Abstract
Objectives:
The comprehensive in silico study aims to figure out the most effective aromatic phytochemical ligands among a number from a library, considering their pharmacokinetic efficacies in blocking “angiotensin-converting enzyme 2 (ACE2) receptor–severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) S protein” complex formation as part of a target-specific drug designing.
Materials and Methods:
A library of 57 aromatic pharmacophore phytochemical ligands was prepared from where the top five ligands depending on Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) and quantitative structure-activity relationship (QSAR)-based pharmacokinetic properties were considered. The selected ligands were optimized for commencing molecular docking and dynamic simulation as a complex with the ACE2 receptor to compare their blocking efficacy with the control drug. The ligand–receptor complexes’ accuracy in preventing the Spike (S) protein of SARS-CoV-2 penetration inside the host cells has been analyzed through hydrogen–hydrophobic bond interactions, principal component analysis (PCA), root mean square deviation (RMSD), root mean square fluctuation (RMSF), and B-Factor. Advanced in silico programming language and bioanalytical software were used for high throughput and authentic results.
Results:
ADMET and QSAR revealed Rhamnetin, Lactupicrin, Rhinacanthin D, Flemiflavanone D, and Exiguaflavanone A as the ligands of our interest to be compared with the control Cassiarin D. According to the molecular docking binding affinity to block ACE2 receptor, the efficiency mountings were Rhinacanthin D > Flemiflavanone D > Lactupicrin > Exiguaflavanone A > Rhamnetin. The binding affinity of the Cassiarin D–ACE2 complex was (−10.2 KJ/mol) found inferior to the Rhinacanthin D–ACE2 complex (−10.8 KJ/mol), referring to Rhinacanthin D as a more stable candidate to use as drugs. The RMSD values of protein–ligand complexes evaluated according to their structural conformation and stable binding pose ranged between 0.1~2.1 Å. The B-factor showed that very few loops were present in the protein structure. The RMSF peak fluctuation regions ranged 5–250, predicting efficient ligand–receptor interactions.
Conclusion:
The experiment sequentially measures all the parameters required in referring to any pharmacophore as a drug, considering which all aromatic components analyzed in the study can strongly be predicted as target-specific medication against the novel coronavirus 2019 infection.
Keywords: Antiviral efficacy, molecular docking, dynamic simulation, principal component analysis, targeted drug designing
Introduction
Coronavirus is a type of zoonotic virus belonging to the Coronaviridae family containing a large positive single-stranded RNA genome of 26~32 kb in size [1]. According to serology and genotype, this family is categorized into four genera: alpha, beta, gamma, and delta coronaviruses [2]. Coronaviruses render severe fatality invertebrates. For instance, alpha and beta coronaviruses affect mostly mammals. In reverse, gamma and delta genera cause diseases in avian species [3]. Up until now, seven members of coronavirus have been detected that provoke illness in humans, among these are OC43, 229E, HKU1, and NL63 that cause mild respiratory sickness in humans [4].
Another three family members are severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), leading to severe respiratory illness in humans for decades [5]. The current century has already faced two highly infectious viruses before, severely influencing human civilization [6]. Starting from 2002, SARS-CoV has outbroken in 30 territories, especially in China, and has taken away about 800 lives with over 8,000 cases. In 2012, MERS-CoV had a outbreak in Saudi Arabia, and till now, about 2,500 cases have been reported, and about 860 deaths have been confirmed from the MERS-CoV invasion [7]. Both SARS-CoV and MERS-CoV manifest interspecies transmission potential and are considered animal-transmitted to humans, using the civet cat and camel as their intermediate hosts [8]. These two coronaviruses show similar flu-type illnesses, prevailing fever, breathing issues, cough, and gastrointestinal problems, which can be fatal with multiple comorbidities [9]. Following SARS-CoV and MERS-CoV, the world has been experiencing a SARS-CoV-2 global pandemic with the novel mutated form of the former SARS-CoV that causes severe pneumonia-like lower respiratory tract illness, firstly experienced in Wuhan in December 2019 [10].
Over 100.9 million people have been found positive with the virus, and more than 2.4 million deaths have been confirmed among all the coronavirus disease 2019 (COVID-19) cases, under the study with nearly 12 months of follow-up [11]. World Health Organization refers to the present circumstance as a global pandemic due to the spreading out of the virus to over 200 nations [12]. SARS-CoV-2 falls in the beta-coronavirus genus like SARS-CoV and MERS-CoV, but is comparatively more contagious than those two viruses [13]. Although SARS-CoV-2 infection shows indistinguishable symptoms (cold, fever, sore throat, and fatigue) like normal flu, such as influenza A/B, the mortality rate is 3–4% higher than influenza infection [14,15]. Around six open reading frame regions in the virus genome encoded by 16 nonstructural proteins (1–16) necessary for the viral replication process along with four proteins means membranes (M), envelope (E), nucleocapsid (N), and spike (S) glycoprotein are for structural fabrication [16]. Angiotensin-converting enzyme 2 (ACE2) is considered as the mother checkpoint of transpassing SARS-CoV-2 inside the host cellular system [17].
ACE2 receptor is a type 1 monocarboxy peptidase glycoprotein that is distributed in most of the body’s organs, especially in the respiratory tract, the esophagus, ileum heart, and kidney [18]. ACE2 gene lying under chromosome Xp22 encodes the ACE2 receptor protein composed of two vital domains: the C and the N terminal domains. ACE2 shares its 40% homology with that domain of ACE via the N terminus region and contains a carboxypeptidase site that plays a vital role in binding SARS-CoV-2 when the C terminal domain helps in anchoring the membrane [19]. SARS-CoV-2 manipulates the S glycoprotein to mediate viral entry to the host like SARS-CoV. Spike glycoprotein is a homo-trimeric protein; it usually exists in a metastable, inactive, and perfusion conformation [20]. This protein’s conformational change occurs at virus–host cell interaction interface, leading through membrane fusion mechanisms [21]. The S glycoprotein cleavage is essential for ACE2 binding, followed by surface membrane penetration and viral entry inside the host [22]. “S glycoprotein” of coronavirus becomes split by the host transmembrane serine protease 2 protein into two subunits known as S1 and S2. The S1 subunit possesses a receptor-binding domain (RBD) region that externally circuits with the N-terminal domain of the ACE2 receptor. Simultaneously, the S2 helps fuse the viral membrane with the host to facilities the viral entrance inside [23].
