Abstract
Efforts are being made worldwide to understand the immune response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for the coronavirus disease 2019 (COVID-19) pandemic, including the impact of T cell immunity and cross-recognition with seasonal coronaviruses. Screening of SARS-CoV-2 peptide pools revealed that the nucleocapsid (N) protein induced an immunodominant response in HLA-B7+ COVID-19-recovered individuals that was also detectable in unexposed donors. A single N-encoded epitope that was highly conserved across circulating coronaviruses drove this immunodominant response. In vitro peptide stimulation and crystal structure analyses revealed T cell-mediated cross-reactivity toward circulating OC43 and HKU-1 betacoronaviruses but not 229E or NL63 alphacoronaviruses because of different peptide conformations. T cell receptor (TCR) sequencing indicated that cross-reactivity was driven by private TCR repertoires with a bias for TRBV27 and a long CDR3β loop. Our findings demonstrate the basis of selective T cell cross-reactivity for an immunodominant SARS-CoV-2 epitope and its homologs from seasonal coronaviruses, suggesting long-lasting protective immunity.
Keywords: SARS-CoV-2, HLA, immunodominant epitope, CD8+ T cell, immune response, cross-reactivity, seasonal coronaviruses
Graphical abstract
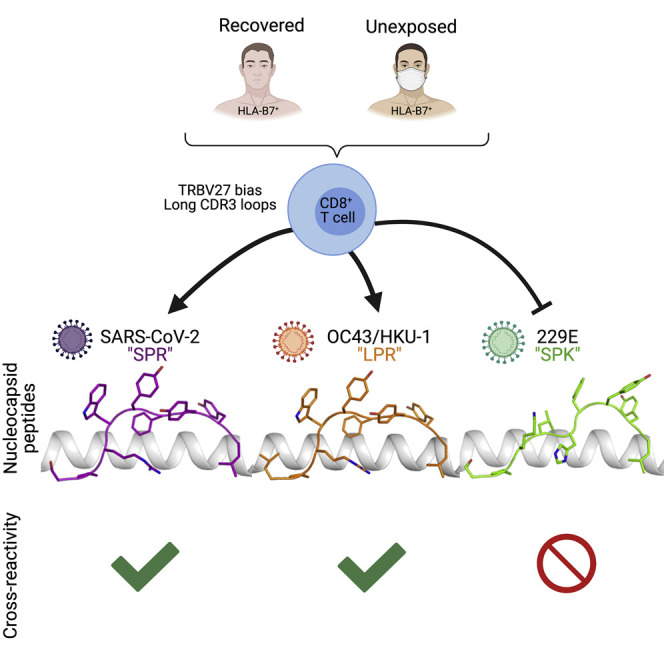
The impact of seasonal coronaviruses on immune responses to SARS-CoV-2 is an active area of research. Lineburg et al. identify CD8+ T cells specific for a conserved and immunodominant SARS-CoV-2 epitope in HLA-B7+ individuals. Furthermore, SARS-CoV-2 epitope-specific CD8+ T cells display cross-reactivity to beta- but not alphacoronaviruses because of distinct peptide-HLA conformations.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an emerging virus responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic. Over 97 million individuals have been infected and over 2 million individuals have succumbed to infection (Dong et al., 2020). Although some vaccines against SARS-CoV-2 are already being administered, and others are in development, many questions remain regarding the immune response toward this virus. Cytotoxic CD8+ T cells are key players in the immune response to viral infections because they participate directly in viral clearance. Among the 26 viral proteins of SARS-CoV-2, some surface proteins, such as the spike protein (S), are more variable, whereas others are internal and more conserved, such as the nucleocapsid protein (N). The sequence conservation of non-surface proteins makes them ideal vaccine targets for activating cytotoxic CD8+ T cells. CD8+ T cells recognize small peptides (typically 8–10 amino acids) together with human leukocyte antigen (HLA) molecule with varying affinity. Although HLA-A2, the most prevalent HLA molecule (∼40% frequency worldwide; Ellis et al., 2000), can present SARS-CoV-2 N-derived peptides (Szeto et al., 2021), they are only weakly immunogenic (Habel et al., 2020). However, it remains unclear whether this is a characteristic specific to the selected N-derived peptides because HLA-A2+ individuals also demonstrate a strong CD8+ T cell response to an S-derived epitope (Shomuradova et al., 2020). Therefore, it is imperative to identify and characterize novel immunogenic CD8+ T cell epitopes against SARS-CoV-2.
Another important aspect of understanding the immune response to this new coronavirus is to understand the properties of protective pre-existing immunity at a population level (Karlsson et al., 2020). It has been proposed that pre-existing immune memory may be generated by previous infection with seasonal coronaviruses (OC43, 229E, NL63, and HKU-1) (Mateus et al., 2020; Ng et al., 2020). However, the identities of the T cell epitopes that give rise to pre-existing immunity are only starting to emerge (Bacher et al., 2020; Braun et al., 2020; Karlsson et al., 2020; Le Bert et al., 2020; Mateus et al., 2020; Nelde et al., 2021; Schulien et al., 2021; Sette and Crotty, 2020; Shomuradova et al., 2020). The SARS-CoV-2 N protein sequence is 90.3% identical to the SARS-CoV-1 N protein but shares only 29% identity with the N proteins of OC43 and HKU-1 and 23% with the NL63 and 229E virus strains. The majority of this sequence homology occurs in the N-terminal domain of the protein, which also contains the immunogenic N105–113 peptide (SPRWYFYYL, hereafter referred to as SPR) (Ferretti et al., 2020; Kared et al., 2020; Peng et al., 2020; Schulien et al., 2021; Sekine et al., 2020; Snyder et al., 2020). The SPR peptide sequence is identical in SARS-CoV-2 and SARS-CoV-1; it differs by only one residue in OC43 and HKU-1 (LPRWYFYYL, hereafter referred to as LPR), three residues in 229E (SPKLHFYYL, hereafter referred to as SPK), and four in NL63 viruses (PPKVHFYYL, hereafter referred to as PPK).
Here we demonstrated that HLA-B7+ individuals who had recently recovered from COVID-19 exhibited a dominant CD8+ T cell response that was highly specific to the SPR epitope of the SARS-CoV-2 N protein. In addition, we identified SPR-specific CD8+ T cells in multiple unexposed HLA-B7+ individuals that showed cross-reactivity with the homologous LPR peptide from the OC43 and HKU-1 strains. However, this same cross-recognition did not extend to the similarly homologous SPK and PPK peptides from the 229E and NL63 viruses, respectively. Using the structural landscape of HLA-B7 bound to SPR and its variants, we delineate the molecular basis for selective T cell cross-reactivity toward specific seasonal coronaviruses.
Results
SARS-CoV-2 N is highly immunogenic in HLA-B7+ COVID-19-recovered individuals
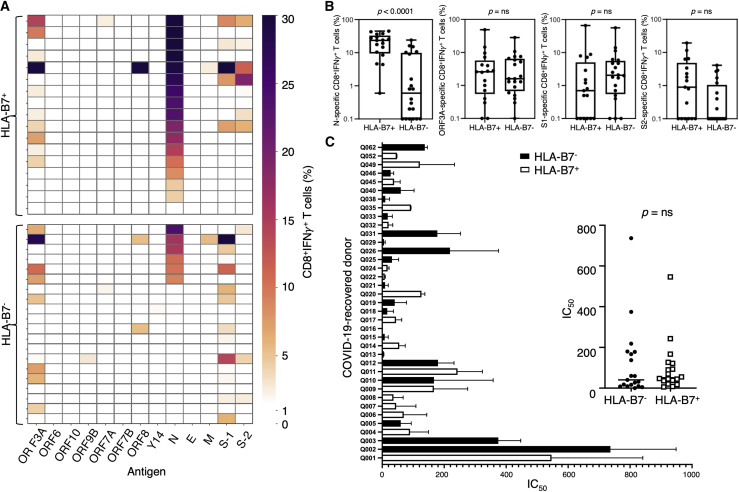
Although many CD8+ T cell studies have focused on different proteins or a limited number of selected peptides (Habel et al., 2020; Kared et al., 2020; Sekine et al., 2020; Shomuradova et al., 2020; Szeto et al., 2021), the specific SARS-CoV-2 proteins that consistently elicit the strongest CD8+ T cell responses remain ill defined (Ferretti et al., 2020; Grifoni et al., 2020; Nelde et al., 2021; Peng et al., 2020). To assess this, we used peripheral blood mononuclear cells (PBMCs) derived from COVID-19-recovered individuals (n = 37) to screen 13 overlapping peptide pools spanning 12 SARS-CoV-2 proteins (Figure 1 ; Tables S1 and S2). Although all 37 COVID-19-recovered donors displayed immunogenicity to SARS-CoV-2 peptide stimulation, 70% of donors or more demonstrated a CD8+interferon γ (IFN-γ)+ response of 5% or higher when stimulated with ORF3A, N, S-1, or S-2 peptide pools (26 of 37). The most consistent CD8+ T cell responses were generated against the N protein-derived peptide pool in conjunction with expression of HLA-B7 (Figure 1A). The majority (76%) of these HLA-B7+ individuals demonstrated a dominant CD8+ T cell response with an average IFN-γ+ frequency of 10% or higher (13 of 17). Conversely, only 25% of HLA-B7– individuals displayed comparable responses (5 of 20). Significant differences in CD8+ T cell responses between HLA-B7+ and HLA-B7– individuals were only observed in response to the N protein (Figure 1B; p < 0.0001).
