Abstract
Hybrids between species often show extreme phenotypes, including some that take place at the molecular level. In this study, we investigated the phenotypes of an interspecies diploid hybrid in terms of protein–protein interactions inferred from protein correlation profiling. We used two yeast species, Saccharomyces cerevisiae and Saccharomyces uvarum, which are interfertile, but yet have proteins diverged enough to be differentiated using mass spectrometry. Most of the protein–protein interactions are similar between hybrid and parents, and are consistent with the assembly of chimeric complexes, which we validated using an orthogonal approach for the prefoldin complex. We also identified instances of altered protein–protein interactions in the hybrid, for instance, in complexes related to proteostasis and in mitochondrial protein complexes. Overall, this study uncovers the likely frequent occurrence of chimeric protein complexes with few exceptions, which may result from incompatibilities or imbalances between the parental proteomes.
Keywords: protein–protein interaction, hybridization, Saccharomyces cerevisiae, Saccharomyces uvarum
Introduction
One of the major goals of evolutionary biology is to understand the molecular basis of phenotypic diversity, which fuels evolution by natural selection. Hybrids between species provide a unique opportunity to investigate the molecular underpinnings of phenotypic diversity. One could expect hybrids to show an intermediate phenotype, in which their characters are midway between parental phenotypes. However, hybrids often show a large spectrum of phenotypes, including extreme values that are unexpected given the parental traits (Rieseberg et al. 1999; Landry et al. 2007; Maheshwari and Barbash 2011; Bar-Zvi et al. 2017). Indeed, previous studies have demonstrated the importance of large-scale regulatory rewiring in hybrids that impacts many processes and molecular phenotypes, such as nucleosome positioning, translation efficiency, protein abundance, methylation, transcription of noncoding RNA, and the replication program (Landry et al. 2007; Tirosh et al. 2010; Tirosh and Barkai 2011; McManus et al. 2014; Zhu et al. 2017; Bar-Zvi et al. 2017; Bamberger et al. 2018; Zhao et al. 2018).
In the context of a cell, protein–protein interactions (PPIs) are central to molecular functions. Stable, nontransient PPIs lead to the formation of protein complexes with diverse functions such as DNA replication, repair and transcription, transport, catalysis, signaling, and many others (Sowmya et al. 2015). As protein complexes have such an important role, variation in their organization impacts the phenotype of organisms. Therefore, examining how PPI networks integrate in interspecific hybrids is of great interest.
Variation in the composition of protein complexes caused by hybridization between species can lead to many qualitative outcomes (as shown in fig. 1A). As hybrids between species that are amenable to genetic studies often do not suffer from dramatic fitness loss, we can broadly expect proteins to interact in the hybrids as they do in the parental species for complexes related to core cellular functions. The complexes could then be of two types: chimeric and parental. Chimeric complexes would result from interlogous (interspecies) and intralogous (intraspecies) PPIs and therefore, they would be a mixture of the two parental proteomes. On the other hand, some parental complexes could assemble from proteins of one parental species only, i.e., complexes would be formed only or majoritarily by intralogous PPIs. Parental complexes in hybrids could preferentially form, for instance, if affinities are higher among proteins from the same parental species than between species, or if the difference in timing of expression in cis between species prevents interlogous PPIs. Lastly, another possibility is that novel complexes could emerge in the hybrids, that is, complexes with interlogous PPIs that do not have equivalent PPIs in the parents.
Fig. 1.
Possible scenarios for the assembly of a protein complex in hybrids and the experimental layout of the comparative proteomics approach. (A) Scenarios depicting the assembly of a three-protein (P1, P2, and P3) complex in hybrid, with intralogous and interlogous PPIs. (B) Experimental layout of the comparative proteomics approach. In the first step, proteins of hybrid and parental strains were labeled with different lysine isotopes (see Materials and Methods and supplementary fig. S2, Supplementary Material online, for details). After extraction, proteins were separated by size-exclusion chromatography (SEC) and the eluted fractions were processed by mass spectrometry. Elution profiles of the proteins were used to infer PPIs and protein complexes by protein correlation profiling (PCP), using dynamic time wrapping (DTW) (supplementary fig. S3, Supplementary Material online), with the principle that two proteins that interact would tend to have similar elution profiles. The peptide counts per protein were used to estimate protein abundance in hybrid and parental species (see Materials and Methods). In the figure, we schematically represent the elution profiles of hypothetical proteins P1, P2 and P3, their putative PPIs, and protein abundances.
The null hypothesis that protein complexes appear to assemble in hybrids as they do in parents is supported by previous studies. For instance, the assembly of the nuclear pore complex and of the RNA polymerase II in yeast hybrids is consistent with the conservation of complexes in hybrids (Leducq et al. 2012), that is, interlogous PPIs are formed in hybrids in a way that reflects the parental PPIs. In fact, it is possible that most proteins between closely related species are capable of interacting this way given that proteins that are as diverged as those of yeast and humans can complement and interact with each other (Kachroo et al. 2015; Zhong et al. 2016). However, cases of incompatibilities and of novel interactions do exist. For instance, a study supported the scenario whereby proteins from two species form only intralogous PPIs when expressed in the same cell: the co-evolution of the proteins belonging to the PCNA (proliferating cell nuclear antigen) complex prevents interlogous PPIs (Zamir et al. 2012). On the other hand, another study found that some complexes contain interlogous PPIs that are not seen in parental species. The authors further linked one of these new hybrid complexes to an enhanced function in tryptophan transport (Piatkowska et al. 2013). Both deleterious and adaptive changes can thus occur in hybrid complexes.
Because previous studies were limited in terms of the coverage of molecular functions, whether hybridization is associated with a global reorganization of protein complexes in the cell is still mostly unexplored. Until recently, we lacked experimental methods that could allow studying a large number of protein complexes in hybrids and parental species. Here, we applied a method that allowed us to survey several protein complexes simultaneously in a yeast hybrid. Several reasons make yeast species excellent experimental models to address the question as to how protein complexes assemble in hybrids. Firstly, hybrids can be readily produced in the laboratory (Krogerus et al. 2018). Second, spontaneous hybridization is common among yeast species and may have an important impact on their performance in nature (Leducq et al. 2016; Barbosa et al. 2016). Third, during industrial processes such as fermentation, hybridization is thought to be an important mechanism of adaptation (González et al. 2006; López-Malo et al. 2013). For example, hybridization leads to a high fermentation capacity with a desirable aroma profile in beer fermentation (Mertens et al. 2015). And finally, the yeast proteome and PPI network have been well characterized (Ito et al. 2001; Krogan et al. 2006; Tarassov et al. 2008), making yeast an ideal model for species comparisons.
We applied size-exclusion chromatography (SEC), protein correlation profiling (PCP), and stable isotope labeling by amino acids in cell culture (SILAC) (SEC–PCP–SILAC) (Kristensen and Foster 2014) to the budding yeast Saccharomyces cerevisiae (denoted as Scer), to S. uvarum (denoted as Suva) (Scannell et al. 2011), and to their F1 diploid hybrid. This approach separates complex mixtures of endogenous proteins into a set of fractions that are analyzed by mass spectrometry. We measured the abundance of ∼400 proteins, with reasonably broad coverage of molecular functions of the proteomes. The profiles of comigrating proteins were clustered to reconstruct PPIs and thus protein complexes (Kristensen et al. 2012). From this data, we inferred the likelihoods of PPIs, which we then used to carry out the comparative analysis between the hybrid and its parental species.
Results and Discussion
Comparative Proteomics Approach to Monitor PPIs in Hybrid and Parental Strains
Scer and Suva are among the most divergent species in the Saccharomyces phylogeny (average distance of 50 My; Kellis et al. 2003), with an overall nucleotide sequence divergence between 30% and 35% (Morales and Dujon 2012) and a 16% divergence at the protein level (supplementary fig. S1A, Supplementary Material online). Scer and Suva strains were used as parental species along with their diploid F1 progeny as hybrid produced in two biological replicates (fig. 1B and supplementary table S1A, Supplementary Material online, for genotypes of the strains). The proteomes of the two parental strains are divergent enough that peptides could in general be distinguished by mass spectrometry (median Jaccard distance of 82.1%, supplementary fig. S1B, Supplementary Material online). The proteomics approach of SEC–PCP–SILAC allowed us to resolve likely interacting proteins (using SEC) and to infer putative PPIs (using PCP), in a comparative manner (using SILAC) (fig. 1B, supplementary fig. S2, Supplementary Material online, and see Materials and Methods for details).
Because some peptides derived from conserved sequences of orthologous proteins cannot be assigned to one parental species or the other in hybrid, only uniquely aligned peptides that could be ascribed to a protein of either Scer or Suva were retained in the analysis. The uniquely aligned peptides amount to ∼70% of the total number of peptides detected (supplementary fig. S1C, Supplementary Material online), representing ∼80% (∼400) of the total number of proteins detected (supplementary fig. S1D, Supplementary Material online). As some proteins were detected by uniquely as well as nonuniquely aligned peptides, only a relatively small fraction of the total number of proteins were removed by the filtering of proteins that were detected exclusively by nonuniquely aligned peptides. Thus, there was not an undue bias toward high sequence divergence in the remaining set of proteins (supplementary fig. S1A, Supplementary Material online). This allowed for a larger coverage of molecular functions, and not only for the proteins that diverge the most between species. Finally, in the PCP step, the elution profiles of proteins (supplementary data S1, Supplementary Material online) were correlated with each other by dynamic time warping (DTW, see Materials and Methods). Similar elution profiles indicate a likely interaction between a given pair of proteins. Therefore, pairwise similarity estimates between all protein pairs of a species provided a putative PPI network for that species (supplementary data S2, Supplementary Material online).
The use of uniquely aligned peptides allowed us to differentiate intralogous and interlogous PPIs with high confidence. In contrast to intralogous PPIs that can occur in parents as well as in the hybrid, interlogous PPIs can only take place in the hybrid. Therefore, for the comparative analysis, as a reference for interlogous PPIs, we inferred the expected interlogous PPIs from the elution profiles obtained for the parental species. Specifically, the elution profiles from the parental species were treated as if they originated from the same cell, called a “theoretical hybrid.” Then, these elution profiles were correlated by DTW to obtain putative PPIs which we hereon refer to as “expected interlogous PPIs.”
