Abstract
Germline variants have a rich history of being studied in the context of cancer risk. Emerging studies now suggest that germline variants contribute not only to cancer risk but to tumor progression as well. In this Opinion article, we discuss the initial discoveries associating germline variants to patient outcome and the mechanisms by which germline variants affect molecular pathways. Germline variants affect molecular pathways through amino acid changes, alteration of splicing patterns or expression of genes, influencing the selection for somatic mutations, and causing genome wide mutational enrichment. These molecular alterations can lead to tumor phenotypes that become clinically apparent such as metastasis, alterations to the immune microenvironment, and modulation of therapeutic response. Overall, the growing body suggests that germline variants play a much larger role in tumor progression than has been previously appreciated and that germline variation holds substantial potential for improving personalized medicine and patient outcomes.
Keywords: Germline Variants, Tumor Progression, Cancer Therapy, Personalized Medicine
The Role of Germline Variants in Tumor Progression
Historically, the study of germline variation in cancer has focused on cancer risk [1, 2]. The study of such germline variants has improved our understanding of basic cellular processes, while also informing clinical screening and treatment guidelines. However, with a few exceptions, germline variants are not used widely for managing cancer after the diagnosis has been made. In contrast, tumor somatic mutations are widely used for cancer treatment decisions [3]. Recent studies suggest that germline variation not only affects the risk of acquiring cancer but also affects the rate of tumor progression and therapeutic responsiveness. These findings suggest that the study of germline variants in the context of cancer progression could further the field’s understanding of molecular processes and identify patients that may benefit from differential clinical management. In this review, we highlight these studies to draw attention to this relatively understudied area of oncology with the potential for improving patient care.
Utility of Germline Variants in the Management of Cancer Patients
While germline variants that increase cancer risk inform cancer screening and prevention guidelines, a broader utilization of germline variants for the management of cancer patients has been challenging (Table 1) [2, 4–8]:
Table 1.
Current challenges and opportunities for further study in understanding how germline variants play a role in shaping the outcome of patients with cancer.
| Category | Challenge | Explanation | Possible Solutions |
|---|---|---|---|
| Cancer Risk, Cancer Progression, and General Problems of Germline Variant Cancer Genomics | Cohort Size | Common variants typically have low effect sizes whereas variants with large effect sizes are often rare, thus requiring large cohorts | Building Large Cohorts or Tests Involving Groups of Variants |
| Integration with Clinical Phenotypes | Datasets in genomics often have minimal clinical annotation | Building datasets with more detailed clinical annotation | |
| Effect Sizes | The effect sizes of individual germline variants must be large to motivate clinical use | Building models with combinations of variants and incorporating variants with large effect sizes, such as pathogenic germline variants | |
| Ethnicity Background | Effects of germline variants may differ based on ethnicity | Perform analyses in ethnically diverse cohorts | |
| Germline-Somatic Interactions | Understanding the molecular basis for the interaction | Associations may be the result of several complex and indirect associations. | Network based computational approaches or experimental perturbation |
| Context Dependency | Interactions may be dependent on specific environmental exposures or may occur in certain genetic backgrounds | Building datasets with more detailed clinical annotation of environmental exposures and performing studies in ethnically diverse cohorts | |
| Drug Responsiveness and Toxicity | Differing Treatment Regimens or Differing Dosages | Patients with the same cancer treated with chemotherapy may undergo different treatment regimens or may be treated with different dosages of chemotherapy drugs | Report detailed treatment information when building genomic datasets and experimental perturbation |
The use of germline variants in managing cancer requires validation in large cohorts because variants with large effect sizes tend to be rare. These rare variants classically exhibit Mendelian inheritance patterns and may be associated with monogenic cancer syndromes. These variants can be studied in the context of familial syndromes. On the other hand, common variants tend to have smaller effect sizes and thus still require large cohorts to validate. There seem to be many common variants that contribute to cancer development and progression and their effects tend to be fairly subtle. Therefore, the discovery of these common variants has shifted our understanding of cancer from a monogenic to multifactorial disease. Common variants are not studied in the context of familial syndromes and are instead studied at the population level [9].
While cancer risk is affected by germline variation, other clinical factors such as environmental exposure and age need to be taken into account to maximize their utility for prediction of cancer risk [10].
The effect sizes of the germline variants must be large enough to alter clinical decisions because of the increased cost and inconvenience to the patient to start screening for cancer earlier.
We suggest in this Opinion piece that there is another use of germline variants in cancer management: they predict tumor progression and outcome.
Germline Variation Shapes the Landscape of Somatic Aberrations in Cancer
The most compelling evidence suggesting that germline variation affects tumor progression initially stemmed from studies that showed germline variation can shape the landscape of somatic aberrations in cancer [11]. Germline variants can interact with somatic aberrations in cancer in multiple ways, such as increasing the likelihood of somatic mutations in the wild type allele of the same gene or genes in the same pathway. Alfred Knudson’s “two-hit hypothesis” published in 1971 predicted this, as his hypothesis suggested that a germline variant that affected the function of RB1 (a tumor suppressor gene) could be followed by a somatic mutation that affected the other allele of that same tumor suppressor gene to cause tumorigenesis (Figure 1A) [12, 13]. A recent study of 429 patients with ovarian carcinoma found that the majority of patients with germline truncating mutations in the tumor suppressor genes BRCA1 and BRCA2 exhibit loss of heterozygosity in their tumor samples in the originally wild type allele [14]. Similarly, genomic studies of myeloproliferative neoplasms identified a germline JAK2 haplotype associated with an increased risk for the development of JAK2V617F somatic mutations---one of the most common and well-characterized drivers of myeloproliferative neoplasms [15–18]. In line with these findings, a previous study identified functional germline variants in the EGFR tyrosine kinase associated with an increased risk for subsequent somatic mutations in EGFR [19].
Figure 1.
