Abstract
Epsilonproteobacteria are a diverse class of eubacteria within the Proteobacteria phylum that includes environmental sulfur-reducing bacteria and the human pathogens, Campylobacter jejuni and Helicobacter pylori. These pathogens infect and proliferate within the gastrointestinal tracts of multiple animal hosts, including humans, and cause a variety of disease outcomes. While infection of these hosts provides nutrients for the pathogenic Epsilonproteobacteria, many hosts have evolved a variety of strategies to either sequester metals from the invading pathogen or exploit the toxicity of metals and drive their accumulation as an antimicrobial strategy. As a result, C. jejuni and H. pylori have developed mechanisms to sense changes in metal availability and regulate their physiology in order to respond to either metal limitation or accumulation. In this review, we will discuss the challenges of metal availability at the host–pathogen interface during infection with C. jejuni and H. pylori and describe what is currently known about how these organisms alter their gene expression and/or deploy bacterial virulence factors in response to these environments.
Keywords: Epsilonproteobacteria, metal, pathogen, Campylobacter, Helicobacter
Graphical Abstract
Graphical Abstract.
This comprehensive review of metal acquisition systems and corresponding regulatory mechanisms provides insight into the importance of nutrient metal homeostasis to the survival of pathogenic Epsilonproteobacteria.
Introduction
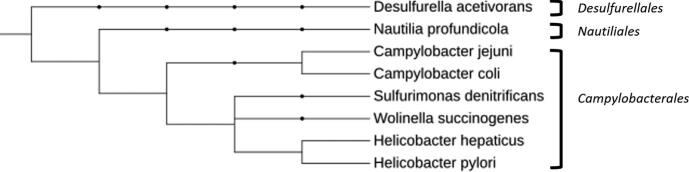
Epsilonproteobacteria are a diverse group of flagellated microorganisms that have evolved into two distinct subgroups based on the environmental niche they inhabit.1 The first subgroup, Nautiliales, includes oceanic thermophiles that are estimated to comprise 66–98% of the microorganisms inhabiting hydrothermal ocean vents and the surrounding area.2 The second subgroup, and the focus of this review, consists of the clinically important Epsilonproteobacteria, Campylobacter spp. and Helicobacter spp., both of which have far-reaching impacts on global health (Fig. 1). Interestingly, Helicobacter pylori was initially considered to be a member of the Campylobacter genus,3 but has since become recognized as a unique genus. Despite the genetic relatedness of Campylobacter and Helicobacter, and the ability of both to cause gastrointestinal disease, the pathogens have diverged to colonize different niches within the gastrointestinal tract and may encounter differing host responses. To combat these responses and successfully colonize, both Campylobacter and Helicobacter have evolved mechanisms to maintain appropriate levels of transition metals from the host environment with some systems being shared and others differing between these two pathogens.
Fig. 1.
Phylogeny of Epsilonproteobacteria. An example phylogenetic tree of Epsilonproteobacteria with example members from the Campylobacterales and Nautiliales orders. A Deltaproteobacteria (Desulfurella acetivorans) has been included as an example from another class. Campylobacter and Helicobacter belong to the same order, but branch into different families with different host niches. Adapted from Waite et al.233 and visualized with iTol.234
Campylobacter spp. are one of the most prevalent causes of bacterial-derived gastroenteritis worldwide4,5 with Campylobacter jejuni being responsible for ∼90% of human campylobacteriosis cases and Campylobacter coli being responsible for ∼10%.6 As a result, C. jejuni is responsible for up to 15% of global cases of diarrhea with an estimated 1.3 million annual cases in the USA.5,7,8 In the developed world, Campylobacter spp. are most often acquired through the consumption of improperly prepared or cross-contaminated food9 primarily due to Campylobacter’s ability to asymptomatically reside within the gastrointestinal tract of agricultural animals, including chickens, cows, pigs, and sheep.10–12 In contrast, the most common source of infection in the developing world is believed to be contaminated water.13 Following ingestion of relatively few viable Campylobacter cells, the organism adheres to the intestinal mucosa and causes inflammation and inhibition of fluid reabsorption.14–17 The pathogen has also been shown to invade host epithelial cells and can survive intracellularly, but campylobacters are generally considered extracellular pathogens18 and are treated as such in colonization studies described in this review, unless otherwise stated. Colonization of a susceptible host typically results in a mild-to-moderate, self-limiting diarrhea that may contain blood, though several persistent post-infectious disorders can occur, including Guillain–Barré syndrome, irritable bowel syndrome, and reactive arthritis.8 Due to this prevalence and severity of infection, both the Centers for Disease Control and Prevention and the World Health Organization have declared Campylobacter a severe threat to global health.9
The Helicobacter group contains several members, including gastric and non-gastric bacteria, but the most impactful, and a focus of this review, is the pathogen H. pylori.19 Similar to C. jejuni, instances of infection and persistent colonization are greater in developing countries, with an estimated 80% of individuals being persistently colonized.19,20 Colonization typically occurs at an early age and persists indefinitely with H. pylori becoming a primary constituent of the gastric microbiota unless the individual undergoes antibiotic treatment.21 Nearly all individuals infected with H. pylori will experience chronic gastritis; however, a small proportion will develop more severe disease such as peptic and duodenal ulcers, neoplasia, hyperplasia, dysplasia, B-cell lymphoma of mucosa-associated lymphoid tissue, and invasive gastric cancer.21 While animal hosts serve as the primary reservoir for Campylobacter leading to human disease, H. pylori lacks an environmental host and is instead thought to be passed between individuals through direct contact.19 Studies have demonstrated the potential of H. pylori to survive intracellularly in host gastric cells, but it was initially considered an extracellular pathogen22 and the systems addressed in this review have been described in extracellular H. pylori unless otherwise stated. Helicobacter pylori is the dominant prokaryote in the gastric niche, and the gastric environment provides numerous nutritional opportunities and challenges, including changes in osmolarity, low pH, innate immune antimicrobial onslaught, and alterations in metal cation concentrations.23–26
Transition metals are vital for all living cells as these ions participate in a variety of critical biological processes, including incorporation into metalloproteins required for respiration, DNA replication, and central metabolism.27–29 To limit growth and dissemination of invading pathogens, the host employs several strategies to bind and sequester essential metals like iron, nickel, and zinc with host proteins like S100A12 in a method termed nutritional immunity.30 In contrast, the host can also use localized increases in metals like copper and bismuth as an antimicrobial strategy due to the toxic effects of elevated metal concentrations on invading pathogens.31 To combat host nutritional immunity or accumulation of metals at antimicrobial levels, C. jejuni and H. pylori have evolved mechanisms to sense changes in metal availability and subsequently alter their physiology to maintain the homeostasis of various metals when invading a susceptible host. The purpose of this review is to briefly examine what is known about the metal challenges at the host–pathogen interface during Campylobacter and Helicobacter infection and what strategies and regulatory systems these bacteria use to counteract those pressures.
Iron
Iron serves a vital role in microbial growth as it is a required cofactor for many enzymes, a catalyst in electron transport processes, and necessary for cellular processes such as the citric acid cycle, DNA synthesis, respiration, and electron transfer.32,33 In addition to the contribution of iron to growth, it can lead to the generation of reactive oxygen species in the presence of oxygen via the Haber–Weiss reaction, which can damage proteins, DNA, and lipids.34–36 To avoid the generation of oxygen-derived free radicals37 while maintaining iron for essential cellular processes, both animal hosts and bacteria have evolved mechanisms to tightly regulate iron concentrations. Due to nutritional immunity, iron levels within the host are also maintained well below the levels necessary for growth of Gram-negative bacteria.38 For example, iron-binding proteins in the intestine are capable of sequestering free ferric iron to a concentration of 10−24 M, while most bacteria require at least 10−7 M iron for normal cell functions.39 As a result of this host sequestration by lactoferrin and other proteins40 and to avoid toxic levels of iron, Epsilonproteobacteria have evolved numerous mechanisms for the efficient uptake and regulation of cellular iron levels (Tables 1 and 2).
Table 1.
Acquisition and efflux mechanisms and regulation of these systems in Campylobacter. List of acquisition systems and the corresponding regulatory mechanism if known for each metal in Campylobacter. The asterisk denotes transport systems or regulatory mechanisms that have been experimentally tested in vitro and superscript A denotes systems investigated in vivo, while those with no superscript have been bioinformatically predicted or have not been experimentally examined
| Metal | Intracellular condition | Transport systems (Campylobacter) | References | Regulation (Campylobacter) | References |
|---|---|---|---|---|---|
| Iron | High | *Fur*PerR | 70, 71 70 | ||
| Low | *AChuZABCD— heme*ACtuA/CfbpABC—transferrin*CfrA/CeuBCDE—FeEnt*AFeoB—ferrous iron*P19 | 32, 50, 51 50, 54, 58 53, 50, 65 47 66, 67 | *Fur*AHeuR | 70, 71 8 | |
| Zinc | High | Zinc conc. | 101, 105 | ||
| Low | *AZnuABC | 101 | PerRFur | 101 101 | |
| Copper | High | *CeuO—detox*CzcD—efflux | 137 55 | *CosR/TatA1Copper conc. | 138 66 |
| Low | *P19 | 66 | |||
| Nickel | High | *Nickel conc. | 152 | ||
| Low | *NikZYXWV | 152, 158, 160 | *HydABCD | 151, 160 | |
| Cadmium | High | *CzcABCD | 194, 195 | *ACadmium conc. | 189 |
| *Cj0612c | 190 | ||||
| Low | |||||
| Cobalt, magnesium, and manganese | High | CzcD | 55 | Cobalt conc. | 201 |
| Low | MntABC | 105 | |||
| Bismuth, molybdenum, and tungsten | High | *ABismuth conc. | 219 | ||
| Low | *ModABC*TupABC | 223 222, 223 | *ModE | 223, 224 |
Table 2.
Acquisition and efflux mechanisms and regulation of these systems in Helicobacter. List of acquisition systems and the corresponding regulatory mechanisms if known for each metal in Helicobacter. The asterisk denotes transport systems or regulatory mechanisms that have been experimentally tested in vitro and superscript A denotes systems investigated in vivo, while those with no superscript have been bioinformatically predicted or have not been experimentally examined
| Metal | Intracellular condition | Transport systems (Helicobacter) | References | Regulation (Helicobacter) | References |
|---|---|---|---|---|---|
| Iron | High | *AFur | 84, 85, 86 | ||
| Pfr | 83 | ||||
| Low | *AFeoB—ferrous iron | 80 | *AFur | 84, 85, 86 | |
| FecAB—ferric dicitrate | 81 | ||||
| FrpB—FeEnt | 82 | ||||
| Zinc | High | CadA—effluxCzcB—efflux*ACznABC—efflux | 118 118 117 | *Zinc conc. | 25, 114 |
| Low | *Fur | 83 | |||
| Copper | High | *CopAP—efflux*CrdAB—efflux*CzcAB—efflux | 118 118 | *CopP*CrdRS*Fur | 144 143 83 |
| Low | |||||
| Nickel | High | *Hpn—sequestration*HspA—sequestration*ACznABC—efflux | 174 174 117 | *ANikRNickel conc. | 175, 176 173 |
| Low | *ANiuBDE | 183 | |||
| Cadmium | High | *CadA—ATPaseCznABC—efflux*Hpn—sequestration*HspA—sequestration | 196, 197 117 174 174 | *Cadmium conc. | 196, 117, 174 |
| Low | |||||
| Cobalt, magnesium, and manganese | High | *CadA | 196 | *PfrCobalt conc. | 218 196 |
| Low | *FecDE*CeuE*NiuBDE*NixA*CorA | 182 182 183 213 217 | |||
| Bismuth, molybdenum, and tungsten | High | *Bismuth conc. | 229 | ||
| Low | ModD | 230 |
Campylobacter
Of the metals discussed in this review, the metalloregulation of iron in Campylobacter has been studied most extensively due to the canonical roles of iron in cellular processes such as DNA synthesis and electron transfer. In addition to the general requirement of iron as a cofactor for metalloenzymes described in other organisms, iron has also been implicated in acid stress tolerance in C. jejuni41 and modulation of chemotaxis through Tlp2, both of which impact colonization potential.42 As previously mentioned, since free iron concentrations are restricted in animal hosts due to nutritional immunity, Campylobacter relies on outcompeting hosts for iron or utilizing iron-associated host molecules as a nutrient source.32 In the case of C. jejuni, several iron acquisition systems have been identified and characterized, including the use of free iron, heme, iron-containing glycoproteins, and siderophores.43–45
Free iron transporters allow pathogens to transport iron encountered during colonization.46 Like the ferrous iron transporter identified in Escherichia coli, FeoAB in Campylobacter transports free ferrous iron across the periplasmic space and into the cytoplasm, especially under iron-limited conditions.47 The FeoAB system is conserved across C. jejuni strains and is important for the persistence of C. jejuni after invasion into intestinal cells.47 Interestingly, differences in colonization potential of feoB mutants in C. jejuni strains 11168, 43431, and 81–176 were observed in a chick colonization model, indicating potential differences in iron transport efficiency between strains encoding the same ferrous iron transport system.47
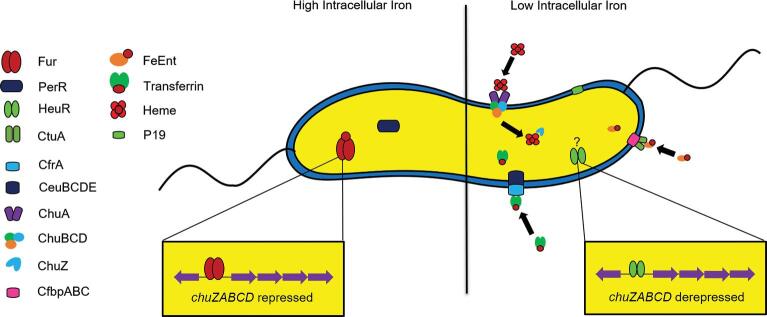
The majority of iron in animal hosts is complexed within heme, a molecule consisting of a porphyrin ring with a centrally located iron atom.48,49 In order for a bacterium to use the iron contained within the heme complex, the heme must be transported into the cytosol via a membrane transport complex.50 In C. jejuni, heme transport is thought to rely on the ChuZABCD system.32 Upon initial binding by the heme receptor, ChuA, heme is translocated into the periplasm in a TonB-dependent manner.45 The ChuBCD transporter then facilitates ATP-dependent movement of heme into the cytosol, but studies indicate transport can still occur without a functional ChuBCD.50,51 Finally, hydrolysis of the porphyrin ring by the heme oxygenase (ChuZ) occurs to release the iron.8,50 Previous transcriptomic work demonstrated chuZABCD was upregulated during colonization of the chicken cecum,52 but no differences in colonization by a chuA mutant were observed, indicating that Campylobacter uses methods other than heme acquisition to obtain iron within the avian host.8,53–55 Expression of chuA increased over 1000-fold during human infection studies, supporting a role for heme as an important source of iron during colonization of a human host.56
The use of host-derived iron-containing glycoproteins, like lactoferrin and transferrin, as iron sources has been described for several pathogens, including those in the Pseudomonadaceae, Pasteurellaceae, Neisseriaceae, and Moraxellaceae families.57–59 It was initially believed that Campylobacter was unable to utilize these glycoproteins,58 but it has since been demonstrated that Campylobacter possesses a transferrin receptor (CtuA) and transporter (CfbpABC), which allows for the use of transferrin as an iron source under iron-limited conditions50,54,58 (Fig. 2). A ctuA mutant exhibited a significant decrease in colonization potential in chickens60 and a somewhat reduced colonization potential in a rabbit model when compared to wild-type C. jejuni,54,60,61 with transcription of both CtuA and CfbpABC upregulated in an iron-limited environment,33,55 indicating transferrin as a potential source of iron during infection.