The SARS-COV-2 RBD of S1 specifically recognizes ACE2 receptors, and the receptor-binding motif mediates the contact between them as part of the RBD. RBDs are slightly exposed to S2 fusion machinery on the virus’s surface and show transiently open and closed states one after another. The open states allow the engagement of receptors, followed by the loosening of S1 and finally membrane fusion by refolding the S2 subunit [24]. The S2 subunit comprises several domains like putative fusion peptide (FP), heptad repeat (HR), transmembrane fusion, and CT, followed by the association of RBD and ACE2 receptor; the FP domain of S2 subunit leads to the conformational change of S2 subunit and the HR domain contains HR1 and HR2, which are expected to lead to viral fusions through forming fusion core [25].
SARS-CoV-2 binds more efficaciously to the ACE2 receptor in comparison with SARS-CoV, and the binding strength of SARS-CoV-2 is 10–20 times greater than its homologous one [10]. SARS-CoV-2 binding activity to different animals, for example pets, farm animals, bats, dogs, pangolins, tigers, lions, and Chinese hamster, allows it to enter the host through the ACE2 proteins for infection [26]. Mainly they act as a putative intermediate host of SARS-CoV-2 due to close contact with humans [27]. The current research has demonstrated that cells remain uninfected by SARS-CoV-2 where no ACE2 receptor is present, but only the cells that have expressed ACE2 on its surface [28].
The viral genome is mutative; therefore, they are exhibiting resistance to antiviral drugs [29]. From the primitive age, many plants are being used to treat diseases as they have potent medicinal properties [30]. Plants that synthesize many phytochemicals show pharmacophore activities against various diseases and exhibit phenomenal bioactive features such as antioxidant, antiviral, antibacterial, antifungal, anti-inflammatory, and anti-cancer properties with a few side effects and toxicity as well [31,32]. Various secondary metabolites, such as flavonoids, terpenes, alkaloids, lignins, tannins, polysaccharides, etc., show significant antiviral activity to fight against many infectious viruses like dengue virus, hepatitis C virus, hepatitis B virus, hemagglutinin type 1 and neuraminidase type 1, porcine epidemic diarrhea virus, human immunodeficiency virus, ebola, retroviruses, and so on [33].
Many pharmaceutical industries are trying hard to invent potential antiviral drugs that can effectively beat coronavirus [34]. Various research articles have revealed that several phytochemicals could efficiently bind to the ACE2 receptor, therefore significantly halting the interaction of viral “S” glycoprotein with this receptor and inhibiting the propagation of the virus [35,36]. An integrated approach of investigation comprising in silico with other in vivo activities is required among the researchers working with COVID-19.
In the current research, a library of phytochemical components was developed depending on their pharmacokinetics to select the best pharmacophore ligands in using effective therapeutics in blocking ACE2 receptor as part of the novel coronavirus 2019 (nCoV) treatment through advanced in silico approaches. Besides, the potentiality of selective phytochemical ligands in using targeted drug modeling for the ACE2 receptor blockage was also studied compared to the control ligands.
Materials and Methods
Construction of the phytochemical library
A library of selective phytochemical compounds was constructed by screening PubChem (authorized by National Center for Biotechnology Information, as part of National Library of Medicine), depending on the established literature. In that case, based on pharmacokinetic properties, a total of 57 pharmacophore ligands were generated.
Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) and Quantitative Structure–Activity Relationship (QSAR) profiling for pharmacokinetic analysis and ligand validation
All the 57 pharmacophore ligands from the constructed library were profiled for understanding their pharmacokinetic properties through ADMET profiling, which illustrates the disposition of a pharmaceutical compound within an organism and measures the drug kinetics of the tissue [37]. In that case, “Molinspiration Cheminformatics” (https://www.molinspiration.com/cgi-bin/properties) along with “Swiss ADME” (http://www.swissadme.ch/index.php) were conducted initially to assess ADME parameters using Lipinski’s rule of violation measurement and drug-likeness features [38]. To understand the high throughput toxicity analysis from the ADMET test, “pkCSM” (http://biosig.unimelb.edu.au/pkcsm/prediction) was considered [39] as a secondary option. Finally, the QSAR testing was conducted for analyzing the efficacy of the ligands used as drugs, which was undertaken through “admetSAR 2” (http://lmmd.ecust.edu.cn/admetsar2/). Considering the ADMET with QSAR profiling, the top five ligands were selected for the molecular docking and dynamics simulation steps from the library of 57 ligands to compare with the control drug Cassiarin D.
Optimization of ligands
The selected ligands were retrieved from the PubChem database in structure data files format to minimize their energy following Gasteiger’s method, where the net charge of the compound was at “zero”, as part of the optimization based on “UCSF Chimera Software Version-1.14” (https://www.cgl.ucsf.edu/chimera/). After minimizing energy, all selected compounds were converted to mol2 format, which is an indispensable part of molecular docking analysis [40].
Extracting targeted macromolecules for optimization
The non-mutated, tertiary micro-crystal structure of the targeted ACE2 Protein Data Bank (PDB ID: 1R4L) was retrieved from the PDB (https://www.rcsb.org/), as the receptor macromolecule of the research. The protein was optimized sequentially in aspects of the protein chain selection, removing unwanted objects like heteroatoms, water molecules, metal ions, ligands, and extra chain subunits interacting with the receptor. Necessary hydrogen atoms were added as part of optimizing the macromolecule for perfect docking. The total optimization was conducted with the tool “UCSF Chimera Software Version-1.14” (https://www.cgl.ucsf.edu/chimera/) [36], and the optimized protein structure was retrieved and conserved for further analysis.
Molecular docking analysis
To predict the efficacy of the optimized ligands mentioned above used as drugs, a protein–ligand docking was conducted targeting the ACE2 receptor as macromolecule through PyRx 0.8 package (based on Auto-Dock Vina configuration). The binding affinity and root mean square deviation (RMSD) values were calculated from the docking results (CSV).
Visualization of the docked file
The two PDBQT output files within the PyRx tool were retrieved after completing the ligands’ dockings on the macromolecule separately. The protein–ligand complex structures were preliminarily studied by the DS Visualizer (64 bit) (http://media.accelrys.com/downloads/visualizer/45/DS45Client.exe). PyMOL Version 2.4.1 (https://pymol.org/2/) was secondarily applied for further structural study and the data were saved as a PDB file.
Analysis of the non-bond interaction and hydrophobicity of the docked file
The final (tertiary) analysis of the non-bonded interactions and the non-covalent interaction (polar and hydrophobic) were conducted by LigPlot+ (V.2.2), based on the java interface (Java SE Runtime Environment 8u271), where only the PDB file obtained from PyMOL was used.
