Abstract
Betel (Piper betle L.) and green tea (Camellia sinensis (L) O. Kuntze) have been used for a long time as traditional medicine. The docking of phytoconstituents contained in the betel plant was evaluated against Mpro, and matcha green tea was evaluated against five target receptors of SARS-CoV-2 as follows: spike ectodomain structure (open state), receptor-binding domain (RDB), main protease (Mpro), RNA-dependent RNA polymerase (RdRp), dan papain-like protease (PLpro). The evaluation was carried out based on the value of binding-free energy and the types of interactions of the amino acids at the receptors that interact with the ligands.
Keywords: Betel, Matcha green tea, Phytoconstituent, In silico, Docking, SARS-CoV-2, Antiviral
Specifications Table
| Subject | Biological sciences |
| Specific subject area | Bioinformatics, in silico analysis, molecular docking |
| Type of data | Tables and Figures |
| How data were acquired | AutoDock Vina and Biovia Discovery Studio Visualizer 2020 |
| Data format | Raw and analyzed Direct URL to the data for betel plant: http://dx.doi.org/10.17632/s72rcpk82b.1 Direct URL to the data for matcha green tea: http://dx.doi.org/10.17632/4dn4svm3jb.1 |
| Parameters for data collection | In the drug discovery setting, Lipinski's rule of 5 predicts that poor absorption or permeation is more likely when there are more than 5 H-bond donors, 10 H-bond acceptors, the molecular weight is greater than 500, and the calculated Log P (CLog P) is greater than 5. The docking score was obtained based on the most negative Gibbs’ free energy of binding generated using autodock Vina. The interactions between receptors’ amino acid residues and the ligands were visualized using Biovia Discovery Studio 2020. |
| Description of data collection | Betel plant phytoconstituents were obtained from GC–MS analysis; Phytoconstituents of matcha were collected from published articles listed in the references. |
| Data source location | The receptors’ structures were retrieved from https://www.rcsb.org/ The ligands’ structures were retrieved from https://pubchem.ncbi.nlm.nih.gov/ |
| Data accessibility | Repository name: Mendeley Data Data identification number for betel plant: http://dx.doi.org/10.17632/s72rcpk82b.1 Data identification number for matcha green tea: http://dx.doi.org/10.17632/4dn4svm3jb.1 Direct URL to the data for betel plant: https://data.mendeley.com/datasets/s72rcpk82b/1 Direct URL to the data for matcha green tea: https://data.mendeley.com/datasets/4dn4svm3jb/1 |
| Related research article | T.E. Tallei, S.G. Tumilaar, A.A. Adam, Fatimawali, Evaluasi potensi polifenol Matcha sebagai agen anti-SARS-CoV-2 menggunakan pendekatan penambatan molekul, in: K. Wikantika, F.M. Dwivany, M.F. Ghazali, L.F. Yayusman, C. Novianti (Eds.), ForMIND Bunga Rampai 2020, ITB Press, Bandung, 2020, pp. 147–155. |
Value of the Data
-
•
The data provide information on the results of GC–MS analysis of various phytoconstituents contained in betel plant (leaf and fruit parts).
-
•
The data provide information on the interactions of various betel leaf and fruit as well as matcha green tea phytoconstituents on important enzyme and proteins of SARS-CoV-2, i.e.: spike ectodomain structure (open state) (PDB code: 6VYB), receptor-binding domain (RDB) (PDB code: 6YLA), main protease (Mpro) (PDB code: 6LU7), RNA-dependent RNA polymerase (RdRp) (PDB code: 6M71), and papain-like protease (PLpro) (PDB code: 6WX4).
-
•
The data may be useful to researchers working on COVID-19 drug discovery and development;
-
•
The data provide promising phytoconstituents for betel and matcha green tea which could serve as potential clues for the development of future therapeutics for COVID-19.
1. Data Description
Plants are sources of phytomedicine which has the potential to be developed as antiviral agents for SARS-C0V-2, as has been reported by previous studies [1], [2]. Betel leaf and fruit contain many phytoconstituents which reveal their uses for various therapeutic purposes. The plant or its parts can be used for the treatment of various disorders in humans such as diabetes, fungal infection, microbial infection, inflammation, antihistaminic, antiulcer, and local anesthetic [3]. Matcha, which is a green tea preparation in powder form [4], is known to have many benefits, including as a source of antioxidants and having antiviral activities [5].
The data described here include the binding free energy value (kcal/mol) of the phytoconstituents contained in betel leaf and matcha green tea which serve as ligands against various targets of SARS-CoV-2. Data on phytoconstituent from betel leaf were obtained from the results of Gas chromatography-mass spectrometry (GC–MS), while information about the phytoconstituent of matcha was obtained through literature searches. The data on the drug-likeness of the ligands based on Lipinski's rule of five are listed in Table 1 for betel leaf and fruit, and Table 2 for matcha green tea. The phytoconstituents of matcha green tea were obtained from the references listed in Table 2. The data on binding free energy resulted from the docking of betel leaf and matcha green tea is presented in Tables 3 and 4, respectively. Tables 5 and 6 show the type of interaction and the interacting amino acids of the receptors with the ligands contained in betel plant and matcha green tea, respectively. The detail of interaction and visualization of the docking results of all phytoconstituents are provided in the supplementary data. The interaction visualization of the best 10 docking results of betel leaf and fruit phytoconstituents is provided in Fig. 1. The visualization of the interaction of matcha green tea with SARS-CoV-2 receptors is available from Supplementary data [6].
Table 1.
