Abstract
Trop-2 is a transmembrane signal transducer that can induce cancer growth. Using antibody targeting and N-terminal Edman degradation, we show here that Trop-2 undergoes cleavage in the first thyroglobulin domain loop of its extracellular region, between residues R87 and T88. Molecular modeling indicated that this cleavage induces a profound rearrangement of the Trop-2 structure, which suggested a deep impact on its biological function. No Trop-2 cleavage was detected in normal human tissues, whereas most tumors showed Trop-2 cleavage, including skin, ovary, colon, and breast cancers. Coimmunoprecipitation and mass spectrometry analysis revealed that ADAM10 physically interacts with Trop-2. Immunofluorescence/confocal time-lapse microscopy revealed that the two molecules broadly colocalize at the cell membrane. We show that ADAM10 inhibitors, siRNAs and shRNAs abolish the processing of Trop-2, which indicates that ADAM10 is an effector protease. Proteolysis of Trop-2 at R87-T88 triggered cancer cell growth both in vitro and in vivo. A corresponding role was shown for metastatic spreading of colon cancer, as the R87A-T88A Trop-2 mutant abolished xenotransplant metastatic dissemination. Activatory proteolysis of Trop-2 was recapitulated in primary human breast cancers. Together with the prognostic impact of Trop-2 and ADAM10 on cancers of the skin, ovary, colon, lung, and pancreas, these data indicate a driving role of this activatory cleavage of Trop-2 on malignant progression of tumors.
Keywords: Trop, Proteolytic processing, Signaling activation, Cell growth, Molecular modeling, Human cancer
Abbreviations: ADAM, a disintegrin and metalloproteinase; LUADs, lung adenocarcinomas; LUSCs, lung squamous cell cancers; mAb, monoclonal antibody; RIP, regulated intramembrane proteolysis; SCC, squamous cell carcinoma; wt, wild-type
Introduction
Trop-2 is a monomeric transmembrane signal transducer [1,2] expressed by epithelial cells at various stages of differentiation [3,4]. The TROP2 gene (TACSTD2; formerly M1S1) [5] is an intronless derivative of the TROP1 gene (EPCAM, TACSTD1; formerly M4S1) [5], [6], [7], [8], which suggests strong evolutionary pressure for conserved functional roles of the Trop molecules.
We and others have shown that Trop-2 is a driver of cancer development and progression [9,10]. Pro-oncogenic Trop-2 signaling occurs through regulation of the expression and activity of cyclin D1, ERK, NFκB, Akt [11], [12], [13], [14], FAK, Rac1, and integrins [9,15]. No Trop-2–activating mutations have been detected in cancers as yet, which indicates that alternative molecular mechanisms are required to account for the functional derangement of Trop-2 [9].
Trop-2 and Trop-1 each contain a GA733 type 1 motif and a thyroglobulin repeat [[6], [7], [8],[16], [17], [18]]. These structural motifs share extended similarities with the corresponding regions of nidogen and IGF-binding proteins [[6], [7], [8],16], and have roles in the homophylic binding of Trop-1/Ep-CAM [19, 20]. We have shown that the proteolytic cleavage of the first loop of the Trop-1 thyroglobulin domain [21] activates the cell-growth stimulatory properties of Trop-1. We then speculated that a similar molecular switch may operate on Trop-2.
Proteolytic enzymes have critical roles in cancer development and progression, and are central to multidirectional signaling networks that regulate processes such as tumor–microenvironment interactions, chemokine/cytokine cross-talk, and angiogenesis. Stoyanova et al. showed that regulated intramembrane proteolysis (RIP) is responsible for the cleavage of Trop-2 that is mediated by ADAM17/TACE at the A187-V188 site, followed by γ-secretase processing at G285-V286 [22]. This induces release of the Trop-2 intracellular domain, which then co-translocates with ß-catenin to the nucleus [22]. Recently, matriptase was reported to cleave Trop-2 at the R87-T88 site [23,24], to induce decreased levels of claudins at the epithelial cell surface. Here, for the first time, we identify ADAM10 as a key interactor of Trop-2 at the cell membrane. ADAM10 is shown to cleave Trop-2 at R87-T88 in cancer cells, to activate Trop-2, and to induce cancer cell growth. Using tumor xenografts of human colon cancer cells that express the R87A-T88A Trop-2 mutant (designated as A87-A88 Trop-2), we demonstrate that Trop-2 proteolytic processing by ADAM10 correspondingly drives metastatic dissemination.
Material and methods
Cells
The human ovarian OVCA-432, mammary MCF-7, pancreatic BxPC3, and colorectal HT-29, HCT-116 and KM12SM [25] cancer cell lines, and the murine myeloma NS-0 cells, were grown in RPMI 1640 medium supplemented with 10% fetal calf serum. The human 293 kidney and the murine MTE 4-14 immortalized thymic epithelium [26] and L fibrosarcoma [27] cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. The cells were transfected with purified DNA [28] using Lipofectamine 2000 or LTX (Invitrogen). Stable transfectants were propagated in complete medium supplemented with 100 μg/mL G418. MTE4-14/Trop-2 transfectants acquire specific features of cancer cells, as they become tumorigenic in vivo, at variance with MTE4-14/vector transfectants [14]. Growth in suspension was obtained by culturing cells in 6-well ultra low attachment plates. Cells were seeded at 106 per well, and analyzed after 5 d.
Tumor patient case series
A human breast cancer case series [29] was analyzed, where eligible patients showed N0, T1/T2 tumors. Snap-frozen material was available for 55 cases. The frozen tissues were ground in liquid nitrogen and processed as for Western blotting. Studies on human tumour samples were approved by the Italian Ministry of Health (RicOncol RF-EMR-2006-361866, 2006).
Antibodies
Balb/c mice were immunized with cells from an ovarian cancer fragment (sample FE). Cell fusion and hybridoma cloning were carried out as previously described [30]. Screening for cell-surface-reactive hybridomas was performed by immunohistochemistry of ovarian cancer cells [31], and by flow cytometry of Trop-2 transfectants. The E1 (IgG2ak by immuno-isotyping), 162-46.2 [27], and T16 [4] anti-Trop-2 [32] hybridomas were grown in serum-free medium. The RS7 anti-Trop-2/EGP-1 ascites was kindly provided by Rhona Stein. Rabbit polyclonal anti-Trop-2 antisera were generated by subcutaneous immunization with the recombinant extracellular domain of human Trop-2 synthesized in bacteria [7], or with KLH-conjugated, N-ter biotinylated peptides corresponding to the cytoplasmic tail of human Trop-2. Anti Trop-2 polyclonal antibodies were purified by affinity chromatography on recombinant Trop-2 conjugated to NHS Sepharose (GE Healthcare) or biotinylated Trop-2 cytoplasmatic tails conjugated to Streptavidin Agarose (Sigma-Aldrich). The AF650 polyclonal goat anti-Trop-2 antibody was from R&D Systems; the rabbit polyclonal anti-ADAM10 antibody was from Merck-Millipore (AB19026); and the goat polyclonal anti-ADAM10 antibody (sc-31853) was from Santa Cruz Biotechnology.
ADAM10 inhibitors
Cells were treated with ADAM10 inhibitors for 24 h at the minimum concentration found to be effective for inhibition of Trop-2 cleavage, and then they were assayed as indicated. The GM6001 metalloprotease family inhibitor (Calbiochem) was dissolved in dimethylsulfoxide and used at 6.5 µM to 13 µM for 24 h. The GI254023X ADAM10 and MMP-9 inhibitor was kindly provided by A. Ludwig and was dissolved in dimethylsulfoxide. The cells received two doses of 10 µM GI254023X every 24 h. Control cells received vehicle alone.
Flow cytometry
Cell staining for flow cytometry was performed as described previously [1, 27, [33], [34], [35]]. Reconstituted mixtures of L cells and Trop-2 transfectants [34] were used for E1 mAb binding and competition studies of E1 with other anti-Trop-2 mAbs, where the cell mixtures were preincubated with 100-fold excess of the indicated ascites.
Immunofluorescence microscopy
Cells grown on glass coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline for 20 min. The cells were permeabilized and blocked in 10% fetal bovine serum, 0.1% saponin [36], and then stained with the 162-46.2 [27], T16 [4], E1 anti-Trop-2 [32], and anti-ADAM10 antibodies. The cells were analyzed by confocal microscopy (LSM-510 META and LSM-800; Zeiss).
Confocal time‐lapse microscopy
Live cells cultured on glass slides were analyzed in Leibovitz's F15 culture medium without phenol red and bicarbonate, supplemented with 10% fetal bovine serum, 100 IU/mL penicillin, 100 µg/mL streptomycin (Euroclone), and 2 mM N‐acetyl‐cysteine (Sigma), to reduce free‐radical damage. The cells were analyzed using a confocal microscope (LSM‐510 META; Zeiss), with images captured at 1 min intervals.
In vitro cell growth assays
MTE 4-14 and KM12SM transfectants were seeded at 1.5-3.0 × 103 cells/well in 96-well plates (5 replica wells per data point). Cell numbers were quantified by staining with crystal violet [37].
Experimental tumors and metastases
Cancer cell lines and TROP2-transfectants were injected subcutaneously into 8-wk-old female athymic Crl:CD1-Foxn1nu mice (Charles River Laboratories). Subcutaneous tumor growth curves were obtained by weekly measurements of tumor volumes (d2 × D/2; shortest diameter2 × longest diameter/2), and computed as described previously [38]. To assess for impact on metastatic dissemination, KM12SM colon cancer cell [25] transfectants were injected into the spleen of 8-wk-old female athymic Crl:CD1-Foxn1nu mice. After 4 wk, the mice were euthanized, and tumor growth and dissemination to the liver and other organs were determined by comparisons of the tumor volumes between the experimental groups. All of the autoptic samples underwent microscopy histopathology analysis to detect minimal tumors and the metastatic burden.
All of the procedures involving animals and their care were conducted in compliance with institutional guidelines, national laws, and international protocols (D.L.No.116, G.U., Suppl.40, Feb. 18, 1992; No.8, G.U., July, 1994; UKCCCR Guidelines for the Welfare of Animals in Experimental Neoplasia; EEC Council Directive 86/609, OJL358. 1, Dec. 12, 1987; Guide for the Care and Use of Laboratory Animals, United States National Research Council, 1996). Preclinical protocols were approved by the Italian Ministry of Health (Prog. 19, 2006) and by the Interuniversity Animal Research Ethics Committee (CEISA) of Chieti-Pescara and Teramo Universities (Prot.26/2011/CEISA/PROG/16).
