Abstract
Meiotic drive, the non-Mendelian transmission of chromosomes to the next generation, functions in asymmetric or symmetric meiosis across unicellular and multicellular organisms. In asymmetric meiosis, meiotic drivers act to alter a chromosome’s spatial position in a single egg. In symmetric meiosis, meiotic drivers cause phenotypic differences between gametes with and without the driver. Here we discuss existing models of meiotic drive, highlighting the underlying mechanisms and regulation governing systems for which the most is known. We focus on outstanding questions surrounding these examples and speculate on how new meiotic drive systems evolve and how to detect them.
Keywords: Meiotic drive, Meiosis, Selfish genetic elements, Germ cells
Germ cells are the only cells in the body that contribute their DNA to the next generation. A feature distinguishing germ cells from somatic cells is their ability to undergo meiosis, the process by which a cell divides to produce haploid gametes (e.g., eggs, sperm and spores) (Fig. 1). The union of gametes leads to the formation of a zygote, which ultimately will develop its own gametes. Meiotic chromosome segregation gives chromosomes a 50% chance of being transmitted to each offspring. However, germ cells are susceptible to hijacking by selfish genetic elements—DNA sequences that increase their associated chromosomes transmission to more than 50% of offspring. The wide variety of selfish genetic elements and their evolutionary dynamics have been reviewed elsewhere [1–4]. This review will focus on meiotic drive, which we define as selfish genetic elements (meiotic drivers) that act to increase chromosome transmission from meiosis to the formation of the zygote, in males, females and fungal spores [5–7].
Fig. 1.
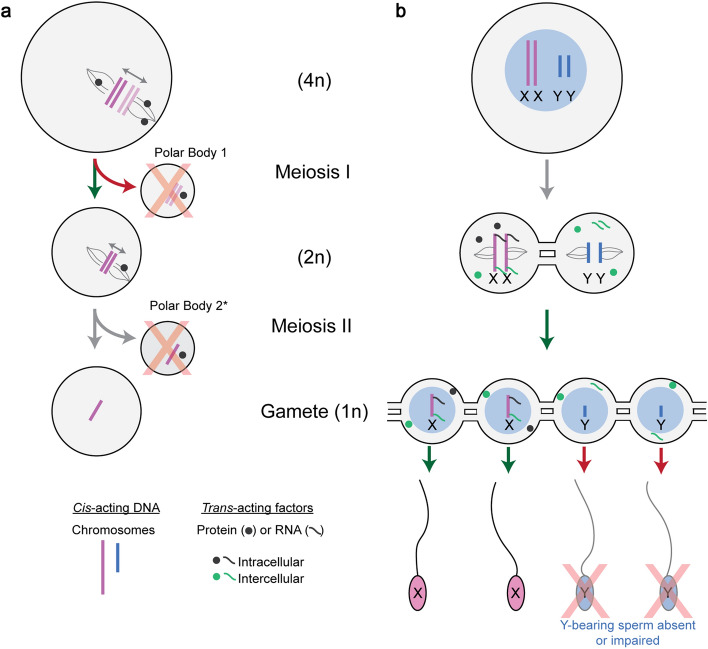
The defining features of meiotic drive in asymmetric and symmetric meiosis. a Females undergo asymmetric meiosis—a single gamete is produced from a single round of meiosis. The driving cis-acting chromosomes (dark pink) biases its retention to the egg by interacting with the inward-facing egg pole (green arrow). The non-driving chromosome (light pink) binds the outward facing cortical pole and is extruded to a polar body (red arrow) which is degraded (red X). * The second polar body is extruded upon fertilization. b Males undergo symmetric meiosis—four gametes are produced from a single round of meiosis. Male meiotic drive systems bias fertilization by increasing the relative abundance of sperm carrying the driving chromosome (green arrow), or by decreasing the fitness of sperm with the non-driving chromosome (red arrow). Gray mRNA and protein represent X-linked trans-acting factors which remain in X-bearing cells (e.g., intracellular). Green mRNA and protein represent X-linked trans-acting factors which are shared and present in Y-bearing cells (e.g., intercellular) via cytoplasmic bridges. Cytoplasmic bridges are established prior to meiosis, in spermatogonia, and connect cells throughout meiosis and after meiosis. For simplicity, a single meiotic cell and the products of meiosis are shown.
Meiotic drivers benefit themselves while typically conferring negative effects on the rest of the genome. To dampen the negative effects of meiotic drivers, suppressors evolve and are selected for. The dynamics of drivers versus suppressors over evolutionary time can dramatically shape genome evolution and influence the survival of a species. In this review, we will focus on meiotic drive systems in their most simplified form, two component systems, with a meiotic driver and a substrate the meiotic driver utilizes to bias chromosome transmission (Fig. 1). In order to bias transmission of one chromosome over the other, one of the two components must be a trans-acting factor (RNA or protein). The trans-acting factor biases chromosome transmission by interaction with either: (1) a cis-acting element (DNA) (Fig. 1a) or (2) another trans-acting factor (Fig. 1b). In many cases, the two components are in tight genetic linkage (on the same chromosome with low/no recombination between them), allowing the two components to co-evolve at the DNA sequence level without being separated by recombination. A universal requirement for meiotic drivers is a DNA sequence (either cis- or trans-acting) difference that distinguishes the driving chromosome from its wild-type non-driving counterpart. These fundamental principles of meiotic drive systems are shaped by whether a germ cell undergoes symmetric or asymmetric meiosis.
Within a species, meiotic drivers generally function exclusively in the female or male germline, but not both [6, 8]. Specifically, we propose that it is the asymmetry or symmetry of meiosis that defines the biological and mechanistic constraints placed on the evolution of meiotic drivers (Fig. 1). Females undergo asymmetric meiosis, producing a single haploid egg and polar bodies (meiotic haploid products that are not transmitted to the next generation) (Fig. 1). In asymmetric female meiosis, meiotic drivers act to increase segregation of a particular chromosome homolog to the single egg (Fig. 1a). Males and sporulators undergo symmetric meiosis producing four haploid sperm (or spores) during each meiotic division. In symmetric meiosis, meiotic drivers act between cells to establish a fitness differential between the, otherwise equivalent, sperm or spores (Fig. 1b). Here we discuss recent advancements in understanding asymmetric and symmetric meiotic drive with a particular focus on the mechanistic constraints governing these systems.
Asymmetric meiosis
In asymmetric meiosis, chromosome segregation into the egg is spatially determined, producing only one haploid oocyte from meiosis. The remaining meiotic products and chromosomes form additional non-transmissive cells (e.g., polar body). For example, in mouse oogenesis, meiotic spindle poles are initially at the center of the cell and then migrate to the periphery. At the periphery, chromosomes attached to the outward facing spindle (cortical pole) are extruded to a polar body, while those attached to the inward facing spindle (egg pole) are retained in the egg (Fig. 1a) [9]. Meiotic drivers act to bias the orientation of chromosomes toward the egg pole, ensuring its transmission to offspring. To accomplish this phenomenon, two features are needed: (i) a cis-acting DNA sequence difference that distinguishes the chromosomes, and (ii) a trans-acting molecule that distinguishes the egg pole from the cortical pole, to spatially orient the chromosomes [10, 11]. Cis-acting centromeric and pericentromeric DNA sequences act as the attachment point between a chromosome and spindle pole. Consequently, alterations in centromeric sequences impact meiotic drive (also known as “centromeric drive”). Centromeric drive has been observed in mice (Mus hybrids) [11–13], Drosophila [14] and monkeyflower (Mimulus hybrids) [15, 16]. Similar spatial principles also govern chromosome segregation in maize, where differences in cis-acting non-centromeric DNA sequences cause meiotic drive [17–20]. A common theme emerging from both centromeric and non-centromeric drive is the involvement of repetitive sequences. Centromeric drive in mice and non-centromeric drive in maize are the experimental systems with the greatest mechanistic understanding, as discussed below.
Centromeric drive
Centromeric meiotic drive results from cis-acting DNA sequence differences in and around the centromere that differentially establish kinetochore-spindle pole interactions and bias chromosome segregation (Figs. 1a, 2a, b). The differential recruitment of proteins between chromosome pairs affects the segregation of chromosomes to either the egg pole or cortical spindle pole (Fig. 2a, b). The chromosome bound to the egg pole is referred to as the “driving” chromosome, whereas the chromosome bound to the cortical pole is the “non-driving” chromosome. In mouse, there are two well-characterized cases of centromeric drive. Each case has been studied by crossing diverged mouse strains to generate hybrid mice [11–13]. Both cases rely on underlying differences in centromere sequence repeat number (cis-acting elements) and differences in spindle functions (trans-acting factors) to influence a chromosomes orientation and fate (Fig. 2a, b) [10–13]. While both cases utilize chromosome reorientation to facilitate drive, they differ in the timing of this reorientation relative to spindle migration [10]. Additionally, in both cases, the driving chromosomes have accumulated minor satellite repeats (Fig. 2a, b) [10, 11].