Figure 1.
Immunodominant response to N in HLA-B7+ COVID-19-recovered donors
PBMCs from COVID-19-recovered donors were stimulated with overlapping peptides from SARS-CoV-2 antigens, cultured for 2 weeks in the presence of IL-2, and then assessed for IFN-γ production following recall with the cognate pool.
(A) Heatmap representing the frequency of CD8+ IFN-γ-producing cells responding to each antigen from a total of 37 COVID-19-recovered donors (n = 17 HLA-B7+ and n = 20 HLA-B7− donors).
(B) Comparative analysis of T cell responses in HLA-B7+ and HLA-B7− donors in response to different antigens. Data are represented as box-and-whisker plots displaying the median, including minimum to maximum range. p values were calculated using a Mann-Whitney test.
(C) Serum samples were serially diluted and tested for neutralizing activity against SARS-CoV-2. Neutralization curves were generated, and the respective inhibitory concentration at 50% (IC50) was analyzed using a “log (inhibitor) versus normalized response” equation (GraphPad). IC50 values of study subjects were graphed individually (left) and grouped according to their HLA-B7 expression profile (right).
See also Figures S1 and S6 and Tables S1 and S2.
To assess whether CD8+ T cell reactivity to the N protein was associated with the presence of elevated antibody titers, we examined neutralizing anti-S immunoglobulin G (IgG) titers in COVID-19-recovered individuals and did not observe any significant differences between HLA-B7+ and HLA-B7– donors (n = 37) (Figure 1C). Similarly, the strong CD8+ T cell response was comparable between males (n = 14) and females (n = 23) and similar within donors categorized as younger (n = 18) or older (n = 19) than 50 years. The distribution of disease severity reported was comparable between HLA-B7+ and HLA-B7– cohorts (Table S2).
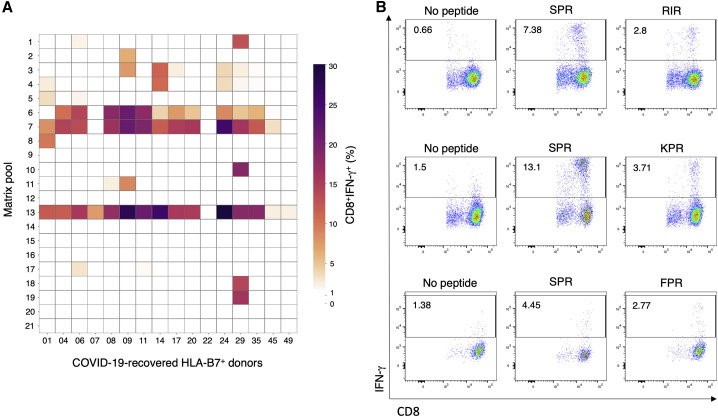
To define the immunogenic peptide(s) responsible for this HLA-B7+ restricted N-specific CD8+ T cell response, we used a matrix of peptide pools to identify which specific peptide elicited the strongest IFN-γ response. Through this process, we identified an immunogenic hotspot (Figure 2 ) corresponding to the same N protein region that contains the known immunogenic HLA-B7-restricted SPR (N105–113) epitope (Table S3; Ferretti et al., 2020; Kared et al., 2020; Schulien et al., 2021; Sekine et al., 2020; Snyder et al., 2020). Eighty percent of HLA-B7+ COVID-19-recovered donors showed a strong CD8+ T cell response to this peptide (14 of 17), consistent with other reports (Ferretti et al., 2020; Kared et al., 2020; Schulien et al., 2021; Sekine et al., 2020; Snyder et al., 2020). Three additional subdominant HLA-B7-restricted epitopes from the N protein were identified: RIRGGDGKM (RIR; N93–101), FPRGQGVPI (FPR; N66–74), and KPRQKRTAT (KPR; N257–265) (Figure 2B; Table S3). Although FPR and KPR are known to be immunogenic in COVID-19-recovered individuals (Schulien et al., 2021), RIR has been predicted but not yet described as an immunogenic SARS-CoV-2 CD8+ T cell epitope (Sekine et al., 2020).
Figure 2.
Peptide matrix analysis of T cell responses to N in COVID-19-recovered donors
(A) T cell cultures from affected individuals showing reactivity to the nucleocapsid (N) were stimulated with matrix pools covering the entire N protein and assessed for cytokine production by ICS. Shown is a heatmap representing the frequency of CD8+ IFN-γ-producing cells responding to each matrix pool from a total of 16 HLA-B7+ COVID-19-recovered donors (the Q052 donor did not show an N protein response and was excluded).
(B) Representative flow plots displaying COVID-19-recovered donors’ CD8+ IFN-γ responses to the SPR, RIR, KPR, and FPR peptides or the no-peptide control.
These data show that the SARS-CoV-2 N protein contains multiple immunogenic epitopes that induce a strong CD8+ T cell response in COVID-19-recovered HLA-B7+ individuals and that the dominant CD8+ T cell response is directed against the SPR peptide.
CD8+ T cells exhibit cross-reactivity between SARS-CoV-2- and OC43/HKU-1-derived N peptides
Given that this dominant SPR-specific CD8+ T cell response has been observed in HLA-B7+ individuals (Ferretti et al., 2020; Kared et al., 2020; Schulien et al., 2021; Sekine et al., 2020; Snyder et al., 2020) and that pre-existing immunity toward this peptide has been suggested for unexposed individuals (Ferretti et al., 2020; Nelde et al., 2021; Schulien et al., 2021), we next sought to identify the basis of the T cell cross-reactivity that elicits the cross-recognition observed against SARS-CoV-2. Sequence alignment comparing SARS-CoV-2 with other coronaviruses demonstrated that, although the entire N protein sequence homology is below 30%, the region containing the SPR peptide is more conserved, displaying 55%–89% sequence homology (Table 1 ). The homologous peptides from SARS-CoV-1, OC43, HKU-1, 229E, and NL63 share a proline at position 2 (P2-P) and a leucine at position 9 (P9-L). These residues are critical anchors for HLA-B7 binding (Sette and Sidney, 1999) and suggest that SPR peptide homologs from these seasonal viral strains might bind to the HLA-B7 molecule. Alignment of 26,158 N protein sequences from SARS-CoV-2 revealed a 100% conservation of the SPR peptide within circulating isolates, including the variants isolated in the United Kingdom (B.1.1.7), South Africa (B.1.351), and Brazil (P.1) (Table S4). In addition, circulating isolates of the homologous peptides (LPR, SPK, and PPK) are entirely conserved within their respective viruses (Table S4), demonstrating their global conservation.
Table 1.
Coronaviruses peptide homologs to SARS-CoV-2 SPR
| Virus | Peptide Sequence | Peptide Name | Peptide Sequence Identity (%)a | N Protein Sequence Identity (%)a | Tm (°C) |
|---|---|---|---|---|---|
| SARS-CoV-2 | SPRWYFYYL | SPR | – | – | 62.8 ± 0.7 |
| SARS-CoV-1 | SPRWYFYYL | SPR | 100 | 90.3 | 62.8 ± 0.7 |
| OC43 | LPRWYFYYL | LPR | 88.9 | 29.2 | 59.8 ± 0.7 |
| HKU-1 | LPRWYFYYL | LPR | 88.9 | 29.6 | 59.8 ± 0.7 |
| 229E | SPKLHFYYL | SPK | 66.7 | 23.2 | 55.7 ± 0.8 |
| NL63 | PPKVHFYYL | PPK | 55.5 | 24.9 | 48.2 ± 1.2 |
Mutations from the SARS-CoV-2 peptide are denoted in italic, and anchor residues are underlined. Tm is determined at 50% of its normalized fluorescence intensity and indicative of the temperature required to unfold 50% of the protein. The Tm value is represented as the mean ± SEM of n = 2.
Sequence identity with SARS-CoV-2-derived SPR peptide.
