Abstract
Thermophilic Campylobacter species are among the major etiologies of bacterial enteritis globally. This study aimed at assessing the antimicrobial resistance (AMR) profiles, virulence genes, and genetic diversity of thermophilic Campylobacter species isolated from a layer poultry farm in South Korea. One hundred fifty-three chicken feces were collected from two layer poultry farms in Gangneung, South Korea. The Campylobacter species were isolated by cultural techniques, while PCR and sequencing were used for species confirmation. Antimicrobial susceptibility testing for six antimicrobials [ciprofloxacin (CIP), nalidixic acid (NAL), sitafloxacin (SIT), erythromycin (ERY), tetracycline (TET), and gentamicin (GEN)] was carried out by broth microdilution. Three AMR and nine virulence genes were screened by PCR. Genotyping was performed by flaA-restriction fragment length polymorphism (RFLP) and multilocus sequence typing (MLST). Of the 153 samples, Campylobacter spp. were detected in 55 (35.9%), with Campylobacter jejuni and Campylobacter coli being 49 (89.1%) and six (10.9%), respectively. High-level resistance was observed for CIP (100%), NAL (100%), and TET (C. jejuni, 93.9%; C. coli: 83.3%). No resistance was observed for SIT. The missense mutation (C257T) in gyrA gene was confirmed by sequencing, while the tet(O) gene was similar to known sequences in GenBank. The rate of multidrug-resistant (MDR) strains was 8.2%, and they all belonged to C. jejuni. All Campylobacter isolates possessed five virulence genes (cdtB, cstII, flaA, cadF, and dnaJ), but none possessed ggt, while the rates for other genes (csrA, ciaB, and pldA) ranged between 33.3 and 95.9%. The flaA-RFLP yielded 26 flaA types (C. jejuni: 21 and C. coli: five), while the MLST showed 10 sequence types (STs) for C. jejuni and three STs for C. coli, with CC-607 (STs 3611) and CC-460 (ST-460) being predominant. Among the 10 STs of C. jejuni, three were newly assigned. The findings of this study highlight the increased resistance to quinolones and TET, the virulence potential, and the diverse genotypes among Campylobacter strains isolated from the layer poultry farm.
Keywords: Campylobacter, quinolones, antimicrobial resistance, flaA RFLP, multilocus sequence typing, poultry, Korea
Introduction
Globally, Campylobacter is the leading cause of bacterial gastroenteritis (Alaboudi et al., 2020). Campylobacter jejuni and Campylobacter coli are the species of clinical significance, being accountable for more than 95% of human campylobacteriosis (Moore et al., 2005; Fitzgerald, 2015). Globally, 96 million cases of diarrhea each year are due to Campylobacter (Havelaar et al., 2015). In Europe, Campylobacter was ranked as the second (next to Salmonella) etiological agent of outbreaks associated with water and food poisoning in 2018 (Klančnik et al., 2020). Contrary to European countries, reports on human campylobacteriosis in Asian countries including South Korea are limited, possibly due to low disease prevalence or the sporadic nature of infections (Kim et al., 2019; Wei et al., 2019).
Human campylobacteriosis requires antimicrobial therapy only in case of complications and in immuno-compromised people (Wieczorek and Osek, 2013; Sproston et al., 2018). Over the years, increasing rates of Campylobacter strains that are resistant to the drugs of choice [fluoroquinolones (FQs) and macrolides] and alternative therapies [gentamicin (GEN) and tetracycline (TET)] have been reported (Blaser and Engberg, 2008; Koolman et al., 2015), making antimicrobial resistant (AMR) Campylobacter strains a public health concern (Mourkas et al., 2019; Windiasti et al., 2019). The resistance to antimicrobials is partly due to their misuse in both human medicine and livestock production (Elisha et al., 2017; Sproston et al., 2018). For instance, different quinolone antibiotics have been extensively used in poultry raising, which led to an accelerated number of quinolone-resistant strains of Campylobacter from chicken and humans (Sproston et al., 2018).
In Korea, FQ use in livestock was banned since 2010, with a prediction to curb the increased resistance in the future (Ku et al., 2011). However, recently, FQ-resistant Campylobacter strains have been isolated from poultry and duck meat (Kim et al., 2019). Due to the increased resistance to quinolones throughout the world (Tang et al., 2017), erythromycin (ERY) has emerged as the recommended drug for treating human campylobacteriosis (Giannatale et al., 2019). Recently, sitafloxacin (SIT), a novel FQ drug, proved to be effective against various FQ-resistant pathogens including Campylobacter (Changkwanyeun et al., 2016; Chen et al., 2020), and could be a promising drug. The persistence of FQ resistance in Campylobacter strains could be linked to the continued use of ciprofloxacin (CIP) in human medicine, international trade, travel, and use of FQs in animal husbandry along with the circulation of resistant isolates among different reservoirs.
Poultry can carry Campylobacter, and chicken intestines are regarded as reservoirs for thermophilic Campylobacter species based on optimal conditions (high body temperature) favoring their growth (Sibanda et al., 2018). Previous reports have associated an increase in human campylobacteriosis cases with the increase in chicken consumption (Oh et al., 2017). Chickens’ ceca alone are usually colonized by C. jejuni to levels above 109 colony-forming unit (CFU)/ml, posing a risk to humans (Humphrey et al., 2007, 2014). Furthermore, Campylobacter can stay in feces and litter for many days, and the use of these byproducts as fertilizers would aggravate the dissemination of the pathogens (Kassem et al., 2016). Campylobacter persistence in chicken farms and slaughterhouses is a hazard to the consumers because it is transmitted along the whole production chain up to the final product (Kim et al., 2019; Ramires et al., 2020). While the literature on broiler chicken is extensive, studies on the epidemiology of Campylobacter species from layer farms are limited (Kassem et al., 2016).
Campylobacter species are equipped with virulome which is used in attachment, establishment, invasion, and production of toxins, contributing to their increased occurrence and epidemiology compared to other enteric bacteria (Bolton, 2015; Otigbu et al., 2018). However, the mechanisms associated with Campylobacter pathogenicity are not fully understood (Nguyen et al., 2016). C. jejuni is known to cause Guillain–Barré syndrome, characterized by acute and progressive neuromuscular paralysis, mediated by sialyltransferases (cstII) (Koga et al., 2006; Humphrey et al., 2007; González-Hein et al., 2013). Sialic acid confers immune avoidance to C. jejuni, as a mutant lacking lipooligosaccharide sialic acid residues showed greater immunoreactivity and decreased serum resistance (Kreling et al., 2020). The CDT complex, another important factor in Campylobacter, codes for the cytolethal distending toxin with cdtB acting as the catalytic site, and in the nucleus, cdtB induces cell cycle arrest and leads to apoptosis of both immune and epithelial cells in the intestines (Jain et al., 2008). It has been reported that C. jejuni cdtB mutants had reduced extra-intestinal invasiveness (Yamasaki et al., 2006) and bowel disturbances (Pokkunuri et al., 2012). A study carried out in Poland showed that Campylobacter strains lacking cdtB and cdtC were non-cytotoxic, confirming their roles in toxin production (Wysok et al., 2020). The presence of ggt contributes to the colonization potential of C. jejuni in chicken and mice intestines (Barnes et al., 2007). The flaA gene contributes to Campylobacter pathogenesis as it is involved in colonization, motility, auto-agglutination, and biofilm formation (Guerry, 2007). Mutation experiments highlighted the role of flaA in chicken colonization (Bolton, 2015). Campylobacter species also possess other genes associated with adhesion (cadF and pldA), invasion (ciaB), thermo-tolerance (dnaJ) (Pillay et al., 2020), and stress response (csrA) (Fields and Thompson, 2008). Studies have shown that Campylobacter strain mutants for cadF and ciaB exhibited a reduced attachment and invasion of INT 407 cell line along with a decline of survival potential (Kreling et al., 2020; Ramires et al., 2020).
Molecular typing methods are important not only in distinguishing bacteria at the species and subspecies levels but also in source attribution of Campylobacter strains (Eberle and Kiess, 2012; Lydekaitiene and Kudirkiene, 2020). Although source attribution aiming at quantifying the contribution of different reservoirs, pathways, exposures, and risk factors to the burden of human illness is difficult (Wagenaar et al., 2013), it is estimated that 80% of human campylobacteriosis are attributed to Campylobacter of poultry origin (Wagenaar et al., 2013; Mulder et al., 2020). Multilocus sequence typing (MLST), based on seven housekeeping genes (HKGs), is the gold-standard method used for epidemiological surveillance (Harrington et al., 2003; Lydekaitiene and Kudirkiene, 2020). Data on MLST of Campylobacter strains in Asia are limited (Nguyen et al., 2016), but previous studies in Korea and Japan have shown the predominance of CC-460, CC-607, CC-21, and CC-45 in poultry and human isolates (Wei et al., 2014; Ozawa et al., 2016; Oh et al., 2017). MLST data are expected to give accurate phylogenic estimation, typing, and strain relatedness (Alaboudi et al., 2020). Whole-genome sequencing (WGS) might become the preferred typing method in the future, but still, there is a need for a consensus upon bioinformatics pipelines and tools for processing WGS data (Duarte et al., 2016b).
Considering the persistence of FQ-resistant Campylobacter strains even in the absence of antimicrobial use and the fact that SIT has a different structure compared to other FQs, we hypothesized that Campylobacter species from chicken are still resistant to both ciprofloxacin and nalidixic acid (NAL) but sensitive to SIT. Furthermore, we think that the same sequence types (STs) are circulating in poultry in Korea and the region. Based on the virulence potential of Campylobacter and the favorable environment offered by chicken, it is most probable that Campylobacter species of poultry origin are hypervirulent and could be of concern. The present study aimed at assessing the antimicrobial resistance profiles, virulence genes, and genetic diversity of thermophilic Campylobacter species isolated from a layer poultry farm in Korea.
