Abstract
Knowledge on genetic structure is key to understand species connectivity patterns and to define the spatiotemporal scales over which conservation management plans should be designed and implemented. The distribution of genetic diversity (within and among populations) greatly influences species ability to cope and adapt to environmental changes, ultimately determining their long-term resilience to ecological disturbances. Yet, the drivers shaping connectivity and structure in marine fish populations remain elusive, as are the effects of fishing activities on genetic subdivision. To investigate these questions, we conducted a meta-analysis and compiled genetic differentiation data (FST/ΦST estimates) for more than 170 fish species from over 200 published studies globally distributed. We modeled the effects of multiple life-history traits, distance metrics, and methodological factors on observed population differentiation indices and specifically tested whether any signal arising from different exposure to fishing exploitation could be detected. Although the myriad of variables shaping genetic structure makes it challenging to isolate the influence of single drivers, results showed a significant correlation between commercial importance and genetic structure, with widespread lower population differentiation in commercially exploited species. Moreover, models indicate that variables commonly used as proxy for connectivity, such as larval pelagic duration, might be insufficient, and suggest that deep-sea species may disperse further. Overall, these results contribute to the growing body of knowledge on marine genetic connectivity and suggest a potential effect of commercial fisheries on the homogenization of genetic diversity, highlighting the need for additional research focused on dispersal ecology to ensure long-term sustainability of exploited marine species.
Keywords: phylogeography, connectivity, dispersal, fisheries, FST
Introduction
Marine ecosystems encompass some of the most productive and biodiverse habitats on the planet. Yet, they face unprecedent changes associated with climate change, pollution, habitat loss, and overexploitation, making them prone to collapse at global scales (Halpern et al. 2007; Hoegh-Guldberg and Bruno 2010). During the last decades, efforts have been put forward to ensure the long-term sustainability of marine biodiversity, including stock management and the implementation of marine protected areas (MPAs). Within this context, the preservation of genetic resources plays a crucial role and is one of the factors that should be taken in consideration to optimize the effectiveness of conservation actions. Genetic diversity greatly influences species’ resilience capacity and their adaptative potential to environmental changes (Pauls et al. 2013). Within each species, genetic resources can be partitioned not only between interbreeding individuals but also among populations. Population genetic differentiation can arise from a myriad of mechanisms related with reproductive isolation, geographic distance (Cunningham et al. 2009), biogeographic barriers (soft boundaries associated with hydrological processes or hard barriers such as land bridges; Cowman and Bellwood 2013), or specific behavioral traits (e.g., fidelity to natal spawning grounds; Boustany et al. 2006). Understanding the patterns of connectivity and the rate of genetic exchange among populations is therefore fundamental from a resource management perspective, as MPAs effectiveness is intrinsically dependent not only on their ability to self-replenish but also on the net export of biomass (i.e., spillover) beyond their boundaries (Harrison et al. 2012).
Species’ dispersal potential is commonly considered to be closely linked with life-history traits such as the duration of planktonic larval stages, egg buoyancy, growth rate, among others. Theoretically, the higher the capability of dispersion, the lower the interpopulational genetic structure of a species. In reef ecosystems, for instance, benthic fish with negatively buoyant eggs have been reported to exhibit greater interpopulational differentiation that pelagic spawning species (Riginos et al. 2014). Likewise, Bradbury et al. (2008) suggest that fish attaining larger sizes during the adult phase tended to display lower FST estimates, potentially due to the increased potential for large-scale, nonpassive dispersal. Yet, although this question has been extensively reviewed, studies are not conclusive and no single trait has been found to explain genetic differentiation consistently across multiple taxa (Riginos et al. 2014). Although it is probably true that marine fish exhibit less populational structure than freshwater species due to the combined effects of longer dispersal capabilities and larger effective populations sizes, recent studies showed that genetic partitioning among populations on the marine realm is significantly higher than previously thought, with the existence of unique genetic diversity at temporal and spatial scales that are relevant to conservation management (Ruzzante et al. 2001; Bradbury et al. 2008, and references therein).
Another factor that remains unclear is the impact of overharvest and selective fishing on populations’ genetic structure. In addition to the removal of large percentages of biomass, there is growing evidence that commercial fishing activities can favor certain life-history or behavioral traits (Alós et al. 2016), a process commonly known as fisheries-induced evolution (Heino et al. 2015). By preferentially targeting individuals with given traits (i.e., larger sizes, bolder behavior, etc.), fishing may exert either direct or indirect selective pressures on exploited populations. Species that evolve in the direction of slow-growing life histories in response to size-selective harvesting, for instance, are subjected to direct human-induced selection (fisheries targeting larger individuals; e.g., Alós et al. 2014). On the other hand, heritable aspects of an individuals’ biology that are covariant with traits under direct selection, such as reproductive or physiological characteristics, might suffer shifts through indirect selection (Heino et al. 2015). It is now accepted that genetic shifts associated with human harvest can occur at contemporary time scales (Audzijonyte et al. 2013) and affect a vast number of exploited fish populations, which can ultimately reduce their genetic diversity (Pinsky and Palumbi 2014). Notwithstanding the importance of these matters, evidence for fisheries-induced evolution remains mostly based on theoretical or experimental, tank-confined studies as it is challenging to disentangle all the variables that may shape populations’ genetic patterns (Balkenhol et al. 2015). These variables range from methodological aspects, to environmental features and past evolutionary events, and can mute any signal of differentiation resulting from organisms’ dispersal or anthropogenic impact.
In order to address this knowledge gap and provide a broader perspective on the patterns shaping genetic structure in marine fish populations, we carried out a global meta-analysis on literature published over the last decade and modeled the effect of several variables on pairwise fixation metrics (FST/ΦST values). We specifically test whether the level of fishing pressure may be reflected on the levels of population divergence and evaluate the importance of isolation-by-distance and life-history traits as factors shaping genetic structure. Furthermore, we investigate at which degree methodological aspects such as genetic marker and fixation statistics may influence the observed trends. Following the rapid advances in molecular biology over the last decades, genetic tools such as mitochondrial DNA (mtDNA), microsatellites (short tandem repeat polymorphisms), and single nucleotide polymorphisms (SNPs) have been widely used in combination with a variety of fixation statistics for the purpose of population genetic inference. However, how their different properties affect the produced estimates remains a controversial topic. The present study provides one of the largest data sets on population genetics of fish species to date and offers further insights into the processes shaping connectivity and genetic structure in marine ecosystems, which may be of paramount importance for stock management and conservation planning.
