Abstract
Ferroptosis is a new form of regulated cell death and closely related to cancer. However, the mechanism underlying the regulation of ferroptosis in lung adenocarcinoma (LUAD) remains unclear. IB, IHC and ELISA were performed to analyze protein expression. RT-qPCR was used to analyze mRNA expression. Cell viability, 3D cell growth, MDA, the generation of lipid ROS and the Fe2+ concentration were measured to evaluate the responses to the induction of ferroptosis. Measurement of luciferase activity and ChIP were used to analyze the promoter activity regulated by the transcriptional regulator. Co-IP assays were performed to identify protein-protein interactions. In the present study, it was revealed that cAMP response element-binding protein (CREB) was highly expressed in LUAD, and knockdown of CREB inhibited cell viability and growth by promoting apoptosis- and ferroptosis-like cell death, concurrently. It was observed that CREB suppressed lipid peroxidation by binding the promoter region of glutathione peroxidase 4 (GPX4), and this binding could be enhanced by E1A binding protein P300 (EP300). The bZIP domain in CREB and the CBP/p300-HAT domain in EP300 were essential for CREB-EP300 binding in LUAD cells. Finally, it was revealed that CREB, GPX4, EP300 and 4-HNE were closely related to tumor size and stage, and tumors with a higher degree of malignancy were more likely to have a low degree of lipid peroxidation. Therefore, targeting this CREB/EP300/GPX4 axis may provide new strategies for treating LUAD.
Keywords: E1A binding protein P300, lipid peroxidation, cell death, promoter, methylation
Introduction
Ferroptosis is a nonapoptotic form of regulated cell death. In recent years, numerous efforts have been made to elucidate the underlying mechanism of ferroptosis. It is considered that the excessive accumulation of lipid peroxides produced by the lipoxygenase family is an important cause of ferroptosis (1,2). This process links ferroptosis to disruption of the redox homeostasis maintained by glutathione and glutathione peroxidase 4 (GPX4) (2). Compounds that inhibit cystine glutamate antiporters (system XC−) and subsequently reduce glutathione (GSH) levels (e.g., erastin) or GPX4 activity (e.g., RSL3) strongly induce ferroptosis (1–3).
In addition to system XC− and GPX4, several other genes have been reported to affect cell sensitivity to ferroptosis, including acyl-CoA synthetase long chain family member 4 (ACSL4), tumor protein p53 (TP53) and glutaminase 2 (GLS2) (4–6). These studies have linked ferroptosis to a variety of cellular processes, such as iron homeostasis, redox homeostasis, lipid metabolism and glutamine decomposition (7,8). Since highly transformed and drug-resistant tumor cells are prone to ferroptosis (9,10), it is important to understand the underlying mechanism of ferroptosis and apply it to personalized anticancer strategies.
Lung cancer is the most common cause of cancer-related deaths in the world, with an estimated 1.8 million deaths per year (11). Approximately 85% of patients are diagnosed with a group of histological subtypes called non-small cell lung cancer (NSCLC), among which lung adenocarcinoma (LUAD) is the most common subtype (12). Studies have revealed that cancer cells grow slowly when cAMP response element-binding protein (CREB), a transcription factor, is knocked down (13–15). However, little is known about the role of CREB in LUAD, and the relationship between CREB and ferroptosis remains unknown.
In the present study, we aimed to analyze CREB expression in LUAD by immunoblotting (IB), immunohistochemistry (IHC) and enzyme-linked immunosorbent assay (ELISA), and investigate how CREB regulates ferroptosis by analyzing cell viability, three-dimensional (3D) cell growth, malondialdehyde (MDA), the generation of lipid reactive oxygen species (ROS) and the Fe2+ concentration. Measurement of luciferase activity and chromatin immunoprecipitation (ChIP) were used to analyze the GPX4 promoter activity regulated by CREB. In addition, co-immunoprecipitation (co-IP) was used to analyze CREB interaction with other GPX4 regulators such as EP300. Targeting CREB-related GPX4 transcription may provide new ferroptotic strategies for LUAD treatment.
Materials and methods
Cells, vectors and patients
The cell lines used in the present study were as follows: 293T cell line, the lung fibroblast cell lines MRC-5 and WI-38, the lung squamous cell carcinoma (LUSC) cell lines: MES-1 and H226, and the LUAD cell lines H358, A549, H1299 and H1650. All cell lines were purchased from Fuheng Biotechnology (Shanghai, China), and validated by short tandem repeat (STR) analysis. All the cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; HyClone, Cytiva) and 1% penicillin and streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). Ferrostatin-1 (Fer-1), ZVAD-FMK, necrostatin-1 (Nec-1), erastin and apoptozole (all from Sigma-Aldrich; Merck KGaA) were used to treat cells. For vectors, CREB-HA and CREB-short hairpin (sh)1/sh2 were acquired from previous studies (15,16) which were constructed by our laboratory (Shanghai Institute of Thoracic Oncology, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shangai, China). EP300-Myc was purchased from Addgene, Inc. Empty vector, GFP-sh and EP300-sh1/sh2 cells were purchased from Shanghai Biolink Co., Ltd. CREB-Del-KID, CREB-Del-bZIP, EP300-Del-KIX, EP300-Del-Bromo and EP300-Del-CBP/p300-HAT were constructed using overlapping PCR and cloned into the pcDNA3.1(+) vector. All the vectors were transfected at a final concentration of 2 µg per 6-well plate using Lipofectamine™ 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.). 293T cells were transfected with FBS-free DMEM for 6 h and cultured in FBS-containing medium for another 24 h. Then, target cells were infected with lentivirus-containing-DMEM for 24 h, and the follow-up experiments were performed. All the aforementioned procedures were conducted at 37°C and 5% CO2. All primers are summarized in Table SI. Tumorous and adjacent lung tissues of patients (mean age ± SD, 63.86±14.12 years; age range, 27.2–88.4 years; 158 males and 138 females) were recruited at the Shanghai Chest Hospital (Shanghai, China) from September 2013 to March 2018. The diagnosis of lung cancer was confirmed by computed tomography and histological analyses by doctors from Shanghai Chest Hospital. Informed written consent was obtained from all patients. The study was approved by the institutional Ethics Committee of Shanghai Chest Hospital.
IB
IB was performed using conventional protocols that have been previously described (17). The primary antibodies used were: Anti-CREB (product nos. 9197 and 9104) and anti-GAPDH (product nos. 5174 and 51332) all from Cell Signaling Technology, Inc. (CST); anti-GPX4 (product code ab125066), anti-cysteinyl-tRNA synthetase (CARS) (product code ab126714), anti-nuclear factor, erythroid 2 like 2 (NRF2) (product code ab62352), anti-heat shock protein family B small member 1 (HSPB1) (also known as Hsp27; product code ab109376), anti-spermidine/spermine N1-acetyltransferase 1 (SAT1) (product code ab105220) all from Abcam; anti-Myc (product nos. 2276 and 2278), anti-HA (product nos. 2367 and 3724) all from CST; and anti-EP300 (product codes ab54984 and ab275378) and anti-SET domain bifurcated histone lysine methyltransferase 2 (SETDB2) (product code ab5517) all from Abcam. The secondary antibodies were anti-rabbit IgG, HRP-linked antibody (product no. 7074; CST) or anti-mouse IgG, HRP-linked antibody (product no. 7076; CST).
