Abstract
The Ros3 protein is a component of the MT-Ros3 transporter complex, considered as the main route of miltefosine entry in Leishmania. L. braziliensis clinical isolates presenting differences in miltefosine susceptibility and uptake were previously shown to differentially express ros3. In this work, we showed that the ros3 gene copy number was increased in the isolate presenting the highest rates of miltefosine uptake and, thus, the highest susceptibility to this drug. The role of the ros3 gene dosage in miltefosine susceptibility was then investigated through a modulation of the gene copy number using two distinct approaches: through an overexpression of ros3 in a tolerant L. braziliensis clinical isolate and in L. major and by generating mono- and diallelic knockouts of this gene in L. major using clustered regularly interspaced short palindromic repeats (CRISPR) Cas9 (Cas = CRISPR-associated). Although the levels of ros3 mRNA were increased at least 40-fold in overexpressing clones, no significant reduction in the half-maximal effective concentration (EC50) for miltefosine was observed in these parasites. The partial or complete deletion of ros3 in L. major, in turn, resulted in a significant increase of 3 and 20 times, respectively, in the EC50 to miltefosine. We unequivocally showed that the ros3 copy number is one of the factors involved in the differential susceptibility and uptake of miltefosine.
Keywords: Leishmania braziliensis, miltefosine, drug resistance, clinical isolates, CRISPR/Cas9, treatment
Leishmania (Viannia) braziliensis is the main etiological agent of tegumentary leishmaniasis in Brazil.1,2 Infections caused by this species predominately manifest as localized cutaneous lesions but can also cause a severe mucosal disease or disseminated cutaneous manifestations.3 Therefore, a systemic treatment in these cases is mandatory, and the limitations of the treatment options in use in Brazil become even more alarming.4 The current therapy for leishmaniasis in Brazil relies on pentavalent antimonial and amphotericin B, both of them highly toxic and parenterally administered.5 Moreover, in some regions of Brazil the cure rates for cutaneous leishmaniasis (CL) upon treatment with meglumine antimoniate have drastically dropped to ∼50%.6,7 For all these reasons, alternative therapies are highly needed in order to overcome these limitations.
Miltefosine (MF) is currently the most effective oral drug available for leishmaniasis treatment.8 In Brazil, recommendations for the use of MF for CL treatment were issued in 2018, but the drug is still not available for clinical use.9 Two clinical trials employing MF for CL treatment in patients infected with the Leishmania (Viannia) species of two different regions of Brazil showed cure rates of ∼70%.6,10 Although MF is not devoid of side effects, those are mostly milder than the side effects of antimony and amphotericin.11 Being the only oral drug in clinical use, MF is of great importance for leishmaniasis chemotherapy. However, the significant drop in cure rates observed in visceral leishmaniasis patients in India, together with the recent isolation of resistant parasites from patients previously treated with MF, raised an alarm on the possible loss of this drug due to the selection of resistance.12−16 Moreover, recently data characterizing the in vitro susceptibility of L. infantum Brazilian clinical isolates recovered from patients enrolled in a clinical trial with MF for VL treatment revealed an alarming correlation between treatment failure and the intrinsic parasite susceptibility to MF.17
Miltefosine’s entry into the Leishmania parasites relies on a P4-ATPase membrane transporter called miltefosine transporter (MT), which has as its main function the transport of phospholipids from the extracellular environment through the cell membrane.18 It is well-known that in vitro MF-selected parasites present a significant reduction in drug accumulation due to mutations in the MT gene, which leads to a defective MF transport machinery.19,20 However, this reduced drug uptake was also shown to be present in parasites naturally less susceptible to MF without significant differences in the MT gene sequence and/or expression of this transporter.21,22
Another protein plays a key role in MF transport, the MT’s beta subunit Ros3, which belongs to the Lem3p/CDC50 family.23 Together, they form the MT-Ros3 complex, and both of them are indispensable for the complex functionality.24 The Ros3 subunit has been shown to play a key role in phospholipid transport and susceptibility of yeast to MF and edelfosine.25 In Leishmania parasites, it has already been demonstrated that the absence or defects in Ros3 cause the retention of the whole MT-Ros3 complex in the endoplasmic reticulum and consequently resistance to MF.24 Moreover, polymorphisms in both MT and ros3 genes were described as responsible for the reduced susceptibility to MF in an L. infantum clinical isolate.15
We previously reported that L. braziliensis clinical isolates from Brazilian patients exhibited differences in susceptibility to MF,26 as a result of differences in drug uptake.21 The levels of ros3 mRNA were found to be decreased in tolerant isolates compared to a sensitive one, suggesting that a low abundance of this component of the MF transport complex could be the cause of a reduced susceptibility observed in tolerant isolates.21
Since gene and/or chromosome copy number variations have been implicated in the mechanisms of drug resistance and variation of susceptibility to drugs in Leishmania,27,28 we hypothesized that the differences in MF susceptibility, drug uptake, and ros3 transcript abundance in these isolates might be the result of a differential ros3 gene dosage in these parasites. Thus, in this work we investigated the copy number of the ros3 gene in these L. braziliensis clinical isolates and the consequences of a differential ros3 gene dosage for MF susceptibility by overexpressing and knocking out this gene in Leishmania.
Results
Differential ros3 DNA Abundance among L. braziliensis Clinical Isolates
It was previously reported by our group that L. braziliensis clinical isolates obtained in Brazil presented variable susceptibility to MF.26 Analysis of these isolates through transcriptome sequencing and quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) allowed the demonstration that the ros3 mRNA abundance was upregulated in the most susceptible isolate (S) when compared to the M2903 reference strain (RS) and the two more tolerant isolates (T1 and T2)21 (Table 1).
Table 1. Susceptibility to MF and ros3 Transcript Abundance in L. braziliensis Clinical Isolates.
| EC50 ± SEMa (μM) | |||||
|---|---|---|---|---|---|
| abbreviationb | identification codec | promastigotes | amastigotes | FC RNaseqd (p-value) | FC real-timee (p-value) |
| RS | MHOM/BR/1975/M2903 | 53.5 ± 6.6 | 2.7 ± 0.2 | ND | –1.85 (0.0993) |
| S | MHOM/BR/2005/LTCP16012 | 22.9 ± 3.7 | 0.8 ± 0.1 | 1.0 | 1.0 |
| T1 | MHOM/BR/2006/LTCP 16907 | 101.2 ± 6.0 | 3.3 ± 0.4 | –2.04 (0.0069) | –3.97 (0.0198) |
| T2 | MHOM/BR/2009/LTCP 19446 | 90.4 ± 5.2 | 4.2 ± 0.2 | –2.10 (0.0146) | –3.93 (0.0114) |
EC50 ± SEM of MF for promastigotes and amastigotes of L. braziliensis clinical isolates and M2903 reference strain. Data previously published in Espada et al.26
Code used for strain and isolates used in this study.
International code of each isolate.