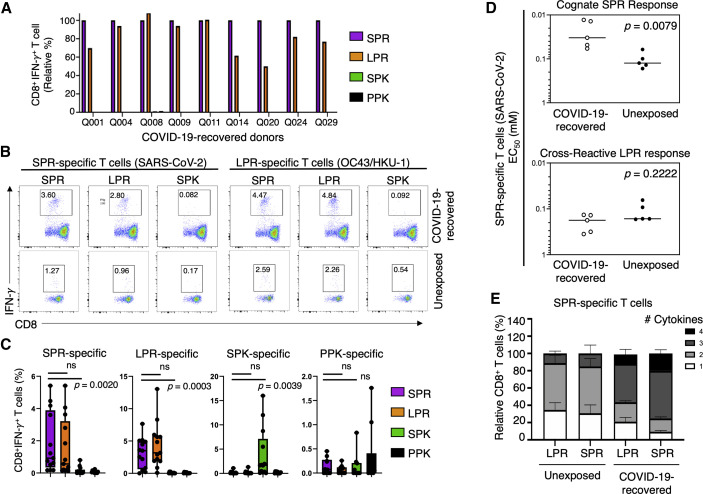
Considering the high degree of conservation between SPR and these homologous peptides (Table 1), we investigated whether SARS-CoV-2-specific CD8+ T cells were able to cross-recognize any of the other conserved homologous peptides. We first expanded SPR-specific T cells from PBMCs isolated from COVID-19-recovered individuals (Figure 3 ). Following re-stimulation of the SPR-specific CD8+ T cells with the SPR peptide, a high level of specificity for the SPR peptide was detected (Figure 3A), with an average CD8+IFN-γ+ response of 23.13%. The same CD8+ T cells were able to cross-react when restimulated with the LPR peptide (Figure 3A), resulting in a similar CD8+IFN-γ+ T cell response of 18.86%. Conversely, we observed little to no reactivity when the SPR-specific CD8+ T cells were restimulated with the SPK or PPK peptide (Figure 3A; Figure S1A).
Figure 3.
CD8+ T cell responses toward N peptides derived from seasonal and pandemic coronaviruses
PBMCs from unexposed and COVID-19-recovered donors were stimulated with SPR (SARS-CoV-2), LPR (OC43/HKU-1 betacoronavirus), SPK (229E alphacoronavirus), or PPK (NL63 alphacoronavirus) peptides, cultured for 2 weeks in the presence of IL-2, and then assessed for IFN-γ production following recall with their cognate peptide or each of the homologous peptides listed above.
(A) CD8+IFN-γ+ responses of SPR-specific T cells expanded from the PBMCs of COVID-19-recovered donors following recall stimulation with the SPR, LPR, SPK, or PPK peptide in an ICS.
(B) Representative flow plots of CD8+IFN-γ+ responses in SPR-specific and LPR-specific T cells expanded from COVID-19-recovered and unexposed donors’ PBMCs, displaying cross-recognition between SPR and LPR peptides following cognate peptide stimulation.
(C) Graph displaying the frequency of CD8+IFN-γ+ T cell responses in SPR, LPR, SPK, and PPK-specific T cells following recall stimulation on day 14 of culture. Each peptide-specific T cell line was re-stimulated individually with its cognate peptide or one of the homologous peptides (SPR, LPR, SPK, or PPK), and their IFN-γ+ response was measured by ICS. Data are represented as median, displaying minimum to maximum range. p values were calculated using a Mann-Whitney test.
(D) Graphs displaying the avidity at the effective concentration used to induce half maximal response (EC50) of SPR-specific T cells in response to cognate SPR peptide and cross-presentation of the LPR peptide, with the median response indicated by a line. Statistical analysis was performed using a Mann-Whitney test, comparing COVID-19-recovered (n = 5) and unexposed (n = 5) donors.
(E) Graph displaying the polyfunctionality of CD8+ SPR-specific T cells from unexposed and COVID-19-recovered donors following re-stimulation with SPR or LPR peptide. Data are represented as relative frequency (percent) of total CD8+ T cells.
See also Figures S1 and S2 and Tables S1 and S2.
Because it was evident that CD8+ T cells were able to cross-recognize the variant peptides from selected seasonal coronaviruses, we next assessed the level of cross-reactive CD8+ T cells in SARS-CoV-2-unexposed individuals. We expanded peptide-specific CD8+ T cells against each of the four peptide variants using PBMCs from unexposed donors (n = 13 for SPR, LPR, and SPK lines and n = 10 for PPK lines). After 10–14 days of expansion, we re-stimulated with each of the individual peptide variants and assessed the ability of CD8+ T cells to produce IFN-γ using an intracellular cytokine staining (ICS) assay (Figures 3B and 3C; Figure S1). Peptide-specific CD8+ T cells generated against SPR, LPR, and SPK peptides were most specific for their cognate peptide (Figure 3C; Figure S1); however, T cells generated against the PPK peptide demonstrated limited specificity (Figure 3C; Figure S1E). Peptide-specific CD8+ T cells generated in response to SPR or LPR peptide stimulation displayed cross-reactivity in the majority of unexposed donors without a significant difference between the two peptides (Figure 3C). Conversely, those generated in response to SPK or PPK stimulation displayed little to no cross-reactivity. Functional avidity analysis confirmed comparable recognition of SPR and LPR peptides by T cells generated from unexposed individuals; it also indicated that SPR-specific T cells generated by COVID-19-recovered donors display significantly higher avidity (Figure 3D; Figure S2A), similar to that reported in CD4+ T cells (Bacher et al., 2020). A higher magnitude of SPR-specific CD8+ T cells was observed from COVID-19-recovered donors (median, 16%) than from unexposed donors (median, 0.4%) (Figure S2B).
To assess the polyfunctionality of activated CD8+ T cells in response to peptide recall, we measured the production of IFN-γ, tumor necrosis factor (TNF), and interleukin-2 (IL-2) as well as CD107a expression. SPR-specific CD8+ T cells from COVID-19-recovered individuals exhibited broader polyfunctionality than those from unexposed donors, with the majority expressing three or four functions (IFN-γ, TNF, CD107a ± IL-2) (Figure 3E; Figure S3A). In comparison, responses of unexposed individuals, irrespective of the peptide used, were dominated by CD8+ T cells generating one or two cytokines that included IFN-γ, TNF, and/or CD107a but not IL-2 (Figure 3E; Figure S3).
These data demonstrate that exposed and unexposed individuals can generate SPR-specific CD8+ T cells that efficiently cross-recognize the homologous LPR peptide from circulating betacoronaviruses.
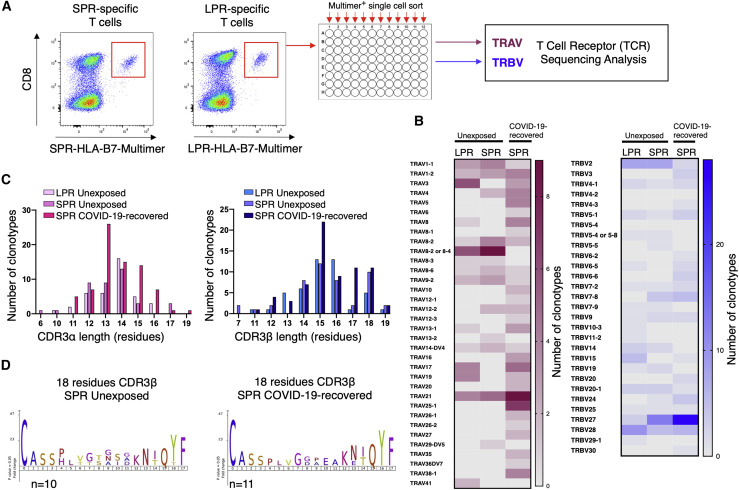
SPR-specific CD8+ T cells share biased TRBV27 gene use and long CDR3β in unexposed and COVID-19-recovered individuals
To further characterize these cross-reactive CD8+ T cells, we next assessed their TCR repertoires. SPR-specific CD8+ T cells from COVID-19-recovered donors were stained with the HLA-B7-SPR multimer, and single cells were isolated by fluorescence-activated cell sorting (FACS) (Figure 4 A). Additionally, SPR- and LPR-specific T cells from unexposed individuals were stained individually with the HLA-B7-SPR or HLA-B7-LPR multimer, and single cells were sorted (Figure S2C). The TCR repertoire was determined using multiplex RT-PCR (Grant et al., 2018; Wang et al., 2012). A total of 767 T cells were sequenced from COVID-19-recovered (n = 5) and unexposed (n = 7) individuals. This analysis revealed that clonotypic diversity underpins the response to SPR and LPR peptides, with a total of 209 clonotypes isolated and 80 unique αβ TCR pairs (Table S5). The TCR repertoires for each individual were private, with no shared (public) TCRs identified between individuals. This suggests that the number of distinct clonotypes capable of responding to the SPR epitope likely underpins the consistent immunodominant response observed in convalescent participants. Cross-reactive CD8+ T cell clones recognizing the SPR and LPR peptides were observed in the majority of unexposed individuals (n = 6 of 7). However, a unique clonotypic profile was evident in all donors’ PBMCs following SPR and LPR stimulation. This suggests that subtle differences in peptide recognition by different TCRs not only influence clonal expansion but also affect the efficiency and avidity of T cell cross-recognition toward homologous peptides (Figure S2A). Comparison of the SPR-specific T cell αβTCR sequences identified a strong T cell receptor beta variable region (TRBV) use bias, with 30% and 40% of these clonotypes expressing the TRBV27 gene in unexposed and COVID-19-recovered individuals, respectively, whereas LPR-specific T cells displayed a bias for TRBV28 (22%; Figure 4B). In comparison, α chain sequencing revealed limited shared use between COVID-19-recovered and unexposed individuals (Figure 4B). A use bias for TRAV8-2 in SPR- and LPR-specific clonotypes was seen in unexposed individuals, and shared TRAV21 use (∼10%–12%) in unexposed and COVID-19-recovered donors (Figure 4B; Table S5). Although we observed a typical distribution in CDR3α length, a preference for long CDR3β was seen in all populations, with the majority of identified sequences greater than 15 amino acids in length (Figure 4C). Although analysis of the CDR3 loop sequences did not reveal a shared motif in the α or β chains, reflecting the private nature of the TCR repertoires (Figures S4), a shared 108PxxGx[P/G/A]x114 motif (where x is any of V/T/S/L/G) was observed in the 18-residue-long CDR3β loops for the SPR-specific TCRs from COVID-19-recovered and unexposed donors (Figure 4D).