Fig. 2.
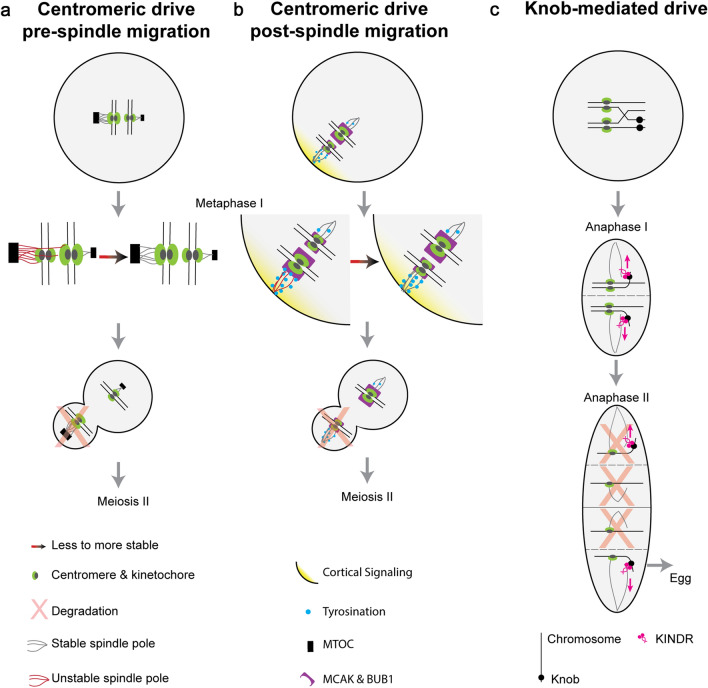
Models of Asymmetric Meiotic Drive. This figure depicts the mechanistic understanding of two examples of centromeric drive (a and b) and one example of non-centromeric drive (c). a An example of female centromeric drive in hybrid mice where cis-acting chromosome reorientation occurs before trans-acting spindle migration. In this system, larger MTOCs (black box) give rise to denser spindle poles (gray line) which preferentially interact with larger kinetochores (green oval). If this favorable interaction is not initially established (red line), then proteins involved in fixing erroneous microtubule attachments are recruited and chromosomes reorient to the more favorable interaction (red–black arrow). Spindles migrate to the periphery, and the outward-facing larger kinetochore is extruded to the polar body and degraded (red X). b Another example of female centromeric drive in hybrid mice where chromosomes reorientation occurs after spindle migration. Cortical signaling (yellow gradient) leads to an enrichment of tyrosination (blue circle) on cortical spindle poles. Spindle pole tyrosination is less stable (red lines) on larger kinetochores (green) with more BUB1 kinase and MCAK (purple). If not in the more stable orientation, then proteins involved in fixing erroneous microtubule attachments cause chromosomes to reorient to the stable orientation (red–black arrow), causing the smaller kinetochore to be extruded to the polar body (red X). c Maize knob-mediated drive requires the kinesin KINDR (pink) which binds knob repeats (black circles) and migrates along the spindle microtubules toward the outward spindle poles (pink arrow), including the distal cell which becomes the egg
In one case of mouse centromeric drive, chromosomes reorient prior to spindle migration [11]. Centromeric sequence differences differentially establish the kinetochores of driving versus non-driving chromosomes. The cis-acting centromeric sequence differences are coupled to differentially established trans-acting factors at the spindle poles [11]. Specifically, the microtubule organizing centers (MTOCs) differ in their density, suggesting differences in protein abundance. Because MTOCs give rise to the spindle poles, the spindles themselves also exhibit different densities (Fig. 2a). However, it is unclear how MTOCs of different densities are established as they are composed of trans-acting proteins [11]. The centromeric minor satellite repeats are twofold higher on the driving chromosome in hybrid mice, and the pericentromeric major satellite repeats are almost undetectable [11]. These sequence differences between centromeres in hybrid mice are not known, but they differentially recruit trans-acting proteins and result in distinguishable kinetochores (Fig. 2a) [11]. Interactions between the spindle poles and kinetochores are most stable when proportional—in other words, when the larger kinetochore is attached to the denser spindle pole (Fig. 2a) [11]. When disproportionate, the chromosomes reorient in an aurora kinase-dependent manner [11]. The correction of erroneous microtubule interactions enables the chromosomes to reorient, and then form more stable kinetochore–spindle interactions during prometaphase (Fig. 2a) [11]. When the spindle poles migrate to the cortex, the denser spindle pole preferentially faces the cortex causing the chromosome with the larger kinetochore to be extruded into a polar body (Fig. 2a) [11]. Clearly, additional research must target understanding the molecular basis of: (1) the instability between the smaller kinetochore and dense spindle pole and (2) how the denser spindle pole is preferentially directed to the cortex.
In the second mouse centromere drive system, the chromosomes reorient after the spindles have migrated to the cortex [10]. Similar to the previous example of centromeric drive, cis-acting centromeric and pericentromeric sequence differences exist and differentially establish trans-acting kinetochores. In this case, the spindle poles are differentiated by cortical signaling which results in an enrichment of tyrosinated α-tubulin on the cortical spindle microtubules [10]. While tyrosination is known to be dependent on CDC42 cortical signaling, the proteins linking CDC42 and tubulin tyrosine kinase ligase (TTL), which catalyzes tyrosination of α-tubulin, remain unknown. Smaller kinetochores interact more stably with tyrosinated microtubules than larger kinetochores (Fig. 2b) [10]. This difference in stability occurs because larger kinetochores recruit more BUB1 kinase and consequently more microtubule destabilizing proteins, including MCAK (mitotic centromere-associated kinesin) (Fig. 2b) [21]. MCAK, a microtubule depolymerase, acts more efficiently on microtubules with tyrosination. Therefore, larger kinetochores with more MCAK form less stable interactions with the tyrosinated cortical microtubules and reorient to the egg pole, promoting their segregation to the egg (Fig. 2b) [21, 22]. A greater understanding of this model could be gained by determining: (1) the underlying centromeric sequence differences by long-read DNA sequencing and (2) identifying the factors involved in TTL recruitment [23, 24]. It is likely that other cases of centromeric drive exist both within the Mus-lineage and across mammals, and these may use distinct molecular mechanisms [11, 13, 21].
Non-centromeric drive
We define other examples of female asymmetric meiotic drive as non-centromeric drive as they involve cis-acting sequences outside of the native centromere and pericentromeric region. Examples of non-centromeric drive include mouse R2d2 [25, 26], HSR (homogeneously stained region) [27] and Om (ovum mutant) [28, 29] and, in rice, S5 [30, 31]. However, the non-centromeric drive system with the greatest mechanistic understanding is knob-mediated meiotic drive in maize. A variant form of maize chromosome 10 (Ab10) contains additional repetitive heterochromatic sequences outside the centromere, known as knobs, which function as cis-acting neocentromeres and interact with spindle microtubules independently of a kinetochore (Fig. 2c) [19]. Similar to mammalian oogenesis, a single egg is ultimately produced during meiosis in maize. However, instead of polar bodies, meiosis results in four meiotic products, where only the distal cell ultimately becomes the egg. Chromosomes with heterochromatic knobs migrate faster along the spindle pole towards the outer spindle pole, promoting their segregation to the distal cell (Fig. 2c) [17, 20]. Knob-mediated meiotic drive is dependent on a trans-acting kinesin protein known as Kinesin driver (KINDR), that is encoded on the Ab10 chromosome and localizes to cis-acting 180 bp knob repeats, on both Ab10 and other chromosomes [18]. KINDR binds knob sequences and, subsequently, moves knob-containing chromosomes towards the outer spindle pole in the direction of chromosome segregation (Fig. 2C) [18]. The established model of knob-mediated meiotic drive segregates knobs to the distal and proximal cells of the meiotic tetrad aided by a recombination event between the centromere and knob (Rhoades Model, Fig. 2c) [32]. The factors that cause KINDR to move knob-containing chromosomes towards the outer spindle poles, as opposed to the inner spindle poles, are not known. While the KINDR encoding sequences and the knob 180 bp repeats are genetically linked, KINDR acts in trans and, therefore, other chromosomes containing the 180 bp repeat are differentially segregated. Maize meiotic drive has resulted in the evolutionarily recent acquisition of ~500 Mb of repetitive sequence in its genome [18]. Maize also contains TR-1 neo-centromeric repeats, which are functionally distinct from the 180-bp repeat mechanism, and may serve to suppress 180-bp drive [17, 18, 33, 34]. Genetic screens have identified suppressors of Ab10 meiotic drive and highlight the importance of tight genetic linkage. Genetic linkage with knobs is necessary to ensure the suppressor functions following the recombination event between the centromere and knobs (Fig. 2c) [35]. Through a similar logic, enhancers of drive would also reside in tight genetic linkage to the initial meiotic driver.
