Abstract
The Coronavirus Disease 2019 (COVID-19) pandemic has forced the scientific community to rapidly develop highly reliable diagnostic methods in order to effectively and accurately diagnose this pathology, thus limiting the spread of infection. Although the structural and molecular characteristics of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were initially unknown, various diagnostic strategies useful for making a correct diagnosis of COVID-19 have been rapidly developed by private research laboratories and biomedical companies. At present, rapid antigen or antibody tests, immunoenzymatic serological tests and molecular tests based on RT-PCR are the most widely used and validated techniques worldwide. Apart from these conventional methods, other techniques, including isothermal nucleic acid amplification techniques, clusters of regularly inter-spaced short palindromic repeats/Cas (CRISPR/Cas)-based approaches or digital PCR methods are currently used in research contexts or are awaiting approval for diagnostic use by competent authorities. In order to provide guidance for the correct use of COVID-19 diagnostic tests, the present review describes the diagnostic strategies available which may be used for the diagnosis of COVID-19 infection in both clinical and research settings. In particular, the technical and instrumental characteristics of the diagnostic methods used are described herein. In addition, updated and detailed information about the type of sample, the modality and the timing of use of specific tests are also discussed.
Keywords: SARS-CoV-2, COVID-19, diagnosis, viral detection, RT-PCR, rapid test, immunoenzymatic assay, ddPCR, isothermal amplification technique, CRISPR-Cas, molecular methods
1. Introduction
Europe and the entire world have faced the second wave of Coronavirus Disease 2019 (COVID-19) pandemic which was characterized by an increased number of infections and related deaths worldwide, thus still highlighting critical issues in the management of this health emergency (1,2). At the time of the writing of the present review article, 113,523,131 laboratory-confirmed COVID-19 cases and 2,519,454 COVID-19-related deaths have been recorded worldwide, highlighting the impressive impact of this pandemic globally (3).
Despite the optimism deriving from the approval of two new mRNA vaccines and of one recombinant vaccine against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections by the Food and Drug Administration (FDA) and the European Medicines Agency (EMA) (5,6), a third wave of infections, already observed in the United States, the United Kingdom and other countries with greater proportions than the one just concluded, is expected in the coming months (3,6,7). In this regard, several governments worldwide have already begun to adopt social distancing measures and the lockdown of collective activities in order to avoid a drastic increase in the number of infections (8,9). In addition, a great concern is also represented by the need to differentiate COVID-19 cases from seasonal flu that could clog hospital emergency services, slowing down the diagnostic and therapeutic procedures for patients with COVID-19 (10). These reasons have led the scientific community to question which diagnostic strategies are optimal in order to efficiently combat the imminent increase in COVID-19 infections, as well as to perform differential diagnoses between COVID-19 infections and seasonal flu.
In this context, population screening strategies have been proposed and are currently being implemented for the effective monitoring of the COVID-19 epidemiological curve and to screen the immunized population; however, it is not yet clear which is the most effective method for these surveillance programs (11-14).
Since the beginning of the pandemic, immense efforts have been made for the development of effective diagnostic strategies which may be used to accurately identify SARS-CoV-2-infected patients, thus limiting the risk of contagion and promptly treating any respiratory symptoms, avoiding serious consequences for individuals (15).
At present, several diagnostic methods have been approved by regulatory agencies worldwide; however, there is still confusion regarding the correct tests to be used based on the patient's medical history or the investigation purpose (16). As regards Europe, there are 365 different commercialized devices CE-IVD- and FDA-approved or used for research purposes. Of these devices, 168 are immunoassays, 192 are PCR-based methods, three are NGS-based methods and two commercialized tools are based on different technologies (17). Therefore, it is evident that the selection of the optimal diagnostic test can be difficult.
On these bases, the present review aimed to provide the rationale for the correct use of SARS-CoV-2 diagnostic tools currently available by setting out the decision-making principles useful for the correct choice of the most appropriate test.
As will be discussed in the following paragraphs, several diagnostic tests are currently available for the early identification of SARS-CoV-2 infection, for the monitoring of the presence of any infections among healthcare workers, for the monitoring the incidence rates and the severity of the infection and for the evaluation of the complete remission of patients with COVID-19 (18-20).
Several parameters should be considered for the selection of the optimal diagnostic test. A diagnostic test must have good sensitivity and specificity rates; however, these parameters are not the only features to be considered. Indeed, in the case of the COVID-19 pandemic, an effective test should be rapid, repeatable, based on technologies available at numerous centers and keep costs limited in order to be carried out on a large fraction of the population (21).
Therefore, it is evident that the selection of the diagnostic test should be performed taking into account the clinical or surveillance purpose of the investigation as well as the possibility to repeat the test several times until the patients are no longer positive.
For all these clinical and epidemiological needs, three main types of tests for COVID-19 diagnosis are available: i) Molecular RT-PCR swab tests; ii) serological tests; iii) rapid antigen or antibody tests.
2. The right test, on the right sample, at the right time
Although the molecular and structural characteristics of SARS-CoV-2 were initially unknown, in a very short period of time, research laboratories and biomedical companies studied the main features of the virus, thus assisting researchers world-wide in developing various diagnostic solutions for a correct diagnosis of COVID-19 (22-24). Among such solutions, the most commonly used and validated methodologies are rapid antigen or antibody tests, immunoenzymatic serological tests and RT-PCR-based molecular tests. Each of these three types of diagnostic tests can be applied at a precise moment of infection. Of note, only kits, reagents and molecular probes validated by the Centers for Diseases Control and Prevention (CDC) and the World Health Organization (WHO) and approved at the American and European level by the FDA and by the EMA can be used for diagnostic purposes (25).
Although immunoenzymatic (either classic or rapid methods) and molecular methods are the most widely used techniques for the diagnosis of COVID-19, other approaches were used in the early phase of the pandemic to identify positive patients and the etiological agent of infection. Among these approaches, viral culture and next-generation sequencing (NGS) methods were crucial for the identification of the novel coronavirus and for the characterization of its molecular structure. These two techniques have made it possible to fully characterize the genome and the viral protein structure, allowing the understanding of the viral mechanisms of action, the mode of transmission, the clinical impact and the development of any therapeutic strategies and diagnostic tools (26,27).
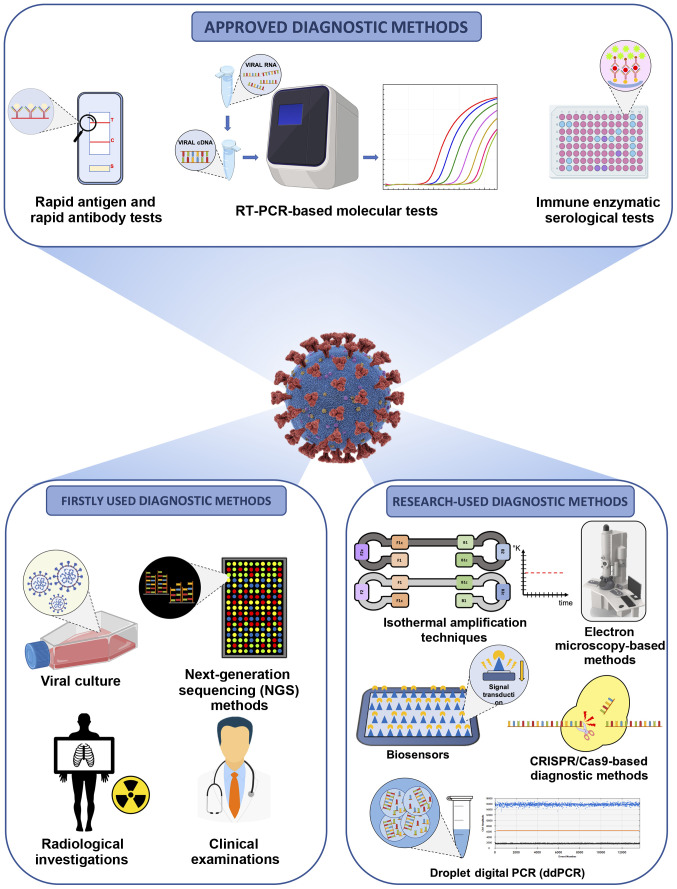
Apart from these conventional strategies, other diagnostic methods are under development and validation or are currently finding application in research contexts. Among these methods, digital PCR, isothermal amplification techniques, clusters of regularly interspaced short palindromic repeats/Cas (CRISPR/Cas)-based diagnostic methods, biosensors, electron microscopy-based methods, etc., represent the armamentarium to effectively diagnose COVID-19 infection and to effectively assess the epidemiological spreading of the pandemic (15,28). Notably, clinical investigations and radiological imaging have represented valid diagnostic alternatives, particularly during the early stages of the pandemic when there were still no validated molecular and serological tests available (29) (Fig. 1).
Figure 1.
Overview of the available clinical, diagnostic and research strategies for the effective diagnosis of COVID-19 infection.
Despite the availability of all these diagnostic techniques, a correct diagnosis of COVID-19 infection can only be established considering the test to be used, the type of sample to be analyzed and the timing of the test itself. Therefore, it is necessary to perform the correct test, at the correct time in the correct biological sample (30,31).
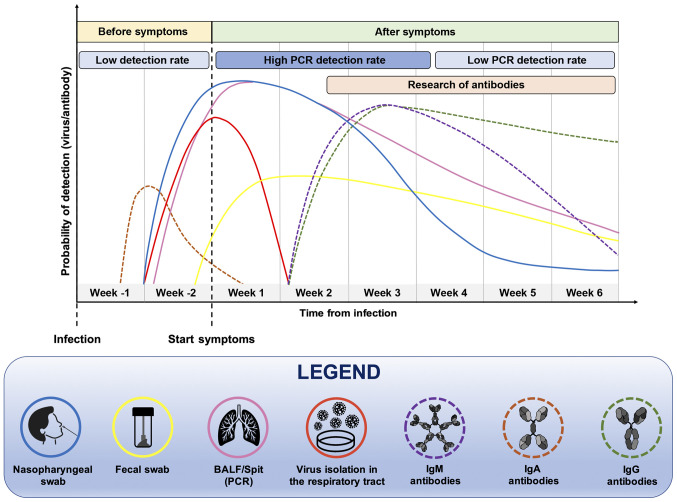
In particular, it is important to take into account the moment of the suspected infection, the patient's medical history, the symptoms and general clinical picture for a successful outcome of the diagnostic test. Furthermore, the positivity of a diagnostic test strongly depends on the moment at which it is carried out (Fig. 2). Both serological and molecular tests are not useful during the first week of the supposed infection because the virus is still in its incubation period and there are not yet sufficient copies of viral RNA in circulation neither antibodies nor viral proteins identifiable by serological tests (32,33). Therefore, before the onset of symptoms, the probability of correctly determining the presence of the virus, particularly using molecular tests, remains low (32).
Figure 2.
Timing and type of samples that should be analyzed for the effective detection of SARS-CoV-2 RNA or anti-SARS-CoV-2 human antibodies.
At two weeks after the presumed infection, and in parallel with the onset of symptoms, molecular tests carried out on nasopharyngeal swabs or bronchoalveolar lavage fluid (BALF) samples will have a greater chance of being positive, as the virus is actively replicating. However, this probability is reduced over time, as a result of the elimination of the virus and the remission of the disease. In the case of molecular tests, the probability of a positive or negative result should be considered, as no commercial diagnostic tools have a sensitivity of 100%, particularly in cases of patients with low viral load as asymptomatic or paucisymptomatic individuals (32,34).
As regards serological tests, the search for IgM and IgG antibodies begins to yield positivity at approximately one month after the presumed infection and the levels of these immunoglobulins remain high for long periods of time. Of note, the IgM serum levels decrease significantly after six weeks from the onset of symptoms (35). The detection of viral proteins and mucosal IgA antibodies is also very important. Indeed, these markers increase in the early stages of infection and can be used for the early diagnosis of COVID-19 infection (36). In summary, all these data suggest that for a correct diagnosis of COVID-19 infection to be made, it is necessary to use the right test, at the right time.
As already mentioned, the selection and the correct handling of samples for both molecular and serological analyses are fundamental for the diagnosis of COVID-19 infection. In particular, the collection of samples and the pre-analytical phases are crucial for the positive outcome of the diagnostic procedure (37,38). Different studies have demonstrated that the positivity rate of molecular tests significantly depends on the quality of the starting sample influenced by sample collection, poor quality material, wrong transport or storage, the presence of inhibitors, etc. (38). Technical troubleshooting will be discussed in the following chapters.
The chief samples used for molecular analyses are obtained from the respiratory tract. In particular, both oropharyngeal and nasopharyngeal swabs represent good samples for viral RNA extraction and amplification through RT-PCR; however, previous studies have highlighted that BALF is the most appropriate sample for SARS-CoV-2 molecular detection (39,40). Apart from these commonly analyzed respiratory tract samples, other specimens may also be collected, including nasal mid-turbinate swabs and nasal or nasopharyngeal wash/aspirate (41). Of note, the permanence of the virus in the respiratory tract is only temporary, with a peak of positivity in the first three days of infection, followed by a constant decrement of the rate of positivity for molecular analyses until the 10th week following symptom onset (42). To ascertain SARS-CoV-2 positivity after a long period of time, other biological samples are used. Among these, fecal samples represent the optimal material to evaluate the permanence of viral RNA in patients with negative nasopharyngeal swabs, but with clinical symptoms attributable to COVID-19 infection (43). Indeed, a greater persistence of SARS-CoV-2 in the gastrointestinal tract has been demonstrated, allowing the detection of SARS-CoV-2 RNA even after more than a month from infection (44).
Other studies have proposed diagnostic screening based on the analysis of saliva and urine samples through serological and molecular tests. However, the collection of these samples is generally carried out by the patients themselves without the surveillance of a healthcare professional, resulting in a possible non-representative sample. In addition, the presence of interfering substances or substances that degrade viral RNA or human antibodies represents a considerable bias that significantly limits the use of these samples in clinical practice (30).
All the types of biological samples used for diagnostic purposes are presented in Table I. A detailed description of the timing of use and the technique for which specific samples are collected is also provided (Table I).
Table I.