Because for Scer protein abundance and PPIs are well characterized, we used them as reference to assess the accuracy of the SEC–PCP–SILAC data. In order to estimate the likelihood of PPIs between pairs of proteins, we applied DTW to calculate the distance between the elution profiles of the proteins. Using the elution profiles of Scer proteins, we first scanned through a range of window sizes used for DTW and a range of thresholds applied on the distance between pairs of elution profiles to obtain optimal parameters (supplementary fig. S3, Supplementary Material online, see Materials and Methods) that provided the best match to the known PPIs from databases (fig. 2A). Also, upon comparing the protein abundance scores obtained from our proteomics data (supplementary data S3, Supplementary Material online) with the reference proteomics data obtained from PAXdb (Wang et al. 2015), we obtained a significant positive correlation (fig. 2B). Overall, for both the likelihood of PPIs and protein abundance, the Scer proteomics data shows high similarity with known reference values for Scer in databases.
Fig. 2.
Primary validation of the proteomics data. SEC-PCP-SILAC data for Scer was compared with reference data obtained from databases (panels A–E). The panels showing the analysis with Scer (A–C) are indicated with the logo of Scer, whereas those showing the analysis with the proteasome protein complex in Scer (panel D and E) are indicated with the logo representing a proteasome inside Scer. (A) Comparison of experimentally determined interaction scores of Scer’s PPIs with the association of interactors in protein complexes obtained from databases (see Materials and Methods). AUC, area under curve; TPR, true positive rate; FPR, false positive rate. (B) Comparison of experimentally determined protein abundance (shown on x-axis) of Scer proteins with reference protein abundance data (y-axis) obtained from PAXdb (Wang et al. 2015) and PeptideAtlas (Desiere et al. 2006). rs, Spearman’s correlation coefficient. The color bar indicates the counts of proteins in the hexbins. (C) Comparison of interaction scores of Scer PPIs that are part of protein complexes with those that are between random sets of proteins (of equal size and number as for protein complexes). The random sets of proteins are not part of any protein complexes known for Scer . P-value from two-sided Mann–Whitney U tests is shown on the plot. On the violin plots, the medians of the distributions are shown by a horizontal black line and quartiles by a vertical thick black line. (D) Comparison of interaction scores of parental Scer PPIs that are within and between subcomplexes of the proteasome. Compositions of subcomplexes were obtained from the Saccharomyces Genome Database (SGD) (Cherry et al. 2012). P-value from two-sided Mann–Whitney U tests is shown . On the violin plots, the medians of the distributions are shown by a horizontal black line and quartiles by a vertical thick black line. (E) Correlation between the interaction scores of parental Scer PPIs within the proteasome complex with distances between the subunits. The dashed line shows the fitted values from a lowess regression fit. Known direct PPIs within the proteasome were obtained from BioGRID (Chatr-aryamontri et al. 2017). The distances between the interactors were obtained from Chrétien et al. (2018). rs, Spearman’s correlation coefficient. (F) Relationship between dosage balance and interaction scores for different classes of PPIs within parental species and the hybrid. For clarity, dosage balance values are divided into bins of equal sizes (x-axis). The average interaction score per bin is shown on the y-axis, with vertical bars indicating 95% confidence interval. Dosage balance is calculated as 1-((|p1-p2|)/(p1+p2)), where p1 and p2 are the protein abundances of the interactors.
In order to further assess whether the inferred PPIs are indicative of the associations between proteins, we compared the average interaction scores for known protein complexes (obtained from Complex Portal database; Meldal et al. 2019) from Scer data with randomly drawn sets of proteins that are not known to be part of complexes (of equal size and number as for protein complexes) (fig. 2C). The interaction scores for protein complexes are significantly higher than the ones for the random sets of proteins (P < 1e−2, two-sided Mann–Whitney U test), indicating that interaction scores indeed capture associations among proteins. For example, in the Scer proteasome protein complex, we find significantly higher interaction scores for PPIs within subcomplexes, compared with PPIs between subcomplexes (P < 1e−2, two-sided Mann–Whitney U test, as shown in fig. 2D; see supplementary fig. S4, Supplementary Material online, for the PPI network within the proteasome complex). We also find that interaction scores significantly correlate with the interprotein distances within the complex (fig. 2E), indicating that empirically obtained interaction scores are sensitive to the physical distances between the proteins and thus that they are a good proxy for association between proteins. Moreover, this analysis for the proteasome complex also suggests that the higher values of interaction scores are likely to indicate direct PPIs.
As the abundance of interacting proteins is often balanced within protein complexes (Ge et al. 2001; Taggart and Li 2018), we tested, for all comparisons, whether the likelihoods of PPIs are positively related with the dosage balance of proteins. From the proteomics data, we captured an expected positive relationship for almost all intralogous PPIs (fig. 2F). Following the relationship between the interaction score and dosage balance, we found that interactors within complexes (annotated for Scer) show significantly higher interaction scores than interactors that are not contained in a protein complex (supplementary fig. S5A, Supplementary Material online). They also maintain greater dosage balance within complexes (supplementary fig. S5B, Supplementary Material online), corroborating earlier reports (Ge et al. 2001; Veitia et al. 2008; Ishikawa et al. 2017). Overall, the biological significance of the empirical data, as revealed from its conformity with benchmarks and other known trends, led us to the further comparative analysis of PPIs between the hybrid and parental species.
Strong Similarity between Parental and Hybrid PPIs and Chimeric Complexes in Hybrid
We compared the interaction scores of intralogous and interlogous PPIs to examine how PPIs form in the hybrid relative to parental species. In each case, we estimated the overall extent of the similarity between PPIs using rank correlation coefficients (fig. 3A). Intralogous PPIs in hybrid show greater similarity with the intralogous PPIs in the parents (fig. 3A, left). This trend holds true for both Scer and Suva intralogous PPIs (fig. 3A, middle) and even in the case of interlogous PPIs relative to the expected interlogous PPIs in the theoretical hybrid (fig. 3A, right). In support of this analysis, a similar trend of conservation was observed upon clustering based on interaction scores and protein abundance (supplementary fig. S6, Supplementary Material online). Interestingly, intralogous PPIs in the hybrid are more similar to those in the parents than for any other comparisons, suggesting that pairs of proteins from the same species tend to interact similarly in hybrids, as they do in the parents. This may reflect a slight preference for intralogous PPIs in hybrids. However, as discussed below, we find limited additional support for this.
Fig. 3.
General conservation of PPIs in the hybrid and formation of chimeric complexes. (A) Putative PPIs in hybrid and parents were classified as intralogous if both interactors belong to the same species and as interlogous otherwise. Similarities between the interaction scores of different classes of PPIs in hybrid and parents (see text), estimated in terms of Spearman’s correlation coefficient (rs), are shown in the plot. The thicknesses of the lines connecting the classes are in scale with rs. (B) Schematic representing the model in which likelihood of interlogous PPIs in hybrid is an additive effect of the intralogous PPIs. (C) Contour plot representing the relationship between the interaction scores of Scer-intralogous (on x-axis) and Suva-intralogous PPIs (on y-axis) in the hybrid, with that of interlogous PPIs (on z-axis). The fitting equation of multiple linear regression between the x and y variables is shown at the top of the plot. The interaction scores of two possible interlogous PPIs between a given pair of two interactors (each from a species) were averaged, as noted in the text. (D) Similar plot as in C, except that interaction scores of intralogous PPIs in the parents instead of the hybrid are plotted on x- and y-axes. (E) The correlations between the empirically estimated interaction scores for interlogous PPIs in hybrid (x-axis) with those predicted by multiple linear regression using interaction scores of intralogous PPIs in parents (orange) and in hybrid (green). rs, Spearman’s correlation coefficient. (F) Protein complexes detected in the hybrid. Scer proteins are highlighted with a orange border, whereas Suva proteins are highlighted with a blue border.
In order to further quantify the similarity of the hybrid PPIs compared with the parental ones, we tested whether the likelihoods of intralogous PPIs in parents can predict the likelihood of interlogous PPIs in hybrid (fig. 3B). This analysis considers that the likelihood of an interlogous PPI is the average of PPI scores of two possible interlogous PPIs, that is, interaction score between P1 of Scer and P2 of Suva and interaction score between P2 of Scer and P1 of Suva are averaged. The positive relationship between the likelihoods of interlogous PPIs in the hybrid with likelihoods of intralogous PPIs in the hybrid (fig. 3C) and in parents (fig. 3D) is clearly evident on the contour maps. Moreover, the fitting of multiple linear regression suggests that the likelihoods of intralogous PPIs contribute nearly equally toward predicting the likelihood of interlogous PPIs. We obtain a stronger association for prediction from intralogous PPIs in hybrid compared with that from intralogous PPIs in parents (fig. 3E), which is expected if the PPI profile in hybrids is overall slightly different from the parental ones. These results indicate that the likelihood of interlogous PPIs in hybrid can at least partially be additively predicted from the likelihoods of intralogous PPIs, either in the parents or in the hybrid, again supporting an overall conservation of PPIs in hybrids.
We next looked directly at known protein complexes that have been characterized in Scer. We considered protein complexes for which at least one PPI was captured in the proteomics data, resulting in 35 protein complexes in the hybrid (fig. 3F), 21 in Scer parent, and 25 in Suva parent. The variation in the number of protein complexes detected is due to the variation in the number of proteins detected and of PPIs inferred in each case. The smaller number of protein complexes in parental Scer and Suva is caused by the smaller number of proteins detected (405 Scer proteins and 401 Suva proteins) as compared with the hybrid (418 Scer proteins and 421 Suva proteins), which consequently led to a relatively smaller number of PPIs inferred in parental species (intralogous PPIs only) as compared with the hybrid (intralogous and interlogous PPIs).
