Abstract
The Red Queen hypothesis depicts evolution as the continual struggle to adapt. According to this hypothesis, new genes, especially those originating from nongenic sequences (i.e., de novo genes), are eliminated unless they evolve continually in adaptation to a changing environment. Here, we analyze two Drosophila de novo miRNAs that are expressed in a testis-specific manner with very high rates of evolution in their DNA sequence. We knocked out these miRNAs in two sibling species and investigated their contributions to different fitness components. We observed that the fitness contributions of miR-975 in Drosophila simulans seem positive, in contrast to its neutral contributions in D. melanogaster, whereas miR-983 appears to have negative contributions in both species, as the fitness of the knockout mutant increases. As predicted by the Red Queen hypothesis, the fitness difference of these de novo miRNAs indicates their different fates.
Keywords: Red Queen hypothesis, de novo gene, microRNA, evolution, male reproduction
Introduction
Organisms evolve to adapt to changing environments via the acquisition of adaptive traits. Adaptations are generally transient because continual changes are needed to keep pace with a moving world, as the Red Queen advises Alice (Carroll 1893). Van Valen (1973) emphasized biotic factors as the driving selective force, which has subsequently been expanded to include the coevolution of host/parasite, prey/predator, male/female (sexual selection), as well as antagonistic evolution (Tobler and Schlupp 2005; King et al. 2009; Brockhurst 2011; Liow et al. 2011; Morran et al. 2011; Brockhurst et al. 2014; Nordbotten and Stenseth 2016; Greenspoon and Mideo 2017). Nevertheless, the essence of this metaphor is continual adaptation, which could be driven by biotic, abiotic, or both factors. Van Valen (1973) first reported that genera and species, regardless of their age, often become extinct when they cannot sufficiently adapt. We proposed to test the Red Queen hypothesis by studying the evolution of new genes, which can be compared with those in younger taxa. If the adaptive landscape shifts rapidly, as posited by the Red Queen hypothesis, this may suggest that newly emerging genes have been under strong pressure to evolve continually or face elimination.
New genes fall in two broad categories: those that originated from reshuffling of existing gene components and those emerging de novo from nongenic sequences (Long et al. 2013; Andersson et al. 2015; Schlotterer 2015; VanKuren and Long 2018). The majority of new genes belong to the former, comprising duplicated, (retro-)transposed, and chimeric genes (Chen et al. 2013). In comparison, de novo genes are more appropriate for testing the Red Queen hypothesis because their products are truly new in the cellular environment. The most common source of de novo genes may be small noncoding loci represented by microRNAs (miRNAs) (Lu, Shen, et al. 2008; Lyu et al. 2014). Given the ease with which miRNAs can be formed de novo (Meunier et al. 2013; Lyu et al. 2014), many could have emerged adaptively (Lu, Fu, et al. 2008; Mohammed et al. 2013, 2014, 2018; Lyu et al. 2014) although the majority may be functionless (Lu, Shen, et al. 2008; Berezikov et al. 2010), or dead-on-arrival (Petrov et al. 1996; Petrov and Hartl 1998). Following the molecular evolutionary analysis performed by Lyu et al. (2014), we use the term “de novo miRNAs” to specifically refer to the first group of adaptive miRNAs. We hypothesize that “de novo miRNAs” must continuously evolve and survive in different fitness landscapes or face elimination, as inferred by the Red Queen hypothesis.
In order to test this possibility, experiments must be conducted in close sibling species, as their divergence time is sufficiently short such that rapidly evolving genes remain identifiable. More importantly, they provide distinct fitness landscapes, and thus de novo genes might end up with different fates. Therefore, we performed functional and molecular analysis on two typical de novo miRNA genes (miR-983 and miR-975; see Materials and Methods) in closely related Drosophila species that diverged about 5–50 Ma (Kumar et al. 2017).
Results
Sequence and Expression Divergence of miR-983 and miR-975 in Drosophila
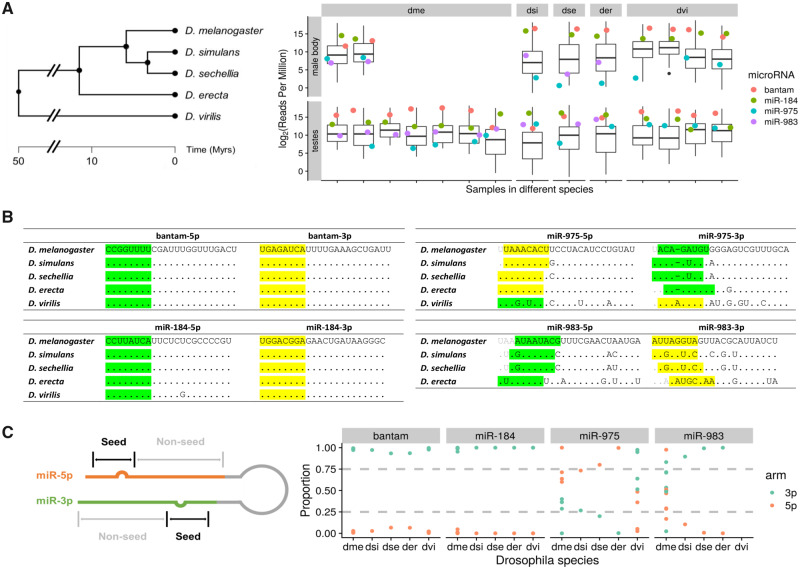
We collected and sequenced 23 small RNA libraries from the testes and male bodies of five Drosophila species to survey the sequence and expression patterns of the de novo miRNAs (fig. 1A). miR-975 is present in all these Drosophila species, whereas miR-983 is absent in D. virilis. Similar to most young miRNA genes, these two de novo miRNAs have rapidly evolved across species, as divergence can be seen in both the seed and nonseed regions compared with conserved miRNAs (e.g., bantam and miR-184) (fig. 1B). Previous study by Lyu et al. (2014) generated DNA sequences from 42 D. melanogaster and 25 D. simulans lines and found both dme-mir-983-1/-2 and dsi-mir-983 show significantly higher divergence/polymorphism ratio compared with its flanking region. The number of substitutions in these miRNAs is far higher than can be sustained by the neutral process, suggesting adaptive evolution by McDonald–Kreitman test (Lyu et al. 2014). In contrast, bantam and miR-184 are highly conserved across Drosophila species, even in nonseed regions, a common trend among older conserved miRNAs.