Materials and Methods
Sample Collection
Fresh chicken fecal samples were purposively collected from two layer poultry farms located in Gangneung city, Republic of Korea in June 2020. The first farm uses an intensive poultry farming system with around 800 1-year-old chickens dispatched into battery cages inside a closed house. The second farm is a small one that is not for commercial purposes, where around 30 1-year-old chickens are enclosed in a cage subdivided into two blocks by a fence. A total of 133 (from the first farm) and 20 (from the second farm) pen floor fecal samples were collected using sterile swabs and transported to the laboratory under refrigeration (ice) within 1 h.
Campylobacter Isolation and Antimicrobial Susceptibility Testing
Upon arrival at the laboratory, the feces were inoculated onto modified charcoal cefoperazone deoxycholate agar (mCCDA) (Oxoid Ltd., Basingstoke, Hampshire, England) containing the Campylobacter mCCDA-selective supplement, SR155E (Oxoid Ltd, Basingstoke, Hampshire, England). Incubation was done as previously described (Kurekci et al., 2012) at 37°C for 48 h under microaerophilic conditions generated by CampyGenTM gas sachets (Oxoid, Basingstoke, England, United Kingdom). Typical colonies of Campylobacter, which are grayish, flat, moistened, and with a tendency to spread (Al-Edany et al., 2015), were sub-cultured on Mueller Hinton Agar supplemented with 5% defibrinated horse blood (MHS) and incubated at 37°C for 48 h under microaerophilic conditions (Kurekci et al., 2012). Campylobacter isolates were preserved at −80°C in Mueller Hinton broth containing 25% glycerol (v/v).
Antimicrobial susceptibility testing was performed by broth microdilution. The isolates were tested against quinolones, namely ciprofloxacin (CIP), NAL (0.25–512 μg/ml), SIT (0.03–16 μg/ml); macrolide, ERY (0.06–64 μg/ml); and aminoglycoside, GEN (0.06–64 μg/ml), and TET (0.125–1,024 μg/ml). Apart from SIT purchased from AdooQ BioScience (Irvine, CA, United States), the other antimicrobials were supplied by Sigma-Aldrich (St. Louis, MO, United States). CIP and ERY were dissolved in 0.1 N HCl and 70% ethanol, respectively. GEN and TET were dissolved in water, while NAL and SIT were dissolved in dimethyl sulfoxide (DMSO). Except for the antibiotics dissolved in DMSO, other solutions of antibiotics were filter-sterilized before being used.
Preserved Campylobacter isolates were inoculated onto MHS (Oxoid Ltd, Basingstoke, Hampshire, England) and incubated at 37°C for 48 h under microaerophilic conditions (Kurekci et al., 2012). A sub-culture was performed on the same media and the same conditions to get well-grown pure colonies free from glycerol. For antimicrobial susceptibility assays, suspensions corresponding to 0.5 McFarland standard (1.5 × 108 CFU/ml) were prepared using normal saline, and the final concentration in a 96-well plate was 2–5 × 106 CFU/ml. The minimal inhibitory concentration (MIC) was determined by checking the absorbance at A600 nm on a microplate reader (Synergy HT; BioTek Instruments Inc., Winooski, VT, United States) and confirmed by the addition of iodonitrotetrazolium chloride to 96-well plates as previously described (Klančnik et al., 2009). The MIC was designated as the lowest concentration of the antimicrobial leading to a significant decrease (>90%) in inoculum viability after 48 h as previously described with modification on incubation time (Burt, 2004). The minimal bactericidal concentration (MBC) was determined as previously described (Dholvitayakhun and Trachoo, 2012; Duarte et al., 2016a). The concentration at which no bacterial growth was noticed after 48 h of incubation was regarded as MBC. The MIC values were interpreted according to the standards of the European Committee for Antimicrobial Susceptibility Testing1, except for SIT which lacks international cutoff values. The MIC values were CIP ≤ 0.5 μg/ml, NAL ≤ 16 μg/ml, ERY ≤ 4 for C. jejuni and ≤8 μg/ml for C. coli, and GEN and TET ≤ 2 μg/ml. However, all Campylobacter strains were sensitive to SIT (MIC ≤ 2 μg/ml) according to the literature (Huang et al., 2015; Xu et al., 2018).
DNA Extraction, PCR Confirmation of Species, and Detection of AMR and Virulence Genes
The extraction of genomic DNA from pure colonies was carried out by using the Qiagen QIAamp PowerFecal Kit (Qiagen, Hilden, Germany) as per the manufacturer’s instructions. Then, a multiplex PCR was conducted using genus-specific primers (C412F and C1228R), cj0414 gene primers (C1 and C3) for C. jejuni, and ask gene primers (CC18F and CC519R) for C. coli (Yamazaki-Matsune et al., 2007). The primers were selected based on the specificity in identifying the genus and species of Campylobacter (Linton et al., 1997; Pajaniappan et al., 2008). The PCR mixture (25 μl) contained 12.5 μl of 2X Master Mix (Thermo Fisher Scientific, Seoul, South Korea), 1 μl of primer (10 μM), 1.5 μl of template DNA (20 μg/ml), and 7 μl of sterile deionized water. The cycling conditions were one cycle of 95°C for 5 min, 35 cycles each of 94°C for 30 s, 55°C for 45 s, and 72°C for 45 s, and a final extension at 72°C for 7 min using MiniAmp Plus Thermal Cycler (Applied Biosystems, MA, United States). The PCR products were held at 4°C before analysis.
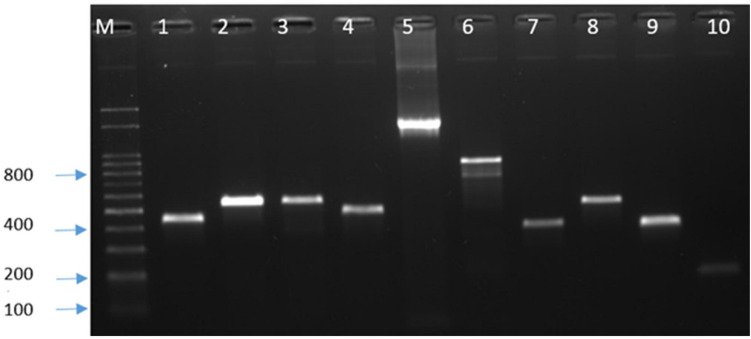
For the genes associated with antibiotic resistance [tet(O), gyrA, and cmeB] and virulence (cstII, cdtB, flaA, ggt, csrA, cadF, ciaB, pldA, and dnaJ), the PCR was performed using specific primers (Supplementary Table 1). After electrophoresis, bands of PCR products (Figure 1) were observed on a Dual UV Transilluminator (Core Bio System, Huntington Beach, CA, United States) under ultraviolet light. The bands of the amplification products were compared to the 100-bp DNA ladder (Dyne bio, Seongnam-si, Republic of Korea). The PCR products of antibiotic resistance genes were purified with AMPure XP beads (Beckman Coulter, Fullerton, CA, United States) and sequenced by the Sanger method at SolGent (Solutions for Genetic Technologies, Daejeon, Republic of Korea).
FIGURE 1.
PCR detection of antimicrobial resistance and virulence genes. M, marker; 1, gyrA; 2, tet(O); 3, cstII; 4, cdt B; 5, flaA; 6, csrA; 7, cadF; 8, ciaB; 9, pldA; and 10, dnaJ.
flaA-Restriction Fragment Length Polymorphism
Genetic diversity was first analyzed by flaA-restriction fragment length polymorphism (RFLP) using 25-μl PCR reactions (Harrington et al., 2003; Wieczorek and Osek, 2008). The flaA amplicon (1.7 kb) was digested for 6 h at 37°C using HpyF3I restriction enzyme (Thermo Scientific, Waltham, MA, United States), and the fragments were separated using 2.5% agarose gel (Lonza Inc., Rockland, ME, United States) in Tris–acetate–EDTA buffer at 90 V for 90 min. The bands were photographed with iBrightTM CL1000 Imaging System (Thermo Fisher Scientific, Seoul, Republic of Korea). The dyne 100-bp and 1-kb DNA ladders (Dyne bio, Seongnam-si, Republic of Korea) were used as standards for molecular size determination.
Multilocus Sequence Typing Analysis
Multilocus sequence typing was performed as previously described (Dingle et al., 2001; Giannatale et al., 2019) using primers available from the Campylobacter MLST website2. Briefly, the seven HKGs aspA (aspartase), glnA (glutamine synthetase), gltA (citrate synthase), glyA (serine hydroxymethyltransferase), pgm (phosphor glucomutase), tkt (transketolase), and uncA (ATP synthase) were PCR-amplified from genomic DNA. For C. jejuni, two rounds of PCR were performed (nested PCR), while for C. coli one set of primers was used. The PCR conditions were denaturation at 94°C for 5 min, 35 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 45 s, and then a final extension at 72°C for 5 min. The purification of amplicons was performed by AMPure XP beads (Beckman Coulter, Fullerton, CA, United States) as per the manufacturer’s recommendations and sequenced by the Sanger method at SolGent (Solutions for Genetic Technologies, Daejeon, Republic of Korea).
