Abstract
A diverse portfolio of SARS-CoV-2 vaccine candidates is needed to combat the evolving COVID-19 pandemic. Here, we developed a subunit nanovaccine by conjugating SARS-CoV-2 Spike protein receptor binding domain (RBD) to the surface of oxidation-sensitive polymersomes. We evaluated the humoral and cellular responses of mice immunized with these surface-decorated polymersomes (RBDsurf) compared to RBD-encapsulated polymersomes (RBDencap) and unformulated RBD (RBDfree), using monophosphoryl lipid A-encapsulated polymersomes (MPLA PS) as an adjuvant. While all three groups produced high titers of RBD-specific IgG, only RBDsurf elicited a neutralizing antibody response to SARS-CoV-2 comparable to that of human convalescent plasma. Moreover, RBDsurf was the only group to significantly increase the proportion of RBD-specific germinal center B cells in the vaccination-site draining lymph nodes. Both RBDsurf and RBDencap drove similarly robust CD4+ and CD8+ T cell responses that produced multiple Th1-type cytokines. We conclude that multivalent surface display of Spike RBD on polymersomes promotes a potent neutralizing antibody response to SARS-CoV-2, while both antigen formulations promote robust T cell immunity.
INTRODUCTION
COVID-19, the disease caused by the novel coronavirus SARS-CoV-2, emerged in late 2019 and was declared a pandemic by the World Health Organization in March 2020. Since its emergence, researchers across the world have sought to rapidly develop vaccine candidates, some of which have received Emergency Use Authorization by the U.S. Food and Drug Administration1,2. While the first vaccines that entered the clinic were based on nucleic acid technologies, subunit vaccines are gaining attention and have also shown promise in clinical trials3,4. The primary antigens used in preclinical and clinical vaccine candidates are the Spike protein and its constituent receptor-binding domain (RBD). The RBD of the Spike protein binds to the ACE-2 receptor on host cell surfaces, enabling viral entry into the host cell5,6.
Several highly potent neutralizing antibodies have been isolated that target RBD and prevent viral binding and uptake, making it an attractive vaccine target7–10. Since RBD is smaller (~25 kDa) and more stable than the full homotrimeric Spike fusion protein (~180 kDa), it is also advantageous from a manufacturing and distribution perspective11. However, RBD has been shown to have lower immunogenicity than the full Spike protein or its RBD-containing S1 domain12,13. Materials science and engineering approaches, particularly strategies involving nanotechnology, may improve RBD immunogenicity and thus aid in the development of next-generation vaccines14–16. Indeed, several approaches of self-assembling RBD into virus-like particles have resulted in potent neutralizing antibody responses17–20.
In order to offer robust protection from infection, cellular in addition to humoral responses are needed21–23. Almost all convalescent individuals show T cell immunity, and the majority have both CD4+ and CD8+ SARS-CoV-2-specific T cells24–27. Conversely, severe disease is associated with lymphopenia and reduced T cell function28–30. Furthermore, T cell immunity may be more durable than humoral responses, and T cells are expected to play an important role in immune memory23,28,31. Therefore, the goal of this study was to improve both humoral and cellular immunogenicity of RBD and compare the efficacy of engineered nanoparticle formulations in order to inform the design of next-generation nanovaccines.
We have previously reported the development of polymersomes (PS) that self-assemble from the oxidation-responsive block copolymer poly(ethylene glycol)-bl-poly(propylene sulfide) (PEG-PPS)32 and shown their efficacy in delivering antigen and adjuvant to dendritic cell endosomes33. In endolysosomal compartments, the PPS block becomes oxidized, which initiates the restructuring of the PS into micelles and concurrent release of encapsulated payload33,34. These vaccine nanocarriers have been shown to activate dendritic cells, induce robust T cell immunity, and elicit high antibody titers with broad epitope coverage33,35,36.
In this study, we hypothesized that we could further improve the humoral responses elicited by PS while retaining their ability to induce T cell immunity by engineering them to mimic the physical form of a viral particle through multivalent surface display of antigen. We envisaged that multivalent surface display of RBD would result in enhanced crosslinking and clustering of B cell receptors (BCRs) and subsequent production of neutralizing antibodies. Here, we report on the development and preclinical evaluation of PS displaying surface-bound RBD (RBDsurf) and PS encapsulating RBD (RBDencap) adjuvanted with monophosphoryl lipid A-encapsulated PS (MPLA PS). We show that mice vaccinated with RBDsurf in combination with MPLA PS in a prime-boost schedule develop high titers of SARS-CoV-2-neutralizing antibodies with robust germinal center responses as well as CD4+ and CD8+ T cell immunity, thus meeting our design criteria.
RESULTS
Formulated polymersomes exhibit long-term stability and in vitro activity
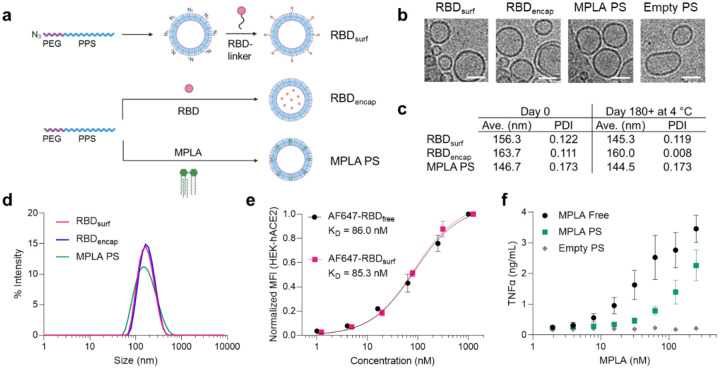
Having previously encapsulated antigen into PS as nanovaccines33, here we developed a conjugation strategy to attach antigens to their surface. To create a modular platform that could be generalized to any antigen, we synthesized N3-PEG-PPS (Suppl. Fig. S1), which, when formulated into PS, yields particles displaying clickable surface moieties (Fig. 1a). Upon the addition of RBD conjugated to a DBCO-containing linker, we generated PS decorated with RBD (RBDsurf, Suppl. Fig. S2). We also synthesized PEG-PPS (Suppl. Fig. S3) and formulated PS encapsulating RBD (RBDencap) or adjuvant (MPLA PS, Fig. 1a). The loading capacities of RBDsurf and RBDencap were 1.57% and 1.75%, respectively, comparable to previous reports of encapsulated ovalbumin33,35, while the loading capacity of MPLA PS was 6.46% (Suppl. Table S1). We confirmed the vesicular structure of PS through cryo-electron microscopy (cryoEM) and demonstrated that the different formulations have similar sizes and morphologies (Fig. 1b, Suppl. Fig. S4). According to dynamic light scattering (DLS) measurements, the average PS diameter is around 150 nm (Fig. 1c,d), which is similar to the reported size of SARS-CoV-2 particles (60–140 nm)37.The polydispersity index (PDI) of each formulation was < 0.2, indicative of a relatively homogenous population of nanoparticles. As indicated by their consistent size and PDI, in addition to the absence of free RBD released into solution, PS remain stable at 4 °C for at least 180 days, which can be beneficial for distribution and shelf-life considerations (Fig. 1c, Suppl. Fig. S5).
Figure 1 |. RBD and MPLA are formulated into stable, biologically active polymersomes (PS).
a, Schematic of formulation of PS. RBD was conjugated to the surface (RBDsurf) or encapsulated inside (RBDencap) of PS, and MPLA was encapsulated in the vesicle membrane (MPLA PS) due to its hydrophobicity. b, Representative cryo-electron microscopy images of PS, depicting vesicle structure. Scale = 50 nm. c, Size and polydispersity index (PDI) from dynamic light scattering (DLS) measurements of PS upon formulation and after > 6 months at 4 °C. d, Representative DLS curves of PS. e, Normalized mean fluorescence intensity (MFI) of AF647 conjugated to free RBD or RBDsurf by flow cytometry showing concentration-dependent binding to HEK-293 cells that express human ACE-2 (HEK-hACE2). Nonlinear regression was used to model data assuming specific binding to one site to determine equilibrium dissociation constants. f, Dose-dependent secretion of TNFα from cultured murine bone marrow-derived dendritic cells (BMDCs) stimulated by free MPLA, MPLA PS, or empty PS. Data represent mean ± SD for n = 2 (e) or 3 (f) replicates.
