euvesicatoria and X. citri pv. fuscans (Xcf) were reported to colonise the inner seeds of cauliflower, tomato and bean, respectively (Sharma and Agrawal 2014; Darrasse et al. 2018; van der Wolf et al. 2019). Plant to seed transmission has long been speculated to originate from infected reproductive organs. Recent work by van der Wolf and colleagues (2013) has indeed identified a direct link between floral infection and inner seed colonisation. Cauliflower plants were spray inoculated with X. campestris at different developmental stages and seed colonisation was monitored. Symptoms were apparent in plants that were inoculated in all developmental stages but seed colonisation was only seen when plants were inoculated during flowering time (van der Wolf, van der Zouwen and van der Heijden 2013). In a further study using GFP-labelled X. campestris, the group demonstrated that the bacteria can successfully colonise and cause symptoms in siliques and subsequently colonise both the outer seed coat and the endosperm and embryo (van der Wolf et al. 2019). This data lead to the hypothesis that infected reproductive tissues later develop into infected endosperms and embryos. The group also estimated that reproductive tissues are externally infected through water splash or pollinators.
PATHOGENOMICS OF XANTHOMONADS
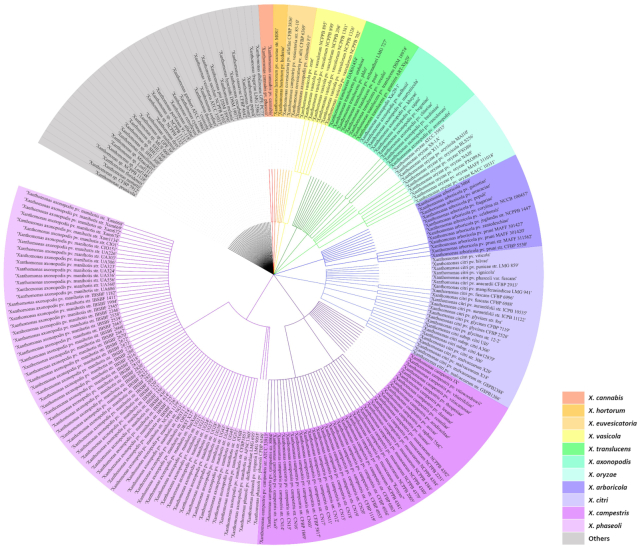
There are ∼1200 genomes of Xanthomonas available on NCBI genome browser, which represents a 21-fold increase compared to the 2011 report (Fig. 2) (Ryan et al. 2011). This wealth of genome data has revolutionised our understanding of bacterial taxonomy and pathogen evolution, as well as factors contributing to virulence and host adaptation. In this section, we discuss case studies/examples that have advanced our understanding of this pathogen.
Figure 2.
Phylogenetic tree of the Xanthomonas genus based on the NCBI taxonomy. The list of taxonomic names obtained from the NCBI Taxonomy Browser filtered using ‘has genome sequences’. The tree was annotated and visualized using iTOL (Interactive Tree Of Life) software.
Taxonomic re-classifications with the knowledge of sequenced genomes
Availability of whole genome sequences for the majority of Xanthomonas spp. has allowed the expansion of our understanding of the overall relationships among different species beyond classically used multilocus sequence typing. Average nucleotide identity among representative strains has been used as a criterion for classifying strains into species (Konstantinidis and Tiedje 2005). Several taxonomic revisions have been proposed in a larger species complex, sp. axonopodis, sp. campestris. While species nomenclature in xanthomonads has been defined based on its host specificity, whole genome comparisons have led to redefinition of some species. For example, Xanthomonas gardneri and Xanthomonas cynarae were recently placed into a single species (Timilsina et al. 2019). Additionally, several species of Xanthomonas (Xanthomonas nasturtii sp. nov. and Xanthomonas floridensis sp. nov.) associated with watercress production in Florida were defined. Sequencing genomes of diverse strains belonging to closely related species infecting tomato and/or pepper indicated the admixture of strains arising due to frequent genetic exchange among different strains of the closely related species (Barak, Koike and Gilbertson 2002; Jibrin et al. 2018). These hybrid strains pose a challenge to the current taxonomic definitions of a species based on average nucleotide identity (ANI). Although their ANI values are above 96%, these strains do differ in many phenotypic tests.
Evolutionary histories of pathogenicity factors
Comparative genomics of the occurrence of the type III secretion system (T3SS) has revealed that the Hrp2 family T3SS (for Hypersensitive Resistance and Pathogenicity) is present in the majority of xanthomonads, with an exception of X. albilineans that in contrast possesses a SPI-1 T3SS system. Four species, Xanthomonas sacchari, Xanthomonas cannabis, Xanthomonas pseudoalbilineans and Xanthomonas maliensis, lack any T3SS and associated effectors. Xanthomonas arboricola strains lacking a T3SS were considered as commensals. Given the relevance of evolution of the T3SS in ecological adaptation of pathogens, Merda et al. (2017) studied evolutionary history of Hrp2 family T3SS and associated effectors (T3Es) in xanthomonads. T3SS loss/acquisition events as well as genomic rearrangements in T3SS-containing region were compared across 82 genome sequences representing the diversity of the Xanthomonas genus (Merda et al. 2017). This study inferred three ancestral acquisitions of this cluster during Xanthomonas evolution followed by subsequent losses in some commensal strains and re-acquisition in some species. Several genes present in the Hrp2 cluster were also influenced by interspecies homologous recombination indicating large fragment recombination in this cluster. Interestingly, HGT of the entire cluster was also observed. It was speculated that loss of T3SS in some of X. arboricola commensals could be due to a high fitness cost associated with maintenance (Merda et al. 2017). These commensals could also act as profiteers given their simultaneous isolation with pathogenic species possessing a T3SS. Interestingly, the species X. arboricola contains strains that possess T3SS and variable numbers of T3Es as well as commensal strains lacking T3SS but possessing four effectors (xopF1, xopM, avrBs2 and xopR). The expansion of genome sequence data in the recent years has challenged the concept of a ‘core effectorome’ of Xanthomonas that comprises 10 T3Es; this ‘core’ is now thought to comprise only four effectors defined as the reduced ancestral core T3SE repertoire (Merda et al. 2017). These four effectors in commensal strains may allow them to overcome basal immune responses. Intriguingly, Merda et al. (2017) also identified a few X. arboricola strains lacking a T3SS but possessing two effectors, avrBs2 and xopR. It remains to be investigated whether these two effectors are secreted into the host and if so, whether these effectors have additional function independent of T3SS. It has been proposed that pathogenic strains that have accumulated other ‘previously defined core’ T3Es (xopL, xopN, xopQ, xopX and xopZ) define a basal step of pathogen emergence since these effectors have been shown to target pattern-triggered immunity (PTI) of plants (Merda et al. 2017).
Host adaptation/host and tissue specificity
Bacteria within the genus Xanthomonas are known for host specificity and the species were defined and classified based on their host range (Dye 1958). Studies involving identification of genetic determinants of host specificity in xanthomonads have been accelerated due to whole genome sequencing of closely related strains/lineages showing differences in phenotypes on certain hosts. Hajri et al. proposed ‘repertoire-for-repertoire hypothesis’ by which repertoires of T3Es in different species of Xanthomonas were proposed as determinants of host specificity (Table S1, Supporting Information) (Hajri et al. 2009). Chen et al. (2018) studied genes encoding transcription activator-like effectors (tal genes) from 17 Xanthomonas strains that cause common bacterial blight of beans, representing the four genetic lineages of Xcf and X. phaseoli pv. phaseoli (Xcp). Two chromosomic tal genes (tal20F and tal18) and two tal genes (tal23A, tal18H) encoded on plasmids were shared among most of phylogenetically distinct Xcf and Xcp strains. Since tal23A homologues genes are conserved in all Xanthomonas strains responsible for the common bacterial blight disease, this gene could be crucial in Xcf and Xcp pathogenicity and adaptation to common bean. These findings support the contention that horizontal transfer of plasmid-borne tal genes between distant lineages contributes to host adaptation (Chen et al. 2018).
Analysis of the complete genome sequence of the X. albilineans pathogenic strain GPE PC73 has allowed identification of several strategies specific to this strain to allow spread within sugarcane xylem vessels (Pieretti et al. 2009). Most of the known pathogenicity/virulence factors from other Xanthomonas strains are conserved in X. albilineans with the exception of the xanthan gum biosynthesis cluster and the T3SS system Hrp 2. The absence of any Hrp cluster can be a result of reductive genome evolution to live and multiply in a dead cell environment (the xylem). The presence of a T3SS belonging to the SP-1 injectisome family may in contrast be involved in the successful colonisation of the leaf surface by the pathogen. A key feature of X. albilineans is the ability to colonise a nutrient poor environment. Pieretti and colleagues identified 35 TonB-dependent transporters encoded by the chromosome of the X. albilineans strain GPE PC73 which could be important in active transport of plant-derived nutrients. In addition, 12 genes that encode for non-ribosomal peptide synthetases (NRPs) comprising 4% of the chromosome were identified. Nine of these genes are present in the genomes of Xanthomonas spp. associated with monocotyledonous species. This suggests a possible interaction between the unknown small molecules synthesized by the NRPS genes and these monocotyledonous plants. X. albilineans possesses a copy of the cbhA gene, which is present only in xylem-associated xanthomonads including Xylella fastidiosa and in Ralstonia solanacearum. This gene is absolutely required for spread in the xylem vessel and the action of the encoded enzyme is possibly to degrade the pit membrane between xylem elements. This X. albilineans strain also encodes two polygalacturonase genes that might be required in pectin degradation to the same purpose (Pieretti et al. 2009). Xanthomonas albilineans strain GPE PC73 does not have gum genes to produce xanthan gum, which may allow pathogen spread inside the xylem vessel without blockage. This may promote pathogen transmission by infected cuttings, since complete obstruction of the xylem vessels would lead to rapid death of the host (Pieretti et al. 2009).
Comparative genome analysis has also shed light on the evolutionary events underpinning the emergence of the three pathotypes (A, A* and Aw) of Xcci and the differences in host range and virulence between them. Pathotype A has the broadest host range in citrus while pathotype A* and Aw have a very limited host range and have only been isolated from citrus species key lime and alemow. Gordon and colleagues (2015) conducted a comparative genomic analysis of 43 strains of Xcci to reveal only extremely low variation in gene content; most of the variant gene content was linked to gene islands and regions of recombination. These findings indicate the role played by horizontal gene transfer (HGT) and recombination for the gene content evolution of these pathotypes in terms of gain and loss of gene content and mutations (Gordon et al. 2015). The avrGf1 gene and its homolog avrGf2 act as a host range restriction factor in the pathotype Aw but the deletion of the avrGf1 gene does not give an A-like host range to this pathotype. This indicates that the presence or absence of other factors is necessary to explain the host range of these pathotypes. The xopAD gene encoding a T3E is found in both A* and Aw where it has undergone pathotype specific recombination. Deletion of xopAD in the strain IAPAR 306 (which is a strain A) had no effect on pathogenicity on different citrus hosts (Escalon et al. 2013). Since xopAD shows unique variations in each pathotype, this gene can be used in future studies to identify pathotype-specific host range.
Ecological genomics
The major sources of inoculum of black rot, which is caused by Xcc, are contaminated seeds or transplants grown from contaminated seeds (Krauthausen et al. 2006) and Xcc-contaminated debris from the previous crop. Black rot-inducing Xcc strains have been isolated from cruciferous weeds (Shepherd's Purse) and contaminated non-cruciferous weeds from black rot infested fields (Krauthausen, Laun and Wohanka 2011). Black rot infested Shepherd's Purse can maintain the pathogenic potential of Xcc for over two successive overwinter periods and that indicates the potential to serve as a source of primary inoculum in the spring. In order to manage black rot it is important to perform crop rotation, apply field sanitation immediately after harvesting and use clean seeds and seedlings.
The existence of multiple lineages of Xanthomonas in the same host plant has been reported: Xanthomonas sp. strain CPBF 424 was isolated from the asymptomatic dormant buds of a diseased walnut tree in Portugal, with typical walnut bacterial blight symptoms (Fernandes et al. 2018). According to the multilocus sequence analysis (MLSA) of some of the housekeeping genes, this new strain is located between the non-pathogenic X. arboricola and X. prunicola clusters. CPBF 424 is divergent from both X. arboricola pv. juglandis (Xaj), the causal agent of walnut bacterial blight, and other X. arboricola pathovars. Since pathogenicity tests confirmed that the CFBF 424 strain is pathogenic to walnut trees, studies of this strain may provide new insights into xanthomonad pathoadaptations (Fernandes et al. 2018).
Comparative genomics of three Xanthomonas pathovars infecting diverse fruit trees has allowed an understanding of their evolutionary origin (Midha and Patil 2014). The maximum likelihood phylogeny indicated a close relationship between Xanthomonas axonopodis pv. citri (Xac) the causal agent of canker in citrus and X. citri pv. mangiferaindicae (Xmi), the causal agent of black spot in mango. Even though these pathogens share common host plants belonging to the order Sapindales, several differences were identified between these two strains, indicating the dynamic nature of genome evolution that these host-specific pathovars are undergoing. Xmi differs from Xac by the presence of a large NRPS/PKS cluster, the absence of CRISPR II, a large deletion in the xanthomonadin cluster and a deletion of the LPS cassette. Interestingly Xac and X. arboricola pv. punicae (Xcp), which do not belong to the same sublineage have similar LPS cassettes and CRISPR elements. These observations indicate the flexibility of these genomes that underpins their evolution for adaptation into different pathosystems (Midha and Patil 2014).
Comparative genomic analysis of pathogenic and non-pathogenic Xanthomonas strains that co-exist in the same host plant can give more information regarding the genomic adaptations of a non-pathogenic strain to a particular environment. A pangenome analysis of Xanthomonas strains isolated from the healthy rice seed identified a number of unique genes that are shared by X. maliensis (that has been previously reported to have a non-pathogenic lifestyle) and rice pathogenic Xanthomonas (Triplett et al. 2015). Even though X. maliensis lacks the T3SS and its associated effectors which are found in pathogenic Xanthomonas spp., it does carry genes for other secretions systems including T1SS, T4SS and T6SS, indicating some features similar to pathogenic Xanthomonas spp. Accordingly, X. maliensis is phylogenetically more related to pathogenic than to non-pathogenic isolates. These observations suggest X. maliensis is a possible intermediate in the evolution of pathogenic from non-pathogenic strains through the acquisition of genes that are usually present in pathogenic isolates (Triplett et al. 2015).
Outbreaks, epidemiology and population structures
Recent advances in high-throughput sequencing with higher speed and output-to-cost ratios have allowed large collections of a single plant pathovar to be sequenced and compared. Bart and colleagues (2012) sequenced 65 Xam strains that cause cassava bacterial blight disease. These 65 strains collected from 12 countries representing three continents and spanning 70 years of evolution, can be considered as an exemplar of sampling diverse strain collection across space and time. Among the studied Xam, strong clustering by geographic origin was observed (Bart et al. 2012). This indicates the significant influence of environmental conditions on pathogen population structure that results in different dominant strains across different geographical regions. Interestingly, no increase in the number of effector genes was observed in strains sampled from the same location during different years. Even though these highly conserved effectors can be subjected to loss or mutation in the face of selection pressure, these can be considered as ideal targets for developing resistance strategies.
In 2012, cotton plants grown in areas centered on Clarksdale, Mississippi were exhibiting symptoms of cotton bacterial blight. The symptoms had been controlled for more than half a century by using resistant germplasm. According to the phylogenic analysis conducted by Phillips et al. (2017), the current outbreak was caused by two strains that cluster with race 18 Xanthomonas citri pv. malvacearum (Xcm) strains. The recent outbreak was not due to an introduction of a new Xcm race, and also was not due to allelic or expression differences among virulence proteins that could cause re-emergence of cotton bacterial blight. Since the newly isolated Xcm was able to trigger the hypersensitive response (HR) on resistant cotton cultivars it was concluded that the recent outbreak of Xcm in the USA was due to re-emergence of race 18 clade Xcm likely due to cultivation of susceptible cultivars (Phillips et al. 2017). Race 18 Xcm might have maintained their populations below detection level on alternative hosts, since resistant cottons were planted in 1990s and early 2000s or might have been re-introduced into the southern USA by contaminated seeds.
Recent outbreaks of Xanthomonas wilt of enset and banana disease, caused by Xvm have been recorded from several major banana-growing countries in East and Central Africa between 2001 and 2008 (Tushemereirwe et al. 2004; Ndungo et al. 2006). Sequence analysis on 20 isolates collected between 1968 and 2005 from the regions where outbreaks had been recorded revealed only few variations (Aritua et al. 2009). Due to this homogenous nature of the Xvm populations with respect to time of isolation, geographic origin or host it is believed that the recent outbreak of Xanthomonas wilt in the Africa was due to the spread of the Xvm that originated in Ethiopia in 1968. Aritua and colleagues were also able to identify a similarity between Xvm isolates and the strains of X. vasicola from sorghum, maize and sugarcane originating in Africa. This musacearum pathovar might have evolved recently to become pathogenic on enset due to the close association with X. vasicola-infected sorghum.
Clustered regularly interspaced short palindromic repeats (CRISPRs) and variable number of tandem repeats (VNTRs) have been identified as high-resolution molecular typing tools that can be used to assess evolutionary changes within closely related microbial isolates by epidemiological typing. The CRISPR/cas system and multiple-locus VNTR analysis (MLVA) system is already used in global surveillance of plant pathogens evolution and epidemiological analysis (Rezzonico, Smits and Duffy 2011; Pruvost et al. 2014). Both of these genotyping methods have been used to study the new outbreaks of strawberry plant pathogen, X. fragariae (Gétaz et al. 2018). In this study, 58 bacterial strains of X. fragariae with miscellaneous geographic origins and years were analysed and two distinct groups that evolved independently before the first X. fragariae isolate was identified.
Type III effector identification and evolution
Machine learning approach combined with in planta translocation assays has been successfully used in recent studies to predict and validate novel T3Es. Previous approaches to predict effectors were limited by the fact that they were based on analysis of single molecular characteristics between effectors and non-effectors. Since a genome-wide machine learning approach scores all the open reading frames (ORFs) based on a large set of features that cover a range of evolutionary, genomics and biochemical characteristics, it can be used to identify T3Es that were missed by previous methods. Using a machine learning approach, Teper et al. (2016) identified seven novel T3Es in X. euvesicatoria also known as X. campestris pv. vesicatoria (Xav). Only three of the identified effectors belong to previously known effector families, while one of the novel effectors (XopAP) was identified as a factor that contributes to the disease development in plants infected with Xav.
A machine learning approach has also been used to detect T3E repertoires in 44 X. arboricola genomes representing 23 commensal and 21 pathogenic strains (Merda et al. 2017). Xanthomonas arboricola group A strain CFBP 2528 was used as the reference strain in this study due to the lower number of contigs of that strain. Based on the prediction, seven ancestral core T3E genes were found and the predicted T3E repertoires were highly variable with some strains with no effector and others with up to 34 effectors. The pan T3E effectome of X. arboricola comprised 57 predicted T3E. Eleven of the predicted effectors were found in both pathogenic and commensal strains, while 46 other effectors were only in pathogenic strains. About 12 novel T3E genes were identified in X. arboricola and six of these showed weak similarity to T3E known in other Xanthomonas species (Merda et al. 2017).
Draft genome sequences have also been used in genome comparisons and to obtain information on genome contents. Peng et al. (2016) obtained a complete genome sequence of X. translucens pv. undulosa (Xtu) strain XT4699 using SMRT sequencing technology. This sequence was compared with Illumina draft genome sequences of nineteen X. translucens strains, which were collected from wheat or barley in different regions and at different times. According to the full genome sequence of XT4699, 8 TAL effector genes that are phylogenetically distinct from other xanthomonad TAL effectors were identified. XT4699-tal7 was conserved among 8 of 12 of the sequenced Xtu strains. From the genomic data of X. translucens strains, 39 putative T3E genes not including TAL effectors were identified (Peng et al. 2016). Genomic comparisons among different pathovars or within pathovars also revealed variations on T3E repertories. It was speculated that these variations might be related to the host range adaptations of these strains.
