Abstract
Recent studies suggest excessive complement activation in severe coronavirus disease-19 (COVID-19). The latter shares common characteristics with complement-mediated thrombotic microangiopathy (TMA). We hypothesized that genetic susceptibility would be evident in patients with severe COVID-19 (similar to TMA) and associated with disease severity. We analyzed genetic and clinical data from 97 patients hospitalized for COVID-19. Through targeted next-generation-sequencing we found an ADAMTS13 variant in 49 patients, along with two risk factor variants (C3, 21 patients; CFH,34 patients). 31 (32%) patients had a combination of these, which was independently associated with ICU hospitalization (p = 0.022). Analysis of almost infinite variant combinations showed that patients with rs1042580 in thrombomodulin and without rs800292 in complement factor H did not require ICU hospitalization. We also observed gender differences in ADAMTS13 and complement-related variants. In light of encouraging results by complement inhibitors, our study highlights a patient population that might benefit from early initiation of specific treatment.
Keywords: COVID-19, SARS-CoV2, Complement, Genetic susceptibility, Eculizumab, Rigorous algorithm
1. Introduction
Coronavirus disease-19 (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has led to unprecedented changes in public health, research, social life, and the world economy [1]. Although many individuals remain asymptomatic, significant morbidity and mortality is observed worldwide. The triumph of SARS-CoV-2 vaccination is expected to overcome many challenges of this pandemic [2]. Nevertheless, it is not likely to address the issue of morbidity and mortality from severe COVID-19 in the short-term.
Despite the plethora of studies in the field, early predictors of outcomes remain to be defined. Clinical factors such as older age, comorbidities, low lymphocyte count and high Radiographic Assessment of Lung Edema (RALE), have been identified as clinical predictors [3]. Lymphocyte subsets have been also extensively studied with regard to the clinical outcome [4,5]. In this context, the role of systemic inflammation is continuously studied, along with potential strategies to mitigate it in patients with severe disease [[6], [7], [8], [9], [10], [11], [12], [13]]. Indeed, severe COVID-19 is a systemic disease associated with small and large vessel thrombosis, renal, cardiac and neurological manifestations [14]. Endothelial dysfunction seems to be a common denominator of this multiorgan pathology. Nevertheless, the pathophysiology of endothelial dysfunction and driver of multiorgan pathology in COVID-19 remains to be elucidated [15].
Studies of previous coronaviruses have shown that blocking C3 activation significantly attenuates the lung-directed proinflammatory sequelae of infections. Both the genetic absence of C3 and blockade of downstream complement effectors, have shown therapeutic promise by containing the detrimental proinflammatory consequences of viral spread mainly via inhibition of monocyte/neutrophil activation and immune cell infiltration into the lungs [16,17]. More importantly, C3 blockade has already provided promising anti-inflammatory results in a small cohort of severe COVID19 patients [18,19]. A recent study also revealed that coronaviruses' proteins (SARS-CoV, MERS-CoV and SARS-CoV-2) bind to a key protein of the lectin pathway (MASP-2/Mannan-binding lectin serine protease 2), leading to complement-mediated inflammatory lung injury [20]. In addition, Magro et al. have detected deposits of C5b-9, C4d, and MAPS-2 in the microvasculature of lung and skin biopsies of severe COVID-19 patients [21]. Furthermore, SARS-CoV-2 spike protein subunits, but not N protein or spike protein from a more benign human coronavirus OC43, have been identified as potent activators of the alternative pathway, withC5 and factor D inhibitors preventing the complement-mediated damage [22]. Interestingly, both C3 and C5aR inhibition have been shown to attenuate thromboinflammation driven by neutrophil extracellular traps (NETs); while the compstatin-based C3 inhibitor Cp40 could disrupt tissue factor expression in NET-releasing neutrophils exposed to COVID-19 sera [23]. Indeed, several studies have shown dysregulation of the classical/lectin, alternative, and terminal pathway activation products in COVID-19 patients [[23], [24], [25], [26]].
Complement activation is a key feature in a number of complement-mediated disorders, also called complementopathies [27]. Among them, complement-mediated thrombotic microangiopathies (TMAs) have been thoroughly investigated, with the prototype disease being atypical hemolytic uremic syndrome (aHUS) [28]. Complement-mediated TMAs present with the typical triad of a TMA (thrombocytopenia, microangiopathic hemolytic anemia and organ damage mostly to the kidneys and central nervous system), caused by excessive complement activation [29]. Although the phenotype of a complementopathy may vary significantly including TMAs, age-related macular degeneration (AMD), catastrophic antiphospholipid antibody syndrome (CAPS) or HELLP (hemolysis, elevated liver enzymes and low platelets), the pathophysiology is common [30].
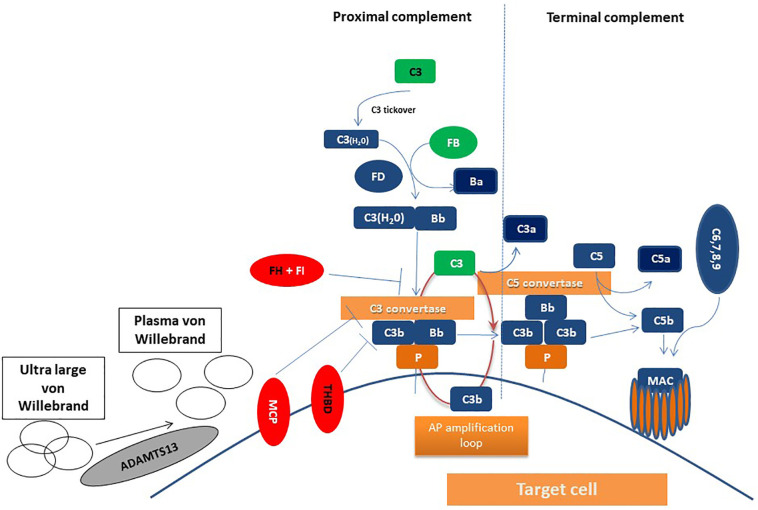
According to the “two-hit hypothesis”, germline mutations in complement-related genes or autoimmune antibodies are the first hit; while complement-amplifying conditions, including inflammation or infections, represent the second hit. Except for complement-mediated TMAs, the other pillar of TMA phenotype is thrombotic thrombocytopenic purpura (TTP), caused by severe ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) deficiency. Genetic variants in ADAMTS13 have been described not only in the hereditary but also in the acquired syndrome [31]. Suggested mechanisms of actions are summarized in Fig. 1 for proteins encoded by TMA-associated genes.
Fig. 1.
Potential interactions of proteins encoded by studied genes in COVID-19. Complement activation leads to C3 activation and C3 convertase formation on C3-opsonized surfaces, culminating in pronounced C3 fragment deposition on complement-targeted surfaces (proximal complement). In the presence of increased surface density of deposited C3b, the terminal complement is triggered, leading to membrane attack complex (MAC) formation on the surface of target cell. The present study focused on complement pathway dysregulation resulting from loss-of-function mutations in regulatory factors (Factor H, I, THBD/thrombomodulin, Membrane Cofactor Protein/MCP or CD46) shown in red, gain-of-function mutations (C3 and Factor B) shown in green, mutations shown in black, indicating the unknown effect on complement cascade. In addition, variants in ADAMTS13 were studied. ADAMTS13 cleaves von Willebrand factor anchored on the endothelial surface, in circulation, and at the sites of vascular injury. Proteins found to be of potential clinical significance (FH, C3, THBD, ADAMTS13) in COVID-19 are shown in black letters. AP: alternative pathway; Properdin: P; MAC: membrane attack complex. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Taken together, these data suggest that severe COVID-19 may resemble TMAs. The importance of this similarity is highlighted by the safety and efficacy of complement inhibitors in these disorders [27], that has been also confirmed by recent results in COVID-19 as summarized by Satyam et al. [32]. Therefore, the “two-hit” hypothesis that has been mentioned above for complement-mediated disorders needs to be further investigated in COVID-19. The second-hit or trigger may be due to inflammatory states, such as diabetes and obesity, or viral infection per se that also activate complement and thus, may exacerbate complement-mediated injury [33]. The first hit is most likely due to a germline predisposition, that has not been confirmed yet.
