Abstract
DNA methyltransferase (Dnmt)3b mediates de novo DNA methylation and modulation of Dnmt3b in respiratory epithelial cells has been shown to affect the expression of multiple genes. Respiratory epithelial cells provide a first line of defense against pulmonary pathogens and play a crucial role in the immune response during pneumonia caused by Pseudomonas (P.) aeruginosa, a gram-negative bacterium that expresses flagellin as an important virulence factor. We here sought to determine the role of Dntm3b in respiratory epithelial cells in immune responses elicited by P. aeruginosa. DNMT3B expression was reduced in human bronchial epithelial (BEAS-2B) cells as well as in primary human and mouse bronchial epithelial cells grown in air liquid interface upon exposure to P. aeruginosa (PAK). Dnmt3b deficient human bronchial epithelial (BEAS-2B) cells produced more CXCL1, CXCL8 and CCL20 than control cells when stimulated with PAK, flagellin-deficient PAK (PAKflic) or flagellin. Dnmt3b deficiency reduced DNA methylation at exon 1 of CXCL1 and enhanced NF-ĸB p65 binding to the CXCL1 promoter. Mice with bronchial epithelial Dntm3b deficiency showed increased Cxcl1 mRNA expression in bronchial epithelium and CXCL1 protein release in the airways during pneumonia caused by PAK, which was associated with enhanced neutrophil recruitment and accelerated bacterial clearance; bronchial epithelial Dnmt3b deficiency did not modify responses during pneumonia caused by PAKflic or Klebsiella pneumoniae (an un-flagellated gram-negative bacterium). Dnmt3b deficiency in type II alveolar epithelial cells did not affect mouse pulmonary defense against PAK infection. These results suggest that bronchial epithelial Dnmt3b impairs host defense during Pseudomonas induced pneumonia, at least in part, by dampening mucosal responses to flagellin.
Author summary
The respiratory epithelium provides a first line of defense against respiratory pathogens. Pseudomonas (P.) aeruginosa is a common causative pathogen in pneumonia and its important virulence factor flagellin is a potent activator of epithelial cells. DNA methyltransferase (Dnmt)3b is an enzyme that mediates de novo methylation of DNA. We here tested the hypothesis that Dnmt3b is involved in the immune response generated in airway epithelial cells upon infection with P. aeruginosa. Using a combination of in vitro investigations with human bronchial epithelial cells and in vivo airway infection models in mice with targeted deletions of the gene encoding Dnmt3b in specific subtypes of airway epithelial cells we demonstrate that Dnmt3b in bronchial but not type 2 alveolar epithelial cells impairs host defense during Pseudomonas induced pneumonia, at least in part by inhibiting mucosal responses to flagellin, by an effect on epithelial cell DNA methylation. We report a thus far unknown role for bronchial epithelial cell Dnmt3b in the innate mucosal immune response to a common respiratory pathogen, providing insight into the regulatory machinery involved in reprograming of epithelial cells during pneumonia.
Introduction
Pseudomonas (P.) aeruginosa is a gram-negative flagellated bacterial pathogen and a common cause of pneumonia in hospitalized patients and those who suffer from chronic lung diseases [1,2]. The emergence of multidrug resistant Pseudomonas strains is a major health care concern, with reported rates of 15–30% in some geographical areas [3].
The respiratory epithelium provides a first line of defense against respiratory pathogens by producing a physical barrier and by releasing antimicrobial peptides, as well as chemotactic mediators [4]. Lung epithelial cells can be activated through a variety of receptors that recognize pathogens or components thereof. P. aeruginosa expresses a flagellum, which is important for its motility and is a major determinant of pathogenicity. Flagellin is the structural component of flagella recognized by Toll-like receptor (TLR) 5, thereby playing a key role in the induction of an innate immune response to infection with Pseudomonas [5,6]. TLR5 is abundantly expressed on the respiratory epithelium and triggering of this receptor results in the activation of the common TLR adaptor myeloid differentiation factor (MyD) 88 and subsequently nuclear factor (NF)-κB [5,6]. We and others previously documented a role for MyD88—dependent signaling in respiratory epithelial cells in boosting host defense during Pseudomonas pneumonia in mice [7,8,9].
The extent of DNA methylation influences gene transcription and chromatin structure, and can change during bacterial infection [10,11]. DNA methyltransferase (Dnmt)3b is one of the main enzymes mediating de novo DNA methylation [12]. Recent studies have suggested that alterations in DNA methylation mediated by Dnmt3b activity may affect transcriptional regulation in a context and cell specific way. The airway epithelium expresses Dnmt3b, which is enhanced by exposure to cigarette smoke [13]. Modulation of airway epithelial cell Dnmt3b expression impacts the expression of multiple genes [14]. Furthermore, exome sequencing associated variants of DNMT3B with community-acquired P. aeruginosa infection in children [15]. Together these results led us to hypothesize that respiratory epithelial Dnmt3b might play a role in host defense against P. aeruginosa infection. To test this hypothesis we evaluated innate immune responses induced in airway epithelial cells with modified Dnmt3b expression in vitro and in mice with airway epithelial cell specific deletion of Dnmt3b in vivo upon exposure to wild-type P. aeruginosa, or isogenic flagellin deficient P. aeruginosa.
Results
Dnmt3b reduces Pseudomonas aeruginosa induced chemokine production by bronchial epithelial cells in vitro
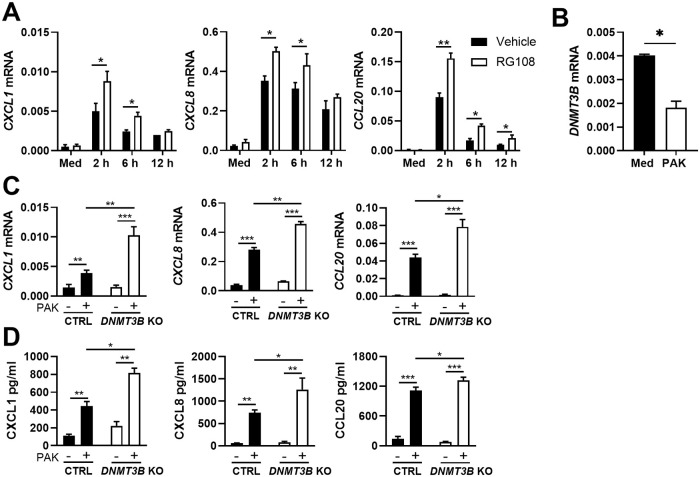
To obtain a first insight into the role of Dnmt3b in the regulation of innate immune responses by respiratory epithelial cells, human bronchial BEAS-2B cells were pretreated with the pan DNA methyltransferase inhibitor RG108 [16] and stimulated with heat killed wild-type P. aeruginosa (PAK). RG108 at doses up to 50 μM did not affect cell viability (S1A Fig), but (at a dose of 10 μM) increased PAK-induced mRNA expression of the chemokines CXCL1, CXCL8 and CCL20 (Fig 1A). PAK stimulation decreased DNMT3B mRNA levels (Fig 1B) without affecting DNMT3A or DNMT1 mRNA expression (S1B Fig). Since P. aeruginosa is a major pathogen in infections of lungs affected by cystic fibrosis, we then examined DNMT expression in the human cystic fibrosis bronchial epithelial cell line CFBE41o- complemented with the wild type cystic fibrosis transmembrane conductance regulator (CFTR) gene after P. aeruginosa infection for 1 hour using publicly available data (GSE30439) [17]. DNMT3B expression was significantly decreased by P. aeruginosa whilst the expression of DNMT3A or DNMT1 was not affected (S1C Fig). To evaluate DNMT expression in 3 dimensional cultures of epithelial cells, we made use of human primary bronchial epithelial cells cultured in air liquid interface exposed to P. aeruginosa PAO1 for 24 hours in experiments published by our laboratory [18]. Consistent with the results obtained in cell lines, PA01 decreased DNMT3B but not DNMT3A or DNMT1 expression (S1D Fig). In mouse primary airway epithelial cells grown in air liquid interface Dnmt3b as well as Dnmt3a and Dnmt1 were decreased upon exposure to P. aeruginosa for 1 or 24 hours (GSE7957) [19] (S1E Fig). Together these data show that DNMT3B expression is decreased in human respiratory epithelial cell lines, as well as in primary human and mouse respiratory epithelial cells after exposure to P. aeruginosa. To determine the role of Dnmt3b in the responsiveness of respiratory epithelial cells to P. aeruginosa, we generated DNMT3B knockout BEAS-2B bronchial epithelial cells using CRISPR-Cas9; control BEAS-2B cells were generated in the same way using a non-targeting control guide RNA. Two confirmed Dnmt3b deficient and two control BEAS-2B clones (S1F Fig) were exposed to heat-killed PAK for 12 hours, after which mRNA and supernatants were harvested. PAK induced a marked upregulation of mRNA and protein levels of CXCL1, CXCL8 and CCL20, which were further increased in Dnmt3b deficient cells (Fig 1C and 1D). In contrast, the expression of genes encoding defensins (β-defensin 1 and 2), barrier function associated proteins (Tight junction protein 1 and 2) and cytokines (IL-1β and TNFα) was not altered by Dnmt3b deficiency (S1G Fig). Therefore, we focused on the regulation of chemokine production by Dnmt3b. Overexpression of Dnmt3b in BEAS-2B cells (S2A Fig) did not influence PAK-induced CXCL1, CXCL8 or CCL20 production (S2B Fig). Hence, these data suggest that endogenous Dnmt3b suppresses chemokine/cytokine production by bronchial epithelial cells upon activation by PAK in vitro.
