Abstract
Integration of a conjugative plasmid into a bacterial chromosome can promote the transfer of chromosomal DNA to other bacteria. Intraspecies chromosomal conjugation is believed responsible for creating the global pathogens Klebsiella pneumoniae ST258 and Escherichia coli ST1193. Interspecies conjugation is also possible but little is known about the genetic architecture or fitness of such hybrids. To study this, we generated by conjugation 14 hybrids of E. coli and Salmonella enterica. These species belong to different genera, diverged from a common ancestor >100 Ma, and share a conserved order of orthologous genes with ∼15% nucleotide divergence. Genomic analysis revealed that all but one hybrid had acquired a contiguous segment of donor E. coli DNA, replacing a homologous region of recipient Salmonella chromosome, and ranging in size from ∼100 to >4,000 kb. Recombination joints occurred in sequences with higher-than-average nucleotide identity. Most hybrid strains suffered a large reduction in growth rate, but the magnitude of this cost did not correlate with the length of foreign DNA. Compensatory evolution to ameliorate the cost of low-fitness hybrids pointed towards disruption of complex genetic networks as a cause. Most interestingly, 4 of the 14 hybrids, in which from 45% to 90% of the Salmonella chromosome was replaced with E. coli DNA, showed no significant reduction in growth fitness. These data suggest that the barriers to creating high-fitness interspecies hybrids may be significantly lower than generally appreciated with implications for the creation of novel species.
Keywords: conjugation, experimental evolution, Escherichia coli, Salmonella Typhimurium, relative fitness, recombination
Introduction
Bacterial genotypes can be altered either by mutation or by the acquisition of foreign genetic material. The great majority of viable mutations are expected to be near-neutral and have a small or even no measurable effect on phenotype (Andersson et al. 2011; Charlesworth and Charlesworth 2018). In contrast, genetic material acquired from another species is often associated with the acquisition of novel phenotypic traits in the recipient but may also be associated with an initial reduction in fitness. In experimental studies where individual essential genes (encoding ribosomal protein S20, translation factor EF-Tu, and dihydrofolate reductase, DHFR) were transferred across species boundaries, fitness defects were common but could in general be compensated by mutations that increased the levels of the respective enzymes (Lind et al. 2010; Bershtein et al. 2015; Kacar et al. 2017). This mechanism of compensatory evolution supported a conclusion that for those genes the reduced fitness was primarily a function of insufficient expression rather than protein functionality. This conclusion is also supported by the results of a recent large-scale cross-phyla gene replacement study involving two nonessential genes, where the major outcome was that mutations upregulating enzyme levels were the most frequent outcome of the experimental evolution to overcome initial fitness defects (Sandberg et al. 2020). In the present study, we have examined the fitness consequences and the potential for compensatory evolution associated with the interspecies replacement of, not one, but many genes simultaneously.
For horizontal genetic transfer (HGT) to have a lasting impact on a bacterial lineage, the acquired genetic material must be stably maintained. Some acquired genetic material encodes the ability to replicate and maintain itself in the new host (for example, plasmids), whereas other genetic material must be recombined into the genome of the new host bacterium to ensure its maintenance. Mobile genetic elements, in particular conjugative plasmids and integrative conjugative elements, are very successful as agents of HGT because they encode the abilities to ensure both transfer and stable maintenance in a new host cell. Such mobile elements, which can be up to several hundred kb in length, play an important role in the transfer of antibiotic resistance and virulence genes between bacteria (Pallen and Wren 2007; Wozniak and Waldor 2010; Johnson and Grossman 2015; Munita and Arias 2016; Diard and Hardt 2017).
Horizontal transfer of chromosomal DNA that is not intrinsically mobile can also occur. This horizontal transfer can be mediated by bacteriophage (Penades et al. 2015; Frazao et al. 2019), by transformation of environmental DNA (Dowson et al. 1989; Ameyama et al. 2002; Domingues et al. 2012), and, the subject of this study, by conjugation mediated by an integrated conjugative plasmid (Baron et al. 1968; McAshan et al. 1999; Hopwood 2006). For example, in Escherichia coli the conjugative F-factor plasmid carries a number of IS elements and can integrate into the chromosome by homologous recombination with similar chromosomal IS elements. The resulting Hfr (high frequency of recombination) strain can conjugate chromosomal DNA to a recipient cell via a conjugative pilus. An important feature of Hfr-mediated HGT, distinguishing it from transduction and transformation, is that the length of DNA transferred into a recipient cell can be very great, in principle, up to the entire length of the donor chromosome (Cavalli-Sforza 1950; Cavalli et al. 1953; Hayes 1953; Davidson et al. 1996; Waksman 2019).
Evidence that conjugative transfer of chromosomal DNA could be clinically important comes from the analysis of pandemic bacterial pathogens. Klebsiella pneumoniae ST258 is a highly virulent and antibiotic-resistant strain that has spread globally since its initial detection in the USA (Kitchel et al. 2009). In ST258, a contiguous segment of DNA, roughly 20% of the chromosome, has been acquired from a distantly related K. pneumoniae strain, ST442 (Chen et al. 2014). The exact nature of ST258’s virulence, its rapid dissemination and whether these features are functionally linked to its hybrid chromosome are not fully understood. Another example is the globally prevalent and highly fluoroquinolone-resistant E. coli ST1193. ST1193 is hypothesized to have been created by the transfer of a single large segment of chromosomal DNA from a distantly related E. coli strain, followed by 11 different homologous recombination events occurring with the recipient’s chromosome (Tchesnokova et al. 2019). This HGT event included the simultaneous transfer of multiple fluoroquinolone resistance mutations located in gyrA and parC, two genes separated by approximately 1 Mb on the chromosome. It is highly unlikely that the acquisition of very long segments of chromosomal DNA as observed in both ST258 and ST1193 could be the result of transformation or phage-mediated transduction and they are more likely due to conjugational transfer of chromosomal DNA.