IHC
IHC was performed using conventional protocols that have been previously described (18). The primary antibodies included anti-CREB (product no. 9197; CST). Immunohistochemical staining was assessed by independent pathologists.
Reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), and the RNA was reverse transcribed into complementary DNA using the PrimeScript RT Reagent Kit (TaKaRa Biotechnology Co., Ltd.) according to the manufacturer's instructions. qPCR was performed using a SYBR Premix Ex Taq (TaKaRa Biotechnology Co., Ltd.) kit for 40 cycles (95°C for 3 sec and 60°C for 30 sec). The relative expression of mRNA level was quantified using the 2−ΔΔCq method (19). The primers are listed in Table SI.
Measurements of cell viability, MDA, 4-HNE and Fe2+
Cells were plated at an initial density of 2×105 cells/well and cultured for 24 h at 37°C and 5% CO2. Cell viability was measured using a Cell Titer-Glo luminescent cell viability assay (cat. no. G7572; Promega Corporation) according to the manufacturer's instructions. MDA, 4-HNE and Fe2+ were measured using kits from Abcam (MDA kit, product code ab118970, 532 nm; 4-HNE kit, product code ab238538, 450 nm; Fe2+, product code ab83366, 593 nm) according to the manufacturer's instructions. The luminance or absorbance was measured by a reader from BioTek Instruments, Inc.
ChIP
ChIP was performed using a kit from Active Motif according to the manufacturer's instructions. Cells (2×107) were fixed using 1% formaldehyde at room temperature for 10 min, washed with PBS and lysed using lysis buffer from the kit. Following sonication, protein-DNA complexes were incubated with antibody-coupled protein G beads at 4°C overnight. The antibodies used were anti-CREB (product no. 9197; CST), anti-IgG (product no. 3900; CST) and anti-hepatocyte nuclear factor 4α (HNF4A; cat. no. PP-H6939-00; R&D Systems, Inc.). On the second day, DNA was eluted in 1% SDS/0.1 M NaHCO3, reverse crosslinked at 65°C, purified via phenol/chloroform extraction and ethanol precipitation, and subjected to qPCR. The primers are listed in Table SI.
Measurement of luciferase activity
Luciferase activities were measured using a dual-luciferase kit (Promega Corporation) according to the the manufacturer's instructions. Wild-type (WT)- and mutant (Mut)-GPX4-promoter luciferase reporters were constructed using the pGL4.21 vector at our laboratory (Shanghai Institute of Thoracic Oncology, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China), and co-transfected into lung cancer cells with Renilla plasmids using Lipofectamine™ 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were transfected with FBS-free DMEM for 6 h and cultured in FBS-containing medium for another 42 h in 37°C and 5% CO2. Then, cells were harvested and then lysed in the passive lysis buffer from the kit. The fluorescence intensity of the luciferase reporters was then examined and normalized to the Renilla luciferase activity.
ELISA
The concentrations of CREB were evaluated by ELISA. The tissue samples were diluted (1:4) in a dilution buffer provided by the manufacturer, and 50 µl of each diluted sample was added to 96-well microtiter plates for analysis. A CREB ELISA kit (cat. no. TX11637) was purchased from Lichen Biotech, Ltd. ELISAs were performed in strict accordance with the manufacturers' guidelines. The signals were determined by measuring the absorbance at 450 nm with a microplate reader (BioTek Instruments, Inc.).
Three-dimensional (3D) cell culture
First, basement membrane extract (BME) was seeded in a 96-well plate at 50 µl/well and warmed at 37°C for 30 min. Then, cells were seeded on top of the plate coated with BME at a density of 10,000 cells/well. After 7 days, cells were stained with SYTOX Green (1 µM; Invitrogen, Thermo Fisher Scientific, Inc.) at 37°C for 30 min. Images were collected using a light microscope, and the relative spheroid size and amount (Φ>30 µm) were counted and calculated.
Co-immunoprecipitation (co-IP)
For co-IP, cell lysates (containing 2×107 cells) were incubated with antibody-conjugated protein A/G magnetic beads (Thermo Fisher Scientific, Inc.) in Western/IP lysis buffer (Beyotime Institute of Biotechnology) at 4°C overnight. Immunoprecipitates isolated with the magnetic beads were washed five times with Western/IP lysis buffer before being subjected to IB. The antibodies used for co-IP were: Anti-Myc (product no. 2276; 1:100; CST), anti-HA (product no. 2367; CST, 1:100), anti-CREB (product no. 9104; 1:100; CST), anti-EP300 (product code ab54984; 1:100; Abcam) and anti-SETDB2 (product code ab13712; 1:50; Abcam).
Lipid reactive oxygen species (ROS) measurement
Lipid ROS generation was measured by adding C11-BODIPY (Invitrogen; Thermo Fisher Scientific, Inc.) to a final concentration of 1.5 µM for 20 min before cell harvest. Lipid ROS-positive cells were finally assessed by a BD FACSCanto II flow cytometer.
Bioinformatic analysis
Data concerning CREB expression in 515 LUAD and 59 normal lung specimens were obtained from The Cancer Genome Atlas (TCGA) and further analyzed using UALCAN database (http://ualcan.path.uab.edu) (20). CREB binding motif was acquired from JASPAR database (21). Potential CREB-binding methyltransferase/acetyltransferase was detected using the STRING database (22). Uniprot database was used to analyzed the domains in CREB and EP300 proteins (23).
Statistical analysis
The differences between groups were examined using Student's t-test, one-way ANOVA followed by Bonferroni's post hoc test, Fisher's exact test, χ2 test and Spearman rank-correlation analysis. P<0.05 was considered to indicate a statistically significant difference. The statistical analysis was performed using Graphpad Prism 8 (GraphPad Software, Inc.) or SPSS version 21 (IBM Corp.).