In the hybrid, the high likelihoods of interlogous PPIs suggest frequent assembly of chimeric complexes (e.g., in proteasome and prefoldin complexes as shown in supplementary figs. S7 and S8 respectively, Supplementary Material online). Assessing the relative proportion of complexes with a chimeric (those that contain interlogous PPIs) versus a parental composition (those that contain only intralogous PPIs, as shown in fig. 1A) is challenging. Indeed, the number of possible chimeric assemblies of protein complexes in the hybrid increases exponentially with the number of subunits because each position in a complex can be occupied by either one of the two parental proteins. To assess the proportion of complexes in these two categories, we considered likelihoods of interlogous PPIs in hybrid as a proxy for the number of protein complexes with chimeric composition, and likelihoods of intralogous PPIs in hybrid as a proxy for protein complexes with parental composition. Because protein complexes with chimeric composition would also contain intralogous PPIs, the proxy for the number of protein complexes with chimeric composition is an underestimate. Using median interaction scores as thresholds, we segmented each type of PPI into “low” (interaction score < median) and “high” (interaction score > median) likelihood. Here, the protein complexes in the “high” class are considered to assemble in the hybrid. A similar number of complexes are represented in the two categories (supplementary fig. S9, Supplementary Material online), suggesting that protein complexes with chimeric composition are generally at least as abundant as those with parental composition in the hybrid. This trend supports the absence of significant bias in the preference for either intralogous or interlogous PPIs in general (supplementary fig. S10, Supplementary Material online). Considering that the proxy for the number of chimeric protein complexes was an underestimate, the chimeric protein complexes are very likely to be more frequent in the hybrid than pure parental complexes.
We also looked for the presence of putative novel complexes in the hybrid by clustering the PPIs with significantly high interaction scores in hybrid but significantly low interaction scores in parents. This analysis revealed a set of proteins, consisting of four Suva proteins (Ahp1, Pcm1, Scw4, and Tsa1) and a Scer protein (Adh4), that do not have any overlap with known protein complexes. The molecular function of this putative protein complex is challenging to infer. It might be related to thioredoxin peroxidase activity as two of the four Suva proteins (Ahp1 and Tsa1) are involved in that molecular function. Interestingly, however, the subunits of the putative protein complex are not known to colocalize. Therefore, the possibility of formation of a novel complex needs further experimental validation. Overall, the lack of power in the data limited the in-depth analysis for the identification of novel protein complexes.
One of the complexes that appear to be chimeric based on these analyses is the prefoldin complex, which we next used as a model to validate the assembly of chimeric complexes in the hybrid (supplementary fig. S8, Supplementary Material online). Prefoldin is a hexameric protein complex that acts as a molecular co-chaperone involved in cytoplasmic folding of actin and tubulin monomers during cytoskeleton assembly (Millán-Zambrano and Chávez 2014). We probed PPIs between pairs of proteins in the prefoldin complex using dihydrofolate reductase protein-fragment complementation assay (DHFR-PCA, see fig. 4A for experimental layout, and supplementary fig. S11, Supplementary Material online, and Materials and Methods for more details) (Tarassov et al. 2008; Michnick et al. 2010; Freschi et al. 2013). DHFR-PCA provides a quantitative signal of interaction strength for direct and near-direct physical interactions (Schlecht et al. 2012; Freschi et al. 2013; Levy et al. 2014; Diss et al. 2017; Diss and Lehner 2018), allowing quantitative comparisons with the proteomics data. From the DHFR-PCA experiments, we obtained relative strengths of PPIs (supplementary data S4, Supplementary Material online) within the complex in the hybrid (fig. 4B) and in parents (supplementary fig. S12, Supplementary Material online).
Fig. 4.
Orthogonal validation for the presence of chimeric PPIs. (A) Experimental layout of the DHFR-PCA. Individual yeast strains harboring different combinations of two interacting proteins in the prefoldin complex (each protein tagged with a complementary fragment of the DHFR) (see supplementary tables S1A and S1C, Supplementary Material online, and Materials and Methods for details) allowed testing for pairwise PPIs: intralogous in parents and both intralogous and interlogous in hybrid. (B) PPI network within the prefoldin complex in hybrid, obtained from DHFR-PCA. The widths of the edges are in scale with interaction strengths. (C) Similarities between the interaction strengths of PPIs within the prefoldin complex belonging to different classes of PPIs in hybrid and parents. The interaction strengths were measured by the DHFR-PCA. The similarities were estimated in terms of Spearman’s correlation coefficient (rs). The thickness of the lines connecting the classes is in scale with rs. (D) Relationship between the strength of intralogous and interlogous PPIs in the hybrid. The colors of the points are scaled according to the scores representing the strengths of interlogous PPIs. The fitted multiple linear regression equation is shown at the top of the plot. (E) Similar analysis as in panel D, except that the interaction scores of the intralogous PPIs in parental species are lined on the x- and y-axes. (F) Correlation between the actual strengths of interlogous PPIs in hybrid with the ones predicted from intralogous PPIs in hybrid (green) and in parents (orange). Spearman’s correlation coefficient (rs) is denoted for each case in the plot legend.
Because the DHFR-PCA experiment detected physical PPIs within complexes between Scer and Suva proteins in the hybrid, it confirms the existence of interlogous complex assembly. Also, the strength of the interlogous PPIs in hybrid (fig. 4B) confirms the chimeric composition of the prefoldin complex observed above. As generally observed in the proteomics data (fig. 3A), intralogous PPI scores in parents are more similar to the intralogous PPIs in the hybrid than when comparing the two parental species to each other (fig. 4C). Furthermore, we tested whether likelihoods of intralogous PPIs can predict the likelihood of PPIs in the hybrid. Again, the strengths of the interlogous PPIs were found to be correlated with intralogous PPIs in the hybrid (fig. 4D). Moreover, they are also correlated with the intralogous PPIs in parents (fig. 4E), corroborating the results from the proteomics data (fig. 3C and D). The intralogous PPIs in parents were found to be a stronger predictor than the intralogous PPIs in hybrid. This exception to the overall trend observed in the case of the proteomics data (fig. 3E) may be because of the sampling of the relatively small number of PPIs in the prefoldin network and of other methodological differences. Overall, the strengths of interlogous PPIs could be predicted from the intralogous PPIs in the hybrid as well as from those in parents (fig. 4F), confirming that chimeric PPIs occur as they are in parental species.
Unexpected Changes in PPIs in the Hybrid
We found a high similarity of PPIs and protein abundance between the hybrid and its parents, as well as patterns consistent with the frequent assembly of chimeric complexes. In this section, we examine how likelihoods of PPIs in some protein complexes deviate from these expectations while keeping in mind our limited statistical power. We first examined the relative preference for either intralogous or interlogous PPIs in hybrid, which as noted earlier (supplementary fig. S10, Supplementary Material online), suggests that both types of PPIs occur in hybrid and that many proteins show no clear preference for either type. Upon stratifying PPIs based on the relative preferences (z-score ≤ −1: prefer intralogous PPIs; z-score ≥ 1: prefer interlogous PPIs), we find that PPIs that do not have a clear preference occur between proteins that have less diverged orthologs (supplementary fig. S13, Supplementary Material online). On the other hand, when PPIs have a higher relative preference for either intralogous or interlogous PPIs, we observe a higher divergence between orthologs (two-sided Mann–Whitney U test, P-value < 1e-4 for intralogous PPIs and P-value < 1e-3 for interlogous PPIs). This led us to examine whether the deviation of the interlogous PPIs from the predicted interlogous PPIs (from the parental intralogous PPIs, as shown in fig. 3E) depends on the divergence of protein sequences between species. We found that orthologs that diverge more in sequence tend to deviate from their expected behavior in hybrids (fig. 5A). Highly diverged orthologs are also more likely to participate in the interlogous PPIs with lower interaction scores than the intralogous PPIs in the hybrid and parents, suggesting the presence of incompatibilities within interlogous PPIs (supplementary fig. S14, Supplementary Material online). However, this effect might be limited to a small number of protein complexes where the interlogous PPIs have very low interaction scores and therefore, the incompatibility is more pronounced. Additionally, because of the positive relationship between protein abundance and sequence conservation (Drummond et al. 2005), such incompatibilities would likely be correlated also with the abundance of the proteins. Overall, although the signal is weak, these results suggest that proteins that diverge in sequence tend to assemble differently in hybrids, which may result from amino acid differences that affect binding directly or other processes that may affect the assembly of complexes in hybrids with specific functions that are less conserved at the protein level.
Fig. 5.
