Abstract
Here, we reveal that the regulation of Drosophila odorant receptor (OR) expression during the pupal stage is permissive and imprecise. We found that directly after hatching an OR feedback mechanism both directs and refines OR expression. We demonstrate that, as in mice, dLsd1 and Su(var)3-9 balance heterochromatin formation to direct OR expression. We show that the expressed OR induces dLsd1 and Su(var)3-9 expression, linking OR level and possibly function to OR expression. OR expression refinement shows a restricted duration, suggesting that a gene regulatory critical period brings olfactory sensory neuron differentiation to an end. Consistent with a change in differentiation, stress during the critical period represses dLsd1 and Su(var)3-9 expression and makes the early permissive OR expression permanent. This induced permissive gene regulatory state makes OR expression resilient to stress later in life. Hence, during a critical period OR feedback, similar to in mouse OR selection, defines adult OR expression in Drosophila.
This study reveals that the regulation of odorant receptor expression during the Drosophila pupal stage is permissive and imprecise; olfactory sensory neuron activity directly after hatching both directs and refines odorant receptor expression. Hence, during a critical period, activity feedback defines adult odorant expression in Drosophila, as happens in mouse.
Introduction
Olfactory sensory neurons (OSNs) in most vertebrates and insects are specified to express a single odorant receptor (OR) from a large repertoire of OR genes in the genome [1–4]. Two OR gene regulatory models have been described: the vertebrate probabilistic selection model and the invertebrate predetermined instructive model.
The vertebrate OR regulatory model depends on chromatin state changes—from a repressed state to an active state and back again to a general repressed state [5,6]. In mice, non-expressed OR genes are embedded in constitutive heterochromatin marked by histone H3 lysine 9 trimethylation (H3K9me3) [5,7]. According to a mathematical model of OR regulation, a yet-to-be-identified H3K9me3 demethylase sporadically opens the constitutive heterochromatin at a single OR locus and initiates expression [6,8]. Lsd1 erases histone H3 lysine 9 dimethylation (H3K9me2), which further opens the chromatin and establishes OR expression [6]. The expressed OR then induces several feedback loops that downregulate Lsd1 and induce heterochromatin formation, blocking the additional initiation of OR expression [6,9–11]. Unknown transcription factors (TFs) restrict the expression of each mouse OR to a stereotyped region in the olfactory epithelium [12].
Drosophila OR expression is generally viewed as a developmentally predetermined and non-plastic process [13–15]. There are several reasons for this assumption. OR expression is stereotypically organized [1], and Drosophila OSNs are specified in a lineage-dependent manner [13]. Notch signaling splits OSNs into 2 subgroups with defined projection patterns and OR expression [16]. Defined TF combinations both drive and restrict OR expression [15,17–19].
Nevertheless, the odor environment and odor exposure early in life can modulate Drosophila OR expression and odor responses [20–22]. Thermal stress and starvation induce plasticity in Drosophila adult OR expression [23]. H3K9me2, which marks OR promoters in vertebrates, also marks OR genes in Drosophila OSNs [19]. G9a, which produces H3K9me2, restricts OR expression in Drosophila [24]. Su(var)3-9, which produces H3K9me3 and induces constitutive heterochromatin, suppresses spurious OR expression [19,23]. The OR cis regulatory regions support cooperative TF interactions that oppose heterochromatin and limit stress-induced plasticity [23,25]. Thus, a heterochromatin-regulated OR expression plasticity that is in some ways similar to that found in vertebrates also seems to exist in Drosophila.
Here, we further address the role of heterochromatin in Drosophila OR regulation. We first demonstrate that OR gene regulation stringency increases after a restricted time of heightened plasticity and a stress-sensitive period of early fly development. We show that dLsd1 and Su(var)3-9 initiate and maintain OR expression stringency in Drosophila. The expressed OR regulates dLsd1 and Su(var)3-9 expression, creating a feedback loop that restricts and balances OR expression. Stress during this period inhibits the feedback loop and produces permanent changes in OR expression.
Results
Drosophila chemoreceptor expression matures during the first few days of adult life
We and others have observed that OR reporter expression varies between OSNs in day-old flies, rising to the uniform high level observed in adult flies after a few days (Fig 1A) [26]. To further investigate OR expression dynamics, we performed RNA sequencing (RNA-seq) analyses comparing antennae from flies 1 day (newly hatched), 4 days (around the point of uniform OR expression), and 14 days (mature) post-eclosion. For each time point biological triplicates were analyzed. Strikingly, all 30 of the 34 adult antennal ORs as well as the olfactory co-receptor Orco increased significantly in expression between 1 and 4 days post-eclosion (DPE) (Fig 1B; S1 Data), but stopped increasing after day 4. The expression of the ionotropic receptors (IRs) and gustatory receptors (GRs) also increased during the first 4 DPE (Fig 1B; S1 Data). We found that 13 of 22 antennal IRs and 8 of 10 GRs expressed in OSNs increased 1-fold or more. As with the ORs, any changes in IR and GR expression after day 4 were minor, without any discernible pattern (Fig 1B; S1 Data). Thus, chemoreceptor expression in general seems to mature during the first 4 DPE.
Fig 1. OR expression matures and OSN development continues after eclosion.
(A) Whole-mount brain and antenna staining shows the Or59b reporter GFP expression (green) in 1- and 7-DPE flies. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). Scale bar denotes 3.5 μm. Below each merged image, the GFP expression in the antenna is shown as the white channel. Note the increased expression and uniform level of expression between OSNs in the 7-day flies. (B–E) Degree of change in RNA sequencing read counts observed at 4 DPE relative to 1 DPE (left) and at 14 DPE relative to 4 DPE (right). Normalized logarithmic counts (log10 size-factor-normalized counts) for each gene from the respective sample were scatter-plotted. Data and statistics are in S1 Data. The raw sequencing data are available on ArrayExpress (#E-MTAB-9805). The code is available on Github (https://github.com/henriksson-lab/mattias-or). Genes shown in grey except (B) ORs (green), GRs (blue), and IRs (red); (C) synapse genes (red); (D) IFT and BBS genes (red); and (E) OBPs (red). The line is the reference at which gene expression is the same between conditions. BBS, BBSome; DPE, days post-eclosion; GR, gustatory receptor; IFT, intraflagellar transport; IR, ionotropic receptor; OBP, odorant binding protein; OR, odorant receptor; OSN, olfactory sensory neuron; RNA-seq, RNA sequencing; TF, transcription factor.
