Abstract
Background
Given the observed olfactory and gustatory dysfunctions in patients with COVID-19 and recent findings on taste receptors possible important activities in the immune system, we elected to estimate the correlation between COVID-19 mortality and polymorphism of a particular type of bitter taste receptor gene called TAS2R38, in a worldwide epidemiological point of view.
Methods
Pooled rate of each of the rs713598, rs1726866, rs10246939, and PAV/AVI polymorphisms of the TAS2R38 gene was obtained in different countries using a systematic review methodology and its relationship with the mortality of COVID-19. Data were analyzed by the comprehensive meta-analysis software and SPSS.
Results
There was only a significant reverse Pearson correlation in death counts and PAV/AVI ratio, p = 0.047, r = −0.503. Also, a significant reverse correlation of PAV/AVI ratio and death rate was seen, r = −0.572 p = 0.021. rs10246939 ratio had a significant positive correlation with death rate, r = 0.851 p = 0.031. Further analysis was not significant. Our results showed that the higher presence of PAV allele than AVI, and a higher rate of G allele than A in rs10246939 polymorphism in a country, could be associated with lower COVID-19 mortality. While assessing all three polymorphisms showed a huge diversity worldwide.
Conclusion
Due to extraoral activities of bitter taste receptor genes, especially in mucosal immunity, this gene seems to be a good candidate for future studies on COVID-19 pathophysiology. Also, the high worldwide diversity of TAS2R38 genes polymorphism and its possible assassination with mortality raises concerns about the efficiency of vaccine projects in different ethnicities.
Keywords: COVID-19, Polymorphism, TAS2R38 gene, Mortality
1. Introduction
The human body function facing difficult conditions such as diseases is largely being modified by two main factors of genetics and environmental factors. The human genome, inherited from previous generations, is one of the most important determinants of the body's response to conditions such as infectious diseases. During the process of modern human evolution out of Africa, various human communities have been expanded around the planet with worldwide genetic variations [1]. In recent years, genetic mapping of human genome genes has attracted the attention of many researchers [2]. Genetic studies and comparisons of different communities have led to the identification of genes that show the evolutionary history of humans, to the extent that some of these genes are referred to as evolution signatures [3]. In an evolutionary view, when a particular selective event contributing to natural selection is distributed over multiple populations, the same widespread families of genes within populations that are acquired from the same parent will respond similarly in all populations. Recognizing these common selective events may recognize significant genomic characteristics of humans in recent global history [3].
The world is currently experiencing the most tangible pandemic of the century due to a virus called n-SARS-CoV, which could be assumed as a very good sample of a global selective event. Coronavirus 2019 disease (COVID-19) is spreading rapidly around the world and is responsible for many deaths worldwide. COVID-19 patients may show mild to severe clinical manifestations of pneumonia.
However, the disease has had a variety of manifestations including cough, body aches, headaches, anosmia, etc. There is currently no definitive or effective treatment for COVID-19 [4]. In our previous study, we showed that in a study of 48,758 people of different ethnicities, polymorphism of a specific type of gene in the Renin-angiotensin system called ACE I was associated with a better improvement ratio of COVID-19 patients in each country [5]. Olfactory and gustatory dysfunctions have also been observed in patients with COVID-19 [6]. Some researchers have suggested a possible link between the Olfactory and gustatory genes and COVID-19 disease [7]. Taste receptors, especially G protein-coupled receptors, which detect different tastes, respond to a wide range of food stimuli [8]. Some of these taste receptors are involved not only in sense but also in immunity and inflammatory processes. Bitter Taste Receptors (TAS2Rs) are a group of taste receptors that in addition to their role in the gustatory system, also have very important activities in the immune system [9]. From the same group, the bitter taste receptor T2R38 is expressed in ciliated cells of the human upper airways [10], where the new coronavirus appears to replicate [11]. T2R38 also regulates the innate defense settings of the human upper respiratory tract by producing nitric oxide (NO), which stimulates mucus secretion and has direct immunological effects [12]. Besides, common polymorphisms of the TAS2R38 gene, based on taste sensitivity to phenylthiocarbamide (PTC), a bitter molecule stimulant, make significant differences in susceptibility of the upper respiratory tract to respiratory infection [13]. Therefore, it is very likely that genetic changes in the TAS2R38 function may cause individual differences in how airway cells act against infectious agents [14]. So in the present study, the TAS2R38 gene was chosen as a suitable factor for mapping its differences in the genome of people around the world. T2R38 is encoded by the TAS2R38 gene, which has two common polymorphisms in different populations around the world. The functional form of the receptor contains proline, alanine, and valine (PAV), while the non-functional form of the receptor contains alanine, valine, and isoleucine (AVI). Therefore, the active haplotype in this field is called PAV and the inactive haplotype is called AVI. Various other polymorphisms have also been suggested for this gene. The TAS2R38 gene is encoded by three non-synonymous coding SNPs (rs713598 - G145C, Ala49Pro; rs1726866 - T785C, Val262Ala; rs10246939 - A886G, Ile296Val) [15].
The present study sought to investigate the possible relationship between genetic differences in bitterness receptors in different communities against COVID-19. Comparison of epidemiological data on the occurrence of Covid-19 disease in different ethnicities can provide a scientific basis for future research to implement protocols related to the treatment and vaccination of COVID-19.
2. Methods
2.1. COVID-19 data
The COVID-19 death rate in each country was retrieved from the WHO Coronavirus Disease Dashboard [https://covid19.who.int/WHO-COVID-19-global-data.csv]. WHO database has provided statics of positive cases and death cases in a monthly manner. The mortality rate on the 31 August of 2020 was very low in lots of countries, so many countries were excluded. The countries with available studies reporting TAS2R38 polymorphism included in our study.