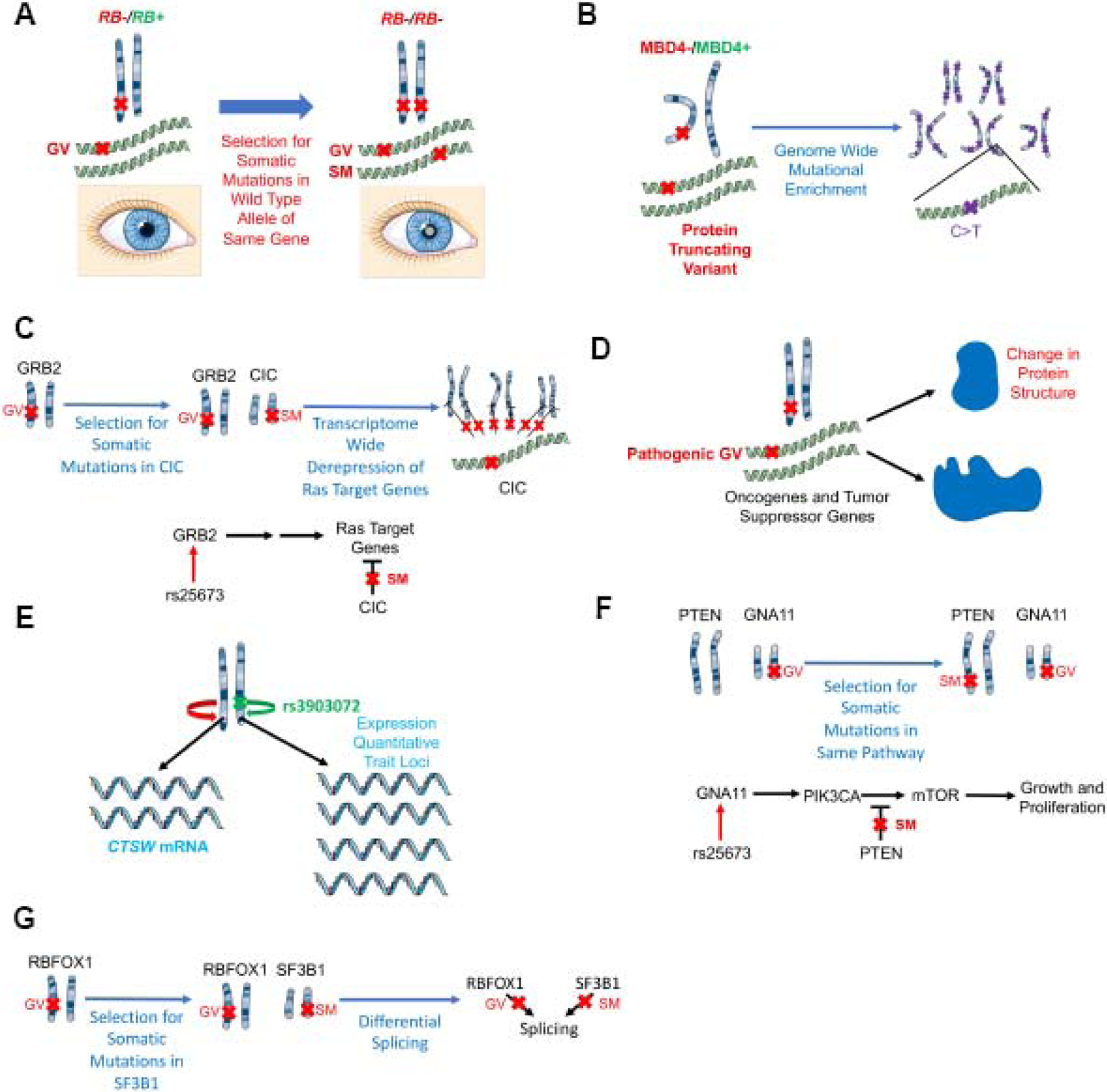
Molecular mechanisms by which germline variants may associate with outcome.
(A) Germline variants increase the likelihood of a somatic mutation in the wild type allele of the same gene: a germline variant in RB1 increases the likelihood of a somatic mutation in the other allele of RB1 leading to retinoblastoma (shown here by the clinical sign of leukocoria).
(B) Germline variants increase specific types of genome wide mutations: germline variants in MBD4 are associated with elevated C>T somatic mutations at CpG dinucleotides, consistent with the role of MBD4 in correcting G:T mismatches.
(C) Germline variants are associated with specific transcriptome alterations: a germline variant in the Ras signaling gene GRB2 is associated with upregulation of Ras target genes.
(D) Molecularly, germline variants may alter the function of tumor suppressor genes or oncogenes through amino acid changes.
(E) Germline variants act as expression quantitative trait loci (eQTL) in cis: a germline variant in the putative regulatory element of the tumor suppressor gene CTSW is associated with increased expression of this tumor suppressor gene.
(F) Germline variants increase the likelihood of additional somatic mutations in the same pathway: germline variants in GNA11, an activator of mTOR signaling, promote the acquisition of somatic mutations in PTEN, a tumor suppressor that suppresses mTOR signaling.
(G) Germline variants perturb splicing machinery: germline variants in RBFOX1, an RNA binding protein involved in splicing, are associated with increased somatic mutations in SF3B1, another component of the spliceosome, leading to varied splicing patterns.
A large study by Carter et al. analyzed the interaction between inherited polymorphisms and somatic aberrations in almost 6,000 tumors across 22 different cancer types [11]. They identified and validated 412 genetic interactions between germline variants and somatic aberrations. The somatic aberrations were sometimes in genes that were not the same as the ones perturbed by the germline variant. For example, some germline variants were associated with increased risk for somatic mutations in other genes in the same pathway. These findings suggest that more complex computational methods are necessary to attain an integrated understanding of the varied factors, including germline variants, that influence which somatic mutations will occur, or be selected for, in a tumor [11].
