Abstract
Childhood cancer survivors treated with radiation therapy (RT) and osteotoxic chemotherapies are at increased risk for fractures. However, understanding of how genetic and clinical susceptibility factors jointly contribute to fracture risk among survivors is limited. To address this gap, we conducted genome-wide association studies of fracture risk after cancer diagnosis in 2,453 participants of European ancestry from the Childhood Cancer Survivor Study (CCSS) with 930 incident fractures using Cox regression models (i.e., time-to-event analysis) and prioritized sex- and treatment-stratified genetic associations. We performed replication analyses in 1,417 survivors of European ancestry with 652 incident fractures from the St. Jude Lifetime Cohort Study (SJLIFE). In discovery, we identified a genome-wide significant (P<5×10−8) fracture risk locus, 16p13.3 (HAGHL), among female CCSS survivors (N=1,289) with strong evidence of sex-specific effects (Psex-heterogeneity<7×10−6). Combining discovery and replication data, rs1406815 showed the strongest association (HR=1.43, P=8.2×10−9; N=1,935 women) at this locus. In treatment-stratified analyses in the discovery cohort, the association between rs1406815 and fracture risk among female survivors with no RT exposures was weak (HR=1.22, 95% CI: 0.95–1.57, P=0.11), but increased substantially among those with greater head/neck RT doses (any RT: HR=1.88, 95% CI: 1.54–2.28, P=2.4×10−10; >36 Gray only: HR=3.79, 95% CI: 1.95–7.34, P=8.2×10−5). These head/neck RT-specific HAGHL SNP effects were replicated in female SJLIFE survivors. In silico bioinformatics analyses suggest these fracture risk alleles regulate HAGHL gene expression and related bone resorption pathways. Genetic risk profiles integrating this locus may help identify female survivors who would benefit from targeted interventions to reduce fracture risk.
Keywords: Genome-wide association studies, GWAS, fracture risk, childhood cancer survivors, osteoporosis
INTRODUCTION
Childhood cancer survivors are at increased risk for developing bone-related late effects. Treatment with osteotoxic chemotherapies (e.g., corticosteroids, methotrexate) may adversely affect normal bone metabolism and skeletal development, while radiation therapy (RT) can induce bone tissue damage and endocrinopathies that influence bone loss.(1,2) Other factors contributing to bone fragility risk include malignancy-related pathologies (e.g., leukemia) and deficiencies in childhood physical activity and nutrition.(1,2) These clinical factors are hypothesized to disrupt the acquisition of sufficient peak bone mass during childhood and adolescence, elevating risk for early onset osteoporosis and subsequent fractures in survivors.(1,2)
In the general population, bone mineral density (BMD) deficits(3,4) and biological sex(5,6) are critical determinants of fracture risk. In a recent study of clinically ascertained late effects in adults treated for childhood cancer (N=1,713),(7) the prevalence of BMD deficits consistent with a diagnosis of osteoporosis (Z-score≤−2.5 SD) was estimated to be ~10% among survivors with a median age of 32 years, which is similar to the prevalence of osteoporosis among adults in the United States aged 60–80 years.(8) Prevalence estimates of BMD deficits in smaller studies of long-term survivors have been reported to be as high as 30% to 50%, varying by diagnostic group, time from diagnosis, and treatment exposures.(1,2,9–12) While broader studies of incident fractures after childhood cancer diagnosis are limited, studies of acute lymphoblastic leukemia survivors have reported higher incident fracture rates during and immediately after treatment: one study observed fracture rates to be ~six-fold higher during the three-year follow-up period after diagnosis relative to healthy controls,(13) while another reported a four-year vertebral fracture cumulative incidence of ~26% after diagnosis.(14) Recent studies of survivors have also reported differential risk for BMD deficits(9,15) and fractures(16) by sex and treatment exposures, which may reflect increased sex-specific vulnerabilities to certain therapeutic agents during childhood and adolescence.(9)
Risk for BMD deficits and fractures after treatment for childhood cancer varies substantially among survivors with similar treatment histories, suggesting genetic susceptibility factors may also play an important role.(1) In the general population, genome-wide association studies (GWAS) of BMD(17,18) and fracture risk(17,19) have shown that these complex bone phenotypes are heritable and offer hundreds of candidate loci for further study. However, there is emerging evidence that top genetic associations identified in GWAS conducted in the general population may not be generalizable to survivors as a consequence of previous cancer treatment exposures,(20) suggesting independent GWAS in survivors are needed. Indeed, a recent genome-wide analysis of BMD in survivors of acute lymphoblastic leukemia identified complex genetic variants (epistatic interactions) including novel SNPs that potentially modify the effects of specific cancer therapies on BMD.(21) To our knowledge, no fracture risk GWAS have been conducted in childhood cancer survivors. Therefore, we performed GWAS of incident fracture risk (i.e., time-to-event analysis) after diagnosis among survivors of European ancestry, using data from the Childhood Cancer Survivor Study (CCSS, N=2,453) for discovery and St. Jude Lifetime Cohort Study (SJLIFE, N=1,417) for replication. To identify genetic loci with sex- and treatment-specific effects on fracture risk in survivors, we selected genome-wide significant (P<5×10−8) SNPs from sex-specific discovery GWAS with evidence of replication for further interrogation in targeted treatment-stratified analyses.
MATERIALS AND METHODS
Survivor cohort study designs and fracture definition
For discovery analyses, we evaluated data from CCSS, the largest multi-institutional cohort study of long-term (≥5 years) survivors of childhood cancer in North America. Survivors were diagnosed before age 21 years between 1970 and 1986, with prospective follow-up of late effects through longitudinal surveys querying health conditions, health-related behaviors, and healthcare care use.(22–24) In this study, we included CCSS survivors of European ancestry with DNA genotype data who did not have bone tumor primary diagnoses. Among the 4,713 CCSS participants meeting these criteria, 62.7% (N=2,955) provided detailed lifetime fracture histories as a part of a larger follow-up questionnaire. Qualifying incident first fractures after primary cancer diagnosis included any fracture at any skeletal site. Covariate data were abstracted from medical records or self-reported in surveys. Participants with allogeneic stem cell transplantation history and incomplete cancer treatment data were excluded.
Replication analyses were performed with data from survivors of European ancestry from SJLIFE,(25,26) a retrospectively-constructed cohort study of 5-year survivors treated for pediatric cancer at St. Jude Children’s Research Hospital (SJCRH) with prospective medical assessment of late effects. Criteria applied to exclude participants and define qualifying fractures in discovery were applied in replication analyses. Most (84.3%) fracture histories were taken from medical history interviews conducted by clinicians at SJCRH visits; otherwise, self-reported responses to fracture prompts identical to the CCSS questionnaires were used. Data for other covariates were clinically assessed during SJCRH visits or abstracted from medical records.
All CCSS and SJLIFE study protocols and contact documents were approved by the institutional review boards of participating study institutions. All study participants provided informed consent. A flow diagram summarizing inclusion criteria for discovery and replication study participation is provided in Supplemental Figure 1. Details regarding phenotype/covariate data collection and processing in CCSS and SJLIFE are provided in the Supplemental Methods.
Genotype data
Methods used to generate genotype data in CCSS and SJLIFE have been described extensively elsewhere.(27–30) In brief, DNA was genotyped using the Illumina HumanOmni5Exome array and imputed using Minimac3(31) for CCSS samples, while whole genome sequencing was performed using the Illumina HiSeq X10 platform with an average coverage per sample of 36.8X in SJLIFE. Stringent sample and variant quality control was applied to autosomal variant data in CCSS and SJLIFE; all discovery and replication analyses were restricted to participants of European genetic ancestry, based on principal components analysis. Discovery analyses in CCSS were performed with ~5.4 million SNPs with minor allele frequency ≥5% and high imputation quality scores (r2≥0.8). Additional details describing genotype data quality control and ancestry ascertainment are given in the Supplemental Methods.
Power calculation
We estimated the power to detect SNP associations for a range of effect allele carrier probabilities (i.e., probability of carrying at least one effect allele under effect allele frequencies or EAFs from 0.05 to 0.3, or 2[EAF][1-EAF]+[EAF]2) and hazard ratios (HRs) comparable in size to reported odds ratios in the fracture risk GWAS literature (HRs up to 2.0). These power estimates used the time-to-event analysis approach(32) and assumed the observed fracture cumulative incidences and sample sizes in male and female CCSS survivors separately, and a type I error probability of 0.05.
Statistical analysis
Previously published GWAS in CCSS and SJLIFE(27,29,30,33) have identified novel genetic loci whose associations with various late effects health conditions (e.g., breast cancer, stroke, premature menopause) are modified by specific cancer therapies in modestly-sized survivor cohorts (N=3,000–6,000) using targeted analyses stratified by relevant treatment exposures. In the current study, we used a similar strategy to identify genetic loci with sex- and treatment-specific effects on fracture risk in survivors by: (1) conducting sex-specific discovery GWAS in CCSS; (2) performing sex-specific replication analyses in SJLIFE; and (3) evaluating treatment-stratified genetic associations with fracture risk, exclusively among SNPs with genome-wide significant (P<5×10−8) fracture risk associations in sex-specific discovery analyses that also showed evidence of replication. In brief, we used discovery and replication analyses to effectively filter SNP candidates for further investigation. Because we do not know which SNPs would have sex-and treatment-specific effects a priori, we adopted this strategy to control the type I error probability while maximizing power for discovery in small survivor cohorts (unlike traditional SNP-treatment interaction analyses). Additional details describing each of these analytic steps are provided below.