2.2. TAS2R38 polymorphism
Preferred Reporting Items for Systematic Studies and Meta-Analysis (PRISMA) statement was considered for collecting studies that had reported the TAS2R38 polymorphisms. Our search strategy was querying through ISI, PubMed, Scopus, and Google Scholar databases with keywords of as TAS2R38 gene, Polymorphism, rs713598, rs1726866, rs10246939, and PAV/AVI from the from eternity to 2020. At the step of title checking, 654 studies were detected. 96 duplicated studies were excluded. Checking the abstracts, 255 irrelevant studies were excluded. Evaluating the full-text of studies, studies not having a proper methodology of polymorphism assessment, having a sample size of patients with genetic disorders, and not reporting the number of subjects, were excluded. In the end, 22 articles were included in our study.
2.3. Static analysis
The collected data of death rate and polymorphisms rate were analyzed using the Comprehensive Meta-Analysis (CMA) software version 2 based on the random-effects model. The I2 index was utilized to detect heterogeneity in studies. Since the I2 index of the combined polymorphisms ratio was 99, 98, 87, 100% percent for rs713598, rs1726866, rs10246939, and PAV/AVI random effect model was used and the inverse method was applied for variance and weight estimation for each study. Grouping was done based on the country. Meta-regression of the COVID-19 mortality rate with polymorphisms ratio was conducted using CMA. There was no evidence of study bias based on egger's test (p > 0.05).
3. Results
In the present study, 21 studies reporting different types of TAS2R38 gene polymorphisms in healthy individuals were included in the study (Table 1 ). These studies reported data on 19,997 people living in 22 different countries.
Table 1.
Frequent study of different types of TAS2R38 gene polymorphisms in healthy individuals.
| Study | Country | n | rs713598 |
rs1726866 |
rs10246939 |
PAV | AVI | |||
|---|---|---|---|---|---|---|---|---|---|---|
| G | C | T | C | A | G | |||||
| Vinuthalakshmi K [16] | India | 100 | 144 | 56 | 124 | 74 | 114 | 84 | 74 | 114 |
| Carrai M [22] | Czech | 576 | 619 | 533 | 663 | 481 | 640 | 540 | 449 | 569 |
| Carrai M [22] | German | 679 | 795 | 563 | 569 | 801 | 739 | 633 | – | – |
| Choi JH [17] | Korea | 3567 | – | – | – | – | 2942 | 4192 | – | – |
| Schembre SM [18] | Native Hawaiians | 236 | – | – | – | – | – | – | 298 | 148 |
| Schembre SM [18] | Japanese Americans | 383 | – | – | – | – | – | – | 423 | 343 |
| Choi JH [19] | Korea | 1361 | – | – | – | – | – | – | 1576 | 1122 |
| Adappa ND [20] | USA | 70 | – | – | – | – | – | – | 50 | 90 |
| Yamaki M [21] | Japanese | 79 | – | – | – | – | – | – | 222 | 150 |
| Barragán R [23] | Spain | 949 | 1137 | 761 | – | – | – | – | – | – |
| Sandell M [24] | Finland | 1860 | 2443 | 1277 | 2331 | 1389 | 2331 | 1389 | 1693 | 3135 |
| Perna S [25] | Italy | 118 | 129 | 107 | – | – | – | – | – | – |
| Deshaware S [27] | India | 393 | 545 | 241 | 533 | 253 | 520 | 266 | 238 | 520 |
| Hayes JE [28] | USA | 198 | – | – | – | – | – | – | 175 | 179 |
| Shen Y [26] | UK | 136 | – | – | – | – | – | – | 118 | 154 |
| Ooi SX [29] | Malaysia | 100 | 122 | 78 | – | – | – | – | – | – |
| Khataan NH [30] | Canada | 911 | 971 | 851 | – | – | – | – | – | – |
| Melis M [31] | Italy | 192 | – | – | – | – | – | – | 348 | 366 |
| Sandell MA [32] | Spain | 23 | – | – | – | – | – | – | 22 | 22 |
| Sandell MA [32] | Finland | 22 | – | – | – | – | – | – | 21 | 23 |
| Sandell MA [32] | UK | 35 | – | – | – | – | – | – | 38 | 32 |
| Lambert JD [33] | UK | 6250 | – | – | – | – | – | – | 374 | 512 |
| Robino A [34] | Georgia | 116 | – | – | – | – | – | – | 282 | 127 |
| Robino A [34] | Armenia | 47 | – | – | – | – | – | – | 95 | 41 |
| Robino A [34] | Azerbaijan | 91 | – | – | – | – | – | – | 198 | 87 |
| Robino A [34] | Uzbekistan | 57 | – | – | – | – | – | – | 127 | 56 |
| Robino A [34] | Kazakhstan | 80 | – | – | – | – | – | – | 174 | 76 |
| Robino A [34] | Tajikistan | 105 | – | – | – | – | – | – | 227 | 98 |
| Wang K [35] | china | 673 | – | – | – | – | – | – | – | – |
| Gorovic N [36] | Denmark | 490 | 627 | 353 | 590 | 392 | 586 | 390 | 327 | 559 |
| Siasi E [37] | Iran | 100 | – | – | 76 | 124 | – | – | – | – |
Studies examined four different types of TAS2R38 gene polymorphisms, including rs713598, rs1726866, rs10246939, and PAV/AVI. To calculate the overall ratio of alleles in different polymorphisms, the random-effect model was used to cumulatively calculate the ratio of alleles for each subgroup (country). In the study of rs713598 polymorphism, data from 10 countries; in rs1726866 assessment, data from 6 countries; in rs10246939 assessment, 6 countries and in the PAV/AVI polymorphism assessment, 16 countries were studied. The results are shown in Table 2 .