Fig. 2.
Metalloregulatory mechanisms expressed during high and low intracellular iron concentrations in Campylobacter. High levels of intracellular iron induce activity of the holo (iron-bound) form of transcriptional regulators Fur and PerR. Low levels of intracellular iron result in the apo form of these regulators and the derepression of genes previously repressed. Low iron also induces increased expression of transporters for the uptake of iron molecules, including ferric–enterobactin (CtuA), transferrin (CeuBCDE/CfrA), and heme (ChuZABCD). Positive regulators of these systems, such as HeuR, also become active under iron-limited conditions.
Fungi and Gram-negative bacteria commonly utilize an iron sequestration process that involves the energy-dependent transport of iron chelators, termed siderophores.62 Importantly, Campylobacter does not encode genes to synthesize siderophores, but it can use exogenous siderophores produced by other microorganisms including those produced by the host gastrointestinal microbiome.32,38 For example, C. jejuni has the ability to take up the siderophore enterobactin (Ent)53,63 using ferric–enterobactin (FeEnt) uptake systems.32,64 To transport FeEnt across the outer membrane, Campylobacter encodes either one or both of the TonB-dependent receptors, CfrA53 and CfrB.65Campylobacter jejuni then uses the CeuBCDE transport system to translocate FeEnt into the cytosol, where it is hydrolyzed through an unknown mechanism releasing iron into a usable form50 (Fig. 2). In addition, a high-affinity iron transporter protein (P19) and a co-localized iron transporter (CJJ81176_1649–1655) are induced during periods of iron limitation and required for optimal growth in media supplemented with human fecal extracts, suggesting a dependence on these transporters in a nutrient-limited environment like the human gut.66,67 Together, these systems represent conserved and effective strategies used by Campylobacter to acquire iron and infect susceptible hosts.38,60,65,68,69
Iron homeostasis must be tightly regulated to ensure adequate amounts are present for biological processes while avoiding toxic levels. As a result, many iron acquisition systems in Campylobacter are controlled by both positive and negative regulators. To prevent excess intracellular iron and respond to oxidative stress, C. jejuni possesses two iron-activated members of the ferric uptake regulator (Fur) family of transcriptional regulators, Fur and PerR (Fig. 2). These regulators are responsible for controlling iron homeostasis and the response to peroxide stress, respectively.70Campylobacter jejuni Fur is homologous to those of other Gram-negative organisms, functioning primarily as an iron-responsive transcriptional regulator and repressing transcription of iron uptake systems when intracellular levels of iron are high; however, C. jejuni Fur can also act as a transcriptional activator and in an iron-dependent manner.7,71 Interestingly, the role of PerR as a sensor for oxidative stress in Campylobacter is unique among Gram-negative organisms because it uses intracellular iron levels to detect oxidative stress.70 Since C. jejuni is a microaerophile, survival under aerobic conditions necessitates the concerted control of oxidative stress response systems,72 including iron cofactored superoxide dismutase and peroxidases, and heme cofactored catalase.72 Thus, the importance of Fur and PerR regulation, directly, indirectly, or co-dependently, is not limited to nutrient iron acquisition, but to environmental persistence and survival.47,60,72–75
Transcriptomic studies have determined the number of genes controlled by Fur or PerR can be as high as 200–300,70,67 which suggests iron homeostasis is particularly important in Campylobacter.32,60,76 Additionally, transcriptomic analysis of a C. jejuni strain mutated for a putative regulatory protein, subsequently termed HeuR (heme utilization regulator), identified the protein as a positive regulator of chuZABCD expression and other iron acquisition systems8 (Fig. 2). This mutant exhibited decreased colonization of the chicken gut when compared to wild-type C. jejuni and was unable to utilize heme as a source of iron in an iron-restricted environment.8 Since heme utilization has been shown not to be involved in chicken colonization, the reduction in colonization potential may be due to HeuR regulating other iron homeostasis systems; however, interactions between HeuR and Fur, which negatively regulates chuZABCD promoter activity, are currently unknown.32,60,70,76
Helicobacter
Helicobacter pylori also has a strict nutritional requirement for iron.77 Within the gastric niche, H. pylori competes with the vertebrate host for dietary iron and has evolved the ability to utilize alternate sources of nutrient iron, including hemoglobin, heme, transferrin, and lactoferrin.78,79 Because of this nutritional flexibility and changes in iron availability within the gastrointestinal tract, H. pylori has evolved elegant strategies to sense and respond to changes in iron concentrations. Helicobacter pylori encodes several iron transporters, including a homolog of the ferrous iron transporter FeoB found in Campylobacter and E. coli, although no homolog to FeoA has been identified.80 Expression of a functional FeoB has been found to be important for successful murine colonization by H. pylori and FeoB mutants possess less cell-associated iron.80 Additionally, H. pylori encodes a fecAB homolog, which may allow for the import of ferric dicitrate molecules, but homologs of the regulatory components found in E. coli are not present in H. pylori.81 The FrpB outer membrane receptor is also present in H. pylori, which may allow for binding and transport of enterobactin during iron limitation.82 In addition to these iron uptake systems, H. pylori contains the ferritin-like protein, Pfr, which helps prevent toxic iron accumulation by sequestering intracellular iron.83
Similar to Fur described in Campylobacter, H. pylori Fur exists in a holo-iron-bound form (holo-Fur) or in an unbound apo-form (apo-Fur) that imparts flexible functionality as a global regulator for gene expression in response to minor changes in iron availability.84–86Helicobacter pylori holo-Fur has been demonstrated to repress and activate gene transcription and apo-Fur has the capacity to regulate gene expression as well.84–86 Upon encountering iron ions, Fur dimers bind to the target sequence within the promoter, effectively repressing transcription of the downstream genes. Many of these genes are involved in iron acquisition and include high-affinity iron transporters such as fecA1, fecA2, frpB1, frpB2, frpB4, feoB, fecD, yaeE, and the exbB–exbD–tonB operon.82,87,88
In addition to its role regulating central metabolism and iron acquisition, Fur also regulates a variety of virulence factors to promote pathogenesis. For example, holo-Fur has been shown to directly activate expression of genes involved in motility such as fliY, flgK, flaB, and cheA.89 Additionally, Fur has been implicated in regulating the cytotoxin-associated gene, cagA, which encodes an oncogenic cytotoxin. Secretion of CagA by the cag-type IV secretion system (cag-T4SS) into host gastric cells results in a variety of changes in host cell signaling, morphology, and immunomodulation.90,91 Translocation of CagA into host cells perturbs host cell polarity and apical release of transferrin, turning the cell surface into a replicative niche for H. pylori.91,92 Interestingly, conditions of low iron availability induce production of cag-T4SS at the host–pathogen interface and enhance the activity of the cag-T4SS.93–95 The importance of iron and Fur in vivo has been studied using the Mongolian gerbil model of chronic H. pylori infection. Results indicate that Fur is required for full colonization of the gerbil stomach at early points in infection.96 Furthermore, interrogation of the Mongolian gerbil model of infection under conditions of varying iron concentration intake reveal that low dietary iron consumption exacerbates H. pylori-dependent carcinogenesis, a result that was not seen in iron-replete animals.93
Zinc
Zinc is an essential trace element necessary for various cellular functions in bacteria, including the proper structure and functioning of multiple metalloproteins involved in DNA replication, transcription, and other enzymatic processes.97–100 To maintain appropriate levels of intracellular zinc, many bacteria employ both zinc uptake and efflux systems that aid in the ability of the organism to acquire exogenous zinc from the environment while preventing the accumulation of zinc to toxic levels.101 For bacterial pathogens, zinc acquisition is often necessary to compete against residents of the host microbiota and allow for colonization of host tissues during infection.54,98,101,102 Susceptible hosts can also use zinc toxicity as a weapon against invading pathogens by preventing the bacterial cell from taking up other necessary metals during pathogenesis.103 As a result, high levels of intracellular zinc must be avoided to prevent zinc outcompeting other necessary metals within the active sites of metalloproteins.101
Vertebrate hosts use both intracellular and extracellular strategies to limit access to zinc by invading pathogens.104 Intracellular examples include the use of transporters to move zinc from the lysosome into the cytoplasm,105 as well as intracellular sequestration of zinc by metallothionein,106 to inhibit zinc-dependent bacterial processes. Hosts also use extracellular strategies, like S100 proteins, to sequester zinc and other metals making them unavailable to bacterial pathogens during infection.101,105,107 These molecules can also serve as immune signaling molecules for the recruitment of neutrophils and monocytes during gastrointestinal infection.104 In humans, the majority of zinc absorption occurs in the small intestines, severely limiting zinc availability to gastrointestinal pathogens like Campylobacter.101 To combat host defenses and maintain zinc levels, both Campylobacter and Helicobacter have evolved several metalloregulatory mechanisms (Tables 1 and 2).
Campylobacter
To survive and proliferate in the zinc-limited environments within the host, Campylobacter uses efficient zinc uptake systems, including the ZnuABC transporter.98,101,105,108 This system belongs to the ATP-binding cassette (ABC) family of transporters, with ZnuA serving as a periplasmic metallochaperone, ZnuB as the membrane permease, and ZnuC as the ATPase subunit.101,109,110 ZnuA from C. jejuni has been shown to bind an average of 3.56 atoms of zinc per molecule and was required for successful colonization of chickens by C. jejuni.101 Interestingly, it was subsequently demonstrated that ZnuA is not required for colonization of germ-free chicks, indicating the primary role of this system in chickens is to compete with the intestinal microbiota for zinc.98 ZnuA was also found to be important for growth when competing for zinc in the presence of host-derived sequestration proteins such as S100A12.104 As with other metals, an overabundance of zinc is toxic to Campylobacter, so the organism uses the conserved ZntA efflux protein to export excess zinc and other metals, keeping concentrations within the tight intracellular range necessary for survival.111
To maintain appropriate levels of zinc, the bacterium must also have regulatory mechanisms to detect and control expression of these uptake systems. Escherichia coli and other bacteria encode the zinc uptake regulator (Zur) that belongs to the Fur family of metalloregulatory proteins, to repress znuABC transcription. Though znuABC in C. jejuni was found to be upregulated in low-zinc environments,101,105 BLAST analysis indicates that Campylobacter lacks a Zur homolog. Due to this absence, it has been suggested that znuABC is cross-regulated by PerR, Fur, or another unidentified regulator.101 Support for PerR regulating znuABC comes from crystallization studies of C. jejuni PerR, where zinc and manganese binding were observed near the PerR active site, but direct interaction of PerR with the znuABC promoter has not been investigated.112 In addition, Campylobacter encodes a putative cobalt–zinc–cadmium efflux protein, CzcD, which has been found to be important for Campylobacter colonization of the murine gut,55 although regulation of these systems beyond response to presence of metals is currently unknown.
Helicobacter
Helicobacter pylori has a strict nutritional requirement for zinc in order to grow and proliferate.77 During infection, H. pylori encounters decreased zinc concentrations within the inflamed gastric mucosa as a result of sequestration by host-derived S100A family proteins.113–116 Conversely, humans consume large quantities of transition metals such as zinc in their diet, so it is likely that H. pylori is exposed to micromolar concentrations of this ion in the lumen of the stomach.117 To persist in the gastric environment, H. pylori has evolved molecular mechanisms to sense and adapt to these wide variations in zinc concentration and availability.
While no zinc-specific import systems have been identified, several efflux systems have been studied in H. pylori. A homolog of the CzcABC efflux pump is encoded by H. pylori, although CzcC is absent and two copies of CzcB are present in the genome,118 aiding in the prevention of toxic levels by exporting excess zinc. Another similar efflux system for zinc detoxification is CadA, which also prevents zinc accumulation by efflux of cobalt, cadmium, and zinc.118 Similar to these two export systems, cznABC in H. pylori encodes a third efflux system to export zinc.117 This transporter effluxes excess cadmium, zinc, and nickel as necessary, and is required for successful colonization of a host as demonstrated in the Mongolian gerbil model.117 These systems function together to prevent toxic effects of metal accumulation, but the regulatory mechanisms of each beyond metal binding have not been studied.
Zinc regulates H. pylori virulence in several ways.113,114,119 Conditions where zinc availability is low, such as those imposed by the presence of calgranulin proteins including S100A12, lead to repression of activity and assembly of major virulence factors such as the cag-T4SS.113,114 Additionally, conditions of low zinc availability imposed by calgranulin proteins (such as calprotectin) also induce lipid modification, biofilm formation, and increased resistance to calgranulin antimicrobial activity.119
Although the molecular mechanism of these virulence properties remains unknown, H. pylori has evolved several ways to respond to varying conditions of zinc availability. Chelation of zinc can result in disruption of the Fur dimeric structure into monomers without associated zinc, impacting the ability of Fur to act in a regulatory manner.120,121 Iron starvation, as well as medium supplementation with zinc, results in repressed synthesis of ferritin (at the mRNA level) in the wild-type strain but not in the H. pylori fur mutant, indicating zinc-associated fur-dependent regulation of H. pylori metal homeostasis occurs, and that Fur is a global metal-dependent regulator.83
Copper
The majority of environmental copper (80%) is found in shale and mining sites, with humans consuming an average of 2–4 mg of copper per day through diet and copper plumbing.122 Copper is important to bacterial growth and pathogens will use host copper as a metal cofactor for enzymes involved in iron uptake and maintenance of the cellular oxidative state.123,124 In addition to the use of copper in metalloenzymes, high levels of copper have been used for biocontrol due to its toxicity.125 This toxicity may be the result of Fenton-like reactions that produce hydroxyl radicals126 or the displacement of other metals, such as iron–sulfur complexes by copper ions.127 As a result, not only is copper sequestered by the host to limit colonization, it may also be used in excess to combat bacterial pathogens.128 Copper in a human host is primarily localized to the brain, kidneys, and liver, and is quickly sequestered by ceruloplasmin to maintain homeostatic levels before excretion in bile.122,128 Innate immune cells, such as macrophages, load their phagosomes with up to millimolar concentrations of copper to intoxicate bacteria.129–131 The host also deploys copper-binding innate immune proteins such as S100A12, which can bind and sequester copper as an antimicrobial strategy.114 As a result of copper requirements for growth, but the potential for toxicity, the ability to tightly regulate levels of copper is a requirement of bacterial pathogens, including Campylobacter and Helicobacter (Tables 1 and 2).
