Abstract
In photoacoustic tomography (PAT), a tunable laser typically illuminates the tissue at multiple wavelengths, and the received photoacoustic waves are used to form functional images of relative total haemoglobin (rHbT) and blood oxygenation saturation (%sO2). Due to measurement errors, the estimation of these parameters can be challenging, especially in clinical studies. In this study, we use a multi-pixel method to smooth the measurements before calculating rHbT and %sO2. We first perform phantom studies using blood tubes of calibrated % sO2 to evaluate the accuracy of our %sO2 estimation. We conclude by presenting diagnostic results from PAT of 33 patients with 51 ovarian masses imaged by our co-registered PAT and ultrasound system. The ovarian masses were divided into malignant and benign/normal groups. Functional maps of rHbT and %sO2 and their histograms as well as spectral features were calculated using the PAT data from all ovaries in these two groups. Support vector machine models were trained on different combinations of the significant features. The area under ROC (AUC) of 0.93 (0.95%CI: 0.90–0.96) on the testing data set was achieved by combining mean %sO2, a spectral feature, and the score of the study radiologist.
Keywords: feature extraction, ovarian cancer diagnosis, oxygen saturation (%sO2), photoacoustic tomography
Graphical Abstract

1 |. INTRODUCTION
Ovarian cancer remains the deadliest of all the gynaecological malignancies. According to the American Cancer Society, 21 750 women in the United States will be diagnosed with ovarian cancer in 2020, and 13 940 deaths will be reported [1]. Due to a lack of effective screening tools, only 20% to 25% of ovarian cancers are diagnosed early. A recent update from the U.S. Preventive Services Task Force concluded that major trials of promising ovarian cancer screening tools have null findings to date among healthy average-risk women, and there are considerable harms associated with screening, which include major surgical complications in women found to have no cancer [2]. Women with a screening abnormality generally undergo prophylactic bilateral salpingo-oophorectomy [3–5], which can result in morbidity and mortality from premature menopause, including accelerated bone loss and cardiovascular death [6]. Thus, there is an urgent need to develop better and more sensitive tools to effectively evaluate the ovary.
Current screening modalities include bimanual pelvic examination and transvaginal ultrasound (US), as well as assessment of cancer antigen 125 (CA125) serum marker levels, which are associated with a high false-positive rate [2, 7]. Conventional MRI and diffusion-weighted MRI are useful follow-up modalities for investigating sonographically indeterminate adnexal masses [8]. Using MRI to measure oxygen saturation for ovarian tumour assessment has not been well studied and is potentially valuable for future research. CT has been historically preferred for the pre-treatment evaluation of ovarian cancer to determine both the extent of the disease and the optimal surgical treatment [9]. FDG-PET/CT can measure residual or recurrent disease and also help select the surgical treatment [10]. However, FDG-PET has limited value in localizing lesions in the early stages of ovarian cancer [11].
In the past decade, with advances in lasers, US transducers, and tomographic reconstruction techniques, photoacoustic imaging or photoacoustic tomography (PAT) has seen an immense growth, providing high spatial resolution same as US and functional information at depths ranging from several millimetres up to several centimetres [12–18]. PAT is a hybrid imaging technology that uses a short-pulsed laser to excite the tissue. Acoustic (or photoacoustic) waves are then generated from thermoelastic expansion due to a transient temperature rise and are measured by US transducers. PAT image contrast is related to tissue optical absorption properties and therefore tumour vasculature or tumour angiogenesis. Oncologic targets of PAT to date include breast cancer [19–27], prostate cancer [28, 29], skin cancer [30], thyroid cancer [31], colon cancer [32], and ovarian cancers [33, 34]. Our limited data have indicated that high grade stage I invasive ovarian cancers show a higher haemoglobin or vasculature content and a lower blood oxygen saturation (%sO2) [33].
%sO2, an indicator of tumour metabolism and therapeutic response, is one important diagnostic parameter measured by PAT. To compute %sO2, PAT data are acquired at multiple wavelengths, then used to solve linear equations that relate the PAT signals to the oxy and deoxyhaemoglobin concentrations at each pixel in the region of interest (ROI), simultaneously obtaining the concentrations of these chromophores at that pixel. This method of calculating %sO2, named “linear unmixing”, has been utilized in many studies [35–44]. In this study, for 49 ovarian masses from 33 patients, we present %sO2 maps computed using this method. However, to mitigate the effect of the measurement errors (eg, spatial and temporal variations in light fluence, system noise, and motion), we replace the value of each pixel with the average of the values of 100 pixels of 1.0 mm by 1.0 mm in size around it before solving the linear equations. The estimated %sO2 maps calculated using this method, along with the relative total haemoglobin (rHbT) mean value and PAT spectral features, are used to develop support vector machine (SVM) models. We demonstrate with a considerable patient population that a combination of PAT features with the radiologists’ diagnostic score, can significantly improve diagnostic accuracy, achieving an area under the receiver operating characteristic curve (ROC) of 0.93 on the testing data set.
2 |. MATERIALS AND METHODS
2.1 |. Co-registered PAT/US imaging system
Our co-registered PAT/US imaging system is described in detail in Ref. [33]. Briefly, the system consists of a commercial ultrasound system (EC-12R, Alpinion Medical Systems, Republic of Korea), a transvaginal US probe (6 MHz, 80% bandwidth), and a tunable Ti-sapphire laser. The light delivery system consists of four optical fibres which are coupled with the US transducer. The system can be programmed to collect data at optical wavelengths in the range of 690 to 900 nm. However, due to time limitations in practical use, we selected four wavelengths of 730, 780, 800, and 830 nm for the patient studies. At each wavelength, the data acquisition time for both US and PAT is about 3 seconds. A total of 3 more seconds is used by the laser system for wavelength tuning from 730 to 830 nm. Thus, the total data acquisition time for all four selected optical wavelengths is 15 seconds.