Fig. 1.
Sequence and expression divergence of miR-975 and miR-983 in Drosophila species. (A) The phylogenetic tree of five Drosophila sibling species is shown on the left. Adopted from TimeTree (Kumar et al. 2017), scale in million years (My). Expression levels (reads per million, RPM) of miR-975 and miR-983 in testes and male bodies are shown on the right. Boxplot represents the expression level of all detected miRNAs, and dots represent the expression of selected miRNAs annotated with different colors. miR-5p and miR-3p expression levels are summed together for comparison. dme, Drosophila melanogaster; dsi, D. simulans; dse, D. sechellia; der, D. erecta; dvi, D. virilis. (B) Sequences of two highly expressed conserved miRNAs, bantam and miR-184, across five Drosophila species are shown on the left. Sequences of miR-975 and miR-983 in the same species are depicted on the right. Seed regions of major and minor miRs are highlighted in yellow and green, respectively. (C) Arm switching of miR-975 and miR-983 in different Drosophila species. Relative expression levels of miR-5p and miR-3p are shown across species (right panel), with an annotation sketch of the miRNA structure (left panel).
We then examined the expression of these miRNAs in different species and multiple lines of D. melanogaster and D. virilis as available (supplementary table S1, Supplementary Material online). Figure 1A shows the expression of bantam, miR-184, miR-975, and miR-983 relative to all miRNAs detected in the testes and male bodies. Bantam and miR-184 are highly expressed and almost always ranked in the first quantile among the evaluated Drosophila species for all samples. In contrast, miR-975 and miR-983, like other adaptive miRNAs (supplementary fig. S1, Supplementary Material online), generally show lower expression in male bodies compared with testes, indicating they are more enriched in testes in other Drosophila species, consistent with what previous studies suggested in D. melanogaster (Lyu et al. 2014; Mohammed et al. 2014). In testes, miR-975 shows substantial variation between and even within species. In D. simulans, D. erecta, and D. virillis, its expression is high and comparable with bantam and miR-184, whereas in D. sechellia, its expression drops below the average level. In D. melanogaster, miR-975 expression is generally low or even undetectable, ranking in or close to the fourth quantile in six of seven samples. Indeed, when considering the 136 canonical miRNAs (supplementary fig. S1, Supplementary Material online; see also Materials and Methods), only miR-983 among the adaptive miRNAs (one out of 14) is consistently expressed above the median expression of all these miRNAs across different samples in D. melanogaster and D. simulans testes, whereas nearly 30% (28 out of 94) conservative miRNAs are expressed above the median across the same species, suggesting the expressions of adaptive miRNAs are more variable.
Each miRNA locus, either new or old, can potentially generate two mature products, which are usually referred as the -5p and -3p arm, respectively (Ameres and Zamore 2013). The starting position of each arm may shift, resulting in distinct mature miRNAs. For example, miR-983-3p has four distinct seeds in four sibling species, and miR-975-5p has three distinct seeds in five sibling species (fig. 1B), which determines the repertoire of target genes and, presumably, their functions. We also observed arm switching for de novo miRNAs among different species (Berezikov 2011; Kim et al. 2020) (supplementary fig. S2A and B, Supplementary Material online), which is less frequent for conserved miRNAs (supplementary fig. S2C and D, Supplementary Material online). Figure 1C shows that the conserved miRNAs (bantam and miR-184) predominantly express one product. In contrast, the expression levels of miR-983 and miR-975 are divergent among Drosophila species. For example, expression of miR-975-3p is strong in D. virilis but rather weak in the remaining species (fig. 1C). These sibling species show arm-switching vis-à-vis D. virilis, with the 5p miR being the major form. We should also note that when sequencing data of multiple lines are available in D. melanogaster and D. virilis, the coexpressions of both miR-5p and miR-3p can be observed for both de novo and conserved miRNAs (supplementary fig. S2, Supplementary Material online), which might be relevant to the general role miRNAs playing in stabilizing gene regulatory network (Zhao et al. 2017, 2018; Ambros 2019; Chen et al. 2019). miRNAs showing coexpression (i.e., minor miRNA accounts for over 25% of total expression) or arm switching in more than two libraries are summarized in table 1 (evolutionary mode is extracted from Lyu et al. [2014]).
Table 1.
miRNAs with Coexpression of Both Matures or Arm Switching in Testes.
| miRNA | Evolutionary Mode | Coexpression | Arm Switching |
|---|---|---|---|
| miR-2498 | Adaptive | √ | √ |
| miR-2499 | √ | √ | |
| miR-973 | √ | ||
| miR-974 | √ | ||
| miR-975 | √ | √ | |
| miR-983 | √ | √ | |
| miR-992 | √ | ||
| miR-10 | Conservative | √ | √ |
| miR-100 | √ | √ | |
| miR-14 | √ | √ | |
| miR-190 | √ | ||
| miR-276a | √ | √ | |
| miR-279 | √ | √ | |
| miR-281-1 | √ | √ | |
| miR-281-2 | √ | √ | |
| miR-282 | √ | ||
| miR-283 | √ | ||
| miR-304 | √ | ||
| miR-305 | √ | √ | |
| miR-306 | √ | ||
| miR-6-3 | √ | √ | |
| miR-92a | √ | √ | |
| miR-958 | √ | √ | |
| miR-965 | √ | √ | |
| miR-981 | √ | √ | |
| miR-988 | √ | √ |
In short, the mature products of de novo miRNAs are extremely variable in sequence, expression, and biogenesis among species, which indicates their rapid evolution across different species. These features are representative of de novo miRNAs and related to their age (fig. 1 and supplementary fig. S2, Supplementary Material online).