Campylobacter
Copper may serve to facilitate iron uptake and sense osmotic stress in Campylobacter123 by acting as an iron-binding protein cofactor, increasing oxygen uptake via oxidation of periplasmic copper, and efflux of copper out of the cell under iron limitation.66,123,124 However, copper can quickly become toxic at higher concentrations as previously described. Animal studies have demonstrated that supplementation of pig feed with exogenous copper sulfate and carbadox decreased shedding of Campylobacter132 and biofilm formation decreased on a surface containing copper with an ∼2-fold decrease in viable cells (CFU/ml).133 These results indicate that increased levels of exogenous copper can be detrimental to Campylobacter growth and that there is potential therapeutic value for copper combating Campylobacter infection. As such, regulating cellular copper levels is likely important to Campylobacter physiology.
The periplasmic protein P19 (mentioned above) also aids in the ability of Campylobacter to use copper and prevent toxicity.66,134 Similar to iron chelation studies conducted with wild-type and P19 mutants, chelation of copper with bathocuproine disulfonic acid under iron-replete conditions resulted in decreased growth of both wild-type and mutant strains,66 indicating a potential role of copper in iron uptake by Campylobacter. In addition, copper also regulates intracellular oxygen via the cytochrome c oxidase complex.123 A homolog of E. coli CueO, the multicopper oxidase in C. jejuni exhibits similar activity to CueO by coupling oxidation of copper-containing substrates to the reduction of oxygen to water.123,124Campylobacter jejuni CueO acts in conjunction with a CopA homolog to remove and detoxify copper from Cu+ in the cytoplasm to Cu2+ in the periplasm.123,124,135,136 While regulation of these genes has not been investigated in depth, the twin arginine translocase TatA1 was shown to be required for correct periplasmic localization of Campylobacter CueO137 and the Campylobacter oxidative stress regulator (CosR) has been implicated as a negative regulator of cueO in a CosR knockdown study138; however, much is still unknown regarding the exact mechanisms regulating these systems.
Helicobacter
Helicobacter pylori encounters micromolar concentrations of copper in the gastric niche, largely derived from the host diet and present at the lumen of the stomach.139 Copper has been shown to influence bacterial cellular movement and acts as a chemotactic repellent for H. pylori motility.140,141 Copper also promotes H. pylori colonization of the rodent stomach, indicating this transition metal is important at the host–pathogen interface.140 Additionally, copper is required for cellular respiration and is a critical cofactor in the terminal oxidase in the H. pylori respiratory chain, a cbb3-type cytochrome oxidase that possesses a heme–copper binuclear center similar to the cytochrome aa3-type oxidases.142
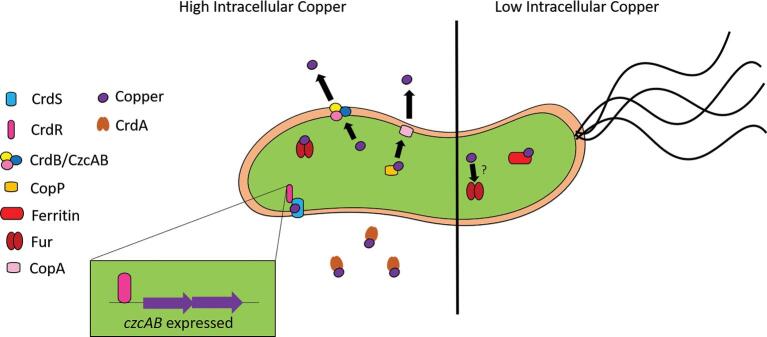
Copper induces the expression of numerous H. pylori genes involved in ion transport, including exbD, crcB, HP1516, HP0733, and the crdAB, czcAB, and copAP operons.118 The exbD gene encodes the energetic component of the iron and nickel transport system (ExbB–ExbD–TonB). Helicobacter pylori employs a two-component system crdRS that encodes a sensor kinase (CrdS) and a response regulator (CrdR) that comprise a phosphorelay that is activated in the presence of copper.143 CrdR induces the expression of crdAB and czcAB operons that encode copper resistance determinants such as secreted copper-binding molecule CrdA, and a copper efflux pump comprised of CrdB, CzcB, and CzcA (Fig. 3). Because evading copper intoxication is clearly a critical strategy for H. pylori, it also has functional redundancy with numerous copper export systems, such as the copAP operon, which encodes a cytoplasmic copper-binding regulator (CopP) that is orthologous to CopZ from E. coli. CopP promotes expression of copA, which encodes a protein that promotes H. pylori resistance to copper.144 Copper exposure alters expression of genes implicated in several metabolic pathways, such as inducing expression of trpA that encodes tryptophan synthase and hpylM that encodes a DNA methyltransferase.118
Fig. 3.
Metalloregulatory mechanisms expressed during high and low intracellular copper concentrations in Helicobacter. High levels of intracellular copper induce activity of the response regulators CrdR, CopP, and Fur. The induction of czcAB expression by CrdR is shown as an example of transcriptional changes in response to copper. Increased levels of copper induce the expression of the CrdB and CzcAB efflux system, as well as the CopA efflux system, and the secretion of the copper-binding protein CrdA to help maintain homeostatic levels of copper. In low levels of intracellular copper, ferritin binds available copper, in addition to other metals, but this sequestration is reversed when high levels of copper are available.
Helicobacter pylori has evolved elegant mechanisms to sense and respond to copper levels and deploy copper detoxification strategies.117,143Helicobacter pylori alters its transcriptional profile in response to copper exposure by reducing the expression of the pfr gene that encodes the metal storage protein, ferritin. Interestingly, this copper-induced downregulation of ferritin is Fur dependent, indicating Fur is a global metal stress regulator in H. pylori.83 In addition to ferritin, numerous additional genes are regulated by copper exposure, including those involved in ion transport, cellular motility, transferase activity, and metabolic flexibility. Copper exposure also results in the upregulation of fliS that encodes a putative flagellin chaperone.118 Additionally, these transcriptional changes may be the indirect result of copper displacing iron ions, as observed in E. coli.127 Thus, copper can influence H. pylori motility at both the chemotactic and transcriptional levels.
Nickel
Nickel is widespread throughout the natural environment at very low levels.145 Small amounts of nickel (3–4 nM) are required for cellular processes in bacteria and humans, but due to the ability of nickel to displace other metal cofactors, much research has focused on the toxic effects of nickel.145–148 The majority of host nickel is located in organs, such as the lungs and adrenal glands, at incredibly low concentrations (<5 ppm).147,149,150 These low levels are maintained in part because dietary nickel is either eliminated in the feces or excreted in the urine.147,150 The host can also utilize metal sequestering molecules, such as S100A8/A9, to bind residual nickel and aid in further limitation of available nickel to invading bacterial pathogens.149,151 Due to both the necessity of nickel and its potential toxicity, sensitive transport and regulatory systems are vital to bacterial survival during infection (Tables 1 and 2).
Campylobacter
Nickel is essential for various cellular functions in Campylobacter. For example, nickel is involved in hydrogenase activity,152 with C. jejuni [Ni–Fe]-hydrogenase catalyzing the reversible oxidation of H2.153 It was previously thought that Campylobacter did not possess a nickel transporter like the nikABCDE transport system in E. coli, and instead used enzymes dependent on nickel cofactors,154,155 but the nikZYXWV (Cj1584c–Cj1580c) gene cluster encoding an outer membrane transporter has since been described in C. jejuni.152 The periplasmic binding protein component of this system (NikZ) is similar to NikA of E. coli,152,156,157 but they exhibit drastically different binding specificities. While purified Campylobacter NikZ was shown to only bind free or uncomplexed nickel, NikA can bind complexed forms including Ni chelated with EDTA.152,158–160 Once inside the periplasm, nickel is bound by the HydABCD complex (Cj1267–Cj1264 in 11168) resulting in the oxidation of hydrogen to release protons and facilitate the proton-motive redox loop with the uptake of protons during reduction of menaquinone.152,160
Since HydABCD activity is dependent on nickel availability in the periplasm,155 its function is impacted by the abundance of the Nik protein. For example, NikZ can be detected in the periplasm of C. jejuni cells under normal conditions, but following supplementation with exogenous nickel, NikZ was unable to be detected in the periplasm, suggesting that nikZYXWV expression is controlled by nickel availability.152 Most strikingly, when grown under the low-nickel conditions like those Campylobacter may encounter in a host, mutation of NikZ resulted in an ∼22-fold decrease in cell-associated nickel and hydrogenase activity was lost. Despite this mutation, hydrogenase activity could be restored when supplemented with exogenous nickel indicating that additional nickel transporters may exist.152
Regulation of these systems beyond the impacts of nickel concentrations either is poorly understood or has not been investigated. Several nickel-responsive regulators such as the Fur-like transcription factor, Nur, and the nik gene regulator, NikR, have been identified in other Gram-negative organisms, but homologs of these regulators either do not exist or are not widely distributed throughout the Campylobacter genus and have regulatory abilities that have not been investigated.152,161 In the 13826 strain of Campylobacter concisus, a putative NikR (13826_0355) was identified based on genome sequence, but the ability of this proposed NikR homolog to regulate the NikZYXWV system has not been investigated.162
Helicobacter
Similar to Campylobacter, H. pylori requires nickel for certain cellular processes like virulence, but also can be intoxicated by this transition metal.163 Two critical H. pylori enzymes require nickel as a cofactor: urease and [Ni–Fe]-hydrogenase. Both urease and [Ni–Fe]-hydrogenase require the protein HypB for the metalation-dependent maturation of these enzymes.164 Urease is an essential colonization factor that facilitates H. pylori survival in the acidic conditions encountered in the lumen of the gastric mucosa.163Helicobacter pylori urease is a nickel-containing enzyme that catalyzes the hydrolysis of urea to ammonium carbonate.165–168Helicobacter pylori requires nickel for the full maturation of the urease complex.169–172 Specifically, expression of urease genes is increased in response to nickel exposure. This likely evolved to repress expression when the critical cofactor for this enzyme is low.173
In conditions of nickel stress or toxicity, H. pylori expresses the Hpn and HspA proteins that participate in metal storage and protect the cells from higher concentrations of external metal concentrations, including nickel.174 Together, these residues bind metal ions with high affinity, to sequester them within the cytoplasm of the cell and overcome the challenge of metal toxicity. Nickel exposure also induces expression of the cznABC loci that encode components of a putative nickel efflux pump.117 This CznABC transport system is critical for circumnavigating nickel stress and is required for colonization of the vertebrate host, indicating nickel toxicity is a challenge in the gastric niche.117
In addition to Fur-dependent regulation of urease gene expression in response to nickel, the dominant transcriptional regulator associated with nickel response is NikR. NikR can bind nickel or copper with picomolar affinity, in what is likely a conserved square-planar site.175 NikR is autoregulatory, binding to the promoter sequence upstream of the gene that encodes it in the presence of nickel.175,176 Comprehensive mapping of the NikR regulon by RNA-seq and ChIP-seq indicates 194 differentially expressed genes between the wild-type and nikR mutant strains, but <50 differentially regulated in response to nickel between these two strains (20 were identified by RNA-seq and 46 by ChIP-seq), indicating NikR regulates a variety of genes in a nickel-independent fashion.177
Holo-NikR (the nickel-bound form of NikR) represses HcpC, HopV, and HopW, which are all outer membrane or secreted proteins involved in host–pathogen interactions.178,179 Additionally, holo-NikR represses expression of the DvnA-RS03305 and MccABC toxin–antitoxin systems, Fur, and the ScoB and AtoE proteins involved in metabolic processes.179 Interestingly, holo-NikR induces expression of urease subunits, which utilize nickel as a cofactor for activity. In addition to urease, the other critical enzyme regulated by holo-NikR is the [Ni–Fe]-hydrogenase. [Ni–Fe]-hydrogenase is a nickel-containing enzyme that enables H. pylori to utilize hydrogen gas within the gastric niche as an energy source.164 NikR has also been implicated in regulating biofilm formation by H. pylori as mutational inactivation of nikR, fur, and the acid-responsive two-component regulatory system encoding ArsRS in double and triple mutants enhances biofilm formation.180 Interestingly, NikR has been implicated as a critical regulator of virulence and isogenic nikR mutants exhibit a colonization defect in rodent models of H. pylori infection.181
Other genes that are regulated by NikR in response to nickel stimulation include FecDE, CeuE, FrpB2, FrpB3, and FecA3, which are involved in metal ion transport and are repressed by holo-NikR.177 It is interesting to note that the FrpB4/FrpB2, FecDE ABC transporter, CeuE transporter, and a TonB1–ExbB–ExbD system contribute to nickel acquisition as well as iron acquisition, underscoring the promiscuity of these critical transporters.182 Furthermore, NixA, a nickel permease required for nickel transport from the periplasm into the cytoplasm, is repressed by holo-NikR.177 Recently, a novel H. pylori nickel transport system, NiuBDE, was discovered that participates in nickel acquisition and nickel-dependent urease activation and is essential for colonization of the mouse stomach.183
Cadmium
Cadmium is a non-essential trace metal that can occur naturally,184,185 although industrial cadmium is ubiquitous in the environment due to industrial contamination.186 At elevated concentrations, cadmium can be toxic due to its ability to replace other divalent cationic metals, such as zinc and calcium.186,187 In both animals and humans, a role for cadmium has not been identified and cadmium has been shown to induce cancer.188 As a result, it is necessary for both hosts and pathogens to tightly regulate the amount of cadmium to prevent toxicity (Tables 1 and 2).
Campylobacter
While it does not appear that Campylobacter uses cadmium for any biological process, cadmium has been shown to reduce Campylobacter colonization of the murine gut189 and cadmium chloride susceptibility was proposed as a means to differentiate Campylobacter spp. from other enteric pathogens.189 Levels as low as 0.1 mM cadmium chloride significantly decreased growth of C. jejuni, with concentrations of 1 mM required to eliminate viability.190 Supplementation of media with either blood or fluid thioglycolate rescued cells from these toxic effects.191 The basis for cadmium toxicity in Campylobacter is not well understood, but several mechanisms for decreased cadmium toxicity have been proposed, including increased intracellular Zn2+ concentrations, decreased Cd2+ uptake, increased expression of metallothionein (a protein that sequesters cadmium), and binding of cadmium ions by other metal sequestering proteins.190,192,193 A change in extracellular cadmium concentrations on a micromolar scale is enough to elicit a bacterial response, where transcripts involved in metal uptake such as bacterioferritin (cj1534c) and an ATP-binding protein (cj1663) were downregulated in the presence of cadmium, while those involved in metal storage like ferritin (cj0612c) were upregulated.190 The CzcABCD efflux system may also help promote cadmium resistance in Campylobacter, similar to the role of cadmium efflux pumps identified in other organisms.194,195 However, the mechanisms regulating these systems are currently unknown.