Results
CREB is highly expressed in LUAD
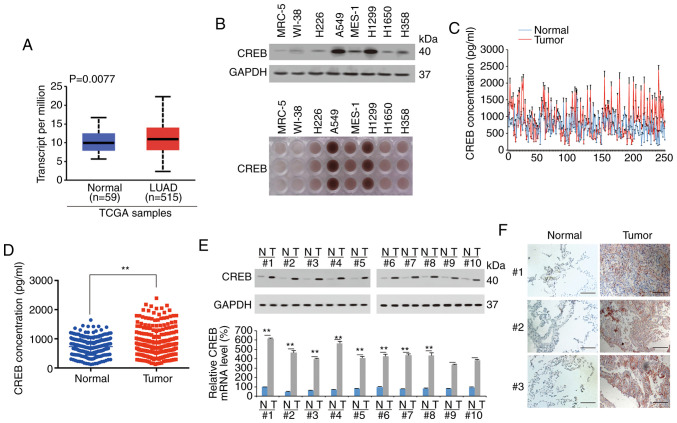
After screening TCGA data using the UALCAN database, it was revealed that CREB was significantly upregulated in 515 LUAD specimens compared to 59 normal lung specimens (Fig. 1A). By using IB and IHC, it was revealed that compared to that of the lung fibroblasts MRC-5 and WI-38, and the LUSC cell lines MES-1 and H226, CREB was highly expressed in LUAD cell lines, especially in A549 and H1299 cells (Fig. 1B). After detecting the expression of CREB in 250 paired samples of LUAD patients by ELISAs, it was determined that the expression of CREB in the tumor tissues was higher than that in adjacent normal tissues (Fig. 1C and D). The tissues of patients 1–10 with the most significant increase in CREB according to the ELISA data of Fig. 1C and D, were also selected to analyze their CREB expression. It was revealed that CREB expression was significantly higher in the LUAD tissues than in the adjacent normal tissues; in addition, the upregulated degree of CREB expression in tumor tissue in Fig. 1E was more obvious than that in Fig. 1A. A similar phenomenon was observed in immunohistochemical analysis by measuring the tissues of no. 1–3 patients (Fig. 1F). CREB expression was also analyzed in squamous cell lung cancer (LUSC) and small-cell lung cancer (SCLC) tissues by ELISAs. It was revealed that CREB was not significantly increased in the LUSC and SCLC tissues compared to their adjacent tissues (Fig. S1). These data indicated that CREB was highly expressed in LUAD tissues.
Figure 1.
CREB is upregulated in LUAD. (A) UALCAN database was used to analyze TCGA data of CREB expression between LUAD (n=515) and normal lung (n=59) tissues. (B) CREB expression in lung fibroblast, LUSC and LUAD cell lines, as measured by IB and IHC. (C and D) CREB expression in LUAD and adjacent normal tissues was measured by ELISA, as presented in a (C) line plot and (D) scatter plot. (E) CREB expression in 10 pairs of LUAD tissues with the most significant increase of CREB according to the ELISA data of C and D, as measured by IB and qPCR. (F) CREB expression in 3 pairs of LUAD tissues, as measured by IHC (scale bar, 800 µm). The data are presented as the mean ± SD from three biological replicates (including IB). **P<0.01 indicates statistical significance. Data from A were analyzed using an independent-sample Student's t-test, D and E were analyzed using paired Student's t-test. CREB, cAMP response element-binding protein; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas; LUSC, lung squamous cell carcinoma; IB, immunoblotting; IHC, immunohistochemistry; ELISA, enzyme-linked immunosorbent assay.
CREB negatively regulates ferroptosis in LUAD
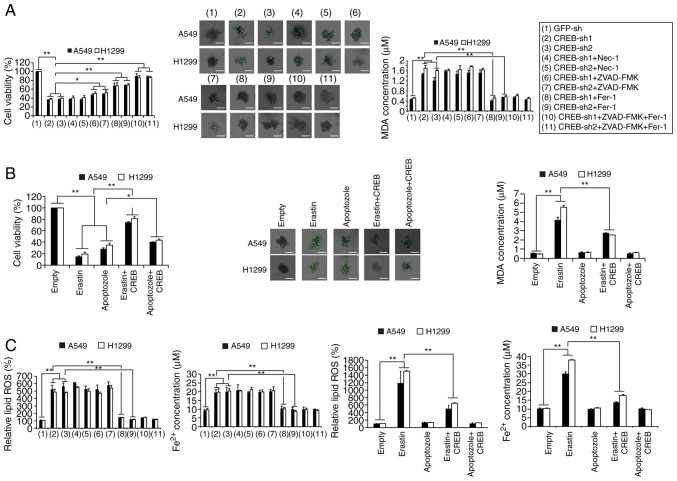
In LUAD, CREB was reported to be a stimulator of cell growth (24,25). However, whether CREB is a regulator of cell death has not been investigated in detail. It was observed that knockdown of CREB inhibited cell viability, and 3D cell growth, whereas these effects could be reversed by the apoptotic inhibitor ZVAD-FMK and the ferroptotic inhibitor Fer-1. However, the CREB knockdown-induced effects could not be regulated by the necrotic inhibitor necrostatin-1 (Nec-1) (Fig. 2A). Compared to ZVAD-FMK, Fer-1 reversed the decrease of cell viability and 3D cell growth induced by CREB knockdown to a greater extent (Fig. 2A). It was also confirmed that CREB knockdown upregulated the level of the lipid peroxidation product MDA and the ferroptotic biomarkers lipid ROS and Fe2+, and these effects could not be reversed by ZVAD-FMK and Nec-1 (Fig. 2A and C). Moreover, it was revealed that ectopically expressed CREB could reverse the apoptotic stimulus Apoptozole- and ferroptosis stimulus erastin-induced decrease of cell viability and 3D cell growth (Fig. 2B), as well as erastin-induced MDA, Fe2+ increase and lipid ROS generation (Fig. 2B and C). These data indicated that knockdown of CREB concurrently induced apoptosis- and ferroptosis-like cell death.
Figure 2.
CREB knockdown inhibits cell growth via stimulating apoptosis- and ferroptosis-like cell death concurrently. (A) Cell viability, 3D cell with SYTOX Green staining and MDA were measured in CREB-knockdown cells with additional treatment using Nec-1 (20 µM), ZVAD-FMK (20 µM) or Fer-1 (1 µM) (scale bar, 50 µm). (B) Cell viability, 3D cell with SYTOX Green staining and MDA were measured in cells treated with erastin (10 µM) or apoptozole (10 µM) with or without ectopically expressed CREB (scale bar, 50 µm). (C) Fe2+ and lipid ROS generation were measured in CREB-knockdown cells with additional treatment using Nec-1 (20 µM), ZVAD-FMK (20 µM) or Fer-1 (1 µM) or cells treated with erastin (10 µM) or apoptozole (10 µM) with or without ectopically expressed CREB. The data are presented as the mean ± SD from three biological replicates. *P<0.05, **P<0.01 indicates statistical significance. Data from A, B and C were analyzed using a one-way ANOVA followed by Bonferroni's post hoc test. CREB, cAMP response element-binding protein; MDA, malondialdehyde; NEC-1, necrostatin-1; Fer-1, ferrostatin-1; ROS, reactive oxygen species; sh, short hairpin.