We next characterized the biological activity of the PS formulations in vitro. To confirm that RBD structure is not substantially altered upon conjugation to the PS surface, we quantified its ability to bind to HEK-293 cells that express human ACE-2 (HEK-hACE2, Fig. 1e). The normalized mean fluorescence intensity (MFI) versus RBD concentration curves were used to calculate the equilibrium dissociation constants (KD) for free RBD and RBDsurf conjugated to AF647 (AF647-RBDfree and AF647-RBDsurf, respectively). The curves and KD values are in excellent agreement, indicating that surface conjugation to PS did not impact ACE-2 binding of RBD. Empty PS conjugated to AF647 did not bind to HEK-hACE2, and neither PS formulation bound to HEK-293 cells lacking hACE-2 (Suppl. Fig. S6). Next, we confirmed that MPLA retained its ability to serve as a TLR4 agonist upon formulation in PS with a HEK-Blue™ TLR4 reporter cell line (Suppl. Fig. S7). To further validate MPLA PS activity in a more physiologically-relevant model, we stimulated murine bone marrow-derived dendritic cells (BMDCs) with free MPLA, MPLA PS, or empty PS, and we measured the subsequent secretion of the pro-inflammatory cytokines TNFα, IL-6, IL-1α, and IL-1β (Fig. 1f, Suppl. Fig. S8). For each cytokine, there was a dose-dependent increase in secretion for free MPLA and MPLA PS with only background levels of secretion for empty PS, indicating that MPLA PS successfully activated antigen presenting cells (APCs) in vitro. Thus, we successfully synthesized two RBD formulations of PS in addition to MPLA PS and showed that they are homogenous vesicular structures with long-term stability and in vitro biological activity.
All adjuvanted formulations elicit RBD-specific IgG responses
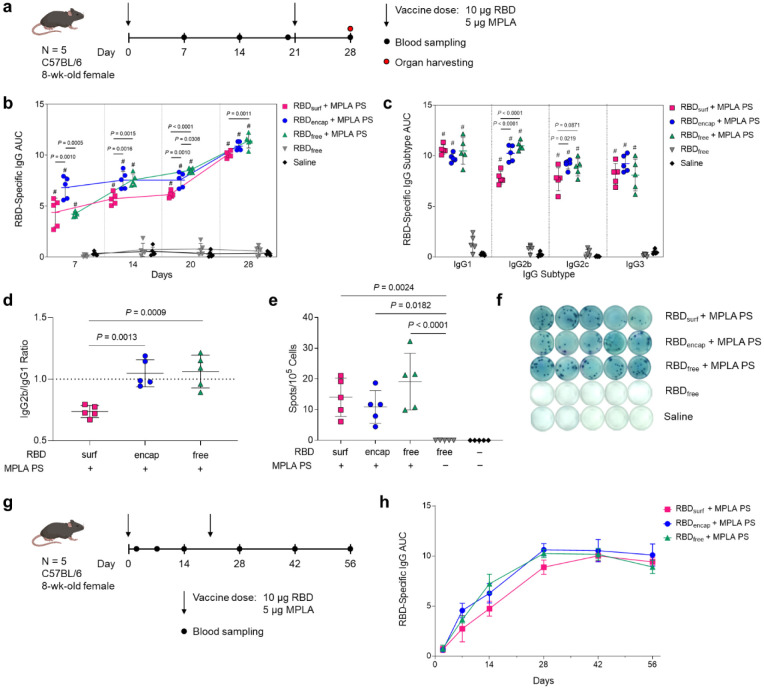
Having confirmed that antigen- and adjuvant-loaded PS exhibit their expected bioactivity in vitro, we next evaluated their ability to enhance humoral and cellular immunity in mice compared to RBDfree. We immunized mice via s.c. injection in the hocks in a prime-boost schedule 3 weeks apart and monitored antibody titers weekly (Fig. 2a). The total RBD-specific IgG is represented by the area under the log-transformed ELISA absorbance curves (AUC), starting at a plasma dilution of 10−2 (see Methods, Suppl. Fig. S9). All adjuvanted groups had significant anti-RBD binding antibody responses within a week after their first dose, with RBDencap stimulating the highest responses (Fig. 2b). The antibody responses in adjuvanted groups either increased gradually or remained constant until a week after the booster, when the mean AUC increased 1.3- to 1.6-fold.
Figure 2 |. High levels of RBD-specific IgG antibodies are produced upon PS vaccination.
a, Vaccination schedule consisting of a priming dose followed by a booster 3 weeks later. b, Total RBD-specific IgG antibodies over time reported as the area under the log-transformed curve (AUC) of absorbance vs. dilution. c, Comparison of RBD-specific IgG isotypes (IgG1, IgG2b, IgG2c, IgG3) on day 28. d, Ratio of AUCs of IgG2b:IgG1 isotypes. e, Quantification of RBD-specific IgG+ antibody secreting cells by ELISpot of splenocytes (Dunn’s post-test compared to unadjuvanted RBDfree). f, Representative ELISpot wells from (e). Data plotted as mean ± SD and represent 1 of 2 experiments with n = 5 mice each. Symbols represent individual mice. g, Vaccine and blood sampling schedule of long-term kinetics study. h, Total RBD-specific IgG antibodies over time for the vaccination schedule in (g). Data represent mean ± SD for n = 5 mice. Comparisons were made using one-way ANOVA with Tukey’s post-test unless stated otherwise. # P < 0.0001 compared to unadjuvanted RBDfree.
In order to explore the humoral response in further detail, we then evaluated IgG subtypes of induced antibodies at the study endpoint (d28, 1 week post-boost). While plasma antibody levels of all adjuvanted groups were similar for IgG1 and IgG3, RBDsurf elicited significantly lower IgG2b and IgG2c antibody responses (Fig. 2c). The ratio of IgG2b/IgG1 was then taken as an indication of a Th1/Th2-mediated response38. While RBDencap and RBDfree + MPLA PS have a ratio of around 1, indicating a balanced Th1/Th2 response, RBDsurf shows a lower ratio of IgG2b to IgG1 indicating a slightly Th2-skewed response (Fig. 2d). Since IgA is important for combating respiratory viruses at the mucosal sites, we also measured plasma levels of RBD-specific IgA antibodies39,40. Although we did not expect high titers of circulating IgA from a s.c. administered vaccine, there were detectable anti-RBD IgA antibodies in all adjuvanted groups, which were significantly higher than the background-level responses elicited by unadjuvanted RBDfree (Suppl. Fig. S10).
Next, to determine if the higher antibody responses of adjuvanted groups stemmed from an expanded number of RBD-specific antibody secreting cells (ASCs), we performed an ex vivo RBD enzyme-linked immunosorbent spot (ELISpot) assay with splenocytes harvested 1 week post-boost. All groups receiving adjuvanted RBD showed significantly higher RBD-specific IgG+ ASCs compared to unadjuvanted RBDfree, consistent with plasma antibody levels (Fig. 2e,f).
Finally, we evaluated the kinetics and durability of the humoral response to demonstrate the persistence of elicited antibodies (Fig. 2g). The RBD-specific IgG AUC for all adjuvanted groups increased until 1 week post-boost and then remained constant over the subsequent 4 weeks, indicating that the antibody responses stimulated by these vaccine formulations persist for at least 2 months in mice after the initial dose. Taken together, MPLA PS-adjuvanted RBDsurf, RBDencap, and RBDfree all stimulated persistent anti-RBD antibodies and increased the frequencies of RBD-specific ASCs in the spleen.