On these grounds, we hypothesized that genetic susceptibility might be present in severe COVID-19 patients at the level of complement-related genes and ADAMTS13. Furthermore, we investigated the association of genetic variants with clinical outcomes aiming to characterize a group of high-risk patients that might benefit from complement inhibition.
2. Materials and methods
2.1. Study population
We prospectively studied consecutive adult Caucasian patients hospitalized with COVID-19 in our referral centers during the first and second wave of the pandemic (April–May and September–October 2020). Diagnosis was confirmed by reverse-transcriptase polymerase chain reaction (RT-PCR). COVID-19 severity was assessed based on World Health Organization's (WHO) criteria into moderate and severe disease. Additional data on patients' history and course were recorded by treating physicians that followed patients up to discharge or death. The final analysis included only patients with available data on clinical course and outcome. The study was approved by Institutional Review Boards (IRBs) of recruiting centers (G Papanicolaou and Attikon Hospital) and conducted according to the Declaration of Helsinki.
2.2. Genetic analysis
Genomic DNA was isolated from peripheral blood samples and analyzed using next generation sequencing (NGS) with a customized complement-related gene panel (complement factor H/CFH, CFH-related, CFI, CFB, CFD, C3, CD55, C5, CD46, thrombomodulin/THBD), including TMA-associated ADAMTS13 (A Disintegrin and Metalloproteinase with Thrombospondin motifs). Probes were designed using DesignStudio (Illumina, San Diego, California) to cover all exons spanning 15 bases into the intronic regions (98% coverage). We used 10 ng of initial DNA material. Libraries were quantified using Qubit and sequenced on a MiniSeq System in a 2 × 150 bp run (Illumina, San Diego, California). Both Ensembl and Refseq resources were used for annotation of the output files. Variants clinical significance was based on ClinVar and the current version of the Complement Database [[34], [35], [36], [37]].
2.3. Statistical analysis
Data were analyzed using the statistical program SPSS 22.0 (IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp). Continuous variables were described as median and range and categorical variables as frequencies. Variables were compared using chi-square statistics for categorical variables and Mann-Whitney for continuous variables. Multivariate analysis was performed using binary logistic regression.
2.4. Proposed algorithm
To find the variants that predict severe disease we developed a collaboration of four international computational centers (Iran, Italy, Malaysia, Greece). This multi-disciplinary approach was necessary to address the multidimensional aspects of COVID-19 infection by established collaborations [[38], [39], [40]]. Through this collaboration, a rigorous algorithm, based on laws and rules of logic, has been developed and trained for the disease risk prediction/assessment of patients with a confirmed infection caused by COVID-19. To this end, the optimum pattern/combination of genetic variants associated with thrombotic microangiopathy which can justify the different clinical response among infected patients has been investigated. This was a challenging task mainly due to their infinite combinations (more than yotta = 1024). To this end experts, from data science and computational mechanics as well as large computer clusters were required in the framework of a multi-Institutional and multidisciplinary approach. In order to implement the proposed rigorous algorithm, a computer program has been developed in Matlab environment, at the Computational Mechanics Laboratory, School of Pedagogical and Technological Education, Athens, Greece. The proposed algorithm is implemented in five distinct steps, described as follows:
Step 1: Obtaining the genetic profile of each Covid-19 infected patient, with emphasis on the genetic variants in her/his genome as identified in the laboratory. Based on the work presented herein, patients exhibit heterogeneous variant profiles including pathogenic, benign, likely benign, and variants of unknown significance.
Step 2: Developing the variants' list (vector) using the list of genetic variants for the entire population of Covid-19 patients under study, as identified in the laboratory.
Step 3: Developing the database registering each patient's genetic variants. This database is defined as a two-dimensional matrix (PGD –Patients Genome Data) of dimension m × n, where m is the number of variants identified in the entire population of patients, and n is the number of patients under study. Each element, denoted as PGD(i,j) where i is the row index and j is the column index of the matrix, can take binary values, either 0 or 1. Specifically, PGD(i,j) = 0 signifies that a patient (corresponding to column with index j) does not exhibit the genetic variant which corresponds to row index i. On the other hand, PGD(i,j) = 1 signifies that a patient (corresponding to column with index j) exhibits the genetic variant which corresponds to row index i.
Step 4: Identification of the optimal combination of genetic variants, identified in the total population of patients (appearing in the relevant list of step 2). The optimal combination categorizes the impact of the disease on the patient's health, in two classes, namely, (i) the patient's needed entry to intensive care unit (ICU), and (ii) no needed entry to ICU. An additional classification can be considered, that is, (a) the combination can be found in the patient's genome, and (b) the combination cannot be found in the patient's genome. According to these classifications, four rules were considered for the genetic variants combinations in the patients cohort under study.
Rule I. Investigation of genetic variants combination, existing in the genome of a Covid-19 patient for whom ICU entry is necessary.
Rule II. Investigation of genetic variants combination, existing in the genome of a Covid-19 patient, not leading to ICU entry.
Rule III. Investigation of genetic variants combination, not existing in the genome of a Covid-19 patient for whom ICU entry is necessary.
Rule IV. Investigation of genetic variants combination, not existing in the genome of a Covid-19 patient, not leading to ICU entry.
Step 5: Identification of the optimal combination of genetic variants (from all combinations studied in the previous step). The optimal combination categorizes the impact of the disease on patient's health, and should take into account additional findings obtained from clinical studies, such as gender differences in ICU cases [41].
3. Results
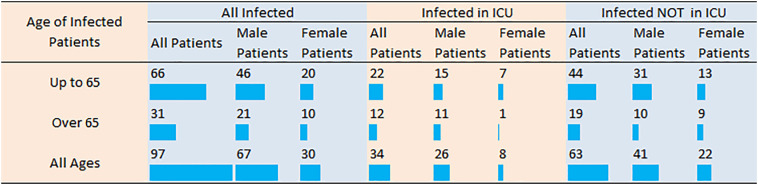
We studied 100 patients hospitalized for COVID-19. Among them, we analyzed only patients with available genetic and clinical data: 63 with moderate disease hospitalized in COVID-19 general ward (GW) and 34 with severe disease hospitalized in intensive care units (ICU). Fig. 2 presents in detail the statistics of the infected patients based on their age, gender, and severity of their disease. Additional patients characteristics are shown in Table 1 . Among them, 12 patients succumbed due to COVID-19 disease (p = 0.229).
Fig. 2.
Number of COVID-19 patients categorized by age, gender and disease severity (requiring hospitalization in intensive care unit or not). ICU: intensive care unit.
Table 1.