Statistical analysis
Student t tests were used for comparisons between mean protein levels in the control and TROP2 transfectants in the antibody microarrays. The normality of the distributions of the assay values was verified (www.graphpad.com). Spearman nonparametric correlation coefficients were computed for protein expression levels in human cancer samples. Two-way ANOVA and post-hoc Bonferroni's t tests were used for growth curve comparisons. The data were analyzed using SigmaStat (SPSS Science Software UK Ltd.) (www.spss.com/software/science/sigmastat/) and GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA) (www.graphpad.com).
Results
Anti‐Trop‐2 reactivity of the E1 monoclonal antibody
Post-translational processing is a key activation step for several tumor growth inducers and adhesion molecules [39], [40], [41], [42]. As Trop-2 activating mutations have not been detected as yet in cancers, we hypothesized that post-translational processing of the molecule might be specifically induced, to provide cancer cells with growth advantage over their normal counterparts. We have previously shown that Trop-1 undergoes activator cleavage at R80-R81 [21]. We explored here the existence and functionality of a corresponding activator mechanism for Trop-2.
To investigate Trop-2 post-translational processing, we generated anti-Trop-2 monoclonal antibodies (mAbs) through immunization and screening procedures aimed at obtaining efficient recognition of Trop-2 under native conditions in living cells. To this end, mAb-producing hybridomas [30] were screened for immunohistochemistry reactivity [31] of cell culture supernatants on ovarian cancer cells [43]. Flow cytometry analysis of live Trop-2 transfectants was then carried out, and anti-Trop-2 mAbs were selected for recognition of native Trop-2 in tumors and cell lines.
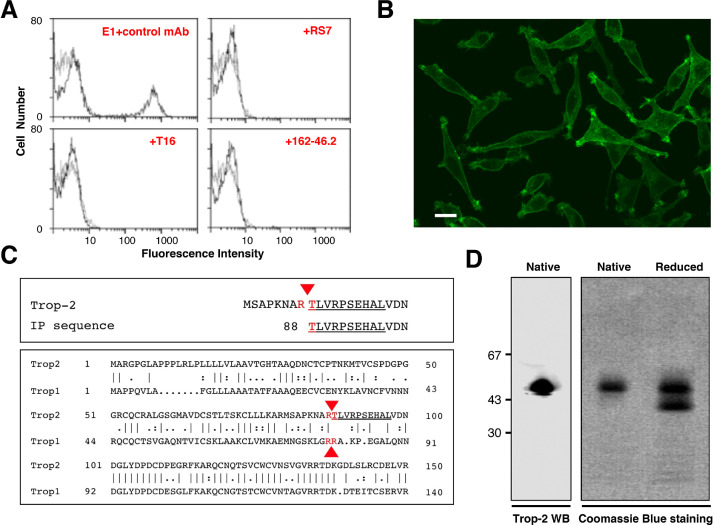
The E1 mAb was shown to mostly bind Trop-2–expressing cancer cells of epithelial origin (Table S1), such as mammary carcinomas, ovarian cancers, lung adenocarcinomas, colorectal cancers, pancreatic adenocarcinomas, prostate cancers, and choriocarcinomas. Specific binding of a Trop-2–negative L-cell murine fibrosarcoma transfected with genomic TROP2 or TROP2 cDNA (including a single-residue polymorphic cDNA clone obtained from the FE ovarian cancer) (Fig. 1A, B, D), provided formal proof for specific recognition of Trop-2. E1 recognized Trop-2 in Western blotting assays under non-denaturing conditions (Fig. 1D), but it did not react with reduced Trop-2.
Fig. 1.
Purification, sequencing and analysis of Trop-2. (A) Reconstituted mixtures of 70% parental L cells and 30% Trop-2/L cells transfectants [34] were used for competition studies of E1 with other anti-Trop-2 mAbs. Cell mixtures were stained with the FITC-E1 mAb. Competition of E1 with other anti-Trop-2 mAbs was performed by incubating cell mixtures with 100-fold excess of the indicated ascites (red). Successful competition was revealed by the disappearance of the peak of stained cells, indicating that the E1 binding site is the same as, or is in close proximity to, that of the competing Abs. E1 was efficiently competed-out by the anti-Trop-2 162-46.2 antibody [80], the T16 mAb [13] and the RS7-3G11 mAb, from which the humanized anti-Trop-2 therapeutic IMMU132 was derived [47]. (B) Reactivity of the E1 mAb with Trop-2. Immunofluorescence microscopy analysis of FITC-E1-stained TROP2/L cell transfectants. (C, top) Alignment of the amino-acid sequence obtained by Edman degradation (underlined) with the canonical Trop-2 sequence. Red arrowhead: cleavage site. (C, bottom) Alignment of the proteolytic sites of Trop-2 and Trop-1 [21] (red arrows). The Edman degradation sequence of E1/Trop-2 is underlined. (D, left) Western blotting of Trop-2, as immunoprecipitated from ovarian cancer cells and purified by affinity chromatography over Sepharose–E1 mAb. Purified Trop-2 was run in SDS-PAGE under native (nonreducing) conditions. (D, right) Coomassie blue staining of purified Trop-2 protein run in SDS-PAGE gradient gel under native or reducing conditions. (Colored version of figure is available online.)
Cross-blocking in competition binding assays showed that the E1 antibody binding site coincides with, or is in close proximity to, that of the main anti-Trop-2 mAbs that have been generated to date (Fig. 1A and [44]), including T16 [4], 162-46.2 (ATCC HB-187), AR47A6.4.2 [45], 77220 (R&D Systems,MN, USA), and the RS7 murine mAb, that has been developed into the anti-Trop-2 antibody–SN-38 drug conjugate [46] Sacituzumab govitecan-hziy (Trodelvy) [47]. Mapping using Trop-2 deletion mutants identified this immunodominant epitope in the extracellular region spanning D146-R178 [44].
Trop‐2 purification and sequencing
We investigated the post-translational processing of Trop-2 using the E1 mAb to immunoprecipitate Trop-2 from ovarian cancer cells. Purification of Trop-2 was then performed by affinity chromatography over Sepharose–E1 mAb columns. Western blotting analysis indicated an essential homogeneity of the isolated Trop-2 (>95% purity) (Fig. 1D).
N-terminal sequences were obtained from the E1-immunoreactive material by Edman degradation. The N-terminal Trop-2 sequence started at T88 (Fig. 1C, top), which first indicated a potential, endogenous cleavage of Trop-2 between R87 and T88 in the thyroglobulin domain. The unprocessed Trop-2 N-terminus was blocked, and did not generate any amino-acid sequence. Post-translational cleavage at R87-T88 was expected to result in a large conformational rearrangement, leading to the generation of a two-chain molecule with a ∼10-kDa fragment bound to the ∼40-kDa membrane-bound segment by a single disulfide bridge between Cys73 and Cys108 (Fig. S1). Consistent with this, reduction of the disulfide bridges led to the loss of a ∼10-kDa fragment from full-length Trop-2 (Fig. 1D).
Trop‐2 immunoprecipitation and mass spectrometry analysis
The Trop-2 cleavage site was confirmed by immunoprecipitation/PAGE/mass spectrometry (MS) peptide mass fingerprinting. The list of the peak fragments of Trop-2 is given in Table S2. The MS spectra were scanned for peptide sequences that did not fit trypsin-cleavage consensus sites, to determine the endogenous proteolytic cleavage sites of Trop-2. Four independent peptides were identified that showed cleavage at R87-T88, which confirmed the N-terminal Edman sequencing data (Fig. 1C, top; Table S2). Of note, the R87-T88 Trop-2 cleavage site shows exact correspondence to the activator cleavage site of the paralogous Trop-1 molecule [21] (Fig. 1C, bottom), suggesting a similar impact on an evolutionarily conserved function.
Three‐dimensional modeling of the Trop‐2 thyroglobulin domain
We generated a three-dimensional model of the thyroglobulin domain of Trop-2 using the crystal structure of the thyroglobulin domain of the p41 splice variant of the major histocompatibility complex class II-associated invariant chain [48] as a template (Fig. S1). Independent prediction of the secondary structure using the PHD program (≥80% accuracy; www.predictprotein.org) provided the corresponding subsegment identification, which supported its overall prediction accuracy. Ramachandran plot analysis allocated 79% of the residues of the Trop-2 model into the core region, and 17% into the allowed region of the plot. A corresponding analysis of the p41 crystal structure positioned 88% of the residues in the core region and 12% in the allowed region of the plot [48], in good correspondence with the Trop-2 model. The Trop-2 thyroglobulin domain showed comparable main-chain parameters versus the p41 crystal. The root mean square deviation between the Trop-2 thyroglobulin domain and the p41 template was 0.399 Å, supporting the reliability of the Trop-2 model [49]. Superimposing the Trop-2 model on the p41 template showed that the Trop-2 and p41 thyroglobulin domains have a relatively flat, wedge-shaped structure, with three loops that are largely aligned on a plane that extends from the three conserved disulfide bridges. The region with the highest structural divergence was between the first and second cysteine, where Trop-2 showed a much longer loop than p41. This region contains the proteolytic cleavage site. PROCHECK and MOLMOL (PaintSurface) residue accessibility routines confirmed high solvent accessibility of the proteolytic cleavage region. Cleavage of the first loop of Trop-2 generated free amino-acid chains of 14 and 20 residues, and removed a rigid constraint to a planar configuration of the thyroglobulin domain (Fig. S1, Movie S1). Favorable main-chain parameters (i.e., Ramachandran plot quality, peptide bond planarity, bad nonbonded interactions, c-a tetrahedron distortion, main chain hydrogen bond energy, overall G-factor) and root mean square deviation were reached, thus allowing reliable analytical insight on the Trop-2 structure.
The first p41 thyroglobulin repeat loop binds the active site of cathepsin L [48]. On the other hand, Trop-2 showed a much longer loop at this position (Fig. S1D, E). Hence, the cathepsin L enzymatic pocket appeared too small to fit the first Trop-2 loop, thus suggesting cleavage by a different protease.