Figure 4.
Diverse TCR repertoires are utilized for recognition of SPR and LPR peptides in unexposed and COVID-19-recovered individuals
PBMCs from COVID-19-recovered or unexposed individuals were stimulated with the SPK or LPR peptides and cultured for 10–14 days in the presence of IL-2. CD8+ T cell lines were stained with the SPK or LPR multimer. Multimer+ cells were single cell sorted, and the TCR repertoire was determined by multiplex PCR.
(A) Schematic displaying representative multimer staining for single-cell isolation and TCR sequencing of the SPR and LPR-specific T cells in HLA-B7+ unexposed donors.
(B) A heatmap displaying preferred TRAV (left panel) and TRBV (right panel) use of SPR and LPR-specific TCRs in unexposed and COVID-recovered individuals.
(C) Summary of CDR3α (left panel) and CDR3β (right panel) lengths for distinct peptide-specific TCR clonotypes.
(D) Motif of the 18-amino-acid-long CDR3β loops from unexposed (left panel) and COVID-19-recovered (right panel) donors.
See also Figures S2 and S4 and Table S5.
Although not definitive, the presence of this shared motif in clonotypes from exposed and unexposed donors suggests that the pre-existing immunity in HLA-B7+ individuals favored clonal expansion.
Distinct epitope presentation drives selective T cell cross-reactivity in HLA-B7+ individuals
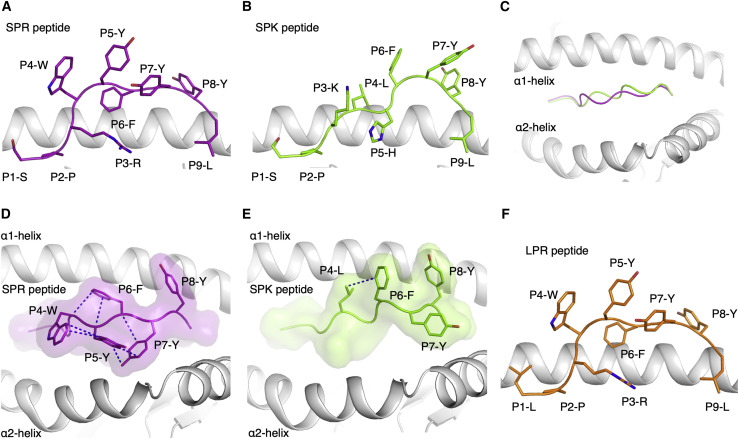
To determine the molecular basis of this selective CD8+ T cell cross-reactivity, we assessed the ability of the SPR and homologous peptides to form a stable complex with HLA-B7. Each peptide refolded efficiently with the HLA-B7 molecule, with the exception of the PPK peptide (poor yield). Next, the thermostability of each peptide-HLA (pHLA) complex was assessed (Figure S5A). The most stable complexes identified were HLA-B7-SPR and HLA-B7-LPR (melting temperature, Tm ≥ 60°C), followed by HLA-B7-SPK (Tm ≈ 56°C), whereas HLA-B7-PPK was less stable than the others (Tm ≈ 48°C) (Table 1). Considering that the SPR and SPK peptides demonstrated similar thermostability in complex with HLA-B7 but that CD8+ T cells were not able to cross-recognize both peptides, we next determined the crystal structures of these peptides bound to the HLA-B7 molecule (Figure 5 ).
Figure 5.
A structural basis for selective T cell cross-reactivity
(A and B) Crystal structures of the (A) SPR (purple stick) and (B) SPK (green stick) peptides presented by the HLA-B7 molecule (white cartoon), represented in the same orientation.
(C) Superposition of the HLA-B7-SPR (purple) and HLA-B7-SPK (green) structures represented as a cartoon from a top-down view of the antigen binding cleft.
(D) Top view of the HLA-B7-SPR structure, with a surface representation of the SPR peptide in purple with transparency and the solvent-exposed residues as sticks. The blue dashed lines represent intra-peptide interactions between the bulky aromatic and solvent-exposed residues of the SPR peptide.
(E) Top view of the HLA-B7-SPK complex in the same orientation as in (D). The SPK peptide is represented as green sticks for the surface-exposed residues and as a green surface. The blue dashed lines represent the intra-peptide interaction.
(F) Model of the structure of the HLA-B7 (white cartoon) molecule presenting the LPR peptide (orange stick), based on modeling from the HLA-B7-SPR complex.
The crystal structures of HLA-B7-SPR and HLA-B7-SPK were solved at 2.88 Å and 1.97 Å, respectively (Table S6). The electron density was well defined for both peptides (Figure S5), showing a stable conformation of the peptides in the cleft of HLA-B7 (Figure 5). Comparison of both structures revealed that different conformations were adopted by SPR and SPK peptides. The arginine (R) at position (P) three (P3-R) in the SPR peptide acted as a secondary anchor residue (Figure 5A), whereas for SPK, the lysine at P3 was pushed out of the cleft by the histidine at P5 that acted as a secondary anchor in the cleft (Figure 5B). The sequence differences at P3 and P5 of the peptides altered the SPK conformation compared with the SPR conformation (Figure 5C) despite the shared 6FYYL9 motif. Four of the five aromatic residues of the SPR peptide formed a network of interaction, stabilizing each other on the surface of the HLA-B7 cleft (Figure 5D). This formed a compact and large binding surface for potential interactions with the TCR. The large binding surface might explain the expansion of T cells with a long CDR3β loop (>16 residues) observed in 34% and 37% of the clonotypes in COVID-19-recovered and unexposed donors, respectively (Figure 4C), which would be able to contact a greater surface area of the peptide because of their length. In comparison, the conformation adopted by the SPK peptide only exposed three aromatic residues located at the C-terminal part of the peptide (6FYY8) (Figure 5E). The LPR peptide only differs from SPR at P1, and this single substitution is unlikely to change the peptide conformation, given that HLA-B7 accommodates a wide range of residues at P1, based on previously reported structures (Brennan et al., 2012; Chan et al., 2018; Du et al., 2016; Rowntree et al., 2018, 2020). Therefore, because of the high sequence identity, the LPR peptide is likely to adopt a conformation similar to the SPR peptide (Figure 5F), forming the basis of the CD8+ T cell cross-reactivity observed in HLA-B7+ individuals. Despite sharing 67% sequence identity, the different structures adopted by the SPR (SARS-CoV-2) and SPK (229E) peptides provide a basis for a low, if any, level of CD8+ T cell cross-reactivity between these peptides, highlighting the fine specificity of CD8+ T cells.