Results
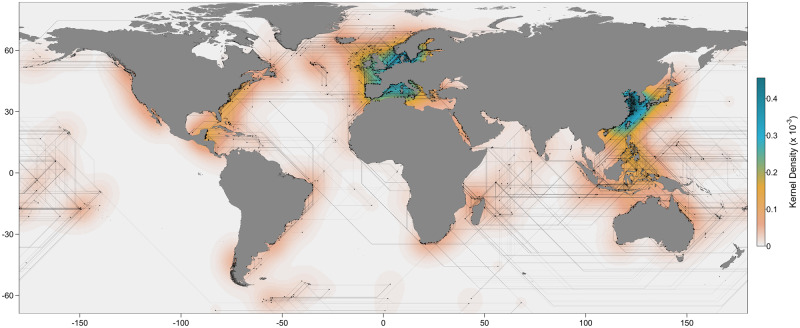
A total of 239 suitable studies were obtained from the literature search, corresponding to a data set of 2,382 samples, 15,234 pairwise differentiation scores, and 188 marine fish species after all data cleaning procedures (supplementary fig. S1, Supplementary Material online). Of the 188 species, 51 (27%) were classified as highly commercial, 63 (34%) as commercial, 44 (23%) as minor commercial, and 30 (16%) as subsistence/noncommercial. Pairwise FST values ranged from −0.23 to 1.00, whereas ΦST values ranged between −0.50 and 0.99 (supplementary fig. S2, Supplementary Material online). Sampled populations were separated by an average distance of approximately 1,780 km, ranging from −67° to 80° in latitude and covering most of world’s oceans (fig. 1).
Fig. 1.
Heat map of analyzed sampling locations (n = 2,382) and illustration of the least-cost trajectories calculated to estimate pairwise geographic distances.
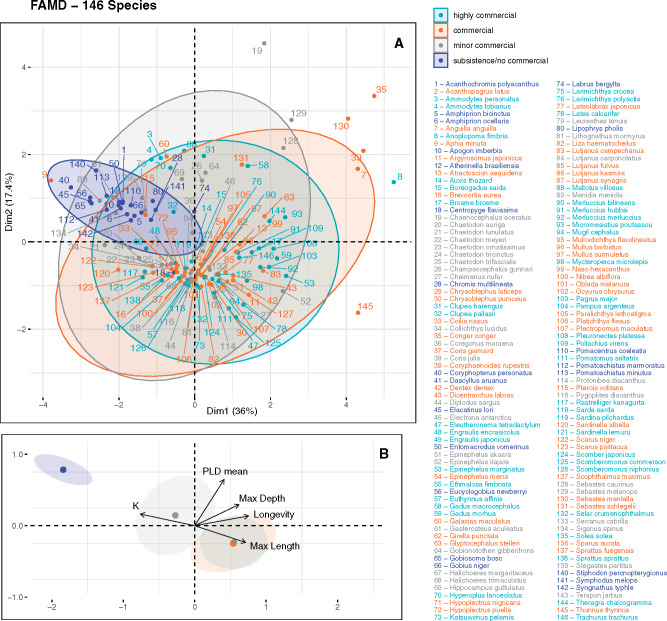
No significant study biases were found, regarding neither reported values (supplementary fig. S2, Supplementary Material online) nor sample size (supplementary fig. S3, Supplementary Material online). Factor analysis of mixed data (FAMD) results showed reduced levels of similarity between species subjected to identical exploitation levels, excepting for species of subsistence fisheries or with noncommercial interest, which formed a tighter cluster on the 2D space (i.e., considering the first two principal components; fig. 2A). Notwithstanding the large overlaps between groups (i.e., concentration ellipses), cluster centroids based on commercial interest were found to be sequentially distributed, from noncommercial to highly commercial species (fig. 2B). No significant biases concerning sample size, number of sampled sites, and spatial scale were identified in relation with different commercial importance (supplementary fig. S4, Supplementary Material online). Nevertheless, some heterogeneity in the geographic distribution of sampled populations was identified when plotting all sampled locations independently per commercial level (supplementary fig. S5, Supplementary Material online).
Fig. 2.
FAMD results based on the life-history traits of the analyzed species, color-coded per commercial importance. (A) Distribution of the included species within the two first principal components, together with concentration ellipses generated assuming multivariate normal distributions. The proportion of variation explained by each of the two principal components is indicated on both axes. (B) Contribution of quantitative variables and commercial importance clusters using 95% confidence intervals around group means.
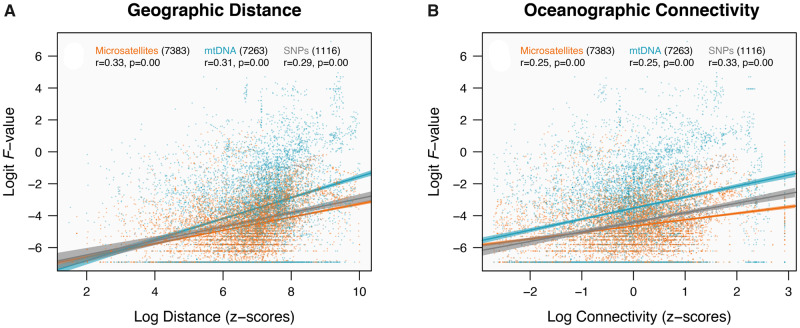
Geographic distance was found to be significantly correlated with pairwise fixation (fig. 3), with moderate Pearson’s coefficients being obtained for all genetic markers (r = 0.33 for microsatellites, r = 0.31 for mtDNA, and r = 0.29 for SNPs). Likewise, significant positive linear relationships were found for oceanographic connectivity estimates, yet with lower correlation coefficients for microsatellites and mtDNA (r = 0.25; SNPs with r = 0.33; fig. 3).
Fig. 3.
Interaction of transformed pairwise fixation indices (logit F-values) with standardized geographic distances (A) and oceanographic connectivity estimates (B), per marker type. Total number of points included is indicated in parenthesis next to each marker and Pearson’s correlation coefficients are displayed below, together with P-values.