Fold-change (FC) in ros3 transcript abundance in tolerant isolates relative to the abundance in the sensitive isolate and adjusted p-value evaluated by three biological replicates of each isolate and M2903 reference strain. (ND) Not differentially expressed in this transcriptome. Data previously published in Espada et al.21
Normalized expression of ros3 and adjusted p-value relative to S isolate assessed by quantitative real-time RT-PCR. Data previously described in Espada et al.21
Since the regulation of gene expression in trypanosomatids does not generally occur at a transcriptional level, a differential expression is mostly a result of differences in gene copy number or post-transcriptional regulation.28,29 Copy number variation (CNV) was assessed in these isolates by a quantification of the ros3 DNA abundance by real-time PCR. Two housekeeping genes, gapdh and tbp, were chosen for normalization in these experiments after they showed a consistent expression between these isolates in the RNaseq data (data not shown).21
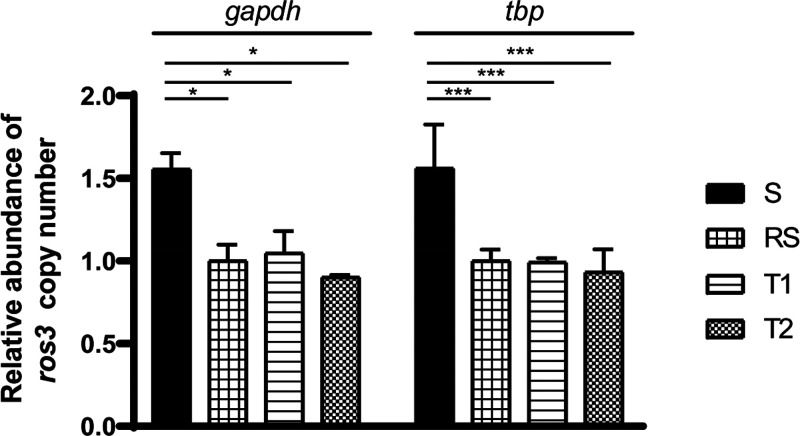
Significant differences in the ros3 DNA abundance were observed between S and T1/T2 isolates using both gapdh and tbp as normalizers (Figure 1). The most susceptible isolate (S) presented a significant 0.5-fold increased abundance of ros3 DNA compared to the tolerant isolates in experiments employing different normalizer genes. The abundance profile of ros3 DNA molecules correlated with the abundance of transcripts of this gene previously reported by Espada et al.21 and is highlighted in Table 1. Interestingly, the DNA quantification did not reveal differences between the reference and T isolates, in spite of the previously noted changes in half-maximal effective concentration (EC50) and mRNA abundance.
Figure 1.
Relative abundance of ros3 DNA in L. braziliensis clinical isolates and reference strain (RS). Each bar represents the mean ros3 DNA molecules number in each isolate relative to the molecule numbers in RS. Relative abundance was assessed by real-time PCR in two independent experiments using two different normalizer genes (gapdh and tbp). Three independent biological replicates and three technical replicates were employed in each experiment. Statistical significance was determined using One-way ANOVA and Tukey’s multiple comparison test. (*) p < 0.05 and (**) p < 0.001.
Overexpressing ros3 in the Tolerant Isolate Does Not Increase MF Susceptibility
The increased abundance of ros3 DNA in the isolate S led us to investigate whether the addition of more copies of this gene in tolerant parasites would lead to an increase in susceptibility to MF. To test this hypothesis, we overexpressed the ros3 gene in the isolate T2, which presented the highest EC50 to MF. The ros3 coding sequence was amplified from the T2 genome, cloned downstream to an L. tarentolae adenine phosphoribosyl (aptr) and upstream to a L. tarentolae calmodulin (camCB) untranslated region (UTR). The generated SR construct was then linearized and delivered by electroporation for integration in the SSU locus of this isolate. Recipient parasites were the T2 isolate and L. major FV-1 (Lm), included to verify if these findings would be similar for another Leishmania species. After a selection with hygromycin B in solid M199 seven clones of T2 and eight clones of Lm were screened by PCR for the presence and correct integration of the SR insert (Figure S1).
Three random T2 and three Lm SR clones presenting the SR cassette integrated in the right orientation were selected for an ros3 mRNA abundance quantification. The relative abundance of the ros3 mRNA in each clone relative to the WT parasites was assessed through real-time qPCR using tbp as a normalizing gene for T2 and Lm lines (Figure 2). An increase in transcript abundance was observed in both species. In L. braziliensis T2 SR clones, levels of ros3 mRNA were increased 44- to 50-fold when compared to WT parasites (Figure 2A). In L. major SR clones, the increase in ros3 mRNA levels varied from 30.8- to 49.0-fold compared to the WT parasite (Figure 2B).
Figure 2.
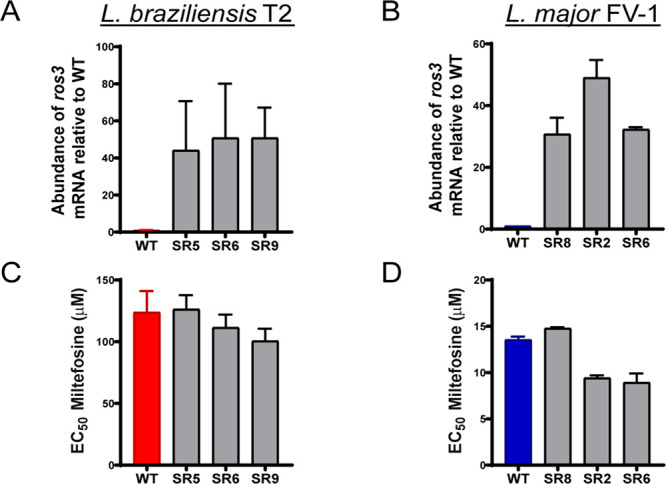
Relative abundance of the ros3 gene mRNA and phenotypic characterization in SR clones. Abundance of the ros3 mRNA was quantified in each SR clone and WT parasites of L. braziliensis T2 (A) and L. major FV-1 (B) by real-time qPCR. Normalization was done using tbp gene Ct values, and each bar represents the mean ± SEM abundance of the ros3 transcript relative to WT parasites obtained in two independent experiments using two biological and three technical replicates (A, B). Susceptibility to MF was evaluated in SR clones and WT parasites of L. braziliensis T2 (C) and L. major FV-1 (D) by an MTT assay. Each bar represents the mean ± SEM EC50 to MF obtained in three independent experiments performed in triplicate.
The effects of ros3 overexpression were investigated by an evaluation of log-phase promastigotes MF susceptibility in SR clones of L. braziliensis by 3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) (MTT). Compared to the EC50 determined for Lb WT (123.9 ± 17.08 μM), a nonstatistically significant decrease of 18% was observed for the clone SR9 (EC50 = 100.6 ± 9.94 μM) (Figure 2C).
In Lm SR clones the EC50 reduction was more pronounced and significant for two out of three clones. A 30% decrease in EC50 values was observed for clone SR8 (8.93 ± 0.97 μM) as compared to the EC50 calculated for the WT parasite (13.54 ± 0.34 μM) (Figure 2D). However, no significant correlation between the ros3 mRNA abundance and MF susceptibility was found for either L. braziliensis or L. major ros3 overexpressor clones (r = −0.800 and p = 0.333; r = −0.600 and p = 0.4167, Spearman’s correlation test, respectively).
Increasing the hygromycin selection pressure from 32 to 128 μg/mL did not lead to a more pronounced reduction in EC50 values in either species. In T2 SR clones, the EC50 of SR2 and SR5 decreased from 111.53 and 126.64 μM to 102.3 and 119.5 μM, respectively. In Lm SR clones the EC50 of SR2 and SR8 under increased hygromycin B pressure changed from 10.8 and 14.7 to 13.5 and 16.5 μM, respectively.
Partial or Complete Removal of ros3 Reduces Susceptibility and Uptake of MF in L. major
As an alternative strategy to evaluate the role of the ros3 gene dosage effect on MF susceptibility, an Lm strain constitutively expressing Cas9 and T7 RNA polymerase (Lm Cas9/T7) was used to generate mono (partial) and diallelic (complete) knockouts for ros3 using CRISPR/Cas9.
For that, small guide RNAs (sgRNAs) coding template and donor DNAs were generated by PCR in vitro and delivered to the parasite, driving the Cas9-mediated break and incorporation of the donor DNA containing a blasticidin resistance (BlastR) gene by homologous recombination, replacing the complete ros3 open read frame (ORF).