Fig 1. Dnmt3b inhibition in BEAS-2B bronchial epithelial cells promotes Pseudomonas aeruginosa induced chemokine production in vitro.
BEAS-2B cells were pretreated with the DNMT inhibitor RG108 at 10 μM/ml (open bars) or vehicle (black bars) for 12 hours and then stimulated with heat killed PAK (MOI = 50) or medium control for 2, 6 or 12 hours. CXCL1, CXCL8 and CCL20 mRNA expression was measured by RT-qPCR (A); DNMT3B mRNA levels in BEAS-2B cells incubated with heat killed PAK or medium control for 12 hours (B); Dnmt3b deficient (knock out, DNMT3B KO; open bars) and control (Ctr; black bars) BEAS-2B cells were stimulated with heat killed PAK (+) or medium control (-) for 12 hours. CXCL1, CXCL8 and CCL20 mRNA levels were measured by RT-qPCR (C) and corresponding protein levels in the supernatant by ELISA (D). Data are presented as means ± SEM (n = 4) and representative of two to three independent experiments and for two DNMT3B KO and control clones. *p < 0.05, **p < 0.01, *** p < 0.001.
Dnmt3b inhibits Pseudomonas aeruginosa induced chemokine production by bronchial epithelial cells in vitro independent of flagellin expression
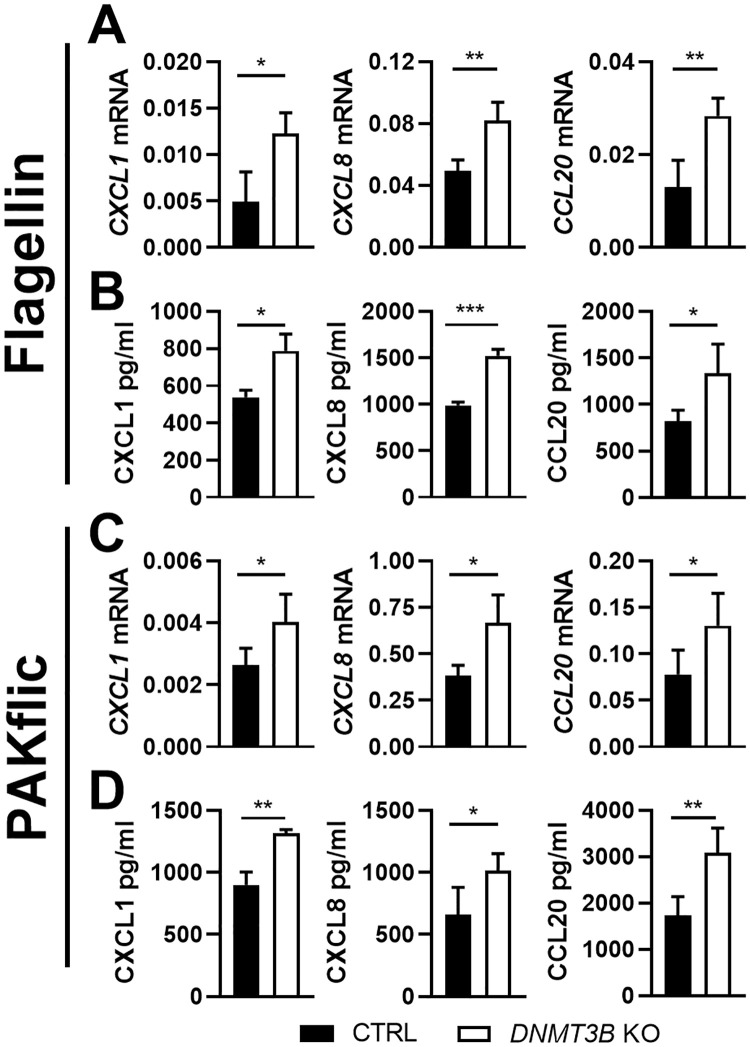
Flagellin is an important virulence factor expressed by PAK and a potent activator of respiratory epithelial cells [6]. Flagellin induced CXCL1, CXCL8 and CCL20 mRNA and protein expression by BEAS-2B cells, which was enhanced in Dnmt3b deficient cells (Fig 2A and 2B). To determine whether flagellin contributes to the regulatory function of Dnmt3b in respiratory epithelial cells activated by PAK, we stimulated BEAS-2B cells with flagellin deficient PAK (PAKflic). Similar to results obtained with wild-type PAK and purified flagellin, PAKflic induced more CXCL1, CXCL8 and CCL20 mRNA expression and protein production by Dnmt3b deficient BEAS-2B cells when compared with control BEAS-2B cells (Fig 2C and 2D). Overexpression of Dnmt3b in BEAS-2B cells did not influence CXCL1, CXCL8 or CCL20 production induced by flagellin or PAKflic (S2C Fig). The heating process used to kill PAK and PAKflic slightly reduced the biological activity of purified flagellin toward BEAS-2B cells, whereas trypsin totally abolished flagellin activity (S3 Fig), which is consistent with the reported heat stability of flagellin [20]. Together, these results suggest that although Dnmt3b influences flagellin responses, the role of Dnmt3b in PAK-induced chemokine production by BEAS-2B cells does not dependent on the presence of flagellin in this bacterium.
Fig 2. Dnmt3b deficiency in BEAS-2B bronchial epithelial cells promotes Pseudomonas aeruginosa induced chemokine production in vitro independent of flagellin.
Dnmt3b deficient (knock out, DNMT3B KO; open bars) and control BEAS-2B cells (black bars) were stimulated with flagellin at final contrition of 1 μg/ml (panels A and B) or heat-killed flagellin-deficient PAK (PAKflic, MOI = 50, panels C and D) for 12 hours. CXCL1, CXCL8 and CCL20 mRNA levels were measured by RT-qPCR (A, C) and corresponding protein levels in supernatants by ELISA (B, D). Data are expressed as the mean ± SEM (n = 4) and representative of two to three independent experiments and for two DNMT3B KO and control clones. *p < 0.05, **p < 0.01, *** p < 0.001.