Data Analysis
GraphPad Prism 8.4.0 (GraphPad Software, La Jolla, CA, United States) was used to compute the descriptive statistics (detection rate, proportions, and frequencies of different attributes). The flaA restriction profiles were analyzed by pairwise comparisons and cluster analysis using the Dice correlation coefficient and the unweighted pair group method with arithmetic mean clustering algorithm in BIONUMERICS V8.0 (Applied Maths, Sint-Martens-Latem, Belgium). The optimization and position tolerance (1%) for band analysis and a cutoff of 100% were used. BioEdit software (version 7.2.6.1) was used to edit, align, and analyze the DNA chromatograms (Hall, 1999). A BLAST search was performed to compare consensus sequences [gyrA and tet(O)] with those from the GenBank database. Then, our sequences were submitted to get the corresponding accession numbers. Standard sensitive strains (L04566.1 and U63413.1) and resistant strains (KX982339.1 and MT176401.1) for gyrA were used for comparison. For the gyrA gene, the comparison was performed with Clustal Omega (Madeira et al., 2019). Amino acid sequences were deduced from the DNA sequences using the ExPASyTranslate tool (Gasteiger et al., 2003). Alleles, STs, and clonal complexes (CCs) were assigned by submitting the sequence data to the MLST database (see text footnote 2) (Maiden, 2006). A minimum spanning tree of C. jejuni and C. coli STs was created from MLST allelic profiles using BIONUMERICS 8.0 (Applied Maths NV, Saint-Martens-Latem, Belgium).
Results
Out of 153 fecal samples, the detection rate of Campylobacter spp. was 35.9% (55), with C. jejuni and C. coli being 89.1% (49) and 10.9% (six), respectively. None of the 20 fecal samples from the second farm was positive for Campylobacter.
Antimicrobial Susceptibility Testing
All Campylobacter isolates were screened for antimicrobial susceptibility to six antimicrobials, and they showed high-level resistance to CIP, NAL, and TET. Resistance to CIP and NAL was 100%, while resistance to TET was 93.9% for C. jejuni and 83.3% for C. coli (Table 1). Four (8.2%) C. jejuni strains were multidrug-resistant (MDR), but none of the C. coli was MDR. Of the four MDR isolates, two were resistant to CIP, NAL, TET, and ERY, while the other two were resistant to CIP, NAL, TET, and GEN. The presence of tet(O) and mutation in gyrA were confirmed by PCR (Figure 1), but all strains did not show bands for the multidrug efflux pump gene (cmeB). Sequencing revealed the presence of a missense mutation (C257T) in the quinolone resistance determining region of gyrA gene, causing resistance to quinolones along with other silent mutations. There was 100% sensitivity to SIT, while 4.1% of the C. jejuni isolates were resistant to both ERY and GEN (Table 1). The MBC values were as follows: CIP, 64–256 μg/ml; NAL, 128–512 μg/ml; TET, 2–1,024 μg/ml; SIT, 0.25–1 μg/ml; ERY, 1–32 μg/ml; and GEN, 1–256 μg/ml.
TABLE 1.
Antimicrobial resistance data for both Campylobacter jejuni and Campylobacter coli species.
| Anti-microbial | Class | Species | Resistance (%) | Number of isolates at the indicated minimal inhibitory concentration (μg/ml) |
|||||||||||||
| 0.06 | 0.13 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | ||||
| C. jejuni | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 36 | 10 | 2 | 0 | 0 | ||
| CIP | C. coli | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2 | 0 | 0 | 0 | |
| Fluoroquinolone | C. jejuni | 0 | 2 | 33 | 11 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| SIT | C. coli | 0 | 0 | 0 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| C. jejuni | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 25 | 17 | 6 | 0 | ||
| NAL | Quinolone | C. coli | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 0 | 0 |
| C. jejuni | 4.1 | 0 | 0 | 1 | 38 | 3 | 2 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| ERY | Macrolide | C. coli | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aminoglycoside | C. jejuni | 4.1 | 0 | 1 | 11 | 27 | 8 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | |
| GEN | C. coli | 0 | 0 | 0 | 2 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Tetracycline | C. jejuni | 93.9 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 3 | 10 | 22 | 10 | 1 | |
| TET | C. coli | 83.3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 0 | |
Upon submission of the gyrA and tet(O) sequences to the GenBank database, the following accession numbers have been assigned: MW067325–MW067370 (Table 2). The main mutation in gyrA gene is the missense mutation (C257T) associated with the codon change from ACA to ATA (C. jejuni) and ACT to ATT (C. coli) leading to T86I substitution. However, silent mutations were also found. It was noticed that TET-resistant strains possessed the tet(O) gene, which was confirmed by sequencing. The BLAST search showed similarity with known tet(O) gene sequences in GenBank.
TABLE 2.
Accession numbers for DNA gyrA and tet(O) resistance genes.
| Isolate | DNA gyrA accession number | tet(O) accession number |
| CJ5 | MW067325 | MW067326 |
| CJ8 | MW067327 | MW067328 |
| CJ17 | MW067329 | MW067350 |
| CJ21 | MW067330 | MW067351 |
| CJ28 | MW067331 | MW067352 |
| CJ32 | MW067332 | MW067353 |
| CJ37 | MW067333 | MW067354 |
| CJ42 | MW067334 | MW067355 |
| CJ46 | MW067335 | MW067356 |
| CJ48 | MW067336 | MW067357 |
| CJ50 | MW067337 | MW067358 |
| CJ51 | MW067338 | MW067359 |
| CJ52 | MW067339 | MW067360 |
| CJ53 | MW067340 | MW067361 |
| CJ54 | MW067341 | MW067362 |
| CJ55 | MW067342 | MW067363 |
| CJ56 | MW067343 | MW067364 |
| CJ60 | MW067344 | MW067365 |
| CJ71 | MW067345 | MW067366 |
| CC13 | MW067346 | MW067367 |
| CC45 | MW067347 | MW067368 |
| CC47 | MW067348 | MW067369 |
| CC2 | MW067349 | MW067370 |
| CC1 | MT947449 | MT967270 |
CJ, Campylobacter jejuni; CC, Campylobacter coli.
Virulence Genes
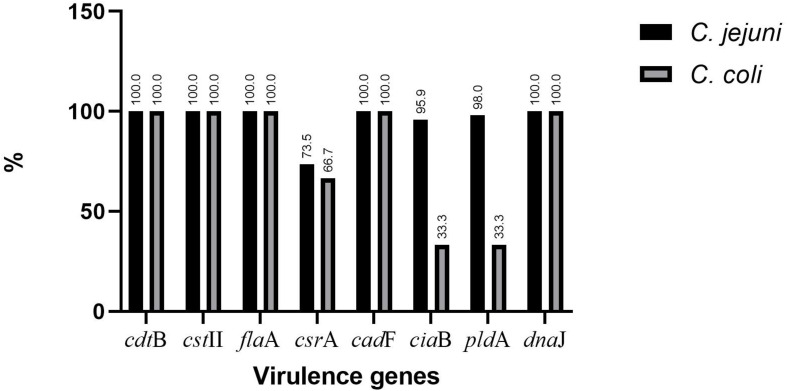
The presence of selected virulence genes (cstII, cdtB, flaA, ggt, csrA, cadF, ciaB, pldA, and dnaJ) was checked by PCR (Figure 1). We found that all isolates possessed cdtB, cstII, flaA, cadF, and dnaJ, but none showed the presence of ggt. The percentages for csrA, ciaB, and pldA were 73.5, 95.9, and 98% for C. jejuni and 66.7, 33.3, and 33.3% for C. coli, respectively (Figure 2).
FIGURE 2.
Distribution of virulence genes in Campylobacter jejuni and Campylobacter coli.
The flaA Polymorphism
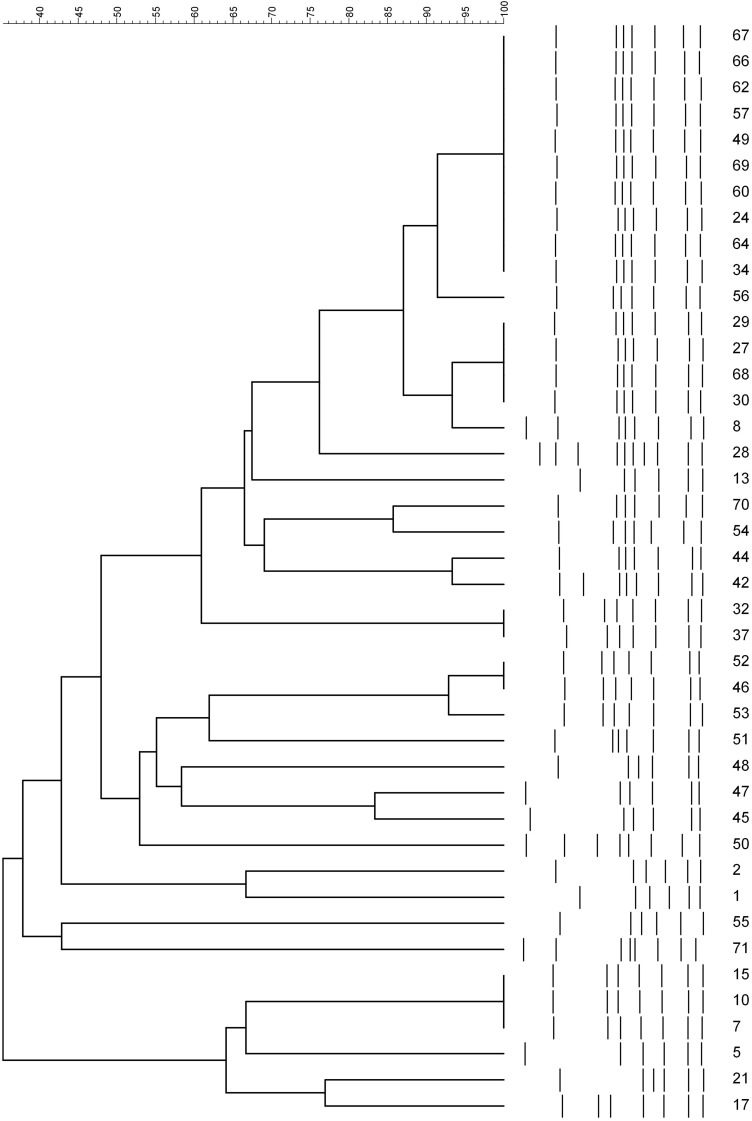
Campylobacter jejuni (n = 37) and C. coli (n = 5) were digested with HpyF3I, yielding six to 10 fragments of DNA. There were 26 fla-A types (C. jejuni: 21 and C. coli: five). For C. jejuni, there was a predominant cluster at the top of the dendrogram. For C. coli, isolate numbers 45 and 47 clustered together; the other isolates had different patterns (Figure 3).