CREB specifically promotes GPX4 expression
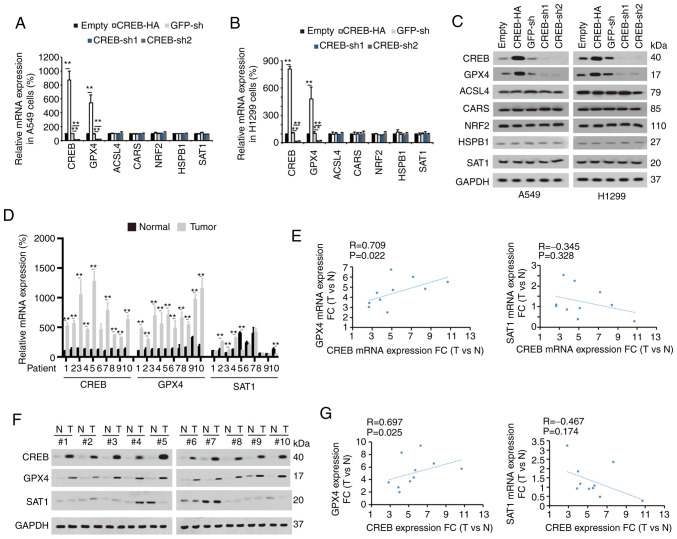
To confirm the target of CREB in ferroptotic regulation and to explore whether their levels were regulated by CREB, several factors (including GPX4, ACSL4, CARS, NRF2, HSPB1 and SAT1) (4,26–30) were selected, which have been recently reported to exert important roles during the ferroptotic process in cancer. It was observed that among these factors, only the mRNA and protein levels of GPX4 were positively regulated by CREB in A549 and H1299 cells (Fig. 3A-C). In the tissues of LUAD patients 1–10, it was observed that the mRNA and protein levels of CREB and GPX4 were upregulated in the LUAD tissues compared to the adjacent normal tissues, and their levels in the LUAD tissues were significantly correlated with each other (Fig. 3D-G). However, parallel experiments revealed that the levels of SAT1 were not significantly increased in the LUAD tissues (Fig. 3D and F). The aforementioned data indicated that GPX4 expression was positively regulated by CREB.
Figure 3.
CREB positively regulates GPX4. (A and B) Indicated mRNA levels were measured in (A) A549 and (B) H1299 cells with or without CREB overexpression or knockdown. (C) Indicated protein expression levels were measured in A549 or H1299 cells with or without CREB overexpression or knockdown. (D) CREB, GPX4 and SAT1 mRNA levels were measured in 10 paired of LUAD tissues. (E) Correlation between GPX4 and CREB mRNA as well as SAT1 and CREB mRNA was calculated. (F) CREB, GPX4 and SAT1 protein expressions were measured in 10 paired of LUAD tissues. (G) Correlation between GPX4 and CREB protein level as well as SAT1 and CREB protein level was calculated. The data are presented as the mean ± SD from three biological replicates (including IB). **P<0.01 indicates statistical significance. Data from A and B were analyzed using a one-way ANOVA followed by Bonferroni's post hoc test. Data from D were analyzed using a paired Student's t-test. Data from E and G were analyzed using Spearman's rank correlation coefficient. CREB, cAMP response element-binding protein; GPX4, glutathione peroxidase 4, SAT1, spermidine/spermine N1-acetyltransferase 1; LUAD, lung adenocarcinoma; IB, immunoblotting; CARS, cysteinyl-tRNA synthetase; NRF2, nuclear factor, erythroid 2 like 2; HSPB1, heat shock protein family B small member 1; ACSL4, acyl-CoA synthetase long chain family member 4; sh, short hairpin; N, normal; T, tumor.
CREB directly binds the promoter of GPX4
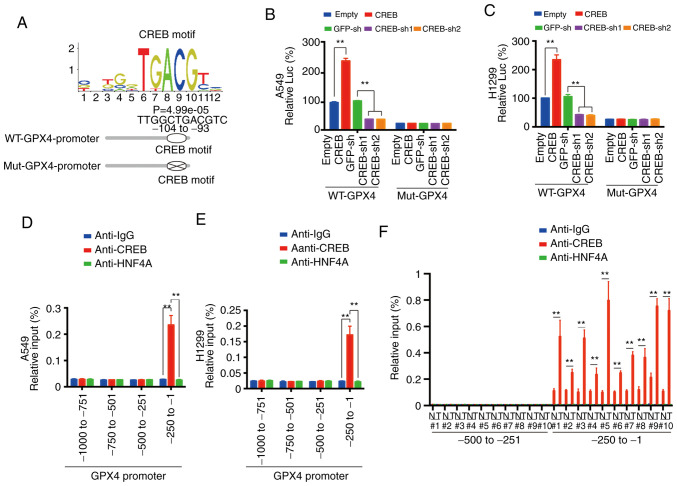
A CREB motif (image from JASPAR database) was observed in the ~-104 to −93 promoter region of GPX4 and two luciferase reporters named WT-GPX4-promoter (containing the CREB motif) and Mut-GPX4-promoter (with the CREB motif deleted) were constructed (Fig. 4A). Via dual-luciferase experiments using these two reporters, it was observed that the promoting effect of CREB for GPX4 promoter was dependent on the CREB motif (Fig. 4B and C). ChIP experiments revealed that CREB bound to the ~-250 to −1 region of the GPX4 promoter (Fig. 4D and E), and the parallel experiments indicated that CREB could not bind to the ~-1000 to −250 region of the GPX4 promoter, and HNF4A and control IgG could not bind to the ~-250 to −1 region of the GPX4 promoter (Fig. 4D and E). As for tissues from no. 1 to no. 10 patients, it was revealed that CREB bound to the ~-250 to −1 region of the GPX4 promoter in both the LUAD and adjacent normal tissues. However, the binding intensity in the LUAD tissues was significantly higher than that in the adjacent normal tissues (Fig. 4F). These data indicated that CREB directly bound to a CREB motif in the GPX4 promoter.
Figure 4.
CREB directly binds the promoter region of GPX4. (A) CREB motif (searched in the JASPAR database) location in GPX4 promoter. (B and C) WT- or Mut-GPX4-Promoter Luc was measured in (B) A549 and (C) H1299 cells with CREB overexpression or knockdown. (D-F) The enrichment of CREB in (D) A549, (E) H1299 cells and (F) 10 paired LUAD tissues at indicated regions of GPX4 promoter was calculated as the percentage of input chromosomal DNA via ChIP using the corresponding antibodies. Anti-IgG and anti-HNF4A were used as the parallel controls. The data are presented as the mean ± SD from three biological replicates. **P<0.01 indicates statistical significance. Data from B-E were analyzed using a one-way ANOVA followed by Bonferroni's post hoc test. Data from F were analyzed using a paired Student's t-test. CREB, cAMP response element-binding protein; GPX4, glutathione peroxidase 4; WT, wild-type; MUT, mutant; LUAD, lung adenocarcinoma; ChIP, chromatin immunoprecipitation; HNF4A, hepatocyte nuclear factor 4α; sh, short hairpin.