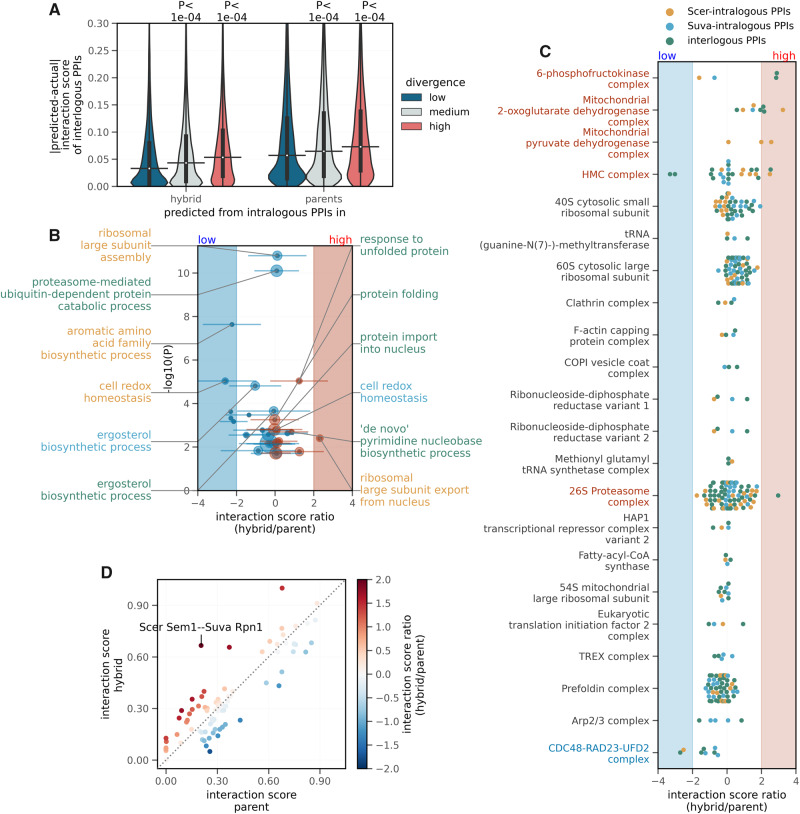
Differences in the PPIs in the hybrid compared with the parental species. (A) Dependence of the predictabilities of the interlogous PPI interaction scores (y-axis) on the interactors protein sequence divergence. Based on the interactors average divergence with respect to their orthologs, interlogous PPIs were categorized into “low,” “medium,” or “high” divergence groups, using first and third quartiles as thresholds. Predictabilities of interlogous PPI interaction scores from intralogous PPIs in hybrid and parents (x-axis) are measured in terms of the absolute difference between predicted and actual scores. P-values from two-sided Mann–Whitney U tests are shown. On the violin plots, the medians of the distributions are shown by a horizontal black line and quartiles, by a vertical thick black line. (B) Volcano plot showing the enrichment of GO terms from the biological processes aspect. The median values of z-score normalized hybrid/parent ratios of interaction scores for PPIs belonging to a given gene set are shown on the x-axis and −log10(P-value) of the enrichment (hypergeometric test, FDR corrected by Benjamini and Hochberg method) is shown on the y-axis. Each point represents sets of Scer-intralogous (orange), Suva-intralogous (blue), or interlogous (green) PPIs. Horizontal bars across the points indicate standard deviation of the ratios, for the PPIs belonging to the gene set. For clarity, only the six most significantly low (on left) and high (on right) enrichment sets are annotated on the plot. The size of the points is proportional to the number of genes per gene set. The region shaded in blue indicates significantly low ratios (z-score ≤ −2), whereas the region shaded in red indicates significantly high ratios (z-score ≥ 2). (C) Distributions of the z-score normalized hybrid/parent ratios of interaction scores (x-axis) per protein complex (y-axis) are shown. Each point represents the ratio for a given PPI. The gene sets are sorted based on the median value of the ratios per protein complex. The region shaded in blue indicates a significantly low ratio (z-score ≤ −2), whereas the region shaded in red indicates a significantly high ratio (z-score ≥ 2). Protein complexes with significantly low or high ratios are shown in blue and red, respectively (y-axis). (D) Comparison of the interaction scores of PPIs in hybrid (on y-axis) with those in parents (on x-axis), in the case of the proteasome complex. Interaction scores of the intralogous PPIs in hybrid are compared with intralogous PPIs in parents, whereas interaction scores of the interlogous PPIs in hybrid are compared with interlogous PPIs in the theoretical hybrid. Colors of the points are scaled according to the z-score normalized hybrid/parent ratios of interaction scores. The PPI with a significantly different ratio (|z-score| ≥ 2) is indicated on the plot.
In order to examine which biological functions may be particularly affected at the level of PPIs in hybrids, we looked at biological processes that have an enrichment of PPIs whose scores in hybrids are higher or lower than in the parents (see Materials and Methods). Remarkably, the gene set enrichment analysis revealed several significant changes in proteostasis-related gene sets (fig. 5B). For example, gene sets related to protein folding and response to unfolded protein are enriched for PPIs with significantly high likelihoods of interlogous PPIs in the hybrid compared with those expected in the theoretical hybrid (P-value = 6e−04 and P-value = 9e−06, respectively, hypergeometric test, false discovery rate (FDR) corrected by Benjamini and Hochberg method). On the other hand, a gene set related to the proteasome-mediated ubiquitin-dependent protein catabolic process contains PPIs with significantly low relative likelihoods of interlogous PPIs in hybrid (P-value = 8e−11). Also, gene sets related to export of the large subunit of the ribosome from nucleus contain PPIs with significantly high likelihoods of interaction in hybrid (P-value = 4e−03), whereas gene sets related to the assembly of the large subunit of the ribosome contain few PPIs with significantly high and few PPIs with significantly low relative likelihoods (P-value = 7e−03 and P-value = 2e−11, respectively). Proteostasis-related changes are also detected when we look at protein complexes (supplementary fig. S15A, Supplementary Material online), where we find that the HMC protein complex (involved in protein folding) contains PPIs with significantly high relative likelihoods of Scer-intralogous PPIs in hybrid (P-value = 1e-03), the proteasome (protein degradation) contains PPIs with significantly high relative likelihoods of interlogous PPIs in hybrid (P-value = 5e−02), and the CDC48–RAD23–UFD2 complex (protein degradation) contains PPIs with significantly low relative likelihoods of interlogous and Scer-intralogous PPIs in hybrid (P-value = 3e−04 and P-value = 2e−04, respectively). Additionally, the gene set related to chaperone binding (molecular function) contains PPIs with significantly low likelihoods of Scer-intralogous PPIs in hybrid compared with parents (P-value = 3e-03, shown in supplementary fig. S15B, Supplementary Material online). Interestingly, an increase in the abundance of complexes associated with proteostasis has been observed in hybrids between species of Drosophila (Bamberger et al. 2018), suggesting that hybrids between species may be particularly prone to changes in proteostasis.
Apart from the proteostasis related genes, among biological processes, gene sets related to the biosynthesis of aromatic amino acid, ergosterol, and pyrimidine nucleobase also contain PPIs with significant changes in likelihoods in the hybrid compared with parents. Some of the metabolic complexes that also show significantly stronger PPIs in hybrids are mitochondrial protein complexes that may reveal incompatibilities between the nuclear and mitochondrial genomes. This is known to take place among pairs of Saccharomyces species (Špírek et al. 2014). Among other examples, the interlogous PPI between the alpha and beta subunit of the 6-phosphofructokinase, a complex involved in glycolysis, is found to be stronger, potentially affecting the function of the protein complex in the hybrid (supplementary fig. S16, Supplementary Material online).
Across protein complexes, however, the distributions of the hybrid/parent ratios are centered at zero (fig. 5C), indicating that the likelihoods of PPIs in hybrid are similar to those in parents or those expected in a theoretical hybrid. Subtle exceptional changes in the likelihoods of PPIs are evident in the PPI networks of the significantly enriched protein complexes as shown in supplementary figures S7 and S16–S20, Supplementary Material online. We illustrate such changes by focusing on the proteasome complex in the hybrid (fig. 5D). In this case, some interlogous PPIs seem favored over intralogous PPIs, suggesting the existence of a certain degree of heterogeneity in the assembly of the complex. For instance, Scer Sem1p has a significantly higher likelihood of interaction with Rpn1p of Suva, compared with the expected interaction in the theoretical hybrid (z-score ≥ 2). The potential changes in the assembly of the proteasome raise interesting links with proteostasis as the proteasome is the main complex involved in protein degradation. Testing whether the particular changes in the assembly of the proteasome is one of the causes or the consequences of the perturbation of proteostasis in hybrids will require further investigation.
Conclusion
Given the functional importance of protein complexes in the cell, variation in their organization may play a central role in the cellular phenotypes of hybrids. In this study, we investigated the molecular basis differentiating hybrids from parents by looking directly at their PPIs. Using the proteomics approach of SEC–PCP–SILAC (Havugimana et al. 2012; Kristensen et al. 2012) (fig. 1B), we obtained likelihood of physical associations between proteins for S. cerevisiae × S. uvarum F1 diploid hybrid and their parents. We investigated the assembly of a few dozen protein complexes in the hybrid in a comparative manner. In general, the accurate identification of interacting protein pairs in data sets resulting from comigration experiments is known to be challenging (Skinnider et al. 2018). The relatively small number of proteins identified for each species was one of the limiting factors of this study. Because of the technical constraints related to the proteomics method, in order to confidently ascribe the parental origin of the proteins, we resorted to one of the most diverged pairs of parental species in the Saccharomyces phylogeny and could only utilize the uniquely aligned peptides for resolving the species origin for the proteins in hybrid. However, a lack of unduly bias for highly divergent proteins among the detected proteins suggests that protein sequence divergence of the orthologs was not the only factor limiting the number of proteins in the data set. The resulting subsampling of the proteome due to the technical constraints related to the proteomics approach limited the extent of whole proteome-wide extrapolations from our study. For a broader coverage of the proteome, among other things, the use of additional labeled amino acids could be advantageous. Further investigation on the dependence of the assembly of chimeric complexes on the divergence between parental species might also require direct protein interaction assays.
The overall equally likely intralogous and interlogous PPIs in hybrids suggests frequent assembly of chimeric protein complexes in hybrid and thus that the subunits of complexes are generally replaceable at this phylogenetic distance, consistent with the results of (Leducq et al. 2012). However, our experimental analysis is biased toward large, abundant and thus possibly more essential and stable complexes, which might be less sensitive to protein divergence for their assembly. Yeast F1 hybrids rarely show a reduction of fitness compared with parents in terms of growth rate (Bernardes et al. 2017; Charron and Landry 2017), although defects of sporulation and spore survival are common (Lee et al. 2008; Greig 2009; Charron et al. 2014). It may therefore be expected that protein complexes involved in the core cellular machinery such as the ones studied here assemble correctly in hybrids. Additionally, the analysis is based on the well-annotated protein complexes of Scer, with the assumption that the composition of protein complexes is mostly similar between Scer and Suva. Although the high overall similarity in likelihoods of the PPIs supports this assumption, it might have limited the detection of subtle differences in the assembly of protein complexes. We also could not thoroughly explore the possibility of novel complexes in hybrids because we had limited technical power for such discovery. Another element to consider is that the strains were grown in synthetic media and differences between hybrids and parents could be manifested in specific conditions such as during stress responses, which may trigger the activity of proteins and pathways that have diverged between species.