Lipinski's rule of five value of betel leaf and fruit phytoconstituents.
| Compound name | Molecular weight | No. H-bond acceptors | No. H-bond donors | log P | Molar refractivity | No. of violations |
|---|---|---|---|---|---|---|
| (5ß)Pregnane-3,20ß-diol, 14a,18a-[4-methyl-3-oxo-(1-oxa-4-azabutane-1,4-diyl)]-, diacetate | 489 | 6 | 0 | 5.962 | 144.653 | 2 |
| N1-Benzyl-N2(bezylidenyl-benzylamino)- | 403 | 0 | 1 | 4.276 | 117.477 | 0 |
| 25-Norisopropyl-9,19-cyclolanostan-22-en-24-one, 3-acetoxy-24-phenyl-4,4,14-trimethyl- | 516 | 3 | 0 | 8.118 | 166.353 | 3 |
| Milbemycin B, 6,28-anhydro-15‑chloro-25-isopropyl-13-dehydro-5-O-demethyl-4-methyl- | 590 | 7 | 1 | 7.752 | 169.584 | 3 |
| 1H-2,8a-Methanocyclopenta [a]cyclopropa[e]cyclodecen-11-one, 1a,2,5,5a,6,9,10,10a-octahydro-5,5a,6-trihydroxy-1,4-bis(hydroxymethyl)−1,7,9-trimethyl-, [1S-(1a,1aa,2a,5ß,5aß,6ß,8aa,9a,10aa)]- |
364 | 6 | 3 | 3.593 | 100.307 | 0 |
| 1H-2,8a-methanocyclopenta[a] cyclopropa[e]cyclodecen-6-yl ester, [1aR-(1aa,2a,5ß,5aß,6ß,8aa,9a,10aa)]- |
430 | 6 | 1 | 5.048 | 122.754 | 1 |
| 2,4,6-Decatrienoic acid, 1a,2,5,5a,6,9,10,10a-octahydro-5,5a-dihydroxy-4-(hydroxymethyl)−1,1,7,9-tetramethyl-11-oxo- | 496 | 6 | 2 | 6.142 | 142.969 | 2 |
| (2,3-Diphenylcyclopropyl)methyl phenyl sulfoxide, trans- | 332 | 1 | 0 | 4.417 | 94.863 | 0 |
| 2-Naphthalenemethanol, decahydro-a,a,4a-trimethyl-8-methylene-, [2R-(2a,4aa,8aß)]- | 240 | 2 | 2 | 3.833 | 81.209 | 0 |
| benzene, 1,1′,1′'-[5-methyl-1-pentene-1,3,5-triyl]tris- | 312 | 0 | 0 | 5.123 | 98.352 | 1 |
Table 2.
Lipinski's rule of five value of the matcha green tea phytoconstituents.
| Compound name | References | Molecular weight | No. H-bond acceptors | No. H-bond donors | log P | Molar refractivity | No. of violations |
|---|---|---|---|---|---|---|---|
| (-)-epicatechin 3,5-di-O-digallate (EC35G) | [7] | 594 | 14 | 9 | 3.16 | 141.986 | 4 |
| Rutin | [8] | 610 | 16 | 10 | −1.88 | 137.495 | 4 |
| (-)- epigallocatechin gallate (EGCG) | [9] | 458 | 11 | 8 | 2.23 | 108.921 | 2 |
| Apigenin glycoside | [10] | 578 | 12 | 6 | 1.86 | 144.558 | 4 |
| Flavonol 3-O-d-glucoside (FOG) | [11] | 400 | 8 | 4 | 0.45 | 99.615 | 0 |
| Myricetin 3-glucoside (M-G) | [12] | 480 | 13 | 9 | −1.01 | 107.939 | 1 |
| (-)- epigallocatechin (EGC) | [13] | 306 | 7 | 6 | 1.25 | 74.288 | 1 |
| Kaempferol | [14] | 286 | 6 | 4 | 2.31 | 72.386 | 0 |
| (-)-epicatechin gallate (ECG) | [9] | 442 | 10 | 7 | 2.53 | 107.256 | 1 |
| (+)-catechin | [9] | 290 | 6 | 5 | 1.55 | 72.623 | 1 |
| (-)-epicatechin (EC) | [9] | 290 | 6 | 5 | 1.55 | 72.623 | 1 |
| Myricetin | [14] | 318 | 8 | 6 | 1.72 | 75.715 | 1 |
| Kaempferol-3-O-glucoside | [10] | 448 | 11 | 7 | −0.44 | 104.609 | 2 |
| Quercetin | [14] | 302 | 7 | 5 | 2.01 | 74.050 | 1 |
| (-)-Epigallocatechin 3-(3-methyl-gallate) (3″Me-EGCG) | [15] | 472 | 11 | 7 | 2.54 | 113.808 | 2 |
| Caffeoylquinic acid (CQA) | [15] | 354 | 9 | 6 | −0.65 | 82.519 | 1 |
| Chlorogenic acid | [8] | 354 | 9 | 6 | −0.65 | 82.519 | 1 |
| Coumaric acid | [8] | 164 | 3 | 2 | 1.49 | 44.776 | 0 |
| Caffeic acid | [8] | 180.16 | 4 | 3 | 1.20 | 46.441 | 0 |
| Gallic Acid | [9] | 170.12 | 5 | 4 | 0.50 | 38.395 | 1 |
| Caffeine | [9] | 194.19 | 5 | 0 | 0.06 | 49.100 | 0 |
Table 3.
Binding free energy of betel leaf phytoconstituents against SARS-CoV-2 Mpro (6LU7).