A study of the interaction between germline variation and somatic aberrations in cancers using whole genome sequencing data from 2,658 patients across 38 tumor types by the Pan-Cancer Analysis of Whole Genomes Consortium found that germline variation is predictive of somatic mutational processes across cancers [20]. For example, germline variants at the 22q13.1 locus were associated with decreased APOBEC mutagenesis in cancer. Rare variants in BRCA1 and BRCA2 were associated with a higher abundance of small somatic structural variant deletions and tandem deletions, consistent with a role of these proteins in error-free homologous recombination directed repair of double-strand breaks. Germline variants in MBD4, a gene that codes for a protein that binds to methylated CpGsCPGs and corrects G:T or G:U mismatches in their vicinity, were associated with an elevated rate of C>T somatic mutations at CpG dinucleotides (Figure 1B). Finally, they identified 114 germline source L1 elements that were capable of active somatic retrotransposition. Overall, their results also suggest that germline variation can shape somatic processes at a genome-wide scale [20].
Numerous additional studies have explored the link between germline variation and somatic aberrations [21–23]. A study from Chatrath et al. analyzed sequencing data from patients with lower grade gliomas (LGG) and identified a germline variant in GRB2, an adaptor protein in the Ras signaling pathway, that was associated with increased expression of genes in the Ras signaling pathway and a doubling of somatic mutations in Capicua transcriptional repressor (CIC), a tumor suppressor gene that negatively regulates the Ras target genes and is mutated in about 21.4% of LGG (Figure 1C) [24].
Understanding the relationship between germline variation and somatic aberrations is particularly promising from a clinical perspective [21]:
A relationship between germline variation and somatic genetic aberration suggests that the aggressiveness of a tumor can be predicted based on the germline status.
Germline variation has begun to be used to improve the selection of clinical therapy as part of this new era of personalized medicine [25–28]. The recent large-scale sequencing of tumors and cancer cell lines has helped to identify the genomic determinants of drug sensitivity [29, 30]. The existence of an interaction between germline variation and somatic aberrations suggests that chemotherapy responsiveness can be predicted using the status of germline variants.
Germline variants in the mismatch repair genes predict microsatellite instability, which increases the chance of producing neo-antigens and is associated with an improved response to immune checkpoint blockade therapy [31–33].
Identification of Germline Variants Associated with Outcome
The idea that there is a link between germline variation and somatic events in cancer implies that germline variation may also affect tumor progression and could be used to predict the prognosis of patients with cancer. Studies in this area have identified germline variants predictive of patient outcome in genes with well-characterized driver roles in cancer. The single nucleotide polymorphism (SNP) rs9939049 in the gene cadherin 1 (CDH1) has been associated with increased risk of colon cancer, along with poor outcome (HR=1.44) [34]. The SNP rs869330 in the MTAP gene was previously associated with increased risk of cutaneous melanoma and is also found to be associated with prolonged relapse-free survival (HR=0.800) and overall survival (HR=0.760) [35]. rs10932384 in ERBB4 was found to be associated with prolonged time to recurrence (HR=0.52) and overall survival (HR=0.50) in patients with renal cell carcinoma [36]. rs187155 in CD44 was associated with increased time to recurrence (HR=0.67) and rs13347 in CD44 was associated with reduced risk of death (HR=0.61) in patients with colon cancer [37]. rs966423 was associated with increased risk of differentiated thyroid cancer and also decreased overall survival (HR=1.89) [38]. Finally, a study of 2,060 patients with colorectal cancer found the germline variant rs863221 in the mismatch repair gene MSH3 to be associated with favorable outcome (HR=0.59) [39].
Recent unbiased screens have also identified associations between patient outcome and germline variants in genes that had not been previously tied to tumorigenesis. In patients with Chronic Lymphocytic Leukemia (CLL), 12 germline variants associated with overall survival were discovered in a variety of genes with moderate to high effect sizes (HR=3.97–43.14) [40]. A study of 1365 patients with multiple myeloma found rs2235013 in ABCB1 (HR=1.52) and rs4148388 in ABCC2 (HR=2.15) to be associated with decreased overall survival [41]. A study by Chatrath et al. of patients with lower grade gliomas identified two germline variants associated with poor outcome, one in the oncogene GRB2 (HR=20.4) and the other in the tumor suppressor gene ANKDD1a (HR=1.73) [24]. A subsequent study extended this approach to all 33 cancers in The Cancer Genome Atlas and characterized the landscape of prognostic germline variants using genomic sequencing data from approximately 10,000 patients [42]. The results suggest that germline variation is associated with patient outcome across all cancers and that germline variation affects tumor progression. Nearly half of the prognostic germline variants were in genes with previously reported roles as oncogenes or tumor suppressor genes. The other half of the genes with prognostic germline variants are of unknown function and require further study [42].
Clinical criteria such as grade and stage of a tumor and selected molecular criteria (e.g. Isocitrate dehydrogenase (IDH) mutation in gliomas) are already being used to predict patient outcome. Such prognostication helps identify patients who are unlikely to respond well to current therapy and for whom newer therapies need to be devised. Adding the status of prognostic germline variants to current prognostic clinical and molecular criteria significantly improve outcome prediction and could therefore potentially be used for patient management decisions [24, 42].
The Molecular Basis for Germline Variants Being Associated with Outcome
Recent studies of the molecular functions of germline variants have suggested multiple mechanisms explaining how germline variants can affect tumorigenesis and progression, including some that have already been described (Figure 1A–C). In this section, we describe the intrinsic impact of germline variability on the tumor itself at the molecular level.
Two types of genes, oncogenes and tumor suppressor genes, have been implicated in cancer development. Oncogenes are genes that have the potential of causing cancer when they acquire gain-of-function mutations or are highly expressed (eg, RAS, MYC, TRK). Gain-of-function mutations are mutations that increase the activity of a protein or enable the protein to perform a novel function. Tumor suppressor genes are genes that normally prevent the development of cancer. Loss-of-function mutations or decreased expression of these genes can contribute to the development of cancer. Loss-of-function mutations alter the proteins coded for by tumor suppressor genes in such a way that they can no longer perform their normal cellular functions. Rare, pathogenic germline variants can result in amino acid changes in oncogenes or tumor suppressor genes resulting in dysfunction of these genes (Figure 1D). While the dysfunction of these genes was originally studied in the context of tumorigenesis, these same molecular mechanisms are likely contributing to differences in tumor progression as well [2].