Discovery in CCSS
To estimate the additive effects of each SNP allele on first fracture risk following diagnosis, we used Cox proportional hazards models (time-to-event analysis). We chose a Cox regression modeling approach that used age as the time scale(34) to adjust for the strong effects of age on fracture risk(5,6) non-parametrically and reduce residual confounding by age. These models were adjusted for potential population stratification and cryptic population substructure and relatedness (first 10 European ancestry principal components), sex, attained height and weight, premature menopause status, and treatments determined to be relevant through univariate association testing (Supplemental Methods): exposure to corticosteroids; intravenous (IV) methotrexate dose; intrathecal (IT) methotrexate dose; and maximum tumor dose (maxTD) from RT to any of seven major body regions (head, neck, chest, abdomen, pelvis, arm, leg).(35) Associations with P<5×10−8 from two-sided tests were considered to be genome-wide significant.
Descriptive cumulative incidence curves among the CCSS survey respondents were examined to compare unadjusted fracture risk by years of follow-up across SNP-genotype groups. Genomic region plots surrounding genome-wide significant SNPs were generated with LocusZoom software.(36) We examined the extent to which genome-wide significant SNPs at each locus were in linkage disequilibrium (LD) in the 1000 Genomes European (1000G EUR) reference panel to assess the relative strength of statistical evidence supporting the observed fracture risk associations. Testing for sex-heterogeneous effects was conducted with GWAMA v2.2.2(37) for suggestively significant (P<1×10−5) SNPs from discovery. GWAMA computes an asymptotically X2-distributed test statistic using summary statistics to test for allelic effect differences between sexes. A Bonferroni-corrected p-value threshold for SNPs with suggestively significant associations in sex-specific discovery analyses (P=0.05/[number of SNPs with P<1×10−5]) was used to assess statistical significance.
Replication in SJLIFE and meta-analysis
We examined all genome-wide significant SNP associations identified in the sex-specific discovery analyses in the SJLIFE replication cohort using the same statistical model from discovery analyses. Associations with replication P≤0.05 (two-sided) with association directions consistent with discovery were considered replicated. For replicated SNP associations, summary effect estimates combining CCSS and SJLIFE association results were computed using the fixed-effects inverse variance-weighted meta-analysis method with GWAMA v2.2.2.(37) The Cochran’s Q statistic and I2 inconsistency index were assessed for effect heterogeneity.
Cancer therapy effect modification
SNPs with genome-wide significant associations in CCSS discovery analyses and evidence of replication in SJLIFE were filtered for further investigation in treatment-stratified analyses to determine whether their genetic effects on fracture risk were modified by specific cancer therapies. We considered three composite treatment definitions in stratified analyses: head/neck RT (maxTD for head or neck), trunk RT (maxTD for chest, abdomen, or pelvis), and chemotherapy (any corticosteroid exposure and IV or IT methotrexate dose). Given the high prevalence of endocrinopathies after RT to the cranial, hypothalamic-pituitary, or neck regions,(38) we evaluated estimates stratified by head/neck RT dose separately from trunk RT. For each SNP meeting the described filtering criteria, we compared adjusted SNP associations in strata with no, any, >medium, and >high dose exposures for these treatment definitions, with medium and high dose corresponding to median and 3rd quartile doses in CCSS, respectively.
Functional/regulatory annotation of SNPs in credible sets
Adopting an annotation procedure similar to Gaulton et al.(39), we constructed 99% credible intervals or SNP sets with 99% probability of containing the causal variant using a Bayesian approach(39–41) for each replicated locus (Supplemental Methods) for annotation, given that: (a) the most strongly associated SNP may not directly influence fracture risk; (b) >1 causal variant may be present at a locus; and (c) the signal could reflect the effects of complex genetic variation, e.g., haplotypes. We interrogated credible-set SNP associations in the Musculoskeletal Knowledge Portal,(42) recent GWAS of bone-related phenotypes(17,18,43) and phenome-wide association studies(44) (PheWAS), and functional/regulatory annotations using external genomic data resources (Supplemental Methods). Lastly, we tested whether credible-set SNPs were likely to drive fracture risk signals in survivors through promoter regulatory mechanisms in specific cell types by using an enrichment test procedure.(39) Specifically, we compared the observed mean posterior probability of credible-set SNPs directly overlapping promoter regions(45) to its null distribution generated with 100,000 randomly-shifted promoter region annotations across cell types specified a priori for relevance to fracture risk in survivors and comparison cell types from the Encyclopedia of DNA Elements(46) (ENCODE) Project (Supplemental Methods).
RESULTS
Discovery: Genetic variants at HAGHL, CD86 loci are associated with fracture risk in female survivors
The major demographic and clinical characteristics of the CCSS discovery cohort (N=2,453) are provided in Table 1. Median age at follow-up survey completion was 42 years (IQR=36–48 years). Post-diagnosis fracture events were reported by 37.9% of survivors in the discovery cohort at follow-up (930 incident fractures); by sex, the cumulative incidence of fracture events was 33.3% (429 events) and 43.0% (501 events) in female and male survivors, respectively (Table 2). Male survivors had significantly greater unadjusted risk of fracture after diagnosis (P=2.0×10−7; Supplemental Figure 2). Limb fractures accounted for the majority (>60%) of post-diagnosis fractures (Supplemental Table 1). While cancer treatment exposures were symmetrically distributed between sexes (Table 1), increases in post-diagnosis fracture risk were associated with increasing IV and IT methotrexate dose in male survivors and higher RT dosages in female survivors (Supplemental Table 2; Supplemental Figure 3).
Table 1:
Demographic and clinical characteristics of childhood cancer survivors in discovery and replication cohorts, split by sex
| Characteristic | Discovery cohort (CCSS) | Replication cohort (SJLIFE) | ||||
|---|---|---|---|---|---|---|
| Sex-combined (N=2,453) | Female (N=1,289) | Male (N=1,164) | Sex-combined (N=1,417) | Female (N=646) | Male (N=771) | |
| % (N) or median (IQR) | % (N) or median (IQR) | % (N) or median (IQR) | % (N) or median (IQR) | % (N) or median (IQR) | % (N) or median (IQR) | |
| Sex | ||||||
| Female | 52.5% (1,289) | 45.6% (646) | ||||
| Male | 47.5% (1,164) | 54.4% (771) | ||||
| Attained age (years) | 42 (36–48) | 42 (36–48) | 43 (37–48) | 31 (26–39) | 31 (26–39) | 32 (26–38) |
| Attained height (cm) | 168 (163–178) | 163 (157–168) | 178 (170–183) | 169 (162–177) | 162 (157–167) | 176 (170–181) |
| Attained weight (kg) | 77 (64–91) | 68 (59–82) | 84 (75–96) | 79 (65–95) | 70 (60–86) | 86 (73–100) |
| Age at cancer diagnosis (years) | 5 (2–12) | 5 (2–12) | 6 (3–12) | 6 (3–12) | 6 (3–13) | 7 (3–12) |
| Primary cancer diagnosis | ||||||
| Leukemia | 35.6% (874) | 38.9% (501) | 32.0% (373) | 35.1% (497) | 35.6% (230) | 34.6% (267) |
| Hodgkin lymphoma | 15.0% (367) | 15.9% (205) | 13.9% (162) | 12.5% (177) | 13.3% (86) | 11.8% (91) |
| Kidney tumors | 12.6% (309) | 14.7% (190) | 10.2% (119) | 7.3% (104) | 9.6% (62) | 5.4% (42) |
| Soft tissue sarcoma | 9.7% (237) | 9.0% (116) | 10.4% (121) | 7.5% (106) | 7.1% (46) | 7.8% (60) |
| Central nervous system tumors | 9.2% (226) | 5.7% (74) | 13.1% (152) | 14.3% (203) | 12.4% (80) | 16.0% (123) |
| Neuroblastoma | 9.1% (224) | 10.9% (141) | 7.1% (83) | 4.7% (66) | 4.8% (31) | 4.5% (35) |
| Non-Hodgkin lymphoma | 8.8% (216) | 4.8% (62) | 13.2% (154) | 7.5% (106) | 5.4% (35) | 9.2% (71) |
| Other | -- | -- | -- | 11.2% (158) | 11.8% (76) | 10.6% (82) |
| Chemotherapy receipt (any) | ||||||
| IV methotrexate | 18.5% (454) | 18.1% (233) | 19.0% (221) | 29.2% (414) | 28.2% (182) | 30.1% (232) |
| IT methotrexate | 38.4% (941) | 37.9% (488) | 38.9% (453) | 38.3% (543) | 37.6% (243) | 38.9% (300) |
| Glucocorticoids | 47.2% (1,158) | 47.0% (606) | 47.4% (552) | 48.3% (685) | 46.9% (303) | 49.5% (382) |
| Methotrexate dosea (in mg/m2) | ||||||
| IV methotrexate | 3,051 (805 – 6,058) | 3,120 (596 – 6,550) | 2,951 (923 – 5,510) | 1,567 (211 – 2,952) | 1,681 (370 – 2,922) | 1,515 (185 – 3,153) |
| IT methotrexate | 126 (71 – 222) | 132 (72 – 223) | 120 (68–222) | 158 (93 – 233) | 171 (84 – 235) | 150 (96 – 233) |
| Radiation therapy receiptb (any) | ||||||
| Any site | 63.0% (1,545) | 61.0% (786) | 65.2% (759) | 48.2% (683) | 48.9% (316) | 47.6% (367) |
| Radiation to head regionc | 45.9% (1,125) | 43.2% (557) | 48.8% (568) | 38.5% (545) | 37.3% (241) | 39.4% (304) |
| Radiation to trunk regiond | 37.0%% (908) | 37.4% (482) | 36.6% (426) | 25.7% (364) | 26.6% (172) | 24.9% (192) |
| Radiation to limb regionse | 1.4% (34) | 1.7% (22) | 1.0% (12) | 3.7% (52) | 4.3% (28) | 3.1% (24) |
| Radiation therapy dosef (in cGy) | ||||||
| Any site | 2,400 (2,000 – 3,900) | 2,400 (1,800 – 3,600) | 2,500 (2,000 – 4,100) | 2,600 (2,100 – 4,500) | 2,600 (2,100 – 4,000) | 2,600 (2,100 – 5,070) |
| Head regions | 2,400 (1,800 – 3,800) | 2,400 (1,800 – 3,500) | 2,400 (2,000 – 4,200) | 2,600 (2,100 – 4,500) | 2,600 (2,100 – 3,700) | 2,600 (2,100 – 5,300) |
| Trunk regions | 3,000 (2,000 – 3,900) | 2,900 (2,000 – 4,000) | 3,000 (2,000 – 3,800) | 2,600 (2,100 – 3,500) | 2,600 (2,175 – 3,600) | 2,600 (2,100 – 3,500) |
| Limb regions | 4,750 (3,625 – 5,900) | 4,700 (3,000 – 5,850) | 4,750 (4,275 – 5,725) | 2,650 (2,000 – 4,600) | 2,700 (2,000 – 4,522) | 2,650 (2,000 – 3,500) |
Dose distributions only include survivors who received any IV or IT methotrexate.