Biological samples and methods used for an effective diagnosis of COVID-19 infection.
| Type of specimens | Collection devices | Transport conditions | Diagnostic methods
|
Comments | |
|---|---|---|---|---|---|
| Approved diagnostic methods | Research-used diagnostic methods | ||||
| NP swab, OP swab, and NP aspirate | Dacron or VTM flocked swabs | Within 5 days, 4°C; >5 days, −70°C | Real-time PCR | NGS - CRISPR/Cas PCR - isothermal amplification techniques −ddPCR - viral culture | |
| Sputum | Sterile vial | Within 48 h, 4°C; >48 h, −70°C | Real-time PCR | NGS - CRISPR/Cas PCR - isothermal amplification techniques - ddPCR - viral culture | |
| Bronchial washing | Sterile vial | Within 48 h, 4°C; >48 h, −70°C | Real-time PCR | NGS - CRISPR/Cas PCR - isothermal amplification techniques - ddPCR - viral culture | Pathogens may be diluted; however, the specimen can be used for diagnostic testing |
| Tracheal aspirate and transtracheal aspirate | Sterile vial | Within 48 h, 4°C; >48 h, −70°C | Real-time PCR | NGS - CRISPR/Cas PCR - isothermal amplification techniques - ddPCR - viral culture | |
| Lung biopsy | Sterile vial | Within 48 h, 4°C; >48 h, −70°C | Real-time PCR | NGS - CRISPR/Cas PCR - isothermal amplification techniques - ddPCR - viral culture | |
| Serum, plasma | Serum/plasma collection tube: Adults, 3-5 ml; infants, 1 ml | Within 5 days, 4°C; >5 days, −70°C | Rapid serological test; immune enzymatic test | Biosensors | For the immune enzymatic test, two samples are collected. The first sample is collected between 1-7 days after symptom onset and the second is collected 14 days after the onset of symptoms |
Overall, the type of sample should be collected considering the timing of the infection. Collecting different samples from different sites may be useful to avoid misdiagnosis of asymptomatic patients negative for molecular tests. Otherwise, it is possible to carry out repeated nasopharyngeal swabs in two or three consecutive days in order to overcome the window period of SARS-CoV-2 incubation thus being able to correctly diagnose a patient infected with SARS-CoV-2 (45).
3. RT-PCR-based molecular tests
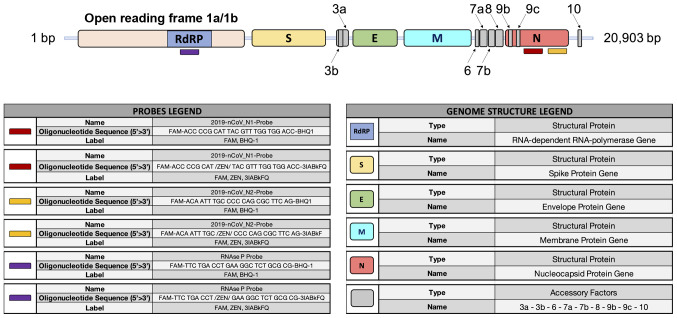
RT-PCR-based molecular methods represent the gold standard techniques used worldwide to make a confirmatory diagnosis of COVID-19 infection (46). Since the complete sequencing of the SARS-CoV-2 genome (26), researchers of different countries have begun to design molecular primers and probes specific to SARS-CoV-2 RNA sequences in order to perform differential diagnosis between COVID-19 infections and other pathologies with similar symptoms, such as seasonal flu or bacterial infections (47,48). Of note, the whole genome sequence of SARS-CoV-2 is 29,903 bp in length containing the following functional elements: A poly-A cap of 50 bases, the open reading frame 1/ab (ORF1/ab) containing the coding sequences for the RNA-dependent RNA polymerase (RdRP), the spike proteins, the envelope proteins, the membrane and nucleocapsid proteins, and a poly-A tail of 30 bp (49).
At present, several portions of the SARS-CoV-2 genome are used for the design of specific primers and probes, including the genomic portions encoding for the RdRP gene, for proteins constituting the nucleocapsid (N gene) and spike molecules (S gene), for proteins of the envelope (E gene), for the membrane, etc. (Fig. 3).
Figure 3.
SARS-CoV-2 genome structure and probes for the molecular detection of SARS-CoV-2 RNA in human samples.
The CDC has made available a list of approved and validated kits and reagents to clinically diagnose COVID-19 infection (50). Similarly, the CDC has also made available a list of three primer pairs that can be used for research purposes only, each working with two different molecular probes, specific for two portions of the N gene and the gene coding for viral RdRP, respectively (51).
RT-PCR-based molecular tests are considered the optimal diagnostic option for wide surveillance strategies due to the relatively low costs of the entire viral RNA extraction, reverse transcription and amplification procedure, and the availability of RT-PCR thermal cyclers in hospitals, research institutes and private laboratories (52). Other advantages of RT-PCR methods compared to other diagnostic techniques are the timesaving of the procedure, the easy execution of the technique and the non-necessity of highly trained personnel (53). In addition, a number of the available RT-PCR kits are based on one-step amplification methods, where the buffer of the nasopharyngeal swab is inserted into the plate and the machine autonomously provides for the extraction, reverse transcription, amplification and analysis of the samples. These procedures ensure fast results guaranteeing excellent reproducibility and standardization of the data obtained which are less influenced by operator bias (54).
As already mentioned, the EMA has approved 192 PCR-based methods while the FDA has approved 235 different molecular tests for both RT-PCR and the rapid detection of SARS-CoV-2 RNA (17,55).
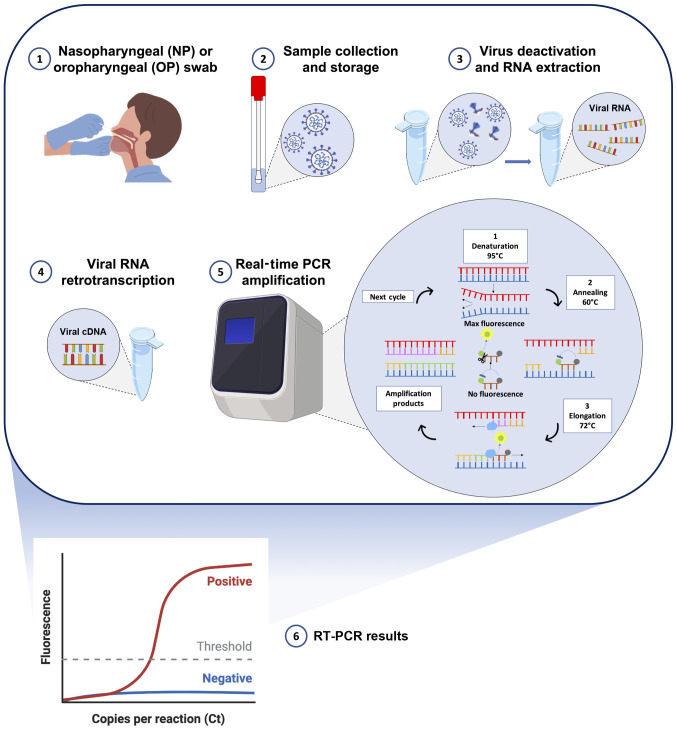
All these approved molecular tests detect two or three fragments of SARS-CoV-2 RNA mainly using multiplex assays based on standard RT-PCR protocols. Briefly, following the collection of nasopharyngeal swabs from individuals with suspected COVID-19 infection, the viral RNA is extracted using commercial kits and lysing solutions with manual or automated extraction protocols. Subsequently, the obtained RNA, containing both human and viral RNA, can be directly analyzed by RT-PCR in the one-step protocols or must be retrotranscribed into complementary DNA (cDNA) before RT-PCR amplification. After obtaining the cDNA, the SARS-CoV-2 targets are amplified through RT-PCR using TaqMan probes specific for the two-three viral targets. In particular, the exonuclease activity of the Taq polymerase (5′→3′ direction) cleaves the probes annealed with the viral targets allowing the emission and real-time detection of fluorescent signals. In this manner, the intensity of the fluorescent signal is proportional to the total amount of the amplified targets. However, all the existing RT-PCR protocols are only qualitative and not quantitative (Fig. 4) (56,57).
Figure 4.
Schematic workflow of RT-PCR-based diagnostic methods. 1) Collection of samples from suspected COVID-19 patient through nasopharyngeal or oropharyngeal swab; 2) storage and handling of the swab to preserve viral RNA integrity; 3) SARS-CoV-2 heat inactivation and RNA extraction through custom or commercial protocols; 4) viral RNA retro-transcription into double strand cDNA; 5) RT-PCR amplification and real-time fluorescent signal detection; 6) interpretation of amplification signals and setting of the positivity threshold.
The entire analytical procedure is completed in 4-8 h, based on the type of RT-PCR protocol used (one-step or two-steps). This makes it possible to establish the positivity of an individual in a relatively short amount of time, allowing the initiation of quarantine protocols that limit the spread of infections (58,59). In addition, the majority of the automated or semi-automated systems available are based on 48- or 96-well platforms in order to process a series of samples, thus reducing the execution times, the costs of plastics and consumables and procedural errors that may occur by analyzing the samples individually (60).
RT-PCR-based methods ensure also a low limit of detection (LoD) of SARS-CoV-2 RNA (61). Specifically, during the early stages of the pandemic, when diagnostic techniques had not yet been optimized and standardized, a significant fraction of COVID-19-positive patients were identified as false-negative due to the low sensitivity of the primers and probes used or the inaccuracy of the whole RT-PCR procedure (false-negative rates ranging from 38% at the day of symptoms onset to 67% before one day from the onset of symptoms or to 66% after 16 days from the onset of symptoms) (62). As will be better described at the end of this chapter, in the case of asymptomatic or paucisymptomatic patients, generally characterized by a low viral load, the RT-PCR investigations could mistakenly yield negative results. Currently, the commercially available RT-PCR kits have partially solved the problem of the low sensitivity of RT-PCR. Indeed, the methods available today have a theoretical LoD that varies from 0.3 copies/µl to 100 copies/µl, depending on the diagnostic system used (63). However, it should be considered that this limit is only theoretical; therefore, in clinical practice, procedural errors or reaction interferers raise the LoD by 10-fold (64).
Although RT-PCR represents the gold standard method for the diagnosis of COVID-19 infection, this method is subject to several limitations and criticisms that can lead to false-positive or false-negative results, thus affecting the correct management of the pandemic. As already mentioned, one of the main limits of RT-PCR is its low sensitivity in correctly diagnosing samples with low viral load, including swabs taken incorrectly or obtained from asymptomatic or paucisymptomatic individuals (65). In addition, RT-PCR is affected by contaminants and interferers contained in the sample or introduced by the operator capable of inhibiting the reaction (65). Other limits also concern the execution time of the analysis, which in non-automated systems, can take up to 24 h to obtain a result that can be communicated to the patient. Finally, RT-PCR is a method that is profoundly affected by pre-analytical and analytical bias regarding the collection, storage and handling of samples, therefore, careful attention should be paid during the collection and management of samples (66).
In order to reduce the errors in the interpretation of molecular tests, the WHO has published specific recommendations useful to make a correct diagnosis of COVID-19. In particular, a molecular test must be conducted in two different biological matrices, for example, a nasopharyngeal swab and a fecal swab, or be performed on two consecutive nasopharyngeal swabs in order to obtain reliable results. The approved tests include the analysis of three different viral genes or two viral genes and a human control gene and the use of specific negative and positive controls useful to ascertain the presence of contamination or evaluate the inhibition of the reaction (67).
Overall, RT-PCR represents the gold standard and most widely used method worldwide to make an accurate diagnosis of COVID-19 infection due to the rapidity of the method and the availability of instruments in public and private hospitals and laboratories. In addition, the RT-PCR tests currently available on the market ensure good sensitivity and specificity rates for the diagnosis of COVID-19 infection. The main RT-PCR diagnostic systems currently available on the market, illustrating their main technical features are presented in Table SI.
4. Rapid antigen and rapid antibody tests
With the increase in the number of individuals with a suspected COVID-19 infection, it became necessary to adopt more rapid and low-cost diagnostic strategies to carry out extensive surveillance campaigns (68-70). To cope with this emergency, various rapid tests have been developed to detect viral antigens or anti-SARS-CoV-2 human antibodies in salivary, nasal or oropharyngeal swabs and blood samples. These tests are currently adopted for the frequent monitoring of personnel operating in at-risk environments such as schools or hospitals or to carry out extensive screening strategies on populations where a new outbreak of infection is suspected (70,71).
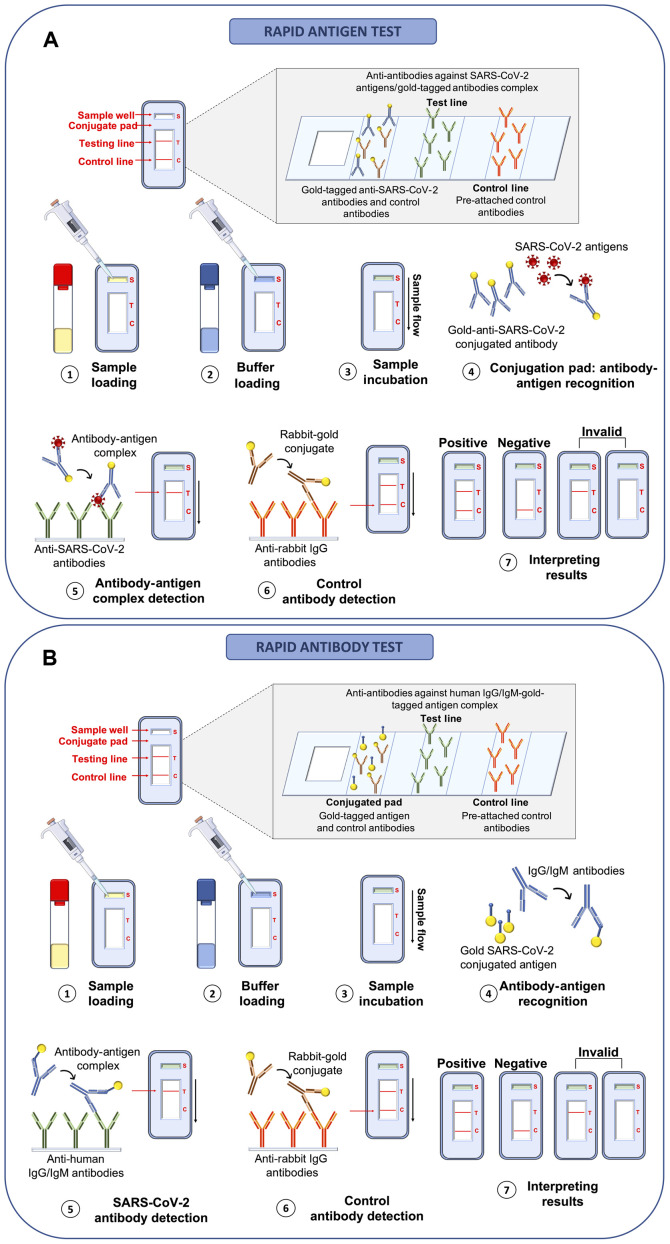
Compared to RT-PCR-based methods, rapid antigenic and rapid antibody tests are characterized by more rapid execution times of ~15-30 min, a lower cost and an easier procedure that does not require the presence of highly trained personnel (72). These tests are mainly built on platforms based on the principle of lateral flow immunoassay (LFIA) for the direct detection of viral proteins (rapid antigen tests) or human antibodies against SARS-CoV-2 antigens (rapid antibody tests). As regards rapid antigen tests, these allow the identification of COVID-19-positive individuals through the detection of SARS-CoV-2 nucleocapsid or Spike proteins (viral antigens) in swabs collected from the upper airways of the subject with suspected infection. The principle behind these LFIA rapid antigen tests is very simple and based on the bond between antigens and antibodies that occurs on the surface of a porous membrane where the swab buffer flows by capillarity. Briefly, the swab buffer is loaded in the sample well of the cartridge and flows by capillarity at the level of the conjugation pad containing control rabbit antibodies and specific antibodies against SARS-CoV-2 antigens both linked to detector molecules (conjugated antibodies). In the case of a positive sample, the link between viral antigens and conjugated antibodies take place at the level of the conjugation pad. Subsequently, the buffer containing conjugated control rabbit antibodies and the antigen-conjugated antibody complex flows to the test line where other antibodies specific for viral antigens are immobilized. In the case of a positive sample, the binding between antibodies immobilized in the test line and the antigen-conjugated antibody complex gives rise to a colorimetric reaction indicating the positivity of the sample. Finally, the buffer flows further to the control line where anti-antibodies specific for the conjugated control rabbit antibodies are immobilized; if the test is performed correctly, the reaction between these two molecules also gives rise to a colorimetric reaction. To be trusted, the test control line must always be positive (73,74) (Fig. 5A).