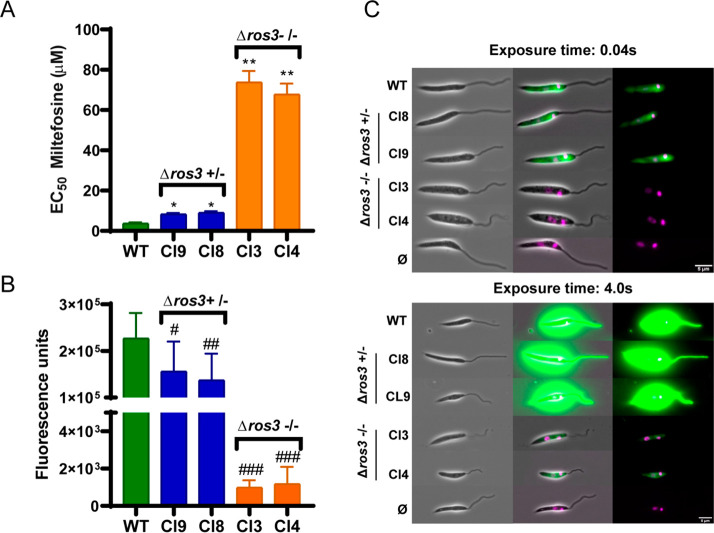
After the selection with blasticidin, single and complete knockouts (KOs) for the ros3 gene were confirmed by PCR in the heterogeneous parasite population that was previously cloned by serial dilution (Figure 3). To verify the presence or absence of ros3 in the clones recovered from ros3 KO experiments, primers annealing outside the ORF were used. The presence of the ros3 gene was identified by the presence of a 1.4 kb fragment, whereas its substitution by a Blast-R gene resulted in the amplification of a 1.9 kb fragment (Figure 3A). Six clones with incomplete and six clones with complete deletions of ros3 (Figure 3) were identified. Two incomplete (Cl8 and Cl9) and two complete (Cl3 and Cl4) ros3-deficient mutants were selected for MF susceptibility and uptake characterizations.
Figure 3.
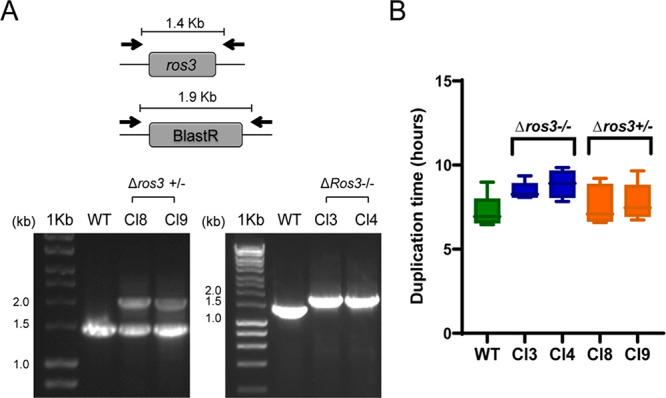
ros3 knockout verification in recovered clones. (A) Schematic representation of the strategy used for ORF KO verification in blasticidin-resistant clones. In diallelic knockouts, Blast-R was amplified and resulted in a 1.9 kb fragment, whereas in monoallelic ros3 KOs both Blast-R and ros3 are amplified resulting in two bands of 1.9 and 1.4 kb, respectively. (B) Parasite doubling time for each incomplete (Δros3 +/–±) and complete (Δros3 −/−) KO clones and for WT L. major Cas9/T7. Each box represents the mean ± SEM of the duplication time evaluated during 4 d.
To exclude the possible interference of growth rates between the mutants in the viability assays, the doubling time of the generated lines in parallel with the WT parasites was determined as described in the Methods Section. Although a mild increase in the doubling time was observed in complete knockouts (Cl3 and Cl4), this difference was not significant [analysis of variance (ANOVA) and Tukey’s multiple comparisons test] (Figure 3). Other features including size, shape, or motility were visually inspected under a bright field-inverted microscope. None of the clonal mutant population cultures presented differences detectable to the human eye (Figure 4C).
Figure 4.
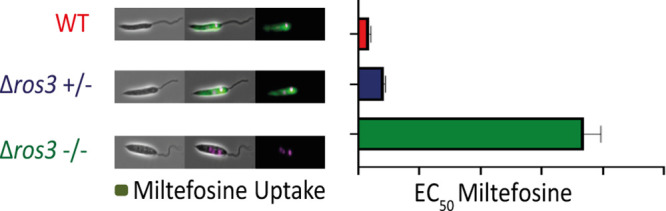
Susceptibility and uptake of MF in L. major ros3 mono- and diallelic KOs. (A) The EC50 of MF was determined for the L. major Cas9/T7 strain and for mono- and diallelic knockouts by MTT. Mean and SEM results of three or more independent experiments were calculated. (*) p < 0.005. (**) p 0.0001. (B) Uptake of MT-EtBDP in monoallelic (Cl8 and Cl9) and diallelic (Cl3 and Cl4) ros3 knockouts. (#) p < 0.01; (##) p < 0.001; (###) p < 0.0001. Uptake of MT-EtBDPY was evaluated by flow cytometry. A reduction in fluorescence (FLH-1) inside parasites was observed in ros3 incomplete knockouts. In complete knockouts, the reduction was even higher. (C) Evaluation of MT-EtBDP uptake in WT (Lm Cas9/T7) and in ros3 monoallelic (Cl8 and Cl9) and diallelic (Cl3 and Cl4) knockouts by fluorescence microscopy. Different exposure times of 0.04 and 4 s were used to confirm the presence of the labeled MF inside parasites. As a background, the control WT Cas9/T7 incubated without MT-EtBDPY was exposed to the same conditions for image acquisition.
The susceptibility of partial and complete knockouts to MF was determined by MTT. Initially, the susceptibility of L. major Cas9/T7 was compared to the L. major WT line (the background in which L. major Cas9/T7 was generated) in order to evaluate if the modified parasite could behave differently regarding MF susceptibility. No significant differences were observed between Lm WT (EC50 = 6.57 ± 0.89 μM) and the Lm WT Cas9/T7 strain (EC50 = 3.46 ± 0.48 μM) (non-parametric t-test).
When compared to Lm Cas9/T7 WT parasites, a significant approximately threefold reduction (p > 0.002) in EC50 values for MF was observed for monoallelic deleted ros3 mutants (Table 2). When the two alleles of ros3 were removed (diallelic deleted ros3 mutants), a more pronounced reduction in the susceptibility to MF (∼20-fold reduction) was detected (p < 0.0001) (Table 2 and Figure 4A).
Table 2. Susceptibility of L. major ros3 Nono- and Diallelic Knockouts to MF.
| sample | EC50 ± SEMa (μM) | activity indexb (AI) |
|---|---|---|
| Lm WT | 6.57 ± 0.89 | |
| Lm WT Cas9/T7 | 3.46 ± 0.48 | |
| Δros3 ± Cl8 | 10.22 ± 1.27 | 2.95 |
| Δros3 ± Cl9 | 9.66 ± 1.43 | 2.79 |
| Δros3 −/– Cl3 | 73.82 ± 5.53 | 21.33 |
| Δros3 −/– Cl4 | 71.47 ± 5.49 | 20.65 |
EC50 ± SEM of MF for promastigotes determined by MTT assay.
Activity index (AI) was calculated by dividing the EC50 of each clone by the EC50 of the reference parasite L. major Friedlin Cas9/T7.
The uptake of MF in mono- and diallelic knockouts was determined by flow cytometry using MF labeled with 11-(4′,4′-difluoro-6′-ethy[11-(4′,4′-difluoro-6′-ethyl-1′,3′,5′,7′-tetramethyl-4′-bora-3′a,4′a-diaza-s-indacen-2′-yl)-undecylphosphocholine] (MT-EtBDPY) and compared to Lm Cas9/T7 WT. In the absence of one allele of ros3, the amount of fluorescence inside the parasite presented a mild but significant reduction. However, in ros3 diallelic knockouts, the uptake is reduced 100-fold compared to that of WT Cas9/T7. These data suggested that, in the absence of one or two copies of ros3, the MF entry in L major is impaired (Figure 4B).
The drug uptake variation was also demonstrated by fluorescence microscopy (Figure 4C). With 0.04 s of exposure, no fluorescence was observed inside Ros3 null mutants, while positive labeling was seen in WT parasites and incomplete knockouts. However, after longer exposures (4 s), a weak MF fluorescence signal was observed in complete knockouts, demonstrating that some MF uptake happened even in the complete absence of ros3 (Figure 4C).