Received more than high scatter doses of radiation therapy.
Head region refers to the brain, neck, or other head region.
Trunk region refers to the chest, abdomen, or pelvis region.
Limb regions refer to arm or leg regions.
Maximum cumulative dosimetry dose; dose distributions only include survivors who received >high scatter doses.
Abbreviations: IQR, inter-quartile range; cm, centimeters; kg, kilograms; IV, intravenous; IT, intrathecal; cGy, centigray.
Table 2:
Characteristics of first fracture events after primary cancer diagnosis in childhood cancer survivors in discovery and replication cohorts, split by sex
| Discovery (CCSS, N=2,453) | Replication (SJLIFE, N=1,417) | |||
|---|---|---|---|---|
| Characteristics | Female (N=1,289) | Male (N=1,164) | Female (N=646) | Male (N=771) |
| Total number of first fractures after diagnosis | 429 | 501 | 246 | 406 |
| Total follow-up (in person-years) | 36,005 | 29,234 | 12,288 | 12,799 |
| Median age at first fracture (IQR), in years | 18 (11–31) | 16 (11–25) | 16 (10–25) | 16 (11–22) |
Our power estimates (Supplemental Figure 4) suggested the sex-specific discovery analyses were sufficiently powered (≥80%) to find common GWAS fracture risk variants with modest effect sizes, e.g., detecting allelic effects of HR≥1.3 for variants with EAF≥0.2. Analyses of the ~5.4 million common autosomal SNPs in the sex-combined and male-specific CCSS discovery samples yielded no genome-wide significant (P<5×10−8) associations with post-diagnosis fracture risk (Supplemental Figure 5). In the female-specific CCSS discovery sample (N=1,289), we identified three SNPs at the CD86 locus (3q13.33) and four SNPs at the HAGHL locus (16p13.3) with fracture risk associations meeting the genome-wide significance threshold (Table 3). The genomic inflation factor across all analyses was 1.02 – 1.03, indicating adequate control of potential population stratification and cryptic population structure and relatedness. Global comparisons of SNP associations with post-diagnosis fracture risk by sex are provided in side-by-side Manhattan plots (Figure 1) and quantile-quantile (QQ) plots (Supplemental Figure 6). We observed no overlap between genome-wide significant variants in CCSS and previously reported fracture risk susceptibility loci, with all discovered SNPs located >1 Mb from lead SNPs in published fracture GWAS (Figure 1, Supplemental Table 3).
Table 3:
All genome-wide significant (P<5×10−8) SNP associations with post-diagnosis fracture risk identified among female survivors in the discovery cohort, compared to male survivors
| Female survivors, discovery cohort (CCSS, N=1,289) | Male survivors, discovery cohort (CCSS, N=1,164) | Sex heterogeneitya | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP | Chr | BP | EA | NEA | EAF | HR (95% CI) | P | EAF | HR (95% CI) | P | Effects | Psex-het |
| 3q13.33 (CD86) | rs4315642 | 3 | 121836049 | C | T | 0.28 | 0.64 (0.54 to 0.75) | 4.1×10−8 | 0.29 | 1.08 (0.95 to 1.24) | 0.24 | −+ | 8.1×10−7 |
| rs2681399 | 3 | 121835908 | G | A | 0.28 | 0.64 (0.54 to 0.75) | 4.7×10−8 | 0.29 | 1.09 (0.95 to 1.25) | 0.23 | −+ | 7.9×10−7 | |
| rs2681400 | 3 | 121837377 | T | C | 0.28 | 0.64 (0.54 to 0.75) | 4.7×10−8 | 0.29 | 1.08 (0.95 to 1.24) | 0.25 | −+ | 9.6×10−7 | |
| 16p13.3 (HAGHL) | rs12448432 | 16 | 778820 | A | G | 0.19 | 1.55 (1.33 to 1.81) | 1.2×10−8 | 0.20 | 0.91 (0.78 to 1.07) | 0.26 | +− | 2.2×10−6 |
| rs1406815 | 16 | 778158 | G | C | 0.19 | 1.55 (1.33 to 1.80) | 1.5×10−8 | 0.21 | 0.92 (0.78 to 1.07) | 0.28 | +− | 3.0×10−6 | |
| rs9928077 | 16 | 784765 | T | C | 0.19 | 1.54 (1.32 to 1.79) | 2.6×10−8 | 0.21 | 0.92 (0.78 to 1.07) | 0.28 | +− | 3.9×10−6 | |
| rs12597563 | 16 | 787738 | C | G | 0.17 | 1.57 (1.34 to 1.84) | 2.8×10−8 | 0.18 | 0.91 (0.77 to 1.08) | 0.29 | +− | 4.1×10−6 | |
For sex heterogeneity testing, a Bonferroni-corrected p-value threshold was used (P<0.05/[41 evaluated SNPs with suggestive significance in sex-specific discovery analyses]=1.2×10−3) to assess statistical significance. The direction of the fracture risk associations by sex are provided in the “Effects” column, with results in female survivors presented first [left] followed by results in male survivors [right]; “-“ corresponds to decreasing risk and “+” corresponds to increasing risk.
Abbreviations: SNP, single nucleotide polymorphism; Chr, chromosome; BP, base position, GRCh37 (hg19) build; EA, effect (risk) allele; NEA, non-effect (reference) allele; EAF, effect allele frequency; HR, hazard ratio; CI, confidence interval; Psex-het, sex heterogeneity test p-value.
