Abstract
INTRODUCTION:
The study of Alzheimer’s disease (AD) has revealed biological pathways with implications for disease neuropathology and pathophysiology. Pathway-level effects may be mediated by covariates such as age or sex.
METHODS:
Gene set enrichment methods test hypotheses at the level of biological pathways. We introduce a method for quantifying gene set enrichment ( gsLRT ), which accounts for covariate effects. We test for age and sex interactions with protein expression in the context of AD and compare results between human and mouse species.
RESULTS:
Gene sets identified by gsLRT are validated by previous AD studies. Differences between gsLRT results on mouse and human datasets are observed.
DISCUSSION:
Characterizing biological pathways involved in AD builds on important work involving single gene drivers. Our method highlights commonalities and differences between human AD and mouse models, which may inform the development of higher fidelity models for the study of AD.
Keywords: Genetics of Alzheimer’s disease, Proteomics, Alzheimer’s disease mouse model development, Biostatistics
1. Background
Strategies for translational research in Alzheimers disease have moved from the characterization of individual variants (SNPs, metabolites, proteins) to the understanding of larger-scale genomic pathways that form the molecular basis of the disease [1–6]. A pathway model of late-onset Alzheimers Disease (LOAD) must integrate numerous genetic, clinical, and environmental factors to more accurately represent the etiology of human LOAD. Relevant factors include: (1) genetic risk factors, e.g. APOE genotype or sex, (2) environmental factors including education level and diet, (3) heterogeneity of biological-behavioral symptoms, and (4) prolonged degenerative processes with pathophysiology beginning years before the onset of detectable symptoms [7] and with accompanying adaptive changes. The motivation for this study is to propose a pathway-based statistical approach to AD that accounts for these factors while informing the development and validation of clinically-relevant mouse models for late-onset or sporadic Alzheimers disease (LOAD).
The use of model systems to identify, describe, and design treatments for human diseases has expanded dramatically over the past 30 years. This increase has kept pace with the technological advancements that have permitted a wide range of advanced approaches to the study of human disease mechanisms. While critical to improving human health, the use of model systems comes with many assumptions. Of primary importance is the assumption that the model system’s pathophysiological pathways are either identical or closely related to those of humans. This assumption leads to the belief that experimental phenomena observed in an animal model, such as a mouse, will forecast similar phenomena in humans. However, clear exceptions exist, which impact the translation of animal model data to humans with disease. These exceptions may require re-evaluation of data generated in mouse studies, redesigning of mouse models, or the use of statistical methodologies that permit evaluation of the differences. In this study, we have developed and tested such a methodology, which permits pathway-level evaluation of differences between mouse and human responses to normal and disease conditions.
Our method uses gene-level data (proteomics, transcriptomics, imaging) collected from human or mouse models to generate significance scores at the level of biological pathways. Informally, the significance scores report the novelty of the observed experimental outcome under the assumption that there is no difference between normal and disease states. Moreover, our approach generates these scores while accounting for age-specific, sex-specific and APOE genotype-specific differences in the human subjects or model systems. It can also test for the significance of the relationship between pathway-level interactions with each of these factors and presence of AD. We validate this method using human proteomic data on Alzheimers disease development and test for interactions with the factors of age, sex and APOE genotype. By applying the same approach to CVN-mouse proteomics data, we are able to compare the pathway significance scores from mouse models with a human-like immune background to those obtained from human Alzheimers patients.
2. Methods
The driver of our pathway-based approach to understanding LOAD in humans and mice is a novel approach to gene set enrichment. Gene set enrichment methods are popular tools for addressing the challenge of finding biologically relevant relationships between sample phenotypes and underlying genomic factors. These methods condense gene-level statistics into gene set scores, each of which corresponds to a group of genes with a common annotation. Gene set annotations may be derived from experiment, computation, or literature-search, though the gene set annotations considered in this article are based on membership to signaling pathways.
Many gene set methods assume that the only piece of information known about a sample is its class label, for instance normal versus AD. However, experimental datasets often include additional sample information, which may be relevant to the particular biological mechanism through which a complex diesease manifests. Motivated by documented age- and sex-driven differences in Alzheimer’s Disease (AD) presentation, we developed a framework for gene set enrichment that takes this information into account. Unlike the method proposed by [8] (ROAST), which relies on the assumption of normally-distributed gene expression profiles, our method (gsLRT) generates likelihood ratio statistics from a pair of nested logistic regression models. The procedure is flexible enough to test for a wide number of complicated gene set hypotheses while adjusting for variation explained by the covariates included in both models.
2.1. Enrichment statistics and p-values
Existing methods that use linear models for gene set enrichment treat each gene expression profile as the response variable in a linear model [8, 9]. They assume that each gene expression profile is normally distributed within categories determined by the diagnosis and experimental design. We remove the assumption of normality by instead treating the diagnosis as the response variable for each of our models. Specifically, we assume a matrix of measurement values (e.g. mRNA expression) for m genes and n samples, a matrix of covariates for the samples, and a binary vector , which codes for the diagnosis (e.g. cognitively normal or AD) for each sample. For each gene j = 1,…,m, we fit the nested models
| (1) |
| (2) |
and find the maximum likelihood estimators of the model coefficients , , .
We define statistics measuring the additional explanatory power of gene measurement profile gj over that of the fixed covariate matrix X as a difference in model log-likelihoods operating at the gene level, which we define as
| (3) |
where ℓ(·) denotes the log-likelihood of the logistic model. We define enrichment score for gene set γk as
| (4) |
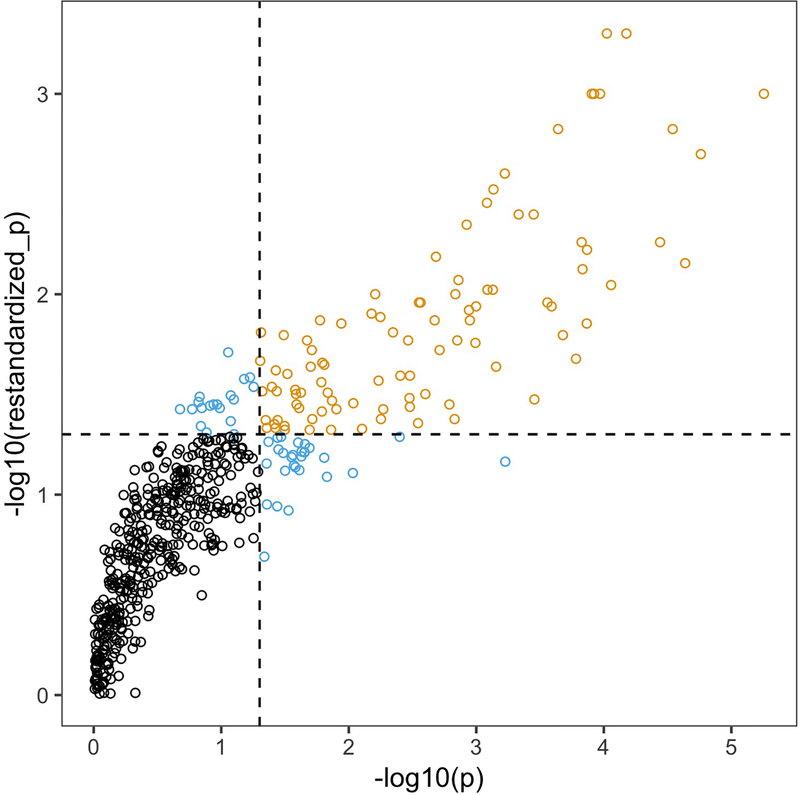
Obtaining appropriate p-values for enrichment scores has been a matter of intense discussion in the literature on gene set testing [10–13]. Most of the discussion has centered around the choice of null hypothesis concerning the relationship among genes. While the assumption that the gene-level statistics Λj are independent leads to a computationally efficient testing procedure, correlation among gene expression profiles in a gene set—possibly due to a violation of independence—will give the corresponding enrichment statistic higher variance, producing overly optimistic levels of significance (Figure 1). We choose to report more conservative p-values obtained by permutation sampling, which is computationally expensive. In practice, one may want to use the independence assumption on larger data sets when computational speed and ranking of gene sets are priorities over type I error control.
Figure 1:
Comparison of p-values obtained from the independence assumption test (x-axis) and the permutation test (y-axis) run on 542 gene sets in Alzheimer’s proteomics data (Section 5). Black color indicates that neither test rejects, gold indicates that both tests reject, and light blue indicates that only one test rejects. Note the difference in ranges on the axes: the permutation test is more conservative than the independence assumption test.
The enrichment scores produced by gsLRT quantify the degree to which including the terms in alters the model log-likelihood. They capture how much the inclusion of effects from the genes within a gene set improves the ability to discriminate between diagnoses. They are directionless in the sense that the likelihood ratio test will reject when a significant improvement in the model likelihood is made, regardless of whether the expression profiles within a gene set are positively or negatively associated with the diagnosis.
The permutation p-values report a significance level with respect to the second null hypothesis described by [10], or equivalently the focused testing described by [8]. They represent the significance of the cumulative improvement in model log-likelihood with respect to the genes in a gene set without reference to other genes in the full collection. As such, permutation p-values from a focused test are particularly appropriate for assigning significance to gene sets when many genes in a dataset have a putative association with the diagnosis of interest, as is the case in our Alzheimer’s study. Gene sets with low permutation p-values are unlikely to contain only genes whose measurement levels have no association to the diagnosis.
2.2. Description of datasets used
After conducting simulation benchmarks (Appendix B), we applied gsLRT to the ADNI Biomarkers Consortium CSF Proteomics MRM data set. These data were obtained from the Alzheimers Disease Neuroimaging Initiative (ADNI1) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a publicprivate partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimers disease (AD). The ADNI study was conducted at multiple sites within North America with retrospective assessment of imaging (MRI and FDG-PET) and biomarkers from cognitively normal controls (CN), MCI subjects, and subjects with AD. MCI subjects defined by the ADNI protocol included subjects with MMSE scores >23, isolated memory impairment based on education adjusted memory scores on the Wechsler Memory Scale Logical Memory III scale and a Clinical Dementia Rating (CDR) [14] global score of 0.5. Study participants with AD were diagnosed using standard criteria (NINCDS/ADRDA criteria for probable AD [15]) and were of mild severity at enrollment, with MMSE scores between 20–26 and CDR scores between 0.5 and 1.0. The cohort in our study was comprised of 152 individuals, 86 classified as cognitively normal and 66 with a diagnosis of AD (following ADNI protocol of NINCDS/ADRDA criteria for probable AD).
Experimental details for the multiplexed proteomics assay including quality control criteria, normalization and the statistical analysis plan are described in [16]. The assay was designed to determine the ability of a panel of peptides in the cerebrospinal fluid (CSF) to discriminate among disease states (normal cognition, MCI and AD), using a targeted mass spectrometry-based assay to qualify the candidate AD biomarkers. The proteomics panel consisted of 567 peptides representing 222 proteins with 2 mass transitions monitored per peptide. The Biomarkers Consortium completed data processing, peptide quantification, mapping of peptides to proteins and normalization [16]. We used the normalized protein intensity data in our analysis.
The cohort of 12 mice used in our cross-species analysis contained 6 wildtype mice and 6 APPSwDI/Nos2−/− (CVN) mice. Each cohort had 3 males and 3 females with a mean age across cohorts of 56.3 months, SD 4.6. Numerous mouse models have been generated to recreate the brain pathology observed in humans with Alzheimers disease. The most common approach to modeling has been to recreate the dominant AD pathologies, aggregated Abeta peptides and amyloid deposits and tau pathology. To generate these models, others have incorporated into mice mutations in specific genes that lead to amyloid or phospho tau accumulation in human brains. This approach clearly shows the accumulation of AD-like pathological deposits in mouse brain. Unfortunately, recreating human amyloid deposition has not led to the loss of neurons leading to brain volume changes in mice. These critical pathologies of human AD are not observed in the widely used AD mouse models. Thus, the commonly used available mouse models do not recreate human brain disease pathology and are unlikely to provide critical information to understand the underlying pathology of AD in humans.