The creation of interspecies hybrids by conjugation could potentially contribute to the emergence of novel clinical pathogens and might also have a general evolutionary significance in the creation of new species. Interspecies hybrids have been created using an E. coli Hfr strain as donor and Salmonella enterica serovar Typhimurium (S. Typhimurium) as recipient (Rayssiguier et al. 1989; Rayssiguier et al. 1991). These studies showed that hybrid creation occurred by RecABCD-dependent homologous recombination and identified mismatch repair (MMR) activity as a major barrier to hybrid strain construction. The resulting hybrid chromosomes were analyzed using gene mapping for the presence or absence of discrete chromosomal markers and the results were consistent with most hybrid strains having incorporated a single continuous segment of donor DNA into the recipient chromosome (Matic et al. 1994). However, very little is known about the detailed genetic architecture of such interspecies hybrids: whether they actually incorporate contiguous segments of DNA (like ST258) or a series of discontinuous segments throughout a chromosomal region (like ST1193), the nature of their recombination joint-points, and whether they suffer significant reductions in growth fitness as a result of mixing genetic elements from different genomes. To address these questions, we created by conjugation interspecies hybrids between E. coli and S. Typhimurium, carrying different lengths of donor and recipient DNA, and used genome sequencing to analyze their genetic architecture. We examined the hybrids in terms of growth fitness, and used experimental evolution to address whether and by which mechanisms hybrid strains could ameliorate their fitness costs.
Results and Discussion
Genetic Architecture of Transconjugants
Chromosomal hybrids between E. coli and S. Typhimurium were created by conjugating an E. coli Hfr donor and a S. Typhimurium recipient (Materials and Methods) on antibiotic-free rich medium for 3, 6, or 12 h. Cells were then resuspended and plated on selective agar medium containing two antibiotics, one selecting a donor chromosomal marker and the other selecting a recipient chromosomal marker. The frequency of colonies on the selective plates ranged from <3.0 × 10−9 up to 9.6 × 10−6, increasing as a function of the amount of time allowed for conjugation, and decreasing as a function of the distance of the selected donor marker from the Hfr origin of transfer (supplementary table S1, Supplementary Material online). Colonies picked from selective plates were streak-purified on the same medium. PCR was used to assess the presence of the selected donor (TET, KAN, or RIF) and recipient (SPT or CHL) antibiotic resistance gene markers to make an initial confirmation of the validity of putative interspecies hybrid strains. To analyze the genetic architecture of the hybrids we determined their whole-genome sequences (WGS).
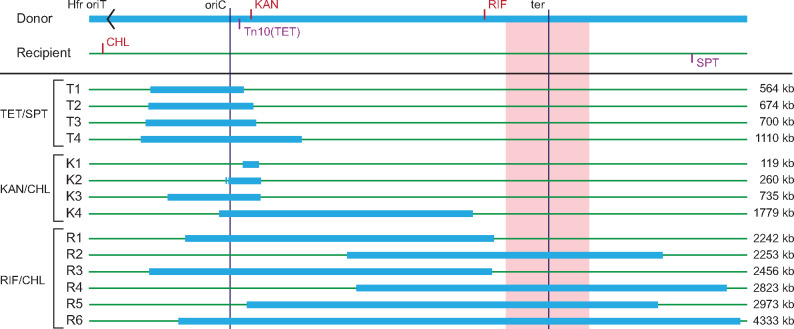
WGS analysis was made on the parental strains and on 14 hybrid strains: Eight strains selected for the transfer of proximal markers (T1–T4 selected with TET, K1–K4 selected with KAN), and six strains (R1–R6) selected for the more distant RIF marker (fig. 1). Analysis of the sequences revealed that in each strain S. Typhimurium chromosomal DNA had been replaced by E. coli chromosomal DNA, creating hybrid interspecies chromosomes (fig. 1). The precise genetic locations of the chromosomal replacements are given in supplementary table S2, Supplementary Material online. In 13 of the 14 hybrid strains, a single contiguous E. coli sequence replaced an equivalent region of Salmonella chromosome. The length of acquired DNA ranged from approximately 119 kb up to 4,333 kb, equivalent to replacing 90% of the Salmonella chromosome with E. coli DNA (fig. 1). In one hybrid strain (K2) the acquired E. coli DNA was in two segments (2.4 and 257 kb) separated by 7 kb of Salmonella DNA. This two-segment recombinant is located very close to the Salmonella origin of replication. None of the 14 hybrids showed replacement of S. Typhimurium DNA closer than 130 kb to the position of the Hfr in the E. coli donor. An inversion of approximately 558 kb around the terminus of replication represents a striking difference between the genomes of the E. coli and S. Typhimurium strains and none of the hybrids had donor DNA recombined within this inversion. Replacements in this region either stopped before the inversion or crossed the inverted region and replaced it completely (fig. 1).
Fig. 1.
Genome structure of the fourteen hybrid strains. Donor DNA (Escherichia coli) is shown in blue and recipient DNA (S. Typhimurium) in green. Drug resistance markers and the Hfr origin of transfer (oriT) are indicated in the parental genomes (top). The E. coli region around the terminus of replication (ter) that is inverted relative to the equivalent region in Salmonella is indicated in red. Selective conditions during hybrid formation are shown to the left and the total size of donor DNA that is inserted into the recipient chromosome is shown to the right.
