Abstract
Cardiolipin (CL) is a lipid that is found in the membranes of bacteria and the inner membranes of mitochondria. CL can increase the activity of integral membrane proteins, in particular components of respiratory pathways. We here report that CL activated detergent-solubilized cytochrome bd, a terminal oxidase from Escherichia coli. CL enhanced the oxygen consumption activity ~ twofold and decreased the apparent KM value for ubiquinol-1 as substrate from 95 µM to 35 µM. Activation by CL was also observed for cytochrome bd from two Gram-positive species, Geobacillus thermodenitrificans and Corynebacterium glutamicum, and for cytochrome bo3 from E. coli. Taken together, CL can enhance the activity of detergent-solubilized cytochrome bd and cytochrome bo3.
Subject terms: Enzymes, Chemical biology
Cardiolipin (CL) is an anionic phospholipid that consists of two phosphatidyl groups connected by a glycerol moiety. CL is important for optimal function of various eukaryotic and prokaryotic membrane protein complexes1–5. CL can interact with bacterial respiratory complexes from phylogenetically diverse species, such as Mycobacterium phlei6, Rhodobacter sphaeroides7 and Escherichia coli8. Among E. coli respiratory chain complexes, CL was shown to activate purified, detergent-free cytochrome bo39 and was the most efficient phospholipid for activation of detergent-solubilized NADH dehydrogenase10 and of liposome-reconstituted nitrate reductase11. Defined binding sites for CL have been determined in crystal structures of E. coli formate dehydrogenase N12, succinate dehydrogenase13, and nitrate reductase11.
The respiratory chain in Escherichia coli features a heme-copper-type terminal oxidase, cytochrome bo3, which transfers electrons from quinol-type substrates onto molecular oxygen. Next to this energetically efficient terminal oxidase, E. coli utilizes cytochrome bd as an alternative branch of the respiratory chain. Cytochrome bd oxidizes quinols, like ubiquinol or menaquinol, coupled with reduction of molecular oxygen to water (Fig. 1A)14,15. Cytochrome bd is particularly important under conditions of stress, such as O2-limitation16, in the presence of nitric oxide17,18 hydrogen peroxide19–21 and hydrogen sulfide22,23. Lack of cytochrome bd in uropathogenic E. coli strains led to attenuation in mouse infection models24.
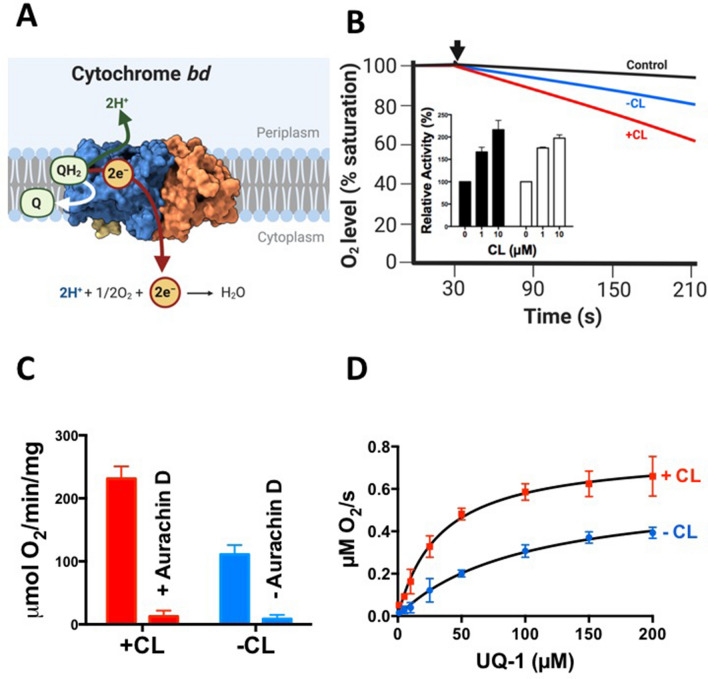
Figure 1.
Activation of E. coli cytochrome bd by CL. (A) Enzymatic function of cytochrome bd (PDB ID: 6RKO42, figure created with BioRender52). (B) The effect of CL (10 μM) on oxygen consumption activity by cytochrome bd (final conc. 2 nM) purified from E. coli was determined using a Clark-type electrode. The reaction was initiated by addition of ubiquinone-1 + DTT (arrow), the negative control contained ubiquinone-1 and DTT, but no cytochrome bd. Inset: Dependency of activation on the pre-incubation time at the indicated CL concentrations. The enzyme was incubated with CL prior to starting the reaction for either 3 min (black bars) or 60 min (white bars). (C) Impact of the inhibitor aurachin D (400 nM) on oxygen consumption by cytochrome bd in the presence or absence of 10 µM CL. (D) Effect of CL on the KM value of E. coli cytochrome bd. Curve fit was done with a simple Michaelis–Menten analysis; R2 values in the absence and presence of CL were 0.978 and 0.969, respectively. Average values were calculated from at least two biological replicates at 37 °C; error bars represent standard deviations.
