Abstract
TEA domain family members (TEADs) play important roles in tumor progression. Till now, the genomic status of TEADs in patients with glioma has not been well investigated. To confirm whether the genomic status of TEADs could affect the prognosis of patients with glioma, the copy number variation (CNV), mutation and expression data of glioma cohorts in The Cancer Genome Atlas, Gene Expression Omnibus and Chinese Glioma Genome Atlas were comprehensively analyzed. Results showed that TEAD CNV frequency in lower grade gliomas (LGGs) was higher than in glioblastoma multiforme (GBM). Multivariate cox regression analysis showed that TEAD4 CNV increase was significantly associated with overall survival (OS) and disease-free survival (DFS) in LGGs (OS p = 0.022, HR = 1.444, 95% CI: 1.054–1.978; DFS p = 0.005, HR = 1.485, 95% CI: 1.124–1.962), while not in GBM. Patients with TEAD4 CNV increase showed higher expression level of TEAD4 gene. In LGG patients with IDH mutation, those with higher TEAD4 expression levels had shorter OS and DFS. Integrating TEAD4 CNV increase, IDH mutations, TP53 mutation, ATRX mutation and 1p19q co-deletion would separate patients with LGG into four groups with significant differences in prognosis. These study results suggested that TEAD4 variations were independent predictive biomarkers for the prognosis in patients with LGG with IDH mutation.
Keywords: low-grade glioma, copy number variation, TEAD4, prognosis, gene expression
1. Introduction
Malignant glioma is a primary brain tumor with extremely high mortality in adults [1,2,3]. Glioblastoma multiforme (GBM; World Health Organization grade IV) is notorious for resistance to therapy and has mean survival of less than 15 months [4,5]. Diffuse low-grade and intermediate-grade gliomas together make up the LGGs (lower grade gliomas), including World Health Organization grades II and III [6]. Majority of the patients with LGGs are sensitive to therapy and experience extended survival depending on the molecular subtype, such as IDH mutant and 1p19q co-deletion [7,8]. While the current curative effect and the prognosis of LGG varies greatly (survival time ranging from 1 to 15 years) due to individual differences, a certain number of patients could not gain satisfactory prognosis [9,10].
Central nervous system tumor diagnosis has entered the molecular era since 2016, which is defined by both histology and molecular features. The molecular parameters contain IDH mutation, ATRX loss, TP53 mutation, etc., and these more precisely defined the entities that are expected to improve therapeutic efficacy, clinical trials and more specific classification [11]. Also, more followed studies focus on searching molecular markers for objective diagnosis and accurate clinical outcomes. Xiao et al. constructed a CD44-related four-gene signature that would well predict the prognosis and effectively distinguish high- and low-risk patients with LGGs [12]. Nevertheless, the signatures associated with stratification of prognosis in patients with LGG remain finite, and identifying novel biomarkers is still essential for improving the diagnostic accuracy and therapeutic efficacy.
Transcription enhancer factors are the most important DNA-binding partner in Hippo and Wnt pathways, and four proteins have been identified, which are named TEAD1–4. When YAP/TAZ, the key molecules in the downstream of Hippo/Wnt pathway, translocate into the nucleus, TEADs directly interact with them and mediate the main transcriptional output of the Hippo/Wnt pathway and then drive cancer cell survival, proliferation, invasive migration and metastasis [13,14]. Hippo and Wnt/β-catenin pathways have been reported as pivotal signaling pathways, regulating cell proliferation and differentiation, immune response and subsequently facilitating tumorigenesis [15,16,17,18,19,20,21]. Meanwhile, YAP/TAZ-TEAD activation may also confer resistance to chemotherapy, radiotherapy or immunotherapy [22,23,24,25]. Therefore, TEADs might be a crucial target for glioma therapy.
Till now, the genomic status of TEADs in patients with glioma was not well investigated. Xu et al. discovered that the overexpression of TEAD4 correlated with poor prognosis of glioma, but they ignored the impact of recognized factors such as IDH mutation and 1p19q co-deletion on the prognosis of glioma and had not considered the difference in outcomes between LGGs and GBM [26]. Simultaneously, Wang et al. conducted a comprehensive study to explore the molecular characterization of the Hippo-signaling pathway in 33 cancers [27]. They found that TEAD2–4 were significantly correlated with LGG survival, but the detailed information of the relationship between TEADs and glioma patients’ outcome was not clearly elaborated. In this study, we are going to take the public data for comprehensively analyzing the relationship between TEADs and glioma prognosis and expecting to provide a novel strategy for individualized medicine for glioma in the future.
2. Methods and materials
2.1. Study samples
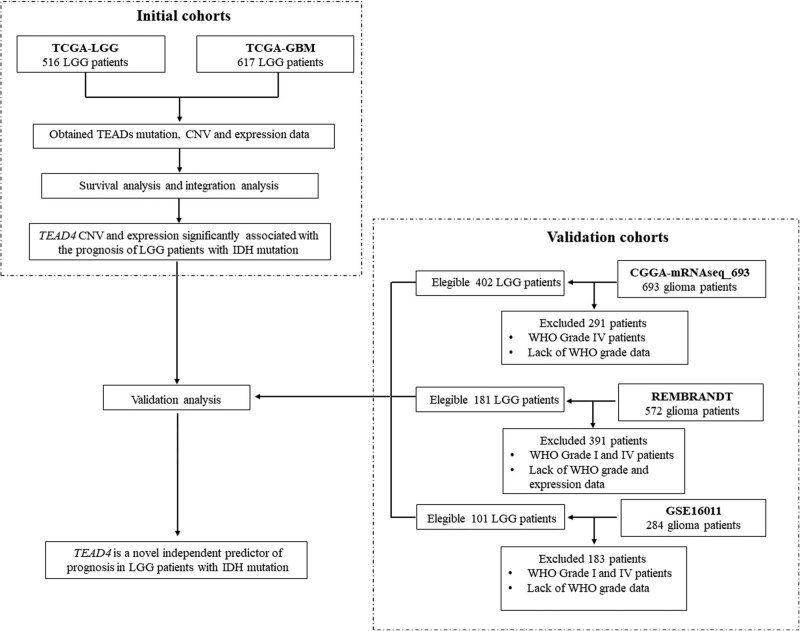
The glioma data set used in this study included TCGA-LGG, TCGA-GBM, CGGA-mRNAseq_693, Rembrandt and GSE16011. Data of somatic mutation, copy number variation (CNV), gene expression and clinical phenotypic in TCGA-LGG and TCGA-GBM data sets were obtained from cbioportal (http://www.cbioportal.org) and the GDC database (https://portal.gdc.cancer.gov/). Data of isocitrate dehydrogenase (IDH) mutations, 1p19q co-deletion status, gene expression and clinical phenotypes in CGGA-mRNAseq_693 were derived from the CGGA database (http://www.cgga.org.cn/). Both of the Rembrandt and GSE16011 data set used Affymetrix gene chip technology to detect gene expression, and the data were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The LGG samples of this study refer to samples of WHO II and WHO III levels [6]. Figure 1 displays the flow diagram of the study patients.