Figure 5.
Rapid antigen and rapid antibody tests. (A) Analytical workflow of rapid antigen test for the rapid detection of SARS-CoV-2 viral antigens through lateral flow immunoassay. (B) Analytical workflow of rapid antibody test for the rapid detection of human IgA, IgG or IgM antibodies against SARS-CoV-2 antigens through lateral flow immunoassay.
Similarly, rapid antibody tests use the same LFIA principle; however, human IgA or IgG and IgM against SARS-CoV-2 antigens are searched for. In particular, the blood (or saliva) sample is loaded inside the sample well and flows to the conjugated pad containing gold-tagged viral antigens and gold-tagged control antibodies. In this test, anti-SARS-CoV-2 human antibodies (IgA, IgG or IgM) bind the gold-tagged antigens and the sample flows to the test line where anti-human IgA, IgG or IgM antibodies are pre-attached to the membrane. In the case of a positive sample, the IgA, IgG or IgM-gold-tagged antigen complex binds to the human anti-antibodies immobilized in the test line determining a colorimetric reaction which indicates the positivity of the sample. Finally, the gold-tagged control rabbit antibodies flow to the control line binding anti-rabbit antibodies thus giving rise to a confirmatory colorimetric reaction (75,76) (Fig. 5B).
Both rapid antigen and rapid antibody tests yield results readable to the naked eye in a very short period of time and can be performed at the 'point-of-care' without any specific instruments or sample processing. Of note, these tests yield qualitative, but not quantitative results; therefore, it is only possible to establish whether the individual is positive or not for COVID-19 infection without assessing the viral load. In addition, in the case of rapid antibody tests, it is possible to establish whether the patient is carrying anti-SARS-CoV-2 antibodies; however, it is not possible to establish whether the patient has an active SARS-CoV-2 infection or already resolved disease (77,78).
Overall, among the main advantages of rapid antigen and rapid antibody tests are the low cost, the possibility of carrying out the test directly at the point-of-care and the high speed and easy execution that ensure a positive or negative result in ~30 min. However, these tests suffer from important limitations mainly related to a low sensitivity and specificity of 56.2 and 99.5%, respectively (79). Indeed, a recent review of the literature demonstrated that rapid antigen tests had a false negative rate of 27.9%, while no false-positive results were observed (68). As regards rapid antibody tests, a recent study comparing three different kits demonstrated a false-positive rate ranging from 51.6 to 28.1%, and a false-negative rate ranging from 0 to 4.2% (80).
As regards rapid antibody tests, these are very straight-forward and can be quickly performed, and provide useful information about the stage of infection. Indeed, in the case of negativity, only the control band is colored. In the case of an ongoing infection, the IgM and control bands are stained. In the case of a recent infection, both the IgM and IgG bands are stained, since both antibodies are present in the bloodstream. Following the remission of the disease, for a period of time that varies from patient to patient, only the IgG band is positive together with the control one, and in the case of re-infection, all the three bands are positive again (Fig. S1). The test is considered invalid when none of the lines stain or when the test lines are stained but the control line is not (81).
The limited sensitivity and the related high false-negative results are mainly related to the low viral load and the low antibody response observed in some patients; however, as already mentioned, the probability of obtaining a positive test depends also on the time of the presumed infection and on the test execution time. Indeed, although the viral antigens are found in the samples after a short time from infection, their permanence and stability in the biological sample are limited; therefore, it is not always possible to correctly identify these proteins (82). Similarly, rapid antibody tests are mainly designed to identify IgM and IgG antibodies, which are not produced by the body immediately, but begin to be found in the bloodstream after the third week of the suspected infection. Therefore, it is important to use the most appropriate test considering the time of the presumed infection. More recently, the use of rapid tests for the detection of IgA to accelerate the diagnosis of COVID-19 infection has been proposed (83).
Overall, rapid antigen and rapid antibody tests are widely used for screening strategies on large portions of the population (70,84); however, they do not ensure a precise diagnosis of COVID-19. Therefore, these tests should be always confirmed by RT-PCR analyses. Details of rapid antigen and rapid anti-body tests currently approved by international agencies are reported in Table SII.
5. Immunoenzymatic serological tests
Most of the immunoenzymatic serological tests used for COVID-19 investigations are based on the principle of indirect enzyme-linked immunosorbent assay (ELISA). Of note, ELISA is a colorimetric, chemiluminescent or fluorescent microwell plate-based assay used for the quantitation and detection of human proteins, immunoglobulins, antigens and other peptides through the binding between the target protein and a specific antibody that results in a detectable signal (85). This technique allows researchers to obtain highly specific and sensitive results in a relatively short time ranging from 1 to 5 h (85).
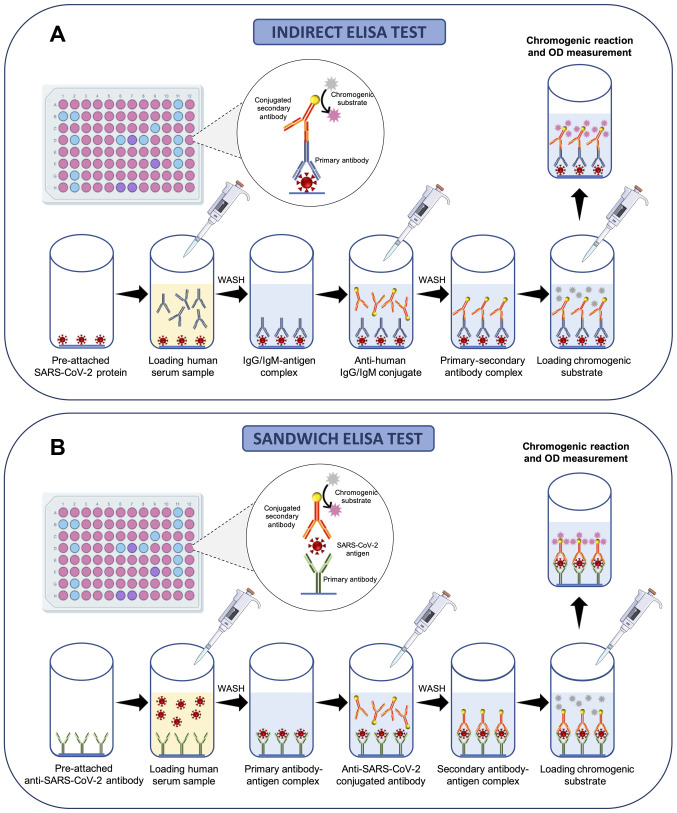
Briefly, 96-well commercial COVID-19 ELISA indirect tests contain immobilized viral antigens at the bottom of each well that are recognized and bound by anti-SARS-CoV-2 antibodies present in the serum of patients following proper dilutions. A series of washes is then made to remove the serum and unbounded antibodies; subsequently, a conjugated anti-body specific for human immunoglobulins is added to each well. Following further washing, a chromogenic (or fluorescent or chemiluminescent) substrate is finally added and metabolized in the presence of antibodies against SARS-CoV-2 and enzyme-conjugated anti-human antibodies. This process gives rise to a colorimetric reaction easily detectable by optical densitometry (or fluorescence or chemiluminescence) whose intensity is indicative of the quantity of IgG, IgM or IgA anti-body presents in the sample (Fig. 6A).
Figure 6.
ELISA-based immune enzymatic serological tests. (A) Analytical workflow of indirect ELISA for the effective detection of human IgA, IgG or IgM antibodies against SARS-CoV-2 antigens. (B) Analytical workflow of sandwich ELISA for the effective detection of SARS-CoV-2 antigens.
Similarly, 96-well commercial COVID-19 ELISA sandwich tests contain immobilized antibodies against SARS-CoV-2 antigens at the bottom of each well able to bind antigens contained in the serum samples of patients. Subsequently, a series of washes is performed to remove the serum and unbounded antigens and a conjugated antibody specific for SARS-CoV-2 antigens is added to each well. Finally, following further washing, a chromogenic substrate is added and metabolized in the case of positive samples giving rise to a colorimetric reaction (Fig. 6B).
As regards the management of the COVID-19 pandemic, ELISAs are currently used for the detection of IgM and IgG antibodies (86) specific for SARS-CoV-2 antigens or for the identification of viral Spike proteins (87). More recently, other ELISAs have been developed for the detection of human IgA antibodies whose determination is of fundamental importance as they are the first antibodies to be produced following exposure to the virus (86).
In a very short period of time, several COVID-19 ELISAs have been developed and some of these have been approved by international agencies (Table SIII). These tests are mainly adopted to monitor the immunological status of patients or for the immunosurveillance of specific work categories, such as healthcare workers or school personnel (88,89). In particular, ELISA tests for the detection of IgG and IgM are often performed on COVID-19 patients who obtained a negative result in molecular tests conducted on nasopharyngeal swabs in order to ascertain the seroconversion of patients and the acquisition of immunocompetence against COVID-19 infection (90). Contrariwise, ELISAs for the detection of viral proteins and anti-SARS-CoV-2 human IgA can be used for diagnostic purposes or for large screening strategies, as these molecules are rapidly founded in clinical samples (83,91). Of note, ELISAs for the detection of viral antigens are based on sandwich ELISA instead of indirect ELISA (92).
Several ELISAs have been approved for the management of COVID-19 infection, allowing the identification of individuals exposed to the SARS-CoV-2 virus (Table SIII); however, these tests are not able to confirm the infectious status of the subject. Indeed, in the case of an ELISA-positive result (either IgA, IgM, IgG or viral antigens), it is necessary to perform molecular confirmatory analyses on nasopharyngeal swabs (93). However, ELISA-based serological tests are much more reliable than rapid antigen or antibody tests; the sensitivity and specificity values of these tests range from 75.6 to 100%, and 85.7 to 100%, respectively, albeit important variations in these values may be related to both the manufacturer or the human antibody or viral antigen tested (94). Indeed, generally, the search for IgG is more accurate compared to that of IgM or IgA (this latter is less sensitive) (94). Of note, both sensitivity and specificity depend on the timing of infection and the timing of test execution.
Overall, SARS-CoV-2 ELISAs represent a good clinical option for large screening and surveillance campaigns mainly adopted for specific work categories due to the rapidity of this method, the possibility of analyzing multiple samples in one round and the availability of automated or semi-automated systems that allow a precise quantitation of viral antigens or human antibodies.
6. Alternative methods for the effective diagnosis of COVID-19 infection
The COVID-19 pandemic has prompted research groups worldwide to develop novel technologies for the diagnosis of COVID-19 infection or to readapt existing diagnostic systems according to the characteristics of the new SARS-CoV-2 virus.
When the infection had not yet assumed the dimension of a global pandemic, traditional culture methods, clinical investigations and NGS techniques were among the first methods used to diagnose COVID-19 infection. Apart from these approaches, other methods have been developed, including biosensors, CRISPR/Cas-based tests, nucleic acid isothermal amplification methods, electron microscopy, etc. (25,95).
Viral culture and electron microscopy
Viral culture has represented the fundamental method that allows the identification of SARS-CoV-2 as a novel causative agent of human pneumonia (96). Despite the difficult realization and the long period of times necessary to obtain a viral culture in vitro, viral isolates represent a milestone for the discovery of novel viral infections (96,97). As regards SARS-CoV-2 infection, viral culture was fundamental in the initial phase of the outbreak prior to the development of other diagnostic assays. Zhu et al (96) (2020) were the first to obtain SARS-CoV-2 viral isolates from clinical specimens and to observe cytopathic effects using transmission electron microscopy. Briefly, the authors of that study inoculated 150 µl of BALF supernatant obtained from a COVID-19 patient into pathogen-free human airway epithelial cells. Following 2 h of incubation at 37°C, infected epithelial cells were washed with phosphate-buffered saline and incubated at 37°C for a long time period. To assess the efficacy of infection and the production of novel viral particles, the authors of that study collected cells supernatant every two days for molecular analyses and observed cytopathic effects under light microscopy. Finally, infected cells were prepared for electron microscopy observation (96).
Subsequently, following the study by Zhu et al (96), other research groups isolated SARS-CoV-2 with an aim to study the structural features and molecular interaction with infected cells (98,99). For these purposes, other cell lines have been used, including the Vero and LLC-MK2 cell lines; by using electron microscopy and cells infected with clinical specimens obtained from COVID-19 patients, it was possible to identify the ultrastructural details of the virus, the interaction between virus and cells and the resulting cytopathic effects (100).
Of note, electron microscopy is one of the pioneering methods for the discovery of novel pathogens, allowing the identification of their structural features. As regards viral infections, two main applications of electron microscopy exist: Solid-phase immune electron microscopy (SPIEM) and immunolabeling electron microscopy (IEM), which are based on the observation of cells blocked in the surface of a grid and on the observation of antibody-antigen complex occurring in infected cells, respectively (101,102).
Overall, viral culture and electron microscopy are important techniques used to observe the main characteristics of the virus. In the case of SARS-CoV-2, these two methods allowed the identification of the typical structure of coronaviruses characterized by a nucleocapsid enclosed within a crown-like envelope composed of spike proteins. As regards the cytopathic effects, both methods displayed a broad range of cellular alterations mainly represented by the formation of plaques characterized by a net-like structure or fused cells. These plaques, composed of multinucleated syncytial cells, also present deformed cilia with a granular formation and a disordered polarity. SARS-CoV-2-infected cells also exhibited double-membrane vesicles and degraded mitochondria. Finally, viral infections also led to the expansion of the endoplasmic reticulum and an increased number of secretory vesicles (96).
Overall, these techniques have made it possible to establish the main characteristics of the novel SARS-CoV-2 virus, which then allowed researchers to develop the diagnostic systems currently used, as well as to propose the first therapeutic approaches for the treatment of COVID-19 infection. Despite the importance of both viral culture and electron microscopy, both methods present some issues that limit their use in clinical settings. Indeed, viral culture is time-consuming and requires specific equipment and high biosafety levels. For these reasons, the CDC recommends SARS-CoV-2 viral culture only for research studies carried out in laboratories equipped with level 3 biosafety cabinets (103). On the other hand, electron microscopy is not widely used as it requires costly instruments and highly trained personnel with specific skills in sample preparation and electron microscopy image analysis. In addition, this technique is characterized by low diagnostic sensitivity and specificity and optimal results can be obtained only if appropriate viral cultures are available (104).