Figure 1:
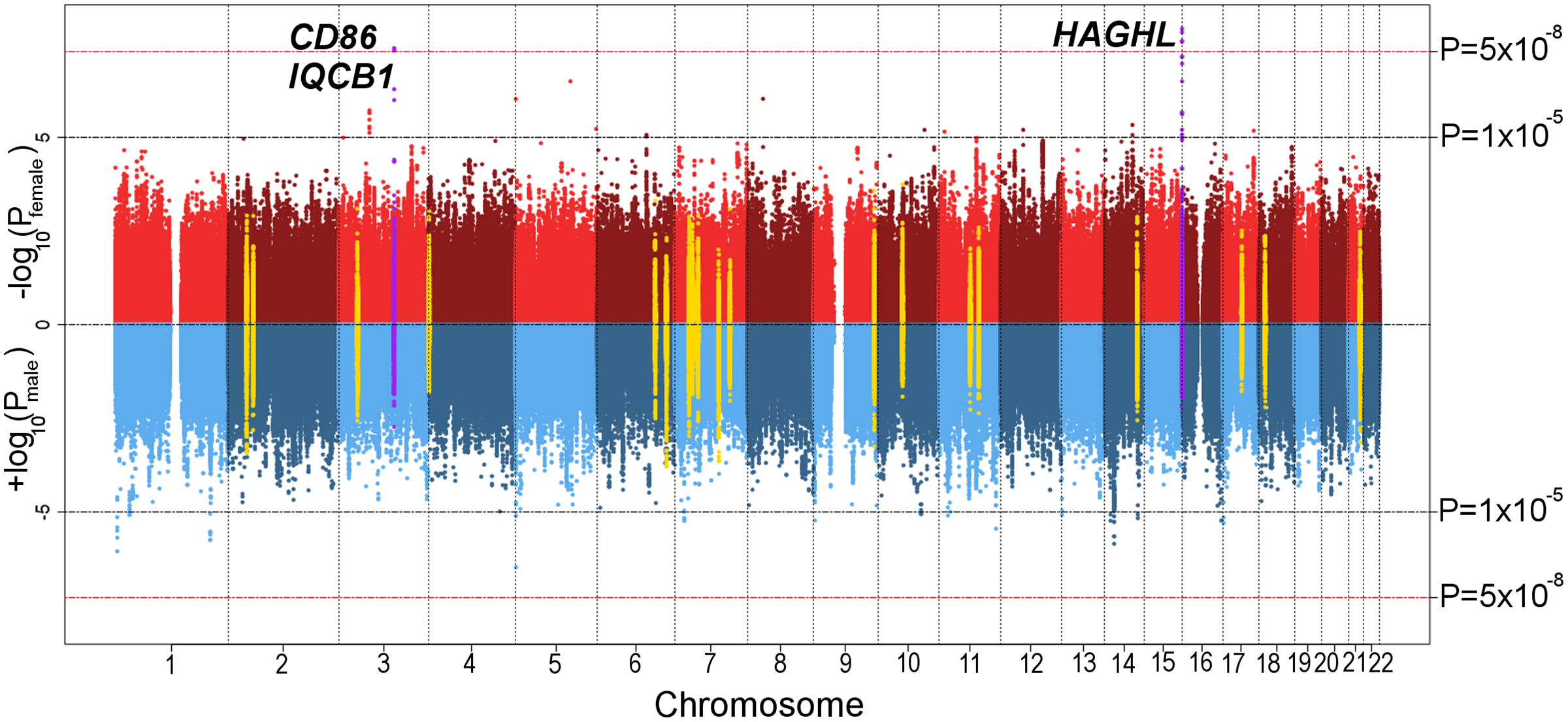
Sex-specific plots of CCSS discovery analysis p-values for autosomal SNP associations with post-diagnosis fracture risk in 2,453 childhood cancer survivors. Depicted p-values are from two-sided Wald tests. On top (red) is the Manhattan plot of –log10 p-values (y-axis) by SNP genomic position (x-axis) from the genome-wide association analysis in 1,289 female survivors. On bottom (blue) is the inverted Manhattan plot (+log10 p-values) from the corresponding analysis in 1,164 male survivors. The red dashed horizontal line signifies the genome-wide significance threshold (P<5×10−8). Log10 p-values for SNPs with previously reported genome-wide significant associations with fracture risk and nearby SNPs (50-kb window) are depicted in yellow. Sex-specific log10 p-values at genome-wide significant loci in female survivors are shown in purple.
Regional plots of SNP associations with subsequent fracture risk at the CD86 and HAGHL loci in female survivors in the CCSS discovery cohort are provided in Supplemental Figures 7–8. At the CD86 locus, rs4315642 had the strongest association with fracture risk (effect allele frequency or EAF=0.28; HR=0.64, 95% CI: 0.54–0.75, P=4.1×10−8), which was supported by two other genome-wide significant SNPs at the CD86 locus in high LD (r2>0.99, 1000G EUR). The most significantly associated SNP at the HAGHL locus was rs12448432 (EAF=0.20; HR=1.55, 95% CI: 1.33–1.81, P=1.2×10−8), whose association with fracture risk was corroborated by three other genome-wide significant SNPs in high LD at this locus (r2≥0.84, 1000G EUR). All seven genome-wide significant SNPs (across the two independent loci) were characterized by significant allelic effect heterogeneity by sex (Bonferroni-corrected threshold P<0.05/[41 evaluated SNPs]=1.2×10−3; Table 3).
Replication: Female-specific HAGHL SNP associations with fracture risk replicated
Following our analytic strategy (see Methods), we evaluated all genome-wide significant SNP associations in an independent sample of survivors from SJLIFE (N=1,417). Demographic, clinical, and fracture event characteristics for survivors in the replication cohort are presented in Tables 1 and 2. Among female SJLIFE survivors (N=646), 38.0% reported post-diagnosis fracture events. We replicated three of the four HAGHL SNP associations with increased post-diagnosis fracture risk in SJLIFE female survivors (HR=1.23–1.24, P≤0.05, Table 4). Meta-analysis combining results from discovery and replication female survivor cohorts (N=1,935) revealed rs1406815 (chr16:778158, GRCh37) had the strongest association with fracture risk (HR=1.43, 95% CI: 1.27–1.62, P=8.2×10−9, Table 4). Among male survivors in the CCSS discovery cohort, non-significant protective fracture risk associations with HAGHL SNPs were seen; consistent with this observation, HAGHL SNPs showed protective fracture risk effects in male survivors from the SJLIFE replication cohort (N=771, P<0.01). CD86 locus SNPs did not replicate, showing the opposite direction of association from discovery.
Table 4:
Replication results for genome-wide significant SNP associations with fracture risk identified in discovery analyses and meta-analysis of replicated (P≤0.05) associations
| Discovery (CCSS; N=1,289 women) | Replication (SJLIFE; N=646 women) | Meta-analysis: Discovery and replication cohorts, combined (CCSS and SJLIFE; N=1,935 women) | Replication in male survivors (SJLIFE; N=771 men) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP | Chr | BP | EA | NEA | EAF | HR (95% CI) | P | EAF | HR (95% CI) | P | EAF | HR (95% CI) | P | Phet | I2 | EAF | HR (95% CI) | P |
| 3q13.33 (CD86) | rs4315642 | 3 | 121836049 | C | T | 0.28 | 0.64 (0.54 to 0.75) | 4.1×10−8 | 0.28 | 1.24 (1.01 to 1.52) | 0.04 | -- | -- | -- | -- | -- | 0.26 | 0.91 (0.77 to 1.07) | 0.26 |
| rs2681399 | 3 | 121835908 | G | A | 0.28 | 0.64 (0.54 to 0.75) | 4.7×10−8 | 0.28 | 1.26 (1.02 to 1.55) | 0.03 | -- | -- | -- | -- | -- | 0.26 | 0.91 (0.78 to 1.08) | 0.28 | |
| rs2681400 | 3 | 121837377 | T | C | 0.28 | 0.64 (0.54 to 0.75) | 4.7×10−8 | 0.28 | 1.25 (1.02 to 1.54) | 0.03 | -- | -- | -- | -- | -- | 0.26 | 0.90 (0.76 to 1.06) | 0.22 | |
| 16p13.3 (HAGHL) | rs12448432 | 16 | 778820 | A | G | 0.19 | 1.55 (1.33 to 1.81) | 1.2×10−8 | 0.21 | 1.23 (1.00 to 1.51) | 0.05 | 0.20 | 1.43 (1.27 to 1.62) | 9.1×10−9 | 0.08 | 0.68 | 0.19 | 0.78 (0.64 to 0.93) | 0.01 |
| rs1406815 | 16 | 778158 | G | C | 0.19 | 1.55 (1.33 to 1.80) | 1.5×10−8 | 0.21 | 1.24 (1.01 to 1.53) | 0.04 | 0.20 | 1.43 (1.27 to 1.62) | 8.2×10−9 | 0.09 | 0.65 | 0.20 | 0.74 (0.62 to 0.90) | 1.9×10−3 | |
| rs9928077 | 16 | 784765 | T | C | 0.19 | 1.54 (1.32 to 1.79) | 2.6×10−8 | 0.21 | 1.23 (1.00 to 1.52) | 0.05 | 0.20 | 1.43 (1.26 to 1.61) | 1.6×10−8 | 0.09 | 0.65 | 0.20 | 0.75 (0.63 to 0.91) | 2.8×10−3 | |
| rs12597563 | 16 | 787738 | C | G | 0.17 | 1.57 (1.34 to 1.84) | 2.8×10−8 | 0.18 | 1.17 (0.94 to 1.46) | 0.17 | -- | -- | -- | -- | -- | 0.18 | 0.76 (0.63 to 0.93) | 0.01 | |
Abbreviations: SNP, single nucleotide polymorphism; Chr, chromosome; BP, base position, GRCh37 (hg19) build; EA, effect (risk) allele; NEA, non-effect (reference) allele; EAF, effect allele frequency; HR, hazard ratio; CI, confidence interval; Phet, p-value from Cochran’s Q test for effect heterogeneity; I2, inconsistency index.
Bolded variant identifies the sentinel SNP with the strongest fracture risk association after meta-analysis for the locus.