EP300 enhances CREB-induced GPX4 transcription
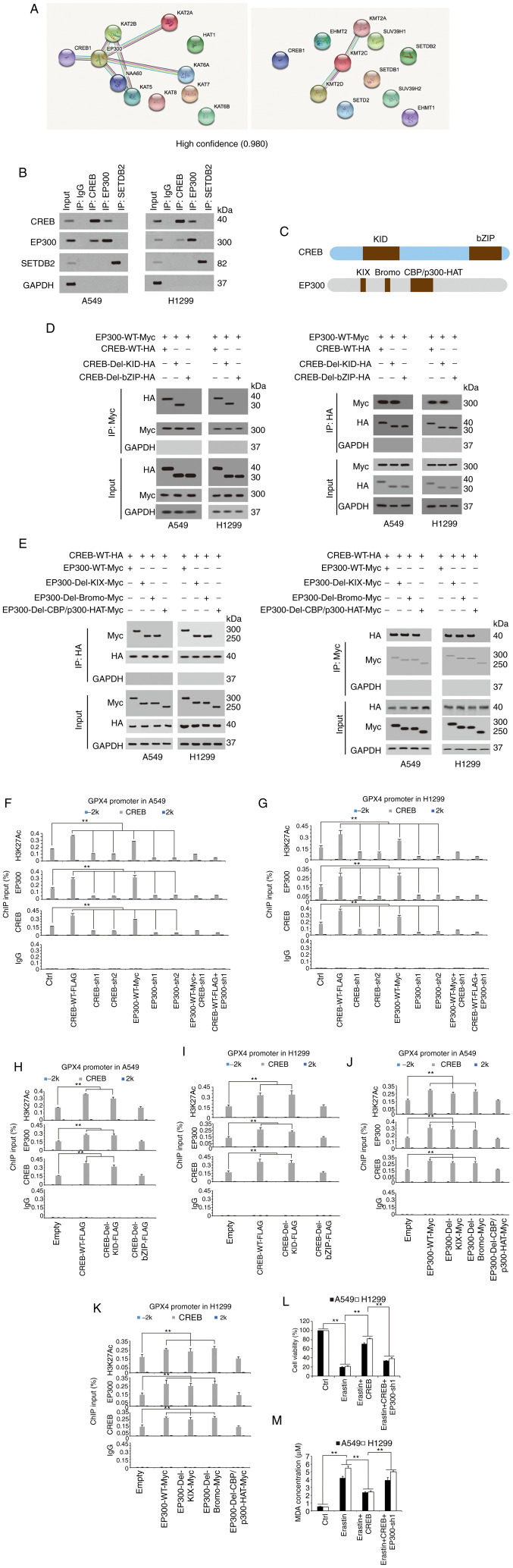
Since histone modification, especially methylation and acetylation, is critical for transcription (31,32), it was further investigated whether methyltransferase or acetyltransferase played important roles in CREB-induced GPX4 transcription. After a STRING analysis detecting potential CREB-binding methyltransferase/acetyltransferase, it was revealed that EP300 had the highest possibility of binding with CREB (Fig. 5A). The co-IP experiments confirmed that CREB could interact with EP300, whereas an obvious interaction was not detected between CREB and SETDB2 (Fig. 5B). The UniProt database revealed (https://www.UniProt.org) that the CREB protein contains two important domains, KID and bZIP, and three important domains, KIX, Bromo and CBP/300-HAT, are located in the EP300 protein (Fig. 5C). Reciprocal co-IP experiments revealed that deletion of the bZIP or CBP/300-HAT domain completely abolished the interaction between CREB and EP300, suggesting that these two domains are essential for the CREB-EP300 interaction (Fig. 5D and E). Additionally, ChIP experiments revealed that CREB or EP300 knockdown (EP300 knockdown efficiency is presented in Fig. S2) reduced the H3K27Ac levels [a hallmark of open chromatin related to EP300 (33)] and inhibited the enrichment of CREB and EP300 around the CREB motif in the GPX4 promoter, and these effects could not be reversed by ectopically expressed EP300 or CREB (Fig. 5F and G), implying that CREB and EP300 were both critical for GPX4 transcription. Furthermore, it was observed that ectopically expressed CREB or EP300 increased the H3K27Ac levels, and stimulated the enrichment of CREB and EP300 around the CREB motif in the GPX4 promoter, whereas these effects could be blocked by deletion of the bZIP domain and CBP/p300-HAT domain, respectively (Fig. 5H-K). In addition, EP300 was knocked down in CREB-overexpressing cells and the cell viability and MDA levels were analyzed. It was determined that CREB could reverse the erastin-induced decrease in cell viability and the MDA increase; however, these effects were abolished by further knocking down EP300 (Fig. 5L and M). Collectively, these data demonstrated that EP300 was essential and had a promoting role in CREB-induced GPX4 transcription.
Figure 5.
EP300 stimulates CREB-dependent GPX4 transcription. (A) STRING analysis revealed CREB interacted with methyltransferase and acyltransferase (confidence=0.980). (B) Co-immunoprecipitation experiments performed in A549 cells using indicated antibodies, and further analysis of CREB, EP300 and SETDB2 expression by IB. (C) Schematic diagram shows the domains in CREB and EP300 protein. (D and E) Co-immunoprecipitation experiments performed using anti-HA or anti-CREB in A549 and H1299 cells with indicated CREB or EP300 plasmids overexpressed, and further analysis of Myc and HA levels by IB. (F and G) The enrichments of H3K27Ac, EP300 and CREB at −2 k, CREB motif or 2 k regions of GPX4 promoter were calculated as the percentage of input chromosomal DNA via ChIP using the corresponding antibodies in (F) A549 and (G) H1299 cells with CREB or EP300 overexpressed or knocked down. Anti-IgG was used as the parallel control. (H-K) The enrichments of H3K27Ac, EP300 and CREB at −2 k, CREB motif or 2 k regions of GPX4 promoter were calculated as the percentage of input chromosomal DNA via ChIP using the corresponding antibodies in (H and J) A549 and H1299 (I and K) cells with WT or mutant (H and I) CREB or (J and K) EP300 overexpressed. Anti-IgG was used as the parallel control. (L and M) Cell viability and MDA, respectively, were measured in A549 and H1299 cells treated with erastin (10 µM) with or without ectopically expressed CREB with or without EP300 knockdown. The data are presented as the mean ± SD from three biological replicates (including IB). **P<0.01 indicates statistical significance. Data from F-M were analyzed using a one-way ANOVA followed by Bonferroni's post hoc test. EP300, E1A binding protein P300; CREB, cAMP response element-binding protein; GPX4, glutathione peroxidase 4; SETDB2, SET domain bifurcated histone lysine methyltransferase 2; IB, immunoblotting; ChIP, chromatin immunoprecipitation; WT, wild-type; MDA, MDA, malondialdehyde; sh, short hairpin; Ctrl, control.