Dnmt3b inhibits Pseudomonas aeruginosa induced NF-ĸB p65 binding to the CXCL1 promoter in bronchial epithelial cells in vitro
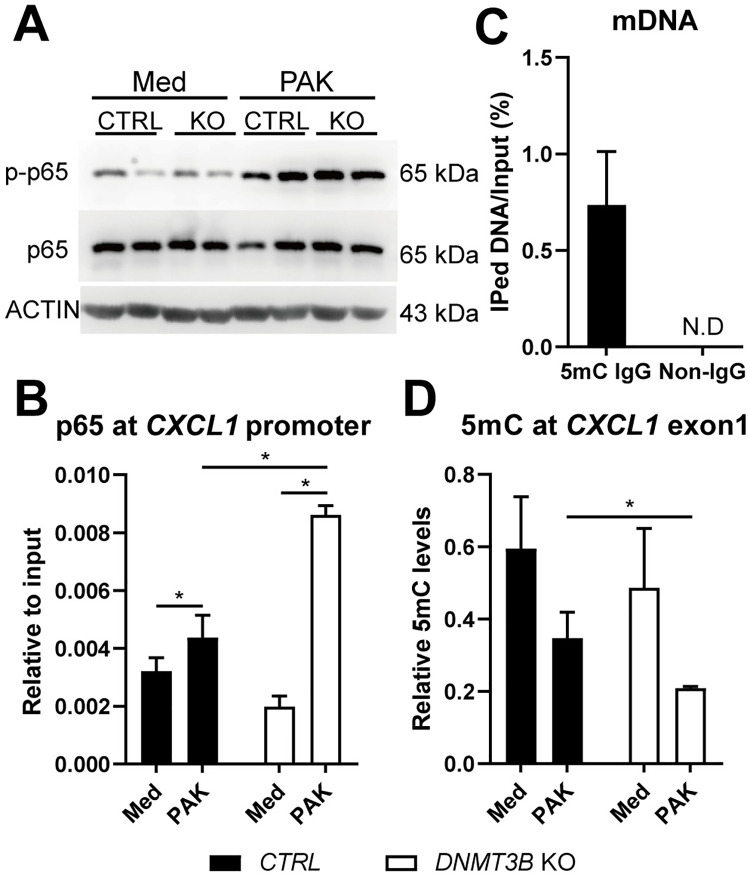
The expression of genes modified by Dnmt3b deficiency in the experiments described above are highly regulated by NF-κB [21]. PAK induced NF-κB activation in BEAS-2B cells as indicated by increased phosphorylation of the NF-κB subunit p65, but this was not altered by Dnmt3b deficiency (Fig 3A). This led us to hypothesize that Dnmt3b might affect chemokine production downstream of NF-κB signaling. To this end, we performed chromatin immunoprecipitation (ChIP) to measure the binding of NF-κB to the promoter region of CXCL1, which is a necessary step to elicit NF-κB target gene expression [22]. PAK induced the binding of NF-κB to the promoter region of CXCL1, and deletion of DNMT3B further potentiated the effect (Fig 3B). It is well documented that DNA methylation regulates gene expression by changing chromatin accessibility to transcriptional factors [23], and Dnmt3b promotes de novo DNA methylation [12]. DNA methylation of the p65 binding motif at the promoter of CXCL1 decreases CXCL1 expression by reducing p65 binding [24]. To evaluate whether Dnmt3b influenced p65 binding via regulating DNA methylation of the binding sites at the regulatory elements of CXCL1, we performed methylated DNA immunoprecipitation (MeDIP) using a 5-methylcytosine antibody (Fig 3C). While control methylated DNA was successfully precipitated in this experiment, we could not detect a signal at the CXCL1 promoter, suggesting low levels of DNA methylation in this region. Dnmt3b deficiency tended to reduce DNA methylation in exon 1 of CXCL1 in unstimulated BEAS-2B cells; PAK induced a decrease in DNA methylation in this region, which was further reduced in Dnmt3b deficient cells (Fig 3D).
Fig 3. Dnmt3b inhibits Pseudomonas aeruginosa induced NF-ĸB p65 binding to the CXCL1 promoter in BEAS-2B bronchial epithelial cells in vitro.
Dnmt3b deficient (knock out, DNMT3B KO) and control BEAS-2B cells (CTRL) were stimulated with heat killed PAK at MOI of 50 for 30 min, total protein was extracted from the activated cells for detection of RelA/NF kappa B (NF-κB) p65 and Phospho-p65 (S536) by western blot (A). DNMT3B deficient (knock out, DNMT3B KO; open bars) and control BEAS-2B cells (CTRL; black bars) were stimulated with heat killed PAK (MOI = 50) for 1 hour, NF-κB p65 binding to CXCL1 promoter was evaluated by ChIP (B). Control methylated DNA (mDNA) was used as control input for testing the efficiency of MeDIP and the specificity of 5-mC IgG, N.D = no signal detected (C). Dnmt3b deficient (knock out, DNMT3B KO) and control BEAS-2B cells (CTRL) were stimulated with heat killed PAK (MOI = 50) for 12 hours, MeDIP was performed to measure DNA methylation (5mC) levels at CXCL1 exon1 (D). Data are expressed as the mean ± SEM (n = 4) and representative of two independent experiments. *p < 0.05.
Bronchial epithelial deficiency of Dnmt3b promotes CXCL1 production by bronchial epithelial cells in the early phase during Pseudomonas pneumonia in vivo
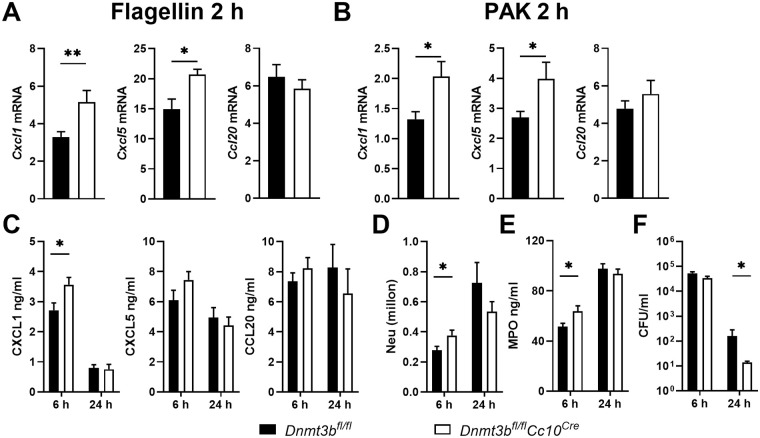
To investigate the role of Dnmt3b in bronchiolar epithelial cells during Pseudomonas pneumonia in vivo, we crossed mice in which the Dnmt3b gene is flanked by two lox-P sites (Dnmt3bfl/fl mice) with mice expressing Cre-recombinase under the control of the club cell 10 kD (CC10) promoter (Cc10Cre mice) to generate Dnmt3bfl/flCc10Cre mice. Our laboratory previously showed that Cre-recombinase is specifically active in bronchiolar epithelial cells in Cc10Cre mice [25]; thus, the cellular distribution of Dnmt3b deficiency in Dnmt3bfl/flCc10Cre mice corresponds with the in vitro studies using Dnmt3b deficient bronchial epithelial cells described above. We first sought to establish whether Dnmt3b influences flagellin induced chemokine expression in bronchial epithelial cells in vivo. Bronchial brushes highly expressed mRNAs encoding the epithelial cell marker CD326 and the bronchial epithelial cell marker CC10, while leukocytes collected by bronchoalveolar lavage (BAL) expressed high levels of mRNA encoding CD45 (S4 Fig), indicating a high purity of respiratory epithelial cells in the brushes. Dnmt3bfl/flCc10Cre mice, relative to Dnmt3bfl/fl Cre-negative littermate control mice, showed significantly enhanced Cxcl1 and Cxcl5 expression in bronchial brushes harvested 2 hours after flagellin treatment, while Ccl20 expression was unaffected (Fig 4A). To determine the impact of bronchial epithelial Dnmt3b on host defense during Pseudomonas pneumonia, Dnmt3bfl/flCc10Cre and Dnmt3bfl/fl control mice were infected with (wild-type) PAK via the airways and euthanized 2, 6 or 24 hours thereafter for analysis. In agreement with the results obtained after flagellin administration, bronchial brushes collected from Dnmt3bfl/flCc10Cre mice 2 hours after PAK infection expressed higher Cxcl1 and Cxcl5 mRNA levels compared to those from Dnmt3bfl/fl control mice, while Ccl20 mRNA were similar between groups (Fig 4B). PAK elicited high levels of CXCL1, CXCL5, and CCL20 in BAL fluid (BALF) at 6 hours post-infection, with lower concentrations at 24 hours (Fig 4C). Dnmt3bfl/flCc10Cre mice showed higher levels of CXCL1 when compared with littermate control mice at 6 hours after infection; this difference was not present anymore at 24 hours. CXCL5 levels tended to be higher in BALF of Dnmt3bfl/flCc10Cre mice relative to control mice at 6 hours post-infection (P = 0.07); BALF CCL20 concentrations were similar in Dnmt3bfl/flCc10Cre and control mice (Fig 4C).
Fig 4. Bronchial epithelial deficiency of Dnmt3b promotes CXCL1 production by bronchial epithelial cells at the early phase of Pseudomonas pneumonia in vivo.
Dnmt3bfl/flCc10Cre and control Dnmt3bfl/fl mice received flagellin (1 μg) purified from P. aeruginosa intranasally. Cxcl1, Cxcl5 and Ccl20 mRNA expression in bronchial brushes collected at 2 hours (A). Dnmt3bfl/flCc10Cre and control Dnmt3bfl/fl mice were infected with PAK (5 x 106 CFU) intranasally. Cxcl1, Cxcl5 and Ccl20 mRNA expression in bronchial brushes collected at 2 hours (B). CXCL1, CXCL5 and CCL20 (C), neutrophil counts (D), MPO (E) and bacterial counts (F) in BALF were determined in BALF harvested after 6 or 24 hours. Data are presented as means ± SEM of 8 mice per group at each time point. * p < 0.05.
