Abstract
Background and Objectives:
Resistance to methicillin in methicillin resistant strains of Staphylococcus aureus (MRSA) is due to the presence of mec-A gene, which encodes a low affinity penicillin binding protein (PBP)-2a or PBP2. Accurate and rapid identification of MRSA in clinical specimens is essential for timely decision on effective treatment. The aim of the study was to compare three different methods for detection of MRSA namely cefoxitin disc diffusion, CHROM agar MRSA and VITEK-2 susceptibility with PCR which is the gold standard reference method and to find the antibiotic susceptibility pattern of these isolates by VITEK-2.
Materials and Methods:
A Total of 100 non-duplicate S. aureus isolates were collected from different clinical samples among both outpatient and inpatients. Detection of MRSA among these isolates was done by cefoxitin disc diffusion, VITEK-2, CHROM agar MRSA and PCR.
Results:
The sensitivity and specificity of cefoxitin disc diffusion and Vitek was found to be 97.2% and 100%, while that of CHROM agar was found to be 100% and 78.6%. The overall prevalence of MRSA in our study by PCR was 72%.
Conclusion:
Based on the findings in our study, isolates which show cefoxitin zone diameter < 22 mm can be reported as MRSA. However, those isolates which have a zone diameter between 22–24 mm, should ideally be confirmed by PCR.
Keywords: Methicillin resistant Staphylococcus aureus, Cefoxitin, Polymerase chain reaction, Chrom agar, Vitek
INTRODUCTION
Staphylococcus aureus is one of the most common bacterial pathogens causing a wide variety of clinical manifestations. Initially infections were being managed with penicillin but gradually the bacteria developed resistance to penicillin (1). Methicillin, a semisynthetic beta-lactam drug was introduced in the UK in 1959 to treat patients infected with pencillin resistant staphylococci (2). The first case of methicillin resistant Staphylococcus aureus (MRSA) was described in 1961 by Jevons et al. (3). Till the mid 1990s, MRSA infections were limited to the hospitals. However within the last 20 years, MRSA were reported in the healthy individuals without association to health care insitutions. These were due to new strains of MRSA known as CA-MRSA (community acquired-methicillin resistant S. aureus) (4). Rate of infections caused by MRSA has been on a rapid rise and the prevalence in India varies from 25% in western India to 50% in South India (5, 6).
Methicillin resistance in MRSA is due to the presence of mec-A gene, which encodes a low affinity penicillin binding protein (PBP)-2a or PBP2’ (7). Other genes—such as femA, femB can also contribute to methicillin resistance in MRSA (8). Detection of MRSA has become extremely complicated due to many reasons. Methicillin resistance in S. aureus is heterogenous in majority of the isolates and these strains appear phenotypically sensitive to methicillin (9). Addition of sodium chloride or sucrose to culture medium, incubation at 30°C or passage in the presence of beta-lactam antibiotics enhances the expression of resistance (10). Accurate and rapid detection of MRSA results in effective antimicrobial therapy, immediate patient isolation and appropriate disinfection measures (11, 12). Methicillin resistance implies that the organism is resistant to pencillins, 1st–4th generation cephalosporins and carbapenems. Vancomycin has become the drug of choice for treatment of MRSA. False positive results of MRSA has lead to widespread use of vancomycin which inturn resulted in the emergence of VISA (vancomycin intermediate S. aureus) and VRSA (vancomycin resistant S. aureus) (13).
Conventional MRSA detection methods included oxacillin disc diffusion, oxacillin MIC (minimum inhibitory concentration) and oxaciilin screen agar methods. Oxacillin disc diffusion method is no longer used. CLSI (Clinical and Laboratory Standards Institute) recommends the use of the cefoxitin for MRSA detection as it is a better inducer of PBP-2a encoding mec-A gene (14). Other methods in the detection of MRSA include latex agglutination assay, CHROM agar, susceptibility testing by VITEK (15,–17). Detection of mec-A gene by PCR has become the gold standard method in the detection of MRSA. However this method is expensive, time consuming and will not detect novel resistance mechanisms such as mecC (18).
The objectives of this study were: i) to compare three different methods namely, cefoxitin disc diffusion, CHROM agar MRSA, VITEK-2 susceptibility with PCR which is the gold standard reference method and ii) to find the antibiotic susceptibility pattern of these isolates by VITEK-2.