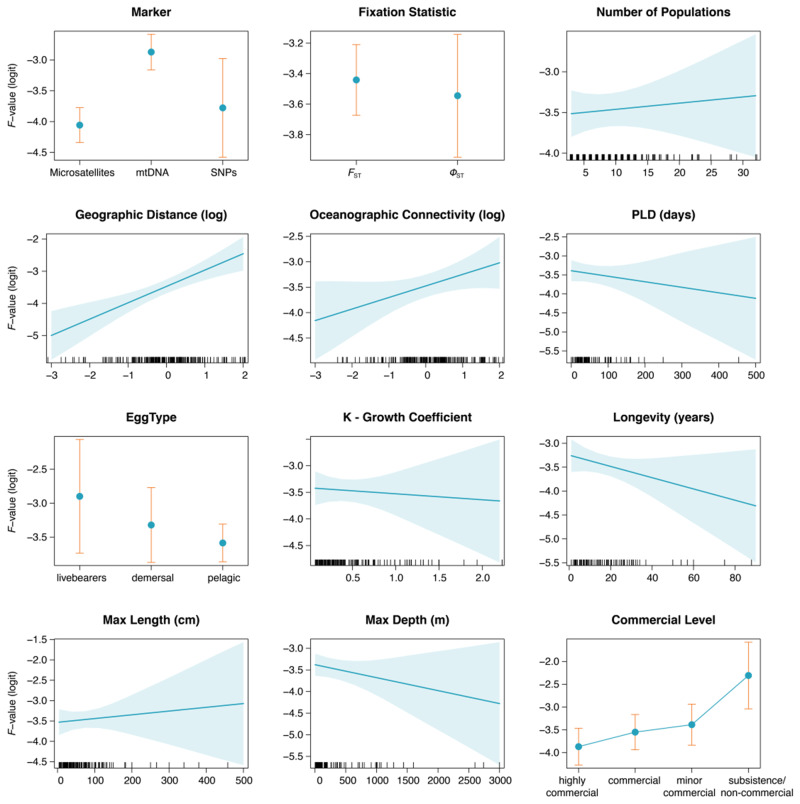
These results were corroborated by the fitted generalized linear mixed models (GLMMs). In both beta and normally distributed models, the response variable was found to be significantly influenced by both marker type and geographic distance (χ2 = 35.53 and χ2 = 17.85 in beta model, χ2 = 34.77 and χ2 = 17.62 in gaussian model, respectively; table 1). Studies conducted using mtDNA markers reported higher FST/ΦST estimates than those using microsatellites or SNPs (fig. 4 and supplementary table S3, Supplementary Material online), and populations separated by larger distances had higher likelihood of being more differentiated. Similar results were obtained when simplifying the models to increase the number of points included. Moreover, fixation indices also showed some degree of association with oceanographic connectivity estimates, although differences were not statistically significant at an α level of 0.05 (table 1). None of the remaining methodological variables (fixation metric and number of populations sampled) had a significant effect on differentiation. Similarly, neither of the analyzed life-history traits consistently explained the response estimates (all P > 0.10), with the exception of maximum depth which exhibited a significance negative association with fixation in the simplified models (P < 0.01; table 1). Commercial importance had a statistically significant association with genetic differentiation in all models (χ2 ranging from 12.44 to 32.06; table 1), with noncommercial or subsistence fisheries species exhibiting higher differentiation than those of minor-commercial, commercial, or highly commercial importance (supplementary table S3, Supplementary Material online). The three latter, although not accounting for enough variability to be considered significantly distinct, displayed a consistent gradient in differentiation estimates, exhibiting a positive trend from the highest to the lowest level of fishing pressure (fig. 4).
Table 1.
GLMM Fixed Effects’ Significance Tests.
| Predictors | Number of Obs: 248; Species: 152 |
Number of Obs: 279; Species: 178 |
||||||
|---|---|---|---|---|---|---|---|---|
|
F-value Mean
a
|
F-value Logit
b
|
F-value Mean
a
|
F-value Logit
b
|
|||||
| χ2 | P-value | χ2 | P-value | χ2 | P-value | χ2 | P-value | |
| Methodological | ||||||||
| Marker | 35.53 | <0.001*** | 34.77 | <0.001*** | 40.55 | <0.001*** | 36.92 | <0.001*** |
| Fixation statistic | 0.15 | 0.702 | 0.27 | 0.643 | 0.01 | 0.911 | 0.03 | 0.877 |
| Number of populations | 0.11 | 0.745 | 0.28 | 0.635 | 0.69 | 0.405 | 1.03 | 0.319 |
| Environmental | ||||||||
| Geographic distance (log) | 17.85 | <0.001*** | 17.62 | <0.001*** | 17.54 | <0.001*** | 15.80 | <0.001*** |
| Oceanographic connectivity (log) | 3.21 | 0.073• | 3.74 | 0.069• | 3.07 | 0.080• | 2.93 | 0.093• |
| Species traits | ||||||||
| PLD mean | 0.42 | 0.519 | 0.64 | 0.475 | — | — | — | — |
| Egg type | 1.01 | 0.603 | 3.28 | 0.245 | — | — | — | — |
| K | 0.88 | 0.347 | 0.10 | 0.780 | 1.20 | 0.273 | 0.40 | 0.528 |
| Longevity | 0.86 | 0.355 | 2.23 | 0.167 | 0.84 | 0.359 | 1.69 | 0.198 |
| Max length | 0.03 | 0.868 | 0.27 | 0.644 | 0.16 | 0.686 | 0.09 | 0.772 |
| Max depth | 1.55 | 0.214 | 1.40 | 0.274 | 8.23 | 0.004** | 10.33 | 0.002** |
| Commercial | ||||||||
| Commercial importance | 13.40 | 0.004** | 12.44 | 0.009** | 32.06 | <0.001*** | 27.67 | <0.001*** |
Note.—P-values of beta models (∼FST mean) obtained through likelihood ratio tests and Gaussian models (∼FST logit) tested using parametric bootstrap and 10,000 iterations.
Model fitted with a beta distribution (glmmTMB package).
Model fitted with normal distribution (lme4 package).
P < 0.001,
P < 0.01,
P < 0.05, •P < 0.10.
Fig. 4.
Effect plots of all fixed factors included in the normally distributed GLMM (using logit F-values as the response variable). Confidence intervals are displayed around fitted lines and value distributions are indicated as marks along the x-axes for the numeric variables.
Discussion
Our results confirm that genetic structure in marine fish species may result from an interplay of different factors, not only related with geographic distance and methodological aspects but potentially also related with fishing exploitation. Most of the life-history traits analyzed were not associated with the observed patterns of population structure across the assemblage of species analyzed. Yet, we found a relationship between species’ commercial importance and the level of population genetic differentiation, with exploited fish tending to exhibit lower FST/ΦST values than those of noncommercial importance or captured by subsistence fisheries.