It has been observed that mouse and human immune responses were different, both in the induction of the NOS2 gene and in the generation of NO by the NOS2 protein. Thus, we used a mouse strain that more closely represented human NOS2 activation and human NO production [17]. When this novel mouse strain was crossed to mice expressing a mutation in APP to generate the APPSwDI/mNos2−/− (CVN-AD) line, this relatively limited, but striking difference in NOS2 and NO production impacted immune-regulated metabolic profiles in the brain [18–20]. These changes lead to loss of neurons, loss of regional connectivity and loss of regional brain volumes, which has been shown using Magnetic Resonance Imaging [21, 22]. Thus, brain changes in the CVN-AD mice closely mimic critical human AD pathology, which other commonly used models do not, that is, neuronal loss leading to significant brain volume changes, and loss of regional connectivity. Because of these structural and pathological commonalities between the APPSwDI/mNos2−/− mice and human AD, we have pushed further in our generation of mouse models by replacing the mouse NOS2 gene with the Human NOS2 gene which recreates a similar profile as our previous model.
The use of the APPSwDI/mNos2−/− model in this study was to compare mice that expressed AD-like pathology in the brain with available human data sets of the same kind. The importance was to uncover functional gene/protein networks that were significantly different from humans as well as to find those areas that were notably similar. The analysis methodology developed here and presented in this manuscript has enabled us to see where this mouse mimics corresponding human data sets and where it fails to mimic them. The data have pointed out very clearly that we have accomplished some of our modeling goals, but are missing other key elements. It shows us where to test and where to modify the genetic/protein landscape further. Overall, at the least, this combined approach of innovative modeling brings us closer to having in hand a more useful, testable model for drug development and translation of drug discovery to humans with AD.
We have used the human CSF for comparison with the mouse brain because CSF is robust surrogate of brain chemistry. The brains interstitial fluid is in direct contact with the CSF and the chemical composition of the CSF reflects the cellular chemical environment in the CNS, including metabolites that are entering or leaving. Because the CSF directly receives metabolites from the entire brain, its chemical composition is an average of all the different brain regions, but as we used whole brain extracts, this is not a major concern for our study. In confirmation of our arguments, [23] obtained comparable results in brain and CSF metabolomes from APPswe/PS1deltaE9 mice, validating the use of CSF as an accurate surrogate of the human brain for use in this study. Moreover, future studies will use analytical methods developed in this publication to compare different stages of disease, using CSF from living human subjects for comparison with animal models. Preparation of mouse brain tissue and subsequent proteomics analysis are each described in Appendix C.
3. Results
We first used gsLRT to test for AD gene set enrichment, including each individual’s age, sex, and APOE genotype as covariates, but not including any interactions between these covariates and protein expression levels. We then tested for AD gene set enrichment with respect to protein expression interactions with sex and age, respectively. For these latter tests, the reference models in the nested logistic regressions were those fitted with only main effects for the covariates and the protein expression profile. The gene sets considered in our analysis were downloaded from MSigDB [24, 25], a repository of annotated gene sets. The gene set collections used were the Hallmark, C2, and C5 collections, which include GO [26, 27], KEGG [28], and REACTOME [29] pathways, among others. We then limited our reference set to include only those gene sets with an overlap of greater than 5 genes and fewer than 100 genes with the ADNI data set, yielding a total of 542 gene sets. Our results therefore extend only to the genes that appear in the filtered gene sets. Below we report the gene set annotations as they appeared in their original collections, but our interpretation focuses on the member genes that overlapped with those included in our study.
The study of sex-specific differences in the pathophysiology and epidemiology of AD has generated a large body of research [30–38]. We are particularly interested in how female biological sex interacts with age and APOE ϵ4 genotypes to affect susceptibility to AD. This is a contentious topic [35, 38], and we believe that addressing the interaction between these factors will help resolve the controversy, and possibly lead to strategies for AD prevention and treatment. We hypothesized that female biological sex and APOE genotype interact at an underlying, immune-regulated metabolic pathway in the brain. Immunity, like sex, age, and APOE genotype, is a primary disease factor in AD and impacts both the onset and pathological features of neurodegenerative events.
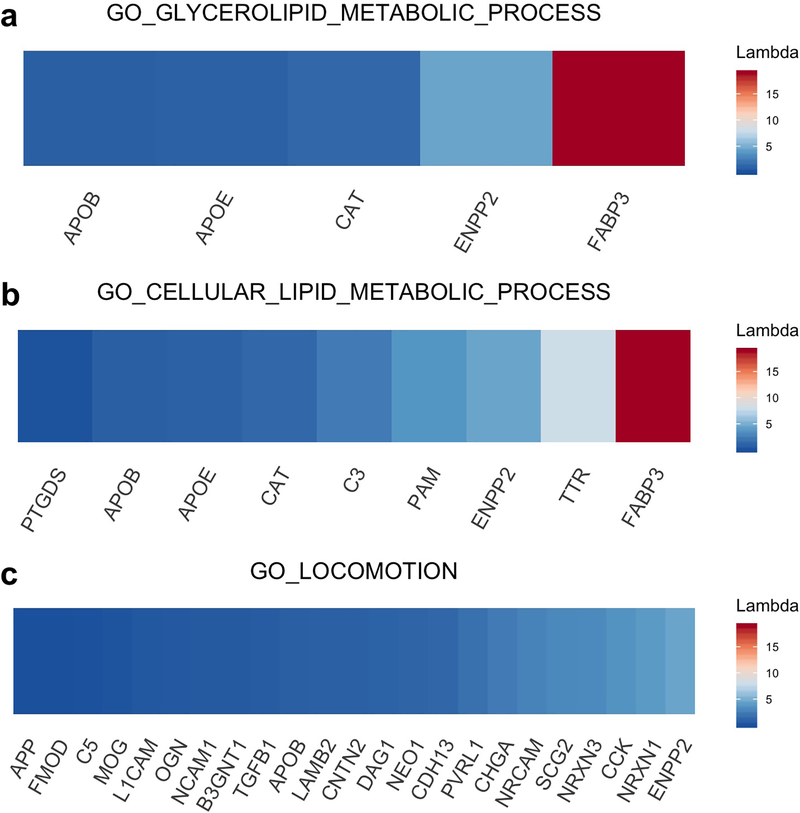
The five gene set enrichment results with the lowest permutation p-values for the class of models that did not include interaction terms for age, sex or APOE genotype are reported in Table 1. Importantly, because of the dependence among gene set tests with overlapping membership, the p-values reported here and in the following tables are not adjusted for multiple testing, and therefore do not have theoretical guarantees for control of False Discovery Rate (FDR). A group of related protein signatures involving lipid metabolism show the strongest enrichment. Most of these signatures include several apolipoproteins (APOE, APOB, APOD) and fatty acid binding protein (FABP3) (Figure 2). The signatures range in size from 5–14 proteins, constituting relatively small to moderate size biological networks. Several metabolic signatures are not identified as significantly enriched for this model. These signatures include glucose metabolism (p=0.34), metabolism of carbohydrates (p=0.37) and heme metabolism (p=0.38). The Gene Ontology pathway for locomotion is a good negative control since this pathway is not identified as significantly enriched (p=0.20) despite inclusion of the APOB and APP proteins in the pathway. This pathway consists of 14 genes and the lack of a strong or moderate difference in protein concentration between the AD and control groups for neuronal and neural cell adhesion proteins (NRCAM, NCAM1), L1 cell adhesion (L1CAM) and neurexins (NRXN1, NRXN3) accounts for the lack of significance at the pathway level.
Table 1:
gsLRT results on ADNI data with no interaction term
| Gene Set | p |
|---|---|
| GO_GLYCEROLIPID_METABOLIC_PROCESS | 0.0009995 |
| GO_CELLULAR_LIPID_METABOLIC_PROCESS | 0.0019990 |
| GO_REGULATION_OF_LIPID_BIOSYNTHETIC_PROCESS | 0.0029985 |
| GO_LIPID_CATABOLIC_PROCESS | 0.0029985 |
| GO_REGULATION_OF_LIPID_METABOLIC_PROCESS | 0.0044978 |
Figure 2:
Gene-level enrichment statistics for two gene sets with significant enrichment (a, b) and one gene set with no significant level of enrichment (c). The inclusion of FABP3 is one of the primary drivers of significance.
Protein set enrichment results for the model that included an interaction term for sex and protein level are reported in Table 2. The p-values for results including this interaction term assesses the statistical significance of the model coefficient that allows for sex-specific differences in the association between the protein expression levels and the disease state. The gene sets with the strongest association contain complement proteins (C2, C3, C4A, C5, C6, C8B, CFB). Several signatures associated with immune function are identified as statistically significant (p<0.05) for this model including immune effector process (p=0.0075) and the innate immune system (p=0.012). For the models that included interaction terms for age, no gene sets were identified as significantly associated (p<0.05) with the AD phenotype.
Table 2:
gsLRT results on ADNI data with sex interaction
| Gene Set | p |
|---|---|
| GO_IMMUNE_EFFECTOR_PROCESS | 0.0074963 |
| GO_REGULATION_OF_PROTEIN_MATURATION | 0.0084958 |
| REACTOME_COMPLEMENT_CASCADE | 0.0089955 |
| HALLMARK_COAGULATION | 0.0094953 |
| REACTOME_INITIAL_TRIGGERING_OF_COMPLEMENT | 0.0094953 |
3.1. Significance of gene sets across species
In order to compare mouse proteomics pathway signatures to those of humans, we applied gsLRT to data from a select population of mice that are useful models of human Alzheimers disease. We applied gsLRT to both the ADNI and mouse data adjusting for age and sex. We then compared the gene set significance results obtained from both datasets, limiting our analysis to only those gene sets corresponding to the intersection of the proteins contained in each dataset.
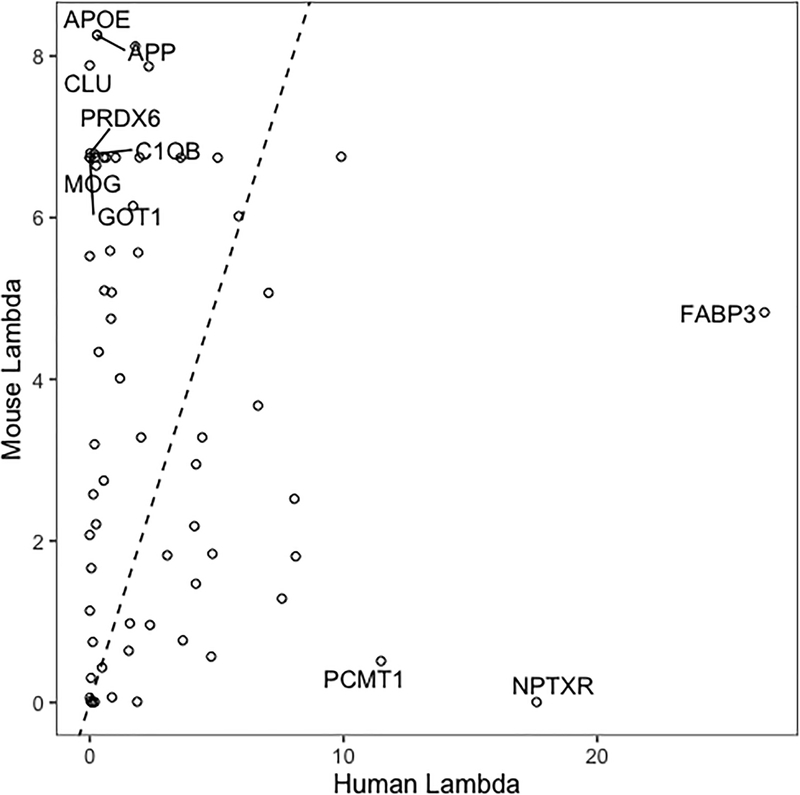
Interestingly, the mouse proteomics dataset yielded more significant pathways overall than the ADNI dataset. As shown in Figure 3, this may be explained by the fact that the human gene set results were driven by a select few proteins with very large Λj values—including Fatty Acid Binding Protein 3 (FABP3) and Neuronal Pentraxin Receptor (NPTXR)—while those for the mouse data were driven by a larger collection of more modest scores for a group of common AD drivers like APOE, APP, CLU, and PRDX6. Hence, pathways involving lipid metabolism were consistently observed as significantly enriched for both species and included regulation of lipid biosynthetic processes, cellular lipid metabolic process, lipid localization and lipid binding, while pathways enriched only in the mouse dataset included those corresponding to immune system processes, aging, and neuronal development. The full table of results is provided as Supplemental Table 2.