ADAM10 is a candidate Trop‐2 protease at the R87‐T88 site
We carried out immunoprecipitation of Trop-2, followed by MS analysis of its binding partners. This revealed four independent ADAM10 peptides across multiple identification procedures (Table S3), thus indicating ADAM10 as a Trop-2 interactor. ADAM10 is a member of the ‘a disintegrin and metalloproteinase’ (ADAM) family of membrane-bound zinc-dependent metalloproteases [50, 51], and it is involved in processing of several adhesion molecules and transmembrane receptors [39, 40, [52], [53], [54], [55], [56]]. Peptide library screening combined with crystal structure analysis of known ADAM10 substrates [57] revealed preference for Pro at P5 (Calsyntenin, proTGFα, TNFα, cleaved peptides [57]), Arg at P1 (EGF, N-Cadherin, CD23), and Thr at P1’ (amyloid β/A4 protein, HB-EGF) in target site subsets (Fig. S2). ADAM10 was also shown to be prone to cleave within the loop segments of target molecules (Figure S2). All of these sequence/structure features are present in the Trop-2 cleavage site (Fig. 1C; Table S2), making ADAM10 a candidate Trop-2-cleaving protease at the Trop-2 R87-T88 site. ADAM10 is expressed at high levels in most human cancer types, and is up-regulated/activated in cancers of the stomach [58], liver [59], colon [60], uterus, ovary [61], oral cavity [62], pancreas [63], lung, and in triple-negative breast cancers [56], as well as in melanoma and glioblastoma [56, 64]. Next-generation sequencing profiling of tumor transcriptomes of almost 8000 cancer patients in the TCGA dataset (www.proteinatlas.org) identified significant association of ADAM10 overexpression with worse prognosis for pancreas (Figure S3A) and lung (Fig. S3B) cancers. Subgroup analysis [65] showed that the negative prognostic impact of ADAM10 mainly occurred on lung adenocarcinomas (LUADs) (P= 0.0013), and only marginally on lung squamous cell cancers (LUSCs) (P = 0.013). Similar data were obtained by meta-analysis of DNA microarray data from 2437 NSCLC patients through the KMPlot database (www.kmplot.com) (Fig. S3B). This closely paralleled our earlier analysis of the prognostic impact of Trop-2 on LUADs versus LUSCs [66]. Taken together, these data prompted us to further investigate the interplay between ADAM10 and Trop-2.
ADAM10 interacts with Trop‐2 at the cell membrane
Previous subcellular localization analysis had revealed that a minor fraction of mature ADAM10 is located in the endoplasmic-reticulum/plasma-membrane–enriched fractions [67], which suggested that most ADAM10 activity is located at the plasma membrane [68]. Here, immunofluorescence (Fig. S4A) and time-lapse fluorescence microscopy (Movie S2) revealed full colocalization between ADAM10 and Trop-2 in 72.2% of the cells analyzed, with partial colocalization in 22.2%, and no colocalization in 5.5% (n = 54). The specificity of the colocalization was verified by immunofluorescence analysis of cells infected with the pLVTHM-GFP lentivirus that co-expresses ADAM10 or scramble shRNA. The antibody used for recognition of ADAM10 did not stain the GFP+ cells co-expressing ADAM10 shRNA. On the other hand, uniform membrane staining for ADAM10 was observed in GFP+ cells that co-express the scramble shRNA (Fig. S4B). We further showed that expression and membrane localization of Trop-2 were not affected by ADAM10 inhibition (Fig. S4B).
Trop‐2 proteolysis is impaired by ADAM10 inhibition
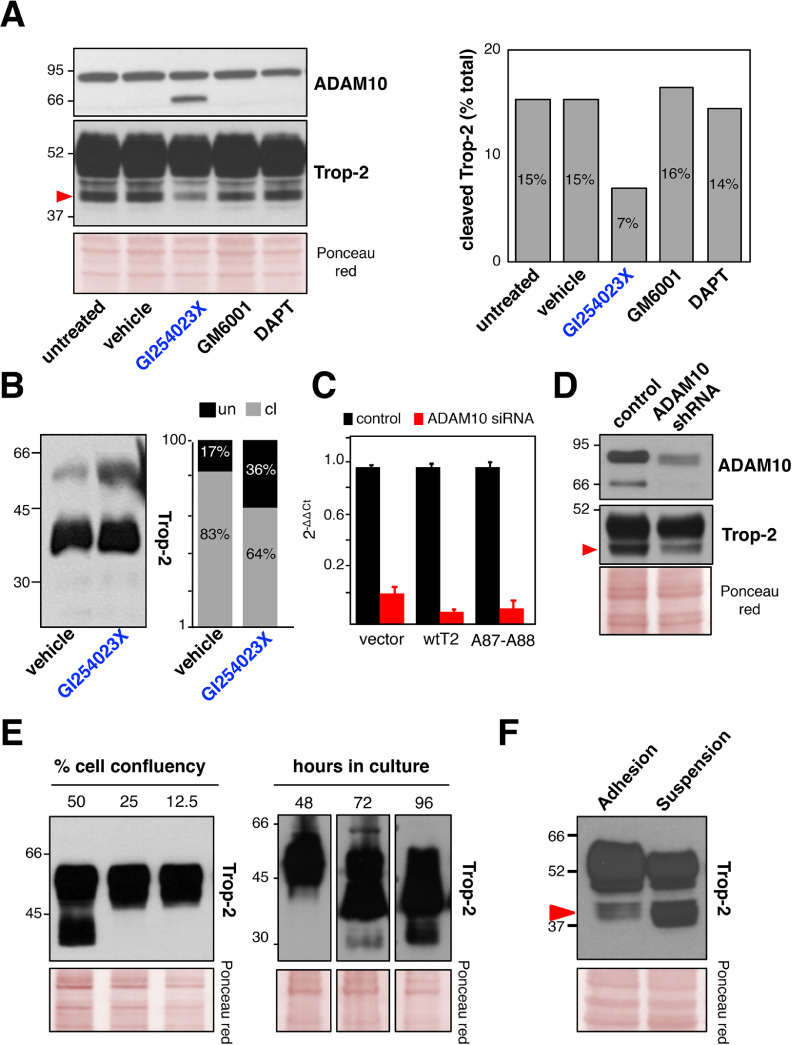
We questioned whether ADAM10 mediates the proteolytic processing of Trop-2 in vivo. We assessed Trop-2 cleavage upon inhibition of ADAM10 activity and expression. Treatment of Trop-2 expressing BxPC3 pancreatic cancer cells or MTE4-14/Trop-2 transfectants with the ADAM10 inhibitor GI254023X [69] led to a strong reduction in the 40-kD cleavage band, while parallel treatment with nonspecific protease inhibitors had no such effect (Fig. 2A, B). Accumulation of the ADAM10 mature form was seen in the GI254023X-treated cells (Fig. 2A), as a result of this inhibition of ADAM10 enzymatic activity [70]. Consistent with the Trop-2 cleavage reduction seen upon chemical inhibition of ADAM10 activity, RNAi-mediated knock-down of ADAM10 expression resulted in a dramatic reduction of the Trop-2 cleavage (Fig. 2C, D). Taken together, our findings indicate that ADAM10 is an effector protease of Trop-2 cleavage at R87-T88.
Fig. 2.
Trop-2 cleavage by ADAM10. (A) Inhibition of ADAM10 activity by treatment of BxPC3 cells with chemical inhibitors. Western blot detections of ADAM10 (top) and Trop-2 (bottom) show accumulation of the mature ADAM10 and corresponding reduction of the 40-kD Trop-2 cleavage band only upon treatment with GI254023X (blue). Quantification of the 40-kD Trop-2 cleavage band upon treatment with protease inhibitors is shown on the right, as percentage of total Trop-2. Ponceau red staining, control of protein loading. (B) Reduction of Trop-2 cleavage upon treatment of MTE4-14/Trop-2 transfectants with the ADAM10 inhibitor GI254023X (blue). Quantification of the Trop-2 cleavage band was carried out, and the ratio between uncleaved (un, black) versus cleaved (cl, gray) Trop-2 is shown on the right panel. (C) ADAM10 mRNA levels (red) 48 h upon treatment with ADAM10 siRNA or control siRNA (human CD133) (black). Vector: vector-alone transfectants; A87-A88: mutagenized Trop-2 at the cleavage site. The 2−ΔΔCT algorithm was used to calculate the relative changes in gene expression. RNA levels of ADAM10 in Vector, wt Trop-2 and A87-A88 Trop-2 cells are shown here for cell group cross-comparison, and are intended as reference data for the functional assays shown in the next figures. (D) ADAM10 protein inhibition by shRNA (top). Corresponding reduction of the Trop-2 cleavage is shown (mid). Ponceau red staining, control of protein loading (bottom). (E) Western blotting of Trop-2 cleavage in MTE 4-14/Trop-2 transfectants. (left) Cells were seeded at the indicated fraction of full confluency, as normalized to 100%, or (right) cells were seeded at equal confluency and then lysed at different subsequent time points. Ponceau red staining, control of protein loading. MW markers are indicated. (F) Western blotting of MCF7 human breast cancer cells grown in adhesion or in suspension and lysed 5 d after seeding for analysis of the Trop-2 cleavage. Red arrow shows cleaved Trop-2. Ponceau red staining, control of protein loading. MW markers are indicated. (Color version of figure is available online.)