HAGHL SNP effects on fracture risk increase with previous head/neck radiation therapy
The three HAGHL SNPs with genome-wide significant, female-specific fracture risk associations in discovery with evidence of replication in female survivors were evaluated for treatment-specific effects on fracture risk. In treatment-stratified analyses, we found that strata with increasing doses of head/neck RT showed corresponding increases in post-diagnosis fracture risk associations with all three selected HAGHL SNPs in both the discovery and replication survivor cohorts (shown in Figure 2; detailed results behind Figure 2 are provided in Supplemental Table 4), while strata with increasing trunk RT and composite chemotherapy doses did not (Supplemental Table 4). As an illustrative example, we describe detailed results for rs1406815, the HAGHL locus SNP with the strongest association with fracture risk after meta-analysis. Among female survivors with no exposure to head/neck radiation in the CCSS discovery cohort, the association between rs1406815 and fracture risk was weak (N=501; per effect allele, HR=1.22, 95% CI: 0.95–1.57, P=0.11); in comparison, rs1406815 showed a considerably higher fracture risk association in CCSS female survivors with any head/neck RT exposure, with little overlap in confidence intervals for the stratum-specific HRs (N=788; HR=1.88, 95% CI: 1.54–2.28, P=2.4×10−10). The magnitude of association per effect allele for rs1406815 was appreciably greater among those with higher head/neck radiation exposures (>3rd quartile dose or 36 Gy stratum, N=117; HR=3.79, 95% CI: 1.95–7.34, P=8.2×10−5). Similar magnitudes of association between rs1406815 and fracture risk were seen among female survivors in the SJLIFE replication cohort (no head/neck radiation, N=331; HR=1.38, 95% CI: 1.03–1.85, P=0.03; >36 Gy head/neck RT, N=61; HR=3.08, 95% CI: 1.09–8.74, P=0.03). In the discovery cohort, the estimated cumulative incidence of fracture was 37.1% and 60.0% at 15 years and 30 years post-diagnosis, respectively, among female survivors with any head/neck RT and homozygous rs1406815 risk alleles (Figure 3); no comparable increases in fracture risk was observed among male survivors with identical genetic and treatment risk profiles (Figure 3).
Figure 2:
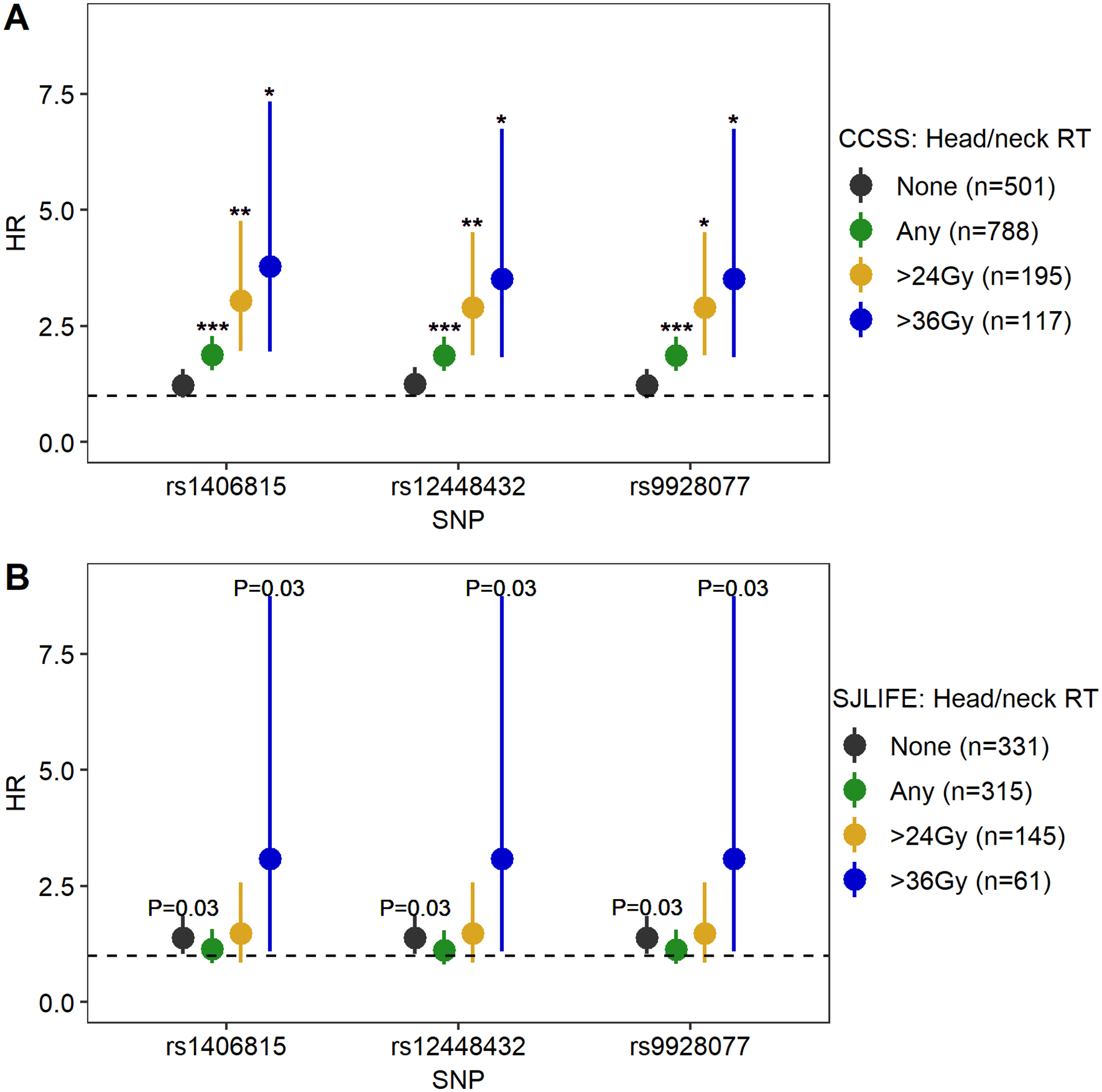
Head/neck radiation therapy-stratified associations between replicated HAGHL locus SNPs and post-diagnosis fracture risk in female survivors from CCSS and SJLIFE. Plots show head/neck radiation therapy (RT) exposure stratum-specific HRs (dots) and 95% confidence intervals (whiskers) for associations between post-diagnosis fracture risk and risk alleles for each of the three replicated HAGHL locus SNPs (rs1406815, rs12448432, rs9928077) in CCSS female survivors (panel 2A, discovery cohort, N=1,289) and SJLIFE female survivors (panel 2B, replication cohort, N=646). Statistical significance thresholds for p-values from two-sided Wald tests for genome-wide (annotated as ***, for P<5×10−8), suggestive (**, for 5×10−8≤P<5×10−5), and nominal (*, for 5×10−5≤ P<0.001) significance are provided for stratum-specific HRs. Actual p-values for 0.001≤P<0.10 are provided in the figure. The dashed line signifies HR=1. The number of survivors in each treatment stratum are provided in plot legends.
Figure 3:
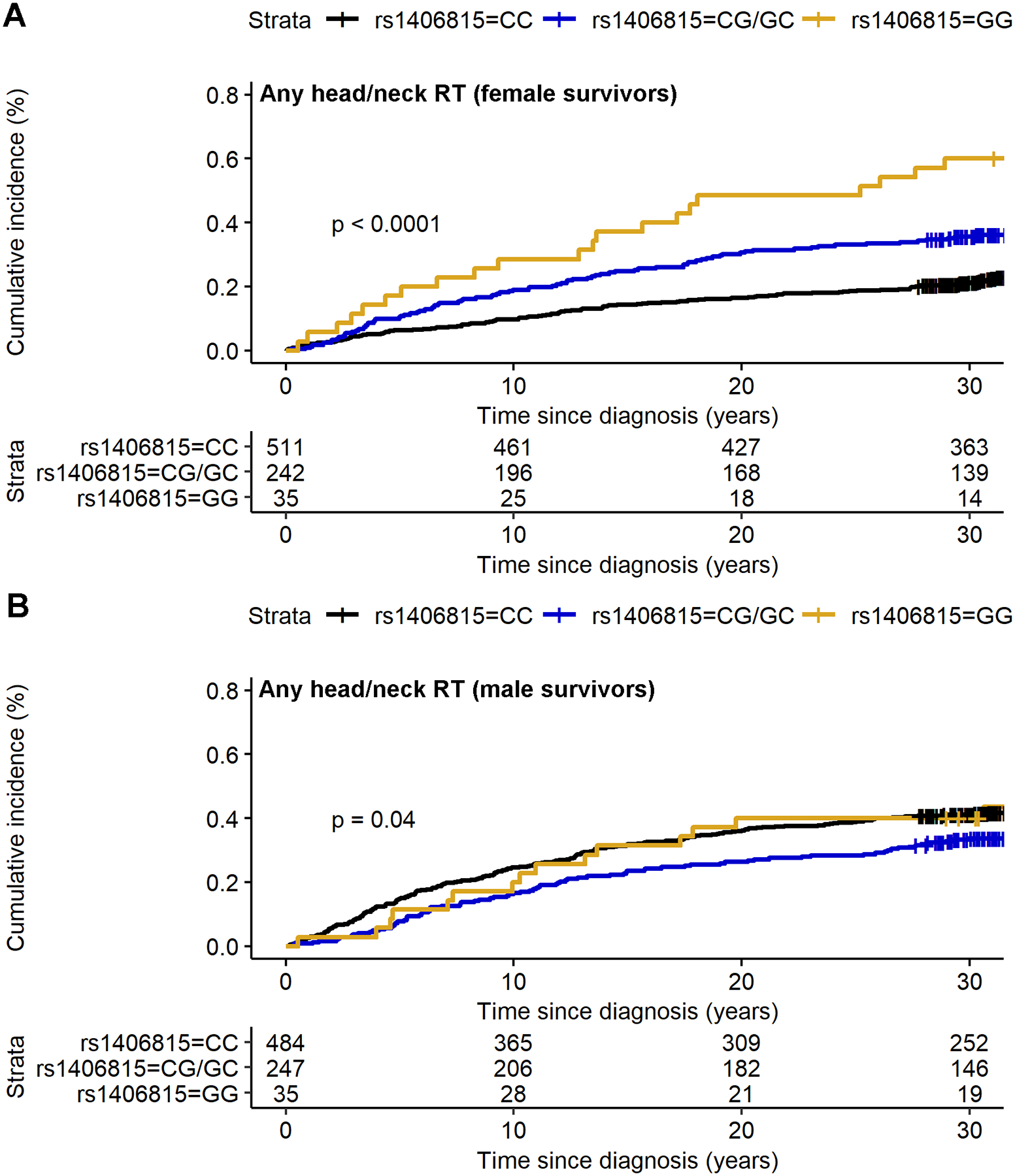
Cumulative incidence curves of post-diagnosis fracture in the CCSS female and male survivors with any exposure to head/neck radiation therapy by HAGHL locus SNP rs1406815 genotype profiles. Panel A shows cumulative incidence curves for fracture by SNP genotype among female survivors with any head/neck radiation therapy (RT; N=788) while panel B is the corresponding figure among males (N=766). The fracture risk allele for SNP rs1406815 is allele G. The p-value from the two-sided log-rank test comparing the fracture risk probability distributions by genotype is provided in the lower left corner.