Bronchial epithelial deficiency of Dnmt3b promotes early neutrophil recruitment and bacterial clearance during Pseudomonas pneumonia in vivo
Neutrophils are attracted to the airways by locally released CXC chemokines such as CXCL1 and play a key role in host defense during pneumonia [26]. In agreement with higher local CXCL1 concentrations, Dnmt3bfl/flCc10Cre mice had higher neutrophil numbers in their BALF when compared with control mice at 6 hours after infection with PAK (Fig 4D). Additionally, Dnmt3bfl/flCc10Cre mice had higher concentrations of the neutrophil degranulation product myeloperoxidase in BALF when compared with control mice (Fig 4E). Neutrophils are important for clearance of Pseudomonas from the airways [27,28]. In agreement with increased neutrophil recruitment to the site of infection, Dnmt3bfl/flCc10Cre mice demonstrated an accelerated bacterial clearance as reflected by lower bacterial burdens in BALF at 24 hours after infection when compared with control mice (Fig 4F). The extent of PAK induced lung pathology, as determined by semi-quantitative scoring of hematoxylin and eosin (H&E) stained lung slides, did not differ between Dnmt3bfl/flCc10Cre and control mice (S5A and S5B Fig). Likewise, bronchial Dnmt3b deficiency did not impact IL-1β and TNFα release in BALF during Pseudomonas pneumonia (S5C Fig).
The enhanced pulmonary response of mice with bronchial epithelial Dnmt3b deficiency to Pseudomonas depends on bacterial expression of flagellin
We and others have previously shown that activation of respiratory epithelial cells by flagellin drives protective innate immunity during Pseudomonas pneumonia through the induction of TLR5-MyD88 dependent signaling [7,8,9]. To determine a role for flagellin in Dnmt3b-mediated inhibition of the immune response in bronchiolar epithelial cells and bacterial clearance during Pseudomonas infection in vivo, we infected Dnmt3bfl/flCc10Cre and control mice with PAKflic via the airways. Unlike after infection with wild-type PAK in vivo (Fig 4) or stimulation with PAKfilc in vitro (Fig 2), Dnmt3bfl/flCc10Cre mice infected with PAKflic showed no differences with regard to local chemokine release, neutrophil recruitment or bacterial clearance (Fig 5). Bronchial epithelial Dnmt3b deficiency also did not influence local IL-1β or TNFα release, or the extent of lung pathology (S6 Fig). Taken together, these results suggest that bronchial epithelial Dnmt3b dampens mucosal immunity during Pseudomonas airway infection resulting in a reduced bacterial clearance in mice, which is dependent on Dnmt3b mediated modulation of respiratory epithelial responses induced by the Pseudomonas component flagellin.
Fig 5. Bronchial epithelial deficiency of Dnmt3b does not modify pulmonary responses during pneumonia caused by flagellin-deficient Pseudomonas.
Dnmt3bfl/flCc10Cre and control Dnmt3bfl/fl mice were infected with flagellin-deficient PAK (PAKflic, 5 x 106 CFU) intranasally. CXCL1, CXCL5 and CCL20 were determined in BALF harvested after 6 or 24 hours (A). Neutrophil counts (B), MPO (C) and bacterial counts (D) in BALF were determined. Data are presented as means ± SEM of 8 mice per group at each time point.
Bronchial epithelial Dnmt3b deficiency does not affect the host response during pneumonia caused by Klebsiella pneumoniae
To further examine the role of flagellin in Dnmt3b regulated functions during bacterial pneumonia we infected Dnmt3bfl/flCc10Cre and control mice with Klebsiella pneumoniae, an un-flagellated gram-negative bacterium. CXCL1 levels, neutrophil counts, MPO concentrations and bacterial burdens in BALF were not affected by Dnmt3b deficiency in bronchial epithelial cells after infection with Klebsiella (S7A, S7B, S7C and S7D Fig). Moreover, bacterial loads in extrapulmonary organs such liver and spleen, and in blood were comparable between groups (S7E Fig). These data provide further support for a role for flagellin in Dnmt3b mediated mucosal immune responses in the infected airways.
Type II alveolar epithelial cell Dnmt3b does not influence the pulmonary response during Pseudomonas pneumonia in vivo
Type II alveolar epithelial cells (AEC2) have been implicated in host defense during P. aeruginosa infection [29]. To determine whether the role of Dnmt3b in the host response during Pseudomonas pneumonia is restricted to bronchiolar epithelial cells, we crossed Dnmt3bfl/fl mice with mice expressing Cre-recombinase under the control of the surfactant protein C promoter (SpCcre mice) to generate AEC2 specific Dntm3b deficient (Dnmt3bfl/fl SpCcre) mice. We previously showed that in SpCcre mice Cre-recombinase is specifically active in AEC2 [25]. In contrast to Dnmt3bfl/flCc10Cre mice, Dnmt3bfl/flSpCcre mice did not differ from littermate control mice with regard to local release of CXCL1 (Fig 6A), neutrophil recruitment (Fig 6B), BALF MPO levels (Fig 6C) or bacterial clearance (Fig 6D) after infection with wild-type PAK via the airways. Furthermore, Dnmt3bfl/flSpCcre and control mice showed similar CXCL5, CCL20 (Fig 6A), IL-1β and TNF-α BALF levels upon infection with PAK (S8 Fig). These data suggest that Dnmt3b expressed in AEC2 does not influence innate immune mechanisms during Pseudomonas pneumonia.
Fig 6. Type II alveolar epithelial cell Dnmt3b has no role in the pulmonary response during Pseudomonas pneumonia in vivo.
Dnmt3bfl/flSpCCre and control Dnmt3bfl/fl mice were infected with PAK (5 x 106 CFU) intranasally. CXCL1, CXCL5 and CCL20 were determined in BALF harvested after 6 or 24 hours (A). Neutrophil counts (B), MPO (C) and bacterial counts (D) in BALF were determined. Data are presented as means ± SEM of 8 mice per group at each time point.
Discussion
Here we report that deficiency of DNMT3B, the gene encoding a key enzyme involved in de novo DNA methylation, in human bronchial epithelial cells in vitro results in increased production of chemokines implicated in mucosal immunity, including CXCL1, in response to the common human pathogen P. aeruginosa. This role of Dnmt3b in epithelial CXCL1 production was reproduced in vivo, using mice with bronchial epithelial cell specific deletion of Dnmt3b infected with P. aeruginosa via the airways. In these mice, increased CXCL1 release into the airways was associated with enhanced neutrophil recruitment and an accelerated bacterial clearance, which depended on expression of flagellin by Pseudomonas. Mechanistically, DNMT3B deficiency was shown to decrease DNA methylation levels at NF-κB binding regions in the promoter region of CXCL1, allowing increased NF-κB binding and transcription of NF-kB response genes upon exposure to Pseudomonas.
Epithelial cells are the first line of defense in lung innate immunity during pulmonary infection [4]. One of their key functions is to produce chemokines that recruit innate immune cells such as neutrophils to control the infection. Previous studies documented that the Dnmt inhibitor 5′-azacytidine increased CCL20 expression in human gingival epithelial cells in response to Fusobacterium nucleatum [30], as well as Cxcl1 expression in mouse MLE-12 respiratory epithelial cells stimulated with IL-17 [31]. Our current finding that the Dnmt inhibitor RG108 enhanced CXCL1, CXCL8 and CCL20 expression in BEAS-2B cells stimulated with P. aeruginosa further supports a role for Dnmt’s in the induction of chemokines by respiratory epithelial cells. The effect of RG108 could be reproduced by elimination of DNMT3B in BEAS-2B cells, and Dnmt3b deficient BEAS-2B cells also produced more CXCL1, CXCL8 and CCL20 upon stimulation with the Pseudomonas component flagellin. The expression of chemokines is partly regulated by NF-κB and binding of the NF-κB subunit p65 to chemokine promoter regions drives their gene transcription [32]. The ability of NF-κB to bind DNA and to trigger inflammatory gene transcription is determined at least in part by the extent of DNA methylation [33]. DNMT3B was downregulated after activation of BEAS-2B cells by PAK, which coincided with a decrease in methylation at exon 1 of CXCL1. Deletion of DNMT3B in BEAS-2B cells further decreased methylation at exon 1 of CXCL1 while enhancing p65 binding to the CXCL1 promoter, providing insight into the mechanism by which Dnmt3b regulates the expression of CXCL1. Noteworthy, DNA methylation can affect other types of epigenetic regulation, such as that mediated by Polycomb-Repressive Complexes (PRCs) [34]. Although PRCs are particularly associated with development, differentiation, and stem cell renewal [35], some studies suggested that PRCs regulate the expression of chemokines, such as CXCL9 and CXCL10 [36]. Therefore, we cannot rule out the possible involvement of PRCs in the regulation of CXCL1 expression in Dnmt3b deficient cells. Additionally, Dnmt3a and Dnmt1 are involved in de novo DNA methylation and maintaining methylation, respectively [23]. Dnmt3b has been reported to work synergistically with Dnmt3a, and their combined effects on de novo methylation have been demonstrated during the epigenetic regulation of hematopoietic stem cell fate decisions [37]. Therefore, further investigations are needed to determine whether complementary mechanisms mediated by Dnmt3a occur in Dnmt3b deficient cells during bacterial infection. The existence of such complementary mechanisms could explain the fact that Dnmt3 deficiency only affected the early response to Pseudomonas infection of the airways. Other factors might also be involved in the transient effect of Dntm3b deficiency, including later induction of other host response mediators by Pseudomonas that overrule the effect of Dnmt3b deficiency and the use of a “single-injury” model with clearance of bacteria (i.e., possibly, repeated injuries might reveal a more sustained role for Dnmt3b).