Discussion
The susceptibility to MF was found to be variable among L. braziliensis clinical isolates, raising the concern of intrinsic tolerance in isolates circulating in Brazil.26 A further investigation of the mechanisms behind the differential susceptibility to MF in these isolates revealed differences in drug uptake and in the abundance of the ros3 transcript, an essential component of the MF transport machinery.21,24 Considering the nature of gene expression regulation in Leishmania,30 the high genome plasticity,31,32 and the previous association of gene copy number variation (CNV) with drug resistance in these organisms,33 in this work we investigated whether or not a differential ros3 mRNA abundance in L. braziliensis clinical isolates was related to variability in ros3 gene dosage. Furthermore, using different DNA manipulation approaches we evaluated if susceptibility to MF could be modulated by the addition or removal of ros3 gene copies, mimicking a CNV condition.
A 0.5-fold increase in the ros3 DNA abundance was found in the L. braziliensis susceptible isolate when compared to tolerant isolates and the reference strain. This indicated the presence of an extra copy of ros3, the most likely reason for the increased abundance of ros3 mRNA observed. The ros3 extra copy may represent an isolated event of gene duplication or a chromosome 32 tetrasomy, but further investigation about the chromosome content in these isolates has not been performed yet.
These findings then led us to investigate whether an alteration in the ros3 gene dosage was enough to modulate the susceptibility to MF. On the one hand, the integration of an extra ros3 copy in the genome significantly increased the mRNA abundance in overexpressing clones. However, the accumulation of ros3 transcripts did not lead to significant changes in the MF susceptibility in these parasites, suggesting that the increase of ros3 transcripts alone was not capable of modulating the MF susceptibility in L. braziliensis T2 and L. major. On the other hand, the generation of mono- and diallelic ros3 knockouts using CRISPR/Cas9 in L.major led to 2-fold and 20-fold increases in the EC50 to MF, respectively.
The observation of the unchanged susceptibility to MF in L. braziliensis T2 and in L. major was surprising. The expression system employed for ros3 overexpression herein (pLEXSY) is capable of inducing the expression of exogenous and endogenous genes in different Leishmania species, including L. braziliensis.34−37 Moreover, differential ros3 and MT expressions have already been shown by others to play a role in the susceptibility of Leishmania to MF.22 An L. braziliensis Peruvian isolate and the Brazilian reference strain M2904 were shown to be 6–10-fold less susceptible to MF when compared to L. donovani due to a reduced expression of Ros3 in the plasma membrane of L. braziliensis.22 This is the same range of variation in susceptibility to MF encountered among Brazilian L. braziliensis clinical isolates.26 Additionally, the same study showed that L. braziliensis overexpressing ros3 demonstrated a 3.5-fold reduction in MF EC50. Importantly, they have shown that, in this context of ros3 overexpression in Leishmania, there is an increase not only in Ros3 protein abundance in plasma membrane but also of MT protein, suggesting that ros3 overexpression triggers an endogenous MT overexpression, since the complete MT-Ros3 complex is essential for MF uptake.22 It is possible therefore that the susceptibility of Ros3 overexpressor mutants was unchanged because they lacked the necessary MT to compound the transporter complex MT-Ros3.
However, one important limitation in this study is the lack of a demonstration of an increased abundance of the Ros3 protein in the overexpressing mutants. Various attempts of immunodetection and protein tagging did not produce clear-cut results, so this remains to be achieved. Therefore, we must consider biological factors that could explain the lack of phenotype in the overexpressor mutants. An overexpression was achieved using the L. braziliensis T2 ros3 coding sequence upstream to a heterologous UTR element. The lack of ros3 UTR elements may have hampered a proper mRNA processing and translation.38 Where overexpression in L. major is concerned, the limited identity (72%) between L. braziliensis and L. major ros3 coding sequences could potentially lead to interference in Ros3 folding, interaction with MT, and membrane insertion when expressed in L. major.23
Moreover, besides ros3, another 35 genes were shown to be differentially expressed between sensitive and tolerant isolates,21 and those could represent indispensable partners for an effective change in the MF uptake. Therefore, increasing the ros3 transcript abundance through the methodology employed herein was not enough to sensitize L. braziliensis T2 and L. major to MF.
However, results obtained employing a loss of function approach revealed that the knockout of ros3 modulates the susceptibility and uptake of MF in a gene-dosage-dependent way. The generation of complete and incomplete ros3 knockout in L. major caused a significant decrease in uptake and in the susceptibility to MF, suggesting that the presence of this gene is critical for the susceptibility to MF. Similar results were observed in L. donovani, which presented a reduction in the susceptibility to MF of 1.7- and 14.2-fold in ros3 mono- and diallelic knockouts, respectively, suggesting that the MF tolerance phenotype caused by the reduction in ros3 gene dosage is not a species-specific phenotype. Interestingly, these values are also comparable to the reduction of 1.9- and 13.7-fold observed in the context of MT mono- and diallelic knockouts, which reinforces the codependence of both proteins.24
In addition, the complete removal of ros3 did not abolish MF internalization completely, as residual fluorescent MF was observed inside the diallelic knockouts, suggesting that other routes for the internalization of MF, such as endocytosis or diffusion after incorporation into cell membranes, may be involved, even if poorly.39 If in the absence of the MT-Ros3 complex, diffusion through the membrane occurs in significant levels, variations in the composition and structure of plasma membrane in Leishmania parasites might also play a role in a differential susceptibility to MF.
To the best of our knowledge, this is the first demonstration of ros3 gene dosage described for Leishmania clinical isolates associated with a differential susceptibility and uptake of miltefosine. However, the dependence of the MT-Ros3 complex for MF transport has been repeatedly shown as the Achille’s heel of MF efficacy in Leishmania parasites either by acquisition of inactivating mutations in these genes15,19,20,40,41 or by a differential expression of this complex in Leishmania plasma membrane.22
Taken together, these results reinforced the role of the Ros3 subunit as a limiting factor for the MF uptake in Leishmania parasites and demonstrated for the first time that the ros3 gene dosage plays a role in a differential susceptibility to MF not only in L. braziliensis isolates never exposed to MF but also in L. major.
The high cure rates,42,43 together with the high intracellular concentrations of MF achieved during therapy44,45 and the low number of cases of resistant parasites recovered after treatment with MF,13,15 are good indicatives that the variations observed in these L. braziliensis isolates are not enough to cause a treatment failure. However, it is important once again to highlight that MT-Ros3 is repeatedly being described as the cause of susceptibility reduction, not only in parasites selected in vitro under drug pressure but also in an isolate recovered after VL treatment with MF failure.15 In this scenario, our findings highly encourage the search for new drug therapy schemes, such as drug combinations that could enhance the MF activity, or even modifications in MF molecule that could promote the entrance of the drug by an alternative route in an attempt to avoid selection of resistant parasites and loss of the only effective oral drug for leishmaniasis treatment.
Conclusions
Being the only oral drug currently in use for leishmaniasis treatment, preventing a loss of MF due to resistance is a necessary effort. Our study reinforce the role of MT-Ros3 machinery in MF resistance by showing the direct effect of ros3 gene dosage in MF susceptibility and uptake not only in long-term laboratory-cultured Leishmania reference strains or parasites selected for resistance but also in L. braziliensis clinical isolates not previously exposed to MF. Our results encourage the search for new variants of MF molecule, different drug-delivery systems, or even coadministration with other molecules that could enhance MF transport and overcome the stringent dependence of active transport through the MT-Ros3 complex.
Methods
Chemical Compounds
Miltefosine and MTT were purchased from Sigma-Aldrich and diluted in sterile water and phosphate-buffered solution (PBS), respectively. The BODIPY-labeled MF MT-EtBDP was kindly donated by Dr. A. U. Acuña (Instituto de Química-Física “Rocasolano”, CSIC) and prepared as described.46 Hygromycin B and blasticidin S hydrochloride were purchased from Melford Laboratories Ltd.
