Abstract
Background
Iron is crucial for survival and growth of microbes. Consequently, limiting iron availability is a human antimicrobial defense mechanism. We explored iron and iron-related proteins as potential biomarkers in community-acquired pneumonia and hypothesized that infection-induced changes in these potential biomarkers differ between groups of pathogens and could predict microbial etiology.
Methods
Blood samples from a prospective cohort of 267 patients with community-acquired pneumonia were analyzed for hepcidin, ferritin, iron, transferrin, and soluble transferrin receptor at admission, clinical stabilization, and a 6-week follow-up. A total of 111 patients with an established microbiological diagnosis confined to 1 microbial group (atypical bacterial, typical bacterial, or viral) were included in predictive analyses.
Results
High admission levels of ferritin predicted atypical bacterial versus typical bacterial etiology (odds ratio [OR], 2.26; 95% confidence interval [CI], 1.18–4.32; P = .014). Furthermore, hepcidin and ferritin predicted atypical bacterial versus viral etiology (hepcidin: OR = 3.12, 95% CI = 1.34–7.28, P = .008; ferritin: OR = 2.38, 95% CI = 1.28–4.45, P = .006). The findings were independent of C-reactive protein and procalcitonin.
Conclusions
Hepcidin and ferritin are potential biomarkers of microbial etiology in community-acquired pneumonia.
Keywords: biomarkers, ferritin, hepcidin, iron, pneumonia
In this prospective cohort of community-acquired pneumonia patients, levels of the iron-related proteins hepcidin and ferritin were higher in atypical bacterial etiology, especially compared with viral etiology. Hepcidin and ferritin merge as potential biomarkers of microbial etiology in community-acquired pneumonia.
Almost all organisms depend on iron to thrive, and the amount of iron available to microbes affects survival and growth and, thus, their pathogenic potential [1]. The human immune system has evolved various defense mechanisms to limit iron availability, resulting in an evolutionary battle for iron between microbes and host. Systemic iron homeostasis is predominantly orchestrated by hepcidin, a peptide hormone mainly produced in the liver [2]. Production of hepcidin changes rapidly and over a wide dynamic range in response to multiple stimuli [3]. Hepcidin is suppressed in response to increased physiological iron requirements and, conversely, induced by increased iron levels to prevent iron accumulation. In addition, as part of the inflammatory response to invading microbes, the hepcidin level rapidly increase in response to inflammatory cytokines, particularly interleukin 6 (IL-6) [4]. Still, how these opposing and converging signals are integrated to regulate hepcidin expression and maintain iron homeostasis is incompletely understood [3].
Hepcidin acts by inducing degradation of the ferroportin iron transporter, causing iron entrapment intracellularly and restricting access of iron to various microbes [5]. This may inhibit replication of typical extracellular bacteria, but it may also provide a nutritional advantage for atypical bacteria with intracellular growth [6]. Induction of hepcidin has also been demonstrated in viral infections, although the contribution to pathogenesis is largely unknown [5]. Knowledge of the various mechanisms involved and the interplay between pathogen and host is emerging, yet a comprehensive overview of the iron metabolism during infections is lacking. Specific differences in host responses to pathogens need to be explored both on individual and group levels, including intracellular versus extracellular pathogens [7].
Whereas hepcidin is a key iron regulator in plasma, ferritin is the major iron storage protein and transferrin the main iron transport protein. As transferrin in plasma exceeds its iron binding capacity (assessed by transferrin saturation; s-iron/s-total iron binding capacity), iron binds to citrate, other organic acids, and albumin, collectively termed nontransferrin-bound iron (NTBI) [8]. Cellular uptake of transferrin-bound iron is mediated by transferrin receptor 1 via endocytosis according to cellular iron requirements [9]. Serum levels of the soluble form (s-transferrin receptor) are correlated to erytropoietic activity and tissue iron content [10].
During inflammation, various cytokines induce synthesis of ferritin, particularly in macrophages [7, 11]. Increased iron storage as ferritin in these cells reduces the labile iron pool intracellularly, which is believed to reduce the growth of intracellular pathogens. However, some pathogens have evolved strategies to overcome this defense mechanism [12]. In addition, ferritin has been hypothesized to leak from damaged cells during the inflammatory process, leading to higher systemic levels. Thus, it may serve as a marker of cellular damage [13].
Because infections induce changes in levels of iron and iron-related proteins that may differ among groups of pathogens, we explored the ability of these potential biomarkers to predict microbial etiology as well as their temporal dynamics in a cohort of hospitalized adult community-acquired pneumonia (CAP) patients.
MATERIALS AND METHODS
In this prospective cohort, adults (≥18 years) with suspected CAP admitted to Drammen hospital (Vestre Viken Hospital Trust, Norway) were recruited over a 3-year period (January 1, 2008–January 31, 2011).