Recombination Junctions Are Nonrandomly Distributed
Previous studies have shown that the formation of hybrid chromosomes between E. coli and Salmonella occurs by RecABCD-dependent homologous recombination and is strongly inhibited by the activity of the MutSLH MMR system (Rayssiguier et al. 1989). We hypothesized that this mechanism would favor hybrids with recombination junctions in chromosomal sequences with higher than average interspecies nucleotide identity. We determined the average nucleotide identity (ANI) between the chromosomes of the E. coli donor and S. Typhimurium recipient and compared this with the ANI in a region centered on each of the 30 recombination junctions in the 14 recombinants. ANIs were calculated for sliding windows of 500 bp and 1 kb (supplementary fig. S1, Supplementary Material online). We observed a strong bias in favor of higher-than-ANI around recombination junctions, consistent with the homologous recombination hypothesis (500 bp: 88.2% vs. 83.9%, P < 0.001; 1 kb: 86.1% vs. 81.9%, P < 0.001). Interestingly, despite the increased overall nucleotide identity around the recombination junction there was very little perfect sequence homology at the point where S. Typhimurium changed to E. coli sequence. Based on single nucleotide polymorphisms (SNPs) between the two species, the median length of perfect sequence identity was 21 bp but could be as short as 4 bp, (supplementary fig. S2, Supplementary Material online). Such short homologies are generally considered to be too short for RecA-dependent homologous recombination (Watt et al. 1985; Shen and Huang 1986; Brandis et al. 2018). We therefore conclude that recombination is initiated by the formation of an imperfect heteroduplex between the S. Typhimurium and E. coli chromosomes (supplementary fig. S3 steps a–c, Supplementary Material online). This hypothesis is consistent with the observation that the MutSLH MMR system inhibits hybrid formation (Rayssiguier et al. 1989). The mismatches within the chromosomal heteroduplex could be repaired by the MMR system or be maintained until the next round of replication, similar to the models for intrachromosomal homologous recombination and gene conversion between the heterologous sequences of tufA and tufB in Salmonella (Abdulkarim and Hughes 1996; Hughes 2000). Either case would lead to the formation of a clear transition between S. Typhimurium and E. coli DNA (supplementary fig. S3 step d, Supplementary Material online). Another possibility is that the MMR system only repairs a section of the heteroduplex or that the base excision repair system (BER) repairs single mismatches (Krokan and Bjoras 2013; Modrich 2016). After the next round of replication, partial MMR and BER can lead to transitions that alternate between S. Typhimurium and E. coli DNA (supplementary fig. S3 steps e–h, Supplementary Material online). We analyzed the chromosomal region around the recombination junctions in all fourteen transconjugants and found that three junctions (3 out of 30 junctions, 10%) displayed alternating transitions (supplementary fig. S4, Supplementary Material online). In one case, the alternating section was 1,736 bp away from the main junction indicating that recombination was initiated with a formation of a heteroduplex of almost 2 kb in length (supplementary fig. S4B, Supplementary Material online). We conclude that formation of hybrid chromosomes between E. coli and Salmonella is initiated by heteroduplex formation of up to and potentially >1.7 kb length. This initial step in the recombination process is most likely responsible for the observed bias of recombination junctions to be located in regions with higher-than-ANI.
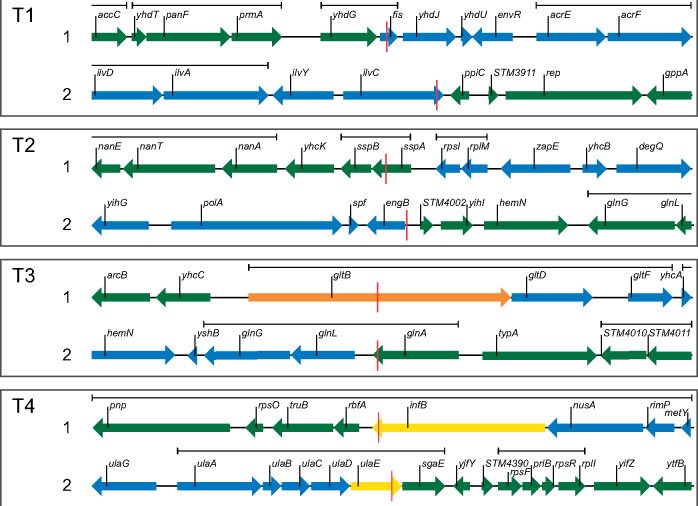
We also examined the consequences of recombination on individual genes and operons by focusing on a window of 10 kb centered around each recombination junction. We expected that most junctions would create hybrid operons (because interoperon space is relatively small) but that junctions within protein coding sequences might be underrepresented due to selection to maintain protein functionality. We observed that recombination junctions frequently occurred within operons (21 out of 30 junctions, 70%), suggesting, at least for these operons, that hybrid operon structures are tolerated (fig. 2 and supplementary fig. S5, supplementary Material online). Half of the recombination junctions were either located outside of protein coding sequences (5/30) or did not create a hybrid protein sequence (10/30). A further seven produced proteins with >99% amino acid identity to the respective protein in S. typhimurium or E. coli whereas the remaining eight had from 86% to 99% identity. The distribution of amino acid identities of proteins encoded by hybrid coding sequences formed during conjugation is significantly higher than that associated with simulated random hybrid protein formation between Salmonella and E. coli (MeanObserved = 97.8%, MeanRandom = 95.4%, P = 0.02, see Materials and Methods). This bias towards higher protein identity could be the result of selection during the conjugation experiment or a consequence of the strong bias in favor of higher-than-ANI around recombination junctions. The experimental conditions during conjugation do not impose any selection for bacterial fitness but only for survival (essential proteins must remain functional). Therefore, it is more likely that the observed high degree of protein identity is a function of recombination at regions of higher-than-ANI.
Fig. 2.
Analysis of recombination junctions. Chromosome structure around each recombination junction in a 10-kb window for hybrids T1–T4. S. Typhimurium genes are shown in green and Escherichia coli genes are shown in blue. Genes with a hybrid coding sequence are colored based on their protein identity. Hybrid genes that encode proteins identical to the protein in either species are shown in green or blue, genes with a protein identity >99% to either species are shown in yellow, and genes with a protein identity <99% to either species are in orange. The red line indicates the first nucleotide that does not match the Salmonella typhimurium sequence. Operon membership as indicated by the EcoCyc database (Keseler et al. 2017) is indicated above the genes. Junctions in hybrid strains K1–K4 and R1–R6 are shown in supplementary figure S5, Supplementary Material online.