Summary of laboratory characteristics.
| Characteristics | General ward patients | Intensive care unit patients | p-value |
|---|---|---|---|
| Total lymphocyte count | 1.0[1.0] | 1.1[0] | 0.172 |
| Neutrophil-to-lymphocyte ratio | 3.1[2.0] | 6.4[8] | <0.001 |
| Platelets (×103/mm3) | 222 ± 103 | 322 ± 189 | 0.016 |
| Alanine aminotransferase (U/L) | 47[74] | 52[95] | 0.111 |
| Asparate aminotransferase (U/L) | 32[48] | 46[85] | 0.676 |
| Direct bilirubin (mg/dl) | 0.19[0.06] | 0.55[0.54] | 0.208 |
| Lactate dehydrogenase (U/L) | 272 ± 98 | 417 ± 172 | <0.001 |
| Creatinine (mg/dl) | 0.8[0.2] | 1.1[1.4] | 0.627 |
| Activated partial thromboplastin time (sec) | 34.9 ± 6.9 | 36.7 ± 5.0 | 0.287 |
| D-dimer (ng/mL) | 875[542] | 2115[2312] | 0.011 |
| C-reactive protein (mg/dl) | 49 ± 44 | 118 ± 69 | <0.001 |
| Procalcitonin (ng/ml) | 0.40 ± 1.24 | 0.49 ± 0.76 | 0.794 |
| Ferritin (ng/ml) | 594 ± 597 | 1055 ± 829 | 0.024 |
Values are presented as mean ± standard deviation for normal variables or median[interquartile range] for variables without normal distribution. Results were measured at the first day of hospitalization in General Ward or Intensive Care Unit.
Using steps 1 and 2 of the outlined methodology, we developed the database indicating which specific mutation of the genome was available in each patient. Based on the available statistics of the database, a total of 347 variants were identified, while the number of variants per infected patient ranged from 47 to 101 with an average value of 71 and standard deviation 12.
We found a variant of ADAMTS13 (rs2301612, missense) in 49 (51%) of patients, previously described in congenital TTP [42]. We also detected two missense risk factor variants, previously described in complement-related diseases: rs2230199 in C3 (21 patients) [43]; and rs800292 in CFH (34 patients) [44]. Among them, 31 (32%) patients had a combination of these characterized variants (18 in the discovery and 13 in the validation cohort). This combination was significantly associated with severe disease that required intensive care (p = 0.042), as well as low lymphocyte counts (p = 0.049) and high neutrophil-to-lymphocyte ratio (p = 0.038). Therefore, we performed a multivariate model (binary logistic regression) to understand which of the above-mentioned factors was associated with this combination of variants. ICU hospitalization (p = 0.022) was independently associated with double heterozygocity in these variants. Furthermore, one patient had a rare germline missense variant in CFI (rs112534524), previously detected in aHUS [45]. This patient suffered from severe disease but survived after long-term ICU hospitalization. Interestingly, 11 (11.5%) patients showed a likely protective missense variant in CFB (rs641153) [46]; while 9 of them did not require ICU hospitalization. Two patients with a protective variant that did require ICU hospitalization had a concomitant combination of ADAMTS13 and C3 variants as mentioned above.
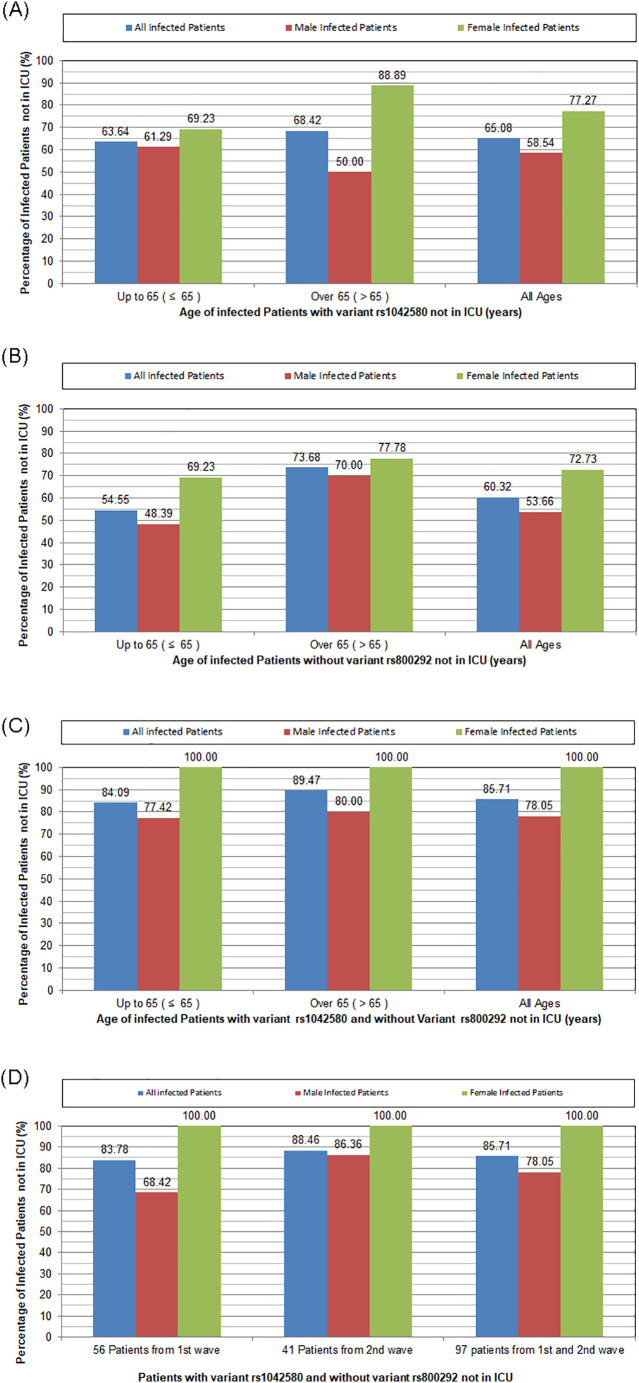
To understand the genetic differences of patients that develop severe COVID-19, we developed a rigorous algorithm, based on laws and rules of logic, trained for disease risk prediction/assessment of patients with confirmed COVID-19. Statistical analysis of all the possible combinations of variants showed that among COVID-19 infected patients, patients with THBD variant rs1042580 and without CFH variant rs800292 were not admitted in ICU. The opposite was true for ICU patients. Fig. 3 shows that this condition was true for 100% of female patients not admitted in ICU during the first and second wave of the pandemic. Interestingly, the percentage of male patients with variant rs1042580 and without variant rs800292 not in ICU significantly increased during the second wave (p = 0.032). This finding might be attributed to a more stringent selection of patients requiring ICU due to lower number of available units during the second wave of the pandemic.
Fig. 3.
Graphs categorizing patients according to gender, age, and percentage of variants predicting ICU hospitalization. A. Percentage of infected patients with THBD variant rs1042580 not in ICU: The majority of patients (78% with age up to 65 years and 89% over 65 years) with this variant did not require ICU hospitalization. Frequencies tend to be higher in female compared to male patients (p = 0.080). B. Percentage of infected patients without CFH variant rs800292 not in ICU. The majority of patients (67% with age up to 65 years and 78% over 65 years) without this variant did not require ICU hospitalization. C. Percentage of infected patients with THBD variant rs1042580 and without CFH variant rs800292 not in ICU. The combination of these variants predicted ICU hospitalization in all female patients (100%), compared to 78% of male patients (p = 0.003). D. Percentage of infected patients with THBD variant rs1042580 and without CFH variant rs800292 not in ICU during the first and the second wave. The percentage of male patients with variant rs1042580 and without variant rs800292 not in ICU significantly increased during the second wave (p = 0.032). The combination of these variants predicted ICU hospitalization in all female patients (100%). ICU: intensive care unit.