Figure 1.
Flow diagram of the study patients.
2.2. Statistical analysis
In this study, Kaplan–Meier (KM) analysis was used to perform univariate survival analysis to determine the effects of TEAD mutations, CNV and gene expression on the overall survival (OS) or disease-free survival (DFS) in patients with LGG. Multivariate survival analysis was performed through cox regression analysis. To select the covariates included in the multivariate survival analysis, backward stepwise cox regression analysis was conducted. The candidate covariates contained IDH mutation, TP53 mutation, ATRX mutation, 1p19q co-deletion, tumor grade and age. Mann–Whitney analysis was utilized to test the relationship between TEAD CNVs and their expression. In CNV analysis, the patients with high-level threshold values of 2 or −2 (calculated by GISTIC 2.0) were considered to have a copy number change. For gene expression data, we took the median value as the grouping basis for the high expression group and the low expression group. All the analyses in this study were performed by SPSS20 and R 3.6.1. GraphPad Prism 6.0 software was utilized for drawing the survival curves and histograms. R package maftools were used to conduct mutation interaction analysis.
2.3. Coexpression and gene ontology (GO) enrichment analysis
We conducted coexpression analysis in carriers of IDH mutation and wild-type patients in both TCGA-LGG and CGGA-mRNAseq_693 LGG data sets. And then filtered the genes that significantly correlated with TEAD4 based on Pearson correlation method (Pearson |r| > 0.4; Bonferroni corrected p < 0.05). The screened genes were finally used to perform GO analysis by David 6.8 (https://david.ncifcrf.gov/tools.jsp).
2.4. Immune infiltration analysis
To test the relationship between TEAD4 expression and immune infiltration, TIMER database (http://timer.cistrome.org/) was utilized. Infiltration scores of six types of immune cells in patients with LGG were calculated. Then the correlation of TEAD4 expression and these scores were tested.
3. Results
3.1. Basic characteristics of the study data sets
The TCGA-LGG data set included 516 patients with glioma, and the data of RNA sequencing, CNV and whole-genome somatic mutations were available for all 506 patients. The TCGA-GBM data set included 617 glioblastoma samples, among them 401 patients had somatic mutation information, 599 patients obtained whole-genome CNV data and 521 patients got whole-genome expression information by Affymetrix U133 microarray. OS and DFS data of all the patients in TCGA were available. The CGGA-mRNAseq_693 data set contained 693 patients, including 402 patients with LGG. The RNA sequencing data, IDH mutation data, 1p19q co-deletion status and OS information were available in this data set. Rembrandt data set contained 572 patients, and 181 were LGG. GSE16011 contained 284 patients including 109 with LGG. Both of Rembrandt data set and GSE16011 used Affymetrix microarray to obtain the whole-genome expression data of patients with glioma. OS was recorded in Rembrandt and GSE16011 data sets. Table 1 summarizes the basic characteristics of each study sample.
Table 1.
Basic characteristics of the study data sets
| Characteristics | TCGA-LGG | TCGA-GBM | CGGA-mRNAseq_693 | Rembrandt | GSE16011 |
|---|---|---|---|---|---|
| Sample size | 516 | 617 | 693 | 572 | 284 |
| Expression detection platform | Illumina | Affymetrix | Illumina RNA | Affymetrix | Affymetrix |
| RNA seq | Microarray | RNA seq | Microarray | Microarray | |
| CNV data | Available | Available | NA | NA | NA |
| IDH mut data | Available | Available | Available | NA | NA |
| 1p19q codel data | Available | Available | Available | NA | NA |
| TP53 and ATRX mutation data | Available | Available | NA | NA | NA |
| Prognostic phenotype | OS, DFS | OS, DFS | OS | OS | OS |
3.2. Mutation and CNV frequency analysis
TCGA data set showed that the incidence of somatic mutations in TEADs in gliomas was extremely low, as only 0.3% (3/812) of the patients was carrying the mutations. Meanwhile TEAD CNV occurred in LGG with a frequency of 11.7% (60/513), and in GBM with a frequency of 2.9% (17/577). Interestingly, the frequency of TEAD CNVs in LGG was significantly higher than that in GBM (p = 1.83 × 10−8). TEAD4 was the main CNV of all TEAD CNVs, accounting for 48.3% (29/60) in LGG and 58.8% (10/17) in GBM. The TEAD4 CNV occurrence in LGG was also significantly higher than that of GBM (p = 5.06 × 10−4). The CNV increase in TEAD4 was the main form of TEAD4 CNV in both LGG and GBM (28/29 in LGG and 9/10 in GBM).
3.3. Survival analysis of patients with TEAD CNV
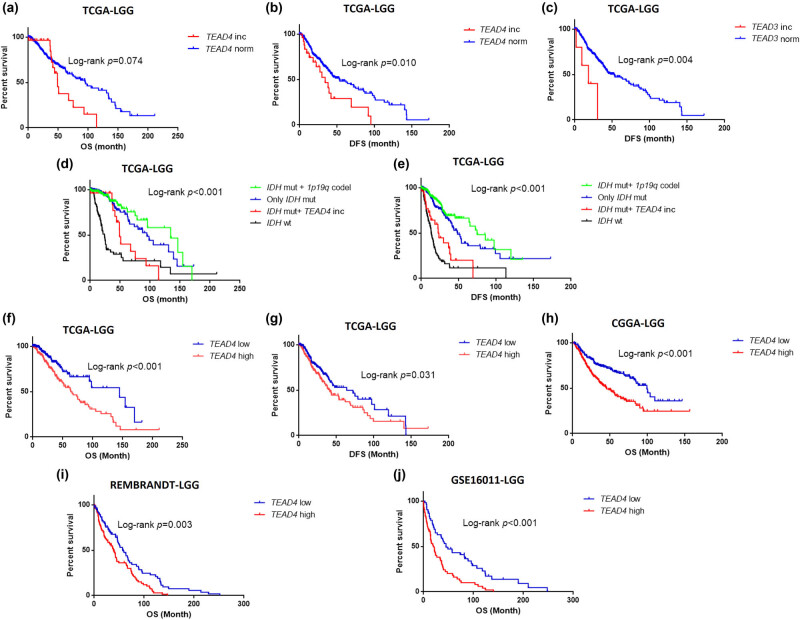
Survival analysis showed that TEAD4 CNV was strongly related to OS and DFS in LGG. The median OS of patients with TEAD4 CNV was obviously shorter than that of patients without TEAD4 CNV (p = 0.074, HR = 1.288, 95% CI: 0.973–1.703) (Figure 2a). And meanwhile, the median DFS for patients without TEAD4 CNV was significantly longer than the patients carrying TEAD4 CNV (p = 0.010, HR = 1.382, 95% CI: 1.074–1.778) (Figure 2b). In GBM, TEAD4 CNV was uncorrelated with OS and DFS (OS p = 0.815, DFS p = 0.463) (Figure A1b and c). Similarly, TEAD3 CNV was also significantly associated with DFS in LGG (p = 0.004, HR = 1.961, 95% CI: 1.190–3.230) (Figure 2c) but not OS (p = 0.918) (Figure A1a). Only one TEAD3 CNV carrier was found in patients with GBM. In contrast, TEAD1 and TEAD2 showed no association with glioma prognosis (Table 2).