Helicobacter
Cadmium is not nutritionally required for H. pylori growth,77 but similar to C. jejuni, H. pylori is quite sensitive to cadmium (Cd2+) toxicity. Helicobacter pylori mitigates cadmium stress by elaborating a putative transition metal ATPase, CadA,196,197 and a metal efflux pump, CznABC,117 which aid in resistance to Cd2+. Additionally, Hpn and HspA proteins from H. pylori participate in metal storage and protect the cells from higher concentrations of external metal ions like cadmium.174
Cobalt, magnesium, and manganese
Campylobacter
Cobalt is a required component of the B12 coenzyme complex, which plays a role in methyl group transfer and can interact with RNA regulatory elements to impact gene expression.154,198 There is little known regarding the import of cobalt in Campylobacter, but a putative cobalt–zinc–cadmium efflux system, CzcD,55 and a cytoplasmic transport protein, CorA,152,199,200 with the potential to bind cobalt have been identified in the C. jejuni genome. Cobalt has been investigated as an antimicrobial coating with visible light-activated cobalt nanoparticles exhibiting bactericidal activity against C. jejuni and other Gram-negative pathogens, which opens the door for its use as a future antimicrobial surface treatment.201
Magnesium is necessary for colonization of the host gut by Campylobacter, but the majority of magnesium within the host is either sequestered in bone or complexed with nucleic acids.202 Therefore, Campylobacter has developed methods to acquire non-sequestered magnesium from the host.203 CorA is the primary transporter of magnesium in C. jejuni, which is a homolog of CorA transporters in other bacteria, including E. coli.203 While little is known about the mechanism of magnesium transport, CorA mutants have been shown to require exogenous magnesium when grown under magnesium-deplete conditions, but these mutants have not been studied in vivo.203 Additionally, CorA is thought to transport iron by acting as a low-affinity transporter under conditions of low magnesium.204 Despite the requirement of magnesium for growth, the introduction of elevated levels of exogenous magnesium oxide to the environment is detrimental to Campylobacter more than most other bacteria since it is particularly sensitive to membrane damage.205
Manganese is a transition metal used by Campylobacter during colonization for the proper functioning of multiple proteins, including pyruvate kinases and catalases,105 and is also predicted to be necessary for PerR activity during periods of iron limitation.76 In humans, the majority of manganese is acquired through the diet and is present in the gastrointestinal tract.206,207 Because of the importance of this metal for both host and bacterial cellular functions, humans employ several methods of manganese limitation, including sequestration by S100A8/A9208 and removal of manganese from the phagosome by natural resistance-associated macrophage protein 1.207,209 To compete with these host mechanisms, Campylobacter uses the MntABC transporter, which is similar to the ZnuABC zinc transporter described previously.105 A regulatory role for PerR in MntABC expression has been identified in Neisseria gonorrhoeae,210 but the role of PerR in mntABC regulation in Campylobacter has not been investigated. Beyond MntABC, other manganese-specific transporters have yet to be identified, although other metal transport systems and binding proteins can interact with manganese nonspecifically.105,200,207,211
Helicobacter
Cobalt is a micronutrient that is nonessential for H. pylori growth,77 but is required for the activity of important enzymes such as arginase. Arginase is a metalloenzyme that hydrolyzes arginine to ornithine and urea. Most arginases are binuclear and utilize Mn2+ as a cofactor; however, the H. pylori arginase enzyme is selective for cobalt.212 Cobalt acquisition is mediated in Helicobacter spp. by the FecDE ABC transporter, CeuE transporter, and a TonB1 ortholog.182 NiuBDE also transports cobalt into the bacterial cell183 and NixA has been implicated in cobalt uptake.213 Additionally, cobalt competes with nickel in metal acquisition pathways indicating cobalt and nickel transporters are promiscuous for these two transition metals.214 In addition to being an important cofactor in enzymes, cobalt can be bactericidal against H. pylori.214 In response to cobalt exposure, H. pylori deploys efflux determinants such as CadD. Plasmid-based expression of cadA in wild-type or cadA mutant strains of H. pylori enhanced resistance to cobalt (Co(II)).196
Magnesium is a strict nutritional requirement for H. pylori growth and viability77 and is essential for H. pylori colonization in the porcine model of colonization and disease.215 Interestingly, systemic magnesium levels are significantly lower in H. pylori-infected individuals compared to uninfected individuals.216 The uptake of magnesium in H. pylori is mediated by the transporter CorA,217 which is essential for the viability of H. pylori. Manganese is another transition metal that is dispensable for H. pylori growth,77 but can be toxic to H. pylori cells. Interestingly, inactivation of the gene pfr that encodes H. pylori ferritin results in enhanced sensitivity to manganese toxicity, indicating metal storage is critical for H. pylori to overcome metal toxicity.218
Bismuth, molybdate/molybdenum, and tungstate/tungsten
Campylobacter
While bismuth has been used for many years to treat stomach ulcers by reducing Helicobacter colonization in humans, little is understood about the effects on Campylobacter.219 However, bismuth has recently been shown to reduce the ability of Campylobacter to colonize the chicken gut.219 Upon dosing chickens with either 0, 50, or 200 ppm, it was determined bismuth administered at 200 ppm for 10 days lowered concentrations of C. jejuni in the chicken gut,219 but the specific mechanisms by which bismuth affects Campylobacter have yet to be determined.
Campylobacter is exceptional among other microorganisms in that it encodes cofactor proteins and transporters for both molybdenum and tungsten.50 Molybdenum and tungsten are transition metals of similar atomic conformations and present in the environment in the usable forms of molybdate and tungstate,220 which can be used interchangeably, though the relative abundance of molybdenum is higher in relation to tungsten.221,222 In Campylobacter, molybdenum and tungsten are transported by ModABC and TupABC transporters, respectively.50,223 Interestingly, the ModA transporter can transport either metal while TupA is believed to be tungsten-specific, since TupA-like transporters in other organisms exhibit high specificity into the picomolar range.222 Expression of these transport systems in Campylobacter is controlled by a regulatory protein similar to ModE in other microbes.50,223 Due to the lack of a specific metal-binding domain, it is thought that Campylobacter ModE regulates both metal uptake systems based on protein–protein interactions, rather than direct interaction of ModE and molybdate or tungstate.50,224
After transport, the respective metals are used by either the molybdenum cofactor (Moco), which is involved in nitrate reductase activity, or the tungsten cofactor (Woco), which acts as a cofactor for formate dehydrogenase during energy metabolism.50 In addition, several other enzymatic cofactors utilize molybdenum, including the periplasmic nitrate reductase NapA.222,225 While the use of tungsten is less understood, it has been shown to be required for proper functioning of formate dehydrogenase50 and can inhibit some molybdenum enzymatic cofactors, though the mechanism remains to be elucidated.222
Helicobacter
Bismuth is considered a common first-line therapy for patients with penicillin allergies suffering from H. pylori infection.226 Bismuth, in combination with other antimicrobial compounds such as rabeprazole, cefuroxime, levofloxacin, esomeprazole, or clarithromycin, is effective at eradicating H. pylori in chronically infected patients.226,227 Standard triple therapy in combination with bismuth results in complete eradication of H. pylori in 90% of patients.228 Bismuth is not required as a nutrient for H. pylori growth,77 but can be toxic to H. pylori cells, underscoring its importance as an antimicrobial therapy. Bismuth can inactivate important transcriptional regulators required for H. pylori colonization and pathogenesis, such as NikR.229
Molybdenum is a rare element essential to nearly all organisms that catalyzes two-electron-transfer oxidation–reduction reactions. Growth analyses demonstrate that molybdenum is dispensable for H. pylori growth and viability.77 However, some H. pylori strains encode a putative ABC transporter, ModD, involved in molybdenum transport from the periplasm into the cytosol.230 Phylogenetic analyses of H. pylori strains from East Asian, Amerindian, European, and African individuals have revealed that all known functions associated with molybdenum appeared to be inactivated in East Asian strains, further underscoring the dispensability of this metal ion within the genus.
Tungsten is not required for H. pylori growth and viability.77 However, tungsten in the form of polyoxotungstates exhibits potent antibacterial activity against H. pylori, an effect that was superior to metronidazole and clarithromycin, both standard therapies for this pathogen.231 Additionally, polyoxotungstates were effective against metronidazole-resistant and clarithromycin-resistant strains of H. pylori, indicating the potential of these molecules as metal-based therapeutics.
Conclusions
Iron
The importance of iron acquisition for survival of bacterial pathogens like Campylobacter and Helicobacter is evidenced by the array of iron uptake systems present in these organisms. Ranging from free iron transport systems to those transporting iron-bound siderophores produced by other bacteria, pathogenic Epsilonproteobacteria can obtain iron from various sources and in numerous forms. The ability to use iron in various forms is reflective of the specific niches occupied by Campylobacter and Helicobacter. With the ability to survive in environmental niches ranging from aquatic sources to the human gut, Campylobacter must employ numerous systems to obtain iron from a variety of potential sources. Similarly, Helicobacter uses multiple uptake systems, but within a very specific environment in the gastric niche. The induction of host responses, such as inflammation during infection, may generate a preference for certain iron acquisition systems over others, such as those that can bind and take up heme from blood released into the lumen during Campylobacter infection. Additionally, iron abundance in the environment and varying levels of oxidative stress encountered during colonization likely impact preferences for certain regulatory systems over others.
Understanding the molecular mechanisms and conditions impacting the regulation of these iron uptake systems can contribute significantly to our understanding of pathogenic Epsilonproteobacteria physiology. Previous studies have indicated a vital role for the global response regulators Fur and PerR, in the transcriptional response of Campylobacter and Helicobacter iron transport systems in response to environmental levels of iron. While these global regulators are important for regulation of virulence factors during infection, other regulators identified recently, such as HeuR in Campylobacter, are worth investigating further. The abundance of iron acquisition systems in these pathogens and the ability of some systems to bind and transport other metals indicate the potential for previously unidentified or uncharacterized regulatory mechanisms to work in conjunction with global regulators like Fur. By focusing on these potential regulatory systems beyond Fur and PerR, we can gain a better understanding of the complex interactions of iron acquisition systems and the ability of Campylobacter and Helicobacter to successfully colonize susceptible hosts.
Zinc
Competition for zinc within the host gut during infection is of interest, as the gut is a major site of zinc absorption in humans. Due to this absorption by the host and sequestration by the native microflora, pathogens must be able to compete with the host and the host microbiome to scavenge available zinc existing at a very low abundance. These environmental pressures encountered in the host have resulted in the ability of Campylobacter and Helicobacter to use efficient zinc acquisition and transport systems to help combat zinc sequestration by host-derived molecules such as S100A8/A9. In addition to zinc acquisition, investigation of these systems demonstrated how some may be able to transport iron or other essential metal nutrients as well, allowing the pathogens to circumvent host nutritional immunity based on metal availability.
Understanding how these pathogens can combat host defenses and sequester zinc is increasingly important due to the employment of zinc treatment as a nutritional supplement in the developing world. Supplementation of zinc in the diet of humans in developing countries may lead to an increase in available zinc levels in the gastric and intestinal niches, resulting in heightened host susceptibility to infection by Helicobacter or Campylobacter, respectively. While some zinc uptake systems have been characterized in C. jejuni, such as ZnuABC, the promiscuity of transport systems for multiple metals and limited known regulatory mechanisms have led to knowledge gaps in the field. Helicobacter differs from Campylobacter as no zinc importers have been identified, but several zinc efflux systems are important to combat potential zinc toxicity in the gastric niche. However, regulation of these systems is not understood beyond the global response to zinc availability. Because of this, it is worth determining whether zinc transport occurs in a hierarchical manner and what changes occur in the response of transport systems under differing conditions, and which systems are most important, in addition to gaining a better understanding of the regulatory mechanisms that control these systems. This would provide insight into potential methods for prevention and treatment, specifically within susceptible hosts residing in the developing world.
Copper
Unlike iron that requires high intracellular concentrations before reaching deleterious levels, copper can quickly become toxic due to the production of reactive radicals or protein mis-metallation.232 Prevention of copper accumulation may explain the pairing of iron- and copper-binding sites within a single transporter. This dual binding in certain pumps can facilitate the efflux of copper while maintaining homeostatic levels of iron; however, this may lead to problems if mutations occur within these efflux systems and intracellular copper accumulates. A better understanding of how these efflux pumps function in copper homeostasis and how copper competes with other nutrient metals for binding sites could lead to exploitation of these mechanisms to combat infection due to the toxic effects of copper accumulation.
Work investigating the global impact of copper on genes in Helicobacter has been done, but research into understanding the effects of copper on Campylobacter gene expression is lacking. The impact of copper on Campylobacter has not been a major focus of metalloregulatory work, but using available knowledge on copper toxicity and the impact on Helicobacter virulence to inform future research can lead to a better understanding of how Campylobacter regulates copper levels via finely tuned transport mechanisms. We can also gain a better understanding of the interplay between copper and other metals and how the availability of these metals during colonization can impact transcription of virulence genes within invading pathogens.
Nickel
Although nickel is necessary for various cellular functions within Campylobacter and Helicobacter, little is known regarding regulation of nickel transport and utilization by these organisms when compared to the dearth of knowledge available for other metals like iron. While it has been demonstrated that Campylobacter requires nickel, a nickel-specific transport system similar to the NikABCDE system in E. coli and H. pylori was only recently described in Campylobacter. While nickel-specific transporters have been identified for both organisms, the promiscuity of nickel binding is not well understood beyond studies indirectly confirming nickel may bind to other metal transporters. Additionally, the toxic potential of nickel based on the ability of nickel to bind to other enzymatic metal-binding sites is worth noting. As seen in other mechanisms involving transition metals such as zinc, metal regulatory systems in Epsilonproteobacteria can often bind more than one type of metal. This lack of specificity may be important to combat host nutritional immunity and compete with the native microflora, so investigating these identified mechanisms further and characterizing other proposed systems would provide useful insights into the ability of pathogens to regulate metal transport through potentially interconnected systems. The impact of nickel on other metal transport systems is worth investigating as novel systems that have yet to be identified may exist in these organisms, allowing for the ability to successfully evade host nutritional immunity in previously uncharacterized ways.
Cadmium
Cadmium is unique among the transition metals described in this review because there is no biological requirement for intracellular cadmium in Epsilonproteobacteria and toxic effects are often experienced by cells due to cadmium accumulation. As a result, these organisms have evolved several methods for dealing with exogenous sources of cadmium, although these processes have not been studied in depth and understanding of the regulatory mechanisms is severely lacking. It is worth noting the impact of cadmium on the regulation of proteins related to the uptake and storage of other metals such as iron and zinc. This sequestration of other metals to avoid uptake of cadmium would be an interesting route to investigate as the accumulation of these other metals can be toxic to the cell. The ability to regulate intracellular levels of certain metals to avoid the uptake of others like cadmium showcases the intertwined metalloregulatory mechanisms of Epsilonproteobacteria and lends support to hypotheses that these regulatory systems work in tandem to maintain homeostasis, especially during infection.