NGS
Apart from viral culture and electron microscopy, NGS has represented a key method for the identification of SARS-CoV-2 and for the development of almost all of the currently adopted molecular diagnostic methods. Through NGS, it was possible to fully characterize the entire genome of SARS-CoV-2, thus establishing that it belongs to the β-coronavirus genus (49). The de novo sequencing of SARS-CoV-2 was performed using the nanopore technology through a sequence-independent single-primer amplification approach (105,106).
At present, NGS is not used for diagnostic purposes, but for molecular epidemiology and for the discovery of novel molecular variants. Indeed, its diagnostic application is limited due to the high costs of the analysis, the requirement of expensive technologies and the need for highly trained personnel with molecular and bioinformatics skills (25). Despite the high procedural costs, some companies have proposed commercial tests for the sequencing of SARS-CoV-2 through NGS platforms. In particular, the commercial kits available are mainly based on NGS techniques coupled with hybrid capture methods (BioCat, Arbor Biosciences and Swift tests). These platforms are built with biotinylated RNA probes that hybridize SARS-CoV-2 RNA fragments. Subsequently, the biotinylated probes are amplified through PCR using streptavidin-coated beads (107,108).
Apart from these platforms, more complex NGS tools have been developed to detect mutations in the sequence of SARS-CoV-2 genome, thus identifying novel emerging strains important from an epidemiological point of view and for the development of novel vaccines. Among these tools, amplicon-based metagenomic sequencing represents the most powerful approach with which to rapidly identify and comprehensively characterize SARS-CoV-2 and other pathogens. In particular, metagenomic sequencing allows the identification of the normal microbiome of patients, while amplicon-based sequencing allows the amplification and the subsequent sequencing of SARS-CoV-2 viral RNA. Together, amplicon-based sequencing and metagenomic sequencing are able to correctly diagnose COVID-19 infection, thus also identifying secondary infections due to other pathogens aggravating the health status of patients (109).
As already mentioned, amplicon-based and metagenomic approaches based on the sequence-independent single primer amplification (SISPA) method are able to detect mutations occurring in the sequence of SARS-CoV-2 and potentially associated with vaccine inefficacy or resistance to antiviral therapies (110,111). Examples of this type of sequencing technique are the MinION and IDbyDNA platforms produced by Oxford Nanopore Technologies and Illumina, respectively (112). As regards the IDbyDNA platform, it ensures the collection of >13 million reads of which >8 million are unique reads with an average length of ~75 bp and an in-depth coverage. This shotgun sequencing allows the generation of high-quality library score and Q score ensuring the correct identification of single variants in the SARS-CoV-2 sequence (113).
Similarly, MinION technology ensures the collection of millions of short and ultra-long reads (4,000 bp in length) obtaining output data up to 30 Gb. This technology is based on a portable platform that allows the real-time analysis of clinical samples with limited costs (114). This technology was effectively used for the analysis of the SARS-CoV-2 genome by using primers for 16 conserved binding sites of coronavirus allowing the reconstruction of the whole genome of SARS-CoV-2 through the generation of 1,000 bp reads with overlapping regions each other (75).
Overall, NGS whole-genome sequencing is the most powerful method for the molecular characterization of SARS-CoV-2, for the identification of novel variants during genomic surveillance screening and for the development of genome-based therapeutic approaches (115-117).
Clinical investigations and imaging techniques
During the early stages of the infection, when the causative agent was still unknown, the diagnosis of COVID-19 was predominantly clinical based on the observation of the patient's respiratory and extra-respiratory symptoms and on the use of radiological imaging techniques (118,119).
Of note, a significant fraction (~50-75%) of COVID-19 patients is asymptomatic or paucisymptomatic, presenting mild symptoms for a limited period of time, while other patients (~10%) present severe respiratory symptoms resulting in acute respiratory distress syndrome (ARDS) responsible for dyspnea, interstitial pneumonitis, multiorgan dysfunctions and in some cases, even death (120,121). Such severe manifestations are often observed in patients with pre-existing comorbidities, such as diabetes, cardiovascular diseases, hypertension and cancer, and are mediated by specific host cell entry mediators (122-126).
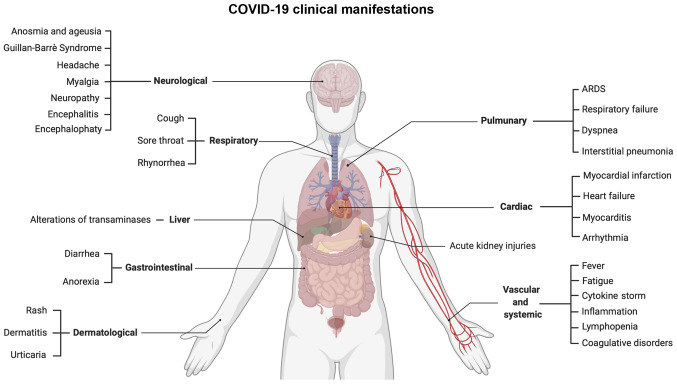
Among the COVID-19 symptoms, there are not only respiratory manifestations, but also other systemic symptoms. According to a recent comprehensive review, the main respiratory symptoms include, but are not limited to dyspnea (19-64%), cough (69-82%), rhinorrhea (4-24%) and a sore throat (5-14%). Other common symptoms are fever (44-98%), headaches (5-14%) and diarrhea (2-5%) (127). However, the most common symptoms reported by patients with COVID-19 are anosmia and ageusia, together with fatigue (128,129).
Apart from these commonly reported symptoms, a plethora of clinical manifestations has been reported, ranging from cardiovascular to neurological disorders and from gastrointestinal to systemic symptoms (Fig. 7). In particular, SARS-CoV-2 exhibits a neuroinvasive behavior via the retrograde transsynaptic invasion of the central nervous system. SARS-CoV-2 is able to bind specific receptors in the cells constituting the olfactory bulb or pulmonary and airways sensorial receptors. Among the most frequent neurological symptoms, there are headaches, neuropathy, myalgia, encephalitis, encephalopathy, etc. (130).
Figure 7.
Respiratory and extra-respiratory clinical manifestations of COVID-19 infection.
Other common symptoms are related to systemic inflammation responsible for the alteration of coagulative and hematological parameters and for the so-called 'cytokine storm' observed in patients with COVID-19 with severe respiratory symptoms. In fact, inflammation leads to the increase of fibrin/fibrinogen debris and D-dimer associated with coagulopathy. In rare cases, such alterations may induce disseminated intravascular coagulopathy that requires anticoagulant prophylactic or curative treatments (131). In addition, inflammation is also responsible for lymphopenia and T-cell exhaustion (132). Strictly related to these hematological alterations are cardiovascular disorders, such as acute myocardial injury, coronary syndrome, cardiomyopathy, myocarditis, arrhythmias, etc. (133).
Among gastrointestinal disorders, COVID-19 infection is responsible for nausea, vomiting, weight loss, anorexia and more frequently, diarrhea. Among these symptoms, hepatic manifestations mainly represented by increasing levels of transaminases are also included (134). Finally, other symptoms affect the kidneys, skin, endocrine system, etc. (135).
Overall, COVID-19 symptoms are mild in 80-90% of positive individuals; however, a small fraction of patients experiences severe symptoms that require hospitalization. Approximately 5% of patients develop interstitial pneumonia associated with respiratory failure, cytokine storm and multiorgan failure that could lead to patient death (127,136).
Therefore, the careful clinical evaluation of all these symptoms, together with radiological and laboratory data, helps the clinicians to correctly diagnose COVID-19 infection, and thus to the timely commencement of effective therapeutic protocols (24).
As already mentioned, radiological investigations help clinicians to correctly diagnose COVID-19 infection in the case of a suspicious case of pneumonia. Among radiological imaging techniques, chest X-ray (CXR) and computed tomography (CT) are the most powerful methods for the diagnosis of COVID-19 pneumonia (137,138).
CXR is usually used for the detection of pulmonary abnormalities following lung injuries due to infectious or neoplastic diseases (139). During the first phase of the COVID-19 outbreak, CXR was widely used to detect multifocal opacities affecting mainly lung interstitial space and alveoli in patients with COVID-19-related symptomatology (140). In particular, CXR is mainly adopted for patients suspected of COVID-19 infection with moderate or severe symptomatology who usually exhibit interstitial opacities (71.7%), or alveolar opacities (60.5%) frequently affecting both lungs (64.5%) (141). These radiological findings become more severe over time with the progression of symptoms and are mostly found in elderly patients with previous pulmonary parenchyma alterations (such as patients with chronic obstructive pulmonary disease) who exhibit both bilateral interstitial and alveolar opacities (142).
Despite the low cost and the rapid radiological findings obtained through CRX, some pulmonary abnormalities are not clearly detectable through this technique. Therefore, besides CRX, CT scan is frequently adopted to better display lung abnormalities mainly represented by bilateral interstitial ground-glass opacities (143). In particular, the CT scan has a great resolution power with a sensitivity of ~95-100%, although the specificity is very low, as this method does not allow for the discrimination of pulmonary alterations associated with other etiological agents other than SARS-CoV-2 (144).
The CT scan has been demonstrated to be an important technique for the diagnosis of asymptomatic patients with COVID-19 negative for molecular tests due to low viral load (145,146). In fact, the CT scan is able to discriminate the presence of morphological abnormalities in the lung already during the early stages of infection; however, this technique is not effective in correctly diagnosing a COVID-19 infection characterized by the absence of respiratory symptoms or where there is no involvement of the lung parenchyma (147). Other limits of the CT scan are represented by the needs of two radiologists' evaluations and the risk of cumulative radiations. This latter issue is addressed through the use of low-dose CT scan or ultra-low-dose CT scan often used for the long-term monitoring of COVID-19 patients with severe respiratory symptoms (148).
Overall, the radiological hallmarks of COVID-19 infection are the bilateral interstitial ground-glass opacities that may also affect the peripheral areas and the alveolar parenchyma. These alterations together with clinical symptoms, lead clinicians to perform a diagnosis of COVID-19 infection.
Biosensor COVID-19 testing techniques
To effectively diagnose COVID-19 infection rapidly and directly at the point-of-care, several electrochemical biosensing systems have been developed. These biosensor-based tools use the principle of impedance and electrochemical reactions occurring when viral RNA or proteins bind specific probes or antibodies. Different types of biosensor platforms are currently available for the diagnosis of COVID-19. These include electrochemical biosensors, colorimetric biosensors, fluorescence-based biosensors, surface-enhanced Raman scattering (SERS) biosensors, quartz crystal microbalance (QCM) biosensors, localized surface plasmon resonance (LSPR) and other platforms that ensure the accurate and fast detection of SARS-CoV-2 particles (149-152).
Among these platforms, the most commonly used are electrochemical biosensors and SERS adopted as point-of-care platforms due to the limited size of instruments, the low cost and the easy execution of the procedure (153).
Electrochemical biosensors can be used for the detection of SARS-CoV-2 proteins or for the detection of viral RNA. Seo et al (154) (2020) described an innovative biosensor for the detection of a low concentration of SARS-CoV-2 spike protein (LoD, 1 fg/ml) built with a graphene sensor with immobilized anti-SARS-CoV-2 spike antibodies. This type of field-effect transistor biosensor (FET) allowed the effective identification of SARS-CoV-2 in different types of samples (PBS, transport medium, viral culture medium, etc.) through the evaluation of electrical performance after S protein-antibody interaction suggesting its application in clinical settings (154).
As regards electrochemical systems for viral RNA detection, Zhao et al (155) (2020) developed an ultrasensitive electrochemical biosensor built with calixarene functionalized graphene oxide containing specific probes for SARS-CoV-2 RNA. This system allows the detection of viral RNA without nucleic acid amplification systems through the use of capture probes and label probes specific for SARS-CoV-2 RNA that through a calixarene substrate are able to detect electrochemical mediators, including toluidine blue and gold nanoparticles, generated by the binding between probes and the viral RNA. The device is portable and the analysis of data can be performed on smartphone apps as a point-of-care testing (155).
Several other types of biosensors have been produced; however, their detailed description is beyond the scope of the present review. Overall, these biosensors exhibited a sensitivity ranging from 86.43 to 93.75%, and a specificity of 90.63-100% depending on the platform used (156). As already mentioned, biosensors present several advantages mainly represented by high sensitivity and specificity, low costs of analysis, rapid execution time, the optimal LoD and the possibility of developing miniaturized platforms used directly in the point-of-care.
Loop-mediated isothermal amplification (LAMP) COVID-19 testing methods
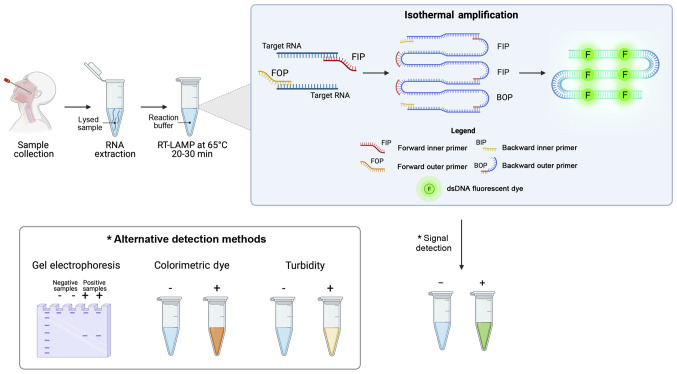
LAMP systems are among the most commonly used alternative nucleic acid amplification methods for the diagnosis of infectious diseases. These techniques are characterized by an easy execution of the assay that does not require specific equipment, a rapid and highly sensitive detection of targets and an easy interpretation of results (157). As regards SARS-CoV-2, several RT-LAMP systems have been developed for the effective detection of viral RNA, highlighting how these methods can be coupled with other diagnostic techniques including NGS, digital detection systems, biosensors, etc. (158).
The SARS-CoV-2 RT-LAMP detection systems are based on autocycling strand displacement DNA synthesis-mediated Bst DNA polymerase and six primers, which work at isothermal temperatures ranging from 60 to 65°C, thus avoiding the use of a costly PCR thermocycler. RT-LAMP amplification is able to amplify RNA fragments up to 106-109 copies in ~30-60 min. The final amplification products can be visualized by agarose gel electrophoresis, by fluorescence labeling, turbidity, or colorimetry for an immediate readout of the data obtained and a prompt diagnosis of COVID-19 infection (Fig. 8).
Figure 8.
Schematic workflow of nucleic acids isothermal amplification methods for the diagnosis of COVID-19.
Overall, SARS-CoV-2 RT-LAMP tests have a high sensitivity and specificity and some of these have been approved for diagnostic purposes by different national and international agencies. Some studies have reported a sensitivity of ~100% for the detection of ORF1ab SARS-CoV-2 gene, highlighting a higher diagnostic accuracy compared to other validated RT-PCR diagnostic methods. In addition, the specificity rate is higher than that obtained through RT-PCR, as the use of more than six primers in the RT-LAMP ensures the correct diagnosis of SARS-CoV-2 infection (25).