Overcoming host resistance and rapid emergence of virulent strains
Rice contains at least 30 bacterial blight resistant genes that have been used in conventional breeding and this has put intense selection pressure on X. oryzae strains to overcome existing resistance (Liu et al. 2014). Rapid emergence of these strains has been studied extensively in the recent years. India is a major region of diversity of rice as well as its pathogen, Xoo. Indian Xoo strains belonging to lineage L-I and L-III can overcome resistance mediated by either xa5 or xa13 but not both, suggesting that lineage L-I and L-III strains acquired the ability to overcome either of these resistances independently (Midha et al. 2017).
Genome dynamics/plasticity-studying evolutionary processes shaping pathogen populations
Comparisons of the complete genome sequence of Xoo strain PXO99A with those of strains MAFF311018 (MAFF) and KACC10331 (KACC), which are highly similar to one another, have provided direct evidence that the Xoo genome is highly plastic and rapidly evolving (Salzberg et al. 2008). The PXO99A genome encodes19 TAL effectors and has at least 10 major chromosomal rearrangements relative to KACC and MAFF strains, resulting in 29 distinct syntenic blocks. Most of these rearrangements appear to be mediated by a set of transposable elements. PXO99A contains fewer classes of elements, but more copies of ISXo8, IS1114/ISXoo4 and ISXo2 while MAFF and KACC have nearly identical numbers of IS elements. Sequences unique to PXO99A include a 38 kbp region that includes several IS elements and is flanked by direct repeats of ISXo5, indicating that the IsXo5 element was involved in the genomic rearrangement that led either to loss of the locus from MAFF and KACC or gain of the locus in PXO99A. The PXO99A strain also has a near-perfect tandem duplication of 212 kb and this repeat is flanked by insertion of ISXo5 at each end and between the two copies. The 212 kb-segment occurs once in the MAFF and KACC sequences. Since these two regions are 100% identical (except for a single base difference in one IS copy), this duplication represents a remarkably recent event. A rapidly evolving CRISPR region contains phage infections unique to the PXO99A lineage and this provides one of the most unique records of differentiation among PXO99A, MAFF and KACC. PXO99A has the largest CRISPR region with 75 spacer elements while MAFF and KACC contain just 48 and 59 spacers, respectively. The majority of the spacers are unique to each strain, indicating the rapid evolution of these regions.
Xanthomonas citri str. Xc-03–1638-1–1 (Xc-A44) is an A group strain that has two plasmids: a 294kb-copper resistance gene-harbouring plasmid and 99kb-pathogenicity plasmid. The size of the pathogenicity plasmid was approximately equal to the combined size of two plasmids in the X. citri. str. 306. Gochez and colleagues sequenced this plasmid using PacBio sequencing to reveal the presence of 4 TALEs. Xc-03–1638-1–1 is the only X. citri strain with all four copies of TALEs carried on a single plasmid. Interestingly none of these TALEs were 100% identical to the TALEs from X. citri strain LM180, another Cu resistant strain that was isolated simultaneously with Xc-03–1638-1–1 (Gochez et al. 2018). The association of Tn-3 like transposon and of repetitive elements suggests that alternative structures of this pathogenicity plasmid exist in nature.
Recombination-mediated evolution of bacterial plant pathogens is important for the colonisation in novel hosts and new disease emergence. Jibrin et al. (2018) compared two newly sequenced Nigerian Xanthomonas strains (N1 and N38) with 70 previously published X. perforans and X. euvesicatoria strains. Recombination was identified as the major driving force of evolution of X. perforans and X. euvesicatoria strains, with X. perforans showing more evidence for recombination than X. euvesicatoria (Jibrin et al. 2018). The analysis showed that both X. perforans and X. euvesicatoria have open and highly dynamic pangenomes. Recombination had also affected the hrp genes, the hrp conserved (hrc) genes and hrp-associated (hpa) genes of X. euvesicatoria and X. perforans lineages. They observed recombination generated intraspecific variation in T3SS alleles, as indicated by genealogies that are incongruent among T3SS genes and with the core genome tree.
An integrative and conjugative element (ICE) was found in Xaj str. CFBP 7179 (Xaj-ICE), isolated from vertical oozing canker (VOC) infected walnut trees in France (Cesbron et al. 2015). This ICE shows 100% gene similarity to the genomic island (GI) found in bacteria from different genera, specifically Pseudomonas aeruginosa strains and Stenotrophomonas maltophilia strain D457, which belongs to the Xanthomonadaceae family. The ICE carried genes that are involved in copper resistance (copA, copB, copC, copD, copF, copG, copK) and most of these strains were confirmed to be copper resistant. The presence of this ICE in the Xaj str. CFBP 7179 might be an indication of the expansion of bacterial genomes due to the extensive use of copper in disease management. Since Xaj-ICE is the first known ICE identified in Xanthomonas, it might have been acquired by lateral transmission from a strain belonging to a different genus (Cesbron et al. 2015).
XANTHOMONAS FACTORS CONTRIBUTING TO DISEASE
The determination of the genome sequence of several species and pathovars of Xanthomonas has facilitated functional analyses aimed at understanding the molecular basis of virulence and adaptation. These functional studies have utilized different infection models and inoculation techniques (spraying, leaf clipping, mesophyll infiltration and vascular inoculation) to identify factors that play a significant role in the different phases of the disease cycle. In the following section, we will discuss recent advances in the study of virulence of Xcc and drawn parallels with other Xanthomonas spp. to demonstrate the adaptable and environmentally responsive nature of these pathogens during infection. In particular, we will highlight cell–cell signalling, second messenger signalling, two component systems, TonB-dependent outer membrane transporters and type III-secreted effectors (Table S2, Supporting Information).
Cell–cell signalling and associated pathways
The synthesis of particular virulence factors in Xanthomonas spp. is controlled by cell–cell signalling (quorum sensing) mediated by molecules of the DSF family, which are cis-2-unsaturated fatty acids (Table S2, Supporting Information) (Ryan et al. 2015; Zhou et al. 2017).
The predominant DSF family signals in Xcc are cis-11-methyl-2-dodecenoic acid (called DSF) and cis-2-dodecenoic acid (BDSF) (Ryan et al. 2015; Zhou et al. 2017). Synthesis and perception of the DSF signal require products of the rpf gene cluster. The synthesis of DSF is dependent on RpfF, a member of the crotonase superfamily with both desaturase and thioesterase activity that generates the signal molecule from a hydroxylated fatty-acyl ACP. A two-component system comprising the sensor kinase RpfC and regulator RpfG is implicated in DSF perception. Recent work has implicated residues in the extreme N-terminal of RpfC, which is located in the periplasm, in the recognition of DSF, although it is likely that other residues in the membrane-associated sensory input domain are also involved (Cai et al. 2017). Signal binding is believed to activate autophosphorylation of RpfC and phosphotranfer to RpfG, a two-component response regulator with an HD-GYP cyclic phosphodiesterase domain; phosphorylation of RpfG activates the protein for cyclic di-GMP degradation, which affects a range of cellular processes (discussed below).
In Xcc, the rpfF gene is linked to rpfB that encodes a fatty acid CoA ligase; rpfB is expressed as an operon with rpfF, although rpfF also has its own promoter. RpfB has been proposed to have a role in the mobilization of saturated free fatty acids generated by the thioesterase action of RpfF, allowing these free fatty acid derivatives to be used in phospholipid biosynthesis (Hu et al. 2018). However, more recent work has shown that rpfB mutants have elevated levels of DSF family signals, leading to the suggestion that RpfB is involved in signal degradation (Hu et al. 2018). This is intriguing since isolated RpfB has no apparent ligase activity against BDSF in vitro.
Comparative transcriptome analysis of wild type and rpfF, rpfC and rpfG mutants has suggested additional complexities in DSF signal perception and transduction (An et al. 2013a, 2014). Specifically, the effects of these mutations suggested alternative sensors for DSF and alternative regulators that interact with RpfC rather than a direct pathway (An et al. 2013a, 2014). Although alternative regulators that interact with RpfC have yet to be defined XC_2579 (RpfS) was identified as a second sensor for DSF in Xcc (An et al. 2013a, 2014). RpfS is a histidine sensor kinase containing an N-terminal PAS domain (PAS_4 domain). This PAS domain binds DSF and is required for regulation. Within the genes controlled by DSF, RpfS controls expression of a subset of genes and functions during epiphytic of Xcc.
DSF signalling has also been implicated in facilitating the entry of Xanthomonas spp. into plants, controlling synthesis of a factor that acts to reverse stomatal closure in Arabidopsis (Gustavo et al. 2009; Kakkar et al. 2015). This plant response is part of the innate immune system and can be induced by bacteria, bacterial components and the plant hormone abscisic acid. The Xanthomonas spp. factor responsible has not been identified but can be extracted from culture supernatants with ethyl acetate. The plant target is equally unknown but may be a signalling pathway involving the guard cell-specific Arabidopsis Mitogen-Activated Protein Kinase3 (Gustavo et al. 2009; Kakkar et al. 2015). Whether this Xcc factor similarly affects the pores of the hydathodes is not known. Although these pores resemble stomata, they do not respond to established elicitors of plant innate immunity (Melotto, Underwood and He 2008). Although the ethyl acetate extract would also contain DSF and derivatives, genetic evidence suggests that it is not the DSF signal molecule per se that acts to reverse stomatal closure. Furthermore, DSF acts to induce plant defences in the leaves of Arabidopsis, Nicotiana benthamiana and rice (Gustavo et al. 2009; Kakkar et al. 2015).
Xanthomonas spp. synthesises a second diffusible factor called DF, whose synthesis depends upon XanB2, which is encoded within the pig locus that directs synthesis of the characteristic yellow pigment xanthomonadin (Zhou et al. 2013a, 2013b). DF is associated with regulation of xanthomonadin biosynthesis, cell viability, epiphytic colonization and systemic invasion (Zhou et al. 2013a, 2013b). XanB2 is a unique bifunctional enzyme with three putative domains that hydrolyzes chorismate to produce 3-hydroxybenzoic acid (3-HBA) and 4-hydroxybenzoic acid (4-HBA). 3-HBA is associated with the xanthomonadin biosynthesis whereas 4-HBA is associated with CoQ8 biosynthesis and antioxidant activity. DF could be considered a metabolite cue rather than a genuine cell–cell signal molecule, as 3-HBA can be incorporated into xanthomonadin. Nevertheless, there is an increasing appreciation of the importance of transfer of metabolites between cells particularly within bacterial communities.
Intracellular signalling mediated by nucleotide second messengers
To date, a regulatory role for the nucleotide intracellular signals cyclic di-GMP (c-di-GMP) and cyclic GMP (cGMP) has been described for Xanthomonas spp. (Table S2, Supporting Informtion) (Ryan 2013; An, Tang and Dow 2017). The cellular level of these signals results from a balance between synthesis and degradation. For c-di-GMP three protein domains are implicated in these processes: the GGDEF domain catalyses synthesis of c-di-GMP from 2 molecules of GTP whereas EAL and HD-GYP domains catalyse hydrolysis of c-di-GMP, first to the linear nucleotide pGpG and then at different rates to GMP (recently reviewed by Dahlstrom and O'Toole 2017; Jenal, Reinders and Lori 2017). The Xcc 8004 genome encodes 37 proteins with GGDEF, EAL or HD-GYP domains. Although many of these are conserved in other Xanthomonas spp., the number of proteins progressively decreases from Xcc to Xca through to X. oryzae pv. oryzicola (Xoc) and Xoo. The effects of deletion/mutation of the genes encoding some of these proteins have been reported to effect virulence and biofilm formation (Wei et al. 2016; Yang et al. 2016; Su et al. 2016b; Li et al. 2018; Xue et al. 2018). The majority of these proteins contain further domains that have a role in recognition of different environmental cues or comprise regulatory elements of two-component systems, such as the HD-GYP domain protein RpfG introduced above. A comprehensive mutational analysis has shown a role for a number of these proteins in virulence, as measured by leaf clipping, and in virulence factor production (Ryan et al. 2007; An, Tang and Dow 2017). A body of work has now identified a role for c-di-GMP in regulation of a wide range of functions in Xcc that include adhesion, biofilm formation, motility, synthesis of polysaccharides and synthesis of extracellular enzymes (Ryan et al. 2007; An, Tang and Dow 2017). The outcome of these experiments suggest that a number of c-di-GMP signalling proteins are organized as a network that allows integration of information from diverse environmental inputs to modulate particular functions such as extracellular enzyme synthesis. In contrast, other c-di-GMP signalling systems appear to be dedicated to alternative specific tasks. Whether different subsets of proteins are important in different phases of the disease cycle remains to be determined.
C-di-GMP exerts a regulatory action by binding to a range of receptors or effectors, such as small ‘adaptor’ proteins with a PilZ domain, transcription factors and riboswitches (Chin et al. 2010; 2012; Guzzo et al. 2013). The cyclic AMP receptor-like protein Clp links Rpf/DSF signalling (and alteration in c-di-GMP) to the expression of genes encoding extracellular enzymes. C-di-GMP binding to Clp prevents the binding of the transcriptional activator to the promoters of several genes encoding extracellular enzymes (Chin et al. 2010, 2012; Guzzo et al. 2013). Lowering of the c-di-GMP levels, for example, by activation of DSF cell–cell signalling, reverses this effect. Clp also regulates biofilm dynamics in response to changes of the level of c-di-GMP. Mutation of clp causes downregulation of manA that encodes an endomannanase involved in biofilm dispersal and upregulation of xag genes, which synthesize a polysaccharide associated with biofilm formation. The nature of this polymer remains to be established.
All Xanthomonas genomes encode four proteins that have a PilZ domain (Yang et al. 2015). In Xcc strain 8004 these are designated XC_0965, XC_2249, XC_2317 and XC_3221 (Chin et al. 2010, 2012; Guzzo et al. 2013). Mutational analysis indicates that XC_0965, XC_2249 and XC_3221 make a significant contribution to the virulence in Chinese radish. A few details of the regulatory action of these proteins (or their orthologs in other Xanthomonas spp.) have emerged (Chin et al. 2010, 2012; Guzzo et al. 2013). XC_0965 and XC_2249 have canonical PilZ domains and hence are predicted to bind c-di-GMP. Both of these proteins positively regulate extracellular enzyme production, whereas XC_2249 additionally affects bacterial motility. In contrast, XC_3221 has a non-canonical type II PilZ domain, does not effectively bind c-di-GMP, but does influence pilus-dependent motility. XC_3221 exerts this effect by interaction with an enzymatically inactive GGDEF-EAL domain protein, called FimX (XC_1824 in Xcc strain 8004), and the PilB pilus motor protein. Although FimX is enzymatically inactive, it can bind c-di-GMP via the EAL domain and then forms a complex with XC_3221. This complex interacts with PilB to influence pilus biogenesis. XC_3221 probably has other cellular functions that contribute to virulence since mutation of pilA, which abolishes motility, has a lower effect on virulence than mutation of XC_3221. Recently, a new class of c-di-GMP effector of the YajQ family has been identified in Xcc (An et al. 2013b; Zhao et al. 2016). This protein (XC_3703) acts to influence the transcription of genes that contribute to virulence in plants and biofilm formation. The available evidence suggests that XC_3703 exerts its action through protein–protein interactions with the LysR family regulator XC_2801, an interaction that is negatively regulated by c-di-GMP (An et al. 2013b; Zhao et al. 2016). Comparative transcriptome profiling indicates that YajQ is involved in further regulatory pathways not involving XC_2801, but these are currently undefined. In silico analysis has predicted riboswitch candidates in Xcc that potentially bind c-di-GMP suggesting that further classes of c-di-GMP effector remain to be discovered (Alkhateeb et al. 2016).
Evidence has recently been provided for a cGMP-mediated pathway in Xcc that acts in the regulation of virulence, biofilm formation and the transcription of specific virulence genes (An et al. 2013b; Ryu et al. 2015). This study is one of the first descriptions of the role of cGMP in bacterial biofilm formation and virulence of a bacterial pathogen. The synthesis of cGMP in Xcc depends upon XC_0250, which is a guanylate cyclase with a class III nucleotidyl cyclase domain attached to a domain of uncharacterized function with tetratricopeptide repeats. The action of cGMP in Xcc depends in part on XC_0249, which has a cGMP-binding domain attached to a GGDEF domain active in c-di-GMP synthesis (An et al. 2013b; Ryu et al. 2015).
Environmental sensing (other than cell–cell signalling) and the role of two-component systems and TonB-dependent outer membrane transporters
Several systematic studies have examined the role of sensing systems in Xanthomonas, in particular in Xcc that respond to environmental or cellular stimuli in the control of virulence, cellular behavior and gene expression (Table S2, Supporting Information; Fig. 3) (Li et al. 2014; Cui et al. 2018). The Xcc genome encodes a number of two component systems including RpfC/RpfG discussed above. Several of these systems have been implicated in regulation of virulence factor synthesis, biofilm formation or resistance to oxidative stress (Li et al. 2014; Cui et al. 2018). In some cases, the nature of the signal or cue to which the system responds has been determined (Table S2, Supporting Information; Fig. 3). A key pathway is that involving the transcriptional regulator HrpG, which directly regulates the expression of HrpX, an AraC-type transcriptional regulator controlling expression of the T3SS (Li et al. 2014; Cui et al. 2018). HrpG is an orphan regulator, as hrpG has no linked gene encoding a sensor. Mutational analysis and protein–protein interaction studies have identified HpaS as a putative sensor (Li et al. 2014). It is proposed that HpaS responds to plant-derived molecules and other environmental stimuli. HpaS may interact with further regulators, in particular HpaR2, which is encoded by a linked gene (Li et al. 2014).
Figure 3.
Xanthomonas employs multiple systems to link sensing of diverse environmental signals to regulation of appropriate responses. The Xcc genome encodes a number of two-component and other systems thought to be involved in environmental sensing and regulation. In a relatively small number of cases the signals that activate these pathways have been established, to include the DSF cell–cell signal, oxygen tension, iron and other metal ions, cytokinin and other plant-derived molecules to include sucrose and light. In addition to two-component regulators, a number of transcription factors have been implicated in downstream signalling pathways that lead to activation of functions associated with virulence and other environmental adaptations. These include the cyclic di-GMP responsive Clp, the type III secretion regulators Zur, FhrR PhoP, HpaR, HrpX, HrpG and HpaR1. Interestingly, VgrR although be shown to be involved in iron uptake also contributes to type III secretion regulation. Code Blue sensor proteins, Green regulatory proteins
Recent work has also established that light is an important environmental cue influencing Xcc interaction with host plants (Bonomi et al. 2016; Otero et al. 2016). Xcc produces a bathy-type photoreceptor called XccBphP that photoconverts between red-absorbing (Pr) and far-red-absorbing (Pfr) states. Upon sensing light, the XccBphP elicits a transcriptional response that leads to down-regulation of xanthan production and biofilm formation and reduced virulence. It has been proposed that XccBphP acts to co-ordinate enhanced virulence factor synthesis to conditions of low light under which the plant tissues exhibit increased susceptibility (Bonomi et al. 2016).