Figure 2.
Survival curve of TEAD3 and TEAD4 in patients with LGG. (a)–(e) Survival curve for CNVs (TEAD3 and TEAD4) in TCGA-LGG cohort. (f)–(j) Survival curve for TEAD4 expression. Blue lines represent gene normal copy number (norm) or TEAD4 low expression, and red lines represent gene copy number increase (inc) or TEAD4 high expression. OS means overall survival and DFS means disease-free survival.
Table 2.
Association analysis results between TEAD CNVs and prognosis in glioma
| Gene | LGG | GBM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| INC | NOR | DEC | OS p | DFS p | INC | NOR | DEC | OS p | DFS p | |
| TEAD1 | 2 | 497 | 14 | 0.165 | 0.988 | 0 | 575 | 1 | NA | NA |
| TEAD2 | 1 | 496 | 16 | 0.791 | 0.132 | 3 | 573 | 1 | 0.857 | 0.882 |
| TEAD3 | 5 | 508 | 0 | 0.918 | 0.004 | 2 | 576 | 0 | NA | NA |
| TEAD4 | 28 | 484 | 1 | 0.074 | 0.010 | 9 | 567 | 1 | 0.815 | 0.463 |
Note: p value was calculated by log-rank method, the p values less than 0.1 were in bold. If the sample size of a group was less than 3, the group was deleted. INC represents CNV increase; NOR represents normal; DEC represents CNV decrease.
Backward stepwise cox regression analysis selected 1p19q co-deletion, TP53 mutation, tumor grade and age as the covariates for OS; and IDH mutation, 1p19q co-deletion and age as the covariates for DFS. After adjusting for covariates, TEAD4 CNV increase significantly remains associated with OS (p = 0.022, HR = 1.444, 95% CI: 1.054–1.978) and DFS (p = 0.005, HR = 1.485, 95% CI: 1.124–1.962) in patients with TCGA-LGG (Table 3). These results indicated that TEAD4 CNV increase was an independent predictor of LGG prognosis.
Table 3.
Univariate and multivariate analysis results of TEAD4 CNV in TCGA-LGG cohort
| OS | DFS | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||
| p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | |
| TEAD4 CNV | 0.077 | 1.288 (0.973–1.703) | 0.022 | 1.444 (1.054–1.978) | 0.012 | 1.382 (1.074–1.778) | 0.005 | 1.485 (1.124–1.962) |
| Age | 6.39 × 10−15 | 1.058 (1.043–1.074) | 1.02 × 10−11 | 1.056 (1.040–1.073) | 4.35 × 10−5 | 1.026 (1.013–1.039) | 0.001 | 1.023 (1.010–1.037) |
| Grade | 1.58 × 10−9 | 3.340 (2.258–4.940) | 6.99 × 10−5 | 2.264 (1.513–3.386) | 0.003 | 1.608 (1.174–2.202) | ||
| IDH mutation | 2.22 × 10−22 | 0.156 (0.108–0.227) | 2.54 × 10−21 | 0.178 (0.124–0.254) | 5.19 × 10−11 | 0.245 (0.161–0.373) | ||
| TP53 mutation | 0.039 | 0.690 (0.485–0.982) | 2.42 × 10−7 | 0.294 (0.185–0.468) | 0.791 | 1.042 (0.767–1.417) | ||
| ATRX mutation | 0.070 | 0.714 (0.497–1.028) | 0.802 | 1.040 (0.764–1.416) | ||||
| 1p19q co-deletion | 1.21 × 10−4 | 0.411 (0.261–0.646) | 3.41 × 10−11 | 0.166 (0.098–0.282) | 4.64 × 10−6 | 0.408 (0.278–0.599) | 0.008 | 0.557 (0.362–0.856) |
Integrated analysis of TEAD4 CNV increase, IDH mutations, TP53 mutation, ATRX mutation and 1p19q co-deletion showed that the patients with LGG would divide into four groups with different prognosis. Group 1 (n = 167) included the patients with both IDH mutation and 1p19q co-deletion, and they had the best outcomes. Group 2 (n = 203) included the patients with IDH mutation but without either TEAD4 CNV increase or 1p19q co-deletion, and their outcomes were only second to group 1. Most of the group 2 patients carried TP53 mutation and/or ATRX mutation, and only two patients had neither TP53 mutation nor ATRX mutation. Group 3 (n = 27) included patients with both IDH mutation and TEAD4 CNV increase but not 1p19q co-deletion, whose prognosis was significantly worse than the patients with LGG having IDH mutations while without TEAD4 CNV increase (OS p = 0.016, HR = 1.465, 95% CI: 1.065–2.016; DFS p = 0.014, HR = 1.405, 95% CI: 1.067–1.850). All of the group 3 patients carried at least one of these two mutations (TP53 mutation and ATRX mutation). Group 4 (n = 96) included the patients with IDH wild-type LGG, and they had significantly worse outcomes than group 3 patients (Figure 2d–e).
3.4. Co-occurrence analysis of TEAD4 CNV, IDH mutation, TP53 mutation, ATRX mutation and 1p19q co-deletion
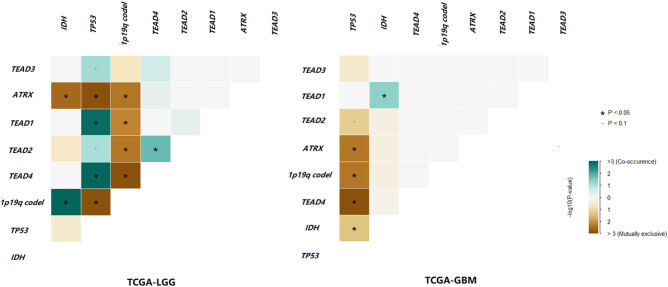
To find the reasons for the diverse clinical effects of TEAD4 CNV increase on LGG and GBM, we conducted a series of explorations, including mutation co-occurrence analysis. Results showed that TEAD4 CNV increase and TP53 mutations were significantly mutually exclusive in GBM, while it was opposite in LGG (Figure 3). In addition, TEAD4 CNV increase and 1p19q co-deletion were significantly mutually exclusive in LGG, while this phenomenon did not occur in GBM. Moreover, 28/29 TEAD4 CNV increase carriers simultaneously had IDH mutation in LGG, while only 1/9 TEAD4 CNV increase carriers had IDH mutation in GBM.
Figure 3.
Interactions of TEAD CNVs, isocitrate dehydrogenase (IDH) mutation, TP53 mutation, ATRX mutation and 1p19q co-deletion in TCGA-LGG and TCGA-GBM cohorts. Green represents co-occurrence and brown represents exclusive.