2.2 |. Phantoms
To evaluate the accuracy of our %sO2 estimation, we prepared five blood tube phantoms, with %sO2 values of 24.9%, 44.2%, 64.9%, 83.9%, and 97.6%. The red blood cells (RBCs) were collected from healthy volunteers and were mixed with 60% saline water inside a hypoxia chamber with a temperature/humidity control. Each blood sample with the desired %sO2 value was produced by controlling the amount of oxygen and nitrogen in the chamber. The %sO2 value of each blood sample was calibrated using an ABL90 FLEX Radiometer. This value was considered as the calibrated %sO2 value in the phantom study.
After each blood sample was prepared, it was injected into a 3 mm diameter tube, which was then tightly capped in the chamber in order to prevent the room oxygen from affecting the %sO2 of the blood sample. Each tube was sequentially positioned at a progression of depths from 1 to 5 cm, with a step size of 0.5 cm, in a homogeneous Intralipid solution with calibrated absorption and reduced scattering coefficients of 0.02 and 4 cm−1, similar to the optical properties of soft tissue. The PAT/US system was used to collect data from each blood tube at each depth and at wavelengths of 730, 780, 800, and 830 nm. To prevent motion during the data acquisition, the transducer was fixed to a post. The acquired PAT data were employed to calculate the %sO2 maps inside the blood tubes, using linear unmixing (described in 2.5). Finally, the mean %sO2 value at each depth was calculated by taking an average over all the pixels with %sO2 values greater than zero.
2.3 |. Patients
The study protocol was approved by the institutional review board and was compliant with the Health Insurance Portability and Accountability Act. A total of 40 patients with ovarian masses gave informed consent and participated in this study from February 2017 to November 2018. Out of these patients, 7 were excluded from the data analysis: 3 patients due to PAT/US system problems, 2 patients without any ovarian masses found at the time of imaging, and 2 patients with deep ovarian masses in the range of 6 to 7 cm from the probe. In 18 of the remaining 33 patients, both ovaries were imaged. Among the other 15 patients, only one suspicious ovary (with pathological evaluation available) was imaged and surgically resected. Thus, the total number of the imaged ovaries was 51, out of which two ovaries were not considered in our analysis because pathology data was not available for them (Figure 1).
FIGURE 1.
Flowchart for inclusion and exclusion of study participants, including reasons for exclusion
In Table 1, the imaged ovaries in this study have been divided into four main categories of invasive epithelial ovarian cancer, other neoplasm, benign ovaries and normal ovaries. On the right side of the table, each mass type has been divided into more subgroups. The number of ovaries belonging to each subgroup as well as the information about the mass size and depth is also provided in the table.
TABLE 1.
Lesion characteristics (33 patients with 49 ovaries: average age of 56 years, range 33–87 years)
| Lesion characteristics | |
|---|---|
| Invasive epithelial ovarian cancer | High grade serous carcinoma (n = 7), endometrioid carcinoma (n = 3) (average size, 9 cm; size range, 2.8–20 cm; average depth, 3 cm; depth range, 2.2–4.3 cm) |
| Other neoplasm | Serous borderline (n = 2) tumour, Sertoli-Leydig cell tumour (n = 1) (average size, 11.1 cm; size range, 4.5–19.2 cm, average depth, 3.6 cm; depth range, 2.6–5.5 cm) |
| Benign ovaries | Fibrothecoma (n = 1; size, 14 cm; depth, 1.9 cm), mature teratoma (n = 1; size, 6 cm; depth, 3.1 cm), serous or mucinous cystadenoma or cystic endometriosis (n = 17; average size, 10 cm; size range, 1.8–3.7 cm; average depth, 2.7 cm; depth range, 0.7–7.4 cm), complex or simple cysts (n = 10; average size, 4.3 cm; size range, 3–7.6 cm; average depth, 2.7 cm; depth range, 0.6–5.5 cm), benign leiomyoma (n = 1; size, 5.5 cm; depth, 1.8 cm), fibrosis and cyst (n = 1; size, 4 cm; depth, 1.9 cm) |
| Normal ovaries | No histopathological abnormalities (n = 5; average size, 2.5 cm; size range, 2.1–2.8 cm; average depth, 1.6 cm; depth range, 0.6–2.6 cm) |
2.4 |. Ovarian mass ranking
For each patient, one attending radiologist evaluated all available images of X-ray CT, MRI, prior US from the patient’s medical record as well as the real-time US images. Based on this information, the radiologist provided a score in a range of 1 to 5 for each ovarian mass before the co-registered photoacoustic and US exam. Scores 1 and 2 are normal, 3 is likely benign, 4 is suspicious for malignancy, score 5 is highly suspicious for malignancy. For the data analysis, we categorized scores 1 and 2 as normal, score 3 as benign, and 4 and 5 as malignant. This mapped feature is referred to as the radiology ranking (Rad rank) in this manuscript.
2.5 |. PAT functional features
To calculate the PAT functional features, that is, rHbT and %sO2, we need to estimate relative oxy- and deoxyhaemoglobin concentrations, and , which can be obtained using the linear unmixing approach [35–44]. Briefly, we find a solution at each pixel by solving a linear least square problem with the constraint of non-negative solutions as:
| (1) |
where g represents measurements at four wavelengths used, H is the known matrix consists of extinction coefficients of the four wavelengths, and f represents the and that need to be estimated.
The subscripts 730, 780, 800, and 830 in the above equations denote the wavelengths used in this study. The rHbT is then simply calculated by summing them. Furthermore, %sO2 is estimated by dividing the relative oxyhaemoglobin by rHbT.