Proteolytic processing of Trop‐2 is required for cell growth stimulation
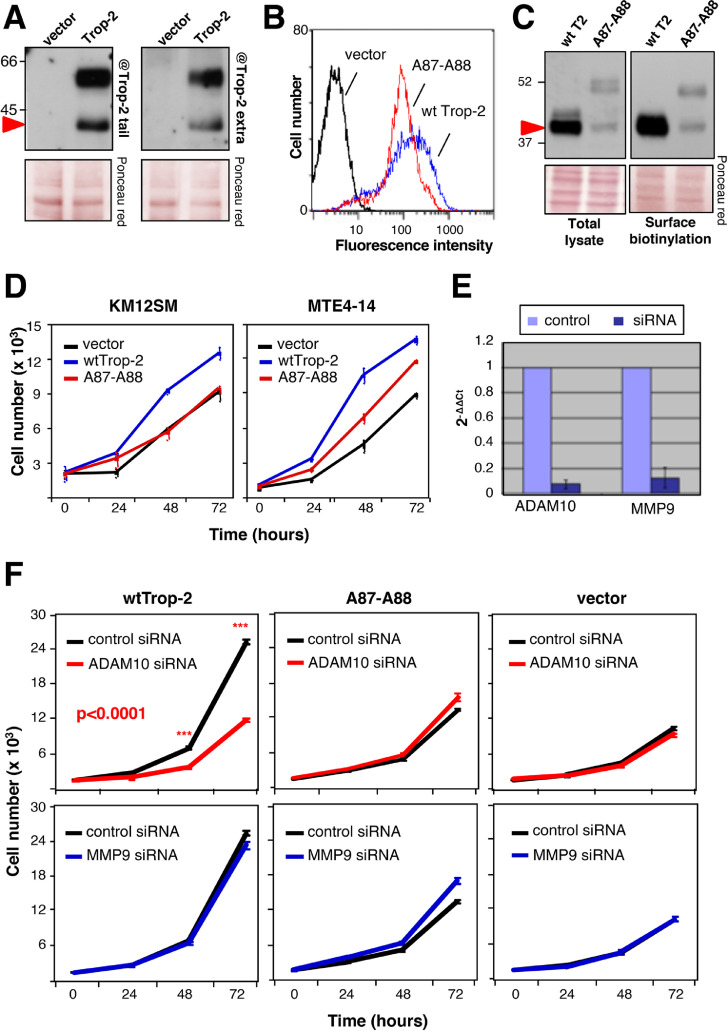
MTE4-14/Trop-2 transfectants were shown to acquire traits of malignant transformation upon Trop-2 expression, such as the ability to form tumors in immunosuppressed animals [14] and to grow at high cell density. This was paralled by increasing levels of Trop-2 cleavage at increasing cell densities (Fig. 2E). Growth under reduced adherence to a substrate is a corresponding hallmark of malignancy [71]. Consistent with this, cells grown in suspension induced Trop-2 cleavage (Fig. 2F). Cell-surface biotinylation/pull-down assays were used to reveal the status of Trop-2 at the cell membrane. This showed that wild type (wt)Trop-2 molecules were cleaved/activated at the cell surface (Fig. 3A). Further, we detected cleaved Trop-2 using antibodies that targeted either the extracellular domain or the intracytoplasmic domain of Trop-2 (Fig. 3A). This demonstrated that the R87-T88–cleaved Trop-2 still contained the cytoplasmic tail, thus indicating that R87-T88 cleavage precedes TACE cleavage, RIP, and release of the Trop-2 extra-cytoplasmic and intra-cytoplasmic segments [22] along the Trop-2 activation cascade. The Trop-2 cleavage site was mutagenized by switching the R87-T88 residues to alanine. Flow cytometry analysis on live cells showed that A87-A88 Trop-2 was efficiently transported to the cell membrane, where it retained wt Trop-2–like expression pattern (Fig. 3B and Fig. S4). Under high cell density and prolonged culture conditions, the A87-A88 Trop-2 showed considerably reduced cleavage compared with wt Trop-2 (Fig. 3C). We then asked whether the cleavage at R87-T88 acted as an activator switch for the Trop-2 growth-stimulatory functions. In colon cancer KM12SM and murine MTE4-14 transfectants, the cleavage-impaired A87-A88 Trop-2 mutant did not stimulate cell growth over baseline, at variance with the cell growth enhancement induced by wt Trop-2 (Fig. 3D).
Fig. 3.
Trop-2 proteolytic processing-defective mutant versus wild-type. (A) Western blotting analysis of KM12SM/Trop-2 transfectants was carried out using anti-Trop-2 antibodies against the cytoplasmic tail and extracellular domain, as indicated. The Trop-2 bands were recognized by both antiextracellular domain and anticytoplasmic tail antibodies, indicating that the Trop-2 signal belongs to transmembrane forms. Red arrow, cleaved Trop-2. Vector, vector-alone transfectants. Ponceau red staining, control of protein loading. MW markers are indicated. (B) Flow cytometry analysis of MTE 4-14/Trop-2 transfectants using the T16 anti-Trop-2 antibody. The A87-A88 Trop-2 mutant (red) and wt Trop-2 (blue) are expressed at comparable levels. Vector: vector-alone transfectants. (C) Trop-2 cleavage at the cell surface. Trop-2 cleavage was assessed upon cell-surface biotinylation on KM12SM metastatic colon cancer cells. Pull-down after cell-surface biotinylation was carried out to reveal the status of Trop-2 at the cell membrane. (left) total cell lysate, (right) cell membrane pulled-down material. Western blotting analysis was carried out with a rabbit polyclonal antibody directed against the extracellular domain of Trop-2, and shows that wt Trop-2 is completely cleaved. Quantitative cleavage reduction was observed for the A87-A88 proteolytic site mutant. Red arrow, cleaved Trop-2. Ponceau red staining, control of protein loading. MW markers are indicated. (D) Growth curves in vitro of KM12SM human colon cancer cells and MTE4-14 murine cells transfected with wt Trop-2 versus A87-A88 Trop-2 mutant. Bars, standard errors of the mean (SEM). (E) ADAM10 (left) and MMP9 (right) reduced RNA levels by siRNAs, as measured by real-time RT-PCR. (F) Cell growth curves of wt Trop-2, A87-A88 Trop-2 or vector-alone transfectants upon ADAM10 (red), MMP9 (blue) or control (black) siRNA-mediated inhibition. Bars, SEM. P value (ADAM10, left panel): ANOVA analysis. Stars indicate post-hoc Bonferroni's t test P values (***, P ≤ 0.001). (Color version of figure is available online.)
We then performed in vitro proliferation assays for cells treated with control siRNAs or ADAM10 siRNAs (Fig. 3E). We here found that ADAM10 inhibition suppressed the growth of wt Trop-2-expressing cells (Fig. 3F). On the other hand, ADAM10 inhibition had no impact on the A87-A88 Trop-2 transfectants, as well as on the negative control transfectants. Of note, the growth rate of wt Trop-2 transfectants treated with ADAM10 siRNAs was reduced to the growth rate of the A87-A88 Trop-2 transfectants, which indicated a mandatory role for ADAM10 in Trop-2 cleavage/activation for induction of cell growth. The ADAM10 inhibitor GI254023X can also inhibit MMP9 [69]. Hence, we functionally tested the role of MMP9 in Trop-2 cleavage by using specific siRNA (Fig. 3E). MMP9 inhibition had no impact on the growth rate of either wt Trop-2 or the A87-A88 mutated Trop-2 cell transfectants (Fig. 3F), which supported the specific involvement of ADAM10 in Trop-2 processing and activity for tumor growth.
Trop‐2 proteolytic processing at R87‐T88 only occurs in cancer cells
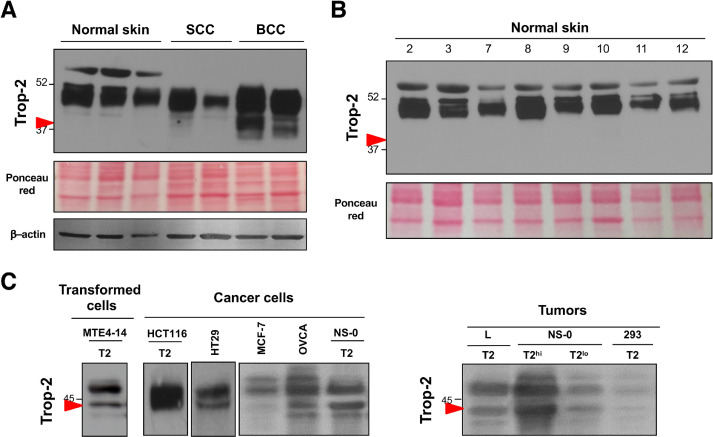
As wt Trop-2 is strongly upregulated in cancer and no cancer-related mutations have been identified in the corresponding gene (TACSTD2) as yet [13], we hypothesized that this Trop-2 proteolytic processing acts as an activator switch, and might be a key activating step for Trop-2 in transformed cells. Human epidermis expresses high levels of Trop-2 under native conditions [4, 13], which allows immediate comparisons of Trop-2 processing in normal epidermal cells versus skin cancers (Fig. 4, Fig. S5). Here, Trop-2 was not cleaved in normal human keratinocytes, whereas its cleavage occurred in the basal cell carcinomas. Minimal, if any, cleavage at R87-T88 was detected in squamous cell carcinomas (Fig. 4A). We speculate that this was due to the prevalent expression of Trop-2 in well-differentiated, keratinizing regions of the tumors, thus suggesting retention of differentiation features of normal human epidermis (Fig. S5).
Fig. 4.
Proteolytic processing of Trop-2 in tumor cells. (A) Western blotting analysis of Trop-2 cleavage in normal epidermal cells and skin tumors (SCC, squamous cell carcinoma; BCC, basal cell carcinoma). Red arrow, cleaved Trop-2. MW markers are indicated. Ponceau red staining and western blotting for β-actin were used as controls of protein loading. (B) Western blotting analysis of Trop-2 cleavage in normal skin samples. Red arrow, absence of Trop-2 cleavage at R87-T88. MW markers are indicated. Ponceau red staining, control of protein loading. (C) Trop-2 cleavage patterns in vitro and in vivo. Western blotting of transformed MTE4-14/Trop-2 transfectants and cancer cells either transfected with or endogenously expressing Trop-2 as indicated, grown in culture (left) or as tumors in nude mice (right). T2: TROP2-transfected cells; T2hi, T2lo: TROP2-transfected NS-0 cells selected to express TROP2 at high or low levels, respectively. Red arrow, cleaved Trop-2. (Color version of figure is available online.)
This analysis was extended to include an indipendent set of normal human skin samples, where absence of Trop-2–activating cleavage was confirmed (Fig. 4B). On the other hand, Trop-2 cleavage was detected in most of the Trop-2-transformed and cancer cells analyzed (i.e., ovarian, breast and colon carcinoma cells; transfectants of carcinoma, myeloma, fibrosarcoma cells) (Fig. 4C). The same cleavage patterns and extent were detected in vivo and in vitro, and across different ranges of cell surface expression (Fig. 4C), suggesting tight regulation of Trop-2 processing.