The role of bronchial epithelial Dnmt3b in limiting Pseudomonas induced CXCL1 transcription in human epithelial cells in vitro was confirmed in Dnmt3bfl/flCc10Cre mice in vivo, in which enhanced CXCL1 mRNA levels in bronchial brushes (2 hours) associated with elevated CXCL1 levels in BALF early (6 hours) after infection. In Dnmt3bfl/flCc10Cre mice Cre-mediated deletion of Dnmt3b is restricted to CC10+ cells, which are the major cell type located in mouse upper airways, trachea and bronchial regions [38]. We showed that bronchial brushes harvested from mice are highly enriched for CC10+ cells, confirming previous reports [39,40]. CC10+ cells are an important source of chemokines [40], which can explain why increased Cxcl1 transcription in bronchial brush cells can result in increased CXCL1 protein levels in BALF. Notably, although bronchial CXCL5 mRNA levels were higher in Dnmt3bfl/flCc10Cre than in control mice infected with PAK, CXCL5 protein levels in BALF were not significantly increased. Moreover, at 24 hours after infection, CXCL1 and CXCL5 concentrations in BALF were similar in both mouse strains. In addition to epithelial cells, these chemokines can be produced by myeloid cells, which likely can obscure possible differences in chemokine release by epithelial cells at later stages during the infection, i.e., after influx of neutrophils. Nonetheless, the enhanced CXCL1 release early after infection was accompanied by a faster influx of neutrophils into the alveolar compartment, which is a likely explanation for a subsequent accelerated bacterial clearance [27,28]. Indeed, neutrophils are among the first cells recruited to infectious sites and are crucial for controlling P. aeruginosa infection through various mechanisms including degranulation, phagocytosis, and the generation of neutrophil extracellular traps [41].
We used BEAS-2B cells to study the function of Dnmt3b in human respiratory epithelial cells. While this cell line is frequently used for mechanistic studies considering its convenience for genetic modification, it is worth noting that BEAS-2B cells have several limitations including their inability to form barriers [42] and to produce mucus [43]. Primary respiratory epithelial cells are not easily accessible for genetic manipulation. We here do provide evidence that the expression of DNMT3B (Dnmt3b) is regulated in the same way (reduced) in human and mouse primary epithelial cells, grown in air-liquid interface, as in BEAS-2B cells upon exposure to Pseudomonas. The data obtained with Dnmt3b deficient BEAS-2B cells and Dnmt3bfl/flCc10Cre mice differed in some aspects. First, expression of CCL20 induced by PAK was affected by Dnmt3b deficiency in BEAS-2B cells but not in bronchial brushes from Dnmt3bfl/flCc10Cre mice, which could be due to differences relating to species (human versus mouse), cell type (cell line versus primary cells) and/or stimulation (heat-killed PAK stably present in cell culture medium versus a gradually decreasing burden of viable bacteria). Importantly, whereas flagellin was not required for the role of Dnmt3b in Pseudomonas induced CXCL1 expression in BEAS-2B cells in vitro, Dnmt3bfl/flCc10Cre mice only demonstrated an enhanced innate immune response after infection with WT PAK but not after infection with flagellin deficient PAKflic. While flagellin activates TLR5, intact P. aeruginosa can also activate TLR2, 4 and 9 [44]; these TLRs are expressed by BEAS-2B cells [45] and activation of different TLRs induces a common set of genes including CXCL1 downstream of NF-κB signaling [46,47]. In BEAS-2B cell cultures, cells were continuously exposed to PAK for 10 hours, likely allowing stimulation of multiple TLRs, which may compensate for the lack of flagellin-TLR5 signaling upon exposure to PAKflic. In mice, however, the recognition of flagellin by TLR5 is required for airway epithelial cells to sense P. aeruginosa and to clear the infection [9,48,49,50,51], which may explain the prominent role of flagellin in the effect of bronchial epithelial Dnmt3b in regulating the mucosal immune response during Pseudomonas infection in vivo. In line with this, we found that the number of PAKflic CFU was higher than that of PAK at 6 hours after infection, which might explain higher IL-1β and TNFα in BALF of PAKflic infected mice as compared to mice infected with PAK. Bronchial epithelial Dnmt3b played no role in CXCL1 release, neutrophil recruitment or anti-bacterial defense during K. pneumoniae infection, which–considering that this gram-negative pathogen does not express flagellin–further supports a role for flagellin in Dnmt3b mediated functions in the respiratory epithelium. An additional explanation for dissimilarities between BEAS-2B cells in vitro and mice in vivo could be species differences; in this context studies comparing the responsiveness of mouse respiratory epithelial cells to PAK and PAKflic would be of interest.
Dnmt3b ablation in mouse AEC2 did not impact CXCL1 production, neutrophil influx or bacterial clearance after infection with Pseudomonas. The differential roles of Dnmt3b in bronchial epithelial (Cc10) cells and AEC2 (SPC cells) could have several mutually non-exclusive explanations. First, AEC2 likely are less important for the production of chemoattractant mediators like CXCL1, but rather are crucial for the synthesis of surfactant lipids and proteins required for the reduction of surface tension in order to prevent collapse of the lungs [4]. Thus, AEC2 produced CXCL1 likely accounts for only a minor part of CXCL1 released in BALF and a possible effect of Dnmt3b on CXCL1 production by AEC2 might not impact overall CXCL1 production. Second, Dnmt3b might function differently in these two epithelial cell types. Notably, whilst the cell-specific expression of the Cc10 (bronchial) and Spc (AEC2) promoters has been documented by several laboratories including ours [25], a small population of epithelial cells named bronchioalveolar stem cells, located at the bronchioalveolar-duct junctions, express both Cc10 and Spc [52,53]. Our data do not exclude a role for Dnmt3b in this cell population during Pseudomonas pneumonia.
Our data do not provide insight into the potential importance of Dnmt3b as a therapeutic target during Pseudomonas pneumonia. Translation of the current respiratory epithelial cell-specific results on Dnmt3b function to a possible therapeutic targeting this enzyme requires additional investigations. This also holds true for further studies on the use of flagellin, administered via the airways as a possible immune enhancing strategy in the treatment of respiratory tract infections, and the role of Dnmt3b herein [6].
Changes in DNA methylation in respiratory epithelia have been implicated in several inflammatory lung diseases, including asthma, chronic obstructive pulmonary disease, cystic fibrosis and idiopathic pulmonary fibrosis [54,55,56]. Knowledge of a potential role of epithelial DNA methylation modifications in the host response to respiratory pathogens is highly limited. This study provides evidence for a role of Dnmt3b, an enzyme mediating de novo DNA methylation, in bronchial epithelial cells in regulating the early innate immune response during airway infection caused by P. aeruginosa.
Methods
Ethics statement
All mouse experiments were approved by the Institutional Animal Care and Use Committee of the University of Amsterdam.
Mice
Homozygous Dnmt3bfl/lf mice (RBRC03733, RIKEN BRC, Tsukuba, Japan) [57] were crossed with mice expressing cre recombinase under the control of the club cell 10-kD promoter (Cc10Cre mice) [25] to generate bronchiolar epithelial–specific Dnmt3b-deficient (Dnmt3bfl/flCc10Cre) mice or with mice expressing cre recombinase under the control of the surfactant protein C promoter (SpCCre mice) [9] to generate type II alveolar epithelial–specific Dnmt3b-deficient (Dnmt3bfl/flSpCCre) mice. Dnmt3bfl/fl Cre–negative littermates were used as controls in all experiments. All genetically modified mice were backcrossed at least eight times to a C57Bl/6 background and age and sex matched when used in experiments. Mice were used at 8–12 weeks of age.
Bacterial strains and culture conditions
Wild-type P. aeruginosa PAK, flagellin deficient P. aeruginosa PAKflic [58] and Klebsiella pneumoniae serotype 2 (American Type Culture Collection no. 43816) were cultured as described previously [9,59]. Briefly, PAK and PAKflic were grown to mid-logarithmic phase in Luria broth at 37°C with shaking and Klebsiella pneumoniae was cultured to mid-logarithmic phase in Tryptic Soy Broth. Bacteria were then harvested by centrifugation at 3,000 rpm for 10 minutes. After washing twice with pyrogen-free 0.9% NaCl, bacteria were suspended in 10 ml of 0.9% NaCl, the number of bacteria was determined by serial dilution in sterile isotonic saline and culture on blood agar plates. Bacteria were diluted to 108 colony-forming units (CFU)/ml for later use.