Patient Selection and Design
Community-acquired pneumonia diagnosis was established based on the following criteria within the first 48 hours of admission: (1) identification of a new pulmonary infiltrate on chest x-ray, (2) rectal temperature ≥38.0°C, and (3) at least 1 of the following: productive or nonproductive cough, dyspnea, respiratory chest pain, crackles, or reduced respiratory sounds. Discharge from hospital within 2 weeks before admission or x-ray findings of other noninfectious pulmonary disease led to exclusion.
Criteria for clinical stabilization were evaluated daily for the first 12 days upon admission. C-reactive protein (CRP) and leucocytes were measured every second day. Clinical stabilization was defined by at least 3 of the following 4 criteria: (1) unchanged antibiotic treatment the last 2 days, (2) improvement of general condition, (3) morning rectal temperature <38°C, and (4) >25% decrease in CRP levels or leucocyte count. The participants were invited to a 6-week follow-up including blood sampling and a renewed chest x-ray.
Patient Consent Statement
All patients signed an informed consent form. The study was approved by the Regional Committee for Medical and Health Research Ethics in South-Eastern Norway (reference number S-06266a) and registered in ClinicalTrials.gov (NCT01563315).
Data Collection and Definitions
Clinical information was recorded on paper case records forms and extracted from medical records. All chest x-rays were re-examined by an experienced study radiologist blinded to the clinical data. Microbial etiology was established through an extensive microbiological investigation as described elsewhere [14] and categorized as pure typical bacterial, pure atypical bacterial, pure viral, mixed, or no established etiology. Pure etiology was defined as 1 or more pathogens identified within a particular group. Severity of illness was assessed by the clinical scoring tool CURB-65 (confusion, urea, respiratory rate, blood pressure, age ≥65 years) [15] and by intensive care unit (ICU) admissions during the hospital stay. A CURB-65 score ≤2 was defined as low risk, and a score ≥3 was defined as high risk. Further details are available in previous publications [14, 16].
Sample Collection and Analysis
Blood samples were collected at admission (within 48 hours), clinical stabilization, and 6-week follow-up. Serum was allowed to clot for at least 30 minutes, and further preparation of serum and plasma was carried out within 60 minutes after sampling by refrigerated centrifugation at 2000 ×g for 12 minutes. Several aliquots were stored at −80°C.
Analysis were performed on all samples given sufficient material. Analysis of plasma hepcidin was performed by liquid chromatography coupled with tandem mass spectrometry, which selectively measures the biological active isoform of hepcidin, hepcidin-25. This method has been validated for clinical applications at Stavanger University Hospital (Stavanger, Norway) according to Eurachem guidelines [17] and later revalidated as part of work in the Working Group on Clinical Quantitative Mass Spectrometry Proteomics of the International Federation of Clinical Chemistry. Samples used for hepcidin analysis had been through 1 freeze-thaw cycle before analysis. All samples were handled equally, and differences in serum hepcidin over 4 freeze-thaw cycles have been reported to be of minor importance [18]. An international standard for hepcidin is not yet universally applied [19], hence comparisons between groups are more relevant than the absolute values. Levels of CRP, ferritin, iron, and transferrin were all analyzed in serum on Architect ci16200 (Abbott Diagnostics, Abbott Park, IL). Serum soluble transferrin receptor was analyzed on BN ProSpec (Siemens Healthineers, Marburg, Germany). Serum procalcitonin ([PCT] BRAHMS; Thermo Fisher Scientific, Hennigsdorf, Germany) was analyzed on ADVIA Centaur (Siemens Healthineers, Tarrytown, NY). C-reactive protein and PCT analysis are previously reported [20]. HFE gene variants were detected by polymerase chain reaction amplification and melt curve analysis on Lightcycler 480 (Roche Diagnostics, Oslo, Norway) using in vitro diagnostics kit from TIB MOLBIOL (Berlin, Germany).
Data Analysis
Categorical data are presented as n (%). Normally distributed continuous variables are presented as mean (standard deviation), whereas skewed data are presented as median (25–75 percentile). Multiple group comparisons were performed using one-way analysis of variance for normally distributed continuous variables. Multiple groups of categorical data were compared (1) by Pearson χ 2 or (2) by Fisher’s exact test if the expected count was <5 in >20% of the cells. Wilcoxon signed-rank test was used to compare levels at admission and 6-week follow-up. In statistical analysis predicting microbial etiology, patients with unknown microbial etiology or copathogens across etiological groups were excluded. Ln-transformation was applied to achieve normally distributed variables for logistic regression analysis. Predictive analysis was performed using bivariate logistic regression of the ln-transformed, standardized variables to estimate odds ratios (ORs) with 95% confidence intervals (CIs). A 2-sided significance level of 0.05 was used in all analysis. Statistical analysis was performed using SPSS (version 25; IBM, Chicago, IL).