Fitness of Chromosomal Hybrids
Escherichia coli and S. enterica have been evolving independently for at least 100 My (Ochman and Groisman 1994). It seems reasonable to assume that their individual genomic contents and complex regulatory networks are under selection for high physiological fitness within each species. Accordingly, we hypothesize that replacing part of the genome of one species with DNA from the other species is likely to reduce fitness because it will mix genetic/physiological elements that have not co-evolved to work efficiently together. In particular, we hypothesized that relative fitness should progressively decrease as the ratio of genomic content from each species approaches equivalence in the hybrids, with 50% from E. coli and 50% from Salmonella.
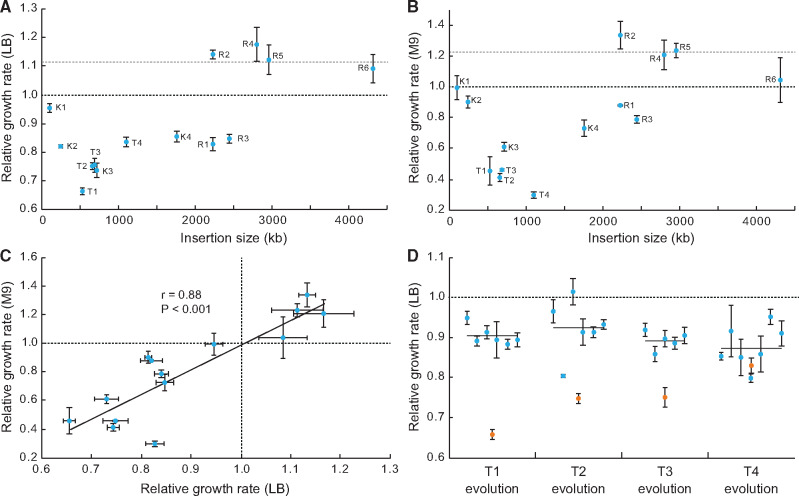
To determine whether the hybrid genomes were associated with a fitness cost we measured the exponential growth rate of each hybrid strain, and of the two parental strains, in rich medium (LB) and in minimal medium (M9) (fig. 3A and B, supplementary table S3, Supplementary Material online). We made two striking observations. Firstly, the genome of the hybrid with the lowest fitness contained only 12% E. coli sequence rather than the expected 50%. More importantly, two of the hybrid strains (R1 and R2) have respectively 49% and 53% remaining Salmonella chromosome (fig. 1) and are accordingly a good test of the null hypothesis that a 50% hybrid should have the lowest fitness. One of these hybrids (R1) has a relatively low fitness (0.82 in LB and 0.87 in M9 relative to the Salmonella parent) whereas the other (R2) has very high fitness (1.14 in LB and 1.33 in M9 relative to the Salmonella parent). The large difference in relative fitness between R1 and R2 despite each having approximately the same amount of native and foreign chromosomal DNA invalidates the null hypothesis and suggests instead that relative fitness is more likely a consequence of the functional complementarity of the respective hybrid genome contents rather than a being directly related to the amount of foreign DNA.
Fig. 3.
Growth rate analysis of hybrid strains. Relative growth rate of the fourteen hybrids as a function of insertion size in rich (A) and minimal (B) growth medium. (C) Correlation of relative growth rate in rich and minimal medium for all hybrids. (D) Relative growth rate of evolved (blue) and unevolved parental (orange) hybrids T1–T4. The black line indicates the average relative growth rate of the evolved hybrids. Error bars in each panel display the standard deviation of four independent measurements. The dashed lines indicate the relative fitness of the parental recipient Salmonella strain (black, defined as 1) and the donor Escherichia strain (gray).
Secondly, although most of the hybrid strains suffered a significant loss of fitness in one or both of the growth media, four of the hybrids did not suffer any growth rate reduction in either medium (fig. 3C). In these four hybrid strains (R2, R4, R5, R6) where 45–90% of the Salmonella chromosome has been replaced with an equivalent region of the E. coli chromosome, the growth rate is equal to or greater than that of the parental Salmonella and similar to the parental E. coli (with the exception of R6 which is less fit in M9 medium relative to E. coli) (fig. 3A and B). A feature common to these four “high-fitness” hybrids is that they have each acquired the chromosomal region around the E. coli terminus of replication that is inverted relative to the equivalent region in Salmonella (fig. 1). The significance of this correlation is uncertain because these four hybrids have also acquired other sequences outside of the inverted region. For each of the 14 hybrid strains, the relative growth rates were broadly similar in both rich and minimal medium (r = 0.88, P < 0.001, fig. 3C) suggesting that the observed reductions in growth rate are not media-specific but are likely to reflect a general loss of fitness, for these hybrids.
Experimental Evolution Can Reduce Hybrid Fitness Costs
Because most hybrid strains suffered a large relative reduction in growth fitness (fig. 3A–C) we next addressed whether, and by which mechanisms, the fitness costs could be ameliorated. We chose four strains (T1–T4) in which growth rate was 20–30% lower in LB relative to the Salmonella parent, and in which the chromosome replacement size ranged from 564 to 1,110 kb (fig. 1). At least five independent culture lineages of each strain were serially passaged in LB. After 500 generations of experimental evolution (50 cycles of 10 generations each) a single colony was isolated from each culture and its exponential growth rate measured. For all of the hybrid strains (T1–T4) some or all of the evolved lineages showed a significant increase in growth rate, in one case equaling that of the parental strain (fig. 3D, supplementary table S4, Supplementary Material online).