During the same period, OSN connectivity is also maturing [26–28]. Analysis of the synaptic gene network showed a slight decrease but no uniform change in expression of the genes in the synaptic network from day 1 to day 4 (Fig 1C; S1 Data). This indicates that a limited set of genes or separate post-transcriptional mechanisms are responsible for refining OSN synapses. We next expanded the analysis further to include other OSN gene networks. Sensory neurons are the only ciliated cells in Drosophila, and the ciliary transport machinery (e.g., intraflagellar transport [IFT] and BBSome [BBS]) is important for the ciliary localization of the chemoreceptors [29]. In OSNs, olfactory transduction levels are affected by OR levels as more ORs are transported into the cilia [29]. Interestingly, we found increasing expression of the IFT and BBS genes during the first 4 DPE and no further change after the fourth day (Fig 1D; S1 Data). OSNs also express high levels of another auxiliary set of olfactory proteins required for specific odor responses, the odorant binding proteins (OBPs) [30]. The expression of most OBP genes either remains steady or increases from day 1 to day 4 (Fig 1E; S1 Data), lending further support to the idea that OSNs continue to develop and sensory transduction continues to change after the pupal stage.
Stress modulates the maturation of OR expression
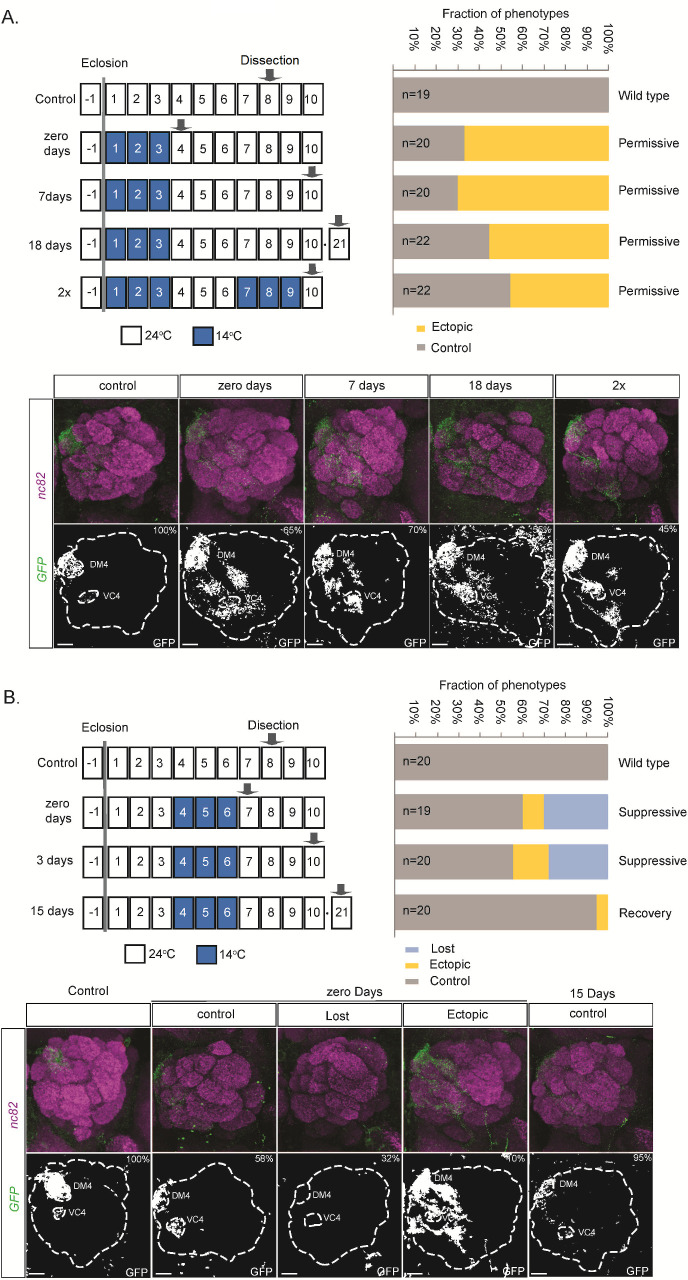
We have previously observed that starvation and thermal stress increase OR expression plasticity [23] and that cooperative TF interactions in the cis regulatory region stabilize the OR expression. In these studies, we focused on adult (5–7 DPE) flies, but the stress-induced plasticity suggested that stress could change also the OR expression maturation process. To visualize stress-induced modulation of OR expression at all stages, we used the Or59b minimal enhancer (Or59bME), an Or59b reporter that lacks the cooperative regulation region required to resist stress-induced changes [23]. After dissection and whole-mount staining of the brain, we analyzed the innervation of the antenna lobe. At room temperature (24°C), Or59bME behaves just like the endogenous Or59b gene, with its expression restricted to the ab2a OSN class [1,2,23] (Fig 2A). Reducing the temperature alters Or59bME reporter expression, but the timing of the temperature shift dictates the resulting phenotype (Fig 2). We found that shifts during the first 3 DPE led to stereotype ectopic Or59bME expression in several OSN classes, as evidenced by the appearance of multiple GFP-positive glomeruli in the antennal lobe (Fig 2A). But all temperature shifts after day 3 produced loss-of-expression phenotypes (Fig 2B). This sharp transition suggests a drastic change in OSN gene regulation. It also suggests stress may alter terminal OSN differentiation.
Fig 2. Permanent OR gene regulation changes following environmental stress during the OR expression maturation.
The schematic drawing shows the time points of thermal stress treatments and sample preparation. The graphs show the fraction of flies with control, lost, or ectopic Or59bME expression after 3 days of thermal stress initiated on (A) day 1 or (B) day 4. The recovery time at ambient temperature is denoted in days (S2 Data). The antennal lobes represent the analyzed phenotypes. GFP expression (green) is driven by the Or59b minimal reporter. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). The percentages show the fraction of the phenotype that is presented in that panel. Scale bar denotes 3.5 μm. Note the persistent ectopic expression after 18 days of recovery or after a second exposure to low temperature (2×). The loss phenotype reverted to single OSN class expression after 14 days of recovery at room temperature. OR, odorant receptor; OSN, olfactory sensory neuron.
If stress modulates terminal OSN differentiation, the ectopic expression phenotypes we observed with early temperature shifts could be expected to become permanent. Indeed, when we returned Or59bME flies that underwent early temperature shifts to room temperature, we found that the stress-induced ectopic Or59bME reporter expression pattern persisted throughout a 7-day recovery period (Fig 2A). It even remained similar after a prolonged 18-day recovery period (Fig 2A). If the process of OR expression maturation is the final stage of OSN differentiation, then temperature shifts after maturation is complete should be reversible. Consistent with this hypothesis, we found that shifts back to room temperature for those exposed to thermal stress after day 3 led to a restoration of the expression pattern to a single OSN class (Fig 2B). This indicates that the OR expression state was already fixed when the flies were subjected to the temperature shift. To address this further, we subjected flies carrying Or59bME to 2 cold shifts, one during the critical period and another after a 3-day recovery period. As expected, the resulting Or59bME ectopic expression pattern for flies subjected to shifts was similar to that of flies subjected to a single early shift (Fig 2A). Together, these results indicate that stress during the maturation phase switches adult OR expression from a stress-sensitive, refined expression pattern to a potentially less refined but stress-resilient expression pattern.
