ABSTRACT
Accumulated studies have revealed the critical role of long non-coding RNAs (lncRNAs) in the carcinogenesis and progression of various cancers. LncRNA TDRG1 has been reported to exhibit oncogenic potential in some cancers. However, its underlying mechanism regulating breast cancer (BC) remains obscure. QRT-PCR was used to measure the relative expression of mRNAs, and western blot was used to detect protein expression levels. CCK8 and CFSE assays were utilized to testify cell proliferation ability. Flow cytometry assay was used for cell apoptosis ability investigation. Transwell and tube formation assays were implemented to test cell migrating and invasive abilities. Relevant mechanism experiments were implemented to determine the molecular mechanism. TDRG1 was remarkably overexpressed in BC cell lines. TDRG1 knockdown suppressed cell proliferation, migration and invasion, but enhanced BC cell apoptosis. Mechanistically, TDRG1 acted as a miR-214-5p sponge to up-regulate CLIC4 expression. MiR-214-5p inhibition or CLIC4 overexpression could revive the tumor-suppressing effects induced by TDRG1 knockdown. TDRG1 promoted cell proliferation, migration, and invasion in BC, suggesting that TDRG1 could promisingly be a therapeutic target for BC.
KEYWORDS: TDRG1, miR-214-5p, clic4, breast cancer
Introduction
Breast cancer (BC), with a rising incidence rate, is the most common malignant tumor and a major reason for cancer-associated deaths among female during 2019.1 Improved diagnosis methods and clinical management have been utilized in recent years, yet the morbidity and mortality of invasion-associated BC are still increasing and remain unoptimistic.2 Thus, exploring the underlying molecular mechanisms behind the carcinogenesis of BC may contribute to providing novel prediction biomarkers and therapeutic target for BC patients.
Long noncoding RNAs (lncRNAs), characterized by more than 200 nucleotides in length, are a cluster of RNAs in lack of protein-coding potential.3 Increasing studies have discovered diversified biological functions of lncRNAs in physiological processes, including the tumorigenesis of disease.4 LncRNAs could play a significant role in biological activities, such as cell proliferation, apoptosis, migration, and invasion during tumorigenesis.5–7 More importantly, with the close implication in the intricate pathogenesis processes of cancers, various lncRNAs have been identified as valuable prediction biomarker or therapeutic targets for various malignant tumors, such as colon cancer and small-cell lung cancer.8,9 In BC, several lncRNAs have been found as potential therapeutic biomarkers. For instance, Yao et al discovered that lncRNA NONHSAT101069 was dramatically up-regulated in BC specimens and cell lines, and identified it as a potential biomarker in BC for its oncogenic properties.10 Therefore, targeting lncRNAs could provide profound implication for the prediction and treatment of BC.
LncRNA TDRG1 has been proved to exert oncogenic properties in some cancers, such as cervical cancer11 and ovarian cancer.12 Furthermore, TDRG1 depletion significantly suppressed cellular malignant behaviors in cervical cancer and ovarian cancer. However, the molecular mechanisms and functions of TDRG1 in BC remain unstudied. Therefore, we selected TDRG1 as candidate lncRNA.
Competing endogenous RNA (ceRNA) network has revealed a new layer of the crucial molecular role of lncRNAs. LncRNAs can function as ceRNA and counteract the effects of miRNA on the expression and biological functions of downstream target genes.13 For example, lncRNA H19 was found to affect cell growth, migration and invasion by regulating miR-138/SOX4 axis in breast cancer.14 However, whether TDRG1 could serve as a ceRNA to regulate BC progression remains unclear. This study aimed to study whether and how TDRG1 affects the biological progresses of BC.
Material and methods
Cell culture
BC cell lines (BT-549, CAL-148, MCF-7, and MDA-MB-231) and human non-tumor breast cell line (MCF-10A) were acquired from ATCC (Rockville, Maryland) and were cultured continuously in DMEM (Invitrogen, Carlsbad, CA, USA) that contained 10% fetal bovine serum (FBS; Invitrogen) as well as 1% penicillin. All cells were incubated in humid air at 37°C with 5% CO2.
Cell transfection
The designed shRNAs for TDRG1 (sh-TDRG1#1/#2), CLIC4 (sh-CLIC4#1/#2), and their corresponding negative controls (sh-NCs) were severally cloned into vector pLKO.1, and transfected into MDA-MB-231 and MCF-7 cells utilizing Lipofectamine 2000 (Invitrogen). The pcDNA3.1/ClIC4 and the empty pcDNA3.1 vector were accordingly constructed by GenePharma. The miR-214-5p mimics, miR-214-5p inhibitor and corresponding NC mimics, NC inhibitor were synthesized by GenePharma (Shanghai, China). 48 h after transfection with these shRNA constructs or pcDNA3.1/CLIC4, the transfected cells were put in 6-well plates at a ratio of 1:10 for passage. Then, stable transfected cells were selected by puromycin treatment.
QRT-PCR
Total RNA was extracted utilizing TRIzol solution (Invitrogen). RNAs were reversely transcribed into cDNA employing Reverse Transcription Kit (Takara, Dalian, China). QRT‐PCR was conducted in Bio-Rad CFX96 System (Takara) by utilizing SYBR Premix Ex Taq II (Takara). Relative RNA expression level was calculated by 2−ΔΔCt approach and normalized to GAPDH/U6.