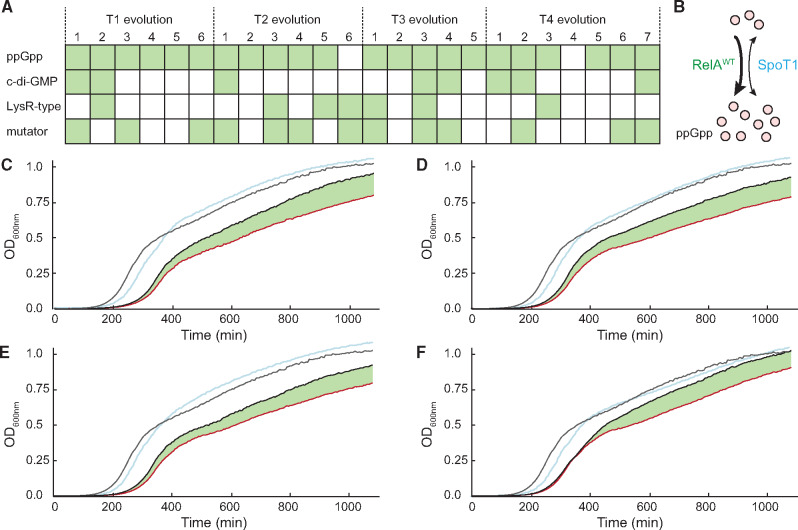
We used whole-genome sequencing to determine the genetic changes associated with the improved growth rates in each of the 24 evolved clones (supplementary table S5, Supplementary Material online). Strikingly, we noted a small number of gene classes where mutations arose in multiple evolved lineages (fig. 4A): 1) genes involved in regulating and reprogramming global transcriptional patterns (relA, cyclic-di-GMP-associated genes, LysR family genes); 2) genes of the MMR system that regulate mutation rates (mutS, mutL, mutH, mutM) and lead to an accumulation of point mutations in the evolved lineages (supplementary tables S5–S7, Supplementary Material online); and 3) genes where mutations have previously been shown to cause adaptation to the growth medium (glpT, treB, arcB, nudE, fliY, fimW, ompF, yjeP) (Knöppel et al. 2018; Garoff et al. 2020). Media adaptation mutations were expected in all experimental evolution studies and are probably not selected specifically to ameliorate the fitness costs of strains with hybrid chromosomes. On the other hand, the high frequency of selection of mutations in genes that regulate global transcriptional patterns, and genes that increase the global mutation rate, strongly suggests that there is no easy or direct mutational route to reduce the fitness costs of these hybrids. Mutations in relA were selected in almost all lineages (22/24), and most of them were clearly gene-inactivating mutations (18 IS10 insertions and 1 premature stop codon). RelA (ppGpp synthase) works in concert with SpoT (dual action ppGpp synthase/hydrolase) to modulate the cellular levels of ppGpp (fig. 4B) in response to perceived carbon or amino acid starvation (Hauryliuk et al. 2015). There is a significant difference between the donor and recipient strains in their respective RelA-SpoT systems (supplementary tables S2 and S8, Supplementary Material online). The E. coli donor is a double mutant (relA1, spoT1) producing an inactive RelA (relA A87 ins IS2) and a SpoT with a missense mutation in the synthetase domain (spoT H255Y) and a two amino acid insertion in the hydrolase domain (spoT D84 ins QD), that potentially makes it less active. In contrast, the Salmonella recipient carries wild-type copies of both genes (relAWT, spoTWT). In the hybrid strains, 9/14 acquired spoT1 from E. coli whereas retaining relAWT from Salmonella (supplementary table S8, Supplementary Material online) creating a hybrid ppGpp regulatory system that potentially causes inappropriate global transcription regulation responses (fig. 4B). We reasoned that this hybrid regulatory system might be reducing growth fitness and hypothesized that the inactivation of relA during experimental evolution was selected because it recreated a system equivalent to that present in the E. coli donor. To test this hypothesis, we inactivated relA in the hybrid strains. The growth characteristics in LB of strains in which relA was deleted were compared with those of the unevolved hybrid strains, T1–T4, and the parental S. Typhimurium. Exponential growth rates of hybrids with relA deletions were indistinguishable from the parental strains but in each case the deletion of relA was sufficient to cause a significant increase in stationary phase culture density (fig. 4C–F, and supplementary table S9, Supplementary Material online). We concluded that inactivation of relA contributes significantly to the improvement in growth fitness of the hybrid strains. The frequent selection in the experimental evolution of mutations in genes associated with reprogramming transcription, or increasing global mutation rates, is indicative of the low-fitness hybrid strains having no simple or direct genetic route to ameliorate their fitness costs. Rewiring of transcriptional networks has been independently observed as one of the frequent response mechanisms in bacteria adapting to gene replacements (Sandberg et al. 2020).
Fig. 4.
Analysis of the fitness compensatory evolution experiment. (A) Overview over acquisition of mutations that effect the production of ppGpp or c-di-GMP, mutations within LysR-type transcriptional regulators, and mutations in genes that can cause a mutator phenotype. A detailed overview over all acquired mutations is given in supplementary table S5, Supplementary Material online. (B) Potential model for the RelA-SpoT interaction within the hybrids. Acquisition of the mutated donor spoT1 gene (spoT D84 ins QD, H255Y) leads to an overproduction of ppGpp due to reduced ppGpp hydrolysis and a premature entry into stationary phase. Deletion of the relA gene reduces cellular ppGpp levels and restores bacterial fitness. (C–F) Growth curves of the S. Typhimurium recipient strain (TH6767, gray), the four unevolved hybrids T1 (C), T2 (D), T3 (E), and T4 (F) (red), a representative evolved hybrid (T1-1, T2-5, T3-1, and T4-7, blue) and unevolved hybrids with deleted relA gene (black). The improvement in growth caused by the deletion of relA is indicated in green.
Conclusions
Escherichia coli and S. Typhimurium are members of two related bacterial genera that have been evolving separately for >100 My (Ochman and Wilson 1987; Doolittle et al. 1996). Despite their independent evolution the two species have maintained a broadly similar gene order on their respective chromosomes (Ochman and Groisman 1994). Previous research has shown that viable interspecies hybrids can be created by conjugation between E. coli and S. Typhimurium (Rayssiguier et al. 1989) but relatively little is known of the details of the genomic organization of hybrid strains (Matic et al. 1994) and the relative fitness and evolvability of such hybrids has not been studied. Recent discoveries showing that clinically important strains of pathogenic bacteria are intraspecies chromosomal hybrids between different sequence types has made understanding the genetics and evolutionary biology of hybrid strains increasingly important (Pitout et al. 2015; Tchesnokova et al. 2019).