Xanthomonas spp. have much larger numbers of TonB-dependent receptors (TBDRs) than many eubacteria, as determined by genome analysis (Blanvillain et al. 2007; Déjean et al. 2013). These proteins are present in the outer membrane of Gram-negative bacteria and transport iron-siderophore complexes and vitamin B12 into the periplasm. The inner membrane energy-coupling TonB-ExbB-ExbD protein complex provides the energy required for transport. Recent evidence has suggested that some Xcc TBDRs might transport plant-derived molecules and may have roles in bacterial virulence or survival (Blanvillain et al. 2007; Déjean et al. 2013). This action was revealed by the definition of specific CUT systems (Carbohydrate Utilization systems containing TonB-dependent transporters) that scavenge plant carbohydrates. One CUT system using N-acetylglucosamine from plant origin has been implicated in virulence whereas a second system involved in the mobilization of xylan contributes to phyllosphere fitness (Blanvillain et al. 2007; Déjean et al. 2013; Boulanger et al. 2014).
Most sequenced Xanthomonas genomes encode multiple ORFs characterized as σ factor. In Xcc, there are 15 such ORFs, of which 10 belong to the extracytoplasmic function (ECF) subfamily, which act under stress conditions to co-ordinate expression of specific genes (Cheng et al. 2008; Bordes et al. 2011). Recent work has shown that these ECF factors are required for the survival of Xcc under stress and also mediate the synthesis of virulence factors. RpoE1 is the best-described ECF σ factor in Xcc, where it plays a role in the heat shock response, adaptation to membrane-perturbing stresses, stationary-phase survival, resistance to cadmium and T3SS expression (Cheng et al. 2008; Bordes et al. 2011b; Yang et al. 2018). Currently little or nothing is known of the mechanisms by which these different ECF σ factors are activated or of any regulatory interplay with other systems involved in sensing. It is clear, however, that the array of such sensing systems allows Xcc to monitor many aspects of the bacterial environment and to respond in an appropriate fashion.
Type III effector biology and action
Xanthomonad repertoires of T3Es are diverse and aid virulence and possibly host specificity (Table S1, Supporting Information). This aspect of Xanthomonad biology has been reviewed extensively recently so we just summary key findings (please see Jacques et al. 2016). To date, a total of 52 effector families have been identified along with three harpin proteins, which are helper or accessory proteins that assist in effector translocation (www.xanthomonas.org).
The majority of sequenced Xcc genomes contain a core set of nine effector genes (xopR, avrBs2, xopK, xopL, xopN, xopP, xopQ, xopX, xopZ) (Table S1, Supporting Information). However, considerable variation in effector complement is seen between Xcc strains and between Xcc and other X. campestris pathovars attacking brassicas. Mutation of genes encoding some core effectors leads to reduced virulence and fitness of Xcc but this can depend on the strain. It has been suggested that there may be substantial functional redundancy among Xcc effectors.
Xcc appears to econtain several effector genes that are only found associated with this species: xopAL1, xopAC, xopAD, xopAH and xopAL2 are unique to Xcc strains. Whether these genes affect host specificity requires further functional testing. Furthermore, Xcc strains appear to encode a specific set of transcription activator-like (TAL) effectors when compared to other Xanthomonas spp. and pathovars (Table S1, Supporting Information) (Denancé et al. 2018). These effectors are considered intriguing candidates for determinants of host specificity and tissue preference.
Effectors from several Xcc strains do affect restriction of host range. Avirulence factors typically determine specificity at the pathogen race/host cultivar level by triggering defence responses in hosts with a corresponding specific resistance gene. AvrXccC and AvrXccE1 determine host specificity for Xcc on mustard and Chinese cabbage, respectively, and AvrBs1 determines non-host resistance on pepper ECW10R (He et al. 2007).
Effectors may also determine tissue specificity: AvrAC has been shown to trigger resistance specifically in the vascular system of Arabidopsis thaliana Col-0 but has no discernible effect on growth in mesophyll tissue (Xu et al. 2008). It is not clear whether this reflects the tissue-specific expression of the corresponding resistance gene. The method of inoculation used in these studies bypasses the hydathodes, which are the usual route of entry into the vascular system and have long been suggested to contribute to resistance. Recent work has shown that bacteria enter the hydathode without apparent restriction but subsequently activate plant immune responses that limit growth in the epithem (Cerutti et al. 2017). Suppression of these defence responses, which is required to allow disease, depends on the type III secretion system and associated effectors. Nevertheless, avrAC acts to restrict pathogen growth upon hydathode infection in Arabidopsis Col-0, consistent with the earlier study (Cerutti et al. 2017). Identification of conserved effector or avirulence genes is a major step towards the cloning of the matching resistance genes, which may be found not only in brassicas but also in non-host plants, and their subsequent deployment in agriculture.
XANTHOMONAS EPS AND LPS
The EPS xanthan
The EPS xanthan is a characteristic component of all xanthomonads with the exception of those with reduced genomes where the xanthan biosynthetic genes (the gum cluster) have been lost (Becker and Vorhölter 2009; Pieretti et al. 2009). The polysaccharide encloses the outer membrane, to which it is not covalently linked. (Becker and Vorhölter 2009). Xanthan has a cellulose-like (1→4)-β-D-glucose polymer backbone, where every second glucose residue of the main chain carries a (1→3)-linked trisaccharide side chain of β-d-mannose-(1→4)-β-D-glucuronic acid-(1→2)-α-D-mannose (Fig. 4). While this carbohydrate structure is highly conserved, there are additional non-sugar decorations, with the terminal mannose residues being acetylated or pyruvylated at varying degrees while mannose residues proximal to the main chain are acetylated, again to varying extents.
Figure 4.
Polysaccharide biosynthesis and selected lipopolysaccharide structures in xanthomonads. (A), An overview indicates how the biosynthesis of polysaccharides and glycoconjugates is embedded in the Xanthomonas metabolism. Main stages of polysaccharide biosynthesis are marked in red. (B), In an inlay, the structure of a pentasaccharide repeat unit of the exopolysaccharide xanthan is given. Two glucose residues contribute to the xanthan main chain, with a trisaccharide side chain attached to every second glucose residue. The mannose residues of the side chains can be modified at varying degrees. The proxymal mannose close to the main chain is depicted with an acetyl group, while the terminal mannose carries a pyruvyl group. Alternatively, this mannose can be acetylated. (C), LPS structures are indicated for X. campestris pv. campestris 8004, which have been partly confirmed for strain B100, for X. translucens pv. translucens DSM-18 974, for X. oryzae pv. oryzicola BLS303, and for X. fragariae NCPPB 1469. Main constituents are identified in the graphical legend. In several cases, side chains are linked non-stoichiometrically. Such non-stoichiometric linkages are marked in red. For X. fragariae NCPPB 1469, part of the monosaccharide side chains were linked in irregular intervals to the main chain.
The role of xanthan in pathogenesis may be complex and vary among Xanthomonas strains and host plants. Xcc mutant strains deficient in xanthan biosynthesis have reduced aggressiveness against host plants (Katzen et al. 1998). Xanthan has been described to suppress induced innate immunity by calcium chelation (Aslam et al. 2008) and to induce plant susceptibility to Xcc by suppressing callose deposition (Yun 2006). For Xac xanthan was reported as not essential for pathogenicity in citrus canker but to contribute to epiphytic survival (Dunger et al. 2007) while in another experiment a xanthan-deficient mutant was reduced in growth and survival on leaf surfaces and provoked reduced disease symptoms (Rigano et al. 2007a). Xanthan pyruvylation has an influence on the virulence of Xcc (Bianco et al. 2016). For Xoo the O-antigen component of LPS was shown to induce rice defence responses, which were suppressed by xanthan, resulting in a modulation of the defence response of the host plant (Girija et al. 2017). Xanthan is also a key component of the biofilm matrix in Xanthomonas (Dow et al. 2003). Besides its pathogenicity-related features, xanthan is relevant as a commercially produced thickening agent. Xanthan is commercially produced by fermentation in large scale (García-Ochoa et al. 2000) and employed in industry, in particular as a component of food, cleansing materials, pharmaceutics and in drilling for oil and natural gas.
Xanthan biosynthesis originates from the interconvertible metabolic intermediates glucose 6-phosphate and fructose 6-phosphate, which are linked to UTP and GTP to obtain the nucleotide sugars UDP-glucose, UDP-glucuronic acid and GDP-mannose in the first main step of xanthan biosynthesis. The sugar residues of these compounds are transferred to a polyprenol lipid carrier (LC, most likely undecaprenol diphosphate), thereby forming a lipid-linked linear glucose-glucose-mannose-glucuronic acid-mannose pentasaccharide in the second main step of xanthan biosynthesis. This pentasaccharide reflects the repeat unit of the xanthan molecules. Consistent with the Wzy-dependent model for heteropolysaccharide biosynthesis of Enterobacteriaceae (Raetz and Whitfield 2002; Whitfield 2006), the LC-linked pentasaccharide is assumed to be subsequently flipped to the outer surface of the inner membrane, where it is polymerized and exported via a specific channel in the outer membrane (Vorhölter et al. 2008). Pyruvylation and acetylation reactions of the mannose residues occur at the LC-linked pentasaccharides, but the exact sequence of the reactions is unclear. Acetyl and pyruvyl groups are donated from the central metablic intermediats acetyl-coenzyme A and phosphoenolpyruvate, respectively.
While genes for the synthesis of the xanthan precursors UDP-glucose and GDP-mannose, which are likely to be also LPS precursors (Vorhölter, Niehaus and Pühler 2001), are clustered with additional LPS biosynthetic genes (Vorhölter, Niehaus and Pühler 2001), the genes that code for the generation of lipid-linked intermediates and the subsequent polymerisation and export of xanthan form a cluster of 12 successive genes with an operon-like identical direction of transcription, gumBto gumM. Mutagenesis has revealed the functions of most of the gum gene products, identifying GumD, GumM, GumH, GumK and GumI as glycosyltransferases that build the pentamer repeating units of xanthan at the LC, and GumF, GumG and GumL as acetyltransferases and pyruvyltransferase, respectively. Similarities to model proteins from Enterobacteriacea paved the way for a model that suggested GumJ as a flippase, which transferred the LC-linked pentasaccharides to the periplasmic face of the inner membrane where it may be polymerized by GumE. The first two genes of the gum cluster, gumB and gumC, are likely to encode components of a channel similar to the Wza/Wzc complex (Collins et al. 2007) of E. coli that spans the outer membrane and the periplasm and facilitates xanthan export. These GumB and GumC proteins (Jacobs et al. 2012; Galván et al. 2013; Bianco et al. 2014) and two of the gum gene products coding for glycosyltransferases, gumI (Salinas et al. 2011) and gumK (Barreras et al. 2008; Salinas et al. 2016), have been characterised in more detail over the past years. Unfortunately, there is little experimental evidence available regarding the putative flippase GumJ and the suggested polymerase GumE.
The phosphorylated sugars that are requisitioned to synthesise cell surface constituents such as EPS and LPS can either be imported, obtained from glycogen breakdown, or be synthesised from citrate cycle intermediates by means of gluconeogenesis (Tang et al. 2005). Xanthomonads have an astonishing repertoire of sugar importers in their inner membrane (Vorhölter et al. 2008), plus a vast variety of outer membrane importers, in particular TonB-dependent receptors (Blanvillain et al. 2007; Serrania et al. 2008; Déjean et al. 2013; Boulanger et al. 2014) that may also be involved in trans-envelope signalling (Wiggerich and Pühler 2000; Vorhölter et al. 2012). The phosphorylated sugars used to synthesize EPS and LPS are also intermediates in pathways leading to the synthesis of glycogen, in catabolism to obtain energy or as building blocks that are ultimately utilised to generate biomass. Xanthomonads mainly employ the Entner–Doudoroff pathway and to a lesser extent the pentose-phosphate pathway for the catabolic utilisation of such intermediates (Schatschneider et al. 2014). The Embden–Meyerhof–Parnas pathway (for glycolysis) was recently revealed as an additional catabolic route in xanthomonads (Schatschneider et al. 2013). This pathway involves a non-canonical pyrophosphate-dependent phosphofructokinase (Frese et al. 2014). Flux analysis produced clear evidence for a remarkable flexibility of the central metabolism, where a major redistribution of resources from xanthan to biomass can occur (Schatschneider et al. 2013). However, only minimal flux was observed for glycolysis under standard laboratory growth conditions (Schatschneider et al. 2014).
Xanthomonas LPS
LPS are main components of the outer leaflet of the outer membrane. Usually, LPS are tripartite glycoconjugates with a glycolipid part called lipid A that carries a core oligosaccharide, and a polysaccharide termed O-specific antigen or O-antigen. LPS that lack the O-antigen component are called lipooligosaccharide (LOS) or rough-type LPS. LPSs play a vital role for bacterial growth, providing a barrier to antimicrobial compounds and protection against stresses as well as being important for the structural properties of the outer membrane. In addition, LPSs have an involvement in adhesion and host recognition (Silipo et al. 2010). LPS from different Xanthomonas species have been shown to induce defence-related responses in both host and non-host plants (Meyer, Pühler and Niehaus 2001; Newman et al. 2002). The acylation and phosphorylation pattern of X. campestris lipid A influences its ability to trigger the innate immune response of its host plant Arabidopsis thaliana (Silipo et al. 2008). The Xoc O-antigen was required for full virulence and type III protein secretion (Wang, Vinogradov and Bogdanove 2013) while the O-antigen component of Xoo LPS-induced rice defence responses during infection which were suppressed by xanthan (Girija et al. 2017).
While lipid A is rather conserved among many Gram-negative bacteria, there is more variation among core oligosaccharides and O-antigen structures may vary even within an individual Xanthomonas spp. Only a limited number of LPS structures has been determined for xanthomonads so far. A selection of these structures is displayed in Fig. 4. For Xcc, the structure of the O-antigen repeating unit (Molinaro et al. 2003) as well as the LOS structure (Silipo et al. 2005) have been determined for the strains 8004 and B100 (Braun et al. 2005; Kaczyński et al. 2007). Attached to the acylated diglucosamine of the lipid A, the Xcc 8004 core oligosaccharide consists of an invariable 3-deoxy-D-manno-oct-2-ulosonic acid (KDO)-mannose-glucose trisaccharide that carries at the KDO a galacturonyl residue that is linked via a phosphate group. An additional galacturonyl phosphate is linked to the mannose residue, which can alternatively be substituted by a phosphoramide group. Finally, the glucose can non-stoichiometrically carry a mannose disaccharide. The complete LOS structure is also available for the X. translucens pv. translucens (Xtt) type strain (Steffens et al. 2017) while its total LPS monosaccharide composition indicates rhamnose and xylose as O-antigen constituents. The LOS of Xoo was found to share the core structure with Xcc (di Lorenzo et al. 2016), but the authors do not mention the phosphoramide found in Xcc LOS. In the LOS of Xoc, no galacturonylphosphate side chain was attached to the KDO, while a galacturonamide was linked via a phosphate group to the mannose residue adjacent to the KDO (Wang, Vinogradov and Bogdanove 2013). While all these core structures agree largely, longer core oligosaccharides have recently been determined for Xac strain Xcc99–1330, where the LOS was characterised as an acylated diglucosamine with an oligosaccharide that consists of two KDO, three galacturonic acids, five hexoses, four rhamnoses and one Fuc3NAc residue, bearing two phosphate groups while to O-antigen was a rhamnose polymer (Casabuono et al. 2011). In addition, some lipid A structures are available (Silipo et al. 2004). Exact O-antigen repeating unit structures were elucidated for the Xcci strains NCPPB 3562 and CFBP 2911 (di Lorenzo et al. 2017), for X. oryzae pv. oryzycola BLS303 (Wang, Vinogradov and Bogdanove 2013) as well as for several other xanthomonads (Molinaro et al. 2000a, 2000b, 2001). A common theme of Xanthomonas O-antigen repeat units appears to be a polyrhamnan main chain with monosaccharide side chains in many strains that are in some cases attached to the main chain in an irregular pattern.
Although the mechanism of LPS biosynthesis is particularly well understood for Enterobacteriaceae (Raetz and Whitfield 2002; Whitfield and Trent 2014), the data available for xanthomonads is as yet highly fragmentary. Nevertheless, the available information indicates that concepts accepted for Enterobactericaeae are by and large valid for LPS biosynthesis in xanthomonads.
NDP-sugar precursors are provided in stage 1 of the LPS biosynthesis. The rmlABCD genes have been identified to code for dTDP-L-rhamnose (Vorhölter, Niehaus and Pühler 2001). This requires glucose 1-phosphate as starting material, which is generated using the xanA gene product, and thereby linked to the synthesis of xanthan. In mutant strains affected in rmlA and rmlD, the rhamnose monosaccharides obtained by acid hydrolysis of LPS were reduced by more than 90%. DNA sequence similarity pointed to two additional genes, gmd and rmd, that were annotated to encode the synthesis of GDP-D-rhamnose from the xanthan precursor GDP-mannose (Vorhölter, Niehaus and Pühler 2001). This second rhamnose source might explain the remaining quantity of rhamnose in the rmlA and rmlD mutants. The biosynthetic pathway of dTDP-3-acetamido-3-deoxyfucose was reconstructed based on DNA sequence analysis; this pathway originates from a dTDP-L-rhamnose precursor (Vorhölter, Niehaus and Pühler 2001). This novel pathway was subsequently confirmed experimentally for similar gene products of Aneurinibacillus thermoaerophilus (Pfoestl et al. 2003). In addition, there is biochemical evidence for the epimerisation of UDP-glucuronic acid to UDP-galacturonic acid, while genome data revealed the gene for this enzymatic conversion (Vorhölter et al. 2008). Genome analysis revealed also the genes that drive the synthesis of UDP-N-acetylglucosamine (UDP-GlcNAc) and CMP-KDO (Vorhölter et al. 2008), probably precursors of the lipid A and LPS core of xanthomonads, respectively.
As with other Gammaproteobacteria, the lipid A and the LPS core are expected to be synthesized at the inner face of the inner membrane to obtain LOS molecules. The LOS molecules are then exported to the outer face of the inner membrane by means of a specific ABC family exporter termed MsbA (Raetz and Whitfield 2002). Unfortunately, there is little available evidence regarding the biosynthesis of these structures in xanthomonads. However, in X. campestris a distinct LC-linked galacturonide was detected, with UDP-galacturonic acid as a likely precursor (Baldessari, Ielpi and Dankert 1990). It is tempting to speculate that the galacturonyl phosphate residues of the LPS core are transferred in a distinct biosynthetic step from these LC-linked intermediates by a mechanism that resembles the lipid A modification in E. coli where the unusual monosaccharide 4-amino-4-deoxy-L-arabinose is added (Raetz et al. 2007).
Subsequently, the O-antigen part of the LPS is expected to be added to the LOS by means of an enzyme termed LPS ligase. As a prerequisite, mature O-antigen is required at the outer face of the inner membrane. The O-antigen is synthesised and exported beforehand in a separate process. The architecture of a channel-forming O-antigen exporter has been determined recently, pointing to a processive O-antigen translocation mechanism (Bi et al. 2018). Based on a first approximation of genome data, the Wzm/Wzt ABC-transporter model developed for Enterobacteriaceae may fundamentally describe O-antigen biosynthesis and export in xanthomonads (Vorhölter, Niehaus and Pühler 2001), where functions important for LPS biosynthesis were related to the wxc gene cluster. LPS biosynthetic gene clusters were also found in other xanthomonads in a conserved genomic location flanked by the etfAB and metB genes (Patil and Sonti 2004). However, the gene content of different strains was quite different, with only the wzm and wzt genes being conserved (Lu et al. 2008). In the immediate genomic vicinity of the Xcc wxc gene cluster are the rmlABCD genes that code for enzymes required to synthesize the LPS precursor dTDP-L-rhamnose, the xanA and xanB genes that code for NDP-sugar precursors of the EPS xanthan which are also LPS precursors (Vorhölter, Niehaus and Pühler 2001), and the lpsI and lpsJ genes that were later renamed as wxcI and wxcJ and that code for 3-oxoacid CoA-transferase subunits which are involved in LPS biosynthesis.