FIGURE 3.
PCR–restriction fragment length polymorphism profiles of flaA gene digested with HpyF31 from strains of Campylobacter species. Numbers represent the laboratory code for isolates.
Multilocus Sequence Typing
Twenty-four isolates (C. jejuni: 19; C. coli: five), selected based on flaA RFLP profiles to maximize the diversity, were genotyped by MLST. C. jejuni isolates were matched with 10 STs grouped into five CCs. However, three C. jejuni isolates had new combinations of previously described alleles but could not be matched with any of the existing STs. Upon submission to the database (see text footnote 2) for ST assignment, the isolates CJ42 (id: 106369), CJ52 (id: 106370), and CJ71 (id: 106371) were assigned to ST-10645, ST-10647, and ST-10648, respectively. Of the 10 STs, ST-3611 was the main one with five isolates, followed by ST-460 with four isolates. The CCs with a higher number of isolates were CC-607 with nine isolates and CC-460 with five isolates. For C. coli (five isolates), three STs were found, with ST-5935 being the most prevalent (three isolates), and all the five isolates belonged to CC-1150 (Table 3).
TABLE 3.
Distribution of sequence types and clonal complexes among Campylobacter strains from chicken (n = 24).
| Species | Isolate ID | Sequence type | Total number | Clonal complex |
| Campylobacter | CJ5; CJ50 | 51 | 2 | 443 |
| jejuni | ||||
| CJ8; CJ51; CJ54; CJ60 | 460 | 4 | 460 | |
| CJ56 | 10613 | 1 | ||
| CJ17; CJ32; CJ37; CJ46; CJ53 | 3611 | 5 | 607 | |
| CJ21; CJ48 | 607 | 2 | ||
| CJ55 | 6238 | 1 | ||
| CJ52 | 10647 | 1 | ||
| CJ28 | 8994 | 1 | 52 | |
| CJ42 | 10645 | 1 | ||
| CJ71 | 10648 | 1 | 446 | |
| Campylobacter | CC1; CC13; CC45 | 5935 | 3 | 1150 |
| coli | CC2 | 8164 | 1 | |
| CC47 | 1121 | 1 | ||
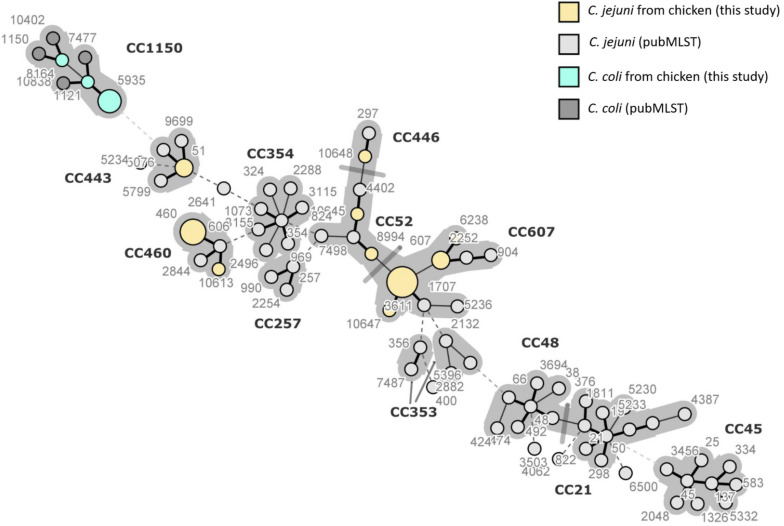
The minimum spanning tree of C. jejuni and C. coli STs was created from MLST allelic profiles using BIONUMERICS 8.0 (Applied Maths NV, Saint-Martens-Latem, Belgium). The tree shows 78 STs grouped into 12 previously characterized CCs, including both STs identified in this study (13) and 65 STs reported in the literature as predominant in Campylobacter strains of human and poultry origins. ST-460 and ST-10613, belonging to CC-460, are clustered together, and they share six of the seven HKGs, with the only difference being in glnA allele. The STs 6238, 607, and 3611 form another cluster belonging to the same CC-607 at the center of the tree. ST-51 belonging to CC-443 is located far from the other STs identified in this study. Of the three newly assigned STs, it can be concluded that ST-10645 is closely related to ST-8994, both belonging to CC-52, while ST-10647 is closely related to ST-3611, both belonging to CC-607. We also included STs that have been previously identified in Campylobacter strains isolated from poultry, human, and cattle fecal samples from the database (see text footnote 2). It can be concluded that CCs of this study are closely related to other CCs (257, 353, and 354) and distanced from CC-45 and CC-21 commonly reported in Campylobacter strains of poultry origin in Asia and elsewhere. The STs for C. coli from this study all belong to CC-1150 along with other STs obtained from the database (Figure 4).
FIGURE 4.
Minimum spanning tree of Campylobacter jejuni and Campylobacter coli sequence types (STs) created from multilocus sequence typing allelic profiles. Each ST is represented as a circle, with the size of the circle proportional to the number of isolates of that ST for the isolates obtained in this study (STs colored in yellow and green). The figure also includes STs commonly reported in Korea and elsewhere from poultry and humans which were retrieved from pubMLST. The branch length and thickness represent the allelic distance: a thick short line connects single-locus variants, a thin longer line connects double-locus variants, and a dashed line connects STs separated by three or more allelic differences. Background shading highlights clonal complexes at a distance of two alleles in this dataset.
Discussion
Chicken contamination or infection by Campylobacter at the farm level usually affects the whole poultry production chain from farm to fork (Giannatale et al., 2019; Tang et al., 2020), and suitable interventions have to be adopted to reduce transmission from poultry to humans (Alter et al., 2011). The detection rate of Campylobacter was 35.9%, with C. jejuni being predominant (89.1%) compared to C. coli (10.9%). The detection rate was slightly higher compared to the rate previously reported for layers in the United States (Rama et al., 2018) but lower than the rates reported in the Netherlands (Schets et al., 2017) and Sri Lanka (Kalupahana et al., 2013). C. jejuni is reported to be the predominant species causing human campylobacteriosis, and our results concur with the literature (Han et al., 2007; Wei et al., 2014). However, exceptions have been reported, where C. coli was the predominant or the only isolated species (Marinou et al., 2012; Wieczorek et al., 2020a).
Campylobacter species exhibited high-level resistance to some antimicrobials (CIP, NAL, and TET), which concurs with findings previously reported in Korea (Kim et al., 2010; Lee et al., 2017; Oh et al., 2017) and elsewhere (Elhadidy et al., 2018; Zhang et al., 2020). In Korea, the use of ciprofloxacin was banned in 2010 (Ku et al., 2011), but mass medication of poultry with FQs, especially enrofloxacin, is still allowed (Seo and Lee, 2020). We characterized the gyrA gene, and the point mutation (C257T) confirmed the phenotypic results. The (C257T) mutation leads to increased resistance to FQs (Frasao et al., 2015) often used in livestock (Perrin-Guyomard et al., 2020) and in humans for treating undiagnosed diarrhea cases (Sproston et al., 2018). Surprisingly, quinolone-resistant Campylobacter strains have been reported in Australia in the absence of their use (Abraham et al., 2020). Although the use of FQs is banned in several countries, resistant strains are maintained in bacterial populations, which may explain their continued occurrence in humans and animals (Sproston et al., 2018). The association of using FQs in animal husbandry and the increased occurrence of AMR pathogens differ depending on the poultry production system, surveillance programs, and geographic location (Hao et al., 2016). It is hypothesized that the increased rates of human campylobacteriosis in Asia, Europe, and America are partly driven by a widespread prevalence of C. jejuni strains resistant to quinolones via poultry (Oh et al., 2017).
There was high resistance to TET, which concurs with previous reports in Korea (Wei et al., 2014; Lee et al., 2017), China (Zhang et al., 2020), and India (Kabir et al., 2015). TET is often used in poultry and pig industries based on their low cost and easy administration to animals through drinking water (Jonker and Picard, 2010). The sequencing and BLAST search showed the similarity of tet(O) gene sequences with known plasmid-mediated tet(O) gene sequences in the GenBank (Elhadidy et al., 2018; Lynch et al., 2020).
All isolates were susceptible to SIT (MIC values: 0.125–1 μg/ml) which possesses specific features lacking in other quinolones like a cyclopropyl ring with fluorine at R-1 and a chloride substituent at R-8 (Changkwanyeun et al., 2016), which may explain its high effectiveness. Our results corroborate previous reports with a MIC of 0.25 μg/ml (Yabe et al., 2010; Changkwanyeun et al., 2015). SIT is a candidate for clinical trials on campylobacteriosis (Changkwanyeun et al., 2015), and it is used for the eradication of multi-drug resistant Helicobacter pylori (Pohl et al., 2019). This could be a breakthrough as the alternative therapies for severe campylobacteriosis are very limited (Pavlova et al., 2020).
The low resistance to ERY and GEN shown by the isolates of this study has also been reported in different countries like Korea, Vietnam, and China (Carrique-Mas et al., 2014; Wei et al., 2014; Zhang et al., 2020). Resistance to ERY has been low and stable in China (Zhang et al., 2020), the United States, and across Europe (Tang et al., 2017). The limited resistance to ERY may be partly explained by a slower process of developing resistant strains when exposed to ERY and reduced survival of resistant strains (Luangtongkum et al., 2009, 2012). Resistance to GEN has also been relatively low as it is used for treating systemic infections (Lynch et al., 2020). Prudent use of existing antimicrobials and efforts to discover new alternative treatment options would help in curbing the AMR trend.