In our previous studies [33,34], we calculated the functional features at each single pixel by solving Equation (1). We then applied a spatial filter to improve the % sO2 image quality. However, due to the measurement errors arise from spatial and temporal variations in light fluence caused by tissue optical absorption and scattering heterogeneities, system noise and motion, solving a linear equation at each single pixel might not be a robust method for finding the functional features. To mitigate this problem, we replaced the value of the envelope data in each pixel by the mean of the envelope at an area around it with a size of 10 × 10 pixels. We named it multi-pixel method. Thus, for each pixel, the area of 1.0 mm × 1.0 mm is chosen so that the smaller pixel is in the centre of the 10 × 10 pixels. Details on how to select the range of pixels for spatial filtering are provided in Appendix A.
We also investigated using Gaussian and Lorentzian algorithms as spatial filters to smooth the PAT data before calculating the %sO2 maps. The size of each filter was fixed at 1.0 mm × 1.0 mm (10 pixels by 10 pixels), the same as the size in the multi-pixel method. To take advantage of the spatial filter’s ability to mitigate measurement errors without significantly impairing the spatial resolution, we had to assign appropriate values to the parameters of the filters. The filter parameters that we adjusted were the SD of the Gaussian filter and the full width at half maximum (FWHM) of the Lorentzian filter, both indicated by “sigma” in this manuscript. Small SDs of the Gaussian filters or small FWHMs of the Lorentzian filters give too much weight to the pixels in the centre of each kernel, neglecting the contributions of the pixels around it. The resulting %sO2 maps are undesirably similar to maps without any spatial filters. On the other hand, if a very large SD or Lorentzian parameter is chosen, all the pixels in the kernel will have almost the same weight. In this case, there will be no difference between the generated %sO2 maps calculated using these filters and those estimated using sliding multi-pixels. Because our multi-pixel kernel size is 10 pixels, we decided to assign half of this value (5) to the SD of the Gaussian filter and Lorentzian filter parameter. Appendix A provides more information about the effect of the parameters of these filters on the estimated %sO2 and residual norm maps.
After finding the %sO2 map, we constructed its histogram and calculated the histogram features (mean, SD, skewness, kurtosis, energy, and entropy). Among these features, three (mean, skewness, and energy) showed significant differences between the benign and malignant ovarian groups.
To compare the robustness of the different smoothing methods, we calculated the value of the squared norm of the normalized residuals (for simplicity, called the normalized residual throughout this manuscript) at that pixel. To find this parameter at each pixel, after solving the linear optimization of Equation (1), we normalized each element of (g − Hf) by the summation of all four wavelengths of g at that pixel, thereby removing the dependency of the residuals on the magnitude of g. Then we calculated the normalized residual at that pixel as:
| (2) |
where superscript T indicates the transpose of the matrix, and subscript n means the matrix has been normalized pixel by pixel after optimization. The better the data fits the model, the lower the value of the normalized residual will be. Thus, we use this parameter to evaluate %sO2 robustness.
2.6 |. PAT spectral features
As explained in the previous section, the envelope of the PAT beam-formed data was employed to calculate the functional features of the tissue. It has been shown in the literature that the spectra of the PAT beam-formed data also contain useful microstructural information of the imaged tissue [45–47].
To calculate the spectral features in this study, the spectrum of each beamline in the ROI is calculated using the fast-Fourier transformation (FFT). The average spectrum is then calculated by taking an average over spectra of all the beamlines in the ROI. Then a line is fitted to this average spectrum. The slope of this line (SS), its intercept with 0.5 MHz frequency line (SI) and its value in the middle of the frequency range (MBF) provide useful information about the micro-scale particles in the ROI. Examples of estimating these features in a malignant and a benign ovary are presented in Figure 4D,H, respectively. The readers are referred to Appendix B for further explanations about these features.
FIGURE 4.
Comparison of photoacoustic tomography (PAT) functional and spectral features of a malignant ovary (A-D) with a benign case (E-H). A and E are the co-registered ultrasound (US) and rHbT maps for the two types of ovarian masses. B and F show the co-registered US and %sO2 map calculated in the region of interest (ROI) indicated by the rectangles in A and E, respectively. The histogram of the %sO2 maps are shown in C and G. The mean spectra of the beamlines in the ROIs and their fitted lines are shown in D and H
2.7 |. Statistical analysis, feature selection, and classification
Each investigated ovarian mass in this study was included in either the benign/normal group (36 ovaries) or the malignant group (13 ovaries). To evaluate the difference between these two groups, a two-tailed t-test with the assumption of an unequal variance between the two groups was performed on each available feature. A feature with a P-value equal to or less than 0.05 was considered significant. Moreover, a random forest model was employed to rank the importance of each feature. The importance of each feature is calculated by averaging the Gini impurity at each node of the forest where that feature is used. Scikit-Learn, a module in Python, was used to calculate the features importance. Further information about feature selection using random forest models is found in Ref. [48].
Support vector machine (SVM) models were trained to distinguish benign/normal ovaries from those with malignant tumours. Models were developed for three subsets of uncorrelated significant features extracted from all the ovaries: radiology ranking (Rad rank) alone; uncorrelated PAT features (%sO2 mean, and rHbT mean, spectral intercept (SI)) alone; and the combination of uncorrelated PAT features and the radiologist’s ranking (%sO2 mean, rHbT mean, SI, and Rad rank). Each model was trained 100 times, each time using a random selection of 2/3 of the available dataset for training, and the rest of the data was reserved for testing. At each iteration the ROC of the training and testing data sets were computed. The mean receiver operating characteristic (ROC) curve and the area under the curve (AUC) over all iterations were then calculated for the training and testing data sets. The model which provided the highest AUC value for the testing data set was considered as the best model. Note that, the AUC of the training data set was found just to monitor the classifiers for overfitting (overfitting occurs when training AUC increases, but the testing AUC does not).
The mean AUC values were also used to calculate the 95% confidence interval (95% CI) for each constructed model using a binomial formula.