Discussion
T cell mediated cross-reactivity in response to seasonal coronaviruses has the potential to limit the development and severity of COVID-19. Here we demonstrated that a robust CD8+ T cell response in HLA-B7+ individuals is primarily directed toward a single immunodominant epitope, SPR, encoded by the N protein of SARS-CoV-2, recognized by more than 80% of tested COVID-19-recovered donors. We also identified SPR-specific CD8+ T cells in more than 90% of tested unexposed HLA-B7+ individuals who had not been exposed to SARS-CoV-2. Although comparative analysis of circulating seasonal coronaviruses revealed a high level of conservation in the homologous peptide sequences, including conservation of the critical proline residue at P2 of the epitope, detailed analysis revealed that T cell responses in unexposed volunteers were driven by cross-reactive CD8+ T cells specific for the LPR peptide from the OC43 and HKU-1 seasonal coronaviruses. It is possible that these cross-reactive memory CD8+ T cells facilitate the strong, polyfunctional, high avidity CD8+ T cell response toward the SPR peptide observed in T cells expanded from COVID-19-recovered individuals. In contrast, despite sharing the C-terminal 6FYY8 motif and the HLA-B7-favored anchor residues (P2-P and P9-L) with the PPK and SPK peptides from the 229E and NL63 viruses, respectively, there was no CD8+ T cell cross-reactivity between these and the SPR or LPR peptides. Considering that an average 4.1% of the global population is HLA-B7+ and it is the sixth most frequent HLA-B molecule worldwide (frequency 0%–20%, depending on ethnicity’ Gonzalez-Galarza et al., 2020; Solberg et al., 2008), these results may have profound implications for T cell-mediated protection against COVID-19. However, they also demonstrated the fine specificity of T cells for their cognate antigen, which may prevent cross-recognition of less conserved epitopes.
Studies have provided tantalizing evidence that individuals not exposed to SARS-CoV-2 may harbor T cells capable of cross-recognizing epitopes presented by HLA class I and class II molecules (Bacher et al., 2020; Braun et al., 2020; Karlsson et al., 2020; Le Bert et al., 2020; Nelde et al., 2021; Schulien et al., 2021; Shomuradova et al., 2020). However, several factors, including lack of sequence conservation between several of the reported cross-reactive epitopes, the non-physiological quantities of antigen used in detection assays, and the limited number of unexposed donors assessed, may contribute to overestimation of the prevalence of pre-existing T cell immunity when it comes to defining truly cross-reactive T cells that could protect against COVID-19. It is therefore critical to undertake a detailed analysis of the molecular characteristics that promote or limit T cell cross-reactive immunity (Karlsson et al., 2020; Sette and Crotty, 2020). It is well established from studies of other human viral infections, including HIV, cytomegalovirus (CMV), and influenza, that even a single amino acid change, particularly in a TCR contact region, can have a profound effect on cross-reactivity (Geldmacher et al., 2009; Goulder et al., 1997; Gras et al., 2009, 2010; Iglesias et al., 2011; Kløverpris et al., 2015; Smith et al., 2014). Here we revealed different conformations of the SPR and SPK peptides in complex with HLA-B7, providing a basis for the observed low or lack of T cell cross-reactivity. This shows that, despite the sequence similarity and conserved C-terminal motif between the peptides, T cell cross-reactivity will not occur toward all seasonal coronaviruses because of fine TCR specificity. Although we were unable to solve the structure of the LPR epitope in complex with HLA-B7, our modeling revealed significant structural homology with minimal changes induced by the S-to-L substitution at P1. Similarly, SPR- and LPR-specific T cells efficiently cross-recognized both peptides, displaying similar functional avidity and polyfunctionality. TCR sequence analysis of SPR- and LPR-specific T cells revealed that, although some biased gene use was observed (TRAV8-2, TRBV2, TRBV27, and TRBV28), the TCR repertoires were entirely private with no public TCR shared between individuals. However, we observed key TCRs able to cross-react toward the SPR and LPR peptides in unexposed individuals. This likely contributes to the strong response observed from COVID-19-recovered donors’ expanded T cells and may suggest a level of pre-existing immunity. Although the TCR repertoire was private, we observed biased TRBV27 gene use with long CDR3β loops expressed preferentially for SPR-specific TCRs in unexposed and COVID-19-recovered individuals. Despite sequence diversity in the TCR repertoire, the bias for long length of CDR3 loops might reflect a common docking mechanism between the clonotypes and facilitate recognition of the numerous large aromatic residues of the SPR peptide.
It is clear that, although pre-existing immunity toward SARS-CoV-2 exists and may provide an advantage to certain individuals, this same advantage might be limited to a few epitopes specific to certain HLA molecules and dependent on previous encounters with certain coronaviruses. In addition, the presence of pre-existing immunity suggests that memory T cells could be activated upon SARS-CoV-2 infection, important for long-lasting protection that will be critical for vaccination (Cañete and Vinuesa, 2020; Dan et al., 2020; Jarjour et al., 2021). Those cross-reactive T cells could also be used as biomarkers to help predict the severity of disease or the efficacy of a vaccine. The discovery of defined epitopes that elicit strong CD8+ T cell responses, particularly those that are pre-existing, is important because these epitopes could be exploited to boost or prime the immune response in a vaccination strategy and therefore could provide significant protection of the global population.
Limitations of study
Our study established that one single epitope dominates the response toward the SARS-CoV-2 N protein in HLA-B7+ individuals who recovered from COVID-19. Although HLA-B7 is the most common HLA-B molecule in populations of European decent, it is the sixth most frequent HLA-B molecule worldwide and is expressed at a frequency of 4% globally. Therefore, although generalization at the global population level is not possible because of our focus on a single allele, our study does provide a potential mechanism for the immunodominant response observed in HLA-B7+ individuals via cross-recognition by seasonal coronavirus T cells. Given the conservation between SARS-CoV-2 and seasonal coronaviruses, especially with betacoronaviruses, it is likely that pre-existing and cross-reactive T cell responses occur for other peptides restricted to diverse HLAs. However, our study also demonstrates how even a few changes in amino acid sequence can affect T cell recognition, suggesting that the presence of immunodominant cross-reactive CD8+ T cells could be restricted to specific pHLA complexes. More detailed and precise research in this direction is critical to determine the actual presence of cross-reactive and pre-existing T cells between SARS-CoV-2 variants and seasonal coronaviruses and their potential role in protection against COVID-19.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD107a-FITC | BD Biosciences/eBioscience | Cat# 555800; RRID:AB_396134 |
| CD107a-AF488 | Thermofisher | Cat# 53-1079-42; RRID:AB_2016657 |
| CD8-PerCP-Cy5.5 | BD Biosciences/eBioscience | Cat# 565310; RRID:AB_2687497 |
| CD4-PE-Cy7 | BD Biosciences | Cat# 560649; RRID:AB_1727475 |
| CD4-Pacific Blue | BD Biosciences | Cat# 558116; AB_397037 |
| CD4-BUV395 | BD Biosciences | Cat# 563550; AB_2738273 |
| Live/Dead Fixable Near-IR Dead Cell Stain | Life Technologies | Cat# L34975 |
| IFN-γ-AF700 | BD Biosciences | Cat# 557995; RRID:AB_396977 |
| IFN-γ-PE | BD Biosciences | Cat# 554701; RRID:AB_395518 |
| IFN-γ-V421 | BD Biosciences | Cat# 562988; 2737934 |
| CD14-APCH7 | BD Biosciences | Cat# 560180 RRID:AB_1645464 |
| CD19-APCH7 | BD Biosciences | Cat# 560727; RRID:AB_1727437 |
| SARS-CoV/SARS-CoV-2 Nucleocapsid Antibody, Rabbit mAb | Sino Biological | 40143-R001; RRID:AB_2827974 |
| Goat anti-Rabbit IgG (H+L) Cross-adsorbed Secondary Ab, HRP | Thermo Fischer | A16104; RRID:AB_2534776 |
| Bacterial and virus strains | ||
| E. coli BL21 | ATCC | N/A |
| SARS-CoV-2 virus | Qld Health | hCoV-19/Australia/QLD02/2020 |
| Biological samples | ||
| Donor-derived PBMCs | QIMR, QLD, Australia | N/A |
| Human Sera samples (heat-treated) | Mater Hospital, QLD, Australia | N/A |
