To the Editor,
We read with great interest the paper by Kashiwagi et al. [1], showing that 7/16 saliva samples resulted positive by RT-PCR test using N2 probe according to the manual provided by Japan National Institute of Infectious Diseases; among these, only 4 showed the presence of SARS-CoV-2 antigen using the ESPLINE® SARS-CoV-2 (Fujirebio Inc., Tokyo) [2]. The Authors infer that the sensitivity of the antigen test depends on the RNA-copy concentrations and is lower than that previously reported [[3], [4], [5]].
We report our experience on the ESPLINE® SARS-CoV-2 antigen test that was obtained on a higher number of saliva samples and, indeed, the results obtained in our laboratory indicate for this sample matrix a clinical sensitivity even lower than that reported in this paper.
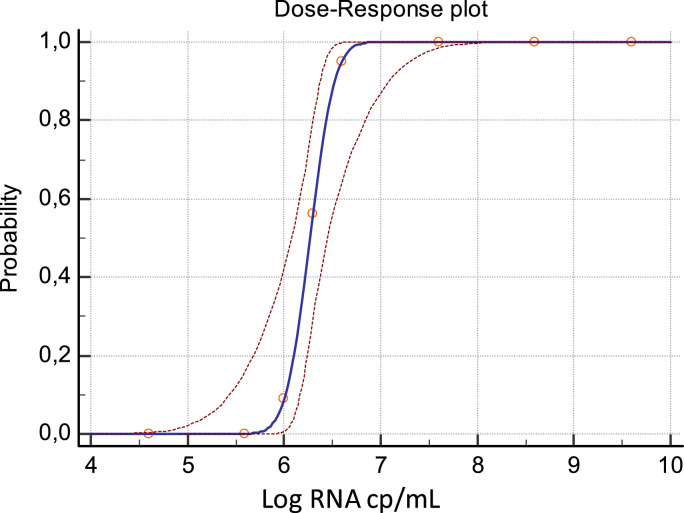
We first evaluated the analytical sensitivity of ESPLINE® SARS-CoV-2 assay by using a pool of fresh saliva samples, collected by passive drooling from healthy donors and spiked with known concentrations of 2019-nCoV/Italy-INMI1 isolate [5,6]. Data obtained from multiple replicates of serial dilutions of the isolate were used to calculate the low limit of detection (LOD) of the assay by Probit analysis using the MedCalc statistical software (MedCalc Software Ltd, Ostend, Belgium). The LOD resulting from this analysis was 2.99 (CI: 2.83–3.69) TCID50/mL, corresponding to 6.60 (CI 6.43–7.29) Log RNA cp/mL (Fig. 1 ).
Fig. 1.
Probit analysis of ESPLINE® SARS-CoV-2 assay applied to saliva samples spiked with 2019-nCoV/Italy-INMI1 isolate. Results are expressed as Log RNA cp/mL.
The clinical sensitivity of the ESPLINE® SARS-CoV-2 antigen assay was evaluated on 136 saliva samples from patients admitted to the National Institute for Infectious Diseases “L. Spallanzani” (INMI) in Rome with suspected COVID-19 infection. The Simplexa™ COVID-19 Direct assay was used as reference molecular test [5]. Among the 136 analyzed samples, 62 resulted positive for SARS-CoV-2 RNA with Simplexa™ COVID-19 Direct assay, and only 5 of the latter were also positive for the presence of antigen, thus showing for the antigen test a sensitivity of 8.1% and a specificity of 100.0%, with slight agreement between the two assays (κ = 0.087; 95% CI = 0.013–0.161), (Table 1 ). All 62 samples resulted positive by molecular assay were from symptomatic patients.
Table 1.
Comparison of ESPLINE® SARS-CoV-2 data vs molecular reference test (Simplexa™ COVID-19 Direct assay) on saliva samples.
| ESPLINE® SARS-CoV-2 |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Simplexa™ COVID-19 Direct | Positive | 5 | 57 | 62 |
| Negative | 0 | 74 | 74 | |
| Total | 5 | 131 | 136 | |
| Proportion# | Percentage (95% CI) | |||
| Sensitivity | 5/62 | 8.1% (2.7% - 17.8%) | ||
| Specificity vs RT-PCR reference test | 74/74 | 100.0% (95.1% - 100.0%) | ||
However, when stratifying samples into groups based on ranges of RT-PCR cycle threshold (Ct, an indirect indicator of viral RNA concentration), the antigen-positives samples were mostly associated with low Ct values, therefore high viral loads (Table 2 ).
Table 2.
Percentage of positivity of saliva samples according to the Ct range of the molecular test.
| Ct Ranges | Ag positive samples N°/total positive PCR | Positivity % with ESPLINE® SARS-CoV-2 |
|---|---|---|
| <20 | 3/4 | 75% |
| 20–25 | 1/14 | 7.14% |
| 25,01-30 | 1/23 | 4.35% |
| >30 | 0/21 | 0.0% |
In our study saliva specimens were collected with the same method used in the study by Kashiwagi et al. but the clinical evaluation was performed on a larger number of samples. In addition, we compared the ESPLINE® SARS-CoV-2 results with Simplexa™ COVID-19 Direct assay, which is the only molecular assay CE licensed for the use of these specimens to our knowledge, thus obtaining a very lower sensitivity 8.1%.
It is to be underlined that saliva is a complex matrix, as it is prone to relevant individual differences in viscosity and other factors, such as pH, presence of spurious materials, etc., which could influence immunochromatographic migration, making this sample problematic for carrying out the test on the reaction cassettes, generating less regular antigenic results than those, observed on swab, if measures are not taken to alleviate these drawbacks.
Funding
This research was supported by funds to National Institute for Infectious Diseases ‘Lazzaro Spallanzani’ IRCCS from Ministero della Salute (Ricerca Corrente, linea 1; COVID-2020-12371817), the European Commission – Horizon 2020 (EU project 101003544 – CoNVat; EU project 101003551 – EXSCALATE4CoV; EU project 101005111-DECISION; EU project 101005075-KRONO) and the European Virus Archive – GLOBAL (grants no. 653316 and no. 871029).
Declaration of competing interest
The authors report no declarations of interest.
References
- 1.Kashiwagi K., Ishii Y., Aoki K., Yagi S., Maeda T., Miyazaki T., et al. Immunochromatographic test for the detection of SARS-CoV-2 in saliva. J Infect Chemother. 2021 Feb;27(2):384–386. doi: 10.1016/j.jiac.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamakawa K., Fujimoto A., Miyamoto K., Ohshima T., Suzuki T., Nagata N. Development of rapid lmmunochromatographic enzyme lmmunoassay for SARS-CoV-2 nucleocapsid protein. Jpn J Med Pharm Sci. 2020;77:937–944. [In Japanese)] [Google Scholar]
- 3.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. Medrxiv. 2020 doi: 10.1101/2020.04.16.20067835. 2020. [DOI] [Google Scholar]
- 4.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/jcm.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordi L., Sberna G., Lalle E., Piselli P., Colavita F., Nicastri E., et al. Frequency and duration of SARS-CoV-2 shedding in oral fluid samples assessed by a modified commercial rapid molecular assay. Viruses. 2020;12(10):1184. doi: 10.3390/v12101184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capobianchi M.R., Rueca M., Messina F., Giombini E., Carletti F., Colavita F., et al. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clin Microbiol Infect. 2020;26:954–956. doi: 10.1016/j.cmi.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]