OR feedback refines OR expression
In mosquitoes, ectopic OR expression suppresses endogenous OR expression [31]. To determine whether OR expression level or function acts in a feedback mechanism on OR expression in Drosophila as well, we expressed an OR in all OSNs with Peb-Gal4 and monitored Or59b-CD8:GFP reporter expression. With the exception of the male pheromone receptor Or47b, most Drosophila ORs have low spontaneous activity [32]. We found that about half of the flies with ectopic Or47b expression lost the Or59b reporter expression (Fig 3A and 3B), indicating that high spontaneous OR activity can suppress OR expression. Interestingly, ectopic expression of Or42b, an OR with lower spontaneous activity compared to Or47b, induced Or59b reporter expression loss in only 11% of the resulting flies (Fig 3A and 3B). To determine whether odor responses induce this negative feedback, we exposed flies to ethyl propionate (EP; diluted 10−4), a strong Or42b ligand. Flies with ectopic Or42b expression exposed to EP showed a slightly larger but still insignificant loss of Or59b reporter expression (11% versus 18%; Fig 3B). EP exposure of control flies (without ectopic Or42b expression) did not affect Or59b reporter expression (Fig 3B). Together, these results indicate OR expression level can feed back on and shape OR gene expression.
Fig 3. OR activity regulates OR expression.
Or59b reporter GFP expression is shown in green, and synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). Below each merged image, the GFP channel is shown. Antennal lobe and labeled glomeruli are marked. Control flies were crossed to w1118. Percentage denotes the proportion of flies with the depicted phenotype (S2 Data). Scale bar denotes 3.5 μm. Ectopic expression of Or47b (A) or Or42b (B) inhibits Or59b reporter expression. The loss of GFP expression is greater when flies with ectopic Or42b expression are exposed to the Or42b-specific odor ligand (EP). (C–E) Degree of change in RNA sequencing read counts observed between 1 and 4 DPE. Normalized logarithmic counts (log10 size-factor-normalized counts) for each gene from the respective sample were scatter-plotted. Genes shown in grey except ORs (green), for (C) control, (D) Peb-Gal4;UAS-Or65a, and (E) Orco−/−. Note that the increase in OR expression between day 1 and 4 shifts to suppression in olfactory sensory neurons with over-activity (D) and lost activity (E). The line is the reference at which gene expression is the same between conditions. Statistics for the figure are in S3 Data. The raw sequencing data are available on ArrayExpress (#E-MTAB-9805). The code is available on Github (https://github.com/henriksson-lab/mattias-or). CPM, counts per million; DPE, days post-eclosion; EP, ethyl propionate; OR, odorant receptor.
Next, we performed an RNA-seq experiment on antennae from flies with ectopic Or65a expression (Fig 3C and 3D). For 30 out of 34 antennal ORs, OR expression decreased in the flies with ectopic Or65a expression between 1 and 4 DPE (Fig 3D; S3 Data). Comparing OR expression with age-matched controls showed that the timing of the feedback regulation of OR expression depended on OSN lineage. In day-old (1 DPE) flies with ectopic Or65a expression, most trichoid-related ORs increased in expression (8/12; S3 Data; S1 Fig), whereas basiconic-related OR expression changes were minor. After OR expression maturation (4 DPE), basiconic-related OR expression was down-regulated, and the trichoid-related OR expression changes were less penetrant (S1 Fig; S2 Data), suggesting that OR feedback establishes trichoid-related OR expression during the pupal stage and restricts basiconic-related OR expression post-eclosion.
To further address whether OSN activity is required for OR expression, we performed an RNA-seq experiment on antennae from Orco mutant flies, in which most of the OSNs lack OR activity [33]. We found a drastic reduction in OR expression in Orco mutant flies 1–4 DPE (Fig 3E). This suggests Orco, and likely OR, function is important for this feedback regulation of OR expression. Consistent with our ectopic expression results in day-old Orco mutant flies, most trichoid-related ORs were up-regulated, while basiconic-related ORs showed no consistent directionality in the changes (S1 Fig). At 4 DPE, the expression of most trichoid and all basiconic ORs was reduced in the Orco mutant compared to controls, indicating that OR feedback post-eclosion is required in non-stress conditions to establish OR expression.
The balance between dLsd1 and Su(var)3-9 refines OR expression
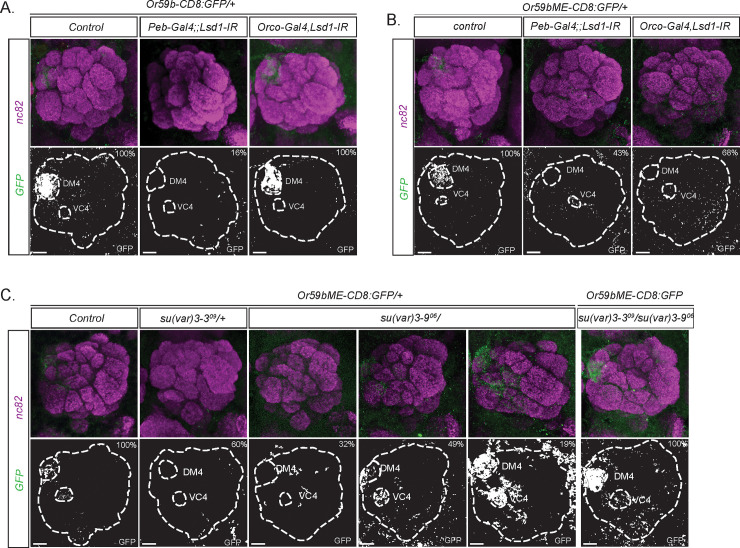
The similarity of the OR feedback we observed to the vertebrate OR choice mechanism suggested a conserved OR regulatory mechanism. In mouse OSNs, Lsd1 catalyzes the demethylation of H3K9me2, opening heterochromatin to initiate OR expression [6]. To determine whether dLsd1 (Su(var)3-3) is also important for Drosophila OR expression, we used Peb-Gal4 to express a UAS-IR (“IR” for “inverted repeats”) line specific to dLsd1 in all OSNs. We found that 16% of the dLsd1-depleted flies showed loss of Or59b reporter expression (Fig 4A), suggesting that dLsd1 is important in the establishment of OR expression in Drosophila. Interestingly, we found that many more dLsd1-depleted flies (43%) showed loss of the Or59bME reporter (Fig 4A and 4B), which lacks cooperative regulation, than loss of the Or59b reporter. This suggests TF cooperativity and dLsd1 are both important for OR expression in flies. In mice, one of the few TFs known to regulate vertebrate OR expression, Lhx2 [34], requires cooperativity to maintain OR expression and counteract heterochromatin formation in OSNs [35].
Fig 4. dLsd1 balances Su(var)3-9 and establishes Or59b expression.