In spite of this large-scale conservation of PPIs in hybrids, we identified some biological functions that seem to be altered specifically in hybrids, including the ones related to proteostasis and metabolism. This could be an effect of or a response to the misregulation of protein proteostasis in hybrids, as has been observed for many other hybrid molecular traits (Landry et al. 2007; Bar-Zvi et al. 2017). Protein complexes involved in proteostasis such as the proteasome could be altered in hybrids, resulting in perturbation of the associated functions. Another possibility is that there could be some imbalance among protein subunits of complexes in the hybrids or marginal incompatibilities among subunits, which would put an additional burden on the protein quality control machinery because the regulation of the stoichiometry of large complexes often occurs through excess protein degradation (Taggart and Li 2018). This hypothesis is supported by a weaker dosage balance for interlogous interacting protein pairs in the hybrid compared with intralogous ones in parents. Proteostasis has also been noticed as being enhanced in hybrids of Drosophila (Bamberger et al. 2018), raising the interesting possibility that protein–protein or gene expression incompatibilities in hybrids are of weak effects but distributed among many proteins, imposing a general proteomic stress. In conclusion, protein complexes appear to be generally robust to protein divergence between species but small differences in their assembly or abundance could accumulate to a point where they perturb the overall protein physiology of the cell. An interesting avenue of research would be to examine how complexes accommodate the evolution of the hybrid genomes with time, for instance, through recombination and loss of heterozygosity (Zhang et al. 2020), which may further enhance the imbalance of complexes.
Materials and Methods
Generation of Yeast Hybrids for the SEC–PCP–SILAC Experiment
The hybrids generated in this study are described in supplementary tables S1A and S1B, Supplementary Material online, along with the replicates that were performed (supplementary table S1C, Supplementary Material online). The parental strains used to generate the hybrids are derived from the reference strains S288C (S. cerevisiae) and CBS7001 (S. uvarum). Using a microneedle (SporePlay micromanipulator, Singer Instruments), a single haploid cell of the first strain was put in physical contact with a single haploid cell of the second strain on a yeast extract, peptone, dextrose (YPD) plate (supplementary table S2, Supplementary Material online). After 3 days of growth at 30 °C, flow cytometry (Millipore Guava easyCyte Flow Cytometry System, MilliporeSigma) was used to determine the relative ploidy of colonies, as reported in Gerstein et al. (2006). Diploid hybrid clones were then validated by polymerase chain reaction (PCR) on the mating-type locus (Huxley et al. 1990). Colony PCRs were performed on single colonies as follows: A small amount of fresh colonies was resuspended in 40 μl of NaOH 20 mM and incubated for 20 min at 95 °C for cell lysis. Cells were centrifuged for 5 min at 4,000 rpm. For each PCR reaction, the mixture contained 2.5 μl of 10× Buffer (BioShop Canada Inc.), 2 μl of the supernatant of the lysed cells, 1.5 μl of MgCl2 25 mM, 0.5 μl of dNTP mix 10 mM, 0.5 μl of each primer at 10 μM, and 0.15 μl of Taq DNA Polymerase (5 U/μl, BioShop Canada Inc.), in a final volume of 25 μl. PCR reactions were carried out in a thermocycler (MasterCycler ProS, Eppendorf) with the following steps: 5 min at 95 °C; 35 cycles of 30 s at 94 °C, 30 s at 55 °C, and 1 min at 72 °C; and a final extension of 3 min at 72 °C. PCR products were then size-verified on an agarose gel. Primers used for verification of mating-types are described in supplementary table S3, Supplementary Material online. After selection of diploid cells, the strains involved in the crosses were validated by analysis of the restriction profiles obtained after DNA digestion with AccI enzyme: Quick DNA extraction was performed on single diploid yeast colonies (Lõoke et al. 2011) followed by PCR amplification using universal primers for Saccharomyces species in the POP2 gene (supplementary table S3, Supplementary Material online). PCR reaction mixture was prepared as described above and incubated following these steps: 5 min at 95 °C; 35 cycles of 30 s at 95 °C, 30 s at 52 °C, and 2 min at 72 °C; and a final extension of 10 min at 72 °C. Each PCR amplification product was digested overnight at 37 °C with 2 μl of CutSmart Buffer (New England Biolabs) and 0.2 μl of AccI enzyme (10 U/ml, New England Biolabs). Restriction profiles specific to each species (Scer or Suva) were identified on agarose gel.
SILAC Labeling
SILAC labeling was performed as described by Fröhlich et al. (2013). All yeast strains used in this study were auxotrophic for lysine. SC (synthetic complete) -lys medium (supplementary table S2, Supplementary Material online) was prepared and enriched by adding 30 mg/l of the following isotopes: 1) L-Lysine (Sigma-Aldrich) for “light” (L) labeled cells, 2) D4 L-Lysine (Cambridge Isotope Laboratories) for “medium” (M) labeling, and 3) 13C6 15N2 L-Lysine (Cambridge Isotope Laboratories) for “heavy” (H) labeling. The strains were precultured overnight in 5 ml of medium containing L, M, or H lysine at 25 °C. Two 50 ml cultures (L samples corresponding to parent Scer [50 ml] and parent Suva [50 ml]) and two 100 ml cultures (M and H samples) of medium with corresponding isotope were inoculated from the precultures to A600 = 0.001. For comparison between a hybrid and a parent, M sample corresponded to one parent, either Scer or Suva (100 ml), and H sample corresponded to one biological hybrid (100 ml). For the comparison between parents, M sample corresponded to parent Scer (100 ml) and H sample corresponded to the other parent Suva (100 ml). Cells were grown to a final A600 = 0.7 corresponding to more than ten doublings. The labeling efficiency was found to be between 85% and 88% (ratio of the number of proteins with M or H modifications on total number of proteins). Summary of the labeled samples can be found in supplementary figure S2, Supplementary Material online.
Cell Lysis
After SILAC labeling, the two parental L samples were pooled to obtain the same final volume as M and H samples (100 ml). Cells were harvested via centrifugation and washed twice with cold Tris-buffered saline solution (50 mM Tris and 150 mM NaCl at pH 7.5). Cells were resuspended in 500 μl of SEC lysis buffer (50 mM Tris, 50 mM NaOAc, and 50 mM KCl at pH 7.2) including Halt Protease and Phosphatase Inhibitor Cocktail (100×) (ThermoFisher Scientific) and then quickly frozen as droplets into liquid nitrogen. The cells were lysed by grinding with a cold mortar and pestle in liquid nitrogen. After lysis, 2.5 ml of SEC lysis buffer with protease inhibitors was added to each lysate. To enrich soluble and cytosolic complexes, the obtained volume was clarified by ultracentrifugation (100,000 rcf for 15 min at 4 °C). A protein concentration assay was performed on supernatant (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific) to inject the same amount of proteins for each sample (L, M, H) in the SEC column. The minimum total amount of proteins injected into the SEC column was 700 μg and the maximum amount was 1,000 μg. Proteins and protein complexes were then extracted and the efficiency of protein extraction was validated with Sodium Dodecyl Sulphate–Polyacrylamide Gel Electrophoresis (SDS-PAGE) and native gels for both hybrids and parents (supplementary fig. S21A–C, Supplementary Material online).
Size-Exclusion Chromatography
The SEC samples were divided into two technical replicates. As described by Kristensen et al. (2012), to reduce the volume and to enrich for high-molecular weight complexes, samples were concentrated using ultrafiltration (100,000 MWCO, Sartorius Stedim). The L sample was concentrated to 200 μl (two technical replicates), whereas M/H samples were combined just before loading into the SEC column (100 + 100 μl) (two technical replicates). Note that technical replicates started from the same cell cultures but samples were divided into two just before injection onto the SEC machine. Biological replicates are experiments starting from independent cultures and different hybrid crosses (supplementary fig. S2, Supplementary Material online). The fractions from the L samples served as an internal standard and were separated by SEC independently from the M/H samples. Samples were loaded into a 300 × 7.8 nm BioSep4000 column (Phenomenex) and separated into 80 fractions by a 1200 Series semipreparative High-Performance Liquid Chromatography (HPLC) (Agilent Technologies) at a flow rate of 0.5 ml/min at 8 °C. The collected volume of the first 20 fractions was 250 μl per fraction and decreased to 125 μl per fraction for fractions 21–80. After protein extraction, SEC separation was performed on the L sample on one side, and M/H samples on the other side. Elution profiles were used as indicators of proper protein complexes separation without aggregates or protein complex dissociation (supplementary fig. S21D, Supplementary Material online). In total, 20 samples were separated by SEC. The information regarding the technical replicates and the samples is provided in supplementary table S1C, Supplementary Material online.
Protein Digestion
Individual SEC–PCP–SILAC samples were prepared for digestion as described in Scott et al. (2015). The first five fractions were skipped as they likely contain the void volume and protein aggregates. The last fractions (66–80) were also skipped to keep only the most abundant cytosolic complexes and to eliminate smaller-sized proteins (most probably noninteracting proteins). A total of 60 fractions (6–65) were thus used in the following steps. Urea (6 M) and thiourea (2 M) were added to each M/H fraction. To generate the SEC–PCP–SILAC reference, fractions 6–65 of the L SEC-separated samples were pooled together and added to each of the M/H fractions at a volume of 1:1. Ammonium bicarbonate (50 mM) was added to the fractions to stabilize the pH. Disulfide reduction was performed by incubating each fraction with 1 μg dithiothreitol for 30 min at room temperature. Samples were then alkylated with 5 μg iodoacetamide in the dark for 20 min at room temperature. LysC was added at a ratio of 1:50 and samples were incubated again overnight at 37 °C. Samples were acidified to a pH <2.5 with 20% trifluoroacetic acid. Peptides were then purified using self-made stop-and-go-extraction tips (StageTips; Rappsilber et al. 2003) made with C18 Empore material packed into 200-μl pipette tips. Stage Tips were first conditioned with 100 μl of methanol and equilibrated with 1% trifluoroacetic acid. Peptides were loaded into the column and then washed twice by adding 100 μl of 0.1% formic acid followed by centrifugation. Peptides were finally eluted with 100 μl of 0.1% formic acid and 80% acetonitrile. Samples were dried down using a vacuum concentrator and stored at 4 °C.