The effects of germline variants are frequently attributed to differences in the expression of nearby genes (expression quantitative trait loci, eQTL, in cis) [43]. Germline variants can thus be associated with tumor progression because they are eQTLs for nearby genes. In fact, these differences in gene expression can occur in non-tumor cells and modulate tumor phenotypes. For example, the germline variant rs3903072 is associated with decreased risk of breast cancer and prolonged survival in patients with breast cancer. This germline variant is found in a putative regulatory element of the tumor suppressor gene CTSW and is associated with its increased expression. Although the function of CTSW is not entirely known, CTSW is specifically expressed in natural killer and T-cells and is believed to play a role in T-cell cytolytic activity. Therefore, this germline variant seems to be associated with differences in cell survival through its association with immune cell function (Figure 1E) [44].
We have described above that germline variants can affect tumor progression by increasing the incidence of somatic mutations elsewhere in the genome (Figure 1A, 1C). As another example, the germline variant rs25673 is associated with increased expression of GNA11, which activates mTOR signaling. Patients with this germline variant are more likely to acquire somatic mutations in PTEN, a tumor suppressor gene, which suppresses mTOR signaling activity (Figure 1F) [11].
Germline variants can have effects on the transcriptome and splicing patterns independent of their roles as eQTLs. Some mechanisms involve selection for mutations elsewhere in the genome, as with Capicua Transcriptional Repressor (CIC) gene deletions in patients with germline variant rs25673 in the oncogene GRB2, an adaptor protein in the Ras signaling pathway, leading to upregulation of Ras target genes (Figure 1C) [24]. As another example, patients with germline variants in RBFOX1, which encodes an RNA binding protein involved in splicing, exhibit increased incidence of somatic mutations in SF3B1, encoding a component of the spliceosome. These patients exhibited varied splicing patterns (Figure 1G) [11].
Germline Variation Affects Drug Responsiveness and Toxicity
Germline variation may also be predictive of patient outcome by modulating responsiveness and toxicity to therapy. These topics have recently been highlighted in other recent articles [25–28].
Effect Sizes of Germline Variants Associated with Outcome
To examine the trend of effect sizes in germline variants that have been identified until now as affecting tumor progression, we compiled the germline variants associated with outcome and plotted their effect size, namely the absolute value of the natural log of the hazard ratio, against the alternate allele frequency (Figure 2). Individually, germline variants with low hazard ratios have minimal clinical significance as their effects are unlikely to be large enough to guide clinical decision-making at the individual level. Instead, the effects of germline variants on clinical decision-making will likely be derived either from individual germline variants with large effect sizes or groups of germline variants pooled together in multifactorial models.
Figure 2.
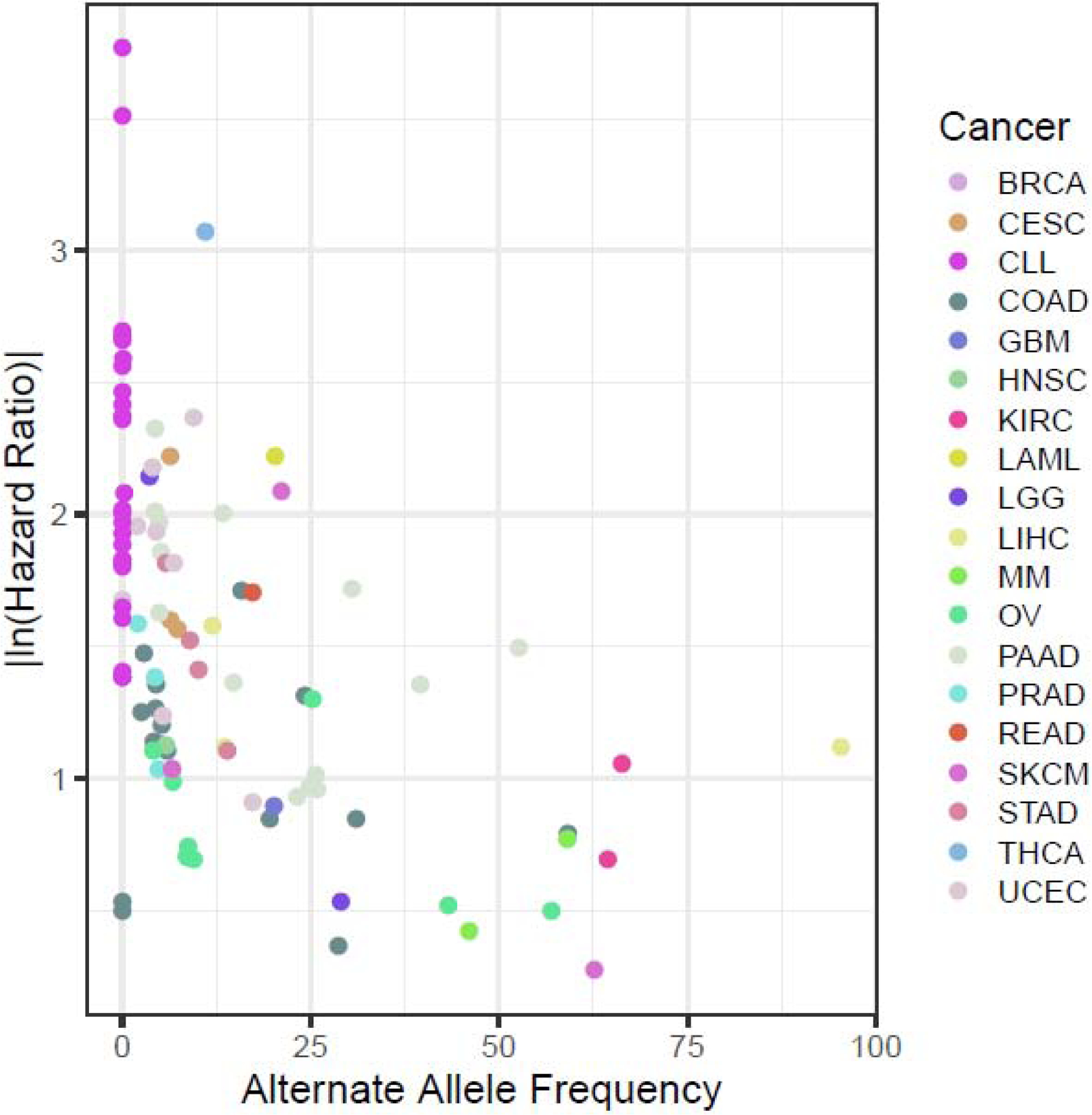
A compilation of germline variants associated with outcome discussed in this review shows that the absolute value of the natural log of the hazard ratio (HR) (effect size) is negatively correlated with the frequency of the alternate allele in the population.