Cobalt, magnesium, and manganese
Cobalt, magnesium, and manganese are among the less well studied enzymatic cofactors in Epsilonproteobacteria, but they encode transporters for either uptake or efflux of each metal to maintain a healthy equilibrium. Although cobalt can serve as a cofactor for some enzymes, the ability to combat cobalt toxicity is necessary to survive in the host environment. Additional metals like magnesium and manganese have not been investigated as thoroughly as iron and zinc, but Epsilonproteobacteria seem to diverge in the ability to tolerate these metals. While Campylobacter requires both to serve as cofactors for different metalloenzymes and utilizes transporters to acquire these metals, Helicobacter does not require manganese for growth and can only tolerate very low levels of the metal. Further research is required to tease apart the mechanisms, but the ability to utilize magnesium and manganese may be a distinguishing characteristic between members of the Epsilonproteobacteria. Beyond the transport systems associated with these metals, our understanding of the regulatory mechanisms impacting these systems is sorely lacking and could be the focus of future metalloregulatory work in Epsilonproteobacteria.
Bismuth, molybdate/molybdenum, and tungstate/tungsten
Unlike the more common transition metals discussed in this review, very little is understood about the biological role of bismuth in Epsilonproteobacteria. Bismuth has been used historically to treat Helicobacter infections, but the mechanistic impact of this metal is poorly understood. Campylobacter is also negatively impacted by bismuth, but the molecular mechanisms behind this detrimental effect are not known though it could be a potential therapeutic agent. Interestingly, the reverse is true for molybdate/molybdenum and tungstate/tungsten in Epsilonproteobacteria, especially Campylobacter. Transporters with high specificity for both metals have been identified in Campylobacter, while we only have a general understanding of those in Helicobacter. The specificity of these transporters in Campylobacter seems unique among Gram-negative organisms, so the characterization of these systems may provide insights into new systems in other pathogenic organisms. Although the identity of these metal transporters is known and a general regulatory mechanism has been identified, understanding of the exact roles of molybdenum and tungsten within Epsilonproteobacteria is lacking.
During the infection state, hosts sequester transition metals as a means of nutritional immunity. To combat the host defenses and obtain nutrient metals, pathogenic Epsilonproteobacteria must employ different mechanisms to transport metals and maintain homeostasis. Taken together, these conclusions demonstrate that although numerous regulatory systems have been identified in Epsilonproteobacteria, many gaps still exist in our understanding of the global impact of metals on the physiology of Campylobacter and Helicobacter. Additionally, our understanding of how these organisms are able to finely regulate these systems to avoid metal toxicity is limited and worth further investigation. The focus of this review was at the host–pathogen interface, but the role of the native microbiome may also have an impact on the ability of these pathogens to acquire and transport metals, and should be taken into account in future studies. The potential for additional metal transport systems that remain to be characterized and the divergence of Epsilonproteobacteria from other more well-studied organisms, such as E. coli, lend support to further investigate the systems in these organisms since we will likely gain novel insights into strategies used by pathogenic bacteria to acquire and regulate metals.
Acknowledgements
This work has been funded by a Career Development Award IK2BX001701 (to JAG) from the Office of Medical Research, Department of Veterans Affairs, and by Vanderbilt University Medical Center's Digestive Disease Research Center supported by NIH grant P30DK058404 and Vanderbilt Institute for Clinical and Translational Research program supported by the National Center for Research Resources, Grant UL1 RR024975-01, and the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Contributor Information
Brittni R Kelley, Department of Microbiology, University of Tennessee, Knoxville, TN, USA.
Jacky Lu, Department of Pathology, Microbiology and Immunology, Vanderbilt University, Nashville, TN, USA.
Kathryn P Haley, Department of Biology, Grand Valley State University, Grand Rapids, MI, USA.
Jennifer A Gaddy, Department of Pathology, Microbiology and Immunology, Vanderbilt University, Nashville, TN, USA; Tennessee Valley Healthcare Systems, Department of Veterans Affairs, Nashville, TN, USA; Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
Jeremiah G Johnson, Department of Microbiology, University of Tennessee, Knoxville, TN, USA.
Author contributions
BRK, JL, KPH, JAG, and JGJ conceived, designed, wrote, and edited this manuscript for critical content. All authors approved this manuscript for content integrity and accuracy.
Funding
None declared.
Conflicts of interest
There are no conflicts of interest to declare.
Data availability
No data was produced for this review.
References
- 1. Porcelli I., Reuter M., Pearson B. M., Wilhelm T., van Vliet A. H., Parallel evolution of genome structure and transcriptional landscape in the Epsilonproteobacteria, BMC Genomics, 2013, 14 (1), 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vetriani C., Voordeckers J. W., Crespo-Medina M., O'Brien C. E., Giovannelli D., Lutz R. A., Deep-sea hydrothermal vent Epsilonproteobacteria encode a conserved and widespread nitrate reduction pathway (Nap), ISME J., 2014, 8 (7), 1510–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vandamme P., Falsen E., Rossau R., Hoste B., Segers P., Tytgat R., De Ley J., Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov., Int. J. Syst. Evol. Microbiol., 1991, 41 (1), 88–103. [DOI] [PubMed] [Google Scholar]
- 4. Blaser M. J., Wells J. G., Feldman R. A., Pollard R. A., Allen J. R., Campylobacter enteritis in the United States: a multicenter study, Ann. Intern. Med., 1983, 98 (3), 360–365. [DOI] [PubMed] [Google Scholar]
- 5. Friedman C. R., Hoekstra R. M., Samuel M., Marcus R., Bender J., Shiferaw B., Reddy S., Ahuja S. D., Helfrick D. L., Hardnett F., Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites, Clin. Infect. Dis., 2004, 38, S285–S296. [DOI] [PubMed] [Google Scholar]
- 6. Meunier M., Guyard-Nicodème M., Hirchaud E., Parra A., Chemaly M., Dory D., Identification of novel vaccine candidates against Campylobacter through reverse vaccinology, J. Immunol. Res., 2016, 2016, 5715790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Askoura M., Sarvan S., Couture J.-F., Stintzi A., The Campylobacter jejuni ferric uptake regulator promotes acid survival and cross-protection against oxidative stress, Infect. Immun., 2016, 84 (5), 1287–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson J. G., Gaddy J. A., DiRita V. J., The PAS domain-containing protein HeuR regulates heme uptake in Campylobacter jejuni, mBio, 2016, 7 (6), e01691–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silva J., Teixeira P., Tackling Campylobacter: a review, Am. J. Adv. Food Sci. Technol., 2015, 3 (2), 107–124. [Google Scholar]
- 10. Saint-Cyr M. J., Guyard-Nicodème M., Messaoudi S., Chemaly M., Cappelier J.-M., Dousset X., Haddad N., Recent advances in screening of anti-Campylobacter activity in probiotics for use in poultry, Front. Microbiol., 2016, 7, 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kreuder A. J., Schleining J. A., Yaeger M., Zhang Q., Plummer P. J., RNAseq reveals complex response of Campylobacter jejuni to ovine bile and in vivo gallbladder environment, Front. Microbiol., 2017, 8, 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Viswanathan M., Pearl D., Taboada E., Parmley E., Mutschall S., Jardine C., Molecular and statistical analysis of Campylobacter spp. and antimicrobial-resistant Campylobacter carriage in wildlife and livestock from Ontario farms, Zoonoses Public Health, 2017, 64 (3), 194–203. [DOI] [PubMed] [Google Scholar]
- 13. Platts-Mills J. A., Kosek M., Update on the burden of Campylobacter in developing countries, Curr. Opin. Infect. Dis., 2014, 27 (5), 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Backert S., Boehm M., Wessler S., Tegtmeyer N., Transmigration route of Campylobacter jejuni across polarized intestinal epithelial cells: paracellular, transcellular or both?, Cell Commun. Signal., 2013, 11 (1), 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Samuelson D. R., Eucker T. P., Bell J. A., Dybas L., Mansfield L. S., Konkel M. E., The Campylobacter jejuni CiaD effector protein activates MAP kinase signaling pathways and is required for the development of disease, Cell Commun. Signal., 2013, 11, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaakoush N. O., Castaño-Rodríguez N., Mitchell H. M., Man S. M., Global epidemiology of Campylobacter infection, Clin. Microbiol. Rev., 2015, 28 (3), 687–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson T. J., Shank J. M., Johnson J. G., Current and potential treatments for reducing Campylobacter colonization in animal hosts and disease in humans, Front. Microbiol., 2017, 8, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Campana R., Baffone W., Intracellular survival and translocation ability of human and avian Campylobacter jejuni and Campylobacter coli strains, in Donelli G. (ed.), Advances in Microbiology, Infectious Diseases and Public Health, Vol. 14, Springer, Switzerland, 2020, p. 115. [DOI] [PubMed] [Google Scholar]
- 19. Gilbreath J. J., Cody W. L., Merrell D. S., Hendrixson D. R., Change is good: variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter, Microbiol. Mol. Biol. Rev., 2011, 75 (1), 84–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Atherton J. C., Blaser M. J., Coadaptation of Helicobacter pylori and humans: ancient history, modern implications, J. Clin. Invest., 2009, 119 (9), 2475–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cover T. L., Blaser M. J., Helicobacter pylori in health and disease, Gastroenterology, 2009, 136 (6), 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang Y., Wang Q.-L, Cheng D.-D., Xu W.-T., Lu N.-H., Adhesion and invasion of gastric mucosa epithelial cells by Helicobacter pylori, Front. Cell. Infect. Microbiol., 2016, 6, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Algood H. M. S., Cover T. L., Helicobacter pylori persistence: an overview of interactions between H. pylori and host immune defenses, Clin. Microbiol. Rev., 2006, 19 (4), 597–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Bernard M., Josenhans C., Pathogenesis of Helicobacter pylori infection, Helicobacter, 2014, 19, 11–18. [DOI] [PubMed] [Google Scholar]
- 25. Gaddy J. A., Haley K. P., Metalloregulation of Helicobacter pylori physiology and pathogenesis, Front. Microbiol., 2015, 6, 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bik E. M., Eckburg P. B., Gill S. R., Nelson K. E., Purdom E. A., Francois F., Perez-Perez G., Blaser M. J., Relman D. A., Molecular analysis of the bacterial microbiota in the human stomach, Proc. Natl Acad. Sci. USA, 2006, 103 (3), 732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hutner S. H., Provasoli L., Schatz A., Haskins C., Some approaches to the study of the role of metals in the metabolism of microorganisms, Proc. Am. Philos. Soc., 1950, 94 (2), 152–170. [Google Scholar]
- 28. Pinchbeck B. J., Soriano-Laguna M. J., Sullivan M. J., Luque-Almagro V. M., Rowley G., Ferguson S. J., Roldán M. D., Richardson D. J., Gates A. J., A dual functional redox enzyme maturation protein for respiratory and assimilatory nitrate reductases in bacteria, Mol. Microbiol., 2019, 111 (6), 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mildvan A. S., Loeb L. A., Wu C.-W., The role of metal ions in the mechanisms of DNA and RNA polymerase, CRC Crit. Rev. Biochem., 1979, 6 (3), 219–244. [DOI] [PubMed] [Google Scholar]
- 30. Crawford A., Wilson D., Essential metals at the host–pathogen interface: nutritional immunity and micronutrient assimilation by human fungal pathogens, FEMS Yeast Res., 2015, 15 (7), fov071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Porcheron G., Garénaux A., Proulx J., Sabri M., Dozois C. M., Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence, Front. Cell. Infect. Microbiol., 2013, 3, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hardy S. L., Glycosylation of Campylobacter iron transport systems and the role of host stress hormones, University of Leicester, 2013. [Google Scholar]
- 33. Butcher J., Stintzi A., The transcriptional landscape of Campylobacter jejuni under iron replete and iron limited growth conditions, PLoS One, 2013, 8 (11), e79475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller R. A., Britigan B. E., Role of oxidants in microbial pathophysiology, Clin. Microbiol. Rev., 1997, 10 (1), 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ishikawa T., Mizunoe Y., Kawabata S.-I., Takade A., Harada M., Wai S. N., Yoshida S.-I., The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni, J. Bacteriol., 2003, 185 (3), 1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oh E., Andrews K. J., Jeon B., Enhanced biofilm formation by ferrous and ferric iron through oxidative stress in Campylobacter jejuni, Front. Microbiol., 2018, 9, 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koppenol W. H., The Haber–Weiss cycle—70 years later, Redox Rep., 2001, 6 (4), 229–234. [DOI] [PubMed] [Google Scholar]
- 38. Zeng X., Lin J., Characterization of high affinity iron acquisition systems in Campylobacter jejuni, in Butcher J., Stintzi A. (eds), Campylobacter jejuni, Springer, New York, 2017, pp. 65–78. [DOI] [PubMed] [Google Scholar]
- 39. Braun V., Hantke K., Koester W., Bacterial iron transport: mechanisms, genetics, and regulation, Met. Ions Biol. Syst., 1998, 35, 67–146. [PubMed] [Google Scholar]
- 40. Haley K. P., Skaar E. P., A battle for iron: host sequestration and Staphylococcus aureus acquisition, Microbes Infect., 2012, 14 (3), 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Askoura M., Youns M., Hegazy W. A. H., Investigating the influence of iron on Campylobacter jejuni transcriptome in response to acid stress, Microb. Pathog., 2020, 138, 103777. [DOI] [PubMed] [Google Scholar]
- 42. Chandrashekhar K., Srivastava V., Hwang S., Jeon B., Ryu S., Rajashekara G., Transducer-like protein in Campylobacter jejuni with a role in mediating chemotaxis to iron and phosphate, Front. Microbiol., 2018, 9, 2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ketley J. M., Konkel M. E., Campylobacter: Molecular and Cellular Biology, Horizon Bioscience, Norfolk, UK, 2005. [Google Scholar]