Rapid Functional Evolution of miR-983 and miR-975
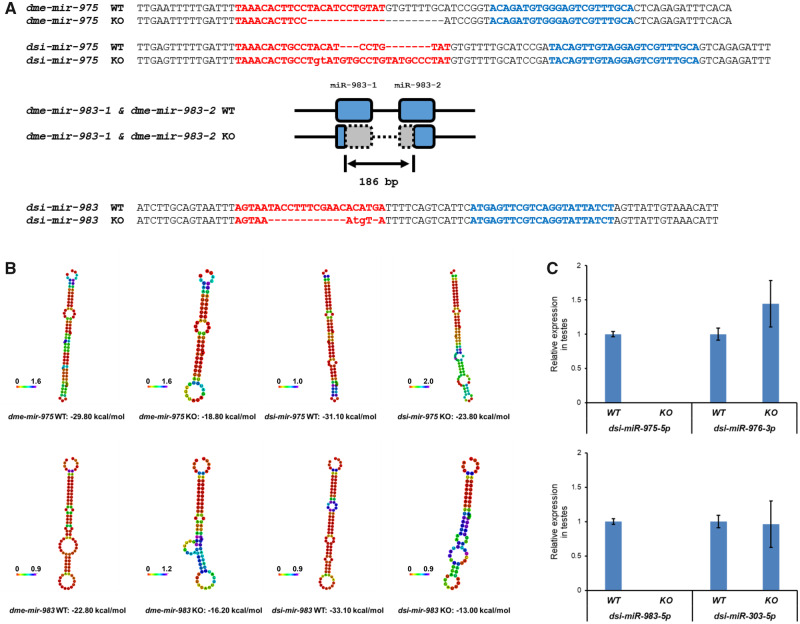
We then surveyed the functional evolution of miR-983 and miR-975 by constructing loss-of-function mutants. For a direct comparison with our previous constructs in D. melanogaster (Lu et al. 2018), we used TALEN (transcription activator-like effector nuclease) to construct miRNA mutants in D. simulans, a nonmodel organism (supplementary fig. S3, Supplementary Material online). Mutants were confirmed by PCR followed by DNA sequencing (fig. 2A and supplementary fig. S4, Supplementary Material online). miRNA harpins are likely to be disrupted after mutant since the minimum free energy increases substantially (fig. 2B). In addition, the absence of mature miRNA expression was further validated by RT–qPCR in testes (fig. 2C).
Fig. 2.
Construction of miR-975 and miR-983 mutants in Drosophila melanogaster and D. simulans. (A) miR-975 and miR-983 mutant sequences in D. melanogaster and D. simulans. miR-5p and miR-3p are highlighted in red and blue, respectively. miR-983 has two sequence identical paralogs in D. melanogaster: dme-mir-983-1 and dme-mir-983-2. A 186-bp deletion disrupts both copies. WT, wildtype; KO, knockout. (B) The optimal secondary structure of miRNA precursors with minimum free energy shown on the bottom. (C) dsi-miR-975-5p is undetectable in dsi-mir-975 KO flies, whereas the adjacent miRNA, dsi-miR-976-3p as a control, is expressed. dsi-miR-983-5p is undetectable in dsi-mir-983 KO flies, whereas the adjacent miRNA, dsi-miR-303-5p as a control, is expressed.
These KO lines were then subjected to a series of analyses focusing on the typical fitness components associated with male reproductive functions, including male mating success, male ability to repress female remating, sperm competition, and male fertility (see Materials and Methods). Male mating success was evaluated by presenting virgin and nonvirgin females to the miRNA KO males and calculating the male mating success rate (Lu et al. 2018). We measured a male’s ability to repress female remating according to the remating rejection rate of the female (Lu et al. 2018). Sperm competition was analyzed using by both offense and defense assays (Yeh et al. 2013; Civetta and Ranz 2019), and we measured male fertility by mating males to two batches of virgin females in series, which aims to exhaust sperm reservoirs in order to obtain an accurate estimate of sperm production (Liufu et al. 2017).
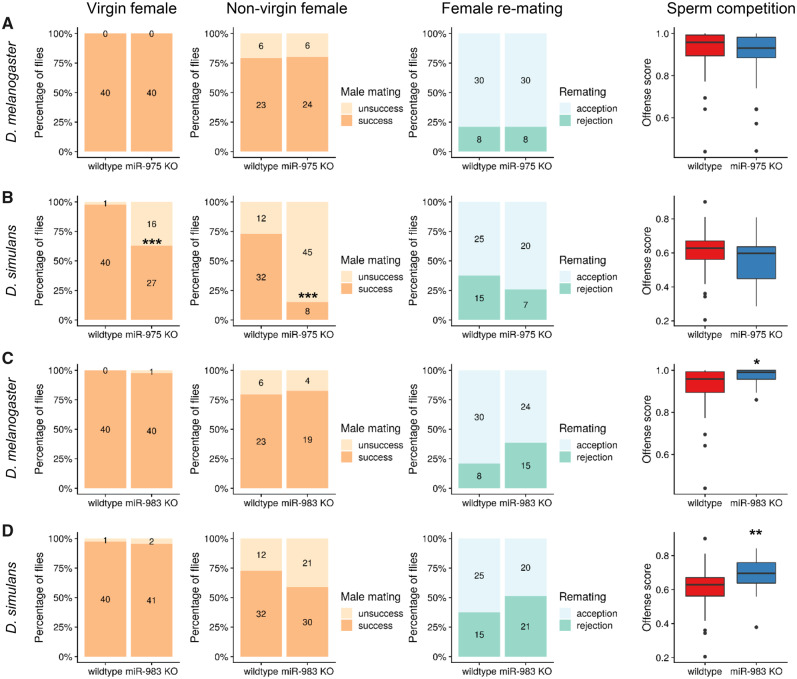
We compared miR-975 KO in D. simulans versus D. melanogaster, which shows no defects in all surveyed components (fig. 3A; also see Lu et al. [2018]). Surprisingly, miR-975 KO in D. simulans resulted in severe defects in male mating success (fig. 3B). More than one-third (16 of 43) of mutant D. simulans males failed to mate with virgin female, compared with only one of 41 wild type males (Fisher’s exact test, P value <0.001). The mating defect was even more pronounced when miR-975 KO males were presented with nonvirgin females, in which the mating success rate drops to only 15% (8 of 53) compared with 73% (32 of 44) in wild type flies (Fisher’s exact test, P value <0.001). This loss of mating success would substantially decrease fitness due to fewer progeny produced in wild. We also found that miR-975 KO males appeared to have reduced ability to repress female remating, but this did not reach statistical significance, perhaps due to the limited numbers tested. According to these data, miR-975 KO seems to have a neutral effect in D. melanogaster but deleterious in D. simulans, which echoes its low expression in D. melanogaster while much higher expression in D. simulans (fig. 1A). Compared with the distinct phenotypic effects of miR-975 KO, effects of miR-983 KO were more similar between the two species (fig. 3C and D). Intriguingly, we found that miR-983 KO males had higher offense scores, indicating that the sperm of miR-983 KO males are more competitive. This enhanced performance is offense-specific because we did not observe an elevated score in the defense assay (supplementary fig. S5, Supplementary Material online). In addition, we observed that miR-983 KO males likely have a stronger ability to repress female remating (fig. 3C and D), and miR-983 KO males produce slightly more progeny in D. melanogaster (supplementary fig. S6, Supplementary Material online). This observation is surprising because the absence of a highly expressed miRNA might be expected to reduce rather than improves fitness (see further discussion below).