Cytochrome bd is present in of a broad variety of Gram-positive and Gram-negative bacteria and archea, but not in the respiratory chain of eukaryotes14. Purified active cytochrome bd has been prepared from several bacterial species, including E. coli25,26, Azotobacter vinelandii27, Corynebacterium glutamicum28 and Geobacillus thermodenitrificans29,30. However, to our knowledge there are no data available concerning the effect of CL on cytochrome bd activity.
In this report, we investigated the influence of CL on detergent-purified cytochrome bd. We found that CL activated the enzymatic activity of cytochrome bd from E. coli, G. thermodenitrificans and C. glutamicum. We then extended our experimentation and also assessed the impact of CL on the activity of purified cytochrome bo3 from E. coli.
Results
CL enhances the activity and decreases the KM value of purified E. coli cytochrome bd
Cytochrome bd was purified from E. coli strain MB43 using streptactin affinity chromatography and β-D-dodecyl-maltoside (DDM) as detergent, as performed earlier32. The purity and spectroscopic properties of the isolated protein were comparable to previous results31,32 (data not shown), Blue-Native PAGE showed that the sample was devoid of large-scale aggregation (Suppl. Figure 1). In line with earlier data21,31,32, the purified enzyme showed a specific oxygen consumption activity of ~ 110 μmol O2*mg−1*min−1 in buffer containing 0.025% DDM, using ubiquinol-1 as substrate. We examined the effect of CL and observed ~ twofold activation of the oxygen consumption activity (Fig. 1B). The activity of cytochrome bd in both the absence and the presence of CL was strongly suppressed by aurachin D (Fig. 1C), an inhibitor of E. coli cytochrome bd33.
We then investigated whether activation of cytochrome bd by CL is only observed at saturating substrate concentrations or if the KM value changes as well. In the absence of CL, cytochrome bd showed a KM value of 95 ± 16 µM for ubiquinol-1 as substrate, in line with previously published results25,26,34. In the presence of 10 μM CL, the KM value decreased to 35 ± 4 μM (Fig. 1D). These results show that CL can influence enzymatic parameters of DDM-solubilized E. coli cytochrome bd.
CL activates purified cytochrome bd from Gram-positive bacteria
Next, we evaluated if activation by CL can also be found for cytochrome bd purified from other bacteria. Genetic classification analyses indicated that two basic types of cytochrome bd can be distinguished, based on the length of a hydrophilic loop (Q-loop) close to the substrate binding site14,35–37. Whereas E. coli cytochrome bd displays a long Q-loop, cytochrome bd from Gram-positive bacteria harbors a short version14,35–37. Previously, purification of cytochrome bd from the two Gram-positive strains Geobacillus thermodenitrificans (formerly called Bacillus stearothermophilus)29,30 and Corynebacterium glutamicum28 was described. As observed above for the E. coli enzyme, purity and spectroscopic properties of these isolated proteins were comparable to previous results28–30 (data not shown) and the samples were devoid of large-scale aggregation (Suppl. Figure 1).
We examined the oxygen consumption activity of purified cytochrome bd from both strains with the same protocol as for E. coli cytochrome bd, except for using menaquinol-1 instead of ubiquinol-1 as substrate, as these Gram-positive bacteria use menaquinone as main constituent of the quinone pool. Cytochrome bd from G. thermodenitrificans showed lower oxygen consumption activity (~ 18 μmol O2*mg−1*min−1 in the initial phase) as compared to the E. coli enzyme, consistent with previous data38. After the initial phase of the reaction, time-dependent inactivation was observed (Fig. 2A). CL significantly increased the activity of cytochrome bd from this strain (Fig. 2A). As observed above for the E. coli enzyme, the activity of G. thermodenitrificans cytochrome bd was sensitive to inhibition by aurachin D in the presence and absence of CL (Fig. 2B).