- 44. Stintzi A., van Vliet A. H., Ketley J. M., Iron metabolism, transport, and regulation, in Nachamkin I, Szymanski C. M., Blaser M. J. (eds), Campylobacter, 3rd edn, American Society of Microbiology, Washington, DC, 2008, pp. 591–610. [Google Scholar]
- 45. Miller C. E., Williams P. H., Ketley J. M., Pumping iron: mechanisms for iron uptake by Campylobacter, Microbiology, 2009, 155 (10), 3157–3165. [DOI] [PubMed] [Google Scholar]
- 46. Bullen J., The significance of iron in infection, Clin. Infect. Dis., 1981, 3 (6), 1127–1138. [DOI] [PubMed] [Google Scholar]
- 47. Naikare H., Palyada K., Panciera R., Marlow D., Stintzi A., Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival, Infect. Immun., 2006, 74 (10), 5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Runyen-Janecky L. J., Role and regulation of heme iron acquisition in Gram-negative pathogens, Front. Cell. Infect. Microbiol., 2013, 3, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Richard K. L., Kelley B. R., Johnson J. G., Heme uptake and utilization by Gram-negative bacterial pathogens, Front. Cell. Infect. Microbiol., 2019, 9, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stahl M., Butcher J., Stintzi A., Nutrient acquisition and metabolism by Campylobacter jejuni, Front. Cell. Infect. Microbiol., 2012, 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ridley K. A., Rock J. D., Li Y., Ketley J. M., Heme utilization in Campylobacter jejuni, J. Bacteriol., 2006, 188 (22), 7862–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taveirne M. E., Theriot C. M., Livny J., DiRita V. J., The complete Campylobacter jejuni transcriptome during colonization of a natural host determined by RNAseq, PLoS One, 2013, 8 (8), e73586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Naikare H., Butcher J., Flint A., Xu J., Raymond K. N., Stintzi A., Campylobacter jejuni ferric–enterobactin receptor CfrA is TonB3 dependent and mediates iron acquisition from structurally different catechol siderophores, Metallomics, 2013, 5 (8), 988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hofreuter D., Defining the metabolic requirements for the growth and colonization capacity of Campylobacter jejuni, Front. Cell. Infect. Microbiol., 2014, 4, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gao B., Vorwerk H., Huber C., Lara-Tejero M., Mohr J., Goodman A. L., Eisenreich W., Galán J. E., Hofreuter D., Metabolic and fitness determinants for in vitro growth and intestinal colonization of the bacterial pathogen Campylobacter jejuni, PLoS Biol., 2017, 15 (5), e2001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crofts A. A., Poly F. M., Ewing C. P., Kuroiwa J. M., Rimmer J. E., Harro C., Sack D., Talaat K. R., Porter C. K., Gutierrez R. L., Campylobacter jejuni transcriptional and genetic adaptation during human infection, Nat. Microbiol., 2018, 3 (4), 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cornelissen C. N., Transferrin–iron uptake by Gram-negative bacteria, Front. Biosci., 2003, 8, d836–d847. [DOI] [PubMed] [Google Scholar]
- 58. Miller C. E., Rock J. D., Ridley K. A., Williams P. H., Ketley J. M., Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni, J. Bacteriol., 2008, 190 (6), 1900–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alexander D. B., Vogel H. J., Tsuda H., Lactoferrin researchers descend on Nagoya Castle, Biochem. Cell Biol., 2017, 95 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- 60. Palyada K., Threadgill D., Stintzi A., Iron acquisition and regulation in Campylobacter jejuni, J. Bacteriol., 2004, 186 (14), 4714–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stintzi A., Marlow D., Palyada K., Naikare H., Panciera R., Whitworth L., Clarke C., Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni, Infect. Immun., 2005, 73 (3), 1797–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ghosh M., Miller P. A., Möllmann U., Claypool W. D., Schroeder V. A., Wolter W. R., Suckow M., Yu H., Li S., Huang W., Targeted antibiotic delivery: selective siderophore conjugation with daptomycin confers potent activity against multidrug resistant Acinetobacter baumannii both in vitro and in vivo, J. Med. Chem., 2017, 60 (11), 4577–4583. [DOI] [PubMed] [Google Scholar]
- 63. Indikova I., Humphrey T. J., Hilbert F., Survival with a helping hand: Campylobacter and microbiota, Front. Microbiol., 2015, 6, 1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Richardson P. T., Park S. F., Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system, Microbiology, 1995, 141 (12), 3181–3191. [DOI] [PubMed] [Google Scholar]
- 65. Xu F., Zeng X., Haigh R. D., Ketley J. M., Lin J., Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter, J. Bacteriol., 2010, 192 (17), 4425–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chan A. C., Doukov T. I., Scofield M., Tom-Yew S. A., Ramin A. B., MacKichan J. K., Gaynor E. C., Murphy M. E., Structure and function of P19, a high-affinity iron transporter of the human pathogen Campylobacter jejuni, J. Mol. Biol., 2010, 401 (4), 590–604. [DOI] [PubMed] [Google Scholar]
- 67. Liu M. M., Boinett C. J., Chan A. C., Parkhill J., Murphy M. E., Gaynor E. C., Investigating the Campylobacter jejuni transcriptional response to host intestinal extracts reveals the involvement of a widely conserved iron uptake system, mBio, 2018, 9 (4), e01347–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guerry P., Perez-Casal J., Yao R., McVeigh A., A genetic locus involved in iron utilization unique to some Campylobacter strains, J. Bacteriol., 1997, 179 (12), 3997–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zeng X., Xu F., Lin J., Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni, Infect. Immun., 2009, 77 (12), 5437–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Butcher J., Handley R. A., van Vliet A. H., Stintzi A., Refined analysis of the Campylobacter jejuni iron-dependent/independent Fur- and PerR-transcriptomes, BMC Genomics, 2015, 16, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. van Vliet A. H., Ketley J. M., Park S. F., Penn C. W., The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense, FEMS Microbiol. Rev., 2002, 26 (2), 173–186. [DOI] [PubMed] [Google Scholar]
- 72. Handley R. A., Mulholland F., Reuter M., Ramachandran V. K., Musk H., Clissold L., Le Brun N. E., van Vliet A. H., PerR controls oxidative stress defence and aerotolerance but not motility-associated phenotypes of Campylobacter jejuni, Microbiology, 2015, 161, 1524–1536. [DOI] [PubMed] [Google Scholar]
- 73. Zeng X., Mo Y., Xu F., Lin J., Identification and characterization of a periplasmic trilactone esterase, Cee, revealed unique features of ferric enterobactin acquisition in Campylobacter, Mol. Microbiol., 2013, 87 (3), 594–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Flint A., Sun Y.-Q., Butcher J., Stahl M., Huang H., Stintzi A., Phenotypic screening of a targeted mutant library reveals Campylobacter jejuni defenses against oxidative stress, Infect. Immun., 2014, 82 (6), 2266–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Oh E., Jeon B., Role of alkyl hydroperoxide reductase (AhpC) in the biofilm formation of Campylobacter jejuni, PLoS One, 2014, 9, e87312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Holmes K., Mulholland F., Pearson B. M., Pin C., McNicholl-Kennedy J., Ketley J. M., Wells J. M., Campylobacter jejuni gene expression in response to iron limitation and the role of Fur, Microbiology, 2005, 151, 243–257. [DOI] [PubMed] [Google Scholar]
- 77. Testerman T. L., Conn P. B., Mobley H. L., McGee D. J., Nutritional requirements and antibiotic resistance patterns of Helicobacter species in chemically defined media, J. Clin. Microbiol., 2006, 44 (5), 1650–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dhaenens L., Szczebara F., Van Nieuwenhuyse S., Husson M.-O., Comparison of iron uptake in different Helicobacter species, Res. Microbiol., 1999, 150 (7), 475–481. [DOI] [PubMed] [Google Scholar]
- 79. Senkovich O., Ceaser S., McGee D. J., Testerman T. L., Unique host iron utilization mechanisms of Helicobacter pylori revealed with iron-deficient chemically defined media, Infect. Immun., 2010, 78 (5), 1841–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Velayudhan J., Hughes N. J., McColm A. A., Bagshaw J., Clayton C. L., Andrews S. C., Kelly D. J., Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter, Mol. Microbiol., 2000, 37 (2), 274–286. [DOI] [PubMed] [Google Scholar]
- 81. Ge R., Sun X., Iron trafficking system in Helicobacter pylori, Biometals, 2012, 25 (2), 247–258. [DOI] [PubMed] [Google Scholar]
- 82. Delany I., Pacheco A. B. F., Spohn G., Rappuoli R., Scarlato V., Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein, J. Bacteriol., 2001, 183 (16), 4932–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bereswill S., Greiner S., van Vliet A. H., Waidner B., Fassbinder F., Schiltz E., Kusters J. G., Kist M., Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori, J. Bacteriol., 2000, 182 (21), 5948–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pich O. Q., Carpenter B. M., Gilbreath J. J., Merrell D. S., Detailed analysis of Helicobacter pylori Fur-regulated promoters reveals a Fur box core sequence and novel Fur-regulated genes, Mol. Microbiol., 2012, 84 (5), 921–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gancz H., Censini S., Merrell D. S., Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori, Infect. Immun., 2006, 74, 602–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Carpenter B. M., Gilbreath J. J., Pich O. Q., McKelvey A. M., Maynard E. L., Li Z.-Z., Merrell D. S., Identification and characterization of novel Helicobacter pylori apo-Fur-regulated target genes, J. Bacteriol., 2013, 195 (24), 5526–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ernst F., Bereswill S., Waidner B., Stoof J., Mader U., Kusters J. G., Kuipers E. J., Kist M., van Vliet A., Homuth G., Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression, Microbiology, 2005, 151, 533–546. [DOI] [PubMed] [Google Scholar]
- 88. Danielli A., Romagnoli S., Roncarati D., Costantino L., Delany I., Scarlato V., Growth phase and metal-dependent transcriptional regulation of the fecA genes in Helicobacter pylori, J. Bacteriol., 2009, 191 (11), 3717–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Danielli A., Roncarati D., Delany I., Chiarini V., Rappuoli R., Scarlato V., In vivo dissection of the Helicobacter pylori Fur regulatory circuit by genome-wide location analysis, J. Bacteriol., 2006, 188 (13), 4654–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tummuru M. K., Sharma S. A., Blaser M. J., Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells, Mol. Microbiol., 1995, 18 (5), 867–876. [DOI] [PubMed] [Google Scholar]
- 91. Odenbreit S., Püls J., Sedlmaier B., Gerland E., Fischer W., Haas R., Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion, Science, 2000, 287 (5457), 1497–1500. [DOI] [PubMed] [Google Scholar]
- 92. Tan S., Noto J. M., Romero-Gallo J., Peek R. M. Jr, Amieva M. R., Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface, PLoS Pathog., 2011, 7 (5), e1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Noto J. M., Khizanishvili T., Chaturvedi R., Piazuelo M. B., Romero-Gallo J., Delgado A. G., Khurana S. S., Sierra J. C., Krishna U. S., Suarez G., Helicobacter pylori promotes the expression of Krüppel-like factor 5, a mediator of carcinogenesis, in vitro and in vivo, PLoS One, 2013, 8, e54344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Noto J. M., Piazuelo M. B., Chaturvedi R., Bartel C. A., Thatcher E. J., Delgado A., Romero-Gallo J., Wilson K. T., Correa P., Patton J. G., Strain-specific suppression of microRNA-320 by carcinogenic Helicobacter pylori promotes expression of the antiapoptotic protein Mcl-1, Am. J. Physiol. Gastrointest. Liver Physiol., 2013, 305 (11), G786–G796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Haley K. P., Blanz E. J., Gaddy J. A., High resolution electron microscopy of the Helicobacter pylori Cag type IV secretion system pili produced in varying conditions of iron availability, J. Vis. Exp., 2014, e52122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Miles S., Piazuelo M. B., Semino-Mora C., Washington M. K., Dubois A., Peek R. M., Correa P., Merrell D. S., Detailed in vivo analysis of the role of Helicobacter pylori Fur in colonization and disease, Infect. Immun., 2010, 78 (7), 3073–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Berg J. M., Shi Y., The galvanization of biology: a growing appreciation for the roles of zinc, Science, 1996, 271 (5252), 1081–1085. [DOI] [PubMed] [Google Scholar]
- 98. Gielda L. M., DiRita V. J., Zinc competition among the intestinal microbiota, mBio, 2012, 3 (4), e00171–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Si M., Wang Y., Zhang B., Zhao C., Kang Y., Bai H., Wei D., Zhu L., Zhang L., Dong T. G., The type VI secretion system engages a redox-regulated dual-functional heme transporter for zinc acquisition, Cell Rep., 2017, 20 (4), 949–959. [DOI] [PubMed] [Google Scholar]
- 100. Moulin P., Patron K., Cano C., Zorgani M. A., Camiade E., Borezée-Durant E., Rosenau A., Mereghetti L., Hiron A., The Adc/Lmb system mediates zinc acquisition in Streptococcus agalactiae and contributes to bacterial growth and survival, J. Bacteriol., 2016, 198 (24), 3265–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Davis L. M., Kakuda T., DiRita V. J., A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization, J. Bacteriol., 2009, 191 (5), 1631–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Diaz-Ochoa V., Jellbauer S., Klaus S., Raffatellu M., Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis, Front. Cell. Infect. Microbiol., 2014, 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Djoko K. Y., Cheryl-lynn Y. O., Walker M. J., McEwan A. G., The role of copper and zinc toxicity in innate immune defense against bacterial pathogens, J. Biol. Chem., 2015, 290 (31), 18954–18961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shank J. M., Kelley B. R., Jackson J. W., Tweedie J. L., Franklin D., Damo S. M., Gaddy J. A., Murphy C. N., Johnson J. G., The host antimicrobial protein calgranulin C participates in the control of Campylobacter jejuni growth via zinc sequestration, Infect. Immun., 2018, 86 (6), e00234–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kehl-Fie T. E., Skaar E. P., Nutritional immunity beyond iron: a role for manganese and zinc, Curr. Opin. Chem. Biol., 2010, 14 (2), 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tapiero H., Townsend D., Tew K., Trace elements in human physiology and pathology. Copper, Biomed. Pharmacother., 2003, 57 (9), 386–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Weinberg E. D., Iron availability and infection, Biochim. Biophys. Acta: Gen. Subj., 2009, 1790 (7), 600–605. [DOI] [PubMed] [Google Scholar]
- 108. Hantke K., Bacterial zinc uptake and regulators, Curr. Opin. Microbiol., 2005, 8 (2), 196–202. [DOI] [PubMed] [Google Scholar]