RESULTS
A total of 267 patients ≥18 years admitted to hospital with suspected CAP were included out of 320 screened patients as detailed previously [14]. Median time from admission to clinical stabilization was 4 days (range, 1–72). Among the survivors at the 6-week follow-up, 90% attended. For biomarker assessment, there were at the most 35, 61, and 43 missing values at admission, clinical stabilization, and 6-week follow-up (missing due to deaths included).
Baseline Characteristics
Baseline characteristics are presented according to microbial etiology in Table 1, and an overview of the identified microbes is provided in Supplementary Table 1. An etiological diagnosis was established in 167 patients (63%) in which 69 patients (41%) had more than 1 detected microbe, as previously published [14]. A total of 111 patients had 1 etiological diagnosis only, either pure typical bacterial, pure atypical bacterial, or pure viral. Within the typical bacterial group, Streptococcus pneumoniae was detected in a majority of the patients (82%). The only significant difference between the etiological groups in baseline characteristics was the overall count of comorbid conditions, with the lowest proportion in the atypical bacterial group.
Table 1.
Baseline Characteristics According to Microbial Etiology
| Characteristicsa | All Patients (n = 267) | Pure Typical Bacterial (n = 51) | Pure Atypical Bacterial (n = 19) | Pure Viral (n = 41) | Mixedb (n = 56) | No Established Etiology (n = 100) | P Value |
|---|---|---|---|---|---|---|---|
| Demographics | |||||||
| Age in years, mean (SD) | 64.4 (17.6) | 64.7 (19.0) | 55.5 (20.3) | 66.5 (17.2) | 63.3 (16.0) | 63.6 (17.9) | .18 |
| Female gender, n (%) | 127 (47.6) | 26 (51.0) | 8 (42.1) | 20 (48.8) | 24 (42.9) | 49 (49.0) | .90 |
| Active smoker, n (%) | 65 (24.4) | 16 (32.0) | 3 (15.8) | 7 (17.1) | 18 (32.1) | 21 (21.0) | .20 |
| Nursing home resident, n (%) | 4 (1.5) | 1 (2.0) | 0 | 2 (4.9) | 0 | 1 (1.0) | .34 |
| Comorbid Conditions, n (%) | 172 (64.4) | 32 (62.7) | 6 (31.6) | 31 (75.6) | 39 (69.6) | 64 (64.0) | .018 |
| Cardiovascular diseasec | 74 (27.7) | 16 (31.4) | 5 (26.3) | 11 (26.8) | 16 (28.6) | 26 (26.0) | .97 |
| COPD | 60 (22.5) | 14 (27.5) | 3 (15.8) | 10 (24.4) | 12 (21.4) | 21 (21.0) | .83 |
| Immunocompromisedd | 48 (18.0) | 10 (19.6) | 2 (10.5) | 8 (19.5) | 9 (16.1) | 19 (19.0) | .90 |
| Autoimmune diseasee | 34 (12.7) | 10 (19.6) | 0 | 4 (9.8) | 7 (12.5) | 13 (13.0) | .26 |
| Diabetes mellitus | 33 (12.4) | 6 (11.8) | 1 (5.3) | 7 (17.1) | 9 (16.1) | 10 (10.0) | .56 |
| Renal disease | 32 (12.0) | 6 (11.8) | 2 (10.5) | 4 (9.8) | 11 (19.6) | 9 (9.0) | .42 |
| Neurological diseasef | 19 (7.1) | 1 (2.0) | 0 | 4 (9.8) | 2 (3.6) | 12 (12.0) | .087 |
| Dementia | 15 (5.6) | 4 (7.8) | 2 (10.5) | 2 (4.9) | 1 (1.8) | 6 (6.0) | .47 |
| Liver disease | 4 (1.5) | 1 (2.0) | 0 | 0 | 1 (1.8) | 2 (2.0) | 1.00 |
| Severity of Illness, n (%) | |||||||
| CURB-65 ≥3 | 97 (37.9) | 17 (34.7) | 8 (42.1) | 14 (35.9) | 24 (44.4) | 34 (35.8) | .81 |
| ICU admission | 48 (18.0) | 7 (13.7) | 5 (26.3) | 7 (17.1) | 9 (16.1) | 20 (20.0) | .74 |
Abbreviations: COPD, chronic obstructive pulmonary disease; CURB-65, confusion, urea, respiratory rate, blood pressure, age ≥65 years; ICU, intensive care unit; SD, standard deviation.
a<5% missing data.
bMix of at least 2 of the groups: typical bacterial, atypical bacterial, and viral.
cHeart failure, coronary heart disease, cerebrovascular disease, peripheral artery disease.
dPrimary or secondary immunodeficiency, active malignancy, immunosuppressive drugs.
eRheumatoid arthritis, systemic lupus erythematosus, inflammatory bowel disease, autoimmune hepatitis, Sjogren’s disease, psoriasis.
fCentral nervous disease, neuromuscular disease.