3.5. TEAD4 CNV was associated with TEAD4 expression
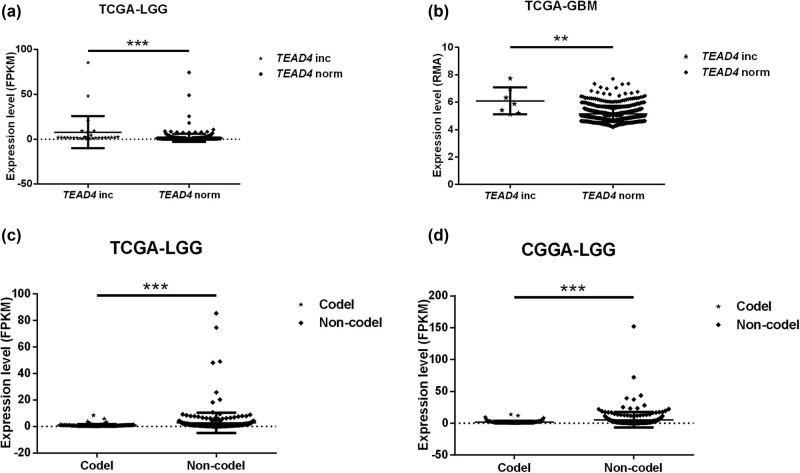
Differential expression analysis showed that carriers with TEAD4 CNV increase had a higher TEAD4 expression level than the normal TEAD4 copy number carriers (LGG p = 8.27 × 10−7; GBM p = 0.004) (Figure 4a and b; FPKM means the expression data were normalized by FPKM method; and RMA means the expression data were normalized by RMA method [28,29]). On the contrary, the TEAD3 CNV was not significantly associated with the expression of TEAD3 (LGG p = 0.077; GBM p is unavailable).
Figure 4.
Distribution of TEAD4 expression according to its CNV and 1p19q co-deletion. (a) and (c) TCGA-LGG cohort. (b) TCGA-GBM cohort. (d) CGGA-mRNAseq_693 LGG cohort. Norm represents TEAD4 normal copy number, inc represents TEAD4 copy number increase. Codel represents 1p19q co-deletion and noncodel represents lack of 1p19q co-deletion. FPKM means the expression data were normalized by FPKM method, while RMA means the expression data were normalized by RMA method.
The relationship between TEAD4 expression and IDH mutation, TP53 mutation, ATRX mutation or 1p19q co-deletion was also tested. We found that the expression of TEAD4 was significantly downregulated in both TCGA-LGG and CGGA-mRNAseq_693 LGG cohorts when 1p19q co-deletion occurred (Figure 4c and d). The TEAD4 expression level was irrelevant to IDH mutation in patients with both of TCGA-LGG and CGGA-mRNAseq_693 LGG. In addition, the expression of TEAD4 was also found to be significantly upregulated in TP53 mutation or ATRX mutation carriers in TCGA-LGG (Figure A2). None of TP53 mutation and ATRX mutation information was provided in CGGA-mRNAseq_693 data sets.
3.6. TEAD4 expression was an independent prognosis predictor for patients with LGG carrying IDH mutation
Survival analysis showed that TEAD4 expression was significantly associated with OS and DFS in TCGA-LGG samples (Figure 2f and g). The median survival time of the high TEAD4 expression group was dramatically shorter than that of the low TEAD4 expression group (63.500 months vs 144.940 months; p = 7.71 × 10−5, HR = 2.113, 95% CI: 1.446–3.088). Similarly, the median DFS time was markedly shorter in patients with high TEAD4 expression than the low TEAD4 expression group (41.060 months vs 72.170 months; p = 0.022, HR = 1.431, 95% CI: 1.050–1.949). Similar results were found in CGGA mRNAseq_693-LGG, Rembrandt-LGG and GSE16011-LGG (Figure 2h and j). The expression of TEAD4 was irrelevant to OS and DFS in patients with TCGA-GBM (OS p = 0.815, DFS p = 0.890).
To confirm whether TEAD4 expression level was an independent factor for the prognosis of LGG, multivariate cox regression analysis was conducted in TCGA-LGG and CGGA mRNAseq_693 LGG data sets. After adjusting for covariates, the expression level of TEAD4 was only significantly associated with OS in CGGA mRNAseq_693 LGG population (Table 4).
Table 4.
Survival analysis of TEAD4 expression in IDH mutant and IDH wild-type patients
| Data set | IDH mutation status | N | Adjusted HR (95% CI) | Adjusted p |
|---|---|---|---|---|
| OS | ||||
| TCGA-LGG | ALL | 505 | 1.325 (0.867–2.024) | 0.193* |
| IDH mut. | 409 | 2.226 (1.260–3.935) | 0.006** | |
| IDH wt. | 96 | 0.869 (0.473–1.596) | 0.651*** | |
| CGGA-mRNAseq_693 | ALL | 402 | 1.666 (1.201–2.311) | 0.002# |
| LGG | IDH mut. | 306 | 1.805 (1.216–2.679) | 0.003 ## |
| IDH wt. | 96 | 1.218 (0.730–2.031) | 0.450### | |
| DFS | ||||
| TCGA-LGG | ALL | 505 | 1.066 (0.766–1.3482) | 0.706& |
| IDH mut. | 409 | 1.128 (0.759–1.677) | 0.550&& | |
| IDH wt. | 96 | 0.824 (0.455–1.492) | 0.523&&& | |
Note: *Adjusted by age, 1p19q co-deletion, TP53 mutation and tumor grade; **adjusted by age, 1p19q co-deletion and tumor grade; ***adjusted by age and tumor grade; #adjusted by 1p19q co-deletion, IDH mutation and tumor grade; ##adjusted by 1p19q co-deletion and tumor grade; ###adjusted by tumor grade; &adjusted by age, 1p19q co-deletion and TP53 mutation; &&adjusted by 1p19q co-deletion; &&&adjusted by age and ATRX mutation. All covariates were selected by backward stepwise cox regression analysis. The p values reaching to the edge of a significant level (<0.1) were in bold.
Based on stratified survival analysis of TEAD4 expression according to IDH mutation status, we found that TEAD4 expression significantly affected the OS in patients with LGG carrying IDH mutants after adjusting for covariates. On the other hand, in IDH wild-type LGG patients, TEAD4 expression had no correlation with the outcomes. These results were validated in CGGA mRNAseq_693 population (Table 4). The results suggested that TEAD4 expression might be an independent predictor of prognosis for patients with LGG carrying IDH mutation, and the TEAD4 gene function might have a synergistic effect with the IDH mutation.
3.7. Coexpression and GO enrichment analysis
A total of 91 genes were found significantly coexpressing with TEAD4 in patients with both TCGA-LGG and mRNAseq_693 with IDH mutations. All of these genes were positively correlated with TEAD4. In carriers of IDH wild-type, 420 genes were found significantly coexpressing with TEAD4, and all genes were positively correlated with TEAD4. Among these genes, 41 coexpressed with TEAD4 only in IDH mutation carriers, while 370 coexpressed with TEAD4 only in IDH wild-type patients (Tables S1 and S2).