Symmetric meiosis
Symmetric meiosis leads to four meiotic products, sperm or spores, half of which contain the meiotic driver. To bias transmission in a symmetric meiotic drive system, gametes with and without the driver must be molecularly distinct from each other (e.g., carrying an X or Y chromosome, Fig. 1b). Differences between gametes with and without the selfish genetic element manifest in one of two ways. First, functional differences such as reduced motility of sperm. Second, failure to develop to maturity leading to differential representation, such as sperm- or spore-killing. Fundamental to both scenarios are molecular differences (DNA, RNA, and/or protein) distinguishing the drive and non-drive containing gametes (Figs. 1b, 3, 4).
Fig. 3.
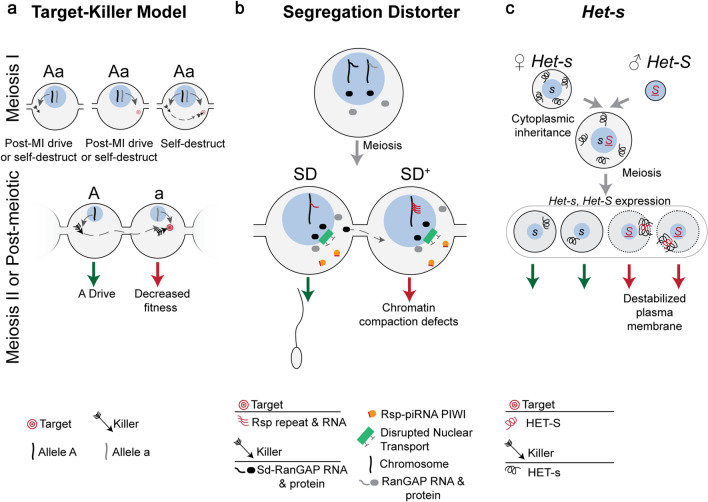
Symmetric Meiotic Drive—Target–Killer. This figure depicts the theoretical importance of intracellular versus intercellular regulation on target–killer meiotic drive systems (a), and the mechanisms underlying two target–killer meiotic drivers (b and c). a The killer protein (black arrow, encoded on the “A” haplotype) targets, directly or indirectly, the target (red bullseye, encoded on the “a” haplotype). During meiosis I (MI), expression of only the killer before or during MI can result in post-MI drive, if the target is a trans-acting factor expressed during or after meiosis II (MII). If the target is a cis-acting sequence, expression of the killer before or during MI could cause the driver to self-destruct. If only a trans-acting target is expressed during MI, post-MI drive occurs if the killer is expressed during or after MII, assuming the target acts intracellularly. However, if the target is partitioned into all MII cells and is therefore intercellular, then expression of the killer will result in the system self-destructing. If a trans-acting killer is expressed during MI and a cis-acting or trans-acting target are accessible, then the system will self-destruction. The target and killer are present in different cells and on different haplotypes in order to not self-destruct, as would be the case if both were expressed in meiotic cells. Expression of the target or killer following homologous chromosome separation in MII prevents self-destruction and results in meiotic drive (green and red arrows). Post-meiosis II expression of the killer necessitates sharing (dashed gray arrow) through cytoplasmic bridges, the target should not be shared in order to prevent self-destruction. b Drosophila melanogaster Segregation Distorter (SD) encodes the killer Sd-RanGAP. One model proposes that Sd-RanGAP mislocalization disrupts piRNA-based silencing (orange) of the target, the high copy number Rsp (red lines) resulting in chromatin compaction defects and SD drive. c The female strain of P. anserina encodes the killer allele, Het-s, and HET-s protein (black curly line) is therefore present upon gamete fusion as the cytoplasm (gray surrounding blue nucleus) is maternally inherited. The male strain’s Het-S allele and female Het-s are expressed following meiosis where they complex in Het-S spores and integrate and destabilize the plasma membrane (red arrow), resulting in Het-s drive (green arrow). Cells are contained within an ascus (large gray oval)
Fig. 4.
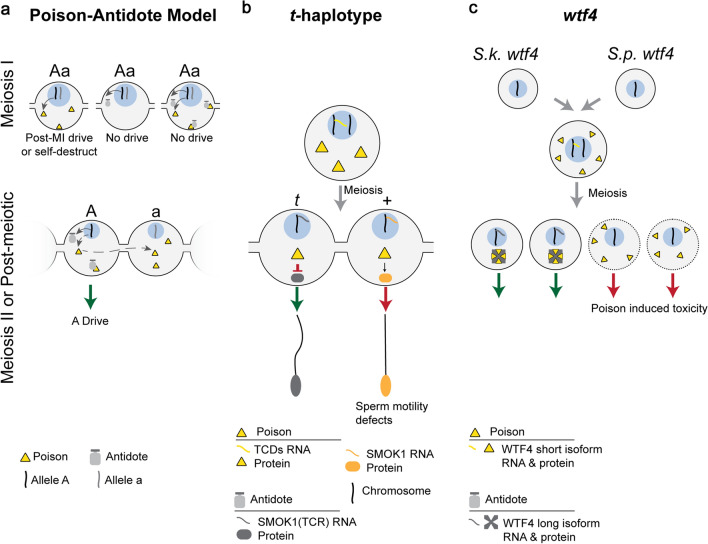
Symmetric Meiotic Drive—Poison-Antidote. This figure depicts the theoretical importance of intracellular versus intercellular regulation on poison–antidote meiotic drive systems (a), and the mechanisms underlying two poison-antidote meiotic drivers (b and c). a The poison protein (yellow triangle) and antidote (gray shape) are encoded by the same haplotype (“A”) in close genetic proximity. The antidote acts intracellularly (“A” haplotype) to spare driving chromosomes from the effects of the intercellular poison. Expression of the poison before or during MI can result in post-MI drive, if the poison acts specifically in MII or post-meiosis. However, if the poison acts on a biological pathway present before or during MI, expression before or during MI leads to self-destruction. Expression of the antidote before or during MI would result in it being partitioned into and consequently sparing all daughter cells, preventing drive. Expression of both the poison and antidote before homologous chromosomes separate would result in the antidote neutralizing the poison and preventing drive. Meiosis II or later expression of the poison requires that the poison is shared through cytoplasmic bridges (dashed gray arrow) to poison the other cell containing the “a” haplotype, resulting in A drive (green arrow). b In the mouse t-haplotype, the poisons, TCDs (yellow triangle), are expressed before meiosis and disrupt the post-meiotically expressed SMOK1 (orange oval) resulting in flagellar dysregulation (red arrow). The antidote, SMOK1(TCR) (gray oval), is a dominant negative variant of SMOK1 and is insensitive to TCD induced dysregulation, resulting in t-haplotype drive (green arrow). c In Schizosaccharomyces, the wtf4 gene encoded both the pre-meiotically expressed poison (yellow triangle) and the post-meiotically expressed antidote (gray shape). These represent distinct isoforms of wtf4. The antidote sequesters poison into distinct subcellular regions preventing its toxicity and results in drive (green arrow)
Symmetric meiotic drive systems rely on the presence of molecular differences; consequently, they function after homologous chromosomes have been separated in meiosis I. In mammalian spermatogenesis, however, meiotic and post-meiotic germ cells are connected by cytoplasmic bridges that enable the sharing of trans-acting factors (RNA and protein gene products) [36–39]. Gene product sharing makes genetically distinct cells (e.g., X- versus Y-bearing sperm) phenotypically or functionally diploid, mitigating molecular distinctions at the RNA and protein level [36, 37, 40]. Therefore, the generation and maintenance of phenotypic differences between sperm (e.g., with or without a meiotic driver) implies there is a spatial regulation among connected cells. This spatial regulation between cells is governed by the restriction of trans-acting factors transit via cytoplasmic bridges. We therefore define trans-acting factors as intercellular when they are shared through cytoplasmic bridges and are present in the cells where they are not transcribed/translated (Fig. 1b, green mRNA and protein). In contrast, we use intracellular to define trans-acting factors which are not shared across cytoplasmic bridges (Fig. 1b, gray mRNA and protein). Despite the prevailing view that cytoplasmic bridges facilitate trans-acting factor sharing, certain factors are not shared, indicating that sharing through cytoplasmic bridges can be a regulated process [41–45]. We highlight both a male and spore-producer for two models of symmetric meiotic drive (target–killer and poison–antidote), placing particular focus on the mechanism of meiotic drive and the intracellular versus intercellular regulation of trans-acting factors.