Cell counting kit-8 (CCK-8) assay
1 × 103 MDA-MB-231 and MCF-7cells were plated in 96-well plates. Cell vitality was detected utilizing CCK-8 assay (Dojindo, Tokyo, Japan). After cultivation over specific time points, the CCK-8 agent was placed to culture for another 4 h. The absorbance at 450 nm was finally evaluated under a microscope reader (Bio-Tek Instruments, Hopkinton, MA, USA).
Carboxyfluorescein succinimidyl ester (CFSE) assay
CFSE assay was implemented in BC cells utilizing the CellTrace™ CFSE Cell Proliferation Kit protocol (Invitrogen) according to the instructions. After being stained, the cells were placed in 6-well plates with 1.0 × 105 cells in each well and cultivated for 72 h. The subsequent analysis was conducted with a flow cytometer (BD Biosciences, San Jose, CA).
Flow cytometry analysis
After 48 h of transfection, the cells were cultured in 6-well plates, and were washed in PBS (Solarbio, Beijing, China). Annexin V-FITC and PI were employed for double staining. Subsequently, apoptosis rate was assessed using a flow cytometer.
Transwell assay
Transwell assays were finalized with transwell chambers (BD Biosciences, San Jose, CA, USA). For migration assay, the upper compartments were placed with transfected MDA-MB-231 and MCF-7 cells, and the bottom compartments were filled with medium containing 10% FBS. 24 h later, cells were fixed with methanol (Sigma-Aldrich) and stained with crystal violet (Sigma-Aldrich). Finally, cells in five randomly chosen fields were counted using a microscope (Olympus). Invasion assay, with the upper compartments matrigel-coated, and has almost the same procedures with migration assay.
Tube formation assay
Human umbilical vein endothelial cells (HUVECs, Corning Incorporated, N.Y., USA), MCF-7 cells and MDA-MB-231 cells were cultured in DMEM with 10% FBS, separately. The latter two types of cells were centrifuged after 48 h of transfection, and then collected with the supernatant. Matrigel was placed in a 96-well plate and cultured in a 37°C incubator for 30 min. After coagulation, HUVEC suspension and tumor-conditioned medium were co-cultured in the plate coated with matrigel. Tube number was figured; images were acquired from four represent chosen fields with a microscope (Olympus).
Subcellular fractionation
The isolation and purification of cytoplasmic and nuclear RNAs were achieved by the utilization of Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Ontario, Canada). Relative expression levels of TDRG1, GAPDH (cytoplasmic control) and U6 (nuclear control) were detected by qRT-PCR analysis.
Fluorescence in situ hybridization (FISH) assay
LncRNA expression was assessed using FISH Kit (RiboBio, Guangzhou, China). After cleaning and fixation, fresh tissue sections were blocked with prehybridization buffer for 60 min. Then, the sections were incubated with 20 μmol/L fluorescence-conjugated TDRG1 Probe Mix overnight. Besides, FISH preparations were counterstained with Hoechst. Finally, the relative TDRG1 expression level was evaluated by Leica Qwin V3 image analysis software.
Luciferase reporter assays
The wild-type (WT) and mutant (MUT) binding sites of miR-214-5p in TDRG1 sequence or CLIC4 3ʹUTR were sub-cloned into pmirGLO luciferase vectors to construct TDRG1-WT/MUT or CLIC4-WT/MUT, respectively. MCF-7 and MDA-MB-231 cells were co-transfected with the recombinant plasmids with miR-214-5p mimics or NC mimics, severally. 48 h later, dual-luciferase Reporter Assay System (Promega, USA) was used to investigate the luciferase activity.
RNA pull-down assay
The TDRG1-WT, TDRG1-MUT, and NC were biotin-labeled into Biotin TDRG1-WT, Biotin TDRG1-MUT and Biotin NC. Then, the biotin-labeled RNA was incubated for a night with cell lysis. Later, magnetic beads containing streptomycin were added and incubated for 48 h. The pulled-down complex was centrifuged and washed in lysis buffer. Finally, qRT-PCR analysis was adopted to detect relative expression level of miR-214-5p.
RNA immunoprecipitation (RIP)
RIP experiment was performed with EZ-Magna RNA-binding protein immunoprecipitation kit (Millipore). MCF-7 and MDA-MB-231 cells were gathered via centrifugation and then lysed in RIP lysis buffer (Invitrogen). Cell lysates were cultivated with immunoprecipitation in the presence of anti-Ago2 (Millipore) or IgG (Millipore). QRT-PCR analysis was utilized to detect relative enrichment of miR-214-5p, TDRG1, and CLIC4.
Xenograft tumor model
Female nude mice were bought from Shi Laike Company (Shanghai, China). MDA-MB-231 cells stably transfected with sh-NC, sh-TDRG1#1 or sh-TDRG1#1+ pcDNA3.1/CLIC4 were inoculated subcutaneously into the hip back of each mice. Tumor volume measurement was performed every 4 days. Euthanasia was performed on the mice for weight measurement 4 weeks later. This study was approved by the Ethics Committee of Qilu Hospital of Shandong University.
Statistical analysis
Data of triplicate independent biological repeats were manifested as mean ± SD. Data analysis was assessed by means of GraphPad Prism 7 software (Graph-Pad Software, La Jolla, CA, USA). One-way ANOVA analysis and Student’s t-test were utilized for the determination of data difference of two or more groups. P < .05 exhibited statistical significance.