| Ligands | Chemical formula | PubChem ID | Binding affinity (kcal/mol) |
|---|---|---|---|
| (5ß)Pregnane-3,20ß-diol, 14a,18a-[4-methyl-3-oxo-(1-oxa-4-azabutane-1,4-diyl)]-, diacetate | C28H43NO6 | 537,242 | −11.5 |
| N1-Benzyl-N2(bezylidenyl-benzylamino)- | C28H25N3 | 562,008 | −8.5 |
| 25-Norisopropyl-9,19-cyclolanostan-22-en-24-one, 3-acetoxy-24-phenyl-4,4,14-trimethyl- | C35H48O3 | 5,373,661 | −8.1 |
| Milbemycin B, 6,28-anhydro-15‑chloro-25-isopropyl-13-dehydro-5-O-demethyl-4-methyl- | C33H47ClO7 | 5,367,225 | −8 |
| 1H-2,8a-Methanocyclopenta[a]cyclopropa[e]cyclodecen-11-one, 1a,2,5,5a,6,9,10,10a-octahydro-5,5a,6-trihydroxy-1,4-bis(hydroxymethyl)−1,7,9-trimethyl-, [1S-(1a,1aa,2a,5ß,5aß,6ß,8aa,9a,10aa)]- | C20H28O6 | 119,057,278 | −7.9 |
| 1H-2,8a-methanocyclopenta[a]cyclopropa[e]cyclodecen-6-yl ester, [1aR-(1aa,2a,5ß,5aß,6ß,8aa,9a,10aa)]- | C24H34O6 | 6,918,670 | −7.9 |
| 2,4,6-Decatrienoic acid, 1a,2,5,5a,6,9,10,10a-octahydro-5,5a-dihydroxy-4-(hydroxymethyl)−1,1,7,9-tetramethyl-11-oxo- | C30H40O6 | 5,367,323 | −7.8 |
| (2,3-Diphenylcyclopropyl)methyl phenyl sulfoxide, trans- | C22H20OS | 562,543 | −7.8 |
| 2-Naphthalenemethanol, decahydro-a,a,4a-trimethyl-8-methylene-, [2R-(2a,4aa,8aß)]- | C15H28O | 165,258 | −7.8 |
| benzene, 1,1′,1′'-[5-methyl-1-pentene-1,3,5-triyl]tris- | C24H24 | 20,138,399 | −7.6 |
| 3-[3-Bromophenyl]−7‑chloro-3,4-dihydro-10‑hydroxy-1,9(2H,10H)-acridinedione | C19H13BrClNO3 | 536,420 | −7.4 |
| (22S)−21-Acetoxy-6a,11ß-dihydroxy-16a,17a-propylmethylenedioxypregna-1,4-diene-3,20-dione | C27H36O8 | 544,325 | −7.4 |
| 2(1H)-Pyrimidinone, 5‑chloro-4,6-diphenyl- | C16H11ClN2O | 624,638 | −7.3 |
| Benz[e]azulene-3,8‑dione, 5-[(acetyloxy)methyl]−3a,4,6a,7,9,10,10a,10b-octahydro- | C19H2406 | 540,437 | −7.3 |
| Alpha-phenyl-alpha-tropylacetaldehyde tosylhydrazone | C22H22N2O2S | 9,602,323 | −7.3 |
| Pregnan-20-one, 3-(acetyloxy)−5,6:16,17-diepoxy-, (3ß,5a,6a,16a)- | C23H32O5 | 265,665 | −7.2 |
| Isoaromadendrene epoxide | C15H24O | 534,398 | −7.1 |
| 2-[4-methyl-6-(2,6,6-trimethylcyclohex-1-enyl)hexa-1,3,5-trienyl]cyclohex-1-en-1-carboxaldehyde | C23H32O | 5,363,101 | −7.1 |
| 5a-Pregn-16-en-20-one, 3ß,12a-dihydroxy (22S)−6a,11ß,21-Trihydroxy-16a,17a-propyl | C25H36O5 C25H34O7 | 1,756,337 | −7.1 |
| Neointermedeol | C15H26O | 11,877,394 | −7 |
| 6-Chloro-3-(2-nitro-1-phenylethyl)−3,4-dihydro-1H-naphthalen-2-one | C18H16ClNO3 | 586,644 | −7 |
| Ethyl isoallocholate | C26H44O5 | 6,452,096 | −7 |
Table 4.
Binding free energy of matcha phytoconstituents against several SARS-CoV-2 receptors.
| Ligands | PubChem CID | SARS-CoV-2 Receptors PDB ID |
||||
|---|---|---|---|---|---|---|
| 6VYB | 6YLA | 6LU7 | 6M71 | 6WX4 | ||
| Binding free energy (kcal/mol) | ||||||
| (-)-epicatechin 3,5-di-O-digallate (EC35G) | 467,299 | −9.7 | −10.0 | −9.1 | −9.2 | −8.8 |
| Rutin | 5,280,805 | −9.9 | −10.1 | −8.8 | −8.8 | −7.2 |
| (-)-epigallocatechin gallate (EGCG) | 65,064 | −9.2 | −9.4 | −8.2 | −8.5 | −7.6 |
| Apigenin glycoside | 44,257,854 | −9.2 | −10.1 | −8.7 | −9.1 | −7.5 |
| Flavonol 3-O-d-glucoside (FOG) | 11,953,828 | −9.0 | −8.1 | −7.8 | −7.8 | −6.7 |
| Myricetin 3-glucoside (M-G) | 44,259,426 | −8.8 | −9.2 | −8.8 | −8.0 | −6.6 |
| (-)-Epigallocatechin (EGC) | 72,277 | −8.6 | −8.4 | −7.1 | −7.5 | −6.7 |
| Kaempferol | 5,280,863 | −8.6 | −7.9 | −7.7 | −7.1 | −6.7 |
| (-)-epicatechin gallate (ECG) | 107,905 | −8.5 | −9.0 | −8.2 | −8.3 | −7.4 |
| (+)-catechin | 9064 | −8.4 | −8.2 | −7.2 | −6.8 | −6.6 |
| (-)-epicatechin (EC) | 72,276 | −8.4 | −7.8 | −7.1 | −7.0 | −6.7 |
| Myricetin | 5,281,672 | −8.4 | −8.4 | −7.3 | −7.3 | −7.1 |
| Kaempferol-3-O-glucoside | 5,282,102 | −8.4 | −8.9 | −8.4 | −7.9 | −6.6 |
| Quercetin | 5,280,343 | −8.3 | −8.4 | −7.4 | −7.6 | −7.0 |
| (-)-Epigallocatechin 3-(3-methyl-gallate) (3″Me-EGCG) | 9,804,842 | −8.3 | −8.1 | −7.6 | −8.6 | −7.6 |
| Caffeoylquinic acid (CQA) | 10,155,076 | −8.1 | −8.0 | −7.2 | −7.0 | −6.6 |
| Chlorogenic acid | 1,794,427 | −7.9 | −7.3 | −7.6 | −6.9 | −7.0 |
| Coumaric acid | 9,840,292 | −6.7 | −7.0 | −7.2 | −5.3 | −6.4 |
| Caffeic acid | 689,043 | −6.7 | −5.9 | −5.7 | −5.3 | −5.3 |
| Gallic acid | 370 | −6.3 | −6.1 | −5.5 | −5.6 | −5.2 |
| Caffeine | 2519 | −6.1 | −6.0 | −5.2 | −5.1 | −5.2 |
Table 5.