Molecular dynamic simulation
In this research, the receptor (ACE2) was dynamically simulated for 10 ns without any ligand bound to it before commencing the protein–ligand docking step to figure out the protein behavior changes over time interacting with the surrounding water molecules and ions. In that case, CABS-flex 2.0 (http://biocomp.chem.uw.edu.pl/CABSflex2/) molecular dynamics simulator got preference. Finally, the protein–ligand docked files commenced with molecular dynamics simulations (MDS) up to 1,100 ps (1.1 ns) via using LARMD molecular dynamics simulator (http://chemyang.ccnu.edu.cn/ccb/server/LARMD/index.php) to analyze their PCA, RMSD, root mean square fluctuation (RMSF), and B-factor results of each protein–ligand interaction separately [41].
Statistical analysis
The necessary statistical analysis of the dynamic simulation-generated data were carried out using “R programming” (version R-4.0.2) [42,43] and “GraphPad Prism” (version 8.0.1) [44,45].
Results
Pharmacokinetics analysis
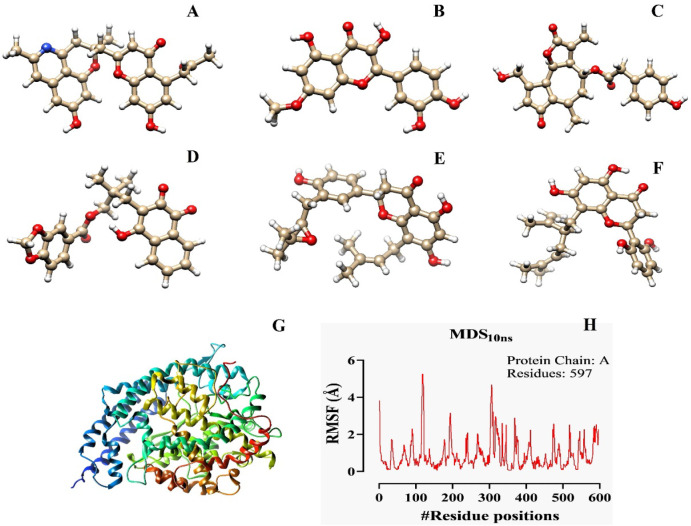
In this research, six ligands (one used as a standard) were selected out of 57 from the initial pharmacophore database according to the ADMET and QSAR analyses. The ligands revealed a better excretion rate from the body after metabolism, showing maximum tolerance at the doses range between 0.438 and 0.506 (log mg/kg/day) (Table 1). All ligands and macromolecules’ crystal structures are shown in Fig. 1.
Table 1. Complete pharmacokinetics profile of six phytochemicals.
| Ligand name | MW | H-Ac | H-Do | Log P | NRB | IA | TC | LD50 | HT | AT | MTD | NLV | DL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cassiarin D | 445.471 | 7 | 2 | 4.12862 | 4 | 100 | −0.295 | 2.694 | Yes | No | 0.155 | 0 | Yes |
| Rhamnetin | 316.265 | 7 | 4 | 2.291 | 2 | 80.214 | 0.473 | 2.453 | No | No | 0.56 | 0 | Yes |
| Lactupicrin | 410.422 | 7 | 2 | 1.782 | 4 | 100 | 0.317 | 2.029 | No | No | −0.72 | 0 | Yes |
| Rhinacanthin D | 408.406 | 7 | 1 | 3.723 | 5 | 93.396 | 0.118 | 1.908 | No | No | 0.52 | 0 | Yes |
| Flemiflavanone D | 424.493 | 6 | 3 | 4.7385 | 5 | 95.506 | 0.198 | 2.454 | No | No | −0.361 | 0 | Yes |
| Exiguaflavanone A | 424.493 | 6 | 4 | 5.3066 | 6 | 81.719 | 0.367 | 2.178 | No | No | 0.221 | 0 | Yes |
MW = molecular weight (g/mol); H-Ac = No. of hydrogen bond acceptor; H-Do = No. of hydrogen bond donor; LogP = Predicted octanol/water partition coefficient; NRB = No. of rotatable bonds; IA = Intestinal absorption (% absorbed); TC = Total clearance (log ml/min/kg); LD50 = Oral rat acute toxicity; HT = Hepatotoxicity; AT = AMES toxicity; MTD = Maximum tolerated dose for human (log mg/kg/day); NLV = No. of Lipinski’s rule violations; DL = Drug-likeness (Lipinski’s rule).
Figure 1. Illustration of the optimized ligands’ crystal structure (A–F) for docking with the ACE2 optimized receptor (G). The molecular dynamic simulation of the ligand-free ACE2 macromolecule has also been mentioned to understand the behaviors of the ACE2 in interacting with the circulating ions and solvents for more than 10 ns (H). The crystal structure of the six optimized phytochemicals are Cassiarin D (A), Rhamnetin (B), Lactupicrin (C), Rhinacanthin D (D), Flemiflavanone D (E), and Exiguaflavanone A (F).
Molecular docking and virtual screening analysis
Molecular docking was carried out to uncover the best candidates among the phytochemicals based on their docking scores which were primarily screened through the PyRx platform. In the PyRx system, the highest binding affinity score determines the best docking interaction between protein and ligand, and the binding score of all phytochemicals is included in Table 2. In this investigation, Cassiarin D is considered as a control ligand that poses the binding affinity at −10.6 KJ/mol with the grid box dimension of X × Y × Z at 35.9128 Å, 33.0428 Å, 25.0000 Å, and the grid center X = 38.8536. Y = 4.8318, Z = 22.4911, respectively. Rhinacanthin D poses the highest binding affinity at −10.8 with the same grid parameters. Besides, at the same grid box sizes, Flemiflavanone D and Lactupicrin pose the binding score at −10.2. Exiguaflavanone A and Rhamnetin posed docking scores at −9.3 and −9.1, respectively (Table 2). Depending on the binding affinities Rhamnetin, Lactupicrin, Rhinacanthin D, Flemiflavanone D, and Exiguaflavanone A were selected for the next steps.