Results
TDRG1 is prominently up-regulated in BC cells and knockdown of TDRG1 impedes malignant behaviors of BC progression in vitro
QRT-PCR was firstly conducted to evaluate TDRG1 expression in BC cell lines (BT-549, CAL-148, MCF-7, and MDA-MB-231) and non-tumor breast epithelial cell line (MCF-10A). It was indicated that the expression level of TDRG1 in BC cell lines was dramatically elevated compared with that in normal breast cell line (Figure 1a). This phenomenon suggested that TDRG1 might play a carcinogenic role in BC progression.
Figure 1.
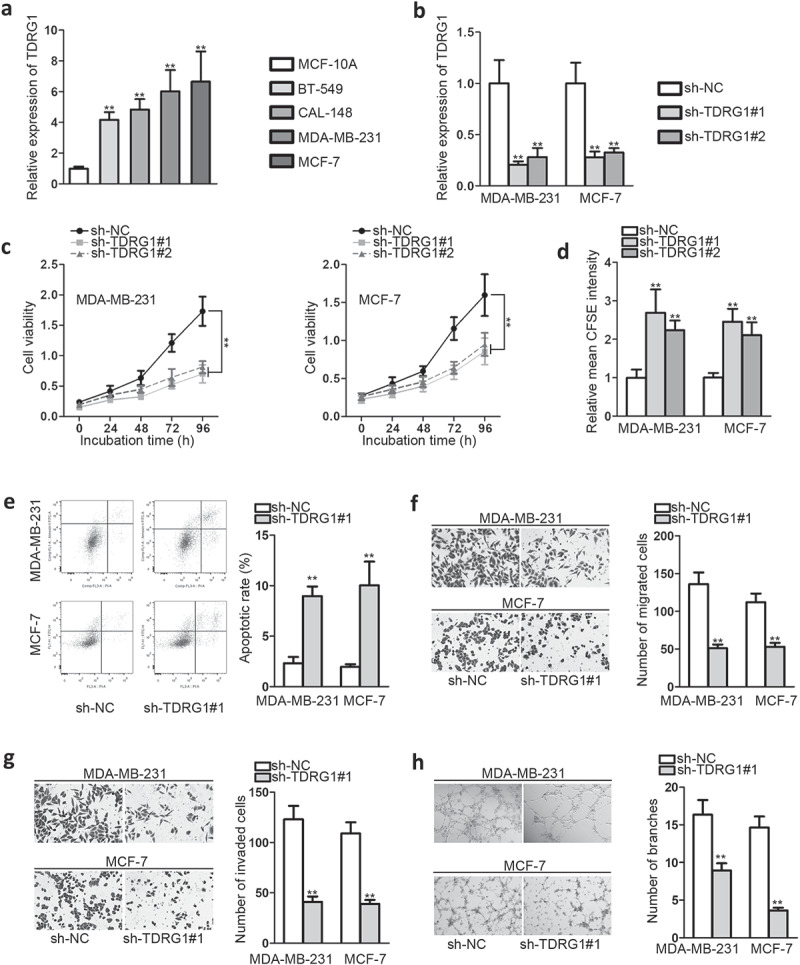
TDRG1 is prominently up-regulated in BC and knockdown of TDRG1 impedes malignant behaviors of BC progression in vitro
A.QRT-PCR analysis was utilized to evaluate the TDRG1 expression level in BC cell lines and normal epithelial cell line (MCF-10A). B. The efficiency of TDRG1 silencing was ensured by qRT-PCR analysis. C-D. CCK8 and CFSE assays were conducted to test cell proliferation ability after knockdown of TDRG1. E. Flow cytometry was utilized to assess apoptosis after knockdown of TDRG1. F-G. Transwell assays were implemented to assess the number of migrated and invaded cells after knockdown of TDRG1. H. Tube formation assay was implemented to detect angiogenesis after knockdown of TDRG1 in HUVECs. **P < .01.
Next, a series of loss-of-function assays were implemented to explore the biological functions of TDRG1 in BC. As shown in Figure 1b, sh-TDRG1#1 and sh-TDRG1#2 were proved to be effective to knock down TDRG1 expression in MDA-MB-231 cells and MCF-7 cells (Figure 1b). On this basis, CCK-8 was conducted and it was shown that knockdown of TDRG1 prominently inhibited BC cell vitality (Figure 1c). Consistently, the relative mean CFSE intensity was significantly enhanced after knockdown of TDRG1, which indicated that sh-TDRG1 weakened cell proliferation ability (Figure 1d). Flow cytometry assay indicated that silencing TDRG1 greatly strengthened BC cell apoptosis capacity (Figure 1e). Transwell assays manifested a significant reduction of migrated and invaded cells after depleting TDRG1, indicating weakened BC migration and invasion abilities (Figure 1(f,figure 1g)). As angiogenesis is a crucial basis for tumor migration, tube formation assay was performed in HUVECs that were cultured in the conditional medium (CM) of MDA-MB-231 and MCF-7 to explore the effects of TDRG1 on angiogenesis. We observed that sh-TDRG1 group showed less tube branches than control group (Figure 1h), which further proved that knockdown of TDRG1 could suppress BC cell migration and invasion abilities.