MATERIALS AND METHODS
Study design and bacterial isolates.
This was a prospective cross-sectional study conducted in the Department of Microbiology, Government TD Medical College, Alleppey from September 2018 to February 2019. A Total of 100 non-duplicate S. aureus isolates were collected from different clinical samples among both outpatient and inpatient like blood, urine, tracheal aspirate, sputum, wound swab, pleural fluid, peritoneal fluid. S. aureus was identified by characteristic haemolytic colonies on blood agar, Gram stain showing Gram-positive cocci in clusters and positive by catalase, slide and tube coagulase methods (19).
Ethical approval.
The study was approved by the Institutional Ethical committee of Government Medical College, Alleppey.
Cefoxitin disc diffusion method.
All 100 isolates of S. aureus were subjected to testing with cefoxitin (30 μg) discs (BD BBL Sensi-Disc, Becton, Dickinson and Company, U.S.A.). A 0.5 McFarland standard suspension of the isolate was made and lawn cultures were made on Muller Hinton agar (MHA) plates (Himedia, New Delhi, India). The zone of inhibition was measured after incubation at 35°C for 16–18 hrs. Zone size was interpreted according to CLSI 2019 criteria. Isolates which showed an inhibition zone ≤ 21 were considered to be MRSA, whereas isolates which showed an inhibition zone ≥ 22 were considered to be MSSA (methicillin sensitive S. aureus) (20). A standard strain of MRSA (ATCC 43300) and MSSA (ATCC 29213) were used as positive and negative controls respectively.
CHROM agar.
CHROMagar (Hicrome™ Rapid MRSA Agar Plate-MP1974, Himedia, New Delhi, India) is a new chromogenic medium for the identification of MRSA. It is a ready made media which contains chromogenic mix, MRSA selective supplement and cefoxitin. The chromogenic mixture incorporated in the medium is specifically cleaved by MRSA to give green coloured colonies. For each isolate, a bacterial suspension adjusted to 0.5 McFarland was made and a swab was dipped into the suspension and streaked onto a CHROMagar plate. The growth of any green colony after incubation for 48 hrs was considered to be positive, indicating MRSA. A standard strain of MRSA (ATCC 43300) and MSSA (ATCC 29213) were used as positive and negative controls respectively.
Vitek-2 susceptibilty system.
All strains were subcultured on Blood agar before testing. A bacterial suspension equivalent to a 0.5 McFarland standard was prepared after 18 to 24 h of incubation on a blood agar plate. Vitek 2-AST-P628 cards (bioMe′rieux, Marcy l’Etoile, France) were inoculated according to the manufacturer’s instructions. Isolates for which cefoxitin screen was positive and oxacillin MIC was ≥ 4 μg/ml were regarded as MRSA as per CLSI 2019 (20). Isolates for which cefoxitin screen was negative and oxacillin MIC was ≤ 2 μg/ml were regarded as MSSA as per CLSI 2019 (20). Antibiotic susceptibility pattern of the isolates was also interpreted from VITEK-2 system.
DNA extraction.
Bacterial DNA was extracted from all the isolates grown on blood agar plate by using the Rapid lysis procedure with Lysostaphin (21, 22).
PCR.
PCR was performed to detect the presence of mec-A gene. The reaction mixture consisted of 5 μl of the 10× reaction buffer; 3 μl of 25 mM MgCl; 1 μl of 2.5 mMdNTPs (Promega); 1 μl mecA1 primer 20 pmol/μl; 1 μl mecA2 primer 20 pmol/μl; 0.2 μl Taq polymerase 5 U/μl (Promega); 10 μl DNA; and 28.8 μl H O. The reaction mixture and the primers used for detection of the mecA gene were F 5’TGGCTATCGTGTCACAATCG 3’ (positions 885 to 905) and R 5’ CTGGAACTTGTTGAGCAGAG 3’ (positions 1174 to 1194) producing a 304-bp amplicon as described by Vannuffel et al. (23).
Amplification was performed as follows, initial denaturation for 5 minutes at 94°C followed by 30 cycles at 94°C for 1 minute, at 54°C for 1 minute, then at 72°C for 1 minute. Final annealing was set at 72°C for 7 minute (23). The PCR products (5 μl) were subjected to electrophoresis in agarose 3% and the band size was assessed by direct comparison with a 100-bp DNA marker (Takara). A standard strain of MRSA (ATCC 43300) and MSSA (ATCC 29213) were used as positive and negative controls respectively.