3 |. RESULTS
3.1 |. Ranking by attending radiologist
A boxplot of the ovarian mass ranking is shown in Figure 5A. Note that our expert radiologist successfully detected all malignant ovaries, but a few benign ovaries were misclassified as malignant.
FIGURE 5.
Box plots of the most significant features and rHbT. The P-value from a t-test on each feature is shown in the associated plot. The number of samples in each group is also shown below the x-axis of each plot. The %sO2 maps were calculated in 10 × 10 multi-pixels. The three-digit numbers that follow SI or SS in the plots indicate the optical wavelength at which the data was acquired
3.2 |. %sO2 calculation for blood tube phantoms
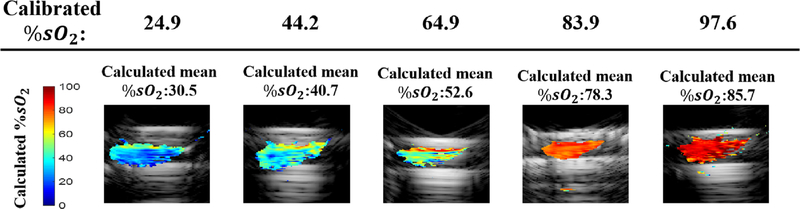
In Figure 2, we demonstrate the accuracy of our %sO2 calculations when the medium was filled with homogeneous Intralipid solution. The multi-pixel method was used before linear unmixing. Each column in this figure shows the result for a different blood tube, with its associated calibrated %sO2 value indicated at the top of the column. The images are from experiments in which the tube was located at a depth of 2.5 cm. Figure 3 summarizes the calculated mean %sO2 values at different depths. Each box plot in this figure shows the calculated mean % sO2 value at depths from 1 to 5 cm, with a step size of 0.5 cm. The mean absolute estimation errors at different depths for the blood tubes with the calibrated %sO2 of 24.9, 44.2, 64.9, 83.9 and 97.6 are 5.6%, 3.8%, 9.8%, 8.7% and 17.7%, respectively.
FIGURE 2.
The co-registered ultrasound (US) and %sO2 maps for different blood tubes located at the depth of 2.5 cm in intralipid. Each column indicates the %sO2 maps for a blood tube with the calibrated %sO2 value specified at the top of the column. The mean of the calculated %sO2 is above each sub image
FIGURE 3.
Calculated %sO2 values vs calibrated values. Each blood vessel was placed in intralipid at depths from 1 to 5 cm below the probe surface, in nine successive steps of 0.5 cm each. At each depth, the mean %sO2 value was calculated. Each box plot summarizes the mean calculated %sO2 at these 9 different depths
3.3 |. PAT features for patients
In Figure 4, the rHbT and %sO2 maps and PAT spectral features are compared for a malignant (A-D) and a benign (E-H) ovarian mass. Multi-pixel method is used before linear unmixing. In these figures, the lesion regions are indicated by the dashed rectangles in the overlaid rHbT (colour-scale image) and background US (grey-scale) images. The malignant ovary has a stronger and more concentrated rHbT map than the benign mass. This difference is also shown in Figure 5H, where the box plot of the mean rHbT for all the benign/normal masses is compared with that of the malignant cases. A significant difference was not observed between these two groups, in terms of the mean of rHbT maps (P = .06), however, the trend toward significant difference can be seen.
The histogram for each %sO2 map is shown below in Figure 4C,G. As can be seen, the mean %sO2 is lower for the malignant group. Also, the histogram of the malignant ovary is skewed toward the lower values, while the benign ovary has a %sO2 histogram skewed toward the higher values. Furthermore, Figure 5B–D verify the expected conclusion that in terms of skewness, mean, and energy, the %sO2 histograms (E and F) show a significant difference between the benign/normal and malignant groups. Because the %sO2 calculation is very sensitive to the PA signal to noise ratio (SNR), we have excluded four ovaries with a poor SNR in the final analysis. For these ovaries, the maximum signal levels at all wavelengths (elements of matrix g in Equation (1)) in the ROI were around the noise level of our system (60 mV). Therefore, performing linear unmixing on these noisy data led to a non-robust estimation of %sO2 in the ROI.
Finally, in Figure 4D,H, the mean spectra of the PAT beamlines in the ROI along with their fitted lines are presented for a malignant and a benign ovary, respectively. The malignant ovaries show a smaller value of SS (more negative) and a larger value of SI (less negative). The box plots in Figure 5E–G verify these observations. As demonstrated in Ref. [49], the lower SS in the malignant ovaries are related to the larger size of absorbers that are present in this type of masses, and the higher SI is associated to the larger size of the absorbers as well as a higher concentration of them in malignant ovaries.
3.4 |. Spatial filtering methods
We investigated the effect of the three smoothing filters on the calculated %sO2 and residual norm maps. Multi-pixel smoothing used a 10× 10 pixel window. The SD of the Gaussian filter and the Lorentzian parameter of the Lorentzian filter were fixed at sigma = 5 (see details in Appendix A). An example of the calculated %sO2 and normalized residual maps using different smoothing methods is shown in Figure 6 for the malignant ovary in Figure 4. As can be seen, the mean values of the calculated %sO2 maps and mean values of the normalized residuals do not change much when different spatial filter methods are used.
FIGURE 6.
The %sO2 (A-C) and the corresponding normalized residual (D-F) maps calculated using different smoothing methods for the malignant ovary in Figure 4. The multi-pixel size or sigma for each smoothing method is in the parenthesis in the image title. MP, GS and LZ in these plots represent multi-pixel, Gaussian, and Lorentzian smoothing methods, respectively
We calculated the histogram features of the %sO2 maps using different smoothing filters (Figure 7). The box plots of the malignant and benign/normal ovaries are located separately in these plots, and the P-values calculated by two-tailed t-tests between the benign/normal and malignant ovaries are shown in each plot. In terms of P-values, the different smoothing methods generate similar results, except for the energy of the %sO2 maps. For this parameter, unlike the multi-pixel and Gaussian filters, the Lorentzian filter does not show a statistically significant difference between the benign/normal and malignant groups (P > .05). Moreover, the box plots of the benign ovaries in the %sO2-skew and %sO2-energy plots calculated using Gaussian method are more spread out than the associated boxplots for the features calculated using the multi-pixel method. Therefore, for our data, the multi-pixel method is a better option than using the other two spatial filters, and it is used in data analysis throughout this manuscript.