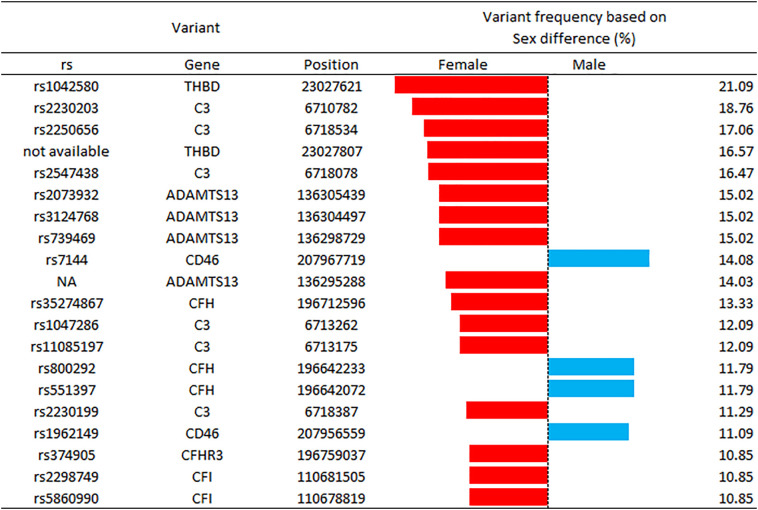
More importantly, Fig. 3 highlights the gender differences in patients with THBD rs1042580 and without CFH rs800292 variant. The combination of these variants predicted ICU hospitalization in all female patients (100%), compared to 78% of male patients (p = 0.003). These results support the existing notion that the clinical response of COVID-19 infected patients is strongly gender-dependent. To further characterize differences between genders, Fig. 4 highlights that variant THBD rs1042580 is by 21.09% likely more present in the genome of women, and also presents variants with different ratios in male and female participants.
Fig. 4.
Graph of variants with a higher than 10% difference in frequency between gender. Bars represent the percentage difference between genders. Red bar is used for variants with a higher percentage in female patients, while blue bar for variants with a higher percentage in male patients. THBD: thrombomodulin; CFH: complement factor H; ADAMTS13: a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13; CFD: complement factor D. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
We found for the first time variants associated with complementopathies and TMAs in patients with severe COVID-19. We were also able to generate a rigorous algorithm that detected a combination of variants predicting ICU hospitalization in all female (100%) and 78% of male patients. Patients with a variant in THBD (rs1042580) and without a variant in CFH (rs800292) did not require ICU hospitalization. Interestingly, we also observed gender differences in several complement-related and ADAMTS13 variants that might contribute to increased morbidity in male COVID-19 patients.
Our study sheds light on the genomic landscape in COVID-19 that is generally under-recognized, although familial clusters have been identified from the early stages of the pandemic [47]. Genetic susceptibility in COVID-19 has so far focused on the role of ABO blood group, angiotensin-converting enzyme 2 (ACE2) polymorphisms and interferons. The genome wide association study involving 1980 patients with COVID-19 and severe disease (defined as respiratory failure) has identified a 3p21.31 gene cluster as a genetic susceptibility locus in patients with COVID-19 and respiratory failure confirming a potential involvement of the ABO blood group system [48]. In addition, variants of ACE2 were found in different populations indicating that some individuals may be less susceptible to SARS-CoV-2 infection than others [49]. Furthermore, loss-of-function rare variants at the 13 human loci known to govern type I interferon immunity to influenza virus were found in 3.5% of adult patients with COVID-19 [50]. Epigenetic dysregulation of ACE2 and interferon-regulated genes also suggested increased COVID-19 susceptibility and severity in lupus patients [51]. Focusing on prediction of severe disease, a most recent study in a large cohort from the United Kingdom identifies and replicated genome-wide associations, on genes encoding antiviral restriction enzyme activators (OAS1, OAS2, OAS3), tyrosine kinase 2 (TYK2), dipeptidyl peptidase 9 (DPP9), and interferon receptor gene IFNAR2 [52]. Despite the delicate methods and analysis of this study, it should be noted that severe COVID-19 patients were compared only to non-COVID-19, non-hospitalized controls. Furthermore, recent clinical studies have shown conflicting results of anti-inflammatory treatment in COVID-19 [53,54]. Regarding complement variants, they have been only detected in a comprehensive genetic and transcriptional analysis of complement and coagulation variants [55]. This study confirms susceptibility to complement activation in large COVID-19 cohorts, although markers of severe disease were not included in the analysis.
Another question remains unanswered during this pandemic: gender differences. Indeed, men present with an increased case fatality rate worldwide [56]. Although this feature also suggests a germline susceptibility to severe infections for male patients, differences have been so far attributed to social and economical factors, along with different expression of ACE2 between genders [57]. A most recent meta-analysis confirmed that men have almost three times the odds of requiring ICU hospitalization and higher odds of death [41]. The authors discuss that known gender differences in the innate and adaptive immunity might be responsible. Our study is the first to describe such differences in germline variants. Interestingly, these differences in complement-related variants may enhance the already observed higher complement activity in healthy male compared to female individuals [58].
Our study is also the first to show that a combination of risk factor variants in complement-related genes (CFH or C3) with a variant in another TMA-associated gene (ADAMTS13) is independently associated with severe disease. A recent experimental study in the field of TMAs provides a rationale for this association, suggesting a synergistic effect of severe ADAMTS13 deficiency and complement activation in TMA pathogenesis [59]. Relevant reports are rare in humans because testing for ADAMTS13 and complement-related genes in the same panel is limited [60]. In addition, the traditional analysis of variants in these patients is only limited to rare variants. In line with this study, previous reports from our group have also suggested that more common variants are also associated with a TMA phenotype, especially where two or more variants are combined [61]. The potential interactions of proteins encoded by genes studied in the present report for patients with COVID-19 is shown in Fig. 1.
Our main finding of variants in THBD and CFH is also a very interesting one. Taking into account that in misssense variants a single nucleotide change results in a codon that codes for a different amino acid, a plethora of studies in other disorders characterized by endothelial dysfunction support our findings. Interestingly, THBD participates in both complement and hemostatic system, potentially proliferating the vicious cycle of thromboinflammation [62]. One of the first studies has found rs1042580 along with other THBD single nucleotide polymorphisms (SNPs) in patients with the model disease of complement-mediated TMA, aHUS [63]. In accordance with the clinical manifestations of COVID-19, rs1042580 has been associated with increased mortality in patients with acute respiratory distress syndrome (ARDS) [64]. Interestingly, the same variant has been also reported to be associated with endothelial dysfunction syndromes post allogeneic hematopoietic cell transplantation [65,66]. On the other hand, the CFH variant rs800292 is characterized a risk factor variant for a complement-related disease, AMD [44]. In line with our data, COVID-19 patients with AMD have been recently shown at significantly increased risk of severe disease and death [55]. Interestingly, COVID-19 spike protein binds heparan sulfate and blocks factor H binding to perturb factor H function, at the same location as the AMD variants [22]. Our results are in line with a recent study, showing decreased deposition of CFH in the COVID-19 infected lung tissue; therefore, speculating genetic dysregulation in patients who do not fare well in response to infection with SARS-Cov-2 [26]. It should be also noted that the detected frequency of variants in our study is similar to the frequency reported in the European population, confirming the validity of our results.
The importance of our findings is underlined by the long-term efficacy and safety of complement inhibition in complement-mediated TMAs [27]. Off-label use of the terminal complement inhibitor eculizumab has been also reported in COVID-19. Diurno et al. treated four severe COVID-19 patients with eculizumab. They observed an immediate reduction of C reactive protein levels and clinical improvement leading to successful disease outcomes [67]. Further supporting evidence has been shown by eculizumab treatment in three cases of critical COVID-19 [68]. Given the promising preclinical data and the severity of COVID-19 infections, eculizumab is currently studied in patients with severe COVID-19 infections. Eculizumab has also the advantage of safety in the pediatric population [69]. In contrast to frequent intravenous infusions every two weeks that are required for eculizumab treatment, the C5 inhibitor ravulizumab has the advantage of 4-fold longer half-life. Using ravulizumab, a single intravenous dose should be sufficient in patients with COVID-19 infections.