Statistical analysis.
Statistical analysis was done using IBM SPSS 20. (SPSS Inc, Chicago, USA). Diagnostic measures such as sensitivity, specificity & positive and negative predictive values of each test and accuracy was computed. K2 test and kappa concordance measures were used for evaluating association & levels of concordance of the data respectively.
RESULTS
Among the 100 isolates of S. aureus, 72 were positive for mec-A gene by PCR. The overall prevalence of MRSA in our study was 72% by PCR, which is regarded as the gold standard method. Remaining 28 isolates were negative for mec-A gene and were regarded as MSSA. Prevalence of MRSA by cefoxitin disc diffusion, CHROM agar MRSA, Vitek-2 were 70%, 78%, 70% respectively. Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of the different methods is shown in Table 1. Antimicrobial susceptibility pattern of the S. aureus isolates (MRSA and MSSA) is shown in Table 2. PCR showing the presence of mec-A gene is shown in Fig. 1. Chrom agar showing green coloured colonies of MRSA are shown in Fig. 2.
Table 1.
Comparison of Phenotypic methods for detection of MRSA
| Method | No of False Positives | No of False Negatives | Sensitivity (%) | Specificity (%) | PPV | NPV | Concordance with PCR (%) |
|---|---|---|---|---|---|---|---|
| Cefoxitin disc diffusion | 0 | 2 | 97.2 | 100 | 100 | 93.3 | 98 |
| Vitek | 0 | 2 | 97.2 | 100 | 100 | 93.3 | 95 |
| Chrom Agar | 6 | 0 | 100 | 78.6 | 92.3 | 100 | 94 |
PPV- Positive Predictive Value, NPV- Negative Predictive Value
Table 2.
Antibiotic susceptibility pattern of S. aureus strains (n=100) by Vitek-2
| Antibiotics tested | Susceptibility of MSSA (%) (n=28) | Susceptibility of MRSA (%) (n=72) |
|---|---|---|
| Ampicillin | 21.4 | NR |
| Cefazolin | 100 | NR |
| Cefotaxime | 100 | NR |
| Cefepime | 100 | NR |
| Gentamicin | 100 | 78.6 |
| Ciprofloxacin | 28.6 | 38.6 |
| Erythromycin | 46.4 | 21.4 |
| Clindamycin | 85.7 | 40 |
| Linezolid | 100 | 100 |
| Vancomycin | 100 | 100 |
| Daptomycin | 100 | 100 |
| Teicoplanin | 100 | 100 |
| Tigecycline | 100 | 100 |
| Rifampicin | 100 | 100 |
| Cotrimoxazole | 89.3 | 65.7 |
NR-Not Required.
Fig. 1.
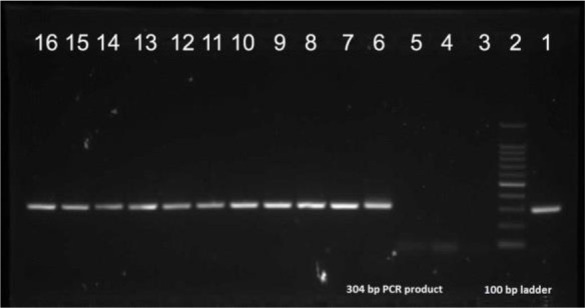
Image of electrophoresis gel of S. aureus. Lane 1.
Positive control strain ATCC 43300
Lane 2. PCR Ladder (100–1500 bp)
Lanes: 3, 4, 5 - Negative results
Lanes: 6–16 - Positive results (MRSA)
Fig. 2.