FIGURE 7.
Box plots of histogram features calculated by using different smoothing methods. (A) Shows the mean of the %sO2 maps, and (B,C) are the skewness and energy of these maps, respectively. MP, GS and LZ indicate the multi-pixel, Gaussian, and Lorentzian smoothing methods. The P-values for each method are in the lower right of each plot
We also calculated the mean residual norm for all the ovarian masses, shown in Figure 8. The average and SD of the mean normalized residual maps using each method are shown on the right of each plot. Overall, % sO2 maps with a slightly lower normalized residual error are achieved by using the multi-pixel method. Therefore, this method was used for estimation of %sO2 maps in this manuscript. For a detailed discussion about choosing the size for each spatial filter, see Appendix A.
FIGURE 8.
Box plots of the means of the normalized residual error maps of all ovaries, calculated using MP, GS and LZ smoothing. The decimal numbers on the right are the mean ± SD of all the samples in each box
3.5 |. Feature ranking
As mentioned before, the significance of each extracted feature was evaluated using the Student’s t test. These features were then ranked in ascending order, meaning that features with lower P-values (shorter bar length in Figure 9A) were located higher in the ranking. Figure 9A shows the first eight most important features based on the P-value ranking. These eight features were ranked using a random forest model (explained Section 2.7) as well and presented in Figure 9B. In this plot, unlike the P-value ranking, the longer bars represent more important features.
FIGURE 9.
Feature ranking based on (A) P-value. (B) Random forest importance. In (A), more significant features have shorter bar length and located higher in the ranking. In (B), more significant features have longer bars and located higher in the ranking
Note that Rad rank is the most important feature in both rankings. Moreover, although rHbT is located on the bottom of the ranking in Figure 9A, the random forest model has ranked it as the second most important feature. Another interesting point about these plots is that three of the first four features in Figure 9A were highly correlated (%sO2-mean, %sO2-skew, %sO2 -energy), while in the random forest ranking (Figure 9B), no correlation was found between each pair of the first 4 features (The correlation between each two features was evaluated using Spearman’s method). This is an important consideration when developing classifiers as highly correlated features lead to overfitting our model. A second point to keep in mind when designing classifiers to decrease the chance of overfitting is that the number of features that is used for training should be less than one-tenth of the training size [50]. The training size in this study was , so we did not use more than four features. As the first four features in the random forest ranking are not correlated, designing the classifiers based on this ranking seems to reduce the chance of overfitting.
3.6 |. Classification
We developed SVM classifiers to distinguish between “malignant” and “benign/normal” groups. In this case, the data set was randomly divided into two groups. The first group included two-thirds of the data used for training, and the rest of the data was employed for testing the classifiers. To lower the chance of overfitting, this process was repeated 100 times.
Four SVM models were trained in this study. The first model was trained using the single radiology feature (Rad rank). The second one was trained using three most important PAT features based on the random forest ranking (rHbT, %sO2 mean, and SI 730). The third model was trained using the combination of the features used in the last two models. Finally, in the last model, rHbT was removed from the features set. For each model, the 95% confidence of interval (CI) was computed. Moreover, the mean ROC of each of the training and testing data sets for these four models were calculated and are shown in Figure 10. In the legend of each plot, the left-hand side of each equation represents the features set employed to train the classifiers and the decimal numbers on the right-hand side are the mean AUC values and the 95% CI associated with the ROC of that classifier. As can be seen on the plots, when just radiology ranking was used to train the SVM classifier, a mean AUC value of 0.85 (95% CI: 0.81–0.89) was achieved. On the other hand, the performance of the classifier was not impressive when just PAT features were used to train the classifiers (mean testing AUC = 0.77 (95%CI: 0.73–0.81). By combining the radiology and PAT features, a superior AUC value of 0.92 (95%CI: 0.89–0.95) on the testing data set was achieved. However, when rHbT was removed from the features set, the performance of the SVM classifier on the testing data set improved to 0.93 (95% CI: 0.90–0.96).
FIGURE 10.
The mean receiver operating characteristic curves (ROCs), area under the curves (AUCs), and 95% CI of the four SVM models developed to classify normal/benign ovaries from malignant ovaries for training (top) and testing (bottom) data sets
Two of the thirteen malignant ovaries imaged in this study were serous borderlines, and another one was Sertoli-Leydig cell tumour. These types of malignancies are considered low-grade cancers. The estimated %sO2 maps for these masses were similar to those for high-grade cancers. However, their rHbT maps and spectral features were close to the ovaries in the benign group. Without these outliers, performing the t-test on rHbT resulted in a P-value of 0.03. However, including these outliers, P-value between all malignant lesions and the benign group is marginal significant (P= .06). As these ovaries have some features similar to the malignant group and some others close to the benign group, performing a multi-class classification to divide the ovaries into three groups of high-grade cancers, low grade cancers, and benign ovaries might be a better choice. However, as we currently have a very limited number of the low-grade cancers (n = 3), it is not feasible to train such a model at this time.
To evaluate the contribution of other significant %sO2 features (skewness and energy), we combined these features with other types of extracted features and constructed new models. Each model was developed once by including rHbT in, and a second time, by excluding this feature from the features set. The performances of different classifiers are given in Table 2. By comparing the testing AUC values, we concluded that rHbT did not improve the performance of any of the constructed models which could be due to the inclusion of the three low-grade tumours in the cancer group.