Lipid peroxidation state may be associated with tumor progression
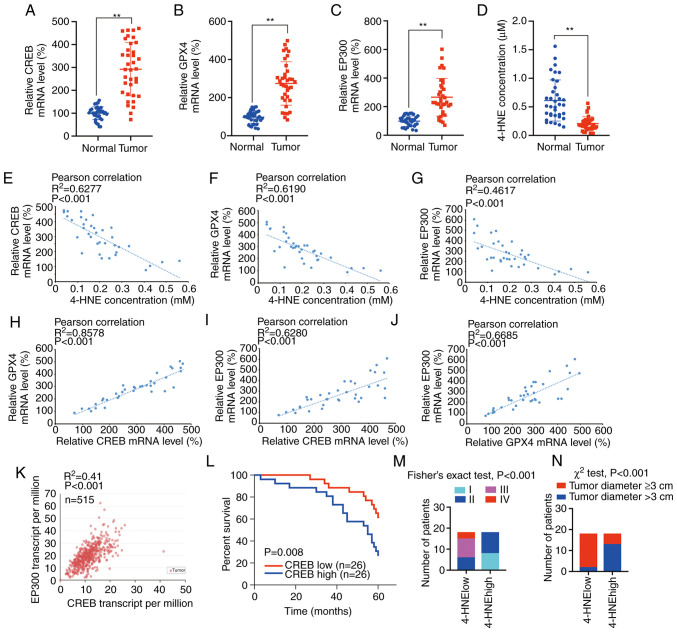
Other paired LUAD tissues were selected to investigate the correlation among CREB, GPX4, EP300 and 4-HNE, a reactive breakdown product of the lipid peroxides that execute ferroptosis. It was observed that the mRNA levels of CREB, GPX4, and EP300 were significantly higher in the tumor tissues than in the normal tissues, while the level of 4-HNE was significantly higher in the normal tissues than in the tumor tissues (Fig. 6A-D). In addition, the level of 4-HNE was negatively associated with the CREB, EP300 and GPX4 levels, whereas significant positive correlations were observed between GPX4 and CREB, EP300 and CREB, and EP300 and GPX4 (Fig. 6E-J).
Figure 6.
Clinical significance of CREB, EP300, GPX4 and 4-HNE. (A-D) Relative (A) CREB, (B) GPX4, (C) EP300 mRNA levels and (D) 4-HNE concentration in 36 paired LUAD tissues. (E-G) Correlations between 4-HNE concentration between (E) CREB, (F) GPX4 and (G) EP300 mRNA levels in 36 paired LUAD tissues. (H-J) Correlations of mRNA levels between (H) GPX4 and CREB, (I) EP300 and CREB, as well as (J) EP300 and GPX4. (K) TCGA data of CREB and EP300 co-expression in 515 LUAD specimens from UALCAN database. (L) Survival in LUAD patients with CREB high (n=26) or low (n=26) expression. (M and N) Correlations between 4-HNE levels and (M) tumor stage and (N) tumor diameter. The data are presented as the mean ± SD from three biological replicates. **P<0.01 indicates statistical significance. Data from A-D were analyzed using a paired Student's t-test. Data from E-K were analyzed using Spearman's rank correlation coefficient. Data from L were analyzed using the log rank analysis. Data from M were analyzed using Fisher's exact test. Data from N were analyzed using χ2 test. CREB, cAMP response element-binding protein; EP300, E1A binding protein P300; GPX4, glutathione peroxidase 4; 4-HNE, 4-hydroxynonenal; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.
Through the TCGA data from the UALCAN database, it was revealed that the CREB level was positively associated with the EP300 level in 515 LUAD specimens (P<0.001) (Fig. 6K). Furthermore, high expression of CREB was significantly associated with poor prognosis in 52 LUAD patients (P=0.008) (Fig. 6L). It was also determined that low 4-HNE levels were associated with more advanced tumor stages and larger tumor diameters (Fig. 6M and N), whereas high CREB, GPX4 and EP300 levels were associated with more advanced tumor stages and larger tumor diameters (Tables I and II). It was also revealed that high levels of CREB, GPX4 and EP300 were associated with advanced N factors (Table II). Furthermore, CREB, GPX4 and EP300 were not associated with patient age, sex or smoking habits (Tables I and II). Collectively, CREB, GPX4, EP300 and 4-HNE, which are related to lipid peroxidation, were closely related to tumor size and stage, and the tumors with a high degree of malignancy were more likely to have a low degree of lipid peroxidation.
Table I.
Associations between mRNA levels of CREB, EP300, GPX4 and clinicopathological parameters including age, sex and tumor stage.
| Sex | Tumor stage | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mRNA levels | Age (years) | P-value (Independent-sample Student's t-test) | Male | Female | P-value (χ2 test) | I | II | III | IV | P-value (Fisher's exact test) |
| CREB high | 65.44 | 0.152 | 8 | 10 | 0.180 | 0 | 7 | 8 | 3 | <0.001 |
| CREB low | 61.39 | 12 | 6 | 8 | 9 | 1 | 0 | |||
| GPX4 high | 65.94 | 0.072 | 7 | 11 | 0.044 | 1 | 6 | 8 | 3 | <0.001 |
| GPX4 low | 60.89 | 13 | 5 | 7 | 10 | 1 | 0 | |||
| EP300 high | 62.11 | 0.361 | 10 | 8 | 0.999 | 0 | 9 | 6 | 3 | <0.001 |
| EP300 low | 64.72 | 10 | 8 | 8 | 7 | 3 | 0 | |||
Data were analyzed by independent-sample Student's t-test, Fisher's exact test and χ2 test.
Table II.
Associations between mRNA levels of CREB, EP300, GPX4 and clinicopathological parameters including smoking habit, tumor diameter as well as N factor.
| Smoking habit | Tumor diameter | N factor | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mRNA levels | Smoking | Non-smoking | P-value (χ2 test) | ≥3 cm | <3 cm | P-value (χ2 test) | N0 | N1 | N2 | N3 | P-value (Fisher's exact test) |
| CREB high | 5 | 13 | 0.725 | 15 | 3 | 0.002 | 3 | 8 | 5 | 2 | 0.001 |
| CREB low | 7 | 11 | 6 | 12 | 12 | 5 | 1 | 0 | |||
| GPX4 high | 6 | 12 | 1 | 15 | 3 | 0.002 | 3 | 8 | 5 | 2 | 0.001 |
| GPX4 low | 6 | 12 | 6 | 12 | 12 | 5 | 1 | 0 | |||
| EP300 high | 4 | 14 | 0.289 | 14 | 4 | 0.018 | 5 | 9 | 3 | 1 | <0.001 |
| EP300 low | 8 | 10 | 7 | 11 | 10 | 4 | 3 | 1 | |||
Data were analyzed by Fisher's exact test and χ2 test.
Discussion
CREB is a ubiquitous transcription factor that activates the transcriptional activity of various promoters through its binding (34). In NSCLC, most studies reported that CREB directly binds to the promoters of proto-oncogenes to exert a cancer promoting effect. For instance, in NSCLC, loss of serine/threonine kinase 11 (LKB1) induced CREB-regulated transcription coactivator (CRTC)-CREB complex activation; the increased enrichment of the CRTC-CREB complex was revealed in the promoter region of LINC00473, and this LINC00473 was essential for the NSCLC cell growth and survival (35). Moreover, the CRTC-CREB complex also induced transcription of inhibitor of DNA binding 1 (ID1), which is associated with stimulated tumor growth and poor prognosis in NSCLC (36). In the present study, it was revealed that CREB could directly bind to the promoter region of GPX4, to stimulate the viability of LUAD cells and inhibit the potential ferroptosis. In summary, CREB is an important transcription factor in NSCLC, that can promote tumor growth by activating a wide range of proto-oncogenes.