The signaling mechanism driven by Trop‐2–activating cleavage
The signaling mechanisms that are triggered by activation cleavage of Trop-2 were explored using proteomic and phosphoproteomic chip analysis (Table S4A, B). The wt Trop-2 versus vector-alone group and the A87-A88 Trop-2 versus vector-alone group were analyzed for concordant and discordant changes in the expression or activity of various signaling effectors (Table S4C, D). This allowed to reveal signaling steps that were specifically activated by wt Trop-2. Protein modifications that operated as central nodes of cleaved/activated Trop-2 were shown to encompass Src, RSK1/2, glycogen synthase, MEK1, CDK9, p21, and BAD as main downstream effectors (Table S4D).
Trop‐2 cleavage induces tumor growth
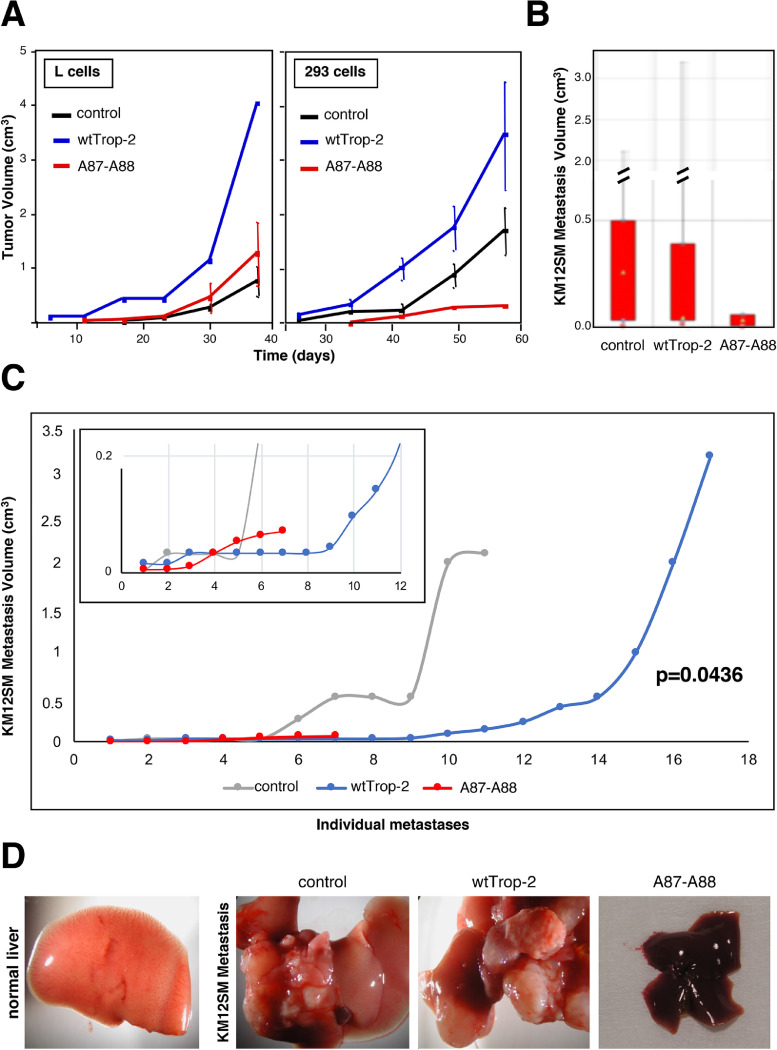
Our findings had indicated that Trop-2 proteolytic activation stimulates cell growth in vitro. We thus assessed the impact of this Trop-2 proteolytic activation on the growth of tumors xenotransplanted in murine models. Tumorigenic L fibrosarcoma and transformed HEK293 cells were transfected with the A87-A88 Trop-2 mutant and injected subcutaneously into immunosuppressed mice. The cells transfected with wt Trop-2 and with the corresponding empty vector were used as controls. Comparison of the tumor volumes between these groups indicated that while transfection of wt Trop-2 resulted in increased growth of both L fibrosarcoma and transformed HEK293 cells as tumors in vivo, the R87A-T88A mutagenesis led to complete loss of this growth-inducing activity (Fig. 5A). Hence, proteolytic processing at R87-T88 is mandatory for triggering the Trop-2 growth-inducing activity.
Fig. 5.
Tumor growth and metastasis dissemination require Trop-2 cleavage. (A) Tumor growth curves of L murine fibrosarcoma and 293 transformed human kidney cells, transfected with wt Trop-2 or A87-A88 Trop-2. Bars, SEM. N = 4 per experimental group. Subcutaneous tumor growth curves were obtained by weekly measurements of tumor volumes (d2 x D/2), followed by normalization on a group-by-group basis, and computation. (B) Boxplot analysis of KM12SM liver metastasis volume. The lowest median in the mutant set of data, together with the lowest max and minimum values of metastasis volume in mutant, processing-less Trop-2 are shown. N = 37 mice were injected with KM12SM/vector control cells, N=34 were injected with the KM12SM/Trop-2 transfectants and N = 13 were injected with the KM12SM/A87-A88 Trop-2 transfectants, i.e. expressing the cleavage-resistant R87-T88 Trop-2 mutant. (C) Individual KM12SM metastasis volume distribution curves. Gray: control metastases; Blue: wt Trop-2 metastases; Red: proteolytic mutant metastases. The distribution and correlated mutant liver metastases versus vector alone control cells and wt Trop-2 transfectants were statistically assessed by Mann-Whitney non-parametric test. This showed a significant reduction of volumes in the mutant transfectant metastases versus wt Trop-2 (P value = 0.0436), consistent with the loss of the Trop-2 activation path. (D) Macroscopic assessment of normal liver, control KM12SM liver metastasis, and of metastasis by wt Trop-2 or proteolytic mutant Trop-2 colon cancer transfectants.
Trop‐2 cleavage induces metastasis
Cancer progression involves tumor growth in the primary site followed by metastatic dissemination to close and distant sites. Hence, we assessed whether Trop-2 cleavage is an activation step for induction of a pro-metastatic activity. To this end, KM12SM colon cancer cells were transfected with the empty vector, wt Trop-2, or A87-A88 Trop-2 and injected into the spleen of immunosuppressed mice. Dissemination to the liver was then determined by comparison of the tumor and metastasis volumes between these groups. All tissue samples from the three groups of xenografts underwent systematic histopathology analysis, to include detection of minimal tumor and metastasis burden and volumes. The individual metastasis volume distribution curves showed that the A87-A88 Trop-2 mutant invariably reduced, or outright abolished, metastasis growth (Fig. 5B-D). Boxplot distribution analysis showed that the dataset of A87-A88 mutant Trop-2 had the lowest median, together with the lowest maximum and minimum values of metastasis volume. The dimensions and volume distributions of mutant liver metastases versus vector-alone control cells and wt Trop-2 transfectants were assessed. These showed that the mutant transfectant metastases volumes were significantly reduced versus wt Trop-2 (nonparametric Mann-Whitney P = 0.0436), due to this loss of the required metastasis activation pathway.
Trop‐2 activation in primary breast cancers
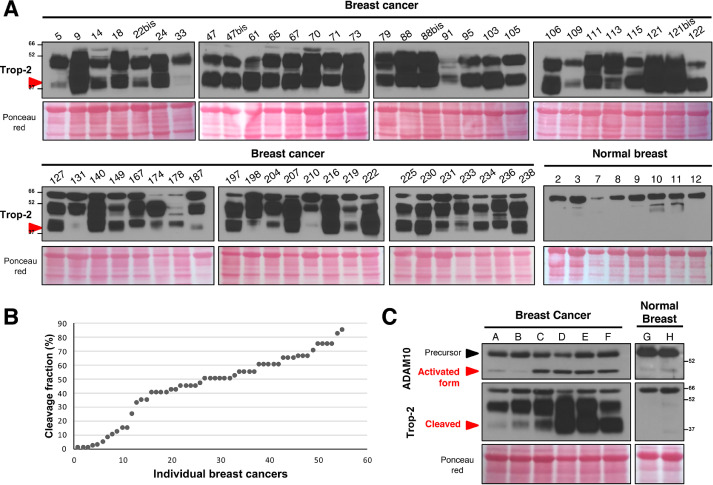
We then assessed whether this Trop-2–activating mechanism operates in primary tumors in cancer patients. Cleavage of Trop-2 was assessed by western blotting in 55 breast cancer samples from a N0, T1/T2 case series [29]. Trop-2 expression was detected in all of the tumor samples, with high intensity (i = 3) in 44 samples (80.0%), low intensity (i = 1) in 3 samples (5.5%), and intermediate intensity (i = 2) in 8 samples (14.5%). Image analysis revealed broadly different levels of Trop-2 cleavage, which ranged from 85% to 1%–2%. However, no primary tumor showed an absence of Trop-2 cleavage, at variance with normal breast samples from control individuals, where we did not observe any cleaved Trop-2 (Fig. 6A). In 20% of the breast cancer samples, there were low levels of cleavage (≤15% of Trop-2 molecules). The majority of cases (67.3%) showed between 18% and 66% cleaved Trop-2 molecules, with high levels of proteolytic processing (67%–85% of Trop-2 molecules) in 12.7% of samples (Fig. 6A, B). These results extended and confirmed similar data obtained in samples of normal skin (Fig. 4A, B). We assessed the expression of ADAM10 in normal breast and in breast cancer samples (Fig. 6C). For best sensitivity, we pooled the cancer samples based on the cleavage pattern of Trop-2 observed in the individual samples. We then quantified global levels of ADAM10 versus levels of activated/secreted form of ADAM10 [72]. This strategy allowed observing a tight direct correlation between the levels of the mature/active form of a secreted ADAM10 in breast cancer and the extent of Trop-2 cleavage.
Fig. 6.