Interacting amino acids of the main protease (6LU7) with the 10 best ligands of betel leaf and fruit.
|
PubChem CID |
Binding free energy (kcal/mol) |
No. of bonds |
Interacting residues and H-bond formation |
|---|---|---|---|
| 537,242 | −11.5 | 18 | Van der Waals: ASN(A142), GLY(A143) CYS(A145), HIS(A164), ASP(A187), MET(A49), TYR(A54), ARG(A188), PRO(A168), LEU(A167), THR(A190), GLN(A189), GLU(A166), MET(A165); conventional H-bond: GLN(A192); unfavorable positive-positive: HIS(A41); attractive charge and pi-anion: HIS(A41); pi-sigma: HIS(A41). |
| 562,008 | −8.5 | 21 | Van der Waals: GLU(A166), MET(A49), THR(A24), THR(A25), THR(A26), GLY(A143), ASN(A142), ARG(A188), ASP(A187), HIS(A164), LEU(A141), GLN(A189), HIS(A163), HIS(A172), PHE(A140); unfavorable positive-positive: HIS(A41); pi-cation: HIS(A41); pi-pi t-shaped: HIS(A41); pi-alkyl: CYS(A145), LEU(A27), MET(A165). |
| 5,373,661 | −8.1 | 19 | Van der Waals: THR(A26), THR(A25), ASN(A142), GLY(A143), HIS(A41), CYS(A145), SER(A144), LEU(A141), GLU(A166), ARG(A188), THR(A190), GLN(A192), GLN(A189), HIS(A164), MET(A49), THR(A24); conventional H-bond: THR(A45), SER(A46); pi-alkyl: MET(A165). |
| 5,367,225 | −8 |
14 | Van der Waals: THR(A25), LEU(A27), MET(A49), GLN(A189), CYS(A145), HIS(A41), SER(A144), MET(A165), PHE(A140), LEU(A141), GLU(A166); conventional H-bond: THR(A26), GLY(A143); pi-alkyl: HIS(A163). |
| 119,057,278 | −7.9 | 14 | Van der Waals: MET(A165), GLN(A189), ASN(A142), SER(A144), GLY(A143), HIS(A172), PHE(A140); conventional H-bond: GLU(A166), 2HIS(A163), LEU(A141); unfavorable positive-positive: 2HIS(A41); alkyl: CYS(A145) |
| 6,918,670 | −7.9 | 16 | Van der Waals: ASN(A142), GLN(A189), HIS(A164), ASP(A187), ARG(A188), MET(A165), LEU(A141), PHE(A140), LEU(A167), PRO(A168); conventional H-bond: 3GLU(A166), HIS(A172); unfavorable positive-positive: HIS(A163); pi-alkyl: HIS(A41). |
| 5,367,323 | −7.8 | 13 | Van der Waals: GLY(A143), HIS(A172), PHE(A140), ASN(A142), MET(A165), PRO(A168), LEU(A167); conventional H-bond: GLU(A166), HIS(A163); unfavorable positive-positive: GLN(A189); pi-alkyl: 3LEU(A141) |
| 562,543 | −7.8 | 15 | Van der Waals: LEU(A141), PHE(A140), GLU(A166), HIS(A163), HIS(A172), HIS(A164), ASP(A187), TYR(A54), ARG(A188), GLN(A189), CYS(A145); conventional H-bond: HIS(A41); pi-cation: HIS(A41); pi-sulfur: MET(A49); pi-pi stacked & pi-alkyl: MET(A165). |
| 165,258 | −7.8 | 12 | Van der Waals: ASP(A187), ARG(A188), GLN(A189), MET(A165), HIS(A164), MET(A49), LEU(A27), GLY(A143), ASN(A142), GLU(A166); conventional H-bond: HIS(A41); alkyl: CYS(A145 |
| 20,138,399 | −7.6 | 15 | Van der Waals: LEU(A141), PHE(A140), HIS(A172), HIS(A163), HIS(A164), ASP(A187), ARG(A188), TYR(A54), THR(A190), GLN(A189), GLU(A166); pi-sulfur: MET(A165), CYS(A145); pi-pi stacked: HIS(A41); pi-alkyl: MET(A49). |
Table 6.
Hydrogen bond interaction of the amino acids of the receptors with phytoconstituents in matcha. The remaining interaction data are available from Tallei et al. [6].
| Interacting amino acids |
|||||
|---|---|---|---|---|---|
|
Receptors |
Ligands |
Conventional H-bond |
Carbon H-bond | Pi-donor H-bond | Pi-carbon H-bond |
| 6VYB | (-)-epicatechin 3,5-di-O-digallate (EC35G) | ASN(C1108), LYS(C1038) | GLY(C910), TYR(A904) | ||
| Rutin | ARG(B1039), ARG(C1039), ARG(A1039), 2ASN(C1023), ARG(A1019) | ||||
| (-)-epigallocatechin gallate (EGCG) | 2SER(B94), ASN(B99), ARG(B190) | ||||
| Apigenin glycoside | ARG(A1039), SER(B1030), THR(B1027), ASP(A1041) | ||||
| Flavonol 3-O-D-glucoside (FOG) | THR(B998), TYB(B756), THR(A998), 2ASP(A994), THR(C998) | ||||
| Myricetin 3-glucoside (M-G) | GLN(A954), GLN(A1010) | GLY(B769), 2GLN(A954) | |||