As is the case with germline variants associated with cancer risk, the alternate allele frequency is negatively correlated with effect size (Spearman’s rho=−0.492, p=1.321E-7). This result is consistent with previous results and follows the general trend of germline variants with deleterious phenotypes [42, 45].
Surprisingly, even though they are not directly comparable, the hazard ratios (effect size) of the germline variants estimated in these studies are surprisingly high, in comparison to the odds ratios determined in studies of cancer risk [45]. Consistent with this, the germline variants associated with outcome were identified in cohorts that are generally much smaller than those of studies of cancer risk. It is not immediately obvious exactly why this is the case, but below are a few hypotheses that need further investigation:
Studies of cancer progression are focused on a trait with a quantitative component (such as time to survival or progression) as opposed to the binary variable used in studies of cancer risk (cancer or no cancer). As a result, studies of cancer progression may be better powered than studies of cancer risk when the cohort size is the same and the follow-up time is sufficiently long.
Common variants may play a larger role than environmental factors in tumor progression than in cancer risk. Generally speaking, at the population level, environmental factors seem to explain a greater proportion of the risk of cancer than germline genetics [1].
The time to development of adult cancers is often longer than the time to progression or time to death after a patient has acquired cancer. As a result, there is less of an opportunity for environmental factors to play a role in shaping the progression of disease, and researchers may be better able to control for confounding factors in studies of tumor progression than studies of cancer risk.
Regardless of the explanation for why germline variants affecting cancer progression can be identified with a smaller number of patients, the smaller cohort requirement will make it easier to conduct studies associating germline variants with cancer progression and patient outcome.
Future Directions: Understanding the Molecular Basis of the Associations with Outcome
Understanding the mechanisms by which germline variants are associated with patient outcome and modulate tumor progression is challenging due to several factors and is likely to be a rich area of future inquiry. Most available datasets are limited to exonic regions and therefore miss potentially important germline variants in introns and intergenic regions, though this is quickly changing. A variant that is associated with prognosis in these datasets may not be the variant responsible for the effect on the outcome. Instead, it could be in genetic linkage with the variant that actually affects the outcome. In addition, germline variants are present in every tissue in the body, and so could affect outcome through effects on non-tumor cells such as through immune system cells or changes in the tumor microenvironment. As a result, determining which nucleotide and which tissue is responsible for the observed phenotype has made understanding the exact molecular mechanisms by which germline variants act quite difficult. Nevertheless, research in this area has suggested possible roles by which the variants may be acting, such as through perturbation of protein structure and function and modulation of gene expression [46–48]. It is most likely that the mechanism by which a given germline variant affects outcome will most easily be elucidated for variants that affect protein sequence of, or are eQTLs for genes that function in the tumor cell itself to directly affect the cancer phenotype.
Future Directions: The Hallmarks of Germline Variants in Cancer
To organize our understanding of the molecular mechanisms by which germline variants could be associated with tumor progression, we were motivated by the hallmarks of cancer originally presented by Hanahan et al. to present the hallmarks (phenotypes) likely affected by germline variants regulating cancer outcome. The hallmarks that we have presented are based on the hallmarks organized and presented by Hanahan et al. (Figure 3) [49]. Broadly, the effects of germline variants on outcome can be divided into two categories: intrinsic effects on the tumor itself (genome instability, signaling, metastasis, cell death resistance, modulation of angiogenesis of the immune microenvironment) and extrinsic effects dependent on interaction with the environment (response to chemotherapy and side effects, radiotherapy, and immune checkpoint blockade).
Figure 3.
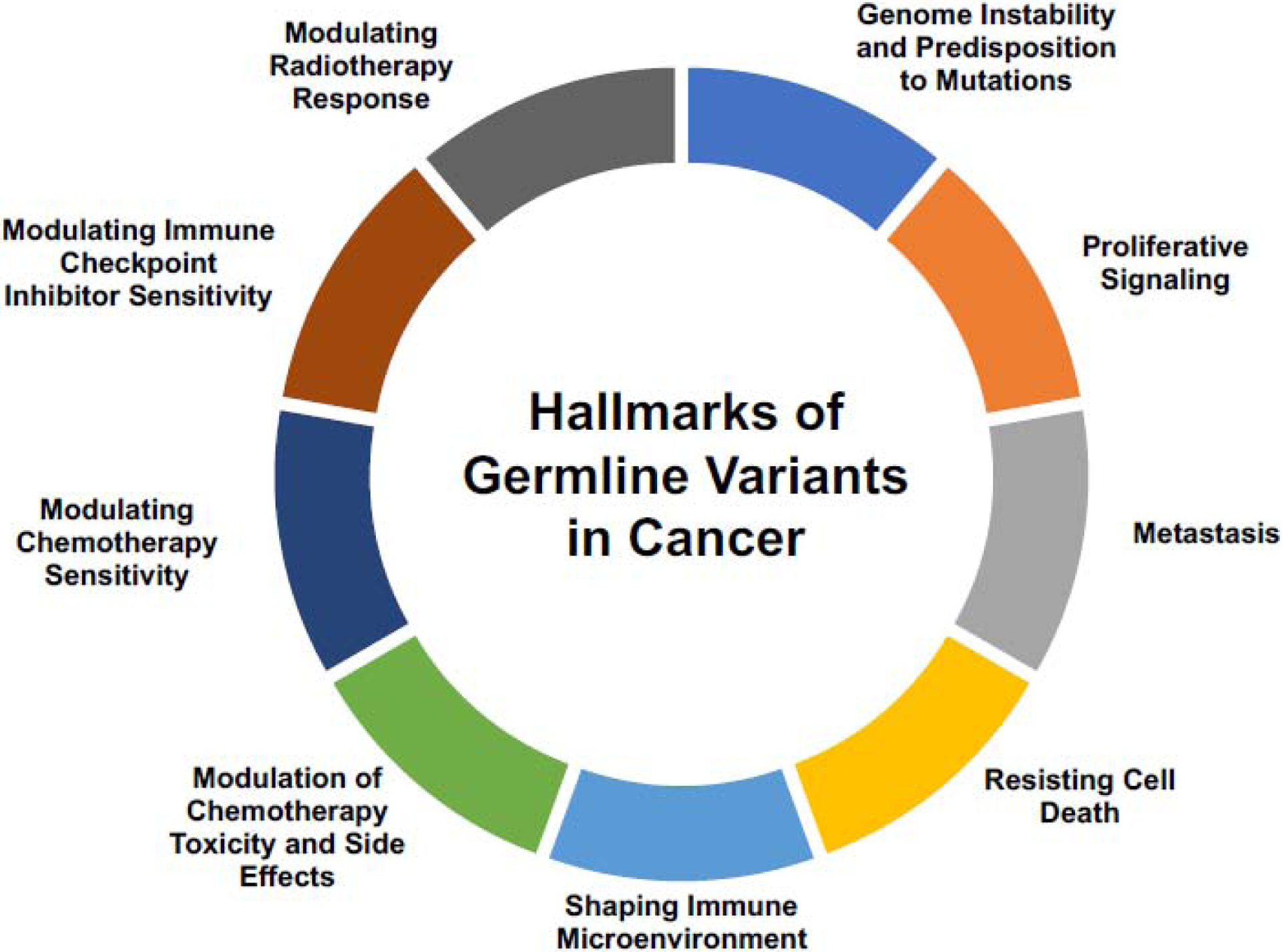
The hallmarks of germline variants in cancer. The molecular mechanisms described in Figure 1 lead to larger phenotypic changes that drive the differences in clinical outcome.