(A) Or59b reporter and (B) Or59bME reporter GFP expression in flies with dLsd1 knock-down before initiation (Peb-Gal4) or after odorant receptor expression (Orco-Gal4). GFP expression is shown in green. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). (C) Or59bME reporter expression in heterozygote Su(var)3−906, heterozygote dLsd109, and double heterozygote flies. Note that the expression changes of the Or59bME reporter in single heterozygote flies were rescued in Su(var)3−906 and dLsd109 heterozygote flies. Control flies were crossed to w1118. Percentage denotes the fraction of flies with the depicted phenotype (S2 Data). Scale bar denotes 3.5 μm.
During Drosophila development, dLsd1 erases H3K4 dimethylation and promotes heterochromatin formation [36,37]. In both Drosophila and mice, Su(var)3-9 methylates H3K9me2 to form H3K9me3, a marker of heterochromatin [38–40]. Or59bME reporter expression in heterozygous Su(var)3-9 mutant flies shows a complex phenotype [23], with 19% of the flies showing ectopic expression, 32% showing loss of expression, and the rest showing single-class expression. Or59bME expression is also lost in 60% of heterozygous Su(var)3−309 (dLsd1 mutant) flies (Fig 4C). Combining the dLsd1 and Su(var)3-9 heterozygotes resets the balance and rescues reporter expression (Fig 4C). This suggests not only that the opening and closing of heterochromatin controls OR expression, but also that dLsd1 promotes open heterochromatin in Drosophila OSNs to support OR expression.
To determine whether dLsd1 initiates or maintains OR expression, we knocked down dLsd1 using Orco-Gal4, which drives expression in most OSNs after OR expression has already begun [33,41]. In these late knock-down flies, Or59b reporter expression was unperturbed (Fig 4A), indicating dLsd1 is required only during the initiation of OR expression. Interestingly, however, when we repeated the late dLsd1 knock-down experiment with the Or59bME reporter, we found a strong loss-of-expression phenotype (Fig 4B), showing that dLsd1 is required continuously to support OR expression.
Kdm4b initiates OR expression
Some mathematical models predict an as-yet-unknown factor that functions at individual OR loci in vertebrates to open constitutive heterochromatin by erasing H3K9me3 [6,8]. There are 2 genes encoding H3K9me3 demethylases in the Drosophila genome, Kdm4a (Kdm4B in vertebrates) and Kdm4b (Kdm4A, -C, -D, -E in vertebrates) [42]. We found, via knock-down of these 2 H3K9me3 demethylases in OSNs, that Kdm4b but not Kdm4a is required for Or59b expression (Fig 5A). Kdm4b is the major H3K9 demethylase in Drosophila [43], which is consistent with the hypothesis that the opening of heterochromatin is required for Or59b expression. We next asked whether Kdm4b is required for continuous Or59b expression by knocking down Kdm4b after OR initiation. Because OR expression begins in the mid-pupal stage and Orco expression begins shortly before eclosion, we decided to use Orco-Gal4 [33] for this knock-down experiment. We found that Orco-Gal4-driven Kdm4b knock-down had no effect on Or59b expression (Fig 5B), indicating that Kdm4b is important for Or59b expression initiation rather than maintenance.
Fig 5. Kdm4b initiates Or59b expression.
Whole-mount brain staining shows Or59b reporter expression of GFP (green). Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). Scale bar denotes 3.5 μm. (A) Loss of expression of the Or59b reporter is observed in the knock-down of Kdm4b but not Kdm4a. Control flies were crossed to w1118. (B) Orco-Gal4 knock-down of Kdm4b after the initiation of odorant receptor expression. Percentage denotes the proportion of flies with the depicted phenotype. Control flies were crossed to w1118 (S2 Data).
OR feedback regulates Kdm4b, dLsd1, and Su(var)3-9 expression
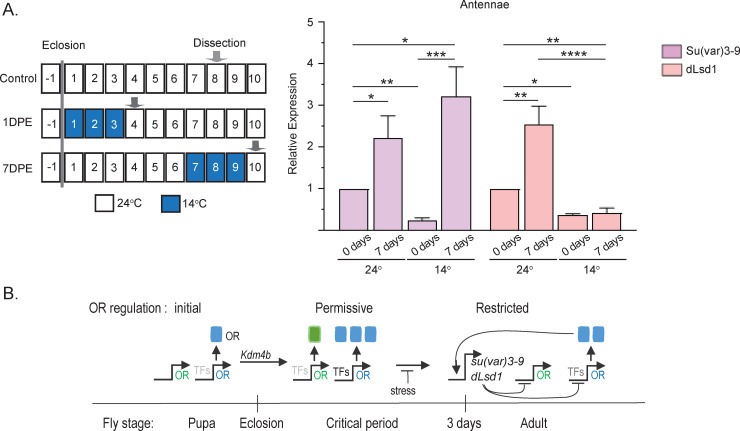
Thus far, our results have revealed that the maturation of OR expression comprises a shift from a developmentally permissive state to a more restrictive state in adults. This suggests that expression levels of dLsd1 and Su(var)3-9 are dynamic. We therefore analyzed antennal expression of Su(var)3-9 and dLsd1 and found that expression increased from low levels in newly eclosed flies to adult levels 3 days later (Fig 6A). Kdm4b expression, in contrast, decreased over the same period (Fig 6B), suggesting that the maturation of OR expression involves a reduction in the initiation and manifestation of OR expression. A more detailed dLsd1 and Su(var)3-9 expression analysis showed that the main increase in mRNA levels for these 2 enzymes occurred during the first hours post-eclosion (Fig 6C). Orco mutant flies that lack OR activity also showed reduced dLsd1 and Su(var)3-9 mRNA levels (Fig 6D), suggesting that OR function or increased expression may induce dLsd1 and Su(var)3-9 expression. Interestingly, we found that Kdm4b mRNA levels also increased in Orco mutants 3 DPE compared to controls (Fig 6D). This suggests that the absence of OR expression and its feedback suppression of Kdm4b likely increases OR expression initiation. Next, we over-expressed Or47b in a heterozygous Su(var)3-9 mutant background. We found, consistent with the hypothesis that increased OR expression or OR activity induces heterochromatin formation, that the heterozygote mutant reduction of Su(var)3-9 balanced the effect of Or47b ectopic expression and rescued the loss of Or59b reporter expression (Fig 6E).
Fig 6. Dynamic expression of chromatin modulators regulates odorant receptor expression.
(A) The graph shows Su(var)3-9 and dLsd1 mRNA levels in antenna at 1, 3, and 7 DPE (*p < 0.05; **p < 0.01; ***p < 0.001; error bars represent SEM (S2 Data). (B) The graph shows the Kdm4b mRNA levels in the antenna at 1 and 7days post-eclosion (DPE). Note that Su(var)3-9/dLsd1 shows contrasting regulation to Kdm4b after eclosion. (C) This graph shows the mRNA levels of Su(var)3-9 and dLsd1 in the antenna at 1 hour and 7 hours after eclosion. Note that the expression levels increase to almost double at 7 hours post-eclosion. (D) The graph compares control (w1118) and Orco mutant mRNA levels of Su(var)3-9, dLsd1, and Kdm4b in the antenna at 4 DPE. Note that the expression levels are lower for Su(var)3-9 and dLsd1 and higher for Kdm4b in Orco mutant flies. (E) GFP expression (green) driven by the Or59b reporter. Note that the loss of Or59b reporter expression in flies with Or47b ectopic expression is rescued in a Su(var)3−906 heterozygote background. Synaptic neuropil is labeled with the presynaptic marker nc82 (magenta). Control flies were crossed to w1118 (S4 Data).