| (-)-Epigallocatechin (EGC) | LEU(A861), LYS(A733), GLY(A1059) | PRO(A1057) | |||
| Kaempferol | 2THR(A549), ASN(B978), MET(B740), TYR(B714), ARG(B1000) | GLY(A541) | |||
| (-)-epicatechin gallate (ECG) | GLY(C744), TYR(C741), 3ARG(31,000), LEU(C977) | ||||
| (+)-catechin | GLN(C1036), HIS(B1048) | ||||
| (-)-epicatechin (EC) | TYR(A741), MET(A740). | ||||
| Myricetin | LYS(1038), GLY(B908), HIS(B104) | TYR(B1047) | |||
| Kaempferol-3-O-glucoside | ALA(C1020), THR(C1027), PHE(C1042), ARG(B1039; | THR(A1027) | |||
| Quercetin | 2LYS(A1038), HIS(A1048), GLY(A1048) | VAL(A1040) | |||
| (-)-Epigallocatechin 3-(3-methyl-gallate) (3″Me-EGCG) | 2IHK(B1027), ARG(B1029) | IHK(C1027) | |||
| Caffeoylquinic acid (CQA) | GLN(A672), ARG(A675), ARG(C1014), ARG(A1019), GLU(A773), GLN(C954) | ||||
| Chlorogenic acid | THR(A961), GLU(A1017), GLU(B773), GLN(A954) | GLY(B769) | |||
| Coumaric acid | SER(A514), TYR(B200), PHE(A515), THR(A430) | ||||
| Caffeic acid | HIS(A1048), 2GLN(B1036) | GLY(B1035) | |||
| Gallic acid | GLN(A1005), GLN(B1002), THR(B1006) | ||||
| Caffeine | GLU(A166), GLY(A143), SER(A144) | CYS(A142) | |||
| 6YLA | (-)-epicatechin 3,5-di-O-digallate (EC35G) | TYR(L:93), LYS(H:43), ALA(H:172), 2GLU(H:152), THR(H:116), GLY(L:47) | |||
| Rutin | GLY(H:112), THR(H:114), GLU(H:152), ALA(H:92) VAL(H:115) | GLY(L:47) | |||
| (-)-epigallocatechin gallate (EGCG) | SER(C:62), MET(H:2), GLU(L:61), THR(H:0), THR(B:0), ASP(B:107), GLN(B:1) | ||||
| Apigenin glycoside | SER(C:174), GLN(C:172),SER(H:75); | SER(H:75), SER(C:174) | |||
| Flavonol 3-O-d-glucoside (FOG) | 2GLN(L:48) | GLN(H:39) | |||
| Myricetin 3-glucoside (M-G) | THR(E:385), THR(H:0), ASP(H:107), SER(L:62), THR(B:0), GLN(B:1), ASP(B107) | ||||
| (-)-Epigallocatechin (EGC) | GLN(H:39) | ||||
| Kaempferol | LYS(L:45), GLN(L:44) | ||||
| (-)-epicatechin gallate (ECG) | LYS(C:213), 2ASN(C:216), 4GLU(C:219), GLY(C:218), LYS(B:218) | PRO(C:125), SER(B:132), PHE(C:122) | |||
| (+)-catechin | GLU(H:152), GLN(L:44), LYS(L:45), GLN(H:39) | ||||
| (-)-epicatechin (EC) | LYS(L:45), GLN(L:44) | ||||
| Myricetin | ILE(H:93), GLN(H:39), GLN(L:44), LYS(L:45) | GLY(L:47) | |||
| Kaempferol-3-O-glucoside | MET(H:2), LYS(A:386), GLN(H:3) | ||||
| Quercetin | LYS(L:45), 2GLN(L:48), ILE(H:93) | ||||
| (-)-Epigallocatechin 3-(3-methyl-gallate) (3″Me-EGCG) | VAL(C:64), GLY(C:63), LYS(E:528), ASP(A:389). 2LYS(A:529), GLU(E:327) | ||||
| Caffeoylquinic acid (CQA) | 2GLN(L:48), GLY(H:42), 2VAL(H:115), ALA(H:92) | PRO(H:41), GLN(H:39) | |||
| Chlorogenic acid | GLY(H:28), 2ASN(H:77), ILE(H:30) | ||||
| Coumaric acid | ASP(E:389), LYS(E:386), GLY(C:63), TYR(E:369), ASN(E:370), VAL(C:64), SER(E:366), 2ASP(C:66) | ||||
| Caffeic acid | LYS(A:528), ASP(L:66), ASN(A:370) | ||||
| Gallic acid | ASN(A:388), TYR(A:369), GLU(L:61), VAL(L:64), ASP(A:364) | ||||
| Caffeine | ARG(H:59), TYR(L:31), SER(H:103) | PRO(E:412), TYR(L:98), TYR(L:31) | |||
| 6LU7 | (-)-epicatechin 3,5-di-O-digallate (EC35G) | THR(A24), THR(A26), THR(A46), HIS(A163), HIS(A164), MET(A165) | GLN(A189) | ||
| Rutin | THR(A26), PHE(A140), LEU(A141), ASN(A142), GLY(A143), HIS(A163), GLU(A166), THR(A190) | ||||
| (-)-epigallocatechin gallate (EGCG) | PHE(A140), HIS(A164), MET(A165) | ||||
| Apigenin glycoside | LEU(A141), 2SER(A144), CYS(A145), HIS(A163), GLU(A166) | CYS(A145) | |||
| Flavonol 3-O-d-glucoside (FOG) | LEU(A141), GLY(A143), SER(A144), CYS(A145) | MET(A165), GLU(A166) | |||
| Myricetin 3-glucoside (M-G) | LEU(A141), ASN(A142), GLY(A143) | ||||
| (-)-Epigallocatechin (EGC) | HIS(A41) | ||||
| Kaempferol | TYR(A54), ASP(A187); | GLU(A166) | |||
| (-)-epicatechin gallate (ECG) | PHE(A140), HIS(164), MET(A165) | ||||
| (+)-catechin | GLU(A166), THR(A190); | GLN(A192) | |||