Germline variants perturbing tumor suppressor genes can lead to genome instability that predisposes to further somatic aberrations, a mechanism of tumor progression that is well understood [2, 50]. Similarly, pathogenic germline variants in oncogenes or germline variants that upregulate oncogenic pathways can lead to increased proliferative signaling [2]. A previous analysis found the germline variant rs1800795 in the promoter of Interleukin 6 (IL-6), a pro-inflammatory cytokine, to be associated with increased risk of metastasis in patients with breast cancer, suggesting that metastasis is another area of tumor progression that may be influenced by germline variants that has only begun to be explored.[51] A study by Chatrath et al. found that patients with a truncating germline variant in MAP2K3 exhibited favorable outcome and decreased apoptotic signaling [42]. Additionally, Lim et al. surveyed 24 human cancer types and showed that germline variation accounts for variable immune responses across patients, suggesting that germline variants play an important role in shaping the tumor microenvironment and the immune system’s response to the tumor [52].
Finally, the growing body of work in the area of pharmacogenomics suggests that germline variants modulate responsiveness to chemotherapy, radiotherapy, immune checkpoint inhibitors, and drug toxicity and side effects, and could thus affect tumor progression [25–28].
Closing Remarks
Germline variation has been studied extensively in the context of cancer risk, and this has been beneficial for basic science and clinical practice. A growing body of work now suggests that germline variation also contributes to tumor progression. Germline variants influence tumor progression by causing amino acid changes, altering the expression of nearby genes, and increasing the probability of somatic mutations in the same gene or same pathway. Germline variants have been tied to larger-scale transcriptomic differences, mutational signature enrichments, and splice isoform alterations in tumors. These molecular mechanisms feed into clinically apparent phenotypes, such as increased genome instability, predisposition to further somatic mutations, proliferative signaling, metastasis, resistance to cell death, differences in the immune microenvironment, and modulation of chemotherapy sensitivity, immune checkpoint inhibitor sensitivity, radiotherapy, drug toxicity, and drug side effects.
The study of germline variation in the context of tumor progression has only just begun and there are a large number of outstanding questions and opportunities in the field that require further investigation (Outstanding Questions). The study of germline variation will undoubtedly yield new molecular insights about basic cellular processes in cancer and we expect the prognostic variants will be widely adopted to inform clinical decisions related to cancer prognosis and therapeutic decisions.
Outstanding Questions.
How can germline variants that affect patient outcome be better integrated with other data types to inform the management of cancer patients?
Do the effects of germline variants differ based on environment, race and other covariates?
What are the molecular mechanisms by which germline variants predispose to specific somatic aberrations in cancer?
What are the molecular mechanisms by which germline variants affect tumor progression and patient outcome?
Highlights.
The differences in somatic aberrations between tumors can partially be explained by a patient’s germline variants, suggesting that germline variants influence the somatic mutational landscape of cancer.
Germline variants are associated with patient outcome across a large array of cancers. This suggests that germline variation is clinically relevant in the context of tumor progression.
Molecularly, germline variants can affect tumor progression through amino acid changes, alteration of splicing patterns or expression of genes, influencing the selection for somatic mutations, and causing genome wide mutational enrichment.
Germline variants affect molecular and clinical phenotypes, such as genome instability, proliferative signaling, metastasis, alterations to the immune microenvironment, and modulation of therapeutic response and side effects.
The smaller patient cohorts required to identify germline variants that affect tumor progression and patient outcome provide an outstanding opportunity to accelerate the identification and validation of clinically useful germline variants.
Acknowledgements
Some templates for Figure 1 were retrieved from https://smart.servier.com/ and used under a Creative Commons Attribution 3.0 License. This work was supported by grants from the NIH R01 CA166054, R01 CA60499, and T32 GM007267 (AC).