Figure 3:
Gene-level enrichment statistics for genes that appear both in the mouse and human proteomics data. The 10 proteins with greatest deviation from equality (dotted line) between species are labeled.
4. Discussion
Application of gsLRT for gene set enrichment in the absence of additional interaction terms (Table 1, Supp. Table 1) confirm known, highly relevant biological signatures and pathways involved in the development of Alzheimers disease [39–43]. Lipid metabolic changes in Alzheimers disease pathophysiology are well replicated. Genes involved in lipid metabolism are identified in large, well-powered genome-wide association studies [42, 43]. Studies of genetic pleiotropy have identified substantial overlap for genes involved with the regulation of serum lipids (LDL, HDL) and Alzheimers disease [44] or cognitive impairment [45]. The gene sets identified as not significantly associated with Alzheimers in Model 1 are not among the strongest genetic effects identified with disease pathophysiology. By contrast, there is compelling phenotypic evidence that these signatures are involved in the disease, most notable for glucose metabolism [46–48] and to a lesser extent for carbohydrate and heme metabolism [49, 50]. If the proteins (or their peptide fragments) that are linked with these signatures are leaked into the CSF, they may appear below the limits of detection in the targeted MS assays.
The inclusion of the protein FABP3 in many of the signatures identified as statistically significant is noteworthy. Increased FABP3 levels have been reported in patients with AD [51], and FABP3 mRNA was increased in microglia from 5XFAD mice compared with C57Bl/6 mice [52]. A specific FABP3 peptide was identified as the most significant peptide differentially expressed between AD and cognitively normal samples identified in the reference study of this proteomic dataset [16]. Lipids serve a variety of roles, from structural to metabolic to signaling [53, 54], and this effect is mediated in part by FABPs [55, 56]. It also has been suggested that one or more of the three FABPs in the brain (FABP3, FABP5, FABP7) may be involved in hippocampal neurogenesis, with a functional defect contributing to hippocampal loss in AD [57]. The lipid metabolism results correlate well with previously reported changes in specific AD biomarkers in the CSF including beta amyloid and tau [58, 59].
It is important to note that our analysis is based on a data set that measures differential protein expression in the CSF. Protein changes in the CSF may not mirror changes in the brain and, specifically, changes associated with glucose metabolism are specific to brain regions and protein changes in metabolism of carbohydrates and heme may be observed in serum [50] in addition to brain [47]. Identification of appropriate controls is both important and challenging for gene set enrichment analysis. Since the proteins chosen for the study had prior support for a role in the development of AD, any significant gene sets will necessarily contain AD-related proteins. However, the finding that a set of lipid metabolic gene sets, specifically (Figure 2a,b), demonstrates enrichment in contrast to the GO locomotion gene set (Figure 2c) suggests that our method of gene set testing confirms a significant role for lipid metabolic signatures involving specific sets of proteins and pathways reported in prior genetic and biochemical studies.
The observation that significant gene set enrichment results were observed for the class of models that included a sex interaction term (Table 2) suggests that the association between AD diagnosis and a subset of protein intensities differs among males and females. The involvement of the complement pathway in AD has strong precedent [60–62]. Prior studies have demonstrated significant sex related differences in complement levels and immune response in healthy individuals [61] and in mice [62]. For example, the clusterin protein is a member of the immune effector process signature, which has the strongest association in the model that includes the sex interaction. The association between clusterin and Alzheimers disease risk is reported in genetic studies [63–66] and in CSF protein level studies [67, 68]. A sex-specific effect of clusterin has been reported as a link between overall decreased bioenergetic metabolism and increased amyloid-related dyshomeostasis in female brains compared with male brains [37].
Since age is a significant risk factor for Alzheimers disease, our prior hypothesis was that we would observe significant enrichment results for the class of models including age interactions. However, the lack of significant results suggests that the interaction between age and protein expression levels does not yield substantial additional explanatory evidence for AD diagnosis beyond that provided by the marginal association between age and AD diagnosis. There are aspects of the study design that may explain the lack of a statistically-significant interaction between age and protein expression. First, The ADNI study includes participants ages 55 to 90 years of age which limits study of changes in dynamic biomarkers such as proteins that occur earlier in life. Second, although the sample size would provide sufficient power for moderate effect size differences between AD and cognitively normal groups, the cohort size may be limiting to detect age-protein concentration level differences of smaller effect sizes. Generalization of the gsLRT statistical analysis to support longitudinal outcome measures would allow more comprehensive assessment of age-specific effects that occur later in life. Prior studies have examined proteomic changes in response to age. One study identified 30 out of 300 proteins in CSF with >20% change in concentrations between older and younger individuals [69], while another study found 248 out of 800 proteins where age differences were greater than twofold over background [70]. The age ranges for these studies were larger than the ADNI study, 22–85 years and 21–85 years respectively. Separating age and disease specific changes in dynamic biomarkers is a challenging problem, requiring large sample sizes to detect interactions between factors.
4.1. Conclusion
Here, we present a statistical approach to derive enrichment scores for biological pathways while accounting for sample covariates in the context of AD. A proof of concept study illustrates cross-species comparisons at the level of pathways or signatures by contrasting the gene-set enrichment results that allowed for interactions between key covariates (age, sex APOE genotype) and protein expression. These pathway-level analyses confirm known genetic interactions and provide unique and informative insights into the relationship between a mouse model of AD and humans with AD. It is clear that overlap in the statistical relevance of specific metabolic pathways can be found. However, it is also clear that, in the data considered for our study, certain pathways clarify critical differences between mouse and human brain. The proof of concept study illustrates that considering the interaction between the covariates and specific analytes that define molecular signatures is important in the development of mouse models that are reliable and consitute a useful model of human neurodegenerative diseases.
There are several caveats and cautions on interpreting the differences between the mouse and human pathway analysis. The genes included in the studies from which these data were drawn focused on specific hypotheses and did not involve an agnostic sampling of the proteome. Measurements were made in two different tissues, brain for the mouse sample and CSF for humans. Sample sizes for the mouse experiments were not adequately powered for stratified analysis by sex or by APOE genotype. The p-values need to be interpreted with caution since there was not a random sampling by the classification factors and since there were multiple comparisons for the number of pathways. The small sample size of the mouse experiments limits the power to detect all but the largest effect sizes. However, there was a substantial overlap (260) in the number of pathways covered by the human and mouse gene sets, supporting an adequate collection of background differences between the classification factors and statistical support for specific signatures is increased by coordinated coregulation or disease-specific differences for a set of proteins, rather than a single protein. For the human data, to be consistent with prior publications based on ADNI data and this specific proteomic dataset, we used the standard diagnostic criteria for AD (NINCDS/ADRDA criteria for probable AD), however future studies may be designed around changes in specific biomarkers including beta amyloid, tau and neurodegenerative markers in accordance with the ATN NIAAA framework [71] and to enable specific comparisons with mouse models that reflect these changes in neuropathology.
Interpretation of dynamic biomarkers such as protein concentrations is complex and cross-species comparisons where there is considerable heterogeneity in the datasets introduces more complexity. As noted in this proof of concept study, there are mechanistic relationships between brain changes and plasma/blood/CSF metabolites that introduce complexities in the human/mouse comparison. Moreover, these studies are cross-sectional, rather than longitudinal; the latter type of study would potentially elucidate age-specific changes and timing/sequencing patterns as a function of disease progression in the proteins concentration. While our study focuses on interactions between critical covariates and the biological signatures, other groups have reported on approaches to compare human and mouse model data, for example to identify of shared cell types across individuals, species, and multiple modalities (gene expression, epigenetic, or spatial data) where non-negative matrix factorization is performed to define a low-dimensional space in which each cell is defined by one set of dataset-specific factors, or metagenes [72] or to examine conserved epigenomic signals in mice and humans in the development of Alzheimers disease [73].
Our future studies will continue to test hypotheses that involve the interactions of four critical factors: age, sex, APOE genotype, and immunity. How these critical factors interact to initiate or accelerate LOAD pathology remains unknown and the approach that we have developed is well-suited to address this question by the field at large. It also facilitates evaluation and comparisons across species, potentially improving the use of animal models to uncover mechanisms of disease and the more rapid development of effective therapeutics for the human population.
Supplementary Material
Table 3:
gsLRT results on ADNI data with age interaction
| Gene Set | p |
|---|---|
| HALLMARK_ESTROGEN_RESPONSE_LATE | 0.0569715 |
| GO_SERINE_HYDROLASE_ACTIVITY | 0.1429285 |
| GO_COFACTOR_METABOLIC_PROCESS | 0.1504248 |
| GO_BLOOD_MICROPARTICLE | 0.1559220 |
| GO_REGULATION_OF_CELLULAR_RESPONSE_TO_STRESS | 0.1739130 |
Highlights.
Development of novel gene set enrichment method (gsLRT) to discover biological pathways relevant to AD.
Gene set enrichment analysis of ADNI proteomics data.
Test for interactions with covariates including age and sex, which may mediate pathway activity.
Comparison of gene set enrichment between human AD and mouse models.
Research in Context.
1. Systematic review:
The authors reviewed literature describing biological pathways involved with AD. Pathway activity related to AD may be mediated by factors like age or sex.
2. Interpretation:
The manuscript proposes a novel statistical approach to derive enrichment scores for biological pathways while accounting for covariates like age and sex in the context of AD. The pathway-level analyses performed on proteomics data confirm known genetic interactions and provide informative insights into the relationship between a mouse model of AD and humans with AD.
3. Future directions:
Future studies will develop methods to integrate several genomics data modalities to identify animal models with optimal pathway-specific correspondence to human AD, while accounting for relevant covariates.
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimers Association; Alzheimers Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimers Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Funding
Funding provided by NIH R56AG057895, RF1 AG057895, and R01 AG066184.
A. Implementation of gsLRT
A.1. Nested logistic regressions
For each gene in our dataset, we use the ‘glm’ function from the base R package ‘stats’ to fit the logistic regression models described above. We apply the Bartlett correction using the correction factor reported by [74]. The statistic used for testing is then
| (5) |
where
| (6) |
| (7) |
Here denotes the diagonal matrix of leverage statistics for the alternative model and denotes the diagonal matrix of leverage statistics for the null model. For a given model and corresponding design matrix , the diagonal leverage matrix has entries defined by
| (8) |
A.2. Permutation
Let ESk be an enrichment statistic computed for gene set k using the original (non-permuted) data. Let the total number of permutations be B. Then the permutation p-value is given by
| (9) |
where is the value of the enrichment statistic upon permutation of the covariate to be tested. For testing main effects, the gene expression profile is the “covariate” that gets permuted. For testing interaction terms, the column containing the product of the gene expression profile and the interacting covariate is permuted.
B. Simulation study
B.1. Dataset simulation
We used a simulation procedure to produce datasets of the same dimension as the proteomics dataset that we used in our Alzheimer’s investigation. For each simulation trial, we generated gene expression data with 152 samples and 142 genes. The genes were split into 20 disjoint gene sets of random sizes. The expression values for each gene and sample gj in gene set γk were drawn independently from the multivariate normal with covariance matrix drawn from an inverse-Wishart distribution
where the entries of were set equal to 1 on the diagonal and set equal to uk, a random number on the unit interval, on the off-diagonal. This construction yields a random correlation structure for genes within gene sets. For each trial, we also generated a covariate matrix X with three continuous covariates drawn i.i.d. from a standard normal distribution and three binary covariates drawn i.i.d. from a Bernoulli distribution with success probability 1/2. Three gene sets were randomly chosen to be associated with the disease outcome through the non-linear relationship
where expit(x) = ex/(1 + ex) and denotes the set of genes included in the 3 enriched gene sets. Note that this means that each gene in the selected gene sets has some association to the disease outome, with magnitude depending on the size of the sampled coefficients α. The coefficients β and α were drawn i.i.d. from a standard normal distribution, which produced simulated disease outcome vectors with an average disease prevalence of 47%, comparable to the 43% observed in the ADNI dataset.