The main observations from our study of hybrids made between E. coli and Salmonella by conjugative transfer of chromosomal DNA are summarized below:
Most hybrid genomes (13/14) contain a single contiguous fragment of E. coli DNA replacing a genetically similar region and length of Salmonella DNA.
Recombination joint points are preferentially located in sequences of higher-than-ANI, as expected for a process that occurs via homologous recombination.
Most hybrids suffered reduced growth rate in both rich and minimal medium.
Two recombinants, R1 and R2, with 50% hybrid genomes but very different fitness values, show that it is the specifics of the genetic replacement rather than the length of the replacement that is most important for determining fitness.
Evolution to restore fitness involved increased mutation rates and transcriptional rewiring.
The most striking observation is the surprisingly high frequency of hybrids with large chromosomal replacements (4/14) that showed no reduction in growth fitness in either rich or minimal medium relative to the parental strains (fig. 3C).
In summary, this study shows that interspecies hybrids between E. coli and Salmonella with high growth fitness can be created at a high frequency by conjugation, suggesting that the physiological and genetic barriers to creating such hybrids could be surmounted. Given the enormity of bacterial populations, the prevalence of conjugative elements, and the ongoing processes that could create novel environmental niches, it would not be surprising if interspecies hybridization, including hybrid chromosomes contribute to the creation of novel species.
Materials and Methods
Bacterial Strains and Genetic Constructions
Bacterial strains are derivatives of S. enterica serovar Typhimurium LT2 (referred to as S. Typhimurium) and E. coli K-12 MG1655. Strain genotypes are shown in supplementary table S2, Supplementary Material online. Lambda Red recombineering using the pSIM6 plasmid (Sharan et al. 2009) was used to place antibiotic-resistance gene cassettes into the S. Typhimurium gene ygdI (TH10684), and into the E. coli genes ymdA, yiiF, and queE (CH6941). The relA gene coding sequence was deleted from hybrid strains T1–T4 (strains CH7193–CH7196), to create CH9713–CH9716 using the DIRex method (Näsvall 2017).
Media, Growth Conditions, and Antibiotic Concentrations
LB (1% tryptone, 0.5% yeast extract, 1% NaCl) was used as rich growth medium. M9 (35 mM Na2HPO4, 22 mM KH2PO4, 7 mM NaCl, 19 mM NH4Cl, 0.1 mM CaCl, and 1 mM MgSO4) supplemented with 0.2% glucose, 0.1% Thiamine, 50 µM FeCl3, and seven amino acids (l-proline, l-methionine, l-leucine, l-valine, l-isoleucine, l-lysine, l-tryptophan) each at a final concentration of 40 mg/l, was used as the defined minimal growth medium. Solid media (Luria agar, LA) contained in addition 1.5% agar. Liquid cultures were grown on a rotary shaker at 180–200 rpm. All cultures, liquid and solid, were incubated at 37 °C for 18 h unless otherwise stated. Selections for the loss of sacB were made on salt-free LA containing 5% sucrose (Ellis et al. 2001). Antibiotics were used at the following final concentrations in media: tetracycline (TET) 15 mg/l; spectinomycin (SPT) 100 mg/l; kanamycin (KAN) 50 mg/l; rifampicin (RIF) 100 mg/l; chloramphenicol (CHL) 60 mg/l; ampicillin (AMP) 100 mg/l.
Hfr Conjugation Protocol
Hfr donor (E. coli) and recipient (S. Typhimurium) strain pairs in conjugation experiments were either CH6459 and TH6767 (selecting transconjugants on TET+SPT), or CH6941 and TH10684, selecting transconjugants on KAN+CHL or RIF+CHL. Liquid cultures of donor and recipient strains were grown individually overnight in LB (final density 2–4 × 109 CFU/ml). For each conjugation experiment an equal volume of donor and recipient culture was mixed and 100 µl of the mixture was spread onto a nitrocellulose filter (pore size 0.22 μm, diameter 25 mm; Sigma Aldrich) placed on an antibiotic-free LA plate incubated at 37 °C for 3 h, 6 h, or 12 h (supplementary table S1, Supplementary Material online). Filters were removed using sterile tweezers, transferred to a microfuge tube containing 1 ml PBS, bacteria were resuspended by vortexing, and 250 µl aliquots were spread onto LA plates containing TET+SPT, CHL+KAN, or CHL+RIF, as appropriate to select transconjugants. Selective plates were incubated at 37 °C for 48 h and colonies were picked and pure-streaked onto the same selective media. Transconjugants with hybrid chromosomes were validated using PCR to identify drug resistance cassettes specific to donor and the recipient chromosomes. Hybrid strains were stored in LB containing 10% glycerol at –80 °C.
Polymerase Chain Reaction
PCR amplification of drug-resistance cassettes for use in Lambda red recombineering was done using VeraSeq 2.0 High-Fidelity DNA Polymerase (Enzymatics, MA), according to the manufacturer’s protocol. Prior to recombineering PCR products were purified using QIAquick PCR Purification Kit (Qiagen, Germany). Resistance cassettes in transconjugants were identified based on the size of the PCR product after amplification across a cassette-insertion site using primers homologous to DNA sequences on either side of the site, using Taq PCR Master Mix (Thermo Scientific, Waltham). Oligonucleotides used for PCR are listed in supplementary table S10, Supplementary Material online.
Whole-Genome Sequencing and Analysis
Genomic DNA was prepared using the MasterPure DNA and RNA Purification Kit (Epicentre, Chicago) from liquid overnight cultures or from cells scraped directly off an agar plate. Extracted DNA was suspended in Elution Buffer (Qiagen, Germany; 10 mM Tris–Cl, pH 8.5). DNA concentrations were measured in a Qubit 3.0 Fluorometer (Invitrogen, California). DNA was diluted in milliQ water (Merck KGaA, Germany) to a final concentration of to 0.2 ng/ml. Diluted samples were prepared for whole-genome sequencing according to the Nextera XT DNA Library Preparation Guide (Illumina Inc., Wisconsin). Before sequencing, the fragment size distribution of the prepared DNA samples was assessed using the Agilent High Sensitivity D1000 ScreenTape System (Agilent Technologies, California). Sequencing was performed in a MiSeq desktop sequencer, according to the manufacturer’s instructions (Illumina Inc., Wisconsin). Sequence data were aligned and analyzed using version 11.0.1 of the CLC Genomics Workbench (CLCbio, Qiagen, Denmark). Recombination junctions were defined as the first nucleotide that does not match the S. typhimurium sequence.