Table 2. Representation of the binding affinity of the ligands of our interest with the active sites of the ACE2 receptor as compared to the control.
| Ligand names | Binding affinity (Kcal/mol) | Amino acid involved interactions | |
|---|---|---|---|
| Hydrogen bond interactions | Hydrophobic bond interactions | ||
| Cassiarin D | −10.6 | His345(2.95 Å), Glu406(2.70 Å), and Arg518(3.03 Å) | Asn149, Asp269, Phe274, Pro346, Asp367, Asp368, Glu375 |
| Rhamnetin | −9.1 | His345 (3.27 Å), Ala348(3.09 Å), His374(3.24 Å), and Glu402 (2.86 Å) | Pro346, Thr347, Thr371, Glu375, Tyr515 |
| Lactupicrin | −10.2 | Asp269 (2.80 Å) and Arg518(2.88 Å and 3.09 Å) | Arg273, Phe274, His345, Asp367, Glu406 |
| Rhinacanthin D | −10.8 | His345 (3.02 Å), Ala348 (3.14 Å), and Arg518 (2.80 Å and 3.12 Å) | with Pro346, Thr347, Asp367, Thr371, His374, Glu375, Tyr515 |
| Flemiflavanone D | −10.2 | Gly268 (3.17 Å), Asn277 (3.14), Cys344 (2.98 Å), and His345 (2.83 Å) | Glu145, Asn149, Asp269, Trp271, Phe274, Lys363, Asp367 |
| Exiguaflavanone A | −9.3 | Asn149 (2.90 Å), Gly268 (3.02 Å), and Asp368 (3.01 Å) | Thr371, Arg273, Trp271, Phe274, Thr276, Asp367, Asp269, Ala153, Cys344, Glu145, Lys363 |
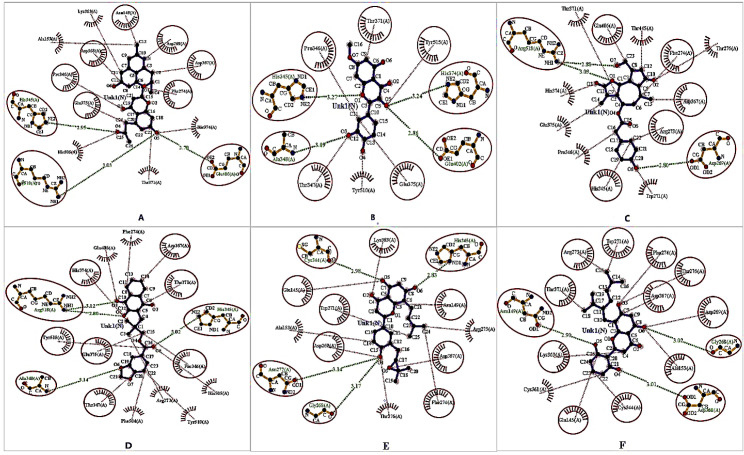
Analysis of the non-covalent interactions of the selected six phytochemicals, where one was standard bound to the ACE2 receptor, reveals the best fitting status, and Ligplot+ V.2.2 was employed to understand their fitness. Cassirin D stabilized via the three H-bonds and seven hydrophobic interactions with the cell membrane receptor protein. On the contrary, four H-bonds and five hydrophobic interactions were displayed by Rhamnetin against the receptor. Lactupicrin forms three H-bonds and five hydrophobic interactions. Rhinacanthin D and Flemiflavanone D were strengthened by four H-bonds and seven hydrophobic interactions, confirming their best binding pose. Exiguaflavanone A forms three H-bonds and 11 hydrophobic interactions with the ACE2 receptor (Fig. 2).
Figure 2. The docked conformation of the ACE2–lead molecule complexes represents the possible hydrogen and hydrophobic interactions, where hydrogen bonds are shown as olive green dotted lines with a specific distance (Å) that are illustrated in red color oval-shaped structures. The hydrophobic interactions are indicated with thin red lines with ellipses in protein residues via the red circle. The red color spiked arc ellipses indicate the protein residues that are mainly equivalent in 3D positions to the residues. Cassiarin D–ACE2 complex (A), Rhamnetin–ACE2 complex (B), Lactupicrin–ACE2 complex (C), Rhinacanthin D–ACE2 complex (D), Flemiflavanone D–ACE2 complex (E), and Exiguaflavanone A–ACE2 complex (F).
“ACE2–Rhamnetin docked complex” possess the four hydrogen bond interactions [with His345 (3.27 Å), Ala348 (3.09 Å), His374 (3.24 Å), and Glu402 (2.86 Å)], six hydrophobic interactions, but mostly hydrophobic interacted residues (Pro346, Thr347, Thr371, Glu375, and Tyr515) were observed (Fig. 2). We noted that the “ACE2–Lactupicrin complex” stabilized by the three hydrogen bond interactions [with Asp269 (2.80 Å), Arg518 (2.88 Å and 3.09 Å)], amino acid residues, and the most interacted hydrophobic bonds among the residues are Arg273, Phe274, His345, Asp367, and Glu406. Additionally, we observed that four hydrogen bond interactions [with His345 (3.02 Å), Ala348 (3.14 Å), and Arg518 (2.80 Å and 3.12 Å)] and multiple non-bonded interactions (hydrophobic interaction) (with Pro346, Thr347, Asp367, Thr371, His374, Glu375, and Tyr515) during the study of “ACE2–Rhinacanthin D” docked complex. However, Cassiarin D formed hydrogen bond interactions with His345 (2.95 Å), Glu406 (2.70 Å), and Arg518 (3.03 Å) residues of ACE2, whereas the amino acid residue of ACE2 involved in the hydrophobic interactions with Cassiarin D are Asn149, Asp269, Phe274, Pro346, Asp367, Asp368, and Glu375 (Table 2). When Flemiflavanone D was docked with the ACE2 receptor active site, we found several binding interaction modes like non-bonded and/or hydrogen bond interaction; the amino acid residues Gly268 (3.17 Å), Asn277 (3.14), Cys344 (2.98 Å), and His345 (2.83 Å) involved in the hydrogen bond interactions and the hydrophobic interacted residues are Glu145, Asn149, Asp269, Trp271, Phe274, Lys363, and Asp367. Asn149 (2.90 Å), Gly268 (3.02 Å), and Asp368 (3.01 Å) are three amino acid residues of ACE2 that are engaged in an H-bond with Exiguaflavanone A, whereas the “ACE2–Exiguaflavanone A” docked complex possesses the hydrophobic interactions among the residues which are Thr371, Arg273, Trp271, Phe274, Thr276, Asp367, Asp269, Ala153, Cys344, Glu145, and Lys363 (Table 2).