NOTE: Bold indicates P < .05.
Dynamics of Iron and Iron-Related Proteins During the Course of Community-Acquired Pneumonia
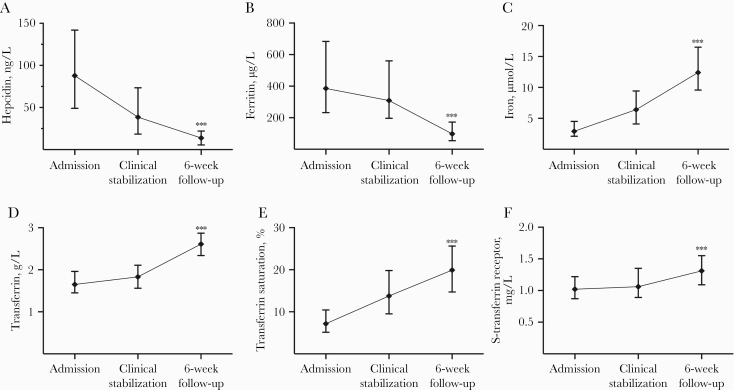
All potential biomarkers showed statistically significant differences in the acute-phase compared with the 6-week follow-up (Figure 1). Whereas serum levels of iron, transferrin, transferrin saturation, and soluble transferrin receptor increased during follow-up, serum levels of hepcidin and ferritin were highest at admission (6.3- and 3.9-fold change compared with the 6-week follow-up).
Figure 1.
Levels of iron and iron-related proteins during the course of pneumonia. Data are shown as median and 25–75 percentiles. Comparison of level at admission and 6-week follow-up by Wilcoxon signed-rank test. ***, P < .001. Reference ranges in parenthesis: (A) hepcidin (morning levels; women, <0.3–32 ng/mL; men, 0.9–34 ng/mL); (B) ferritin (women, 15–200 μg/L; men, 30–400 μg/L); (C) iron (9–34 µmol/L); (D) transferrin (2.0–3.3 g/L); (E) transferrin saturation (women 18–49 years, 10%–50%; women ≥50 years, 15%–50%; men, 15%–57%); (F) soluble transferrin receptor (0.76–1.76 mg/L).
Association Between the Potential Biomarkers and Microbial Etiology
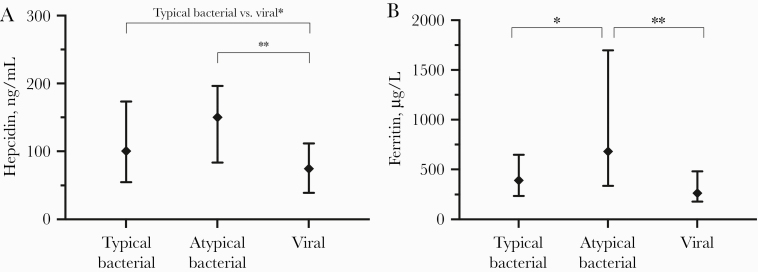
Comparing the 2 bacterial groups, admission levels of ferritin were significantly higher in patients with atypical bacterial etiology (Figure 2). Both hepcidin and ferritin levels were significantly higher in patients with atypical bacterial etiology compared with viral etiology. Hepcidin levels were also higher in typical bacterial etiology compared with viral etiology.
Figure 2.
Levels of iron and iron-related proteins at admission according to microbial etiology. Data are displayed as median and 25–75 percentile. Two-group comparison by Mann-Whitney U test. *, P < .05; **, P < .01. (A) Hepcidin; (B) ferritin.
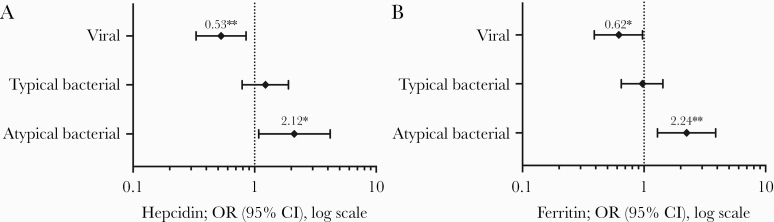
The ability of hepcidin and ferritin to predict microbial etiology is displayed in Figure 3 by comparing 1 etiological group to the 2 other groups combined. High levels of both hepcidin and ferritin were associated with a higher risk of atypical bacterial etiology (hepcidin: OR = 2.12, 95% CI = 1.08–4.18, P = .030; ferritin: OR = 2.24, 95% CI = 1.29–3.89, P = .004) and a lower risk of viral etiology (hepcidin: OR = 0.53, 95% CI = 0.33–0.85, P = .009; ferritin: OR = 0.62, 95% CI = 0.39–0.97, P = .035).