GO enrichment analysis suggested that the top 20 GO terms enriched by 41 genes were mainly related to immune and membrane, such as T-cell receptor-signaling pathway, MHC class II protein complex, plasma membrane and so on (Table S3). However, the top 20 GO terms enriched by 370 genes were mainly correlated with binding, such as protein binding, integrin binding, actin filament binding and so on (Table S4).
3.8. Immune infiltration analysis
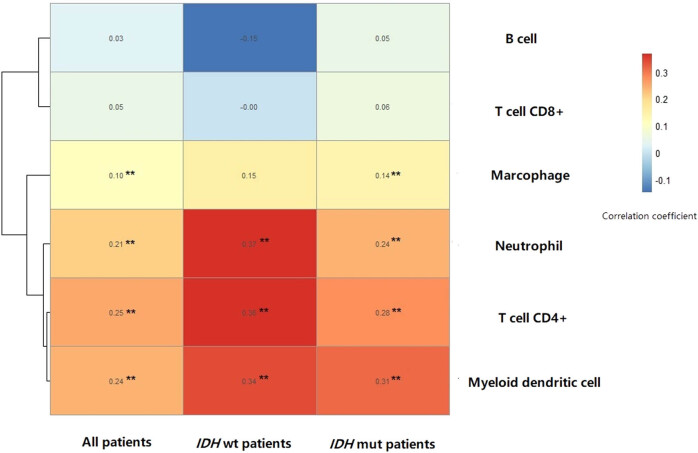
The expression level of TEAD4 was significantly positively correlated with immune infiltration scores of myeloid dendritic cell, T-cell CD4+, neutrophil and macrophage in patients with TCGA-LGG. The same results were found in patients with TCGA-LGG carrying IDH mutation. In patients with TCGA-LGG carrying wild-type IDH mutation, except for macrophage, the immune infiltration scores of myeloid dendritic cell, T-cell CD4+ and neutrophil significantly positively correlated with TEAD4 expression (Figure 5). No significant difference was observed between IDH mutation and IDH wild-type patients.
Figure 5.
Heat map of the correlation coefficient of immune infiltration scores and TEAD4 expression.
4. Discussion
In this study, we found that TEAD CNVs had a higher incidence in LGG than in GBM. Additionally, TEAD4 CNV which strongly regulated TEAD4 expression was significantly associated with the outcomes of patients with LGG. Interestingly, we’ve discovered that both TEAD CNVs and TEAD expressions were taking effect only in patients with LGG, but not in patients with GBM. Meanwhile, we also found that TEAD4 CNV increase and IDH mutations might be mutually exclusive in GBM; while in LGG, these two mutations occur synergistically. Moreover, integrating TEAD4 CNV, TP53 mutation, ATRX mutation, IDH mutations and 1p19q co-deletion would separate patients with LGG into four groups with different prognosis, which might provide a new biomarker for developing new therapeutic regimens to improve the outcomes in patients with LGG carrying IDH mutations but suffering poor prognosis.
In our study, we discovered that TEAD4 CNV increase might lead to poor prognosis in patients with LGG carrying IDH mutations, but not in patients with LGG not carrying IDH mutation or the patients with GBM. This compelling phenomenon might most probably be due to the IDH mutation that could promote the synthesis of 2-hydroxyglutarate and then lead to hypermethylation phenotype in cells which would regulate lots of gene expression levels in various pathways [30,31,32]. So there might be significant differences in the activity of many signaling pathways including Hippo and Wnt between IDH mutation and wild-type individuals.
Meanwhile, we conducted TEAD4 coexpression analysis in IDH mutant and IDH wild-type patients, and pathway analysis was performed for the relating genes. The results suggested enormous difference in TEAD4 coexpression genes between IDH mutation and wild-type patients. In IDH mutation carriers, the TEAD4 correlating genes mainly enriched in immune response–related pathways; while in IDH wild-type patients, the genes concentrated in other biological pathways. The reported IDH mutant could alter the tumor immunological microenvironment in LGGs, and the immune system gene signature could predict the prognosis of glioma [32,33]. Several studies found that YAP/TAZ expression could regulate the cross-talk between immune cells and tumor cells in the tumor microenvironment through binding to TEADs and then suppressing the T-cell viability and triggering tumor immune evasion [34,35,36].
Based on these, the expression of TEAD4 would take on different roles in glioma prognosis depending on whether the patient is carrying IDH variation. As we know, IDH mutation carriers in LGG tend to have longer survival than other types of gliomas [37]. Nevertheless, some of these patients are still suffering poor prognosis, while the reasons were unknown yet. The influence of CNV on clinical outcome is ubiquitous in various malignant tumors including gliomas [38]. The level of total genomic CNV inversely correlated with both PFS and OS in IDH-mutant LGG (grades II and III) [39,40,41]. Our results suggested that TEAD4 CNV increases and the high expression level would dramatically aggravate outcomes in patients with LGG carrying IDH mutations, which might partly explain why these patients are experiencing poorer prognosis.
In the meantime, our study discovered that 1p19q co-deletion would downregulate TEAD4 expression in glioma. However, TEAD4 CNV and expression level could affect the prognosis in LGG independent of 1p19q. TEAD4 is located on chromosome 12, suggesting that its decreased expression is not caused by the deletion of chromosome 1p or 19q directly. The specific reason for the correlation of 1p19q co-deletion with TEAD4 expression is not clear at present, and further functional studies are needed to determine it.
Our research results were somewhat different from those reported by Xu et al. as mentioned in the Introduction section [26]. Our study found that CNV and overexpression of TEAD4 only affected the prognosis in patients with LGG carrying IDH mutation. The reasons for this difference might be that Xu et al. did not distinguish the discrepancy of prognosis and genes expression between LGG and GBM, and they had not considered the effects of IDH mutation, TP53 mutation, ATRX mutation and 1p19q on the prognosis of glioma. In addition, Xu et al. found that the expression of TEAD4 was negatively correlated with IDH1 mutation; while our study found that TEAD4 expression level was irrelevant to IDH mutation in patients with LGG [26]. This might because they did not take into account that the frequency of IDH mutation was extremely low in GBM and the relatively higher level of TEAD4 expression in GBM. So it appeared that IDH1 mutation was significantly negatively related with TEAD4 expression.
Nevertheless, our study has several limitations. First of all, in addition to TEAD4, the frequency of other TEAD CNV was very low, and thus we could not confirm the influence of TEAD1–3 in prognosis of patients with LGG. Second, the frequency of IDH mutation in GBM was very low, hence we could not explore whether the interaction between TEAD4 CNVs and IDH mutation in GBM is the same as in LGG. Finally, the results in this study were acquired from clinical data analysis; therefore, further experiments were required for validation.
5. Conclusion
This study discovered that CNV and gene expression status of TEAD4 were closely related to the prognosis of patients with LGG carrying IDH mutation. Incorporating TEAD4 CNVs might better stratify the patients with LGG, which would provide new biomarkers for establishing new molecular classification systems for further precision medicine in glioma.
Appendix
Figure A1.