Breast cancer case series. (A) Frozen samples from a consecutive breast cancer case series were analyzed for Trop-2 expression and cleavage by Western blotting. Western blot lanes bear the original patient number in the case series. The latin “bis” indication refers to a second sampling of the same tumor case, upon local relapse. Red arrow: cleaved Trop-2. Ponceau red staining, control of protein loading. MW markers are indicated. The bottom right panel shows a Western blotting analysis of Trop-2 expression and cleavage in normal breast tissue samples from control individuals. (B) Distribution of the fraction of cleaved Trop-2 molecules in individual breast cancer samples, ordered from low to high Trop-2 cleavage. (C) Western blotting analysis of ADAM10 expression and of Trop-2 cleavage in samples from breast cancer and normal breast tissues. The samples were pooled from the individual cases shown in the panel A as based on the extent of Trop-2 cleavage. The pools are as follows: A = 5+22+78+109+187; B = 113+131+174+187+204; C = 78+131+198+231+233; D = 9+18+24+47+61; E = 65+67+70+73+88; F = 95+103+106+115+122; G = 2+8+12; H = 3+7+9. ADAM10 was detected as bands at 55-50 kDa, designated as precursor and activated forms of the protein. Red arrow: presence/absence of Trop-2 cleavage at R87-T88. (Color version of figure is available online.)
Discussion
Better knowledge of the mechanisms underlying cancer cell growth and metastasis is an urgent need for cancer care. In this study, we set out to investigate the activation mechanisms of Trop-2, a signal transducer that is involved in tumor growth and metastasis.
The TROP2/TACSTD2 gene is an intronless derivative of TROP1/EPCAM [7, 8, 73]. The extracellular domain of the Trop molecules contains a GA733 type 1 motif and a thyroglobulin repeat, which together have 12 conserved cysteine residues [5, 7, 8]. Both the GA733 type 1 and thyroglobulin domains have been proposed to be involved in homophilic intra-membrane and inter-membrane interactions of the paralogous Trop-1/EpCAM [20, 73], which suggested a corresponding function for Trop-2.
We previously reported that proteolytic cleavage occurs between R80 and R81 in the first loop of the Trop-1 thyroglobulin domain [21], and that this activates the cell-growth stimulatory properties of Trop-1. We hypothesized that Trop-1 and Trop-2 may share the same activation mechanism. Here we have shown that this is indeed the case, as Edman degradation sequencing and peptide mass fingerprinting of Trop-2 purified to homogeneity revealed that the cleavage of the molecule occurs in the thyroglobulin domain, between R87 and T88. This site precisely aligns to the corresponding cleavage site of Trop-1. Cleavage at R87-T88 implies a large conformational rearrangement and potential swiveling of a 10 kDa subunit over the Trop-2 backbone, suggesting a large impact on the interactions of Trop-2 with binding partners.
Cleavage of the extracellular domain(s) of transmembrane signal transducers is frequently catalyzed by MMPs. ADAM10 is a member of this family, and it has been shown to induce proteolytic processing of many type I and II transmembrane proteins, including cadherins [53, 55], CD44 [52], L1 [54], Her2 [74], HB-EGF [75], [76], [77], TNF-α [76], and APP [78].
Peptide library screening, combined with crystal structure analysis of known ADAM10 substrates [57] indicated preference for a consensus target site that includes Arg at P1 and Thr at P1’, which are both present in the Trop-2 candidate cleavage site. Similarly to Trop-2, ADAM10 is expressed by most human cancer types, and is upregulated in many of them [[58], [59], [60], [61], [62], 79]. We have shown here direct interaction and dynamic colocalization of ADAM10 and Trop-2, in agreement with previous reports that signaling activation enhances the association of ADAM10 with its substrates, such as for HB-EGF and CD9, thereby modulating tumor invasion [75]. Inhibition of ADAM10 expression or activity in Trop-2–expressing cells was then shown to markedly reduce Trop-2 processing. These findings indicated that ADAM10 is effector protease at Trop-2 R87-T88.
Recently, matriptase was reported to recognize the R87-T88 Trop-2 cleavage site [23, 24], which raises the possibility of a finely regulated, multipronged post-translational processing at this position. Stoyanova et al showed that TACE mediates proteolysis and ectodomain shedding of Trop-2, which is followed by RIP through γ-secretase/presenilins, and nuclear translocation of the intracellular domain of Trop-2 upon release from the cell membrane [22]. Our membrane biotinylation/pull-down assays revealed that R87-T88–cleaved Trop-2 molecules are still anchored to the cell membrane. Furthermore, this R87-T88–cleaved Trop-2 remains linked to the cytoplasmic tail, which demonstrates that the Trop-2 cleavage at R87-T88 precedes TACE cleavage and RIP in the Trop-2 activation cascade.
R87A-T88A mutagenesis was shown here to lead to complete loss of wt Trop-2 pro-growth activity in vitro. Striking correspondence was observed also in vivo, where the A87-A88 Trop-2 mutant failed to stimulate tumor growth in both murine xenografts of L and HEK-293 cells.
We have previously shown that Trop-2 is highly expressed in most metastatic cancer cells and represents a key driver of metastasis. The proteolytic cleavage of Trop-2 at the R87-T88 site was shown to be a requirement for activation of Trop-2 for induction of the complete cascade from growth to metastasis in vivo.
Analysis of the Trop-2 processing in human samples here reveals that Trop-2 is not cleaved in normal epidermidis and in squamous cell carcinomas, where Trop-2 could associate with retention of differentiation, whereas proteolytic cleavage occurs in basal cell carcinomas.
Correspondingly, no Trop-2–activating cleavage was detected in normal human breast samples. On the other hand, the majority of the breast tumors analyzed showed the Trop-2 cleavage, and a tight correlation between the extent of Trop-2 cleavage and the expression levels of the mature/active form of a secreted ADAM10 was observed. Together with the prognostic impact of Trop-2/ADAM10 on cancers of the lung and pancreas, this indicates a driving role for this Trop-2 activatory cleavage on the progression of malignant human tumors.
Conclusions
We have demonstrated here that Trop-2 recruits ADAM10 and is activated by ADAM10-mediated cleavage to trigger molecular pathways for tumor growth and metastasis. Although wt Trop-2 is strongly up-regulated in cancers, no cancer-related mutations have been as yet identified in the corresponding human gene (TACSTD2) [13]. Our findings here show that post-translational processing of Trop-2 acts as the Trop-2 activator, to drive tumor growth and metastatic dissemination.
Our findings that the ADAM10-mediated cleavage of Trop-2 at the R87-T88 site is necessary to activate the transformed growth stimulatory activity of Trop-2 and promote its metastasis driver function now pave the way for the development of new therapeutic precision strategies. Many antibody-based therapeutic approaches that target Trop-2 are currently being developed; these include use of the humanized anti-Trop-2 antibody–SN-38 drug conjugate Sacituzumab govitecan-hziy (Trodelvy) in metastatic triple-negative breast cancers [47]. Inhibition of Trop-2 proteolytic processing may correspondingly open new perspectives for such specific therapies to control tumor growth and metastatic dissemination through targeting the Trop-2 activation steps in cancer cells.
Author contributions
Marco Trerotola: Conceptualization, Investigation, Formal analysis, Writing – Original draft, Visualization, Funding acquisition. Emanuela Guerra: Conceptualization, Investigation, Formal analysis, Writing – Original draft, Visualization. Zeeshan Ali: Investigation, Formal analysis. Anna Laura Aloisi: Investigation, Formal analysis. Pasquale Simeone: Investigation, Formal analysis, Data curation. Angela Acciarito: Investigation, Formal analysis. Paola Zanna: Investigation, Formal analysis. Giovanna Vacca: Investigation, Formal analysis. Martina Ceci: Investigation, Formal analysis, Data curation. Antonella D'Amore: Investigation, Formal analysis. Khouloud Boujnah: Investigation, Formal analysis, Data curation. Valeria Garbo: Investigation, Formal analysis, Data curation. Antonino Moschella: Investigation, Formal analysis, Data curation. Rossano Lattanzio: Resources, Investigation, Formal analysis. Saverio Alberti: Conceptualization, Formal analysis, Writing – Original draft, Visualization, Supervision, Project administration, Funding acquisition.
Competing interests
All authors declare that they have no competing financial or nonfinancial interests that might have influenced the performance or presentation of the work described in this manuscript.
Funding
The research was supported by Fondazione of the Cassa di Risparmio della Provincia di Chieti, the Italian Association for Cancer Research (AIRC), ABO Foundation (Grant VE01D0019 and CH01D0081). The Programma Per Giovani Ricercatori “Rita Levi Montalcini”, Italian Ministry of University and Research (Grant PGR12I7N1Z) to M.T. is gratefully acknowledged.
Acknowledgments
We thank L. Zardi for help over the course of this study, and R. Stein for providing RS7 antibodies. The support of Fondazione of the Cassa di Risparmio della Provincia di Chieti, the Italian Association for Cancer Research (AIRC), ABO Foundation (Grant VE01D0019 and CH01D0081), Fondazione Compagnia di San Paolo, Italian Ministry of Health (RicOncol RF-EMR-2006-361866) and Marie Curie Transfer of Knowledge Fellowship – EC VI Framework Programme (Contract 014541) is gratefully acknowledged. The Programma Per Giovani Ricercatori “Rita Levi Montalcini”, Italian Ministry of University and Research (Grant PGR12I7N1Z) to M.T. is gratefully acknowledged. We also thank C. Berrie for language editing of the manuscript.
The sponsors had no role in the design and conduct of this study, nor in the collection, analysis, and interpretation of the data, or in the preparation, review, or approval of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.03.006.