TDRG1 exerts oncogenic properties in BC via negatively regulating miR-214-5p
CeRNA mechanism implies that cytoplasm lncRNAs may compete with mRNAs for the binding of shared miRNAs, which showcased profound implication in current anti-cancer research.
To determine the possible role of TDRG1 in BC, we began with subcellular fractionation assay to detect its subcellular situation. It was found that TDRG1 was predominantly located in cytoplasm (Figure 2a). Additionally, FISH assay further proved it (Figure 2b). These experiments suggested that the ceRNA mechanical role of TDRG1 in BC seemed possible. Then, we detected five microRNAs with putative binding sites with TDRG1 via starBase (http://starbase.sysu.edu.cn/). Among the five microRNAs, only miR-214-5p expression was significantly elevated after knockdown of TDRG1 from qRT-PCR analysis (Figure 2c). Furthermore, we found that miR-214-5p expression was dramatically down-regulated in BC cells compared within normal cells, which was contrary to TDRG1 expression (Figure 2d). Given these, it was preliminarily judged that miR-214-5p might be negatively regulated by TDRG1. Next, the putative miR-214-5p binding sites with TDRG1 were explored (Figure 2e), and dual luciferase reporter assay was conducted to investigate the physical association. As a result, miR-214-5p mimics could attenuate the luciferase activity of TDRG1-WT, but exert no impact on that of TDRG1-MUT (figure 2f). To further certify this association, RNA pull-down assay was followed, and it manifested that miR-214-5p was markedly enriched by biotinylated TDRG1-WT, which further elucidated the interaction between miR-214-5p and TDRG1 (Figure 2g). Take together, TDRG1 was verified as a sponge of miR-214-5p in BC.
Figure 2.
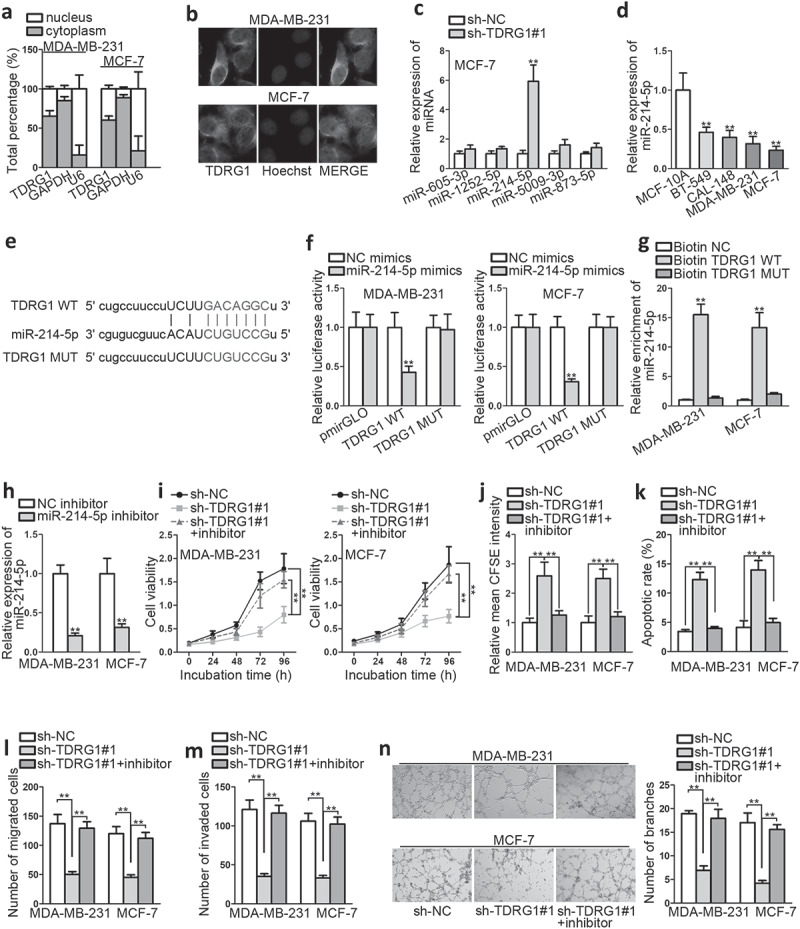
TDRG1 exerts oncogenic properties in BC via negatively regulating miR-214-5p
A-B. Subcellular fractionation and FISH assay were conducted to determine the location of TDRG1. C. QRT-PCR analysis was implemented to investigate miRNA expression when TDRG1 was silenced. D. QRT-PCR analysis was performed to detect the expression profile of miR-214-5p in cell lines. E. The binding sites of miR-214-5p with TDRG1 were predicted. F. Wild and mutant TDRG1 sequences were sub-cloned into luciferase reporter. Luciferase activities were normalized to renilla luciferase. G. RNA pull down was performed to explore the mechanical relationship between miR-214-5p and TDRG1. H. The inhibiting efficiency of miR-214-5p inhibitor was evaluated by qRT-PCR. I-N. Rescue experiments were performed to evaluate the impact of miR-214-5p inhibitor on sh-TDRG1induced BC biological activities. **P < .01.