Cultivation of Leishmania Parasites
The cell lines used in this work were: the parental strain of L. major Friedlin FV-1 (MHOM/IL/1980/Friedlin) (Lm) and the modified Cas9/T7-expressing L. major Friedlin FV-1 (Lm Cas9/T7);47 the L. braziliensis reference strain (RS) (MHOM/BR/1975/M2903) and three L. braziliensis Brazilian clinical isolates, namely, MHOM/BR/2005/LTCP16012 (named S, for Sensitive), MHOM/BR/2006/LTCP16907 (T1, for Tolerant 1), and MHOM/BR/2009/LTCP19446 (T2, for Tolerant 2). The susceptibility of these isolates to MF was previously reported26 (Table 1).
Promastigotes of Leishmania were cultivated at 28 °C in M199 medium (Sigma-Aldrich) supplemented with 2.2 g/L NaHCO3, 0.005% hemein, 40 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), pH 7.4, and 10% heat-inactivated fetal calf serum (FCS). For L. braziliensis, 2% male urine was added to the culture. Transfectants were maintained in MM199 media, which was made by diluting M199 media powder and supplementing with 2.2 g/L NaHCO3, 0.0025% hemein, 40 mM HEPES, pH 7.4, 0.1 mM adenine hemisulfate, 1.2 μg/mL biopterin, and 20% FCS. The appropriate selection drug was added at 32 μg mL–1 hygromycin B or 5 μg mL–1 blasticidin S hydrochloride.
Generation of Parasites Overexpressing of ros3 Gene
For the ros3 overexpression in the L. braziliensis T2 isolate and L. major Friedlin FV-1 strain, the open reading frame of the gene ros3 (LbrM.32.0580) was amplified from L. braziliensis T2 total DNA using the primer pair SR_BglII-Fow and SR_NotI-Rev (Table S1) and cloned into the plasmid pLEXSY-hyg2 (Jena Biosciences). In this expression vector, the gene of interest is under the regulation of L. tarentolae adenine phosphoribosyl transferase (aprt) UTR at 5′ and calmodulin (camCB) UTR at 3′.48 The generated construct (SR) was then linearized with the restriction enzyme SwaI, and purified cassette pLEXSY-hyg2-SR was delivered to L. braziliensis T2 isolate and L. major Friedlin FV-1 parasites for integration in the small subunit of rDNA (SSU) by electroporation, as previously described.49 Transgenic parasites overexpressing the ros3 gene (SR clones) were selected in a semisolid M199 medium supplemented with 32 μg/mL hygromycin B as described.50 Recovered clones were then maintained in liquid M199 in the presence of 32 μg/mL hygromycin B. Genomic DNA was extracted using the protocol described by Rotureau et al.,51 and integration of the SR cassette into the SSU rDNA locus was confirmed by PCR using the primers provided with pLEXSY-hyg2 F3001, A264, F3002, and A384 (Jena Bioscience) (Table S1).
Quantitative Real-Time PCR
The abundance of ros3 mRNA and DNA in SR clones and isolates was quantified by real-time RT-PCR (qPCR) using total RNA or genomic DNA as templates, respectively. For the mRNA quantification, cDNA was synthesized from total RNA using MuLV Reverse Transcriptase (Applied Biosystems). Briefly, 6 μg of DNase-treated RNA was incubated with 1 μg of random primers (Thermo Fischer Scientific) for 10 min at 70 °C. After this period 1X MulV-RT buffer, 0.01 mM dithiothreitol (DTT), 5.5 mM MgCl2, and 1 mM dNTPs were added to the system and incubated at 42 °C for 2 min. Reverse Transcriptase was then added to the RT+ tubes but not the RT– tubes (control of DNA absence), and the reaction was incubated at 25 °C for 10 min, followed by incubations at 48 °C for 30 min and at 95 °C for 5 min according to the manufacturer’s instructions.
One hundred nanograms of in vitro synthesized cDNA was used a as template for qPCR, which was performed in a StepOne Plus System (Applied Bios Systems) using SYBR Green PCR Master Mix (Thermo Fisher Scientific). The following program was used: 95 °C for 10 min followed by 40 cycles at 95 °C for 15s, 60 °C for 60 s, and 72 °C for 20 s. The 163 base pair (bp) ros3 gene fragment was amplified using the primer pair LbLm_Ros3-F and LbLm_Ros3-R (Table S1). The housekeeping glyceraldehyde 3-phosphate dehydrogenase (gapdh) and tata-box-binding protein (tbp) coding genes were used for normalization and amplified using the primer pair gapdh-F and gapdh-R and Lb_tbp-F/Lm_tbp-F and LbLm_tbp-R, respectively (Table S1). Three biological replicates and three technical replicates of each sample were evaluated for ros3 and gapdh/tbp mRNA and DNA abundance determination. The threshold cycle (Ct) obtained for ros3 in each sample was normalized by the Ct of the gapdh/tbp genes. The 2–ΔΔCt equation was used to determine the expression of ros3 genes relative to RS, in the case of isolates, or to the wild type (WT) parasites when SR clones were characterized, respectively.52 2–ΔΔCt values were then plotted on GraphPad Prism 6, and statistical analyses were performed using a one-way ANOVA analysis followed by Tukey’s multiple comparison tests.
For quantifying the copy number of the ros3 gene, the samples were submitted to real-time PCR together with a standard curve using the pGEM-T-Ros3-M2903 plasmid (previously available in the laboratory).21 A linear regression of Ct values and ros3 molecule number was constructed based on the data obtained for the standard curve and employed to determine the number of molecules in each sample. After normalization by tbp or gapdh molecule values, the relative abundance of ros3 was calculated by dividing the normalized amount of ros3 in each isolate by the amount in RS.
Generation of Mono- and Diallelic Knockouts for the ros3 Gene
Knockout mutants of this study were generated by CRISPR/Cas9 technology and the LeishGEdit toolkit on the background of L. major overexpressing Cas9/T7.53 Primer sequences for the PCR generation of sgRNA templates and donor DNAs for the ros3 gene ID (LmjF.32.0510) were selected using LeishGEdit (http://www.leishgedit.net/).53
The sgRNA templates for target gene cleavage were generated by PCR reactions using the G00 primer together with the 5′ (LmRos3_5′sgRNA) or 3′sgRNA (LmRos3_3′sgRNA) LeishGEdit primers in individual tubes (Table S1). Donor DNA for generation of ros3 knockouts was also obtained by PCR reactions using the pTBlast_v1 plasmid as a DNA template and Upstream Forward Primer (LmRos3_UFP) and Downstream Reverse Primer (LmRos3_DRP) initiators (Table S1). Detailed protocols used for PCR reactions are described in Beneke and Gluenz.53
The delivery of sgRNA templates and donor DNA to 1 × 107 Lm Cas9/T7 log-phase promastigotes was done by a transfection with Amaxa Nucleofector using program X-001 in a transfection buffer as previously described.53 After transfections, parasites were added to flasks containing MM199, and after 6 h, 5 μg/mL blasticidin was added to the culture.53 After two splits in 1:100 proportion, the recovered population was cloned in 96-well plates in three different proportions 0.1, 1.0, and 10 promastigotes/ml. Population was considered to be clonal when no more than 30% of the wells in each dilution presented growth.
Monoallelic (single) and diallelic (double) knockouts were verified through PCR reactions using primers LmRos3_UTR-F and LmRos3_UTR-R, which anneal outside the ros3 ORF. The presence of WT ros3 results in the amplification of a 1.4 kb fragment, whereas the substitution by the Blast-R gene would result in the amplification of a 1.9 kb fragment. DNA obtained from the ros3 transfectant population and of Lm Cas9/T7 were used as positive controls for partial KO and target gene presence, respectively.