Appendix. Supplementary materials
References
- 1.Ripani E, Sacchetti A, Corda D, Alberti S. The human Trop-2 is a tumor-associated calcium signal transducer. Int J Cancer. 1998;76:671–676. doi: 10.1002/(sici)1097-0215(19980529)76:5<671::aid-ijc10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 2.Basu A, Goldenberg DM, Stein R. The epithelial/carcinoma antigen EGP-1, recognized by monoclonal antibody RS7-3G11, is phosphorylated on serine 303. Int J Cancer. 1995;62:472–479. doi: 10.1002/ijc.2910620419. [DOI] [PubMed] [Google Scholar]
- 3.Klein CE, Hartmann B, Schön MP, Weber L, Alberti S. Expression of 38-kD cell-surface glycoprotein in transformed human keratinocyte cell lines, basal cell carcinomas, and epithelial germs. J Invest Dermatol. 1990;95:74–82. doi: 10.1111/1523-1747.ep12873988. [DOI] [PubMed] [Google Scholar]
- 4.Alberti S, Miotti S, Stella M, Klein CE, Fornaro M, Ménard S, Colnaghi MI. Biochemical characterization of Trop-2, a cell surface molecule expressed by human carcinomas: formal proof that the monoclonal antibodies T16 and MOv-16 recognize Trop-2. Hybridoma. 1992;11:539–545. doi: 10.1089/hyb.1992.11.539. [DOI] [PubMed] [Google Scholar]
- 5.Calabrese G, Crescenzi C, Morizio E, Palka G, Guerra E, Alberti S. Assignment of TACSTD1 (alias TROP1, M4S1) to human chromosome 2p21 and refinement of mapping of TACSTD2 (alias TROP2, M1S1) to human chromosome 1p32 by in situ hybridization. Cytogenet Cell Genet. 2001;92:164–165. doi: 10.1159/000056891. [DOI] [PubMed] [Google Scholar]
- 6.Linnenbach AJ, Seng BA, Wu S, Robbins S, Scollon M, Pyrc JJ, Druck T, Huebner K. Retroposition in a family of carcinoma-associated antigen genes. Mol Cell Biol. 1993;13:1507–1515. doi: 10.1128/mcb.13.3.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Sewedy T, Fornaro M, Alberti S. Cloning of the murine Trop2 gene: conservation of a PIP2-binding sequence in the cytoplasmic domain of Trop-2. Int J Cancer. 1998;75:324–330. doi: 10.1002/(sici)1097-0215(19980119)75:2<324::aid-ijc24>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 8.Fornaro M, Dell'Arciprete R, Stella M, Bucci C, Nutini M, Capri MG, Alberti S. Cloning of the gene encoding TROP-2, a cell-surface glycoprotein expressed by human carcinomas. Int J Cancer. 1995;62:610–618. doi: 10.1002/ijc.2910620520. [DOI] [PubMed] [Google Scholar]
- 9.Trerotola M, Jernigan D, Liu Q, Siddiqui J, Fatatis A, Languino L. Trop-2 promotes prostate cancer metastasis by modulating β1 integrin functions. Cancer Res. 2013;73:3155–3167. doi: 10.1158/0008-5472.CAN-12-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu E-C, Rice MA, Bermudez A, Marques FJG, Aslan M, Liu S, Ghoochani A, Zhang CA, Chen Y-S, Zlitni A. Trop2 is a driver of metastatic prostate cancer with neuroendocrine phenotype via PARP1. Proc Natl Acad Sci. 2020;117:2032. doi: 10.1073/pnas.1905384117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra E, Trerotola M, Aloisi AL, Tripaldi R, Vacca G, La Sorda R, Lattanzio R, Piantelli M, Alberti S. The Trop-2 signalling network in cancer growth. Oncogene. 2013;32:1594–1600. doi: 10.1038/onc.2012.151. [DOI] [PubMed] [Google Scholar]
- 12.Guerra E, Trerotola M, Dell’ Arciprete R, Bonasera V, Palombo B, El-Sewedy T, Ciccimarra T, Crescenzi C, Lorenzini F, Rossi C. A bi-cistronic CYCLIN D1-TROP2 mRNA chimera demonstrates a novel oncogenic mechanism in human cancer. Cancer Res. 2008;68:8113–8121. doi: 10.1158/0008-5472.CAN-07-6135. [DOI] [PubMed] [Google Scholar]
- 13.Trerotola M, Cantanelli P, Guerra E, Tripaldi R, Aloisi AL, Bonasera V, Lattanzio R, de Lange R, Weidle UH, Piantelli M. Up-regulation of Trop-2 quantitatively stimulates human cancer growth. Oncogene. 2013;32:222–233. doi: 10.1038/onc.2012.36. [DOI] [PubMed] [Google Scholar]
- 14.Guerra E, Trerotola M, Tripaldi R, Aloisi AL, Simeone P, Sacchetti A, Relli V, A DA, La Sorda R, Lattanzio R. Trop-2 induces tumor growth through Akt and determines sensitivity to Akt inhibitors. Clin Cancer Res. 2016;22:4197–4205. doi: 10.1158/1078-0432.CCR-15-1701. [DOI] [PubMed] [Google Scholar]
- 15.Trerotola M, Li J, Alberti S, Languino LR. Trop-2 inhibits prostate cancer cell adhesion to fibronectin through the β1 integrin-RACK1 axis. J Cell Physiol. 2012;227:3670–3677. doi: 10.1002/jcp.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linnenbach AJ, Wojcierowski J, Wu SA, Pyrc JJ, Ross AH, Dietzschold B, Speicher D, Koprowski H. Sequence investigation of the major gastrointestinal tumor-associated antigen gene family, GA733. Proc Natl Acad Sci USA. 1989;86:27–31. doi: 10.1073/pnas.86.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong JM, Speicher DW. Determination of disulfide bond assignments and N-glycosylation sites of the human gastrointestinal carcinoma antigen GA733-2 (CO17-1A, EGP, KS1-4, KSA, and Ep-CAM. J Biol Chem. 2001;276:5804–5813. doi: 10.1074/jbc.M008839200. [DOI] [PubMed] [Google Scholar]
- 18.Strnad J, Hamilton AE, Beavers LS, Gamboa GC, Apelgren LD, Taber LD, Sportsman JR, Bumol TF, Sharp JD, Gadski RA. Molecular cloning and characterization of human adenocarcinoma/epithelial cell-surface-antigen-complementary DNA. Cancer Res. 1989;49:314–317. [PubMed] [Google Scholar]
- 19.Trebak M, Begg GE, Chong JM, Kanazireva EV, Herlyn D, Speicher DW. Oligomeric state of the colon carcinoma-associated glycoprotein GA733-2 (Ep-CAM/EGP40) and its role in GA733-mediated homotypic cell-cell adhesion. J Biol Chem. 2001;276:2299–2309. doi: 10.1074/jbc.M004770200. [DOI] [PubMed] [Google Scholar]
- 20.Balzar M, Briaire-de Bruijn IH, Rees-Bakker HAM, Prins FA, Helfrich W, Leij LD, Riethmuller G, Alberti S, Warnaar SO, Fleuren GJ. Epidermal growth factor-like repeats mediate lateral and reciprocal interactions of Ep-CAM molecules in homophilic adhesions. Mol Cell Biol. 2001;21:2570–2580. doi: 10.1128/MCB.21.7.2570-2580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schön MP, Schön M, Mattes MJ, Stein R, Weber L, Alberti S, Klein CE. Biochemical and immunological characterization of the human carcinoma- associated antigen MH 99/KS 1/4. Int J Cancer. 1993;55:988–995. doi: 10.1002/ijc.2910550619. [DOI] [PubMed] [Google Scholar]
- 22.Stoyanova T, Goldstein AS, Cai H, Drake JM, Huang J, Witte ON. Regulated proteolysis of Trop2 drives epithelial hyperplasia and stem cell self-renewal via beta-catenin signaling. Genes Dev. 2012;26:2271–2285. doi: 10.1101/gad.196451.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu CJ, Lu M, Feng X, Nakato G, Udey MC. Matriptase cleaves EpCAM and TROP2 in keratinocytes, destabilizing both proteins and associated claudins. Cells. 2020;9 doi: 10.3390/cells9041027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamble PR, Rane S, Breed AA, Joseph S, Mahale SD, Pathak BR. Proteolytic cleavage of Trop2 at Arg87 is mediated by matriptase and regulated by Val194. FEBS Letters. 2020;594:3156–3169. doi: 10.1002/1873-3468.13899. [DOI] [PubMed] [Google Scholar]
- 25.Morikawa K, Walker SM, Jessup JM, Fidler IJ. In vivo selection of highly metastatic cells from surgical specimens of different primary human colon carcinomas implanted into nude mice. Cancer Res. 1988;48:1943–1948. [PubMed] [Google Scholar]
- 26.Naquet P, Lepesant H, Luxembourg A, Brekelmans P, Devaux C, Pierres M. Establishment and characterization of mouse thymic epithelial cell lines. Thymus. 1989;13:217–226. [PubMed] [Google Scholar]
- 27.Alberti S, Herzenberg LA. DNA methylation prevents transfection of genes for specific surface antigens. Proc Natl Acad Sci USA. 1988;85:8391–8394. doi: 10.1073/pnas.85.22.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alberti S, Fornaro M. Higher transfection efficency of genomic DNA purified with a guanidinium-thiocyanate-based procedure. Nucleic Acids Res. 1990;18:351–353. doi: 10.1093/nar/18.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambrogi F, Fornili M, Boracchi P, Trerotola M, Relli V, Simeone P, La Sorda R, Lattanzio R, Querzoli P, Pedriali M. Trop-2 is a determinant of breast cancer survival. PLoS One. 2014;9:e96993. doi: 10.1371/journal.pone.0096993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zardi L, Carnemolla B, Siri A, Santi L, Accolla RS. Somatic cell hybrids producing antibodies specific to human fibronectin. Int J Cancer. 1980;25:325–329. doi: 10.1002/ijc.2910250304. [DOI] [PubMed] [Google Scholar]
- 31.Carnemolla B, Neri D, Castellani P, Leprini A, Neri G, Pini A, Winter G, Zardi L. Phage antibodies with pan-species recognition of the oncofoetal angiogenesis marker fibronectin ED-B domain. Int J Cancer. 1996;68:397–405. doi: 10.1002/(SICI)1097-0215(19961104)68:3<397::AID-IJC20>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Alberti S, Nutini M, Herzenberg LA. DNA methylation prevents the amplification of TROP1, a tumor associated cell surface antigen gene. Proc Natl Acad Sci USA. 1994;91:5833–5837. doi: 10.1073/pnas.91.13.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dell'Arciprete R, Stella M, Fornaro M, Ciccocioppo R, Capri MG, Naglieri AM, Alberti S. High-efficiency expression gene cloning by flow cytometry. J Histochem Cytochem. 1996;44:629–640. doi: 10.1177/44.6.8666748. [DOI] [PubMed] [Google Scholar]
- 34.Alberti S, Bucci C, Fornaro M, Robotti A, Stella M. Immunofluorescence analysis in flow cytometry: better selection of antibody-labeled cells after fluorescence overcompensation in the red channel. J Histochem Cytochem. 1991;39:701–706. doi: 10.1177/39.5.1901878. [DOI] [PubMed] [Google Scholar]
- 35.Alberti S, Parks DR, Herzenberg LA. A single laser method for subtraction of cell autofluorescence in flow cytometry. Cytometry. 1987;8:114–119. doi: 10.1002/cyto.990080203. [DOI] [PubMed] [Google Scholar]