Figure 3.
Forest plots; each etiological group is compared with the other 2 groups. Odds ratios (ORs) are calculated on basis of ln-transformed and standardized values. *, P < .05; **, P < .01. (A) Hepcidin; (B) ferritin. CI, confidence interval.
We further explored predictive comparisons between single etiological groups (Table 2). Ferritin had the ability to discriminate atypical bacterial from both typical bacterial and viral etiology but not typical bacterial from viral etiology. Hepcidin discriminated both atypical bacterial and typical bacterial etiology from viral etiology but not between the 2 bacterial groups. In contrast, iron, transferrin saturation, and soluble transferrin receptor were not, or only modestly, associated with microbial etiology of CAP.
Table 2.
Prediction of Microbial Etiology by Logistic Regression of Admission Levels of Potential Biomarkers
| Atypical Bacterial vs Typical Bacterial OR (95% CI), P Value | Atypical Bacterial vs Viral OR (95% CI), P Value | Typical Bacterial vs Viral OR (95% CI), P Value | ||||
|---|---|---|---|---|---|---|
| Levels at Admission | Univariate | Multivariatea | Univariate | Multivariatea | Univariate | Multivariatea |
| Hepcidin | 1.64 (0.82–3.28), .16 | 2.64 (0.99–7.02), .053 | 3.12 (1.34–7.28), .008 | 3.22 (1.20–8.64), .020 | 1.68 (1.01–2.77), .044 | 1.16 (0.65–2.06), .62 |
| Ferritin | 2.26 (1.18–4.32), .014 | 3.50 (1.42–8.61), .006 | 2.38 (1.28–4.45), .006 | 2.41 (1.15–5.02), .019 | 1.35 (0.84–2.17), .21 | 0.97 (0.57–1.66), .91 |
| Iron | 0.87 (0.50–1.52), .63 | 0.71 (0.34–1.48), .37 | 0.62 (0.31–1.23), .17 | 0.74 (0.35–1.56), .43 | 0.78 (0.51–1.21), .27 | 1.03 (0.63–1.70), .90 |
| Transferrin | 0.83 (0.48–1.45), .52 | 0.83 (0.44–1.57), .57 | 0.50 (0.26–0.99), .045 | 0.55 (0.26–1.17), .12 | 0.68 (0.44–1.05), .079 | 0.88 (0.55–1.43), .62 |
| Transferrin saturation | 0.94 (0.54–1.65), .83 | 0.84 (0.44–1.57), .58 | 0.86 (0.47–1.60), .64 | 0.97 (0.51–1.85), .93 | 0.94 (0.62–1.42), .76 | 1.07 (0.68–1.69), .76 |
| S-Transferrin receptor | 1.16 (0.67–1.98), .60 | 1.07 (0.59–1.93), .83 | 0.79 (0.42–1.46), .45 | 0.70 (0.35–1.38), .30 | 0.73 (0.48–1.10), .13 | 0.92 (0.59–1.44), .71 |
| CRP | 0.92 (0.59–1.45), .73 | NA | 1.39 (0.80–2.42), .25 | NA | 1.51 (1.01–2.24), .043 | NA |
| Procalcitonin | 0.46 (0.25–0.84), .011 | NA | 1.04 (0.59–1.85), .89 | NA | 2.24 (1.41–3.56), .001 | NA |
Abbreviations: CI, confidence interval; CRP, C-reactive protein; NA, not applicable; OR, odds ratio.
aAdjusted for CRP and procalcitonin.
NOTES: ORs are calculated on basis of ln-transformed and standardized values. Bold indicates P < .05.
Multivariate analysis with adjustment for CRP and PCT was included, because these are the most studied biomarkers in clinical use for this purpose [21]. Our main findings were largely unchanged after adjustment for CRP and PCT in the logistic regression analysis (Table 2). However, the borderline significant ability of hepcidin to discriminate between typical bacterial and viral microbial etiology in univariate analysis did not sustain.
Association Between the Potential Biomarkers and HFE Genotype
Because genetic variation in the HFE gene may influence levels of hepcidin and ferritin, we also analyzed 2 single-nucleotide variants in the HFE gene, Cys282Tyr (C282Y) and His63Asp (H63D), to assess the ability of the potential biomarkers to predict HFE genotype during acute-phase pneumonia (Supplementary Table 2). Increased levels of serum iron and transferrin saturation were associated with the 7 cases of potential disease-related genotypes: C282Y/C282Y and C282Y/H63D. However, we did not find any association between hepcidin or ferritin levels and the most common heterozygous HFE genotypes, C282Y/WT and H63D/WT.