Target–killer
In the target–killer model, the driving chromosome encodes a trans-acting killer that acts intercellularly on a target in the non-drive containing cell to disrupt a cellular process (e.g., developmental arrest, perturbed motility, apoptosis, etc.). The cis-acting or trans-acting target is encoded on the non-driving chromosome and must exist intracellularly. To prevent the killer from self-destructing, the killer-encoding and target-encoding sequences must be on distinct haplotypes (Fig. 3a). To prevent the killer-encoding and target-encoding sequences from being on the same haplotype, the two loci must be in tight genetic linkage, making recombination events infrequent (Fig. 3a). The killer component can be expressed before meiosis as long as the target is not present and vice versa (Fig. 3a). If the target is a gene product expressed before meiosis, it must remain associated with the non-driving chromosome and expression of the killer must occur after meiosis I to enable the killer to exclusively target the non- driving chromosome (Fig. 3a). Below we will discuss two target–killer systems, the Drosophila melanogaster Segregation Distorter (SD) [46] and the fungus Podospora anserina Het-s [47–49].
Drosophila melanogaster—segregation distorter
Segregation Distorter (SD) is an autosomal meiotic drive system present on chromosome 2 in Drosophila melanogaster. Males heterozygous for SD and wild-type chromosome 2 (denoted SD/SD+) transmit the SD chromosome up to ~ 95% of the time. Meiotic drive results from the disrupted development of sperm carrying the wild type SD+ chromosome 2 [46]. The SD locus encodes an intercellular trans-acting killer. The SD gene product is a truncated duplicate of wild-type Ran GTPase activating protein that is expressed before meiosis and mislocalizes to the nucleus, Sd-RanGAP (Fig. 3b) [50, 51]. The truncated Sd-RanGAP protein acts intercellularly by targeting the intracellular cis-acting 120-bp pericentric repeat, Responder (Rsp) [52–54]. Though Rsp sequences are present on both SD and SD+ chromosomes, the sensitivity of Rsp correlates with copy number. Wild-type SD+ chromosomes contain many copies and are sensitive (> 700 copies, RspS allele), whereas SD chromosomes are insensitive, with few copies (< 20, RspI allele) (Fig. 3b) [46, 52–54]. Both the proximity of SD to the centromere and the presence of inversions suppress recombination between the killer locus (Sd-RanGAP) and the target locus (RspS), keeping Sd-RanGAP and RspI physically linked and preventing the killer from targeting itself [46].
How Sd-RanGAP disrupts SD+ cellular development is unknown, and likewise, it is unclear whether Sd-RanGAP interacts with RspS directly or indirectly [46]. Multiple models for SD have been proposed, and all assume a disruption in nuclear transport arising from a disrupted RAN protein gradient, normally established by the cytoplasmic localization of wild-type RanGAP and nuclear RanGEF (Fig. 3b) [6, 46, 55]. Sd-RanGAP mislocalizes to the nucleus, and nuclear mislocalization of wild-type RanGAP is sufficient to cause drive. This result implicates the nuclear localization of the intercellular Sd-RanGAP in the mechanism underlying SD drive [56]. Phenotypically, SD+ sperm fail to undergo chromatin compaction, a process necessary for proper sperm development (Fig. 3b) [57]. One possible mechanism linking these two observations is that Rsp-derived piRNAs (trans-acting) are needed to silence the Rsp locus (cis-acting) and allow proper chromatin compaction, and disruptions in nuclear transport prevent production and/or localization of piRNAs silencing complexes [6, 56]. However, it is unclear whether piRNA production and trafficking rely on the RAN gradient. If piRNA production is dependent on a RAN gradient, then it is surprising that more widespread problems do not arise as piRNAs play a critical role in transposon silencing [58]. Sd-RanGAP protein likely affects all sperm, as it is expressed before meiosis. Disrupted Rsp silencing, therefore, would have greater effects on high copy number RspS than low copy number RspI. Supporting this mechanism, small Rsp RNAs are associated with the germline-specific piRNA silencing proteins [59]. Additionally, mutations in the PIWI protein, Aubergine, enhance the SD phenotype, implicating piRNA-based silencing as a component of the SD mechanism [60].
Podospora anserina—Het-s
Het-s drive in the fungus Podospora anserina utilizes two different alleles, at the same locus, that produce two variants of the same trans-acting proteins, HET-s (killer) and HET-S (target) [47]. The two alleles (Het-s and Het-S) become heterozygous when two different strains of P. anserina fuse [47]. The HET-s protein product, which acts intercellularly, can undergo a conformational switch to form a prion (killer) that is toxic when it interacts with its intracellular target protein product, HET-S (Fig. 3c) [47]. It is not known what causes HET-s protein to switch from the wild type to prion form. The prion-forming interaction causes the HET-S protein to integrate into and destabilize the plasma membrane of het-S spores (Fig. 3c) [48]. Given the similarity in sequence between Het-s and Het-S, it is surprising that HET-s is not toxic by itself. One possibility for evading self-toxicity is that the transmembrane domain of HET-s is less hydrophobic and therefore less able to permeate the plasma membrane. Furthermore, meiotic drive only occurs when the Het-s genotype comes from the “female” parental strain, since fused cytoplasm is almost entirely maternally inherited and thus constitutes intracellular HET-s protein (Fig. 3c) [47]. Since Het-s and Het-S are transcriptionally silenced until after meiosis when individual spores are formed [49], the maternal transmission of the cytoplasmic contents ensures HET-s segregates to all resulting spores to function intercellularly. When Het-s and Het-S are then expressed in individual spores, Het-S spores produce HET-S protein which is intracellular and complexes with the maternally transmitted HET-s prion, specifically killing Het-S spores (Fig. 3c) [49].
Poison-antidote
In the poison–antidote model, the driving chromosome encodes both a poison and an antidote in tight genetic linkage. The poison acts intercellularly and is toxic to all cells. The antidote neutralizes the effects of poison but must act intracellularly to preferentially spare the cell containing the driving chromosome. Tight genetic linkage between the poison and antidote prevents recombination, and the driver from poisoning itself. The poison can either be: (1) shared across cytoplasmic bridges following meiosis, or (2) expressed during meiosis I and persist into non-driving cells after meiosis. Importantly, if expressed before or during meiosis I, the poison cannot function until after homologous chromosomes have separated, otherwise the poison will self-destruct (Fig. 4a). In contrast, the antidote is expressed after the driving and non-driving chromosome separate (meiosis I). Passage of the antidote through cytoplasmic bridges must be prevented to ensure selective killing (Fig. 4a). Poison–antidote meiotic drive systems include: the mouse t-haplotype [43, 61–67], Neurospora intermedia Sk-2 and Sk-3 [68–70], Podospora anserina Spok genes [71, 72], Schizosaccharomyces pombe wtf genes [73–77] and Oryza sativa qHMS7 [78]. Below we discuss the male t-haplotype and yeast wtf poison–antidote systems.
Mouse t-haplotype
The mouse t-haplotype is a well-studied mammalian poison–antidote meiotic drive system on chromosome 17. The t-haplotype contains four inversions that suppress recombination with wild-type chromosome 17, and keep the antidote and poisons in tight genetic linkage. Males heterozygous for the t-haplotype (t) and wild-type chromosome 17 ( +) can transmit the t-haplotype to up to 99% of offspring [79]. Preferential transmission of the t-haplotype arises from multiple intercellular poisons which collectively dysregulate cellular signaling pathways and disrupt sperm motility [61–67]. Specifically, poison-induced dysregulation is thought to converge on post-meiotically expressed intracellular wild-type SMOK1 (sperm motility kinase 1), which results in disrupted sperm motility. In contrast, t-haplotype SMOK1 (TCR) functions intracellularly as an antidote (Fig. 4b), sparing t-haplotype carrying sperm (Fig. 4b) [43, 67]. It remains to be determined whether the motility defects are specific to sperm carrying wild-type chromosome 17 [80].