Next, functional rescue experiments were implemented to explore the impacts of miR-214-5p in TDRG1-regulated BC progression. Transfection of miR-214-5p inhibitor into BC cell lines was proved to down-regulate miR-214-5p (Figure 2h); miR-214-5p inhibition was found to significantly abrogate the molecular effects induced by sh-TDRG1, including cell proliferation, apoptosis, migration, and invasion abilities, as shown in Figure 2(i,figure 2j,figure 2k,figure 2l,figure 2m). Additionally, the angiogenesis ability reduced by silencing TDRG1 was counteracted by miR-214-5p inhibitor in HUVECs (Figure 2n). Thus, we identified miR-214-5p as the direct molecule downstream of TDRG1 in BC development. In addition, TDRG1 exerted oncogenic properties in BC via down-regulating miR-214-5p.
TDRG1 exerted positive effects on CLIC4 expression by down-regulating miR-214-5p
To probe into the downstream target of miR-214-5p, we screened combinable mRNA by searching on starBase and limiting the binding qualifications (clade: mammal; genome: human; assembly: hg19; miRNA: miR-214-5p; program number: 2; Degradome data: medium stringency; CLIP data: high stringency;). As a result, DDX5, CLIC4, MBNL1, SPEN, KCNC4, RPS26, and POMGNT1 were chosen as qualified candidates. After ensuring the overexpression transfection efficiency of miR-214-5p (Figure 3a), we performed qRT-PCR to determine the change of mRNA expression. It was found that CLIC4 depleted much more significantly than other candidate mRNAs in MDA-MB-231 after transfecting with miR-214-5p mimics (Figure 3b). Meanwhile, CLIC4 was remarkably elevated in BC cell lines than in normal ones, just contrary to the situation of miR-214-5p (Figure 3c). Thus, CLIC4 was selected for further study, and its putative binding sites with miR-214-5p were predicted via starBase (Figure 3d). Subsequently, we conducted dual-luciferase reporter assays and observed a decrease in the luciferase activity of CLIC4-WT by co-transfecting miR-214-5p mimics (Figure 3e). Consistently, RIP assay indicated significant enrichment of miR-214-5p, CLIC4 as well as TDRG1 in anti-Ago2 group compared within the IgG control group (figure 3f).
Figure 3.
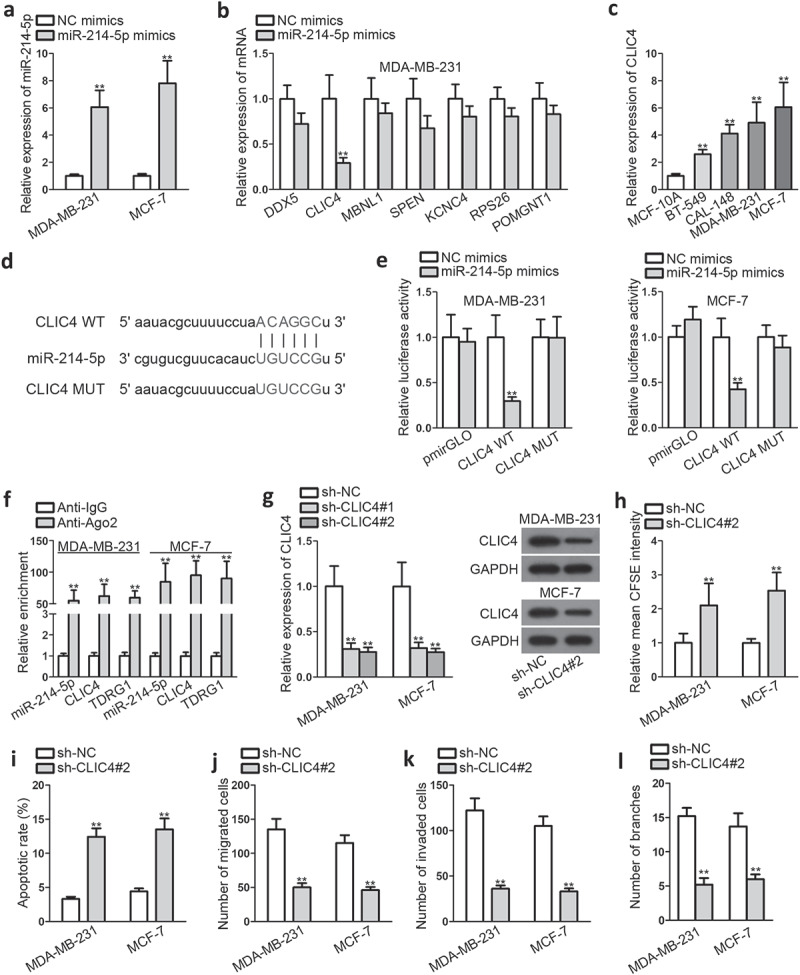
TDRG1 exerts positive effects on CLIC4 expression by negatively regulating miR-214-5p
A. QRT-PCR was utilized to evaluate the transfection efficiency of miR-214-5p mimics plasmid. B. MiRNA candidates’ expression was examined by qRT-PCR analysis after overexpressing miR-214-5p. C. QRT-PCR was performed to evaluate miR-214-5p expression in BC cell lines and normal one. D. Putative binding sites between miR-214-5p and CLIC4 and. E. Wild and mutant CLIC4 sequences were sub-cloned into luciferase reporter, and dual luciferase reporter was utilized to determine the relationship between CLIC4 and miR-214-5p. F. RIP assay was performed to verify the association of miR-214-5p, CLIC4, and TDRG1. G. The inhibiting efficiency of sh-CLIC4 plasmid was examined by qRT-PCR. H-L. Functional experiments were conducted to investigate CLIC4’s role in BC cellular activities. **P < .01.