Table 2.
Polymorphisms of TAS2R38 gene in different countries.
| Country | n | rs713598 |
rs1726866 |
rs10246939 |
PAV/AVI |
|---|---|---|---|---|---|
| G/C | T/C | A/G | |||
| Canada | 911 | 1.141011 | – | – | – |
| China | 673 | 2.167059 | – | – | – |
| Czech | 576 | 1.161351 | 1.378378 | 1.185185 | 0.789104 |
| Denmark | 490 | 1.776204 | – | – | – |
| Finland | 1860 | 1.913078 | 1.678186 | 1.678186 | 0.542749 |
| German | 679 | 1.412078 | 0.710362 | 1.167457 | – |
| Denmark | 491 | – | 1.505102 | 1.502564 | 0.584973 |
| Italy | 118 | 1.205607 | – | – | 0.95082 |
| Malaysia | 100 | 1.564103 | – | – | – |
| Spain | 949 | 1.494087 | – | – | 1 |
| India | 492 | 2.319865 | 2.009174 | 1.811429 | 0.492114 |
| Iran | 100 | – | 0.612903 | – | – |
| Korea | 3567 | – | – | 0.701813 | 1.404635 |
| Japan | 79 | – | – | – | 1.48 |
| USA | 198 | – | – | – | 1.254197 |
| UK | 6421 | – | – | – | 0.75 |
| Georgia | 116 | – | – | – | 2.220472 |
| Armenia | 47 | – | – | – | 2.317073 |
| Azerbaijan | 91 | – | – | – | 2.275862 |
| Uzbekistan | 57 | – | – | – | 2.267857 |
| Kazakhstan | 80 | – | – | – | 2.289474 |
| Tajikistan | 105 | – | – | – | 2.316327 |
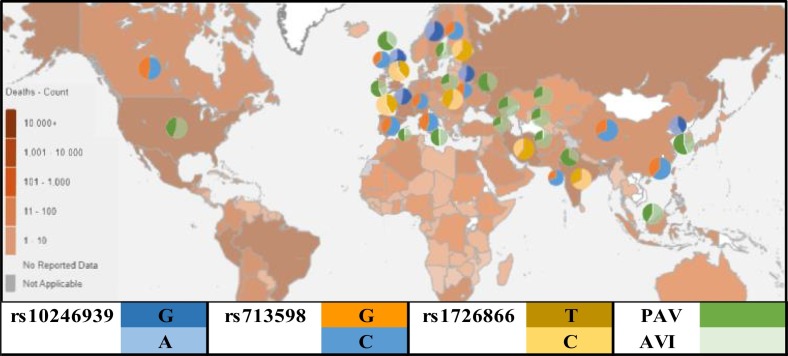
Data on the prevalence and mortality of COVID-19 in different countries were extracted from the WHO website. As shown in Fig. 1 , the disease has spread to most parts of the world, with the highest mortality rates (brown colored) in the USA, Brazil, various parts of Europe, and Asian countries such as India, Japan, and Iran, and so on. The ratio of allele's distribution for different polymorphisms is shown in Fig. 1. The interpretation of geographical differences of polymorphisms was difficult due to wide changes between different countries. But the data was not available in other parts of the world.
Fig. 1.
Image of the Earth based on the COVID-19 mortality rate from the WHO website (https://covid19.who.int/explorer).
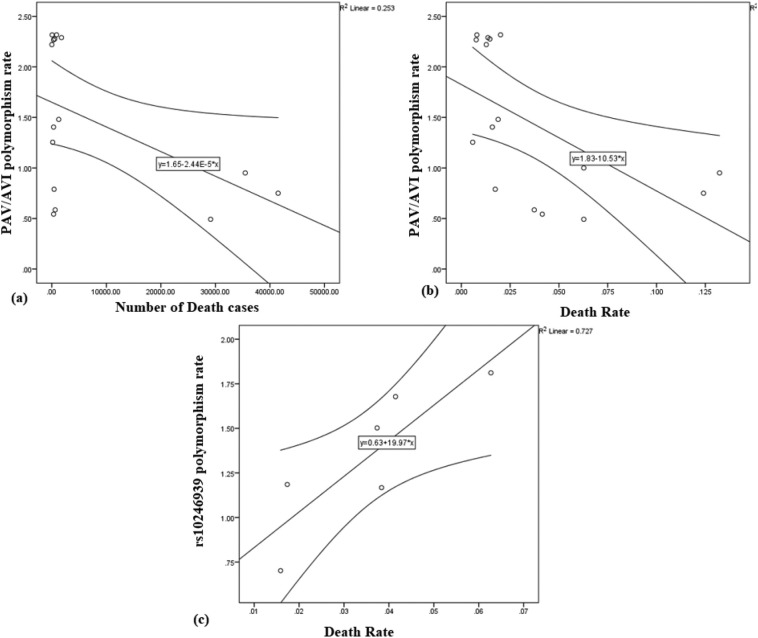
Then correlation of the calculated point estimate for the polymorphism rate of each country with reported cases and deaths of COVID-19 was estimated. There was only a significant reverse Pearson correlation in death counts and PAV/AVI ratio, p = 0.047, r = −0.503 (Fig. 2 .a). Also, a significant reverse correlation of PAV/AVI ratio and death rate was seen, r = −0.572 p = 0.021 (Fig. 2.b). rs10246939 ratio had a significant positive correlation with death rate, r = 0.851 p = 0.031 (Fig. 2.c). Further analysis was not significant.
Fig. 2.