Three distinct regions were distinguished in the Xcc wxc gene cluster (Vorhölter, Niehaus and Pühler 2001). While gene products encoded in region 2 were similar to NDP-sugar biosynthetic enzymes, genes from region 1 coded for a Wzm/Wzt ABC-exporter in addition to glycosyltransferases and proteins of unclear function. Mutant strains deficient in these genes were affected in O-antigen biosynthesis (Hötte et al. 1990; Vorhölter, Niehaus and Pühler 2001). The functions of the products of some other genes in region 2, for example WxcB, are poorly understood. In X. euvesicatoria, a wxcB knock-out mutant had reduced virulence while effects on the LPS were minimal (Park, Jung and Han 2014). In contrast in an Xcc mutant deficient in wxcB the O-antigen was missing completely (Steffens et al. 2016). Another unusual gene of region 2 was wxcD, where a mutant strain (Vorhölter, Niehaus and Pühler 2001) produced LPS with fragmentary O-antigen (Braun et al. 2005), relating this gene possibly to O-antigen primer or adapter biosynthesis. Mutagenesis of the genes wxcK and wxcN (Steffens et al. 2016) in region 3 of the wxc gene cluster indicates a distinct biosynthetic mechanism for the synthesis of the O-antigen side chain.
Cyclic glucans
Xanthomonads produce low-molecular-weight β-glucans including a cyclic β-glucan that is sequestered in the periplasmic space (Miller and Salama 2018). These molecules have been classified as membrane-derived oligosaccharides (MDOs). The cyclic β-glucan structures of X. campestris pv. glycines (Xcg) PDDCC 7414 and Xac have been confirmed as a β-(1→2)-linked glucan with 15 β-(1→2)-linkages and one α-(1→2)-linkage (York 1995). The cyclic β-(1→2)-glucan of Xcc has a role in the systemic suppression of plant immune responses (Rigano et al. 2007a) and may also protect bacteria against iron-induced toxicity (Javvadi et al. 2017).
Regulation of cell surface carbohydrate biosynthesis
While the biosynthesis of cyclic glucan in Xcc appears to be regulated by the DSF system (Vojnov et al. 2001b), both DSF and DF signalling systems have an influence on xanthan biosynthesis. As outlined previously, the DSF system influences the levels of the second messenger cyclic di-GMP via several rpf gene products (O'Connell et al. 2013). The CAP-like protein (CLP), binds directly to the promoter region upstream of gumB in a manner modulated by the nucleotide second messenger (Chen et al. 2010a). It is not known how DSF regulates cyclic glucan synthesis or how DF signalling influences xanthan. There are additional regulatory effectors of gum expression: the GntR family regulator HpaR1 is an activator of gum gene expression that interacts directly with the gumB promoter (Su et al. 2016a) and CysB was recently identified as another transcriptional regulator that is likely to modulate gum gene expression (Schulte et al. 2019). These effects of DNA-binding transcriptional regulators are likely complemented by antisense transcripts that have been detected with several start sites, most outstanding for the promoter region upstream of gumB (Alkhateeb et al. 2016). In addition, proteomics data revealed that besides transcriptional regulation the post-translational modification of key enzymes may influence xanthan biosynthesis, in particular at the level of NDP-sugar synthesis from central metabolic intermediates (Musa et al. 2013). More research will be required to better understand this complex of regulatory influences.
XANTHOMONAS DISEASE MANAGEMENT AND CONTROL
A suite of strategies for prevention and suppression of Xanthomonas-caused diseases have been developed; these have involved inhibition of pathogen dispersal, use of chemical and biocontrol agents and development and deployment of resistant varieties.
Application of bactericides and biocontrol agents
For more than a century, copper and copper-based antimicrobial compounds were the main chemical control agents of Xanthomonas caused diseases. Currently, copper applications are widely used in the field and the compound is still considered as the most efficient means to control bacterial diseases (Lamichhane et al. 2018). However, in the past few decades many concerns have arisen regarding the sustainability and environmental effects of copper leading to the search for other control methods (Lamichhane et al. 2018). Resistance to copper has become more and more prevalent within Xanthomonas field isolates from different crops and locations. This resistance is attributed to the acquisition of copper resistance gene cluster through HGT (Bender et al. 1990; Cooksey 1994; Behlau et al. 2008). Copper resistance is usually plasmid encoded, and the plasmid is thought to be readily conjugated into other bacteria (Basim et al. 1999). Different strategies have been deployed to provide an efficient and sustainable substitute for copper and several products are now commercially available as biocontrol agents for Xanthomonas caused diseases (Table 2).
Table 2.
Examples of commercially available biocontrol agents used for Xanthomonas-induced diseases.
| Biocontrol agent | Commercial name/s | Diseases that can be controlled by the product A | Approved for use | Comments | |||
|---|---|---|---|---|---|---|---|
| USAB | EUC,F | BrazilD | ChinaE | ||||
| Acibenzolar-s-methyl (ASM) | ACTIGARDTM (Syngenta) | Broad range: Bacterial spot of tomato and pepper (X. euvesicatoria, X. perforans, X. gardneri, X. vesicatoria), Citrus canker (X. citri pv. citri), black rot of crucifers (X. campestris pv. campestris), bacterial spot of cucurbits (X. cucurbitae) | √ | √ | √ | unknown | |
| BIONR (Syngenta) | |||||||
| CGA 245704TM (Syngenta) | |||||||
| Bactericides | |||||||
| Bismerthiazol | Bacterial blight of rice (X. oryzae pv. oryzae), bacterial streak of rice (X. oryzae pv. oryzicola) | √ | Reported as a thyroid toxicant (Zhang, Wang and Zhu 2009), not approved use in multiple countries due to possible toxicity | ||||
| Streptomycin | FireWall TM (Agrosource) | Bacterial spot of tomato and pepper (X. euvesicatoria, X. perforans, X. gardneri, X. vesicatoria), citrus canker (X. citri pv. citri) | √ | √ | High resistance reported in tomato infecting Xanthomonas strains (Sundin and Wang 2018) | ||
| Oxytetracycline | FireLineTM (Agrosource) | Bacterial spot of peaches and nectarines (X. arboricola pv. pruni) | √ | ||||
| MycoshieldTM (Nufarm Americas) | |||||||
| Kasugamycin | Kasumin 2 L, (Arysta) | In USA: walnut blight (X. arboricola pv. juglandis) In China: black rot of crucifers (X. campestris pv. campestris), citrus canker (X. citri pv. citri), bacterial spot of peaches and nectarines (X. arboricola pv. pruni) |
√ | √ | |||
| Bacteriophages | |||||||
| AgriPhageTM tomato spot & speck (omnilytics) | Bacterial spot of tomato and pepper (X. euvesicatoria, X. perforans, X. gardneri, X. vesicatoria) | √ | |||||
| AgriPhage™ citrus canker (omnilytics) | Citrus canker (X. citri pv. citri) | √ | |||||
| Bacterial control agents | |||||||
| Bacillus pumilus QST 2808 | Sonata (Wilbur-Ellisa®) | Broad range | √ | √ | √ | ||
| Bacillus amyloliquefaciens QST 713 | SerenadeASO (AgraQuest®) | √ | √ | √ | |||
| Bacillus amyloliquefaciens D747 | CX 9030 (Agrobase) | √ | √ | √ | |||
| Bacillus amyloliquefaciens Strain FZB24 | TAEGRO® (Syngenta) | Bacterial spot of tomato and pepper (X. euvesicatoria, X. perforans, X. gardneri, X. vesicatoria) | √ | √ | |||
| Bacillus amyloliquefaciens MBI 600 | Serifel® Biofungicide (BASF) | Black rot of crucifers (X. campestris pv. campestris) | √ | ||||
| Bacillus amyloliquefaciens B7900 | Broad range | √ | |||||
| Bacillus amyloliquefaciens PQ21 | √ | ||||||
| Bacillus amyloliquefaciens B1619 | √ | ||||||
| Bacillus amyloliquefaciens LX-11 | Bacterial leaf streak of rice (X. oryzae pv. oryzicola), broad range | √ | |||||
| Streptomyces lydicus WYEC 108 | Actinivate (Novoenzymes) | Walnut blight (X. arboricola pv. juglandis), bacterial spot of tomato (X. perforans), citrus canker (X. citri pv. citri), angular leaf spot of strawberries (X. fragariae) | √ | √ | |||
According to company specification sheet.
Approved by United States Environmental Protection Agency, US EPA.
Approved by EU according to EU commission—EU Pesticides database.
Approved by the Brazilian Health Regulatory Agency (Anvisa).
Approved by ministry of Agriculture of the People's Republic of China. We thank Drs. Xiaoyong Yuan, Zhigang Ouyang, Mingqing Shang, and Shengqi Chi for their help in collecting information.
Some products approved by EU commission but is not yet authorised for use on a national level for each country.
Several types of antimicrobials were approved to be used for control against certain bacterial diseases (Table 2). Regulations of which compound can be used vary between countries and introduction of new antimicrobial agents in the field is subject to extensive testing prior to approval. Different antibiotics have been used against Xanthomonas caused diseases with some success. Streptomycin was first introduced as an agricultural antimicrobial agent in the 1950’s for treatment of bacterial diseases in various crops. The application of streptomycin, however, is soon followed by high frequency of resistance to the compound in different Xanthomonas spp. (Sundin and Wang 2018). Oxytetracycline is currently used for treatment of bacterial spot of peach caused by X. arboricola pv. pruni. This antibiotic treatment is effective for disease control in peach and no resistance to the treatment has been reported. The quinolone antibiotic oxolinic acid was reported to be successful in initial field trials against X. frageria and other bacterial pathogens such as Erwinia amylovora and Burkholderia glumae (Kim et al. 2016). However, this treatment failed to sustain its effect in long-term application because of the occurrence of resistant E. amylovora and B. glumae strains (Stockwell and Duffy 2012). Kasugamaycin had demonstrated some positive effect on Xanthomonas bacterial spot of pepper and tomato (Vallad et al. 2010) and was recently approved as a pesticide in the USA (Table 2). Field trials have been conducted to examine the effectiveness of the compound against other Xanthomonas caused diseases.
The potential application of antimicrobial peptides (AMPs) as a biocontrol agent for Xanthomonas treatments is currently under investigation. AMPs naturally occur in all forms of life and play a role in defence or competition against or between microorganisms (Hancock and Diamond 2000). AMPs biochemical properties give them semi-differential but usually not very specific toxic activity against multitude of microorganisms. Several AMPs are currently subjected to clinical trials for potential medicinal uses (Mahlapuu et al. 2016). Multiple studies have tested the potential application of natural and synthetic AMPs against bacterial and fungal pathogens, including Xanthomonas spp. and showed promising preliminary data. Studies have demonstrated high toxicity of AMPs on various Xanthomonas spp. in vitro while showing minimal effect on plant or mammalian cells (Choi and Moon 2009; Zeitler et al. 2013; Inui Kishi et al. 2018; Camó et al. 2019). Peptides were also tested for inhibition of plant diseases in greenhouse conditions in a few studies (Choi and Moon 2009; Shi et al. 2016).
Metal ions such as Zn2+ or Ag+ were previously reported to have an antimicrobial effect. However, the amount required for antimicrobial activity is high and will potentially have a negative environmental effect. Recent advances in nanoparticle technology should enable effective modification resulting in a more potent product. Accordingly, different metal-ion-based nanoparticles have been successfully utilised as antimicrobial compound in greenhouse and field trials against tomato infecting Xanthomonas strains (Ocsoy et al. 2013; Paret et al. 2013; Makarovsky et al. 2018; Liao et al. 2019).
Bacteriophages
Bacteriophages are bacterial viruses that occur in almost all bacteria and usually have high host specificity. Soon after their discovery in the beginning of the 20th century the potential use of these viruses as antimicrobial agents was suggested; greenhouse-based studies displayed effective prevention of plant bacterial diseases as early as the mid-1920s (Abedon et al. 2011). An interest in utilising bacteriophages for biocontrol as an alternative to copper has arisen again in the past two decades. Many studies have been dedicated to the identification of bacteriophages in different Xanthomonas spp. and the utilisation of them against the bacterial pathogens in the field (Buttimer et al. 2017). Bacteriophages were indeed found to be effective in suppressing Xanthomonas caused diseases in tomato, citrus and onion in field conditions (Obradovic et al. 2004; Lang, Gent and Schwartz 2007; Balogh et al. 2008). While promising, there are limiting factors to bacteriophage application in the field to include their stability in the different environments and sensitivity to UV radiation.
Systemic acquired resistance inducers
Several chemicals identified to actively induce systemic acquired resistance (SAR) have been utilised for management of diseases through activation of plant defence responses. The application of these inducers has a broad effect against many types of pathogens and in particular can effectively inhibit Xanthomonas diseases in multiple crops in the field (Thakur and Sohal 2013; Bektas and Eulgem 2015). A range of chemicals can induce SAR, although the exact mechanism of induction is not completely understood in most cases (Thakur and Sohal 2013; Bektas and Eulgem 2015). The main chemical compounds identified as SAR inducers are abicenzolar-S-mehtyl (ASM), Probenazole, β-amino butyric acid (BABA) and bismerthiazol. ASM has been commercialized (Table 2) and has been shown to be effective against many Xanthomonas-induced diseases (Obradovic et al. 2004; Gent and Schwartz 2005; Francis et al. 2009).
Biocontrol through beneficial microorganisms
Naturally occurring bacteria from the plant rhizosphere and phyllosphere have been shown to affect plant pathogens and disease development (Parnell et al. 2016). A lot of work has been dedicated towards identifying isolates with antimicrobial-producing or SAR-inducing activity and utilising these bacteria to combat Xanthomonas-induced diseases. Plant-associated microorganisms exhibiting antimicrobial activity are attractive candidates for field biocontrol agents. Bacillus and Streptomyces spp. in particular, are considered as good candidates because of established antimicrobial attributes and sporulation capabilities. Indeed, many Bacillus and Streptomyces strains have been shown to have a broad range biocontrol effect (Shafi, Tian and Ji 2017; Vurukonda, Giovanardi and Stefani 2018) and several have been commercialised (Table 2). Numerous studies have demonstrated that foliar treatment with commercial or naturally isolated bacilli and Streptomyces strains significantly reduced Xanthomonas caused diseases in multiple crops in field trials (Van Hop et al. 2014; Thapa and Babadoost 2016; Daranas et al.2019).
Many bacterial isolates were reported to exert biocontrol through SAR induction. Repeated applications of P. fluorescens by foliar spray suppressed the disease severity of bacterial blight in repeated field trials and improved yield (Vidhyasekaran et al. 2001). Several rhizosphere isolates of Burkholderia and Stenotrophomonas geniculata with SAR-inducing traits successfully inhibited X. citri infection of grapefruit through soil drench treatment in greenhouse experiments (Riera et al. 2018).
Biocontrol of pathogenic Xanthomonas through non-pathogenic or attenuated Xanthomonas strains has also been examined. The general hypothesis behind using these non-pathogenic or attenuated strains is that pathogenic and non-pathogenic strains occupy same niches, but non-pathogenic strains induce SAR or utilise species specific bacteriocins against the virulent strains. Field trials utilising mutants in the type III secretion system that potentially elicit SAR indeed displayed strong biocontrol inhibition of X. euvesicatoria and X. oryzae on tomato and rice, respectively (Dardick et al. 2003; Moss et al. 2007). Non-pathogenic isolates of X. campestris that occur naturally have been shown to exert biocontrol activity against X. arabicola pv. pruni (Xap) in peach under greenhouse conditions (Kawaguchi, Inoue and Inoue 2014). While displaying high biocontrol capability during co-inoculation, the population of the non-pathogenic strains was not sustained in the host for long periods of time, questioning the sustainability of these strains in the field (Kawaguchi, Inoue and Inoue 2014). To overcome the inability to sustain an in planta population of non-pathogenic strains and type III secretion system mutants, Hert et al. attempted to use a bacteriocin-producing attenuated X. perforans strain to control bacteriocin sensitive X. euvesicatoria strains in tomato. The assumption here was that the attenuated strain would sustain a stable population that would be insufficient to induce disease symptoms while nevertheless inhibiting the pathogenic strain (Hert et al. 2009). Although the pathogenic strain was inhibited, it was not completely eradicated, however, and multiple applications of the biocontrol strain were required to sustain inhibition (Hert et al. 2009).
Generation of resistant varieties
Generation of disease resistant varieties is one of the most efficient, sustainable, environmentally friendly and desired disease control approaches. Plant resistance or susceptibility to certain pathogens have been demonstrated to be a hereditable trait. Many cultivar specific Xanthomonas resistance loci were identified in different plants. The number of resistance genes identified in domesticated varieties varies greatly between plant species. For instance, dozens of Xanthomonas resistance associated loci have been identified in rice (Khan, Naeem and Iqbal 2014) while to date none have been identified in other crops, such as banana, peach or citrus,.
Resistance to Xanthomonas spp. can be multiple quantitative trait loci (QTL) or can originate from a single dominant or recessive locus. In most cases, a dominant locus represents active recognition of pathogen determinant by a resistance (R) or pattern recognition receptor (PRR) proteins while recessive loci are usually associated with the modulation of pathogen susceptibility genes. One of the well characterised recessive resistance loci against Xanthomonas was found to induce the expression of the SWEET transporter family genes, which are named susceptibility (S) genes, in rice by X. oryzea TAL effectors (Hutin et al. 2015a; Ji, Wang and Zhao 2018). The effector-binding element (EBE) of SWEET genes is mutated in rice R loci xa13, xa25 and xa41 (Chu et al. 2006; Yang, Sugio and White 2006; Liu et al. 2011; Hutin et al. 2015b). In addition, reduced affinity of TAL effectors to transcriptional machinery was identified as the functional mechanism of xa5 resistance locus (Yuan et al. 2016).
A handful of dominant resistance genes against Xanthomonas spp. were cloned from pepper, rice, tomato and maize and the bacterial determinants were identified in some of them. The majority of such genes were identified to be intracellular NB-LRR receptor genes (pepper Bs2, maize Rxo1, rice Xa1 and tomato Bs4) or outer-membrane receptor genes (rice Xa21, Xa3/Xa26 and Xa4) (Table S3, Supporting Information). Dominant executor R genes (pepper Bs3 and Bs4C and rice Xa10, Xa23, and Xa27) target specific TAL effectors. Executor genes encode hypersensitive-response inducing proteins under a promoter region that contains the EBE of virulent TALs (Zhang, Yin and White 2015) (Table S3, Supporting Information). Even though considerable effort has been dedicated to the identification and characterisation of hereditable resistance traits, the majority of genes conferring resistance are unknown. In many cases, a resistance trait is identified and even mapped to a certain chromosomal region but the resistance gene itself is not identified (Khan, Naeem and Iqbal 2014) (Table S4, Supporting Information). Many virulence determinants, mainly T3Es, have been shown to dictate cultivar and even species host specificity but the plant R genes responsible for the resistance have not been cloned (Table S4). Identification, cloning and characterisation of these unknown R genes can greatly contribute to the development of new Xanthomonas resistant varieties that will increase yield in the field.
Generation of resistant varieties through traditional breeding
Resistance to pathogens is a major trait to be considered during breeding. Substantial efforts are being made to identify and introduce resistant or tolerant accessions for breeding against disease. However, in many cases, the screened resistant lines are either significantly different from the main commercial lines or are wild germplasms of a domesticated crop. The introduction of resistant genes into desired domesticated crops requires significant amount of crossings to produce near isogenic lines, a task that is not completely realistic in perennial tree plants or hybrid lines that are vegetatively propagated such as most domesticated citrus varieties (Wu et al. 2018). Nevertheless, genes conferring resistance against Xanthomonas have been introduced through traditional breeding to produce near-isogenic lines of rice, pepper and tomatoes (Stall, Jones and Minsavage 2009) (Chukwu et al. 2019). Cross-breeding procedures are currently ongoing for many annual crops such as strawberries, maize, cassava and wheat (Roach et al. 2016; Soto Sedano et al. 2017; Wen et al. 2018).