Among the most commonly used RT-LAMP methods there is The ID NOW™ COVID-19 assay (Abbott Laboratories), which has been approved by FDA with Emergency Use Authorization (EUA). This RT-LAMP-based system ensures high-sensitive results in ~5 min through the identification of the SARS-CoV-2 RdRp gene. Abbott ID NOW™ and other similar systems represent effective point-of-care testing, which have improved the management of COVID-19 infection thanks to their rapid results and limited costs (159).
Through the development of novel integrated diagnostic tools, RT-LAMP has been coupled with other diagnostic systems improving their applicability in clinical settings. In particular, Broughton et al (160) developed an RT-LAMP system combined with CRISPR/Cas12 lateral flow assay, which ensures a better display of the results and a significant reduction of the execution times.
Similarly, other LAMP systems have been combined with recombinase polymerase amplification (RPA) techniques, resulting in a high-sensitive method termed as RAMP. In particular, RAMP consists of two consecutive isothermal enzymatic steps that are RPA and LAMP. Firstly, RPA is performed at 37°C for 20 min in order to amplify all viral targets; subsequently, RPA amplicons are detected through isothermal LAMP performed for 30 min at 65°C. The amplification data are finally displayed by using colorimetric dyes or are monitored in real-time by using intercalating fluorescent dyes, such as SYBR-Green (161). At present, a RAMP detection system is used in research contexts for the effective detection of the SARS-CoV-2 ORF1ab gene with a sensitivity of 100% (162). If adequately validated, both RT-LAMP and RAMP may represent an effective alternative to nucleic acid amplification systems (163-165).
CRISPR/Cas-based COVID-19 testing methods
The CRISPR/Cas system is a bacterial nucleoprotein complex that confers resistance to external plasmids or phages nucleic acids acting as a type of prokaryotic immune system. The CRISPR/Cas system has revolutionized the field of molecular biology allowing the development of genome editing, a technology that has received the Nobel Prize 2020 for Chemistry (166,167). The applications and potentialities of CRISPR/Cas systems are almost unlimited, ranging from genome editing to diagnostics and from therapies to gene regulation (168).
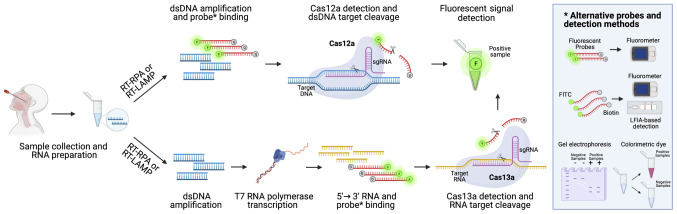
As mentioned in above, CRISPR/Cas technology has been used for the effective diagnosis of COVID-19 associated with LAMP isothermal amplification (169). The technology behind the SARS-CoV-2 diagnostic CRISPR/Cas systems is very simple and based on the use of CRISPR sequences with guide RNAs (gRNAs) specific for SARS-CoV-2 viral RNA, Cas nuclease proteins, target nucleic acid fragments and fluorescent probes or colorimetric dyes. At present, two main CRISPR/Cas technologies are used for the effective detection of the viral genome, i.e., CRISPR/Cas13a and CRISPR/Cas12a that are able to bind and cleave RNA and DNA targets, respectively. Other systems, not yet approved, but with a higher level of accuracy, use also FnCas9 nuclease (170).
Following viral RNA extraction, the sample is retrotranscribed into cDNA using RT-RPA or RT-LAMP, as described in above [please see section entitled 'Loop-mediated isothermal amplification (LAMP) COVID-19 testing methods']. Subsequently, the obtained cDNA can be directly analyzed through CRISPR/Cas12a detection system or can be transcribed into RNA using T7 transcriptase. In this latter case, the RNA obtained is then processed through the CRISPR/Cas13a nucleoprotein complex able to cleave labeled targeted viral RNA, thus allowing the detection of a positive signal in case of COVID-19 infection. If the CRISPR/Cas12a detection system is used, the double-strand DNA bound by a probe labeled with a fluorescent reporter and a quencher is cleaved by Cas12a generating a readable signal. The data generated by using both CRISPR/Cas13a or CRISPR/Cas12a systems can be observed via different methods by using electrophoresis gel, lateral flow strips or instruments able to detect real-time fluorescent signals occurring when probes are cleaved by Cas proteins (159) (Fig. 9).
Figure 9.
Schematic workflow of CRISPR/Cas-based methods for the diagnosis of COVID-19 infection. CRISPR/Cas12a systems effectively detect SARS-CoV-2 viral RNA after cDNA synthesis through the recognition of a specific SARS-CoV-2 sequence that is cleaved by Cas12a activity resulting in the collateral cleavage of fluorescent probes. In the same manner, CRISPR/Cas13a systems effectively detect SARS-CoV-2 RNA that is cleaved by Cas13a activity resulting in the collateral cleavage of fluorescent probes. Apart from fluorescence, alternative detection methods are based on the use of colorimetric dyes, electrophoresis gel and LFIA cartridge. CRISPR/Cas, clusters of regularly interspaced short palindromic repeats/Cas; LFIA, lateral flow immunoassay.
Some SARS-CoV-2 CRISPR/Cas detection systems have been already approved by international agencies and can be used as point-of-care testing due to their smart execution that does not require specific laboratory equipment, the rapidity of the procedure and the low cost of the assay (171).
Among the most commonly used systems, there is the Sherlock™ CRISPR SARS-CoV-2 assay (Sherlock Biosciences) that has been approved by the US FDA for emergency use. This system based on Cas13a nuclease activity is composed of two different stages. The first one is an RT-LAMP, where viral RNA is reverse transcribed into cDNA and amplified through a strand-displacing DNA polymerase (LAMP). The second step consists of RNA transcription and collateral cleavage mediated by the CRISPR/Cas13a complex able to target a specific SARS-CoV-2 sequence. The cleavage of SARS-CoV-2 RNA results in a fluorescent signal easily detectable (172,173). This system has exhibited a more rapid execution time compared with RT-PCR, with 100% sensitivity and specificity to correctly diagnose COVID-19 in clinical samples, a minimal footprint of instruments and a LoD of 6.75 copies/µl (174).
Other CRISPR/Cas-based systems have proven similar accuracy. Hou et al (175) (2020) developed a CRISPR/Cas13a system able to produce high-sensitive results within 40 min with a LoD of 7.5 copies per reaction. Similarly, Curti et al (176) (2020) developed an ultrasensitive, rapid, and portable CRISPR/Cas12a-based assay for the effective detection of SARS-CoV-2 RNA demonstrating a LoD of 10 copies/µl.
Overall, as shown for RT-LA MP methods, CRISPR/Cas-based assays (176) represent a promising alternative for the effective diagnosis of COVID-19, ensuring high sensitivity and specificity rates, and low costs of analysis.
Digital PCR COVID-19 testing methods
Digital PCR represents a technological evolution of the currently adopted RT-PCR methods (177). Digital PCR has been widely used for different applications, including mutational analysis, viral load detection, microbiological investigations, copy number variant analysis, single-cell analysis, analysis of liquid biopsy samples and detection of low expressed targets (177-183).
Different digital PCR platforms have been developed by different companies. Among these, droplet digital PCR (ddPCR) represents one of the most accurate systems currently available for the effective detection of viral infections. Of note, ddPCR (and other digital PCR systems in general) was initially developed to find application in microbiological diagnostics, especially in that of the virus, where it is of fundamental importance to assess the viral load of an infected individual in order to establish its contagiousness and prognosis (177). Subsequently, ddPCR was applied to various clinical fields and currently represents one of the most sensitive and accurate methods for the diagnosis of numerous pathologies, including COVID-19 infection (178,184,185).
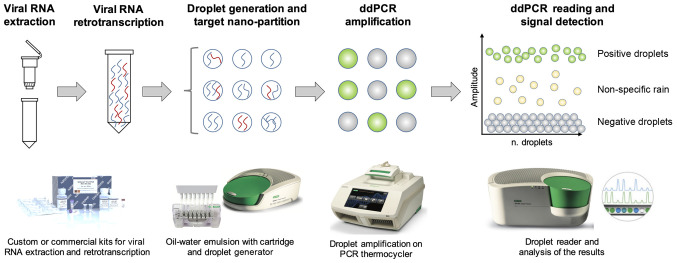
Of note, digital PCR uses the principles of the sample micro-partitions and DNA ultra-dilutions performed on solid supports or through the water-oil emulsion of the reaction mix. This latter example is the principle of ddPCR, where the reaction mix is nano-partitioned in >20,000 oil-water droplets containing the target nucleic acid, specific primers and probes, Taq polymerase and the amplification buffer necessary for the PCR amplification. The workflow is similar to that observed for RT-PCR; following sample collection, viral RNA is extracted through custom or commercial protocols. Subsequently, viral RNA can be directly processed in ddPCR by using one-step protocols or needs a previous RT step to obtain cDNA. The ddPCR protocol consists of the preparation of the reaction mix and the subsequent generation of oil-water emulsion droplets performed through specific cartridges and a droplet generator. The thousands of droplets thus generated are subsequently amplified through a classic RT-PCR amplification protocol which amplifies target DNA contained in each individual droplet resulting in hundreds of separate amplification reactions in a single well. Finally, the amplified droplets are read through a droplet reader which uses capillary tubes where the droplets flow separately and are excited by a laser beam that in case of positivity will generate a fluorescent emission signal detected by a CCD camera (186) (Fig. 10).
Figure 10.
Schematic workflow of the ddPCR detection system. ddPCR, droplet digital PCR.
As regards SARS-CoV-2 RNA detection, ddPCR ensures a greater sensitivity, specificity and accuracy compared to other molecular systems, including RT-PCR (178,187). Indeed, the elevated false-negative rate and the lack of availability of quantitative RT-PCR methods significantly limit the diagnostic potential of RT-PCR. Conversely, ddPCR is able to accurately assess SARS-CoV-2 viral load, thus allowing clinicians to effectively diagnose COVID-19 infection, particularly in asymptomatic or paucisymptomatic patients (188). In addition, ddPCR may be used for the monitoring of COVID-19 patients, thus allowing the discharge of patients who have resolved the infection and to guarantee correct social isolation measures useful for limiting the spread of infections (189).
Apart from the advantages in terms of sensitivity and specificity, ddPCR has been proven to have a higher robustness compared to RT-PCR, also in terms of stability and reproducibility of the method. Indeed, RT-PCR efficiency is affected by several pre-analytical and analytical biases due to the source of specimens, the storage of samples, the sampling timing, the sampling modalities, the extraction protocols and the nucleic acid quality (38,64). Conversely, ddPCR has exhibited a lower LoD compared to RT-PCR and it is also less susceptible to PCR interferers (178,190).
The better accuracy of the ddPCR translates into the optimal performance in terms of LoD. Indeed, in an experimental setting ddPCR has an LoD of 2.1 and 1.8 copies/reaction for ORF1ab and N SARS-CoV-2 genes, respectively, compared to RT-PCR, which has an LoD of 1,039 copies/reaction for ORF1ab and 873.2 copies/reaction for N gene. Overall, ddPCR has a 500-fold greater sensitivity compared to RT-PCR (188).
These data suggest how ddPCR may be considered a turning point method for the effective diagnosis of COVID-19 infection, due to its greater sensitivity, specificity and accuracy compared to other existing molecular methods.
7. Conclusions
The COVID-19 pandemic has highlighted the importance of laboratory diagnostics in the management of a health emergency that has deeply affected the social, economic and health fabric of the entire world. At present, nucleic acid amplification methods represent the gold standard for the diagnosis of COVID-19 infection with several RT-PCR-based tests approved by different national and international regulatory agencies. These tests, together with clinical and radiological investigations, have significantly improved the ability to correctly and rapidly diagnose a COVID-19 infection compared to the first diagnostic methods mainly represented by viral culture analysis and de novo sequencing of the SARS-CoV-2 genome. However, despite the high sensitivity of RT-PCR, patients with a low viral load are not often correctly diagnosed. In addition, this technique requires confirmatory analyses, trained personnel and expensive instruments and reagents that limit its application, especially in low-income countries. For these purposes, other methods have been developed to overcome the limitations of RT-PCR. Among these methods, CRISPR/Cas-based assays and isothermal amplification methods represent promising low-cost diagnostic strategies that can be used for the effective diagnosis of COVID-19 infection in middle- and low-income countries. Moreover, more sensitive molecular methods, including ddPCR and biosensors, are beginning to be approved by international agencies and used for the diagnosis of COVID-19 infection, as well as for the monitoring of viral load in hospitalized patients or quarantined individuals.
Apart from these widely used diagnostic strategies, other techniques have been developed for the screening of SARS-CoV-2 infection in the population or in high-risk environments, such as hospitals and schools. Among these, rapid antigen and rapid antibody tests and immunoenzymatic serological tests represent the most used techniques for the monitoring of COVID-19 infection spreading. Of note, despite the low-cost of such techniques, the low sensitivity and specificity of LFIAs and ELISAs do not allow clinicians to formulate a precise diagnosis of COVID-19 infection that should be confirmed with other techniques. However, the use of such point-of-care testing made it possible to implement effective health surveillance systems that allowed the effective management of the COVID-19 pandemic thus limiting the number of infections.
At present, other techniques are essential for the genomic surveillance of the pandemic. Indeed, NGS techniques and SARS-CoV-2 whole-genome sequencing are fundamental for the precise characterization of novel genetic variants of SARS-CoV-2. However, the high costs of these methods limit their application in clinical practice.
Overall, in a short period of time, the scientific community has developed several methods useful for correctly diagnosing a suspected case of COVID-19 infection. However, in order to perform a correct diagnosis of COVID-19, it is necessary to take into account, not only the test to be used, but also the patient's medical history, the time of the suspicious SARS-CoV-2 exposure, the type of sample to be collected and analyzed and how to interpret the result. Only by integrating all these elements, it will be possible to formulate a correct diagnosis of COVID-19 infection and effectively manage the COVID-19 pandemic.
Supplementary Data
Acknowledgments
Not applicable.
Funding Statement
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The original contributions/references of studies presented in the study are publicly available. These data can be found at: www.pubmed.com and https://www.who.int.
Authors' contributions
LF, DAS and ML conceptualized the study. LF and GG wrote and prepared the draft of the manuscript. AT, DAS and ML provided critical revisions. LF and GG drew the figures and prepare the tables. LF and ML confirm the authenticity of all the raw data. All authors contributed to manuscript revision and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article. The other authors declare that they have no competing interests.