Fig. 3.
Functional divergence of miR-975 and miR-983 in Drosophila melanogaster and D. simulans. (A) Phenotypic assays for miR-975 KO in D. melanogaster. Four fitness measurement are shown from left to right: male mating success with virgin and nonvirgin females, Fisher’s exact test was used for significance, *** represents P ≤ 0.001; male ability to repress female remating; offense score for sperm competition, Mann–Whitney U test was used for significant test, * represents P ≤ 0.05 and ** represents P ≤ 0.01. (B) Phenotypic assays for miR-975 KO in D. simulans. (C) Phenotypic assays for miR-983 KO in D. melanogaster. (D) Phenotypic assays for miR-983 KO in D. simulans. Phenotypic assays for miR-975 and miR-983 in D. melanogaster are adopted from Lu et al. (2018).
Collectively, these functional assays demonstrate the diverse contributions of the studied miRNAs to male reproduction functions in different species, suggesting the rapid functional evolution of these de novo miRNAs.
Transcriptome Divergence between Species
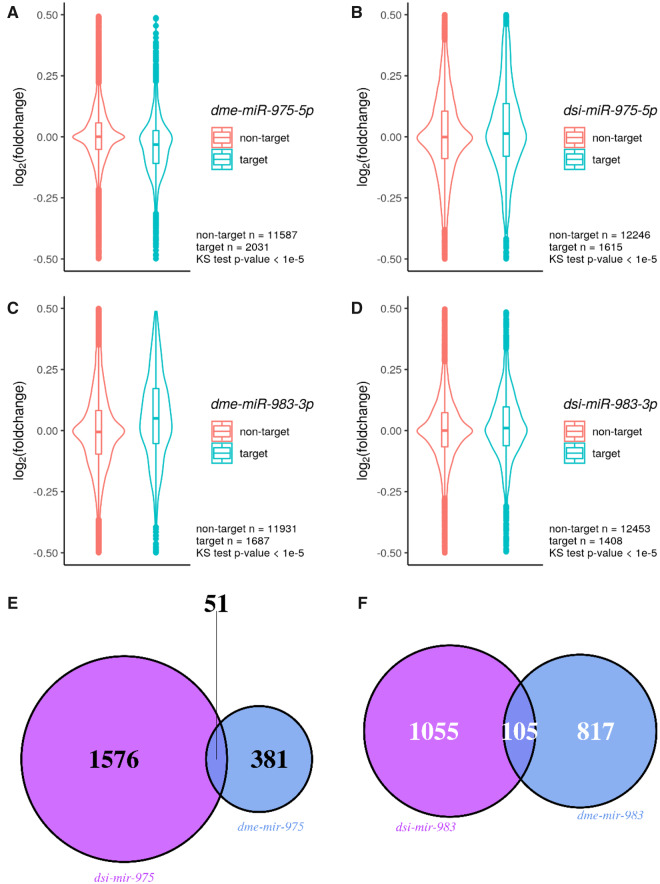
The phenotypic consequences of the de novo miRNA deletions (miR-983 KO and miR-975 KO) between D. melanogaster and D. simulans should be a manifestation of the underlying transcriptomic dysregulation in testes. We next compared the number of dysregulated genes in the two species upon miRNA knockout. For miR-975, we found only 432 genes that were significantly dysregulated in D. melanogaster (supplementary fig. S7A, Supplementary Material online) whereas 1,843 genes were dysregulated in D. simulans (supplementary fig. S7B, Supplementary Material online), a 4-fold difference. This discrepancy, similar to the differences observed in phenotypic assays, might be due to the weak expression of miR-975 in D. melanogaster compared with a much higher expression level in D. simulans (fig. 1A). Indeed, when we surveyed the target expression changes upon miRNA KO, we found dsi-mir-975 KO causes modest but significant target derepression, whereas dme-mir-975 KO does not (fig. 4A and B). In contrast, for miR-983, which has similar expressions in the two species, the magnitude of target derepression (fig. 4C and D) and transcriptomic changes were much closer, with 922 genes dysregulated in D. melanogaster (supplementary fig. S7C, Supplementary Material online) and 1,333 in D. simulans (supplementary fig. S7D, Supplementary Material online). We found the overlap among these dysregulated genes to be small between the two species (fig. 4E and F). Further permutation tests show the observed number of overlapping gene for miR-983 is higher than the mean of the permuted distribution (105 vs. 83, at the 99.6% quantile) but less than 2-fold, which suggests miR-983 is retaining some of its regulations in the two species. However, the observed number of overlapping genes for miR-975 is lower than the mean of the permuted distribution (51 vs. 53, at the 43% quantile), which suggests fast evolving in its regulations. Lastly, we also performed gene ontology analysis and found dysregulated genes are enriched in distinct terms (supplementary table S2, Supplementary Material online), indicating significant divergence in target gene regulation between the two species, which results in downstream effects on the entire transcriptome.