The Campylobacter species virulome contributes to their pathogenicity (Han et al., 2019). In this study, cstII, cdtB, flaA, cadF, and dnaJ were detected in all isolates. The detection rates were similar to a previous report in Korea (Oh et al., 2017) but higher than those reported in South Africa and Chile (González-Hein et al., 2013; Otigbu et al., 2018; Pillay et al., 2020). cadF seems to be a prerequisite for the invasion of epithelial cells by any bacterial pathogen (Ramires et al., 2020). The outer membrane phospholipase A (pldA) was observed more in C. jejuni than in C. coli (Figure 2), which corroborates the study in South Africa (Pillay et al., 2020), while for ciaB, our findings are above those of a previous report in Korea (Oh et al., 2017) and contrast with the study in South Africa (Pillay et al., 2020). The presence of cadF and ciaB facilitates the adhesion and internalization of Campylobacter in cellular models (Ramires et al., 2020). The detection rate for csrA in C. jejuni was slightly higher than the rate in C. coli, but csrA was lacking in Campylobacter strains from South Africa (Otigbu et al., 2018). None of our isolates expressed ggt. The latter was reported to be only 5.5% in Chile (González-Hein et al., 2013), but our findings contrast with the high values (30.9–43.2%) reported in Finland (Gonzalez et al., 2009). The difference could be associated with the complexity of the colonization process involving several genes and the use of strains from a single sampling site (González-Hein et al., 2013). Furthermore, MDR and virulent C. jejuni strains have been found more in summer than in winter, suggesting the role played by climate in the expression of some genes (Kim et al., 2019). We also evoke that several virulence genes are plasmid-mediated, which may affect their presence in different strains (Oh et al., 2017). The virulence genes reported in this study have been previously reported in Campylobacter strains isolated from humans (González-Hein et al., 2013; Oh et al., 2017), highlighting the potential virulence of these Campylobacter strains in causing human infections.
The flaA-RFLP typing showed a considerable diversity of Campylobacter strains despite being from the same farm. In our study, 26 types were found by flaA typing of 42 Campylobacter isolates. In another study in Korea, 30 flaA types were reported for 100 C. jejuni from chicken (Han et al., 2007), while 19 flaA types were reported for 100 C. jejuni from Grenada, Puerto Rico, and Alabama (Behringer et al., 2011). The flaA typing is suitable for laboratories dealing with a small number of isolates as it is cheaper and reproducible (Behringer et al., 2011). However, the drawback of the flaA typing is increased recombination events in the flaA gene, which modifies RFLP profiles (Harrington et al., 1997). Furthermore, the flaA typing focuses only on a single gene from a considerable genome, and there is a lack of inter-laboratory comparison of obtained results (Nguyen et al., 2016) due to the absence of flaA database. Therefore, a combination of several typing methods is recommended (Wieczorek and Osek, 2008).
Molecular typing techniques showed that 80% of human campylobacteriosis is associated with Campylobacter of poultry origin (Newell et al., 2011). In this study, the MLST revealed 10 STs for C. jejuni and three STs for C. coli. Three STs were new but could fit in existing clonal complexes. The predominant STs (3611 and 460) have been previously isolated in C. jejuni from poultry, while ST-51 was found in C. jejuni of both chicken and human origins in various regions of Korea (Oh et al., 2017). In Korea, in addition to the CCs identified in this study, other CCs including CC-48, CC-21, and CC-45 (Figure 4) have been recovered from Campylobacter isolates of poultry and human origins (Wei et al., 2014; Oh et al., 2017). The predominance of these clonal complexes and STs could be associated with environmental factors (geography and climate) and increasing poultry consumption in Korea (Oh et al., 2017). Globally, CC-45 has been found in various hosts, including poultry, cattle, dogs, wild birds, penguins, and it was also isolated from environmental samples (Cody et al., 2012; Shin et al., 2013), while CC-607 is largely associated with chicken and humans as reported from the United Kingdom, France, Canada, Thailand, and Uruguay (Cody et al., 2012; Duarte et al., 2014; Guyard-Nicodème et al., 2015). ST-45 and ST-50, predominant in Korea, are also common in C. jejuni from chickens in Europe (Wieczorek et al., 2020b). In 2019, ST-21, ST-50, and ST-137 have been isolated from wild ducks, indicating their widespread distribution (Wei et al., 2019). Three of the 10 STs (607, 443, and 51) were found in Japan and China (Ozawa et al., 2016; Ma et al., 2017), while CC-460 and CC-607 have also been reported in samples from humans and cattle, indicating their threat to public health (Ozawa et al., 2016; Kiatsomphob et al., 2019; Wei et al., 2019). CC-460 and CC-607 are thought to be virulent as they both possess many virulence genes. In Israel, type VI secretion system (T6SS), implicated in virulence, metabolism, AMR and contributing to host adaptation, has been found in both CCs (Rokney et al., 2018). In Japan, CC-21 is known as the prevalent clonal complex in human-derived C. jejuni isolates sharing a common genetic background and similar antimicrobial susceptibilities with C. jejuni strains from chicken (Ohishi et al., 2017). CC-21 (ST-50 and ST-21) could be of interest in the region as it has been found in Korea, Japan, and China from humans, poultry, and cattle (Ozawa et al., 2016; Oh et al., 2017; Kiatsomphob et al., 2019; Wei et al., 2019; Zhang et al., 2020). However, further studies are needed to confirm this hypothesis. Except for ST-257, other STs (ST-21, ST-48, and ST-353) identified in Korea have been recovered from C. jejuni of human origin in the United Kingdom, Brazil, and China (Gomes et al., 2019). It is known that the predominance of certain genotypes of C. jejuni depends on several factors such as animal reservoirs, zoonotic transmission, rates of recombination events, food source, and the first time when a given genotype is recorded in the country (Rokney et al., 2018).
Although resistance to quinolones and TET was not a particularity of certain STs, a strong correlation between CC-460, CC-607, CC-45, CC-48, CC-21, and resistance to both TET and quinolones in C. jejuni strains is not new (Cody et al., 2012; Shin et al., 2013; Guyard-Nicodème et al., 2015; Zhang et al., 2020). In Korea, C. jejuni strains of human origin with ST-607, ST-137, ST-45, ST-21, and ST-48 were found to be MDR (Shin et al., 2013). Two isolates of C. jejuni belonged to CC-443, which is suggested to harbor antibiotic-resistant and pathogenic C. jejuni strains (Kim et al., 2019). There is limited literature associating virulence genes to specific CCs and STs; thus, this study highlights the virulence potential of the presented Campylobacter isolates. For C. coli, ST-1121 has been reported from broilers in China (Tang et al., 2020). We suggest carrying out WGS to have a detailed picture of the pathogenicity by analyzing all the housekeeping, virulence, and AMR genes. The WGS would also enlighten the evolutionary pathways of the Campylobacter spp. used in this study to inform better practices that may lead to a reduction of campylobacteriosis cases.
The limitation of this study was the restricted access to various poultry farms across the city which would have given a broad picture of the prevalence, AMR profiles, and genotypes associated with Campylobacter in Gangneung. Campylobacter strains from the current study proved to be highly diverse considering the number of obtained STs and CCs. This is in agreement with other published studies reporting the possibility of getting several genotypes in a single poultry flock, suggesting different exposure sources via horizontal transmission and/or genetic drifts within the Campylobacter population (Alter et al., 2011; El-Adawy et al., 2013).
Conclusion
This study highlights the role of layers as a reservoir of Campylobacter spp., harboring various AMR and virulence genes. The genotyping highlighted that C. jejuni isolates were more diverse than C. coli as analyzed by MLST. The MLST revealed that CC-607 (ST-3611) and CC-460 (ST-460) were the predominant ones, while three STs were newly assigned. ERY, GEN, and SIT need to be appropriately used to prevent or delay the increasing resistance in Campylobacter species. The isolates of this study may present a potential hazard to public health based on their AMR profiles, virulence genes, and genotyping data.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Ethical review and approval was not required for the animal study.
