Abstract
Curcuma longa, C. wenyujin and C. phaeocaulis are important herbal medicine which of rhizomatous herbaceous perennial plant of the family Zingiberaceae. This study generated a complete chloroplast genome sequence of three medicinal species were characterized by de novo assembly with whole genome sequencing data. The length of complete chloroplast genome were 162,180 bp (C. longa), 162,266 bp (C. wenyujin), and 162,133 bp (C. phaeocaulis), respectively, with four structures that were included in large single copy region (87,001 bp, 87,042 bp, and 87,013 bp), small single copy region (15,681 bp, 15,710 bp, and 15,622 bp), and duplicated inverted regions (29,749 bp, 29,757 bp and 29,749 bp of each). Based on phylogenetic trees, C. longa, C. wenyujin, and C. phaeocaulis were grouped by high bootstrap value with Curcuma species. This result approved that C. longa, C. wenyujin and C. phaeocaulis were comprised in Alpinia and Wurfbainia. Therefore, this chloroplast genome data firstly generated valuable genetic resource in discrimination of herbal materials, phylogeny and development DNA marker.
Keywords: Chloroplast, genome sequencing, C. longa, C. wenyujin, C. phaeocaulis
Curcuma longa, C. wenyujin, and C. phaeocaulis are perennial plants belonging to the family Zingeraceae. The genus Curcuma consists of about 50 species in Southeast Asia, of which more than 12 species are found in China (Wu and Larsen 2000), widely cultivated in the subtropical regions of the world (Li et al. 2011).
These plants part of rhizoma have commonly used as spice, natural food dye, and medicine (Curcumae Longae Rhizoma and Zeodariae Rhizoma), widely used in Asia (Sigrist et al. 2011). In Ayurveda medicine, it is used as a treatment for inflammatory conditions; and, in traditional medicine, it is used as stimulant, aspirant, carminative, cordeal, emenagogue, astringent, detergent, and diuretic effects (Sasikumar 2005; Remadevi et al. 2007; Jurenka 2009). In this study, we determined the complete chloroplast genome sequences of three medicinal species, C. longa, C. wenyujin, and C. phaeocaulis. These sequences will be a valuable genetic resource for further molecular study and phylogenetic analysis of Curcuma species with other species in the family Zingeraceae.
The plant sample of C. longa, C. wenyujin, and C. phaeocaulis were supplied from National Institute of Horticultural and Herbal Science in Korea (Eumseong-gun, Chungcheongbuk-do, 36°56’39.6”N 127°45’17.3”E 36.944327, 127.754793). A voucher specimen (C. longa; CUR16, C. wenyujin; CUR09, and C. phaeocaulis; CUR16) was deposited at the herbarium, in the National Center of Herbal Medicinal Resources at OK-chen (National Institute of Food and Drung Safety Evaluation, Korea).
The genomic DNA was extracted from dried leaf were stored at −70 °C refrigerator in the laboratory of the NIFDS (National Institute of Food and Drug Safety Evaluation), Herbal Medicine Research Division and assembled to an Illumina paired-end library. Genomic sequencing was performed using Illumina Hiseq 2000 instrument (Illumina, San Diego, CA) and assembled by CLC genomic assembler (v. beta 4.6, CLC Inc., Rarhus, Denmark).
The total size of the assembled chloroplast genome of C. longa, C. wenyujin, and C. phaeocaulis were 162,180 bp, 162,266 bp, and 162,133 bp, respectively (accession No. MK109018, MK109019, and MK109020), and genome reads were 215 X of average coverage. The three medicinal plant genome structure typically consisted of four parts that were large single copy region (LSC) of 87,001 bp, 87,042 bp, and 87,013 bp, small single copy region (SSC) of 15,681 bp, 15,710 bp, and 15,622 bp, and a pair of inverted regions (IRa and IRb) of 29,749 bp, 29,757 bp, and 29,749 bp each. In genome, a total 114 coding regions, which were comprised 80 protein-coding regions, four rRNA genes, and 30 tRNA genes, were predicted through OGDraw (http://ogdraw.mpimp-golm.mpg.de, Lohse et al. 2007) and manual curation using BLAST.