Stress regulates Su(var)3-9 expression differently during and after OR expression maturation
To determine whether dLsd1 and Su(var)3-9 expression are sensitive to stress, we analyzed their expression in flies shifted to low temperature at different time points (Fig 7). Flies subjected to a temperature shift at eclosion (1 DPE) showed a 2-fold reduction in dLsd1 and Su(var)3-9 expression (Fig 7). The balanced reduction is consistent with a continuous permissiveness. Interestingly, reduction in copy number of both Su(var)3-9 and dLsd1 produced single-class expression, whereas stress produced ectopic expression, suggesting that additional stress signals enhance the permissive state. After a similar shift in adult flies (7 DPE), dLsd1 expression fell to the level found at eclosion, whereas Su(var)3-9 expression showed no significant change (Fig 7). The resulting imbalance between Su(var)3-9 and dLsd1 is consistent with the loss of Or59b expression we observed when we exposed adult heterozygous dLsd1 mutant flies to stress (Fig 4C).
Fig 7. dLsd1 regulation differs following environmental stress during or after the critical period.
(A) Schematic showing the time points of thermal stress treatment and sample preparation. The graph shows the Su(var)3-9 and dLsd1 mRNA levels in the antenna after 3 days of thermal stress (14°C) ending at 4 DPE or 10 DPE, compared to flies maintained at ambient temperature (24°C) (S4 Data). (B) Schematic model of the progression of OR gene expression regulation from initiation to adult mature OR expression. OR proteins depicted as rectangles. DPE, days post-eclosion; OR, odorant receptor; TF, transcription factor.
Discussion
Here, we show that Drosophila OR expression matures and that OSNs become terminally differentiated after OR expression initiation. Our results show that OR expression matures in 3 steps: initiation, establishment, and refinement.
OR expression initiation: Predetermined versus stochastic
Models of vertebrate OR expression suggest the existence of a heterochromatin switch that initiates expression [6,8]. We found Kdm4b, an H3K9me3 demethylase, induces OR expression. In a direct instructive model, a predetermined differentiation path produces TFs that recruit Kdm4b to open an OR locus. In a more stochastic model, Kdm4b opens chromatin at an OR locus, and if the necessary factors are available, the locus is kept open. A recent study revealed that low OR expression precedes the initiation step [41], which, together with our results, favors a model in which Kdm4b is recruited to the OR locus or even attracted by low OR expression, and in the presence of high OR expression inhibits OR initiation and expression at other OR loci.
A deeply conserved OR maturation mechanism
The mechanism that establishes OR expression was first identified in mice [44]. Perhaps the most striking point of conservation is the unique OSN-specific function of Lsd1. In most Drosophila and vertebrate cells, Lsd1 erases H3K4 methylation and dimethylation and induces heterochromatin formation [36,37]. Our results and several vertebrate OR choice studies [6,45,46] show that Lsd1 opposes Su(var)3-9 and constitutive heterochromatin formation in OSNs. The enzyme that forms H3K9me2, G9a, also restricts OR expression in both Drosophila and mice [24,47], making it clear that H3K9me2 lies at the center of OR gene regulation across phyla. The cis regulatory regions have also evolved to balance heterochromatin formation in similar ways between Drosophila and mice. TF cooperativity opposes heterochromatin formation and stabilizes Or59b expression [23]. In mice, Lhx2, one of the few TFs known to regulate vertebrate OR expression [34], also requires cooperativity to block heterochromatin formation and establish OR expression [35].
Also, the OR feedback loop was first described in mice [44,48]. The vertebrate feedback mechanisms build on the folding of [6,9] or signaling from [10,11] an expressed OR to inhibit the expression of other ORs. With the many levels of conserved features in OR regulation, it is possible that vertebrate-like OR feedback mechanisms link the expressed OR and dLsd1 and Su(var)3-9 expression.
After maturation, the OR expression mechanisms differ between Drosophila and mice. In Drosophila, both OR alleles are expressed in each OSN [49]. In mice, 1 OR allele is selected and expressed continuously by what is likely a separate mechanism. Another difference in Drosophila is that dLsd1 activity balances Su(var)3-9 activity after maturation, whereas in mice, Lsd1 is down-regulated after maturation [44]. We found that it is only Lsd1 that is suppressed after maturation, suggesting that the memory mechanism that maintains strict monogenic OR expression is an inflexible Su(var)3-9 expression that produces a defined heterochromatin level and sets the OR expression baseline. It remains unclear if such a memory mechanism is conserved, given the differences in regulation after OR expression maturation.
A critical period mechanism controls OR expression
The restricted duration of OR expression maturation suggests that the period of gene regulation plasticity may be a bona fide critical period [50,51]. OR regulation does fulfill the criteria. First, a critical period should have a restricted duration, and OR expression maturation ends after a very sharp transition in gene regulation 2 DPE. Second, the plasticity of a critical period should be sensitive to activity in the circuit, and we found that feedback from an expressed OR can refine OR expression. Third, the phenotype changing in a critical period should be refined through competition, and we show that ectopic OR expression can outcompete endogenous OR expression. Fourth, the plasticity in a critical period should be sensitive to external stress, and we show that stress can dramatically alter OR expression during and after the relevant period. Fifth, the phenotype developing during a critical period becomes permanent after the period has passed, and we show that adult OR expression reaches a permanent state, indicating that OSN differentiation ends as the critical period closes. The conserved nature of the mechanisms, and the fact that immature vertebrate OSNs also show a low frequency of OR co-expression [52,53], suggests vertebrate OSN differentiation closes with a critical period as well.
Differences in OR feedback regulation between OSN classes
We found that the timing of the OR feedback regulation depends on the OSN lineage. According to our results, most trichoid-related OR expression regulation changes take place in the pupal stage, whereas basiconic-related OR expression refinement seems to take place after eclosion. Interestingly, this difference in regulation relates to olfactory function because basiconic-related ORs respond more to food-related odorants while trichoid-related ORs respond more to pheromones for the sake of social interactions. When a fly emerges from its pupal case, it does so in the vicinity of the food it lived on as a larva but not necessarily close to other flies. Consistent with this, pheromone responses increase as social interactions increase post-maturation [54]. These response increases come, at least in part, from the sensitization of Or47b OSNs rather than from changes in Or47b expression, suggesting a separate mechanism. Thus, early trichoid-related OR gene regulation supports OR expression even in the absence of stimuli and allows for plasticity even after the OSNs have matured. For basiconic-related OR expression, the ORs’ late regulation provides more tuning possibilities in a dynamic food odor environment.