Cells
The human bronchial epithelial cell line BEAS-2B [60] was obtained from the American Type Culture Collection (ATCC, Rockville, MD). Cells were cultured in DMEM (InvivoGen, San Diego, CA) supplemented with 10% FBS and 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, San Diego, CA) and kept at 37°C in an incubator with a humidified atmosphere containing 5% CO2. BEAS-2B cells were stimulated with heat-killed (65 °C for 15 minutes) wild-type P. aeruginosa (PAK), isogenic flagellin deficient P. aeruginosa (PAKflic) (both at multiplicity of infection (MOI) = 50) or flagellin purified from P. aeruginosa (tlrl-pafla, Invivogen, San Diego, CA; 1 μg/ml) for 12 hours. Before stimulation cells were plated in 24-well plates overnight. A lentivirus mediated CRISPR/Cas9 system was used to generate DNMT3B knockout cells as described [61]. Briefly, the single guide (sg)RNA targeting sequences were designed using an online gRNA design tool (Crispr.mit.edu). The sgRNA sequence 5-ATCCGCACCCCGGAGATCAG-3 was chosen to target DNMT3B and cloned into lentiCRISPR v2 (Addgene #52961, http://n2t.net/addgene:52961; RRID:Addgene_52961) [62]. Control cells were generated using the same method with a non-targeting sgRNA sequence ACGGAGGCTAAGCGTCGCAA. Two independent Dnmt3b deficient or control cell lines were selected for further experiments. Overexpression of Dnmt3b in BEAS-2B cells was performed using human DNMT3B ORF clone lentiviral particle (Vigene Biosciences, Rockville, MD) according to manufacturer’s instruction. Clones with confirmed Dnmt3b overexpression (by Western blot: see below) were selected for experiments; control clones were generated by transducing a lentiviral particle carrying the same vector skeleton used for Dnmt3b overexpression. For some experiments, cells were pretreated with the Dnmt inhibitor RG108 at 10 μM/ml [16] (Sigma, Zwijndrecht, Nederland) or vehicle DMSO for 12 hours, and then stimulated with heat killed bacteria at MOI = 50. The effect of RG108 treatment on cell viability was determined using flow cytometry after straining of cells with fixable viability dye eFluor 780 (Invitrogen, Carlsbad, CA).
Western blot
Total protein was extracted from BEAS-2B cells, separated by 10% SDS gel electrophoresis and transferred to a PVDF membrane (Millipore, Billerica, MA). Membranes were blocked for 1 hour in 5% milk in tris-buffered saline with 0.1% Tween 20 (TBST) buffer (TBST) and incubated overnight with (primary) antibodies against phospho-NF-κB p65 (Ser536) (1: 1000, #3033; Cell Signaling Technology, Leiden, The Netherlands), NF-κB p65 (1: 1000, #3034N; Cell Signaling Technology, Leiden, The Netherlands), Dnmt3b (1:250, ab2851; Abcam, Cambridge, UK) or beta-actin (1: 1000, 4967L; Cell Signaling Technology) at 4°C. After incubation with horseradish peroxidase (HRP)-conjugated secondary antibody against rabbit IgG (1: 2000, #7074; Cell Signaling Technology) for 1 hour at room temperature, blots were imaged using Lumilight plus ECL substrate (Roche, Almere, The Netherlands) on an ImageQuant LAS 4000 biomolecular imager (GE Healthcare, Buckinghamshire, UK). For quantification, densitometry was performed with ImageJ (National Institutes of Health, Bethesda, MD; https://imagej.nih.gov/ij/) using the histogram function in a selected area of mean gray value for each band.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA from BEAS-2B cells, mouse bronchial brushes and BALF cells was isolated with NucleoSpin columns (Bioke, Leiden, The Netherlands) according the manufacturer’s recommendations. All RNA samples were quantified by spectrophotometry and stored at -80°C until further analysis. cDNA was prepared using AMV Reverse Transcriptase (Promega, Leiden, The Netherlands) according to manufacturer’s instructions. Gene expression analysis was performed using a Roche LightCycler 480 thermocycler with SensiFAST Real-time PCR kit (#CSA-01190; Bioline, London, UK) using the gene specific primers listed in S1 Table. For qPCR of bronchial brush samples, primers specific for Epcam (encoding the epithelial cell marker CD326), Ptprc (hematopoietic cell marker CD45) and Pecam1 (endothelial cell marker CD31) were used to quantify the enrichment of epithelial cells in the brushes; primers for Scgb1a1 (cube cell marker CC10) were used to evaluate the relative enrichment of bronchial epithelial cells in the brushes. Data was analyzed with LinRegPCR based on PCR efficiency values derived from amplification curves [63]. All results were normalized to Hprt expression levels.
Enzyme-linked immunosorbent assay (ELISA)
Human chemokine (C-X-C motif) ligand (CXCL)1, CXCL8, and chemokine (C-C motif) ligand (CCL) 20, as well as murine CXCL1, CXCL5, CCL20, tumor necrosis factor (TNF)-α, IL-1β and myeloperoxidase (MPO) were measured by species specific commercially available ELISA’s (R&D Systems, Minneapolis, MN) according to manufacturer’s description (protocols can be found at www.rndsystems.com).
Methylated DNA immunoprecipitation (MeDIP)
MeDIP analysis was performed using Methylamp Methylated DNA Capture Kit (Epigentek, Farmingdale, NY) following the manufacturer’s instructions. BEAS-2B cells were stimulated with heat killed PAK for 12 hours. Cells were collected for DNA purification using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Prior to immunoprecipitation, genomic DNA was sonicated with a Diagnode BioRuptor to obtain DNA fragments ranging in size from 200 to 1000 bp, with a mean fragment size of around 300 bp. Methylated DNA was captured using Methylamp Methylated DNA Capture Kit. In total 100 ng of fragmented DNA was applied in every antibody-coated well and incubated at room temperature on a horizontal shaker for 2 hours. The immunoprecipitated DNA was released by proteinase K. The DNA was eluted and adjusted to a final volume of 100 μl with nuclease-free water. For each sample, an input vial was included using total sonicated DNA as loading control. Total DNA and immunoprecipitated DNA were used to perform qPCR with CXCL1 promoter primer pairs that included a p65 binding site (S2 Table).
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed using ChIP-IT Express kit (Active Motif, Carlsbad, CA) following the manufacturer’s instructions. BEAS-2B cells were stimulated with heat killed PAK for 60 minutes and subsequently were cross-linked with 1% paraformaldehyde for 10 minutes at room temperature. Extracted chromatin was sheared by sonication using a Diagnode BioRuptor (10 pulses of 20 seconds each, with a 30 second rest between each pulse) into fragments of 200–800 bp length and immunoprecipitated using 3 μg NF-κB p65 antibody (Sigma-Aldrich, St. Louis, MO) at 4°C overnight. The samples were reverse cross-linked, and then proteins were digested with proteinase K and participated DNA was purified by NucleoSpin Gel and PCR Clean-up kit (Bioke, Leiden, The Netherlands) after reversal of cross-linking. qPCR amplification using 3 μl of total DNA and immunoprecipitated DNA was performed with the same primer pairs used in the MeDIP experiment (S2 Table).
Induction of pneumonia and sampling of organs
To induce pneumonia, mice were inoculated with viable PAK, PAKflic (5 × 106 CFU) or K. pneumoniae (1 × 104 CFU) or flagellin purified from P. aeruginosa (1 μg, tlrl-pafla, Invivogen) intranasally. At predefined time points mice were euthanized by heart puncture after injection of ketamine/medotomidine as described [59,64]. Briefly, the right lung was used for bronchoalveolar lavage (BAL) by instilling 2 × 0.5 ml of sterile phosphate-buffered saline; the left lung was preserved for histopathology after fixation in 10% formalin. Bronchial brushing was performed after BAL to collect bronchial epithelial cells as described [40]; BALF was serially diluted and plated on blood agar plates for measurements of bacterial numbers. Cell counts in BALF were determined using a hemocytometer (Beckman Coulter, Fullerton, CA). BALF supernatants and bronchial brushes were stored at −20°C until further analysis. In some experiments BALF was pelleted and cells were lysed for RNA isolation. For all animal experiments, littermate controls and conditional knockout were mixed in cages (i.e. randomized) and procedures were done without knowledge of the genotype (blinded). Readout parameters were thereby determined in a double-blinded manner.