RBD-surface-decorated polymersomes, but not RBD-encapsulated polymersomes, induce neutralizing antibodies
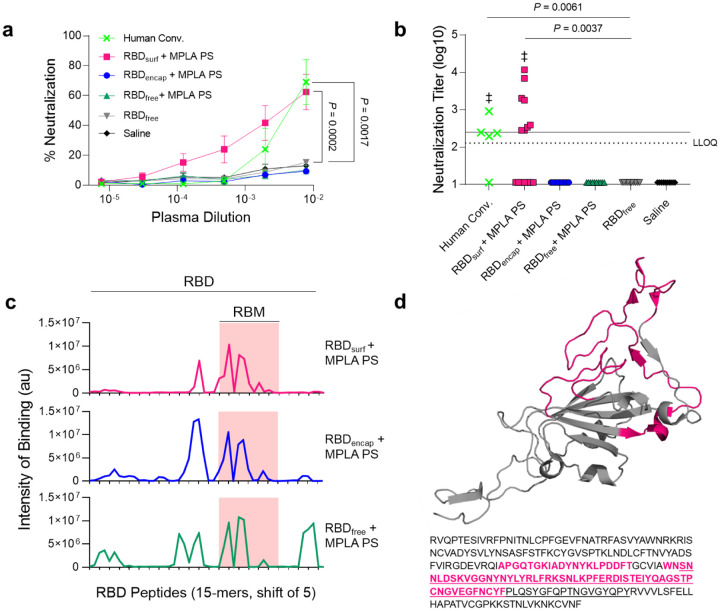
After analyzing the quantity of RBD-specific antibodies produced by the vaccine candidates, we next sought to determine their neutralizing capacity and breadth of epitope recognition. Neutralizing antibodies were assessed against SARS-CoV-2 infection of Vero E6 cells in vitro. Although all adjuvanted groups elicited similarly high titers of RBD-binding IgG antibodies (105–107, Suppl. Fig. S11a), only RBDsurf neutralized the virus to a greater extent than unadjuvanted RBDfree at a plasma dilution of 10−2.11 (Fig. 3a). We then quantified the viral neutralization titer (VNT) as the dilution at which 50% of SARS-CoV-2-mediated cell death is neutralized. There was no significant difference between VNTs of human convalescent individual samples and RBDsurf plasma, and both groups induced higher VNTs compared to unadjuvanted RBDfree (Fig. 3b). Furthermore, the median VNT elicited by RBDsurf was 2.45, which falls within the FDA classification of “high titer” for convalescent plasma therapy (> 2.40)41. To ensure reproducibility, the neutralization assay was repeated with 3 different cohorts of n = 5 mice each, and no significant differences were observed between the resulting VNTs (Suppl. Fig. S11b).
Figure 3 |. Antibodies induced by vaccination with RBDsurf + MPLA PS are neutralizing and localized to the receptor binding motif.
a, Plasma from mice 1 week post-boost was tested for its ability to neutralize SARS-CoV-2 infection of Vero E6 cells in vitro. Percent neutralization for multiple plasma dilutions normalized to cells without virus (100%) or without plasma (0%). Data plotted as mean ± SEM for n = 5 convalescent human samples (human conv.) or 10–15 mice. Comparisons to unadjuvanted RBDfree were made for lowest dilution (10−2.11) using one-way ANOVA with Dunnett’s post-test. b, Viral neutralization titers, representing the plasma dilution at which 50% of SARS-CoV-2-mediated cell death is neutralized. Dashed line: lower limit of quantification (LLOQ = 2.11). For values below the LLOQ, LLOQ/2 values were plotted. Solid line: FDA recommendation for “high titer” classification (= 2.40). Comparisons were made using Kruskal-Wallis nonparametric test with Dunn’s post-test or Wilcoxon signed rank test (‡ ns, P > 0.05 compared to hypothetical value of 2.40). Symbols represent individual mice. c, Pooled plasma was then tested for antibody binding to linear epitopes using overlapping 15-amino-acid peptides with 5-amino-acid offsets, spanning the entire RBD sequence. X-axis represents sequential peptide number within the RBD amino acid sequence, and y-axis represents average luminescence for each peptide epitope. d, 3D structure of RBD with the receptor binding motif (RBM) underlined and main peptide sequences recognized by mouse plasma highlighted in pink (Protein Data Bank Entry 7DDD).
We next aimed to determine whether differences in neutralizing ability resulted from the epitope recognition breadth elicited by each vaccine formulation. To test this, we mapped the epitopes recognized by vaccine-elicited antibodies using a linear peptide array from the full-length RBD sequence. While IgG antibodies elicited by RBDsurf primarily recognized linear epitopes concentrated within the receptor binding motif of RBD (RBM; aa 438–508), RBDencap and RBDfree + MPLA PS exhibited broader linear epitope diversity (Fig. 3c,d, Suppl. Fig. S12). Within RBD, the RBM is particularly critical for interacting with ACE-2, so antibodies specific for this region may have potent neutralizing potential42,43.
Because RBDsurf appeared to offer the advantage of improved neutralizing activity, while RBDencap offered epitope diversity, we asked if co-administration would synergize to further enhance protection. To test this, we mixed RBDsurf and RBDencap (with total RBD dose remaining constant) with MPLA PS and treated mice using the same vaccination schedule. As an additional control, we also investigated the humoral response of RBDfree adjuvanted with free MPLA. While both groups produced high RBD-specific IgG AUCs, neither exhibited neutralizing potential against the SARS-CoV-2 virus above background levels (Suppl. Fig. S13a,b). Analysis of the peptide arrays for these groups shows the presence of high-intensity-binding antibodies outside of the RBM (Suppl. Fig. S13c).
In summary, while all adjuvanted groups elicit high titers of RBD-specific antibodies, only RBDsurf generated neutralizing antibodies against SARS-CoV-2 at titers comparable to human convalescent plasma. Additionally, these antibodies uniquely bound to linear epitopes localized within the RBM, while all other groups produced antibodies with greater epitope breadth.
All adjuvanted formulations increase Tfh and B cell activation in the dLN
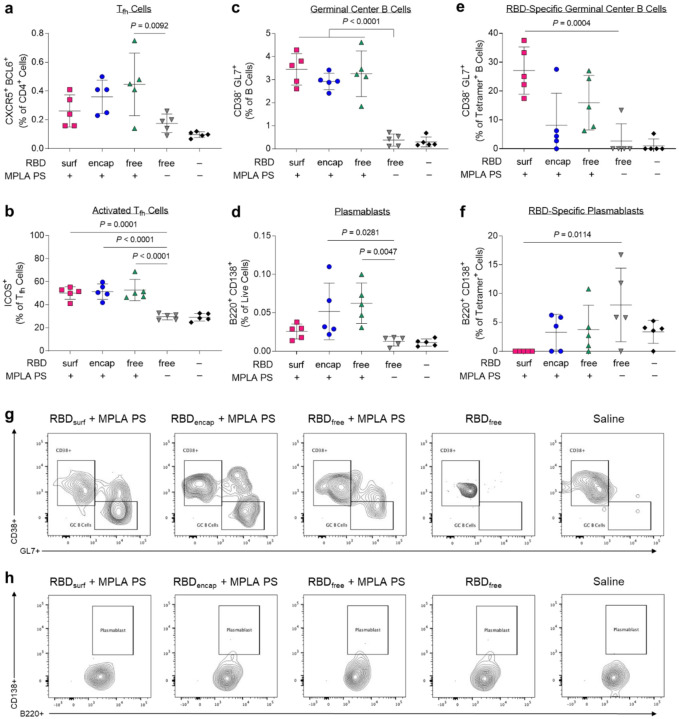
Given the differences in antibody responses and neutralizing activity elicited by RBDsurf versus RBDencap vaccination, we further investigated the phenotypes of the B and T cells involved in the initiation of the humoral immune response. All adjuvanted groups showed trends of higher frequencies of T follicular helper cells (Tfh; CD4+CXCR5+BCL6+) in the injection site-draining lymph nodes (dLNs) 1 week post-boost compared to unadjuvanted RBDfree (Fig. 4a, Suppl. Fig. S14), although differences were only statistically significant for RBDfree + MPLA PS. Interestingly, a greater percentage of Tfh cells in all adjuvanted groups highly upregulated expression of ICOS, a co-stimulatory receptor important in Tfh activation and maintenance (Fig. 4b)44.