Transcriptional activation is regulated by histone modifications, such as methylation and acetylation (37,38). The interaction between CREB and EP300 has been reported previously (39,40), yet the specific domains responsible for the interaction in LUAD have not been elucidated. In the present study, it was observed that the bZIP domain in CREB and the CBP/p300-HAT were essential for the interaction between CREB and EP300. The bZIP domain was revealed to be involved in CREB dimerization and DNA-binding and contributed to CREB transactivation by recruiting the coactivator TORC (41). The CBP/p300-HAT domain was revealed to be critical for the interaction of EP300 with histones or the transcription factor AP-2 alpha (TFAP2A) (42,43). Therefore, the bZIP and CBP/p300-HAT domains are important for protein interactions.
Ferroptotic therapy may be a favorable selection for cancer treatment because ferroptosis exhibits greater induction in certain types of tumor cells than in normal cells (44). In addition, ferroptotic treatment specifically targets cells with a high degree of malignancy, such as cells with a high metastatic tendency or cisplatin resistance (9,45). In contrast, a strong antioxidant system exists in tumor cells, which maintains ROS at an appropriate level, stimulates the proliferation of tumor cells, and does not cause cell death due to excessive stress (46). In LUAD, both SLC7A11 and NRF2 produce high levels of GSH to protect tumor cells from lipid peroxidation damage (47,48). In the present study, it was also revealed that CREB is an important component of the antioxidant system and plays an antioxidant role by stimulating transcription of GPX4. According to the present data, targeting CREB inhibited LUAD cell proliferation and promoted cell lipid peroxidation. Therefore, CREB may be a suitable drug target in LUAD therapy.
There are two main ways to stimulate ferroptosis: Blocking the cystine/glutamate transporter system XC− (49) or directly inhibiting the GSH-dependent antioxidant enzyme GPX4 (50). Studies have reported that treating lung cancer cells using system XC inhibitors such as sorafenib or temozolomide can inhibit cell growth and cause cell death (51,52). However, drugs directly targeting GPX4 have not exhibited potential for clinical application. For example, tumor cells exhibit high tolerance to RSL3 treatment in vivo (53). In the present study, it was observed that knockdown of CREB caused ferroptotic-like effects in LUAD cells by inhibiting GPX4 transcription. Therefore, in the future, a combination of drug treatment and gene knockout to inhibit both system XC− and GPX4 may produce improved therapeutic effects for cancer treatment.
In conclusion, it was revealed that CREB inhibited ferroptosis by stimulating the transcription of GPX4 in the absence of EP300. Targeting this CREB/EP300/GPX4 axis may be a new strategy for treating LUAD.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant nos. 81822029, 81871907, 81672332, 81902869, 81902315 and 81774291), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine (grant no. 20191834), the Shanghai Rising Star Program (grant no. 18QA1403400), the Shanghai Municipal Commission of Health and Family Planning (grant no. 2017YQ024), the Grant Support ‘Chen Guang’ project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (grant no. 18CG16), the Shanghai Sailing Program (grant no. 19YF1444800), and the Nurture Projects for Basic Research of Shanghai Chest Hospital (grant no. 2018YNJCQ06).
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81822029, 81871907, 81672332, 81902869, 81902315 and 81774291), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine (grant no. 20191834), the Shanghai Rising Star Program (grant no. 18QA1403400), the Shanghai Municipal Commission of Health and Family Planning (grant no. 2017YQ024), the Grant Support ‘Chen Guang’ project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (grant no. 18CG16), the Shanghai Sailing Program (grant no. 19YF1444800), and the Nurture Projects for Basic Research of Shanghai Chest Hospital (grant no. 2018YNJCQ06).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The results published and presented in Fig. 1A are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Authors' contributions
ZW and XZ researched, analyzed data and wrote the manuscript. XT constructed the plasmids. YYa and LM researched and analyzed data. JW and YYu designed the study and revised the manuscript for important intellectual content. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Informed written consents were obtained from all patients. The study was approved by the institutional Ethics Committee of Shanghai Chest Hospital (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, et al. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Minikes AM, Gao M, Bian H, Li Y, Stockwell BR, Chen ZN, Jiang X. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature. 2019;572:402–406. doi: 10.1038/s41586-019-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, You JH, Kim MS, Roh JL. Epigenetic reprogramming of epithelial-mesenchymal transition promotes ferroptosis of head and neck cancer. Redox Biol. 2020;37:101697. doi: 10.1016/j.redox.2020.101697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melosky B, Juergens R, McLeod D, Leighl N, Brade A, Card PB, Chu Q. Immune checkpoint-inhibitors and chemoradiation in stage III unresectable non-small cell lung cancer. Lung Cancer. 2019;134:259–267. doi: 10.1016/j.lungcan.2019.05.027. [DOI] [PubMed] [Google Scholar]
- 12.Pinto R, Petriella D, Lacalamita R, Montrone M, Catino A, Pizzutilo P, Botticella MA, Zito FA, Del Bene G, Zonno A, et al. KRAS-Driven lung adenocarcinoma and B Cell infiltration: Novel insights for immunotherapy. Cancers (Basel) 2019;11:1145. doi: 10.3390/cancers11081145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitton B, Chae HD, Hsu K, Dutta R, Aldana-Masangkay G, Ferrari R, Davis K, Tiu BC, Kaul A, Lacayo N, et al. Small molecule inhibition of cAMP response element binding protein in human acute myeloid leukemia cells. Leukemia. 2016;30:2302–2311. doi: 10.1038/leu.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chae HD, Mitton B, Lacayo NJ, Sakamoto KM. Replication factor C3 is a CREB target gene that regulates cell cycle progression through the modulation of chromatin loading of PCNA. Leukemia. 2015;29:1379–1389. doi: 10.1038/leu.2014.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Ma L, Weng W, Qiao Y, Zhang Y, He J, Wang H, Xiao W, Li L, Chu Q, et al. Mutual interaction between YAP and CREB promotes tumorigenesis in liver cancer. Hepatology. 2013;58:1011–1020. doi: 10.1002/hep.26420. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Tang X, Weng W, Qiao Y, Lin J, Liu W, Liu R, Ma L, Yu W, Yu Y, et al. The membrane protein melanoma cell adhesion molecule (MCAM) is a novel tumor marker that stimulates tumorigenesis in hepatocellular carcinoma. Oncogene. 2015;34:5781–5795. doi: 10.1038/onc.2015.36. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, Zhu G, Liu Y, Bian Z, Xu W, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9:1091. doi: 10.1038/s41419-018-1132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Sun F, Qiao Y, Zheng W, Liu Y, Chen Y, Wu Q, Liu X, Zhu G, Chen Y, et al. TFCP2 is required for YAP-Dependent transcription to stimulate liver malignancy. Cell Rep. 2017;21:1227–1239. doi: 10.1016/j.celrep.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Balasubramanya SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu W, Li L, Zheng F, Yang W, Zhao S, Tian C, Yin W, Chen Y, Guo W, Zou L, Deng W. β-Catenin Cooperates with CREB binding protein to promote the growth of tumor cells. Cell Physiol Biochem. 2017;44:467–478. doi: 10.1159/000485013. [DOI] [PubMed] [Google Scholar]