| Donor-derived PBMCs | Monash University, VIC, Australia | N/A |
| Donor-derived PBMCs | Blood bank, VIC, Australia | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI-1640 | Thermofisher | Cat# 21870092 |
| MEM nonessential amino acid solution | Thermofisher | Cat# 11140050 |
| HEPES | Life Technologies | Cat# 15630080 |
| l-glutamine | GIBCO | Cat# 25030149 |
| penicillin/streptomycin | Life Technologies | Cat# 15140122 |
| FCS | Scientifix | Cat# SFBS-NZ |
| IL-2 | BD Biosciences | Cat# 559334 |
| GolgiPlug Protein Trnsp Inhb | BD Biosciences | Cat# 555029 |
| Cytofix/Cytoperm W/Golgi Stop Kit | BD Biosciences | Cat# 554715 |
| Paraformaldehyde (BD cytofix) | BD Biosciences | Cat# 554655 |
| Triton X-100 | Astral | Cat# 786-514 |
| ExoSAP | Life Technologies | Cat# 78201.1.ML |
| Urea | Thermofisher | Cat# AJA817-5KG |
| L-Arginine | Sigma | Cat# A5131-1KG |
| Tris-HCl | Thermofisher | Cat# FSBBP152-5 |
| EDTA | BDH | Cat# BDH9232-500G |
| glutathione reduced | Goldbio | Cat# G-155-500 |
| glutathione oxidised | Goldbio | Cat# G-060-25 |
| SYPRO orange | Thermofisher | Cat# S6650 |
| NaCl | Thermofisher | Cat# CHE700/NACL-25KG |
| NH4SO4 | Astral | Cat# BIOADB0060-500 g |
| PEG3350 | Sigma | Cat# 88276-1KG-F |
| Potassium Iodide | Sigma | Cat # 221945-500G |
| Ethylene glycol | Sigma | Cat# 293237-1L |
| Minimum Essential Media | GIBCO | 11095080 |
| FCS | GIBCO | 16000044 |
| PBS | GIBCO | 003002 |
| Triton X-100 | Sigma Aldrich | T8787 |
| Tween-20 | Sigma | 003005 |
| 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt | Sigma Aldrich | A1888 |
| SARS-CoV-2 antigen overlapping peptide pools POOLS CoV1-5 contain: (AP31, NS6, ORF10, ORF9B, NS7A, NS7B, NS8, Y14, NCAP, VEMP, VME1, Spike Glycoprotein) | JPT | PM-WCPV-AP3A-1 PM-WCPV-NS6-1 PM-WCPV-ORF10-1 PM-WCPV-ORF9B-1 PM-WCPV-NS7A-1 PM-WCPV-NS7B-1 PM-WCPV-NS8-1 PM-WCPV-Y14-1 PM-WCPV-NCAP-1 PM-WCPV-VEMP-1 PM-WCPV-WME1-1 PM-WCPV-S-1 |
| SPR, SPK, PPR, LPR peptides | Genscript | N/A |
| Critical commercial assays | ||
| VILO cDNA synthesis kit | Life Technologies | Cat# 11754250 |
| Deposited data | ||
| Crystal structure of HLA-B7-SPR | This paper | 7LGD (PDB code) |
| Crystal structure of HLA-B7-SPK | This paper | 7LGT (PDB code) |
| TCR sequences deposited in Mendeley Data | This paper | https://doi.org/10.17632/p34mzy8jfx.1 |
| Experimental models: cell lines | ||
| Vero cells | ATCC | ATCC® CRL-1586 |
| Oligonucleotides | ||
| Primer for TCR alpha and beta chain sequencing | Sigma | N/A |
| Recombinant DNA | ||
| Human beta 2 microglobulin | genscript | N/A |
| HLA-B∗07:02 soluble fraction (1-275 residues) | genscript | N/A |
| Software and algorithms | ||
| FlowJo software (TreeStar) | FlowJo, LLC | N/A |
| FACSDiva software | BD Biosciences | N/A |
| FinchTV | Geospiza | N/A |
| IMGT software | IMGT | N/A |
| GraphPad Prism 8 (version 8.4.2) | Graphpad | N/A |
| XDS | XDS | N/A |
| PHASER | CCP4 | N/A |
| CCP4 suite | CCP4 | N/A |
| COOT | Coot | N/A |
| BUSTER | BUSTER | N/A |
| Pymol | Schrödinger | N/A |
| Nonlinear regression analysis, log (inhibitor) versus normalized response | Graphpad | N/A |
Resource availability
Lead contact
Further information and requests for resources and materials should be directed to the Lead Contact, Prof. Stephanie Gras (S.Gras@latrobe.edu.au)
Materials availability
Materials are available upon reasonable request.
Data and code availability
The final crystal structure models for the peptide-HLA-B∗07:02 complexes have been deposited to the Protein Data Bank (PDB) under the following accession codes: 7LGD for HLA-B7-SPR and 7LGT for HLA-B7-SPK.
Experimental model and subject details
Study participants
This study was performed according to the principles of the Declaration of Helsinki. Ethics approval to undertake the research was obtained from the QIMR Berghofer Medical Research Institute Human Research Ethics Committee and Monash University Human Research Ethics Committee. COVID-19-recovered donors were over the age of 18, had been clinically diagnosed by PCR with SARS-CoV-2 infection, and had subsequently been released from isolation following resolution of symptomatic infection. A total of 37 participants were recruited in May and June 2020 from the south-east region of Queensland, Australia. The majority of participants were returned overseas travelers. Participants ranged in age from 20 to 75, 14 were male and 23 were female, and were a median of 62 (46 – 124) days post-initial diagnosis. Blood samples were collected from all participants to isolate peripheral blood mononuclear cells (PBMCs) to assess SARS-CoV-2 immunity. Healthy donors over the age of 18, with no known COVID-19 infection or exposure, were recruited. These donors are referred to as unexposed throughout the manuscript. A total of 17 unexposed donors were recruited, ranging in age from 19 to 56 (average of 33 years old), 9 were male, 7 were female and 1 was undetermined. Informed consent was obtained from all participants. The HLA typing was performed by AlloSeq Tx17 (CareDx Pty Ltd, Fremantle, Australia), or Australian Red Cross Victorian Transplant and Immunogenetics Service (Melbourne, Australia), or PathWest Laboratory Medicine, Fiona Stanley Hospital using AllType NGS high resolution typing on the IonTorrent NGS platform, and these details are provided in Table S1.
Method details
Peripheral blood mononuclear cells
PBMCs were separated from whole blood or buffy coats using density gradient centrifugation. PBMCs were used fresh or were cryogenically stored until use.
Generation of peptide-specific CD8+ T cell lines
CD8+ T cell lines were generated as previously described (Grant et al., 2018; Lineburg et al., 2020). Briefly, PBMCs were incubated with 10 μM SARS-CoV-2 overlapping peptide pools, or 10 μM of individual peptides and cultured for 10-14 days in RPMI-1640 supplemented with 2 mM MEM nonessential amino acid solution (Sigma), 100 mM HEPES (Sigma), 2 mM l-glutamine (Sigma), penicillin/streptomycin (Life Technologies), 50 mM 2-ME (Sigma) and 10% heat-inactivated (FCS; Scientifix). Cultures were supplemented with 10IU IL-2 2-3 times weekly. CD8+ T cell lines were used freshly harvested, or were cryogenically stored for subsequent analysis.
Intracellular cytokine assay
CD8+ T cell lines were stimulated with cognate SARS-CoV-2 antigen overlapping peptide pools, or 10 μM individual peptides and were incubated for 4-5 hours in the presence of GolgiPlug (BD Biosciences), GolgiStop (BD Biosciences) and anti-CD107a-FITC or -AF488 (BD Biosciences/eBioscience). Following stimulation, cells were surface stained for 30 mins with anti-CD8-PerCP-Cy5.5 (eBioscience/BD Biosciences), anti-CD4-PE-Cy7 or -Pacific Blue or -BUV395 (all BD Biosciences) and Live/Dead Fixable Near-IR Dead Cell Stain (Life Technologies). Cells were fixed and permeabilised using BD Cytofix/Cytoperm solution (BD Biosciences) and then intracellularly stained with anti-IFN-γ-AF700 or -PE or -V421 (all from BD Biosciences) as well as anti-TNF-PE-Cy7, and IL2-PE (all BD Biosciences) for a further 30 minutes. Cells were acquired on a BD LSRFortessa with FACSDiva software. Post-acquisition analysis was performed using FlowJo software (TreeStar). Cytokine detection levels identified in the no-peptide control condition were subtracted from the corresponding test conditions in all summary graphs to account for non-specific, spontaneous cytokine production.
Multimer staining
CD8+ T cell lines were multimer stained for 1 hr at room temperature. Cells were washed and surface stained with anti-CD8-PerCP-Cy5.5 (BD Biosciences), anti-CD4-BUV395 (BD Biosciences) anti-CD14-APCH7, anti-CD19-APCH7 and Live/Dead Fixable Near-IR Dead Cell Stain (Life Technologies). Cells were either fixed with 1% paraformaldehyde and acquired on the BD LSR Fortessa, or were directly single-cell sorted into PCR plates (Eppendorf) using a BD Aria Fusion. Plates were centrifuged, and stored at −80°C until use.
Single-cell multiplex PCR
Single-cell multiplex PCR was carried out as previously described (Grant et al., 2018; Wang et al., 2012). Briefly, cDNA was generated using the VILO cDNA synthesis kit (Invitrogen) at 1/20 of the manufacturer’s recommendation with 0.1% Triton X. Nested PCR comprising 40 α- and 27 β-chains was subsequently undertaken. PCR products were purified using ExoSAP (GE Healthcare) and were sequenced at AGRF (Melbourne, Australia) or The QIMR Berghofer Sequencing Facility (Brisbane, Australia). Sequences were analyzed using FinchTV (Geospiza) and IMGT software (Brochet et al., 2008; Giudicelli et al., 2011). CDR3 sequences shown are all productive (no stop codons).