Flow cytometry
Neutrophils in BALF were determined by flow cytometry as described [65]. Briefly, BALF cells were resuspended FACS buffer (5% BSA, 0.35 mM EDTA, 0.01% NaN3) and stained with fixable viability dye eFluor 780, rat anti mouse-CD45 PE-eFluor610 (30-F11), rat anti-mouse CD11b PE-Cy7 (clone M1/70), rat anti-mouse Siglec-F Alexa Fluor 647 (clone E50-2440), rat anti-mouse Ly-6C Alexa Fluor 700 (clone AL-21) (all from BD Biosciences) and rat anti-mouse Ly-6G FITC (clone 1A8; Biolegend, San Diego, CA) before loading on FACS Calibur (Becton Dickinson, Franklin Lakes, NJ). Data were analyzed using FlowJo software (Becton Dickinson). Neutrophils were identified as CD45+/Siglec-F-/CD11b+/Ly6C+/Ly6G+ cells. Examples of the gating strategy for BALF neutrophils are depicted in S9 Fig.
Pathology scores
The left lung lobe was fixed in 10% formaldehyde solution and embedded into paraffin blocks. Sections were stained with Hematoxylin and eosin (H&E). Slides were coded and scored from 0 (absent) to 4 (severe) for the following parameters: interstitial inflammation, endothelialitis, bronchitis, edema, thrombi, pleuritis, and percentage of the lung surface demonstrating confluent (diffuse) inflammatory infiltrate by a pathologist blinded for groups. The total “lung inflammation score” was expressed as the sum of the scores for each parameter [66].
Statistical analysis
All statistical analyses were performed using GraphPad Prism 8 software (GraphPad software, San Diego, CA). Significance was evaluated using two tailed unpaired t tests or non-parametric Mann-Whitney U tests where appropriate. Mouse experiments were done with 8 mice per group at each time point. Using a group size of 8 animals with a standard deviation of 35%, we are able to show a difference of >50% between two groups with a power of 80%. Results with a P-value of less than 0.05 were considered significant. ns: not significant.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
(DOCX)
Acknowledgments
We acknowledge M.S. ten Brink for helping with the animal experiments.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
WQ is supported by the State Scholarship Fund from China Scholarship Council (CSC #201606170115). XB is supported by a grant from the Netherlands Organization for Health Research and Development (ZonMW #50-53000-98-139. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis. 2010; 51 Suppl 1:S81–7. 10.1086/653053 [DOI] [PubMed] [Google Scholar]
- 2.Fujitani S, Sun HY, Yu VL, Weingarten JA. Pneumonia due to Pseudomonas aeruginosa: part I: epidemiology, clinical diagnosis, and source. Chest. 2011; 139:909–19. 10.1378/chest.10-0166 [DOI] [PubMed] [Google Scholar]
- 3.Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, et al. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin Microbiol Rev. 2019; 32. 10.1128/CMR.00031-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol. 2015; 16:27–35. 10.1038/ni.3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riquelme SA, Ahn D, Prince A. Pseudomonas aeruginosa and Klebsiella pneumoniae Adaptation to Innate Immune Clearance Mechanisms in the Lung. J Innate Immun. 2018; 10:442–54. 10.1159/000487515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijayan A, Rumbo M, Carnoy C, Sirard JC. Compartmentalized Antimicrobial Defenses in Response to Flagellin. Trends Microbiol. 2018; 26:423–35. 10.1016/j.tim.2017.10.008 [DOI] [PubMed] [Google Scholar]
- 7.Feuillet V, Medjane S, Mondor I, Demaria O, Pagni PP, Galán JE, et al. Involvement of Toll-like receptor 5 in the recognition of flagellated bacteria. Proc Natl Acad Sci U S A. 2006; 103:12487–92. 10.1073/pnas.0605200103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mijares LA, Wangdi T, Sokol C, Homer R, Medzhitov R, Kazmierczak BI. Airway epithelial MyD88 restores control of Pseudomonas aeruginosa murine infection via an IL-1-dependent pathway. J Immunol. 2011; 186:7080–8. 10.4049/jimmunol.1003687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anas AA, van Lieshout MH, Claushuis TA, de Vos AF, Florquin S, de Boer OJ, et al. Lung epithelial MyD88 drives early pulmonary clearance of Pseudomonas aeruginosa by a flagellin dependent mechanism. Am J Physiol Lung Cell Mol Physiol. 2016; 311:L219–28. 10.1152/ajplung.00078.2016 [DOI] [PubMed] [Google Scholar]
- 10.Abiko Y U O, Fukumoto S O T. Epigenetics of oral infection and inflammatory diseases—DNA methylation changes in infections and inflammation diseases. J Oral Biosci. 2014: 105–9. [Google Scholar]
- 11.Pérez-Novo CA, Bachert C. DNA methylation, bacteria and airway inflammation: latest insights. Curr Opin Allergy Clin Immunol. 2015; 15:27–32. 10.1097/ACI.0000000000000130 [DOI] [PubMed] [Google Scholar]
- 12.Gagliardi M, Strazzullo M, Matarazzo MR. DNMT3B Functions: Novel Insights From Human Disease. Front Cell Dev Biol. 2018; 6:140. 10.3389/fcell.2018.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, Killian JK, Yang M, Walker RL, Hong JA, Zhang M, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010; 29:3650–64. 10.1038/onc.2010.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teneng I, Tellez CS, Picchi MA, Klinge DM, Yingling CM, Snider AM, et al. Global identification of genes targeted by DNMT3b for epigenetic silencing in lung cancer. Oncogene. 2015; 34:621–30. 10.1038/onc.2013.580 [DOI] [PubMed] [Google Scholar]
- 15.Asgari S, McLaren PJ, Peake J, Wong M, Wong R, Bartha I, et al. Exome Sequencing Reveals Primary Immunodeficiencies in Children with Community-Acquired Pseudomonas aeruginosa Sepsis. Front Immunol. 2016; 7:357. 10.3389/fimmu.2016.00357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rondelet G, Fleury L, Faux C, Masson V, Dubois J, Arimondo PB, et al. Inhibition studies of DNA methyltransferases by maleimide derivatives of RG108 as non-nucleoside inhibitors. Future Med Chem. 2017; 9:1465–81. 10.4155/fmc-2017-0074 [DOI] [PubMed] [Google Scholar]
- 17.Hampton TH, Ballok AE, Bomberger JM, Rutkowski MR, Barnaby R, Coutermarsh B, et al. Does the F508-CFTR mutation induce a proinflammatory response in human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2012; 303:L509–18. 10.1152/ajplung.00226.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramirez-Moral I, Yu X, Butler JM, van Weeghel M, Otto NA, Ferreira BL, et al. mTOR-driven glycolysis governs induction of innate immune responses by bronchial epithelial cells exposed to the bacterial component flagellin. Mucosal Immunol. 2021. 10.1038/s41385-021-00377-8 [DOI] [PubMed] [Google Scholar]
- 19.Kassim SY, Gharib SA, Mecham BH, Birkland TP, Parks WC, McGuire JK. Individual matrix metalloproteinases control distinct transcriptional responses in airway epithelial cells infected with Pseudomonas aeruginosa. Infect Immun. 2007; 75:5640–50. 10.1128/IAI.00799-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sierro F, Dubois B, Coste A, Kaiserlian D, Kraehenbuhl JP, Sirard JC. Flagellin stimulation of intestinal epithelial cells triggers CCL20-mediated migration of dendritic cells. Proc Natl Acad Sci U S A. 2001; 98:13722–7. 10.1073/pnas.241308598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017; 2:e17023. 10.1038/sigtrans.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Dea E, Hoffmann A. The regulatory logic of the NF-kappaB signaling system. Cold Spring Harb Perspect Biol. 2010; 2:a000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schübeler D. Function and information content of DNA methylation. Nature. 2015; 517:321–6. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- 24.Atsumi T, Suzuki H, Jiang JJ, Okuyama Y, Nakagawa I, Ota M, et al. Rbm10 regulates inflammation development via alternative splicing of Dnmt3b. Int Immunol. 2017; 29:581–91. 10.1093/intimm/dxx067 [DOI] [PubMed] [Google Scholar]
- 25.Anas AA, Claushuis T, Mohan RA, Christoffels VM, Aidinis V, Florquin S, et al. Epithelial Myeloid-Differentiation Factor 88 Is Dispensable during Klebsiella Pneumonia. Am J Respir Cell Mol Biol. 2017; 56:648–56. 10.1165/rcmb.2016-0190OC [DOI] [PubMed] [Google Scholar]