Author Contributions
NG, C-HP, and EK conceived the study. NG carried out the experiments, analyzed and interpreted the data, and wrote the manuscript. D-GS and K-YY substantially contributed to the analysis of the results. LM, MM, DM, and RA substantially revised the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was financially supported by the Ministry of Oceans and Fisheries, South Korea (grant no. 20170488) and the Partnership for Skills in Applied Sciences, Engineering, and Technology – Regional Scholarship and Innovation Fund (PASET-RSIF) in collaboration with the government of the Republic of Korea, and the KIST intramural grant (2Z06483).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.622275/full#supplementary-material
References
- Abraham S., Sahibzada S., Hewson K., Laird T., Abraham R., Pavic A., et al. (2020). Emergence of fluoroquinolone-resistant Campylobacter jejuni and Campylobacter coli among Australian chickens in the absence of fluoroquinolone use. Appl. Environ. Microbiol. 86:e02765-19. 10.1128/AEM.02765-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaboudi A. R., Malkawi I. M., Osaili T. M., Abu-Basha E. A., Guitian J. (2020). Prevalence, antibiotic resistance and genotypes of Campylobacter jejuni and Campylobacter coli isolated from chickens in Irbid governorate, Jordan. Intern. J. Food Microbiol. 327:108656. 10.1016/j.ijfoodmicro.2020.108656 [DOI] [PubMed] [Google Scholar]
- Al-Edany A. A., Khudor M. H., Radhi L. Y. (2015). Isolation, identification and toxigenic aspects of Campylobacter jejuni isolated from slaughtered cattle and sheep at Basrah city. Microbiology 14 316–327. [Google Scholar]
- Alter T., Weber R. M., Hamedy A., Glünder G. (2011). Carry-over of thermophilic Campylobacter spp. between sequential and adjacent poultry flocks. Vet. Microbiol. 147 90–95. 10.1016/j.vetmic.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Barnes I. H. A., Bagnall M. C., Browning D. D., Thompson S. A., Manning G., Newell D. G. (2007). γ-Glutamyl transpeptidase has a role in the persistent colonization of the avian gut by Campylobacter jejuni. Microb. Pathog. 43 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer M., Miller W. G., Oyarzabal O. A. (2011). Typing of Campylobacter jejuni and Campylobacter coli isolated from live broilers and retail broiler meat by flaA-RFLP, MLST, PFGE and REP-PCR. J. Microbiol. Methods 84 194–201. 10.1016/j.mimet.2010.11.016 [DOI] [PubMed] [Google Scholar]
- Blaser M. J., Engberg J. (2008). “Clinical aspects of Campylobacter jejuni and Campylobacter coli infections,” in Campylobacter, 3edn Edn, eds Nachamkin I., Szymanski C. M., Blaser M. J. (Washington, DC: ASM Press; ), 99–121. 10.1128/9781555815554.ch6 [DOI] [Google Scholar]
- Bolton D. J. (2015). Campylobacter virulence and survival factors. Food Microbiol. 48 99–108. [DOI] [PubMed] [Google Scholar]
- Burt S. (2004). Essential oils: their antibacterial properties and potential applications in foods—a review. Intern. J. Food Microbiol. 94 223–253. 10.1016/j.ijfoodmicro.2004.03.022 [DOI] [PubMed] [Google Scholar]
- Carrique-Mas J. J., Bryant J. E., Cuong N. V., Hoang N. V. M., Campbell J., Hoang N. V., et al. (2014). An epidemiological investigation of Campylobacter in pig and poultry farms in the Mekong delta of Vietnam. Epidemiol. Infect. 142 1425–1436. 10.1017/S0950268813002410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changkwanyeun R., Usui M., Kongsoi S., Yokoyama K., Kim H., Suthienkul O., et al. (2015). Characterization of Campylobacter jejuni DNA gyrase as the target of quinolones. J. Infect. Chemother. 21 604–609. 10.1016/j.jiac.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Changkwanyeun R., Yamaguchi T., Kongsoi S., Changkaew K., Yokoyama K., Kim H., et al. (2016). Impact of mutations in DNA gyrase genes on quinolone resistance in Campylobacter jejuni. Drug Test. Analys. 8 1071–1076. 10.1002/dta.1937 [DOI] [PubMed] [Google Scholar]
- Chen C. K., Cheng I., Chen Y. H., Lai C. C. (2020). Efficacy and safety of sitafloxacin in the treatment of acute bacterial infection: a meta-analysis of randomized controlled trials. Antibiotics 9 1–9. 10.3390/antibiotics9030106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody A. J., McCarthy N. M., Wimalarathna H. L., Colles F. M., Clark L., Bowler I. C. J. W., et al. (2012). A longitudinal 6-Year study of the molecular epidemiology of clinical Campylobacter isolates in Oxfordshire, United Kingdom. J. Clin. Microbiol. 50 3193–3201. 10.1128/JCM.01086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dholvitayakhun A., Trachoo N. (2012). Antibacterial activity of ethanol extract from Some Thai medicinal plants against Campylobacter jejuni. Nat. Product Res. 6 113–116. [DOI] [PubMed] [Google Scholar]
- Dingle K. E., Colles F. M., Wareing D. R. A., Ure R., Fox A. J., Bolton F. E., et al. (2001). Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39 14–23. 10.1128/JCM.39.1.14-23.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A., Luís Â, Oleastro M., Domingues F. C. (2016a). Antioxidant properties of coriander essential oil and linalool and their potential to control Campylobacter spp. Food Control 61 115–122. 10.1016/j.foodcont.2015.09.033 [DOI] [Google Scholar]
- Duarte A., Santos A., Manageiro V., Martins A., Fraqueza M. J., Caniça M., et al. (2014). Human, food and animal Campylobacter spp. isolated in Portugal: high genetic diversity and antibiotic resistance rates. Intern. J. Antimicrob. Agents 44 306–313. 10.1016/j.ijantimicag.2014.06.012 [DOI] [PubMed] [Google Scholar]
- Duarte A., Seliwiorstow T., Miller W. G., De Zutter L., Uyttendaele M., Dierick K., et al. (2016b). Discriminative power of Campylobacter phenotypic and genotypic typing methods. J. Microbiol. Methods 125 33–39. 10.1016/j.mimet.2016.03.004 [DOI] [PubMed] [Google Scholar]
- Eberle K. N., Kiess A. S. (2012). Phenotypic and genotypic methods for typing Campylobacter jejuni and Campylobacter coli in poultry. Poult. Sci. 91 255–264. 10.3382/ps.2011-01414 [DOI] [PubMed] [Google Scholar]
- El-Adawy H., Hotzel H., Tomaso H., Neubauer H., Taboada E. N., Ehricht R., et al. (2013). Detection of genetic diversity in Campylobacter jejuni isolated from a commercial turkey flock using flaA typing, MLST analysis and microarray assay. PLoS One 8:e051582. 10.1371/journal.pone.0051582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhadidy M., Miller W. G., Arguello H., Álvarez-Ordóñez A., Duarte A., Dierick K., et al. (2018). Genetic basis and clonal population structure of antibiotic resistance in Campylobacter jejuni isolated from broiler carcasses in Belgium. Front. Microbiol. 9:1014. 10.3389/fmicb.2018.01014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisha I. L., Botha F. S., McGaw L. J., Eloff J. N. (2017). The antibacterial activity of extracts of nine plant species with good activity against Escherichia coli against five other bacteria and cytotoxicity of extracts. BMC Complem. Altern. Med. 17:133. 10.1186/s12906-017-1645-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields J. A., Thompson S. A. (2008). Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 190 3411–3416. 10.1128/JB.01928-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald C. (2015). Campylobacter. Clin. Lab. Med. 35 289–298. 10.1016/j.cll.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Frasao B., da S., Medeiros V., Barbosa A. V., de Aguiar W. S., dos Santos F. F., et al. (2015). Detection of fluoroquinolone resistance by mutation in gyrA gene of Campylobacter spp. isolates from broiler and laying (Gallus gallus domesticus) hens, from Rio de Janeiro State, Brazil. Ciência Rural 45 2013–2018. 10.1590/0103-8478cr20141712 [DOI] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31 3784–3788. 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannatale E. D., Calistri P., Donato G. D., Decastelli L., Goffredo E., Adriano D., et al. (2019). Thermotolerant Campylobacter spp. in chicken and bovine meat in Italy: prevalence, level of contamination and molecular characterization of isolates. PLoS One 14:e0225957. 10.1371/journal.pone.0225957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C. N., Frazão M. R., Passaglia J., Duque S. S., Medeiros M. I. C., Falcão J. P. (2019). Molecular epidemiology and resistance profile of Campylobacter jejuni and Campylobacter coli strains isolated from different sources in Brazil. Microb. Drug Resist. 26 1516–1525. 10.1089/mdr.2019.0266 [DOI] [PubMed] [Google Scholar]
- Gonzalez M., Hakkinen M., Rautelin H., Hänninen M. L. (2009). Bovine Campylobacter jejuni strains differ from human and chicken strains in an analysis of certain molecular genetic markers. Appl. Environ. Microbiol. 75 1208–1210. 10.1128/AEM.01879-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Hein G., Huaracán B., García P., Figueroa G. (2013). Prevalence of virulence genes in strains of Campylobacter jejuni isolated from human, bovine and broiler. Braz. J. Microbiol. 44 1223–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P. (2007). Campylobacter flagella: not just for motility. Trends Microbiol. 15 456–461. 10.1016/j.tim.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Guyard-Nicodème M., Rivoal K., Houard E., Rose V., Quesne S., Mourand G., et al. (2015). Prevalence and characterization of Campylobacter jejuni from chicken meat sold in French retail outlets. Intern. J. Food Microbiol. 203 8–14. 10.1016/j.ijfoodmicro.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41 95–98. [Google Scholar]
- Han K., Jang S. S., Choo E., Heu S., Ryu S. (2007). Prevalence, genetic diversity, and antibiotic resistance patterns of Campylobacter jejuni from retail raw chickens in Korea. Intern. J. Food Microbiol. 114 50–59. 10.1016/j.ijfoodmicro.2006.10.042 [DOI] [PubMed] [Google Scholar]