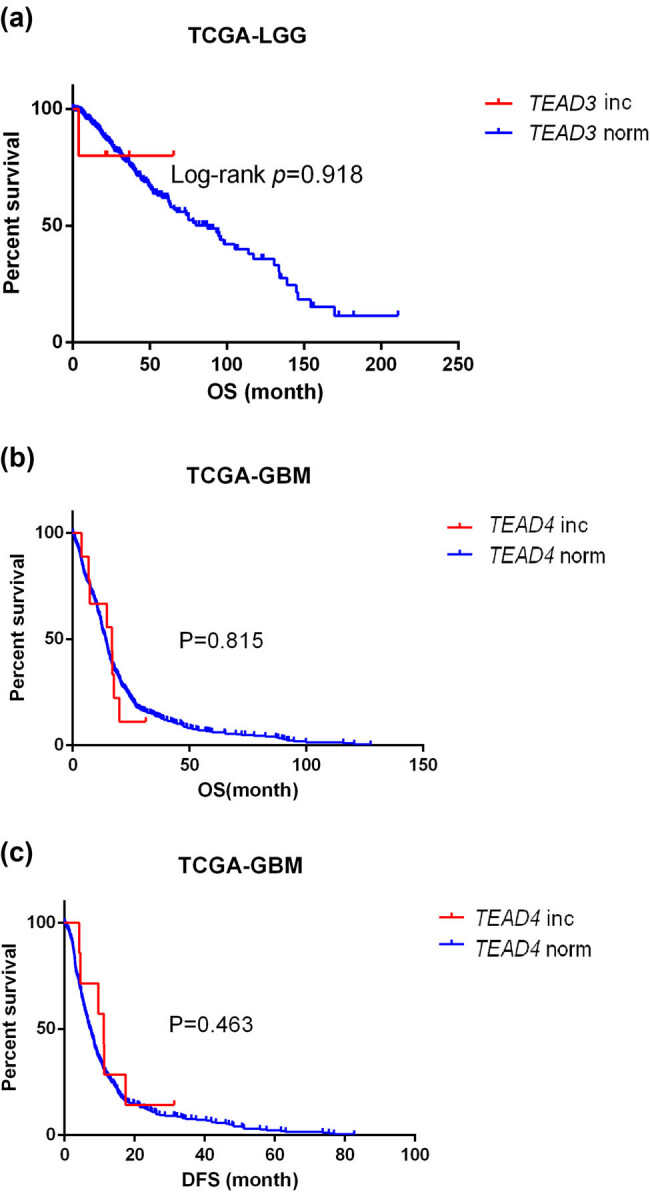
Survival Curve of TEAD3 and TEAD4 CNV in patients with glioma. (a) Overall survival curve for TEAD3 CNV in TCGA-LGG cohort. (b) Overall survival curve for TEAD4 CNV in TCGA-GBM cohort. (c) Disease-free survival curve for TEAD4 CNV in TCGA-GBM cohort. Blue lines represent gene normal copy number (norm), while red lines represent gene copy number increase (inc). OS means overall survival, and DFS means disease-free survival.
Figure A2.
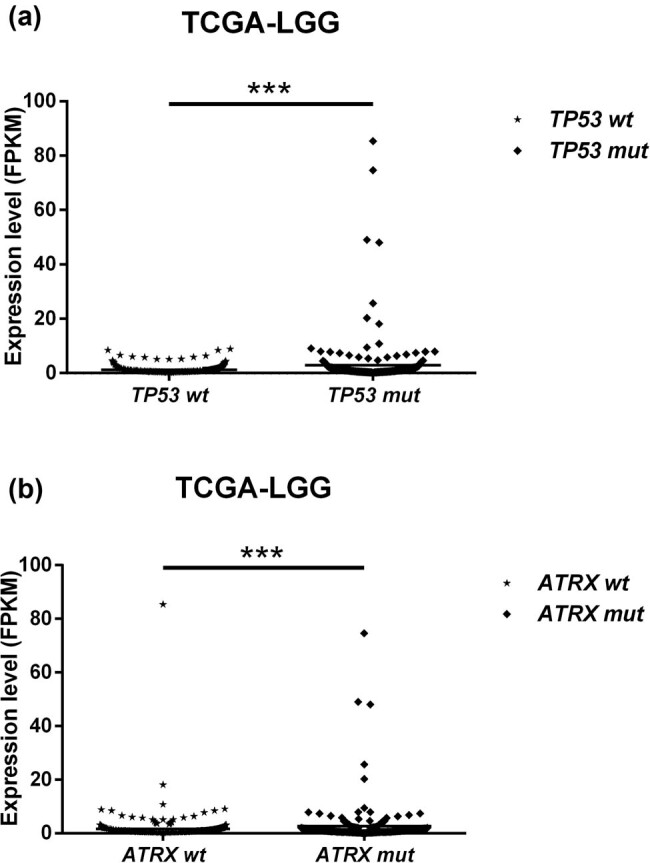
Distribution of TEAD4 expression according to TP53 mutation and ATRX mutation. mut represents mutation, wt represents wild type, FPKM means the expression data were normalized by FPKM method.
Footnotes
Funding information: This project was supported by Chinese National Science Foundation (No. 81803583, No. 81671123), Hunan Provincial Natural Science Foundation of China (2017JJ3480, 2019JJ50854), Open Fund Project of Hunan Universities Innovation Platform (18K006, 17K100) and Fundamental Research Funds for the Central Universities of Central South University (No. 2018zzts903).
Conflict of interest: The authors state no conflicts of interest.