The SEC–PCP–SILAC steps described in this protocol were adapted from Kristensen et al. (2012).
Mass Spectrometry
Prior to the mass spectrometry analysis, samples were resuspended in 30 μl (fractions 6–20) or 15 μl (fractions 21–65) of 0.1% formic acid. Peptides were analyzed using a quadrupole-time of flight mass spectrometer (Impact II, Bruker Daltonics) on-line coupled to an Easy nano LC 1000 HPLC (ThermoFisher Scientific) using a Captive spray nanospray ionization source (Bruker Daltonics) including a 2-cm-long, 100-μm-inner diameter fused silica fritted trap column, a 40-cm-long (average), 75-μm-inner diameter fused silica analytical column with an integrated spray tip (6-8-μm-diameter opening, pulled on a P-2000 laser puller, Sutter Instruments). The trap column is packed with 5 μm Aqua C18 beads (Phenomenex), whereas the analytical column is packed with 1.9-μm-diameter Reprosil-Pur C18-AQ beads (Dr. Maisch, www.Dr-Maisch.com). The analytical column was held at 50 °C by an in-house constructed column heater. Buffer A consisted of 0.1% formic acid in water, and buffer B consisted of 0.1% formic acid in acetonitrile. Peptides were separated from 0% to 40% Buffer B in 90 min, then the column was washed with 100% Buffer B for 20 min before re-equilibration with Buffer A.
The Impact II was set to acquire in a data-dependent auto-MS/MS (tandem mass spectrometry) mode with inactive focus fragmenting the 20 most abundant ions (one at a time at 18-Hz rate) after each full-range scan from m/z 200 Th to m/z 2,000 Th (at 5-Hz rate). The isolation window for MS/MS was 2–3 Th depending on parent ion mass to charge ratio and the collision energy ranged from 23 to 65 eV depending on ion mass and charge. Parent ions were then excluded from MS/MS for the next 0.4 min and reconsidered if their intensity increased more than five times. Singly charged ions were excluded because in electrospray ionization (ESI) mode peptides usually carry multiple charges. Strict active exclusion was applied. The nano ESI source was operated at 1,700 V capillary voltage, 0.20 Bar nano buster pressure, 3 l/min drying gas, and 150 °C drying temperature. The mass spectrometry data are available through PRIDE (Perez-Riverol et al. 2019) at https://www.ebi.ac.uk/pride/archive/projects/PXD010136.
Database Searching and Quantification
Tandem mass spectra were extracted from the data files using MaxQuant version 1.6.0.1 (Cox and Mann 2008) and were searched against protein sequences from Scer and Suva retrieved from Saccharomyces online resources (http://www.saccharomycessensustricto.org/) (Scannell et al. 2011), plus common contaminants and reverse database for FDR filtering. Peptide and protein identification was performed using MaxQuant (Tyanova et al. 2016) with the following parameters: carbamidomethylation of cysteine as a fixed modification; oxidation of methionine, acetylation of protein N-terminal and SILAC labeling as variable modifications; lysine/K cleavage with a maximum of two missed cleavages, 0.006 Da precursor mass error tolerance, and 40-ppm fragment ion mass tolerance and requantify option was enabled. The data were filtered for 1% FDR at both peptide and protein levels. The search results and the protein databases are available at https://www.ebi.ac.uk/pride/archive/projects/PXD010136.
Preprocessing of Proteomics Raw Data
From the “proteinGroups” output of MaxQuant, proteins (column name: “Protein Groups”) with commonly occurring contaminant (column name: “Potential contaminant”), proteins identified only by a modification site (column name: “Only identified by site”), and other spurious protein hits (column name: “Reverse”) were discarded. Additionally, protein hits with Andromeda score (column name: “Score”) of less than 0.05 quantile threshold were discarded, to retain only high-quality hits. The filtered protein hits were annotated by gene name/ID based on the proteome reference, and by species of origin based on the SILAC labeling (supplementary fig. S2, Supplementary Material online). Only uniquely aligned peptides were considered while assigning the species of origin, thus avoiding ambiguity in the data. The L labeling served as a reference for the calculation of the ratios of intensities, that is, M/L and H/L. The ratios of intensities along the elution fractions constituted an elution profile for a given peptide. Such peptide-wise data and replicates were aggregated by taking the mean of the ratios of intensities per elution profile, resulting in protein-wise elution profiles. During this procedure, the aggregation of replicates helped in reducing the sparsity of the data. The data for the parental strains labeled with M and H isotopes were considered as biological replicates. Processed elution profiles of the proteins are included in supplementary data S1, Supplementary Material online.
Estimation of Interaction Scores from Proteomics Data
The peak heights in the preprocessed protein-wise elution profiles were first rescaled between 0 and 1, so that the minimum peak height is 0 and the maximum peak height is 1. The rescaled elution profiles were used to calculate pairwise similarity (in “all vs. all” manner) using DTW, implemented through dtaidistance (Wannesm et al. 2019). For the optimization of the window size parameter of the DTW, using the database reported interactions for Scer as a reference, we scanned a continuous range of window sizes (supplementary fig. S3, Supplementary Material online). Reference PPIs were obtained from STRING (release version: 11.0, accessed on November 5, 2019) (Szklarczyk et al. 2019) and HitPredict (accessed on November 8, 2019) (López et al. 2015) databases. Based on the distance scores obtained from the DTW, an upper threshold was selected which marks the “no interaction” range, with the most accuracy. PPIs with distance scores higher than the upper threshold were assigned an interaction score of 0, indicating no interaction. If the distance score was less than the upper threshold, the interaction score was scaled in a way that the highest interaction score was equal to the lowest distance score and vice versa (interaction score = 1 – [distance score/upper threshold of distance score]). After optimizing the window size and the upper threshold of the distance score using parent Scer data, the interaction scores for Suva and hybrid were estimated using the optimized settings. Estimated interaction scores of the proteins are included in supplementary data S2, Supplementary Material online.
Estimation of Protein Abundance from Proteomics Data
From the “proteinGroups” output of MaxQuant, the “Peptide counts (all)” column was used to estimate the protein abundances. The sum of the counts for all peptides of the same protein was used as protein abundance. In order to remove low spurious peptide counts, total counts of less than 3 were discarded. The protein-wise aggregated abundance was transformed with pseudo-log (base 10 with pseudocount of 0.5). Finally, the protein abundances were quantile normalized across species. Note that in the case of the hybrid, protein abundances for Scer as well as Suva proteins were estimated.
As shown in figure 2B, as a validation of the protein abundances estimated from the proteomics data, we compared protein abundance obtained from the proteomics for parental Scer species with the reference protein abundance obtained from proteomics source of PeptideAtlas (Desiere et al. 2006) (March 2013, filename: 4932-PA_201303.txt, accessed on May 23, 2018) available on PAXdb database (Wang et al. 2015).
Dosage balance was measured as 1-((|p1-p2|)/(p1+p2)), where p1 and p2 are protein abundances of two proteins. Protein abundance and the dosage balance scores are included in supplementary data S3, Supplementary Material online.
Comparative Analysis of the Interaction Scores between Hybrid and Parental Species
Comparative analysis of PPIs between hybrid and parental species was carried out in terms of ratios of the interaction scores of interlogous PPIs in hybrid to interaction scores of corresponding intralogous PPIs in parents. These ratios were log2 transformed and z-score normalized. The resulting distribution of z-score normalized ratios was used to stratify the PPIs. The ones with z-score greater than or equal to 2 were classified as significantly “high” relative likelihoods in hybrid and those with z-score less than −2 as with significantly “low” relative likelihoods in hybrid. The ratios of interaction scores are included in supplementary data S5, Supplementary Material online.
Gene Set Enrichment Analysis
Reference gene sets used in the enrichment analysis include sets of protein complexes obtained from the complex portal (Meldal et al. 2019) and GO (Gene Ontology) terms obtained from QuickGO (Binns et al. 2009). In order to avoid redundancy in the set of protein complexes, if a given protein complex possesses different variants (e.g., proteasome, ID: CPX-2262), only the variant carrying the largest number of interactors was considered in the analysis. Among GO terms, gene sets that qualify as “part_of,” “involved_in,” or “enables” from Molecular function (F), Biological Process (P), and cellular component (C) were included in the data set. Only gene sets assigned by either GO_Central (Binns et al. 2009; Gene Ontology Consortium 2019) or SGD (Cherry et al. 2012) were retrieved from QuickGO (accessed on March 17, 2020); link to access the GO terms used in the study: https://www.ebi.ac.uk/QuickGO/annotations? qualifier=part_of, involved_in, enables&assignedBy=GO_Central, SGD&reference=PMID&taxonId=559292&taxonUsage=descendants&geneProductSubset=Swiss-Prot&proteome=gcrpCan, gcrpIso, complete&geneProductType=protein&withFrom=GO, SGD
The sets of PPIs with significantly “high” and “low” interaction scores obtained from the comparative analysis of the hybrid with parents were used as test sets. The significance of the overlap between the reference and test sets was examined using hypergeometric test, and it was corrected for FDR using Benjamini–Hochberg procedure. The results of the gene set enrichment analysis are included in supplementary data S6, Supplementary Material online.
DHFR-PCA Screening of the Prefoldin Complex PPIs
Strain Construction
DHFR-PCA was used to detect PPIs between proteins forming the prefoldin complex in diploid cells. Strains were constructed as follows (supplementary tables S1A and S1C, Supplementary Material online).