| (-)-epicatechin (EC) | THR(A26), HIS(A41), GLN(A189) | ||||
| Myricetin | GLY(A143), SER(A144), ARG(A188) | GLU(A166) | |||
| Kaempferol-3-O-glucoside | THR(A24), THR(A26), THR(A46), HIS(A163), HIS(A164), MET(A165) | GLN(A189) | |||
| Quercetin | TYR(A54), ASP(A187) | GLU(A166) | |||
| (-)-Epigallocatechin 3-(3-methyl-gallate) (3″Me-EGCG) | LEU(A141), 2CYS(A145), THR(A190), ASN(A188) | GLU(A166), | |||
| GLN(A189) | |||||
| Caffeoylquinic acid (CQA) | LEU(A141),GLY(A143), 2SER(A144), CYS(A145), HIS(A163), GLU(A166), 2THR(A190); | MET(A165) | |||
| Chlorogenic acid | ASN(A142), SER(A144), 2THR(A190) | ||||
| Coumaric acid | LEU(A141), GLY(A143), 2SER(A144), CYS(A145), THR(A190) | PRO(A168) | |||
| Caffeic acid | GLU(A166), GLY(A143), SER(A144) | CYS(A145). | |||
| Gallic acid | LEU(A141), GLY(A143), CYS(A145), GLU(A166), GLN(A189) | CYS(A145) | |||
| Caffeine | GLY(A143), GLU(A166) | LEU(A141), 2CYS(A145), GLN(A189) | |||
| 6M71 | (-)-epicatechin 3,5-di-O-digallate (EC35G) | THR(A:710), ASN(A:781) 2SER(A:708), LYS(A:780), SER(A:784), HIS(A:133), ASN(A:138) | LYS(A:780) | ||
| Rutin | LYS(A:47), 2TYR(A:129), SER(A:784), LYS(A:780), ASN(A:138) | ASP(A:135) | |||
| (-)-epigallocatechin gallate (EGCG) | 3ASN(A:781), 2SER(A:709), ALA(A:706), 2SER(A:784) | ||||
| Apigenin glycoside | THR(A:394), ARG(A:349), LEU(A:245), LEU(A:251) | THR(A:319), CYS(A:395) | |||
| Flavonol 3-O-d-glucoside (FOG) | VAL(A:675) | ARG(A:457) | |||
| Myricetin 3-glucoside (M-G) | LYS(A:47) THR(A:710), ASN(A:781), SER(A:709) | 2GLY(A:774) | |||
| (-)-Epigallocatechin (EGC) | LYS(A:47), TYR(A:129), 2SER(A:784) | ||||
| Kaempferol | TYR(A:689), ILE(A:494), ARG(A:569), 2ASN(A:496) | ||||
| (-)-epicatechin gallate (ECG) | 2SER(A:709), HIS(A:133), SER(A:784) | ||||
| (+)-catechin | TYR(A:129), SER(A:784), PHE(A:134), LYS(A:780), SER(A:772) | ||||
| (-)-epicatechin (EC) | TYR(A:129), SER(A:709), GLN(A:778), 2THR(A:710), ASP(A:711), LYS(A:47) | TYR(A:129) | |||
| Myricetin | THR(A:710), 2SER(A:784) | GLY(A:774) | |||
| Kaempferol-3-O-glucoside | ASN(A:781), SER(A:709), ASN(A:138) | TYR(A:129), TYR(A:32) | |||
| Quercetin | ASN(A:628) | ||||
| (-)-Epigallocatechin 3-(3-methyl-gallate) (3″Me-EGCG) | LYS(A:780), SER(A:784), LYS(A:47), TYR(A:32) | ||||
| Caffeoylquinic acid (CQA) | ASP(A:623), CYS(A:622), LYS(A:621), PHE(A:793), SER(A:795), ASP(A:618) | ||||
| Chlorogenic acid | THR(A:206), ASN(A:208) | ||||
| Coumaric acid | LYS(A:47), SER(A:709), 2ASN(A:781), SER(A:784) | ||||
| Caffeic acid | TYR(A:619), GLU(A:811), TRP(A:800) | ||||
| Gallic acid | ASP(A:761), TRP(A:617) | ||||
| Caffeine | TYR(B:135), GLY(B:144), TYR(B:138) | TYR(B:135), ASP(B:148 | |||
| 6WX4 | Rutin | 2GLU(D:252), TYR(D:252), SER(D:212),LEU(D:211) | |||
| (-)-epicatechin 3,5-di-O-digallate (EC35G) | TYR(D:213), GLU(D:214), LYS(D:306) | ||||
| (-)-epigallocatechin gallate (EGCG) | 164), TYR(D:273), 2GLY(D:163) | PRO(D:248) | |||
| Apigenin glycoside | HIS(D:175), THR(D:74), TYR(D:154) | GLN(D:174) | |||
| Flavonol 3-O-d-glucoside (FOG) | 2SER(D:180), ASN(D:308), GLU(D:124) | ||||
| Myricetin 3-glucoside (M-G) | LEU(D:58), ASN(D:60), ASP(D:61) | GLN(D:30), PHE(D:31 | |||
| (-)-Epigallocatechin (EGC) | LYS(D:306), GLU(D:307), 2GLU(D:214), TYR(D:305) | ||||
| Kaempferol | ASP(D:164) | ||||
| (-)-epicatechin gallate (ECG) | 2GLY(D:163), ASP(D:164), TYR(D:273), ARG(D:166) | ||||
| (+)-catechin | SER(D:212) | ||||
| (-)-epicatechin (EC) | LYS(D:306), 2GLU(D:307), TYR(D:305), TYR(D:213), GLU(D:214) | ||||
| Myricetin | GLY(D:266), THR(D:301), ASP(D:164) | ||||
| Kaempferol-3-O-glucoside | LYS(D:297), SER(D:212), THR(D:210) | ||||
| Quercetin | TYR(D:273) | ||||
| (-)-Epigallocatechin 3-(3-methyl-gallate) (3″Me-EGCG) | THR(D:301), ASP(D:164), TYR(D:273) | ||||