SMOK1 dysregulation occurs by the combined efforts of poisons encoded on the t-haplotype known as t-complex distorters (Tcd) [63–66]. Trans-acting Tcds have been identified as the duplicated Tagap1 (Tcd4), overexpressed Fgd2 (Tcd2a), hypomorphic Nme3 (Tcd2b) and an isoform of Tiam2s (Tcd1) [63–66], which are pre-meiotically expressed and are therefore predicted to function intercellularly in wild-type cells after meiosis I [63–66]. The identification of Tcds indicates disrupted Rho-signaling pathways act to dysregulate SMOK1. However, there are poorly defined components in this drive system including Tcd3 and the proteins that link Tcds to Rho-signaling. Furthermore, TAGAP1 and FGD2 are thought to enhance both inhibitory and activating regulators of Rho-signaling, respectively, but the directionality of SMOK1 dysregulation is not known. Smok1 is present in multiple copies on the t-haplotype and wild-type chromosome 17 [80, 81], but it is unclear how wild-type copies of Smok1 on the t-haplotype fail to sensitize t-haplotype sperm. Clearly, the identification of SMOK1 targets will help identify the flagellar functions impaired in sperm motility defects.
Schizosaccharomyces pombe and Schizosaccharomyces kambucha—wtf4
The wtf meiotic drivers have been uncovered in yeast hybrids between S. pombe and S. kambucha [73–77]. The wtf4 gene, on chromosome 3 in S. kambucha encodes both the poison and antidote, due to alternative transcriptional start sites (Fig. 4c) [73]. The longer isoform encodes the antidote, and the short isoform encodes a poison. The poison (short isoform) acts intercellularly to poison spores that do not encode it, whereas the antidote (long isoform) acts intracellularly. The wtf4 poison is expressed before meiosis which enables wtf4 mRNA/protein to be present in all spores (Fig. 4c). In contrast, the antidote is expressed after meiotic divisions and is present exclusively in spores with the S. kambucha wtf4 allele (Fig. 4c) [73]. These findings indicate that poison-induced toxicity is specific to post-meiotic cells, since pre-meiotic and meiotic cells containing the poison appear unaffected. Recent work indicates that the poison forms dispersed aggregates that are toxic and kill spores, though the mechanism underlying this toxicity is unclear [82]. In cells expressing the antidote allele, the poison and antidote aggregate together near vacuoles and are no longer toxic, indicating the targeting of the complex to the vacuole is an important step (Fig. 4c) [82].
In S. pombe, the wtf gene family has expanded and diverged at the sequence level, resulting in multiple additional dual poison-antidote, and antidote only, wtf genes [73–76, 83]. It remains unclear how wtf systems act independently of one another given their sequence similarity and likely similar mechanism dependent on alternative transcriptional start sites. Interestingly, Spok genes in the fungus P. anserina also constitute both the poison and antidote [71]. Unlike wtf4, which relies on distinct isoforms, a single isoform of Spok appears to simultaneously function as the poison and antidote, though the mechanism and regulation of these functions is unknown [72]. Future studies could investigate the sequence differences between wtf family members that enable them to function as distinct poison-antidote meiotic drivers.
Future considerations for studying meiotic drivers
Meiotic drive systems continue to shape the evolutionary trajectory of genomes and species, yet remain challenging to study [1, 3, 84, 85]. First, meiotic drivers reside in regions of the genome where recombination is suppressed and are therefore not amenable to traditional genetic approaches. Second, meiotic drivers are typically not associated with an easily observable phenotype. Only in rare cases, they are genetically-linked with readily observable phenotypes such as sex ratio distortion or shortened tails associated with the t-haplotype. Third, genetic suppressors readily evolve to silence a driver; consequently, experimental hybrid models separate meiotic drivers into naïve genetic backgrounds lacking suppressors. Lastly, meiotic drivers are often rapidly evolving and in duplicated regions of the genome, making their identification, annotation and characterization more challenging than conserved single-copy sequences. As a result, even within a species, independent mechanisms can govern seemingly similar drive systems, as is the case for female mouse centromeric drive [10, 11, 13, 21].
New meiotic drive systems can also evolve on top of one another by integrating into pre-existing meiotic drive pathways. In maize, TR-1 repeats create neocentromeres which can suppress 180 bp repeat neocentromeres (knobs) [33, 34]. Similarly, two sex chromosome drive systems in Drosophila simulans, Winters and Durham, share a common suppressor [86]. This complex layering of drivers, enhancers and suppressors may be a common theme of meiotic drive. Such layering further complicates their mechanistic characterization, creating systems not easily classifiable under existing models. Greater mechanistic understandings may also reclassify existing drive systems, such as the Sd-RanGAP killer with RspS being the target, maybe Sd-RanGAP as a poison and RspI as the antidote.
Emerging drive models with evolving complexity: the case of Slxl1 and Sly in mice.
In mouse, a sex ratio drive system between X-linked Slxl1 and Y-linked Sly does not fit under existing models. Mechanistically, the ratio of Slxl1 to Sly gene copy number governs the offspring sex ratio [87, 88]. Removal of all Slxl1 gene copies results in male-biased litters (60% male), whereas overexpression of Slxl1 and the related Slx gene results in female-biased litters (60% female) [88]. The poison–antidote model requires both components be on the same chromosome, but Slxl1 and Sly are on opposite sex chromosomes. Under the target–killer model, loss of the killer, Slxl1, would be expected to neutralize meiotic drive, yet loss of Slxl1 results in meiotic drive. Slxl1 and Sly may represent individual poison antidote systems or factors beneficial to X-bearing and Y-bearing sperm, respectively. Under either of these models, the back and forth duplication of Slxl1 and Sly would alter the offspring sex ratio to a female or male bias, respectively. Repeated back and forth duplication may be a common theme of meiotic drivers [88]. Slxl1 and Sly might also be regulators of another drive system as they both interact with the Y-linked massively duplicated SSTY1 and SSTY2 [88, 89]. Given the shared signature of gene duplication and testis expression, it is tempting to speculate that Ssty1/2 are sex ratio meiotic drivers, creating a complex Ssty1/2-Slxl1-Sly network of meiotic drive [88, 90].
Conclusion
In its simplest form, meiotic drive is a two-component system. The two components are either a cis- and trans-acting factor, or two trans-acting factors. For example, the cis-acting Knobs with trans-acting KINDR in maize and the trans-acting factors Slx/Slxl1 versus Sly in mice are potentially two-component meiotic drive systems. New meiotic driver systems likely start as a two-component system. Over time, additional factors may influence drive systems. For example, the t-complex is a more complex drive system with at least four factors influencing chromosome inheritance, known as t-complex distorters [91, 92]. These distorters are harbored within a series of distinct inversions on mouse chromosome 17, which maintains their genetic linkage [91, 92]. Retracing the evolutionary history of multi-component drive systems, like the t-complex, can help reveal the originating two-component system and provide insights into how new factors are layered on existing drive systems.
There are multiple strategies to reveal meiotic drivers. One strategy is to generate hybrid organisms, which “release” the meiotic driver from suppressors. For example, the meiotic drive function of wtf4 in S. kambucha is revealed when S. kambucha is crossed to S. pombe and centromeric drive in females is revealed in hybrid mice. This suggests a plethora of meiotic drive systems could be revealed via hybridization of two species. Moreover, ancestral hybridization events between species, though rare, may have facilitated meiotic drivers to move from one species to another. By comparing closely related species genomes regions of the genome can be scanned for introgression of sequence from one species to another, potentially due to meiotic drive. Another strategy to reveal meiotic drive genes is via precise genetic manipulations. For example, megabase-sized deletions and duplications of the Slx/Slxl1 gene family were used in mice. These genetic manipulations revealed that Slx/Slxl1 versus Sly meiotic drive occurs in a gene dose-dependent manner. Recent advances in genome engineering will enable future studies to test whether candidate genes function as meiotic drivers. Indeed, while comparative sequence analysis helps identify candidate meiotic drivers, functional assessments are essential to determine if they drive.