Correlation of the PAV/AVI polymorphism rate and death cases of each country (a); PAV/AVI polymorphism rate vs. death rate (b); rs10246939 polymorphism rate and death rate (c).
4. Discussion
Variation in the disease spreads in different regions can be attributed to a large number of genetic, cultural, and environmental influences, which has led researchers to investigate the possible link between genetic factors and susceptibility to COVID-19. Anosmia has been reported as a manifestation of COVID-19 and other coronavirus infections of SARS and MERS [38].
Surprisingly it is not accompanied by nasal obstruction or other symptoms of rhinitis. Therefore, this is probably due to the direct damage of the virus to the olfactory and taste receptors [39]. Our study examined the genetic basis of this issue by looking at a gene involved in the olfactory and taste systems, at a worldwide perspective. Our results showed that the higher presence of PAV allele than AVI, and a higher rate of G allele than A in rs10246939 polymorphism in a country, could be associated with lower COVID-19 mortality. So far, no study has examined the relationship between these genetic factors and COVID-19 at the epidemiological level. However, some studies provide laboratory evidence to support this hypothesis. Innate airway immunity depends on the efficient and timely detection of bacteria and viruses by the airway mucosa [40]. Deshpande et al. showed the presence of bitter taste receptors in the human lung and broncho dilation associated with their stimulation [41].
Shah et al. stated that TAS2R expression in the human airway epithelial cells. Also they find that bitter compounds increase intracellular Ca2+ concentration and stimulate ciliary beat frequency. Ciliated cells of the airways help to clear mucus and toxic chemicals from the airways. Viral infections and cigarette smoke have also been shown to damage these cilias and can interfere with the defense mechanism [42]. Recent data suggest that a specific TAS2R (TAS2R38) is involved in the innate immune defense of the upper airways, and changes in this gene contribute to individual differences in susceptibility to respiratory infection [13]. Airway T2R expression was first studied in human bronchial ciliated epithelial cells, where they amplify calcium signals that increase the frequency of ciliary beats [43].
In the lung, T2Rs may be very important in the sinus cavity, which is at the forefront of the respiratory defenses, and are in contact with large volumes and types of pathogenic microbes. Host and pathogen interactions occur continuously with each breath, and when the nasal line of defense is destroyed, bacterial infections are likely to cause lower airway infections or exacerbate airway disease [44].
Immune system components also contribute to the regeneration of damaged tissues by stem cells, which could be important in COVID-19 recovery, especially in taste and loss recovery. Stem cells are also present in the oral cavity. Immune system responses to SARS-COV-2 may also get started from the oral cavity. Research has shown that important molecules for SARS-CoV-2 infection are plentiful throughout the oral cavity [45]. The SARS-CoV-2 receptor, ACE2, and TMPRSS2 that facilitate entry of the SARS-CoV-2 into the human cells were defined as factors of SARS-CoV-2 infection. ACE2 and TMPRSS2 expression were detected in the dorsal part of the tongue, gingiva, saliva, and tongue coating cells from taste buds [46]. The main concern is that the SARS-CoV-2 specifically influences smell perception by contaminating olfactory sensory neurons or passively via disrupting linked cells. Chou et al. described the olfactory epithelium and olfactory bulb cells expressing SARS-CoV-2 cell entry molecules. In COVID-19 patients, non-neuronal forms of cells contribute to anosmia and associated defects of smell sensation. Sustentacular and oral stem cells are probably direct targets of SARS-CoV-2 in the human oral cavity [47]. Oral-derived stem cells are known to have a contribution to injured tissue regeneration and local immunomodulation [48]. Rather than immune responses as being in the first line of the virus confrontation, these stem cells may also be a good candidate for cell therapy due to regenerative potential [49]. Various clinical studies have previously been conducted on mesenchymal stromal/stem cells as a potential therapy for COVID-19. Treatment options for COVID-19 are not completely provided, but stem cell therapies have already shown efficiency in treating inflammatory responses in patients with COVID-19 [50]. Unless the stem cells of the oral cavity are not severely impaired, the olfactory system may have the substantial regenerative ability, contributing to spontaneous odor perception recovery [51]; So, stem cell therapies can be queried for smell and taste perception loss treatment in COVID-19. It is possible to provide a specific technique to recognize signaling pathways that trigger the stimulation and neurogenesis of olfactory stem cells and thus utilize the intrinsic ability of the olfactory resident stem cells for recovery [52]. Novel biotechnology advancements could help us and should be taken to account as our available choices for the COVID-19 researches. Bioimpedance is a technique that is used to detect inflammatory tissues in the oral cavity [53,54]. Also, some researchers have used it to detect inflammatory processes in airways, by analyzing extracellular bioelectrical conductivity [55]. The taste sensation is a bioelectrical process at the level of the taste receptors [56]. Interestingly, human tongue-recorded bio-electrical impulses were shown to be related to the TAS2R38 PAV-AVI genotypes [57]. So, we think that bio-electrical analysis could help in researches which are going to be conducted on the gustatory recovery of COVID-19 [58].
Renin-angiotensin system (RAS) is an important regulator of body fluids and sodium homeostasis. Angiotensin II (AngII) is an important active product of RAS. The RAS system appears to alter individuals' sensitivity to taste through the AngII receptor type 1 (AT1), which is expressed in taste cells. However, the molecular mechanisms involved in these changes have not been identified [59].
On the other hand, the RAS system is involved in modulating the activity of taste cells. Expression of the ACE-2 gene, the major route of new coronavirus entry into cells, in taste cells provides a possible explanation for taste disorders in patients with COVID-19. That is, SARS-CoV-2 may enter taste cells via ACE-2: as a result, the normal function of these sensory cells is disrupted, leading to a change or loss of taste perception [60].