MDS analysis
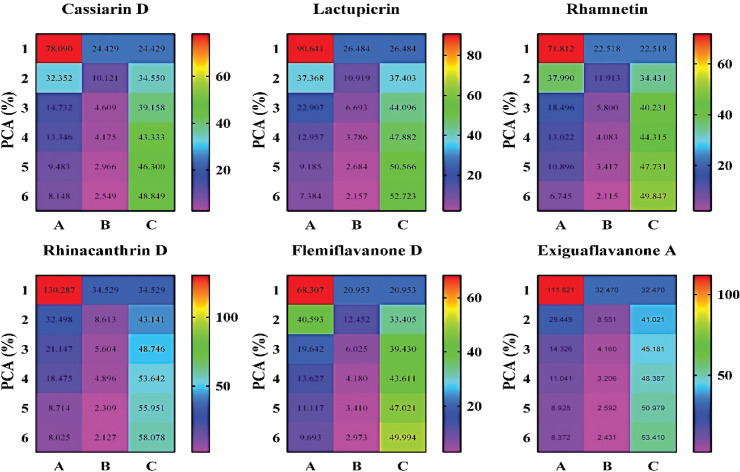
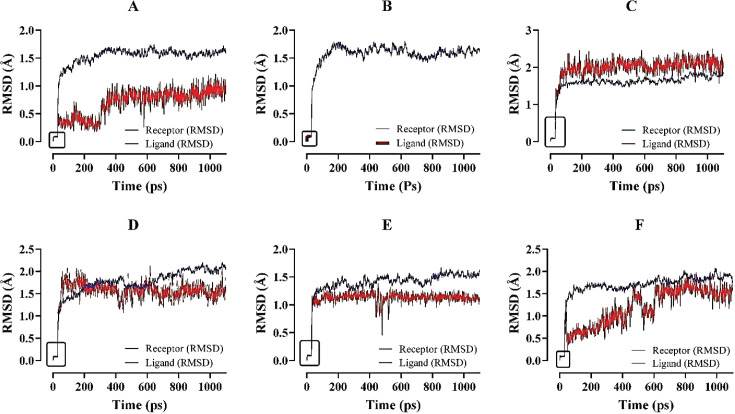
During the MDS process, the PCA, RMSD, RMSF, B-factor, and so on criteria to determine the best candidates were revealed. Principal component analysis (PCA) determines the lead molecule’s cluster conformations and evaluates the ligand’s best stability by analyzing the MDS parameters. So, the PCA result primarily displayed the best stable ligand from the selected compounds during the MDS method (Fig. 3). Besides, RMSD data analysis of MDS results covers the structural conformation and regular binding pose of lead and ACE2 enzymes. RMSD values range between 0.1~2.1 Å (Fig. 4).
Figure 3. The demonstration of the PCA of the selected ligands as compared to Cassiarin D control pharmacophore. Cassiarin D showed a lower eigenvalue, whereas the other five compounds exhibited the highest eigenvalues. Most importantly, Rhinacanthrin D and Exiguaflavanone A revealed the highest eigenvalues, resulting in more stable compounds against ACE2. Besides, the cumulative variance parameter described the positive results of the ligands than the phytochemical Cassiarin D. Eigenvalue (A), variance (B), cumulative variance (C), and principal component analysis (PCA).
Figure 4. The individual representation of the complex stability analysis assessing the RMSD values of ACE2–ligand complexes through molecular dynamic simulation for 1,100 ps. The protein and ligand fluctuation represent the following order: (A) ACE2 with Cassiarin D; (B) ACE2 with Rhamnetin; (C) ACE2 with Lactupicrin; (D) ACE2 with Rhinacanthin D; (E) ACE2 with Flemiflavanone D; and (F) ACE2 with Exiguaflavanone A. ACE2–Cassiarin D complex (A); ACE2–Rhamnetin complex (B); ACE2–Lactupicrin complex (C); ACE2–Rhinacanthin D complex (D); ACE2–Flemiflavanone D complex (E); and ACE2–Exiguaflavanone A complex (F). The grid box refers to the starting point of the simulation of both the ligands and the macromolecules.
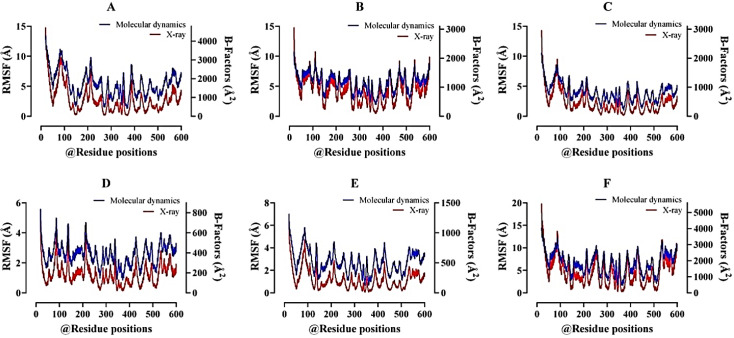
The complete RMSF profile was displayed for each residue dynamics of the target receptor and showed consistent atomic-pattern fluctuations at MDS. The peak fluctuation regions of 5–250 Å were observed and predicted that most compounds were binding at this region of the ACE2 receptor. On the other hand, B-factors of resulting trajectories were calculated with significantly fewer loops present at the protein structure (Fig. 5).
Figure 5. Analysis of the root mean square fluctuations (RMSF) plot with B-factor values, where most data represent the protein’s highest fluctuation rate for the determination of possible binding residues with that. A better B-factor value represents that there is a lesser amount of loop present in the protein structure. All the data have been found to be authentic for the lead compound selection. ACE2–Cassiarin D complex (A); ACE2–Rhamnetin complex (B); ACE2–Lactupicrin complex (C); ACE2–Rhinacanthin D complex (D); ACE2–Flemiflavanone D complex (E); and ACE2–Exiguaflavanone A complex (F).
Discussion
COVID-19 has been considered a global health emergency with a significant number of deaths around the world caused by nCoV from the Wuhan outbreak in December 2019. ACE2 is the receptor of interest for pathogenic entry and invasion of COVID-19 inside the cell [46]. Despite the researchers’ endeavors to find out the noble therapeutic agents for preoccupying the entry points of the 2019-nCoV, no established drugs are prescribed. In silico drug design has become one of the fundamental approaches in modern drug discovery that accelerates drug development by analyzing pharmacophore profiling, molecular docking, post-docking binding capability, molecular dynamics simulations, and prediction of the noble compound(s) [47]. Here six documented phytochemicals were used as a control (Fig. 1) and the remaining five compounds were functionally active against the cell surface receptor ACE2 by following several screening methods.
Pharmacokinetics study is an important method to predict the test drugs’ intended activities, compare with other prevalent drugs, and even suggest new assays [48]. These six phytochemicals’ pharmacokinetics were studied using the pkCSM–pharmacokinetics and Swiss ADME online server [39]. The drug mimicing properties, for example absorption, distribution, metabolism, excretion, and toxicity factors, were mainly analyzed. At first, we studied the “Lipinski’s rule of 5” which included molecular weight, H-bond donors and acceptors, number of violations, and consensus log P; most of the compounds fulfill our demand as a drug-likeness property. Besides all the compounds resulted AMES toxicity and hepatotoxicity negatively without standard compound, parameters recommended that selected phytochemicals are not carcinogenic (Table 1). More importantly, these compounds also support the LD50 (oral rat acute toxicity) criteria and follow the better absorption in the human intestinal mucosal cell line.