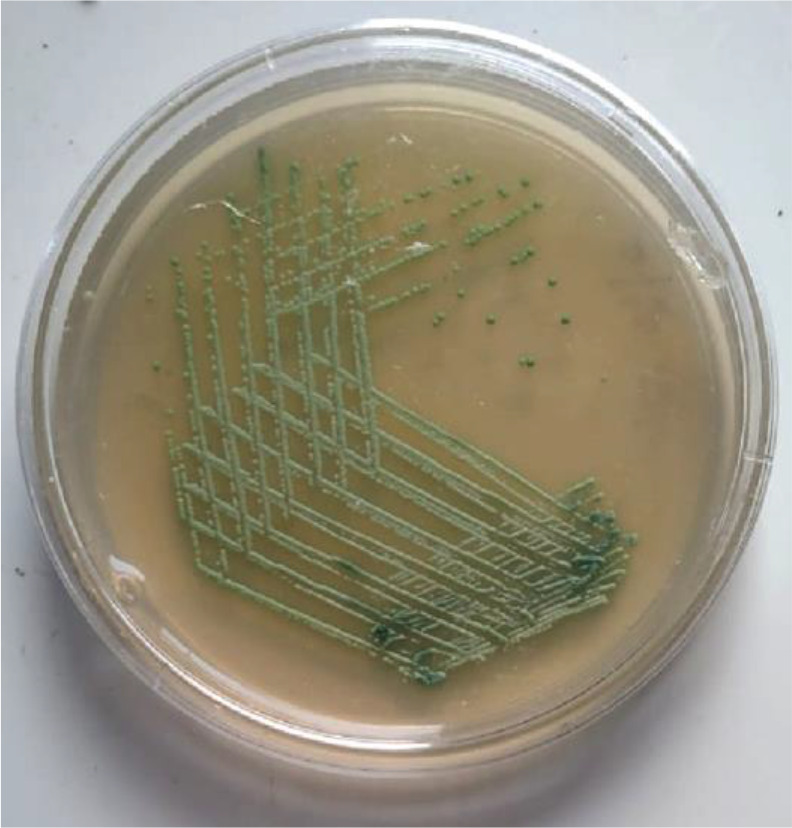
CHROM agar showing green coloured colonies of MRSA
DISCUSSION
Accurate and early detection of methicillin resistance is of immense importance in the prognosis of infections caused by S. aureus. Correct identification of MRSA by conventional methods is quite difficult, as isolates which appear sensitive to methicillin by one method may appear resistant by another method (24). Methicillin resistance is difficult to detect because mec-A positive strains differ in their level of expression of resistance. The resistance is usually heterogenous, with only a few cells (one in 104 or 106) expressing the phenotype. Accurate identification is needed not only for choosing the appropriate antibiotic but also to control the endemicity of MRSA (25). Eventhough detection of mecA gene by PCR is the gold standard method for identification of MRSA, use of molecular methods for identification may not be feasible in a resource poor setting. Therefore it is necessary to implement an accurate, rapid and cost effective phenotypic method for detection of MRSA (26).
In our study among the 100 isolates, 72 were identified as MRSA by PCR. Disc diffusion by cefoxitin and oxacillin are the most commonly used phenotypic methods in laboratory for the detection of MRSA. Oxacillin disc diffusion is no longer recommended by CLSI for MRSA. Disc diffusion by cefoxitin was used in our study, which showed a sensitivity of 97.2% and specificity of 100%. High sensitivity of this method is attributed to the increased expression of mecA encoded protein PBP2a, cefoxitin being a potent inducer of mecA gene (27). Our finding is concordant with other studies around the world, which have also reported that disc diffusion by cefoxitin has high sensitivity and specificity (28–30). In our study prevalence of MRSA by cefoxitin disc diffusion was 70% and there were 2 isolates which showed false negative results. The two isolates which showed false negative results had a zone diameter of 23 and 24 mm respectively which is just above the cut off zone. However, both the isolates which gave false negative results by cefoxitin disc diffusion where found to be MRSA by Vitek and CHROM agar. CHROM agar was used in our study for the identification of MRSA and the sensitivity, specificity was found to be 100% and 78.6% respectively. Eventhough this method was highly sensitive, it gave six false positive results, resulting in a low specificity. These six isolates were false positive after 24 hrs of incubation and the sensitivity, specificity did not increase after 48 hrs of incubation. High sensitivity of this medium has also been reported by Diederen et al. and Datta et al. (16, 31). False positive results while using chromogenic medium for the detection of MRSA was also described by Stoakes et al. who reported three false positive results (32). The six false positive isolates by CHROM agar were found to be MSSA by Vitek and cefoxitin disc diffusion. Prevalence of MRSA by CHROM agar was 78%.