Figure 4 |. CD4+ T follicular helper cell (Tfh) and B cells are activated by PS vaccine 1 week post-boost in the injection site-draining lymph nodes (dLN).
a, Tfh cells (CXCR5+ BCL6+) of vaccinated mice quantified via flow cytometry as a percent of live CD4+ T cells. b, Highly activated ICOS+ Tfh cells quantified as percent of Tfh cells. c, Germinal center B cells (CD38− GL7+) quantified as a percentage of total B cells (B220+ CD19+). d, Plasmablasts (B220+ CD138+) quantified as a percentage of total dLN cells. e, Germinal center B cells quantified as a percentage of RBD-specific B cells. f, Plasmablasts quantified as a percentage of RBD-specific dLN cells. g-h, Concatenated flow cytometry contour plots for n = 5 mice/group showing RBD-specific GC B cells (g) or plasmablasts (h). Data plotted as mean ± SD and represent 1 of 2 experiments with n = 5 mice each. Symbols represent individual mice. Comparisons to unadjuvanted RBDfree were made using one-way ANOVA with Dunn’s post-test.
Following activation by Tfh cells, naïve B cells can either undergo a germinal center (GC)-dependent response in which they become GC B cells and undergo cycles of class-switching and somatic hypermutation (SHM) before differentiation into long-lived plasma cells and class-switched memory B cells, or they can differentiate into short-lived plasmablasts or IgM memory cells in a GC-independent response45. A stronger GC response is valuable in vaccination because it results in higher affinity and longer-lived antibody production46. Overall frequencies of B cells (CD19+B220+) were unaffected across groups, but there were significantly lower frequencies of naïve IgD+ B cells in the adjuvanted groups compared to unadjuvanted RBDfree (Suppl. Figs. S15, S16). All adjuvanted groups generated GC responses, characterized by increased frequencies of GC B cells (CD38−GL7+) in the dLN compared to unadjuvanted RBDfree (Fig. 4c). Both RBDencap and RBDfree + MPLA PS formulations, but not RBDsurf, significantly increased the frequencies of plasmablasts (B220+CD138+) in the dLN compared to unadjuvanted RBDfree (Fig. 4d).
To determine the antigen-specificity of the B cells, we developed a set of fluorescent RBD protein tetrameric probes. To ensure selectivity, B cells were considered RBD-specific if they were double-positive for both PE- and APC-conjugated RBD-tetramers and negative for the non-specific control protein tetramer (Suppl. Fig. S17). RBDfree + MPLA PS was the only formulation to significantly increase the frequency of RBD-specific B cells in the dLN (Suppl. Fig. S18). We next sought to further determine the phenotype of these RBD-specific B cells. RBDsurf, unlike the other adjuvanted formulations, led to a significantly higher fraction of RBD-specific B cells with GC B cell phenotype, suggesting a more robust GC response to RBD (Fig. 4e). RBDsurf was also the only formulation with a significantly lower fraction of plasmablasts within the RBD-specific B cell subset compared to unadjuvanted RBDfree (Fig. 4f). These differences are also visually apparent in pooled flow cytometry plots for RBD-specific GC B cells (Fig. 4g) and plasmablasts (Fig. 4h). In summary, all adjuvanted formulations of RBD increased activation of Tfh cells and GC B cells in the dLN, but within the RBD-specific B cell population, only RBDsurf generated a higher fraction of GC B cells and a lower fraction of plasmablasts.
Vaccination with polymersome-formulated RBD generates RBD-specific Th1 T cell responses
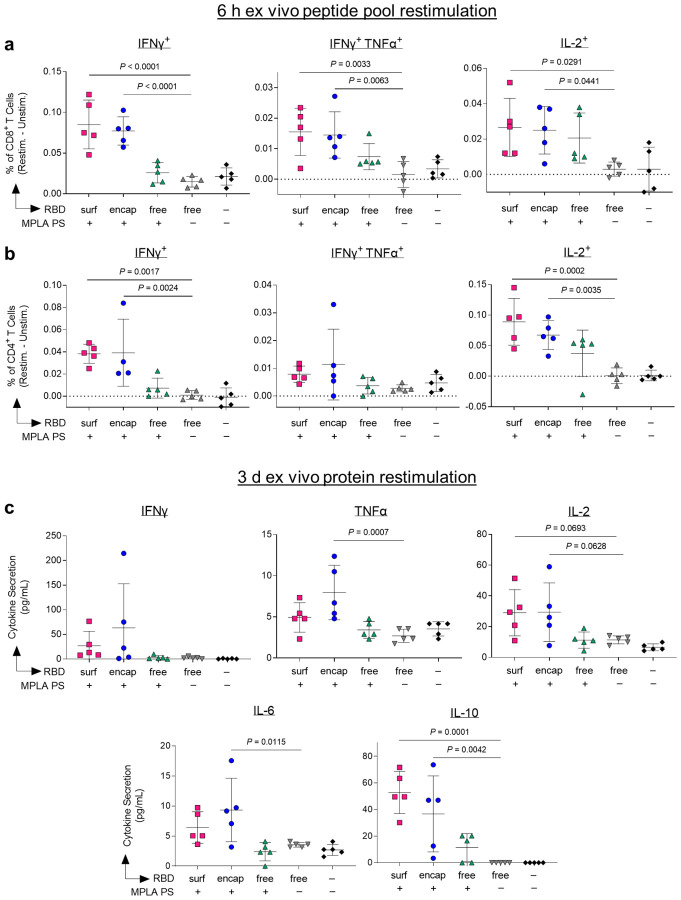
Having demonstrated that our PS vaccines can generate strong humoral responses, we next sought to determine their capacity to generate robust CD8+ and CD4+ T cell immunity. In order to assess the RBD-specific T cell response, we isolated cells from the dLNs of vaccinated mice 1 week post-boost. Prior to intracellular staining, cells were restimulated with RBD peptide pools for 6 hours. The RBD-specific response was quantified by subtracting the signal from cells incubated in media alone from those incubated with RBD peptide pools (Suppl. Fig. S19). Only PS-formulated RBD groups RBDsurf and RBDencap generated significantly higher frequencies of IFNγ+, bifunctional IFNγ+TNFα+, and IL-2+ secreting CD8+ T cells compared to unadjuvanted RBDfree (Fig. 5a). Similar trends were seen in the CD4+ T cell compartment. Treatment with RBDsurf and RBDencap but not RBDfree + MPLA PS led to significantly higher frequencies of IFNγ+ and IL-2+ secreting CD4+ T cells, although the increase in bifunctional IFN γ+TNFα+ was not statistically significant (Fig. 5b). The production of these cytokines implies a Th1 T cell response, which is correlated with less severe cases of SARS-CoV infection21.
Figure 5 |. Vaccination with polymersome-formulated RBD improves antigen-specific T cell responses in mice.
Cells isolated from the injection-site draining lymph nodes of PS-vaccinated mice 1 week post-boost were restimulated ex vivo for 6 h with RBD peptide pools or full RBD protein and analyzed by flow cytometry or multiplexed ELISA, respectively. a-b, percentages of cytokine-positive CD8+ T cells (a) and CD4+ T cells (b) in response to RBD peptide pools, subtracting unstimulated controls. c, Pro-inflammatory cytokine levels released into the supernatant measured after 3 d restimulation with whole RBD protein. Data plotted as mean ± SD and represent 1 of 2 experiments with n = 5 mice each. Symbols represent individual mice. Comparisons to unadjuvanted RBDfree were made using one-way ANOVA with Dunn’s post-test.
For further validation of the cytokine response, cells from the dLN isolated from the same vaccinated mice were restimulated with full RBD protein ex vivo for 3 days, followed by quantification of cytokines in the supernatant. The RBD-specific response was quantified by subtracting unstimulated signal from stimulated signal as above. The levels of Th1-type cytokines detected were consistent with intracellular staining data. Levels of IFNγ and IL-2 were modestly but not significantly increased in the RBDsurf and RBDencap groups compared to the RBDfree group, while cells from RBDencap-treated mice produced TNFα at significantly higher levels than unadjuvanted RBDfree (Fig. 5c). RBDencap also led to increased production of IL-6 and IL-10, which are pleiotropic cytokines known to be secreted during Th1 responses (Fig. 5c)47,48. Levels of secreted Th2-type cytokines were also measured. More IL-4 was produced in the supernatant of samples treated with RBDsurf and RBDencap compared to RBDfree, albeit at an overall low concentration. There was no significant elevation of IL-5 secretion across any of the treatment groups compared to the saline control and no detectable levels of IL-13 in any sample (Suppl. Fig. S20). In summary, vaccination with RBD delivered via polymersome formulations generated stronger RBD-specific Th1-type CD8+ and CD4+ T cell responses than unadjuvanted RBDfree, while RBDfree + MPLA PS did not.