On the other hand, one of the most important interpretations of our current study and previous study on ACE I polymorphism's correlation with epidemiological aspects of the COVID-19 is raising cautions about the possible effect of huge ethnicitical differences of communities on the COVID-19 vaccine's efficiency that are going to be available worldwide. Also, there is the possibility of bitter taste receptors' effect on cellular immunity against COVID-19. A study stated that Tas2Rs genes are also being expressed in white blood cells and upregulation of TAS2Rs in human white blood cells is seen in patients with severe refractory asthma [61]. Tran et al. have shown the higher expression of T2R38 in resting and activated human lymphocytes [62]. Identification of bitter receptors role in response to n-SARS-Cov may help to get new therapeutic targets for stimulating mucociliary clearance [12] as a major defect in the elderly making them susceptible to severe COVID-19 [60]. As one of the agonists of TAS2Rs, caffeine is a very good candidate for future studies in this field [63].
4.1. Limitations
However, this study deals with various known and unknown factors affecting global mortality of COVID-19, including demographic characteristics such as patient sex, age, underlying diseases, and different methods for treating patients in different countries.
An important limitation of studies such as our study that examines correlational relationships is the impossibility of confidence to the investigated relationship as a causal relationship. Understanding whether there is a real causal relationship or whether these relationships occur for a variety of mediating reasons requires further study. Because the human taste system is greatly affected by genetic polymorphisms; these genetic changes may be associated with a change in a person's lifestyle. T2R38 gene polymorphisms have been important predictors of behaviors such as alcohol or smoking [64]. Also, concerning other types of genes involved in taste perception, genetic variation in taste receptors has been associated with differences in food salinity perception, which has affected a person's risk of developing hypertension [65]. Blood pressure is also one of the main risk factors for mortality in COVID-19. These countless potential relationships can all exist. Therefore, it is necessary to determine the exact mechanisms in the transmission of T2R38 signals in response to COVID-19 in more detail.
5. Conclusion
Extraoral functions of bitter taste receptor genes, especially in mucosal immunity, suggest it a good candidate for future studies seeking to understand the pathophysiology of COVID-19. Our study showed a high diversity of TAS2R38 genes polymorphism in various parts of the world and its possible assassination with the mortality rate that raises concerns about the efficiency of vaccine projects in different ethnicities. Also, this study suggests further attention to bitter taste receptors and underlying pathways as a potential approach to help COVID-19 patients' improvement.
Ethics approval and consent to participate
There is no human subject participating in this study.
Consent for publication
Not applicable.
Availability of data and material
All data would be available in the manuscript.
Funding
None.
CRediT authorship contribution statement
Shima Parsa: Conceptualization, Methodology, Data curation, Resources, Investigation, Writing - Original draft, Supervision. Vahid Mogharab: Data curation, Investigation, Resources, Writing - Original draft. Mohsen Ebrahimi: Conceptualization, Writing - Original draft, Data curation. Sayyed Reza Ahmadi: Writing - Review & editing, Data curation, Methodology, Supervision. Behzad Shahi: Data curation, Resources. Neema John Mehramiz: Supervision, Investigation. Mahdi Foroughian: Investigation, Data curation. Mohammad Zarenezhad: Data curation, Writing - Original draft. Navid Kalani: Writing - Original draft, Writing - Review & editing, Investigation. Mohammad Hashem Abdi: Investigation. Farshid Javdani: Investigation. Pouyan Keshavarz: Data curation, Resources, Methodology, Writing - Original draft. Naser Hatami: Conceptualization, Methodology, Data curation, Writing - Original draft, Writing - Review & editing, Supervision.
Declaration of competing interest
None.
Acknowledgements
We would like to thank the Clinical Research Development Unit of Peymanieh Educational and Research and Therapeutic Center of Jahrom University of Medical Sciences for providing facilities for this work.
References
- 1.Nielsen R., Akey J.M., Jakobsson M., Pritchard J.K., Tishkoff S., Willerslev E. Tracing the peopling of the world through genomics. Nature. 2017;541(7637):302–310. doi: 10.1038/nature21347. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams K.L. Encyclopedia of Bioinformatics and Computational Biology. Academic Press; 2019. Gene mapping; pp. 242–250. [Google Scholar]