The critical period provides flexibility for OR gene regulation
Predetermined systems of OR gene regulation lack the flexibility that feedback mechanisms can provide. We and others have shown that high odor responses suppress Drosophila OR expression [20,22]. Feedback mechanisms like this could tune responses to environmental odor levels and ensure odor responses fall within physiological limits. Our results further predict that feedback refinement buffers and allows for imperfect gene regulation, reducing the regulatory cost to maintain tight monogenic OR expression. The TFs required to express 1 particular OR can likely even vary with internal state and stress level. Our results also predict that the DNA binding motif locations and cis regulatory mechanisms can be plastic between species.
Stress inhibition of the feedback mechanisms also adds to the flexibility of the system. Stress can induce OR paralog expression, allowing the previously suppressed paralog to contribute to odor responses when the environment changes. In short-lived organisms like Drosophila, stress early in life predicts an insecure future. It therefore follows logically that stress early in life would inhibit OR expression maturation, and if the stress lasts beyond the critical period, the changes become permanent. In this way, the permissiveness built into the system makes OSNs and OR gene regulation more robust and resilient to continued or future episodes of stress.
The stress-altered OR expression also makes the animal more robust to environmental variability. Our results indicate that the feedback systems and the critical period function as a capacitator, silencing the effect of allelic variability, allowing changes in the OR genes and adaptation of olfactory function. This capacitator function hypothesis predicts that in the non-stressed ambient state, OR feedback keeps paralogs and alternate alleles dormant and produces the uniform OR expression observed in adult flies. But when the environment changes, stress blocks feedback suppression and dormant OR alleles or paralogs can be expressed, leading to an individualization of OR expression and odor responses in the population. Interestingly, the OSNs that express ORs also express IR co-receptors in both Drosophila and mosquitoes [55,56], suggesting that ORs and IRs are co-expressed in some OSN classes. Electrophysiology also shows that some OSN classes in Drosophila (ab1b, ab3a, and ab6a) respond to IR odors [57], suggesting that stress can tweak the balance between co-expressed ORs and IRs. Thus, our prediction is that stress accentuates alternative responses and OR allele expression when environmental conditions change, and shifts the system from optimal function to maximal detection.
Materials and methods
Drosophila stocks
The Or59b promoter fusion and Or59b minimal enhancer constructs were described previously [23]. Pebbled-Gal4 (Peb-Gal4) was a kind gift from Liqun Luo (Stanford University, Stanford, CA, US). The Su(var)3−906 and Lsd109 mutants were a kind gift from Anita Öst (Linköping University, Linköping, Sweden). UAS-Or42b was a kind gift of Matthieu Louis, and UAS-Or47b:HA;UAS-Or65a was a kind gift from John Carlson. The following RNA interference (RNAi) lines were obtained from the Transgenic RNAi Project (TRiP; Harvard Medical School, Boston, MA, US; http://www.flyrnai.org): Su(var)3-3 (dLsd1)-IR (36867; 32853, 33726), Kdm4a-IR (34629), and Kdm4b-IR (35676, 57721). The following fly lines were provided by the Bloomington Drosophila Stock Center (Indiana University, Bloomington, IN, US; http://flystocks.bio.indiana.edu): w1118 (38690) and Orco-Gal4 (23909).
RNAi methodology and environmental experiments
Virgin RNAi females were mated with males carrying Pebbled-Gal4, UAS-Dicer2, and the cluster transgenes. The crosses were set up and maintained at 24°C. Then, 2–5 days after eclosion, the flies were dissected, stained, and scored for phenotypes.
For the stress experiments, flies were collected as virgins and raised on standard Drosophila culture medium at 24°C. On the day for the temperature shift, the temperature-stressed flies were transferred to new vials and maintained for 3 days at 14°C, while control flies were maintained at ambient temperature. Further information can be found in the supplemental experiment statics and details (S2 Data).
Immunofluorescence
Immunofluorescence was performed as previously described [15]. The following primary antibodies were used: rabbit anti-GFP (1:2,000, TP-401; Torrey Pines Biolabs) and mouse anti-nc82 (1:100; Developmental Studies Hybridoma Bank). Secondary antibodies were conjugated with Alexa Fluor 488 (1:500; Molecular Probes) and Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Rhodamine Red-X (1:250; Thermo Fisher Scientific). Confocal microscopy images were collected on an LSM 700 (Zeiss) and analyzed using the LSM Image Browser. The numbers of OSNs co-expressing BP104 and GFP for the different constructs were counted in these images. Adobe Photoshop CS4 (Adobe Systems) was used for image processing.
Quantitative PCR
Antennae were obtained with a sieve after freezing the appropriate flies in liquid nitrogen. Total RNA from the antennae was extracted with TRIzol (Invitrogen) and purified with the RNeasy kit (Qiagen). Quantitative PCR was conducted on an Applied Biosystems 7900HT Fast Real-Time PCR System (Life Technologies) using the Power SYBR Green PCR Master Mix (Applied Biosystems, Life Technologies) and primer sets designed using Primer Express Software v3.0.1 (Integrated DNA Technologies). Actin 5c was used as an internal control. To amplify cDNA products and not genomic DNA, primers were designed to join the end of one exon with the beginning of the next exon. Quantitative PCR for each primer set was performed on both control and experimental samples for 40 cycles. Following amplification, melt curve analysis and ethidium bromide agarose gel electrophoresis were performed to evaluate the PCR products. The relative quantification of the fold change in mRNA expression was calculated using the 2−ΔΔCT threshold cycle method.
Library preparation
For RNA-seq experiments, virgin flies were collected, and 50 antennae were handpicked, either immediately or after 4 or 14 days on standard Drosophila culture medium at 24°C. Total RNA was extracted using TRIzol (Invitrogen, cat. no. 15596–018) according to the manufacturer’s instructions. DNA was degraded using the Invitrogen TURBO DNA-free Kit. After DNase treatment, TRIzol RNA extraction was repeated a second time. The concentration and quality of the RNA was determined using a sensitive fluorescent-dye-based Qubit RNA HS Assay Kit and the Agilent HS RNA kit and an Agilent 4200 TapeStation System.
Using 1–5 μg of total RNA for each sample, we performed 2 rounds of mRNA isolation using the NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490) according to the manufacturer’s instructions. Libraries were generated using the NEBNext RNA Ultra II RNA Library Prep Kit. The samples were quality controlled and successfully sequenced on an Illumina NextSeq 500 next-generation sequencing system in mid-output mode via 1 × 100 bp paired-end sequencing.
RNA-seq analysis
The RNA read counts were estimated with Kallisto (version 0.45.1). Differentially expressed genes were estimated by DESeq2 (version 1.26.0) after counts had been rounded to the nearest integer count. The linear model was simply one group versus the other group, e.g., WT day 1 versus day 4, or WT day 1 versus treatment day 1. Plots were made using ggplot2 and R, showing log10 size-factor-normalized read counts.