- 25.Jin X, Di X, Wang R, Ma H, Tian C, Zhao M, Cong S, Liu J, Li R, Wang K. RBM10 inhibits cell proliferation of lung adenocarcinoma via RAP1/AKT/CREB signalling pathway. J Cell Mol Med. 2019;23:3897–3904. doi: 10.1111/jcmm.14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei HM, Zhang KR, Wang CH, Wang Y, Zhuang GL, Lu LM, Zhang J, Shen Y, Chen HZ, Zhu L. Aldehyde dehydrogenase 1A1 confers erlotinib resistance via facilitating the reactive oxygen species-reactive carbonyl species metabolic pathway in lung adenocarcinomas. Theranostics. 2019;9:7122–7139. doi: 10.7150/thno.35729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayano M, Yang WS, Corn CK, Pagano NC, Stockwell BR. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016;23:270–278. doi: 10.1038/cdd.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, Tang D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X, Wang H, Cao L, Tang D. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene. 2015;34:5617–5625. doi: 10.1038/onc.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou Y, Wang SJ, Li D, Chu B, Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc Natl Acad Sci USA. 2016;113:E6806–E6812. doi: 10.1073/pnas.1607152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 32.Kinnaird A, Zhao S, Wellen KE, Michelakis ED. Metabolic control of epigenetics in cancer. Nat Rev Cancer. 2016;16:694–707. doi: 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 33.Ebrahimi A, Sevinc K, Gurhan Sevinc G, Cribbs AP, Philpott M, Uyulur F, Morova T, Dunford JE, Göklemez S, Arı Ş, et al. Bromodomain inhibition of the coactivators CBP/EP300 facilitate cellular reprogramming. Nat Chem Biol. 2019;15:519–528. doi: 10.1038/s41589-019-0264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoeffler JP, Meyer TE, Yun Y, Jameson JL, Habener JF. Cyclic AMP-responsive DNA-binding protein: Structure based on a cloned placental cDNA. Science. 1988;242:1430–1433. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Li JL, Lin S, Cao C, Gimbrone NT, Yang R, Fu DA, Carper MB, Haura EB, Schabath MB, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. 2016;126:2267–2279. doi: 10.1172/JCI85250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodon L, Svensson RU, Wiater E, Chun MG, Tsai WW, Eichner LJ, Shaw RJ, Montminy M. The CREB coactivator CRTC2 promotes oncogenesis in LKB1-mutant non-small cell lung cancer. Sci Adv. 2019;5:eaaw6455. doi: 10.1126/sciadv.aaw6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daskalaki MG, Tsatsanis C, Kampranis SC. Histone methylation and acetylation in macrophages as a mechanism for regulation of inflammatory responses. J Cell Physiol. 2018;233:6495–6507. doi: 10.1002/jcp.26497. [DOI] [PubMed] [Google Scholar]
- 38.Thanh Nha Uyen L, Amano Y, Al-Kzayer LFY, Kubota N, Kobayashi J, Nakazawa Y, Koike K, Sakashita K. PCDH17 functions as a common tumor suppressor gene in acute leukemia and its transcriptional downregulation is mediated primarily by aberrant histone acetylation, not DNA methylation. Int J Hematol. 2020;111:451–462. doi: 10.1007/s12185-019-02799-4. [DOI] [PubMed] [Google Scholar]
- 39.Tong Q, Weaver MR, Kosmacek EA, O'Connor BP, Harmacek L, Venkataraman S, Oberley-Deegan RE. MnTE-2-PyP reduces prostate cancer growth and metastasis by suppressing p300 activity and p300/HIF-1/CREB binding to the promoter region of the PAI-1 gene. Free Radic Biol Med. 2016;94:185–194. doi: 10.1016/j.freeradbiomed.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tung WH, Hsieh HL, Lee IT, Yang CM. Enterovirus 71 modulates a COX-2/PGE2/cAMP-dependent viral replication in human neuroblastoma cells: Role of the c-Src/EGFR/p42/p44 MAPK/CREB signaling pathway. J Cell Biochem. 2011;112:559–570. doi: 10.1002/jcb.22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boer U, Eglins J, Krause D, Schnell S, Schofl C, Knepel W. Enhancement by lithium of cAMP-induced CRE/CREB-directed gene transcription conferred by TORC on the CREB basic leucine zipper domain. Biochem J. 2007;408:69–77. doi: 10.1042/BJ20070796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braganca J, Eloranta JJ, Bamforth SD, Ibbitt JC, Hurst HC, Bhattacharya S. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J Biol Chem. 2003;278:16021–16029. doi: 10.1074/jbc.M208144200. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 44.Friedmann Angeli JP, Krysko DV, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer. 2019;19:405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 45.Lee J, You JH, Shin D, Roh JL. Inhibition of Glutaredoxin 5 predisposes cisplatin-resistant head and neck cancer cells to ferroptosis. Theranostics. 2020;10:7775–7786. doi: 10.7150/thno.46903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chio IIC, Tuveson DA. ROS in cancer: The burning question. Trends Mol Med. 2017;23:411–429. doi: 10.1016/j.molmed.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma L, Chen T, Zhang X, Miao Y, Tian X, Yu K, Xu X, Niu Y, Guo S, Zhang C, et al. The m6A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 2020;38:101801. doi: 10.1016/j.redox.2020.101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galan-Cobo A, Sitthideatphaiboon P, Qu X, Poteete A, Pisegna MA, Tong P, Chen PH, Boroughs LK, Rodriguez MLM, Zhang W, et al. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-Mutant lung adenocarcinoma. Cancer Res. 2019;79:3251–3267. doi: 10.1158/0008-5472.CAN-18-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song X, Zhu S, Chen P, Hou W, Wen Q, Liu J, Xie Y, Liu J, Klionsky DJ, Kroemer G, et al. AMPK-Mediated BECN1 phosphorylation promotes ferroptosis by directly blocking system Xc- Activity. Curr Biol. 2018;28:2388–2399.e5. doi: 10.1016/j.cub.2018.05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaschler MM, Andia AA, Liu H, Csuka JM, Hurlocker B, Vaiana CA, Heindel DW, Zuckerman DS, Bos PH, Reznik E, et al. FINO2 initiates ferroptosis through GPX4 inactivation and iron oxidation. Nat Chem Biol. 2018;14:507–515. doi: 10.1038/s41589-018-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houessinon A, Francois C, Sauzay C, Louandre C, Mongelard G, Godin C, Bodeau S, Takahashi S, Saidak Z, Gutierrez L, et al. Metallothionein-1 as a biomarker of altered redox metabolism in hepatocellular carcinoma cells exposed to sorafenib. Mol Cancer. 2016;15:38. doi: 10.1186/s12943-016-0526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sehm T, Rauh M, Wiendieck K, Buchfelder M, Eyupoglu IY, Savaskan NE. Temozolomide toxicity operates in a xCT/SLC7a11 dependent manner and is fostered by ferroptosis. Oncotarget. 2016;7:74630–74647. doi: 10.18632/oncotarget.11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The results published and presented in Fig. 1A are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.