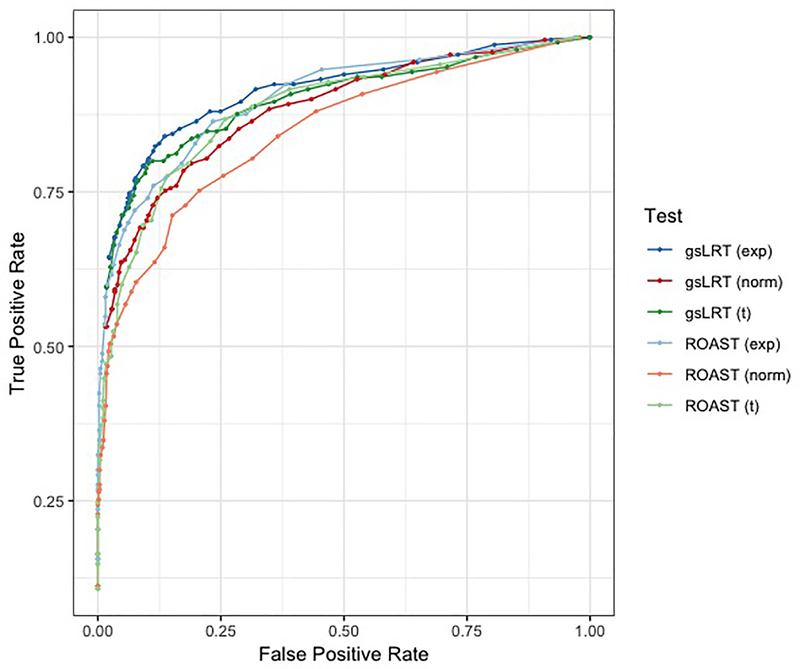
B.2. True and false positive rates compared to ROAST
We performed 50 trials following the procedure above and computed p-values using 200 permutations in each trial. The number of permutations was chosen to limit computation time. The three gene sets per trial that were randomly selected to be associated with the disease outcome were the true positive cases; all other gene sets were considered true negatives. We then computed the average true positive rate (TPR) and false positive rate (FPR) for gsLRT at fixed p-value thresholds. For comparison, we also ran the ROAST methodology [8] using 200 rotations in each trial. ROAST achieved better Type I Error control than gsLRT, but was less sensitive on average (Figure 4). We ran a second and third round of trials using exponentially-distributed gene expression profiles and t-distributed gene expression profiles with five degrees of freedom. For a fixed FPR, gsLRT achieved comparable or modestly higher true positive rates than ROAST in all scenarios. The improvement was most marked in the region of low Type I Error (FPR < 0.1).
Figure 4:
Evaluation of gsLRT’s performance on simulated gene expression data versus that of ROAST methodology. On average, gsLRT (darker colors) achieves a higher true positive rate than ROAST (lighter colors) at each fixed false positive rate when evaluated on normally distributed gene expression profiles (red) exponentially distributed gene expression profiles (blue) and t-distributed gene expression profiles (green).
B.3. Validation under null
We modified the simulation setup as described above in order to sample from the null distribution that gsLRT is designed to test against. Specifically, we ran simulations with the same dimensions as above, but sampled gene expression values for the samples within gene set k from
This reflects the independence assumption assumed by the gsLRT chi squared test (though this is not necessary for the permutation test). We also sampled the disease outcome independently of the gene expression profiles and covariates
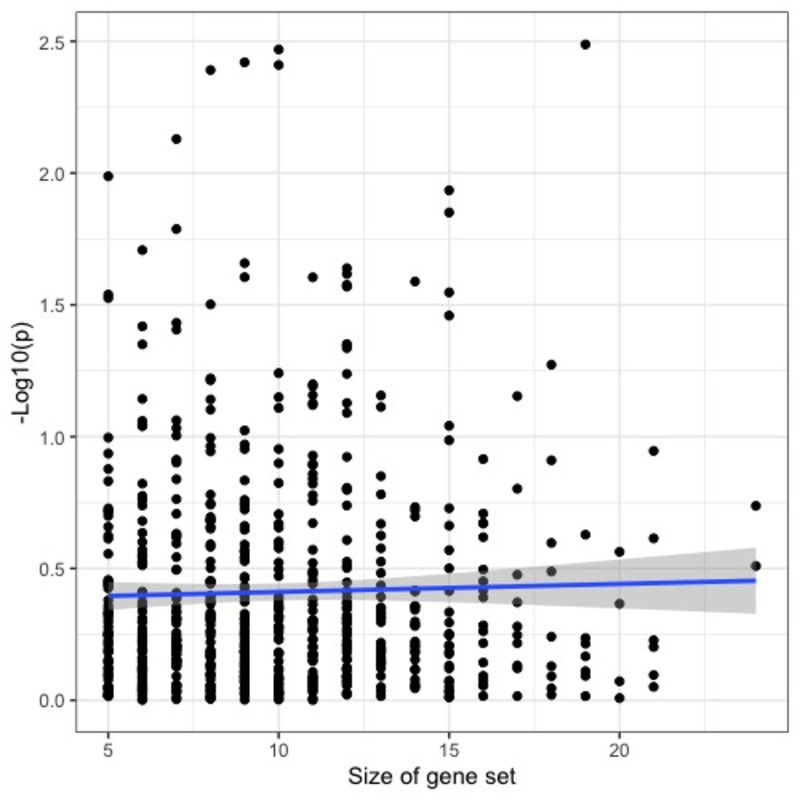
so that the simulated data reflected the null hypothesis that the addition of the gene expression profiles offered no additional explanatory power. The empirical false positive rate at level 0.05 was observed to be 0.0479. In addition, we observed no bias in significance level as relates to the size of gene set (correlation between gene set size and gsLRT p-value was −0.02137, CI = [−0.09835,0.05586]).
Figure 5:
Evaluation of gsLRT’s performance on simulated gene expression data with no relationship to disease outcome (i.e. data conforming to the null model assumed by the gsLRT test). In addition to controlling Type I error at the nominal level (0.0479 vs. 0.05), the gsLRT p-values show no bias towards assigning significance to gene sets of larger size. The blue line shows the line of best-fit, whose slope is proportional to the correlation between the gene set size and −log10(p).
C. Proteomics Methods
C.1. Brain tissue preparation
Brain tissue samples stored in 1.5 mL tubes were delivered to the Duke Proteomics & Metabolomics Core Facility (n = 6 per genotype). 0.5% w/v ALS-1 surfactant in 50 mM ammonium bicarbonate (AmBic) was added to each sample at a volume of 10 uL/mg wet weight of tissue. Tissue homogenization and cell lysis was performed with probe sonication (Misonix) over three pulses at power level 3 for 5 seconds each with cooling on ice between pulses. A 5 uL aliquot of homogenate was diluted 25x in AmBic for determination of protein content by Bradford assay. Based on Bradford results, samples were 0.7±0.2 mg protein / mg tissue. Following normalization (100 μg protein at 1 mg/mL protein in 0.5% ALS-1/AmBic), samples were reduced with 10 mM dithiothreitol (DTT) at 80° C with shaking for 15 minutes, alkylated with 20 mM iodoacetamide (IAA) at room temperature in the dark for 30 minutes, and digested with 2 μg sequencing grade modified trypsin (Promega) overnight at 37° C with shaking. Digestion was stopped with the addition of 12 μL 10/20/70 v/v/v TFA/MeCN/H2O and heating at 60° C for 2 hours. Diluted further with 1/2/97 v/v/v TFA/MeCN/H2O for a final digested protein concentration of 0.5 ug/uL. A pool of all samples (Study Pool QC, SPQC) was created from equal volume of each sample, and analyzed at regular intervals throughout the study to allow observation of any experimental drift.
C.2. Proteomics Analysis
The samples were analyzed using a nanoAcquity UPLC system (Waters) coupled to a Q Exactive HF Orbitrap high-resolution accurate-mass tandem mass spectrometer (Thermo Scientific) via a nanoelectrospray ionization source. Each sample was analyzed once, and the SPQC was analyzed approximately every 6 samples. Briefly, the sample was first trapped and desalted on a Symmetry C18 180 um x 20 mm trapping column (5 uL/min at 99.8/0.1/0.1 v/v water/acetonitrile/formic acid), then the analytical separation was performed using a 1.7 um Acquity HSS T3 C18 75 um x 250 mm column (Waters). The peptides on the column were eluted using a 90-minute gradient of 5–40% acetonitrile with 0.1% formic acid at a flow rate of 400 nanoliters/minute (nL/min) with a column temperature of 55° C. Data collection on the Q Exactive HF mass spectrometer was performed in a data-dependent MS/MS manner, using a 120,000 resolution precursor ion (MS1) scan followed by MS/MS (MS2) of the top 12 most abundant ions at 30,000 resolution. MS1 was accomplished using an automatic gain control (AGC) target of 3e6 ions and mass accumulation time of up to 50 msec. MS2 used AGC target of 5e4 ions, up to 45 msec maxiumum ion accumulation, 1.2 m/z isolation window, 27V normalized collision energy, and 20 sec dynamic exclusion.
Following the analyses, the data was imported into Rosetta Elucidator v 4.0 (Rosetta Biosoftware, Inc.), and all LC-MS files were aligned based on the accurate mass and retention time of detection ions (“features”) using a PeakTeller algorithm (Elucidator). The relative peptide abundance was calculated based on area-under-the-curve (AUC) of aligned features across all runs.
The MS/MS data was searched against a custom built database based on the SwissProt database with Mus musculus taxonomy (downloaded April 28, 2017) with additional proteins, including yeast ADH1_YEAST (surrogate standard), ALBU_BOVIN (contaminant), APOE_HUMAN (genetic substitution), and additional mutated proteins expressed in the mice with sequences provided by the investigators, were also included in the custom database. An equal number of reversed-sequence decoys were appended to this forward DB for false discovery rate determination. A total of 3084 proteins were quantified, and 2118 (69%) proteins were quantified with 2 or more peptides (Supplementary Table 3).
Footnotes
Conflicts of Interest: The authors report no relevant disclosures for this work.
Software availability
Software with the implementation of gsLRT is available in an R package. It may be downloaded from https://github.com/j-g-b.
Data used in preparation of this article were obtained from the Alzheimers Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
ClinicalTrials.gov identifier, NCT00106899
Data availability
ADNI data are available with approved data access request from http://adni.loni.usc.edu/.