| Caffeoylquinic acid (CQA) | THR(D:257), TYR(D:213), TYR(D:251), GLU(D:214) | ||||
| Chlorogenic acid | 2GLU(D:307), 2LYS(D:217), 2LYS(D:306), THR(D:257), TYR(D:251) | ||||
| Coumaric acid | GLU(D:252), LYS(D:217) | THR(D:257) | |||
| Caffeic acid | LYS(D:217), SER(D:212) | ||||
| Gallic acid | GLU(D:214) | ||||
| Caffeine | ARG(D:116), THR(D:301) | ||||
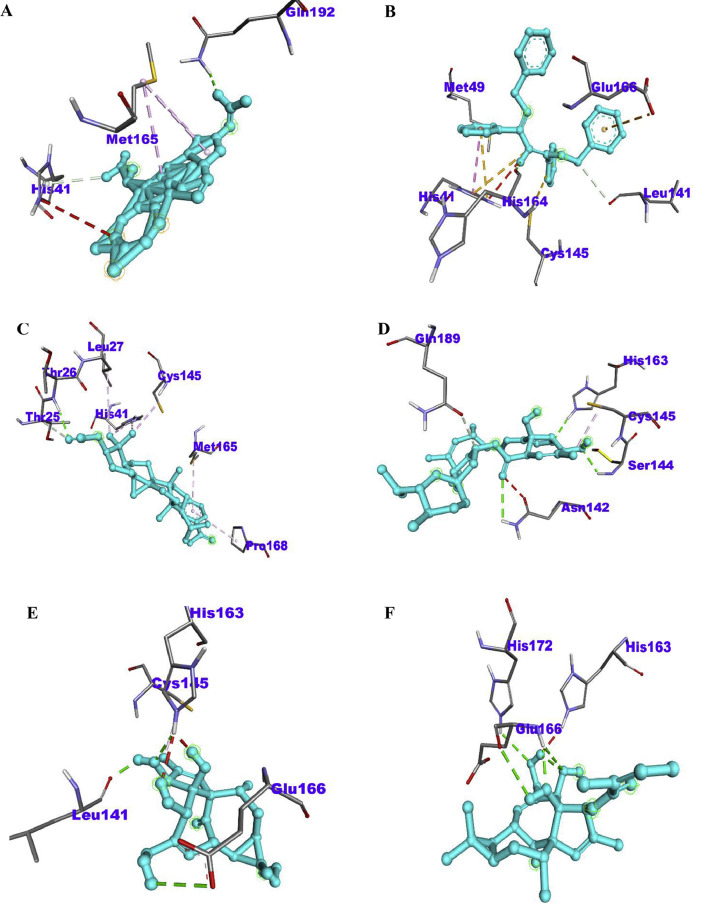
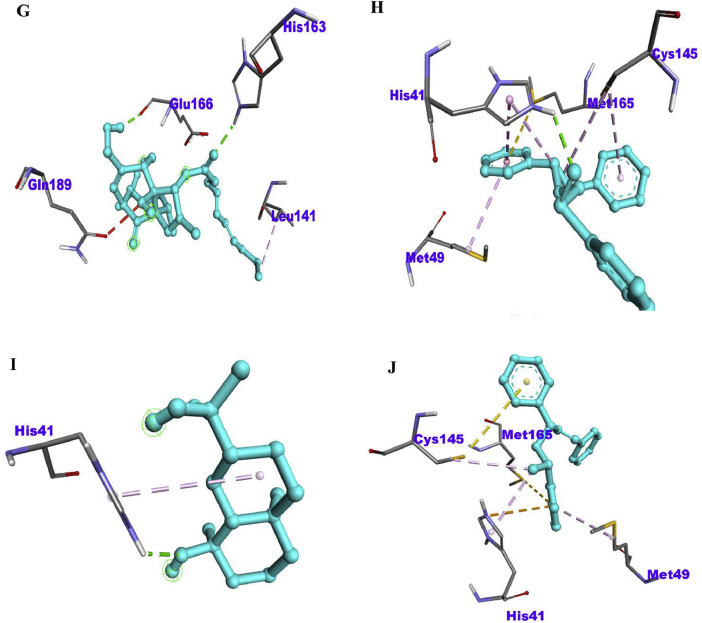
Fig. 1.
The 2D diagram showing the types of amino acid residues involved in the bond between the phytoconstituents in betel plant and the Mpro receptor of Sars-Cov-2. (A) PubChem ID 537,242, (B) PubChem ID 562,008, (C) PubChem ID 5,373,661, (D) PubChem ID 5,367,225, (E) PubChem ID 119,057,278, (F) PubChem ID 6,918,670, (G) PubChem ID 5,367,323, (H) PubChem ID 562,543, (I) PubChem ID 165,258, (J) PubChem ID 20,138,399.
2. Experimental Design, Materials, and Methods
2.1. Receptors and ligands selection
The selection of receptors is based on the information contained in the literature. Five essentials enzyme and proteins of SARS-CoV-2 selected as receptors in this study were spike ectodomain structure (open state) (PDB code: 6VYB), receptor-binding domain (RDB) (PDB code: 6YLA), main protease (Mpro) (PDB code: 6LU7), RNA-dependent RNA polymerase (RdRp) (PDB code: 6M71), and papain-like protease (PLpro) (PDB code: 6WX4). The phytoconstituents of betel leaf which serve as ligands were based on GC–MS data [16]. The GC–MS procedure was carried out following the research conducted by Tumilaar et al. [17]. The phytoconstituents of matcha green tea were selected based on a literature survey as listed in Table 2.
2.2. Receptors and ligands preparation
The structures of the receptor (Mpro) were retrieved from Protein Data Bank (http://www.rcsb.org) and opened in BIOVIA Discovery Studio Visualizer 2020 [18]. After removing the water molecules and native ligands, the receptor was saved in a .pdb format. All the structures of the ligands were retrieved from PubChem (http://pubchem.ncbi.nlm.nih.gov) in .sdf format. The files were converted into a .pdb format using Open Babel [19]. After adjusting the torque, the files were saved in .pdbqt format.
2.3. The docking process and visualization
Autudock Vina [20] was used in the docking analysis. The .pdbqt formats of ligands and receptors were copied into the Vina folder. Vina configuration was typed in notepad and saved as conf.txt. Vina program was performed in a command prompt mode. The most negative Gibbs’ free energy of binding indicated the best pose. The visualization of the interacting amino acids of the receptors with the ligands was performed in Biovia Discovery Studio 2020 [18].
Ethics Statement
The work did not involve the use of endangered species of wild flora.
Supplementary Data
Supplementary data to this article can be found at http://dx.doi.org/10.17632/w8h74c6hsy.1 and https://doi.org/10.17632/4dn4svm3jb.1.
CRediT Author Statement
Fatimawali: Conceptualization, Methodology, Data curtion, Writing - original draft, Writing – review & editing; Rizky Ramadhan Maulana: Software, Validation; Axl Laurens Lukas Windah: Software, Validation; Irma Febrianti Wahongan: Visualization, Investigation; Sefren Geiner Tumilaar: Visualization, Investigation; Ahmad Akroman Adam: Data curtion, Writing – review & editing; Billy Johnson Kepel: Data curtion, Writing – review & editing; Widdhi Bodhi: Visualization, Investigation; Trina Ekawati Tallei: Conceptualization, Methodology, Data curtion, Writing – review & editing, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have or could be perceived to have influenced the work reported in this article.