Average Nucleotide Identity
The ANI between the full genomes of the donor E. coli and the recipient S. Typhimurium genomes was calculated as previously described (Goris et al. 2007) using a sliding 500 bp window (step size: 100 bp, minimal length 350 bp, minimum identity: 70%) and 1-kb window (step size: 200 bp, minimal length 700 bp, minimum identity: 70%). ANI values for the recombination junctions were calculated for the 500-bp and 1-kb windows centered on the first nucleotide mismatch in the hybrid relative to the equivalent sequence in each of the parental strains. ANI values around the recombination junctions were compared with the genome-wide ANI values using a two-tailed Kolmogorov–Smirnov test.
Average Amino Acid Identity
The average amino acid identity (AAI) (Konstantinidis and Tiedje 2005) between the full proteomes of the donor E. coli and the recipient S. Typhimurium genomes was calculated using a minimal alignment length of 20 amino acids with a minimal identity of 70%. The set of homologous proteins identified during this analysis was used to simulate random hybrid protein formation. In total, 1,000 pairs of proteins were randomly chosen from this set (individual protein pairs could be chosen multiple times) and hybrid proteins were formed that contain Salmonella and E. coli sequence at a random proportion (the choice of protein pairs and proportions was made using Excel version 16.16.27). The distribution of AAI values of the 25 hybrid proteins formed during conjugation were compared with the simulated random hybrid proteins using a two-tailed Kolmogorov–Smirnov test.
Growth Rate Measurements
Growth rates in liquid culture were measured using a Bioscreen C machine (Oy Growth Curves Ab Ltd, Finland) recording changes in optical density (OD) as a function of time. Measurements were performed in a 100-well honeycomb plate, each well containing 300 µl culture, with continuous shaking at 37 °C for 18 h (LB growth) or 48 h (M9 growth) with OD recorded every 5 min. Measurements in LB medium were initiated from overnight cultures (biological replicates) each diluted 1:1000 in LB before inoculation into assay wells. For measurements in M9, colonies were scraped from LA plates (two biological replicates) resuspended in 0.9% NaCl to approximately the density of an overnight culture, diluted 1:1000 in M9 (supplemented as described above in Media), and inoculated into assay wells. Each experiment was run in four independent replicates. Doubling times were calculated from the increase in OD over the full growth curve with a sliding window of ten measurement points. Maximum exponential growth rates were defined as the measurement window with the minimal doubling time. Growth fitness was calculated relative to the doubling time of the parental Salmonella isolate and compared using a two-tailed t-test.
Experimental Evolution
Independent lineages of four hybrid strains, T1–T4 (CH7193–CH7196, supplementary table S2, Supplementary Material online) were serially passaged in LB culture for 500 generations by transferring 2 µl of overnight culture into 2 ml of fresh LB every day for 50 days. Cultures were grown at 37 °C on a rotary shaker set at 180 rpm and every 100 generations an aliquot was stored at –80 °C for future analysis.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by grants to D.H. from the Swedish Research Council (Vetenskapsrådet, Grant No. 2017-03953) and the Carl Trygger Foundation (Grant No. CTS17:204). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
The whole-genome sequence data underlying this article (paired-end sequence reads of the parental Salmonella Typhimurium and Escherichia coli strains, interspecies hybrid strains, and evolved hybrids strains) have been deposited at the NCBI Bioproject database with accession number: PRJNA667834, https://www.ncbi.nlm.nih.gov/sra/PRJNA667834.
References
- Abdulkarim F, Hughes D.. 1996. Homologous recombination between the tuf genes of Salmonella typhimurium. J Mol Biol. 260(4):506–522. [DOI] [PubMed] [Google Scholar]
- Ameyama S, Onodera S, Takahata M, Minami S, Maki N, Endo K, Goto H, Suzuki H, Oishi Y.. 2002. Mosaic-like structure of penicillin-binding protein 2 Gene (penA) in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime. Antimicrob Agents Chemother. 46(12):3744–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DI, Hughes D, Roth JR.. 2011. The origin of mutants under selection: interactions of mutation, growth and selection. In: Böck RCI, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, Ussery D, editors. EcoSal-Escherichia coli and Salmonella: cellular and molecular biology. Washington (DC: ): ASM Press. [DOI] [PubMed] [Google Scholar]
- Baron LS, Gemski P, Johnson EM, Wohlhieter JA.. 1968. Intergeneric bacterial matings. Bacteriol Rev. 32(4_Pt_1):362–369. [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Serohijos AW, Bhattacharyya S, Manhart M, Choi JM, Mu W, Zhou J, Shakhnovich EI.. 2015. Protein homeostasis imposes a barrier on functional integration of horizontally transferred genes in bacteria. PLoS Genet. 11(10):e1005612.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandis G, Cao S, Hughes D.. 2018. Co-evolution with recombination affects the stability of mobile genetic element insertions within gene families of Salmonella. Mol Microbiol. 108(6):697–710. [DOI] [PubMed] [Google Scholar]
- Cavalli LL, Lederberg J, Lederberg EM.. 1953. An infective factor controlling sex compatibility in Bacterium coli. J Gen Microbiol. 8(1):89–103. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL. 1950. Sexuality of bacteria. Boll Ist Sieroter Milan. 29(9–10):281–289. [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 2018. Neutral variation in the context of selection. Mol Biol Evol. 35(6):1359–1361. [DOI] [PubMed] [Google Scholar]
- Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN.. 2014. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. mBio 5(3):e01355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson BE, Kordias N, Dobos M, Hillier AJ.. 1996. Genomic organization of lactic acid bacteria. Antonie Van Leeuwenhoek. 70(2-4):161–183. [DOI] [PubMed] [Google Scholar]
- Diard M, Hardt WD.. 2017. Evolution of bacterial virulence. FEMS Microbiol Rev. 41(5):679–697. [DOI] [PubMed] [Google Scholar]
- Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, Nielsen KM.. 2012. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 8(8):e1002837.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF, Feng DF, Tsang S, Cho G, Little E.. 1996. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271(5248):470–477. [DOI] [PubMed] [Google Scholar]