HAGHL locus SNPs have plausible functional and regulatory consequences on fracture risk
We used external genomic annotation resources to interrogate HAGHL locus SNPs in a 99% credible set representing the set of common variants most likely to be responsible for the fracture risk association signal at the HAGHL locus. The 99% credible set for HAGHL locus SNPs consisted of 11 variants spanning a ~14-kb region (Supplemental Table 3).
A PheWAS of UK Biobank phenotypes showed the top (P<1×10−16) associated phenotypes for credible-set SNPs were for height and body composition (Supplemental Table 5). Given the strength of reported associations between HAGHL locus SNPs and height and weight in the UK Biobank, we evaluated whether the observed HAGHL locus SNP fracture risk associations could be biased by adjusting for heritable covariates(47) (i.e., height, weight). The potential impact of collider bias on effect estimates appeared to be negligible, since associations between HAGHL locus SNPs and fracture risk after omitting adjustments for attained height, weight, or both covariates yielded no appreciable changes to effect estimates, and no associations between these SNPs and height or weight were observed in female CCSS survivors (Supplemental Table 6). While not phenome-wide significant, the leading (P<5×10−3) phenotypes for credible-set SNPs in a second PheWAS of ICD9 codes were largely related to musculoskeletal conditions. In published GWAS of bone phenotypes conducted in the general population(17,18,43), we found multiple credible-set SNPs were associated with nominally significant (P<0.05) decreases in BMD; all showed nominally significant increases in femoral neck area and non-significant but directionally consistent increases with fracture risk (Supplemental Table 7).
To determine likely gene targets and cellular contexts, we considered external functional annotations of the credible-set SNPs. Six SNPs mapped to HAGHL transcripts, of which two SNPs were also putative HAGHL coding alleles(48) (rs1406815, encoding p.Arg50Gly; rs12448432, encoding p.[Ala202Thr;Ala94Thr;Ala84Thr;Ala21Thr]) (Supplemental Table 3). All credible-set SNPs’ fracture risk alleles were strongly associated with increased expression of NARFL and HAGHL (FDR≤5%), particularly in thyroid cells(49) (Supplemental Table 3). Risk alleles for multiple credible-set SNPs (7/11) were also significantly associated (FDR<5%) with increased DNA methylation at a CpG site near NARFL and HAGHL (cg27144592) in whole blood.(50) Lastly, among differential expression quantitative trait loci (eQTLs) in human osteoblasts treated with pharmacological agents known to affect bone cells(51), rs12448432 was identified as a cis-eQTL (FDR<5%) for HAGHL in osteoblasts treated with dexamethasone or prostaglandin E2 compared to untreated samples.
We examined chromatin state annotations(45) and found that credible-set SNPs predominantly overlapped putative promoter and transcribed regions (Supplemental Table 3). We therefore assessed whether the credible-set SNPs were likely to drive fracture risk signals in survivors by regulating promoter activity in specific cell types. We examined a set of four cell types likely to be relevant to fracture risk in female survivors exposed to head/neck RT and a comparison set of diverse cell types. We found that credible-set SNPs with high posterior probabilities (>0.2) for being causal fracture-risk SNPs co-localize with the HAGHL promoter region and showed significant selectivity (Bonferroni-corrected threshold P<3.6×10−3) for putative poised/bivalent promoter chromatin states in bone cells (osteoblasts, chondrocytes), female fetal brain tissue, and ovary tissue (Figure 4). No comparison cell type showed statistically significant enrichment of credible-set SNPs in promoter sites.
Figure 4:
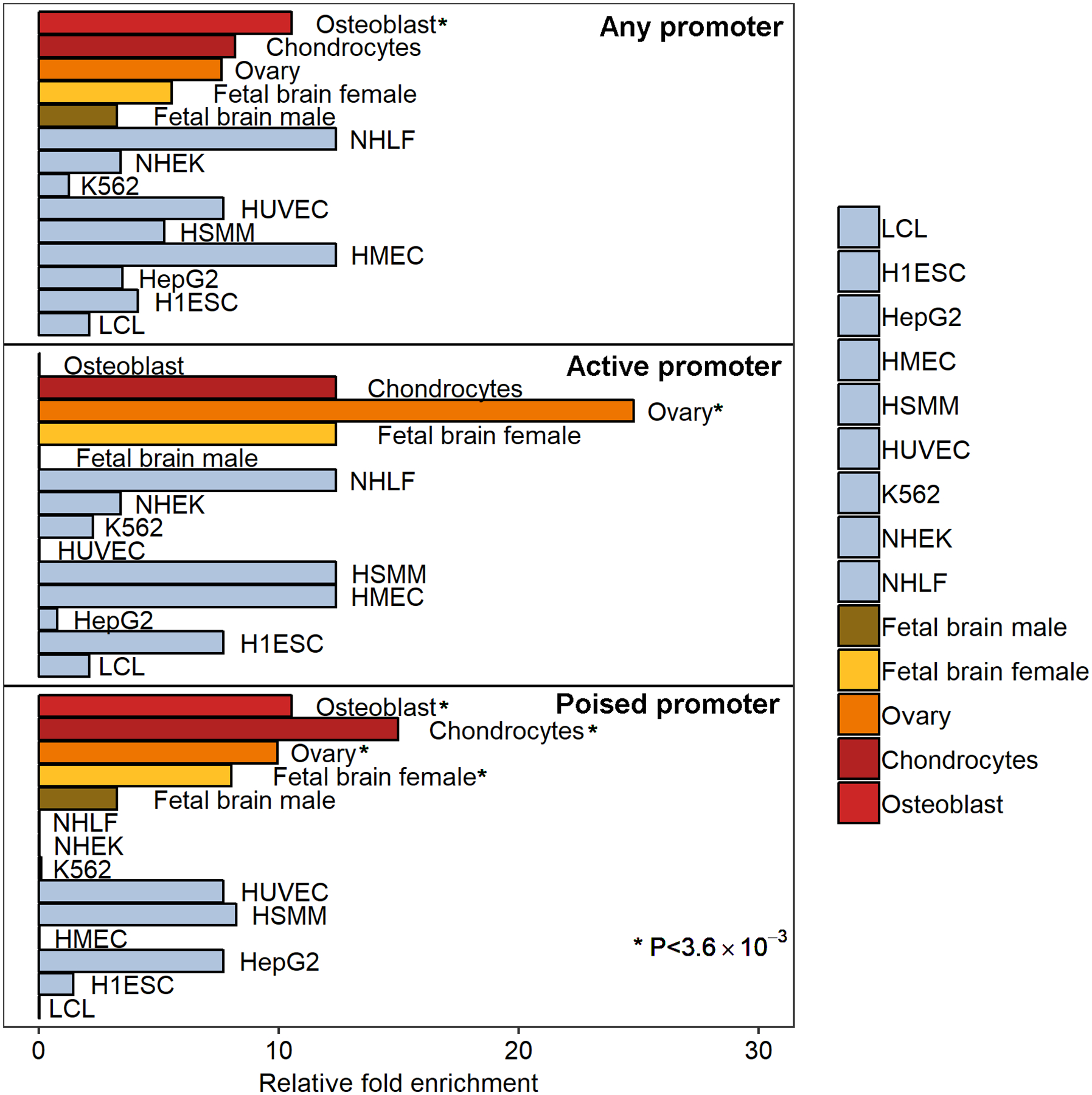
HAGHL/NARFL post-diagnosis fracture risk variants in female survivors overlap promoter epigenetic features in bone, ovary, and female brain cell types. Enrichments of posterior probabilities for credible-set SNPs that overlap promoter chromatin state annotations (25-state ChromHMM) compared to null distribution posterior probabilities are illustrated. Relative fold enrichments in 9 comparison ENCODE cell types are shown (pale blue), along with enrichments in 4 phenotype-relevant cell types: bone osteoblasts and chondrocytes (red); ovary (orange); and female fetal brain cells (yellow). Top, middle, and bottom panels show enrichment results from overlap with any promoter (active/poised promoters), active promoter, and poised promoter states, respectively. Cell types with significant enrichments meeting the Bonferroni-corrected threshold P<0.05/[14 evaluated cell types]=3.6×10−3 from permutation tests are annotated (*) and include: chondrocytes (poised promoter, P=2.0×10−3); female fetal brain (poised promoter, P=3.3×10−3); osteoblasts (any promoter, P=2.9×10−3; poised promoter, P=2.9×10−3); and ovary (active promoter, P=2.0×10−3; poised promoter, P=3.1×10−3). Abbreviations for ENCODE cell types are as follows: GM12878 (B-lymphocyte), K562 (chronic myelogenous leukemia), HepG2 (hepatocellular carcinoma), HSMM (skeletal muscle myoblast), HUVEC (umbilical vein endothelial), NHEK (epidermal keratinocyte), NHLF (lung fibroblast), H1-hESC (embryonic stem cell), HMEC (mammary epithelial]).