Acknowledgments
This research was funded by the Directorate of Research and Community Service, Ministry of Research and Technology / National Research and Innovation Agency under the scheme of Excellent Applied Research in Higher Education (Contract No. 1179/UN12.13/LT/2020) and the COVID-19 Refocusing Research scheme for the fiscal year 2020 (Contract No. 206/SP2H/AMD/LT/DRPM/2020).
Contributor Information
Fatimawali, Email: fatimawali@unsrat.ac.id.
Trina Ekawati Tallei, Email: trina_tallei@unsrat.ac.id.
References
- 1.Tallei T.E., Tumilaar S.G., Niode N.J., Fatimawali F., Kepel B.J., Idroes R., Effendi Y., Sakib S.A., Emran T.B. Potential of plant bioactive compounds as SARS-CoV-2 main protease (Mpro) and spike (S) glycoprotein inhibitors: a molecular docking study. Scientifica (Cairo) 2020 doi: 10.1155/2020/6307457. 2020Article ID 6307457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rakib A., Paul A., Chy M., Sami S.A., Baral S.K., Majumder S.M., Tareq A.M., Amin M.N., Shahriar A., Uddin M.Z., Dutta M., Tallei T.E., Emran T.B., Simal-Gandara J. Biochemical and computational approach of selected phytocompounds from Tinospora crispa in the management of COVID-19. Molecules. 2020;25(17):3936. doi: 10.3390/molecules25173936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sengupta R., Banik J.K. A review on betel leaf (PAN) Int. J. Pharm. Sci. 2013;4(12):4519–4524. doi: 10.13040/IJPSR.0975-8232.4(12).4519-24. [DOI] [Google Scholar]
- 4.Farooq S., Sehgal A. Antioxidant activity of different forms of green tea: loose leaf, bagged and matcha. Curr. Res. Nutr. Food. Sci. J. 2018;6:35–40. doi: 10.12944/CRNFSJ.6.1.04. [DOI] [Google Scholar]
- 5.Jakubczyk K., Kochman J., Kwiatkowska A., Kałduńska J., Dec K., Kawczuga D., Janda K. Antioxidant properties and nutritional composition of matcha green tea. Foods. 2020;9(4):483. doi: 10.3390/foods9040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tallei T.E. 2020. Supplementary Dataset on the Docking of Phytoconstituents of Matcha Green Tea on SARS-CoV-2 . Mendeley Data, V1. </Dataset>. [DOI] [Google Scholar]
- 7.Yao L., Jiang Y., Datta N., Datta N., Singanusong R., Liu X., Duan J., Raymont K., Lisle A., Xu Y. HPLC analyses of flavanols and phenolic acids in the fresh young shoots of tea (Camellia sinensis) grown in Australia. Food Chem. 2004;84(2):253–263. doi: 10.1016/s0308-8146(03)00209-7. [DOI] [Google Scholar]
- 8.Jeszka-Skowron M., Zgoła-Grześkowiak A. Analysis of antioxidant activity, chlorogenic acid, and rutin content of Camellia sinensis infusions using response surface methodology optimization. Food Anal. Meth. 2014;7:2033–2041. doi: 10.1007/s12161-014-9847-1. [DOI] [Google Scholar]
- 9.Kim Y., Lee K.G., Kim, M. K. M.K. Volatile and non-volatile compounds in green tea affected in harvesting time and their correlation to consumer preference. Int. J. Food Sci. Technol. 2016;53(10):3735–3743. doi: 10.1007/s13197-016-2349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savić I.M., Nikolić V.D., Savić I.M., Nikolić L.B., Jović M.D., Jović M.D. The qualitative analysis of the green tea extract using ESI-MS method. Savremene tehnologije. 2014;3(1):30–37. doi: 10.3724/sp.j.1123.2015.04028.10.5937/savteh1401030S. [DOI] [Google Scholar]
- 11.Jiang H., Engelhardt U.H., Thräne C., B Maiwald, Stark J. Determination of flavonol glycosides in green tea, oolong tea and black tea by UHPLC compared to HPLC. Food Chem. 2015;183:30–35. doi: 10.1016/j.foodchem.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z., Sha Y., Yu X., Liang Y. Determination of flavonol glycosides in tea samples by ultra-high performance liquid chromatography-photodiode array detection-tandem mass spectrometry. Se Pu. 2015;33(9):974–980. doi: 10.3724/sp.j.1123.2015.04028. PMID: 26753286. [DOI] [PubMed] [Google Scholar]
- 13.Reygaert W.C. Green tea catechins: their use in treating and preventing infectious diseases. BioMed. Res. Int. 9105261. 2018 doi: 10.1155/2018/9105261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeganathan B., Punyasiri P.A., Kottawa-Arachchi J.D., Ranatunga M.A., Abeysinghe I.S., Gunasekare M.T., Bandara B.M. Genetic variation of flavonols, myricetin, and kaempferol in the Sri Lankan Tea (Camellia sinensis L.) and their health-promoting aspects. Int. J. Food Sci. 2016;6057434 doi: 10.1155/2016/6057434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang K., Cha K.H., Song D., Kim S.M., Lee E.H., Lee H.J., Nho C.W., Pan C. Inhibititory effect of gastrointestinal lipolysis by green tea, coffee, and gomchui (Ligularia fischeri) tea during simulated digestion. FASEBJ. 2012;26 doi: 10.1096/fasebj.26.1_supplement.lb360. lb360-lb360. [DOI] [PubMed] [Google Scholar]
- 16.Fatimawali F., Tallei T.E. Supplementary Dataset of Betel Plant GC-MS and Docking results. Mendeley Data. 2021;V1 doi: 10.17632/s72rcpk82b.1. [DOI] [Google Scholar]
- 17.Tumilaar S.G., Fatimawali F., Niode N.J., Effendi Y., Idroes R., Adam A.A., Rakib A., Emran T.B., Tallei T.E. The potential of leaf extract of Pangium edule Reinw as HIV-1 protease inhibitor: a computational biology approach. J. Appl. Pharm. Sci. 2021;11(01):101–110. doi: 10.7324/JAPS.2021.110112. [DOI] [Google Scholar]
- 18.Systèmes D.B. Discovery Studio Modeling Environment, Release 2020. https://discover.3ds.com/discovery-studio-visualizer-download, 2020 (accessed 10 June 2020).
- 19.O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Com. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]