References
- 1.Bontempi E. The europe second wave of COVID-19 infection and the Italy 'strange' situation. Environ Res. 2021;193:110476. doi: 10.1016/j.envres.2020.110476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cacciapaglia G, Cot C, Sannino F. Second wave COVID-19 pandemics in Europe: A temporal playbook. Sci Rep. 2020;10:15514. doi: 10.1038/s41598-020-72611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) Jonhs Hopkins University (JHU) doi: 10.1016/S1473-3099(22)00434-0. https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6. Accessed February 27, 2021. [DOI] [PMC free article] [PubMed]
- 4.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, C4591001 Clinical Trial Group et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Medicine Agency (EMA): COVID-19 Vaccines. https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines/covid-19-vaccines. Accessed February 27, 2021.
- 6.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pullano G, Di Domenico L, Sabbatini CE, Valdano E, Turbelin C, Debin M, Guerrisi C, Kengne-Kuetche C, Souty C, Hanslik T, et al. Underdetection of cases of COVID-19 in France threatens epidemic control. Nature. 2021;590:134–139. doi: 10.1038/s41586-020-03095-6. [DOI] [PubMed] [Google Scholar]
- 8.O'Dowd A. Covid-19: Avoid indoor mixing over Christmas or risk third wave, warns iSAGE. BMJ. 2020;371:m4832. doi: 10.1136/bmj.m4832. [DOI] [PubMed] [Google Scholar]
- 9.Farsalinos K, Poulas K, Kouretas D, Vantarakis A, Leotsinidis M, Kouvelas D, Docea AO, Kostoff R, Gerotziafas GT, Antoniou MN, et al. Improved strategies to counter the COVID-19 pandemic: Lockdowns vs. primary and community healthcare. Toxicol Rep. 2021;8:1–9. doi: 10.1016/j.toxrep.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S, Pan C. Differentiating diagnosis of COVID-19 or influenza in patients based on laboratory data during flu season. EClinicalMedicine. 2020;26:100511. doi: 10.1016/j.eclinm.2020.100511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Basteiro AL, Chaccour C, Guinovart C, Llupià A, Brew J, Trilla A, Plasencia A. Monitoring the COVID-19 epidemic in the context of widespread local transmission. Lancet Respir Med. 2020;8:440–442. doi: 10.1016/S2213-2600(20)30162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu AHB. Screening the General Population for SARS-CoV-2 Virus and COVID-19 Antibodies: A Counterargument. J Appl Lab Med. 2020;5:1107–1110. doi: 10.1093/jalm/jfaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rostami A, Sepidarkish M, Leeflang MMG, Riahi SM, Nourollahpour Shiadeh M, Esfandyari S, Mokdad AH, Hotez PJ, Gasser RB. SARS-CoV-2 seroprevalence worldwide: a systematic review and meta-analysis. Clin Microbiol Infect. 2021;27:331–340. doi: 10.1016/j.cmi.2020.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wells PM, Doores KJ, Couvreur S, Nunez RM, Seow J, Graham C, Acors S, Kouphou N, Neil SJD, Tedder RS, et al. Estimates of the rate of infection and asymptomatic COVID-19 disease in a population sample from SE England. J Infect. 2020;81:931–936. doi: 10.1016/j.jinf.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji T, Liu Z, Wang G, Guo X, Akbar Khan S, Lai C, Chen H, Huang S, Xia S, Chen B, et al. Detection of COVID-19: A review of the current literature and future perspectives. Biosens Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanyaolu A, Okorie C, Marinkovic A, Ayodele O, Abbasi AF, Prakash S, Ahmed M, Kayode D, Jaferi U, Haider N. Navigating the diagnostics of COVID-19. SN Compr Clin Med. 2020 Jul 25; doi: 10.1007/s42399-020-00408-8. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.European Commission (EC): Current performance of COVID-19 test methods and devices and proposed performance criteria. EC; Brussels: 2020. https://ec.europa.eu/docsroom/documents/40805. Accessed January 2, 2021. [Google Scholar]
- 18.Taleghani N, Taghipour F. Diagnosis of COVID-19 for controlling the pandemic: A review of the state-of-the-art. Biosens Bioelectron. 2021;174:112830. doi: 10.1016/j.bios.2020.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung J, Jang H, Kim HK, Kim J, Kim A, Ko KP. The Importance of Mandatory COVID-19 Diagnostic Testing Prior to Release from Quarantine. J Korean Med Sci. 2020;35:e314. doi: 10.3346/jkms.2020.35.e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winter AK, Hegde ST. The important role of serology for COVID-19 control. Lancet Infect Dis. 2020;20:758–759. doi: 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandenberg O, Martiny D, Rochas O, van Belkum A, Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat Rev Microbiol. 2021;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tromberg BJ, Schwetz TA, Pérez-Stable EJ, Hodes RJ, Woychik RP, Bright RA, Fleurence RL, Collins FS. Rapid scaling up of Covid-19 diagnostic testing in the United States - The NIH RADx Initiative. N Engl J Med. 2020;383:1071–1077. doi: 10.1056/NEJMsr2022263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guglielmi G. The explosion of new coronavirus tests that could help to end the pandemic. Nature. 2020;583:506–509. doi: 10.1038/d41586-020-02140-8. [DOI] [PubMed] [Google Scholar]
- 24.Tsatsakis A, Calina D, Falzone L, Petrakis D, Mitrut R, Siokas V, Pennisi M, Lanza G, Libra M, Doukas SG, et al. SARS-CoV-2 pathophysiology and its clinical implications: An integrative overview of the pharmacotherapeutic management of COVID-19. Food Chem Toxicol. 2020;146:111769. doi: 10.1016/j.fct.2020.111769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alpdagtas S, Ilhan E, Uysal E, Sengor M, Ustundag CB, Gunduz O. Evaluation of current diagnostic methods for COVID-19. APL Bioeng. 2020;4:041506. doi: 10.1063/5.0021554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Erratum. Nature. 2020;580:E7. doi: 10.1038/s41586-020-2202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. China novel coronavirus investigating and research team: a novel coronavirus from patients with pneumonia in China 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar R, Nagpal S, Kaushik S, Mendiratta S. COVID-19 diagnostic approaches: Different roads to the same destination. Virusdisease. 2020;31:97–105. doi: 10.1007/s13337-020-00599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brun AL, Gence-Breney A, Trichereau J, Ballester MC, Vasse M, Chabi ML, Mellot F, Grenier PA. COVID-19 pneumonia: High diagnostic accuracy of chest CT in patients with intermediate clinical probability. Eur Radiol. 2021;31:1969–1977. doi: 10.1007/s00330-020-07346-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez RM. Clinical samples for SARS-CoV-2 detection: Review of the early literature. Clin Microbiol Newsl. 2020;42:121–127. doi: 10.1016/j.clinmicnews.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao GG, Agarwal A, Batura D. Testing times in Coronavirus disease (COVID-19): A tale of two nations. Med J Armed Forces India. 2020;76:243–249. doi: 10.1016/j.mjafi.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarvis KF, Kelley JB. Temporal dynamics of viral load and false negative rate influence the levels of testing necessary to combat COVID19 spread. medRxiv. doi: 10.1101/2020.08.12.20173831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rode OĐ, Kurolt IC, Puljiz I, Čivljak R, Balent NC, Laškaj R, Tiljak MK, Mikulić R, Markotić A. Antibody response and the clinical presentation of patients with COVID-19 in Croatia: The importance of a two-step testing approach. Eur J Clin Microbiol Infect Dis. 2021;40:261–268. doi: 10.1007/s10096-020-04019-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bisoffi Z, Pomari E, Deiana M, Piubelli C, Ronzoni N, Beltrame A, Bertoli G, Riccardi N, Perandin F, Formenti F, et al. Sensitivity, specificity and predictive values of molecular and serological tests for COVID-19: a longitudinal study in emergency room. Diagnostics (Basel) 2020;10:669. doi: 10.3390/diagnostics10090669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krajewski R, Gołębiowska J, Makuch S, Mazur G, Agrawal S. Update on serologic testing in COVID-19. Clin Chim Acta. 2020;510:746–750. doi: 10.1016/j.cca.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chirumbolo S. Swabbing Thoroughly People for COVID-19 Positivity. Insights on the Main Bio-analytical and Microbiology Bias and Concerns. Curr Microbiol. 2020;77:3680–3684. doi: 10.1007/s00284-020-02167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. 2020;58:1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 39.Ora J, Puxeddu E, Cavalli F, Giorgino FM, Girolami A, Chiocchi M, Sergiacomi G, Federici M, Rogliani P. Does bronchoscopy help the diagnosis in COVID-19 infection? Eur Respir J. 2020;56:2001619. doi: 10.1183/13993003.01619-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong Y, Bao A, Chen H, Huang J, Lv Z, Feng L, Cheng Y, Wang Y, Bai L, Rao W, et al. Necessity for detection of SARS-CoV-2 RNA in multiple types of specimens for the discharge of the patients with COVID-19. J Transl Med. 2020;18:411. doi: 10.1186/s12967-020-02580-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McFee DRB. COVID-19 Laboratory Testing/CDC Guidelines. Dis Mon. 2020;66:101067. doi: 10.1016/j.disamonth.2020.101067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Y, Li Y, Guo E, He L, Liu J, Yang B, Li F, Wang Z, Li Y, Xiao R, et al. Dynamics and correlation among viral positivity, seroconversion, and disease severity in COVID-19: a retrospective study. Ann Intern Med. 2020;8:M20–M3337. doi: 10.7326/M20-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mesoraca A, Margiotti K, Viola A, Cima A, Sparacino D, Giorlandino C. Evaluation of SARS-CoV-2 viral RNA in fecal samples. Virol J. 2020;17:86. doi: 10.1186/s12985-020-01359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng SC, Tilg H. COVID-19 and the gastrointestinal tract: More than meets the eye. Gut. 2020;69:973–974. doi: 10.1136/gutjnl-2020-321195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long DR, Gombar S, Hogan CA, Greninger AL, O'Reilly-Shah V, Bryson-Cahn C, Stevens B, Rustagi A, Jerome KR, Kong CS, et al. Occurrence and timing of subsequent severe acute respiratory syndrome coronavirus 2 reverse-transcription polymerase chain reaction positivity among initially negative patients. Clin Infect Dis. 2021;72:323–326. doi: 10.1093/cid/ciaa722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahendiratta S, Batra G, Sarma P, Kumar H, Bansal S, Kumar S, Prakash A, Sehgal R, Medhi B. Molecular diagnosis of COVID-19 in different biologic matrix, their diagnostic validity and clinical relevance: A systematic review. Life Sci. 2020;258:118207. doi: 10.1016/j.lfs.2020.118207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.da Silva SJR, Silva CTAD, Guarines KM, Mendes RPG, Pardee K, Kohl A, Pena L. Clinical and Laboratory Diagnosis of SARS-CoV-2, the Virus Causing COVID-19. ACS Infect Dis. 2020;6:2319–2336. doi: 10.1021/acsinfecdis.0c00274. [DOI] [PubMed] [Google Scholar]
- 48.Li D, Zhang J, Li J. Primer design for quantitative real-time PCR for the emerging Coronavirus SARS-CoV-2. Theranostics. 2020;10:7150–7162. doi: 10.7150/thno.47649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Centers for Disease Control and Prevention (CDC) CDC's Diagnostic Test for COVID-19 Only and Supplies. https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html. Accessed January 5, 2021.
- 51.Centers for Disease Control and Prevention (CDC) Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. Accessed January 5, 2021.
- 52.Dorlass EG, Monteiro CO, Viana AO, Soares CP, Machado RRG, Thomazelli LM, Araujo DB, Leal FB, Candido ED, Telezynski BL, et al. Lower cost alternatives for molecular diagnosis of COVID-19: Conventional RT-PCR and SYBR Green-based RT-qPCR. Braz J Microbiol. 2020;51:1117–1123. doi: 10.1007/s42770-020-00347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neilan AM, Losina E, Bangs AC, Flanagan C, Panella C, Eskibozkurt GE, Mohareb A, Hyle EP, Scott JA, Weinstein MC, et al. Clinical Impact, Costs, and Cost-Effectiveness of Expanded SARS-CoV-2 Testing in Massachusetts. Clin Infect Dis. 2020 Sep 18; doi: 10.1093/cid/ciaa1418. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyrlaki I, Ekman M, Lentini A, Rufino de Sousa N, Papanicolaou N, Vondracek M, Aarum J, Safari H, Muradrasoli S, Rothfuchs AG, et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat Commun. 2020;11:4812. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.U.S Food & Drug: COVID-19 tests and collection kits authorized by the FDA in 2020: Infographic. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/covid-19-tests-and-collection-kits-authorized-fda-2020-infographic. Accessed January 8, 2021.