Molecular docking is a technique of assessing the pharmacophores’ potentiality to act as real drugs when complexed with any receptor, depending on their affinity of bonding scores between them, conducted via PyRx 0.8 virtual screening tool [49]. Molecular docking always takes place at a specific targeted site of the macromolecules of interest. Protein active sites are the main corresponding ligand docking sites to reform a natural H-bond [50]. Based on the scoring value, molecular docking ensures more potentially useful drug candidates among a number. However, the analysis of the several interactions between protein and ligand complexes with LigPlot+ V.2.2 tool has conducted in this study, which precisely configured on java interface to represent protein–ligand interactions with necessary quantitative measurements like atomic distances. Most importantly, Ligplot+ runs only the PDB file that is generated by the PyMOL visualizer tool.
MDS is the only way of investigating biomolecular interactions and has been applied for modern drug discovery. The output data from the dynamic trajectory paves the way to determine the relationship between protein structure and function. MDS output demonstrates the structural dynamics of the receptor upon binding with lead molecule/proposed drug. In this current study, we arranged the online dynamics simulators LARMD to analyze MDS due to posing the results rapidly. The LARMD server is developed based on Amber16 software, which accurately provides reports on PCA, RMSD, RMSF, and B-factor [51]. We analyze our protein–ligand complex file and measure some parameters that determine the relationship between protein structure and ligand via the normal mode analysis (Nor-mod) method. CABS-flex 2.0 is another efficient protein flexibility modeling online web-based service that is automatically analyzed and processed to provide vital information of native protein dynamics within 10 ns. More importantly, PDB ID of native protein structure to submit the CABS-flex 2.0 server to investigate the dynamics simulations [41].
The phytocompounds complexed with ACE2 receptor were studied by molecular dynamics simulation for 1,000 ps (1.0 ns) to investigate the stability confirmations with ionic fluctuations of the complex within a water explicit condition where Na+ and/or Cl− Na+ or Cl− are added as counter ions in the system. During the molecular dynamic simulations, LARMD program was applied for investigating the protein–ligand conformational binding modes basis on the force field of Amber16 that were employing 3,000 steps for conjugated gradient method but during the minimization process used 2,000 steps steepest descent method. All atoms were relaxed in the explicit water [51]. Principle component analysis (PCA) considers practical data analysis and predictive methods, where we can find out the best lead molecule by analyzing the data of MDS. PCA evaluates the quantitative structure–activity relationship (QSAR) of ligand molecule via analyzing the eigenvalue and cumulative variance. Here, eigenvalue determines the stability and vibration analysis of ligand molecule, where Cassiarin D determines the less stability result but other lead compounds potentially stable than the standard molecule. However, cumulative Variance data set are authentic for the selected compounds (Fig. 3). The PCA data confirmed the stable and best binding capability toward the receptor [52,53]. The average RMSD values range between 0.1~2.1 Å. The fluctuated values of ligand 0.1~2.1 Å and proteins 0~2.0 Å that evaluate the complex did not show any undesirable repulsion and precisely identify the ligand–protein interactions (Fig. 4).
On the other hand, the complete RMSF profile displayed each residue dynamics of the target receptor and showed consecutive atomic fluctuations at the MDS. The peak fluctuation regions from 5 to 250 were observed (Fig. 5) and predict that most compounds were binding at this region of the ACE2 receptor. After that, B-factors of resulting trajectories were evaluated with very few loops present at protein structure and showed no significant fluctuations in the atom mean square isotropic displacement [54]. In recent times, bacteriocins are getting equal concerns of the researchers to be used for therapeutic purposes like phytochemical ligands where the milk-derived probiotic microorganisms are given the main attentions [55,56] due to having robust immune simulation against viruses and in boosting secondary immunization [57].
Conclusion
In this present in silico study, five ligands out of 57 phytochemicals have been selected based on their pharmacokinetic properties through ADMET and QSAR analyses. The selected ligands were characterized, optimized, and implemented to molecular docking on ACE2 receptor protein. Molecular dynamic simulation for 1100 ps revealed the RMSD, RMSF, and B-factor values for those targeted ligands complexed with the ACE2 receptor individually and precisely. This current study suggests that Rhamnetin, Lactupicrin, Rhinacanthin D, Flemiflavanone D, and Exiguaflavanone A can interact more efficiently with the common receptor macromolecule (ACE2) of both animals and human beings, suggesting their therapeutic potentiality in treating SARS-CoV-2 infections via blocking the entrance into the cell. To figure out better therapeutic efficacy of the aforementioned experimentally designed ligands prior to clinical trials, integrative in silico and in situ comprehensive research is heartily suggested.
List of abbreviations
SARS-CoV: Severe acute respiratory syndrome coronavirus; MERS-CoV: Middle East respiratory syndrome coronavirus; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; COVID-19: Coronavirus disease 2019; S protein: Spike protein; ACE2: Angiotensin-converting enzyme 2; ADMET: Absorption, distribution, metabolism and excretion; QSAR: Quantitative structure–activity relationship; PCA: Principal component analysis; RMSD: Root mean square deviation; RMSF: Root mean square fluctuation; nCoV: Novel coronavirus; RBD: Receptor-binding domain; FP: Fusion peptide; HR: Heptad repeat; PDB: Protein data bank; MDS: Molecular dynamics simulations; TC: Total clearance; CSV: Comma Separated Values; LARMD: Ligand and Receptor Molecular Dynamics.
Acknowledgments
The authors are genuinely grateful to RPG (Govt. License ID: 05-060-06021) for rendering all types of technical and financial supports under the Grant of TEAM-RPG Project Category: F1 (Project ID. #06-2020/21).
Conflict of interest
The authors declare that they have no conflict of interests.
Authors’ contribution
All the authors participated in the research and performed equally and cordially. Parag Kumar Paul strongly contributed to the molecular docking and dynamic simulation studies. Salauddin Al Azad conducted all the statistical analyses generated from the dynamic simulations. The rest of the authors prepared the manuscript, which was reviewed and edited by each of the authors individually. The total research work was supervised by Salauddin Al Azad (PhD Researcher) and co-supervised by Mohammad Faysal Al Mazid (PhD Researcher).