DISCUSSION
All potential biomarkers changed profoundly during the course of acute-phase pneumonia, reflecting a high degree of systemic inflammation. As expected, serum iron and transferrin saturation were low in the acute phase compared with the 6-week follow-up, and they were far below the lower reference limits. These findings are consistent with, and probably due to, the increase of the main iron regulatory hormone hepcidin [2]. Indeed, both hepcidin and ferritin showed an opposite pattern with the highest levels at admission.
A major finding of this explorative study was that admission levels of ferritin predicted atypical bacterial versus typical bacterial etiology. Furthermore, levels of both hepcidin and ferritin were particularly higher in atypical bacterial etiology compared with viral etiology. Accordingly, both hepcidin and ferritin showed predictive power of atypical bacterial versus viral etiology.
The battle for iron between microbes and host is multifaceted, and on an individual level, microbes utilize different strategies to extract iron [22]. The host strategies for withholding iron and their effect also vary depending on the intra- or extracellular location of the microbes. The increased levels of ferritin and hepcidin in atypical bacterial pneumonia in our study are novel observations, supporting diverse mechanisms of iron regulation depending on pathogen category.
Increased levels of hepcidin is theoretically an indirect measure of intracellular iron entrapment, which has been shown to promote intracellular pathogen survival and growth in vitro [6], a finding supported by studies of cultured macrophages [23]. On the other hand, mice studies indicate a pathogen-induced downregulation of ferroportin expression independent of hepcidin and iron levels [24], and that elevated hepcidin levels may not promote intracellular infections [25]. Possible explanations of such hepcidin-independent involvement are specific microbe-induced iron regulation, or a compartmentalization of iron within the macrophages, thus restricting iron access to intracellular pathogens [8]. Although hepcidin is mainly produced in the liver, mice studies suggest that infection can induce a peripheral hepcidin production by leukocytes, macrophages, and monocytes similar to ferritin that may influence iron availability locally [26–28], but not necessarily change systemic hepcidin levels and iron balance [29]. In general, local regulation may differ considerably from the systemic effects assessed by measurement of biomarkers in serum [7]. Because iron is tightly bound to transferrin throughout the physiological concentration range, it has been argued that the primary defense mechanism of hepcidin induction during infection is mainly to prevent NTBI formation that may occur due to infection-related destruction of tissue and erythrocytes [25]. Nontransferrin-bound iron comprises a heterogeneous group of iron species that may be more easily utilizable to pathogens than transferrin-bound iron and may enhance growth of some microbes [25]. Thus, in addition to withholding iron from invading microbes, the hepcidin response during infection may protect the host from toxic effects of NTBI [1]. Furthermore, in vitro evidence of direct antimicrobial effects of hepcidin has been demonstrated, but supporting evidence in vivo in humans are yet sparse [5]. Finally, we can only speculate whether the hepcidin levels were elevated before or as a consequence of atypical bacterial pneumonia. High baseline hepcidin levels may theoretically promote atypical bacterial infections.
Because hyperferritinemia may be cytokine-induced or related to cellular damage [11, 13], the degree of cellular damage in pneumonia as well as the preferred intracellular or extracellular site of infection may be important. Hypothetically, the higher level of ferritin in atypical bacterial infections could be related to a more profound cellular damage in this group or be linked to its intracellular location. Atypical bacterial infections also differ from other bacterial infections of the lung by the prevalence of extrapulmonary manifestations [30], and ferritin leakage due to secondary organ involvement may contribute to elevated ferritin levels.
It should be noted that the term atypical bacteria is not a clearly defined entity [31–33]. The pathogens we included as atypical bacteria in this study are either predominantly intracellular (Legionella pneumophila, Mycoplasma pneumoniae, Chlamydia pneumoniae) [34] or there is evidence of some degree of intracellular involvement (Bordetella pertussis) [35]. To complicate this distinction, the most common CAP pathogen S pneumoniae may also occasionally reside intracellularly [36]. Nevertheless, we believe that our categorization is sound, and we hypothesize that a key point to the particularly increased levels of hepcidin and ferritin in atypical pneumonia is their dominant intracellular pathophysiology. Viruses are obligate intracellular, but, unlike atypical bacteria, their reproduction relies entirely on the machinery of the host cells [37]. Although the pathophysiological mechanisms resulting in the measured levels of hepcidin and ferritin have yet to be detailed, the finding of lower hepcidin and ferritin levels among patients with viral pneumonia compared with atypical bacterial pneumonia suggest that there are differences in the iron-related immune response between these intracellular pathogen groups that should be further explored.