DISCUSSION
In this study, we developed antigen-decorated, oxidation-sensitive polymersomes that mimic virus particles as next-generation nanovaccines. While all adjuvanted formulations generated long-lived RBD-binding IgG responses, surface conjugation of antigen was necessary to generate neutralizing antibodies against SARS-CoV-2. More generally, this comparison of antigen formulation on the quality of immune response offers valuable insight into vaccine design, demonstrating the benefits of surface antigen display.
The differences in the immune responses elicited by the two PS antigen formulations, RBDsurf and RBDencap, suggest that surface display of antigen leads to stronger GC responses, while PS-encapsulated antigen elicits more predominantly an extrafollicular response. Though RBD-specific GC B cells are present in the dLN after treatment with both formulations, a much higher percentage of the RBD-specific B cells recovered after vaccination with RBDsurf exhibited a GC B cell phenotype. This higher percentage could be due to the induction of higher numbers of RBD-specific GC B cells after vaccination with RBDsurf or an increase in their affinity, leading to easier detection via RBD protein tetramer staining. Both possibilities suggest a more robust GC response, as GC responses are necessary for an efficient SHM leading to increased B cell affinity49. Evidence for affinity maturation due to SHM also includes the relatively few epitopes on the RBD linear peptide array to which IgG from RBDsurf-treated groups were specific compared to the other adjuvanted groups. Clonal bursts in GC responses can lead to rapid expansion of high-affinity SHM variants and loss of overall clonal diversity50. A difference in the affinity of RBD-specific IgG generated by these vaccines may also explain the neutralization ability of the plasma after vaccination with RBDsurf but not RBDencap. Further data that suggest that RBDencap elicits a more extrafollicular response include the fast initial antibody response after priming by RBDencap, which resulted in RBD-specific IgG AUC 1 week after the initial prime that was significantly higher than that of RBDsurf. In an extrafollicular B cell response, B cells can differentiate immediately into plasmablasts and begin secreting antibodies after initial T cell help, whereas B cells that enter GCs delay the antibody response by several days45. The preferential differentiation into plasmablasts rather than GC B cells was also evident in the increased fraction of plasmablasts within the RBD-specific B cell population generated by RBDencap. In vaccine development, the generation of robust GC reactions is preferable to extrafollicular responses because GC formation will usually result in higher affinity B cells, increased memory B cells and increased long-lived plasma cells in the bone marrow.45
The difference in mechanism of B cell activation by these two RBD formulations may be explained by previous studies on multimerization of antigen and use of virus-like particles in vaccination. Virus-like particles are multi-protein supra-molecular structures constructed of many identical protein copies. Their multimeric nature has been associated with the induction of potent antibody responses due to BCR crosslinking in the presence of CD4+ T cell help51. Mechanistic studies by Kato et al. demonstrated that increasing antigen valency can enhance the early activation and proliferation of antigen-specific B cells as well as increase B cell accumulation at the T-B border leading to increased differentiation of antigen-specific B cells into GC B cell and plasma cells52. Furthermore, in contrast to RBDencap, RBDsurf most likely efficiently exposes for BCR interaction the conformational epitopes of RBD reported to be targeted by neutralizing antibodies in plasma of convalescent or vaccinated individuals7–10. Thus, the multimerization of RBD on the PS surface in addition to its increased availability to B cells led to an improved functional quality of humoral response compared to encapsulated or unformulated RBD.
The type of immune response to SARS-CoV-2 may have important implications in how the infection is cleared and should be considered in vaccine design21. Less severe cases of the original SARS-CoV were associated with increased Th1-type cell responses53,54. In contrast, Th2-type responses were associated with increased pathology due to antibody-dependent enhancement, and several vaccine formulations against SARS-CoV tested in animal models showed signs of immunopathology due to Th2 cell-mediated eosinophil infiltration55,56. In this study, we measured the ratio of IgG1/IgG2b and the cytokine profile of dLN cells restimulated with RBD protein in order to characterize the type of immune responses generated by our vaccine formulations. RBDencap and RBDfree + MPLA PS generated IgG2b/IgG1 ratios around 1, suggesting a balance between type 1 and type 2 immunity, whereas RBDsurf generated a response with a slightly IgG1-bias, suggestive of a Th2-skewed response. However, both RBDsurf and RBDencapf generated significant levels of the Th1-type cytokines IFNγ, TNFα and IL-2. This Th1-type response is likely induced by the MPLA PS, as MPLA has been shown to generate strong type-1 immunity57. Although RBDsurf generated a slightly IgG1-biased antibody response, it did not result in an increase in Th2-type cytokines upon restimulation compared to RBDencap. Combined with the evidence of Th1-type cytokine production upon restimulation, we believe the risk of adverse events related to Th2-type responses is low.
The PS-formulated RBD vaccines were able to generate stronger Th1-driven CD4+ and CD8+ T cell responses compared to RBDfree. We previously demonstrated that as an antigen delivery vehicle, PEG-PPS polymersomes could improve cross-presentation to CD8+ T cells33. This increased T cell response likely occurs because of both enhanced APC targeting as well as rapid endosomal antigen release33. APCs are able to accumulate membrane-impermeable nanocarriers such as PS more efficiently than other cell types due to their constitutive macropinocytosis58. Once endocytosed, PEG-PPS polymersomes require only a small amount of oxidation to release encapsulated antigen, and payload delivery to the cytosol is not restricted to endosomal compartments with reductive or acidic conditions33. Unlike antigen-encapsulated PS, however, acidification may be important for proteolytic degradation of RBDsurf and translocation to the cytosol, as evidenced by studies on VLPs59,60. Additionally, peptides derived from large antigen particles have been found to enter the cross-presentation pathways more efficiently than those derived from soluble antigens, which may provide rationale for the enhanced CD8+ response of RBDsurf, whereas no such benefit was seen for adjuvanted RBDfree61. Therefore, antigen formulation using PS to improve T cell responses could be beneficial in the engineering of future vaccines against cancer or other infectious diseases for which T cell immune responses are thought to be necessary for protection, such as herpesviruses, human immunodeficiency virus, and hepatitis C virus62.
In summary, we have demonstrated that a polymersome-based antigen and adjuvant delivery system generates robust humoral immunity and neutralizing antibody titers, as well as T cell responses, against a key SARS-CoV-2 vaccine target, the RBD of Spike. This platform technology is amenable to a wide variety of antigens and formulated or soluble adjuvants. Once the type and dose of adjuvant has been optimized for a given application, a single particle could be used to deliver both antigen and adjuvant to APCs in the injection site-draining lymph node. Additionally, multiple antigens, for example from different viruses or different strains or variants of the same virus, could be conjugated to the same particle as a strategy to induce cross-reactive neutralizing antibodies20. Importantly, both surface-decorated and antigen-encapsulated polymersomes remained stable at 4 °C for at least 6 months, as indicated by consistent particle size and absence of antigen released into solution. Vaccines that exhibit long-term stability without requiring sub-zero temperatures will likely be important for widespread vaccine distribution, for example to rural populations or developing nations with poor cold chain network. The evaluation of RBD-decorated polymersomes presented here could thus provide insight into the next generation of stable formulations of nanovaccines to combat the current COVID-19 pandemic as well as future viral outbreaks.
METHODS
RBD production and purification
For production of the Spike protein RBD (Spike319–541; GenBank: MN908947.3), we obtained expression plasmids on pCAGGS backbone containing mammalian codon-optimized sequences for this gene from Florian Krammer’s laboratory (Icahn School of Medicine at Mount Sinai, New York, NY)63. Suspension-adapted HEK-293F cells were maintained in serum-free Free Style 293 Expression Medium (Gibco). On the day of transfection, cells were inoculated into at a concentration of 1 × 106 cells mL−1. Plasmid DNA (1 mg mL−1) was mixed with linear 25 kDa polyethyleneimine (2 mg mL−1; Polysciences) and co-transfected in OptiPRO SFM medium (4% final concentration; Thermo Fisher). Flasks were cultured in an orbital shaking incubator (135 rpm, 37 °C, 5% CO2) for 7 days. Culture medium was then collected by centrifugation, filtered, and loaded into a HisTrap HP 5 mL column (GE Healthcare) using an ÄKTA pure 25 (GE Healthcare). After washing the column with wash buffer (20 mM NaH2PO4 and 0.5 M NaCl, pH 8.0), protein was eluted using a gradient of 500 mM imidazole in wash buffer. The protein was further purified by size-exclusion chromatography using a HiLoad Superdex 200PG column (GE Healthcare) with PBS as an eluent. Dimers of RBD were reduced by the addition of dithiothreitol (1 mM) which was subsequently dialyzed against PBS. All purification steps were carried out at 4 °C. The expressed proteins were verified to be >90% pure through SDS–PAGE. The purified proteins were tested for endotoxin using a HEK-Blue™ TLR4 reporter cell line (InvivoGen), and the endotoxin levels were confirmed to be below 0.01 EU mL−1. Protein concentration was determined by absorbance at 280 nm using a NanoDrop spectrophotometer (Thermo Scientific). Proteins were stored at a concentration of 4 mg mL−1 at −80 °C until use.