Though CLIC4 has been found to act as an oncogene in some cancers, such as ovarian cancer,15 its function in BC was first studied here by performing loss-of-function experiments. As the inhibiting transfection efficiency of sh-CLIC4#2 was superior to sh-CLIC4#1 (Figure 3g), it was selected for the functional experiments. The outcomes indicated that knockdown of CLIC4 prominently dampened the proliferation, migration, and invasion capacities of BC cells (Figure 3(h,figure 3k,figure 3l)), but accelerated BC cell apoptosis (Figure 3i). Collectively, these findings demonstrated that CLIC4 played an essential oncogenic role in BC malignant behaviors.
TDRG1 expedites cell proliferation, migration and invasion by regulating miR-214-5p/CLIC4 axis
To further determine whether TDRG1 promotes BC by indirectly regulating CLIC4 through sponging miR-214-5p, a series of rescue experiments were conducted. Firstly, we transfected pcDNA3.1/CLIC4 into BC cells to overexpress CLIC4 (Figure 4a). Then, we studied the effects on CLIC4 mRNA and protein expression induced by sh-TDRG1. QRT-PCR results showed that CLIC4 expression was depleted after TDRG1 knockdown, but elevated again in the presence of CLIC4 overexpression (Figure 4b). Western blot showed the identical level change of CLIC4 protein (Figure 4c). Further, we discovered that CLIC4 up-regulation greatly abrogated the anti-tumor ability of TDRG1 knockdown (Figure 4(d,figure 4e,figure 4f,figure 4g,figure 4h, figure 4i)). For further exploration of the tumor abating effects of sh-TDRG1 in BC, we inoculated the transfected MDA-MB-231 cells into nude mice. The results indicated that knockdown of sh-TDRG1 could slower xenograft tumor growth compared with control group, but the impact was counteracted by overexpressing CLIC4 (Figure 4(j,figure 4k)). Meanwhile, the tumor volume and weight were also decreased in sh-TDRG1 group, but increased by overexpressing CLIC4 (Figure 4(l,figure 4m)). These findings suggested that TDRG1 knockdown could inhibit BC xenograft tumor growth in vivo, further indicating that TDRG1 promoted cell proliferation, migration, and invasion via up-regulating CLIC4 through negative regulation of miR-214-5p in BC.
Figure4.
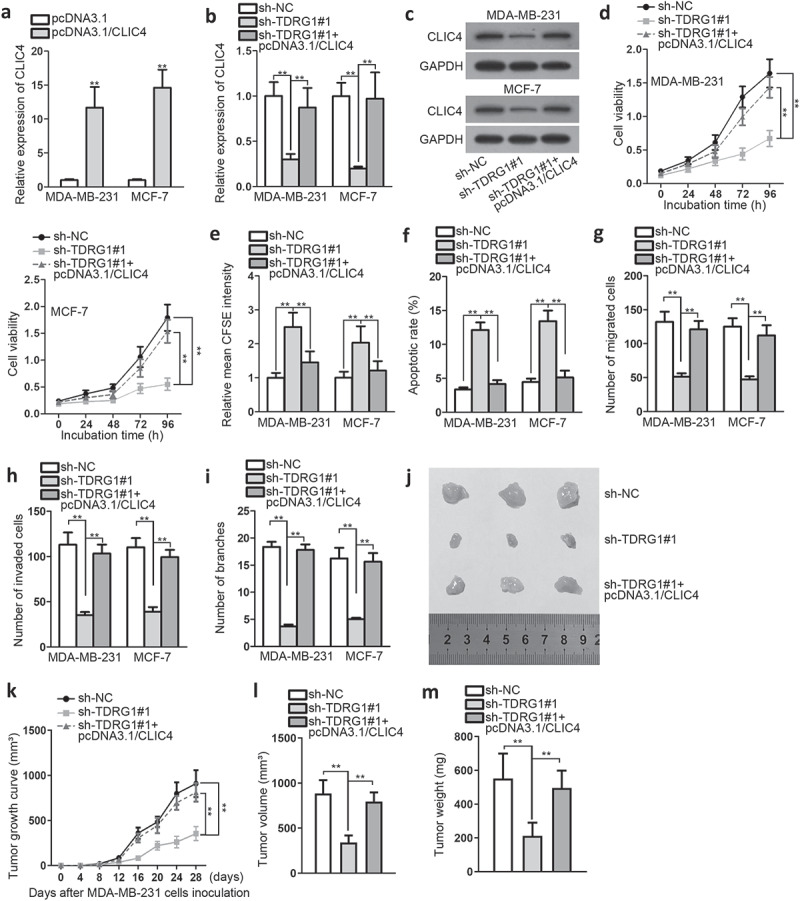
TDRG1 expedites cell proliferation and migration by regulating miR-214-5p/CLIC4 axis
A. QRT-PCR was utilized to determine CLIC4 expression after pcDNA3.1/CLIC4 plasmid transfection. B-C. CLIC4 mRNA expression and protein expression were detected, respectively, by QRT-PCR and western blot. D-I. Functional experiments were conducted to detect the effects of pcDNA3.1/CLIC4 on sh-TDRG1-mediated biological activities in rescue analysis. J. Xenograft tumors shrank after knockdown of TDRG1. K. Xenograft tumor growth curved within four weeks after inoculating MDA-MB-231 cells. L-M. Xenograft tumors were measured and compared in terms of volume and weight. **P < .01.