Single-tagged Haploid Strains
First, Scer haploid MATa (BY4741) and MATα, (BY4742) strains were retrieved from the Yeast Protein Interactome Collection (Tarassov et al. 2008) (except for strain with PFD1 gene tagged with DHFR F[3] which was reconstructed as specified below for Suva). Second, Suva haploid MATa (MG032) and MATα, (MG031) strains were constructed as follows. DHFR fragments and associated resistance modules were amplified from plasmids pAG25-linker-F[1,2]-ADHterm (NAT resistance marker) and pAG32-linker-F[3]-ADHterm (HPH resistance marker) (Tarassov et al. 2008) using oligonucleotides described in supplementary table S3, Supplementary Material online. PCR mixture contained 15 ng of plasmid, 5 μl of 5× Buffer with Mg2+, 0.75 μl of 10 mM dNTPs, 3 μl of each primer at 10 μM, and 0.5 μl of 1 U/µl Kapa HiFi HotStart DNA polymerase (Kapa Biosystems, Inc, A Roche Company) for a total volume of 25 μl. PCR was performed with the following cycling protocol: initial denaturation (5 min, 95 °C), 32 cycles of 1) denaturation (20 s, 98 °C), 2) annealing (15 s, 64°C), and 3) extension (1 min, 72 °C) and one cycle for final extension (5 min, 72 °C). PCR products were then concentrated using an OligoPrep OP120 SpeedVac Concentrator (Savant). Competent cells collected during exponential growth (A600 = 0.7) were transformed with either DHFR F[1,2] (MATa cells) or DHFR F[3] (MATα, cells) modules as described in Tarassov et al. (2008) with the following modifications: heat shock was performed for 20–30 min after adding 5 μl of dimethyl sulfoxide (DMSO), followed by recovery in YPD at 25 °C for 5 h. Cells were plated onto selective NAT (DHFR F[1,2]) or HYG (DHFR F[3]) media (supplementary table S2, Supplementary Material online) and incubated for 5 days at 25 °C. The correct genome integration of DHFR fragment module was validated by colony PCR using primers described in supplementary table S3, Supplementary Material online. Colony PCRs were performed as mentioned previously (mating type verification PCR), with the only difference that we used 2.5 μl of supernatant of lysed cells for each PCR reaction. The product of the PCR reaction was finally Sanger sequenced with O1-50 primer ensuring that no insertion, deletion, or nonsynonymous mutation occurred at the junction between the gene and the DHFR fragment.
Double-tagged Haploid Strains
At this point, we had all the haploid strains with one of the prefoldin complex genes tagged with either DHFR F[1,2] or DHFR F[3] (supplementary table S1A, Supplementary Material online). These strains would be used to test intralogous PPIs in parents and interlogous PPIs in hybrids. However, to test for parental PPIs in hybrids, we needed single haploid strains tagged for both prefoldin genes for which we want to test the interaction, one tagged with DHFR F[1,2] and the other with DHFR F[3] (see supplementary fig. S11, Supplementary Material online).
To achieve that, for Scer strains, we crossed previously described MATa and MATα, haploid tagged strains of interest to obtain all desired combinations. Briefly, cells of opposite mating types were combined into 3 ml of YPD. Cells were incubated overnight at 30 °C and diploid selection was performed on NAT+HygB (supplementary table S2, Supplementary Material online). Cells were then transferred onto enriched sporulation medium and incubated for at least 1 week at room temperature. Following ascus digestion with 200 μg/ml zymolyase 20T (BioShop Canada Inc.), sporulated cultures were put on solid YPD medium. Tetrads were then dissected with a microneedle (SporePlay micromanipulator, Singer Instruments) to isolate single haploid spores. The mating type of these spores was PCR identified following Huxley et al. (1990) procedure described above, and spores were replicated on SC -met and SC -lys (supplementary table S2, Supplementary Material online) to determine auxotrophies. Selected strains for the following experiment are identified in supplementary table S1D, Supplementary Material online.
For Suva strains, we used an alternative approach. We transformed MG032 haploid strains already tagged for one gene with DHFR F[1,2] directly with specific DHFR F[3] modules amplified from a pAG32-linker-F[3]-ADHterm plasmid in which the TEF terminator for the antibiotic resistance was changed for a CYC terminator (pAG32-DHFR[3]-HPHNT1) to avoid unwanted recombination between the resistance markers. We performed the same steps as described above with the following changes: Reverse primers used for plasmid amplification were different and are described in supplementary table S3, Supplementary Material online, whereas forward primers remained the same. After transformations, cells were plated on NAT+HygB. The following steps remained similar, including validation of transformations by colony PCRs and sequencing.
For positive controls, we transformed Scer BY4741 MATa and BY4742 MATα, strains with plasmids p41-ZL-DHFR[1,2] and p41-ZL-DHFR[3]. As described by Leducq et al. (2012), these plasmids express interacting leucine zipper moieties that strongly dimerize and that thus lead to a strong signal in DHFR-PCA. For negative controls, we used plasmids expressing the linkers and DHFR fragments alone, p41-L-DHFR[1,2] and p41-L-DHFR[3], that show no signal in DHFR-PCA.
Screening of PPIs in the Prefoldin Complex
We performed all possible crosses among the constructed strains (see above and supplementary table S1D, Supplementary Material online) for a total of 15 different tested PPIs and 180 independent crosses. MATa and MATα, strains were combined from solid medium into a 96-well deepwell plate with 1 ml of YPD per well. About the same amount of cells for each strain were transferred into each well. Plates were incubated overnight at 25 °C. The next day, cells were resuspended and 6 μl of each cross was deposited on solid SC medium lacking the proper amino acids to ensure diploid selection (supplementary tables S1D and S2, Supplementary Material online). Plates were incubated for at least 2 days at 25 °C. After diploid selection, cells were transferred again into a 96-well deepwell plate with 1 ml of liquid SC medium lacking the proper amino acids and put at 25 °C overnight. The following days, cells were printed and then rearrayed on YPD plates in a way to have in a 1536 format a minimum of six replicates per diploid strains and to include a double border of a control PPI of medium strength (LSM8-DHFR F[1,2]/CDC39-DHFR F[3]). To perform DHFR-PCA, cells were in the end transferred on methotrexate (MTX) and control condition, dimethyl sulfoxide (DMSO) media (supplementary table S2, Supplementary Material online) for two successive rounds of a 4-day incubation at 25 °C. Starting from the rearraying step, all the following steps were done using robotically manipulated pin tools (BM5-SC1, S&P Robotics Inc).
Estimation of Interaction Strengths from DHFR-PCA Experiment Data
Images of agar media plates used for the DHFR-PCA experiment were taken each day of the two selection rounds, with an EOS Rebel T5i camera (Canon). We used images taken after 4 days of growth on the second selection round for analysis of both MTX and DMSO plates. Images were analyzed using gitter (R package version 1.1.1; Wagih and Parts 2014) to quantify colony sizes (supplementary data S6, Supplementary Material online) by defining a square around the colony center and measuring the foreground pixel intensity minus the background pixel intensity. For the estimation of the scores representing interaction strengths, first, log2 transformed ratios of sizes of the colonies on MTX with respect to sizes of the colonies on the DMSO plates were calculated. The ratios calculated from crosses representing the same interaction types (as shown in supplementary fig. S11, Supplementary Material online) were averaged. All the ratios were rescaled between 0 (no interaction) and 1 (strong interaction). In the text, the rescaled ratios are referred to as interaction strengths. These scores are included in supplementary data S4, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the members of the Landrylab for discussions. We thank Anna Fijarczyk, Angel F. Cisneros, Johan Hallin, and Diana I. Ascencio for comments on the manuscript. C.M.B. thanks the teams of L.J.F. and Thibault Mayor for their help in the project and access to equipment. R.D. is funded by Fonds de recherche du Québec-Santé (FRQS), Programme Postdoctoral fellowship, and Human Frontier Science Program (HFSP) (RGP0034/2018) grant to C.R.L. C.M.B. was funded by PROTEO via a Fonds Nature et technologies (FRQNT)/PROTEO International Internship Program. This research was supported by the Natural Sciences and Engineering Research Council (NSERC) Discovery Grant to C.R.L. and by Genome Canada and Genome British Columbia to L.J.F. (Project 214PRO). C.R.L. holds the Canada Research Chair in Cellular Systems and Synthetic Biology. The mass spectrometry infrastructure used was supported by the Canada Foundation for Innovation, the British Columbia Knowledge Development Fund, and the British Columbia Proteomics Network (BCPN).
Author Contributions
C.R.L., I.G.-A., and L.J.F. designed research. C.M.B., R.G.S., I.G.-A., and K.-M.M. performed experiments. R.D. and C.M.B. performed the analyses. R.D., I.G.-A., and C.R.L. wrote the article with input from C.M.B.
Data Availability
Supplementary data files are available on figshare at https://doi.org/10.6084/m9.figshare.12428975.v1. Raw proteomics data are available on PRIDE data repository at https://www.ebi.ac.uk/pride/archive/projects/PXD010136. Computer code to reproduce all the analyses shown in this article is available at https://github.com/Landrylab/dandage_berger_2020.