- 109. Patzer S. I., Hantke K., The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli, Mol. Microbiol., 1998, 28 (6), 1199–1210. [DOI] [PubMed] [Google Scholar]
- 110. Patzer S. I., Hantke K., The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli, J. Biol. Chem., 2000, 275 (32), 24321–24332. [DOI] [PubMed] [Google Scholar]
- 111. Outten C. E., O'Halloran T. V., Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis, Science, 2001, 292 (5526), 2488–2492. [DOI] [PubMed] [Google Scholar]
- 112. Sarvan S., Charih F., Butcher J., Brunzelle J. S., Stintzi A., Couture J. F., Crystal structure of Campylobacter jejuni peroxide regulator, FEBS Lett., 2018, 592 (13), 2351–2360. [DOI] [PubMed] [Google Scholar]
- 113. Gaddy J. A., Radin J. N., Loh J. T., Piazuelo M. B., Kehl-Fie T. E., Delgado A. G., Ilca F. T., Peek R. M., Cover T. L., Chazin W. J., The host protein calprotectin modulates the Helicobacter pylori cag type IV secretion system via zinc sequestration, PLoS Pathog., 2014, 10 (10), e1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Haley K. P., Delgado A. G., Piazuelo M. B., Mortensen B. L., Correa P., Damo S. M., Chazin W. J., Skaar E. P., Gaddy J. A., The human antimicrobial protein calgranulin C participates in control of Helicobacter pylori growth and regulation of virulence, Infect. Immun., 2015, 83 (7), 2944–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Leach S. T., Mitchell H. M., Geczy C. L., Sherman P. M., Day A. S., S100 calgranulin proteins S100A8, S100A9 and S100A12 are expressed in the inflamed gastric mucosa of Helicobacter pylori-infected children, Can. J. Gastroenterol., 2008, 22, 461–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sempértegui F., Diaz M., Mejia R., Rodríguez-Mora O. G., Rentería E., Guarderas C., Estrella B., Recalde R., Hamer D. H., Reeves P. G., Low concentrations of zinc in gastric mucosa are associated with increased severity of Helicobacter pylori-induced inflammation, Helicobacter, 2007, 12, 43–48. [DOI] [PubMed] [Google Scholar]
- 117. Stähler F. N., Odenbreit S., Haas R., Wilrich J., van Vliet A. H., Kusters J. G., Kist M., Bereswill S., The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization, Infect. Immun., 2006, 74 (7), 3845–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Waidner B., Melchers K., Ivanov I., Loferer H., Bensch K. W., Kist M., Bereswill S., Identification by RNA profiling and mutational analysis of the novel copper resistance determinants CrdA (HP1326), CrdB (HP1327), and CzcB (HP1328) in Helicobacter pylori, J. Bacteriol., 2002, 184 (23), 6700–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gaddy J. A., Radin J. N., Cullen T. W., Chazin W. J., Skaar E. P., Trent M. S., Algood H. M., Helicobacter pylori resists the antimicrobial activity of calprotectin via lipid A modification and associated biofilm formation, mBio, 2015, 6 (6), e01349–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dian C., Vitale S., Leonard G. A., Bahlawane C., Fauquant C., Leduc D., Muller C., de Reuse H., Michaud-Soret I., Terradot L., The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites, Mol. Microbiol., 2011, 79 (5), 1260–1275. [DOI] [PubMed] [Google Scholar]
- 121. Vitale S., Fauquant C., Lascoux D., Schauer K., Saint-Pierre C., Michaud-Soret I., A ZnS4 structural zinc site in the Helicobacter pylori ferric uptake regulator, Biochemistry, 2009, 48 (24), 5582–5591. [DOI] [PubMed] [Google Scholar]
- 122. Latorre M., Troncoso R., Uauy R., Biological aspects of copper, in Kerkar N., Roberts E. A. (eds), Clinical and Translational Perspectives on Wilson Disease, Elsevier, Amsterdam, 2019, pp. 25–31. [Google Scholar]
- 123. Hall S. J., Hitchcock A., Butler C. S., Kelly D. J., A multicopper oxidase (Cj1516) and a CopA homologue (Cj1161) are major components of the copper homeostasis system of Campylobacter jejuni, J. Bacteriol., 2008, 190 (24), 8075–8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Silva C. S., Durão P., Fillat A., Lindley P. F., Martins L. O., Bento I., Crystal structure of the multicopper oxidase from the pathogenic bacterium Campylobacter jejuni CGUG11284: characterization of a metallo-oxidase, Metallomics, 2012, 4, 37–47. [DOI] [PubMed] [Google Scholar]
- 125. Shih H.-Y., Lin Y. E., Efficacy of copper–silver ionization in controlling biofilm- and plankton-associated waterborne pathogens, Appl. Environ. Microbiol., 2010, 76 (6), 2032–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Pham A. N., Xing G., Miller C. J., Waite T. D., Fenton-like copper redox chemistry revisited: hydrogen peroxide and superoxide mediation of copper-catalyzed oxidant production, J. Catal., 2013, 301, 54–64. [Google Scholar]
- 127. Macomber L., Imlay J. A., The iron–sulfur clusters of dehydratases are primary intracellular targets of copper toxicity, Proc. Natl Acad. Sci. USA, 2009, 106 (20), 8344–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Weiss G., Carver P., Role of divalent metals in infectious disease susceptibility and outcome, Clin. Microbiol. Infect., 2018, 24, 16–23. [DOI] [PubMed] [Google Scholar]
- 129. Wolschendorf F., Ackart D., Shrestha T. B., Hascall-Dove L., Nolan S., Lamichhane G., Wang Y., Bossmann S. H., Basaraba R. J., Niederweis M., Copper resistance is essential for virulence of Mycobacterium tuberculosis, Proc. Natl Acad. Sci. USA, 2011, 108 (4), 1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Johnson M. D., Kehl-Fie T. E., Klein R., Kelly J., Burnham C., Mann B., Rosch J. W., Role of copper efflux in pneumococcal pathogenesis and resistance to macrophage-mediated immune clearance, Infect. Immun., 2015, 83 (4), 1684–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Neyrolles O., Wolschendorf F., Mitra A., Niederweis M., Mycobacteria, metals, and the macrophage, Immunol. Rev., 2015, 264, 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wells J., Oliver W., Yen J. T., The effects of dietary additives on faecal levels of Lactobacillus spp., coliforms, and Escherichia coli, and faecal prevalence of Salmonella spp. and Campylobacter spp. in US production nursery swine 1, J. Appl. Microbiol., 2010, 108, 306–314. [DOI] [PubMed] [Google Scholar]
- 133. Reeser R. J., Medler R. T., Billington S. J., Jost B. H., Joens L. A., Characterization of Campylobacter jejuni biofilms under defined growth conditions, Appl. Environ. Microbiol., 2007, 73 (6), 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gardner S. P., Olson J. W., Interaction of copper toxicity and oxidative stress in Campylobacter jejuni, J. Bacteriol., 2018, 200 (21), e00208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Grass G., Rensing C., Genes involved in copper homeostasis in Escherichia coli, J. Bacteriol., 2001, 183 (6), 2145–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Outten F. W., Huffman D. L., Hale J. A., O'Halloran T. V., The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli, J. Biol. Chem., 2001, 276 (33), 30670–30677. [DOI] [PubMed] [Google Scholar]
- 137. Liu Y.-W., Hitchcock A., Salmon R. C., Kelly D. J., It takes two to tango: two TatA paralogues and two redox enzyme-specific chaperones are involved in the localization of twin-arginine translocase substrates in Campylobacter jejuni, Microbiology, 2014, 160, 2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hwang S., Kim M., Ryu S., Jeon B., Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni, PLoS One, 2011, 6 (7), e22300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Barceloux D. G., Barceloux D., Copper, J. Toxicol. Clin. Toxicol., 1999, 37 (2), 217–230. [DOI] [PubMed] [Google Scholar]
- 140. Montefusco S., Esposito R., D'Andrea L., Monti M. C., Dunne C., Dolan B., Tosco A., Marzullo L., Clyne M., Copper promotes TFF1-mediated Helicobacter pylori colonization, PLoS One, 2013, 8 (11), e79455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Sanders L., Andermann T. M., Ottemann K. M., A supplemented soft agar chemotaxis assay demonstrates the Helicobacter pylori chemotactic response to zinc and nickel, Microbiology, 2013, 159, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Nagata K., Tsukita S., Tamura T., Sone N., A db-type cytochrome-c oxidase terminates the respiratory chain in Helicobacter pylori, Microbiology, 1996, 142 (7), 1757–1763. [DOI] [PubMed] [Google Scholar]
- 143. Waidner B., Melchers K., Stähler F. N., Kist M., Bereswill S., The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA, J. Bacteriol., 2005, 187 (13), 4683–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Beier D., Spohn G., Rappuoli R., Scarlato V., Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation, J. Bacteriol., 1997, 179 (15), 4676–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. McIlveen W., Negusanti J., Nickel in the terrestrial environment, Sci. Total Environ., 1994, 148, 109–138. [DOI] [PubMed] [Google Scholar]
- 146. Lusi E., Patrissi T., Guarascio P., Nickel-resistant bacteria isolated in human microbiome, New Microbes New Infect., 2017, 19, 67–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Buxton S., Garman E., Heim K. E., Lyons-Darden T., Schlekat C. E., Taylor M. D., Oller A. R., Concise review of nickel human health toxicology and ecotoxicology, Inorganics, 2019, 7 (7), 89. [Google Scholar]
- 148. Nieminen T. M., Ukonmaanaho L., Rausch N., Shotyk W., Biogeochemistry of nickel and its release into the environment, Metal Ions Life Sci., 2007, 2, 1–30. [Google Scholar]
- 149. Maier R. J., Benoit S. L., Role of nickel in microbial pathogenesis, Inorganics, 2019, 7 (7), 80. [Google Scholar]
- 150. Sunderman F. W. Jr, Aitio A., Morgan L. G., Norseth T., Biological monitoring of nickel, Toxicol. Ind. Health, 1986, 2, 17–78. [DOI] [PubMed] [Google Scholar]
- 151. Palmer L. D., Skaar E. P., Transition metals and virulence in bacteria, Annu. Rev. Genet., 2016, 50, 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Howlett R. M., Hughes B. M., Hitchcock A., Kelly D. J., Hydrogenase activity in the foodborne pathogen Campylobacter jejuni depends upon a novel ABC-type nickel transporter (NikZYXWV) and is SlyD-independent, Microbiology, 2012, 158 (6), 1645–1655. [DOI] [PubMed] [Google Scholar]
- 153. Dufour V., Li J., Flint A., Rosenfeld E., Rivoal K., Georgeault S., Alazzam B., Ermel G., Stintzi A., Bonnaure-Mallet M., Inactivation of the LysR regulator Cj1000 of Campylobacter jejuni affects host colonization and respiration, Microbiology, 2013, 159, 1165–1178. [DOI] [PubMed] [Google Scholar]
- 154. Rodionov D. A., Hebbeln P., Gelfand M. S., Eitinger T., Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters, J. Bacteriol., 2006, 188, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Weerakoon D. R., Borden N. J., Goodson C. M., Grimes J., Olson J. W., The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology, Microb. Pathog., 2009, 47, 8–15. [DOI] [PubMed] [Google Scholar]
- 156. Heddle J., Scott D. J., Unzai S., Park S.-Y., Tame J. R., Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli, J. Biol. Chem., 2003, 278 (50), 50322–50329. [DOI] [PubMed] [Google Scholar]
- 157. Cavazza C., Martin L., Laffly E., Lebrette H., Cherrier M. V., Zeppieri L., Richaud P., Carrière M., Fontecilla-Camps J. C., Histidine 416 of the periplasmic binding protein NikA is essential for nickel uptake in Escherichia coli, FEBS Lett., 2011, 585 (4), 711–715. [DOI] [PubMed] [Google Scholar]
- 158. Kirsch F., Eitinger T., Transport of nickel and cobalt ions into bacterial cells by S components of ECF transporters, Biometals, 2014, 27 (4), 653–660. [DOI] [PubMed] [Google Scholar]
- 159. Lebrette H., Brochier-Armanet C., Zambelli B., de Reuse H., Borezée-Durant E., Ciurli S., Cavazza C., Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins, Structure, 2014, 22 (10), 1421–1432. [DOI] [PubMed] [Google Scholar]
- 160. Taylor A. J., Kelly D. J., The function, biogenesis and regulation of the electron transport chains in Campylobacter jejuni: new insights into the bioenergetics of a major food-borne pathogen, in Poole R. K. (ed.), Advances in Microbial Physiology, Vol. 74, Elsevier, Amsterdam, 2019, pp. 239–329. [DOI] [PubMed] [Google Scholar]
- 161. An Y. J., Ahn B.-E., Han A.-R., Kim H.-M., Chung K. M., Shin J.-H., Cho Y.-B., Roe J.-H., Cha S.-S., Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition, Nucleic Acids Res., 2009, 37 (10), 3442–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Benoit S. L., Maier R. J., Site-directed mutagenesis of Campylobacter concisus respiratory genes provides insight into the pathogen's growth requirements, Sci. Rep., 2018, 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. De Reuse H., Vinella D., Cavazza C., Common themes and unique proteins for the uptake and trafficking of nickel, a metal essential for the virulence of Helicobacter pylori, Front. Cell. Infect. Microbiol., 2013, 3, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Sydor A. M., Lebrette H., Ariyakumaran R., Cavazza C., Zamble D. B., Relationship between Ni(II) and Zn(II) coordination and nucleotide binding by the Helicobacter pylori [NiFe]-hydrogenase and urease maturation factor HypB, J. Biol. Chem., 2014, 289 (7), 3828–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Evans D. J. Jr, Evans D. G., Kirkpatrick S. S., Graham D. Y., Characterization of the Helicobacter pylori urease and purification of its subunits, Microb. Pathog., 1991, 10, 15–26. [DOI] [PubMed] [Google Scholar]
- 166. Hawtin P., Delves H., Newell D., The demonstration of nickel in the urease of Helicobacter pylori by atomic absorption spectroscopy, FEMS Microbiol. Lett., 1991, 77, 51–54. [DOI] [PubMed] [Google Scholar]
- 167. Maier R. J., Benoit S. L., Seshadri S., Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori, Biometals, 2007, 20, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Benoit S. L., Maier R. J., Hydrogen and nickel metabolism in Helicobacter species, Ann. N. Y. Acad. Sci., 2008, 1125, 242–251. [DOI] [PubMed] [Google Scholar]
- 169. Yang X., Li H., Lai T.-P., Sun H., UreE–UreG complex facilitates nickel transfer and preactivates GTPase of UreG in Helicobacter pylori, J. Biol. Chem., 2015, 290 (20), 12474–12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Chivers P. T., Nickel recognition by bacterial importer proteins, Metallomics, 2015, 7 (4), 590–595. [DOI] [PubMed] [Google Scholar]
- 171. Johnson R. C., Hu H. Q., Merrell D. S., Maroney M. J., Dynamic HypA zinc site is essential for acid viability and proper urease maturation in Helicobacter pylori, Metallomics, 2015, 7 (4), 674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Jones M., Ademi I., Yin X., Gong Y., Zamble D., Nickel-responsive regulation of two novel Helicobacter pylori NikR-targeted genes, Metallomics, 2015, 7 (4), 662–673. [DOI] [PubMed] [Google Scholar]
- 173. van Vliet A. H., Kuipers E. J., Waidner B., Davies B. J., de Vries N., Penn C. W., Vandenbroucke-Grauls C. M., Kist M., Bereswill S., Kusters J. G., Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level, Infect. Immun., 2001, 69 (8), 4891–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Rowinska-Zyrek M., Witkowska D., Bielinska S., Kamysz W., Kozlowski H., The –Cys–Cys– motif in Helicobacter pylori’s Hpn and HspA proteins is an essential anchoring site for metal ions, Dalton Trans., 2011, 40 (20), 5604–5610. [DOI] [PubMed] [Google Scholar]