Durability is also a significant factor in the introduction of resistant varieties. Many Xanthomonas races within a species are found to infect resistant lines and in many cases quick field adaptation of the pathogen occurs (Stall, Jones and Minsavage 2009; Hutin et al. 2015a). For example, resistance to TAL effectors that is achieved through alterations in the promoters of S gene or utilisation of executor R genes are usually specific to certain pathogen races and can be easily overcome by Xanthomonas pathogens (Hutin et al. 2015a). Xanthomonas oryzae races contain multiple TALs that target different promoter regions in the same S gene or alternatively target a different S gene with similar function (Hutin et al. 2015a).
The majority of non-TAL effectors are functionally redundant and a single effector is usually not significant for pathogenicity (Hajri et al. 2009; Cunnac et al. 2011). For pepper or tomato, Xanthomonas bacteria were able to quickly overcome the introduction of non-TAL recognising resistant genes (pepper Bs1 and tomato Xv3 and rx1, rx2 and rx3) in the field through the loss or inactivation of the avirulence effector and shift in pathogen races (Stall, Jones and Minsavage 2009). On the other hand, the non-TAL effector gene avrBs2 is highly conserved in Xanthomonas spp. (Hajri et al. 2009) and is required for the virulence of many Xanthomonas strains such X. euvesicatoria, Xac and Xam (Li et al. 2015; Medina et al. 2018). Consequently, the pepper Bs2 locus, which confer resistance upon recognition of AvrBs2, serves as a good candidate for durable field resistance. Production of near isogenic lines of pepper containing Bs2 was indeed found to give relatively stable field resistance to bacterial spot, however, few X. euvesciatoria isolates were identified in the resistant lines and mutation in avrBs2 in relatively low rate frequency was identified in Xanthomonas mutant collections (Wichmann et al. 2005).
Pyramiding disease resistance genes against Xanthomonas has been conducted in pepper, cotton and rice (Delannoy et al. 2005; Stall, Jones and Minsavage 2009; Chukwu et al. 2019). Pepper near isogenic lines ECW123 (containing Bs1, Bs2 and Bs3) were generated and indeed showed enhanced resistance to bacterial spot but still could not provide complete protection against the Xanthomonas pathogen (Stall, Jones and Minsavage 2009). Several commercially available rice lines harbouring a combination of multiple resistance genes against bacterial blight caused by Xoo have been produced and introduced into the field (Chukwu et al. 2019). Improved Samba Mahsuri is a near isogenic line containing three bacterial blight resistance genes (Xa21, xa13, and xa5) in 2008 (Sundaram et al. 2009); thus far this line has provided high resistance against bacterial blight in India.
Introduction of resistant varieties through genetic engineering
The introduction of genetically modified (GM) crops has long been suggested as an approach to overcome the obstacles of traditional breeding, in particular to enable modification of crops with long generation times that cannot be bred into near isogenic lines in a realistic time window. Producing GM crops does raise many issues. Regulation of production, consumption and research of GM crops and addressing public concerns regarding the potential health hazards or the economic effect of patenting agricultural products are among the limiting factors that should be considered before proceeding in that direction (Raman 2017; Thomas and De Tavernier 2017; Faure and Napier 2018). With that in mind, multiple approaches are being used for introduction of resistance or tolerance to Xanthomonas in commercial crops.
The most direct means to generate resistant transgenic plants is the introduction of R genes or immune receptors. General induction of immune response was proven efficient in laboratory conditions but is usually associated with other pleiotropic effects on plant physiology that might reduce yield in field conditions. The introduction of immune receptors might overcome such issues, but, similarly to the resistant lines in breeding, it can be associated with relatively fast adaptation of the pathogens. Several R or PRR genes from different plant species have been introduced into commercial crops. For example, introduction of the pepper Bs2 or Arabidopsis ELONGATION FACTOR TU RECEPTOR (EFR) into tomato increases resistance to Xanthomonas bacterial spot in greenhouse and field trials (Horvath et al. 2012; Kunwar et al. 2018). Introduction of other resistance genes like pepper Bs2, rice Xa21 and N. benhtamiana FLAGELLIN SENSITIVE2 (FLS2) into citrus, maize Rxo1 into rice and rice Xa21 into banana inhibited Xanthomonas induced diseases in greenhouse and glasshouse conditions (Zhou et al. 2010; Tripathi et al. 2014; Hao et al. 2015; Sendín et al. 2017; Omar et al. 2018). Durability of these transgenic plants in the field is yet to be determined.
Another strategy for transgene-induced resistance is through promoter engineering of executor R genes. Zeng et al. engineered the promoter region of Xa10 executor R gene to contain EBE recognition site of multiple X. oryzae TALEs and demonstrated that the modified executor R gene, Xa10 (E5), provided broad spectrum resistance against 27 X. oryzae field isolated (Zeng et al. 2015). Using a similar approach, Shantharaj and colleagues introduced the EBE region of the CsLOB1, the canker susceptibility gene induced by X. citri PthA4, into the promoter of the Bs3 gene and replaced Bs3 with avrGf1 which is recognised as an avirulence gene in grapefruit (Shantharaj et al. 2017). Transient expression of the engineered executor R gene in grapefruit leaves leads to the development of HR in response to X. citri and reduced bacterial colonisation (Shantharaj et al. 2017).
Few studies have introduced bacterial genes into crops to combat Xanthomonas diseases in greenhouse conditions. For example, Caserta and colleagues have introduced DSF producing gene rpfF from Xylella into citrus and demonstrated enhanced resistance to X. citri, possibly through misregulation of virulence-associated genes in an improper bacterial quorum (Caserta et al. 2014). In addition, Zhang and colleagues expressed Pantoea albD, which was shown to detoxify the X. albilineans toxin albicidin in sugarcane. This lead to a reduction of X. albilineans leaf scald symptoms with partial success (Zhang, Xu and Birch 1999).
Xanthomonas-insensitive rice and citrus were produced through targeting the EBE of S locus promoter. Li and colleagues utilized TALEN-based gene editing to modify the EBE region, recognized by two X. oryzae TAL effectors, of the rice OsSWEET14 susceptibility gene (Li et al. 2012). Selfing of modified lines resulted in a non-transgenic rice plants with resistance to the X. oryzae strains (Li et al. 2012). Two groups independently disrupted the X. citri PthA4 EBE binding region of the citrus canker susceptibility gene CsLOB1 using CRISPR/Cas9 system in grapefruit and Wangjincheng orange (Jia et al. 2016; Peng et al. 2017). The PthA4 EBE was disrupted in differential ratios in transgenic plants expressing the CRISPR/Cas9 system and, as expected, the plants displayed reduced canker symptoms when inoculated with X. citri (Jia et al. 2016; Peng et al. 2017). A novel technique developed by Murata and colleagues was based on production of cybrid cells containing combinations of nucleus and organelles from different cultivars (Murata et al. 2018). Using somatic hybridisation, the group produced a cybrid grapefruit containing the nucleus of X. citri susceptible grapefruit (Citrus paradisi) and the chloroplast of X. citri tolerant/resistant Meiwa kumquat (Citrus japonica). The cybrids displayed enhanced resistance to X. citri infection, thus demonstrating that organelle genetic background plays an important role in pathogen response (Murata et al. 2018).
CONCLUDING REMARKS
Since the report of the first two Xanthomonas genomes almost 20 years ago, there has been an extensive body of work describing factors that contribute to Xanthomonas disease in their respective host plants. It is now clear that Xanthomonas has developed a range of highly regulated and co-ordinated traits needed to adhere to plant tissue, acquire nutrients, suppress plant defence responses and ultimately cause disease. In spite of these detailed studies of molecular mechanisms underlying the activity of Xanthomonas virulence factors there are still many unanswered questions. Some of the major challenges include the understanding Xanthomonas tissue specificity and preference for portal of entry mechanisms of sensing of plant-derived signals; the elucidation of virulence-associated regulatory systems, which include RNA-binding proteins and small RNAs. It is believed that multidisciplinary approaches (comparative genomics, functional genomics, population genomics, metagenomics, transcriptomics, proteomics and systems biology) will be deployed in the future research to address these issues.
Efforts are dedicated to understanding Xanthomonas ecology, epidemiology, and phytopathology have the ultimate goal of controlling or suppressing plant disease. This knowledge has begun to be translated into various strategies including pathogen chemical inhibition of dispersal, biocontrol agents and utilisation of resistant varieties. In addition to these approaches, the growing appreciation of the role of the plant microbiome in promoting plant health and growth suggests the possibility of manipulation of endophytic/epiphytic communities to achieve some measure of disease control. However, studies of the impact of the microbiome on Xanthomonas colonisation and disease control are currently at an early stage.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Chance Nowak, Prabha Liyanapathiranage and Jeffrey Jones for helpful comments on the manuscript. We apologise to the many authors whose work we were unable to cite because of space limitations and the large scope of this current review. Work in the Tang laboratory is supported by grants from the State Key Laboratory Program SKLCUSA-b201817, National Natural Science Foundation of China (31470237) and the Ba Gui Scholar Program of Guangxi Zhuang Autonomous Region of China (2014A002). Work in the Bleris laboratory is supported by US National Science Foundation (NSF) CAREER grant 1351354 and a Cecil H. and Ida Green Endowment. Work in the Wang laboratory is supported by USDA-NIFA Plant Biotic Interactions Program 2017-67013-26527. Additionally, Shi-qi An is supported by State Key Laboratory Program SKLCUSA-b201817 and BBSRC grant BB/R012415/1.
Contributor Information
Shi-Qi An, National Biofilms Innovation Centre (NBIC), Biological Sciences, University of Southampton, University Road, Southampton SO17 1BJ, UK.
Neha Potnis, Department of Entomology and Plant Pathology, Rouse Life Science Building, Auburn University, Auburn AL36849, USA.
Max Dow, School of Microbiology, Food Science & Technology Building, University College Cork, Cork T12 K8AF, Ireland.
Frank-Jörg Vorhölter, MVZ Dr. Eberhard & Partner Dortmund, Brauhausstraße 4, Dortmund 44137, Germany.
Yong-Qiang He, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, 100 Daxue Road, Nanning 530004, Guangxi, China.
Anke Becker, Loewe Center for Synthetic Microbiology and Department of Biology, Philipps-Universität Marburg, Hans-Meerwein-Straße 6, Marburg 35032, Germany.
Doron Teper, Citrus Research and Education Center, Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, 700 Experiment Station Road, Lake Alfred 33850, USA.
Yi Li, Bioengineering Department, University of Texas at Dallas, 2851 Rutford Ave, Richardson, TX 75080, USA; Center for Systems Biology, University of Texas at Dallas, 800 W Campbell Road, Richardson, TX 75080, USA.
Nian Wang, Citrus Research and Education Center, Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, 700 Experiment Station Road, Lake Alfred 33850, USA.
Leonidas Bleris, Bioengineering Department, University of Texas at Dallas, 2851 Rutford Ave, Richardson, TX 75080, USA; Center for Systems Biology, University of Texas at Dallas, 800 W Campbell Road, Richardson, TX 75080, USA; Department of Biological Sciences, University of Texas at Dallas, 800 W Campbell Road, Richardson, TX75080, USA.
Ji-Liang Tang, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, 100 Daxue Road, Nanning 530004, Guangxi, China.
Conflicts of interest. None declared.
REFERENCES
- Abedon ST, Thomas-Abedon C, Thomas Aet al.. Bacteriophage prehistory: is or is not Hankin, 1896, a phage reference?. Bacteriophage. 2011;1:174–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addis T, Turyagyenda LF, Alemu Tet al.. Garden tool transmission of Xanthomonas campestris pv. musacearum on banana (Musa spp.) and enset in Ethiopia. Acta Hortic. 2010;879:367–72. [Google Scholar]
- Alkhateeb RS, Vorhölter FJ, Rückert Cet al.. Genome wide transcription start sites analysis of Xanthomonas campestris pv. campestris B100 with insights into the gum gene cluster directing the biosynthesis of the exopolysaccharide xanthan. J Biotechnol. 2016;225:18–28. [DOI] [PubMed] [Google Scholar]
- An S-Q, Febrer M, McCarthy Yet al.. High-resolution transcriptional analysis of the regulatory influence of cell-to-cell signalling reveals novel genes that contribute to Xanthomonas phytopathogenesis. Mol Microbiol. 2013a;88:1058–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S-Q, Tang J-L, Dow JM. Probing the role of cyclic di-GMP signaling systems in disease using Chinese radish. Methods Mol Biol. 2017;1657:205–12. [DOI] [PubMed] [Google Scholar]
- An SQ, Allan JH, Mccarthy Yet al.. The PAS domain-containing histidine kinase RpfS is a second sensor for the diffusible signal factor of Xanthomonas campestris. Mol Microbiol. 2014;92:586–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SQ, Chin KH, Febrer Met al.. A cyclic GMP-dependent signalling pathway regulates bacterial phytopathogenesis. EMBO J. 2013b;32:2430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aritua V, Parkinson N, Thwaites Ret al.. Molecular epidemiology of Xanthomonas campestris pv. musacearum, the causal agent of xanthomonas wilt of banana and enset. Acta Hortic. 2009;828:219–26. [Google Scholar]
- Aslam SN, Newman MA, Erbs Get al.. Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol. 2008;18:1078–83. [DOI] [PubMed] [Google Scholar]
- Bae N, Park HJ, Park Het al.. Elucidating functions of fleq in Xanthomonas oryzae pv. oryzae by comparative proteomic and phenotypic analyses. Int J Mol Sci. 2018;19:pii: E3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldessari A, Ielpi L, Dankert MA. A novel galacturonide from Xanthomonas campestris. J Gen Microbiol. 1990;136:1501–8. [Google Scholar]
- Balogh B, Canteros BI, Stall REet al.. Control of citrus canker and citrus bacterial ppot with bacteriophages. Plant Dis. 2008, doi:10.1094/pdis-92-7-1048. [DOI] [PubMed] [Google Scholar]
- Barak JD, Koike ST, Gilbertson RL. Movement of Xanthomonas campestris pv. vitians in the stems of lettuce and seed contamination. Plant Pathol. 2002;51:506–12. [Google Scholar]
- Barreras M, Salinas SR, Abdian PLet al.. Structure and mechanism of GumK, a membrane-associated glucuronosyltransferase. J Biol Chem. 2008;283:25027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R, Cohn M, Kassen Aet al.. High-throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance. Proc Natl Acad Sci. 2012;109:E1972–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basim H, Stall RE, Minsavage GVet al.. Chromosomal gene transfer by conjugation in the plant pathogen Xanthomonas axonopodis pv. vesicatoria. Phytopathology. 1999;89:1044–9. [DOI] [PubMed] [Google Scholar]
- Bayer-Santos E, Lima L dos P, Ceseti L de Met al.. Xanthomonas citri T6SS mediates resistance to Dictyostelium predation and is regulated by an ECF σ factor and cognate Ser/Thr kinase. Environ Microbiol. 2018;20:1562–75. [DOI] [PubMed] [Google Scholar]
- Becker A, Vorhölter F-J. Xanthan biosynthesis by Xanthomonas bacteria: an overview of the current biochemical and genomic data. In: Bernd H. A. Rehm (ed). Microbial Production of Biopolymers and ·polymer Precursors: Applications and Perspectives. UK: Caister Academic Press, 2009, 1–12. [Google Scholar]
- Behlau F, Belasque J, Bergamin Filho Aet al.. Copper sprays and windbreaks for control of citrus canker on young orange trees in southern Brazil. Crop Prot. 2008;27:807–13. [Google Scholar]
- Bektas Y, Eulgem T. Synthetic plant defense elicitors. Front Plant Sci. 2015;5:804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasque J, Parra-Pedrazzoli ALL, Rodrigues Neto Jet al.. Adult citrus leafminers (Phyllocnistis citrella) are not efficient vectors for Xanthomonas axonopodis pv. citri. Plant Dis. 2005;89:590–4. [DOI] [PubMed] [Google Scholar]
- Bender CL, Malvick DK, Conway KEet al.. Characterization of pXV10A, a copper resistance plasmid in Xanthomonas campestris pv. vesicatoria. Appl Environ Microbiol. 1990;56:170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D, Duta S, Yu SMet al.. Taxonomic and functional changes of bacterial communities in the rhizosphere of Kimchi cabbage after seed bacterization with Proteus vulgaris JBLS202. Plant Pathol J. 2018;34:286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Mann E, Whitfield Cet al.. Architecture of a channel-forming O-antigen polysaccharide ABC transporter. Nature. 2018;553:361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco MI, Jacobs M, Salinas SRet al.. Biophysical characterization of the outer membrane polysaccharide export protein and the polysaccharide co-polymerase protein from Xanthomonas campestris. Protein Expr Purif. 2014;101:42–53. [DOI] [PubMed] [Google Scholar]
- Bianco MI, Toum L, Yaryura PMet al.. Xanthan pyruvilation is essential for the virulence of Xanthomonas campestris pv. campestris. Mol Plant Microbe Interact. 2016;29:688–99. [DOI] [PubMed] [Google Scholar]
- Blanvillain S, Meyer D, Boulanger Aet al.. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One. 2007;2:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme G, Dita M, Jacobsen KSet al.. Bacterial diseases of bananas and enset: current state of knowledge and integrated approaches toward sustainable management. Front Plant Sci. 2017;8:1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock CH, Cook AZ, Parker PEet al.. Short-distance dispersal of splashed bacteria of Xanthomonas citri subsp. citri from canker-infected grapefruit tree canopies in turbulent wind. Plant Pathol. 2012;61:829–36. [Google Scholar]
- Bonomi HR, Toum L, Sycz Get al.. Xanthomonas campestris attenuates virulence by sensing light through a bacteriophytochrome photoreceptor. EMBO Rep. 2016;17:1565–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordes P, Lavatine L, Phok Ket al.. Insights into the extracytoplasmic stress response of Xanthomonas campestris pv. campestris: role and regulation of σE-dependent activity. J Bacteriol. 2011, doi:10.1128/JB.00884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger A, Zischek C, Lautier Met al.. The plant pathogen Xanthomonas campestris pv. campestris exploits N-Acetylglucosamine during infection. MBio. 2014;5:e01527–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury JF. Genus II Xanthomonas Dowson 1939. In: Krieg NR, Holt JG (eds). Bergey's Manual of Systematic Bacteriology, Vol. 1. London: Williams and Wilkins, 1984, pp. 199–210. [Google Scholar]
- Braun SG, Meyer A, Holst Oet al.. Characterization of the Xanthomonas campestris pv. campestris Lipopolysaccharide substructures essential for elicitation of an oxidative burst in tobacco cells. Mol Plant-Microbe Interact. 2005;18:674–81. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Garrido-Oter R, Münch PCet al.. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17:392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch AY, Do PT, Sbodio Aet al.. High-level culturability of epiphytic bacteria and frequency of biosurfactant producers on leaves. Appl Environ Microbiol. 2016, doi:10.1128/AEM.01751-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buregyeya H, Kityo R, Kubiriba Jet al.. Role of birds & bats in long distance transmission of banana bacterial wilt in Uganda. Int J Agric Innov Res. 2014;2:636–40. [Google Scholar]