TABLE 2.
Area under the curve (AUC) of models constructed using different %sO2 features, with and without rHbT
| Features (predictors) | Training AUC (0.95% CI) | Testing AUC (0.95% CI) |
|---|---|---|
| %sO2 mean, SI730, Rad rank | 0.95 (0.93–0.97) | 0.93 (0.90–0.96) |
| %sO2 mean, SI730, Rad rank, rHbT | 0.95 (0.93–0.97) | 0.92 (0.89–0.95) |
| %sO2 skew, SI730, Rad rank | 0.95 (0.93–0.97) | 0.92 (0.89–0.95) |
| %sO2 skew, SI730, Rad rank, rHbT | 0.95(0.92–0.97) | 0.92 (0.89–0.95) |
| %sO2 energy, SI730, Rad rank | 0.93 (0.90–0.96) | 0.92 (0.89–0.95) |
| %sO2 energy, SI730, Rad rank, rHbT | 0.93 (0.91–0.96) | 0.90(0.87–0.93) |
4 |. DISCUSSION AND SUMMARY
In this work, we used the linear unmixing method to calculate %sO2 values from phantom and clinical PAT data. In our earlier publication [34], the %sO2 maps were calculated by solving linear equations independently for every single pixel. A spatial filter was then applied to smooth the %sO2 maps. Here, before the linear equations are solved, the value of the envelope data at each pixel is replaced with the average value of the envelope data for 100 surrounding pixels, which makes the results more robust to measurement errors. Additionally, whereas our earlier study computed only the mean %sO2 from each %sO2 map, here we calculated six histogram features of the %sO2 maps (mean, skewness, energy, variance, kurtosis, entropy), and the first three features were statistically significant. Furthermore, our earlier study ranked features based on their p-values estimated from t-tests. Here, a random forest model has been utilized for ranking the features, reducing the chance of overfitting. Finally, the radiology score has been used in this study, which is a significant step toward the clinical translation of the technology.
We demonstrated the accuracy of estimating the %sO2 for different calibrated blood tube phantoms embedded inside a homogeneous Intralipid medium. Whole blood consists of a substantial number of impurities, including enzymes that cause blood coagulation. Therefore, we followed the standard process of centrifuging whole blood in order to collect RBCs. The RBCs were diluted with saline water and placed inside a hypoxia chamber, where oxygen and nitrogen concentrations were controlled to produce the desired %sO2 condition. A 60% saline solution was used to dilute the RBCs and avoid saturation of the oximeter used to calibrate each blood sample. Since the goal was to estimate %sO2 of the blood sample using PAT, diluting the RBCs with 60% saline provided strong PA signals without affecting the results.
We also calculated %sO2 using clinical data. The calculated %sO2 and rHbT maps, along with SI and Rad rank, were then used to construct different SVM models. The highest AUC value on the testing data set was found for the model constructed from %sO2 mean, SI730, and Rad rank. In our earlier publication [34], we developed two classifiers, models A and B. Model A distinguished “benign/normal” masses from those with “epithelial ovarian cancers”. For the testing data, the best AUC of 0.94 was achieved using an SVM classifier. Model B distinguished “benign/normal” lesions from those with “epithelial ovarian cancers and other types of tumours”. With this model, we achieved a best testing AUC of 0.93. In the present manuscript with a larger sample size, we have developed one classifier, model C, which differentiates “benign/normal” masses from “epithelial cancers and other types of tumours”. With the addition of the radiology scores, an AUC of 0.93, closely similar to that of model B, was also achieved. In practice, model C has more translational potential because it integrates conventional imaging with PAT functional imaging features.
Depleted oxygen and glucose in the microenvironment of ovarian cancer limits the potential for cellular metabolic plasticity, which has been appreciated as playing a key role in cancer progression and chemoresistance [51]. In a recent study [52], Qiu et al reported the first evidence that levels of a natural antisense transcript of hypoxia-inducible factor 1 (aHIF) were increased in epithelial ovarian cancer (EOC) tissues and were upregulated by hypoxia in EOC cells. Functional data revealed that aHIF knockdown accelerated cell apoptosis under hypoxia and inhibited EOC tumorigenesis and tumour growth in vivo. PAT provides blood oxygen saturation, which is related to the tumour oxygen microenvironment and may have a greater role in ovarian cancer diagnosis and assessing chemotherapy response.
Our ovarian mass reader evaluation is a single institution experience, and the ovarian mass ranking was performed by two highly experienced radiologists with over 55 years of combined expertise in pelvic US. Their experience and breadth in pelvic imaging does not reflect the wider radiology community and will not be the case as PAT gets integrated in the clinical imaging armamentarium. In fact, we anticipate that the addition of PAT to ovarian mass imaging may bring the confidence and accuracy of a general radiologist into the realm of a specialist.
Currently, there are limitations of the photoacoustic technique. First, the typical penetration depth of PAT is about 5 cm. In cystic ovarian tissue, however, the PAT penetration depth may reach 6 to 7 cm. We were able to image more than 95% of the ovaries at these imaging depths. A second limitation is the assumption of wavelength-independent fluence in the linear unmixing method. Quantitative %sO2 calculation without this assumption has been a hot topic in the field of PAT in recent years, and several methods, including deep learning models and model-based algorithms, have been proposed [53–58]. Nevertheless, although interesting results have been observed for simulation and phantom data, applying these methods to clinical data remains a challenge.
In summary, our study with a considerable patient population has shown for the first time that %sO2 and its distribution can play a significant role in ovarian cancer diagnosis. Combined with the diagnosis of radiologists, the overall accuracy of identifying both high grade epithelial ovarian cancer and low-grade ovarian tumour can achieve an AUC of 0.93. A large clinical study is currently underway to validate these initial findings.