- Dowson CG, Hutchison A, Brannigan JA, George RC, Hansman D, Linares J, Tomasz A, Smith JM, Spratt BG.. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 86(22):8842–8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis HM, Yu D, DiTizio T, Court DL.. 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA. 98(12):6742–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazao N, Sousa A, Lassig M, Gordo I.. 2019. Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. Proc Natl Acad Sci USA. 116(36):17906–17915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff L, Pietsch F, Huseby DL, Lilja T, Brandis G, Hughes D.. 2020. Population bottlenecks strongly influence the evolutionary trajectory to fluoroquinolone resistance in Escherichia coli. Mol Biol Evol. 37(6):1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM.. 2007. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 57(1):81–91. [DOI] [PubMed] [Google Scholar]
- Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K.. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 13(5):298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes W. 1953. The mechanism of genetic recombination in Escherichia coli. Cold Spring Harb Symp Quant Biol. 18:75–93. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. 2006. Soil to genomics: the Streptomyces chromosome. Annu Rev Genet. 40(1):1–23. [DOI] [PubMed] [Google Scholar]
- Hughes D. 2000. Co-evolution of the tuf genes links gene conversion with the generation of chromosomal inversions. J Mol Biol. 297(2):355–364. [DOI] [PubMed] [Google Scholar]
- Johnson CM, Grossman AD.. 2015. Integrative and Conjugative Elements (ICEs): what they do and how they work. Annu Rev Genet. 49(1):577–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacar B, Garmendia E, Tuncbag N, Andersson DI, Hughes D.. 2017. Functional constraints on replacing an essential gene with its ancient and modern homologs. mBio 8(4):e01276-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martínez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, et al. 2017. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 45(D1):D543–D550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG.. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 53(8):3365–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöppel A, Knopp M, Albrecht LM, Lundin E, Lustig U, Näsvall J, Andersson DI.. 2018. Genetic adaptation to growth under laboratory conditions in Escherichia coli and Salmonella enterica. Front Microbiol. 9:756.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM.. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 187(18):6258–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan HE, Bjoras M.. 2013. Base excision repair. Cold Spring Harb Perspect Biol. 5(4):a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Tobin C, Berg OG, Kurland CG, Andersson DI.. 2010. Compensatory gene amplification restores fitness after inter-species gene replacements. Mol Microbiol. 75(5):1078–1089. [DOI] [PubMed] [Google Scholar]
- Matic I, Radman M, Rayssiguier C.. 1994. Structure of recombinants from conjugational crosses between Escherichia coli donor and mismatch-repair deficient Salmonella typhimurium recipients. Genetics 136(1):17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAshan SK, Vergin KL, Giovannoni SJ, Thaler DS.. 1999. Interspecies recombination between enterococci: genetic and phenotypic diversity of vancomycin-resistant transconjugants. Microb Drug Resist. 5(2):101–112. [DOI] [PubMed] [Google Scholar]
- Modrich P. 2016. Mechanisms in E. coli and human mismatch repair (Nobel lecture). Angew Chem Int Ed Engl. 55(30):8490–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Arias CA.. 2016. Mechanisms of antibiotic resistance. Microbiol Spectr. 4:VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näsvall J. 2017. Direct and Inverted Repeat stimulated excision (DIRex): simple, single-step, and scar-free mutagenesis of bacterial genes. PLoS One 12(8):e0184126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Groisman EA.. 1994. The origin and evolution of species differences in Escherichia coli and Salmonella typhimurium. EXS 69:479–493. [DOI] [PubMed] [Google Scholar]
- Ochman H, Wilson AC.. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 26(1–2):74–86. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Wren BW.. 2007. Bacterial pathogenomics. Nature 449(7164):835–842. [DOI] [PubMed] [Google Scholar]
- Penades JR, Chen J, Quiles-Puchalt N, Carpena N, Novick RP.. 2015. Bacteriophage-mediated spread of bacterial virulence genes. Curr Opin Microbiol. 23:171–178. [DOI] [PubMed] [Google Scholar]
- Pitout JD, Nordmann P, Poirel L.. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 59(10):5873–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayssiguier C, Dohet C, Radman M.. 1991. Interspecific recombination between Escherichia coli and Salmonella typhimurium occurs by the RecABCD pathway. Biochimie 73(4):371–374. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C, Thaler DS, Radman M.. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342(6248):396–401. [DOI] [PubMed] [Google Scholar]
- Sandberg TE, Szubin R, Phaneuf PV, Palsson BO.. 2020. Synthetic cross-phyla gene replacement and evolutionary assimilation of major enzymes. Nat Ecol Evol. 4(10):1402–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Thomason LC, Kuznetsov SG, Court DL.. 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 4(2):206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P, Huang HV.. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112(3):441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokova V, Radey M, Chattopadhyay S, Larson L, Weaver JL, Kisiela D, Sokurenko EV.. 2019. Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene loci. Proc Natl Acad Sci USA. 116(29):14740–14748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G. 2019. From conjugation to T4S systems in Gram-negative bacteria: a mechanistic biology perspective. EMBO Rep. 20(2):e47012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt VM, Ingles CJ, Urdea MS, Rutter WJ.. 1985. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci USA. 82(14):4768–4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK.. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 8(8):552–563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome sequence data underlying this article (paired-end sequence reads of the parental Salmonella Typhimurium and Escherichia coli strains, interspecies hybrid strains, and evolved hybrids strains) have been deposited at the NCBI Bioproject database with accession number: PRJNA667834, https://www.ncbi.nlm.nih.gov/sra/PRJNA667834.