Fig. 4.
Derepression of miRNA targets in miRNA KO flies. (A) dme-miR-975-5p targets versus nontargets in dme-mir-975 KO flies. Log2(fold change of expressions between miRNA KO vs. WT) is shown on the y axis. Numbers of target and nontarget, as well as P values for Kolmogorov–Smirnov test are indicated in each violin plot. (B) dsi-miR-975-5p targets versus nontargets in dsi-mir-975 KO flies. (C) dme-miR-983-3p targets versus nontargets in dme-mir-983-1/-2 KO flies. (D) dsi-miR-983-3p targets versus nontargets in dsi-mir-983 KO flies. (E) Overlap of dysregulated genes between dsi-mir-975 KO and dme-mir-975 KO. Dysregulated genes are determined by DEseq2 with FDR < 0.05. (F) Overlap of dysregulated genes between dsi-mir-983 KO and dme-mir-983 KO.
Discussion
Van Valen suggested that lower taxa (e.g., genera) are subjected to more unpredictable selective pressure compared with the higher taxa (e.g., classes or orders). Hence, these lower groups often go extinct, even following a period of adaptive existence. In this study, we demonstrated this scenario from a gene-level perspective by focusing on de novo miRNA genes.
A unique feature of this study is the choice of genes. The selected de novo genes must have been adaptive in the past. Many earlier studies have proposed high turnover rates of new elements in the genome, for both coding (Tautz and Domazet-Loso 2011; Palmieri et al. 2014) and noncoding genes (Lu, Shen, et al. 2008; Meunier et al. 2013; Lyu et al. 2014). As these new elements are mostly nonfunctional, they may belong to a class often characterized as dead-on-arrival (DOA) elements. The high turnover of neutral DOA elements is unsurprising, whereas new genes that arrive in an adaptive manner are expected to remain functional. In addition, new genes originated from existing genes would also continuously evolve as suggested by the Red Queen hypothesis (Innan and Kondrashov 2010; Ross et al. 2013; Wang et al. 2019). Although the diversity of de novo new genes in close-related species will be easier to observe, because the duplicates had avoided their “childhood.”
Our phenotypic assays revealed that miR-975 is functional in D. simulans, as deleting the miRNA substantially decreased male mating success, and hence fitness (fig. 3B). However, when the same gene continuously evolves in D. melanogaster, a distinct fitness landscape, it may end up in a completely different sequence and functional space due to its neutral contributions to fitness components. Indeed, miR-975 in D. melanogaster has lower expression (fig. 1A), loses its ability to repress targets (fig. 4A), and unsurprisingly, being phenotypically neutral for all fitness components we surveyed (fig. 3A). We also observed strong divergence of miR-975 regulations in close sister species (fig. 4C). These observations are direct evidence for the Red Queen hypothesis from a gene-level perspective.
In contrast, our assays showed that knockout of miR-983 increased fitness in general for both species. This negative contribution of miR-983 is unexpected given its abundant expression level and adaptive signal in the McDonald–Kreitman test (Lyu et al. 2014). However, since the signal detected by MK test mainly suggests the adaptive evolution in the past (Hejase et al. 2020), it is not necessarily proving that miR-983 is still advantageous at present in the evolving Red Queen landscape. Similar evolution dynamics were also observed for young protein-coding genes, which are fast evolving and under rapid turnover across different species (Betran 2015). In our previous study, we noted that the total fitness effect of miRNAs may be “quasi-neutral,” which means the miRNA KO flies could perform better in some fitness components and worse in others (Lu et al. 2018). Here, we only measured four components of male reproduction, which may not be sufficient to draw definitive conclusions. Nevertheless, de novo genes die regularly, not only due to complete loss of functionality but also because of potentially opposing positive and negative fitness effects. Our observations indicate that miR-983 may be on an evolutionary death trajectory, as it shows modest negative effects on some fitness components.
The driving force underlying the continual evolution of new genes has been widely discussed (McLysaght and Hurst 2016). Here, we should mention that most new genes are specific to male reproduction (Kaessmann 2010; Schlotterer 2015). The arms race in sexual competition is a never-ending battle (Gage 2004; Brennan and Prum 2015; Perry and Rowe 2015; Morimoto et al. 2019), and selective advantages are often transient. In this sense, sexual selection may very likely operate in a Red Queen landscape. Because new genes, including all the de novo miRNAs analyzed here, are often testis-specific in expression, it is plausible that the evolution of new genes may be subject to the Red Queen effect via sexual selection.
Materials and Methods
Small RNA Analyses
Testes were dissected and collected from 3- to 5-day-old D. erecta (UCSD Stock No.: 14021-0224.01) and D. sechellia (Stock No.: 14021-0248.20) adults. Total RNA was extracted using the TRIzol Reagent. Small RNA libraries were generated using an Illumina Small RNA Sample Preparation kit and subsequently sequenced using an Illumina HiSeq 2000 machine at the Beijing Genomics Institute (Shenzhen).