- 3.Johnson K.E., Voight B.F. Patterns of shared signatures of recent positive selection across human populations. Nature ecology & evolution. 2018;2(4):713–720. doi: 10.1038/s41559-018-0478-6. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J., He W.T., Wang L., Lai A., Ji X., Zhai X., Li G., Suchard M.A., Tian J., Zhou J., Veit M. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol. Med. 2020;26(5):483–495. doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatami N., Ahi S., Sadeghinikoo A., et al. Worldwide ACE (I/D) polymorphism may affect COVID-19 recovery rate: an ecological meta-regression. Endocrine. 2020;68:479–484. doi: 10.1007/s12020-020-02381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiesa-Estomba C.M., Lechien J.R., Portillo-Mazal P., Martínez F., Cuauro-Sanchez J., Calvo-Henriquez C., Saussez S. Olfactory and gustatory dysfunctions in COVID-19. First reports of Latin-American ethnic patients. Am. J. Otolaryngol. Jun 6 2020;41(5) doi: 10.1016/j.amjoto.2020.102605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilinska K., Jakubowska P., Von Bartheld C.S., Butowt R. Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem. Neurosci. 2020;11(11):1555–1562. doi: 10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandrashekar J., Mueller K.L., Hoon M.A., Adler E., Feng L., Guo W., Zuker C.S., Ryba N.J. T2Rs function as bitter taste receptors. Cell. 2000;100(6):703–711. doi: 10.1016/s0092-8674(00)80706-0. Mar 17. [DOI] [PubMed] [Google Scholar]
- 9.Luo M., Ni K., Jing Y., Yu Z., Deng L. Towards the identification of extra-oral TAS2R agonists as drug agents for muscle relaxation therapies via bioinformatics-aided screening of bitter compounds in traditional Chinese medicine. Front. Physiol. 2019;10:861. doi: 10.3389/fphys.2019.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen N.A. The genetics of the bitter taste receptor T2R38 in upper airway innate immunity and implications for chronic rhinosinusitis. Laryngoscope. 2017;127(1):44–51. doi: 10.1002/lary.26198. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan R.W., Chan M.C., Agnihothram S., Chan L.L., Kuok D.I., Fong J.H., Guan Y., Poon L.L., Baric R.S., Nicholls J.M., Peiris J.M. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 2013;87(12):6604–6614. doi: 10.1128/JVI.00009-13. Jun 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee R.J., Cohen N.A. Role of the bitter taste receptor T2R38 in upper respiratory infection and chronic rhinosinusitis. Curr. Opin. Allergy Clin. Immunol. 2015;15(1):14. doi: 10.1097/ACI.0000000000000120. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee R.J., Xiong G., Kofonow J.M., Chen B., Lysenko A., Jiang P., Abraham V., Doghramji L., Adappa N.D., Palmer J.N., Kennedy D.W. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J. Clin. Invest. 2012;122(11):4145–4159. doi: 10.1172/JCI64240. Nov 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim U., Wooding S., Ricci D., Jorde L.B., Drayna D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum. Mutat. 2005;26(3):199–204. doi: 10.1002/humu.20203. Sep. [DOI] [PubMed] [Google Scholar]
- 15.Lalueza-Fox C., Gigli E., de la Rasilla M., Fortea J., Rosas A. Bitter taste perception in Neanderthals through the analysis of the TAS2R38 gene. Biol. Lett. 2009;5(6):809–811. doi: 10.1098/rsbl.2009.0532. Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinuthalakshmi K., Sheik N., Mustak M.S. TAS2R38 gene polymorphism and its association with taste perception, alcoholism and tobacco chewing among the Koraga-a primitive tribal population of Southwest coast of India. Meta Gene. Jun 1 2019;20 [Google Scholar]
- 17.Choi J.H. Variation in the TAS2R38 bitterness receptor gene was associated with food consumption and obesity risk in Koreans. Nutrients. 2019;11(9):1973. doi: 10.3390/nu11091973. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schembre S.M., Cheng I., Wilkens L.R., Albright C.L., Marchand L.L. Variations in bitter-taste receptor genes, dietary intake, and colorectal adenoma risk. Nutr. Cancer. 2013;65(7):982–990. doi: 10.1080/01635581.2013.807934. Oct 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi J.H., Lee J., Oh J.H., Chang H.J., Sohn D.K., Shin A., Kim J. Variations in the bitterness perception-related genes TAS2R38 and CA6 modify the risk for colorectal cancer in Koreans. Oncotarget. 2017;8(13):21253. doi: 10.18632/oncotarget.15512. Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adappa N.D., Zhang Z., Palmer J.N., Kennedy D.W., Doghramji L., Lysenko A., Reed D.R., Scott T., Zhao N.W., Owens D., Lee R.J. InInternational Forum of Allergy & Rhinology. vol. 4(1) Jan 2014. The bitter taste receptor T2R38 is an independent risk factor for chronic rhinosinusitis requiring sinus surgery; pp. 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaki M., Saito H., Isono K., Goto T., Shirakawa H., Shoji N., Satoh-Kuriwada S., Sasano T., Okada R., Kudoh K., Motoi F. Genotyping analysis of bitter-taste receptor genes TAS2R38 and TAS2R46 in Japanese patients with gastrointestinal cancers. J. Nutr. Sci. Vitaminol. 2017;63(2):148–154. doi: 10.3177/jnsv.63.148. [DOI] [PubMed] [Google Scholar]
- 22.Carrai M., Steinke V., Vodicka P., Pardini B., Rahner N., Holinski-Feder E., Morak M., Schackert H.K., Görgens H., Stemmler S., Betz B. Association between TAS2R38 gene polymorphisms and colorectal cancer risk: a case-control study in two independent populations of Caucasian origin. PLoS One. Jun 2 2011;6(6) doi: 10.1371/journal.pone.0020464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barragán R., Coltell O., Portolés O., Asensio E.M., Sorlí J.V., Ortega-Azorín C., González J.I., Sáiz C., Fernández-Carrión R., Ordovas J.M., Corella D. Bitter, sweet, salty, sour and umami taste perception decreases with age: sex-specific analysis, modulation by genetic variants and taste-preference associations in 18 to 80 year-old subjects. Nutrients. 2018;10(10):1539. doi: 10.3390/nu10101539. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandell M., Hoppu U., Mikkilä V., Mononen N., Kähönen M., Männistö S., Rönnemaa T., Viikari J., Lehtimäki T., Raitakari O.T. Genetic variation in the hTAS2R38 taste receptor and food consumption among Finnish adults. Genes Nutr. 2014;9(6):433. doi: 10.1007/s12263-014-0433-3. Nov 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perna S., Riva A., Nicosanti G., Carrai M., Barale R., Vigo B., Allegrini P., Rondanelli M. Association of the bitter taste receptor gene TAS2R38 (polymorphism RS713598) with sensory responsiveness, food preferences, biochemical parameters and body-composition markers. A cross-sectional study in Italy. Int. J. Food Sci. Nutr. 2018;69(2):245–252. doi: 10.1080/09637486.2017.1353954. Feb 17. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y., Kennedy O.B., Methven L. Exploring the effects of genotypical and phenotypical variations in bitter taste sensitivity on perception, liking and intake of brassica vegetables in the UK. Food Qual. Prefer. 2016;50:71–81. Jun 1. [Google Scholar]
- 27.Deshaware S., Singhal R. Genetic variation in bitter taste receptor gene TAS2R38, PROP taster status and their association with body mass index and food preferences in Indian population. Gene. 2017;627:363–368. doi: 10.1016/j.gene.2017.06.047. Sep 5. [DOI] [PubMed] [Google Scholar]
- 28.Hayes J.E., Bartoshuk L.M., Kidd J.R., Duffy V.B. Supertasting and PROP bitterness depends on more than the TAS2R38 gene. Chem. Senses. 2008;33(3):255–265. doi: 10.1093/chemse/bjm084. Mar 1. [DOI] [PubMed] [Google Scholar]
- 29.Ooi S.X., Lee P.L., Law H.Y., Say Y.H. Bitter receptor gene (TAS2R38) P49A genotypes and their associations with aversion to vegetables and sweet/fat foods in Malaysian subjects. Asia Pac. J. Clin. Nutr. 2010;19(4):491. Dec. [PubMed] [Google Scholar]
- 30.Khataan N.H., Stewart L., Brenner D.M., Cornelis M.C., El-Sohemy A. TAS2R38 genotypes and phenylthiocarbamide bitter taste perception in a population of young adults. Lifestyle Genomics. 2009;2(4–5):251–256. doi: 10.1159/000297217. [DOI] [PubMed] [Google Scholar]
- 31.Melis M., Errigo A., Crnjar R., Pes G.M., Barbarossa I.T. TAS2R38 bitter taste receptor and attainment of exceptional longevity. Sci. Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-54604-1. Dec 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandell M.A., Collado M.C. Genetic variation in the TAS2R38 taste receptor contributes to the oral microbiota in North and South European locations: a pilot study. Genes Nutr. 2018;13(1):30. Dec 1. [Google Scholar]
- 33.Lambert J.D., VanDusen S.R., Cockroft J.E., Smith E.C., Greenwood D.C., Cade J.E. Bitter taste sensitivity, food intake, and risk of malignant cancer in the UK Women’s Cohort Study. Eur. J. Nutr. 2019;58(5):2111–2121. doi: 10.1007/s00394-018-1772-4. Aug 1. [DOI] [PubMed] [Google Scholar]
- 34.Robino A., Mezzavilla M., Pirastu N., Dognini M., Tepper B.J., Gasparini P. A population-based approach to study the impact of PROP perception on food liking in populations along the Silk Road. PLoS One. Mar 13 2014;9(3) doi: 10.1371/journal.pone.0091716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K, Pang L, Yin L, Li X, Zhang J, Cui T, Tao Y, Lin HC. AMBN Gene Polymorphism Alters the Caries Susceptibility of Adolescents in South China. 10.21203/rs.2.12670/v1S.
- 36.Gorovic N., Afzal S., Tjønneland A., Overvad K., Vogel U., Albrechtsen C., Poulsen H.E. Genetic variation in the h TAS2R38 taste receptor and brassica vegetable intake. Scand. J. Clin. Lab. Invest. 2011;71(4):274–279. doi: 10.3109/00365513.2011.559553. Jul 1. [DOI] [PubMed] [Google Scholar]
- 37.Siasi E., Aleyasin A., Mowla J. Study of t886c snp in tas2r38 gene association with azoospermia and oligospermia in idiopathic infertile men. International journal of reproductive biomedicine (iranian journal of reproductive medicine) 2011;9(Suppl. 2):53 to 53. spring. [Google Scholar]
- 38.Meng X., Deng Y., Dai Z., Meng Z. COVID-19 and anosmia: a review based on up-to-date knowledge. Am. J. Otolaryngol. 2020;41(5):102581. doi: 10.1016/j.amjoto.2020.102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaira L.A., Salzano G., Fois A.G., Piombino P., De Riu G. Potential pathogenesis of ageusia and anosmia in COVID-19 patients. InInternational forum of allergy & rhinology. 2020;10(9):1103–1104. doi: 10.1002/alr.22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kikkert M. Innate immune evasion by human respiratory RNA viruses. Journal of innate immunity. 2020;12(1):4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deshpande D.A., Wang W.C., McIlmoyle E.L., Robinett K.S., Schillinger R.M., An S.S., Sham J.S., Liggett S.B. Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat. Med. 2010;16(11):1299. doi: 10.1038/nm.2237. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah A.S., Ben-Shahar Y., Moninger T.O., et al. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee R.J., Cohen N.A. Taste receptors in innate immunity. Cell. Mol. Life Sci. 2015;72(2):217–236. doi: 10.1007/s00018-014-1736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hens G., Hellings P.W. The nose: gatekeeper and trigger of bronchial disease. Rhinology. 2006;44(3):179. Sep. [PubMed] [Google Scholar]