- Buttimer C, McAuliffe O, Ross RPet al.. Bacteriophages and bacterial plant diseases. Front Microbiol. 2017;8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Yuan ZH, Zhang Het al.. Fatty acid DSF binds and allosterically activates histidine kinase RpfC of phytopathogenic bacterium Xanthomonas campestris pv. campestris to regulate quorum-sensing and virulence. PLoS Pathog. 2017;13:e1006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camó C, Bonaterra A, Badosa Eet al.. Antimicrobial peptide KSL-W and analogues: promising agents to control plant diseases. Peptides. 2019;112:85–95. [DOI] [PubMed] [Google Scholar]
- Canteros BI, Gochez AM, Moschini RC. Management of citrus canker in argentina, a success story. Plant Pathol J. 2017;33:441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabuono A, Petrocelli S, Ottado Jet al.. Structural analysis and involvement in plant innate immunity of Xanthomonas axonopodis pv. citri lipopolysaccharide. J Biol Chem. 2011;286:25628–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta R, Picchi SC, Takita MAet al.. Expression of Xylella fastidiosa RpfF in citrus disrupts signaling in Xanthomonas citri subsp. citri and thereby its virulence. Mol Plant-Microbe Interact. 2014;27:1241–52. [DOI] [PubMed] [Google Scholar]
- Castiblanco LF, Sundin GW. New insights on molecular regulation of biofilm formation in plant-associated bacteria. J Integr Plant Biol. 2016;58:362–72. [DOI] [PubMed] [Google Scholar]
- Cazorla FM, Codina JC, Abad Cet al.. 62-kb plasmids harboring rulAB homologues confer UV-tolerance and epiphytic fitness to Pseudomonas syringae pv. syringae mango isolates. Microb Ecol. 2008;56:283–91. [DOI] [PubMed] [Google Scholar]
- Cerutti A, Jauneau A, Auriac M-Cet al.. Immunity at cauliflower hydathodes controls systemic infection by Xanthomonas campestris pv campestris. Plant Physiol. 2017;174:700–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesbron S, Briand M, Essakhi Set al.. Comparative genomics of pathogenic and nonpathogenic strains of Xanthomonas arboricola unveil molecular and evolutionary events linked to pathoadaptation. Front Plant Sci. 2015;6:1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoiseau P, Petit-Bourg G, Rott Pet al.. Epiphytic populations of Xanthomonas albilineans and subsequent sugarcane stalk infection are linked to rainfall in guadeloupe. Plant Dis. 2009;93:339–46. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Almeida RPP, Lindow S. Living in two worlds: the plant and insect lifestyles of xylella fastidiosa. Annu Rev Phytopathol. 2008;46:243–71. [DOI] [PubMed] [Google Scholar]
- Chen CH, Lin NT, Hsiao YMet al.. Two non-consensus Clp binding sites are involved in upregulation of the gum operon involved in xanthan polysaccharide synthesis in Xanthomonas campestris pv. campestris. Res Microbiol. 2010a;161:583–9. [DOI] [PubMed] [Google Scholar]
- Chen NWG, Serres-Giardi L, Ruh Met al.. Horizontal gene transfer plays a major role in the pathological convergence of Xanthomonas lineages on common bean. BMC Genomics. 2018;19:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Wu CH, Lin JWet al.. Mutation of the gene encoding a major outer-membrane protein in Xanthomonas campestris pv. campestris causes pleiotropic effects, including loss of pathogenicity. Microbiology. 2010b;156:2842–54. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Shieh SY, Hsu CCet al.. Characterization and transcriptional analysis of an ECF sigma factor from Xanthomonas campestris pv. campestris. FEMS Microbiol Lett. 2008;289:250–7. [DOI] [PubMed] [Google Scholar]
- Chin KH, Kuo WT, Yu YJet al.. Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein. Acta Crystallogr Sect D Biol Crystallogr. 2012;68:1380–92. [DOI] [PubMed] [Google Scholar]
- Chin KH, Lee YC, Tu ZLet al.. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396:646–62. [DOI] [PubMed] [Google Scholar]
- Choi J, Moon E. Identification of novel bioactive hexapeptides against phytopathogenic bacteria through rapid screening of a synthetic combinatorial library. J Microbiol Biotechnol. 2009;19:792–802. [DOI] [PubMed] [Google Scholar]
- Christiano RSC, Dalla Pria M, Jesus Junior WCet al.. Modelling the progress of Asiatic citrus canker on Tahiti lime in relation to temperature and leaf wetness. Eur J Plant Pathol. 2009;124:1–7. [Google Scholar]
- Chu Z, Yuan M, Yao Jet al.. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006;20:1250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukwu SC, Rafii MY, Ramlee SIet al.. Bacterial leaf blight resistance in rice: a review of conventional breeding to molecular approach. Mol Biol Rep. 2019;46:1519–32. [DOI] [PubMed] [Google Scholar]
- Collins RF, Beis K, Dong Cet al.. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc Natl Acad Sci. 2007;104:2390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey DA. Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol Rev. 1994;14:381–6. [DOI] [PubMed] [Google Scholar]
- Crossman L, Dow JM. Biofilm formation and dispersal in Xanthomonas campestris. Microbes Infect. 2004;6:623–9. [DOI] [PubMed] [Google Scholar]
- Cubero J, Gell I, Johnson EGet al.. Unstable green fluorescent protein for study of Xanthomonas citri subsp. citri survival on citrus. Plant Pathol. 2011;60:977–85. [Google Scholar]
- Cui P, Li RF, Zhang DPet al.. HpaP, a novel regulatory protein with ATPase and phosphatase activity, contributes to full virulence in Xanthomonas campestris pv. campestris. Environ Microbiol. 2018;20:1389–404. [DOI] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BHet al.. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci. 2011;108:2975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom KM, O'Toole GA. A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu Rev Microbiol. 2017;71:179–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daranas N, Roselló G, Cabrefiga Jet al.. Biological control of bacterial plant diseases with Lactobacillus plantarum strains selected for their broad-spectrum activity. Ann Appl Biol. 2019;174:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardick C, da Silva FG, Shen Yet al.. Antagonistic Interactions Between Strains of Xanthomonas oryzae pv. oryzae Phytopathology. 2003;93:705–11. [DOI] [PubMed] [Google Scholar]
- Darrasse A, Barret M, Cesbron Set al.. Niches and routes of transmission of Xanthomonas citri pv. fuscans to bean seeds. Plant Soil. 2018;422:115–28. [Google Scholar]
- Darrasse A, Bureau C, Samson Ret al.. Contamination of bean seeds by Xanthomonas axonopodis pv. phaseoli associated with low bacterial densities in the phyllosphere under field and greenhouse conditions. Eur J Plant Pathol. 2007;119:203–15. [Google Scholar]
- Déjean G, Blanvillain-Baufumé S, Boulanger Aet al.. The xylan utilization system of the plant pathogen Xanthomonas campestris pv campestris controls epiphytic life and reveals common features with oligotrophic bacteria and animal gut symbionts. New Phytol. 2013;198:899–915. [DOI] [PubMed] [Google Scholar]
- Delannoy E, Lyon BR, Marmey Pet al.. Resistance of Cotton Towards Xanthomonas campestris pv. malvacearum. Annu Rev Phytopathol. 2005;43:63–82. [DOI] [PubMed] [Google Scholar]
- Delmotte N, Knief C, Chaffron Set al.. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci. 2009;106:16428–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denancé N, Szurek B, Doyle ELet al.. Two ancestral genes shaped the Xanthomonas campestris TAL effector gene repertoire. New Phytol. 2018;219:391–407. [DOI] [PubMed] [Google Scholar]
- Diab S, Bashan Y, Okon Yet al.. Effects of relative humidity on bacterial scab caused by Xanthomonas campestris pv. vesicatoria on pepper. Phytopathology. 1982;72:1257–60. [Google Scholar]
- Di Lorenzo F, Palmigiano A, Silipo Aet al.. The structure of the lipooligosaccharide from Xanthomonas oryzae pv. Oryzae: the causal agent of the bacterial leaf blight in rice. Carbohydr Res. 2016;427:38–43. [DOI] [PubMed] [Google Scholar]
- Di Lorenzo F, Silipo A, Andersen Gersby LBet al.. Xanthomonas citri pv. citri Pathotypes: LPS structure and function as microbe-associated molecular patterns. ChemBioChem. 2017;18:772–81. [DOI] [PubMed] [Google Scholar]
- Dow JM, Crossman L, Findlay Ket al.. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci. 2003;100:10995–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunger G, Guzzo CR, Andrade MOet al.. Xanthomonas citri subsp. citri type IV pilus is required for twitching motility, biofilm development, and adherence. Mol Plant-Microbe Interact. 2014;27:1132–47. [DOI] [PubMed] [Google Scholar]
- Dunger G, Relling VM, Tondo MLet al.. Xanthan is not essential for pathogenicity in citrus canker but contributes to Xanthomonas epiphytic survival. Arch Microbiol. 2007;188:127–35. [DOI] [PubMed] [Google Scholar]
- Dutta B, Schneider RW, Robertson CLet al.. Embryo localization enhances the survival of acidovorax citrulli in watermelon seeds. Phytopathology. 2016;106:330–8. [DOI] [PubMed] [Google Scholar]
- Dye DW. Host Specificity in Xanthomonas. Nature. 1958;182:1813–4. [Google Scholar]
- Escalon A, Javegny S, Vernière Cet al.. Variations in type III effector repertoires, pathological phenotypes and host range of Xanthomonas citri pv. citri pathotypes. Mol Plant Pathol. 2013;14:483–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure J-D, Napier JA. Europe's first and last field trial of gene-edited plants?. Elife. 2018;7:e42379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargier E, Fischer-Le Saux M, Manceau C. A multilocus sequence analysis of Xanthomonas campestris reveals a complex structure within crucifer-attacking pathovars of this species. Syst Appl Microbiol. 2011;34:156–65. [DOI] [PubMed] [Google Scholar]
- Fernandes C, Blom J, Pothier JFet al.. high-quality draft genome sequence of xanthomonas sp. Strain CPBF 424, a walnut-pathogenic strain with atypical features. Microbiol Resour Announc. 2018;7:pii: e00921–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis MI, Redondo A, Burns JKet al.. Soil application of imidacloprid and related SAR-inducing compounds produces effective and persistent control of citrus canker. Eur J Plant Pathol. 2009;124:283–92. [Google Scholar]
- Frese M, Schatschneider S, Voss Jet al.. Characterization of the pyrophosphate-dependent 6-phosphofructokinase from Xanthomonas campestris pv. campestris. Arch Biochem Biophys. 2014;546:53–63. [DOI] [PubMed] [Google Scholar]
- Galván EM, Ielmini MV, Patel YNet al.. Xanthan chain length is modulated by increasing the availability of the polysaccharide copolymerase protein GumC and the outer membrane polysaccharide export protein GumB. Glycobiology. 2013;23:259–72. [DOI] [PubMed] [Google Scholar]
- García-Ochoa F, Santos VE, Casas JAet al.. Xanthan gum: production, recovery, and properties. Biotechnol Adv. 2000;18:549–79. [DOI] [PubMed] [Google Scholar]
- Gent DH, Schwartz HF. Management of xanthomonas leaf blight of onion with a plant activator, biological control agents, and copper bactericides. Plant Dis. 2005;89:631–9. [DOI] [PubMed] [Google Scholar]
- Gétaz M, Krijger M, Rezzonico Fet al.. Genome-based population structure analysis of the strawberry plant pathogen Xanthomonas fragariae reveals two distinct groups that evolved independently before its species description. Microb Genomics. 2018, doi:10.1099/mgen.0.000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanardi D, Biondi E, Ignjatov Met al.. Seed transmission of Xanthomonas vesicatoria and Clavibacter michiganensis subsp. michiganensis in tomato and Xanthomonas euvesicatoria in pepper and implementation of seed disinfection methods. Proc 7th Congr Plant Prot "Integrated Plant Prot - a Knowledge-Based Step Towar Sustain Agric For Landsc Archit Novemb 24–28, 2014, Zlatibor, Serbia2015:65–70.
- Girija AM, Kinathi BK, Madhavi MBet al.. Rice leaf transcriptional profiling suggests a functional interplay between Xanthomonas oryzae pv. oryzae lipopolysaccharide and extracellular polysaccharide in modulation of defense responses during infection. Mol Plant-Microbe Interact. 2017;30:16–27. [DOI] [PubMed] [Google Scholar]
- Gitaitis R, Walcott R. The epidemiology and management of seedborne bacterial diseases. Annu Rev Phytopathol. 2007;45:371–97. [DOI] [PubMed] [Google Scholar]
- Gochez AM, Huguet-Tapia JC, Minsavage GVet al.. Pacbio sequencing of copper-tolerant Xanthomonas citri reveals presence of a chimeric plasmid structure and provides insights into reassortment and shuffling of transcription activator-like effectors among X. citri strains. BMC Genomics. 2018;19:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Lefeuvre P, Escalon Aet al.. Comparative genomics of 43 strains of Xanthomonas citri pv. citri reveals the evolutionary events giving rise to pathotypes with different host ranges. BMC Genomics. 2015;16:1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald TR, Bassanezi RB, Amorim Let al.. Spatial pattern analysis of citrus canker-infected plantings in São Paulo, Brazil, and augmentation of infection elicited by the asian leafminer. Phytopathology. 2007;97:674–83. [DOI] [PubMed] [Google Scholar]
- Gottwald TR, Irey M. Post-hurricane analysis of citrus canker II: predictive model estimation of disease spread and area potentially impacted by various eradication protocols following catastrophic weather events. Plant Heal Prog. 2007, doi:10.1094/php-2007-0405-01-rs. [Google Scholar]
- Gottwald TR, Sun X, Riley Tet al.. Geo-referenced spatiotemporal analysis of the urban citrus canker epidemic in Florida. Phytopathology. 2002;92:361–77. [DOI] [PubMed] [Google Scholar]
- Guo A, Leach J. Examination of rice hydathode water pores exposed to Xanthomonas campestris pv oryzae. Phytopathology. 1989;79:433–6. [Google Scholar]
- Guo Y, Zhang Y, Li J-Let al.. Diffusible signal factor-mediated quorum sensing plays a central role in coordinating gene expression of Xanthomonas citri subsp. citri. Mol Plant-Microbe Interact. 2011;25:165–79. [DOI] [PubMed] [Google Scholar]
- Gustavo E, Gudesblat, Torres PSet al.. Stomata and pathogens: warfare at the gates. Plant Signal Behav. 2009;4:1114–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo CR, Dunger G, Salinas RKet al.. Structure of the PilZ-FimXEAL-c-di-GMP complex responsible for the regulation of bacterial type IV pilus biogenesis. J Mol Biol. 2013;425:2174–97. [DOI] [PubMed] [Google Scholar]
- Hajri A, Brin C, Hunault Get al.. A «repertoire for repertoire» hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS One. 2009;4:e6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock REW, Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8:402–10. [DOI] [PubMed] [Google Scholar]
- Hao G, Pitino M, Duan Yet al.. Reduced susceptibility to Xanthomonas citri in transgenic citrus expressing the FLS2 receptor from nicotiana benthamiana. Mol Plant-Microbe Interact. 2015, doi:10.1094/mpmi-09-15-0211-r. [DOI] [PubMed] [Google Scholar]
- He Y, Munkvold GP. Comparison of seed transmission and survival of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans in common bean seeds. Plant Heal Prog. 2013;14:41. [Google Scholar]
- He YQ, Zhang L, Jiang BLet al.. Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biol. 2007;8:R218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hert AP, Marutani M, Momol MTet al.. Suppression of the bacterial spot pathogen Xanthomonas euvesicatoria on tomato leaves by an attenuated mutant of Xanthomonas perforans. Appl Environ Microbiol. 2009, doi:10.1128/AEM.02399-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockett KL, Burch AY, Lindow SE. Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. PLoS One. 2013;8:e59850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hop D, Phuong Hoa PT, Quang NDet al.. Biological control of Xanthomonas oryzae pv. oryzae causing rice bacterial blight disease by Streptomyces toxytricini VN08-A-12, isolated from soil and leaf-litter samples in Vietnam. Biocontrol Sci. 2014;19:103–11. [DOI] [PubMed] [Google Scholar]
- Horvath DM, Stall RE, Jones JBet al.. Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS One. 2012;7:e42036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotte B, Rath-Arnold I, Puhler Aet al.. Cloning and analysis of a 35.3-kilobase DNA region involved in exopolysaccharide production by Xanthomonas campestris pv. campestris. J Bacteriol. 1990;172:2804–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Dong H, Ma JCet al.. Novel Xanthomonas campestris long-chain-specific 3-oxoacyl- acyl carrier protein reductase involved in diffusible signal factor synthesis. MBio. 2018;9:e00596–18. doi:10.1128/mBio.00596-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeau L, Roumagnac P, Picard Yet al.. Quantitative and molecular epidemiology of bacterial blight of onion in seed production fields. Phytopathology. 2006;96:1345–54. [DOI] [PubMed] [Google Scholar]
- Hutin M, Pérez-Quintero AL, Lopez Cet al.. MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front Plant Sci. 2015a;6:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin M, Sabot F, Ghesquière Aet al.. A knowledge-based molecular screen uncovers a broad-spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 2015b;84:694–703. [DOI] [PubMed] [Google Scholar]
- Inui Kishi RN, Stach-Machado D, Singulani J, de Let al.. Evaluation of cytotoxicity features of antimicrobial peptides with potential to control bacterial diseases of citrus. Dalisay DS (ed.). PLoS One. 2018;13:e0203451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JL, Carroll TL, Sundin GW. The role of pigmentation, ultraviolet radiation tolerance, and leaf colonization strategies in the epiphytic survival of phyllosphere bacteria. Microb Ecol. 2005;49:104–13. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Salinas SR, Bianco MIet al.. Expression, purification and crystallization of the outer membrane lipoprotein GumB from Xanthomonas campestris. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:1255–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques M-A, Arlat M, Boulanger Aet al.. Using ecology, physiology, and genomics to understand host specificity in Xanthomonas. Annu Rev Phytopathol. 2016;54:163–87. [DOI] [PubMed] [Google Scholar]
- Javvadi S, Pandey SS, Mishra Aet al.. Bacterial cyclic β‐(1,2)‐glucans sequester iron to protect against iron‐induced toxicity. EMBO Rep. 2017;19:172–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. 2017;15:271–84. [DOI] [PubMed] [Google Scholar]
- Jha G, Sonti RV. Attack and defense in xanthomonas-rice interactions. Proc indian Natn Sci Acad. 2009;75:49–68. [Google Scholar]
- Ji Z, Wang C, Zhao K. Rice routes of countering xanthomonas oryzae. Int J Mol Sci. 2018;19:E3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Orbovic V, Jones JBet al.. Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol J. 2016;14:1291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jibrin MO, Potnis N, Timilsina Set al.. Genomic inference of recombination-mediated evolution in Xanthomonas euvesicatoria and X. perforans. Appl Environ Microbiol. 2018;84:e00136–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun SR, Sims GE, Wu GAet al.. Whole-proteome phylogeny of prokaryotes by feature frequency profiles: An alignment-free method with optimal feature resolution. Proc Natl Acad Sci U S A. 2010;107:133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczyński Z, Braun S, Lindner Bet al.. Investigation of the chemical structure and biological activity of oligosaccharides isolated from rough-type Xanthomonas campestris pv. campestris B100 lipopolysaccharide. J Endotoxin Res. 2007;13:101–8. [DOI] [PubMed] [Google Scholar]
- Kakkar A, Jain D, Khanna Pet al.. Sarcomatoid carcinoma of lung - A case report and review of epidermal growth factor receptor mutation status. Lung India. 2015;32:533–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F, Ferreiro DU, Oddo CGet al.. Xanthomonas campestris pv. campestris gum mutants: effects on xanthan biosynthesis and plant virulence. J Bacteriol. 1998;180:1607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Inoue K, Inoue Y. Biological control of bacterial spot on peach by nonpathogenic Xanthomonas campestris strains AZ98101 and AZ98106. J Gen Plant Pathol. 2014;80:158–63. [Google Scholar]