- 56.Fomsgaard AS, Rosenstierne MW. An alternative workflow for molecular detection of SARS-CoV-2 - escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill. 2020;25:2000398. doi: 10.2807/1560-7917.ES.2020.25.14.2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, Chen H, Mubareka S, Gubbay JB, Chan WCW. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 58.Ravi N, Cortade DL, Ng E, Wang SX. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wells CR, Townsend JP, Pandey A, Moghadas SM, Krieger G, Singer B, McDonald RH, Fitzpatrick MC, Galvani AP. Optimal COVID-19 quarantine and testing strategies. Nat Commun. 2021;12:356. doi: 10.1038/s41467-020-20742-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bustin SA, Nolan T. RT-qPCR Testing of SARS-CoV-2: A Primer. Int J Mol Sci. 2020;21:3004. doi: 10.3390/ijms21083004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Böger B, Fachi MM, Vilhena RO, Cobre AF, Tonin FS, Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am J Infect Control. 2021;49:21–29. doi: 10.1016/j.ajic.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020;173:262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Afzal A. Molecular diagnostic technologies for COVID-19: Limitations and challenges. J Adv Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev Mol Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan Y, Long L, Zhang D, Yuan T, Cui S, Yang P, Wang Q, Ren S. Potential false-negative nucleic acid testing results for severe acute respiratory syndrome coronavirus 2 from thermal inactivation of samples with low viral loads. Clin Chem. 2020;66:794–801. doi: 10.1093/clinchem/hvaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020;92:903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.World Health Organization (WHO) Recommendations for laboratories testing by PCR for presence of SARS coronavirus-RNA. WHO; Geneva: 2020. https://www.who.int/csr/sars/coronarec-ommendations/en/. Accessed January 26, 2021. [Google Scholar]
- 68.Cerutti F, Burdino E, Milia MG, Allice T, Gregori G, Bruzzone B, Ghisetti V. Urgent need of rapid tests for SARS CoV-2 antigen detection: Evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J Clin Virol. 2020;132:104654. doi: 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olalekan A, Iwalokun B, Akinloye OM, Popoola O, Samuel TA, Akinloye O. COVID-19 rapid diagnostic test could contain transmission in low- and middle-income countries. Afr J Lab Med. 2020;9:1255. doi: 10.4102/ajlm.v9i1.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Augustine R, Das S, Hasan ASA, Abdul Salam S, Augustine P, Dalvi YB, Varghese R, Primavera R, Yassine HM, et al. Rapid antibody-based COVID-19 mass surveillance: relevance, challenges, and prospects in a pandemic and post-pandemic world. J Clin Med. 2020;9:3372. doi: 10.3390/jcm9103372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Albert E, Torres I, Bueno F, Huntley D, Molla E, Fernández-Fuentes MÁ, Martínez M, Poujois S, Forqué L, Valdivia A, et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect. 2021;27:472.e7–472.e10. doi: 10.1016/j.cmi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pilarowski G, Lebel P, Sunshine S, Liu J, Crawford E, Marquez C, Rubio L, Chamie G, Martinez J, Peng J, et al. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. J Infect Dis. 2021 Jan 4; doi: 10.1093/infdis/jiaa802. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JH, Choi M, Jung Y, Lee SK, Lee CS, Kim J, Kim J, Kim NH, Kim BT, Kim HG. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens Bioelectron. 2021;171:112715. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Diao B, Wen K, Zhang J, Chen J, Han C, Chen Y, Wang S, Deng G, Zhou H, Wu Y. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect. 2021;27:289.e1–289.e4. doi: 10.1016/j.cmi.2020.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, Sasso JM, Gregg AC, Soares DJ, Beskid TR, et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang C, Wen T, Shi FJ, Zeng XY, Jiao YJ. Rapid detection of igm antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5:12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mak GC, Cheng PK, Lau SS, Wong KK, Lau CS, Lam ET, Chan RC, Tsang DN. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lanser L, Bellmann-Weiler R, Öttl KW, Huber L, Griesmacher A, Theurl I, Weiss G. Evaluating the clinical utility and sensitivity of SARS-CoV-2 antigen testing in relation to RT-PCR Ct values. Infection. 2020;13:1–3. doi: 10.1007/s15010-020-01542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, Emperador D, Takwoingi Y, Cunningham J, Beese S, et al. Cochrane COVID-19 Diagnostic Test Accuracy Group: Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2020;8:CD013705. doi: 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Montesinos I, Gruson D, Kabamba B, Dahma H, Van den Wijngaert S, Reza S, Carbone V, Vandenberg O, Gulbis B, Wolff F, et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 2020;128:104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prazuck T, Colin M, Giachè S, Gubavu C, Seve A, Rzepecki V, Chevereau-Choquet M, Kiani C, Rodot V, Lionnet E, et al. Evaluation of performance of two SARS-CoV-2 Rapid IgM-IgG combined antibody tests on capillary whole blood samples from the fingertip. PLoS One. 2020;15:e0237694. doi: 10.1371/journal.pone.0237694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jacobs J, Kühne V, Lunguya O, Affolabi D, Hardy L, Vandenberg O. Implementing COVID-19 (SARS-CoV-2) rapid diagnostic tests in Sub-Saharan Africa: a review. Front Med (Lausanne) 2020;7:557797. doi: 10.3389/fmed.2020.557797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.La Rosa Fabián C, Urquizo Briceño L. Anti-SARS-Cov-2 IgA in Current Scenario of IgM and IgG Rapid Test: A New Alternative for the Diagnostic of COVID-19. SN Compr Clin Med. 2020;26:1–3. doi: 10.1007/s42399-020-00551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Canetti D, Dell'Acqua R, Riccardi N, Della Torre L, Bigoloni A, Muccini C, Bruzzesi E, Ranzenigo M, Chiurlo M, Racca S, et al. SARS-CoV-2 IgG/IgM Rapid Test as a Diagnostic Tool in Hospitalized Patients and Healthcare Workers, at a large Teaching Hospital in northern Italy, during the 2020 COVID-19 Pandemic. New Microbiol. 2020;43:161–165. [PubMed] [Google Scholar]
- 85.Alhajj M, Farhana A. StatPearls. StatPearls Publishing; Treasure Island, FL: 2020. Enzyme Linked Immunosorbent Assay. [PubMed] [Google Scholar]
- 86.Tré-Hardy M, Wilmet A, Beukinga I, Favresse J, Dogné JM, Douxfils J, Blairon L. Analytical and clinical validation of an ELISA for specific SARS-CoV-2 IgG, IgA, and IgM anti-bodies. J Med Virol. 2021;93:803–811. doi: 10.1002/jmv.26303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Freeman B, Lester S, Mills L, Rasheed MAU, Moye S, Abiona O, Hutchinson GB, Morales-Betoulle M, Krapinunaya I, Gibbons A, et al. Validation of a SARS-CoV-2 spike protein ELISA for use in contact investigations and serosurveillance. bioRxiv. doi: 10.1101/2020.04.24.057323. [DOI] [Google Scholar]
- 88.Korth J, Wilde B, Dolff S, Anastasiou OE, Krawczyk A, Jahn M, Cordes S, Ross B, Esser S, Lindemann M, et al. SARS-CoV-2-specific antibody detection in healthcare workers in Germany with direct contact to COVID-19 patients. J Clin Virol. 2020;128:104437. doi: 10.1016/j.jcv.2020.104437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alharbi SA, Almutairi AZ, Jan AA, Alkhalify AM. Enzyme-linked immunosorbent assay for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) IgM/IgA and IgG antibodies among healthcare workers. Cureus. 2020;12:e10285. doi: 10.7759/cureus.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stringhini S, Wisniak A, Piumatti G, Azman AS, Lauer SA, Baysson H, De Ridder D, Petrovic D, Schrempft S, Marcus K, et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): A population-based study. Lancet. 2020;396:313–319. doi: 10.1016/S0140-6736(20)31304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vashist SK. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics (Basel) 2020;10:202. doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tanaka Y, Kamimura K, Nakamura R, Ohkoshi-Yamada M, Koseki Y, Mizusawa T, Ikarashi S, Hayashi K, Sato H, Sakamaki A, et al. Usefulness of ultrasonography to assess the response to steroidal therapy for the rare case of type 2b immunoglobulin G4-related sclerosing cholangitis without pancreatitis: A case report. World J Clin Cases. 2020;8:5821–5830. doi: 10.12998/wjcc.v8.i22.5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.AvŞar T, ÇaliŞ Ş, Yilmaz B, Demİrcİ OtluoĞlu G, Holyavkİn C, Kili CT. Genome-wide identification of Chiari malformation type I associated candidate genes and chromosomal variations. Turk J Biol. 2020;44:449–456. doi: 10.3906/biy-2009-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.La Marca A, Capuzzo M, Paglia T, Roli L, Trenti T, Nelson SM. Testing for SARS-CoV-2 (COVID-19): A systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod Biomed Online. 2020;41:483–499. doi: 10.1016/j.rbmo.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sheikhzadeh E, Eissa S, Ismail A, Zourob M. Diagnostic techniques for COVID-19 and new developments. Talanta. 2020;220:121392. doi: 10.1016/j.talanta.2020.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu N, Wang W, Liu Z, Liang C, Wang W, Ye F, Huang B, Zhao L, Wang H, Zhou W, et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jain R, Sarkale P, Mali D, Shete AM, Patil DY, Majumdar T, Suryawanshi A, Patil S, Mohandas S, Yadav PD. Inactivation of SARS-CoV-2 by gamma irradiation. Indian J Med Res. 2020 Nov 27; doi: 10.4103/ijmr.IJMR_2789_20. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park WB, Kwon NJ, Choi SJ, Kang CK, Choe PG, Kim JY, Yun J, Lee GW, Seong MW, Kim NJ, et al. Virus Isolation from the First Patient with SARS-CoV-2 in Korea. J Korean Med Sci. 2020;35:e84. doi: 10.3346/jkms.2020.35.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao J, Zhou H, Huang W, Zhou J, Qiu M, Deng Z, Chen L, Weng Y, Cai L, Gu Y, et al. Cell morphological analysis of SARS-CoV-2 infection by transmission electron microscopy. J Thorac Dis. 2020;12:4368–4373. doi: 10.21037/jtd-20-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Akilesh S, Nicosia RF, Alpers CE, Tretiakova M, Hsiang TY, Gale M, Jr, Smith KD. Characterizing viral infection by electron microscopy: lessons from the coronavirus disease 2019 pandemic. Am J Pathol. 2021;191:222–227. doi: 10.1016/j.ajpath.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, Rota PA, Bankamp B, Bellini WJ, Zaki SR. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bain W, Lee JS, Watson AM, Stitt-Fischer MS. Practical guidelines for collection, manipulation and inactivation of SARS-CoV-2 and COVID-19 clinical specimens. Curr Protoc Cytom. 2020;93:e77. doi: 10.1002/cpcy.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu SY, Yau HS, Yu MY, Tsang HF, Chan LWC, Cho WCS, Shing Yu AC, Yuen Yim AK, Li MJW, Wong YKE, et al. The diagnostic methods in the COVID-19 pandemic, today and in the future. Expert Rev Mol Diagn. 2020;20:985–993. doi: 10.1080/14737159.2020.1816171. [DOI] [PubMed] [Google Scholar]
- 105.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang M, Fu A, Hu B, Tong Y, Liu R, Liu Z, Gu J, Xiang B, Liu J, Jiang W, et al. Nanopore targeted sequencing for the accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. Small. 2020;16:e2002169. doi: 10.1002/smll.202002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Addetia A, Xie H, Roychoudhury P, Shrestha L, Loprieno M, Huang ML, Jerome KR, Greninger AL. Identification of multiple large deletions in ORF7a resulting in in-frame gene fusions in clinical SARS-CoV-2 isolates. J Clin Virol. 2020;129:104523. doi: 10.1016/j.jcv.2020.104523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Di Giallonardo F, Duchene S, Puglia I, Curini V, Profeta F, Cammà C, Marcacci M, Calistri P, Holmes EC, Lorusso A. Genomic Epidemiology of the First Wave of SARS-CoV-2 in Italy. Viruses. 2020;12:1438. doi: 10.3390/v12121438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moore SC, Penrice-Randal R, Alruwaili M, Randle N, Armstrong S, Hartley C, Haldenby S, Dong X, Alrezaihi A, Almsaud M, et al. Amplicon-Based Detection and Sequencing of SARS-CoV-2 in Nasopharyngeal Swabs from Patients With COVID-19 and Identification of Deletions in the Viral Genome That Encode Proteins Involved in Interferon Antagonism. Viruses. 2020;12:1164. doi: 10.3390/v12101164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kames J, Holcomb DD, Kimchi O, DiCuccio M, Hamasaki-Katagiri N, Wang T, Komar AA, Alexaki A, Kimchi-Sarfaty C. Sequence analysis of SARS-CoV-2 genome reveals features important for vaccine design. Sci Rep. 2020;10:15643. doi: 10.1038/s41598-020-72533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Conti P, Caraffa A, Gallenga CE, Kritas SK, Frydas I, Younes A, Di Emidio P, Tetè G, Pregliasco F, Ronconi G. The British variant of the new coronavirus-19 (Sars-Cov-2) should not create a vaccine problem. J Biol Regul Homeost Agents. 2021;35:1–4. [PubMed] [Google Scholar]
- 112.Hourdel V, Kwasiborski A, Balière C, Matheus S, Batéjat CF, Manuguerra JC, Vanhomwegen J, Caro V. rapid genomic characterization of SARS-CoV-2 by direct amplicon-based sequencing through comparison of MinION and Illumina iSeq100TM system. Front Microbiol. 2020;11:571328. doi: 10.3389/fmicb.2020.571328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.illumina: Comprehensive Workf low for Detecting Coronavirus Using Illumina Benchtop Systems - A Shotgun Metagenomics Sequencing Workflow for Effective Detection and Characterization of Coronavirus Strains. https://www.illumina.com/content/dam/illumina-marketing/documents/products/appnotes/ngs-coronavirus-app-note-1270-2020-001.pdf. Accessed January 20, 2021.
- 114.Oxford Nanopore Technologies MinION - Powerful, real-time, long-read sequencing in the palm of your hand. https://nanoporetech.com/sites/default/files/s3/literature/MinION-Mk1C-brochure.pdf. Accessed January 20, 2021.