Since C3 and the lectin pathway have been implicated in the pathophysiology of coronaviruses infections, inhibitors of proximal complement pathways, under clinical development for complement-mediated TMAs, could also be efficacious in COVID-19 infections. The first case report of C3 inhibition with the compstatin-based inhibitor AMY-101 has shown safety and efficacy in severe COVID-19 [18]. Further comparative results of AMY-101 to eculizumab point toward a broader pathogenic involvement of C3-mediated pathways in thromboinflammation [19]. The lectin pathway inhibitor narsoplimab has also reported favorable results in six COVID-19 patients compared to retrospectively studied control groups [70].
Our study has certain limitations. Due to its observational nature, our present findings cannot be considered as definitive, indicating the need for future studies. Furthermore, our study highlights the significant amount of data derived from NGS that are not readily available for clinicians. Further studies are needed to incorporate these data into clinical practice. Translating the genotype to phenotype is also an important aspect of this field that was not available in our patients and might be feasible through novel ex vivo assays to detect complement activation in complementopathies [28,71]. In addition, our study design did not permit incorporation of all related variables in one model. Last but not least, it is evident that a larger population of COVID-19 patients from different centers could provide more information on the clinical importance of genetic susceptibility. Therefore, our multi-institutional and multi-disciplinary team is currently planning to overcome these obstacles in future studies.
In conclusion, variants associated with complementopathies and TMAs were for the first time detected in adult patients with severe COVID-19, confirming our hypothesis of genetic susceptibility. Furthermore, a combination of certain variants led to a high prediction rate of clinical outcomes, highlighting the need for further studies utilizing a multi-disciplinary approach in the field. Genetic testing in high-risk patients might be an important aspect aiming at intensified monitoring and early initiation of specific treatment in patients with genetic susceptibility.
Authors' contributions
E.G. and A.A. conceptualization. S.K., V.K., A.V., K.E., D.C., I.K., M.B., D.B., E.K., M.B., I.K., S.P., C.V. data curation. E.E., M.K., P.P., An.P., Ap.P., F.P., M.C., D.C. investigation. S.G., M.K., L.C., D.A. data analysis. P.A. software. E.G., T.T. P.A. writing-original draft . E.Y., D.S., I.S., R.A.B., S.K. and A.A. writing-review and editing.
Role of the funding source
This research was also supported by an independent investigator-driven grant (Pfizer Pharmaceuticals).
Acknowledgments
Acknowledgments
The authors would like to thank the biologist Maria Spachidou for her technical assistance.
Disclosure of conflicts of interest
R.A.B. is currently serving as an Alexion Pharmaceuticals Scientific Advisory Board member. E.G. is supported by the ASH Global Research Award and has consulted for Omeros Cooperation. The other authors declare no competing financial interest.
References
- 1.Kabanova A., Gavriilaki E., Pelzer B.W., Brunetti L., Maiques-Diaz A. Effect of the COVID-19 pandemic on laboratory and clinical research: a testimony and a call to action from researchers. Hemasphere. 2020;4 doi: 10.1097/HS9.0000000000000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubin E.J., Longo D.L. SARS-CoV-2 vaccination - an Ounce (Actually, Much Less) of Prevention. N. Engl. J. Med. 2020;383(27):2677–2678. doi: 10.1056/NEJMe2034717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciceri F., Castagna A., Rovere-Querini P., De Cobelli F., Ruggeri A., Galli L., Conte C., De Lorenzo R., Poli A., Ambrosio A., Signorelli C., Bossi E., Fazio M., Tresoldi C., Colombo S., Monti G., Fominskiy E., Franchini S., Spessot M., Martinenghi C., Carlucci M., Beretta L., Scandroglio A.M., Clementi M., Locatelli M., Tresoldi M., Scarpellini P., Martino G., Bosi E., Dagna L., Lazzarin A., Landoni G., Zangrillo A. Early predictors of clinical outcomes of COVID-19 outbreak in Milan, Italy. Clin. Immunol. 2020;217 doi: 10.1016/j.clim.2020.108509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y., Wei X., Guan J., Qin S., Wang Z., Lu H., Qian J., Wu L., Chen Y., Chen Y., Lin X. COVID-19 pneumonia: CD8(+) T and NK cells are decreased in number but compensatory increased in cytotoxic potential. Clin. Immunol. 2020;218:108516. doi: 10.1016/j.clim.2020.108516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urra J.M., Cabrera C.M., Porras L., Rodenas I. Selective CD8 cell reduction by SARS-CoV-2 is associated with a worse prognosis and systemic inflammation in COVID-19 patients. Clin. Immunol. 2020;217:108486. doi: 10.1016/j.clim.2020.108486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Rial J., Martinon-Torres F. A strategy targeting monocyte-macrophage differentiation to avoid pulmonary complications in SARS-Cov2 infection. Clin. Immunol. 2020;216:108442. doi: 10.1016/j.clim.2020.108442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vojdani A., Kharrazian D. Potential antigenic cross-reactivity between SARS-CoV-2 and human tissue with a possible link to an increase in autoimmune diseases. Clin. Immunol. 2020;217:108480. doi: 10.1016/j.clim.2020.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J., Xia P., Zhou Y., Liu Z., Zhou X., Wang J., Li T., Yan X., Chen L., Zhang S., Qin Y., Li X. Potential effect of blood purification therapy in reducing cytokine storm as a late complication of critically ill COVID-19. Clin. Immunol. 2020;214:108408. doi: 10.1016/j.clim.2020.108408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vuitton D.A., Vuitton L., Seilles E., Galanaud P. A plea for the pathogenic role of immune complexes in severe Covid-19. Clin. Immunol. 2020;217:108493. doi: 10.1016/j.clim.2020.108493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roncati L., Ligabue G., Fabbiani L., Malagoli C., Gallo G., Lusenti B., Nasillo V., Manenti A., Maiorana A. Type 3 hypersensitivity in COVID-19 vasculitis. Clin. Immunol. 2020;217:108487. doi: 10.1016/j.clim.2020.108487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yager E.J. Antibody-dependent enhancement and COVID-19: moving toward acquittal. Clin. Immunol. 2020;217:108496. doi: 10.1016/j.clim.2020.108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryabkova V.A., Churilov L.P., Shoenfeld Y. Influenza infection, SARS, MERS and COVID-19: cytokine storm - the common denominator and the lessons to be learned. Clin. Immunol. 2021;223:108652. doi: 10.1016/j.clim.2020.108652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dauletova M., Hafsan H., Mahhengam N., Zekiy A.O., Ahmadi M., Siahmansouri H. Mesenchymal stem cell alongside exosomes as a novel cell-based therapy for COVID-19: a review study. Clin. Immunol. 2021;226:108712. doi: 10.1016/j.clim.2021.108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavriilaki E., Brodsky R.A. Severe COVID-19 infection and thrombotic microangiopathy: success does not come easily. Br. J. Haematol. 2020;189:e227–e230. doi: 10.1111/bjh.16783. [DOI] [PubMed] [Google Scholar]