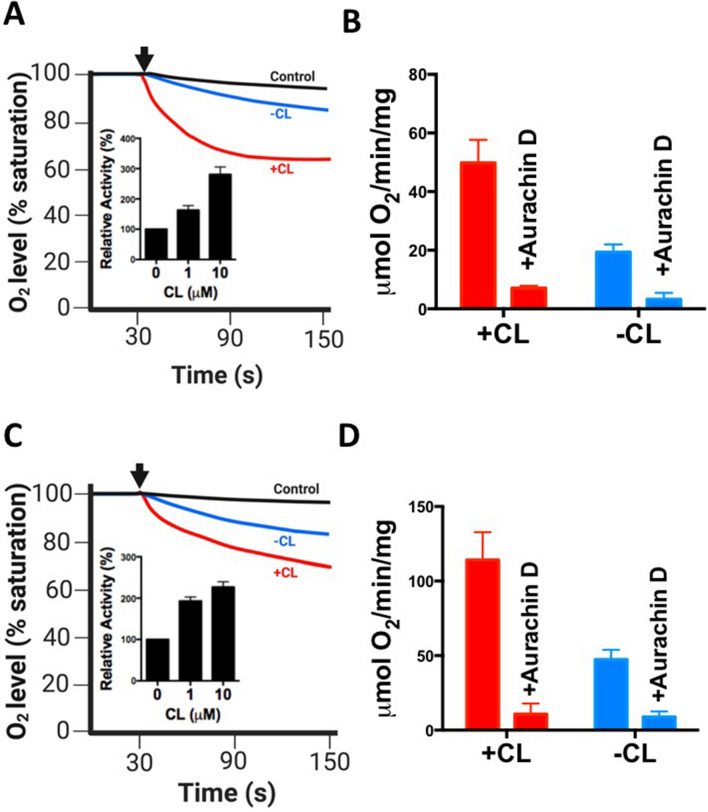
Figure 2.
Activation of cytochrome bd from Gram-positive bacteria by CL. The effect of CL (final conc. 10 μM) on oxygen consumption activity of cytochrome bd purified from G. thermodenitrificans (final conc. 10 nM) (A,B) and C. glutamicum (final conc. 2.8 nM) (C,D) was determined using a Clark-type electrode at 37 °C. The reaction was initiated by addition of menaquinone-1 + DTT (arrow), the negative control contained menaquinone-1 and DTT, but no cytochrome bd. Insets (A,C) Dependency of activation on the CL concentration. (B,D) Impact of the inhibitor aurachin D (400 nM) on the oxygen consumption activity in the presence or absence of 10 µM CL. Average values were calculated from at least two biological replicates; error bars represent standard deviations.
Consistent with previous results28, the oxygen consumption activity of cytochrome bd from C. glutamicum (~ 50 μmol O2*mg1*min−1) was lower than that of the E. coli enzyme, but higher than that of G. thermodenitrificans cytochrome bd. Importantly, the activity was significantly enhanced by CL (Fig. 2C). We confirmed that the observed oxygen consumption activity in the presence and absence of CL was sensitive to inhibition by aurachin D (Fig. 2D). These results reveal that activation by CL is not restricted to cytochrome bd from E. coli, but can also be found for this enzyme isolated from two Gram-positive bacteria.
CL activates enzymatic activity of cytochrome bo3 from E. coli
We then extended our efforts to the second terminal oxidase found in E. coli, cytochrome bo3. Cytochrome bo3 is a heme-copper-type quinol oxidase and evolutionary is not related to cytochrome bd14,39. Cytochrome bo3 was purified from E. coli strain GO105/pJRhisA using DDM as detergent without significant aggregation (Suppl. Figure 1), displaying similar spectroscopic properties as described earlier40,41 (data not shown). Like cytochrome bd, cytochrome bo3 can accept ubiquinol-1 as electron donor and reduces molecular oxygen (Fig. 3A). In the absence of CL, cytochrome bo3 displayed a specific oxygen consumption activity of 47 μmol O2*mg−1*min−1, comparable to previously reported values39,40. Addition of CL caused a pronounced increase in activity (Fig. 3B). Oxygen consumption by cytochrome bo3 in both the absence and in the presence of CL was highly susceptible to the inhibitor potassium cyanide (KCN) (Fig. 3C). The KM value decreased from 56 ± 13 μM in the absence of CL to 38 ± 4 μM in the presence of CL (Fig. 3D). Previously, a KM of 59 μM has been reported for cytochrome bo3 in the presence of CL in the detergent-free state8. Taken together, our results show that CL can activate both terminal oxidases in E. coli.