- 36.Polishchuk RS, Polishchuk EV, Marra P, Alberti S, Buccione R, Luini A, Mironov A. Correlative light-electron microscopy reveals the saccular-tabular ultrastructure of carriers operating between the Golgi apparatus and the plasma membrane. J Cell Biol. 2000;148:45–58. doi: 10.1083/jcb.148.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orsulic S, Li Y, Soslow RA, Vitale-Crosss LA, Gutkind JS, Varmus HE. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell. 2002;1:53–62. doi: 10.1016/s1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi C, Di Lena A, La Sorda R, Lattanzio R, Antolini L, Patassini C, Piantelli M, Alberti S. Intestinal tumour chemoprevention with the antioxidant lipoic acid stimulates the growth of breast cancer. Eur J Cancer. 2008;44:2696–2704. doi: 10.1016/j.ejca.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 39.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 41.Overall CM, Blobel CP. In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol. 2007;8:245–257. doi: 10.1038/nrm2120. [DOI] [PubMed] [Google Scholar]
- 42.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zardi L, Carnemolla B, Siri A, Petersen TE, Paolella G, Sebastio G, Baralle FE. Transformed human cells produce a new fibronectin isoform by preferential alternative splicing of a previously unobserved exon. EMBO J. 1987;6:2337–2342. doi: 10.1002/j.1460-2075.1987.tb02509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda M, Yamaguchi M, Kato K, Nakamura K, Shiina S, Ichikawa-Ando T, Misaka H, Myojo K, Sugimoto Y, Hamada H. Pr1E11, a novel anti-TROP-2 antibody isolated by adenovirus-based antibody screening, recognizes a unique epitope. Biochem Biophys Res Commun. 2015;458:877–882. doi: 10.1016/j.bbrc.2015.02.051. [DOI] [PubMed] [Google Scholar]
- 45.Truong AHL, Feng N, Sayegh D, Mak BC, O'Reilly K, Fung SW, Ceric N, Hahn SH, Pereira D, Findlay H. AR47A6.4.2, a functional naked monoclonal antibody targeting Trop-2, demonstrates in vivo efficacy in human pancreatic, colon, breast and prostate cancer models. Mol Cancer Therap. 2007;6:3334s. [Google Scholar]
- 46.Thomas A, Pommier Y. Targeting topoisomerase I in the era of precision medicine. Clin Cancer Res. 2019;25:6581–6589. doi: 10.1158/1078-0432.CCR-19-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bardia A, Mayer IA, Vahdat LT, Tolaney SM, Isakoff SJ, Diamond JR, O'Shaughnessy J, Moroose RL, Santin AD, Abramson VG. Sacituzumab govitecan-hziy in refractory metastatic triple-negative breast cancer. N Engl J Med. 2019;380:741–751. doi: 10.1056/NEJMoa1814213. [DOI] [PubMed] [Google Scholar]
- 48.Guncar G, Pungercic G, Klemencic I, Turk V, Turk D. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J. 1999;18:793–803. doi: 10.1093/emboj/18.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tramontano A. Homology modeling with low sequence identity. Methods. 1998;14:293–300. doi: 10.1006/meth.1998.0585. [DOI] [PubMed] [Google Scholar]
- 50.Crawford HC, Dempsey PJ, Brown G, Adam L, Moss ML. ADAM10 as a therapeutic target for cancer and inflammation. Curr Pharm Des. 2009;15:2288–2299. doi: 10.2174/138161209788682442. [DOI] [PubMed] [Google Scholar]
- 51.Duffy MJ, McKiernan E, O'Donovan N, McGowan PM. Role of ADAMs in cancer formation and progression. Clin Cancer Res. 2009;15:1140–1144. doi: 10.1158/1078-0432.CCR-08-1585. [DOI] [PubMed] [Google Scholar]
- 52.Nagano O, Murakami D, Hartmann D, De Strooper B, Saftig P, Iwatsubo T, Nakajima M, Shinohara M, Saya H. Cell-matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca(2+) influx and PKC activation. J Cell Biol. 2004;165:893–902. doi: 10.1083/jcb.200310024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maretzky T, Schulte M, Ludwig A, Rose-John S, Blobel C, Hartmann D, Altevogt P, Saftig P, Reiss K. L1 is sequentially processed by two differently activated metalloproteases and presenilin/gamma-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol Cell Biol. 2005;25:9040–9053. doi: 10.1128/MCB.25.20.9040-9053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci U S A. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller MA, Sullivan RJ, Lauffenburger DA. Molecular pathways: receptor ectodomain shedding in treatment, resistance, and monitoring of cancer. Clin Cancer Res. 2017;23:623–629. doi: 10.1158/1078-0432.CCR-16-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J. 2009;424:79–88. doi: 10.1042/BJ20090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang YY, Ye ZY, Li L, Zhao ZS, Shao QS, Tao HQ. ADAM 10 is associated with gastric cancer progression and prognosis of patients. J Surg Oncol. 2011;103:116–123. doi: 10.1002/jso.21781. [DOI] [PubMed] [Google Scholar]
- 59.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gavert N, Conacci-Sorrell M, Gast D, Schneider A, Altevogt P, Brabletz T, Ben-Ze'ev A. L1, a novel target of beta-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J Cell Biol. 2005;168:633–642. doi: 10.1083/jcb.200408051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fogel M, Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Smirnov A, Edler L, Ben-Arie A, Huszar M, Altevogt P. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet. 2003;362:869–875. doi: 10.1016/S0140-6736(03)14342-5. [DOI] [PubMed] [Google Scholar]
- 62.Ko SY, Lin SC, Wong YK, Liu CJ, Chang KW, Liu TY. Increase of disintergin metalloprotease 10 (ADAM10) expression in oral squamous cell carcinoma. Cancer Lett. 2007;245:33–43. doi: 10.1016/j.canlet.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 63.Gaida MM, Haag N, Gunther F, Tschaharganeh DF, Schirmacher P, Friess H, Giese NA, Schmidt J, Wente MN. Expression of A disintegrin and metalloprotease 10 in pancreatic carcinoma. Int J Mol Med. 2010;26:281–288. doi: 10.3892/ijmm_00000463. [DOI] [PubMed] [Google Scholar]
- 64.Simeone P, Trerotola M, Urbanella A, Lattanzio R, Ciavardelli D, Di Giuseppe F, Eleuterio E, Sulpizio M, Eusebi V, Pession A. A unique four-hub protein cluster associates to glioblastoma progression. PLoS One. 2014;9 doi: 10.1371/journal.pone.0103030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Relli V, Trerotola M, Guerra E, Alberti S. Abandoning the notion of non-small cell lung cancer. Trends Mol Med. 2019;25:585–594. doi: 10.1016/j.molmed.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 66.Relli V, Trerotola M, Guerra E, Alberti S. Distinct lung cancer subtypes associate to distinct drivers of tumor progression. Oncotarget. 2018;9:35528–35540. doi: 10.18632/oncotarget.26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, Zentgraf H, Fogel M, Altevogt DP. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 2003;17:292–294. doi: 10.1096/fj.02-0430fje. [DOI] [PubMed] [Google Scholar]
- 68.Saftig P, Hartmann D (2005). ADAM10. A Major Membrane Protein Ectodomain Sheddase Involved in Regulated Intramembrane Proteolysis. (eds.) NMHaUL (ed), pp. 85-121.
- 69.Ludwig A, Hundhausen C, Lambert MH, Broadway N, Andrews RC, Bickett DM, Leesnitzer MA, Becherer JD. Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb Chem High Throughput Screen. 2005;8:161–171. doi: 10.2174/1386207053258488. [DOI] [PubMed] [Google Scholar]
- 70.Brummer T, Pigoni M, Rossello A, Wang H, Noy PJ, Tomlinson MG, Blobel CP, Lichtenthaler SF. The metalloprotease ADAM10 (a disintegrin and metalloprotease 10) undergoes rapid, postlysis autocatalytic degradation. FASEB J. 2018;32:3560–3573. doi: 10.1096/fj.201700823RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 72.Sogorb-Esteve A, García-Ayllón M-S, Gobom J, Alom J, Zetterberg H, Blennow K, Sáez-Valero J. Levels of ADAM10 are reduced in Alzheimer's disease CSF. J Neuroinflammation. 2018;15:213. doi: 10.1186/s12974-018-1255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zanna P, Trerotola M, Vacca G, Bonasera V, Palombo B, Guerra E, Rossi C, Lattanzio R, Piantelli M, Alberti S. Trop-1 are conserved growth stimulatory molecules that mark early stages of tumor progression. Cancer. 2007;110:452–464. doi: 10.1002/cncr.22785. [DOI] [PubMed] [Google Scholar]
- 74.Liu PC, Liu X, Li Y, Covington M, Wynn R, Huber R, Hillman M, Yang G, Ellis D, Marando C. Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells. Cancer Biol Ther. 2006;5:657–664. doi: 10.4161/cbt.5.6.2708. [DOI] [PubMed] [Google Scholar]
- 75.Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J Cell Biol. 2002;158:221–226. doi: 10.1083/jcb.200112026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arduise C, Abache T, Li L, Billard M, Chabanon A, Ludwig A, Mauduit P, Boucheix C, Rubinstein E, Le Naour F. Tetraspanins regulate ADAM10-mediated cleavage of TNF-alpha and epidermal growth factor. J Immunol. 2008;181:7002–7013. doi: 10.4049/jimmunol.181.10.7002. [DOI] [PubMed] [Google Scholar]
- 77.Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164:769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu D, Sharma C, Hemler ME. Tetraspanin12 regulates ADAM10-dependent cleavage of amyloid precursor protein. Faseb J. 2009;23:3674–3681. doi: 10.1096/fj.09-133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Knosel T, Emde A, Schluns K, Chen Y, Jurchott K, Krause M, Dietel M, Petersen I. Immunoprofiles of 11 biomarkers using tissue microarrays identify prognostic subgroups in colorectal cancer. Neoplasia. 2005;7:741–747. doi: 10.1593/neo.05178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lipinski M, Parks DR, Rouse RV, Herzenberg LA. Human trophoblast cell-surface antigens defined by monoclonal antibodies. Proc Natl Acad Sci USA. 1981;78:5147–5150. doi: 10.1073/pnas.78.8.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.