Fishing Pressure
Continuous exploitation over multiple generations might have contributed toward some of the evolutionary changes behind the detected trend. Empirical and theoretical evidences show that size-selective harvesting targeting large individuals, for instance, tends to favor slower growth and earlier age at maturity (Conover and Munch 2002), although not exclusively so (Hilborn and Minte-Vera 2008). Thus, the dispersion potential of individuals, both during the larval and adult life stages, may also be nonrandom relatively to the genotype and, rather, directly dependent on some of these traits (Pukk et al. 2013). Even though a high heterogeneity between species within commercial levels was observed, FAMD results suggested the existence of some traits-related patterns (cluster centroids sequentially ordered on the first two principal components). Noncommercial/subsistence fisheries species exhibited, on average, a K growth coefficient twice as large as the commercial or highly commercial fish. Yet, these differences might have existed prior to human impact and therefore, without considering the temporal nature of these patterns (i.e., changes over time), it is impossible to attribute this trend to fisheries alone. The Clupeidae and Gadidae families, for example, include some of the most commercially important species and are historically highly prolific, often producing large numbers of pelagic eggs during spawning (Dulvy et al. 2003), which can increase their capacity of dispersal. Consequently, the reverse scenario can also be true, that is, species with certain sets of biological characteristics (typically favoring connectivity) can make them more profitable and therefore more heavily fished. Moreover, theoretically, some of the fisheries-induced shifts could occur in the opposite direction, favoring reduced dispersal and thus a higher differentiation between populations. By removing older individuals, fishing might be responsible, for example, for a decrease in the fitness and survivorship of larvae (Birkeland and Dayton 2005), which could also reduce gene flow between allopatric populations. This means that, together with varying selection pressures exerted by different types of fishing gear (Jørgensen et al. 2009), changing environmental conditions (McLean et al. 2018) and density-dependent processes (Hixon and Webster 2002; Lorenzen and Enberg 2002), evolutionary outcomes on species’ dispersal capabilities are inherently difficult to predict.
An alternative and non/exclusive mechanism could be linked with genetic homogenization due to fisheries-induced bottlenecks and consequent population replacement by immigration, even in marine species with putatively large population sizes. Similarly, interbreeding with hatchery-reared stocks through accidental or deliberate introductions can lead to accentuated decreases in genetic differentiation between populations, although this is expected to impact particularly freshwater species (Kenchington 2003). According to the Food and Agriculture Organization of the United Nations, over 30% of the world’s marine fish stocks are currently overexploited (FAO 2018). Considering that the genetic structure of many marine species is still unknown, it is highly probable that the extirpation and extinction of many populations of overfished species could have gone unnoticed due to replacement from neighboring populations (Pukk et al. 2013), a process of shifted genetic baselines (Assis et al. 2013). A large number of populations and subpopulations have either been driven to extinction or are still recovering from past depletions, with as many as 83 species of marine fish in Europe alone being reported to exhibit declining populations sizes (Nieto et al. 2015). Subsequently, individuals dispersing out of neighboring stocks could swamp and homogenize gene pools of these declined/fragmented populations, preventing or disrupting the development of local adaptations. A similar process has been proposed by Hutchinson et al. (2003) to explain genetic changes in a declining North Sea cod population, for example. After enduring severe commercial exploitation, the isolated population started to lose allelic diversity as its effective size was substantially reduced. Yet, following a period of increased divergence due to enhanced genetic drift, the growing impact of occasional immigrant fish from proximate populations has likely led to a gradual genetic replacement and consequent decrease in differentiation estimates. The same reduction in allelic richness and heterozygosity for overfished stocks has been observed by Pinsky and Palumbi (2014), using an analogous meta-analytical approach. Together, these results suggest that fishing activities might have the potential not only to reduce species genetic diversity but also to disrupt spatial genetic structure, which may negatively impact resilience and species’ capacity to withstand environmental changes across their distributional range.
Isolation by Distance
Differently to previous meta-analysis of genetic differentiation on the marine realm, we collected pairwise fixation estimates beyond global FST statistics, which allowed us to more accurately assess the influence of geographic distance and large-scale oceanographic flow patterns in populational genetic structure. Contrarily to what was expected, geographic distance explained better dispersal potential and gene flow than distances estimated through global ocean current patterns. This finding can be associated with the fact that fish, in general, are able to travel large distances, carry out long migrations throughout their lifetimes and cross important oceanographic barriers, which may reduce the importance of larval dispersion in the patterns of genetic connectivity, at least when compared with sessile organisms such as corals, sponges, and macroalgae (Assis, Serrão, et al. 2018). Moreover, most fish larvae have strong navigational capabilities and are able to perform diel vertical movements (Kingsford et al. 2002; Staaterman and Paris 2014) which can affect their distribution and make their dispersal trajectories challenging to explain, even despite the fact that active dispersion was accounted for in the models. Both larval behavior (e.g., geo and phototaxis) and vertical mixing of water masses may result in transport over deeper layers, in which case dispersal discrepancies could be expected (Gilg and Hilbish 2003). It is also likely that the broad temporal and spatial resolution at which sea currents data are available at global scales is not sufficient to capture microscale ocean dynamics that can also drive local transport or retention (Werner et al. 2007). These results suggest that seascape studies should implement higher-resolution circulation models, whenever available, and take into account larval behavior and the spawning period of each species, at each region, when analyzing species connectivity patterns.
Life-History Traits
Our results corroborate the idea that life-history traits are not strong predictors of genetic population structure, with no single variable significantly explaining the observed patterns excepting maximum depth. Similarly to additional studies (Weersing and Toonen 2009; Riginos et al. 2011; Nanninga and Manica 2018) we did not find a particularly strong association between average pelagic larval duration (PLD) and the extent of genetic differentiation, contributing to the growing body of evidence suggesting that this trait is a poor determinant of dispersal in marine species, contrarily to what has been traditionally assumed. It is possible that maximum PLD or larval swimming capacities are better correlated with species’ connectivity potential and population structure as suggested by Weersing and Toonen (2009) and Nanninga and Manica (2018); however, these variables are difficult to measure and therefore still unknown for many species, which renders them useless as proxies for dispersal. In contrast with Bradbury et al. (2008) and Riginos et al. (2011), we did not find an association between FST/ΦST estimates and egg type alone (livebearers vs. demersal vs. pelagic), neither between egg type and PLD, which cast doubts on the hypothesized lower connectivity of species with demersal/fixed eggs advanced by these studies. This lack of association was also reported by Galarza et al. (2009) and Nanninga and Manica (2018), which emphasizes that these early-life traits may be insufficient to estimate gene flow patterns across multispecific assemblages. Admittedly, factors such as temperature-dependent larval growth, intraspecific variability, stochastic mortality, and settlement success may also be important sources of variation and have ecologically meaningful impacts in the observed population structures.
Although deep-sea taxa have been hypothesized to disperse larger distances and exhibit higher connectivity, quantitative evidence supporting this notion is mostly nonexistent (Hilário et al. 2015). In our models, maximum depth was the life-history trait that accounted for the greatest amount of variance, especially when using a higher number of observations (excluding PLD and egg type). Our analyses suggest that this paradigm holds true and that, on average, fish occurring at greater depths might disperse further, corroborating the results obtained by Baco et al. (2016). It is possible that broader areas of contiguous environment together with uniform abiotic conditions (temperature, pressure, etc.) and the absence of strong barriers to dispersal might contribute to the observed levels of connectivity, yet the logistic and economic efforts required to survey deeper habitats make it challenging to investigate these questions. By adopting a meta-analytic approach and controlling for several confounding factors, our results are among the first to identify a significant association between depth occurrence and population structure in marine fish and might be of direct relevance for the design of deep-sea MPA networks and reserves.