- Han X., Guan X., Zeng H., Li J., Huang X., Wen Y., et al. (2019). Prevalence, antimicrobial resistance profiles and virulence-associated genes of thermophilic Campylobacter spp. isolated from ducks in a Chinese slaughterhouse. Food Control 104 157–166. 10.1016/j.foodcont.2019.04.038 [DOI] [Google Scholar]
- Hao H., Sander P., Iqbal Z., Wang Y., Cheng G., Yuan Z. (2016). The risk of some veterinary antimicrobial agents on public health associated with antimicrobial resistance and their molecular basis. Front. Microbiol. 7:1626. 10.3389/fmicb.2016.01626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington C. S., Moran L., Ridley A. M., Newell D. G., Madden R. H. (2003). Inter-laboratory evaluation of three flagellin PCR/RFLP methods for typing Campylobacter jejuni and C. coli: the CAMPYNET experience. J. Appl. Microbiol. 95 1321–1333. 10.1046/j.1365-2672.2003.02101.x [DOI] [PubMed] [Google Scholar]
- Harrington C. S., Thomson-Carter F. M., Carter P. E. (1997). Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35 2386–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelaar A. H., Kirk M. D., Torgerson P. R., Gibb H. J., Hald T., Lake R. J., et al. (2015). World health organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 12:1001923. 10.1371/journal.pmed.1001923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. S., Wang J. T., Sheng W. H., Chuang Y. C., Chang S. C. (2015). Comparative in vitro activity of sitafloxacin against bacteremic isolates of carbapenem resistant Acinetobacter baumannii complex. J. Microbiol. Immunol. Infect. 48 545–551. 10.1016/j.jmii.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Humphrey S., Chaloner G., Kemmett K., Davidson N., Williams N., Kipar A., et al. (2014). Campylobacter jejuni is not merely a commensal in commercial broiler chickens and affects bird welfare. mBio 5:e01364-14. 10.1128/mBio.01364-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T., O’Brien S., Madsen M. (2007). Campylobacters as zoonotic pathogens: a food production perspective. Intern. J. Food Microbiol. 117 237–257. [DOI] [PubMed] [Google Scholar]
- Jain D., Prasad K. N., Sinha S., Husain N. (2008). Differences in virulence attributes between cytolethal distending toxin positive and negative Campylobacter jejuni strains. J. Med. Microbiol. 57(Pt 3), 267–272. 10.1099/jmm.0.47317-0 [DOI] [PubMed] [Google Scholar]
- Jonker A., Picard J. A. (2010). Antimicrobial susceptibility in thermophilic Campylobacter species isolated from pigs and chickens in South Africa. J. South Afric. Vet. Assoc. 81 228–236. [DOI] [PubMed] [Google Scholar]
- Kabir S. L., Asakura M., Shiramaru S., Pal A., Hinenoya A., Yamasaki S. (2015). Molecular identification and antimicrobial resistance profiles of Campylobacter strains of poultry origin in India with special emphasis on fluoroquinolone resistance. Asian J. Med. Biol. Res. 1 1–8. 10.3329/ajmbr.v1i1.25491 [DOI] [Google Scholar]
- Kalupahana R. S., Kottawatta K. S. A., Kanankege K. S. T., Bergen M. A. P., van Abeynayake P., Wagenaar J. A. (2013). Colonization of Campylobacter spp. in broiler chickens and laying hens reared in tropical climates with low-Bbosecurity housing. Appl. Environ. Microbiol. 79 393–395. 10.1128/AEM.02269-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem I. I., Kehinde O., Kumar A., Rajashekara G. (2016). Antimicrobial-resistant Campylobacter in organically and conventionally raised layer chickens. Foodbo. Pathog. Dis. 14 29–34. 10.1089/fpd.2016.2161 [DOI] [PubMed] [Google Scholar]
- Kiatsomphob S., Taniguchi T., Tarigan E., Latt K. M., Jeon B., Misawa N. (2019). Aerotolerance and multilocus sequence typing among Campylobacter jejuni strains isolated from humans, broiler chickens, and cattle in Miyazaki Prefecture, Japan. J. Vet. Med. Sci. 81 1144–1151. 10.1292/jvms.19-0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Hong J., Bae W., Koo H. C., Kim S. H., Park Y. H. (2010). Prevalence, antibiograms, and transferable tet (O) plasmid of Campylobacter jejuni and Campylobacter coli isolated from raw chicken, pork, and human clinical cases in Korea. J. Food Protect. 73 1430–1437. 10.4315/0362-028X-73.8.1430 [DOI] [PubMed] [Google Scholar]
- Kim J., Park H., Kim J., Kim J. H., Jung J. I., Cho S., et al. (2019). Comparative analysis of aerotolerance, antibiotic resistance, and virulence gene prevalence in Campylobacter jejuni isolates from retail raw chicken and duck meat in South Korea. Microorganisms 7:433. 10.3390/microorganisms7100433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klančnik A., Guzej B., Kolar M. H., Abramovič H., Možina S. S. (2009). In Vitro antimicrobial and antioxidant activity of commercial rosemary extract formulations. J. Food Protect. 72 1744–1752. 10.4315/0362-028X-72.8.1744 [DOI] [PubMed] [Google Scholar]
- Klančnik A., Šimunović K., Sterniša M., Ramić D., Smole Možina S., Bucar F. (2020). Anti-adhesion activity of phytochemicals to prevent Campylobacter jejuni biofilm formation on abiotic surfaces. Phytochem. Rev. 1–30. 10.1007/s11101-020-09669-6 [DOI] [Google Scholar]
- Koga M., Gilbert M., Takahashi M., Li J., Koike S., Hirata K., et al. (2006). Comprehensive analysis of bacterial risk factors for the development of Guillain-Barré syndrome after Campylobacter jejuni enteritis. J. Infect. Dis. 193 547–555. [DOI] [PubMed] [Google Scholar]
- Koolman L., Whyte P., Burgess C., Bolton D. (2015). Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 12 424–432. 10.1089/fpd.2014.1883 [DOI] [PubMed] [Google Scholar]
- Kreling V., Falcone F. H., Kehrenberg C., Hensel A. (2020). Campylobacter sp.: pathogenicity factors and prevention methods—new molecular targets for innovative antivirulence drugs? Appl. Microbiol. Biotechnol. 104 10409–10436. 10.1007/s00253-020-10974-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B. K., Kim H. J., Lee Y. J., Kim Y. I., Choi J. S., Park M. Y., et al. (2011). Genetic Characterization and antimicrobial susceptibility of Campylobacter Spp. isolated from domestic and imported chicken meats and humans in Korea. Foodborne Pathog. Dis. 8 381–386. 10.1089/fpd.2010.0680 [DOI] [PubMed] [Google Scholar]
- Kurekci C., Bishop-Hurley S. L., Vercoe P. E., Durmic Z., Al Jassim R. A. M., McSweeney C. S. (2012). Screening of Australian plants for antimicrobial activity against Campylobacter jejuni. Phytother. Res. 26 186–190. 10.1002/ptr.3526 [DOI] [PubMed] [Google Scholar]
- Lee J., Jeong J., Lee H., Ha J., Kim S., Choi Y., et al. (2017). Antibiotic susceptibility, genetic diversity, and the presence of toxin producing genes in Campylobacter isolates from poultry. Intern. J. Environ. Res. Public Health 14 1–11. 10.3390/ijerph14111400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton D., Lawson A. J., Owen R. J., Stanley J. (1997). PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35 2568–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luangtongkum T., Jeon B., Han J., Plummer P., Logue C. M., Zhang Q. (2009). Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Fut. Microbiol. 4 189–200. 10.2217/17460913.4.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luangtongkum T., Shen Z., Seng V. W., Sahin O., Jeon B., Liu P., et al. (2012). Impaired fitness and transmission of macrolide-resistant Campylobacter jejuni in its natural host. Antimicrob. Agents Chemother. 56 1300–1308. 10.1128/AAC.05516-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydekaitiene V. L., Kudirkiene E. (2020). Prevalence and genotyping diversity of C. jejuni isolated from Broilers and their environment using flaA-RFLP typing and MLST analysis. Ann. Anim. Sci. 20 485–501. 10.2478/aoas-2020-0008 [DOI] [Google Scholar]
- Lynch C. T., Lynch H., Burke S., Hawkins K., Buttimer C., Mc Carthy C., et al. (2020). Antimicrobial resistance determinants circulating among thermophilic Campylobacter isolates recovered from broilers in Ireland over a one-year period. Antibiotics 9 1–20. 10.3390/antibiotics9060308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Su Y., Ma L., Ma L., Li P., Du X., et al. (2017). Prevalence and Characterization of Campylobacter jejuni Isolated from Retail Chicken in Tianjin, China. J. Food Protect. 80 1032–1040. 10.4315/0362-028X.JFP-16-561 [DOI] [PubMed] [Google Scholar]
- Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47 W636–W641. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M. C. J. (2006). Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60 561–588. 10.1146/annurev.micro.59.030804.121325 [DOI] [PubMed] [Google Scholar]
- Marinou I., Bersimis S., Ioannidis A., Nicolaou C., Mitrousia-Ziouva A., Legakis N. J., et al. (2012). Identification and antimicrobial resistance of Campylobacter species isolated from animal sources. Front. Microbiol. 3:58. 10.3389/fmicb.2012.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. E., Corcoran D., Dooley J. S. G., Fanning S., Lucey B., Matsuda M., et al. (2005). Campylobacter. Vet. Res. 36 351–382. 10.1051/vetres:2005012 [DOI] [PubMed] [Google Scholar]
- Mourkas E., Florez-Cuadrado D., Pascoe B., Calland J. K., Bayliss S. C., Mageiros L., et al. (2019). Gene pool transmission of multidrug resistance among Campylobacter from livestock, sewage and human disease. Environ. Microbiol. 21 4597–4613. 10.1111/1462-2920.14760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder A. C., Franz E., de Rijk S., Versluis M. A. J., Coipan C., Buij R., et al. (2020). Tracing the animal sources of surface water contamination with Campylobacter jejuni and Campylobacter coli. Water Res. 187:116421. 10.1016/j.watres.2020.116421 [DOI] [PubMed] [Google Scholar]