Protein expression, refold and purification
DNA plasmids encoding HLA-B7 α chain and β-2-microglobulin were transformed separately into a BL21 strain of E. coli. Recombinant proteins were expressed individually, where inclusion bodies were extracted and purified from the transformed E. coli cells. Soluble pHLA complexes were produced by refolding 30 mg of HLA-B7 α chain with 10 mg of β-2-microglobulin and 5 mg of peptide (Genscript) into a buffer of 3M Urea, 0.5 M L-Arginine, 0.1 M Tris-HCl pH 8.0, 2.5 mM EDTA pH 8.0, 5 mM glutathione (reduced), 1.25 mM glutathione (oxidised). The refold mixture was dialysed into 10 mM Tris-HCl pH 8.0 and soluble pHLA was purified using anion exchange chromatography using a HiTrapQ column (GE Healthcare).
Differential scanning fluorimetry
Differential Scanning fluorimetry was performed in a QIAGEN RG6 real-time PCR machine, with pHLA samples heated from 30 to 95°C at a rate of 0.5°C/min using a default excitation and emission channel set to yellow (excitation of ∼530 nm and detection at ∼557 nm). The experiment was set up using two concentrations of pHLA (5 μM and 10 μM), each in duplicate. Each sample was dialysed in 10mM Tris-HCl pH 8.0, 150mM NaCl and contained a final concentration of 10X SYPRO Orange Dye. Fluorescence intensity data was normalized and plotted using GraphPad Prism 8 (version 8.4.2). The Tm value is determined by the temperature when 50% of maximum fluorescence intensity is reached, and summarized in Table 1.
Crystallization and structural determination
Crystals of pHLA complexes were grown via sitting-drop, vapor diffusion method at 20°C with a protein: reservoir drop ratio of 1:1, at a concentration of 7 mg/mL in 10 mM Tris-HCl pH 8.0, 150 mM NaCl. Crystals of HLA-B7 in complex with SARS-CoV-2 N105-113 (SPRWYFYYL) were grown in 2M ammonium sulfate, 0.1M HEPES pH 7.5; or with 229E N105-113 (SPKLHFYYL) were grown in 18% PEG3350, 0.2 M KI. These crystals were soaked in a cryoprotectant containing mother liquor and 20% EG or 30% PEG3350 (w/v) and then flash-frozen in liquid nitrogen. The data were collected on the MX2 beamline at the Australian Synchrotron, part of ANSTO, Australia (Aragão et al., 2018). The data were processed using XDS (Kabsch, 2010) and the structures were determined by molecular replacement using the PHASER program (McCoy et al., 2007) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994) with a model of HLA-B7 without the peptide (derived from PDB ID: 5WMN; Rowntree et al., 2018). Manual model building was conducted using COOT (Emsley et al., 2010) followed by refinement with BUSTER (Bricogne et al., 2011). The final model has been validated using the wwPDB OneDep System with the accession number of 7LGD for HLA-B7-SPR and 7LGT for HLA-B7-SPK structures. The final refinement statistics are summarized in Table S6. All molecular graphics representations were created using PyMOL.
Model building
Model building of the structure of HLA-B7-LPR complex was performed using the crystal structure of HLA-B7-SPR as a starting model. The SPR peptide P1-Ser residue was mutated into a P1-Leu residue using the crystallographic software, COOT (Emsley et al., 2010), where the side chain rotamer was selected based on the least steric clashes, as evaluated using MolProbity.
SARS-CoV-2 microneutralization assay
Vero cells were cultured in 96 well plates. Convalescent serum harvested from COVID-19-recovered patients was heat-treated at 56°C for 1 hour. The sera was then serial diluted with minimum essential medium (MEM) (GIBCO) supplemented with 2% FCS. In physical containment 3 settings, the diluted sera were incubated with the SARS-CoV-2 (QLD/02; MOI 1) for 1 hour at room temperature. The serum-virus mixture was transferred to the cultured vero cells and further incubated for 1 hour at room temperature for infection. The inoculum was then removed and replaced with MEM (GIBCO) supplemented with 2% FCS (GIBCO). The cells were incubated at 37°C for 72 hours. The cells were fixed with 10% formaldehyde for 24 hours.
Cells were permeabilised with PBS containing 0.1% Trition-X-100 (Sigma) for 15mins at room temperature. The plates were blocked using PBS (Sigma) supplemented with 0.1% Tween-20 (Sigma) and 3% skim milk for 1 hour at room temperature. The cells were primarily stained with rabbit anti-NP (1:3000)(Sino Biological) for 1 hour at room temperature. The plates were washed with PBS (Sigma) supplemented with 0.1% Tween-20 (Sigma). The cells were stained using goat anti-rabbit immunoglobulin horse-radish protein (HRP) (Thermo Fischer) (1:3000) for 1 hour at room temperature. The plates were washed with PBS (Sigma) supplemented with 0.1% Tween-20 (Sigma). The plates were incubated with 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid (ABTS) (Thermo Fischer) substrate for color development and incubated for 20 minutes. Measurements were done at 405 nm.
Sequence alignment of SPR and homologs from coronaviruses isolates
Complete full length protein sequences for the SARS-CoV-2 (taxid ID 2697049, 419 amino acids), SARS-CoV-1 (taxid ID 694069, 422 amino acids), OC43 (taxid ID 31631, 448 amino acids), HKU-1 (taxid ID 290028, 441 amino acids), 229E (taxid ID 11137, 389 amino acids) and NL63 (taxid ID 277944, 377 amino acids) coronaviruses were obtained from the NCBI virus database http://www.ncbi.nlm.nih.gov/labs/virus on the 10th of November 2020. Sequences were aligned using https://www.fludb.org/brc/home.spg?decorator=influenza. All sequences were used except a single SARS-CoV-2 North American sequence that was unable to be aligned, and a single SARS-CoV-2 Europe sequence with an unknown amino acid (denoted as X) within the peptide sequence.
Quantification and statistical analysis
Statistical analysis
GraphPad Prism 8.2.1 (San Diego, CA) was used to perform statistical analysis. Statistical comparisons between participant groups (unexposed and COVID-19-recovered) were made using unpaired Mann-Whitney U Wilcoxon rank-sum tests. p < 0.05 was considered statistically significant.
Acknowledgments
The authors would like to thank Queensland Health Forensic & Scientific Services, Queensland Department of Health who provided the SARS-CoV-2 isolate QLD02; Monash facilities (Flow Core, Macromolecular Crystallization Facility, Imaging Facility); VTIS, PathWest Laboratory Medicine and CareDx for HLA Typing; and the MX team for assistance at the Australian Synchrotron. The authors would also like to thank all participants who took part in the study. This work was supported by generous donations from the QIMR Berghofer COVID-19 appeal and financial contributions from Monash University, the Australian Nuclear Science and Technology Organisation (ANSTO; AINSE ECR grants), the Australian Research Council (ARC), the National Health and Medical Research Council (NHMRC, GNT1132519), and the Medical Research Future Fund (MRFF, APP2005654). S.S. and H.S. are supported by an Australian Government Research Training Program scholarship. E.J.G. was supported by an NHMRC C.J. Martin fellowship (1110429) and is supported by an Australian Research Council DECRA (DE210101479). K.R.S. is supported by an Australian Research Council DECRA (DE180100512). S.G. is supported by an NHMRC SRF (1159272).
Author contributions
Conceptualization, K.E.L., E.J.G., S.S., D.S.M.C., C. Smith, and S.G.; formal analysis, K.E.L., E.J.G., S.S., D.S.M.C., H.S., C. Szeto., C. Smith, and S.G.; funding acquisition, K.R.S., C. Smith, and S.G.; investigation and methodology, K.E.L., E.J.G., S.S., D.S.M.C., C. Szeto, H.S., A.P., J.R., P.C., S.R., A.T.N., L.L., M.A.N., Z.W.M.T., D.J., K.Y.C., C.A.L., H.H., J.M.B., K.R.S., and S.G.; project administration, C. Smith and S.G.; resources, W.C., L.D., C. Smith, and S.G.; supervision, K.E.L., E.J.G., S.S., D.S.M.C., C. Smith, and S.G.; validation, K.E.L., E.J.G., S.S., D.S.M.C., C. Smith, and S.G.; visualization, K.E.L., E.J.G., S.S., D.S.M.C., H.S., C. Smith, and S.G. ; writing – original draft, K.E.L., E.J.G., S.S., D.S.M.C., H.S., C. Smith, and S.G. ; writing – review & editing, all authors.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. One or more of the authors of this paper self-identifies as living with a disability. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: April 13, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.04.006.