To determine the expression patterns of de novo miRNAs, we also collected and surveyed additional testes and male body libraries in D. melanogaster, D. simulans, D. sechellia, D. erecta, and D. virilis. Sequencing data were retrieved from the GEO database (http://www.ncbi.nlm.nih.gov/geo/; last accessed December 17, 2020) (Edgar et al. 2002), and accession numbers are shown in supplementary table S1, Supplementary Material online. Fly genomes were retrieved from FlyBase (http://flybase.org/; last accessed December 17, 2020) (Attrill et al. 2016). The genome versions used were: D. melanogaster: r6.34, D. simulans: r2.02, D. sechellia: r1.3, D. erecta: r1.3, and D. virilis: r1.2. miRNA precursor and mature miRNA sequences were retrieved from miRBase (http://www.mirbase.org; last accessed December 17, 2020; release 22.1) (Kozomara and Griffiths-Jones 2014; Kozomara et al. 2019). miRNA expression was measured using the Mapper and Quantifier modules in miRDeep2 (version: 2.0.1.2). Short reads were mapped to genomes using mapper.pl with parameters “-e -h -i -j -l 18 -m -v –n.” Reads matching miRNAs in each library were normalized using all reads mappable to mature sequences and scaled as reads per million (RPM). For coexpression and arm switching analysis, we focus on 136 miRNAs, which is detailed below.
Selection of De Novo miRNAs for Functional Assays
In the previous study (Lyu et al. 2014), Lyu et al. studied all miRNAs in Drosophila melanogaster and found there are 136 highly expressed canonical miRNAs contributing to the majority of miRNA repertoire over long periods of time, which shows particularly interesting evolutionary patterns. Among these miRNAs, 22 of them were newly emerged in the last 30 My, Interestingly, more than half of the 22 new miRNAs are found in three clusters, including miR-972 cluster, miR-310 cluster, and miR-982 cluster (Lyu et al. 2014). In the past few years, we further investigated these clusters using functional assays in D. melanogaster (Liufu et al. 2017; Lu et al. 2018). In this study, for the purpose of comparing functional evolution of de novo miRNAs between Drosophila species, we picked two of the de novo miRNAs we surveyed before, that is, miR-975 and miR-983, as representatives for the de novo miRNAs. miR-983 is from the miR-982 cluster, which only contains new miRNAs (i.e., emerged in the last 30 My), whereas miR-975 is from the miR-972 cluster, containing both old and new miRNAs (Lyu et al. 2014).
miRNA Mutant Fly Construction and Stocks
miRNA mutant flies were generated using a TALEN (Transcription Activator-Like Effector Nuclease) method (Katsuyama et al. 2013). TALEN plasmids were designed and constructed by ViewSolid Biotech Co., Ltd (http://www.v-solid.com/; last accessed December 17, 2020). TALEN binding sites are shown in supplementary table S3, Supplementary Material online. Plasmids were transcribed to mRNA in vitro using the mMESSAGE mMACHINE T7 ULTRA Transcription Kit. w501 embryos were used for microinjection in D. simulans. After microinjection, embryos were kept at 25 °C, and emerging adult flies were crossed to the Sim6 strain in D. simulans (see supplementary fig. S3, Supplementary Material online, for a detailed cross scheme). DNA was extracted from F0 adult flies after 5 days of crossing. PCR was performed to amplify the regions containing miR-983 and miR-975 in D. simulans for mutation detection using the Surveyor Mutation Detection Kit. Once a mutation was detected, single-pair crossing was performed with F1 flies. Sanger sequencing of mutants was performed to determine InDel sequences. Primers used for PCR are shown in supplementary table S4, Supplementary Material online. Flies without miRNA mutations were subjected to the same cross scheme and used as controls in subsequent assays. All procedures using commercial kits were performed following manufacturers’ protocols. Fly stocks used in this study are listed in supplementary table S5, Supplementary Material online.
Quantitative miRNA Analyses by qRT–PCR
We measured relative expression levels of miRNAs in the miRNA knock-out and control strains. Total RNA was extracted from testes of 3- to 5-day-old flies using the TRIzol Reagent (Thermo Fisher Scientific Inc., Catalog No. 15596026). We used stem-loop primers for reverse transcription (Chen et al. 2005), and Taqman PCR analysis was then performed using the miRNA UPL (Roche Diagnostics) probe assay protocol (Varkonyi-Gasic et al. 2007; He et al. 2016). Primers are listed in supplementary table S6, Supplementary Material online. Three biological replicates were conducted for each genotype, and 2s RNA was used as an endogenous control.
Phenotypic Assays
Male Fertility
Two rounds of mating were performed with miR-983 KO and miR-975 KO flies. In D. melanogaster, each 3- to 5-day-old male was mated with five 3- to 5-day-old w1118 virgin females for 2 days, and then transferred to a new vial to mate with another five virgin females for 2 days. In D. simulans, the two mating procedure rounds were the same, except that D. simulans control virgin females were used for mating. All offspring were counted in each of the two consecutive mating rounds. More than 15 males were examined for each genotype in D. melanogaster and D. simulans. Male fertility was measured as the average number of offspring per male. Two-tailed t-test was used to determine the statistical significance of phenotypic differences between genotypes.
Mating Success and Male Ability to Repress Female Remating
Three distinct metrics were measured: male mating success with virgin females, male mating success with premated females, and males’ ability to repress female remating. To survey mating success with virgin females, wild-type virgin females were mated to either wild-type or miRNA knockout males for 2 h. Mating success rates for the males were inferred by whether progeny were produced. This was calculated according to the number of females with progeny divided by the total number of females used in the assay. To survey male mating success with nonvirgin females, wild-type females were first mated to reference males for 2 h. After 2.5 days, these mated females were presented with either wild-type or miRNA knockout males for 12 h. Male mating success with nonvirgin females was calculated by the number of females with both white-eyed and red-eyed progeny (successful fertilization by experimental and reference males, respectively) divided by the total number of females successfully premated with reference males. To survey males’ ability to repress female remating, virgin females were mated to either wild-type or miRNA knockout males for 2 h. After 2.5 days, these mated females were presented with reference males for 12 h. We measured two outcomes: 1) number of females (x) with white-eyed progeny only (rejection of the reference male), and 2) number of females (y) with both white-eyed and red-eyed progeny (acceptance of the reference male). Then, male ability to repress female remating was calculated as x/(x + y). The reference males were from the Sim6 line in D. simulans. P values were calculated using Fisher’s exact test. Drosophila melanogaster data from Lu et al. (2018) were reused for analysis.