It is important to acknowledge, however, that the revised studies comprised a larger proportion of demersal and reef-associated fish (34% and 36%, respectively) comparatively to benthopelagic (12%) and pelagic (19%) species, which might have introduced a small bias on the models. Nonetheless, pelagic fish are expected to show less genetic population differentiation than other classes of fish (Hauser and Ward 1998) and, hence, it is likely that our findings are in fact overconservative.
Genetic Markers and Fixation Indices
Population structure is commonly reported using a variety of different sampling and statistical frameworks. As anticipated, marker type had a significant influence on the obtained FST values, indicating that direct comparisons between studies using different genetic methodological tools should be done with care, as they may lead to incorrect inferences. Microsatellite and SNP markers conveyed significantly lower genetic differentiation than those using mtDNA alleles. An equivalent pattern has been found by Weersing and Toonen (2009) and Medina et al. (2018), with potential explanations ranging from different mutation rates, inheritance processes, and effective population sizes of each marker. The higher FST and ΦST values obtained by mtDNA in relation to the two nuclear markers are in line with the reduced effective population size (Ne) of haploid mitochondrial genes, one-fourth of the Ne of nuclear DNA due to maternal inheritance (assuming balanced sex ratios). This results in faster genetic drift and, therefore earlier lineage sorting, even though microsatellite loci can potentially suffer faster mutation rates. Moreover, in many studies, the high polymorphism of microsatellites loci is not accounted for when computing FST estimates (i.e., standardization based on marker heterozygosity), which together with homoplasy may lead to an underestimation of genetic differentiation (Balloux et al. 2000; Coates et al. 2009).
Regarding fixation metrics, we chose to include both FST and ΦST in the analysis to improve statistical power and to assess the relationship between the indices. FST has been the most widely used descriptive statistics in evolutionary genetics. Yet, following its introduction by Wright, multiple analogous metrics have been implemented (see Holsinger and Weir 2009 and Meirmans and Hedrick 2011 for a more indepth review). Although FST is based on haplotype frequencies, ΦST also takes into account the genetic distances among haplotypes. Thus, the lack of divergence between the two estimators might point toward recently diverged populations, that is, when evolutionary time was not long enough to result in the build-up of genetic differences. These results are further supported by the additional analyses carried out using solely mtDNA samples estimated using both metrics (supplementary fig. S6, Supplementary Material online).
Estimates of population differentiation do not seem to be significantly influenced by the number of samples surveyed contrarily to the findings of Jenkins et al. (2010), that reported an asymptotic relationship between the number of sampled populations and the likelihood of IBD (isolation-by-distance) significance.
Conclusion
By using a meta-analytic framework on a comprehensive data set and controlling for multiple factors potentially affecting connectivity estimates, this works builds on previous findings and reveals for the first time the potential association between genetic structure and commercial fisheries. Contrarily to marked spatial structure, harvested species exhibit lower genetic differentiation. It is likely that their typically large populational sizes might allow them to replenish depleted populations, yet, with the detrimental cost of reducing overall genetic diversity levels and therefore their potential for adaptation and evolution. Given the caveats inherently associated with meta-analysis approaches and the lack of studies focusing on the temporal nature of population structure over ecological meaningful periods, this hypothesis warrants further investigation.
Our review also confirms previous findings on the effect of marker type, with mtDNA loci typically yielding larger differentiation estimates than those measured with microsatellites or SNPs, which underlines the importance of considering their divergent nature when inferring population structure. Moreover, we quantitatively show that early-life-history traits (namely PLD) and broad-scale ocean circulation patterns might be insufficient to accurately predict patterns of connectivity in the absence of further knowledge, and that species occurring at higher depths may be able to sustain higher gene flow between allopatric populations.
Although further analyses are required to accurately characterize and predict connectivity for individual species, the broad-scale perspective here provided offers novel insights into the role of several factors in shaping marine genetic structure. As these patterns of subdivision are influenced by a plethora of processes, further insights could be obtained by analyzing changes in genetic differentiation over consecutive generations and by accounting for additional aspects such as species’ historical demography, environmental continuity, abiotic conditions, and larval behavior. Even though it remains a challenging task, understanding how multiple processes operate together is fundamental to ensure the sustainability of exploited marine species on the long term. Overall, the diversity in connectivity patterns and dispersal pathways described by this and several other studies (Bradbury et al. 2008; Ramesh et al. 2019) highlights the importance of conservation management strategies supported by sound scientific knowledge and argues the need for multilateral, holistic cooperation across national borders.
Materials and Methods
Literature Search and Data Collection
Genetic differentiation data were obtained from a systematic literature search. Studies published over a 10-year period (from 2008 to 2018) were compiled through Web of Science, using the terms: “phylogeography” or “population genetic structure” and “fish” and “marine.” All resulting references were methodically reviewed and selected whenever they met five main criteria: 1) the study included at least one marine fish species, 2) it analyzed genetic differentiation between non co-occurrent populations (i.e., spatial structure analysis), 3) at least three different locations were sampled, 4) pairwise FST or ΦST values were supplied (either in the main article or under the supplementary files), and 5) coordinates of the sampling sites were reported or could be accurately inferred (supplementary fig. S1, Supplementary Material online).
Posteriorly, all available data were collected and organized using a relational database structure, including methodological details such as scientific names, sampling site coordinates and number of fish analyzed, pairwise fixation values together with the corresponding genetic marker and all relevant species traits. These traits were selected based on previous studies and ecological meaningfulness, and included: average PLD, egg type (livebearers vs. demersal vs. pelagic), growth coefficient (K; rate at which an asymptotic length is approached in the von Bertalanffy equation), longevity, maximum length, maximum depth, and commercial importance (supplementary table S1, Supplementary Material online). Commercial importance was grouped into four main levels according to the extent to which the species are exploited: highly commercial, commercial, minor commercial, and subsistence fisheries/noncommercial, as defined in Froese et al. (2000) and based on the Food and Agriculture Organization’s list of commercial species and Sea Around Us fisheries database (http://www.seaaroundus.org, last accessed October 2020). Data manipulation as well as all subsequent analyses were carried out using R v3.6 (R Core Team 2020). Species traits were sourced from FishBase (Froese and Pauly 2019), either using the “rfishbase” package (Boettiger et al. 2012) or through custom web scraping routines, or inferred from closely related species (species within the same genus or family). Whenever values for a given variable were missing, we retrieved the traits of all available species belonging to the same genus. Similarly, if no data were available for the genus-level, we sampled species within the same family. Then, for numeric variables, we computed the average of all the values retrieved; and for categorical variables, we used the most common value. Additionally, some of the categorical variables were reclassified and aggregated in broader classes in order to simplify the analysis and increase sample size (supplementary table S2, Supplementary Material online).