DISCUSSION
In this GWAS of incident fracture risk after diagnosis in long-term survivors of childhood cancer, we identified two independent genetic loci with sex-specific fracture risk effects, CD86 (3q13.33) and HAGHL (16p13.3), among 1,289 female survivors in the CCSS discovery cohort. The female-specific fracture risk susceptibility locus 16p13.3 (HAGHL) was replicated in an independent cohort of 646 female survivors in SJLIFE. Using discovery and replication analyses as a filter to reduce the potential for false negative results and identify candidate loci with sex-specific effects for further investigation, we evaluated whether selected HAGHL locus SNPs also had treatment-specific effects on fracture risk. We found the fracture risk effects of replicated HAGHL locus SNPs increased incrementally in subgroups of female survivors with greater doses of RT to the head or neck, which was not observed among male survivors with comparable genotype and treatment profiles. In general population study samples, HAGHL locus SNPs were observed to have nominally significant associations with BMD and femoral neck area,(18,43) and genome-wide significant associations with phenotypes corresponding to skeletal size (e.g., body height and mass)(18,44). Of particular note was the relatively strong external association observed between increased femoral neck area (P=5.6×10−3) and rs1406815, the HAGHL locus variant with the strongest association with fracture risk in female survivors in our meta-analysis; larger femoral neck area has been reported to have a greater genetic correlation with increased hip fracture risk than femoral neck BMD deficits.(43) These external GWAS results suggest the HAGHL locus plausibly contributes to elevated fracture risk in female survivors.
Current long-term follow-up guidelines for bone density and fracture late effects such as those issued by the Children’s Oncology Group(52) are broad, recommending bone densitometry screening or clinical follow-up of all survivors with any exposure to radiation, antimetabolites, corticosteroids, or hematopoietic cell transplant. Our results suggest that an evaluation of both genetic and clinical risk factors and their interactions, i.e., HAGHL genetic variants modified by sex and varying exposures to head/neck RT, may potentially be more informative in identifying subgroups of childhood cancer survivors at greater risk for fractures after diagnosis than existing follow-up recommendations.
An improved understanding of the biological mechanisms underpinning fracture risk in survivors is warranted. Insight into how HAGHL locus SNPs affect fracture risk in female survivors, particularly those exposed to head/neck RT, may reveal new pathways for bone biology which are potentially useful as future targets for treatments for low bone density and osteoporosis. In silico analyses suggest that the fracture risk alleles at the HAGHL locus are strongly associated with HAGHL gene expression in endocrine tissues and may also play a role in the regulation of a poised/bivalent state at the HAGHL promoter in bone cell types. Notably, these HAGHL locus SNPs have also been associated with differential HAGHL gene expression in osteoblasts treated with dexamethasone and prostaglandin E2 (PGE2). HAGHL encodes a member of the glyoxalase II subfamily of the metallo-β-lactamase protein superfamily; while the exact functions of HAGHL-encoded glyoxalases remain unknown, glyoxalase I and II work in tandem in detoxifying pathways for byproducts of glycolysis and may contribute to the maturation of osteoclasts (bone-resorbing cells).(53) Given that poised/bivalent promoter regions are posited as keeping genes “poised” for rapid activation in response to environmental stimuli, we speculate that HAGHL locus SNPs may increase fracture risk in female survivors by affecting osteoclastogenesis pathways mediated by HAGHL gene expression in response to head/neck radiation. PGE2 levels increase as a part of the local proinflammatory response after irradiation(54) and increase in osteoblasts as an indirect effect of altered levels of thyroid hormones.(55) Since damage to the thyroid gland and consequent thyroid hormone imbalance are common after head/neck RT,(38) head/neck RT may elevate fracture risk in female survivors with HAGHL risk alleles by altering baseline levels of thyroid hormones and PGE2 to influence HAGHL transcription and increase bone resorption.
To our knowledge, this study constitutes the first genome-wide assessment of SNP associations with fracture risk in long-term survivors of childhood cancer. Although our study is relatively underpowered to detect typical fracture risk signals observed in GWAS performed in general population samples, the major strength of our analysis is that we focus on capturing genetic variants with sex- and treatment-specific fracture risk associations in survivors. Among the limitations of our study is that fractures were self-reported and are therefore subject to recall bias. Studying fractures confirmed by radiographic reports would be optimal, but previous studies of fracture risk in survivors suggest the validity of self-reported fractures is high.(16) Because detailed fracture histories were only available for survivors who responded to a specific CCSS follow-up questionnaire, these results may not be generalizable to all 5-year survivors. For example, we found that CCSS survivors who provided detailed fracture histories (~63% response rate) were more likely to be female and older at follow-up; they were also more likely to have been diagnosed with leukemia and less likely to have been exposed to radiation therapy (Supplemental Table 8). Among these factors, the difference in sex was greatest between responders and non-responders; given that our analyses were stratified by sex, the impact of sample sex differences on the generalizability of results was likely mitigated. Temporal data for potential confounders at fracture occurrence were also unavailable, including use of medications and supplements to improve BMD (e.g., hormone replacement therapy, vitamin D, calcium), alcohol use, smoking, exercise, height, and weight in CCSS; consequently, we did not account for these risk factors or used best available proxies (i.e., attained height/weight). Due to the unavailability of well-cleaned variant data for sex chromosomes in CCSS and SJLIFE, analyses were restricted to autosomal variants; an evaluation of variants on sex chromosomes and their associations with fracture risk is needed in survivor cohorts. Another limitation of the CCSS data is that measures of BMD and bone area are unavailable. Assessments of how SNPs, especially HAGHL locus SNPs, affect BMD and bone area in survivors are warranted. Lastly, functional validation for posited biological mechanisms involving HAGHL locus SNPs, head/neck RT, and fracture risk in female survivors is needed.
In summary, we performed GWAS of first fracture risk following primary cancer diagnosis in long-term survivors of childhood cancer. We identified a credible novel genetic locus (HAGHL, 16p13.3) for fracture risk that is both female-specific and sensitive to previous exposures to head/neck RT. Our study demonstrates the importance of interrogating sex-specific SNP effects in survivors, especially for bone phenotypes with differential risk by sex in both the general population, due to sex-specific patterns for bone accretion and loss,(5,6) and survivors, as a consequence of sex-specific vulnerabilities to cancer treatments.(9,15) Because multiple clinical interventions to lessen fracture risk and increase BMD exist, future investigations should be pursued to evaluate whether top genetic associations identified among survivors, including HAGHL genetic variants, and polygenic risk scores based on published BMD/fracture risk GWAS conducted in general population samples can improve fracture risk prediction in survivors.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Cancer Institute (U24 CA55727 to G.T.A., principal investigator, U01 CA195547 to M.M.H. and L.L.R., principal investigators, CA21765 to C. Roberts, Principal Investigator and R01 CA216354 to Y.Y. and J. Zhang principal investigators); Childhood Cancer Survivor Study Career Development Award; and American Lebanese Syrian Associated Charities.
Footnotes
Conflict of interest disclosure statement: The authors declare no conflicts of interest.
Data accessibility statement: The CCSS data used in this study may be accessed from the database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) under accession number phs001327.v1.p1. Data from SJLIFE is available from the St. Jude Cloud (https://www.stjude.cloud/) under accession number SJC-DS-1002.