Sperm Competition
Sperm competition assays were conducted as described in Yeh et al. (2013). Briefly, double-mating experiments were set up for each miRNA KO and wild-type line. In D. simulans, virgin wild-type females were mated to reference males (a wild type red eye strain, Sim6) for 2 h and then mated to experimental males (miRNA KO or wild-type) for 12 h after 56 h to measure offensive ability. Conversely, wild-type females were mated to experimental males for 2 h and then to the reference males for 12 h after 56 h for the defense assay. Each assay used 40–50 replicates. Progeny eye color was used as a marker of mating success and paternity identification. F0 females with both red-eyed and white-eyed progeny had successfully mated with both experimental and reference males. Red-eyed female progenies (y) arose from reference males, whereas white-eyed female progenies (x) were generated from experimental fathers. Sperm competitive ability was estimated using the score P = x/(x + y). Mann–Whitney U test was used to determine the statistical significance of phenotypic differences between genotypes. Drosophila melanogaster data from Lu et al. (2018) were reused for analysis.
Transcriptome Analyses
To detect the effects of miR-983 and miR-975 KO on the transcriptome, we performed RNA-seq analysis. Total RNA was extracted from ∼30 testes of 3- to 5-day-old males using the TRIzol Reagent. PolyA-enriched RNA libraries were constructed and sequenced on an Illumina HiSeq 2000 at GENEWIZ Co. (https://www.genewiz.com/; last accessed December 17, 2020). Three samples with abnormal GC content were removed and not used for further analysis. Drosophila melanogaster (r6.34) and D. simulans (r2.02) genomes were downloaded from FlyBase and used for mapping. Adapters were trimmed using Cutadapt version 2.10. Salmon was used to quantify gene expression (Patro et al. 2017). DEseq2 was used to detect significantly dysregulated genes with FDR < 0.05 (Love et al. 2014). Gene Ontology analysis was performed using the R package topGO (Alexa and Rahnenfuhrer 2020). For the permutation test, we permuted the labels of dysregulated genes in both species 1,000 times and calculated the number of dysregulated genes overlapped. The observed number of overlapped genes is then compared with the distribution of overlapped gene number from permutation.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Professor Chung-I Wu for his original idea of Red Queen hypothesis on new genes and his generous support of this study. We thank Yumei Huang for her help on fly functional assays. This work was supported by National Natural Science Foundation of China (31900417, 31770246, 32070644) and Guangdong Basic and Applied Basic Research Foundation (2019A1515010708 to G.A.L., 2019A1515110387 to J.X.).
Data Availability
The small RNA-seq libraries generated for this study are available in GEO under accession numbers GSM2977400 and GSM2977401. RNA-seq libraries are available in the National Genomics Data Center (https://bigd.big.ac.cn/; last accessed December 17, 2020) under accession number PRJCA002747.
References
- Ameres SL, Zamore PD.. 2013. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 14(8):475–488. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Jerlstrom-Hultqvist J, Nasvall J.. 2015. Evolution of new functions de novo and from preexisting genes. Cold Spring Harb Perspect Biol. 7(6):a017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill H, , Falls K, , Goodman JL, , Millburn GH, , Antonazzo G, , Rey AJ, , Marygold SJ. 2016. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 44(D1):D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E. 2011. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 12(12):846–860. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Liu N, Flynt AS, Hodges E, Rooks M, Hannon GJ, Lai EC.. 2010. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat Genet. 42(1):6–9; author reply 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E. 2015. The “life histories” of genes. J Mol Evol. 80:186–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan PL, Prum RO.. 2015. Mechanisms and evidence of genital coevolution: the roles of natural selection, mate choice, and sexual conflict. Cold Spring Harb Perspect Biol. 7(7):a017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA. 2011. Evolution. Sex, death, and the Red Queen. Science 333(6039):166–167. [DOI] [PubMed] [Google Scholar]
- Brockhurst MA, Chapman T, King KC, Mank JE, Paterson S, Hurst GD.. 2014. Running with the Red Queen: the role of biotic conflicts in evolution. Proc Biol Sci. 281(1797):20141382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll L. 1893. Through the looking-glass, and what Alice found there. New York: T.Y. Crowell & Co. [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33(20):e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M.. 2013. New genes as drivers of phenotypic evolution. Nat Rev Genet. 14(9):645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YX, Shen Y, Lin P, Tong D, Zhao YX, Allesina S, Shen X, Wu CI.. 2019. Gene regulatory network stabilized by pervasive weak repressions: microRNA functions revealed by the May-Wigner theory. Natl Sci Rev. 6(6):1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta A, Ranz JM.. 2019. Genetic factors influencing sperm competition. Front Genet. 10:820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE . 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30(1):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage M. 2004. Evolution: sexual arms races. Curr Biol. 14(10):R378–R380. [DOI] [PubMed] [Google Scholar]
- Greenspoon PB, Mideo N.. 2017. Parasite transmission among relatives halts Red Queen dynamics. Evolution 71(3):747–755. [DOI] [PubMed] [Google Scholar]
- He L, Xie M, Huang J, Zhang T, Shi S, Tang T.. 2016. Efficient and specific inhibition of plant microRNA function by anti-microRNA oligonucleotides (AMOs) in vitro and in vivo. Plant Cell Rep. 35(4):933–945. [DOI] [PubMed] [Google Scholar]
- Hejase HA, Dukler N, Siepel A.. 2020. From summary statistics to gene trees: methods for inferring positive selection. Trends Genet. 36(4):243–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F.. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11(2):97–108. [DOI] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20(10):1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuyama T, Akmammedov A, Seimiya M, Hess SC, Sievers C, Paro R.. 2013. An efficient strategy for TALEN-mediated genome engineering in Drosophila. Nucleic Acids Res. 41(17):e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim J, Yu S, Lee YY, Park J, Choi RJ, Yoon SJ, Kang SG, Kim VN.. 2020. A mechanism for microRNA arm switching regulated by uridylation. Mol Cell. 78(6):1224–1236 e1225. [DOI] [PubMed] [Google Scholar]