- 15.Gavriilaki E., Anyfanti P., Gavriilaki M., Lazaridis A., Douma S., Gkaliagkousi E. Endothelial dysfunction in COVID-19: lessons learned from coronaviruses. Curr. Hypertens. Rep. 2020;22:63. doi: 10.1007/s11906-020-01078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gralinski L.E., Sheahan T.P., Morrison T.E., Menachery V.D., Jensen K., Leist S.R., Whitmore A., Heise M.T., Baric R.S. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9 doi: 10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y., Zhao G., Song N., Li P., Chen Y., Guo Y., Li J., Du L., Jiang S., Guo R., Sun S., Zhou Y. Blockade of the C5a-C5aR axis alleviates lung damage in hDPP4-transgenic mice infected with MERS-CoV. Emerg. Microbes Infect. 2018;7:77. doi: 10.1038/s41426-018-0063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mastaglio S., Ruggeri A., Risitano A.M., Angelillo P., Yancopoulou D., Mastellos D.C., Huber-Lang M., Piemontese S., Assanelli A., Garlanda C., Lambris J.D., Ciceri F. The first case of COVID-19 treated with the complement C3 inhibitor AMY-101. Clin. Immunol. 2020;108450 doi: 10.1016/j.clim.2020.108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mastellos D.C., Pires da Silva B.G.P., Fonseca B.A.L., Fonseca N.P., Auxiliadora-Martins M., Mastaglio S., Ruggeri A., Sironi M., Radermacher P., Chrysanthopoulou A., Skendros P., Ritis K., Manfra I., Iacobelli S., Huber-Lang M., Nilsson B., Yancopoulou D., Connolly E.S., Garlanda C., Ciceri F., Risitano A.M., Calado R.T., Lambris J.D. Complement C3 vs C5 inhibition in severe COVID-19: early clinical findings reveal differential biological efficacy. Clin. Immunol. 2020;220:108598. doi: 10.1016/j.clim.2020.108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao T., Hu M., Zhang X., Li H., Zhu L., Liu H., Dong Q., Zhang Z., Wang Z., Hu Y., Fu Y., Jin Y., Li K., Zhao S., Xiao Y., Luo S., Li L., Zhao L., Liu J., Zhao H., Liu Y., Yang W., Peng J., Chen X., Li P., Liu Y., Xie Y., Song J., Zhang L., Ma Q., Bian X., Chen W., Liu X., Mao Q., Cao C. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv. 2020 2020.2003.2029.20041962 (preprint) [Google Scholar]
- 21.Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J., Yuan X., Chen H., Chaturvedi S., Braunstein E.M., Brodsky R.A. Direct activation of the alternative complement pathway by SARS-CoV-2 spike proteins is blocked by factor D inhibition. Blood. 2020;136(18):2080–2089. doi: 10.1182/blood.2020008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skendros P., Mitsios A., Chrysanthopoulou A., Mastellos D.C., Metallidis S., Rafailidis P., Ntinopoulou M., Sertaridou E., Tsironidou V., Tsigalou C., Tektonidou M., Konstantinidis T., Papagoras C., Mitroulis I., Germanidis G., Lambris J.D., Ritis K. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 2020;130:6151–6157. doi: 10.1172/JCI141374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holter J.C., Pischke S.E., de Boer E., Lind A., Jenum S., Holten A.R., Tonby K., Barratt-Due A., Sokolova M., Schjalm C., Chaban V., Kolderup A., Tran T., Gjolberg T. Tollefsrud, Skeie L.G., Hesstvedt L., Ormasen V., Fevang B., Austad C., Muller K.E., Fladeby C., Holberg-Petersen M., Halvorsen B., Muller F., Aukrust P., Dudman S., Ueland T., Andersen J.T., Lund-Johansen F., Heggelund L., Dyrhol-Riise A.M., Mollnes T.E. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc. Natl. Acad. Sci. U. S. A. 2020;117:25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busch M.H., Timmermans S., Nagy M., Visser M., Huckriede J., Aendekerk J.P., de Vries F., Potjewijd J., Jallah B., Ysermans R., Oude Lashof A.M.L., Breedveld P.H., van de Poll M.C.G., van de Horst I.C.C., van Bussel B.C.T., Theunissen R., Spronk H.M.H., Damoiseaux J., Ten Cate H., Nicolaes G.A.F., Reutelingsperger C.P., van Paassen P. Neutrophils and contact activation of coagulation as potential drivers of COVID-19. Circulation. 2020;142:1787–1790. doi: 10.1161/CIRCULATIONAHA.120.050656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satyam A., Tsokos M.G., Brook O.R., Hecht J.L., Moulton V.R., Tsokos G.C. Activation of classical and alternative complement pathways in the pathogenesis of lung injury in COVID-19. Clin. Immunol. 2021;226:108716. doi: 10.1016/j.clim.2021.108716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavriilaki E., Brodsky R.A. Complementopathies and precision medicine. J. Clin. Invest. 2020;130:2152–2163. doi: 10.1172/JCI136094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gavriilaki E., Yuan X., Ye Z., Ambinder A.J., Shanbhag S.P., Streiff M.B., Kickler T.S., Moliterno A.R., Sperati C.J., Brodsky R.A. Modified ham test for atypical hemolytic uremic syndrome. Blood. 2015;125:3637–3646. doi: 10.1182/blood-2015-02-629683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavriilaki E., Anagnostopoulos A., Mastellos D.C. Complement in thrombotic microangiopathies: unraveling Ariadne’s thread into the labyrinth of complement therapeutics. Front. Immunol. 2019;10:337. doi: 10.3389/fimmu.2019.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaturvedi S., Braunstein E.M., Yuan X., Yu J., Alexander A., Chen H., Gavriilaki E., Alluri R., Streiff M.B., Petri M., Crowther M.A., McCrae K.R., Brodsky R.A. Complement activity and complement regulatory gene mutations are associated with thrombosis in APS and CAPS. Blood. 2020;135:239–251. doi: 10.1182/blood.2019003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jian C., Xiao J., Gong L., Skipwith C.G., Jin S.Y., Kwaan H.C., Zheng X.L. Gain-of-function ADAMTS13 variants that are resistant to autoantibodies against ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. Blood. 2012;119:3836–3843. doi: 10.1182/blood-2011-12-399501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satyam A., Tsokos G.C. Curb complement to cure COVID-19. Clin. Immunol. 2020;221:108603. doi: 10.1016/j.clim.2020.108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shim K., Begum R., Yang C., Wang H. Complement activation in obesity, insulin resistance, and type 2 diabetes mellitus. World J. Diabetes. 2020;11:1–12. doi: 10.4239/wjd.v11.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osborne A.J., Breno M., Borsa N.G., Bu F., Fremeaux-Bacchi V., Gale D.P., van den Heuvel L.P., Kavanagh D., Noris M., Pinto S., Rallapalli P.M., Remuzzi G., Rodriguez de Cordoba S., Ruiz A., Smith R.J.H., Vieira-Martins P., Volokhina E., Wilson V., Goodship T.H.J., Perkins S.J. Statistical validation of rare complement variants provides insights into the molecular basis of atypical hemolytic uremic syndrome and C3 glomerulopathy. J. Immunol. 2018;200:2464–2478. doi: 10.4049/jimmunol.1701695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez E., Rallapalli P.M., Osborne A.J., Perkins S.J. New functional and structural insights from updated mutational databases for complement factor H, Factor I, membrane cofactor protein and C3. Biosci. Rep. 2014;34 doi: 10.1042/BSR20140117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders R.E., Abarrategui-Garrido C., Fremeaux-Bacchi V., Goicoechea de Jorge E., Goodship T.H., Lopez Trascasa M., Noris M., Ponce Castro I.M., Remuzzi G., Rodriguez de Cordoba S., Sanchez-Corral P., Skerka C., Zipfel P.F., Perkins S.J. The interactive Factor H-atypical hemolytic uremic syndrome mutation database and website: update and integration of membrane cofactor protein and Factor I mutations with structural models. Hum. Mutat. 2007;28:222–234. doi: 10.1002/humu.20435. [DOI] [PubMed] [Google Scholar]