Supplementary Material
ACKNOWDELGMENTS
This work was supported by NCI (R01CA151570, R01CA237664). We gratefully acknowledge the efforts of Ruth Holdener and Lynne Lippmann in coordinating the study schedules and identifying and consenting patients to the study. We thank the entire GYN oncology group for helping identify patients, and the Radiology US technologists for helping with US scans. The help of Dr Stephen Rogers and Dr Allan Doctor with blood phantom preparation is also appreciated. Guang Yang acknowledges the support of an Imaging Science Pathway Trainee Fellowship from Washington University. Finally, we sincerely thank James Ballard for manuscript editing.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
Data available based on reasonable request.
REFERENCES
- [1].American Cancer Society, Cancer Facts and Figures, American Cancer Society, Atlanta, GA: 2019. [Google Scholar]
- [2].Henderson JT, Webber EM, Sawaya GF, JAMA 2018, 319 (6), 595. 10.1001/jama.2017.21421. [DOI] [PubMed] [Google Scholar]
- [3].Domchek SM, Friebel TM, Neuhausen SL, Wagner T, Evans G, Isaacs C, Garber JE, Daly MB, Eeles R, Matloff E, Tomlinson GE, Van’t Veer L, Lynch HT, Olopade OI, Weber BL, Rebbeck TR, Lancet Oncol. 2006, 7, 223. [DOI] [PubMed] [Google Scholar]
- [4].Finch A, Beiner M, Lubinski J, Lynch HT, Moller P, Rosen B, Murphy J, Ghadirian P, Friedman E, Foulkes WD, Kim-Sing C, Wagner T, Tung N, Couch F, Stoppa-Lyonnet D, Ainsworth P, Daly M, Pasini B, Gershoni-Baruch R, Eng C, Olopade OI, McLennan J, Karlan B, Weitzel J, Sun P, Narod SA, Hereditary Ovarian Cancer Clinical Study Group, JAMA 2006, 296, 185. [DOI] [PubMed] [Google Scholar]
- [5].Kwon JS, Tinker A, Pansegrau G, McAlpine J, Housty M, McCullum M, Gilks CB, Obstet. Gynecol. 2013, 121, 14. [DOI] [PubMed] [Google Scholar]
- [6].Shuster LT, Gostout BS, Grossardt BR, Rocca WA, Menopause Int. 2008, 14, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mathieu KB, Bedi DG, Thrower SL, Qayyum A, Bast RC Jr., Ultrasound Obstet. Gynecol. 2018, 51(3), 293. 10.1002/uog.17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Addley H, Moyle P, Freeman S, Clin. Radiol. 2017, 72(11), 981. 10.1016/j.crad.2017.07.014. [DOI] [PubMed] [Google Scholar]
- [9].Iyer VR, Lee SI, AJR 2010, 194(2), 311. [DOI] [PubMed] [Google Scholar]
- [10].Mapelli P, Incerti E, Fallanca F, Gianolli L, Picchio M, Nucl QJ. Med. Mol. Imaging 2016, 60(2), 93. [PubMed] [Google Scholar]
- [11].Kumar R, Chauhan A, Jana S, Dadparvar S, Expert Rev. Anticancer Ther. 2006, 6(7), 1033. [DOI] [PubMed] [Google Scholar]
- [12].Oraevsky AA, Karabutov AA, Optoacoustic tomography. in Biomedical Photonics Handbook (Ed: Vo-Dinh T), CRC, Boca Raton, FL: 2003. [Google Scholar]
- [13].Bouchard R, Sahin O, Emelianov S, IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2014, 61(3), 450. [DOI] [PubMed] [Google Scholar]
- [14].Valluru KS, Willmann JK, Ultrasonography 2016, 35(4), 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zackrisson S, van de Ven SMWY, Gambhir SS, Cancer Res. 2014, 74(4), 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Su R, Ermilov S, Liopo A, Oraevsky A, Nucl. Instrum. Methods Phys. Res. A 2013, 720, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang G, Amidi E, Nandy S, Mostafa A, Zhu Q, Photoacoustics 2019, 13, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Luo H, Yang G, Zhu Q, Opt. Lett. 2020, 45(3), 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Weidner N, Semple JP, Welch WR, Folkman J, Engl N. J. Med. 1991, 324(1), 1. [DOI] [PubMed] [Google Scholar]
- [20].Vaupel P, Kallinowski F, Okunieff P, Cancer Res. 1989, 49, 6449. [PubMed] [Google Scholar]
- [21].Kruger RA, Kuzmiak CM, Lam RB, Reinecke DR, del Rio SP, Steed D, Med. Phys. 2013, 40(11), 113301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Heijblom M, Piras D, van den Engh FM, van der Schaaf M, Klaase JM, Steenbergen W, Manohar S, Eur. Radiol. 2016, 26(11), 3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Menke J, Eur. Radiol. 2015, 25(8), 2205. [DOI] [PubMed] [Google Scholar]
- [24].Wong TTW, Zhang R, Hai P, Zhang C, Pleitez MA, Aft RL, Novack DV, Wang LV, Sci. Adv. 2017, 3(5), 1602168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Garcia-Uribe A, Erpelding TN, Krumholz A, Ke H, Maslov K, Appleton C, Margenthaler JA, Wang LV, Sci. Rep. 2015, 5, 15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kruger RA, Lam RB, Reinecke DR, del Rio SP, Doyle RP, Med. Phys. 2010, 37(11), 6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Becker A, Masthoff M, Claussen J, Ford SJ, Roll W, Burg M, Barth PJ, Heindel W, Schäfers M, Eisenblätter M, Wildgruber M, Eur. Radiol. 2018, 28(2), 602. [DOI] [PubMed] [Google Scholar]