Supplemental information
References
- Aragão D., Aishima J., Cherukuvada H., Clarken R., Clift M., Cowieson N.P., Ericsson D.J., Gee C.L., Macedo S., Mudie N., et al. MX2: a high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 2018;25:885–891. doi: 10.1107/S1600577518003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schröder I., Wieters I., Khodamoradi Y., et al. Low-Avidity CD4+ T Cell Responses to SARS-CoV-2 in Unexposed Individuals and Humans with Severe COVID-19. Immunity. 2020;53:1258–1271.e5. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Brennan R.M., Petersen J., Neller M.A., Miles J.J., Burrows J.M., Smith C., McCluskey J., Khanna R., Rossjohn J., Burrows S.R. The impact of a large and frequent deletion in the human TCR β locus on antiviral immunity. J. Immunol. 2012;188:2742–2748. doi: 10.4049/jimmunol.1102675. [DOI] [PubMed] [Google Scholar]
- Bricogne G.B.E., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O.S., Vonrhein C., Womack T.O. Global Phasing Ltd.; 2011. BUSTER version 2.10. [Google Scholar]
- Brochet X., Lefranc M.P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn316. W503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañete P.F., Vinuesa C.G. COVID-19 Makes B Cells Forget, but T Cells Remember. Cell. 2020;183:13–15. doi: 10.1016/j.cell.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.F., Gully B.S., Gras S., Beringer D.X., Kjer-Nielsen L., Cebon J., McCluskey J., Chen W., Rossjohn J. Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide. Nat. Commun. 2018;9:1026. doi: 10.1038/s41467-018-03321-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato K., Hastie K.M., Faliti C.E., Ramirez S.I., Frazier A., Yu E.D., Grifoni A., Rawlings S.A., et al. Immunological memory to SARS-CoV-2 assessed for greater than six months after infection. bioRxiv. 2020 doi: 10.1101/2020.11.15.383323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du V.Y., Bansal A., Carlson J., Salazar-Gonzalez J.F., Salazar M.G., Ladell K., Gras S., Josephs T.M., Heath S.L., Price D.A., et al. HIV-1-Specific CD8 T Cells Exhibit Limited Cross-Reactivity during Acute Infection. J. Immunol. 2016;196:3276–3286. doi: 10.4049/jimmunol.1502411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.M., Henson V., Slack R., Ng J., Hartzman R.J., Katovich Hurley C. Frequencies of HLA-A2 alleles in five U.S. population groups. Predominance Of A∗02011 and identification of HLA-A∗0231. Hum. Immunol. 2000;61:334–340. doi: 10.1016/s0198-8859(99)00155-x. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W., et al. Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldmacher C., Metzler I.S., Tovanabutra S., Asher T.E., Gostick E., Ambrozak D.R., Petrovas C., Schuetz A., Ngwenyama N., Kijak G., et al. Minor viral and host genetic polymorphisms can dramatically impact the biologic outcome of an epitope-specific CD8 T-cell response. Blood. 2009;114:1553–1562. doi: 10.1182/blood-2009-02-206193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V., Brochet X., Lefranc M.P. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb. Protoc. 2011;2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Galarza F.F., McCabe A., Santos E.J.M.D., Jones J., Takeshita L., Ortega-Rivera N.D., Cid-Pavon G.M.D., Ramsbottom K., Ghattaoraya G., Alfirevic A., et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48(D1):D783–D788. doi: 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Phillips R.E., Colbert R.A., McAdam S., Ogg G., Nowak M.A., Giangrande P., Luzzi G., Morgan B., Edwards A., et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- Grant E.J., Josephs T.M., Loh L., Clemens E.B., Sant S., Bharadwaj M., Chen W., Rossjohn J., Gras S., Kedzierska K. Broad CD8+ T cell cross-recognition of distinct influenza A strains in humans. Nat. Commun. 2018;9:5427. doi: 10.1038/s41467-018-07815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gras S., Saulquin X., Reiser J.B., Debeaupuis E., Echasserieau K., Kissenpfennig A., Legoux F., Chouquet A., Le Gorrec M., Machillot P., et al. Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope. J. Immunol. 2009;183:430–437. doi: 10.4049/jimmunol.0900556. [DOI] [PubMed] [Google Scholar]
- Gras S., Kedzierski L., Valkenburg S.A., Laurie K., Liu Y.C., Denholm J.T., Richards M.J., Rimmelzwaan G.F., Kelso A., Doherty P.C., et al. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc. Natl. Acad. Sci. USA. 2010;107:12599–12604. doi: 10.1073/pnas.1007270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., Koutsakos M., Hensen L., Jia X., Chua B., et al. Suboptimal SARS-CoV-2-specific CD8+ T cell response associated with the prominent HLA-A∗02:01 phenotype. Proc. Natl. Acad. Sci. USA. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias M.C., Almeida J.R., Fastenackels S., van Bockel D.J., Hashimoto M., Venturi V., Gostick E., Urrutia A., Wooldridge L., Clement M., et al. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood. 2011;118:2138–2149. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarjour N.N., Masopust D., Jameson S.C. T Cell Memory: Understanding COVID-19. Immunity. 2021;54:14–18. doi: 10.1016/j.immuni.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P., et al. CD8+ T cell responses in convalescent COVID-19 individuals target epitopes from the entire SARS-CoV-2 proteome and show kinetics of early differentiation. bioRxiv. 2020 doi: 10.1101/2020.10.08.330688. [DOI] [Google Scholar]
- Karlsson A.C., Humbert M., Buggert M. The known unknowns of T cell immunity to COVID-19. Sci. Immunol. 2020;5:eabe8063. doi: 10.1126/sciimmunol.abe8063. [DOI] [PubMed] [Google Scholar]
- Kløverpris H.N., Cole D.K., Fuller A., Carlson J., Beck K., Schauenburg A.J., Rizkallah P.J., Buus S., Sewell A.K., Goulder P. A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape. Retrovirology. 2015;12:20. doi: 10.1186/s12977-015-0149-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Lineburg K.E., Srihari S., Altaf M., Swaminathan S., Panikkar A., Raju J., Crooks P., Ambalathingal G.R., Martins J.P., Matthews K.K., et al. Rapid detection of SARS-CoV-2-specific memory T-cell immunity in recovered COVID-19 cases. Clin. Transl. Immunology. 2020;9:e1219. doi: 10.1002/cti2.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Oxford Immunology Network Covid-19 Response T cell Consortium. ISARIC4C Investigators Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowntree L.C., Nguyen T.H.O., Halim H., Purcell A.W., Rossjohn J., Gras S., Kotsimbos T.C., Mifsud N.A. Inability To Detect Cross-Reactive Memory T Cells Challenges the Frequency of Heterologous Immunity among Common Viruses. J. Immunol. 2018;200:3993–4003. doi: 10.4049/jimmunol.1800010. [DOI] [PubMed] [Google Scholar]
- Rowntree L.C., Nguyen T.H.O., Farenc C., Halim H., Hensen L., Rossjohn J., Kotsimbos T.C., Purcell A.W., Kedzierska K., Gras S., Mifsud N.A. A Shared TCR Bias toward an Immunogenic EBV Epitope Dominates in HLA-B∗07:02-Expressing Individuals. J. Immunol. 2020;205:1524–1534. doi: 10.4049/jimmunol.2000249. [DOI] [PubMed] [Google Scholar]
- Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Karolinska COVID-19 Study Group Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- Shomuradova A.S., Vagida M.S., Sheetikov S.A., Zornikova K.V., Kiryukhin D., Titov A., Peshkova I.O., Khmelevskaya A., Dianov D.V., Malasheva M., et al. SARS-CoV-2 Epitopes Are Recognized by a Public and Diverse Repertoire of Human T Cell Receptors. Immunity. 2020;53:1245–1257.e5. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C., Gras S., Brennan R.M., Bird N.L., Valkenburg S.A., Twist K.A., Burrows J.M., Miles J.J., Chambers D., Bell S., et al. Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire. Sci. Rep. 2014;4:3993. doi: 10.1038/srep03993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder T.M., Gittelman R.M., Klinger M., May D.H., Osborne E.J., Taniguchi R., Zahid H.J., Kaplan I.M., Dines J.N., Noakes M.N., et al. Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. medRxiv. 2020 doi: 10.1101/2020.07.31.20165647. [DOI] [Google Scholar]
- Solberg O.D., Mack S.J., Lancaster A.K., Single R.M., Tsai Y., Sanchez-Mazas A., Thomson G. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 2008;69:443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto C., Chatzileontiadou D.S.M., Nguyen A.T., Sloane H., Lobos C.A., Jayasinghe D., Halim H., Smith C., Riboldi-Tunnicliffe A., Grant E.J., Gras S. The presentation of SARS-CoV-2 peptides by the common HLA-A∗02:01 molecule. iScience. 2021;24:102096. doi: 10.1016/j.isci.2021.102096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.C., Dash P., McCullers J.A., Doherty P.C., Thomas P.G. T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci. Transl. Med. 2012;4:128ra42. doi: 10.1126/scitranslmed.3003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The final crystal structure models for the peptide-HLA-B∗07:02 complexes have been deposited to the Protein Data Bank (PDB) under the following accession codes: 7LGD for HLA-B7-SPR and 7LGT for HLA-B7-SPK.