Doubling Time Measurement
For doubling time characterization in mutants and WT parasites, the culture density was adjusted to 1 × 106 promastigotes/mL in M199. After an incubation for 24 h at 28 °C, the cell culture density was determined using a cell counter (CASY model TT, Roche Diagnostics) with a 60 μm capillary and exclusion of particles with a pseudo diameter below 2.0 μm. The cell density was adjusted again to 1 × 106 promastigotes/mL in a new flask. This procedure was repeated for 4 d. The doubling time (DT) was calculated using the following formula.
| 1 |
Susceptibility Assays
The susceptibility to MF was determined by an MTT assay.54 Briefly, 2 × 106 (L. braziliensis) or 2 × 105 (L. major) log-phase promastigotes were incubated in the presence of increasing concentrations of MF for 24 h (SR clones) or 48 h (knockout mutants). MF concentrations employed for L. braziliensis were 400, 280, 240, 200, 140, 120, 100, 70, 60, 50, and 35 μM, and for L. major they were 120, 90, 80, 70, 60, 45, 30, 15, 7.5, 3.75, and 1.875 μM. The cell viability was then determined by an incubation with 5 mg/mL MTT followed by cell lysis with 4% SDS and optical density (OD) measurement at 690 and 595 nm. OD values were then converted in EC50 values by sigmoidal regression curves using GraphPad Prism 6 software. Susceptibility assays were conducted in triplicate, and at least three independent experiments were performed.
Uptake of MT-EtBDPY
The uptake of MT-EtBDPY was evaluated as described in Espada et al.21 Briefly, log-phase Leishmania promastigotes were incubated in HEPES-NaCl buffer (21 mM HEPES, 137 mM NaCl, 5 mM KCl, 0.7 mM NaH2PO4, 6 mM glucose, pH 7.05) supplemented with 0.3% (w/v) bovine serum albumin (BSA) and 500 μM phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich) for 15 min at 28 °C. After this period, 1 μM MT-EtBDP was added, and the incubation was continued for 5 min at 28 °C. Parasites were washed three times with HEPES-NaCl containing 0.3% BSA to remove the noninternalized labeled molecules. The parasites were then suspended in PBS, and the fluorescence intensity was measured using a BD Accuri C6 flow cytometer (BD Biosciences). Statistical analyses were performed using one-way ANOVA followed by Tukey’s multiple comparison tests using Graph Pad Prism 6. Values are reported with the standard error of measure (SEM).
Fluorescence Microscopy
For MT-EtBDP uptake analysis, after noninternalized molecules were washed, a fraction of the parasites was incubated in PBS with 10 μg/mL Hoechst 33342. Parasites were pelleted, suspended in PBS, and then placed on a microscope slide inside a small area marked with a liquid blocker pen. A coverslip was applied, and the living cells were immediately imaged in a Zeiss Axioimager.Z2 microscope with a 63× numerical aperture (NA) 1.40 oil immersion objective and a Hamamatsu ORCA-Flash4.0 camera. The filters used for Hoechst 33342 and MT-EtBDP were 350/461 nm (excitation/emission) and 527/536 nm, respectively. As a background control, WT Cas9/T7 untagged and/or that did not receive the ligands or fluorescent molecules were imaged using the specific filters at the same exposure time (0.04 and 4 s). Images were processed using Fiji.55
Acknowledgments
We want to thank J. K. U. Yokoyama-Yasunaka and H. Banks (supported by Wellcome Trust Grant [104627/Z/14/Z] to K. Gull) for the valuable lab support. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2015/09080-2, 2016/23405-4, 2016/21171-6 and 2018/25299-2). S.R.B.U. is the recipient of a senior researcher scholarship from CNPq. A.C.C. has also, in part, received funding from UK Research and Innovation via the Global Challenges Research Fund under the grant “A Global Network for Neglected Tropical Diseases” (Grant No. MR/P027989/1). V.H. acknowledges the predoctoral Grant FPI (2004-2009) from Ministerio de Educación y Ciencia of Spain (BQU2003-04413) and the Contract RYC-2017-22294 from the Spanish Ministerio de Ciencia e Innovación. A.A.-W. is the recipient of a Marie Sklodowska-Curie Individual Fellowship (transLEISHion-EU FP7, No. 798736) and acknowledges Fundação para a Ciência e a Tecnologia for funds to GHTM (UID/04413/2020). E.G. is a Royal Society University Research Fellow and supported through the WCIP core Wellcome Centre Award No. 104111/Z/14/Z.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00857.
Author Present Address
∇ Caroline R. Espada: Departamento de Biologia Celular e Molecular e Bioagentes Patogênicos, Faculdade de Medicina de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, São Paulo, Brazil.
Author Contributions
C.R.E.: Conceptualization, investigation, formal analysis, methodology, visualization, writing, reviewing and editing. A.A.-W.: Conceptualization, methodology, formal analysis, reviewing, and editing. V.H.: development of MT-EtBDP and reviewing. E.G.: Conceptualization, methodology, formal analysis, reviewing, and editing. A.C.C.: Conceptualization, methodology, writing, reviewing, and editing. S.R.B.U.: Conceptualization, methodology, formal analysis, writing, reviewing, and editing.
The authors declare no competing financial interest.
Supplementary Material
References
- Grimaldi G. Jr.; Tesh R. B.; McMahon-Pratt D. (1989) A review of the geographic distribution and epidemiology of leishmaniasis in the New World. Am. J. Trop. Med. Hyg. 41 (6), 687–725. 10.4269/ajtmh.1989.41.687. [DOI] [PubMed] [Google Scholar]
- Silveira F. T. (2019) What makes mucosal and anergic diffuse cutaneous leishmaniases so clinically and immunopathogically different? A review in Brazil. Trans. R. Soc. Trop. Med. Hyg. 113, 505. 10.1093/trstmh/trz037. [DOI] [PubMed] [Google Scholar]
- Cincurá C.; de Lima C. M. F.; Machado P. R. L.; Oliveira-Filho J.; Glesby M. J.; Lessa M. M.; Carvalho E. M. (2017) Mucosal leishmaniasis: A Retrospective Study of 327 Cases from an Endemic Area of Leishmania (Viannia) braziliensis. Am. J. Trop. Med. Hyg. 97 (3), 761–766. 10.4269/ajtmh.16-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burza S.; Croft S. L.; Boelaert M. (2018) Leishmaniasis. Lancet 392 (10151), 951–970. 10.1016/S0140-6736(18)31204-2. [DOI] [PubMed] [Google Scholar]
- Uliana S. R. B.; Trinconi C. T.; Coelho A. C. (2018) Chemotherapy of leishmaniasis: present challenges. Parasitology 145 (4), 464–480. 10.1017/S0031182016002523. [DOI] [PubMed] [Google Scholar]
- Machado P. R., Ampuero J., Guimaraes L. H., Villasboas L., Rocha A. T., Schriefer A., Sousa R. S., Talhari A., Penna G., and Carvalho E. M.. Miltefosine in the treatment of cutaneous leishmaniasis caused by Leishmania braziliensis in Brazil: a randomized and controlled trial. PLoS neglected tropical diseases 2010, 4 ( (12), ). 10.1371/journal.pntd.0000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado P. R. L.; Prates F. V. O.; Boaventura V.; Lago T.; Guimarães L. H.; Schriefer A.; Corte T. W. F.; Penna G.; Barral A.; Barral-Netto M.; Carvalho E. M. (2020) A double-bind and randomized trial to evaluate Miltefosine and topical GM-CSF in the treatment of cutaneous leishmaniasis caused by Leishmania braziliensis in Brazil. Clin. Infect. Dis. 10.1093/cid/ciaa1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyoto T., Potet J., and Boelaert M.. Why miltefosine-a life-saving drug for leishmaniasis-is unavailable to people who need it the most. BMJ. global health 2018, 3 ( (3), ). 10.1136/bmjgh-2018-000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil, Ministério da Saúde. Secretaria de Ciência, Tecnologia e Insumos Estratégicos. Comissão Nacional de Inocorporação de Tecnologia no SUS Miltefosina para o tratamento da Leishmaniose Tegumentar. http://conitec.gov.br/images/Relatorios/2018/Relatorio_Miltefosina_LeishmanioseTegumentar.pdf (accessed 2021-03-14).