More importantly, the abilities of hepcidin and ferritin to discriminate between atypical bacterial and viral pneumonia were independent of CRP and PCT. Indeed, neither CRP nor PCT discriminated between atypical bacterial and viral causes. However, as expected, CRP discriminated typical bacterial from viral etiology, although by a lower odds ratio than what we found for hepcidin and ferritin in predicting atypical bacterial etiology. Otherwise, CRP failed to separate the etiological groups. Compared with CRP, PCT showed a better ability to discriminate between viral and typical bacterial causes. The use of a combination of iron-related and traditional inflammatory biomarkers may be a future strategy to predict microbial etiology. Because there were no differences in CURB-65 or ICU admissions between the etiological groups, we do not believe that the differences in biomarker levels between etiological groups were related to disease severity.
Although comparative analyses show significant differences, the number of patients with an atypical bacterial infection in the present study is limited. In addition, being a single-center study with specific inclusion and exclusion criteria, the external validity to other populations must be carefully considered. The microbial etiology of CAP was established through extensive microbiological investigations. Still, a definite etiological diagnosis was not obtained for 37% of patients, and undetected copathogens may have biased the etiological groups. Larger studies are needed to validate our findings, preferably with a power to distinguish between single pathogens as well as etiological groups, because pathogens within the atypical bacterial group may also be different. Finally, the statistical analyses in the present study are used to assess associations and cannot prove causal relationships.
CONCLUSIONS
In this prospective cohort of CAP patients, high admission levels of ferritin and hepcidin were in particular associated with a higher risk of atypical bacterial etiology and a lower risk of viral etiology. Hepcidin and ferritin emerge as potential biomarkers of microbial etiology in CAP.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank Ulla Helene Enger (Department of Laboratory Medicine, Drammen Hospital, Vestre Viken Hospital Trust, Drammen, Norway) for technical assistance and guidance in the analysis of HFE gene variants.
Author contributions. L. H., J. C. H., E. H., W. W. S., T. L., P. A., T. U., and K. O. conceptualized and designed the study. C. B., Ø. S., T. L., A. E. M., and K. O. contributed to the acquisition of data. T. U. and K. O. performed statistical analysis. L. H., T. U., P. A., T. S., E. H., J. C. H., and K. O. were central in interpretation of the data. K. O. and L. H. drafted the manuscript. All authors revised it critically for important intellectual content and approved the final version
Financial support. Siemens Healthineers provided reagent kits for s-transferrin receptor analysis.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Ganz T, Nemeth E. Iron homeostasis in host defence and inflammation. Nat Rev Immunol 2015; 15:500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sangkhae V, Nemeth E. Regulation of the iron homeostatic hormone hepcidin. Adv Nutr 2017; 8:126–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sangkhae V, Nemeth E. To induce or not to induce: the fight over hepcidin regulation. Haematologica 2019; 104:1093–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Girelli D, Nemeth E, Swinkels DW. Hepcidin in the diagnosis of iron disorders. Blood 2016; 127:2809–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Michels K, Nemeth E, Ganz T, Mehrad B. Hepcidin and host defense against infectious diseases. PLoS Pathog 2015; 11:e1004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paradkar PN, De Domenico I, Durchfort N, et al. . Iron depletion limits intracellular bacterial growth in macrophages. Blood 2008; 112:866–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nairz M, Dichtl S, Schroll A, et al. . Iron and innate antimicrobial immunity-depriving the pathogen, defending the host. J Trace Elem Med Biol 2018; 48:118–33. [DOI] [PubMed] [Google Scholar]
- 8. Ganz T. Iron and infection. Int J Hematol 2018; 107:7–15. [DOI] [PubMed] [Google Scholar]
- 9. Kawabata H. Transferrin and transferrin receptors update. Free Radic Biol Med 2019; 133:46–54. [DOI] [PubMed] [Google Scholar]
- 10. Harms K, Kaiser T. Beyond soluble transferrin receptor: old challenges and new horizons. Best Pract Res Clin Endocrinol Metab 2015; 29:799–810. [DOI] [PubMed] [Google Scholar]
- 11. Cullis JO, Fitzsimons EJ, Griffiths WJ, et al. ; British Society for Haematology . Investigation and management of a raised serum ferritin. Br J Haematol 2018; 181:331–40. [DOI] [PubMed] [Google Scholar]
- 12. Parrow NL, Fleming RE, Minnick MF. Sequestration and scavenging of iron in infection. Infect Immun 2013; 81:3503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kell DB, Pretorius E. Serum ferritin is an important inflammatory disease marker, as it is mainly a leakage product from damaged cells. Metallomics 2014; 6:748–73. [DOI] [PubMed] [Google Scholar]