Data availability statement: The data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol. 2016;18:v1–75. 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed]; Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C. et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol. 2016;18:v1–75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Liu W, Lv G, Li Y, Li L, Wang B. Downregulation of CDKN2A and suppression of cyclin D1 gene expressions in malignant gliomas. J Exp Clin Cancer Res. 2011;30:76. 10.1186/1756-9966-30-76. [DOI] [PMC free article] [PubMed]; Liu W, Lv G, Li Y, Li L, Wang B. Downregulation of CDKN2A and suppression of cyclin D1 gene expressions in malignant gliomas. J Exp Clin Cancer Res. 2011;30:76. doi: 10.1186/1756-9966-30-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA A Cancer J Clinicians. 2016;66:115–32. 10.3322/caac.21338. [DOI] [PubMed]; Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA A Cancer J Clinicians. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- [4].Batich KA, Reap EA, Archer GE, Sanchez-Perez L, Nair SK, Schmittling RJ, et al. Long-term survival in glioblastoma with cytomegalovirus pp65-targeted vaccination. Clin Cancer Res. 2017;23:1898–909. 10.1158/1078-0432.ccr-16-2057. [DOI] [PMC free article] [PubMed]; Batich KA, Reap EA, Archer GE, Sanchez-Perez L, Nair SK, Schmittling RJ. et al. Long-term survival in glioblastoma with cytomegalovirus pp65-targeted vaccination. Clin Cancer Res. 2017;23:1898–909. doi: 10.1158/1078-0432.ccr-16-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9:717–26. 10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed]; Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9:717–26. doi: 10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–98. 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed]; Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR. et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–98. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Baumert BG, Hegi ME, van den Bent MJ, von Deimling A, Gorlia T, Hoang-Xuan K, et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033–6033): a randomised, open-label, phase 3 intergroup study. Lancet Oncol. 2016;17:1521–32. 10.1016/S1470-2045(16)30313-8. [DOI] [PMC free article] [PubMed]; Baumert BG, Hegi ME, van den Bent MJ, von Deimling A, Gorlia T, Hoang-Xuan K. et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033–6033): a randomised, open-label, phase 3 intergroup study. Lancet Oncol. 2016;17:1521–32. doi: 10.1016/S1470-2045(16)30313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–68. 10.1038/ng.3273. [DOI] [PubMed]; Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y. et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–68. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- [9].van den Bent MJ. Practice changing mature results of RTOG study 9802: another positive PCV trial makes adjuvant chemotherapy part of standard of care in low-grade glioma. Neuro Oncol. 2014;16:1570–4. 10.1093/neuonc/nou297. [DOI] [PMC free article] [PubMed]; van den Bent MJ. Practice changing mature results of RTOG study 9802: another positive PCV trial makes adjuvant chemotherapy part of standard of care in low-grade glioma. Neuro Oncol. 2014;16:1570–4. doi: 10.1093/neuonc/nou297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31:344–50. 10.1200/jco.2012.43.2229. [DOI] [PubMed]; van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY. et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31:344–50. doi: 10.1200/jco.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- [11].Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–20. 10.1007/s00401-016-1545-1. [DOI] [PubMed]; Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK. et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- [12].Xiao Y, Cui G, Ren X, Hao J, Zhang Y, Yang X, et al. A novel four-gene signature associated with immune checkpoint for predicting prognosis in lower-grade glioma. Front Oncol. 2020;10:605737. 10.3389/fonc.2020.605737. [DOI] [PMC free article] [PubMed]; Xiao Y, Cui G, Ren X, Hao J, Zhang Y, Yang X. et al. A novel four-gene signature associated with immune checkpoint for predicting prognosis in lower-grade glioma. Front Oncol. 2020;10:605737. doi: 10.3389/fonc.2020.605737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed]; Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee CK, Jeong SH, Jang C, Bae H, Kim YH, Park I, et al. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science. 2019;363:644–9. 10.1126/science.aav0173. [DOI] [PubMed]; Lee CK, Jeong SH, Jang C, Bae H, Kim YH, Park I. et al. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science. 2019;363:644–9. doi: 10.1126/science.aav0173. [DOI] [PubMed] [Google Scholar]
- [15].Mo Y, Wang Y, Zhang L, Yang L, Zhou M, Li X, et al. The role of Wnt signaling pathway in tumor metabolic reprogramming. J Cancer. 2019;10:3789–97. 10.7150/jca.31166. [DOI] [PMC free article] [PubMed]; Mo Y, Wang Y, Zhang L, Yang L, Zhou M, Li X. et al. The role of Wnt signaling pathway in tumor metabolic reprogramming. J Cancer. 2019;10:3789–97. doi: 10.7150/jca.31166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199–221. 10.1007/s00432-003-0431-0. [DOI] [PMC free article] [PubMed]; Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wu J, Fang J, Yang Z, Chen F, Liu J, Wang Y. Wnt inhibitory factor-1 regulates glioblastoma cell cycle and proliferation. J Clin Neurosci. 2012;19:1428–32. 10.1016/j.jocn.2011.12.023. [DOI] [PubMed]; Wu J, Fang J, Yang Z, Chen F, Liu J, Wang Y. Wnt inhibitory factor-1 regulates glioblastoma cell cycle and proliferation. J Clin Neurosci. 2012;19:1428–32. doi: 10.1016/j.jocn.2011.12.023. [DOI] [PubMed] [Google Scholar]
- [18].Park HW, Kim YC, Yu B, Moroishi T, Mo JS, Plouffe SW, et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell. 2015;162:780–94. 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed]; Park HW, Kim YC, Yu B, Moroishi T, Mo JS, Plouffe SW. et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell. 2015;162:780–94. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed]; Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guo C, Liang C, Yang J, Hu H, Fan B, Liu X. LATS2 inhibits cell proliferation and metastasis through the Hippo signaling pathway in glioma. Oncol Rep. 2019;41:2753–61. 10.3892/or.2019.7065. [DOI] [PMC free article] [PubMed]; Guo C, Liang C, Yang J, Hu H, Fan B, Liu X. LATS2 inhibits cell proliferation and metastasis through the Hippo signaling pathway in glioma. Oncol Rep. 2019;41:2753–61. doi: 10.3892/or.2019.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xu G, Fang P, Chen K, Xu Q, Song Z, Ouyang Z. MicroRNA-362-3p targets PAX3 to inhibit the development of glioma through mediating Wnt/beta-catenin pathway. Neuroimmunomodulation. 2019;26(3):119–28. 10.1159/000499766. [DOI] [PubMed]; Xu G, Fang P, Chen K, Xu Q, Song Z, Ouyang Z. MicroRNA-362-3p targets PAX3 to inhibit the development of glioma through mediating Wnt/beta-catenin pathway. Neuroimmunomodulation. 2019;26(3):119–28. doi: 10.1159/000499766. [DOI] [PubMed] [Google Scholar]