REFERENCES
- 1.Wilson CL, Ness KK. Bone mineral density deficits and fractures in survivors of childhood cancer. Curr Osteoporos Rep. 2013;11(4):329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wasilewski-Masker K, Kaste SC, Hudson MM, Esiashvili N, Mattano LA, Meacham LR. Bone mineral density deficits in survivors of childhood cancer: long-term follow-up guidelines and review of the literature. Pediatrics. Mar 2008;121(3):e705–13. [DOI] [PubMed] [Google Scholar]
- 3.Sambrook P, Cooper C. Osteoporosis. Lancet. 2006;367(9527):2010–8. [DOI] [PubMed] [Google Scholar]
- 4.Johnell O, Kanis J. An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteoporos Int. 2006;17(12):1726–33. [DOI] [PubMed] [Google Scholar]
- 5.Farr JN, Melton III LJ, Achenbach SJ, Atkinson EJ, Khosla S, Amin S. Fracture incidence and characteristics in young adults aged 18 to 49 years: a population‐based study. J Bone Miner Res. 2017;32(12):2347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Court-Brown CM, Caesar B. Epidemiology of adult fractures: a review. Injury. 2006;37(8):691–7. [DOI] [PubMed] [Google Scholar]
- 7.Hudson MM, Ness KK, Gurney JG, Mulrooney DA, Chemaitilly W, Krull KR, et al. Clinical ascertainment of health outcomes among adults treated for childhood cancer. JAMA. 2013;309(22):2371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright NC, Looker AC, Saag KG, Curtis JR, Delzell ES, Randall S, et al. The recent prevalence of osteoporosis and low bone mass in the United States based on bone mineral density at the femoral neck or lumbar spine. J Bone Miner Res. 2014;29(11):2520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Hoed MA, Klap BC, te Winkel ML, Pieters R, van Waas M, Neggers S, et al. Bone mineral density after childhood cancer in 346 long-term adult survivors of childhood cancer. Osteoporos Int. 2015;26(2):521–9. [DOI] [PubMed] [Google Scholar]
- 10.Gurney JG, Kaste SC, Liu W, Srivastava DK, Chemaitilly W, Ness KK, et al. Bone mineral density among long-term survivors of childhood acute lymphoblastic leukemia: results from the St. Jude Lifetime Cohort Study. Pediatr Blood Cancer. July 2014;61(7):1270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang M, Kim SM, Lee YA, Shin CH, Yang S, Lim J. Risk factors for osteoporosis in long-term survivors of intracranial germ cell tumors. Osteoporos Int. 2012;23(7):1921–9. [DOI] [PubMed] [Google Scholar]
- 12.Sala A, Talsma D, Webber C, Posgate S, Atkinson S, Barr R. Bone mineral status after treatment of malignant lymphoma in childhood and adolescence. Eur J Cancer Care. 2007;16(4):373–9. [DOI] [PubMed] [Google Scholar]
- 13.van der Sluis IM, van den Heuvel-Eibrink MM, Hählen K, Krenning EP, de Muinck Keizer-Schrama SM. Altered bone mineral density and body composition, and increased fracture risk in childhood acute lymphoblastic leukemia. J Pediatr. 2002;141(2):204–10. [DOI] [PubMed] [Google Scholar]
- 14.Cummings EA, Ma J, Fernandez CV, Halton J, Alos N, Miettunen PM, et al. Incident vertebral fractures in children with leukemia during the four years following diagnosis. J Clin Endocrinol Metab. 2015;100(9):3408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Atteveld JE, Pluijm SM, Ness KK, Hudson MM, Chemaitilly W, Kaste SC, et al. Prediction of low and very low bone mineral density among adult survivors of childhood cancer. J Clin Oncol. 2019;37(25):2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson CL, Dilley K, Ness KK, Leisenring WL, Sklar CA, Kaste SC, et al. Fractures among long-term survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. Cancer. December 01 2012;118(23):5920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemp JP, Morris JA, Medina-Gomez C, Forgetta V, Warrington NM, Youlten SE, et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat Genet. 2017;49(10):1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai RC, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet. 2019;51(2):258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trajanoska K, Morris JA, Oei L, Zheng H-F, Evans DM, Kiel DP, et al. Assessment of the genetic and clinical determinants of fracture risk: genome wide association and mendelian randomisation study. BMJ. 2018;362:k3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im C, Qin N, Wang Z, Qiu W, Howell CR, Sapkota Y, et al. Generalizability of” GWAS hits” in clinical populations: Lessons from childhood cancer survivors. Am J Hum Genet. 2020;107(4):636–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im C, Ness KK, Kaste SC, Chemaitilly W, Moon W, Sapkota Y, et al. Genome-wide search for higher order epistasis as modifiers of treatment effects on bone mineral density in childhood cancer survivors. Europ J Hum Genet. 2018;26:275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robison LL, Armstrong GT, Boice JD, Chow EJ, Davies SM, Donaldson SS, et al. The Childhood Cancer Survivor Study: a National Cancer Institute–supported resource for outcome and intervention research. J Clin Oncol. 2009;27(14):2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robison LL, Mertens AC, Boice JD, Breslow NE, Donaldson SS, Green DM, et al. Study design and cohort characteristics of the Childhood Cancer Survivor Study: a multi‐institutional collaborative project. Med Pediatr Oncol. 2002;38(4):229–39. [DOI] [PubMed] [Google Scholar]
- 24.Leisenring WM, Mertens AC, Armstrong GT, Stovall MA, Neglia JP, Lanctot JQ, et al. Pediatric cancer survivorship research: experience of the Childhood Cancer Survivor Study. J Clin Oncol. 2009;27(14):2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudson MM, Ness KK, Nolan VG, Armstrong GT, Green DM, Morris EB, et al. Prospective medical assessment of adults surviving childhood cancer: study design, cohort characteristics, and feasibility of the St. Jude Lifetime Cohort study. Pediatr Blood Cancer. May 2011;56(5):825–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson MM, Ehrhardt MJ, Bhakta N, Baassiri M, Eissa H, Chemaitilly W, et al. Approach for classification and severity grading of long-term and late-onset health events among childhood cancer survivors in the St. Jude Lifetime Cohort. Cancer Epidemiol Biomarkers Prev. 2017;26(5):666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morton LM, Sampson JN, Armstrong GT, Chen T-H, Hudson MM, Karlins E, et al. Genome-wide association study to identify susceptibility loci that modify radiation-related risk for breast cancer after childhood cancer. J Natl Cancer Inst. 2017;109(11):djx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Wilson CL, Easton J, Thrasher A, Mulder H, Liu Q, et al. Genetic Risk for Subsequent Neoplasms Among Long-Term Survivors of Childhood Cancer. J Clin Oncol. 2018;36(20):2078–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sapkota Y, Cheung YT, Moon W, Shelton K, Wilson CL, Wang Z, et al. Whole–Genome Sequencing of Childhood Cancer Survivors Treated with Cranial Radiation Therapy Identifies 5p15. 33 Locus for Stroke: A Report from the St. Jude Lifetime Cohort Study. Clin Cancer Res. 2019;25(22):6700–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sapkota Y, Turcotte LM, Ehrhardt MJ, Howell RM, Arnold MA, Wilson CL, et al. Genome-wide association study in irradiated childhood cancer survivors identifies HTR2A for subsequent basal cell carcinoma. J Invest Dermatol. 2019;139(9):2042–5. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chow S-C, Wang H, Shao J. Sample size calculations in clinical research: CRC press; 2007. [Google Scholar]
- 33.Brooke RJ, Im C, Wilson CL, Krasin MJ, Liu Q, Li Z, et al. A high-risk haplotype for premature menopause in childhood cancer survivors exposed to gonadotoxic therapy. J Natl Cancer Inst. 2018;110(8):895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kom EL, Graubard BI, Midthune D. Time-to-event analysis of longitudinal follow-up of a survey: choice of the time-scale. Am J Epidemiol. 1997;145(1):72–80. [DOI] [PubMed] [Google Scholar]
- 35.Howell RM, Smith SA, Weathers RE, Kry SF, Stovall M. Adaptations to a Generalized Radiation Dose Reconstruction Methodology for Use in Epidemiologic Studies: An Update from the MD Anderson Late Effect Group. Radiat Res. 2019;192(2):169–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11(1):288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chemaitilly W, Cohen LE, Mostoufi-Moab S, Patterson BC, Simmons JH, Meacham LR, et al. Endocrine late effects in childhood cancer survivors. J Clin Oncol. 2018;36(21):2153–9. [DOI] [PubMed] [Google Scholar]
- 39.Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Mägi R, Reschen ME, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47(12):1415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wakefield J A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81(2):208–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiel DP, Kemp JP, Rivadeneira F, Westendorf JJ, Karasik D, Duncan EL, et al. The Musculoskeletal Knowledge Portal: making omics data useful to the broader scientific community. J Bone Miner Res. 2020;35(9):1626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Styrkarsdottir U, Stefansson OA, Gunnarsdottir K, Thorleifsson G, Lund SH, Stefansdottir L, et al. GWAS of bone size yields twelve loci that also affect height, BMD, osteoarthritis or fractures. Nat Commun. 2019;10(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.UK Biobank PheWeb. http://pheweb.sph.umich.edu:5000. Accessed February 1, 2020.
- 45.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. February 19 2015;518(7539):317–30. Epub 2015/02/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aschard H, Vilhjálmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet. 2015;96(2):329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348(6235):648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49(1):131. [DOI] [PubMed] [Google Scholar]
- 51.Grundberg E, Adoue V, Kwan T, Ge B, Duan QL, Lam KC, et al. Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet. 2011;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Children’s Oncology Group. http://www.survivorshipguidelines.org/pdf/2018/COG_LTFU_Guidelines_v5.pdf. Accessed June 1, 2020.
- 53.Kawatani M, Okumura H, Honda K, Kanoh N, Muroi M, Dohmae N, et al. The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I. Proc Natl Acad Sci USA. 2008;105(33):11691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Porter RL, Georger MA, Bromberg O, McGrath KE, Frisch BJ, Becker MW, et al. Prostaglandin E2 increases hematopoietic stem cell survival and accelerates hematopoietic recovery after radiation injury. Stem Cells. 2013;31(2):372–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bassett JD, Williams GR. Role of thyroid hormones in skeletal development and bone maintenance. Endocr Rev. 2016;37(2):135–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