Data Analyses
To assess the effect of the metric chosen, we included both FST and ΦST values in the analysis by converting data to long format and storing the type of index (FST or ΦST). Whenever both FST and ΦST values were available for a given record, the entry was duplicated to accommodate the two response variables. Pairwise comparisons between overlapping sampling locations within the same study were discarded due to the increased probabilities of belonging to the same population and to avoid biasing the analyses with temporally structured samples. Moreover, due to the uneven number of data points coming from different genetic markers, only samples resulting from the more commonly used microsatellites, mtDNA, or SNPs markers were included (accounting approximately for 95% of the full data set).
Negative FST/ΦST values were converted to zero, as results lower than zero indicate that population differentiation is negligible (Meirmans 2006; Medina et al. 2018). All coordinates were converted to decimal degrees and checked against the GSHHG (Global Self-consistent, Hierarchical, High-resolution Geography Database; Wessel and Smith 1996) coastline layer, at intermediate resolution. Whenever the reported locations lied within land surfaces, they were relocated to the nearest marine location. Subsequently, geographic distances (i.e., shortest distance by water between pairs of sites) were calculated using a least-cost path analysis (Dijkstra’s algorithm, “gdistance” package; van Etten 2017). Oceanographic connectivity (i.e., relative measure of distance via ocean currents) was calculated using the “rWind” package (Fernández‐López and Schliep 2019). Data on the northward and eastward components of oceanographic currents were retrieved from Bio-ORACLE (Assis, Tyberghein, et al. 2018), a data set providing geophysical, biotic, and environmental data for surface and benthic marine realms at a spatial resolution of 5 arcmin (∼9.2 km at the equator). A conductance matrix was computed based on the direction and intensity of currents, and relative distances were calculated using the same least-cost algorithm. As this transition layer is anisotropic (asymmetric due to the directionality of water flows), the process was repeated twice for each pair of sampling locations with swapped “from” and “to” directions, with only the smaller of the distances being assigned to the pair. Moreover, we accounted for the swimming capabilities of most fish larvae by allowing movements against flow direction, at an increased cost (Felicísimo et al. 2008).
Prior to statistical analyses, we assessed the potential bias in publications related to reported values or sample sizes, to test whether larger differentiation estimates had a higher probability of being published and whether any skew in differentiation values existed in relationship with the number of individuals sampled. The former hypothesis was investigated through histograms of reported FST/ΦST values and the second was assessed using funnel-like plots, that is, scatterplots of fixation values against mean sample size. Furthermore, similarities between biological traits of ray-finned species (class Actinopterygii) within each commercial level were assessed through an FAMD (FactoMineR package; Lê et al. 2008), a principal component method comprising both quantitative and qualitative variables. Finally, to visually assess eventual discrepancies in the distribution of sampling locations (spatial bias) in relation with commercial level, we produced heat maps of sample sites using kernel density estimation.
As a preliminary data analysis and in order to provide visually interpretable patterns of correlation between the considered traits and the levels of genetic structure observed, we plotted all pairwise differentiation values against geographic distance and oceanographic connectivity estimates, per marker and fixation metric, and calculated Pearson’s correlation coefficients. To reduce heteroskedasticity, distances were log-transformed and standardized into a z-score, and fixation values were linearized using a logit transformation.
To test the effect of each variable in the observed differentiation estimates, we averaged the FST/ΦST values (per species, marker and publication) and adjusted GLMMs. Two different sets of models were generated, one using the raw averages of the fixation estimates following a beta distribution (“glmmTMB” package; Brooks et al. 2017), and another using linearized indices (F-index logit) with a normal distribution (“lme4” package; Bates et al. 2015). As missing values are not allowed in the modeling algorithm used, the analysis was conducted exclusively on the subset of species for which all variables could be retrieved (n = 152, ∼80% of all species). In both cases, hierarchical taxonomic structure (Family/Genus/Species) was included as a nested random effect to account for both phylogenetic nonindependence and repetitive measurements (i.e., the cases when multiple publications studied the same species). An amount of 1 × 10−7 was added to all values in the first set of models, because beta distribution parametrizations are not possible with exact zeros. Additionally, as complete life-history traits were not available for all species and GLMMs adopt listwise deletion excluding all incomplete data rows, we repeated the analyses dropping the insignificant terms that had higher percentages of missing values in order to increase the number of observations included.
Before model fitting, all numeric variables were scaled and collinearity between all predictors was assessed through a variance inflation factor analysis (Zuur et al. 2010). Overall significance of the fixed covariates was assessed with ANOVA (analysis of variance) tables, using likelihood ratio tests for the beta-distributed models and parametric bootstrap for the normally distributed models (10,000 iterations). Fixed effects plots with corresponding confidence intervals were generated using the “effects” package (Fox and Hong 2009) and pairwise post hoc analysis was conducted using Tukey’s HSD (honestly significant difference) tests, available through the “emmeans” package (Lenth 2020).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by the European Maritime and Fisheries Fund and the MAR2020 program through project REDAMP (MAR-01.04.02-FEAMP-0015). Additional funding was provided by FCT - Foundation for Science and Technology through the projects UIDB/04326/2020 and PTDC/BIA-CBI/6515/2020, and through the transitional norms DL57/2016/CP1361/CT0035 and DL 57/2016/CP1361/CT0036.
Data Availability
Genetic differentiation data supporting this article are from previously reported studies and data sets, which are cited in supplementary information S2, Supplementary Material online. The processed data will be shared on reasonable request to the corresponding author. Species life-history traits were sourced from FishBase and are available in Figshare: https://figshare.com/s/8d8a73dfc1eedc0fb5e0.