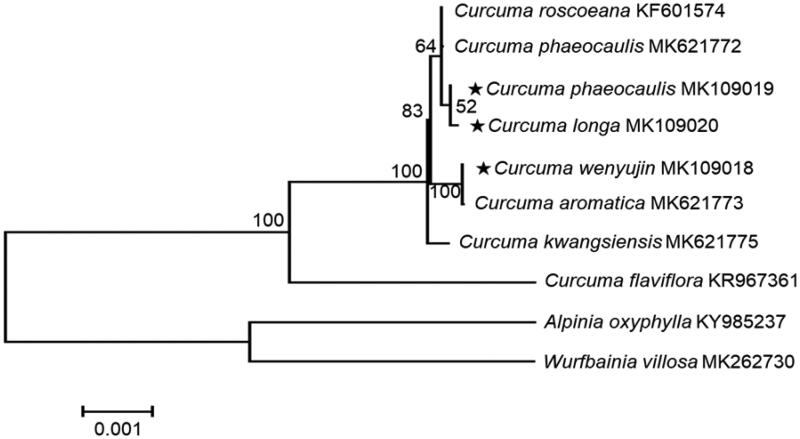
To determine the taxonomic status of C. longa, C. wenyujin, and C. phaeocaulis, phylogenetic analyses based on phylogenetic tree were performed from genome sequences of ten taxa, consisting of genus Curcuma, Alpinia, and Wurfbainia. These sequences were aligned using MAFFT (http://mafft.cbrc.jp/alignment/software/, Katoh and Standley 2013). The Phylogenetic relationship was constructed by eight species, and 1000 bootstrap replications in MEGA6 (Tamura et al. 2013). Phylogenetic analysis was conducted with chloroplast genome sequences of Curcuma longa, C. wenyujin, and C. phaeocaulis with eight species belonging to the family Zingeraceae (Figure 1). This chloroplast genome data could be supported to discriminate between three similar medicinal species for herbal medicine and adulterations, analyze the relation of closely related taxa, and study phylogeny and identification marker.
Figure 1.
Maximum-likelihood analysis of C. longa, C. wenyujin, and C. phaeocaulis with related species in Zingiberaceae based on complete chloroplast genome sequences. Numbers on branch indicated bootstrap values.
Funding Statement
This research was supported by a grant [16171MFDS228] from the Ministry of Food and Drug Safety of the Republic of Korea in 2018.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI, National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov/nuccore/MK109019.1/, https://www.ncbi.nlm.nih.gov/nuccore/MK109020.1/, and https://www.ncbi.nlm.nih.gov/nuccore/MK109018.1/, reference number [accession No. MK109018, MK109019, and MK109020].
References
- Jurenka S. 2009. Anti-inflammatory properties of curcumin, a major constituent of Curcuma longa: a review of preclinical and clinical research. Altern Med Rev. 14:141–153. [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yuan W, Deng G, Wang P, Yang P, Aggarwal B.. 2011. Chemical composition and product quality control of turmeric (Curcuma longa L.). TOPHARMCJ. 5(1):28–54. [Google Scholar]
- Lohse M, Drechsel O, Bock R.. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274. [DOI] [PubMed] [Google Scholar]
- Remadevi R, Surendran E, Kimura T, 2007. Turmeric in traditional medicine. In: Ravindran PN, Nirmal Babu K, Sivaraman K, editors. Turmeric: the genus Curcuma. Boca Raton, London, New York: CRC Press; p. 409–436. [Google Scholar]
- Sasikumar B. 2005. Genetic resources of Curcuma: diversity, characterization and utilization. Plant Genet Resour. 3(2):230–251. [Google Scholar]
- Sigrist MS, Pinheiro JB, Filho JAA, Zucchi MI.. 2011. Genetic diversity of turmeric germplasm (Curcuma longa; Zingiberaceae) identified by microsatellite markers. Genet Mol Res. 10(1):419–428. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Larsen K.. 2000. Curcuma Linnaeus. In: Wu ZY, Raven PH, editors. Flora of China. Beijing: Science Press; p. 359. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in NCBI, National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov/nuccore/MK109019.1/, https://www.ncbi.nlm.nih.gov/nuccore/MK109020.1/, and https://www.ncbi.nlm.nih.gov/nuccore/MK109018.1/, reference number [accession No. MK109018, MK109019, and MK109020].