- King KC, Delph LF, Jokela J, Lively CM.. 2009. The geographic mosaic of sex and the Red Queen. Curr Biol. 19(17):1438–1441. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S.. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47(D1):D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S.. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42(D1):D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Liow LH, Van Valen L, Stenseth NC.. 2011. Red Queen: from populations to taxa and communities. Trends Ecol Evol. 26(7):349–358. [DOI] [PubMed] [Google Scholar]
- Liufu Z, Zhao Y, Guo L, Miao G, Xiao J, Lyu Y, Chen Y, Shi S, Tang T, Wu CI.. 2017. Redundant and incoherent regulations of multiple phenotypes suggest microRNAs’ role in stability control. Genome Res. 27(10):1665–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, VanKuren NW, Chen S, Vibranovski MD.. 2013. New gene evolution: little did we know. Annu Rev Genet. 47(1):307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu GA, Zhao Y, Yang H, Lan A, Shi S, Liufu Z, Huang Y, Tang T, Xu J, Shen X, et al. 2018. Death of new microRNA genes in Drosophila via gradual loss of fitness advantages. Genome Res. 28(9):1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Fu Y, Kumar S, Shen Y, Zeng K, Xu A, Carthew R, Wu CI.. 2008. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol Biol Evol. 25(5):929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI.. 2008. The birth and death of microRNA genes in Drosophila. Nat Genet. 40(3):351–355. [DOI] [PubMed] [Google Scholar]
- Lyu Y, Shen Y, Li H, Chen Y, Guo L, Zhao Y, Hungate E, Shi S, Wu CI, Tang T.. 2014. New microRNAs in Drosophila–birth, death and cycles of adaptive evolution. PLoS Genet. 10(1):e1004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hurst LD.. 2016. Open questions in the study of de novo genes: what, how and why. Nat Rev Genet. 17(9):567–578. [DOI] [PubMed] [Google Scholar]
- Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H.. 2013. Birth and expression evolution of mammalian microRNA genes. Genome Res. 23(1):34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J, Bortolamiol-Becet D, Flynt AS, Gronau I, Siepel A, Lai EC.. 2014. Adaptive evolution of testis-specific, recently evolved, clustered miRNAs in Drosophila. RNA 20(8):1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J, Flynt AS, Panzarino AM, Mondal MMH, DeCruz M, Siepel A, Lai EC.. 2018. Deep experimental profiling of microRNA diversity, deployment, and evolution across the Drosophila genus. Genome Res. 28(1):52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J, Flynt AS, Siepel A, Lai EC.. 2013. The impact of age, biogenesis, and genomic clustering on Drosophila microRNA evolution. RNA 19(9):1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto J, McDonald GC, Smith E, Smith DT, Perry JC, Chapman T, Pizzari T, Wigby S.. 2019. Sex peptide receptor-regulated polyandry modulates the balance of pre- and post-copulatory sexual selection in Drosophila. Nat Commun. 10(1):283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Schmidt OG, Gelarden IA, Parrish RC II, Lively CM.. 2011. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science 333(6039):216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordbotten JM, Stenseth NC.. 2016. Asymmetric ecological conditions favor Red-Queen type of continued evolution over stasis. Proc Natl Acad Sci U S A. 113(7):1847–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri N, Kosiol C, Schlotterer C.. 2014. The life cycle of Drosophila orphan genes. Elife 3:e01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 14(4):417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JC, Rowe L.. 2015. The evolution of sexually antagonistic phenotypes. Cold Spring Harb Perspect Biol. 7(6):a017558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL.. 1998. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol Biol Evol. 15(3):293–302. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Lozovskaya ER, Hartl DL.. 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384(6607):346–349. [DOI] [PubMed] [Google Scholar]
- Ross BD, Rosin L, Thomae AW, Hiatt MA, Vermaak D, de la Cruz AF, Imhof A, Mellone BG, Malik HS.. 2013. Stepwise evolution of essential centromere function in a Drosophila neogene. Science 340(6137):1211–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer C. 2015. Genes from scratch – the evolutionary fate of de novo genes. Trends Genet. 31:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T.. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12(10):692–702. [DOI] [PubMed] [Google Scholar]
- Tobler M, Schlupp I.. 2005. Parasites in sexual and asexual mollies (Poecilia, Poeciliidae, Teleostei): a case for the Red Queen? Biol Lett. 1(2):166–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. 1973. A new evolutionary law. Evol Theory1:1–30. [Google Scholar]
- VanKuren NW, Long M.. 2018. Gene duplicates resolving sexual conflict rapidly evolved essential gametogenesis functions. Nat Ecol Evol. 2(4):705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP.. 2007. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-D, Shao X-J, Bai B, Wang J, Wang X, Cao X, Liu Y-H, Wang X, Yin T-T, Zhang S-J, et al. 2019. Structural variation during dog domestication: insights from gray wolf and dhole genomes. Natl Sci Rev. 6(1):110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh SD, Chan C, Ranz JM.. 2013. Assessing differences in sperm competitive ability in Drosophila. J Vis Exp. (78):e50547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lin P, Liufu Z, Yang H, Lyu Y, Shen X, Wu CI, Tang T.. 2018. Regulation of large number of weak targets-new insights from twin-microRNAs. Genome Biol Evol. 10(5):1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Shen X, Tang T, Wu CI.. 2017. Weak regulation of many targets is cumulatively powerful – an evolutionary perspective on microRNA functionality. Mol Biol Evol. 34(12):3041–3046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The small RNA-seq libraries generated for this study are available in GEO under accession numbers GSM2977400 and GSM2977401. RNA-seq libraries are available in the National Genomics Data Center (https://bigd.big.ac.cn/; last accessed December 17, 2020) under accession number PRJCA002747.