References
- Bamberger C, Martínez-Bartolomé S, Montgomery M, Lavallée-Adam M, Yates JR.. 2018. Increased proteomic complexity in Drosophila hybrids during development. Sci Adv. 4(2):eaao3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa R, Almeida P, Safar SVB, Santos RO, Morais PB, Nielly-Thibault L, Leducq J-B, Landry CR, Gonçalves P, Rosa CA, et al. 2016. Evidence of natural hybridization in Brazilian wild lineages of Saccharomyces cerevisiae. Genome Biol Evol. 8(2):317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Zvi D, Lupo O, Levy AA, Barkai N.. 2017. Hybrid vigor: the best of both parents, or a genomic clash? Curr Opin Syst Biol. 6:22–27. [Google Scholar]
- Bernardes JP, Stelkens RB, Greig D.. 2017. Heterosis in hybrids within and between yeast species. J Evol Biol. 30(3):538–548. [DOI] [PubMed] [Google Scholar]
- Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R.. 2009. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics 25(22):3045–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G, Landry CR.. 2017. No evidence for extrinsic post-zygotic isolation in a wild Saccharomyces yeast system. Biol Lett. 13(6):20170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G, Leducq J-B, Landry CR.. 2014. Chromosomal variation segregates within incipient species and correlates with reproductive isolation. Mol Ecol. 23(17):4362–4372. [DOI] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O'Donnell L, Oster S, Theesfeld C, Sellam A, et al. 2017. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45(D1):D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40(D1):D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien A-È, Gagnon-Arsenault I, Dubé AK, Barbeau X, Després PC, Lamothe C, Dion-Côté A-M, Lagüe P, Landry CR.. 2018. Extended linkers improve the detection of protein-protein interactions (PPIs) by dihydrofolate reductase protein-fragment complementation assay (DHFR PCA) in living cells. Mol Cell Proteomics. 17(3):549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26(12):1367–1372. [DOI] [PubMed] [Google Scholar]
- Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R.. 2006. The PeptideAtlas project. Nucleic Acids Res. 34(90001):D655–D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diss G, Gagnon-Arsenault I, Dion-Coté A-M, Vignaud H, Ascencio DI, Berger CM, Landry CR.. 2017. Gene duplication can impart fragility, not robustness, in the yeast protein interaction network. Science 355(6325):630–634. [DOI] [PubMed] [Google Scholar]
- Diss G, Lehner B.. 2018. The genetic landscape of a physical interaction. Elife 7:e32472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, , Bloom JD, , Adami C, , Wilke CO, , Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 102(40):14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freschi L, Torres-Quiroz F, Dubé AK, Landry CR.. 2013. qPCA: a scalable assay to measure the perturbation of protein-protein interactions in living cells. Mol Biosyst. 9(1):36–43. [DOI] [PubMed] [Google Scholar]
- Fröhlich F, Christiano R, Walther TC.. 2013. Native SILAC: metabolic labeling of proteins in prototroph microorganisms based on lysine synthesis regulation. Mol Cell Proteomics. 12(7):1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Liu Z, Church GM, Vidal M.. 2001. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet. 29(4):482–486. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. 2019. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47(D1):D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Chun H-JE, Grant A, Otto SP.. 2006. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2(9):e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González SS, Barrio E, Gafner J, Querol A.. 2006. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 6(8):1221–1234. [DOI] [PubMed] [Google Scholar]
- Greig D. 2009. Reproductive isolation in Saccharomyces. Heredity (Edinb). 102(1):39–44. [DOI] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. 2012. A census of human soluble protein complexes. Cell 150(5):1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C, Green ED, Dunham I.. 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 6(8):236. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Makanae K, Iwasaki S, Ingolia NT, Moriya H.. 2017. Post-translational dosage compensation buffers genetic perturbations to stoichiometry of protein complexes. PLoS Genet. 13(1):e1006554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y.. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 98(8):4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo AH, Laurent JM, Yellman CM, Meyer AG, Wilke CO, Marcotte EM.. 2015. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348(6237):921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES.. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423(6937):241–254. [DOI] [PubMed] [Google Scholar]
- Kristensen AR, Foster LJ.. 2014. Protein correlation profiling-SILAC to study protein-protein interactions. Methods Mol Biol. 1188:263–270. [DOI] [PubMed] [Google Scholar]
- Kristensen AR, Gsponer J, Foster LJ.. 2012. A high-throughput approach for measuring temporal changes in the interactome. Nat Methods. 9(9):907–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440(7084):637–643. [DOI] [PubMed] [Google Scholar]
- Krogerus K, Preiss R, Gibson B.. 2018. A unique Saccharomyces cerevisiae × Saccharomyces uvarum hybrid isolated from Norwegian farmhouse beer: characterization and reconstruction. Front Microbiol. 9:2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Hartl DL, Ranz JM.. 2007. Genome clashes in hybrids: insights from gene expression. Heredity 99(5):483–493. [DOI] [PubMed] [Google Scholar]
- Leducq J-B, Charron G, Diss G, Gagnon-Arsenault I, Dubé AK, Landry CR.. 2012. Evidence for the robustness of protein complexes to inter-species hybridization. PLoS Genet. 8(12):e1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leducq J-B, Nielly-Thibault L, Charron G, Eberlein C, Verta J-P, Samani P, Sylvester K, Hittinger CT, Bell G, Landry CR.. 2016. Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nat Microbiol. 1(1):15003. [DOI] [PubMed] [Google Scholar]
- Lee H-Y, Chou J-Y, Cheong L, Chang N-H, Yang S-Y, Leu J-Y.. 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135(6):1065–1073. [DOI] [PubMed] [Google Scholar]
- Levy ED, Kowarzyk J, Michnick SW.. 2014. High-resolution mapping of protein concentration reveals principles of proteome architecture and adaptation. Cell Rep. 7(4):1333–1340. [DOI] [PubMed] [Google Scholar]
- Lõoke M, Kristjuhan K, Kristjuhan A.. 2011. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques 50(5):325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Malo M, Querol A, Guillamon JM.. 2013. Metabolomic comparison of Saccharomyces cerevisiae and the cryotolerant species S. bayanus var. uvarum and S. kudriavzevii during wine fermentation at low temperature. PLoS One 8(3):e60135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Y, Nakai K, Patil A.. 2015. HitPredict version 4: comprehensive reliability scoring of physical protein-protein interactions from more than 100 species. Database 2015:bav117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA.. 2011. The genetics of hybrid incompatibilities. Annu Rev Genet. 45(1):331–355. [DOI] [PubMed] [Google Scholar]
- McManus CJ, May GE, Spealman P, Shteyman A.. 2014. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 24(3):422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldal BHM, Bye-A-Jee H, Gajdoš L, Hammerová Z, Horácková A, Melicher F, Perfetto L, Pokorný D, Lopez MR, Türková A, et al. 2019. Complex Portal 2018: extended content and enhanced visualization tools for macromolecular complexes. Nucleic Acids Res. 47(D1):D550–D558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens S, Steensels J, Saels V, De Rouck G, Aerts G, Verstrepen KJ.. 2015. A large set of newly created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl Environ Microbiol. 81(23):8202–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V.. 2010. Chapter 14—A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein–protein interactions in living cells. In: Methods in enzymology. Vol. 470. San Diego (CA): Academic Press. p. 335–368. [DOI] [PubMed] [Google Scholar]
- Millán-Zambrano G, , Chávez S. 2014. Nuclear functions of prefoldin. Open Biol. 4(7):140085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales L, Dujon B.. 2012. Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol Mol Biol Rev. 76(4):721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47(D1):D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkowska EM, Naseeb S, Knight D, Delneri D.. 2013. Chimeric protein complexes in hybrid species generate novel phenotypes. PLoS Genet. 9(10):e1003836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M.. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 75(3):663–670. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK.. 1999. Transgressive segregation, adaptation and speciation. Heredity 83(4):363–372. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT.. 2011. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 1(1):11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht U, Miranda M, Suresh S, Davis RW, St Onge RP.. 2012. Multiplex assay for condition-dependent changes in protein-protein interactions. Proc Natl Acad Sci U S A. 109(23):9213–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott NE, Brown LM, Kristensen AR, Foster LJ.. 2015. Development of a computational framework for the analysis of protein correlation profiling and spatial proteomics experiments. J Proteomics. 118:112–129. [DOI] [PubMed] [Google Scholar]
- Skinnider MA, Stacey RG, Foster LJ.. 2018. Genomic data integration systematically biases interactome mapping. PLoS Comput Biol. 14(10):e1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowmya G, Breen EJ, Ranganathan S.. 2015. Linking structural features of protein complexes and biological function. Protein Sci. 24(9):1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Špírek M, Poláková S, Jatzová K, Sulo P.. 2014. Post-zygotic sterility and cytonuclear compatibility limits in S. cerevisiae xenomitochondrial cybrids . Front Genet. 5:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart JC, Li G-W.. 2018. Production of protein-complex components is stoichiometric and lacks general feedback regulation in eukaryotes. Cell Syst. 7(6):580–589.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Molina MMS, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW.. 2008. An in vivo map of the yeast protein interactome. Science 320(5882):1465–1470. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N.. 2011. Inferring regulatory mechanisms from patterns of evolutionary divergence. Mol Syst Biol. 7(1):530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N.. 2010. Divergence of nucleosome positioning between two closely related yeast species: genetic basis and functional consequences. Mol Syst Biol. 6(1):365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J.. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 11(12):2301–2319. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA.. 2008. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 24(8):390–397. [DOI] [PubMed] [Google Scholar]
- Wagih O, Parts L.. 2014. gitter: a robust and accurate method for quantification of colony sizes from plate images. G3 4(3):547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, Mering C.. 2015. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15(18):3163–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannesm K, Wusai VCT, Ma E.. 2019. Wannesm/dtaidistance v1.2.2 [Internet]. [accessed May 22, 2020]. Available from: https://zenodo.org/record/3276100.
- Zamir L, Zaretsky M, Fridman Y, Ner-Gaon H, Rubin E, Aharoni A.. 2012. Tight coevolution of proliferating cell nuclear antigen (PCNA)-partner interaction networks in fungi leads to interspecies network incompatibility. Proc Natl Acad Sci U S A. 109(7):E406–E414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Bendixsen DP, Janzen T, Nolte AW, Greig D, Stelkens R.. 2020. Recombining your way out of trouble: the genetic architecture of hybrid fitness under environmental stress. Mol Biol Evol. 37(1):167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Tao X, Feng S, Wang L, Hong H, Ma W, Shang G, Guo S, He Y, Zhou B, et al. 2018. LncRNAs in polyploid cotton interspecific hybrids are derived from transposon neofunctionalization. Genome Biol. 19(1):195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Pevzner SJ, Hao T, Wang Y, Mosca R, Menche J, Taipale M, Taşan M, Fan C, Yang X, et al. 2016. An inter‐species protein–protein interaction network across vast evolutionary distance. Mol Syst Biol. 12(4):865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Hu B, Becker C, Doğan ES, Berendzen KW, Weigel D, Liu C.. 2017. Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol. 18(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary data files are available on figshare at https://doi.org/10.6084/m9.figshare.12428975.v1. Raw proteomics data are available on PRIDE data repository at https://www.ebi.ac.uk/pride/archive/projects/PXD010136. Computer code to reproduce all the analyses shown in this article is available at https://github.com/Landrylab/dandage_berger_2020.