- 115.Meredith LW, Hamilton WL, Warne B, Houldcroft CJ, Hosmillo M, Jahun AS, Curran MD, Parmar S, Caller LG, Caddy SL, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: A prospective genomic surveillance study. Lancet Infect Dis. 2020;20:1263–1271. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wen F, Yu H, Guo J, Li Y, Luo K, Huang S. Identification of the hyper-variable genomic hotspot for the novel coronavirus SARS-CoV-2. J Infect. 2020;80:671–693. doi: 10.1016/j.jinf.2020.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lam TT. Tracking the Genomic Footprints of SARS-CoV-2 Transmission. Trends Genet. 2020;36:544–546. doi: 10.1016/j.tig.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Landete P, Quezada Loaiza CA, Aldave-Orzaiz B, Muñiz SH, Maldonado A, Zamora E, Sam Cerna AC, Del Cerro E, Alonso RC, Couñago F. Clinical features and radiological manifestations of COVID-19 disease. World J Radiol. 2020;12:247–260. doi: 10.4329/wjr.v12.i11.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen H, Ai L, Lu H, Li H. Clinical and imaging features of COVID-19. Radiol Infect Dis. 2020;7:43–50. doi: 10.1016/j.jrid.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Al-Qahtani M, AlAli S, AbdulRahman A, Salman Alsayyad A, Otoom S, Atkin SL. The prevalence of asymptomatic and symptomatic COVID-19 in a cohort of quarantined subjects. Int J Infect Dis. 2021;102:285–288. doi: 10.1016/j.ijid.2020.10.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yanes-Lane M, Winters N, Fregonese F, Bastos M, Perlman-Arrow S, Campbell JR, Menzies D. Proportion of asymptomatic infection among COVID-19 positive persons and their transmission potential: A systematic review and meta-analysis. PLoS One. 2020;15:e0241536. doi: 10.1371/journal.pone.0241536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Katopodis P, Kerslake R, Davies J, Randeva HS, Chatha K, Hall M, Spandidos DA, Anikin V, Polychronis A, Robertus JL, et al. COVID 19 and SARS CoV 2 host cell entry mediators: Expression profiling of TMRSS4 in health and disease. Int J Mol Med. 2021;47:64. doi: 10.3892/ijmm.2021.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vivarelli S, Falzone L, Torino F, Scandurra G, Russo G, Bordonaro R, Pappalardo F, Spandidos DA, Raciti G, Libra M. Immune-checkpoint inhibitors from cancer to COVID-19: A promising avenue for the treatment of patients with COVID-19 (Review) Int J Oncol. 2021;58:145–157. doi: 10.3892/ijo.2020.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vivarelli S, Falzone L, Grillo CM, Scandurra G, Torino F, Libra M. Cancer Management during COVID-19 Pandemic: Is Immune Checkpoint Inhibitors-Based Immunotherapy Harmful or Beneficial? Cancers (Basel) 2020;12:2237. doi: 10.3390/cancers12082237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abdi A, Jalilian M, Sarbarzeh PA, Vlaisavljevic Z. Diabetes and COVID-19: A systematic review on the current evidences. Diabetes Res Clin Pract. 2020;166:108347. doi: 10.1016/j.diabres.2020.108347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang S, Wang J, Liu F, Liu J, Cao G, Yang C, Liu W, Tu C, Zhu M, Xiong B. COVID-19 patients with hypertension have more severe disease: A multicenter retrospective observational study. Hyperte Res. 2020;43:824–831. doi: 10.1038/s41440-020-0485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pascarella G, Strumia A, Piliego C, Bruno F, Del Buono R, Costa F, Scarlata S, Agrò FE. COVID-19 diagnosis and management: A comprehensive review. J Intern Med. 2020;288:192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Torre La G, Massetti AP, Antonelli G, Fimiani C, Fantini M, Marte M, Faticoni A, Previte CM, Turriziani O, Pugliese F, et al. Covid-Sapienza Collaborative Group: Anosmia and Ageusia as Predictive Signs of COVID-19 in Healthcare Workers in Italy: A Prospective Case-Control Study. J Clin Med. 2020;9:2870. doi: 10.3390/jcm9092870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tanasa IA, Manciuc C, Carauleanu A, Navolan DB, Bohiltea RE, Nemescu D. Anosmia and ageusia associated with coronavirus infection (COVID-19) - what is known? Exp Ther Med. 2020;20:2344–2347. doi: 10.3892/etm.2020.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pennisi M, Lanza G, Falzone L, Fisicaro F, Ferri R, Bella R. SARS-CoV-2 and the Nervous System: From Clinical Features to Molecular Mechanisms. Int J Mol Sci. 2020;21:5475. doi: 10.3390/ijms21155475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Connors JM, Levy JH. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020;135:2033–2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, Chen L, Li M, Liu Y, Wang G, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Basu-Ray I, Almaddah Nk, Adeboye A, Soos MP. StatPearls. StatPearls Publishing; Treasure Island, FL: 2020. Cardiac Manifestations Of Coronavirus. [PubMed] [Google Scholar]
- 134.Garland V, Kumar AB, Borum ML. Gastrointestinal and hepatic manifestations of COVID-19: evolving recognition and need for increased understanding in vulnerable populations. J Natl Med Assoc. 2020 Aug 15; doi: 10.1016/j.jnma.2020.07.017. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Baj J, Karakuła-Juchnowicz H, Teresiński G, Buszewicz G, Ciesielka M, Sitarz E, Forma A, Karakuła K, Flieger W, Portincasa P, et al. COVID-19: Specific and non-specific clinical manifestations and symptoms: the current state of knowledge. J Clin Med. 2020;9:1753. doi: 10.3390/jcm9061753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Renu K, Prasanna PL, Valsala Gopalakrishnan A. Coronaviruses pathogenesis, comorbidities and multi-organ damage - A review. Life Sci. 2020;255:117839. doi: 10.1016/j.lfs.2020.117839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chang MC, Lee W, Hur J, Park D. Chest Computed Tomography Findings in Asymptomatic Patients with COVID-19. Respiration. 2020;99:748–754. doi: 10.1159/000509334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jędrusik P, Gaciong Z, Sklinda K, Sierpiński R, Walecki J, Gujski M. Diagnostic role of chest computed tomography in coronavirus disease 2019. Pol Arch Intern Med. 2020;130:520–528. doi: 10.20452/pamw.15348. [DOI] [PubMed] [Google Scholar]
- 139.Kang EY, Staples CA, McGuinness G, Primack SL, Müller NL. Detection and differential diagnosis of pulmonary infections and tumors in patients with AIDS: Value of chest radiography versus CT. AJR Am J Roentgenol. 1996;166:15–19. doi: 10.2214/ajr.166.1.8571866. [DOI] [PubMed] [Google Scholar]
- 140.Larici AR, Cicchetti G, Marano R, Merlino B, Elia L, Calandriello L, Del Ciello A, Farchione A, Savino G, Infante A, et al. Multimodality imaging of COVID-19 pneumonia: From diagnosis to follow-up. A comprehensive review. Eur J Radiol. 2020;131:109217. doi: 10.1016/j.ejrad.2020.109217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ippolito D, Maino C, Pecorelli A, Allegranza P, Cangiotti C, Capodaglio C, Mariani I, Giandola T, Gandola D, Bianco I, et al. Chest X-ray features of SARS-CoV-2 in the emergency department: A multicenter experience from northern Italian hospitals. Respir Med. 2020;170:106036. doi: 10.1016/j.rmed.2020.106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li P, Chen L, Liu Z, Pan J, Zhou D, Wang H, Gong H, Fu Z, Song Q, Min Q, et al. Clinical features and short-term outcomes of elderly patients with COVID-19. Int J Infect Dis. 2020;97:245–250. doi: 10.1016/j.ijid.2020.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cui N, Zou X, Xu L. Preliminary CT findings of coronavirus disease 2019 (COVID-19) Clin Imaging. 2020;65:124–132. doi: 10.1016/j.clinimag.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kovács A, Palásti P, Veréb D, Bozsik B, Palkó A, Kincses ZT. The sensitivity and specificity of chest CT in the diagnosis of COVID-19. Eur Radiol. 2020;13:1–6. doi: 10.1007/s00330-020-07347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao W, He L, Tang H, Xie X, Tang L, Liu J. The Relationship Between Chest Imaging Findings and the Viral Load of COVID-19. Front Med (Lausanne) 2020;7:558539. doi: 10.3389/fmed.2020.558539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meng H, Xiong R, He R, Lin W, Hao B, Zhang L, Lu Z, Shen X, Fan T, Jiang W, et al. CT imaging and clinical course of asymptomatic cases with COVID-19 pneumonia at admission in Wuhan, China. J Infect. 2020;81:e33–e39. doi: 10.1016/j.jinf.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mehrabi S, Fontana S, Mambrin F, Nguyen HQ, Righi E, Tacconelli E, Mansueto G. Pitfalls of computed tomography in the coronavirus 2019 (COVID-19) era: a new perspective on ground-glass opacities. Cureus. 2020;12:e8151. doi: 10.7759/cureus.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tofighi S, Najafi S, Johnston SK, Gholamrezanezhad A. Low-dose CT in COVID-19 outbreak: Radiation safety, image wisely, and image gently pledge. Emerg Radiol. 2020;27:601–605. doi: 10.1007/s10140-020-01784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 150.Pandey LM. Design of engineered surfaces for prospective detection of SARS-CoV-2 using quartz crystal micro-balance-based techniques. Expert Rev Proteomics. 2020;17:425–432. doi: 10.1080/14789450.2020.1794831. [DOI] [PubMed] [Google Scholar]
- 151.Maddali H, Miles CE, Kohn J, O'Carroll DM. Optical Biosensors for Virus Detection: Prospects for SARS-CoV-2/COVID-19. ChemBioChem. 2021;22:1176–1189. doi: 10.1002/cbic.202000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cui F, Zhou HS. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Samson R, Navale GR, Dharne MS. Biosensors: frontiers in rapid detection of COVID-19. 3 Biotech. 2020;10:385. doi: 10.1007/s13205-020-02369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 155.Zhao H, Liu F, Xie W, Zhou TC, OuYang J, Jin L, Li H, Zhao CY, Zhang L, Wei J, et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens Acuators B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Choi JR. Development of point-of-care biosensors for COVID-19. Front Chem. 2020;8:517. doi: 10.3389/fchem.2020.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lu R, Wu X, Wan Z, Li Y, Jin X, Zhang C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int J Mol Sci. 2020;21:2826. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yuan X, Yang C, He Q, Chen J, Yu D, Li J, Zhai S, Qin Z, Du K, Chu Z, et al. Current and perspective diagnostic techniques for COVID-19. ACS Infect Dis. 2020;6:1998–2016. doi: 10.1021/acsinfecdis.0c00365. [DOI] [PubMed] [Google Scholar]
- 159.Huang WE, Lim B, Hsu CC, Xiong D, Wu W, Yu Y, Jia H, Wang Y, Zeng Y, Ji M, et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Song J, Liu C, Mauk MG, Rankin SC, Lok JB, Greenberg RM, Bau HH. Two-Stage Isothermal Enzymatic Amplification for Concurrent Multiplex Molecular Detection. Clin Chem. 2017;63:714–722. doi: 10.1373/clinchem.2016.263665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.El-Tholoth M, Bau HH, Song J. A Single and Two-Stage, Closed-Tube, Molecular Test for the 2019 Novel Coronavirus (COVID-19) at Home, Clinic, and Points of Entry. ChemRxiv. doi: 10.26434/chemrxiv.11860137. [DOI] [Google Scholar]
- 163.Smithgall MC, Scherberkova I, Whittier S, Green DA. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the Rapid Detection of SARS-CoV-2. J Clin Virol. 2020;128:104428. doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rhoads DD, Cherian SS, Roman K, Stempak LM, Schmotzer CL, Sadri N. Comparison of Abbott ID Now, DiaSorin Simplexa, and CDC FDA Emergency Use Authorization Methods for the Detection of SARS-CoV-2 from Nasopharyngeal and Nasal Swabs from Individuals Diagnosed with COVID-19. J Clin Microbiol. 2020;58:e00760–e20. doi: 10.1128/JCM.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Basu A, Zinger T, Inglima K, Woo KM, Atie O, Yurasits L, See B, Aguero-Rosenfeld ME. Performance of Abbott ID Now COVID-19 Rapid Nucleic Acid Amplification Test Using Nasopharyngeal Swabs Transported in Viral Transport Media and Dry Nasal Swabs in a New York City Academic Institution. J Clin Microbiol. 2020;58:e01136–e20. doi: 10.1128/JCM.01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Westermann L, Neubauer B, Köttgen M. Nobel Prize 2020 in Chemistry honors CRISPR: A tool for rewriting the code of life. Pflugers Arch. 2021;473:1–2. doi: 10.1007/s00424-020-02497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 168.Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Broughton JP, Deng X, Yu G, Fasching CL, Singh J, Streithorst J, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, et al. Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. medRxiv. doi: 10.1101/2020.03.06.20032334. [DOI]
- 170.Javalkote VS, Kancharla N, Bhadra B, Shukla M, Soni B, Goodin M, Bandyopadhyay A, Dasgupta S. CRISPR-based assays for rapid detection of SARS-CoV-2. Methods. 2020 Oct 9; doi: 10.1016/j.ymeth.2020.10.003. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.van Dongen JE, Berendsen JTW, Steenbergen RDM, Wolthuis RMF, Eijkel JCT, Segerink LI. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens Bioelectron. 2020;166:112445. doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Joung J, Ladha A, Saito M, Segel M, Bruneau R, Huang MW, Kim NG, Yu X, Li J, Walker BD, et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv. doi: 10.1101/2020.05.04.20091231. [DOI] [Google Scholar]
- 174.Food and Drug Administration (FDA): Sherlock™ CRISPR SARS-CoV-2 kit - Instructions for use. https://www.fda.gov/media/137746/download. Accessed February 2, 2021.
- 175.Hou T, Zeng W, Yang M, Chen W, Ren L, Ai J, Wu J, Liao Y, Gou X, Li Y, et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020;16:e1008705. doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Curti L, Pereyra-Bonnet F, Gimenez CA. An Ultrasensitive, Rapid, and Portable Coronavirus SARS-CoV-2 Sequence Detection Method Based on CRISPR-Cas12. bioRxiv. doi: 10.1101/2020.02.29.971127. [DOI]
- 177.Salipante SJ, Jerome KR. Digital PCR-An Emerging Technology with Broad Applications in Microbiology. Clin Chem. 2020;66:117–123. doi: 10.1373/clinchem.2019.304048. [DOI] [PubMed] [Google Scholar]
- 178.Falzone L, Musso N, Gattuso G, Bongiorno D, Palermo CI, Scalia G, Libra M, Stefani S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int J Mol Med. 2020;46:957–964. doi: 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Falzone L, Gattuso G, Lombardo C, Lupo G, Grillo CM, Spandidos DA, Libra M, Salmeri M. Droplet digital PCR for the detection and monitoring of Legionella pneumophila. Int J Mol Med. 2020;46:1777–1782. doi: 10.3892/ijmm.2020.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Crimi S, Falzone L, Gattuso G, Grillo CM, Candido S, Bianchi A, Libra M. Droplet Digital PCR Analysis of Liquid Biopsy Samples Unveils the Diagnostic Role of hsa-miR-133a-3p and hsa-miR-375-3p in Oral Cancer. Biology (Basel) 2020;9:379. doi: 10.3390/biology9110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Salemi R, Falzone L, Madonna G, Polesel J, Cinà D, Mallardo D, Ascierto PA, Libra M, Candido S. MMP-9 as a Candidate Marker of Response to BRAF Inhibitors in Melanoma Patients With BRAFV600E Mutation Detected in Circulating-Free DNA. Front Pharmacol. 2018;9:856. doi: 10.3389/fphar.2018.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Battaglia R, Palini S, Vento ME, La Ferlita A, Lo Faro MJ, Caroppo E, Borzì P, Falzone L, Barbagallo D, Ragusa M, et al. Identification of extracellular vesicles and characterization of miRNA expression profiles in human blastocoel fluid. Sci Rep. 2019;9:84. doi: 10.1038/s41598-018-36452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bizouarn F. Clinical applications using digital PCR. Methods Mol Biol. 2014;1160:189–214. doi: 10.1007/978-1-4939-0733-5_16. [DOI] [PubMed] [Google Scholar]
- 184.Olmedillas-López S, García-Arranz M, García-Olmo D. Current and Emerging Applications of Droplet Digital PCR in Oncology. Mol Diagn Ther. 2017;21:493–510. doi: 10.1007/s40291-017-0278-8. [DOI] [PubMed] [Google Scholar]
- 185.Denis JA, Nectoux J, Lamy PJ, Rouillac Le Sciellour C, Guermouche H, Alary AS, Kosmider O, Sarafan-Vasseur N, Jovelet C, Busser B, et al. Development of digital PCR molecular tests for clinical practice: Principles, practical implementation and recommendations. Ann Biol Clin (Paris) 2018;76:505–523. doi: 10.1684/abc.2018.1372. [DOI] [PubMed] [Google Scholar]
- 186.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84:1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Alteri C, Cento V, Antonello M, Colagrossi L, Merli M, Ughi N, Renica S, Matarazzo E, Di Ruscio F, Tartaglione L, et al. Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One. 2020;15:e0236311. doi: 10.1371/journal.pone.0236311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Suo T, Liu X, Feng J, Guo M, Hu W, Guo D, Ullah H, Yang Y, Zhang Q, Wang X, et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liu C, Shi Q, Peng M, Lu R, Li H, Cai Y, Chen J, Xu J, Shen B. Evaluation of droplet digital PCR for quantification of SARS-CoV-2 Virus in discharged COVID-19 patients. Aging (any NY) 2020;12:20997–21003. doi: 10.18632/aging.104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vasudevan HN, Xu P, Servellita V, Miller S, Liu L, Gopez A, Chiu CY, Abate AR. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification. Sci Rep. 2021;11:780. doi: 10.1038/s41598-020-80715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The original contributions/references of studies presented in the study are publicly available. These data can be found at: www.pubmed.com and https://www.who.int.