- Kemp BP, Horne J, Bryant Aet al.. Xanthomonas axonopodis pv. manihotis gumD gene is essential for EPS production and pathogenicity and enhances epiphytic survival on cassava (Manihot esculenta). Physiol Mol Plant Pathol. 2004;64:209–18. [Google Scholar]
- Khan AK, Naeem M, Iqbal M. Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur J Plant Pathol. 2014;139:27–37. [Google Scholar]
- Kim D-R, Gang G-H, Jeon C-Wet al.. Epidemiology and control of strawberry bacterial angular leaf spot disease caused by Xanthomonas fragariae. plant Pathol J. 2016;32:290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Kim JG, Lee BMet al.. Mutational analysis of the gum gene cluster required for xanthan biosynthesis in Xanthomonas oryzae pv oryzae. Biotechnol Lett. 2009;31:265–70. [DOI] [PubMed] [Google Scholar]
- Knief C, Delmotte N, Chaffron Set al.. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012;6:1378–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiselburd I, Alet AI, Tondo MLet al.. A LOV protein modulates the physiological attributes of xanthomonas axonopodis pv. citri relevant for host plant colonization. PLoS One. 2012;7:e38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthausen H-J, Laun N, Wohanka W. Methods to reduce the spread of the black rot pathogen, Xanthomonas campestris pv. campestris, in brassica transplants. J Plant Dis Prot. 2011;118:7–16. [Google Scholar]
- Krauthausen HJ, Kreiselmaier J, Laun Net al.. Xanthomoans-Adernschwärze des Kohls. Gemüse. 2006;42:24–6. [Google Scholar]
- Kumar R, Shamarao Jahagridar MR, Yenjerappa STet al.. Epidemiology and management of bacterial blight of pomegranate caused by Xanthomonas axonopodis pv. punicae. Acta Hortic. 2006;818:291–6. [Google Scholar]
- Kumar Verma R, Samal B, Chatterjee S. Xanthomonas oryzae pv. oryzae chemotaxis components and chemoreceptor Mcp2 are involved in the sensing of constituents of xylem sap and contribute to the regulation of virulence-associated functions and entry into rice. Mol Plant Pathol. 2018;19:2397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar S, Iriarte F, Fan Qet al.. Transgenic expression of EFR and Bs2 genes for field management of bacterial wilt and bacterial spot of tomato. Phytopathology. 2018;108:1402–11. [DOI] [PubMed] [Google Scholar]
- Lamichhane JR, Osdaghi E, Behlau Fet al.. Thirteen decades of antimicrobial copper compounds applied in agriculture. A review. Agron Sustain Dev. 2018;38:28. [Google Scholar]
- Lang JM, Gent DH, Schwartz HF. Management of Xanthomonas Leaf Blight of Onion with Bacteriophages and a Plant Activator. Plant Dis. 2007;91:871–8. [DOI] [PubMed] [Google Scholar]
- Li H, Xue D, Tian Fet al.. Xanthomonas oryzae pv. oryzae response regulator TriP regulates virulence and exopolysaccharide production via interacting with c-di-GMP phosphodiesterase PdeR. Mol Plant-Microbe Interact. 2018, doi:10.1094/mpmi-09-18-0260-r. [DOI] [PubMed] [Google Scholar]
- Li J, Wang N. Genome-wide mutagenesis of xanthomonas axonopodis pv. citri reveals novel genetic determinants and regulation mechanisms of biofilm formation. PLoS One. 2011a;6:e21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang N. The wxacO gene of Xanthomonas citri ssp. citri encodes a protein with a role in lipopolysaccharide biosynthesis, biofilm formation, stress tolerance and virulence. Mol Plant Pathol. 2011b;12:381–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang N. The gpsX gene encoding a glycosyltransferase is important for polysaccharide production and required for full virulence in Xanthomonas citri subsp. citri. BMC Microbiol. 2012;12:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang N. Foliar application of biofilm formation–inhibiting compounds enhances control of citrus canker caused by Xanthomonas citri subsp. citri. Phytopathology. 2013, doi:10.1094/phyto-04-13-0100-r. [DOI] [PubMed] [Google Scholar]
- Li R-F, Lu G-T, Li Let al.. Identification of a putative cognate sensor kinase for the two-component response regulator HrpG, a key regulator controlling the expression of the hrp genes in X anthomonas campestris pv. campestris Environ Microbiol. 2014;16:2053–71. [DOI] [PubMed] [Google Scholar]
- Li S, Wang Y, Wang Set al.. The type III effector AvrBs2 in Xanthomonas oryzae pv. oryzicola suppresses rice immunity and promotes disease development. Mol Plant Microbe Interact. 2015;28:869–80. [DOI] [PubMed] [Google Scholar]
- Li Y, Moore R, Guinn Met al.. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci Rep. 2012;2:897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y-Y, Strayer-Scherer AL, White Jet al.. Nano-Magnesium oxide: a novel bactericide against copper-tolerant xanthomonas perforans causing tomato bacterial spot. Phytopathology. 2019;109:52–62. [DOI] [PubMed] [Google Scholar]
- Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69:1875–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Carvalhais LC, Schenk PMet al.. Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Sci Rep. 2017;7:41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yuan M, Zhou Yet al.. A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 2011;34:1958–69. [DOI] [PubMed] [Google Scholar]
- Lu H, Patil P, Van Sluys MAet al.. Acquisition and evolution of plant pathogenesis-associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas. PLoS One. 2008;3:e3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Liu J, Triplett Let al.. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol. 2014;52:213–41. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Håkansson J, Ringstad Let al.. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol. 2016;6:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarovsky D, Fadeev L, Salam BBet al.. Silver nanoparticles complexed with bovine submaxillary mucin possess strong antibacterial activity and protect against seedling infection. Appl Environ Microbiol. 2018;84:pii: e02212–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamud F, Torres PS, Roeschlin Ret al.. The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology. 2011;157:819–29. [DOI] [PubMed] [Google Scholar]
- Martínez S, Martínez-Salazar J, Camas Aet al.. Evaluation of the role of recA protein in plant virulence with recA mutants of Xanthomonas campestris pv. campestris. Mol Plant Microbe Interact. 1997;10:911–6. [DOI] [PubMed] [Google Scholar]
- Medina CA, Reyes PA, Trujillo CAet al.. The role of type III effectors from Xanthomonas axonopodis pv. manihotis in virulence and suppression of plant immunity. Mol Plant Pathol. 2018;19:593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, He SY. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol. 2008;46:101–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merda D, Briand M, Bosis Eet al.. Ancestral acquisitions, gene flow and multiple evolutionary trajectories of the type three secretion system and effectors in Xanthomonas plant pathogens. Mol Ecol. 2017;26:5939–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Pühler A, Niehaus K. The lipopolysaccharides of the phytopathogen Xanthomonas campestris pv. campestris induce an oxidative burst reaction in cell cultures of Nicotiana tabacum. Planta. 2001;213:214–22. [DOI] [PubMed] [Google Scholar]
- Midha S, Bansal K, Kumar Set al.. Population genomic insights into variation and evolution of Xanthomonas oryzae pv. oryzae. Sci Rep. 2017;7:40694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midha S, Patil PB. Genomic insights into the evolutionary origin of Xanthomonas axonopodis pv. citri and its ecological relatives. Appl Environ Microbiol. 2014;80:6266–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Salama NR. The gram-negative bacterial periplasm: Size matters. PLoS Biol. 2018;16:e2004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro A, Evidente A, Fiore Set al.. Structure elucidation of the O-chain from the major lipopolysaccharide of the Xanthomonas campestris strain 642. Carbohydr Res. 2000a;325:222–9. [DOI] [PubMed] [Google Scholar]
- Molinaro A, Evidente A, Iacobellis Net al.. Structural determination of the O-Specific polysaccharide from the xanthomonas fragariae lipopolysaccharide fraction. European J Org Chem. 2001;2001:927–31. [Google Scholar]
- Molinaro A, Evidente A, Lanzetta Ret al.. O-specific polysaccharide structure of the aqueous lipopolysaccharide fraction from Xanthomonas campestris pv. vitians strain 1839. Carbohydr Res. 2000b;328:435–9. [DOI] [PubMed] [Google Scholar]
- Molinaro A, Silipo A, Lanzetta Ret al.. Structural elucidation of the O-chain of the lipopolysaccharide from Xanthomonas campestris strain 8004. Carbohydr Res. 2003;338:277–81. [DOI] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. Spatial organization of dual-species bacterial aggregates on leaf surfaces. Appl Environ Microbiol. 2005;71:5484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales G, Moragrega C, Montesinos Eet al.. Effects of leaf wetness duration and temperature on infection of Prunus by Xanthomonas arboricola pv. pruni. PLoS One. 2018a;13:e0193813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales G, Moragrega C, Montesinos Eet al.. Environmental and inoculum effects on epidemiology of bacterial spot disease of stone fruits and development of a disease forecasting system. Eur J Plant Pathol. 2018b;152: doi:10.1007/s10658-018-1507-7. [Google Scholar]
- Moreira LM, Facincani AP, Ferreira CBet al.. Chemotactic signal transduction and phosphate metabolism as adaptive strategies during citrus canker induction by Xanthomonas citri. Funct Integr Genomics. 2015;15:197–210. [DOI] [PubMed] [Google Scholar]
- Moschini RC, Canteros BI, Martinez MIet al.. Quantification of the environmental effect on citrus canker intensity at increasing distances from a natural windbreak in northeastern Argentina. Australas Plant Pathol. 2014;43:652–62. [Google Scholar]
- Moss WP, Byrne JM, Campbell HLet al.. Biological control of bacterial spot of tomato using hrp mutants of Xanthomonas campestris pv. vesicatoria. Biol Control. 2007;41:199–206. [Google Scholar]
- Murata MM, Omar AA, Mou Zet al.. Novel plastid-nuclear genome combinations enhance resistance to citrus canker in cybrid grapefruit. Front Plant Sci. 2018;9:1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa YR, Bäsell K, Schatschneider Set al.. Dynamic protein phosphorylation during the growth of Xanthomonas campestris pv. campestris B100 revealed by a gel-based proteomics approach. J Biotechnol. 2013;167:111–22. [DOI] [PubMed] [Google Scholar]
- Nakato V, Mahuku G, Coutinho T. Xanthomonas campestris pv. musacearum: a major constraint to banana, plantain and enset production in central and east Africa over the past decade. Mol Plant Pathol. 2018;19:525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndungo V, Eden-Green S, Blomme Get al.. Presence of banana xanthomonas wilt (Xanthomonas campestris pv. musacearum) in the Democratic Republic of Congo (DRC). Plant Pathol. 2006;55:294–. [Google Scholar]
- Newman MA, Von Roepenack-Lahaye E, Parr Aet al.. Prior exposure to lipopolysaccharide potentiates expression of plant defenses in response to bacteria. Plant J. 2002;29:487–95. [DOI] [PubMed] [Google Scholar]
- O'Connell A, An S-Q, McCarthy Yet al.. Proteomics analysis of the regulatory role of Rpf/DSF cell-to-cell signaling system in the virulence of Xanthomonas campestris. Mol Plant Microbe Interact. 2013;26:1131–7. [DOI] [PubMed] [Google Scholar]
- Obradovic A, Jones JB, Momol MTet al.. Management of tomato bacterial spot in the field by foliar applications of bacteriophages and SAR inducers. Plant Dis. 2004;88:736–40. [DOI] [PubMed] [Google Scholar]
- Ocimati W, Ssekiwoko F, Karamura Eet al.. Systemicity of Xanthomonas campestris pv. musacearum and time to disease expression after inflorescence infection in East African highland and Pisang Awak bananas in Uganda. Plant Pathol. 2012;62:777–85. [Google Scholar]
- Ocsoy I, Paret ML, Ocsoy MAet al.. Nanotechnology in plant disease management: DNA-directed silver nanoparticles on graphene oxide as an antibacterial against Xanthomonas perforans. ACS Nano. 2013;7:8972–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol. 2015;41:341–52. [DOI] [PubMed] [Google Scholar]
- Omar AA, Murata MM, El-Shamy HAet al.. Enhanced resistance to citrus canker in transgenic mandarin expressing Xa21 from rice. Transgenic Res. 2018;27:179–91. [DOI] [PubMed] [Google Scholar]
- Otero LH, Klinke S, Rinaldi Jet al.. Structure of the full-length bacteriophytochrome from the plant pathogen Xanthomonas campestris provides clues to its long-range signaling mechanism. J Mol Biol. 2016;428:3702–20. [DOI] [PubMed] [Google Scholar]
- Ottesen A, Ramachandran P, Reed Eet al.. Metagenome tracking biogeographic agroecology: phytobiota of tomatoes from Virginia, Maryland, North Carolina and California. Food Microbiol. 2019;79:132–6. [DOI] [PubMed] [Google Scholar]
- Ottesen AR, González Peña A, White JRet al.. Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol. 2013;13:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen AR, Gorham S, Pettengill JBet al.. The impact of systemic and copper pesticide applications on the phyllosphere microflora of tomatoes. J Sci Food Agric. 2015;95:1116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paret ML, Vallad GE, Averett DRet al.. Photocatalysis: effect of light-activated nanoscale formulations of TiO(2) on Xanthomonas perforans and control of bacterial spot of tomato. Phytopathology. 2013;103:228–36. [DOI] [PubMed] [Google Scholar]
- Park HJ, Jung HW, Han SW. Functional and proteomic analyses reveal that wxcB is involved in virulence, motility, detergent tolerance, and biofilm formation in Xanthomonas campestris pv. vesicatoria. Biochem Biophys Res Commun. 2014;452:389–94. [DOI] [PubMed] [Google Scholar]
- Parnell JJ, Berka R, Young HAet al.. From the lab to the farm: an industrial perspective of plant beneficial microorganisms. Front Plant Sci. 2016;7:1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil PB, Sonti RV. Variation suggestive of horizontal gene transfer at a lipopolysaccharide (lps) biosynthetic locus in Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen of rice. BMC Microbiol. 2004;4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A, Chen S, Lei Tet al.. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J. 2017;15:1509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Hu Y, Xie Jet al.. Long read and single molecule DNA sequencing simplifies genome assembly and TAL effector gene analysis of Xanthomonas translucens. BMC Genomics. 2016;17. doi:10.1186/s12864-015-2348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocelli S, Arana MR, Cabrini MNet al.. Deletion of pilA, a minor pilin-like gene, from Xanthomonas citri subsp. citri influences bacterial physiology and pathogenesis. Curr Microbiol. 2016;73:904–14. [DOI] [PubMed] [Google Scholar]
- Petrocelli S, Tondo ML, Daurelio LDet al.. Modifications of Xanthomonas axonopodis pv. citri lipopolysaccharide affect the basal response and the virulence process during citrus canker. PLoS One. 2012;7:e40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfoestl A, Hofinger A, Kosma Pet al.. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-α-D-galactose in Aneurinibacillus thermoaerophilus L420-91 T. J Biol Chem. 2003;278:26410–7. [DOI] [PubMed] [Google Scholar]
- Phillips AZ, Berry JC, Wilson MCet al.. Genomics-enabled analysis of the emergent disease cotton bacterial blight. PLoS Genet. 2017;13:e1007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti I, Royer M, Barbe Vet al.. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics. 2009;10:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplawsky AR, Kawalek MD, Schaad SW. A Xanthomonadin-encoding gene cluster for the identification of pathovars of Xanthomonas campestris. Mol Plant-Microbe Interact. 1993;6:545. [Google Scholar]
- Poplawsky AR, Urban SC, Chun W. Biological role of xanthomonadin pigments in Xanthomonas campestris pv. campestris. Appl Environ Microbiol. 2000;66:5123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan BB, Ranjan M, Chatterjee S. XadM, a novel adhesin of Xanthomonas oryzae pv. oryzae, exhibits similarity to Rhs family proteins and is required for optimum attachment, biofilm formation, and virulence. Mol Plant-Microbe Interact. 2012;25:1157–70. [DOI] [PubMed] [Google Scholar]
- Pruvost O, Magne M, Boyer Ket al.. A MLVA genotyping scheme for global surveillance of the citrus pathogen Xanthomonas citri pv. citri suggests a worldwide geographical expansion of a single genetic lineage. PLoS One. 2014;9:e98129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Reynolds CM, Trent MSet al.. Lipid a modification systems in Gram-Negative Bacteria. Annu Rev Biochem. 2007;76:295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Ranjan M, Pradhan BBet al.. Atypical regulation of virulence-associated functions by a diffusible signal factor in Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 2012;25:789–801. [DOI] [PubMed] [Google Scholar]
- Raman R. The impact of Genetically Modified (GM) crops in modern agriculture: A review. GM Crops Food. 2017;8:195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi G, Sbodio A, Tech JJet al.. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012;6:1812–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezzonico F, Smits THM, Duffy B. Diversity, evolution, and functionality of Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) regions in the fire blight pathogen erwinia amylovora. Appl Environ Microbiol. 2011;77:3819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera N, Wang H, Li Yet al.. Induced Systemic Resistance Against Citrus Canker Disease by Rhizobacteria. Phytopathology. 2018;108:1038–45. [DOI] [PubMed] [Google Scholar]
- Rigano LA, Payette C, Brouillard Get al.. Bacterial cyclic beta-(1,2)-glucan acts in systemic suppression of plant immune responses. Plant Cell. 2007a;19:2077–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigano LA, Siciliano F, Enrique Ret al.. Biofilm Formation, Epiphytic Fitness, and Canker Development in Xanthomonas axonopodis pv. citri. Mol Plant-Microbe Interact. 2007b;20:1222–30. [DOI] [PubMed] [Google Scholar]
- Roach JA, Verma S, Peres NAet al.. FaRXf1: a locus conferring resistance to angular leaf spot caused by Xanthomonas fragariae in octoploid strawberry. Theor Appl Genet. 2016;129:1191–201. [DOI] [PubMed] [Google Scholar]
- Roper C. Pantoea stewartii subsp. stewartii: lessons learned from a xylem-dwelling pathogen of sweet corn. Mol Plant Pathol. 2011;12:628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutikanga A, Night G, Tusiime Get al.. Spatial and temporal distribution of insect vectors of Xanthomonas campestris pv. musacearum and their activity across banana cultivars grown in Rwanda. In: Marcic Det al. (eds). Proceedings of the 7th Congress on Plant Protection. Serbia: Plant Protection Society of Serbia, 2015; , 139–53. [Google Scholar]
- Ryan RP. Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiol (United Kingdom). 2013;159:1286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, qi An S, Allan JHet al.. The DSF family of cell–cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog. 2015;11:e1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JFet al.. Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris. Mol Microbiol. 2007;63:429–42. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Vorhölter FJ, Potnis Net al.. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat Rev Microbiol. 2011;9:344–55. [DOI] [PubMed] [Google Scholar]
- Ryu MH, Youn H, Kang IHet al.. Identification of bacterial guanylate cyclases. Proteins Struct Funct Bioinforma. 2015;83:799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas SR, Bianco MI, Barreras Met al.. Expression, purification and biochemical characterization of GumI, a monotopic membrane GDP-mannose:glycolipid 4 - D-mannosyltransferase from Xanthomonas campestris pv. campestris. Glycobiology. 2011;21:903–13. [DOI] [PubMed] [Google Scholar]