References
- [1].Canchi Saranya, Raao Balaji, Masliah Deborah, Rosenthal Sara Brin, Sasik Roman, Fisch Kathleen M., De Jager Philip L., Bennett David A., and Rissman Robert A.. Integrating Gene and Protein Expression Reveals Perturbed Functional Networks in Alzheimers Disease. Cell Reports, 28(4):1103–1116.e4, July 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Castrillo Juan I. and Oliver Stephen G.. Alzheimers as a Systems-Level Disease Involving the Interplay of Multiple Cellular Networks. In Castrillo Juan I. and Oliver Stephen G., editors, Systems Biology of Alzheimer’s Disease, volume 1303, pages 3–48. Springer New York, New York, NY, 2016. Series Title: Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- [3].Ng Bernard, Casazza William, Patrick Ellis, Tasaki Shinya, Novakovsky Gherman, Felsky Daniel, Ma Yiyi, Bennett David A., Gaiteri Chris, De Jager Philip L., and Mostafavi Sara. Using Transcriptomic Hidden Variables to Infer Context-Specific Genotype Effects in the Brain. The American Journal of Human Genetics, 105(3):562–572, September 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Satizabal Claudia L., Adams Hieab H. H., Hibar Derrek P., White Charles C., Knol Maria J., Stein Jason L., Scholz Markus, Sargurupremraj Muralidharan, Jahanshad Neda, Roshchupkin Gennady V., Smith Albert V., Bis Joshua C., Jian Xueqiu, Luciano Michelle, Hofer Edith, Teumer Alexander, van der Lee Sven J., Yang Jingyun, Yanek Lisa R., Lee Tom V., Li Shuo, Hu Yanhui, Koh Jia Yu, Eicher John D., Desrivières Sylvane, Arias-Vasquez Alejandro, Chauhan Ganesh, Athanasiu Lavinia, Rentería Miguel E., Kim Sungeun, Hoehn David, Armstrong Nicola J., Chen Qiang, Holmes Avram J., den Braber Anouk, Kloszewska Iwona, Andersson Micael, Espeseth Thomas, Grimm Oliver, Abramovic Lucija, Alhusaini Saud, Milaneschi Yuri, Papmeyer Martina, Axelsson Tomas, Ehrlich Stefan, Roiz-Santiañez Roberto, Kraemer Bernd, Håberg Asta K., Jones Hannah J., Pike G. Bruce, Stein Dan J., Stevens Allison, Bralten Janita, Vernooij Meike W., Harris Tamara B., Filippi Irina, Witte A. Veronica, Guadalupe Tulio, Wittfeld Katharina, Mosley Thomas H., Becker James T., Doan Nhat Trung, Hagenaars Saskia P., Saba Yasaman, Cuellar-Partida Gabriel, Amin Najaf, Hilal Saima, Nho Kwangsik, Mirza-Schreiber Nazanin, Arfanakis Konstantinos, Becker Diane M., Ames David, Goldman Aaron L., Lee Phil H., Boomsma Dorret I., Lovestone Simon, Giddaluru Sudheer, Hellard Stephanie Le, Mattheisen Manuel, Bohlken Marc M., Kasperaviciute Dalia, Schmaal Lianne, Lawrie Stephen M., Agartz Ingrid, Walton Esther, Tordesillas-Gutierrez Diana, Davies Gareth E., Shin Jean, Ipser Jonathan C., Vinke Louis N., Hoogman Martine, Jia Tianye, Burkhardt Ralph, Klein Marieke, Crivello Fabrice, Janowitz Deborah, Carmichael Owen, Haukvik Unn K., Aribisala Benjamin S., Schmidt Helena, Strike Lachlan T., Cheng Ching-Yu, Risacher Shannon L., Pütz Benno, Fleischman Debra A., Assareh Amelia A., Mattay Venkata S., Buckner Randy L., Mecocci Patrizia, Dale Anders M., Cichon Sven, Boks Marco P., Matarin Mar, Penninx Brenda W. J. H., Calhoun Vince D., Chakravarty M. Mallar, Marquand Andre F., Macare Christine, Masouleh Shahrzad Kharabian, Oosterlaan Jaap, Amouyel Philippe, Hegenscheid Katrin, Rotter Jerome I., Schork Andrew J., Liewald David C. M., de Zubicaray Greig I., Wong Tien Yin, Shen Li, Sämann Philipp G., Brodaty Henry, Roffman Joshua L., de Geus Eco J. C., Tsolaki Magda, Erk Susanne, van Eijk Kristel R., Cavalleri Gianpiero L., van der Wee Nic J. A., McIntosh Andrew M., Gollub Randy L., Bulayeva Kazima B., Bernard Manon, Richards Jennifer S., Himali Jayandra J., Loeffler Markus, Rommelse Nanda, Hoffmann Wolfgang, Westlye Lars T., Valdés Hernández Maria C., Hansell Narelle K., van Erp Theo G. M., Wolf Christiane, Kwok John B. J., Vellas Bruno, Heinz Andreas, Loohuis Loes M. Olde, Delanty Norman, Ho Beng-Choon, Ching Christopher R. K., Shumskaya Elena, Singh Baljeet, Hofman Albert, van der Meer Dennis, Homuth Georg, Psaty Bruce M., Bastin Mark E., Montgomery Grant W., Foroud Tatiana M., Reppermund Simone, Hottenga Jouke-Jan, Simmons Andrew, Meyer-Lindenberg Andreas, Cahn Wiepke, Whelan Christopher D., van Donkelaar Marjolein M. J., Yang Qiong, Hosten Norbert, Green Robert C, Thalamuthu Anbupalam, Mohnke Sebastian, Pol Hilleke E. Hulshoff, Lin Honghuang, Jack Clifford R., Schofield Peter R., Mühleisen Thomas W., Maillard Pauline, Potkin Steven G., Wen Wei, Fletcher Evan, Toga Arthur W., Gruber Oliver, Huentelman Matthew, Smith George Davey, Launer Lenore J., Nyberg Lars, Jönsson Erik G., Crespo-Facorro Benedicto, Koen Nastassja, Greve Douglas N., Uitterlinden André G., Weinberger Daniel R., Steen Vidar M., Fedko Iryna O., Groenewold Nynke A., Niessen Wiro J., Toro Roberto, Tzourio Christophe, Longstreth William T., Ikram M. Kamran, Smoller Jordan W., van Tol Marie-Jose, Sussmann Jessika E., Paus Tomas, Hervé Lemaître Matthias L. Schroeter, Mazoyer Bernard, Andreassen Ole A., Holsboer Florian, Depondt Chantal, Veltman Dick J., Turner Jessica A., Pausova Zdenka, Schumann Gunter, van Rooij Daan, Djurovic Srdjan, Deary Ian J., McMahon Katie L., Müller-Myhsok Bertram, Brouwer Rachel M., Soininen Hilkka, Pandolfo Massimo, Wassink Thomas H., Cheung Joshua W., Wolfers Thomas, Martinot JeanLuc, Zwiers Marcel P., Nauck Matthias, Melle Ingrid, Martin Nicholas G., Kanai Ryota, Westman Eric, Kahn René S., Sisodiya Sanjay M., White Tonya, Saremi Arvin, van Bokhoven Hans, Brunner Han G., Völzke Henry, Wright Margaret J., van t Ent Dennis, Nöthen Markus M., Ophoff Roel A., Buitelaar Jan K., Fernández Guillén, Sachdev Perminder S., Rietschel Marcella, van Haren Neeltje E. M., Fisher Simon E., Beiser Alexa S., Francks Clyde, Saykin Andrew J., Mather Karen A., Romanczuk-Seiferth Nina, Hartman Catharina A., DeStefano Anita L., Heslenfeld Dirk J., Weiner Michael W., Walter Henrik, Hoekstra Pieter J., Nyquist Paul A., Franke Barbara, Bennett David A., Grabe Hans J., Johnson Andrew D., Chen Christopher, van Duijn Cornelia M., Lopez Oscar L., Fornage Myriam, Wardlaw Joanna M., Schmidt Reinhold, DeCarli Charles, De Jager Philip L., Villringer Arno, Debette Stéphanie, Gudnason Vilmundur, Medland Sarah E., Shulman Joshua M., Thompson Paul M., Seshadri Sudha, and Ikram M. Arfan. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet, 51(11):1624–1636, November 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schadt Eric E., Friend Stephen H., and Shaywitz David A.. A network view of disease and compound screening. Nat Rev Drug Discov, 8(4):286–295, April 2009. [DOI] [PubMed] [Google Scholar]
- [6].Zhang Bin, Gaiteri Chris, Bodea Liviu-Gabriel, Wang Zhi, Joshua McElwee Alexei A. Podtelezhnikov, Zhang Chunsheng, Xie Tao, Tran Linh, Dobrin Radu, Fluder Eugene, Clurman Bruce, Melquist Stacey, Narayanan Manikandan, Suver Christine, Shah Hardik, Mahajan Milind, Gillis Tammy, Mysore Jayalakshmi, MacDonald Marcy E., Lamb John R., Bennett David A., Molony Cliona, Stone David J., Gudnason Vilmundur, Myers Amanda J., Schadt Eric E., Neumann Harald, Zhu Jun, and Emilsson Valur. Integrated Systems Approach Identifies Genetic Nodes and Networks in Late-Onset Alzheimers Disease. Cell, 153(3):707–720, April 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Khachaturian Zaven S., Mesulam M. Marsel, Khachaturian Ara S., and Mohs Richard C.. The Special Topics Section of Alzheimer’s & Dementia. Alzheimer’s & Dementia, 11(11):1261–1264, November 2015. [DOI] [PubMed] [Google Scholar]
- [8].Wu Di, Lim Elgene, Vaillant François, Asselin-Labat Marie-Liesse, Visvader Jane E., and Smyth Gordon K.. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics, 26(17):2176–2182, September 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Oron Assaf and Gentleman Robert. GSEAlm, 2017. [Google Scholar]
- [10].Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, and Park PJ. Discovering statistically significant pathways in expression profiling studies. Proceedings of the National Academy of Sciences, 102(38):13544–13549, September 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Barry WT, Nobel AB, and Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics, 21(9):1943–1949, May 2005. [DOI] [PubMed] [Google Scholar]
- [12].Efron Bradley and Tibshirani Robert. On testing the significance of sets of genes. The Annals of Applied Statistics, 1(1):107–129, June 2007. [Google Scholar]
- [13].Tamayo Pablo, Steinhardt George, Liberzon Arthur, and Mesirov Jill P. The limitations of simple gene set enrichment analysis assuming gene independence. Stat Methods Med Res, 25(1):472–487, February 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Morris JC. The Clinical Dementia Rating (CDR): Current version and scoring rules. Neurology, 43(11):2412–2412, November 1993. [DOI] [PubMed] [Google Scholar]
- [15].McKhann G, Drachman D, Folstein M, Katzman R, Price D, and Stadlan EM. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group* under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology, 34(7):939–939, July 1984. [DOI] [PubMed] [Google Scholar]