Meiotic drivers in asymmetric meiosis converge on similar mechanisms. In the few examples of meiotic drive, the goal of the driver is to preferentially segregate itself and the chromosome encoding it to the egg for transmission to the next generation. As one would expect, considering the role of the centromere in chromosome segregation, centromeric repeats and their attachment to spindles are key components in asymmetric meiosis drive systems. A variation on this theme is the presence of ectopic centromere-like repeats known as Knobs in maize that bias attachment of the spindle and thus chromosome segregation. Recent long-read sequencing technologies have been able to resolve centromeric sequences, providing an opportunity to uncover the differences in the underlying repeat sequence that contribute to drive. These findings may help identify new candidate drive loci in asymmetric meiosis with sequence features resembling centromeric-like or Knob-like repeats.
Meiotic drivers in symmetric meiosis are often simplified into the target–killer or poison–antidote model, though great diversity in the underlying mechanisms exists. For example, Segregation Distorter uses a RanGAP pathway, the t-complex uses Rho-signaling pathway, the HET-s system affects the plasma membrane, and the wtf system results in prion-like aggregates. In each of these cases, the goal of the driver is to incapacitate the non-driver containing haploid cell. Since there is more than one way to incapacitate a cell, we expect future studies will uncover a diversity of meiotic drive mechanisms in symmetric meiosis.
The impact of meiotic drivers on evolution of a species and its genome can be dramatic. Since meiotic drivers are associated with negative fitness costs [93], suppressors capable of silencing the driver evolve and are readily selected for. A back and forth arms race ensues as drive enhancers and suppressors continuously emerge, altering the genome. The evolution of suppressors and enhancers can create new gene functions that reconfigure existing biological pathways. Meiotic drivers are thus a powerful force continuously shaping the genetic architecture of future genomes and biological pathways.
Acknowledgements
We would like to acknowledge M. Arlt, D. Burke, S. Hammoud, S. Kalantry and J. Moran for their comments.
Funding
This work was supported by National Institutes of Health grants HD094736 to JLM, and T32GM007544 and a National Science Foundation Graduate Research Fellowship DGE 1256260 to ANK.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hurst GD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet. 2001;2(8):597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 2.Werren JH. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10863–10870. doi: 10.1073/pnas.1102343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agren JA, Clark AG. Selfish genetic elements. PLoS Genet. 2018;14(11):e1007700. doi: 10.1371/journal.pgen.1007700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurst LD. A century of bias in genetics and evolution. Heredity (Edinb) 2019;123(1):33–43. doi: 10.1038/s41437-019-0194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helleu Q, Gerard PR, Montchamp-Moreau C. Sex chromosome drive. Cold Spring Harb Perspect Biol. 2014;7(2):a017616. doi: 10.1101/cshperspect.a017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courret C, Chang CH, Wei KH, Montchamp-Moreau C, Larracuente AM. Meiotic drive mechanisms: lessons from Drosophila. Proc Biol Sci. 2019;286(1913):20191430. doi: 10.1098/rspb.2019.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyttle TW. Segregation distorters. Annu Rev Genet. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- 8.Bravo Nunez MA, Nuckolls NL, Zanders SE. Genetic villains: killer meiotic drivers. Trends Genet. 2018;34(6):424–433. doi: 10.1016/j.tig.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol. 2008;18(24):1986–1992. doi: 10.1016/j.cub.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 10.Akera T, Chmatal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, Lampson MA. Spindle asymmetry drives non-Mendelian chromosome segregation. Science. 2017;358(6363):668–672. doi: 10.1126/science.aan0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T, Lane SIR, Morgan SL, Jones KT. Spindle tubulin and MTOC asymmetries may explain meiotic drive in oocytes. Nat Commun. 2018;9(1):2952. doi: 10.1038/s41467-018-05338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chmatal L, Gabriel SI, Mitsainas GP, Martinez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2014;24(19):2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmatal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol. 2017;27(15):2365–2373 e2368. doi: 10.1016/j.cub.2017.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei KH, Reddy HM, Rathnam C, Lee J, Lin D, Ji S, Mason JM, Clark AG, Barbash DA. A pooled sequencing approach identifies a candidate meiotic driver in Drosophila. Genetics. 2017;206(1):451–465. doi: 10.1534/genetics.116.197335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322(5907):1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- 16.Fishman L, Willis JH. A novel meiotic drive locus almost completely distorts segregation in mimulus (monkeyflower) hybrids. Genetics. 2005;169(1):347–353. doi: 10.1534/genetics.104.032789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buckler EST, Phelps-Durr TL, Buckler CS, Dawe RK, Doebley JF, Holtsford TP. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics. 1999;153(1):415–426. doi: 10.1093/genetics/153.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawe RK, Lowry EG, Gent JI, Stitzer MC, Swentowsky KW, Higgins DM, Ross-Ibarra J, Wallace JG, Kanizay LB, Alabady M, Qiu W, Tseng KF, Wang N, Gao Z, Birchler JA, Harkess AE, Hodges AL, Hiatt EN. A Kinesin-14 motor activates neocentromeres to promote meiotic drive in maize. Cell. 2018;173(4):839–850 e818. doi: 10.1016/j.cell.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Dawe RK, Reed LM, Yu HG, Muszynski MG, Hiatt EN. A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell. 1999;11(7):1227–1238. doi: 10.1105/tpc.11.7.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu HG, Hiatt EN, Chan A, Sweeney M, Dawe RK. Neocentromere-mediated chromosome movement in maize. J Cell Biol. 1997;139(4):831–840. doi: 10.1083/jcb.139.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akera T, Trimm E, Lampson MA. Molecular strategies of meiotic cheating by selfish centromeres. Cell. 2019;178(5):1132–1144 e1110. doi: 10.1016/j.cell.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16(4):335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain M, Olsen HE, Turner DJ, Stoddart D, Bulazel KV, Paten B, Haussler D, Willard HF, Akeson M, Miga KH. Linear assembly of a human centromere on the Y chromosome. Nat Biotechnol. 2018;36(4):321–323. doi: 10.1038/nbt.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langley SA, Miga KH, Karpen GH, Langley CH. Haplotypes spanning centromeric regions reveal persistence of large blocks of archaic DNA. Elife. 2019 doi: 10.7554/eLife.42989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Didion JP, Morgan AP, Clayshulte AM, McMullan RC, Yadgary L, Petkov PM, Bell TA, Gatti DM, Crowley JJ, Hua K, Aylor DL, Bai L, Calaway M, Chesler EJ, French JE, Geiger TR, Gooch TJ, Garland T, Jr, Harrill AH, Hunter K, McMillan L, Holt M, Miller DR, O'Brien DA, Paigen K, Pan W, Rowe LB, Shaw GD, Simecek P, Sullivan PF, Svenson KL, Weinstock GM, Threadgill DW, Pomp D, Churchill GA, Pardo-Manuel de Villena F. A multi-megabase copy number gain causes maternal transmission ratio distortion on mouse chromosome 2. PLoS Genet. 2015;11(2):e1004850. doi: 10.1371/journal.pgen.1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Didion JP, Morgan AP, Yadgary L, Bell TA, McMullan RC, Ortiz de Solorzano L, Britton-Davidian J, Bult CJ, Campbell KJ, Castiglia R, Ching YH, Chunco AJ, Crowley JJ, Chesler EJ, Forster DW, French JE, Gabriel SI, Gatti DM, Garland T, Jr, Giagia-Athanasopoulou EB, Gimenez MD, Grize SA, Gunduz I, Holmes A, Hauffe HC, Herman JS, Holt JM, Hua K, Jolley WJ, Lindholm AK, Lopez-Fuster MJ, Mitsainas G, da Luz MM, McMillan L, Ramalhinho Mda G, Rehermann B, Rosshart SP, Searle JB, Shiao MS, Solano E, Svenson KL, Thomas-Laemont P, Threadgill DW, Ventura J, Weinstock GM, Pomp D, Churchill GA, Pardo-Manuel de Villena F. R2d2 drives selfish sweeps in the house mouse. Mol Biol Evol. 2016;33(6):1381–1395. doi: 10.1093/molbev/msw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agulnik SI, Agulnik AI, Ruvinsky AO. Meiotic drive in female mice heterozygous for the HSR inserts on chromosome 1. Genet Res. 1990;55(2):97–100. doi: 10.1017/s0016672300025325. [DOI] [PubMed] [Google Scholar]