Supporting information
Degree of change in sequence counts observed between control and the different genotypes at 4 DPE relative to 1 DPE. Normalized logarithmic read counts (log10 size-factor-normalized) for each gene from the respective sample were scatter-plotted. Genes shown in grey except basiconic ORs (green) and trichoid ORs (magenta). The line is the reference at which gene expression is the same between conditions, with increased expression above, and suppression below, the line. Statistics for the figure are in S3 Data.
(TIF)
(XLSX)
(DOCX)
Acknowledgments
We thank John Carlson, Matthieu Louis, Liqun Luo, and Anita Öst for flies, and Najat Dzaki and Jan Larsson for discussion and comments on the manuscript.
Abbreviations
- BBS
BBSome
- DPE
days post-eclosion
- EP
ethyl propionate
- GR
gustatory receptor
- H3K9me2
histone H3 lysine 9 dimethylation
- H3K9me3
histone H3 lysine 9 trimethylation
- IFT
intraflagellar transport
- IR
ionotropic receptor
- OBP
odorant binding protein
- OR
odorant receptor
- OSN
olfactory sensory neuron
- RNA-seq
RNA sequencing
- TF
transcription factor
Data Availability
All the code is available on GitHub (https://github.com/henriksson-lab/mattias-or). The RNA-seq data is available on ArrayExpression with accession #E-MTAB-9805. The quantitative data are in the Supporting Materials.
Funding Statement
The authors have received support from the following funders: Vetenskapsrådet VR grant 2016-0520 to MA and SJ, Vetenskapsrådet VR grant 2016- 06726 to SJ, Vetenskapsrådet VR grant #2016-06598 (https://www.vr.se/) to JH, and National Science Foundation I/UCRC, the Center for Arthropod Management Technologies under Grant No. IIP-1821914 (https://www.iucrc-camtech.org/) (to HY). The computations were performed using resources provided by the Swedish National Infrastructure for Computing (SNIC) through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project SNIC 2020/6-251. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15(17):1535–47. 10.1016/j.cub.2005.07.034 [DOI] [PubMed] [Google Scholar]
- 2.Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15(17):1548–53. 10.1016/j.cub.2005.07.066 [DOI] [PubMed] [Google Scholar]
- 3.Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79(7):1245–55. 10.1016/0092-8674(94)90015-9 [DOI] [PubMed] [Google Scholar]
- 4.Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, et al. Visualizing an olfactory sensory map. Cell. 1996;87(4):675–86. 10.1016/s0092-8674(00)81387-2 [DOI] [PubMed] [Google Scholar]
- 5.Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145(4):555–70. 10.1016/j.cell.2011.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154(2):325–36. 10.1016/j.cell.2013.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyons DB, Magklara A, Goh T, Sampath SC, Schaefer A, Schotta G, et al. Heterochromatin-mediated gene silencing facilitates the diversification of olfactory neurons. Cell Rep. 2014;9(3):884–92. 10.1016/j.celrep.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan LZ, Zong CH, Xie XS. Rare event of histone demethylation can initiate singular gene expression of olfactory receptors. Proc Natl Acad Sci U S A. 2013;110(52):21148–52. 10.1073/pnas.1321511111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155(2):321–32. 10.1016/j.cell.2013.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira T, Wilson SR, Choi YG, Risso D, Dudoit S, Speed TP, et al. Silencing of odorant receptor genes by G protein betagamma signaling ensures the expression of one odorant receptor per olfactory sensory neuron. Neuron. 2014;81(4):847–59. 10.1016/j.neuron.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischmann A, Abdus-Saboor I, Sayed A, Shykind B. Functional interrogation of an odorant receptor locus reveals multiple axes of transcriptional regulation. PLoS Biol. 2013;11(5):e1001568. 10.1371/journal.pbio.1001568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zapiec B, Mombaerts P. The zonal organization of odorant receptor gene choice in the main olfactory epithelium of the mouse. Cell Rep. 2020;30(12):4220–34.e5. 10.1016/j.celrep.2020.02.110 [DOI] [PubMed] [Google Scholar]
- 13.Barish S, Volkan PC. Mechanisms of olfactory receptor neuron specification in Drosophila. Wires Dev Biol. 2015;4(6):609–21. 10.1002/wdev.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray A, van Naters WV, Carlson JR. A regulatory code for neuron-specific odor receptor expression. PLoS Biol. 2008;6(5):e125. 10.1371/journal.pbio.0060125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jafari S, Alkhori L, Schleiffer A, Brochtrup A, Hummel T, Alenius M. Combinatorial activation and repression by seven transcription factors specify Drosophila odorant receptor expression. PLoS Biol. 2012;10(3):e1001280. 10.1371/journal.pbio.1001280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endo K, Aoki T, Yoda Y, Kimura K, Hama C. Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nat Neurosci. 2007;10(2):153–60. 10.1038/nn1832 [DOI] [PubMed] [Google Scholar]
- 17.Tichy AL, Ray A, Carlson JR. A new Drosophila POU gene, pdm3, acts in odor receptor expression and axon targeting of olfactory neurons. J Neurosci. 2008;28(28):7121–9. 10.1523/JNEUROSCI.2063-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komiyama T, Carlson JR, Luo L. Olfactory receptor neuron axon targeting: intrinsic transcriptional control and hierarchical interactions. Nat Neurosci. 2004;7(8):819–25. 10.1038/nn1284 [DOI] [PubMed] [Google Scholar]
- 19.Sim CK, Perry S, Tharadra SK, Lipsick JS, Ray A. Epigenetic regulation of olfactory receptor gene expression by the Myb-MuvB/dREAM complex. Genes Dev. 2012;26(22):2483–98. 10.1101/gad.201665.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koerte S, Keesey IW, Khallaf MA, Llorca LC, Grosse-Wilde E, Hansson BS, et al. Evaluation of the DREAM technique for a high-throughput deorphanization of chemosensory receptors in Drosophila. Front Mol Neurosci. 2018;11:366. 10.3389/fnmol.2018.00366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyengar A, Chakraborty TS, Goswami SP, Wu CF, Siddiqi O. Post-eclosion odor experience modifies olfactory receptor neuron coding in Drosophila. Proc Natl Acad Sci U S A. 2010;107(21):9855–60. 10.1073/pnas.1003856107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von der Weid B, Rossier D, Lindup M, Tuberosa J, Widmer A, Col JD, et al. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat Neurosci. 2015;18(10):1455–63. 10.1038/nn.4100 [DOI] [PubMed] [Google Scholar]