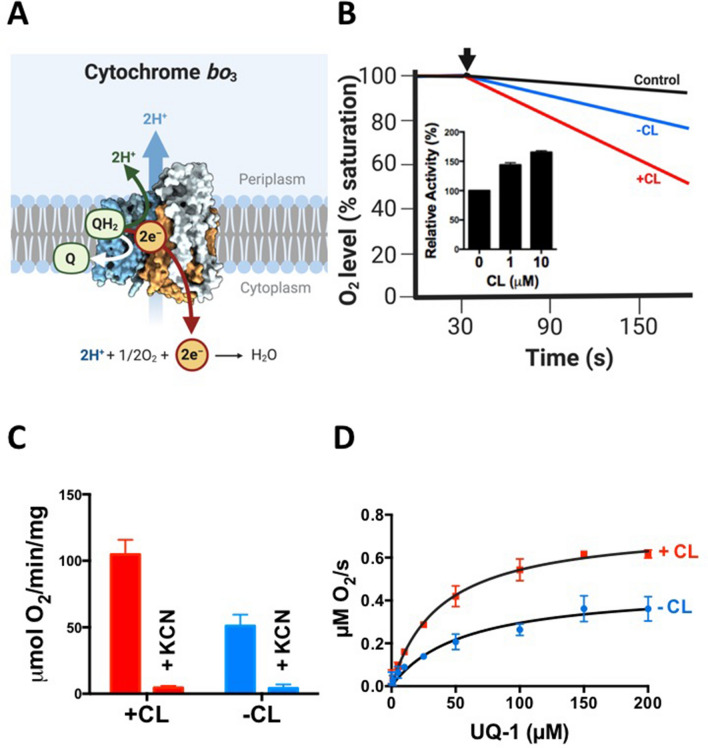
Figure 3.
Activation of E. coli cytochrome bo3 by CL. (A) Enzymatic function of cytochrome bo3 (PDB ID: 1FFT39, figure created with BioRender52) (B) The effect of CL (final conc. 10 μM) on oxygen consumption activity by cytochrome bo3 purified from E. coli (final conc. 5 nM) was determined using a Clark-type electrode. The reaction was initiated by addition of ubiquinone-1 + DTT (arrow), the negative control contained ubiquinone-1 and DTT, but no cytochrome bo3. Inset: dependency of activation on the CL concentration. (C) Impact of KCN (2 mM) on oxygen consumption by cytochrome bo3 in the presence or absence of 10 µM CL. (D) Effect of CL on the KM value of purified E. coli cytochrome bo3. Curve fit was done with a simple Michaelis–Menten analysis, R2 values in the absence and presence of CL were 0.974 and 0.983, respectively. All experiments were carried out at 37 °C. Average values were calculated from at least two biological replicates; error bars represent standard deviations.
Discussion
It has been established that CL can enhance the activity of various bacterial membrane proteins, including complexes of both aerobic and of anaerobic respiration1,2,5,9,11,12. Previously, activation of purified cytochrome bo3 by CL and activation of purified cytochrome bd by asolectin was reported9,26. However, these experiments were carried out in detergent-free state. In the absence of detergent, membrane protein aggregation likely causes a significant decrease in activity, which subsequently is relieved by addition of lipid. In this study, we found that CL enhanced the activity of both terminal oxidases of the E. coli respiratory chain in the detergent-solubilized state. In line with our results, recently high enzymatic activity (889 e−*s−1 ≅ 135 μmol O2*mg−1*min−1) has been reported for E. coli cytochrome bd solubilized in MSP1D1/POPC-containing nano-discs42, likely reflecting the importance of the lipid environment for the performance of this enzyme.
CL can be located at the outer surface of a detergent-solubilized membrane protein, enabling proper vertical positioning of the protein, or it may bind to clefts or cavities on the protein surface1,3. CL may play a structural role, e.g. by binding at the interface between individual subunits, as previously reported for formate dehydrogenase N12. Alternatively, CL may enhance the interaction with the quinol substrate and/or facilitate the electron transfer reaction, as reported for nitrate reductase, where CL binds to a niche near the quinol-binding site11. In the respiratory chain of Saccharomyces cerevisiae CL stabilizes the super-complex formed by the cytochrome bc1 complex and cytochrome c oxidase, binding at the interface of the two components5,43. In case of mitochondrial ATP synthase, CL transiently binds to conserved lysine residues in subunit c, possibly lubricating the motion of this membrane-embedded rotary machine44. As found for DDM-purified cytochrome c oxidase from bovine heart mitochondria, CL can be functionally required for optimal electron transports and proton translocation43,44. Three-dimensional structures are available for cytochrome bd from Geobacillus thermodenitrificans30 and from E. coli42,45, however, the presently achieved resolution might not allow for identification of all bound lipid molecules. The decreased KM value of E. coli cytochrome bd for ubiquinol-1 measured here indicates that CL influences the substrate binding process.