- 45.Aurora A.B., Olson E.N. Immune modulation of stem cells and regeneration. Cell Stem Cell. 2014;15(1):14–25. doi: 10.1016/j.stem.2014.06.009. Jul 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakaguchi W., Kubota N., Shimizu T., Saruta J., Fuchida S., Kawata A., Yamamoto Y., Sugimoto M., Yakeishi M., Tsukinoki K. Existence of SARS-CoV-2 entry molecules in the oral cavity. Int. J. Mol. Sci. 2020;21(17):6000. doi: 10.3390/ijms21176000. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou H.J., Fletcher R.B., Das D. Non-neuronal expression of SARS-CoV-2 entry genes in the olfaory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020;6(31):eabc5801. doi: 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spagnuolo G., Codispoti B., Marrelli M., Rengo C., Rengo S., Tatullo M. Commitment of oral-derived stem cells in dental and maxillofacial applications. Dentistry journal. 2018;6(4):72. doi: 10.3390/dj6040072. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moll G., Drzeniek N., Kamhieh-Milz J., Geissler S., Volk H.D., Reinke P. MSC therapies for COVID-19: importance of patient coagulopathy, thromboprophylaxis, cell product quality and mode of delivery for treatment safety and efficacy. Front. Immunol. 2020;11:1091. doi: 10.3389/fimmu.2020.01091. May 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi R., Goldstein B.J. Olfactory epithelium: cells, clinical disorders, and insights from an adult stem cell niche. Laryngoscope investigative otolaryngology. 2018;3(1):35–42. doi: 10.1002/lio2.135. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mainland J.D., Barlow L.A., Munger S.D., Millar S.E., Vergara M.N., Jiang P., Schwob J.E., Goldstein B.J., Boye S.E., Martens J.R., Leopold D.A. Identifying treatments for taste and smell disorders: gaps and opportunities. Chem. Senses. 2020;45(7):493–502. doi: 10.1093/chemse/bjaa038. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ballini A., Cantore S., Scacco S., Coletti D., Tatullo M. Mesenchymal stem cells as promoters, enhancers, and playmakers of the translational regenerative medicine 2018. Stem Cells Int. 2018;1:2018. doi: 10.1155/2018/6927401. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emran S., Hurskainen M., Tomppo L., Lappalainen R., Kullaa A.M., Myllymaa S. Bioimpedance spectroscopy and spectral camera techniques in detection of oral mucosal diseases: a narrative review of the state-of-the-art. Journal of Medical Engineering & Technology. 2019;43(8):474–491. doi: 10.1080/03091902.2019.1692940. Nov 17. [DOI] [PubMed] [Google Scholar]
- 54.Tatullo M., Marrelli M., Amantea M., Paduano F., Santacroce L., Gentile S., Scacco S. Bioimpedance detection of oral lichen planus used as preneoplastic model. J. Cancer. 2015;6(10):976. doi: 10.7150/jca.11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peroni D.G., Bodini A., Loiacono A., Paida G., Tenero L., Piacentini G.L. Bioimpedance monitoring of airway inflammation in asthmatic allergic children. Allergol. Immunopathol. 2009;37(1):3–6. doi: 10.1016/s0301-0546(09)70243-5. Feb 1. [DOI] [PubMed] [Google Scholar]
- 56.Aroulmoji V., Mathlouthi M., Portmann-Richardson M.O. Solution properties and the masking of unpleasant tastes of nicotine-sweetener-water mixtures. International Journal of Pharmaceutical Sciences and Research. Jun 1 2013;4(6):2190. [Google Scholar]
- 57.Sollai G., Melis M., Pani D., Cosseddu P., Usai I., Crnjar R., Bonfiglio A., Barbarossa I.T. First objective evaluation of taste sensitivity to 6-n-propylthiouracil (PROP), a paradigm gustatory stimulus in humans. Sci. Rep. 2017;7(1):1–2. doi: 10.1038/srep40353. Jan 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shigemura N., Takai S., Hirose F., Yoshida R., Sanematsu K., Ninomiya Y. Expression of renin-angiotensin system components in the taste organ of mice. Nutrients. 2019;11(9):2251. doi: 10.3390/nu11092251. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020;55(4):2000607. doi: 10.1183/13993003.00607-2020. Apr 16. (PMID: 32269085; PMCID: PMC7144260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oncken C., Feinn R., Covault J., Duffy V., Dornelas E., Kranzler H.R., Sankey H.Z. Genetic vulnerability to menthol cigarette preference in women. Nicotine Tob. Res. 2015;17(12):1416–1420. doi: 10.1093/ntr/ntv042. Dec 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noh H., Paik H.Y., Kim J., Chung J. Salty taste acuity is affected by the joint action of αENaC A663T gene polymorphism and available zinc intake in young women. Nutrients. 2013;5(12):4950–4963. doi: 10.3390/nu5124950. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orsmark-Pietras C., James A., Konradsen J.R., Nordlund B., Söderhäll C., Pulkkinen V., Pedroletti C., Daham K., Kupczyk M., Dahlén B., Kere J. Transcriptome analysis reveals upregulation of bitter taste receptors in severe asthmatics. Eur. Respir. J. 2013;42(1):65–78. doi: 10.1183/09031936.00077712. Jul 1. [DOI] [PubMed] [Google Scholar]
- 63.Tran H.T., Herz C., Ruf P., Stetter R., Lamy E. Human T2R38 bitter taste receptor expression in resting and activated lymphocytes. Front. Immunol. 2018;9:2949. doi: 10.3389/fimmu.2018.02949. Dec 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chamoun E., Mutch D.M., Allen-Vercoe E., Buchholz A.C., Duncan A.M., Spriet L.L., Haines J., Ma D.W. Guelph Family Health Study, A review of the associations between single nucleotide polymorphisms in taste receptors, eating behaviors, and health. Crit. Rev. Food Sci. Nutr. 2018;58(2):194–207. doi: 10.1080/10408398.2016.1152229. [DOI] [PubMed] [Google Scholar]
- 65.Bigiani A. Gustatory dysfunctions in COVID-19 patients: possible involvement of taste renin-angiotensin system (RAS) Eur. Arch. Otorhinolaryngol. 2020;20:1. doi: 10.1007/s00405-020-06054-z. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data would be available in the manuscript.