In our study Vitek-2 was used to detect methicillin resistance. All the isolates which were correctly identified as MRSA had oxacillin MIC ≥ 4 μg/ml, whereas isolates which were correctly identified as MSSA had oxacillin MIC ranging from 0.25–1 μg/ml. This method had a sensitivity of 97.2% and a specificity of 100%. By this method two isolates were falsely identified as MSSA and these isolates had oxacillin MIC of 0.5 μg/ml. Both isolates had zone of inhibition of 20 mm by cefoxitin disc diffusion and produced green colonies on CHROM agar. Overall prevalence of MRSA by Vitek-2 was 70%. Roisin et al. in her study also reported high sensitivity and specificity of 97.5% and 100% respectively for Vitek-2 in the detection of MRSA (33). One advantage of Vitek over cefoxitin disc diffusion is that while disc diffusion requires 16–18 hrs incubation, Vitek can classify the isolate as MRSA or MSSA within 8 hrs of growth in culture. Antimicrobial susceptibility pattern of the isolates by Vitek-2 showed that all the isolates were sensitive to vancomycin, teicoplanin, linezolid and rifampicin. Inducible clindamycin resistance was seen in 57.1% of MRSA isolates and in 10.7% of MSSA isolates.
CONCLUSION
This study showed an overall MRSA prevalence of 70% by PCR. Although cefoxitin disc diffusion and Vitek-2 are excellent methods to detect methicillin resistance in S. aureus, it still produced two false negative results. Our study showed that, while CHROM agar had a high sensitivity for MRSA detection, it showed poor specificity. Based on the findings in our study, isolates which show cefoxitin zone diameter < 22 mm can be reported as MRSA. However, those isolates which have a zone diameter between 22–24 mm, should ideally be confirmed by PCR.
REFERENCES
- 1.Kirby WM. Extraction of a highly potent penicillin inactivator from penicillin resistant staphylococci. Science 1944; 99: 452–453. [DOI] [PubMed] [Google Scholar]
- 2.KNOX R. A new penicillin (BRL 1241) active against penicillin-resistant staphylococci. Br Med J 1960;2: 690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jevons MP. “Celbenin” - resistant Staphylococci. Br Med J 1961; 1: 124–125. [Google Scholar]
- 4.Otto M. Community-associated MRSA: what makes them special? Int J Med Microbiol 2013; 303: 324–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopalakrishnan R, Sureshkumar D. Changing trends in antimicrobial susceptibility and hospital acquired infections over an 8 year period in a tertiary care hospital in relation to introduction of an infection control programme. J Assoc Physicians India 2010;58 Suppl:25–31. [PubMed] [Google Scholar]
- 6.Patel AK, Patel KK, Patel KR, Shah S, Dileep P. Time trends in the epidemiology of microbial infections at a tertiary care center in west India over last 5 years. J Assoc Physicians India 2010;58 Suppl:37–40. [PubMed] [Google Scholar]
- 7.Mbah AN, Isokpehi RD. Identification of functional regulatory residues of the β-lactam inducible penicillin binding protein in methicillin-resistant Staphylococcus aureus. Chemother Res Pract 2013;2013: 614670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger-Bächi B, Rohrer S. Factors influencing methicillin resistance in staphylococci. Arch Microbiol 2002; 178: 165–171. [DOI] [PubMed] [Google Scholar]
- 9.Chambers HF. Methicillin-resistant staphylococci. Clin Microbiol Rev 1988; 1: 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman BJ, Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother 1986; 29: 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper BS, Stone SP, Kibbler CC, Cookson BD, Roberts JA, Medley GF, et al. Isolation measures in the hospital management of methicillin resistant Staphylococcus aureus (MRSA): Systematic review of the literature. BMJ 2004;329:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Towner KJ, Talbot DC, Curran R, Webster CA, Humphreys H. Development and evaluation of a PCR based immunoassay for the rapid detection of methicillin resistant Staphylococcus aureus. J Med Microbiol 1998; 47: 607–613. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention (CDC) . Staphylococcus aureus resistant to vancomycin--United States, 2002. MMWR Morb Mortal Wkly Rep 2002; 51: 565–567. [PubMed] [Google Scholar]