- 37.Saunders R.E., Goodship T.H., Zipfel P.F., Perkins S.J. An interactive web database of factor H-associated hemolytic uremic syndrome mutations: insights into the structural consequences of disease-associated mutations. Hum. Mutat. 2006;27:21–30. doi: 10.1002/humu.20268. [DOI] [PubMed] [Google Scholar]
- 38.Kaxiras E., Neofotistos G. Multiple epidemic wave model of the COVID-19 pandemic: modeling study. J. Med. Internet Res. 2020;22 doi: 10.2196/20912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahimi I., Gandomi A.H., Asteris P.G., Chen F. Analysis and prediction of COVID-19 using SIR, SEIQR, and machine learning models: Australia, Italy, and UK cases. Information. 2021;12:109. [Google Scholar]
- 40.Asteris P.-G., Douvika M.-G., Karamani C.-A., Skentou A.-D., Chlichlia K., Cavaleri L., Daras T., Armaghani D.-J., Zaoutis T.-E. A novel heuristic algorithm for the modeling and risk assessment of the COVID-19 pandemic phenomenon. Comput. Model. Eng. Sci. 2020;125:815–828. [Google Scholar]
- 41.Peckham H., de Gruijter N.M., Raine C., Radziszewska A., Ciurtin C., Wedderburn L.R., Rosser E.C., Webb K., Deakin C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020;11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kokame K., Matsumoto M., Soejima K., Yagi H., Ishizashi H., Funato M., Tamai H., Konno M., Kamide K., Kawano Y., Miyata T., Fujimura Y. Mutations and common polymorphisms in ADAMTS13 gene responsible for von Willebrand factor-cleaving protease activity. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11902–11907. doi: 10.1073/pnas.172277399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J., Li S., Hu S., Yu J., Xiang Y. Association between genetic variation of complement C3 and the susceptibility to advanced age-related macular degeneration: a meta-analysis. BMC Ophthalmol. 2018;18:274. doi: 10.1186/s12886-018-0945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.G.S. Hageman, D.H. Anderson, L.V. Johnson, L.S. Hancox, A.J. Taiber, L.I. Hardisty, J.L. Hageman, H.A. Stockman, J.D. Borchardt, K.M. Gehrs, R.J. Smith, G. Silvestri, S.R. Russell, C.C. Klaver, I. Barbazetto, S. Chang, L.A. Yannuzzi, G.R. Barile, J.C. Merriam, R.T. Smith, A.K. Olsh, J. Bergeron, J. Zernant, J.E. Merriam, B. Gold, M. Dean, R. Allikmets, A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration, Proc. Natl. Acad. Sci. U. S. A., 102 (2005) 7227–7232. [DOI] [PMC free article] [PubMed]
- 45.Nilsson S.C., Karpman D., Vaziri-Sani F., Kristoffersson A.C., Salomon R., Provot F., Fremeaux-Bacchi V., Trouw L.A., Blom A.M. A mutation in factor I that is associated with atypical hemolytic uremic syndrome does not affect the function of factor I in complement regulation. Mol. Immunol. 2007;44:1835–1844. doi: 10.1016/j.molimm.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Wang X., Zhang Y., Zhang M.N. Complement factor B polymorphism (rs641153) and susceptibility to age-related macular degeneration: evidence from published studies. Int. J. Ophthalmol. 2013;6:861–867. doi: 10.3980/j.issn.2222-3959.2013.06.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han Y., Jiang M., Xia D., He L., Lv X., Liao X., Meng J. COVID-19 in a patient with long-term use of glucocorticoids: a study of a familial cluster. Clin. Immunol. 2020;214:108413. doi: 10.1016/j.clim.2020.108413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernandez J., Prati D., Baselli G., Asselta R., Grimsrud M.M., Milani C., Aziz F., Kassens J., May S., Wendorff M., Wienbrandt L., Uellendahl-Werth F., Zheng T., Yi X., de Pablo R., Chercoles A.G., Palom A., Garcia-Fernandez A.E., Rodriguez-Frias F., Zanella A., Bandera A., Protti A., Aghemo A., Lleo A., Biondi A., Caballero-Garralda A., Gori A., Tanck A., Nolla A. Carreras, Latiano A., Fracanzani A.L., Peschuck A., Julia A., Pesenti A., Voza A., Jimenez D., Mateos B., Jimenez B. Nafria, Quereda C., Paccapelo C., Gassner C., Angelini C., Cea C., Solier A., Pestana D., Muniz-Diaz E., Sandoval E., Paraboschi E.M., Navas E., Sanchez F. Garcia, Ceriotti F., Martinelli-Boneschi F., Peyvandi F., Blasi F., Tellez L., Blanco-Grau A., Hemmrich-Stanisak G., Grasselli G., Costantino G., Cardamone G., Foti G., Aneli S., Kurihara H., ElAbd H., My I., Galvan-Femenia I., Martin J., Erdmann J., Ferrusquia-Acosta J., Garcia-Etxebarria K., Izquierdo-Sanchez L., Bettini L.R., Sumoy L., Terranova L., Moreira L., Santoro L., Scudeller L., Mesonero F., Roade L., Ruhlemann M.C., Schaefer M., Carrabba M., Riveiro-Barciela M., Basso M.E. Figuera, Valsecchi M.G., Hernandez-Tejero M., Acosta-Herrera M., D’Angio M., Baldini M., Cazzaniga M., Schulzky M., Cecconi M., Wittig M., Ciccarelli M., Rodriguez-Gandia M., Bocciolone M., Miozzo M., Montano N., Braun N., Sacchi N., Martinez N., Ozer O., Palmieri O., Faverio P., Preatoni P., Bonfanti P., Omodei P., Tentorio P., Castro P., Rodrigues P.M., Ortiz A. Blandino, de Cid R., Ferrer R., Gualtierotti R., Nieto R., Goerg S., Badalamenti S., Marsal S., Matullo G., Pelusi S., Juzenas S., Aliberti S., Monzani V., Moreno V., Wesse T., Lenz T.L., Pumarola T., Rimoldi V., Bosari S., Albrecht W., Peter W., Romero-Gomez M., D’Amato M., Duga S., Banales J.M., Hov J.R., Folseraas T., Valenti L., Franke A., Karlsen T.H., G.G. Severe Covid Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schluter A., Le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Al Turki S., Hasanato R., van de Beek D., Biondi A., Bettini L.R., D’Angio M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.E., Keles S., Colkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela S.N., Gallego C.R., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouenan C., Clinicians C.-S., Clinicians C., Imagine C.G., French C.C.S.G., Co V.C.C., Amsterdam U.M.C.C., Biobank C.H.G. Effort, Niaid U., T.C.I. Group, Snow A.L., Dalgard C.L., Milner J., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., Garcia-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R., Zhang S.Y., Gorochov G., Beziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.L. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawalha A.H., Zhao M., Coit P., Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020;215:108410. doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., Furniss J., Richmond A., Gountouna E., Wrobel N., Harrison D., Wang B., Wu Y., Meynert A., Griffiths F., Oosthuyzen W., Kousathanas A., Moutsianas L., Yang Z., Zhai R., Zheng C., Grimes G., Beale R., Millar J., Shih B., Keating S., Zechner M., Haley C., Porteous D.J., Hayward C., Yang J., Knight J., Summers C., Shankar-Hari M., Klenerman P., Turtle L., Ho A., Moore S.C., Hinds C., Horby P., Nichol A., Maslove D., Ling L., McAuley D., Montgomery H., Walsh T., Pereira A., Renieri A., Gen O.I., I. Investigators, C.