PS formulation
PEG-PPS polymersomes were formulated by thin film rehydration as previously described33. In brief, 20 mg of polymer was dissolved in 750 μL dichloromethane (DCM), and DCM was removed by vacuum desiccation overnight. 1 mL of PBS was then added to the vial, which was rotated at room temperature (RT) for 24 h to allow complete dispersal of the polymer. The solution was then sequentially extruded through 0.8, 0.4, 0.2, and 0.1 μm pore membranes (Whatman). To formulate RBD-encapsulated PS, 250 μL of PBS containing 4 mg/mL RBD was added to the polymer thin film for rehydration, and the solution was rotated at 4 °C for 72 h before extrusion as above. After extrusion, RBD-encapsulated polymersomes were passed through a sepharaose size exclusion chromatography (SEC) column to remove unencapsulated free RBD. The RBD content was quantified by SDS-PAGE using mini-protein TGX stain-free precast gels (Bio-Rad). Gels were imaged on a ChemiDoc XRS+ Gel Documentation System (Bio-Rad) and analyzed using ImageJ.
RBD-surface-conjugated PS were synthesized by first formulating empty PS as above consisting of 25% N3-PEG-PPS. RBD was conjugated to a sulfo DBCO-Maleimide linker (Click Chemistry Tools) at the molar ratio of 1:5 (RBD:linker) and reacted for 2.5 hours. Unconjugated linker was removed by Zeba spin desalting columns (7K MWCO; ThermoFisher). The resulting RBD-linker was analyzed by MALDI-TOF MS using an α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) matrix and a Bruker ultrafleXtreme MALDI TOF/TOF instrument. RBD-linker (4 wt% of polymersome) was then incubated with empty N3-PEG-PPS polymersomes overnight at RT. RBD-surface-conjugated PS were then passed through SEC to remove unconjugated RBD-linker. The conjugation was monitored by SDS-PAGE, and the final RBD content was quantified by the CBQCA Protein Quantitation Kit (ThermoFisher). Fluorescently labeled PS were prepared by conjugating AlexaFluor-647 alkyne dye (ThermoFisher) to PS containing 25% N3-PEG-PPS. The dye was mixed with PS at a molar ratio of 1:20 (dye:N3-PEG-PPS), and the solution was stirred overnight at RT. Labeled PS were then passed through SEC to remove unconjugated dye. To make the fluorescently labeled RBDsurf, labeled PS were prepared as above and conjugated with RBD as described, followed by SEC purification. Final RBD content was quantified by the CBQCA Protein Quantitation Kit (ThermoFisher).
MPLA PS were fabricated by flash nanoprecipitation using a 3D printed impingement jets mixer64. 20 mg PEG-PPS and 2 mg MPLA (PHAD®; Avanti Polar Lipids) were dissolved in 500 μL tetrahydrofuran (THF) and loaded into a 1 mL plastic disposable syringe. 500 μL PBS was loaded into a second syringe, and the two solutions were impinged against one another slowly within the mixer by hand. The impinged solution was immediately vortexed to form a homogenous polymersome solution which was then extruded and purified by SEC as described above. MPLA loading was quantified using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) method as previously described65 using PHAD®−504 (Avanti Polar Lipids) as an internal standard on an Agilent 6460 Triple Quad MS-MS with 1290 UHPLC.
PS characterization
The size and polydispersity index (PDI) of all the polymersome formulations were measured by dynamic light scattering (DLS) using a Zetasizer Nano ZS90 (Malvern Instruments). Cryogenic electron microscopy (cryoEM) images were obtained on a FEI Talos 200kV cryoEM dedicated electron microscope.
MPLA PS in vitro activity
To determine TLR4 activation, HEK-Blue™ TLR4 cells (Invivogen) were incubated with increasing concentrations of MPLA PS for 24 h 37 °C in a 5% CO2 incubator. NF-kB-induced SEAP activity was detected using QUANTI-Blue™ (Invivogen) and by reading the OD at 650 nm. For dendritic cell activation experiments, BMDCs were prepared from C57Bl/6 mice (Jackson) as previously described.66 After 9 days of culture, cells were seeded at 2 × 105 cells/well in round-bottom 96-well plates (Fisher Scientific) in IMDM with 10% FBS and 2% penicillin/streptomycin (Life Technologies). Cells were treated with varying concentrations of MPLA PS or free MPLA and incubated for 24 h at 37 °C in a 5% CO2 incubator. After 24 h, the supernatant was collected, and cytokine concentration was measured using a multiplexed mouse Th cytokine panel (BioLegend) according to the manufacturer’s instructions.
RBDsurf in vitro activity
For the cell-based hACE2-binding assay, human embryonic kidney (HEK)-293T cells overexpressing human ACE-2 (HEK-hACE2) were obtained from BEI Resources (NIH NIAID). Fluorescently labeled empty PS, RBDsurf, or RBDfree was incubated at varying concentrations with 5 × 104 HEK-hACE2 cells at 4 °C for 20 min. Cells were then washed three times in PBS with 2% FBS. Binding of RBD to hACE-2 on the cell surface was assessed via the mean fluorescent intensity measured by flow cytometry using BD LSRFortessa (BD Biosciences).
Production of RBD protein tetramers
RBD protein expressed with AviTag was purchased from GenScript. Site-specific biotinylation of the AviTag was performed using BirA Biotin-Protein Ligase Reaction kit (Avidity). Next, unconjugated biotin was removed using Zeba spin desalting columns, 7K MWCO (ThermoFisher). The quantification of reacted biotin was performed using the Pierce Biotin Quantification Kit (ThermoFisher). Biotinylated RBD was incubated with either streptavidin-conjugated PE (Biolegend) or streptavidin-conjugated APC fluorophores (Biolegend) for 20 min on ice at a molar ratio of 4:1 of biotin to streptavidin. Streptavidin-conjugated FITC (BioLegend) was reacted with excess free biotin to form a non-RBD-specific streptavidin probe as a control. Tetramer formation was confirmed using SDS-PAGE gel. Cells were stained for flow cytometry with all three streptavidin probes at the same time as other fluorescent surface markers at a volumetric ratio of 1:100 for RBD-streptavidin-PE and 1:200 for RBD-streptavidin-APC and biotin-streptavidin-FITC.
Mouse vaccination experiments
All experiments were performed in accordance with the Institutional Animal Care and Use Committee at the University of Chicago. Female 8-week-old C57BL/6 mice (Jackson Laboratory) were randomly assigned to cohorts of n = 5 and vaccinated with 10 μg of antigen and 5 μg of adjuvant s.c. in the hock (either 2 or 4 hocks) and boosted on day 21. On day 7, 14, 20, and 28 post vaccination, 100 μL of blood was collected in EDTA-K2-coated tubes (Milian), and plasma was separated by centrifugation and stored at −80 °C until use. On day 28 after initial vaccination, mice were sacrificed. Splenocytes and lymph node cells from draining lymph nodes were collected. Single-cell suspensions of the lymph node were prepared by digestion in collagenase D for 45 min at 37 °C. Splenocytes and lymph node cells were filtered through a 70 μm cell strainer. Splenocytes were then incubated in ACK lysis buffer to remove red blood cells. Lymph node cells were stained for Tfh cells and RBD-specific B cells using fluorescent probes listed in Supplementary Tables 1 and 2, respectively. Samples were acquired on a BD LSR-Fortessa (BD) and analyzed using FlowJo™ software. Representative gating strategies used to identify the cell populations are shown in Supplemental Figures 14 (Tfh cells) and 15 (RBD-specific B cells).