- 28.Pardo-Manual de Villena F, Slamka C, Fonseca M, Naumova AK, Paquette J, Pannunzio P, Smith M, Verner A, Morgan K, Sapienza C. Transmission-ratio distortion through F1 females at chromosome 11 loci linked to Om in the mouse DDK syndrome. Genetics. 1996;142(4):1299–1304. doi: 10.1093/genetics/142.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pardo-Manuel de Villena F, Naumova AK, Verner AE, Jin WH, Sapienza C. Confirmation of maternal transmission ratio distortion at Om and direct evidence that the maternal and paternal "DDK syndrome" genes are linked. Mamm Genome. 1997;8(9):642–646. doi: 10.1007/s003359900529. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Zhao X, Cheng K, Du H, Ouyang Y, Chen J, Qiu S, Huang J, Jiang Y, Jiang L, Ding J, Wang J, Xu C, Li X, Zhang Q. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science. 2012;337(6100):1336–1340. doi: 10.1126/science.1223702. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Ding J, Ouyang Y, Du H, Yang J, Cheng K, Zhao J, Qiu S, Zhang X, Yao J, Liu K, Wang L, Xu C, Li X, Xue Y, Xia M, Ji Q, Lu J, Xu M, Zhang Q. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc Natl Acad Sci USA. 2008;105(32):11436–11441. doi: 10.1073/pnas.0804761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhoades MM, Dempsey E. Further studies on preferential segregation. Maize Genetics: Coop. Maize Genetics Cooperation Newsletter; 1957. [Google Scholar]
- 33.Kanizay LB, Albert PS, Birchler JA, Dawe RK. Intragenomic conflict between the two major knob repeats of maize. Genetics. 2013;194(1):81–89. doi: 10.1534/genetics.112.148882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiatt EN, Kentner EK, Dawe RK. Independently regulated neocentromere activity of two classes of tandem repeat arrays. Plant Cell. 2002;14(2):407–420. doi: 10.1105/tpc.010373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawe RK, Cande WZ. Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proc Natl Acad Sci USA. 1996;93(16):8512–8517. doi: 10.1073/pnas.93.16.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morales CR, Lefrancois S, Chennathukuzhi V, El-Alfy M, Wu X, Yang J, Gerton GL, Hecht NB. A TB-RBP and Ter ATPase complex accompanies specific mRNAs from nuclei through the nuclear pores and into intercellular bridges in mouse male germ cells. Dev Biol. 2002;246(2):480–494. doi: 10.1006/dbio.2002.0679. [DOI] [PubMed] [Google Scholar]
- 37.Braun RE, Behringer RR, Peschon JJ, Brinster RL, Palmiter RD. Genetically haploid spermatids are phenotypically diploid. Nature. 1989;337(6205):373–376. doi: 10.1038/337373a0. [DOI] [PubMed] [Google Scholar]
- 38.Ventela S, Toppari J, Parvinen M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Mol Biol Cell. 2003;14(7):2768–2780. doi: 10.1091/mbc.e02-10-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fawcett DW, Ito S, Slautterback D. The occurrence of intercellular bridges in groups of cells exhibiting synchronous differentiation. J Biophys Biochem Cytol. 1959;5(3):453–460. doi: 10.1083/jcb.5.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caldwell KA, Handel MA. Protamine transcript sharing among postmeiotic spermatids. Proc Natl Acad Sci USA. 1991;88(6):2407–2411. doi: 10.1073/pnas.88.6.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umehara T, Tsujita N, Shimada M. Activation of Toll-like receptor 7/8 encoded by the X chromosome alters sperm motility and provides a novel simple technology for sexing sperm. PLoS Biol. 2019;17(8):e3000398. doi: 10.1371/journal.pbio.3000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, Deng X, Martin-DeLeon PA. Lack of sharing of Spam1 (Ph-20) among mouse spermatids and transmission ratio distortion. Biol Reprod. 2001;64(6):1730–1738. doi: 10.1095/biolreprod64.6.1730. [DOI] [PubMed] [Google Scholar]
- 43.Veron N, Bauer H, Weisse AY, Luder G, Werber M, Herrmann BG. Retention of gene products in syncytial spermatids promotes non-Mendelian inheritance as revealed by the t complex responder. Genes Dev. 2009;23(23):2705–2710. doi: 10.1101/gad.553009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler A, Gordon RE, Gatt S, Schuchman EH. Sperm abnormalities in heterozygous acid sphingomyelinase knockout mice reveal a novel approach for the prevention of genetic diseases. Am J Pathol. 2007;170(6):2077–2088. doi: 10.2353/ajpath.2007.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhutani K, Stansifer K, Ticau S, Bojic L, Villani C, Slisz J, Cremers C, Roy C, Donovan J, Fiske B, Friedman R. Widespread haploid-based gene expression in mammalian spermatogenesis associated with frequent selective sweeps and evolutionary conflict. Biorxiv. 2019;4:120–134. [Google Scholar]
- 46.Larracuente AM, Presgraves DC. The selfish segregation distorter gene complex of Drosophila melanogaster. Genetics. 2012;192(1):33–53. doi: 10.1534/genetics.112.141390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalstra HJ, Swart K, Debets AJ, Saupe SJ, Hoekstra RF. Sexual transmission of the [Het-S] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci USA. 2003;100(11):6616–6621. doi: 10.1073/pnas.1030058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, Riek R. The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol. 2012;10(12):e1001451. doi: 10.1371/journal.pbio.1001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalstra HJ, van der Zee R, Swart K, Hoekstra RF, Saupe SJ, Debets AJ. Non-mendelian inheritance of the HET-s prion or HET-s prion domains determines the het-S spore killing system in Podospora anserina. Fungal Genet Biol. 2005;42(10):836–847. doi: 10.1016/j.fgb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Merrill C, Bayraktaroglu L, Kusano A, Ganetzky B. Truncated RanGAP encoded by the segregation distorter locus of Drosophila. Science. 1999;283(5408):1742–1745. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- 51.Kusano A, Staber C, Ganetzky B. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev Cell. 2001;1(3):351–361. doi: 10.1016/s1534-5807(01)00042-9. [DOI] [PubMed] [Google Scholar]
- 52.Brittnacher JG, Ganetzky B. On the components of segregation distortion in Drosophila melanogaster. IV. Construction and analysis of free duplications for the Responder locus. Genetics. 1989;121(4):739–750. doi: 10.1093/genetics/121.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyttle TW. The effect of novel chromosome position and variable dose on the genetic behavior of the Responder (Rsp) element of the segregation distorter (SD) system of Drosophila melanogaster. Genetics. 1989;121(4):751–763. doi: 10.1093/genetics/121.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu CI, Lyttle TW, Wu ML, Lin GF. Association between a satellite DNA sequence and the responder of segregation distorter in D. melanogaster. Cell. 1988;54(2):179–189. doi: 10.1016/0092-8674(88)90550-8. [DOI] [PubMed] [Google Scholar]
- 55.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 56.Kusano A, Staber C, Ganetzky B. Segregation distortion induced by wild-type RanGAP in Drosophila. Proc Natl Acad Sci USA. 2002;99(10):6866–6870. doi: 10.1073/pnas.102165099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hauschteck-Jungen E, Hartl DL. Defective Histone Transition during Spermiogenesis in Heterozygous segregation distorter Males of D. Melanogaster. Genetics. 1982;101(1):57–69. doi: 10.1093/genetics/101.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seto AG, Kingston RE, Lau NC. The coming of age for piwi proteins. Mol Cell. 2007;26(5):603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 59.Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA (New York, NY) 2010;16(12):2503–2515. doi: 10.1261/rna.2270710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gell SL, Reenan RA. Mutations to the piRNA pathway component aubergine enhance meiotic drive of segregation distorter in Drosophila melanogaster. Genetics. 2013;193(3):771–784. doi: 10.1534/genetics.112.147561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katz DF, Erickson RP, Nathanson M. Beat frequency is bimodally distributed in spermatozoa from T/t12 mice. J Exp Zool. 1979;210(3):529–535. doi: 10.1002/jez.1402100316. [DOI] [PubMed] [Google Scholar]