- Chrusciak-Talhari A.; Dietze R.; Chrusciak Talhari C.; da Silva R. M.; Gadelha Yamashita E. P.; de Oliveira Penna G.; Lima Machado P. R.; Talhari S. (2011) Randomized controlled clinical trial to access efficacy and safety of miltefosine in the treatment of cutaneous leishmaniasis Caused by Leishmania (Viannia) guyanensis in Manaus, Brazil. Am. J. Trop. Med. Hyg. 84 (2), 255–260. 10.4269/ajtmh.2011.10-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorlo T. P.; Balasegaram M.; Beijnen J. H.; de Vries P. J. (2012) Miltefosine: a review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. J. Antimicrob. Chemother. 67 (11), 2576–2597. 10.1093/jac/dks275. [DOI] [PubMed] [Google Scholar]
- Sundar S.; Singh A.; Rai M.; Prajapati V. K.; Singh A. K.; Ostyn B.; Boelaert M.; Dujardin J. C.; Chakravarty J. (2012) Efficacy of miltefosine in the treatment of visceral leishmaniasis in India after a decade of use. Clin. Infect. Dis. 55 (4), 543–550. 10.1093/cid/cis474. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Mishra J., Gupta A. K., Singh A., Shankar P., and Singh S.. Laboratory confirmed miltefosine resistant cases of visceral leishmaniasis from India. Parasites & Vectors 2017, 10 ( (1), ). 10.1186/s13071-017-1969-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal S.; Ostyn B.; Uranw S.; Rai K.; Bhattarai N. R.; Dorlo T. P.; Beijnen J. H.; Vanaerschot M.; Decuypere S.; Dhakal S. S.; Das M. L.; Karki P.; Singh R.; Boelaert M.; Dujardin J. C. (2013) Increasing failure of miltefosine in the treatment of kala-azar in Nepal and the potential role of parasite drug resistance, reinfection, or noncompliance. Clin. Infect. Dis. 56 (11), 1530–1538. 10.1093/cid/cit102. [DOI] [PubMed] [Google Scholar]
- Mondelaers A., Sanchez-Canete M. P., Hendrickx S., Eberhardt E., Garcia-Hernandez R., Lachaud L., Cotton J., Sanders M., Cuypers B., Imamura H., Dujardin J. C., Delputte P., Cos P., Caljon G., Gamarro F., Castanys S., and Maes L.. Genomic and Molecular Characterization of Miltefosine Resistance in Leishmania infantum Strains with Either Natural or Acquired Resistance through Experimental Selection of Intracellular Amastigotes. PloS one 2016, 11 ( (4), ). 10.1371/journal.pone.0154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojean S.; Houze S.; Haouchine D.; Huteau F.; Lariven S.; Hubert V.; Michard F.; Bories C.; Pratlong F.; Le Bras J.; Loiseau P. M.; Matheron S. (2012) Leishmania resistance to miltefosine associated with genetic marker. Emerging Infect. Dis. 18 (4), 704–706. 10.3201/eid1804.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnielli J. B. T.; Monti-Rocha R.; Costa D. L.; Molina Sesana A.; Pansini L. N. N.; Segatto M.; Mottram J. C.; Costa C. H. N.; Carvalho S. F. G.; Dietze R. (2019) Natural Resistance of Leishmania infantum to Miltefosine Contributes to the Low Efficacy in the Treatment of Visceral Leishmaniasis in Brazil. Am. J. Trop. Med. Hyg. 101 (4), 789–794. 10.4269/ajtmh.18-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria F. J.; Gamarro F.; Ouellette M.; Castanys S. (2003) Functional cloning of the miltefosine transporter. A novel P-type phospholipid translocase from Leishmania involved in drug resistance. J. Biol. Chem. 278 (50), 49965–49971. 10.1074/jbc.M308352200. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria F. J.; Castanys S.; Gamarro F. (2003) Leishmania donovani resistance to miltefosine involves a defective inward translocation of the drug. Antimicrob. Agents Chemother. 47 (8), 2397–2403. 10.1128/AAC.47.8.2397-2403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho A. C., Trinconi C. T., Costa C. H., and Uliana S. R.. In Vitro and In Vivo Miltefosine Susceptibility of a Leishmania amazonensis Isolate from a Patient with Diffuse Cutaneous Leishmaniasis. PLoS Negl Trop Dis 2014, 8 ( (7), ). 10.1371/journal.pntd.0002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada C. R.; Magalhaes R. M.; Cruz M. C.; Machado P. R.; Schriefer A.; Carvalho E. M.; Hornillos V.; Alves J. M.; Cruz A. K.; Coelho A. C.; Uliana S. R. B. (2019) Investigation of the pathways related to intrinsic miltefosine tolerance in Leishmania (Viannia) braziliensis clinical isolates reveals differences in drug uptake. Int. J. Parasitol.: Drugs Drug Resist. 11, 139. 10.1016/j.ijpddr.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Canete M. P.; Carvalho L.; Perez-Victoria F. J.; Gamarro F.; Castanys S. (2009) Low plasma membrane expression of the miltefosine transport complex renders Leishmania braziliensis refractory to the drug. Antimicrob. Agents Chemother. 53 (4), 1305–1313. 10.1128/AAC.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sánchez S.; Sánchez-Cañete M. P.; Gamarro F.; Castanys S. (2014) Functional role of evolutionarily highly conserved residues, N-glycosylation level and domains of the Leishmania miltefosine transporter-Cdc50 subunit. Biochem. J. 459 (1), 83–94. 10.1042/BJ20131318. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria F. J.; Sanchez-Canete M. P.; Castanys S.; Gamarro F. (2006) Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J. Biol. Chem. 281 (33), 23766–23775. 10.1074/jbc.M605214200. [DOI] [PubMed] [Google Scholar]
- Hanson P. K.; Malone L.; Birchmore J. L.; Nichols J. W. (2003) Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J. Biol. Chem. 278 (38), 36041–36050. 10.1074/jbc.M305263200. [DOI] [PubMed] [Google Scholar]
- Espada C. R.; Ribeiro-Dias F.; Dorta M. L.; Pereira L. I. A.; Carvalho E. M.; Machado P. R.; Schriefer A.; Yokoyama-Yasunaka J. K. U.; Coelho A. C.; Uliana S. R. B. (2017) Susceptibility to Miltefosine in Brazilian Clinical Isolates of Leishmania (Viannia) braziliensis. Am. J. Trop Med. Hyg 96 (3), 656–659. 10.4269/ajtmh.16-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing T.; Imamura H.; Decuypere S.; Clark T. G.; Coombs G. H.; Cotton J. A.; Hilley J. D.; de Doncker S.; Maes I.; Mottram J. C.; Quail M. A.; Rijal S.; Sanders M.; Schonian G.; Stark O.; Sundar S.; Vanaerschot M.; Hertz-Fowler C.; Dujardin J. C.; Berriman M. (2011) Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 21 (12), 2143–2156. 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprohon P.; Legare D.; Raymond F.; Madore E.; Hardiman G.; Corbeil J.; Ouellette M. (2009) Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 37 (5), 1387–1399. 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Freue G.; Holzer T. R.; Forney J. D.; McMaster W. R. (2007) Global gene expression in Leishmania. Int. J. Parasitol. 37 (10), 1077–1086. 10.1016/j.ijpara.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Clayton C. E. (2016) Gene expression in Kinetoplastids. Curr. Opin. Microbiol. 32, 46–51. 10.1016/j.mib.2016.04.018. [DOI] [PubMed] [Google Scholar]