-H.G. Initiative, I and Me, B. Investigators, Gen C.I., Shen X., Ponting C.P., Fawkes A., Tenesa A., Caulfield M., Scott R., Rowan K., Murphy L., Openshaw P.J.M., Semple M.G., Law A., Vitart V., Wilson J.F., Baillie J.K. Genetic mechanisms of critical illness in Covid-19. Nature. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 53.Stone J.H., Frigault M.J., Serling-Boyd N.J., Fernandes A.D., Harvey L., Foulkes A.S., Horick N.K., Healy B.C., Shah R., Bensaci A.M., Woolley A.E., Nikiforow S., Lin N., Sagar M., Schrager H., Huckins D.S., Axelrod M., Pincus M.D., Fleisher J., Sacks C.A., Dougan M., North C.M., Halvorsen Y.D., Thurber T.K., Dagher Z., Scherer A., Wallwork R.S., Kim A.Y., Schoenfeld S., Sen P., Neilan T.G., Perugino C.A., Unizony S.H., Collier D.S., Matza M.A., Yinh J.M., Bowman K.A., Meyerowitz E., Zafar A., Drobni Z.D., Bolster M.B., Kohler M., D'Silva K.M., Dau J., Lockwood M.M., Cubbison C., Weber B.N., Mansour M.K., B.B.T.T. Investigators Efficacy of tocilizumab in patients hospitalized with Covid-19. N. Engl. J. Med. 2020;383:2333–2344. doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan N.A. Anakinra for severe forms of COVID-19. Lancet Rheumatol. 2020;2:e586–e587. doi: 10.1016/S2665-9913(20)30273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., May B., De Freitas J.K., Glicksberg B.S., Mason C.E., Tatonetti N.P., Shapira S.D. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat. Med. 2020;26:1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gebhard C., Regitz-Zagrosek V., Neuhauser H.K., Morgan R., Klein S.L. Impact of sex and gender on COVID-19 outcomes in Europe. Biol. Sex Differ. 2020;11:29. doi: 10.1186/s13293-020-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kopel J., Perisetti A., Roghani A., Aziz M., Gajendran M., Goyal H. Racial and gender-based differences in COVID-19. Front. Public Health. 2020;8:418. doi: 10.3389/fpubh.2020.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaya da Costa M., Poppelaars F., van Kooten C., Mollnes T.E., Tedesco F., Wurzner R., Trouw L.A., Truedsson L., Daha M.R., Roos A., Seelen M.A. Age and sex-associated changes of complement activity and complement levels in a healthy Caucasian population. Front. Immunol. 2018;9:2664. doi: 10.3389/fimmu.2018.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng L., Zhang D., Cao W., Song W.C., Zheng X.L. Synergistic effects of ADAMTS13 deficiency and complement activation in pathogenesis of thrombotic microangiopathy. Blood. 2019;134:1095–1105. doi: 10.1182/blood.2019001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fidalgo T., Martinho P., Pinto C.S., Oliveira A.C., Salvado R., Borras N., Coucelo M., Manco L., Maia T., Mendes M.J., Del Orbe Barreto R., Corrales I., Vidal F., Ribeiro M.L. Combined study of ADAMTS13 and complement genes in the diagnosis of thrombotic microangiopathies using next-generation sequencing. Res. Practi. Thromb. Haemostas. 2017;1:69–80. doi: 10.1002/rth2.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gavriilaki E., Touloumenidou T., Sakellari I., Batsis I., Mallouri D., Psomopoulos F., Tsagiopoulou M., Koutra M., Yannaki E., Papalexandri A., Taylor P., Nikolousis E., Stamouli M., Holbro A., Baltadakis I., Liga M., Spyridonidis A., Tsirigotis P., Charchalakis N., Tsakiris D.A., Brodsky R.A., Passweg J., Stamatopoulos K., Anagnostopoulos A. Pretransplant genetic susceptibility: clinical relevance in transplant-associated thrombotic microangiopathy. Thromb. Haemostas. 2020;120:638–646. doi: 10.1055/s-0040-1702225. [DOI] [PubMed] [Google Scholar]
- 62.Weitz I.C. Complement the hemostatic system: an intimate relationship. Thromb. Res. 2014;133(Suppl. 2):S117–S121. doi: 10.1016/S0049-3848(14)50020-5. [DOI] [PubMed] [Google Scholar]
- 63.Delvaeye M., Noris M., De Vriese A., Esmon C.T., Esmon N.L., Ferrell G., Del-Favero J., Plaisance S., Claes B., Lambrechts D., Zoja C., Remuzzi G., Conway E.M. Thrombomodulin mutations in atypical hemolytic-uremic syndrome. N. Engl. J. Med. 2009;361:345–357. doi: 10.1056/NEJMoa0810739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sapru A., Liu K.D., Wiemels J., Hansen H., Pawlikowska L., Poon A., Jorgenson E., Witte J.S., Calfee C.S., Ware L.B., Matthay M.A., Network N.A. Association of common genetic variation in the protein C pathway genes with clinical outcomes in acute respiratory distress syndrome. Crit. Care. 2016;20:151. doi: 10.1186/s13054-016-1330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rachakonda S.P., Dai H., Penack O., Blau O., Blau I.W., Radujkovic A., Muller-Tidow C., Kumar R., Dreger P., Luft T. Single nucleotide polymorphisms in CD40L predict endothelial complications and mortality after allogeneic stem-cell transplantation. J. Clin. Oncol. 2018;36:789–800. doi: 10.1200/JCO.2017.76.4662. [DOI] [PubMed] [Google Scholar]
- 66.Rachakonda S.P., Penack O., Dietrich S., Blau O., Blau I.W., Radujkovic A., Isermann B., Ho A.D., Uharek L., Dreger P., Kumar R., Luft T. Single-nucleotide polymorphisms within the thrombomodulin gene (THBD) predict mortality in patients with graft-versus-host disease. J. Clin. Oncol. 2014;32:3421–3427. doi: 10.1200/JCO.2013.54.4056. [DOI] [PubMed] [Google Scholar]
- 67.Diurno F., Numis F.G., Porta G., Cirillo F., Maddaluno S., Ragozzino A., De Negri P., Di Gennaro C., Pagano A., Allegorico E., Bressy L., Bosso G., Ferrara A., Serra C., Montisci A., D'Amico M., Morello S. Schiano Lo, Di Costanzo G., Tucci A.G., Marchetti P., Di Vincenzo U., Sorrentino I., Casciotta A., Fusco M., Buonerba C., Berretta M., Ceccarelli M., Nunnari G., Diessa Y., Cicala S., Facchini G. Eculizumab treatment in patients with COVID-19: preliminary results from real life ASL Napoli 2 Nord experience. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4040–4047. doi: 10.26355/eurrev_202004_20875. [DOI] [PubMed] [Google Scholar]
- 68.Laurence J., Mulvey J.J., Seshadri M., Racanelli A., Harp J., Schenck E.J., Zappetti D., Horn E.M., Magro C.M. Anti-complement C5 therapy with eculizumab in three cases of critical COVID-19. Clin. Immunol. 2020;219:108555. doi: 10.1016/j.clim.2020.108555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahajan R., Lipton M., Broglie L., Jain N.G., Uy N.S. Eculizumab treatment for renal failure in a pediatric patient with COVID-19. J. Nephrol. 2020;33(6):1373–1376. doi: 10.1007/s40620-020-00858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rambaldi A., Gritti G., Mico M.C., Frigeni M., Borleri G., Salvi A., Landi F., Pavoni C., Sonzogni A., Gianatti A., Binda F., Fagiuoli S., Di Marco F., Lorini L., Remuzzi G., Whitaker S., Demopulos G. Endothelial injury and thrombotic microangiopathy in COVID-19: treatment with the lectin-pathway inhibitor narsoplimab. Immunobiology. 2020;152001 doi: 10.1016/j.imbio.2020.152001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan X., Yu J., Gerber G., Chaturvedi S., Cole M., Chen H., Metjian A., Sperati C.J., Braunstein E.M., Brodsky R.A. Ex vivo assays to detect complement activation in complementopathies. Clin. Immunol. 2020;221:108616. doi: 10.1016/j.clim.2020.108616. [DOI] [PMC free article] [PubMed] [Google Scholar]