- 62.Olds-Clarke P, Johnson LR. t haplotypes in the mouse compromise sperm flagellar function. Dev Biol. 1993;155(1):14–25. doi: 10.1006/dbio.1993.1002. [DOI] [PubMed] [Google Scholar]
- 63.Bauer H, Schindler S, Charron Y, Willert J, Kusecek B, Herrmann BG. The nucleoside diphosphate kinase gene Nme3 acts as quantitative trait locus promoting non-Mendelian inheritance. PLoS Genet. 2012;8(3):e1002567. doi: 10.1371/journal.pgen.1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bauer H, Veron N, Willert J, Herrmann BG. The t-complex-encoded guanine nucleotide exchange factor Fgd2 reveals that two opposing signaling pathways promote transmission ratio distortion in the mouse. Genes Dev. 2007;21(2):143–147. doi: 10.1101/gad.414807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bauer H, Willert J, Koschorz B, Herrmann BG. The t complex-encoded GTPase-activating protein Tagap1 acts as a transmission ratio distorter in mice. Nat Genet. 2005;37(9):969–973. doi: 10.1038/ng1617. [DOI] [PubMed] [Google Scholar]
- 66.Charron Y, Willert J, Lipkowitz B, Kusecek B, Herrmann BG, Bauer H. Two isoforms of the RAC-specific guanine nucleotide exchange factor TIAM2 act oppositely on transmission ratio distortion by the mouse t-haplotype. PLoS Genet. 2019;15(2):e1007964. doi: 10.1371/journal.pgen.1007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herrmann BG, Koschorz B, Wertz K, McLaughlin KJ, Kispert A. A protein kinase encoded by the t complex responder gene causes non-mendelian inheritance. Nature. 1999;402(6758):141–146. doi: 10.1038/45970. [DOI] [PubMed] [Google Scholar]
- 68.Rhoades NA, Harvey AM, Samarajeewa DA, Svedberg J, Yusifov A, Abusharekh A, Manitchotpisit P, Brown DW, Sharp KJ, Rehard DG, Peters J, Ostolaza-Maldonado X, Stephenson J, Shiu PKT, Johannesson H, Hammond TM. Identification of rfk-1, a meiotic driver undergoing RNA editing in neurospora. Genetics. 2019;212(1):93–110. doi: 10.1534/genetics.119.302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turner BC, Perkins DD. Spore killer, a chromosomal factor in neurospora that kills meiotic products not containing it. Genetics. 1979;93(3):587–606. doi: 10.1093/genetics/93.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hammond TM, Rehard DG, Xiao H, Shiu PK. Molecular dissection of Neurospora Spore killer meiotic drive elements. Proc Natl Acad Sci USA. 2012;109(30):12093–12098. doi: 10.1073/pnas.1203267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grognet P, Lalucque H, Malagnac F, Silar P. Genes that bias Mendelian segregation. PLoS Genet. 2014;10(5):e1004387. doi: 10.1371/journal.pgen.1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogan AA, Ament-Velasquez SL, Granger-Farbos A, Svedberg J, Bastiaans E, Debets AJ, Coustou V, Yvanne H, Clave C, Saupe SJ, Johannesson H. Combinations of Spok genes create multiple meiotic drivers in Podospora. Elife. 2019 doi: 10.7554/eLife.46454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nuckolls NL, Bravo Nunez MA, Eickbush MT, Young JM, Lange JJ, Yu JS, Smith GR, Jaspersen SL, Malik HS, Zanders SE. wtf genes are prolific dual poison-antidote meiotic drivers. Elife. 2017 doi: 10.7554/eLife.26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bravo Nunez MA, Lange JJ, Zanders SE. A suppressor of a wtf poison-antidote meiotic driver acts via mimicry of the driver's antidote. PLoS Genet. 2018;14(11):e1007836. doi: 10.1371/journal.pgen.1007836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bravo Nunez MA, Sabbarini IM, Eickbush MT, Liang Y, Lange JJ, Kent AM, Zanders SE. Dramatically diverse Schizosaccharomyces pombe wtf meiotic drivers all display high gamete-killing efficiency. PLoS Genet. 2020;16(2):e1008350. doi: 10.1371/journal.pgen.1008350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu W, Jiang ZD, Suo F, Zheng JX, He WZ, Du LL. A large gene family in fission yeast encodes spore killers that subvert Mendel's law. Elife. 2017 doi: 10.7554/eLife.26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zanders SE, Eickbush MT, Yu JS, Kang JW, Fowler KR, Smith GR, Malik HS. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. Elife. 2014;3:e02630. doi: 10.7554/eLife.02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu X, Zhao Z, Zheng X, Zhou J, Kong W, Wang P, Bai W, Zheng H, Zhang H, Li J, Liu J, Wang Q, Zhang L, Liu K, Yu Y, Guo X, Wang J, Lin Q, Wu F, Ren Y, Zhu S, Zhang X, Cheng Z, Lei C, Liu S, Liu X, Tian Y, Jiang L, Ge S, Wu C, Tao D, Wang H, Wan J. A selfish genetic element confers non-Mendelian inheritance in rice. Science. 2018;360(6393):1130–1132. doi: 10.1126/science.aar4279. [DOI] [PubMed] [Google Scholar]
- 79.Chesley P, Dunn LC. The inheritance of taillessness (Anury) in the house mouse. Genetics. 1936;21(5):525–536. doi: 10.1093/genetics/21.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schimenti J. Segregation distortion of mouse t haplotypes the molecular basis emerges. Trends Genet. 2000;16(6):240–243. doi: 10.1016/s0168-9525(00)02020-5. [DOI] [PubMed] [Google Scholar]
- 81.Schimenti J, Vold L, Socolow D, Silver LM. An unstable family of large DNA elements in the center of the mouse t complex. J Mol Biol. 1987;194(4):583–594. doi: 10.1016/0022-2836(87)90235-x. [DOI] [PubMed] [Google Scholar]
- 82.Nuckolls NL, Mok AC, Lange JJ, Yi K, Kandola TS, Hunn AM, McCroskey S, Snyder JL, Bravo Nunez MA, McClain ML, McKinney SA, Wood C, Halfmann R, Zanders SE. The wtf4 meiotic driver utilizes controlled protein aggregation to generate selective cell death. biorXiv. 2020;16:13–54. doi: 10.7554/eLife.55694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eickbush MT, Young JM, Zanders SE. Killer meiotic drive and dynamic evolution of the wtf gene family. Mol Biol Evol. 2019;36(6):1201–1214. doi: 10.1093/molbev/msz052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lindholm AK, Dyer KA, Firman RC, Fishman L, Forstmeier W, Holman L, Johannesson H, Knief U, Kokko H, Larracuente AM, Manser A, Montchamp-Moreau C, Petrosyan VG, Pomiankowski A, Presgraves DC, Safronova LD, Sutter A, Unckless RL, Verspoor RL, Wedell N, Wilkinson GS, Price TAR. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol Evol. 2016;31(4):315–326. doi: 10.1016/j.tree.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 85.Meiklejohn CD, Tao Y. Genetic conflict and sex chromosome evolution. Trends Ecol Evol. 2010;25(4):215–223. doi: 10.1016/j.tree.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin CJ, Hu F, Dubruille R, Vedanayagam J, Wen J, Smibert P, Loppin B, Lai EC. The hnRNP/RNAi pathway is essential to resolve intragenomic conflict in the drosophila male germline. Dev Cell. 2018;46(3):316–326. doi: 10.1016/j.devcel.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cocquet J, Ellis PJ, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 2012;8(9):e1002900. doi: 10.1371/journal.pgen.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kruger AN, Brogley MA, Huizinga JL, Kidd JM, de Rooij DG, Hu YC, Mueller JL. A Neofunctionalized X-Linked Ampliconic Gene Family Is Essential for Male Fertility and Equal Sex Ratio in Mice. Curr Biol. 2019;29(21):3699–3706 e3695. doi: 10.1016/j.cub.2019.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Comptour A, Moretti C, Serrentino ME, Auer J, Ialy-Radio C, Ward MA, Toure A, Vaiman D, Cocquet J. SSTY proteins co-localize with the post-meiotic sex chromatin and interact with regulators of its expression. FEBS J. 2014;281(6):1571–1584. doi: 10.1111/febs.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soh YQ, Alfoldi J, Pyntikova T, Brown LG, Graves T, Minx PJ, Fulton RS, Kremitzki C, Koutseva N, Mueller JL, Rozen S, Hughes JF, Owens E, Womack JE, Murphy WJ, Cao Q, de Jong P, Warren WC, Wilson RK, Skaletsky H, Page DC. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159(4):800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ardlie KG. Putting the brake on drive: meiotic drive of t haplotypes in natural populations of mice. Trends Genet. 1998;14(5):189–193. doi: 10.1016/s0168-9525(98)01455-3. [DOI] [PubMed] [Google Scholar]
- 92.Silver LM. The peculiar journey of a selfish chromosome: mouse t haplotypes and meiotic drive. Trends Genet. 1993;9(7):250–254. doi: 10.1016/0168-9525(93)90090-5. [DOI] [PubMed] [Google Scholar]
- 93.Wong HWS, Holman L. Fitness consequences of the selfish supergene Segregation Distorter. J Evol Biol. 2020;33(1):89–100. doi: 10.1111/jeb.13549. [DOI] [PubMed] [Google Scholar]