- Cupolillo E.; Brahim L. R.; Toaldo C. B.; De Oliveira-Neto M. P.; De Brito M. E.; Falqueto A.; De Farias Naiff M.; Grimaldi G. Jr. (2003) Genetic Polymorphism and Molecular Epidemiology of Leishmania (Viannia) braziliensis from Different Hosts and Geographic Areas in Brazil. J. Clin Microbiol 41 (7), 3126–3132. 10.1128/JCM.41.7.3126-3132.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebede A.; De Doncker S.; Arevalo J.; Le Ray D.; Dujardin J. C. (1999) Size-polymorphism of mini-exon gene-bearing chromosomes among natural populations of Leishmania, subgenus Viannia. Int. J. Parasitol. 29 (4), 549–557. 10.1016/S0020-7519(99)00010-7. [DOI] [PubMed] [Google Scholar]
- Laffitte M. N., Leprohon P., Papadopoulou B., and Ouellette M.. Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000Research 2016, 5. 10.12688/f1000research.9218.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdag E. M.; Cirstea I. C.; Breitling R.; Lukes J.; Blankenfeldt W.; Alexandrov K. (2010) Purification and crystallization of human Cu/Zn superoxide dismutase recombinantly produced in the protozoan Leishmania tarentolae. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 66, 871–877. 10.1107/S1744309110019330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos M. S. e; Souza L. A. d.; Onofre T. S.; Silva Junior A.; Almeida M. R. d.; Bressan G. C.; Fietto J. L. R. (2017) Achievement of constitutive fluorescent pLEXSY-egfp Leishmania braziliensis and its application as an alternative method for drug screening in vitro. Memorias do Instituto Oswaldo Cruz 112 (2), 155–159. 10.1590/0074-02760160237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei Z., Van Reet N., Pouladfar G., Kühne V., Ramezani A., Sarkari B., Pourabbas B., and Büscher P.. Expression of a rK39 homologue from an Iranian Leishmania infantum isolate in Leishmania tarentolae for serodiagnosis of visceral leishmaniasis. Parasites & vectors 2019, 12 ( (1), ). 10.1186/s13071-019-3839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza L. M. B., Carvalho J., Bates M. D., Petterle R. R., Thomaz-Soccol V., and Bates P. A.. Production of a kinesin-related recombinant protein (Lbk39) from Leishmania braziliensis by Leishmania tarentolae promastigotes and its application in the serodiagnosis of leishmaniasis. One health (Amsterdam, Netherlands) 2019, 8. 10.1016/j.onehlt.2019.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas Castro F.; Ruy P. C.; Nogueira Zeviani K.; Freitas Santos R.; Simoes Toledo J.; Kaysel Cruz A. (2017) Evidence of putative non-coding RNAs from Leishmania untranslated regions. Mol. Biochem. Parasitol. 214, 69–74. 10.1016/j.molbiopara.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Fernandes K. S.; de Souza P. E.; Dorta M. L.; Alonso A. (2017) The cytotoxic activity of miltefosine against Leishmania and macrophages is associated with dynamic changes in plasma membrane proteins. Biochimica et biophysica acta. Biochim. Biophys. Acta, Biomembr. 1859 (1), 1–9. 10.1016/j.bbamem.2016.10.008. [DOI] [PubMed] [Google Scholar]
- Coelho A. C., Leprohon P., and Ouellette M.. Generation of Leishmania hybrids by whole genomic DNA transformation. PLoS Negl Trop Dis 2012, 6 ( (9), ). 10.1371/journal.pntd.0001817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw C. D.; Lonchamp J.; Downing T.; Imamura H.; Freeman T. M.; Cotton J. A.; Sanders M.; Blackburn G.; Dujardin J. C.; Rijal S.; Khanal B.; Illingworth C. J.; Coombs G. H.; Carter K. C. (2016) In vitro selection of miltefosine resistance in promastigotes of Leishmania donovani from Nepal: genomic and metabolomic characterization. Mol. Microbiol. 99 (6), 1134–1148. 10.1111/mmi.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar S.; Jha T. K.; Thakur C. P.; Engel J.; Sindermann H.; Fischer C.; Junge K.; Bryceson A.; Berman J. (2002) Oral miltefosine for Indian visceral leishmaniasis. N. Engl. J. Med. 347 (22), 1739–1746. 10.1056/NEJMoa021556. [DOI] [PubMed] [Google Scholar]
- Jha T. K.; Sundar S.; Thakur C. P.; Bachmann P.; Karbwang J.; Fischer C.; Voss A.; Berman J. (1999) Miltefosine, an oral agent, for the treatment of Indian visceral leishmaniasis. N. Engl. J. Med. 341 (24), 1795–1800. 10.1056/NEJM199912093412403. [DOI] [PubMed] [Google Scholar]
- Castro M. D., Gomez M. A., Kip A. E., Cossio A., Ortiz E., Navas A., Dorlo T. P., and Saravia N. G.. Pharmacokinetics of Miltefosine in Children and Adults with Cutaneous Leishmaniasis. Antimicrob. Agents Chemother. 2017, 61 ( (3), ). 10.1128/aac.02198-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorlo T. P.; van Thiel P. P.; Huitema A. D.; Keizer R. J.; de Vries H. J.; Beijnen J. H.; de Vries P. J. (2008) Pharmacokinetics of miltefosine in Old World cutaneous leishmaniasis patients. Antimicrob. Agents Chemother. 52 (8), 2855–2860. 10.1128/AAC.00014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornillos V.; Carrillo E.; Rivas L.; Amat-Guerri F.; Acuna A. U. (2008) Synthesis of BODIPY-labeled alkylphosphocholines with leishmanicidal activity, as fluorescent analogues of miltefosine. Bioorg. Med. Chem. Lett. 18 (24), 6336–6339. 10.1016/j.bmcl.2008.10.089. [DOI] [PubMed] [Google Scholar]
- Beneke T.; Madden R.; Makin L.; Valli J.; Sunter J.; Gluenz E. (2017) A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. Royal Society open science 4, 1–16. 10.1098/rsos.170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T. (2012) Recombinant protein production in the eukaryotic protozoan parasite Leishmania tarentolae: a review. Methods Mol. Biol. (N. Y., NY, U. S.) 824, 307–315. 10.1007/978-1-61779-433-9_15. [DOI] [PubMed] [Google Scholar]
- Coburn C. M.; Otteman K. M.; McNeely T.; Turco S. J.; Beverley S. M. (1991) Stable DNA transfection of a wide range of trypanosomatids. Mol. Biochem. Parasitol. 46 (1), 169–179. 10.1016/0166-6851(91)90210-W. [DOI] [PubMed] [Google Scholar]
- Beverley S. M.; Coburn C. M. (1990) Recurrent de novo appearance of small linear DNAs in Leishmania major and relationship to extra-chromosomal DNAs in other species. Mol. Biochem. Parasitol. 42 (1), 133–141. 10.1016/0166-6851(90)90121-2. [DOI] [PubMed] [Google Scholar]
- Rotureau B.; Gego A.; Carme B. (2005) Trypanosomatid protozoa: a simplified DNA isolation procedure. Exp. Parasitol. 111 (3), 207–209. 10.1016/j.exppara.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Livak K. J.; Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (Amsterdam, Neth.) 25 (4), 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Beneke T.; Gluenz E. (2019) LeishGEdit: A Method for Rapid Gene Knockout and Tagging Using CRISPR-Cas9. Methods Mol. Biol. (N. Y., NY, U. S.) 1971, 189–210. 10.1007/978-1-4939-9210-2_9. [DOI] [PubMed] [Google Scholar]
- Zauli-Nascimento R. C.; Miguel D. C.; Yokoyama-Yasunaka J. K.; Pereira L. I.; Pelli de Oliveira M. A.; Ribeiro-Dias F.; Dorta M. L.; Uliana S. R. (2010) In vitro sensitivity of Leishmania (Viannia) braziliensis and Leishmania (Leishmania) amazonensis Brazilian isolates to meglumine antimoniate and amphotericin B. Tropical Medicine & International Health 15 (1), 68–76. 10.1111/j.1365-3156.2009.02414.x. [DOI] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J. Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 (7), 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.