- [16].Spellman Daniel S., Wildsmith Kristin R., Honigberg Lee A., Tuefferd Marianne, Baker David, Raghavan Nandini, Nairn Angus C., Croteau Pascal, Schirm Michael, Allard Rene, Lamontagne Julie, Chelsky Daniel, Hoffmann Steven, Potter William Z., Alzheimer’s Disease Neuroimaging Initiative, and the Foundation for NIH (FNIH) Biomarkers Consortium CSF Proteomics Project Team. Development and evaluation of a multiplexed mass spectrometry based assay for measuring candidate peptide biomarkers in Alzheimer’s Disease Neuroimaging Initiative (ADNI) CSF. Prot. Clin. Appl, 9(7–8):715–731, August 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Colton Carol A., Wilson Joan G., Everhart Angela, Wilcock Donna M., Puoliväli Jukka, Heikkinen Taneli, Oksman Juho, Jääskeläinen Olli, Lehtimäki Kimmo, Laitinen Teemu, Vartiainen Nina, and Vitek Michael P.. mNos2 Deletion and Human NOS2 Replacement in Alzheimer Disease Models. J Neuropathol Exp Neurol, 73(8):752–769, August 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hoos Michael D., Richardson Brenna M., Foster Matthew W., Everhart Angela, Thompson J. Will, Moseley M. Arthur, and Colton Carol A.. Longitudinal Study of Differential Protein Expression in an Alzheimers Mouse Model Lacking Inducible Nitric Oxide Synthase. J. Proteome Res, 12(10):4462–4477, October 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hoos Michael D, Vitek Michael P, Ridnour Lisa A, Wilson Joan, Jansen Marilyn, Everhart Angela, Wink David A, and Colton Carol A. The impact of human and mouse differences in NOS2 gene expression on the brains redox and immune environment. Mol Neurodegeneration, 9(1):50, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Linnertz Colton, Anderson Lauren, Gottschalk William, Crenshaw Donna, Lutz Michael W., Allen Jawara, Saith Sunita, Mihovilovic Mirta, Burke James R., Welsh-Bohmer Kathleen A., Roses Allen D., and Chiba-Falek Ornit. The cis -regulatory effect of an Alzheimer’s disease-associated poly-T locus on expression of TOMM40 and apolipoprotein E genes. Alzheimer’s & Dementia, 10(5):541–551, September 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Badea Alexandra, Wu Wenlin, Shuff Jordan, Wang Michele, Anderson Robert J., Qi Yi, Johnson G. Allan, Wilson Joan G., Koudoro Serge, Garyfallidis Eleftherios, Colton Carol A., and Dunson David B.. Identifying Vulnerable Brain Networks in Mouse Models of Genetic Risk Factors for Late Onset Alzheimers Disease. Front. Neuroinform, 13:72, December 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Badea Alexandra, Delpratt Natalie A., Anderson RJ, Dibb Russell, Qi Yi, Wei Hongjiang, Liu Chunlei, Wetsel William C., Avants Brian B., and Colton Carol. Multivariate MR biomarkers better predict cognitive dysfunction in mouse models of Alzheimer’s disease. Magnetic Resonance Imaging, 60:52–67, July 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pan Xiaobei, Nasaruddin Muhammad Bin, Elliott Christopher T., McGuinness Bernadette, Passmore Anthony P., Kehoe Patrick G., Hölscher Christian, McClean Paula L., Graham Stewart F., and Green Brian D.. Alzheimer’s diseaselike pathology has transient effects on the brain and blood metabolome. Neurobiology of Aging, 38:151–163, February 2016. [DOI] [PubMed] [Google Scholar]
- [24].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences, 102(43):15545–15550, October 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liberzon Arthur, Birger Chet, Helga Thorvaldsdóttir Mahmoud Ghandi, Mesirov Jill P., and Tamayo Pablo. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Systems, 1(6):417–425, December 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Research, 47(D1):D330–D338, January 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, and Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet, 25(1):25–29, May 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kanehisa Minoru, Sato Yoko, Furumichi Miho, Morishima Kanae, and Tanabe Mao. New approach for understanding genome variations in KEGG. Nucleic Acids Research, 47(D1):D590–D595, January 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fabregat Antonio, Jupe Steven, Matthews Lisa, Sidiropoulos Konstantinos, Gillespie Marc, Garapati Phani, Haw Robin, Jassal Bijay, Korninger Florian, May Bruce, Milacic Marija, Corina Duenas Roca Karen Rothfels, Sevilla Cristoffer, Shamovsky Veronica, Shorser Solomon, Varusai Thawfeek, Viteri Guilherme, Weiser Joel, Wu Guanming, Stein Lincoln, Hermjakob Henning, and D’Eustachio Peter. The Reactome Pathway Knowledgebase. Nucleic Acids Res, 46(D1):D649–D655, January 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fan Chun Chieh, Banks Sarah J., Thompson Wesley K., Chen Chi-Hua, McEvoy Linda K., Tan Chin Hong, Kukull Walter, Bennett David A., Farrer Lindsay A., Mayeux Richard, Schellenberg Gerard D., Andreassen Ole A., Desikan Rahul, and Dale Anders M.. Sex-dependent polygenic effects on the clinical progressions of Alzheimers disease. preprint, Genetics, April 2019. [Google Scholar]
- [31].for the Womens Brain Project and the Alzheimer Precision Medicine Initiative, Ferretti Maria Teresa, Iulita Maria Florencia, Cavedo Enrica, Chiesa Patrizia Andrea, Dimech Annemarie Schumacher, Chadha Antonella Santuccione, Baracchi Francesca, Girouard Hélène, Misoch Sabina, Giacobini Ezio, Depypere Herman, and Hampel Harald. Sex differences in Alzheimer disease the gateway to precision medicine. Nat Rev Neurol, 14(8):457–469, August 2018. [DOI] [PubMed] [Google Scholar]
- [32].Jack Clifford R, Wiste Heather J, Weigand Stephen D, Therneau Terry M, Knopman David S, Lowe Val, Knopman David S, Val Lowe, Vemuri Prashanthi, Mielke Michelle M, Roberts Rosebud O, Machulda Mary M, Senjem Matthew L, Gunter Jeffrey L, Rocca Walter A, and Petersen Ronald C. Age-specific and sex-specific prevalence of cerebral beta-amyloidosis, tauopathy, and neurodegeneration in cognitively unimpaired individuals aged 5095 years: a cross-sectional study. The Lancet Neurology, 16(6):435–444, June 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mielke Michelle M.. Sex and Gender Differences in Alzheimer’s Disease Dementia. Psychiatr Times, 35(11):14–17, November 2018. [PMC free article] [PubMed] [Google Scholar]
- [34].Nebel Rebecca A., Aggarwal Neelum T., Barnes Lisa L., Gallagher Aimee, Goldstein Jill M., Kantarci Kejal, Mallampalli Monica P., Mormino Elizabeth C., Scott Laura, Wai Haung Yu Pauline M. Maki, and Mielke Michelle M.. Understanding the impact of sex and gender in Alzheimer’s disease: A. call to action. Alzheimer’s & Dementia, 14(9):1171–1183, September 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Neu Scott C., Pa Judy, Kukull Walter, Beekly Duane, Kuzma Amanda, Gangadharan Prabhakaran, Wang Li-San, Romero Klaus, Arneric Stephen P., Redolfi Alberto, Orlandi Daniele, Frisoni Giovanni B., Au Rhoda, Devine Sherral, Auerbach Sanford, Espinosa Ana, Boada Mercè, Ruiz Agustín, Johnson Sterling C., Koscik Rebecca, Wang Jiun-Jie, Hsu Wen-Chuin, Chen Yao-Liang, and Toga Arthur W.. Apolipoprotein E Genotype and Sex Risk Factors for Alzheimer Disease: A Meta-analysis. JAMA Neurol, 74(10):1178, October 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Riedel Brandalyn C., Thompson Paul M., and Brinton Roberta Diaz. Age, APOE and sex: Triad of risk of Alzheimers disease. The Journal of Steroid Biochemistry and Molecular Biology, 160:134–147, June 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhao Liqin, Mao Zisu, Woody Sarah K., and Brinton Roberta D.. Sex differences in metabolic aging of the brain: insights into female susceptibility to Alzheimer’s disease. Neurobiology of Aging, 42:69–79, June 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kulminski Alexander M., Arbeev Konstantin G., Culminskaya Irina, Arbeeva Liubov, Ukraintseva Svetlana V., Stallard Eric, Christensen Kaare, Schupf Nicole, Province Michael A., and Yashin Anatoli I.. Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan. PLoS Genet, 10(1):e1004141, January 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Karch Celeste M., Cruchaga Carlos, and Goate Alison M.. Alzheimers Disease Genetics: From the Bench to the Clinic. Neuron, 83(1):11–26, July 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pimenova Anna A., Raj Towfique, and Goate Alison M.. Untangling Genetic Risk for Alzheimers Disease. Biological Psychiatry, 83(4):300–310, February 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].the Alzheimer’s Disease Neuroimaging Initiative, CHARGE consortium, EADI1 consortium, Hollingworth Paul, Harold Denise, Sims Rebecca, Gerrish Amy, Lambert Jean-Charles, Carrasquillo Minerva M, Abraham Richard, Hamshere Marian L, Pahwa Jaspreet Singh, Moskvina Valentina, Dowzell Kimberley, Jones Nicola, Stretton Alexandra, Thomas Charlene, Richards Alex, Ivanov Dobril, Widdowson Caroline, Chapman Jade, Lovestone Simon, Powell John, Proitsi Petroula, Lupton Michelle K, Brayne Carol, Rubinsztein David C, Gill Michael, Lawlor Brian, Lynch Aoibhinn, Brown Kristelle S, Passmore Peter A, Craig David, McGuinness Bernadette, Todd Stephen, Holmes Clive, Mann David, Smith A David, Beaumont Helen, Warden Donald, Wilcock Gordon, Love Seth, Kehoe Patrick G, Hooper Nigel M, Vardy Emma R L C, Hardy John, Mead Simon, Fox Nick C, Rossor Martin, Collinge John, Maier Wolfgang, Jessen Frank, Ruther Eckart, Schurmann Britta, Heun Reiner, Kolsch Heike, van den Bussche Hendrik, Heuser Isabella, Kornhuber Johannes, Wiltfang Jens, Dichgans Martin, Frolich Lutz, Hampel Harald, Gallacher John, Hull Michael, Rujescu Dan, Giegling Ina, Goate Alison M, Kauwe John S K, Cruchaga Carlos, Nowotny Petra, John C Morris Kevin Mayo, Sleegers Kristel, Bettens Karolien, Engelborghs Sebastiaan, De Deyn Peter P, Van Broeckhoven Christine, Livingston Gill, Bass Nicholas J, Gurling Hugh, McQuillin Andrew, Gwilliam Rhian, Deloukas Panagiotis, Al-Chalabi Ammar, Shaw Christopher E, Tsolaki Magda, Singleton Andrew B, Guerreiro Rita, Muhleisen Thomas W, Nothen Markus M, Moebus Susanne, Jöckel Karl-Heinz, Klopp Norman, Wichmann H-Erich, Pankratz V Shane, Sando Sigrid B, Aasly Jan O, Barcikowska Maria, Wszolek Zbigniew K, Dickson Dennis W, Graff-Radford Neill R, Petersen Ronald C, van Duijn Cornelia M, Breteler Monique M B, Ikram M Arfan, DeStefano Anita L, Fitzpatrick Annette L, Lopez Oscar, Launer Lenore J, Seshadri Sudha, Berr Claudine, Campion Dominique, Epelbaum Jacques, Dartigues Jean-François, Tzourio Christophe, Alperovitch Annick, Lathrop Mark, Feulner Thomas M, Friedrich Patricia, Riehle Caterina, Krawczak Michael, Schreiber Stefan, Mayhaus Manuel, Nicolhaus S, Wagenpfeil Stefan, Steinberg Stacy, Stefansson Hreinn, Stefansson Kari, Snædal Jon, Björnsson Sigurbjörn, Jonsson Palmi V, Chouraki Vincent, Genier-Boley Benjamin, Hiltunen Mikko, Soininen Hilkka, Combarros Onofre, Zelenika Diana, Delepine Marc, Bullido Maria J, Pasquier Florence, Mateo Ignacio, Frank-Garcia Ana, Porcellini Elisa, Hanon Olivier, Coto Eliecer, Alvarez Victoria, Bosco Paolo, Siciliano Gabriele, Mancuso Michelangelo, Panza Francesco, Solfrizzi Vincenzo, Nacmias Benedetta, Sorbi Sandro, Bossù Paola, Piccardi Paola, Arosio Beatrice, Annoni Giorgio, Seripa Davide, Pilotto Alberto, Scarpini Elio, Galimberti Daniela, Brice Alexis, Hannequin Didier, Licastro Federico, Jones Lesley, Holmans Peter A, Jonsson Thorlakur, Riemenschneider Matthias, Morgan Kevin, Younkin Steven G, Owen Michael J, O’Donovan Michael, Amouyel Philippe, and Williams Julie. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet, 43(5):429–435, May 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Alzheimer Disease Genetics Consortium (ADGC), The European Alzheimers Disease Initiative (EADI), Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE), Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimers Disease Consortium (GERAD/PERADES),Kunkle Brian W., Grenier-Boley Benjamin, Sims Rebecca, Bis Joshua C., Damotte Vincent, Naj Adam C., Boland Anne, Vronskaya Maria, van der Lee Sven J., Amlie-Wolf Alexandre, Bellenguez Céline, Frizatti Aura, Chouraki Vincent, Martin Eden R., Sleegers Kristel, Badarinarayan Nandini, Jakobsdottir Johanna, Hamilton-Nelson Kara L., Moreno-Grau Sonia, Olaso Robert, Raybould Rachel, Chen Yuning, Kuzma Amanda B., Hiltunen Mikko, Morgan Taniesha, Ahmad Shahzad, Vardarajan Badri N., Epelbaum Jacques, Hoffmann Per, Boada Merce, Beecham Gary W., Garnier Jean-Guillaume, Harold Denise, Fitzpatrick Annette L., Valladares Otto, Moutet Marie-Laure, Gerrish Amy, Smith Albert V., Qu Liming, Bacq Delphine, Denning Nicola, Jian Xueqiu, Zhao Yi, Zompo Maria Del, Fox Nick C., Choi Seung-Hoan, Mateo Ignacio, Hughes Joseph T., Adams Hieab H., Malamon John, Sanchez-Garcia Florentino, Patel Yogen, Brody Jennifer A., Dombroski Beth A., Naranjo Maria Candida Deniz, Daniilidou Makrina, Eiriksdottir Gudny, Mukherjee Shubhabrata, Wallon David, Uphill James, Aspelund Thor, Cantwell Laura B., Garzia Fabienne, Galimberti Daniela, Hofer Edith, Butkiewicz Mariusz, Fin Bertrand, Scarpini Elio, Sarnowski Chloe, Bush Will S., Meslage Stéphane, Kornhuber Johannes, White Charles C., Song Yuenjoo, Barber Robert C., Engelborghs Sebastiaan, Sordon Sabrina, Voijnovic Dina, Adams Perrie M., Vandenberghe Rik, Mayhaus Manuel, Cupples L. Adrienne, Albert Marilyn S., De Deyn Peter P., Gu Wei, Himali Jayanadra J., Beekly Duane, Squassina Alessio, Hartmann Annette M., Orellana Adelina, Blacker Deborah, Rodriguez-Rodriguez Eloy, Lovestone Simon, Garcia Melissa E., Doody Rachelle S., Munoz-Fernadez Carmen, Sussams Rebecca, Lin Honghuang, Fairchild Thomas J., Benito Yolanda A., Holmes Clive, Karamuji-omi Hata, Frosch Matthew P., Thonberg Hakan, Maier Wolfgang, Roschupkin Gena, Ghetti Bernardino, Giedraitis Vilmantas, Kawalia Amit, Li Shuo, Huebinger Ryan M., Kilander Lena, Moebus Susanne, Hernández Isabel, Kamboh M. Ilyas, Brundin RoseMarie, Turton James, Yang Qiong, Katz Mindy J., Concari Letizia, Lord Jenny, Beiser Alexa S., Keene C. Dirk, Helisalmi Seppo, Kloszewska Iwona, Kukull Walter A., Koivisto Anne Maria, Lynch Aoibhinn, Tarraga Lluís, Larson Eric B., Haapasalo Annakaisa, Lawlor Brian, Mosley Thomas H., Lipton Richard B., Solfrizzi Vincenzo, Gill Michael, Longstreth WT, Montine Thomas J., Frisardi Vincenza, Diez-Fairen Monica, Rivadeneira Fernando, Petersen Ronald C., Deramecourt Vincent, Alvarez Ignacio, Salani Francesca, Ciaramella Antonio, Boerwinkle Eric, Reiman Eric M., Fievet Nathalie, Rotter Jerome I., Reisch Joan S., Hanon Olivier, Cupidi Chiara, Uitterlinden A. G. Andre, Royall Donald R., Dufouil Carole, Maletta Raffaele Giovanni, de Rojas Itziar, Sano Mary, Brice Alexis, Cecchetti Roberta, St George-Hyslop Peter, Ritchie Karen, Tsolaki Magda, Tsuang Debby W., Dubois Bruno, Craig David, Wu Chuang-Kuo, Soininen Hilkka, Avramidou Despoina, Albin Roger L., Fratiglioni Laura, Germanou Antonia, Apostolova Liana G., Keller Lina, Koutroumani Maria, Arnold Steven E., Panza Francesco, Gkatzima Olymbia, Asthana Sanjay, Hannequin Didier, Whitehead Patrice, Atwood Craig S., Caffarra Paolo, Hampel Harald, Quintela Inés, Carracedo Angel, Lannfelt Lars, Rubinsztein David C., Barnes Lisa L., Pasquier Florence, Frölich Lutz, Barral Sandra, McGuinness Bernadette, Beach Thomas G., Johnston Janet A., Becker James T., Passmore Peter, Bigio Eileen H., Schott Jonathan M., Bird Thomas D., Warren Jason D., Boeve Bradley F., Lupton Michelle K., Bowen James D., Proitsi Petra, Boxer Adam, Powell John F., Burke James R., Kauwe John S. K., Burns Jeffrey M., Mancuso Michelangelo, Buxbaum Joseph D., Bonuccelli Ubaldo, Cairns Nigel J., McQuillin Andrew, Cao Chuanhai, Livingston Gill, Carlson Chris S., Bass Nicholas J., Carlsson Cynthia M., Hardy John, Carney Regina M., Bras Jose, Carrasquillo Minerva M., Guerreiro Rita, Allen Mariet, Chui Helena C., Fisher Elizabeth, Masullo Carlo, Crocco Elizabeth A., DeCarli Charles, Bisceglio Gina, Dick Malcolm, Ma Li, Duara Ranjan, Graff-Radford Neill R., Evans Denis A., Hodges Angela, Faber Kelley M., Scherer Martin, Fallon Kenneth B., Riemenschneider Matthias, Fardo David W., Heun Reinhard, Farlow Martin R., Kölsch Heike, Ferris Steven, Leber Markus, Foroud Tatiana M., Heuser Isabella, Galasko Douglas R., Giegling Ina, Gearing Marla, Michael Hüll, Geschwind Daniel H., Gilbert John R., Morris John, Green Robert C., Mayo Kevin, Growdon John H., Feulner Thomas, Hamilton Ronald L., Harrell Lindy E., Drichel Dmitriy, Honig Lawrence S., Cushion Thomas D., Huentelman Matthew J., Hollingworth Paul, Hulette Christine M., Hyman Bradley T., Marshall Rachel, Jarvik Gail P., Meggy Alun, Abner Erin, Menzies Georgina E., Jin Lee-Way, Leonenko Ganna, Real Luis M., Jun Gyungah R., Baldwin Clinton T., Grozeva Detelina, Karydas Anna, Russo Giancarlo, Kaye Jeffrey A., Kim Ronald, Jessen Frank, Kowall Neil W., Vellas Bruno, Kramer Joel H., Vardy Emma, LaFerla Frank M., Jöckel Karl-Heinz, Lah James J., Dichgans Martin, Leverenz James B., Mann David, Levey Allan I., PickeringBrown Stuart, Lieberman Andrew P., Klopp Norman, Lunetta Kathryn L., Wichmann H-Erich, Lyketsos Constantine G., Morgan Kevin, Marson Daniel C., Brown Kristelle, Martiniuk Frank, Medway Christopher, Mash Deborah C., Nöthen Markus M., Masliah Eliezer, Hooper Nigel M., McCormick Wayne C., Daniele Antonio, McCurry Susan M., Bayer Anthony, McDavid Andrew N., Gallacher John, McKee Ann C., van den Bussche Hendrik, Mesulam Marsel, Brayne Carol, Miller Bruce L., Riedel-Heller Steffi, Miller Carol A., Miller Joshua W., Al-Chalabi Ammar, Morris John C., Shaw Christopher E., Myers Amanda J., Wiltfang Jens, OBryant Sid, Olichney John M., Alvarez Victoria, Parisi Joseph E., Singleton Andrew B., Paulson Henry L., Collinge John, Perry William R., Mead Simon, Peskind Elaine, Cribbs David H., Rossor Martin, Pierce Aimee, Ryan Natalie S., Poon Wayne W., Nacmias Benedetta, Potter Huntington, Sorbi Sandro, Quinn Joseph F., Sacchinelli Eleonora, Raj Ashok, Spalletta Gianfranco, Raskind Murray, Caltagirone Carlo, Bossù Paola, Orfei Maria Donata, Reisberg Barry, Clarke Robert, Reitz Christiane, Smith A David, Ringman John M., Warden Donald, Roberson Erik D., Wilcock Gordon, Rogaeva Ekaterina, Bruni Amalia Cecilia, Rosen Howard J., Gallo Maura, Rosenberg Roger N., Ben-Shlomo Yoav, Sager Mark A., Mecocci Patrizia, Saykin Andrew J., Pastor Pau, Cuccaro Michael L., Vance Jeffery M., Schneider Julie A., Schneider Lori S., Slifer Susan, Seeley William W., Smith Amanda G., Sonnen Joshua A., Spina Salvatore, Stern Robert A., Swerdlow Russell H., Tang Mitchell, Tanzi Rudolph E., Trojanowski John Q., Troncoso Juan C., Van Deerlin Vivianna M., Van Eldik Linda J., Vinters Harry V., Vonsattel Jean Paul, Weintraub Sandra, Welsh-Bohmer Kathleen A., Wilhelmsen Kirk C., Williamson Jennifer, Wingo Thomas S., Woltjer Randall L., Wright Clinton B., Yu Chang-En, Yu Lei, Saba Yasaman, Pilotto Alberto, Bullido Maria J., Peters Oliver, Crane Paul K., Bennett David, Bosco Paola, Coto Eliecer, Boccardi Virginia, Lleo Alberto, De Jager Phil L., Lleo Alberto, Warner Nick, Lopez Oscar L., Ingelsson Martin, Deloukas Panagiotis, Cruchaga Carlos, Graff Caroline, Gwilliam Rhian, Fornage Myriam, Goate Alison M., Sanchez-Juan Pascual, Kehoe Patrick G., Amin Najaf, Ertekin-Taner Nilifur, Berr Claudine, Debette Stéphanie, Love Seth, Launer Lenore J., Younkin Steven G., Dartigues Jean-Francois, Corcoran Chris, Ikram M. Arfan, Dickson Dennis W., Nicolas Gael, Campion Dominique, Tschanz JoAnn, Schmidt Helena, Hakonarson Hakon, Clarimon Jordi, Munger Ron, Schmidt Reinhold, Farrer Lindsay A., Van Broeckhoven Christine, ODonovan Michael C., DeStefano Anita L., Jones Lesley, Haines Jonathan L., Deleuze Jean-Francois, Owen Michael J., Gudnason Vilmundur, Mayeux Richard, Escott-Price Valentina, Psaty Bruce M., Ramirez Alfredo, Wang Li-San, Ruiz Agustin, van Duijn Cornelia M., Holmans Peter A., Seshadri Sudha, Williams Julie, Amouyel Phillippe, Schellenberg Gerard D., Lambert Jean-Charles, and Pericak-Vance Margaret A.. Genetic meta-analysis of diagnosed Alzheimers disease identifies new risk loci and implicates A-beta, tau, immunity and lipid processing. Nat Genet, 51(3):414–430, March 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].European Alzheimer’s Disease Initiative (EADI), Genetic and Environmental Risk in Alzheimer’s Disease (GERAD), Alzheimer’s Disease Genetic Consortium (ADGC), Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE), Lambert Jean-Charles, IbrahimVerbaas Carla A, Harold Denise, Naj Adam C, Sims Rebecca, Bellenguez Céline, Jun Gyungah, DeStefano Anita L, Bis Joshua C, Beecham Gary W, Grenier-Boley Benjamin, Russo Giancarlo, Thornton-Wells Tricia A, Jones Nicola, Smith Albert V, Chouraki Vincent, Thomas Charlene, Ikram M Arfan, Zelenika Diana, Vardarajan Badri N, Kamatani Yoichiro, Lin ChiaoFeng, Gerrish Amy, Schmidt Helena, Kunkle Brian, Dunstan Melanie L, Ruiz Agustin, Bihoreau Marie-Thérèse, Choi Seung-Hoan, Reitz Christiane, Pasquier Florence, Hollingworth Paul, Ramirez Alfredo, Hanon Olivier, Fitzpatrick Annette L, Buxbaum Joseph D, Campion Dominique, Crane Paul K, Baldwin Clinton, Becker Tim, Gudnason Vilmundur, Cruchaga Carlos, Craig David, Amin Najaf, Berr Claudine, Lopez Oscar L, De Jager Philip L, Deramecourt Vincent, Johnston Janet A, Evans Denis, Lovestone Simon, Letenneur Luc, Morón Francisco J, Rubinsztein David C, Eiriksdottir Gudny, Sleegers Kristel, Goate Alison M, Fiévet Nathalie, Huentelman Matthew J, Gill Michael, Brown Kristelle, Kamboh M Ilyas, Keller Lina, Barberger-Gateau Pascale, McGuinness Bernadette, Larson Eric B, Green Robert, Myers Amanda J, Dufouil Carole, Todd Stephen, Wallon David, Love Seth, Rogaeva Ekaterina, Gallacher John, St George-Hyslop Peter, Clarimon Jordi, Lleo Alberto, Bayer Anthony, Tsuang Debby W, Yu Lei, Tsolaki Magda, Bossù Paola, Spalletta Gianfranco, Proitsi Petroula, Collinge John, Sorbi Sandro, Sanchez-Garcia Florentino, Fox Nick C, Hardy John, Naranjo Maria Candida Deniz, Bosco Paolo, Clarke Robert, Brayne Carol, Galimberti Daniela, Mancuso Michelangelo, Matthews Fiona, Moebus Susanne, Mecocci Patrizia, Del Zompo Maria, Maier Wolfgang, Hampel Harald, Pilotto Alberto, Bullido Maria, Panza Francesco, Caffarra Paolo, Nacmias Benedetta, Gilbert John R, Mayhaus Manuel, Lannfelt Lars, Hakonarson Hakon, Pichler Sabrina, Carrasquillo Minerva M, Ingelsson Martin, Beekly Duane, Alvarez Victoria, Zou Fanggeng, Valladares Otto, Younkin Steven G, Coto Eliecer, Hamilton-Nelson Kara L, Gu Wei, Razquin Cristina, Pastor Pau, Mateo Ignacio, Owen Michael J, Faber Kelley M, Jonsson Palmi V, Combarros Onofre, O’Donovan Michael C, Cantwell Laura B, Soininen Hilkka, Blacker Deborah, Mead Simon, Mosley Thomas H, Bennett David A, Harris Tamara B, Fratiglioni Laura, Holmes Clive, de Bruijn Renee F A G, Passmore Peter, Montine Thomas J, Bettens Karolien, Rotter Jerome I, Brice Alexis, Morgan Kevin, Foroud Tatiana M, Kukull Walter A, Hannequin Didier, Powell John F, Nalls Michael A, Ritchie Karen, Lunetta Kathryn L, Kauwe John S K, Boerwinkle Eric, Riemenschneider Matthias, Boada Mercè, Hiltunen Mikko, Martin Eden R, Schmidt Reinhold, Rujescu Dan, Wang Li-San, Dartigues Jean-François, Mayeux Richard, Tzourio Christophe, Hofman Albert, Nöthen Markus M, Graff Caroline, Psaty Bruce M, Jones Lesley, Haines Jonathan L, Holmans Peter A, Lathrop Mark, Pericak-Vance Margaret A, Launer Lenore J, Farrer Lindsay A, van Duijn Cornelia M, Van Broeckhoven Christine, Moskvina Valentina, Seshadri Sudha, Williams Julie, Schellenberg Gerard D, and Amouyel Philippe. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet, 45(12):1452–1458, December 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Desikan Rahul S., Schork Andrew J., Wang Yunpeng, Thompson Wesley K., Dehghan Abbas, Ridker Paul M, Chasman Daniel I., McEvoy Linda K., Holland Dominic, Chen Chi-Hua, Karow David S., Brewer James B., Hess Christopher P., Williams Julie, Sims Rebecca, ODonovan Michael C., Choi Seung Hoan, Bis Joshua C., Ikram M. Arfan, Gudnason Vilmundur, DeStefano Anita L., van der Lee Sven J., Psaty Bruce M., van Duijn Cornelia M., Launer Lenore, Seshadri Sudha, Pericak-Vance Margaret A., Mayeux Richard, Haines Jonathan L., Farrer Lindsay A., Hardy John, Ulstein Ingun Dina, Dag Aarsland, Fladby Tormod, White Linda R., Sando Sigrid B., Rongve Arvid, Witoelar Aree, Djurovic Srdjan, Hyman Bradley T., Snaedal Jon, Steinberg Stacy, Stefansson Hreinn, Stefansson Kari, Schellenberg Gerard D., Andreassen Ole A., and Dale Anders M.. Polygenic Overlap Between C-Reactive Protein, Plasma Lipids, and Alzheimer Disease. Circulation, 131(23):2061–2069, June 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lutz Michael W., Casanova Ramon, Saldana Santiago, Kuchibhatla Maragatha, Plassman Brenda L., and Hayden Kathleen M.. Analysis of pleiotropic genetic effects on cognitive impairment, systemic inflammation, and plasma lipids in the Health and Retirement Study. Neurobiology of Aging, 80:173–186, August 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu Xia, Wang Wei, Chen Hua-li, Zhang Hai-yan, and Zhang Nai-xia. Interplay between Alzheimers disease and global glucose metabolism revealed by the metabolic profile alterations of pancreatic tissue and serum in APP/PS1 transgenic mice. Acta Pharmacol Sin, May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Swerdlow R, Marcus DL, Landman J, Kooby D, Frey W, and Freedman ML. Brain glucose metabolism in Alzheimer’s disease. Am. J. Med. Sci, 308(3):141–144, September 1994. [DOI] [PubMed] [Google Scholar]
- [48].Szablewski Leszek. Glucose Transporters in Brain: In Health and in Alzheimers Disease. JAD, 55(4):1307–1320, December 2016. [DOI] [PubMed] [Google Scholar]
- [49].Ghosh Chandradeep, Seal Manas, Mukherjee Soumya, and Dey Somdatta Ghosh. Alzheimers Disease: A HemeAbeta Perspective. Acc. Chem. Res, 48(9):2556–2564, September 2015. [DOI] [PubMed] [Google Scholar]
- [50].Mueller Claudius, Zhou Weidong, VanMeter Amy, Heiby Michael, Magaki Shino, Ross Mark M., Espina Virginia, Schrag Matthew, Dickson Cindy, Liotta Lance A., and Kirsch Wolff M.. The Heme Degradation Pathway is a Promising Serum Biomarker Source for the Early Detection of Alzheimer’s Disease. JAD, 19(3):1081–1091, January 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chiasserini Davide, Biscetti Leonardo, Eusebi Paolo, Salvadori Nicola, Frattini Giulia, Simoni Simone, De Roeck Naomi, Tambasco Nicola, Stoops Erik, Vanderstichele Hugo, Engelborghs Sebastiaan, Mollenhauer Brit, Calabresi Paolo, and Parnetti Lucilla. Differential role of CSF fatty acid binding protein 3, alpha-synuclein, and Alzheimers disease core biomarkers in Lewy body disorders and Alzheimers dementia. Alz Res Therapy, 9(1):52, December 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rangaraju Srikant, Dammer Eric B., Syed Ali Raza Tianwen Gao, Xiao Hailian, Betarbet Ranjita, Duong Duc M., Webster James A., Hales Chadwick M., Lah James J., Levey Allan I., and Seyfried Nicholas T.. Quantitative proteomics of acutely-isolated mouse microglia identifies novel immune Alzheimers disease-related proteins. Mol Neurodegeneration, 13(1):34, December 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hotamisligil Gökhan S. and Erbay Ebru. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol, 8(12):923–934, December 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Moore Sherri, Kremer Michael, Sanderlin Edward, Wheeler Michael, and Hines Ian. Emerging Roles for Lipids in the Hepatic Innate Immune Response. Journal of Human Nutrition and Food Science, 1, 2013. [Google Scholar]
- [55].Zhang Yuwen and Li Bing. E-FABP: regulator of immune function. Oncoscience, 1:398, June 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Furuhashi Masato and Hotamisligil Gökhan S.. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov, 7(6):489–503, June 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Matsumata Miho, Inada Hitoshi, and Osumi Noriko. Fatty acid binding proteins and the nervous system: Their impact on mental conditions. Neuroscience Research, 102:47–55, January 2016. [DOI] [PubMed] [Google Scholar]
- [58].Walter Jochen and van Echten-Deckert Gerhild. Cross-talk of membrane lipids and Alzheimer-related proteins. Mol Neurodegeneration, 8(1):34, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Huang Yadong and Mahley Robert W.. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiology of Disease, 72:3–12, December 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, Lemere CA, Selkoe DJ, and Stevens B. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science, 352(6286):712–716, May 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].da Costa Mariana Gaya, Poppelaars Felix, van Kooten Cees, Mollnes Tom E., Tedesco Francesco, Würzner Reinhard, Trouw Leendert A., Truedsson Lennart, Daha Mohamed R., Roos Anja, and Seelen Marc A.. Age and Sex-Associated Changes of Complement Activity and Complement Levels in a Healthy Caucasian Population. Front. Immunol, 9:2664, November 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mangold Colleen A., Wronowski Benjamin, Du Mei, Masser Dustin R., Hadad Niran, Bixler Georgina V., Brucklacher Robert M., Ford Matthew M., Sonntag William E., and Freeman Willard M.. Sexually divergent induction of microglial-associated neuroinflammation with hippocampal aging. J Neuroinflammation, 14(1):141, December 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Foster Evangeline M., Dangla-Valls Adrià, Lovestone Simon, Ribe Elena M., and Buckley Noel J.. Clusterin in Alzheimers Disease: Mechanisms, Genetics, and Lessons From Other Pathologies. Front. Neurosci, 13:164, February 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lee Joseph H., Cheng Rong, Barral Sandra, Reitz Christiane, Medrano Martin, Lantigua Rafael, Jiménez-Velazquez Ivonne Z., Rogaeva Ekaterina, St. George-Hyslop Peter H., and Mayeux Richard. Identification of Novel Loci for Alzheimer Disease and Replication of CLU, PICALM, and BIN1 in Caribbean Hispanic Individuals. Arch Neurol, 68(3), March 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wijsman Ellen M., Pankratz Nathan D., Choi Yoonha, Rothstein Joseph H., Faber Kelley M., Cheng Rong, Lee Joseph H., Bird Thomas D., Bennett David A., Ramon Diaz-Arrastia Alison M. Goate, Farlow Martin, Ghetti Bernardino, Sweet Robert A., Foroud Tatiana M., Mayeux Richard, and The NIA-LOAD/NCRAD Family Study Group. Genome-Wide Association of Familial Late-Onset Alzheimer’s Disease Replicates BIN1 and CLU and Nominates CUGBP2 in Interaction with APOE. PLoS Genet, 7(2):e1001308, February 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yu Jin-Tai and Tan Lan. The Role of Clusterin in Alzheimers Disease: Pathways, Pathogenesis, and Therapy. Mol Neurobiol, 45(2):314–326, April 2012. [DOI] [PubMed] [Google Scholar]
- [67].Nordengen Kaja, Kirsebom Bjørn-Eivind, Henjum Kristi, Selnes Per, Gísladóttir Berglind, Wettergreen Marianne, Torsetnes Silje Bøen, Grøntvedt Gøril Rolfseng, Waterloo Knut K., Aarsland Dag, Nilsson Lars N. G., and Fladby Tormod. Glial activation and inflammation along the Alzheimers disease continuum. J Neuroinflammation, 16(1):46, December 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Slot Rosalinde E.R., Kester Maartje I., Van Harten Argonde C., Jongbloed Wesley, Bouwman Femke H., Teunissen Charlotte E., Scheltens Philip, van der Flier Wiesje M., and Veerhuis Robert. ApoE and clusterin CSF levels influence associations between APOE genotype and changes in CSF tau, but not CSF Abeta42, levels in non-demented elderly. Neurobiology of Aging, 79:101–109, July 2019. [DOI] [PubMed] [Google Scholar]
- [69].Zhang Jing, Goodlett David R., Peskind Elaine R., Quinn Joseph F., Zhou Yong, Wang Qin, Pan Catherine, Yi Eugene, Eng Jimmy, Aebersold Ruedi H., and Montine Thomas J.. Quantitative proteomic analysis of age-related changes in human cerebrospinal fluid. Neurobiology of Aging, 26(2):207–227, February 2005. [DOI] [PubMed] [Google Scholar]
- [70].Baird Geoffrey S., Nelson Sally K., Keeney Tracy R., Stewart Alex, Williams Stephen, Kraemer Stephan, Peskind Elaine R., and Montine Thomas J.. Age-Dependent Changes in the Cerebrospinal Fluid Proteome by Slow Off-Rate Modified Aptamer Array. The American Journal of Pathology, 180(2):446–456, February 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jack Clifford R., Bennett David A., Blennow Kaj, Carrillo Maria C., Feldman Howard H., Frisoni Giovanni B., Hampel Harald, Jagust William J., Johnson Keith A., Knopman David S., Petersen Ronald C., Scheltens Philip, Sperling Reisa A., and Dubois Bruno. A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology, 87(5):539–547, August 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Welch Joshua D., Kozareva Velina, Ferreira Ashley, Vanderburg Charles, Martin Carly, and Macosko Evan Z.. Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell, 177(7):1873–1887.e17, June 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gjoneska Elizabeta, Pfenning Andreas R., Mathys Hansruedi, Quon Gerald, Kundaje Anshul, Tsai Li-Huei, and Kellis Manolis. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimers disease. Nature, 518(7539):365–369, February 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cordeiro Gauss M. and Montenegro Lourdes Coral C.. BARTLETT COR-RECTED LIKELIHOOD RATIO TESTS IN LOCATION-SCALE NON-LINEAR MODELS. Communications in Statistics - Theory and Methods, 30(7):1353–1372, June 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ADNI data are available with approved data access request from http://adni.loni.usc.edu/.