- 26.Lovewell RR, Patankar YR, Berwin B. Mechanisms of phagocytosis and host clearance of Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol. 2014; 306:L591–603. 10.1152/ajplung.00335.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen SM, Cheng DS, Williams BJ, Sherrill TP, Han W, Chont M, et al. The nuclear factor kappa-B pathway in airway epithelium regulates neutrophil recruitment and host defence following Pseudomonas aeruginosa infection. Clin Exp Immunol. 2008; 153:420–8. 10.1111/j.1365-2249.2008.03707.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun L, Guo RF, Newstead MW, Standiford TJ, Macariola DR, Shanley TP. Effect of IL-10 on neutrophil recruitment and survival after Pseudomonas aeruginosa challenge. Am J Respir Cell Mol Biol. 2009; 41:76–84. 10.1165/rcmb.2008-0202OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannan S, Huang H, Seeger D, Audet A, Chen Y, Huang C, et al. Alveolar epithelial type II cells activate alveolar macrophages and mitigate P. Aeruginosa infection. PLoS One. 2009; 4:e4891. 10.1371/journal.pone.0004891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin L, Chung WO. Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol. 2011; 4:409–19. 10.1038/mi.2010.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J, An X, Yao Y, Erb C, Ferguson A, Kolls JK, et al. Epigenetic Regulation of IL-17-Induced Chemokines in Lung Epithelial Cells. Mediators Inflamm. 2019; 2019:9050965. 10.1155/2019/9050965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackwell TS, Christman JW. The role of nuclear factor-kappa B in cytokine gene regulation. Am J Respir Cell Mol Biol. 1997; 17:3–9. 10.1165/ajrcmb.17.1.f132 [DOI] [PubMed] [Google Scholar]
- 33.Campbell KJ, Perkins ND. Regulation of NF-kappaB function. Biochem Soc Symp. 2006: 165–80. 10.1042/bss0730165 [DOI] [PubMed] [Google Scholar]
- 34.Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013; 14:R25. 10.1186/gb-2013-14-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006; 20:1123–36. 10.1101/gad.381706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagarsheth N, Peng D, Kryczek I, Wu K, Li W, Zhao E, et al. PRC2 Epigenetically Silences Th1-Type Chemokines to Suppress Effector T-Cell Trafficking in Colon Cancer. Cancer Res. 2016; 76:275–82. 10.1158/0008-5472.CAN-15-1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014; 15:350–64. 10.1016/j.stem.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasaki A, Foxman EF, Molony RD. Early local immune defences in the respiratory tract. Nat Rev Immunol. 2017; 17:7–20. 10.1038/nri.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin W, Brands X, van ’t Veer C, de Vos AF, Scicluna BP, van der Poll T. Bronchial Epithelial Tet2 Maintains Epithelial Integrity during Acute Pseudomonas aeruginosa Pneumonia. Infect Immun. 2020; 89. 10.1128/IAI.00603-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen K, Eddens T, Trevejo-Nunez G, Way EE, Elsegeiny W, Ricks DM, et al. IL-17 Receptor Signaling in the Lung Epithelium Is Required for Mucosal Chemokine Gradients and Pulmonary Host Defense against K. pneumoniae. Cell Host Microbe. 2016; 20:596–605. 10.1016/j.chom.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thanabalasuriar A, Scott B, Peiseler M, Willson ME, Zeng Z, Warrener P, et al. Neutrophil Extracellular Traps Confine Pseudomonas aeruginosa Ocular Biofilms and Restrict Brain Invasion. Cell Host Microbe. 2019; 25:526–36.e4. 10.1016/j.chom.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Godfrey RW. Human airway epithelial tight junctions. Microsc Res Tech. 1997; 38:488–99. [DOI] [PubMed] [Google Scholar]
- 43.Veranth JM, Kaser EG, Veranth MM, Koch M, Yost GS. Cytokine responses of human lung cells (BEAS-2B) treated with micron-sized and nanoparticles of metal oxides compared to soil dusts. Part Fibre Toxicol. 2007; 4:2. 10.1186/1743-8977-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McIsaac SM, Stadnyk AW, Lin TJ. Toll-like receptors in the host defense against Pseudomonas aeruginosa respiratory infection and cystic fibrosis. J Leukoc Biol. 2012; 92:977–85. 10.1189/jlb.0811410 [DOI] [PubMed] [Google Scholar]
- 45.Sha Q, Truong-Tran AQ, Plitt JR, Beck LA, Schleimer RP. Activation of airway epithelial cells by toll-like receptor agonists. Am J Respir Cell Mol Biol. 2004; 31:358–64. 10.1165/rcmb.2003-0388OC [DOI] [PubMed] [Google Scholar]
- 46.Mayer AK, Muehmer M, Mages J, Gueinzius K, Hess C, Heeg K, et al. Differential recognition of TLR-dependent microbial ligands in human bronchial epithelial cells. J Immunol. 2007; 178:3134–42. 10.4049/jimmunol.178.5.3134 [DOI] [PubMed] [Google Scholar]
- 47.Borghini L, Lu J, Hibberd M, Davila S. Variation in Genome-Wide NF-κB RELA Binding Sites upon Microbial Stimuli and Identification of a Virus Response Profile. J Immunol. 2018; 201:1295–305. 10.4049/jimmunol.1800246 [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z, Louboutin JP, Weiner DJ, Goldberg JB, Wilson JM. Human airway epithelial cells sense Pseudomonas aeruginosa infection via recognition of flagellin by Toll-like receptor 5. Infect Immun. 2005; 73:7151–60. 10.1128/IAI.73.11.7151-7160.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skerrett SJ, Wilson CB, Liggitt HD, Hajjar AM. Redundant Toll-like receptor signaling in the pulmonary host response to Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol. 2007; 292:L312–22. 10.1152/ajplung.00250.2006 [DOI] [PubMed] [Google Scholar]
- 50.Morris AE, Liggitt HD, Hawn TR, Skerrett SJ. Role of Toll-like receptor 5 in the innate immune response to acute P. aeruginosa pneumonia. Am J Physiol Lung Cell Mol Physiol. 2009; 297:L1112–9. 10.1152/ajplung.00155.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raoust E, Balloy V, Garcia-Verdugo I, Touqui L, Ramphal R, Chignard M. Pseudomonas aeruginosa LPS or flagellin are sufficient to activate TLR-dependent signaling in murine alveolar macrophages and airway epithelial cells. PLoS One. 2009; 4:e7259. 10.1371/journal.pone.0007259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu K, Tang M, Liu Q, Han X, Jin H, Zhu H, et al. Bi-directional differentiation of single bronchioalveolar stem cells during lung repair. Cell Discov. 2020; 6:1. 10.1038/s41421-019-0132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005; 121:823–35. 10.1016/j.cell.2005.03.032 [DOI] [PubMed] [Google Scholar]
- 54.Tzouvelekis A, Kaminski N. Epigenetics in idiopathic pulmonary fibrosis. Biochem Cell Biol. 2015; 93:159–70. 10.1139/bcb-2014-0126 [DOI] [PubMed] [Google Scholar]
- 55.Magalhães M, Tost J, Pineau F, Rivals I, Busato F, Alary N, et al. Dynamic changes of DNA methylation and lung disease in cystic fibrosis: lessons from a monogenic disease. Epigenomics. 2018; 10:1131–45. 10.2217/epi-2018-0005 [DOI] [PubMed] [Google Scholar]
- 56.Solazzo G, Ferrante G, La Grutta S. DNA Methylation in Nasal Epithelium: Strengths and Limitations of an Emergent Biomarker for Childhood Asthma. Front Pediatr. 2020; 8:256. 10.3389/fped.2020.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dodge JE, Okano M, Dick F, Tsujimoto N, Chen T, Wang S, et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005; 280:17986–91. 10.1074/jbc.M413246200 [DOI] [PubMed] [Google Scholar]
- 58.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, et al. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003; 50:809–24. 10.1046/j.1365-2958.2003.03740.x [DOI] [PubMed] [Google Scholar]
- 59.Otto NA, de Vos AF, van Heijst J, Roelofs J, van der Poll T. Myeloid Liver Kinase B1 depletion is associated with a reduction in alveolar macrophage numbers and an impaired host defense during gram-negative pneumonia. J Infect Dis. 2020. 10.1093/infdis/jiaa416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, et al. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 1988; 48:1904–9. [PubMed] [Google Scholar]
- 61.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014; 343:84–7. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014; 11:783–4. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003; 339:62–6. 10.1016/s0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- 64.van Lieshout MH, Anas AA, Florquin S, Hou B, van’t Veer C, de Vos AF, et al. Hematopoietic but not endothelial cell MyD88 contributes to host defense during gram-negative pneumonia derived sepsis. PLoS Pathog. 2014; 10:e1004368. 10.1371/journal.ppat.1004368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Porto AP, Liu Z, de Beer R, Florquin S, de Boer OJ, Hendriks RW, et al. Btk inhibitor ibrutinib reduces inflammatory myeloid cell responses in the lung during murine pneumococcal pneumonia. Mol Med. 2019; 25:3. 10.1186/s10020-018-0069-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.García-Laorden MI, Stroo I, Blok DC, Florquin S, Medema JP, de Vos AF, et al. Granzymes A and B Regulate the Local Inflammatory Response during Klebsiella pneumoniae Pneumonia. J Innate Immun. 2016; 8:258–68. 10.1159/000443401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.