To assess antigen-specific cytokine production by T cells, 1 × 106 lymph node cells were incubated with pools of 15-mer peptides overlapping by 10 amino acids covering the N-terminus of SARS-CoV-2 Spike protein up to the furin cleavage site (S1 pool; PepMix SARS-CoV-2 Spike Glycoprotein, JPT) for 6 h at 37 °C with 5% CO2. Monensin (GolgiStop, BD) was added after 2 h of incubation to inhibit cytokine secretion. Cells were stained for surface markers using fluorescent monoclonal antibodies (mAbs). Cells were subsequently fixed and permeabilized using BD Cytofix/Cytoperm™, and intracellular cytokines were stained using fluorescent-mAbs listed in Supplementary Table 3. Samples were acquired on a BD LSR-Fortessa (BD) and analyzed using FlowJo™ software. A representative gating strategy used to identify the cell populations is shown in Supplementary Figure 19. To assess antigen-specific cytokine secretion, lymph node cells were plated at 5 × 105 cells/well were incubated with 100 μg RBD for 3 d at 37 °C with 5% CO2. After 3 d, the supernatant was analyzed for cytokine concentration via a multiplexed mouse Th cytokine panel (BioLegend) according to the manufacturer’s instructions. Samples were aquired on an Attune NxT flow cytometer (ThermoFisher), and analyzed with LEGENDplex v8.0 software.
For long-term experiments, mice were not sacrificed but continued to be bled on day 2, 7, 14 and then every 2 weeks for 80 days.
RBD-binding ELISA
Plasma was assessed for anti-RBD IgG and IgA by ELISA. 96-well ELISA plates (Costar high-bind flat-bottom plates, Corning) were coated with 10 μg/mL RBD in carbonate buffer (50 mM sodium carbonate/sodium bicarbonate, pH 9.6) overnight at 4 °C. The following day, plates were washed three times in PBS with 0.05% Tween 20 (PBS-T) and then blocked with 1x casein (Sigma) for 1 h at RT. Following blocking, wells were washed three times with PBS-T and further incubated with six 10-fold dilutions of plasma in 1x casein for 2 h at RT. Wells were then washed five times with PBS-T and incubated for an additional hour at RT with horseradish peroxide (HRP)-conjugated antibodies against mouse IgG, IgG1, IgG2b, IgG3, or IgA (Southern Biotech). After five washes with PBS-T, bound RBD-specific Ig was detected with tetramethylbenzidine (TMB) substrate. Stop solution (3% H2SO4 + 1% HCl) was added after 18 min of TMB incubation at RT, and the OD was measured at 450 and 570 nm on an Epoch Microplate Spectrophotometer (BioTek). Background signal at 570 nm was subtracted from the OD at 450 nm. Fold-change over the average of blank wells was then calculated and log-transformed. The area under the curve (AUC) of log-transformed fold change versus log-transformed dilution was then calculated.
RBD-binding IgG ELISpot assay
ELISpot plates (Millipore IP Filter plate) were coated with 20 μg/mL RBD in sterile PBS overnight at 4 °C. Plates were then blocked using ELISpot Media (RPMI 1640, 1% glutamine, 10% fetal bovine serum, 1% penicillin-streptomycin) for 2 hours at 37 °C. Splenocytes from vaccinated mice were seeded in triplicate at a starting concentration of 6.75 × 105 cell/well and diluted seven times in 3-fold serial dilutions. Plates were incubated for 18 hours at 37 °C and 5% CO2 after which the cells were washed five times in PBS. Wells were incubated with 100 μL IgG-biotin HU adsorbed (Southern Biotech) for 2 h at RT. Next, plates were washed four times in PBS followed by the addition of 100 μL HRP-conjugated streptavidin/well for 1 h at RT. Plates were washed again and incubated with 100 μL TMB/well for 5 minutes until distinct spots emerged. Finally, plates are then washed three times with distilled water and left to dry completely in a laminar flow hood. A CTL ImmunoSpot Analyzer was used to image plates, count spots, and perform quality control.
SARS-CoV-2 virus neutralization assay
SARS-CoV-2 viruses (400 plaque forming units; strain nCov/Washington/1/2020, provided by the National Biocontainment Laboratory, Galveston TX, USA) were incubated with 4-fold serial dilutions of heat-inactivated plasma from vaccinated or control mice and for 1 h at 37 °C in DMEM with 2% fetal bovine serum, penicillin-streptomycin (100 U/mL penicillin, 100 μg/mL streptomycin), and non-essential amino acids (10 mM, glycine, L-alanine, L-asparagine, L-aspartic acid, L-glutamic acid, L-proline, L-serine; Gibco). The pre-incubated viruses were then applied to Vero-E6 cell monolayers, and the cells were maintained until > 90% cell death for the negative control (4–5 d). Cells were then washed with PBS, fixed with 10% formalin, stained with crystal violet, and quantified with a Tecan infinite m200 microplate reader (excitation/emission 592 nm/636 nm). Neutralization titer is measured as the greatest dilution that inhibits 50% of SARS-CoV-2 induced cell death (EC50). To determine the EC50, data were fit using a least squares variable slope four-parameter model. To ensure realistic EC50 values, we considered a dilution (1/X) of X = 10−1 to be 100% neutralizing and a dilution of X = 108 to be 0% neutralizing and constrained EC50 > 0. Plasma from convalescent human COVID-19 patients were provided by Ali Ellebedy (Washington University School of Medicine, St. Louis, MO; Catalog # NR-53661, NR-53662, NR-53663, NR-53664, and NR-53665).
Peptide array analysis
Antibody specificity to linear epitopes of the spike protein was analyzed using a CelluSpots™ Covid19_hullB Peptide Array (Intavis Peptide Services, Tubingen, Germany) according to the manufacturer’s protocol. The array comprises 254 peptides spanning the full-length sequence of the Spike protein (NCBI GenBank accession # QHD43416.1), with each 15-mer peptide offset from the previous one by 5 amino acids. Briefly, peptide arrays were blocked in 1x casein solution at 4 °C overnight. Arrays were then incubated with pooled serum diluted 1:200 in 1x casein for 6 h at RT on an orbital shaker (60 rpm). After 6 h, arrays were washed four times with PBS-T and incubated for an addition 2 h at RT, 60 rpm with goat anti-mouse IgG conjugated to HRP (Southern Biotech) diluted 1:5000 in 1x casein. Arrays were washed another four times with PBS-T. Spots were detected with Clarity™ Western ECL Substrate (Bio-Rad), and chemiluminescence was measured using a ChemiDoc XRS+ Gel Documentation System (Bio-Rad). Spots were analyzed using Spotfinder software (version v3.2.1).
Software packages and statistical analysis
Statistical analysis was performed using GraphPad Prism 9 (GraphPad). Data were analyzed using one-way ANOVA with Dunn’s or Tukey’s post-hoc correction for multiple hypothesis testing unless otherwise stated. All flow cytometry data were analyzed using FlowJo_v10.7.2 software (FlowJo LLC, BD Biosciences). Figures 1 and 2 were created using BioRender (https://biorender.com) as part of an Academic License through the Chicago Immunoengineering Innovation Center.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for funding support through a pilot project grant from the Chicago Immunoengineering Innovation Center of the University of Chicago, as well as the Chicago Biomedical Consortium COVID-19 Response Award (# CR-002) to M.A.S. In addition, a number of researchers were supported by fellowships via the NIH NHLBI (# T32-HL007605 to L.R.V.), Canadian Institutes of Health Research (#201910MFE-430736-73744 to N.M.), University of Chicago Comprehensive Cancer Center (Sigal Fellowship in Immuno-Oncology to N.M.), NIH NCI (# F30-CA221250 to M.R.S.), NIH NIGMS (# T32-GM007281 to M.R.S.) and NIH NIAID (# T32-AI007090 to T.M.M. and A.C.T.). We are grateful to the laboratory of Florian Krammer (Icahn School of Medicine at Mount Sinai, New York City, NY) for providing plasmids coding for the Spike RBD, produced with support from the NIH NIAID (Contract # HHSN272201400008C). We are also grateful to the groups of Jesse Bloom (Fred Hutchinson Cancer Research Center, Seattle, WA) and Ali Ellebedy (Washington University School of Medicine, St. Louis, MO) for contributing reagents via the NIH NIAID BEI Resources repository. We acknowledge helpful discussions with Patrick C. Wilson, Jenna J. Guthmiller, Anne I. Sperling, and Aaron Esser-Kahn (University of Chicago, Chicago, IL) and with Robert Baker and David Boltz (Illinois Institute of Technology Research Institute, Chicago, IL) that were instrumental to experimental planning and model development. We acknowledge Suzana Gomes and Tera Lavoie for technical assistance. Parts of this work were carried out at the Cytometry and Antibody Technology Core Facility (Cancer Center Support Grant P30CA014599), the Soft Matter Characterization Facility, the Mass Spectrometry Facility (NSF instrumentation grant CHE-1048528), the Nuclear Magnetic Resonance Facility, the Advanced Electron Microscopy Facility (RRID:SCR_019198), and the Human Immunologic Monitoring Facility (RRID:SCR_017916) at the University of Chicago.