- Newell D. G., Elvers K. T., Dopfer D., Hansson I., Jones P., James S., et al. (2011). Biosecurity-based interventions and strategies to reduce Campylobacter spp. On poultry farms. Appl. Environ. Microbiol. 77 8605–8614. 10.1128/AEM.01090-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. N. M., Hotzel H., El-Adawy H., Tran H. T., Le M. T. H., Tomaso H., et al. (2016). Genotyping and antibiotic resistance of thermophilic Campylobacter isolated from chicken and pig meat in Vietnam. Gut Pathog. 8 19. 10.1186/s13099-016-0100-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. Y., Kwon Y. K., Wei B., Jang H. K., Lim S.-K., Kim C. H., et al. (2017). Epidemiological relationships of Campylobacter jejuni strains isolated from humans and chickens in South Korea. J. Microbiol. 55 13–20. 10.1007/s12275-017-6308-8 [DOI] [PubMed] [Google Scholar]
- Ohishi T., Aoki K., Ishii Y., Usui M., Tamura Y., Kawanishi M., et al. (2017). Molecular epidemiological analysis of human- and chicken-derived isolates of Campylobacter jejuni in Japan using next-generation sequencing. J. Infect. Chemother. 23 165–172. 10.1016/j.jiac.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Otigbu A. C., Clarke A. M., Fri J., Akanbi E. O., Njom H. A. (2018). Antibiotic sensitivity profiling and virulence potential of Campylobacter jejuni isolates from estuarine water in the Eastern Cape Province, South Africa. Intern. J. Environ. Res. Public Health 15 1–15. 10.3390/ijerph15050925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., Hiki M., Kawanishi M., Abo H., Kojima A., Asai T., et al. (2016). Molecular typing of fluoroquinolone-resistant Campylobacter jejuni isolated from broilers in Japan using multilocus sequence typing and pulsed-field gel electrophoresis. Foodborne Pathog. Dis. 13 1–7. 10.1089/fpd.2015.1975 [DOI] [PubMed] [Google Scholar]
- Pajaniappan M., Hall J. E., Cawthraw S. A., Newell D. G., Gaynor E. C., Fields J. A., et al. (2008). A temperature-regulated Campylobacter jejuni gluconate dehydrogenase is involved in respiration-dependent energy conservation and chicken colonization. Mol. Microbiol. 68 474–491. 10.1111/j.1365-2958.2008.06161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova M., Alexandrova E., Donkov G., Mitova-Mineva Y., Kantardjiev T., Velev V. (2020). Campylobacter infections among Bulgarian children: molecular characterization and antimicrobial susceptibility. Biotechnol. Biotechnol. Equip. 34 1038–1042. 10.1080/13102818.2020.1817783 [DOI] [Google Scholar]
- Perrin-Guyomard A., Jouy E., Urban D., Chauvin C., Granier S. A., Mourand G., et al. (2020). Decrease in fluoroquinolone use in French poultry and pig production and changes in resistance among E. coli and Campylobacter. Vet. Microbiol. 243:108637. 10.1016/j.vetmic.2020.108637 [DOI] [PubMed] [Google Scholar]
- Pillay S., Amoako D. G., Abia A. L. K., Somboro A. M., Shobo C. O., Perrett K., et al. (2020). Characterisation of Campylobacter spp. isolated from poultry in KwaZulu-Natal, South Africa. Antibiotics 9 1–15. 10.3390/antibiotics9020042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl D., Keller P. M., Bordier V., Wagner K. (2019). Review of current diagnostic methods and advances in Helicobacter pylori diagnostics in the era of next generation sequencing. World J. Gastroenterol. 25 4629–4660. 10.3748/wjg.v25.i32.4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokkunuri V., Pimentel M., Morales W., Jee S.-R., Alpern J., Weitsman S., et al. (2012). Role of cytolethal distending toxin in altered stool form and bowel phenotypes in a rat model of post-infectious irritable bowel syndrome. J. Neurogastroenterol. Motil. 18 434–442. 10.5056/jnm.2012.18.4.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rama E. N., Bailey M., Jones D. R., Gast R. K., Anderson K., Brar J., et al. (2018). Prevalence, persistence, and antimicrobial resistance of Campylobacter spp. from eggs and laying hens housed in five commercial housing systems. Foodborne Pathog. Dis. 15 506–516. 10.1089/fpd.2017.2404 [DOI] [PubMed] [Google Scholar]
- Ramires T., de Oliveira M. G., Kleinubing N. R., Rauber Wurfel S., de F., Mata M. M., et al. (2020). Genetic diversity, antimicrobial resistance, and virulence genes of thermophilic Campylobacter isolated from broiler production chain. Braz. J. Microbiol. 51 2021–2032. 10.1007/s42770-020-00314-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokney A., Valinsky L., Moran-Gilad J., Vranckx K., Agmon V., Weinberger M. (2018). Genomic Epidemiology of Campylobacter jejuni transmission in Israel. Front. Microbiol. 9:2432. 10.3389/fmicb.2018.02432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schets F. M., Jacobs-Reitsma W. F., van der Plaats R. Q. J., Heer L. K. D., van Hoek A. H. A. M., Hamidjaja R. A., et al. (2017). Prevalence and types of Campylobacter on poultry farms and in their direct environment. J. Water Health 15 849–862. 10.2166/wh.2017.119 [DOI] [PubMed] [Google Scholar]
- Seo K. W., Lee Y. J. (2020). Prevalence and characterization of plasmid-mediated quinolone resistance determinants qnr and aac(6’)-Ib-cr in ciprofloxacin-resistant Escherichia coli isolates from commercial layer in Korea. J. Microbiol. Biotechnol. 30 1180–1183. 10.4014/jmb.2003.03058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin E., Oh Y., Kim M., Jung J., Lee Y. (2013). Antimicrobial resistance patterns and corresponding multilocus sequence types of the Campylobacter jejuni isolates from human Diarrheal samples. Microb. Drug Resist. 19 110–116. 10.1089/mdr.2012.0099 [DOI] [PubMed] [Google Scholar]
- Sibanda N., McKenna A., Richmond A., Ricke S. C., Callaway T., Stratakos A. C., et al. (2018). A review of the effect of management practices on Campylobacter prevalence in poultry farms. Front. Microbiol. 9:2002. 10.3389/fmicb.2018.02002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproston E. L., Wimalarathna H. M. L., Sheppard S. K. (2018). Trends in fluoroquinolone resistance in Campylobacter. Microb. Genom. 4:198. 10.1099/mgen.0.000198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Fang L., Xu C., Zhang Q. (2017). Antibiotic resistance trends and mechanisms in the foodborne pathogen, Campylobacter. Anim. Health Res. Rev. 18 87–98. 10.1017/S1466252317000135 [DOI] [PubMed] [Google Scholar]
- Tang Y., Jiang Q., Tang H., Wang Z., Yin Y., Ren F., et al. (2020). Characterization and prevalence of Campylobacter spp. from broiler chicken rearing period to the slaughtering process in Eastern China. Front. Vet. Sci. 7:227. 10.3389/fvets.2020.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar J. A., French N. P., Havelaar A. H. (2013). Preventing Campylobacter at the source: why is it so difficult? Clin. Infect. Dis. 57 1600–1606. [DOI] [PubMed] [Google Scholar]
- Wei B., Cha S. Y., Kang M., Roh J. H., Seo H. S., Yoon R. H., et al. (2014). Antimicrobial susceptibility profiles and molecular typing of Campylobacter jejuni and Campylobacter coli isolates from ducks in South Korea. Appl. Environ. Microbiol. 80 7604–7610. 10.1128/AEM.02469-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B., Kang M., Jang H.-K. (2019). Genetic characterization and epidemiological implications of Campylobacter isolates from wild birds in South Korea. Transbound. Emerg. Dis. 66 56–65. 10.1111/tbed.12931 [DOI] [PubMed] [Google Scholar]
- Wieczorek K., Osek J. (2008). Comparison of two molecular methods for genotyping of Campylobacter Sp. Isolated from poultry. Appl. Environ. Microbiol. 52 523–528. [Google Scholar]
- Wieczorek K., Osek J. (2013). Antimicrobial resistance mechanisms among Campylobacter. Biomed. Res. Intern. 2013 1–12. 10.1155/2013/340605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek K., Bocian Ł, Osek J. (2020a). Prevalence and antimicrobial resistance of Campylobacter isolated from carcasses of chickens slaughtered in Poland - a retrospective study. Food Control 112:107159. 10.1016/j.foodcont.2020.107159 [DOI] [Google Scholar]
- Wieczorek K., Wołkowicz T., Osek J. (2020b). MLST-based genetic relatedness of Campylobacter jejuni isolated from chickens and humans in Poland. PLoS One 15:e0226238. 10.1371/journal.pone.0226238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windiasti G., Feng J., Ma L., Hu Y., Hakeem M. J., Amoako K., et al. (2019). Investigating the synergistic antimicrobial effect of carvacrol and zinc oxide nanoparticles against Campylobacter jejuni. Food Control 96 39–46. 10.1016/j.foodcont.2018.08.028 [DOI] [Google Scholar]
- Wysok B., Wojtacka J., Kivisto R. (2020). Pathogenicity of Campylobacter strains of poultry and human origin from Poland. Intern. J. Food Microbiol. 334:108830. 10.1016/j.ijfoodmicro.2020.108830 [DOI] [PubMed] [Google Scholar]
- Xu N., Wang G., Leng Y., Dong X., Chen F., Xing Q. (2018). Sulbactam enhances the in vitro activity of sitafloxacin against extensively-drug resistant Acinetobacter baumannii. Exp. Therap. Med. 16 3485–3491. 10.3892/etm.2018.6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe S., Higuchi W., Takano T., Razvina O., Iwao Y., Isobe H., et al. (2010). In vitro susceptibility to antimicrobial agents and ultrastructural characteristics related to swimming motility and drug action in Campylobacter jejuni and C. coli. J. Infect. Chemother. 16 174–185. 10.1007/s10156-010-0040-1 [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Asakura M., Tsukamoto T., Faruque S. M., Deb R., Ramamurthy T. (2006). Cytolethal distending toxin (cdt): genetic diversity, structure and role in diarrheal disease. Toxin Rev. 25 61–88. 10.1080/15569540500320938 [DOI] [Google Scholar]
- Yamazaki-Matsune W., Taguchi M., Seto K., Kawahara R., Kawatsu K., Kumeda Y., et al. (2007). Development of a multiplex PCR assay for identification of Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis subsp. hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis. J. Med. Microbiol. 56 1467–1473. [DOI] [PubMed] [Google Scholar]
- Zhang L., Li Y., Shao Y., Hu Y., Lou H., Chen X., et al. (2020). Molecular characterization and antibiotic resistant profiles of Campylobacter species isolated from poultry and diarrheal patients in Southeastern China 2017-2019. Front. Microbiol. 11:1244. 10.3389/fmicb.2020.01244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.