- 14. Holter JC, Müller F, Bjørang O, et al. . Etiology of community-acquired pneumonia and diagnostic yields of microbiological methods: a 3-year prospective study in Norway. BMC Infect Dis 2015; 15:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim WS, van der Eerden MM, Laing R, et al. . Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax 2003; 58:377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holter JC, Ueland T, Jenum PA, et al. . Risk factors for long-term mortality after hospitalization for community-acquired pneumonia: a 5-year prospective follow-up study. PLoS One 2016; 11:e0148741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Magnusson B, Örnemark U, (eds.). Eurachem Guide: The Fitness for Purpose of Analytical Methods—A Laboratory Guide to Method Validation and Related Topics. 2nd ed. 2014. Available from: http://www.eurachem.org [Google Scholar]
- 18. Kemna EH, Tjalsma H, Podust VN, Swinkels DW. Mass spectrometry-based hepcidin measurements in serum and urine: analytical aspects and clinical implications. Clin Chem 2007; 53:620–8. [DOI] [PubMed] [Google Scholar]
- 19. Aune ET, Diepeveen LE, Laarakkers CM, et al. . Optimizing hepcidin measurement with a proficiency test framework and standardization improvement. Clin Chem Lab Med 2020; 59:315–23. [DOI] [PubMed] [Google Scholar]
- 20. Siljan WW, Holter JC, Michelsen AE, et al. . Inflammatory biomarkers are associated with aetiology and predict outcomes in community-acquired pneumonia: results of a 5-year follow-up cohort study. ERJ Open Res 2019; 5:00014-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Savvateeva EN, Rubina AY, Gryadunov DA. Biomarkers of community-acquired pneumonia: a key to disease diagnosis and management. Biomed Res Int 2019; 2019:1701276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Palmer LD, Skaar EP. Transition metals and virulence in bacteria. Annu Rev Genet 2016; 50:67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chlosta S, Fishman DS, Harrington L, et al. . The iron efflux protein ferroportin regulates the intracellular growth of Salmonella enterica. Infect Immun 2006; 74:3065–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Willemetz A, Beatty S, Richer E, et al. . Iron- and hepcidin-independent downregulation of the iron exporter ferroportin in macrophages during salmonella infection. Front Immunol 2017; 8:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stefanova D, Raychev A, Arezes J, et al. . Endogenous hepcidin and its agonist mediate resistance to selected infections by clearing non-transferrin-bound iron. Blood 2017; 130:245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Motley ST, Morrow BJ, Liu X, et al. . Simultaneous analysis of host and pathogen interactions during an in vivo infection reveals local induction of host acute phase response proteins, a novel bacterial stress response, and evidence of a host-imposed metal ion limited environment. Cell Microbiol 2004; 6:849–65. [DOI] [PubMed] [Google Scholar]
- 27. Peyssonnaux C, Zinkernagel AS, Datta V, et al. . TLR4-dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood 2006; 107:3727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Theurl I, Theurl M, Seifert M, et al. . Autocrine formation of hepcidin induces iron retention in human monocytes. Blood 2008; 111:2392–9. [DOI] [PubMed] [Google Scholar]
- 29. Drakesmith H, Prentice AM. Hepcidin and the iron-infection axis. Science 2012; 338:768–72. [DOI] [PubMed] [Google Scholar]
- 30. Cunha BA. The atypical pneumonias: clinical diagnosis and importance. Clin Microbiol Infect 2006; 12:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murdoch DR, Chambers ST. Atypical pneumonia–time to breathe new life into a useful term? Lancet Infect Dis 2009; 9:512–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woodhead M, Blasi F, Ewig S, et al. ; European Respiratory Society; European Society of Clinical Microbiology and Infectious Diseases . Guidelines for the management of adult lower respiratory tract infections. Eur Respir J 2005; 26:1138–80. [DOI] [PubMed] [Google Scholar]
- 33. Mandell LA, Wunderink RG, Anzueto A, et al. ; Infectious Diseases Society of America; American Thoracic Society . Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 2007; 44:S27–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cillóniz C, Torres A, Niederman M, et al. . Community-acquired pneumonia related to intracellular pathogens. Intensive Care Med 2016; 42:1374–86. [DOI] [PubMed] [Google Scholar]
- 35. Martín C, Etxaniz A, Uribe KB, et al. . Adenylate cyclase toxin promotes bacterial internalisation into non phagocytic cells. Sci Rep 2015; 5:13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Subramanian K, Henriques-Normark B, Normark S. Emerging concepts in the pathogenesis of the Streptococcus pneumoniae: from nasopharyngeal colonizer to intracellular pathogen. Cell Microbiol 2019; 21:e13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eisenreich W, Rudel T, Heesemann J, Goebel W. How viral and intracellular bacterial pathogens reprogram the metabolism of host cells to allow their intracellular replication. Front Cell Infect Microbiol 2019; 9:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.