Discussion
Recently, lncRNAs gained increasing attention for its important role in the carcinogenesis and progression of cancers.16–18 TDRG1, as a novel identified lncRNA, was found differentially expressed in several cancers. Here, we found that TDRG1 was significantly elevated in different BC cells (including triple negative CAL-148, MDA-MB-231, BT-549 cells, and ER-positive MCF-7 cells) than in normal breast cell line, which was the same expression profile as in cervical cancer.19 This phenomenon implies that TDRG1 may have a rolein BC of different types, the confirmation of which needs further research.
Functional experiments manifested that TDRG1 knockdown could not only remarkably suppressed the proliferation, migration, invasion of BC cells, but also hampered HUVECs tube formation. It was suggested that TDRG1 was closely associated with BC progression and acted as an oncogene in BC. Interestingly, this oncogenic property of TDRG1 in BC was consensus with its role in the enhancement of endometrial carcinoma tumorigenicity.20
More and more studies have discovered that cytoplasmic lncRNAs could indirectly regulate mRNA expression levels by acting as competing endogenous RNAs through binding to miRNAs.21–23 In the present study, we found that TDRG1 was mainly situated in the cytoplasm, and identified as a molecular sponge for miR-214-5p in BC through multiple channels. Moreover, we verified that miR-214-5p inhibition notably abrogated the biological impacts mediated by silencing TDRG1 in vitro. TDRG1-regulated BC progression via negatively regulating miR-214-5p.
MiRNAs achieved their cellular function via enhancing translational repression or mRNA degradation of target genes.24,25 MiR-214-5p, whose expression was reported to be abnormal in multiple cancers including prostate cancer and hepatocellular carcinoma,26,27 was found to be down-regulated in BC cell lines in this research. And its target gene CLIC4 was identified and the mechanism was investigated. CLIC4, based on the former studies, is an oncogene in some cancers like head and neck squamous carcinoma.28 Here, we determined the oncogenic property of CLIC4 in BC. It was found that both CLIC4 mRNA and protein levels were decreased after knockdown of TDRG1, supporting the ceRNA role of TDRG1. Rescue experiments manifested that CLIC4 up-regulation could reverse the anti-tumor effects of TDRG1 knockdown, elucidating that CLIC4 exerted effects on TDRG1-mediated biological effects in BC. Thus, it could be concluded that overexpression of CLIC4 aggravated the proliferation, migration, and invasion of BC cells but decreased the apoptosis. Taken together, TDRG1 affected biological progresses of BC via modulating CLIC4by binding to miR-214-5p competitively. As for the downstream of CLIC4, several previous reports give some hints about this. Silencing CLIC4 could promote ATP-induced HNCS cell apoptosis through mitochondrial and endoplasmic reticulum pathways.28CLIC4 expression was negatively linked to cytogenetically normal acute myeloid leukemia progression, as it could induce the NF-kB-dependent activation of HIF (hypoxia-inducible factor) and be involved in tumor growth via its micro-environmental function.29
In a nutshell, the upregulation of TDRG1 expression was found in BC cells. Besides, it was also verified that TDRG1, acting as a molecular sponge for miR-214-5p to regulate CLIC4 indirectly, facilitated cell growth, migration, and invasion of BC. Our data indicated that TDRG1 might serve as a potential novel therapeutic target for BC therapy.
Acknowledgments
We are very grateful to all individuals and groups involved in this study.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (81572587); Youth Program of National Natural Science Foundation of China (81602310); Key R & D projects of Shandong Province (2017GSF18114).
Disclosure statement
No potential conflict of interest was reported by the authors.
Ethical statement
The mice experiment was approved by the Ethics Committee of Qilu Hospital of Shandong University.Funding The present study was supported by the National Natural Science Foundation of China (81572587); Youth Program of National Natural Science Foundation of China (81602310); Key R & D projects of Shandong Province (2017GSF18114).