Glossary:
- Germline Variant
a nucleotide change that is inherited and therefore expected to be present in every cell in the body
- Monogenic Diseases
Disorders caused by genetic variation of large effect in a single gene
- Multifactorial Diseases
Disorders caused by genetic variations of variable effects in a large number of genes that together contribute to the disease phenotype
- Somatic Aberration
An acquired (not inherited) genetic abnormality. These genetic abnormalities are limited to the somatic cells that they occurred in (and the progeny of these cells) and are therefore not found in every cell of any organism
- Alfred Knudson’s two-hit hypothesis
A model for understanding the development of cancer in individuals that carry germline variants in tumor suppressor genes. This model suggests that individuals with one “hit” (or germline variant) in a tumor suppressor gene are at a higher risk of acquiring cancer as they only require a single somatic mutation in the other wild type allele of the same gene to impair the function of that gene
- Retinoblastoma Protein (RB1)
Protein encoded by a tumor suppressor gene which regulates cell cycle progression from G1 to S phase of the cell cycle
- Retinoblastoma
a rare cancer that develops from immature cells of the retina. It classically occurs in children. Mutations of the tumor suppressor gene RB1 substantially increase the risk for the development of retinoblastomas
- Tumor suppressor gene
a gene that regulates the cell in many ways to prevent cancer, such as by ensuring genome stability, repairing DNA damage, inhibiting cell cycle progression, and inducing apoptosis. Mutations in tumor suppressor genes increase the risk for cancer development
- Tumorigenesis
The formation of a cancer
- BRCA1 and BRCA2
DNA repair (tumor suppressor) genes involved with the repair of double-strand breaks. BRCA1 and BRCA2 are frequently mutated in breast and ovarian cancers
- Loss of heterozygosity
The loss of function of one allele of a gene due to mutation
- Myeloproliferative Neoplasms
a set of rare blood cancers characterized by the proliferation of red blood cells, white blood cells, or platelets
- Janus Kinase 2 (JAK2)
A non-receptor tyrosine kinase of the JAK/STAT pathway that promotes the growth and proliferation of cells. Frequently mutated in hematologic cancers
- Epidermal Growth Factor Receptor (EGFR)
A transmembrane protein that acts as a receptor for epidermal growth factor (EGF) and promotes cell proliferation, growth, and survival. Frequently mutated in cancer
- MBD4
a gene that codes for a protein that binds to methylated CpGs and corrects G:T or G:U mismatches in their vicinity
- Retrotransposition
The insertion of genetic components called retrotransposons (also known as Class I transposable elements and transposons via RNA intermediates) into areas of the genome through reverse transcription using an RNA transposition intermediate
- Lower grade glioma
A type of grade II or grade III malignant tumor present in the central nervous system that arise from glial cells
- Capicua transcriptional repressor (CIC)
A transcriptional repressor which negatively regulates MAPK signaling
- Microsatellite instability
Widespread genetic instability resulting from the dysfunction of proteins involved with DNA mismatch repair
- Immune checkpoint blockade
a form of cancer immunotherapy which targets immune checkpoints, which suppress the immune response. Immune checkpoint blockade therapy uses antibodies to block these immune checkpoints, thereby encouraging the activation of the immune system and the subsequent targeting of cancer cells by cells of the immune system
- Single Nucleotide Polymorphism (SNP)
A DNA sequence variation present in a sufficiently large fraction of a population that occurs when a single nucleotide in the genome of an individual differs from the reference genome
- The Cancer Genome Atlas
a cancer genomics program which characterized 33 different types of cancers through the generation and analysis of multi-omic data
- Isocitrate Dehydrogenase (IDH)
A gene that normally codes for an enzyme that catalyzes the oxidative decarboxylative of isocitrate. When mutated in the context of cancer, this enzyme acquires a novel function in which it produces (D)-2-hydroxyglutarate from alpha-ketoglutarate
- Oncogenes
Genes that have the potential of causing cancer when they acquire gain-of-function mutations or are highly expressed (eg, RAS, MYC, TRK)
- Gain of function mutations
Mutations that increase the activity of a protein or enable the protein to perform a novel function
- Loss of function mutations
Mutations that alter the functions of proteins coded (typically used in the context of proteins coded for by tumor suppressor genes) in such a way that they can no longer perform their normal cellular functions
- Expression Quantitative Trait Loci
A genetic loci with genetic variants that are associated with variations in the expression levels of mRNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors are employees of the University of Virginia, which has applied for a patent on the use of germline variants for management of cancer patients.