- [22].Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–5. 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed]; Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA. et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–5. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma Y, Yang Y, Wang F, Wei Q, Qin H. Hippo-YAP signaling pathway: a new paradigm for cancer therapy. Int J Cancer. 2015;137:2275–86. 10.1002/ijc.29073. [DOI] [PubMed]; Ma Y, Yang Y, Wang F, Wei Q, Qin H. Hippo-YAP signaling pathway: a new paradigm for cancer therapy. Int J Cancer. 2015;137:2275–86. doi: 10.1002/ijc.29073. [DOI] [PubMed] [Google Scholar]
- [24].Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed]; Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Thompson BJ. YAP/TAZ: drivers of tumor growth, metastasis, and resistance to therapy. Bioessays. 2020;42:e1900162. 10.1002/bies.201900162. [DOI] [PubMed]; Thompson BJ. YAP/TAZ: drivers of tumor growth, metastasis, and resistance to therapy. Bioessays. 2020;42:e1900162. doi: 10.1002/bies.201900162. [DOI] [PubMed] [Google Scholar]
- [26].Xu A, Wang X, Zeng Y, Zhou M, Yi R, Wu Z, et al. Overexpression of TEAD4 correlates with poor prognosis of glioma and promotes cell invasion. Int J Clin Exp Pathol. 2018;11:4827–35. [PMC free article] [PubMed]; Xu A, Wang X, Zeng Y, Zhou M, Yi R, Wu Z. et al. Overexpression of TEAD4 correlates with poor prognosis of glioma and promotes cell invasion. Int J Clin Exp Pathol. 2018;11:4827–35. [PMC free article] [PubMed] [Google Scholar]
- [27].Wang Y, Xu X, Maglic D, Dill MT, Mojumdar K, Ng PK, et al. Comprehensive molecular characterization of the Hippo signaling pathway in cancer. Cell Rep. 2018;25:1304–5. 10.1016/j.celrep.2018.10.001. [DOI] [PMC free article] [PubMed]; Wang Y, Xu X, Maglic D, Dill MT, Mojumdar K, Ng PK. et al. Comprehensive molecular characterization of the Hippo signaling pathway in cancer. Cell Rep. 2018;25:1304–5. doi: 10.1016/j.celrep.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed]; Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed]; Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24:1073–81. 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed]; Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E. et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24:1073–81. doi: 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhou H, Chang K, Bai HX, Xiao B, Su C, Bi WL, et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J Neurooncol. 2019;142:299–307. 10.1007/s11060-019-03096-0. [DOI] [PMC free article] [PubMed]; Zhou H, Chang K, Bai HX, Xiao B, Su C, Bi WL. et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J Neurooncol. 2019;142:299–307. doi: 10.1007/s11060-019-03096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Han S, Liu Y, Cai SJ, Qian M, Ding J, Larion M, et al. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br J Cancer. 2020;122:1580–9. 10.1038/s41416-020-0814-x. [DOI] [PMC free article] [PubMed]; Han S, Liu Y, Cai SJ, Qian M, Ding J, Larion M. et al. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br J Cancer. 2020;122:1580–9. doi: 10.1038/s41416-020-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Qian Z, Li Y, Fan X, Zhang C, Wang Y, Jiang T, et al. Molecular and clinical characterization of IDH associated immune signature in lower-grade gliomas. Oncoimmunology. 2018;7:e1434466. 10.1080/2162402X.2018.1434466. [DOI] [PMC free article] [PubMed]; Qian Z, Li Y, Fan X, Zhang C, Wang Y, Jiang T. et al. Molecular and clinical characterization of IDH associated immune signature in lower-grade gliomas. Oncoimmunology. 2018;7:e1434466. doi: 10.1080/2162402X.2018.1434466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Feng J, Yang H, Zhang Y, Wei H, Zhu Z, Zhu B, et al. Tumor cell-derived lactate induces TAZ-dependent upregulation of PD-L1 through GPR81 in human lung cancer cells. Oncogene. 2017;36:5829–39. 10.1038/onc.2017.188. [DOI] [PubMed]; Feng J, Yang H, Zhang Y, Wei H, Zhu Z, Zhu B. et al. Tumor cell-derived lactate induces TAZ-dependent upregulation of PD-L1 through GPR81 in human lung cancer cells. Oncogene. 2017;36:5829–39. doi: 10.1038/onc.2017.188. [DOI] [PubMed] [Google Scholar]
- [35].Janse van Rensburg HJ, Azad T, Ling M, Hao Y, Snetsinger B, Khanal P, et al. The Hippo pathway component taz promotes immune evasion in human cancer through PD-L1. Cancer Res. 2018;78:1457–70. 10.1158/0008-5472.CAN-17-3139. [DOI] [PubMed]; Janse van Rensburg HJ, Azad T, Ling M, Hao Y, Snetsinger B, Khanal P. et al. The Hippo pathway component taz promotes immune evasion in human cancer through PD-L1. Cancer Res. 2018;78:1457–70. doi: 10.1158/0008-5472.CAN-17-3139. [DOI] [PubMed] [Google Scholar]
- [36].Pan Z, Tian Y, Cao C, Niu G. The emerging role of YAP/TAZ in tumor immunity. Mol Cancer Res. 2019;17:1777–86. 10.1158/1541-7786.MCR-19-0375. [DOI] [PubMed]; Pan Z, Tian Y, Cao C, Niu G. The emerging role of YAP/TAZ in tumor immunity. Mol Cancer Res. 2019;17:1777–86. doi: 10.1158/1541-7786.MCR-19-0375. [DOI] [PubMed] [Google Scholar]
- [37].Tang L, Deng L, Bai HX, Sun J, Neale N, Wu J, et al. Reduced expression of DNA repair genes and chemosensitivity in 1p19q codeleted lower-grade gliomas. J Neurooncol. 2018;139:563–71. 10.1007/s11060-018-2915-4. [DOI] [PMC free article] [PubMed]; Tang L, Deng L, Bai HX, Sun J, Neale N, Wu J. et al. Reduced expression of DNA repair genes and chemosensitivity in 1p19q codeleted lower-grade gliomas. J Neurooncol. 2018;139:563–71. doi: 10.1007/s11060-018-2915-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mirchia K, Snuderl M, Galbraith K, Hatanpaa KJ, Walker JM, Richardson TE. Establishing a prognostic threshold for total copy number variation within adult IDH-mutant grade II/III astrocytomas. Acta Neuropathol Commun. 2019;7:121. 10.1186/s40478-019-0778-3. [DOI] [PMC free article] [PubMed]; Mirchia K, Snuderl M, Galbraith K, Hatanpaa KJ, Walker JM, Richardson TE. Establishing a prognostic threshold for total copy number variation within adult IDH-mutant grade II/III astrocytomas. Acta Neuropathol Commun. 2019;7:121. doi: 10.1186/s40478-019-0778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mirchia K, Sathe AA, Walker JM, Fudym Y, Galbraith K, Viapiano MS, et al. Total copy number variation as a prognostic factor in adult astrocytoma subtypes. Acta Neuropathol Commun. 2019;7:92. 10.1186/s40478-019-0746-y. [DOI] [PMC free article] [PubMed]; Mirchia K, Sathe AA, Walker JM, Fudym Y, Galbraith K, Viapiano MS. et al. Total copy number variation as a prognostic factor in adult astrocytoma subtypes. Acta Neuropathol Commun. 2019;7:92. doi: 10.1186/s40478-019-0746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Richardson TE, Patel S, Serrano J, Sathe AA, Daoud EV, Oliver D, et al. Genome-wide analysis of glioblastoma patients with unexpectedly long survival. J Neuropathol Exp Neurol. 2019;78:501–7. 10.1093/jnen/nlz025. [DOI] [PMC free article] [PubMed]; Richardson TE, Patel S, Serrano J, Sathe AA, Daoud EV, Oliver D. et al. Genome-wide analysis of glioblastoma patients with unexpectedly long survival. J Neuropathol Exp Neurol. 2019;78:501–7. doi: 10.1093/jnen/nlz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Richardson TE, Sathe AA, Kanchwala M, Jia G, Habib AA, Xiao G, et al. Genetic and epigenetic features of rapidly progressing IDH-mutant astrocytomas. J Neuropathol Exp Neurol. 2018;77:542–8. 10.1093/jnen/nly026. [DOI] [PMC free article] [PubMed]; Richardson TE, Sathe AA, Kanchwala M, Jia G, Habib AA, Xiao G. et al. Genetic and epigenetic features of rapidly progressing IDH-mutant astrocytomas. J Neuropathol Exp Neurol. 2018;77:542–8. doi: 10.1093/jnen/nly026. [DOI] [PMC free article] [PubMed] [Google Scholar]