References
- 1.Ahmad A. Breast cancer statistics: recent trends. Adv Exp Med Biol. 2019;1152:1–7. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz RS, Erban JK, Phimister EG. Timing of metastasis in breast cancer. N Engl J Med. 2017;376:2486–2488. doi: 10.1056/NEJMcibr1701388. [DOI] [PubMed] [Google Scholar]
- 3.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 4.Bouckenheimer J, Assou S, Riquier S, Hou C, Philippe N, Sansac C, Lavabre-Bertrand T, Commes T, Lemaitre JM, Boureux A, et al. Long non-coding RNAs in human early embryonic development and their potential in ART. Hum Reprod Update. 2016;23:19–40. doi: 10.1093/humupd/dmw035. [DOI] [PubMed] [Google Scholar]
- 5.Hua Q, Jin M, Mi B, Xu F, Li T, Zhao L, Liu J, Huang G. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol. 2019;12:91. doi: 10.1186/s13045-019-0773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Gu T, Lu Z, Qiu L, Xiao G, Zhu X, Li F, Yu H, Li G, Liu H. Roles of MYC-targeting long non-coding RNA MINCR in cell cycle regulation and apoptosis in non-small cell lung cancer. Respir Res. 2019;20:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu M, Xu X, Pan B, Chen X, Lin K, Zeng K, Liu X, Xu T, Sun L, Qin J, et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer. 2019;18:135. doi: 10.1186/s12943-019-1063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F, Shen F, Zheng Z, Ruan J. The LncRNA XIRP2-AS1 predicts favorable prognosis in colon cancer. Onco Targets Ther. 2019;12:5767–5778. doi: 10.2147/OTT.S215419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng F, Wang Q, Wang S, Liang S, Huang W, Guo Y, Peng J, Li M, Zhu W, Guo L. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene. 20. 20; 39:293-307. [DOI] [PubMed] [Google Scholar]
- 10.Yao N, Fu Y, Chen L, Liu Z, He J, Zhu Y, Xia T, Wang S. Long non-coding RNA NONHSAT101069 promotes epirubicin resistance, migration, and invasion of breast cancer cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene. 2019;38:7216–7233. doi: 10.1038/s41388-019-0904-5. [DOI] [PubMed] [Google Scholar]
- 11.Jiang H, Liang M, Jiang Y, Zhang T, Mo K, Su S, Wang A, Zhu Y, Huang G, Zhou R. The lncRNA TDRG1 promotes cell proliferation, migration and invasion by targeting miR-326 to regulate MAPK1 expression in cervical cancer. Cancer Cell Int. 2019;19:152. doi: 10.1186/s12935-019-0872-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Wang LL, Sun KX, Xiu YL, Zong ZH, Chen X, Zhao Y. The role of the long non-coding RNA TDRG1 in epithelial ovarian carcinoma tumorigenesis and progression through miR-93/RhoC pathway. Mol Carcinog. 2018;57:225–234. doi: 10.1002/mc.22749. [DOI] [PubMed] [Google Scholar]
- 13.Yan J, Jia Y, Chen H, Chen W, Zhou X. Long non-coding RNA PXN-AS1 suppresses pancreatic cancer progression by acting as a competing endogenous RNA of miR-3064 to upregulate PIP4K2B expression. J Exp Clin Cancer Res: CR. 2019;38:390. doi: 10.1186/s13046-019-1379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Si H, Chen P, Li H, Wang X. Long non-coding RNA H19 regulates cell growth and metastasis via miR-138 in breast cancer. Am J Transl Res. 2019;11:3213–3225. [PMC free article] [PubMed] [Google Scholar]
- 15.Singha B, Harper SL, Goldman AR, Bitler BG, Aird KM, Borowsky ME, Cadungog MG, Liu Q, Zhang R, Jean S, et al. CLIC1 and CLIC4 complement CA125 as a diagnostic biomarker panel for all subtypes of epithelial ovarian cancer. Sci Rep. 2018;8:14725. doi: 10.1038/s41598-018-32885-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniue K, Kurimoto A, Sugimasa H, Nasu E, Takeda Y, Iwasaki K, Nagashima T, Okada-Hatakeyama M, Oyama M, Kozuka-Hata H, et al. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc Natl Acad Sci U S A. 2016;113:1273–1278. doi: 10.1073/pnas.1500992113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C, Wang Y, Wang Y, Zhang S, Yu L, Guo C, Xu H. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene. 2017;36:5392–5406. doi: 10.1038/onc.2017.133. [DOI] [PubMed] [Google Scholar]
- 18.Wu XS, Wang F, Li HF, Hu YP, Jiang L, Zhang F, Li ML, Wang XA, Jin YP, Zhang YJ, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18:1837–1853. doi: 10.15252/embr.201744147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H, Hu GM, Wang WL, Wang ZH, Fang Y, Liu YL. LncRNA TDRG1 functions as an oncogene in cervical cancer through sponging miR-330-5p to modulate ELK1 expression. Eur Rev Med Pharmacol Sci. 2019;23:7295–7306. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Wang LL, Sun KX, Liu Y, Guan X, Zong ZH, Zhao Y. LncRNA TDRG1 enhances tumorigenicity in endometrial carcinoma by binding and targeting VEGF-A protein. Biochimica Et Biophysica Acta Molr Basis Dis. 2018;1864:3013–3021. doi: 10.1016/j.bbadis.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng C, Guo K, Chen B, Wen Y, Xu Y. miR-214-5p inhibits human prostate cancer proliferation and migration through regulating CRMP5. Cancer Biomarkers: Section A Dis Markers. 2019;26:193–202. doi: 10.3233/CBM-190128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elemeery MN, Badr AN, Mohamed MA, Ghareeb DA. Validation of a serum microRNA panel as biomarkers for early diagnosis of hepatocellular carcinoma post-hepatitis C infection in Egyptian patients. World J Gastroenterol. 2017;23:3864–3875. doi: 10.3748/wjg.v23.i21.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue H, Lu J, Yuan R, Liu J, Liu Y, Wu K, Wu J, Du J, Shen B. Knockdown of CLIC4 enhances ATP-induced HN4 cell apoptosis through mitochondrial and endoplasmic reticulum pathways. Cell Biosci. 2016;6:5. doi: 10.1186/s13578-016-0070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang S, Huang Z, Chen P, Feng C. Aberrant chloride intracellular channel 4 expression is associated with adverse outcome in cytogenetically normal acute myeloid leukemia. Front Oncol. 2020;10:1648. doi: 10.3389/fonc.2020.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]