References
- 1.Lichtenstein P et al. (2000) Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343 (2), 78–85. [DOI] [PubMed] [Google Scholar]
- 2.Huang KL et al. (2018) Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 173 (2), 355–370.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger MF and Mardis ER (2018) The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol 15 (6), 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chirita-Emandi A et al. (2020) Challenges in reporting pathogenic/potentially pathogenic variants in 94 cancer predisposing genes - in pediatric patients screened with NGS panels. Sci Rep 10 (1), 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sud A et al. (2017) Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer 17 (11), 692–704. [DOI] [PubMed] [Google Scholar]
- 6.Li MM et al. (2017) Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 19 (1), 4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J et al. (2015) Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med 373 (24), 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodmer W and Tomlinson I (2010) Rare genetic variants and the risk of cancer. Curr Opin Genet Dev 20 (3), 262–7. [DOI] [PubMed] [Google Scholar]
- 9.Zhao M et al. (2014) The Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer 120 (15), 2299–307. [DOI] [PubMed] [Google Scholar]
- 10.Tomasetti C and Vogelstein B (2015) Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347 (6217), 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter H et al. (2017) Interaction Landscape of Inherited Polymorphisms with Somatic Events in Cancer. Cancer Discov 7 (4), 410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahman N (2014) Realizing the promise of cancer predisposition genes. Nature 505 (7483), 302–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger AH et al. (2011) A continuum model for tumour suppression. Nature 476 (7359), 163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanchi KL et al. (2014) Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun 5, 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones AV et al. (2009) JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat Genet 41 (4), 446–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilpivaara O et al. (2009) A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet 41 (4), 455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell PJ (2009) Somatic and germline genetics at the JAK2 locus. Nat Genet 41 (4), 385–6. [DOI] [PubMed] [Google Scholar]
- 18.Olcaydu D et al. (2009) A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet 41 (4), 450–4. [DOI] [PubMed] [Google Scholar]
- 19.Liu W et al. (2011) Functional EGFR germline polymorphisms may confer risk for EGFR somatic mutations in non-small cell lung cancer, with a predominant effect on exon 19 microdeletions. Cancer Res 71 (7), 2423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(2020) Pan-cancer analysis of whole genomes. Nature 578 (7793), 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamidi TKK et al. (2019) Mapping the Germline and Somatic Mutation Interaction Landscape in Indolent and Aggressive Prostate Cancers. J Oncol 2019, 4168784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y et al. (2018) Integrated case-control and somatic-germline interaction analyses of melanoma susceptibility genes. Biochim Biophys Acta Mol Basis Dis 1864 (6 Pt B), 2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramroop JR et al. (2019) Germline Variants Impact Somatic Events during Tumorigenesis. Trends Genet 35 (7), 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatrath A et al. (2019) The Germline Variants rs61757955 and rs34988193 Are Predictive of Survival in Lower Grade Glioma Patients. Mol Cancer Res 17 (5), 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Relling MV and Evans WE (2015) Pharmacogenomics in the clinic. Nature 526 (7573), 343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinshilboum RM and Wang L (2017) Pharmacogenomics: Precision Medicine and Drug Response. Mayo Clin Proc 92 (11), 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daly AK (2017) Pharmacogenetics: a general review on progress to date. Br Med Bull 124 (1), 65–79. [DOI] [PubMed] [Google Scholar]
- 28.Romero Lagunes ML and Vera Badillo FE (2019) Design and Implementing Pharmacogenomics Study in Cancer. Adv Exp Med Biol 1168, 43–77. [DOI] [PubMed] [Google Scholar]
- 29.Ghandi M et al. (2019) Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569 (7757), 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tate JG et al. (2019) COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 47 (D1), D941–d947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battaglin F et al. (2018) Microsatellite instability in colorectal cancer: overview of its clinical significance and novel perspectives. Clin Adv Hematol Oncol 16 (11), 735–745. [PMC free article] [PubMed] [Google Scholar]
- 32.Duffy MJ and Crown J (2019) Biomarkers for Predicting Response to Immunotherapy with Immune Checkpoint Inhibitors in Cancer Patients. Clin Chem 65 (10), 1228–1238. [DOI] [PubMed] [Google Scholar]
- 33.Arora S et al. (2019) Existing and Emerging Biomarkers for Immune Checkpoint Immunotherapy in Solid Tumors. Adv Ther 36 (10), 2638–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Summers MG et al. (2020) Comprehensive analysis of colorectal cancer-risk loci and survival outcome: A prognostic role for CDH1 variants. Eur J Cancer 124, 56–63. [DOI] [PubMed] [Google Scholar]
- 35.Marasigan V et al. (2019) Melanoma susceptibility variant rs869330 in the MTAP gene is associated with melanoma outcome. Melanoma Res 29 (6), 590–595. [DOI] [PubMed] [Google Scholar]
- 36.Shu X et al. (2018) Germline genetic variants in somatically significantly mutated genes in tumors are associated with renal cell carcinoma risk and outcome. Carcinogenesis 39 (6), 752–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stotz M et al. (2017) Cancer Stem Cell Gene Variants in CD44 Predict Outcome in Stage II and Stage III Colon Cancer Patients. Anticancer Res 37 (4), 2011–2018. [DOI] [PubMed] [Google Scholar]
- 38.Świerniak M et al. (2016) Association between GWAS-Derived rs966423 Genetic Variant and Overall Mortality in Patients with Differentiated Thyroid Cancer. Clinical Cancer Research 22 (5), 1111–1119. [DOI] [PubMed] [Google Scholar]
- 39.Koessler T et al. (2009) Common germline variation in mismatch repair genes and survival after a diagnosis of colorectal cancer. Int J Cancer 124 (8), 1887–91. [DOI] [PubMed] [Google Scholar]
- 40.Mosquera Orgueira A et al. (2019) The association of germline variants with chronic lymphocytic leukemia outcome suggests the implication of novel genes and pathways in clinical evolution. BMC Cancer 19 (1), 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macauda A et al. (2018) Inherited variation in the xenobiotic transporter pathway and survival of multiple myeloma patients. Br J Haematol 183 (3), 375–384. [DOI] [PubMed] [Google Scholar]
- 42.Chatrath A et al. (2020) The pan-cancer landscape of prognostic germline variants in 10,582 patients. Genome Med 12 (1), 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.(2013) The Genotype-Tissue Expression (GTEx) project. Nat Genet 45 (6), 580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y et al. (2019) The Cancer-Associated Genetic Variant Rs3903072 Modulates Immune Cells in the Tumor Microenvironment. Front Genet 10, 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JH et al. (2011) Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci U S A 108 (44), 18026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Page C et al. (2019) Clinicopathological features of women with epithelial ovarian cancer and double heterozygosity for BRCA1 and BRCA2: A systematic review and case report analysis. Gynecol Oncol. [DOI] [PubMed] [Google Scholar]
- 47.Musa J et al. (2019) Cooperation of cancer drivers with regulatory germline variants shapes clinical outcomes. Nat Commun 10 (1), 4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mastorakos P et al. (2019) Tumor to Cerebellar Peduncle T2-Weighted Imaging Intensity Ratio Fails to Predict Pituitary Adenoma Consistency. J Neurol Surg B Skull Base 80 (3), 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanahan D and Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144 (5), 646–74. [DOI] [PubMed] [Google Scholar]
- 50.Lu C et al. (2015) Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun 6, 10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abana CO et al. (2017) IL-6 variant is associated with metastasis in breast cancer patients. PLoS One 12 (7), e0181725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim YW et al. (2018) Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration. Proc Natl Acad Sci U S A 115 (50), E11701–e11710. [DOI] [PMC free article] [PubMed] [Google Scholar]