Footnotes
COMPETING INTERESTS
M.A.S. and J.A.H. have patents related to the polymersome technology and interests in LantaBio, which has licensed those patents.
REFERENCES
- 1.Le T. et al. Evolution of the COVID-19 vaccine development landscape. Nature Review Drug Discovery 19, 667–668 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Kyriakidis N. C. et al. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines 6, 28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richmond P. et al. Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: a phase 1, randomised, double-blind, placebo-controlled trial. The Lancet 397, 682–694 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keech C. et al. Phase 1–2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. New England Journal of Medicine 383, 2320–2332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lan J. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Cao Y. et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 182, 73–84.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zost S. J. et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi R. et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Liu L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Sternberg A & Naujokat C.. Structural features of coronavirus SARS-CoV-2 spike protein: Targets for vaccination. Life Sciences 257, 118056 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai L. & Gao G. F. Viral targets for vaccines against COVID-19. Nature Reviews Immunology 21, 73–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Wang L., Cao H. & Liu C. SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen. Journal of Medical Virology 93, 892–898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irvine D. J. & Read B. J. Shaping humoral immunity to vaccines through antigen-displaying nanoparticles. Current Opinion in Immunology 65 1–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin M. D. et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nature Nanotechnology 15 646–655 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Singh A. Eliciting B cell immunity against infectious diseases using nanovaccines. Nature Nanotechnology 16, 16–24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kit Tan T. et al. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nature Communications 12, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y.-F. et al. Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. ACS Nano 15, 2738–2752 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Walls A. C. et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen A. A. et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 371, 735–741 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeyanathan M. et al. Immunological considerations for COVID-19 vaccine strategies. Nature Reviews Immunology 20, 615–632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox R. J. & Brokstad K. A. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nature Reviews Immunology 20, 581–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sette A. & Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 184 861–880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z. & John Wherry E. T cell responses in patients with COVID-19. Nature Reviews Immunology. 20 529–536 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekine T. et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 183, 158–168.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grifoni A. et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181, 1489–1501.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng Y. et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nature Immunology 21, 1336–1345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonifacius A. et al. COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses. Immunity 54, 340–354.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caserta S. et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol 11, 827 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen G. et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. Journal of Clinical Investigation 130, 2620–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altmann D. M. & Boyton R. J. SARS-CoV-2 T cell immunity: Specificity, function, durability, and role in protection. Science Immunology. 5 6160 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Napoli A., et al. Oxidation-responsive polymeric vesicles. Nature Materials 3, 183–189 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Scott E. A. et al. Dendritic cell activation and T cell priming with adjuvant- and antigen-loaded oxidation-sensitive polymersomes. Biomaterials 33, 6211–6219 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Cerritelli S. et al. Aggregation Behavior of Poly(ethylene glycol-bl-propylene sulfide) Di-and Triblock Copolymers in Aqueous Solution. Langmuir 25, 11328–11335 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Stano A., et al. Tunable T cell immunity towards a protein antigen using polymersomes vs. solid-core nanoparticles. Biomaterials 34, 4339–4346 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Galan-Navarro C. et al. Oxidation-sensitive polymersomes as vaccine nanocarriers enhance humoral responses against Lassa virus envelope glycoprotein. Virology 512, 161–171 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Zhu N. et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England Journal of Medicine 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens T. L. et al. Regulation of antibody isotype secretion by subsets of antigen-specific helper T cells. Nature 334, 255–258 (1988). [DOI] [PubMed] [Google Scholar]
- 39.Chao Y. X., Rötzschke O. & Tan E.-K. The role of IgA in COVID-19. Brain, Behavior, and Immunity 87, 182–183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renegar K. B., et al. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. The Journal of Immunology 173, 1978–1986 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Hinton D. M. Convalescent Plasma EUA Letter of Authorization. (March 9, 2021).
- 42.Yi C. et al. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cellular and Molecular Immunology 17, 621–630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang J. et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi Y. S. et al. ICOS Receptor Instructs T Follicular Helper Cell versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity 34, 932–946 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akkaya M., Kwak K. & Pierce S. K. B cell memory: building two walls of protection against pathogens. Nature Reviews Immunology 20, 229–238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toyama H. et al. Memory B Cells without Somatic HypermutationAre Generated from Bcl6-Deficient B Cells. Immunity 17, 329–339 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Mocellin S., Marincola F. M. & Young H. A. Interleukin-10 and the immune response against cancer: a counterpoint. Journal of Leukocyte Biology 78, 1043–1051 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Kishimoto T. Interleukin-6: discovery of a pleiotropic cytokine. Arthritis Research & Therapy 8, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cyster J. G. & Allen C. D. C. B Cell Responses: Cell Interaction Dynamics and Decisions. Cell 177 524–540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mesin L., Ersching J. & Victora G. D. Germinal Center B Cell Dynamics. Immunity 45 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohsen M. O. et al. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Seminars in Immunology 34, 123–132 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Kato Y. et al. Multifaceted Effects of Antigen Valency on B Cell Response Composition and Differentiation In Vivo. Immunity 53, 548–563.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh H. et al. Understanding the T cell immune response in SARS coronavirus infection. Emerging Microbes and Infections 1, 1–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin H.-S. et al. Immune Responses to Middle East Respiratory Syndrome Coronavirus During the Acute and Convalescent Phases of Human Infection. Clinical Infectious Diseases 68, 984–992 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yasui F. et al. Prior Immunization with Severe Acute Respiratory Syndrome (SARS)-Associated Coronavirus (SARS-CoV) Nucleocapsid Protein Causes Severe Pneumonia in Mice Infected with SARS-CoV. The Journal of Immunology 181, 6337–6348 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Liu L. et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Casella C. R. & Mitchell T. C. Putting endotoxin to work for us: Monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cellular and Molecular Life Sciences 65, 3231–3240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sallusto F. et al. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. Journal of Experimental Medicine 182, 389–400 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leclerc Víctor C. et al. Endosome-to-Cytosol Pathway Virus-Like Particles Using an In Vivo, Dendritic Cells Can Cross-Present. Journal of Immunology References 171, 2242–2250 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Bachmann M. F. & Jennings G. T. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nature Reviews Immunology 10, 787–796 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Gamvrellis A. et al. Vaccines that facilitate antigen entry into dendritic cells. Immunology and Cell Biology 82 506–516 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Quiroga M. F. et al. Features of effective T Cell-inducing vaccines against chronic viral infections. Frontiers in Immunology 9, 276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amanat F. et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nature Medicine 26, 1033–1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allen S., Osorio O., Liu Y. G. & Scott E. Facile assembly and loading of theranostic polymersomes via multi-impingement flash nanoprecipitation. Journal of Controlled Release 262, 91–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamdy S., Haddadi A., Somayaji V., Ruan D. & Samuel J. Pharmaceutical analysis of synthetic lipid A-based vaccine adjuvants in poly (d,l-lactic-co-glycolic acid) nanoparticle formulations. Journal of Pharmaceutical and Biomedical Analysis 44, 914–923 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Lutz M. B. et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. Journal of Immunological Methods 223, 77–92 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.