References
- [1].Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63’. J Virol. 2012;86(23):12816–25. doi: 10.1128/JVI.00906-12. https://doi.org/10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chan JF, To KK, Tse H, Jin DY, Yuen KY. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21(10):544–55. doi: 10.1016/j.tim.2013.05.005. https://doi.org/10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Phan MV, Ngo Tri T, Hong Anh P, Baker S, Kellam P, Cotten M. Identification and characterization of Coronaviridae genomes from Vietnamese bats and rats based on conserved protein domains. Virus Evol. 2018;4(2):vey035. doi: 10.1093/ve/vey035. https://doi.org/10.1093/ve/vey035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zeng ZQ, Chen DH, Tan WP, Qiu SY, Xu D, Liang HX, et al. Epidemiology and clinical characteristics of human coronaviruses OC43, 229E, NL63, and HKU1: a study of hospitalized children with acute respiratory tract infection in Guangzhou, China. Eur J Clin Microbiol Infect Dis. 2018;37(2):363–9. doi: 10.1007/s10096-017-3144-z. https://doi.org/10.1007/s10096-017-3144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Singh A, Singh RS, Sarma P, Batra G, Joshi R, Kaur H, et al. A comprehensive review of animal models for coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV. Virol Sin. 2020;35(3):290–304. doi: 10.1007/s12250-020-00252-z. https://doi.org/10.1007/s12250-020-00252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):59. doi: 10.3390/v11010059. https://doi.org/10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-15562-9. https://doi.org/10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–34. doi: 10.1016/j.virusres.2014.11.021. https://doi.org/10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Momattin H, Mohammed K, Zumla A, Memish ZA, Al-Tawfiq JA. Therapeutic options for Middle East respiratory syndrome coronavirus (MERS-CoV)–possible lessons from a systematic review of SARS-CoV therapy. Int J Infect Dis. 2013;17(10):e792–e798. doi: 10.1016/j.ijid.2013.07.002. https://doi.org/10.1016/j.ijid.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Uddin M, Mustafa F, Rizvi TA, Loney T, Suwaidi HA, Al-Marzouqi AH, et al. SARS-CoV-2/COVID-19: viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12(5):526. doi: 10.3390/v12050526. https://doi.org/10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Coronavirus Update (Live): Cases and Deaths from COVID-19 Virus Pandemic - Worldometer. (no date)
- [12].Alanagreh L, Alzoughool F, Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9(5):331. doi: 10.3390/pathogens9050331. https://doi.org/10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12(3):e7423. doi: 10.7759/cureus.7423. https://doi.org/10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hsih WH, Cheng MY, Ho MW, Chou CH, Lin PC, Chi CY, et al. Featuring COVID-19 cases via screening symptomatic patients with epidemiologic link during flu season in a medical center of central Taiwan. J Microbiol Immunol Infect. 2020;53(3):459–66. doi: 10.1016/j.jmii.2020.03.008. https://doi.org/10.1016/j.jmii.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pedersen SF, Ho YC. SARS-CoV-2: a storm is raging. J Clin Invest. 2020;130(5):2202–220. doi: 10.1172/JCI137647. https://doi.org/10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–23. doi: 10.1002/jmv.25681. https://https://doi.org/10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12(1):8. doi: 10.1038/s41368-020-0074-x. https://doi.org/10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wiese O, Zemlin AE, Pillay TS. Molecules in pathogenesis: angiotensin converting enzyme 2 (ACE2) J Clin Pathol. 2020 doi: 10.1136/jclinpath-2020-206954. https://doi.org/10.1136/jclinpath-2020-206954. [DOI] [PubMed] [Google Scholar]
- [19].Dalan R, Bornstein SR, El-Armouche A, Rodionov RN, Markov A, Wielockx B, et al. The ACE-2 in COVID-19: foe or friend? Horm Metab Res. 2020;52(5):257. doi: 10.1055/a-1155-0501. https://doi.org/10.1055/a-1155-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sinica. 2020;41(9):1141–9. doi: 10.1038/s41401-020-0485-4. https://doi.org/10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–4. doi: 10.1038/s41586-020-2179-y. https://doi.org/10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sternberg A, Naujokat C. Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination. Life Sci. 2020;257:118056. doi: 10.1016/j.lfs.2020.118056. https://doi.org/10.1016/j.lfs.2020.118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727 LP–34. doi: 10.1073/pnas.2003138117. https://doi.org/10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–20. doi: 10.1038/s41586-020-2180-5. https://doi.org/10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- [25].Sigrist CJ, Bridge A, Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 2020;177:104759. doi: 10.1016/j.antiviral.2020.104759. https://doi.org/10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Leroy EM, Gouilh MA, Brugère-Picoux J. The risk of SARS-CoV-2 transmission to pets and other wild and domestic animals strongly mandates a one-health strategy to control the COVID-19 pandemic. One Health. 2020;10:100133. doi: 10.1016/j.onehlt.2020.100133. https://doi.org/10.1016/j.onehlt.2020.100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhai X, Sun J, Yan Z, Zhang J, Zhao J, Zhao Z, et al. Comparison of SARS-CoV-2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J Virol. 2020;94(15):e00831–20. doi: 10.1128/JVI.00831-20. https://doi.org/10.1128/JVI.00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24(1):1–10. doi: 10.1186/s13054-020-03120-0. https://doi.org/10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ghildiyal R, Prakash V, Chaudhary VK, Gupta V, Gabrani R. Berlin, Germany: Springer; 2020. Phytochemicals as antiviral agents: recent updates, in plant-derived bioactives; pp. 279–295. https://doi.org/10.1007/978-981-15-1761-7_12. [Google Scholar]
- [30].Ben-Shabat S, Yarmolinsky L, Porat D, Dahan A. Antiviral effect of phytochemicals from medicinal plants: applications and drug delivery strategies. Drug Deliv Transl Res. 2020;10(2):1–14. doi: 10.1007/s13346-019-00691-6. https://doi.org/10.1007/s13346-019-00691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kapoor R, Sharma B, Kanwar SS. Antiviral phytochemicals: an overview’. Biochem Physiol. 2017;6(2):7. https://doi.org/10.4172/2168-9652.1000220. [Google Scholar]
- [32].Kabera JN, Semana E, Mussa AR, He X. Plant secondary metabolites: biosynthesis, classification, function and pharmacological properties. J Pharm Pharmacol. 2014;2:377–92. [Google Scholar]