References
- Alós J, Palmer M, Catalan IA, Alonso-Fernández A, Basterretxea G, Jordi A, Buttay L, Morales-Nin B, Arlinghaus R.. 2014. Selective exploitation of spatially structured coastal fish populations by recreational anglers may lead to evolutionary downsizing of adults. Mar Ecol Prog Ser. 503:219–233. [Google Scholar]
- Alós J, Palmer M, Rosselló R, Arlinghaus R.. 2016. Fast and behavior-selective exploitation of a marine fish targeted by anglers. Sci Rep. 6(1):38093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis J, Castilho Coelho N, Alberto F, Valero M, Raimondi P, Reed D, Alvares Serrão E.. 2013. High and distinct range-edge genetic diversity despite local bottlenecks. PLoS One 8(7):e68646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis J, Serrão EÁ, Coelho NC, Tempera F, Valero M, Alberto F.. 2018. Past climate changes and strong oceanographic barriers structured low‐latitude genetic relics for the golden kelp Laminaria ochroleuca. J Biogeogr. 45(10):2326–2336. [Google Scholar]
- Assis J, Tyberghein L, Bosch S, Verbruggen H, Serrão EA, De Clerck O.. 2018. Bio‐ORACLE v2.0: extending marine data layers for bioclimatic modelling. Glob Ecol Biogeogr. 27(3):277–284. [Google Scholar]
- Audzijonyte A, Kuparinen A, Fulton EA.. 2013. How fast is fisheries‐induced evolution? Quantitative analysis of modelling and empirical studies. Evol Appl. 6(4):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baco AR, Etter RJ, Ribeiro PA, Von der Heyden S, Beerli P, Kinlan BP.. 2016. A synthesis of genetic connectivity in deep‐sea fauna and implications for marine reserve design. Mol Ecol. 25(14):3276–3298. [DOI] [PubMed] [Google Scholar]
- Balkenhol N, Cushman S, Storfer A, Waits L.. 2015. Landscape genetics: concepts, methods, applications. West Sussex, UK: John Wiley & Sons. [Google Scholar]
- Balloux F, Brunner H, Lugon-Moulin N, Hausser J, Goudet J.. 2000. Microsatellites can be misleading: an empirical and simulation study. Evolution 54(4):1414–1422. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S.. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw. 67(1):1–48. [Google Scholar]
- Birkeland C, Dayton PK.. 2005. The importance in fishery management of leaving the big ones. Trends Ecol Evol. 20(7):356–358. [DOI] [PubMed] [Google Scholar]
- Boettiger C, Lang DT, Wainwright PC.. 2012. rfishbase: exploring, manipulating and visualizing FishBase data from R. J Fish Biol. 81(6):2030–2039. [DOI] [PubMed] [Google Scholar]
- Boustany AM, Reeb CA, Teo SLH, De Metrio G, Block BA.. 2006. Genetic data and electronic tagging indicate that the Gulf of Mexico and Mediterranean Sea are reproductively isolated stocks of bluefin tuna (Thunnus thynnus). ICCAT SCRS 089. Col Vol Sci Pap. 60(4):1154–1159.
- Bradbury IR, Laurel B, Snelgrove PVR, Bentzen P, Campana SE.. 2008. Global patterns in marine dispersal estimates: the influence of geography, taxonomic category and life history. Proc R Soc B. 275(1644):1803–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks ME, Kristensen K, van Benthem KJ, Magnusson A, Berg CW, Nielsen A, Skaug HJ, Machler M, Bolker BM.. 2017. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 9(2):378–400. [Google Scholar]
- Coates BS, Sumerford DV, Miller NJ, Kim KS, Sappington TW, Siegfried BD, Lewis LC.. 2009. Comparative performance of single nucleotide polymorphism and microsatellite markers for population genetic analysis. J Hered. 100(5):556–564. [DOI] [PubMed] [Google Scholar]
- Conover DO, Munch SB.. 2002. Sustaining fisheries yields over evolutionary time scales. Science 297(5578):94–96. [DOI] [PubMed] [Google Scholar]
- Cowman PF, Bellwood DR.. 2013. Vicariance across major marine biogeographic barriers: temporal concordance and the relative intensity of hard versus soft barriers. Proc R Soc B. 280(1768):20131541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KM, Canino MF, Spies IB, Hauser L.. 2009. Genetic isolation by distance and localized fjord population structure in Pacific cod (Gadus macrocephalus): limited effective dispersal in the northeastern Pacific Ocean. Can J Fish Aquat Sci. 66(1):153–166. [Google Scholar]
- Dulvy NK, Sadovy Y, Reynolds JD.. 2003. Extinction vulnerability in marine populations. Fish Fisheries. 4(1):25–64. [Google Scholar]
- FAO. 2018. The State of World Fisheries and Aquaculture 2018—meeting the sustainable development goals. Rome (Italy: ): FAO. [Google Scholar]
- Felicísimo ÁM, Muñoz J, González-Solis J.. 2008. Ocean surface winds drive dynamics of transoceanic aerial movements. PLoS One. 3(8):e2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐López J, Schliep K.. 2019. rWind: download, edit and include wind data in ecological and evolutionary analysis. Ecography 42(4):804–810. [Google Scholar]
- Fox J, Hong J.. 2009. Effect displays in R for multinomial and proportional-odds logit models: extensions to the effects package. J Stat Softw. 32(1):1–24. [Google Scholar]
- Froese R, Capuli E, Garilao C, Pauly D.. 2000. The SPECIES table. In: Froese R, Pauly D, editors. FishBase 2000: concepts designs and data sources. Los Baños (Philippines: ): ICLARM. p. 76–85. [Google Scholar]
- Froese R, Pauly D.. 2019. FishBase. Available from: http://www.fishbase.org. World Wide Web Electron. Publ. version (12/2019).
- Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF, Rico C.. 2009. The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci U S A. 106(5):1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilg MR, Hilbish TJ.. 2003. The geography of marine larval dispersal: coupling genetics with fine‐scale physical oceanography. Ecology 84(11):2989–2998. [Google Scholar]
- Halpern BS, Selkoe KA, Micheli F, Kappel CV.. 2007. Evaluating and ranking the vulnerability of global marine ecosystems to anthropogenic threats. Conserv Biol. 21(5):1301–1315. [DOI] [PubMed] [Google Scholar]
- Harrison HB, Williamson DH, Evans RD, Almany GR, Thorrold SR, Russ GR, Feldheim KA, van Herwerden L, Planes S, Srinivasan M, et al. 2012. Larval export from marine reserves and the recruitment benefit for fish and fisheries. Curr Biol. 22(11):1023–1028. [DOI] [PubMed] [Google Scholar]
- Hauser L, Ward RD.. 1998. Population identification in pelagic fish: the limits of molecular markers. In: Carvalho GR, editor. Advances in molecular ecology. Amsterdam: IOS Press. p. 191–224. [Google Scholar]
- Heino M, Pauli BD, Dieckmann U.. 2015. Fisheries-induced evolution. Annu Rev Ecol Evol Syst. 46(1):461–480. [Google Scholar]
- Hilário A, Metaxas A, Gaudron SM, Howell KL, Mercier A, Mestre NC, Ross RE, Thurnherr AM, Young C.. 2015. Estimating dispersal distance in the deep sea: challenges and applications to marine reserves. Front Mar Sci. 2:1–14. [Google Scholar]
- Hilborn R, Minte-Vera CV.. 2008. Fisheries-induced changes in growth rates in marine fisheries: are they significant? Bull Mar Sci. 83(1):95–105. [Google Scholar]
- Hixon MA, Webster MS.. 2002. Density dependence in reef fish populations. In: Sale PF, editor. Coral reef fishes: dynamics and diversity in a complex ecosystem. San Diego (CA: ): Academic Press. p. 303–325. [Google Scholar]
- Hoegh-Guldberg O, Bruno JF.. 2010. The impact of climate change on the world’s marine ecosystems. Science 328(5985):1523–1528. [DOI] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS.. 2009. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet. 10(9):639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson WF, Oosterhout C, Rogers SI, Carvalho GR.. 2003. Temporal analysis of archived samples indicates marked genetic changes in declining North Sea cod (Gadus morhua). Proc R Soc Lond B. 270(1529):2125–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins DG, Carey M, Czerniewska J, Fletcher J, Hether T, Jones A, Knight S, Knox J, Long T, Mannino M, et al. 2010. A meta‐analysis of isolation by distance: relic or reference standard for landscape genetics? Ecography 33(2):315–320. [Google Scholar]
- Jørgensen C, Ernande B, Fiksen Ø.. 2009. Size‐selective fishing gear and life history evolution in the Northeast Arctic cod. Evol Appl. 2(3):356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenchington EL. 2003. The effects of fishing on species and genetic diversity. In: Sinclair M, Valdimarsson G, editors. Responsible fisheries in the marine ecosystem. Rome and Wallington: FAO and CAB International. p. 235–253. [Google Scholar]
- Kingsford MJ, Leis JM, Shanks A, Lindeman KC, Morgan SG, Pineda J.. 2002. Sensory environments, larval abilities and local self-recruitment. Bull Mar Sci. 70(1):309–340. [Google Scholar]
- Lê S, Josse J, Husson F.. 2008. FactoMineR: an R package for multivariate analysis. J Stat Softw. 25(1):1–18. [Google Scholar]
- Lenth R. 2020. Emmeans: estimated marginal means, aka least-squares means. R package version 1.4.5. Available from: https://CRAN.R-project.org/package=emmeans.
- Lorenzen K, Enberg K.. 2002. Density-dependent growth as a key mechanism in the regulation of fish populations: evidence from among-population comparisons. Proc R Soc Lond B. 269(1486):49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M, Mouillot D, Auber A.. 2018. Ecological and life history traits explain a climate-induced shift in a temperate marine fish community. Mar Ecol Prog Ser. 606:175–186. [Google Scholar]
- Medina I, Cooke GM, Ord TJ.. 2018. Walk, swim or fly? Locomotor mode predicts genetic differentiation in vertebrates. Ecol Lett. 21(5):638–645. [DOI] [PubMed] [Google Scholar]
- Meirmans PG. 2006. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60(11):2399–2402. [PubMed] [Google Scholar]
- Meirmans PG, Hedrick PW.. 2011. Assessing population structure: FST and related measures. Mol Ecol Resour. 11(1):5–18. [DOI] [PubMed] [Google Scholar]
- Nanninga GB, Manica A.. 2018. Larval swimming capacities affect genetic differentiation and range size in demersal marine fishes. Mar Ecol Prog Ser. 589:1–12. [Google Scholar]
- Nieto A, Ralph GM, Comeros-Raynal MT, Kemp J, García Criado M, Allen DJ, Dulvy NK, Walls RHL, Russell B, Pollard D.. 2015. European Red List of marine fishes. Luxembourg (Luxembourg: ): Publications Office of the European Union. [Google Scholar]
- Pauls SU, Nowak C, Bálint M, Pfenninger M.. 2013. The impact of global climate change on genetic diversity within populations and species. Mol Ecol. 22(4):925–946. [DOI] [PubMed] [Google Scholar]
- Pinsky ML, Palumbi SR.. 2014. Meta‐analysis reveals lower genetic diversity in overfished populations. Mol Ecol. 23(1):29–39. [DOI] [PubMed] [Google Scholar]
- Pukk L, Kuparinen A, Järv L, Gross R, Vasemägi A.. 2013. Genetic and life‐history changes associated with fisheries‐ induced population collapse. Evol Appl. 6(5):749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: https://www.r-project.org/. Accessed January 14, 2020. [Google Scholar]
- Ramesh N, Rising JA, Oremus KL.. 2019. The small world of global marine fisheries: the cross-boundary consequences of larval dispersal. Science 364(6446):1192–1196. [DOI] [PubMed] [Google Scholar]
- Riginos C, Buckley YM, Blomberg SP, Treml EA.. 2014. Dispersal capacity predicts both population genetic structure and species richness in reef fishes. Am Nat. 184(1):52–64. [DOI] [PubMed] [Google Scholar]
- Riginos C, Douglas KE, Jin Y, Shanahan DF, Treml EA.. 2011. Effects of geography and life history traits on genetic differentiation in benthic marine fishes. Ecography 34(4):566–575. [Google Scholar]
- Ruzzante DE, Taggart CT, Doyle RW, Cook D.. 2001. Stability in the historical pattern of genetic structure of Newfoundland cod (Gadus morhua) despite the catastrophic decline in population size from 1964 to 1994. Conserv Genet. 2(3):257–269. [Google Scholar]
- Staaterman E, Paris CB.. 2014. Modelling larval fish navigation: the way forward. ICES J Mar Sci. 71(4):918–924. [Google Scholar]
- van Etten J. 2017. R package gdistance: distances and routes on geographical grids. J Stat Softw. 76:1–21. [Google Scholar]
- Weersing K, Toonen RJ.. 2009. Population genetics, larval dispersal, and connectivity in marine systems. Mar Ecol Prog Ser. 393:1–12. [Google Scholar]
- Werner FE, Cowen RK, Paris CB.. 2007. Coupled biological and physical models: present capabilities and necessary developments for future studies of population connectivity. Oceanography 20(3):54–69. [Google Scholar]
- Wessel P, Smith WHF.. 1996. A global, self‐consistent, hierarchical, high‐resolution shoreline database. J Geophys Res. 101(B4):8741–8743. [Google Scholar]
- Zuur AF, Ieno EN, Elphick CS.. 2010. A protocol for data exploration to avoid common statistical problems. Methods Ecol Evol. 1(1):3–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genetic differentiation data supporting this article are from previously reported studies and data sets, which are cited in supplementary information S2, Supplementary Material online. The processed data will be shared on reasonable request to the corresponding author. Species life-history traits were sourced from FishBase and are available in Figshare: https://figshare.com/s/8d8a73dfc1eedc0fb5e0.