- 14.CLSI (2008). Performance Standards for Antimicrobial Susceptibility Testing, 15th informational supplement, M100-S15. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- 15.Hussain Z, Stoakes L, Garrow S, Longo S, Fitzgerald V, Lannigan R. Rapid detection of mecA-positive and mecA-negative coagulase-negative staphylococci by an anti-penicillin binding protein 2a slide latex agglutination test. J Clin Microbiol 2000; 38: 2051–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diederen B, van Duijn I, van Belkum A, Willemse P, van Keulen P, Kluytmans J. Performance of CHROMagar MRSA medium for detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 2005; 43: 1925–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nonhoff CG, Mascart MJ, Struelens C, Van Den Borre, Denis O. (2004). Detection of hetero-resistant MRSA: controlled comparison of oxacillin/cefoxitin susceptibility testing by disk diffusion, agar screen, Vitek-2 and BD Phoenix automated systems, abstr. P-1630, p. 460. Abstr. 14th European Congress of Clinical Microbiology and Infectious Diseases, Prague, Czech Republic. [Google Scholar]
- 18.Lee JH, Jeong JM, Park YH, Choi SS, Kim YH, Chae- JS, et al. Evaluation of the methicillin-resistant Staphylococcus aureus (MRSA)-Screen latex agglutination test for detection of MRSA of animal origin. J Clin Microbiol 2004; 42: 2780–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baron S. editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. [PubMed] [Google Scholar]
- 20.CLSI . Performance standards for antimicrobial susceptibility testing. 29th ed. Clinical and Laboratory Standards Institute; Wayne, PA: 2019. CLSI supplement M100. [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. (1989). Molecular cloning: a laboratory manual. New York: Cold Spring Harb. Lab. Press. [Google Scholar]
- 22.Unal S, Hoskins J, Flokowitsch JE, Wu CY, Preston DA, Skatrud PL. Detection of methicillin-resistant Staphylococci by using the polymerase chain reaction. J Clin Microbiol 1992; 30: 1685–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wauters G, et al. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J Clin Microbiol 1995; 33: 2864–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baddour MM, AbuElKheir MM, Fatani AJ. Comparison of mecA polymerase chain reaction with phenotypic methods for the detection of Methicillin-resistant Staphylococcus aureus. Curr Microbiol 2007; 55: 473–479. [DOI] [PubMed] [Google Scholar]
- 25.Pramodhini S, Thenmozhivalli PR, Selvi R, Dillirani V, Vasumathi A, Agatha D. Comparison of various phenotypic methods and mecA based PCR for the detection of MRSA. J Clin Diagn Res 2012; 5: 1359–1362. [Google Scholar]
- 26.Krishnan PU, Miles K, Shetty N. Detection of methicillin and mupirocin resistance in Staphylococcus aureus isolates using conventional and molecular methods: a descriptive study from a burns unit with high prevalence of MRSA. J Clin Pathol 2002; 55: 745–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velasco D, Tomas MM, Cartelle M, Beceiro A, Perez A, Molina F, et al. Evaluation of different methods for detecting methicillin (oxacillin) resistance in Staphylococcus aureus. J Antimicrob Chemother 2005; 55: 379–382. [DOI] [PubMed] [Google Scholar]
- 28.Pillai MM, Latha R, Sarkar G. Detection of methicillin resistance in Staphylococcus aureus by polymerase chain reaction and conventional methods: a comparative study. J Lab Physicians 2012; 4: 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pourmand MR, Hassanzadeh S, Mashhadi R, Askari E. Comparison of four diagnostic methods for detection of methicillin resistant Staphylococcus aureus. Iran J Microbiol 2014; 6: 341–344. [PMC free article] [PubMed] [Google Scholar]
- 30.Panda RK, Mahapatra A, Mallick B, Chayani N. Evaluation of genotypic and phenotypic methods for detection of methicillin resistant Staphylococcus aureus in a tertiary care hospital of eastern Odisha. J Clin Diagn Res 2016;10:DC19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datta P, Gulati N, Singhla N, Vasudeva H, Bala K, Chander J, et al. Evaluation of various methods for the detection of methicillin-resistant Staphylococcus aureus (MRSA) and their susceptibility pattern. J Med Microbiol 2011; 60: 1613–1616. [DOI] [PubMed] [Google Scholar]
- 32.Stoakes L, Reyes R, Daniel J, Lennox G, John MA, Lannigan R, et al. Prospective comparison of a new chromogenic medium, MRSASelect, to CHROMagar MRSA and mannitol-salt medium supplemented with oxacillin or cefoxitin for detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 2006; 44: 637–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roisin S, Nonhoff C, Denis O, Struelens MJ. Evaluation of new Vitek 2 card and disk diffusion method for determining susceptibility of Staphylococcus aureus to oxacillin. J Clin Microbiol 2008; 46: 2525–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]