- Salinas SR, Petruk AA, Brukman NGet al.. Binding of the substrate UDP-glucuronic acid induces conformational changes in the xanthan gum glucuronosyltransferase. Protein Eng Des Sel. 2016;29:197–207. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Sommer DD, Schatz MCet al.. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 2008;9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatschneider S, Huber C, Neuweger Het al.. Metabolic flux pattern of glucose utilization by Xanthomonas campestris pv. campestris: prevalent role of the Entner-Doudoroff pathway and minor fluxes through the pentose phosphate pathway and glycolysis. Mol Biosyst. 2014;10:2663–76. [DOI] [PubMed] [Google Scholar]
- Schatschneider S, Persicke M, Watt SAet al.. Establishment, in silico analysis, and experimental verification of a large-scale metabolic network of the xanthan producing Xanthomonas campestris pv. campestris strain B100. J Biotechnol. 2013;167:123–34. [DOI] [PubMed] [Google Scholar]
- Schulte F, Leßmeier L, Voss Jet al.. Regulatory associations between the metabolism of sulfur-containing amino acids and xanthan biosynthesis in Xanthomonas campestris pv. campestris B100. FEMS Microbiol Lett. 2019;366. doi:10.1093/femsle/fnz005. [DOI] [PubMed] [Google Scholar]
- Sena-Vélez M, Redondo C, Graham JHet al.. Presence of extracellular DNA during biofilm formation by Xanthomonas citri subsp. citri strains with different host range. PLoS One. 2016;11:e0156695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendín LN, Orce IG, Gómez RLet al.. Inducible expression of Bs2 R gene from Capsicum chacoense in sweet orange (Citrus sinensis L. Osbeck) confers enhanced resistance to citrus canker disease. Plant Mol Biol. 2017;93:607–21. [DOI] [PubMed] [Google Scholar]
- Serrania J, Vorhölter FJ, Niehaus Ket al.. Identification of Xanthomonas campestris pv. campestris galactose utilization genes from transcriptome data. J Biotechnol. 2008;135:309–17. [DOI] [PubMed] [Google Scholar]
- Shafi J, Tian H, Ji M. Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol Biotechnol Equip. 2017;31:446–59. [Google Scholar]
- Shantharaj D, Römer P, Figueiredo JFLet al.. An engineered promoter driving expression of a microbial avirulence gene confers recognition of TAL effectors and reduces growth of diverse Xanthomonas strains in citrus. Mol Plant Pathol. 2017;18:976–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DK, Agrawal K. Incidence and histopathological study of Xanthomonas axonopodis pv. vesicatoria in tomato (Lycopersicon esculentum Mill.) seeds. Int J Agric Technol. 2014;10:233–42. [Google Scholar]
- Shekhawat GS, Srivastava DN. Mode of infection in bacterial leaf streak of rice and histology of the diseased leaf. J Phytopathol. 1972, doi:10.1111/j.1439-0434.1972.tb04649.x. [Google Scholar]
- Shen CH, Chiang YC, Hsu CHet al.. Identification and characterization of two uvrA genes of Xanthomonas axonopodis pathovar citri. Mol Genet Genomics. 2007;277:149–60. [DOI] [PubMed] [Google Scholar]
- Shi W, Li C, Li Met al.. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl Microbiol Biotechnol. 2016;100:5059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A, Erbs G, Shinya Tet al.. Glycoconjugates as elicitors or suppressors of plant innate immunity. Glycobiology. 2010;20:406–19. [DOI] [PubMed] [Google Scholar]
- Silipo A, Molinaro A, Lanzetta Ret al.. The structures of the lipid A moieties from the lipopolysaccharides of two phytopathogenic bacteria, Xanthomonas campestris pv. pruni and Xanthomonas fragariae. European J Org Chem. 2004;1336–43.. doi:10.1002/ejoc.200300721. [Google Scholar]
- Silipo A, Molinaro A, Sturiale Let al.. The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J Biol Chem. 2005;280:33660–8. [DOI] [PubMed] [Google Scholar]
- Silipo A, Sturiale L, Garozzo Det al.. The acylation and phosphorylation pattern of lipid A from Xanthomonas campestris strongly influence its ability to trigger the innate immune response in arabidopsis. ChemBioChem. 2008;9:896–904. [DOI] [PubMed] [Google Scholar]
- Singh S. Guttation: quantification, microbiology and implications for phytopathology. Progress in Botany. 2013;75:187–214. [Google Scholar]
- Soto Sedano JC, Mora Moreno RE, Mathew Bet al.. Major novel QTL for resistance to cassava bacterial blight identified through a multi-environmental analysis. Front Plant Sci. 2017;8:1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza DP, Oka GU, Alvarez-Martinez CEet al.. Bacterial killing via a type IV secretion system. Nat Commun. 2015;6:6453. [DOI] [PubMed] [Google Scholar]
- Souza RC, Mendes IC, Reis-Junior FBet al.. Shifts in taxonomic and functional microbial diversity with agriculture: how fragile is the Brazilian Cerrado? BMC Microbiol. 2016;16:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stall RE, Jones JB, Minsavage GV. Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu Rev Phytopathol. 2009;47:265–84. [DOI] [PubMed] [Google Scholar]
- Steffens T, Duda K, Lindner Bet al.. The lipopolysaccharide of the crop pathogen Xanthomonas translucens pv. translucens: Chemical characterization and determination of signaling events in plant cells. Glycobiology. 2017;27:264–74. [DOI] [PubMed] [Google Scholar]
- Steffens T, Vorhölter FJ, Giampà Met al.. The influence of a modified lipopolysaccharide O-antigen on the biosynthesis of xanthan in Xanthomonas campestris pv. campestris B100. BMC Microbiol. 2016;16:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell VO, Duffy B. Use of antibiotics in plant agriculture. Rev Sci Tech. 2012;31:199–210. [DOI] [PubMed] [Google Scholar]
- Su HZ, Wu L, Qi YHet al.. Characterization of the GntR family regulator HpaR1 of the crucifer black rot pathogen Xanthomonas campestris pathovar campestris. Sci Rep. 2016a;6:19862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Zou X, Huang Let al.. DgcA, a diguanylate cyclase from Xanthomonas oryzae pv. oryzae regulates bacterial pathogenicity on rice. Sci Rep. 2016b;6:25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhaimi NSM, Goh SY, Ajam Net al.. Diversity of microbiota associated with symptomatic and non-symptomatic bacterial wilt-diseased banana plants determined using 16S rRNA metagenome sequencing. World J Microbiol Biotechnol. 2017;33:168. [DOI] [PubMed] [Google Scholar]
- Sundaram RM, Vishnupriya MR, Laha GSet al.. Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety. Biotechnol J. 2009;4:400–7. [DOI] [PubMed] [Google Scholar]
- Sundin GW, Wang N. Antibiotic resistance in plant-pathogenic bacteria. Annu Rev Phytopathol. 2018;56:161–80. [DOI] [PubMed] [Google Scholar]
- Tang D-J, He Y-Q, Feng J-Xet al.. Xanthomonas campestris pv. campestris possesses a single gluconeogenic pathway that is required for virulence. J Bacteriol. 2005;187:6231–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JL, Liu YN, Barber CEet al.. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. MGG Mol Gen Genet. 1991;226:409–17. [DOI] [PubMed] [Google Scholar]
- Tao F, Swarup S, Zhang LH. Quorum sensing modulation of a putative glycosyltransferase gene cluster essential for Xanthomonas campestris biofilm formation. Environ Microbiol. 2010;12:3159–70. [DOI] [PubMed] [Google Scholar]
- Teper D, Burstein D, Salomon Det al.. Identification of novel Xanthomonas euvesicatoria type III effector proteins by a machine-learning approach. Mol Plant Pathol. 2016;17:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur M, Sohal BS. Role of elicitors in inducing resistance in plants against pathogen infection: a review. ISRN Biochem. 2013;2013:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa S, Babadoost M. Effectiveness of chemical compounds and biocontrol agents for management of bacterial spot of pumpkin Caused by Xanthomonas cucurbitae. Plant Heal Prog. 2016;17:106–13. [Google Scholar]
- Thomas G, De Tavernier J. Farmer-suicide in India: debating the role of biotechnology. Life Sci Soc policy. 2017;13:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Yu C, Li Het al.. Alternative sigma factor RpoN2 is required for flagellar motility and full virulence of Xanthomonas oryzae pv. oryzae. Microbiol Res. 2015;170:177–83. [DOI] [PubMed] [Google Scholar]
- Timilsina S, Kara S, Jacques MAet al.. Reclassification of Xanthomonas gardneri (ex Šutič 1957) Jones et al. 2006 as a later heterotypic synonym of Xanthomonas cynarae Trébaol et al. 2000 and description of X. cynarae pv. cynarae and X. cynarae pv. gardneri based on whole genome analyses. Int J Syst Evol Microbiol. 2019;69:343–9. [DOI] [PubMed] [Google Scholar]
- Tinzaara W, Gold CS, Ssekiwoko Fet al.. Role of insects in the transmission of banana bacterial wilt. African Crop Sci J. 2006;14:105–10. [Google Scholar]
- Tondo ML, Delprato ML, Kraiselburd Iet al.. KatG, the bifunctional catalase of xanthomonas citri subsp. Citri, responds to hydrogen peroxide and contributes to epiphytic survival on citrus leaves. PLoS One. 2016;11:e0151657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi JN, Lorenzen J, Bahar Oet al.. Transgenic expression of the rice Xa21 pattern-recognition receptor in banana (Musa sp.) confers resistance to Xanthomonas campestris pv. musacearum. Plant Biotechnol J. 2014;12:663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett LR, Verdier V, Campillo Tet al.. Characterization of a novel clade of Xanthomonas isolated from rice leaves in Mali and proposal of Xanthomonas maliensis sp. nov. Antonie van Leeuwenhoek, Int J Gen Mol Microbiol. 2015;107:869–81. [DOI] [PubMed] [Google Scholar]
- Tushemereirwe W, Kangire A, Ssekiwoko Fet al.. First report of Xanthomonas campestris pv. musacearum on banana in Uganda. Plant Pathol. 2004;53:802–. [Google Scholar]
- Vallad GE, Pernezny KL, Balogh Bet al.. Comparison of Kasugamycin to Traditional Bactericides for the Management of Bacterial Spot on Tomato. HortScience. 2010;45:1834–40. [Google Scholar]
- Vicente JG, Holub EB. Xanthomonas campestris pv. Campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol Plant Pathol. 2013;14:2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidhyasekaran P, Kamala N, Ramanathan Aet al.. Induction of systemic resistance by Pseudomonas fluorescens Pf1 against Xanthomonas oryzae pv.oryzae in rice leaves. Phytoparasitica. 2001;29:155–66. [Google Scholar]
- Vojnov AA, Slater H, Daniels MJet al.. Expression of the gum operon directing xanthan biosynthesis in Xanthomonas campestris and its regulation in planta. Mol Plant Microbe Interact. 2001a;14:768–74. [DOI] [PubMed] [Google Scholar]
- Vojnov AA, Slater H, Newman MAet al.. Regulation of the synthesis of cyclic glucan in Xanthomonas campestris by a diffusible signal molecule. Arch Microbiol. 2001b;176:415–20. [DOI] [PubMed] [Google Scholar]
- Vojnov AA, Zorreguieta A, Dow JMet al.. Evidence for a role for the gumB and gumC gene products in the formation of xanthan from its pentasaccharide repeating unit by Xanthomonas campestris. Microbiology. 1998;144:1487–93. [DOI] [PubMed] [Google Scholar]
- Vorhölter FJ, Niehaus K, Pühler A. Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: a cluster of 15 genes is involved in the biosynthesis of the LPS O-antigen and the LPS core. Mol Genet Genomics. 2001;266:79–95. [DOI] [PubMed] [Google Scholar]
- Vorhölter FJ, Schneiker S, Goesmann Aet al.. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J Biotechnol. 2008;134:33–45. [DOI] [PubMed] [Google Scholar]
- Vorhölter FJ, Wiggerich HG, Scheidle Het al.. Involvement of bacterial TonB-dependent signaling in the generation of an oligogalacturonide damage-associated molecular pattern from plant cell walls exposed to Xanthomonas campestris pv. campestris pectate lyases. BMC Microbiol. 2012;12:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurukonda SSKP, Giovanardi D, Stefani E. Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int J Mol Sci. 2018;19:E952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Vinogradov EV, Bogdanove AJ. Requirement of the lipopolysaccharide O-chain biosynthesis gene wxocB for type III secretion and virulence of Xanthomonas oryzae pv. Oryzicola. J Bacteriol. 2013;195:1959–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Pierson EA, Setubal JCet al.. The Candidatus liberibacter-host interface: insights into pathogenesis mechanisms and disease control. Annu Rev Phytopathol. 2017;55:451–82. [DOI] [PubMed] [Google Scholar]
- Wei C, Jiang W, Zhao Met al.. A systematic analysis of the role of GGDEF-EAL domain proteins in virulence and motility in Xanthomonas oryzae pv. oryzicola. Sci Rep. 2016;6:23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen A, Jayawardana M, Fiedler Jet al.. Genetic mapping of a major gene in triticale conferring resistance to bacterial leaf streak. Theor Appl Genet. 2018;131:649–58. [DOI] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. [DOI] [PubMed] [Google Scholar]
- Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. [DOI] [PubMed] [Google Scholar]
- Wichmann G, Ritchie D, Kousik CSet al.. Reduced genetic variation occurs among genes of the highly clonal plant pathogen Xanthomonas axonopodis pv. vesicatoria, including the effector gene avrBs2. Appl Environ Microbiol. 2005;71:2418–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggerich HG, Pühler A. The exbD2 gene as well as the iron-uptake genes tonB, exbB and exbD1 of Xanthomonas campestris pv. campestris are essential for the induction of a hypersensitive response on pepper (Capsicum annuum). Microbiology. 2000;146:1053–60. [DOI] [PubMed] [Google Scholar]
- van der Wolf J, Kastelein P, Tadeu AFet al.. Colonization of siliques and seeds of rapid cycling Brassica oleracea plants by Xanthomonas campestris pv. campestris after spray-inoculation of flower clusters. Eur J Plant Pathol. 2019;154:445–61. [Google Scholar]
- van der Wolf J, van der Zouwen P. Colonization of cauliflower blossom (Brassica oleracea) by Xanthomonas campestris pv. campestris, via Flies (Calliphora vomitoria) can result in seed infestation. J Phytopathol. 2010;158:726–32. [Google Scholar]
- van der Wolf J, van der Zouwen P, van der Heijden L. Flower infection of Brassica oleracea with Xanthomonas campestris pv. campestris results in high levels of seed infection. Eur J Plant Pathol. 2013;136:103–11. [Google Scholar]
- Wu GA, Terol J, Ibanez Vet al.. Genomics of the origin and evolution of Citrus. Nature. 2018;554:311–6. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang Y, Zhang Pet al.. The structure and function of the global citrus rhizosphere microbiome. Nat Commun. 2018;9:4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RQ, Blanvillain S, Feng JXet al.. AvrACXcc8004, a type III effector with a leucine-rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col-0. J Bacteriol. 2008;190:343–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Tian F, Yang Fet al.. Phosphodiesterase EdpX1 Promotes Xanthomonas oryzae pv. oryzae Virulence, Exopolysaccharide Production, and Biofilm Formation. Appl Environ Microbiol. 2018, doi:10.1128/aem.01717-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Hu X, Wang N. The novel virulence-related gene nlxA in the lipopolysaccharide cluster of Xanthomonas citri ssp. citri is involved in the production of lipopolysaccharide and extracellular polysaccharide, motility, biofilm formation and stress resistance. Mol Plant Pathol. 2012;13:923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci U S A. 2006;103:10503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Qian S, Tian Fet al.. The GGDEF-domain protein GdpX1 attenuates motility, exopolysaccharide production and virulence in Xanthomonas oryzae pv. oryzae. J Appl Microbiol. 2016;120:1646–57. [DOI] [PubMed] [Google Scholar]
- Yang F, Tian F, Chen Het al.. The Xanthomonas oryzae pv. oryzae PilZ Domain Proteins Function Differentially in Cyclic di-GMP Binding and Regulation of Virulence and Motility. Appl Environ Microbiol. 2015;81:4358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LY, Yang LC, Gan YLet al.. Systematic functional analysis of sigma (s) factors in the phytopathogen Xanthomonas campestris reveals novel roles in the regulation of virulence and viability. Front Microbiol. 2018;9:1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T-C, Leu Y-W, Chang-Chien H-Cet al.. Flagellar biogenesis of Xanthomonas campestris requires the alternative sigma factors RpoN2 and FliA and is temporally regulated by FlhA, FlhB, and FlgM. J Bacteriol. 2009;191:2266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS. A conformational model for cyclic β-(1 → 2)-linked glucans based on NMR analysis of the β-glucans produced by Xanthomonas campestris. Carbohydr Res. 1995;278:205–25. [DOI] [PubMed] [Google Scholar]
- Yu X, Lund SP, Scott RAet al.. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc Natl Acad Sci. 2013;110:E425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Ke Y, Huang Ret al.. A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. Elife. 2016;5:e19605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH. Xanthan induces plant susceptibility by suppressing callose deposition. PLANT Physiol. 2006;141:178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandjanakou‐Tachin M, Fanou A, LeGall Pet al.. Detection, survival and transmission of Xanthomonas axonopodis pv. manihotis and X. axonopodis pv. vignicola, causal agents of Cassava and Cowpea bacterial blight, respectively, in/by insect vectors. J Phytopathol. 2007;115:159–69. [Google Scholar]
- Zarei S, Taghavi SM, Hamzehzarghani Het al.. Epiphytic growth of Xanthomonas arboricola and Xanthomonas citri on non-host plants. Plant Pathol. 2018, doi:10.1111/ppa.12769. [Google Scholar]
- Zeitler B, Herrera Diaz A, Dangel Aet al.. De-novo design of antimicrobial peptides for plant protection. PLoS One. 2013;8:e71687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Melotto M, He SY. Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr Opin Biotechnol. 2010;21:599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Tian D, Gu Ket al.. Genetic engineering of the Xa10 promoter for broad-spectrum and durable resistance to Xanthomonas oryzae pv. oryzae. Plant Biotechnol J. 2015;13:993–1001. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yin Z, White F. TAL effectors and the executor R genes. Front Plant Sci. 2015, doi:10.3389/fpls.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu J, Birch RG. Engineered detoxification confers resistance against a pathogenic bacterium. Nat Biotechnol. 1999;17:1021–4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Callaway EM, Jones JBet al.. Visualisation of hrp gene expression in Xanthomonas euvesicatoria in the tomato phyllosphere. Eur J Plant Pathol. 2009;124:379–90. [Google Scholar]
- Zhao Y, Damicone JP, Bender CL. Detection, survival, and sources of inoculum for bacterial diseases of leafy crucifers in Oklahoma. Plant Dis. 2002;86:883–8. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Wu Z, Zhang Jet al.. Crystal structure of the YajQ-family protein XC_3703 from Xanthomonas campestris pv. campestris. Acta Crystallogr Sect F Struct Biol Commun. 2016;72:720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Huang T-W, Wang J-Yet al.. The Rice Bacterial Pathogen Xanthomonas oryzae pv. oryzae Produces 3-Hydroxybenzoic Acid and 4-Hydroxybenzoic Acid via XanB2 for Use in Xanthomonadin, Ubiquinone, and Exopolysaccharide Biosynthesis. Mol Plant-Microbe Interact. 2013a;26:1239–48. [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang JY, Wang Jet al.. The diffusible factor synthase XanB2 is a bifunctional chorismatase that links the shikimate pathway to ubiquinone and xanthomonadins biosynthetic pathways. Mol Microbiol. 2013b;87:80–93. [DOI] [PubMed] [Google Scholar]
- Zhou L, Zhang LH, Cámara Met al.. The DSF family of quorum sensing signals: diversity, biosynthesis, and turnover. Trends Microbiol. 2017;25:293–303. [DOI] [PubMed] [Google Scholar]
- Zhou Y-L, Xu M-R, Zhao M-Fet al.. Genome-wide gene responses in a transgenic rice line carrying the maize resistance gene Rxo1 to the rice bacterial streak pathogen, Xanthomonas oryzae pv. oryzicola. BMC Genomics. 2010;11:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv C, Zhao Z, Gao YGet al.. Multifunctional roles of plant cuticle during plant-pathogen interactions. Front Plant Sci. 2018;9:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.