- 23.Jafari S, Alenius M. Cis-regulatory mechanisms for robust olfactory sensory neuron class-restricted odorant receptor gene expression in Drosophila. PLoS Genet. 2015;11(3):e1005051. 10.1371/journal.pgen.1005051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alkhori L, Ost A, Alenius M. The corepressor Atrophin specifies odorant receptor expression in Drosophila. FASEB J. 2014;28(3):1355–64. 10.1096/fj.13-240325 [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez A, Jafari S, Zenere A, Alenius M, Altafini C. Thermodynamic model of gene regulation for the Or59b olfactory receptor in Drosophila. PLoS Comput Biol. 2019;15(1):e1006709. 10.1371/journal.pcbi.1006709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golovin RM, Vest J, Vita DJ, Broadie K. Activity-dependent remodeling of Drosophila olfactory sensory neuron brain innervation during an early-life critical period. J Neurosci. 2019;39(16):2995–3012. 10.1523/JNEUROSCI.2223-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devaud JM, Acebes A, Ramaswami M, Ferrus A. Structural and functional changes in the olfactory pathway of adult Drosophila take place at a critical age. J Neurobiol. 2003;56(1):13–23. 10.1002/neu.10215 [DOI] [PubMed] [Google Scholar]
- 28.Sachse S, Rueckert E, Keller A, Okada R, Tanaka NK, Ito K, et al. Activity-dependent plasticity in an olfactory circuit. Neuron. 2007;56(5):838–50. 10.1016/j.neuron.2007.10.035 [DOI] [PubMed] [Google Scholar]
- 29.Sanchez GM, Alkhori L, Hatano E, Schultz SW, Kuzhandaivel A, Jafari S, et al. Hedgehog signaling regulates the ciliary transport of odorant receptors in Drosophila. Cell Rep. 2016;14(3):464–70. 10.1016/j.celrep.2015.12.059 [DOI] [PubMed] [Google Scholar]
- 30.Larter NK, Sun JS, Carlson JR. Organization and function of Drosophila odorant binding proteins. Elife. 2016;5:e20242. 10.7554/eLife.20242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maguire SE, Afify A, Goff LA, Potter CJ. A feedback mechanism regulates odorant receptor expression in the malaria mosquito, Anopheles gambiae. bioRxiv. 2020. July 24. 10.1101/2020.07.23.218586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125(1):143–60. 10.1016/j.cell.2006.01.050 [DOI] [PubMed] [Google Scholar]
- 33.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43(5):703–14. 10.1016/j.neuron.2004.08.019 [DOI] [PubMed] [Google Scholar]
- 34.Kolterud A, Alenius M, Carlsson L, Bohm S. The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development. 2004;131(21):5319–26. 10.1242/dev.01416 [DOI] [PubMed] [Google Scholar]
- 35.Monahan K, Schieren I, Cheung J, Mumbey-Wafula A, Monuki ES, Lomvardas S. Cooperative interactions enable singular olfactory receptor expression in mouse olfactory neurons. Elife. 2017;6:e28620. 10.7554/eLife.28620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr Biol. 2007;17(9):808–12. 10.1016/j.cub.2007.03.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26(1):103–15. 10.1016/j.molcel.2007.02.025 [DOI] [PubMed] [Google Scholar]
- 38.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21(5):1121–31. 10.1093/emboj/21.5.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292(5514):110–3. 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- 40.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–9. 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- 41.McLaughlin CN, Brbić M, Xie Q, Li T, Horns F, Kolluru SS, et al. Single-cell transcriptomes of developing and adult olfactory receptor neurons in Drosophila. Elife. 2021;10:e63856. 10.7554/eLife.63856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–57. 10.1038/nrg3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsurumi A, Dutta P, Shang R, Yan SJ, Li WX. Drosophila Kdm4 demethylases in histone H3 lysine 9 demethylation and ecdysteroid signaling. Sci Rep. 2013;3:2894. 10.1038/srep02894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dalton RP, Lomvardas S. Chemosensory receptor specificity and regulation. Annu Rev Neurosci. 2015;38:331–49. 10.1146/annurev-neuro-071714-034145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coleman JH, Lin B, Schwob JE. Dissecting LSD1-dependent neuronal maturation in the olfactory epithelium. J Comp Neurol. 2017;525(16):3391–413. 10.1002/cne.24259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vyas RN, Meredith D, Lane RP. Lysine-specific demethylase-1 (LSD1) depletion disrupts monogenic and monoallelic odorant receptor (OR) expression in an olfactory neuronal cell line. Mol Cell Neurosci. 2017;82:1–11. 10.1016/j.mcn.2017.04.005 [DOI] [PubMed] [Google Scholar]
- 47.Lyons DB, Lomvardas S. Repressive histone methylation: a case study in deterministic versus stochastic gene regulation. Biochim Biophys Acta. 2014:1839(12):1373–84. 10.1016/j.bbagrm.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 48.Abdus-Saboor I, Fleischmann A, Shykind B. Setting limits: maintaining order in a large gene family. Transcription. 2014;5:e28978. 10.4161/trns.28978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37(5):827–41. 10.1016/s0896-6273(03)00094-1 [DOI] [PubMed] [Google Scholar]
- 50.Burggren WW. Phenotypic switching resulting from developmental plasticity: fixed or reversible? Front Physiol. 2019;10:1634. 10.3389/fphys.2019.01634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hensch TK. Critical period regulation. Annu Rev Neurosci. 2004;27:549–79. 10.1146/annurev.neuro.27.070203.144327 [DOI] [PubMed] [Google Scholar]
- 52.Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, Sun Y, et al. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117(6):801–15. 10.1016/j.cell.2004.05.015 [DOI] [PubMed] [Google Scholar]
- 53.Hanchate NK, Kondoh K, Lu Z, Kuang D, Ye X, Qiu X, et al. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 2015;350(6265):1251–5. 10.1126/science.aad2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sethi S, Lin HH, Shepherd AK, Volkan PC, Su CY, Wang JW. Social context enhances hormonal modulation of pheromone detection in Drosophila. Curr Biol. 2019;29(22):3887–98.e4. 10.1016/j.cub.2019.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Task D, Lin C-C, Afify A, Li H, Vulpe A, Menuz K, et al. Widespread polymodal chemosensory receptor expression in Drosophila olfactory neurons. bioRxiv. 2020. November 8. 10.1101/2020.11.07.355651 [DOI] [Google Scholar]
- 56.Younger MA, Herre M, Ehrlich AR, Gong Z, Gilbert ZN, Rahiel S, et al. Non-canonical odor coding ensures unbreakable mosquito attraction to humans. bioRxiv. 2020. November 8. 10.1101/2020.11.07.368720 [DOI] [Google Scholar]
- 57.de Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30(2):537–52. 10.1016/s0896-6273(01)00289-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Degree of change in sequence counts observed between control and the different genotypes at 4 DPE relative to 1 DPE. Normalized logarithmic read counts (log10 size-factor-normalized) for each gene from the respective sample were scatter-plotted. Genes shown in grey except basiconic ORs (green) and trichoid ORs (magenta). The line is the reference at which gene expression is the same between conditions, with increased expression above, and suppression below, the line. Statistics for the figure are in S3 Data.
(TIF)
(XLSX)
(DOCX)
Data Availability Statement
All the code is available on GitHub (https://github.com/henriksson-lab/mattias-or). The RNA-seq data is available on ArrayExpression with accession #E-MTAB-9805. The quantitative data are in the Supporting Materials.