- 175. Abraham L. O., Li Y., Zamble D. B., The metal- and DNA-binding activities of Helicobacter pylori NikR, J. Inorg. Biochem., 2006, 100, 1005–1014. [DOI] [PubMed] [Google Scholar]
- 176. Evans S. E., Michel S. L., Dissecting the role of DNA sequence in Helicobacter pylori NikR/DNA recognition, Dalton Trans., 2012, 41 (26), 7946–7951. [DOI] [PubMed] [Google Scholar]
- 177. Vannini A., Pinatel E., Costantini P. E., Pelliciari S., Roncarati D., Puccio S., De Bellis G., Peano C., Danielli A., Comprehensive mapping of the Helicobacter pylori NikR regulon provides new insights in bacterial nickel responses, Sci. Rep., 2017, 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Contreras M., Thiberge J. M., Mandrand-Berthelot M. A., Labigne A., Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori, Mol. Microbiol., 2003, 49 (4), 947–963. [DOI] [PubMed] [Google Scholar]
- 179. Ernst F. D., Stoof J., Horrevoets W. M., Kuipers E. J., Kusters J. G., van Vliet A. H., NikR mediates nickel-responsive transcriptional repression of the Helicobacter pylori outer membrane proteins FecA3 (HP1400) and FrpB4 (HP1512), Infect. Immun., 2006, 74 (12), 6821–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Servetas S. L., Carpenter B. M., Haley K. P., Gilbreath J. J., Gaddy J. A., Merrell D. S., Characterization of key Helicobacter pylori regulators identifies a role for ArsRS in biofilm formation, J. Bacteriol., 2016, 198 (18), 2536–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Bury-Moné S., Thiberge J. M., Contreras M., Maitournam A., Labigne A., De Reuse H., Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori, Mol. Microbiol., 2004, 53 (2), 623–638. [DOI] [PubMed] [Google Scholar]
- 182. Stoof J., Kuipers E. J., Klaver G., van Vliet A. H., An ABC transporter and a TonB ortholog contribute to Helicobacter mustelae nickel and cobalt acquisition, Infect. Immun., 2010, 78 (10), 4261–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Fischer F., Robbe-Saule M., Turlin E., Mancuso F., Michel V., Richaud P., Veyrier F. J., De Reuse H., Vinella D., Characterization in Helicobacter pylori of a nickel transporter essential for colonization that was acquired during evolution by gastric Helicobacter species, PLoS Pathog., 2016, 12 (12), e1006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Quezada-Hinojosa R. P., Matera V., Adatte T., Rambeau C., Föllmi K. B., Cadmium distribution in soils covering Jurassic oolitic limestone with high Cd contents in the Swiss Jura, Geoderma, 2009, 150, 287–301. [Google Scholar]
- 185. Liu Y., Xiao T., Baveye P. C., Zhu J., Ning Z., Li H., Potential health risk in areas with high naturally-occurring cadmium background in southwestern China, Ecotoxicol. Environ. Saf., 2015, 112, 122–131. [DOI] [PubMed] [Google Scholar]
- 186. Chakraborty J., Das S., Characterization and cadmium-resistant gene expression of biofilm-forming marine bacterium Pseudomonas aeruginosa JP-11, Environ. Sci. Pollut. Res., 2014, 21 (24), 14188–14201. [DOI] [PubMed] [Google Scholar]
- 187. Sutoo D., Akiyama K., Imamiya S., A mechanism of cadmium poisoning: the cross effect of calcium and cadmium in the calmodulin-dependent system, Arch. Toxicol., 1990, 64 (2), 161–164. [DOI] [PubMed] [Google Scholar]
- 188. Huff J., Lunn R. M., Waalkes M. P., Tomatis L., Infante P. F., Cadmium-induced cancers in animals and in humans, Int. J. Occup. Environ. Health, 2007, 13 (2), 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Kazmi S., Roberson B., Stern N., Cadmium chloride susceptibility, a characteristic of Campylobacter spp., J. Clin. Microbiol., 1985, 21 (5), 708–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Kaakoush N. O., Raftery M., Mendz G. L., Molecular responses of Campylobacter jejuni to cadmium stress, FEBS J., 2008, 275 (20), 5021–5033. [DOI] [PubMed] [Google Scholar]
- 191. Stern N., Kazmi S., Roberson B., Ono K., Juven B., Response of Campylobacter jejuni to combinations of ferrous sulphate and cadmium chloride, J. Appl. Bacteriol., 1988, 64 (3), 247–256. [DOI] [PubMed] [Google Scholar]
- 192. Ron E. Z., Minz D., Finkelstein N., Rosenberg E., Interactions of bacteria with cadmium, in Rosenberg E. (ed.), Microorganisms to Combat Pollution, Springer, Dordrecht, The Netherlands, 1992, pp. 37–46. [Google Scholar]
- 193. Banaś A. M., Bocian-Ostrzycka K. M., Plichta M., Dunin-Horkawicz S., Ludwiczak J., Płaczkiewicz J., Jagusztyn-Krynicka E. K., C8J_1298, a bifunctional thiol oxidoreductase of Campylobacter jejuni, affects Dsb (disulfide bond) network functioning, PLoS One, 2020, 15 (3), e0230366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Große C., Anton A., Hoffmann T., Franke S., Schleuder G., Nies D. H., Identification of a regulatory pathway that controls the heavy-metal resistance system Czc via promoter czcNp in Ralstonia metallidurans, Arch. Microbiol., 2004, 182, 109–118. [DOI] [PubMed] [Google Scholar]
- 195. Jia S., Wang Z., Zhang X.-X., Liu B., Li W., Cheng S., Metagenomic analysis of cadmium and copper resistance genes in activated sludge of a tannery wastewater treatment plant, J. Environ. Biol., 2013, 34, 375. [PubMed] [Google Scholar]
- 196. Herrmann L., Schwan D., Garner R., Mobley H. L., Haas R., Schäfer K. P., Melchers K., Helicobacter pylori cadA encodes an essential Cd(II)–Zn(II)–Co(II) resistance factor influencing urease activity, Mol. Microbiol., 1999, 33 (3), 524–536. [DOI] [PubMed] [Google Scholar]
- 197. Melchers K., Schuhmacher A., Buhmann A., Weitzenegger T., Belin D., Grau S., Ehrmann M., Membrane topology of CadA homologous P-type ATPase of Helicobacter pylori as determined by expression of phoA fusions in Escherichia coli and the positive inside rule, Res. Microbiol., 1999, 150 (8), 507–520. [DOI] [PubMed] [Google Scholar]
- 198. Nahvi A., Barrick J. E., Breaker R. R., Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes, Nucleic Acids Res., 2004, 32, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Hu Y., Huang J., Jiao X.-A., Screening of genes expressed in vivo during interaction between chicken and Campylobacter jejuni, J. Microbiol. Biotechnol., 2014, 24 (2), 217–224. [DOI] [PubMed] [Google Scholar]
- 200. Taylor Z. W., Raushel F. M., Manganese-induced substrate promiscuity in the reaction catalyzed by phosphoglutamine cytidylyltransferase from Campylobacter jejuni, Biochemistry, 2019, 58 (16), 2144–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Ali M., Hussain R., Tariq F., Noreen Z., Toufiq A. M., Bokhari H., Akhtar N., ur Rahman S., Highly effective visible light-activated cobalt-doped TiO2 nanoparticles for antibacterial coatings against Campylobacter jejuni, Appl. Nanosci., 2020, 10 (3), 1005–1012. [Google Scholar]
- 202. Wolf F. I., Cittadini A., Chemistry and biochemistry of magnesium, Mol. Aspects Med., 2003, 24, 3. [DOI] [PubMed] [Google Scholar]
- 203. Effarizah M. E. P., Simon F, Characterization of a magnesium transport in Campylobacter jejuni and the role of CorA in magnesium uptake, Int. J. Curr. Microbiol. Appl. Sci., 2014, 3 (7), 83–92. [Google Scholar]
- 204. Wooldridge K. G., van Vliet A. H., Iron transport and regulation, in Konkel M. E., Ketley J. M. (eds), Campylobacter: Molecular and Cellular Biology, Horizon Bioscience, Wymondham, UK, 2005, pp. 293–310. [Google Scholar]
- 205. He Y., Ingudam S., Reed S., Gehring A., Strobaugh T. P., Irwin P., Study on the mechanism of antibacterial action of magnesium oxide nanoparticles against foodborne pathogens, J. Nanobiotechnol., 2016, 14, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Aschner J. L., Aschner M., Nutritional aspects of manganese homeostasis, Mol. Aspects Med., 2005, 26, 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Juttukonda L. J., Skaar E. P., Manganese and nutritional immunity, in Collins J. F. (ed.), Molecular, Genetic, and Nutritional Aspects of Major and Trace Minerals, Elsevier, Amsterdam, 2017, pp. 377–387. [Google Scholar]
- 208. Zackular J. P., Chazin W. J., Skaar E. P., Nutritional immunity: S100 proteins at the host–pathogen interface, J. Biol. Chem., 2015, 290 (31), 18991–18998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Mantovani A., Cassatella M. A., Costantini C., Jaillon S., Neutrophils in the activation and regulation of innate and adaptive immunity, Nat. Rev. Immunol., 2011, 11 (8), 519–531. [DOI] [PubMed] [Google Scholar]
- 210. Wu H. J., Seib K. L., Srikhanta Y. N., Kidd S. P., Edwards J. L., Maguire T. L., Grimmond S. M., Apicella M. A., McEwan A. G., Jennings M. P., PerR controls Mn-dependent resistance to oxidative stress in Neisseria gonorrhoeae, Mol. Microbiol., 2006, 60 (2), 401–416. [DOI] [PubMed] [Google Scholar]
- 211. Sanchuki H. B., Valdameri G., Moure V. R., Rodriguez J. A., Pedrosa F. O., Souza E. M., Korolik V., Ribeiro R. R., Huergo L. F., Conserved histidine residues at the ferroxidase centre of the Campylobacter jejuni Dps protein are not strictly required for metal binding and oxidation, Microbiology, 2016, 162, 156–163. [DOI] [PubMed] [Google Scholar]
- 212. Srivastava A., Dwivedi N., Samanta U., Sau A. K., Insight into the role of a unique SSEHA motif in the activity and stability of Helicobacter pylori arginase, IUBMB Life, 2011, 63 (11), 1027–1036. [DOI] [PubMed] [Google Scholar]
- 213. Komeda H., Kobayashi M., Shimizu S., A novel transporter involved in cobalt uptake, Proc. Natl Acad. Sci. USA, 1997, 94, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Bruggraber S. F., French G., Thompson R. P., Powell J. J., Selective and effective bactericidal activity of the cobalt(II) cation against Helicobacter pylori, Helicobacter, 2004, 9 (5), 422–428. [DOI] [PubMed] [Google Scholar]
- 215. Koga T., Sato K., Shimada Y., Takahashi K., Kikuchi I., Okazaki Y., Katsuta M., Iwata M., Essential role of magnesium ion in water for colonization of Helicobacter pylori in 2-week-old miniature pigs, Microbiol. Res., 2003, 158, 69–75. [DOI] [PubMed] [Google Scholar]
- 216. Öztürk N., Kurt N., Özgeriş F. B., Baygutalp N. K., Tosun M. S., Bakan N., Bakan E., Serum zinc, copper, magnesium and selenium levels in children with Helicobacter pylori infection, Eurasian J. Med., 2015, 47 (2), 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Pfeiffer J., Guhl J., Waidner B., Kist M., Bereswill S., Magnesium uptake by CorA is essential for viability of the gastric pathogen Helicobacter pylori, Infect. Immun., 2002, 70 (7), 3930–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Bereswill S., Waidner U., Odenbreit S., Lichte F., Fassbinder F., Bode G., Kist M., Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori, Microbiology, 1998, 144 (9), 2505–2516. [DOI] [PubMed] [Google Scholar]
- 219. Farnell M., Donoghue A., de los Santos F. S., Reyes-Herrera I., Cole K., Dirain M., Blore P., Pandya K., Donoghue D., Effect of oral administration of bismuth compounds on Campylobacter colonization in broilers, Poult. Sci., 2006, 85 (11), 2009–2011. [DOI] [PubMed] [Google Scholar]
- 220. Hall D. R., Gourley D. G., Leonard G. A., Duke E. M., Anderson L. A., Boxer D. H., Hunter W. N., The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds, EMBO J., 1999, 18 (6), 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Kletzin A., Adams M. W., Tungsten in biological systems, FEMS Microbiol. Rev., 1996, 18, 5–63. [DOI] [PubMed] [Google Scholar]
- 222. Smart J. P., Cliff M. J., Kelly D. J., A role for tungsten in the biology of Campylobacter jejuni: tungstate stimulates formate dehydrogenase activity and is transported via an ultra-high affinity ABC system distinct from the molybdate transporter, Mol. Microbiol., 2009, 74 (3), 742–757. [DOI] [PubMed] [Google Scholar]
- 223. Xi D., Alter T., Einspanier R., Sharbati S., Gölz G., Campylobacter jejuni genes Cj1492c and Cj1507c are involved in host cell adhesion and invasion, Gut Pathog, 2020, 12, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Taveirne M. E., Sikes M. L., Olson J. W., Molybdenum and tungsten in Campylobacter jejuni: their physiological role and identification of separate transporters regulated by a single ModE-like protein, Mol. Microbiol., 2009, 74 (3), 758–771. [DOI] [PubMed] [Google Scholar]
- 225. Mintmier B., McGarry J. M., Sparacino-Watkins C. E., Sallmen J., Fischer-Schrader K., Magalon A., McCormick J. R., Stolz J. F., Schwarz G., Bain D. J., Molecular cloning, expression and biochemical characterization of periplasmic nitrate reductase from Campylobacter jejuni, FEMS Microbiol. Lett., 2018, 365 (16), fny151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Song Z., Fu W., Zhou L., Cefuroxime, levofloxacin, esomeprazole, and bismuth as first-line therapy for eradicating Helicobacter pylori in patients allergic to penicillin, BMC Gastroenterol., 2019, 19, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Auttajaroon J., Vilaichone R.-K., Chotivitayatarakorn P., Mahachai V., Once-daily rabeprazole, levofloxacin, clarithromycin-MR, and bismuth for Helicobacter pylori eradication: a randomized study of 7 or 14 days (ONCE study), Helicobacter, 2019, 24 (5), e12615. [DOI] [PubMed] [Google Scholar]
- 228. McNicholl A. G., Bordin D. S., Lucendo A., Fadeenko G., Fernandez M. C., Voynovan I., Zakharova N. V., Sarsenbaeva A. S., Bujanda L., Perez-Aisa Á., Combination of bismuth and standard triple therapy eradicates Helicobacter pylori infection in more than 90% of patients, Clin. Gastroenterol. Hepatol., 2020, 18, 89–98. [DOI] [PubMed] [Google Scholar]
- 229. Guo Y., Guan C., Wan H., Zhang Z., Li H., Sun H., Xia W., Inactivation of NikR from Helicobacter pylori by a bismuth drug, J. Inorg. Biochem., 2019, 196, 110685. [DOI] [PubMed] [Google Scholar]
- 230. Zhang Y.-N., Ding S.-G., Huang L.-H., Zhang J., Shi Y.-Y., Zhong L.-J., Comparative proteome analysis of Helicobacter pylori clinical strains by two-dimensional gel electrophoresis, J. Zhejiang Univ. Sci. B, 2011, 12 (10), 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Inoue M., Segawa K., Matsunaga S., Matsumoto N., Oda M., Yamase T., Antibacterial activity of highly negative charged polyoxotungstates, K27 [KAs4W40O140] and K18 [KSb9W21O86], and Keggin-structural polyoxotungstates against Helicobacter pylori, J. Inorg. Biochem., 2005, 99 (5), 1023–1031. [DOI] [PubMed] [Google Scholar]
- 232. Foster A. W., Osman D., Robinson N. J., Metal preferences and metallation, J. Biol. Chem., 2014, 289 (41), 28095–28103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Waite D. W., Vanwonterghem I., Rinke C., Parks D. H., Zhang Y., Takai K., Sievert S. M., Simon J., Campbell B. J., Hanson T. E., Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.), Front. Microbiol., 2017, 8, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: recent updates and new developments, Nucleic Acids Res., 2019, 47, W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was produced for this review.