- [33].da Silva Antonio A, Wiedemann LS, Veiga-Junior VF. Natural products’ role against COVID-19. RSC Adv. 2020;10(39):23379–93. doi: 10.1039/d0ra03774e. https://doi.org/10.1039/D0RA03774E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Abd El-Aziz TM, Stockand JD. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2)-an update on the status. Infect Genet Evol. 2020;83:104327. doi: 10.1016/j.meegid.2020.104327. https://doi.org/10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chojnacka K, Witek-Krowiak A, Skrzypczak D, Mikula K, Młynarz P. Phytochemicals containing biologically active polyphenols as an effective agent against Covid-19-inducing coronavirus. J Funct Foods. 2020;73:104146. doi: 10.1016/j.jff.2020.104146. https://doi.org/10.1016/j.jff.2020.104146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Basu A, Sarkar A, Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci Rep. 2020;10(1):17699. doi: 10.1038/s41598-020-74715-4. https://doi.org/10.1038/s41598-020-74715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kshatriya R, Kambale D, Mali S, Jejurkar VP, Lokhande P, Chaudhari HK, et al. Brønsted acid catalyzed domino synthesis of functionalized 4H-chromens and their ADMET, molecular docking and antibacterial studies. ChemistrySelect. 2019;4(27):7943–8. https://doi.org/10.1002/slct.201901775. [Google Scholar]
- [38].Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. https://doi.org/10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pires DE, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–72. doi: 10.1021/acs.jmedchem.5b00104. https://doi.org/10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, et al. UCSF chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179(3):269–78. doi: 10.1016/j.jsb.2011.09.006. https://doi.org/10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kuriata A, Gierut AM, Oleniecki T, Ciemny MP, Kolinski A, Kurcinski M, et al. CABS-flex 2.0: a web server for fast simulations of flexibility of protein structures. Nucleic Acids Res. 2018;46(W1):W338–43. doi: 10.1093/nar/gky356. https://doi.org/10.1093/nar/gky356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Akter KM, Tushi T, Jahan Mily S, Mohona RA, Anis S, Chakraborty AK, et al. RT-PCR Mediated Identification of SARS-CoV-2 patients from particular regions of Bangladesh and the multi-factorial analysis considering their pre and post infection health conditions. Biotechnol J Int. 2020;24(6):43–56. https://doi.org/10.9734/bji/2020/v24i630121. [Google Scholar]
- [43].Akther T, Rony MKK, Anowar A, Habiba MU, Akhi OJ, Tabassum E, et al. Comparative analysis of the government investments and revenue from different sectors in Bangladesh and its impact on the development of HRM sectors: a 20 years of study. Int J Bus Manage Soc Res. 2020;10(01):553–62. https://doi.org/10.18801/ijbmsr.100120.58. [Google Scholar]
- [44].Rashaduzzaman M, Kamrujjaman M, Islam MA, Ahmed S, Al Azad S. An experimental analysis of different point specific musculoskeletal pain among selected adolescent-club cricketers in Dhaka city. Eur J Clin Exp Med. 2019;17(4):308–14. https://doi.org/10.15584/ejcem.2019.4.4. [Google Scholar]
- [45].Al Azad S, Farjana M, Mazumder B, Abdullah-Al-Mamun M, Haque AI. Molecular identification of a Bacillus cereus strain from Murrah buffalo milk showed in vitro bioremediation properties on selective heavy metals. J Adv Vet Anim Res. 2020;7(1):62–8. doi: 10.5455/javar.2020.g394. https://doi.org/10.5455/javar.2020.g394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dashraath P, Jeslyn WJL, Karen LMX, Min LL, Sarah L, Biswas A, et al. Coronavirus disease 2019 (COVID-19) pandemic and pregnancy. Am J Obstet Gynecol. 2020;222(6):521–31. doi: 10.1016/j.ajog.2020.03.021. https://doi.org/10.1016/j.ajog.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pandey P, Rane JS, Chatterjee A, Kumar A, Khan R, Prakash A, et al. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: an in-silico study for drug development. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1796811. https://doi.org/10.1080/07391102.2020.1796811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Olasupo SB, Uzairu A, Shallangwa G, Uba S. QSAR modeling, molecular docking and ADMET/pharmacokinetic studies: a chemometrics approach to search for novel inhibitors of norepinephrine transporter as potent antipsychotic drugs. J Iran Chem Soc. 2020;17(8):1953–66. https://doi.org/10.1007/s13738-020-01902-5. [Google Scholar]
- [49].Prabhavathi H, Dasegowda KR, Renukananda KH, Lingaraju K, Naika HR. Exploration and evaluation of bioactive phytocompounds against BRCA proteins by in silico approach. J Biomol Struct Dyn. 2020;8:1–15. doi: 10.1080/07391102.2020.1790424. https://doi.org/10.1080/07391102.2020.1790424. [DOI] [PubMed] [Google Scholar]
- [50].Fukunishi Y, Nakamura H. Prediction of ligand-binding sites of proteins by molecular docking calculation for a random ligand library. Protein Sci. 2011;20(1):95–106. doi: 10.1002/pro.540. https://doi.org/10.1002/pro.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yang JF, Wang F, Chen YZ, Hao GF, Yang GF. LARMD: integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of estrogen receptor. Brief Bioinform. 2019;21(6):2206–18. doi: 10.1093/bib/bbz141. https://doi.org/10.1093/bib/bbz141. [DOI] [PubMed] [Google Scholar]
- [52].Steindl TM, Crump CE, Hayden FG, Langer T. Pharmacophore modeling, docking, and principal component analysis-based clustering: combined computer-assisted approaches to identify new inhibitors of the human rhinovirus coat protein. J Med Chem. 2005;48(20):6250–60. doi: 10.1021/jm050343d. https://doi.org/10.1021/jm050343d. [DOI] [PubMed] [Google Scholar]
- [53].Bender A, Jenkins JL, Scheiber J, Sukuru SCK, Glick M, Davies JW. How similar are similarity searching methods? A principal component analysis of molecular descriptor space. J Chem Inf Model. 2009;49(1):108–19. doi: 10.1021/ci800249s. https://doi.org/10.1021/ci800249s. [DOI] [PubMed] [Google Scholar]
- [54].Liu Q, Kwoh CK, Li J. Binding affinity prediction for protein–ligand complexes based on β contacts and B factor. J Chem Inf Model. 2013;53(11):3076–85. doi: 10.1021/ci400450h. https://doi.org/10.1021/ci400450h. [DOI] [PubMed] [Google Scholar]
- [55].Abdullah-Al-Mamun M, Jakir Hasan M, Al Azad S, Giash Uddin M, Shahriyar S, Jyoti Mondal K. Evaluation of potential probiotic characteristics of isolated lactic acid bacteria from goat milk. Biotechnol J Int. 2016;14(2):1–7. https://doi.org/10.9734/BBJ/2016/26397. [Google Scholar]
- [56].Al Azad S, Moazzem Hossain K, Rahman SMM, Al Mazid MF, Barai P, Gazi MS. In ovo inoculation of duck embryos with different strains of Bacillus cereus to analyse their synergistic post-hatch anti-allergic potentialities. Vet Med Sci. 2020;6(4):992–9. doi: 10.1002/vms3.279. https://doi.org/10.1002/vms3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Al Azad S, Shahriyar S, Mondal KJ. Opsonin and its mechanism of action in secondary immune. J Mol Studies Med Res. 2016;1(02):48–56. https://doi.org/10.18801/jmsmr.010216.06. [Google Scholar]