- [28].Dogra VS, Chinni BK, Valluru KS, Joseph JV, Ghazi A, Yao JL, Evans K, Messing EM, Rao NA, J. Clin. Imag. Sci. 2013, 3, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang X, Roberts WW, Carson PL, Wood DP, Fowlkes JB, Biomed. Opt. Express 2010, 1(4), 1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhou Y, Tripathi SV, Rosman I, Ma J, Hai P, Linette GP, Council ML, Fields RC, Wang LV, Cornelius LA, J. Invest. Dermatol. 2017, 137(6), 1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang M, Zhao L, He X, Su N, Zhao CY, Tang H, Hong T, Li W, Yang F, Lin L, Zhang B, Zhang R, Jiang Y, Li C, Biomed. Opt. Express 2017, 8(7), 3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yang G, Amidi E, Chapman W, Nandy S, Mostafa A, Abdelal H, Alipour Z, Chatterjee D, Mutch M, Zhu Q, J. Biomed. Opt. 2019, 24(12), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nandy S, Mostafa A, Hagemann IS, Powell MA, Amidi E, Robinson K, Mutch DG, Siegel C, Zhu Q, Radiology 2018, 289(3), 740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Amidi E, Mostafa A, Nandy S, Yang G, Middleton W, Siegel C, Zhu Q, Biomed. Opt. Express 2019, 10(5), 2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li ML, Oh JT, Xie X, Ku G, Wang W, Li C, Lungu G, Stoica G, Wang LV, Proc. IEEE 2008, 96(3), 481. [Google Scholar]
- [36].Wang X, Xie X, Ku G, Wang LV, Stoica G, J. Biomed. Opt. 2006, 11(2), 24015. [DOI] [PubMed] [Google Scholar]
- [37].Needles A, Heinmiller A, Sun J, Theodoropoulos C, Bates D, Hirson D, Yin M, Foster FS, IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2013, 60(5), 888. [DOI] [PubMed] [Google Scholar]
- [38].Wang LV, Wang X, Ku G, Xie X, Stoica G. High-resolution spectroscopic photoacoustic tomography for noninvasive functional imaging of small-animal brains in vivo. The Second Asian and Pacific Rim Symp. on Biophotonics, 2004. APBP 2004, 246–247 [Google Scholar]
- [39].Hennen SN, Xing W, Shui YB, Zhou Y, Kalishman J, Andrews-Kaminsky LB, Kass MA, Beebe DC, Maslov KI, Wang LV, Exp. Eye Res. 2015, 138, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hochuli R, An L, Beard PC, Cox BT, J. Biomed. Opt. 2019, 24(12), 121914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lavaud J, Henry M, Gayet P, Fertin A, Vollaire J, Usson Y, Coll JL, Josserand V, Int. J. Biol. Sci. 2020, 16(9), 1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li X, Zhang S, Wu J, Huang S, Feng Q, Qi L, Chen W, IEEE Trans. Med. Imaging 2020, 39, 3463. [DOI] [PubMed] [Google Scholar]
- [43].Kim J, Park B, Ha J, Steinberg I, Park EY, Choi W, Hooper S, Gambhir SS, DjLim C Kim. Multispectral photoacoustic assessment of thyroid cancer nodules in vivo. Photons Plus Ultrasound: Imaging and Sensing 2020; 1124004. [Google Scholar]
- [44].Durairaj DA, Agrawal S, Johnstonbaugh K, Chen H, Krishna Karri SP, Kothapalli SR. Unsupervised deep learning approach for photoacoustic spectral unmixing. Photons Plus Ultrasound: Imaging and Sensing 2020; 112403. [Google Scholar]
- [45].Feng T, Perosky JE, Kozloff KM, Xu G, Cheng Q, Du S, Yuan J, Deng CX, Wang X, Opt. Express 2020, 23(19), 25217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Leng X, Chapman W, Rao B, Nandy S, Chen R, Rais R, Gonzalez I, Zhou Q, Chatterjee D, Mutch M, Zhu Q, Biomed. Opt. Express 2018, 9(11), 5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hysi E, Wirtzfeld LA, May JP, Undzys E, Li SD, Kolios MC, Photoacoustics 2017, 2017(5), 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Menze BH, Kelm BM, Masuch R, Himmelreich U, Bachert P, Petrich W, Hamprecht FA, BMC Bioinformatics 2009, 10, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xu G, Dar IA, Tao C, Liu X, Deng CX, Wang X, Appl. Phys. Lett. 2012, 101(22), 221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Harrell F, The PHGLM procedure. in SUGI Supple- Mental Library User’s Guide, SAS Institute, Cary, NC: 1983, p. 267. [Google Scholar]
- [51].Emmings E, Mullany S, Chang Z, Landen CN Jr., Linder S, Bazzaro M, Int. J. Mol. Sci. 2019, 20(1), 229. 10.3390/ijms20010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Qiu JJ, Lin XJ, Zheng TT, Tang XY, Hua KQ, Onco. Targets Ther. 2018, 14(11), 9101. 10.2147/OTT.S173816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cox BT, Laufer JG, Beard PC. The challenges for quantitative photoacoustic imaging. Proc. SPIE 7177, Photons Plus Ultrasound: Imaging and Sensing 2009;717713. [Google Scholar]
- [54].Pulkkinen A, Cox BT, Arridge SR, Kaipio JP, Tarvainen T, Inverse Probl. 2014, 30(6), 65012. [Google Scholar]
- [55].Kirchner T, Gröhl J, Maier-Hein L, J. Biomed. Opt. 2018, 23(5), 56008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li M, Tang Y, Yao J, Photoacoustics 2018, 10, 65. 10.1016/j.pacs.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Cai C, Deng K, Ma C, Luo J, Opt. Lett. 2018, 43(12), 2752. [DOI] [PubMed] [Google Scholar]
- [58].Bench C Towards accurate quantitative photoacoustic imaging: learning vascular blood oxygen saturation in 3D. arXiv: 2005.01089 2020. https://arxiv.org/abs/2005.01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available based on reasonable request.