In our study we investigated cytochrome bd and cytochrome bo3 in the detergent-solubilized state and our results therefore do not clarify if CL has a similar effect on these enzymes in the native membrane. CL as high-curvature lipid is predominantly localized at the poles in rod-shaped bacteria and may thereby influence the cellular localization of membrane protein complexes46, as suggested for the SecYEG translocon47. Previously, for cytochrome bd, a distribution in mobile patches in the E. coli cytoplasmic membrane has been reported48. It needs to be investigated if CL can influence function, localization or dynamics of cytochrome bd or cytochrome bo3 in the native plasma membrane.
Materials and methods
Chemicals
Aurachin D was synthesized as described earlier in Li et al. 201349 and was kindly provided by Dr. Jennifer Herrmann (Helmholtz Centre for Infection Research and Pharmaceutical Biotechnology, Saarbrücken). CL was purchased from Sigma (C1649, from bovine heart, > 80% polyunsaturated fatty acid content, primarily linoleic acid). All other chemicals were bought from Sigma, unless indicated otherwise.
Purification of cytochrome bd
Cytochrome bd from E. coli was purified based on Hoeser et al.31, with modifications as described by Goojani et al.32. Briefly, E. coli MB43 carrying the pET17cydABX-Strep-tag plasmid was grown in Luria–Bertani (LB) medium with 100 µg/ml Ampicillin at 37 °C overnight with shaking at 200 rpm. The bacteria were diluted to OD600 ~ 0.01 in 800 ml LB medium with 100 µg/ml Ampicillin and incubated until reaching OD60 ~ 0.4. Then IPTG (0.45 mM final conc.) was added and the bacteria were incubated again at 37 °C, 200 rpm until reaching OD600 ~ 2.0. Cells were sedimented by centrifugation at 6000 g for 20 min (JA-10 rotor). The pellets were washed by phosphate buffer saline, pH 7.4, and spun down at 6000 g for 20 min. Each 15 g of wet cells were re-suspended with 75 ml of MOPS solution (50 mM 3-N-morpholino-propanesulfonic acid, 100 mM NaCl and protease inhibitor (cOmplete, Roche). The cells were disrupted by passing three times though a Stansted cell homogenizer at 1.8 kb. Unbroken cells were centrifuged at 9500 g (Ja-3050-ti rotor) for 20 min. Subsequently, the supernatant was pelleted by ultracentrifugation 250,000 g (70-ti rotor) for 75 min at 4 °C. The pellet was re-suspended in MOPS solution and the protein concentration was measured using the BCA Protein Assay kit (Pierce) as described by the manufacturer. The concentration was adjusted to 10 mg/ml and incubated in MOPS solution containing 1% DDM (final conc.) at 4 °C for an hour with gentle shaking. Un-solubilized material was sedimented by ultracentrifugation at 250,000 g at 4 °C for 15 min (70-ti rotor). The collected supernatant was applied on streptactin column at 4 °C (cold room) and the flowthrough was collected. The column was washed with washing buffer (50 mM sodium phosphate, 300 mM NaCl, protease inhibitor (cOmplete), containing 0.01% DDM, pH 8.0) to remove unspecific protein binding and the flow-through was collected again. The elution buffer (50 mM sodium phosphate, 300 mM NaCl, protease inhibitor (C0mplete EDTA free), 0.01% DDM, and 2.5 mM desthiobiotin pH 8.0) was added to the column at 4 °C to elute the protein.
Purification of cytochrome bd from Geobacillus thermodenitrificans and from Corynebacterium glutamicum
Cytochrome bd from G. thermodenitrificans was extracted and purified from membrane fractions of G. thermodenitrificans K1041/pSTE-cbdAB recombinant cells with two consecutive column chromatography of DEAE-Toyopearl and hydroxyapatite in the presence of 0.5% (w/v) MEGA9 + 10, as described previously in Arutyunyan et al. 201236. Cytochrome bd from C. glutamicum was extracted and purified from membrane fractions of C. glutamicum ΔctaD/pPC4-cydABDC recombinant cells50 with two consecutive chromatography of hydroxyapatite and then DEAE-Toyopearl in the presence of 0.05% (w/v) DDM.
Purification of E. coli cytochrome bo3
Cytochrome bo3 was extracted and purified from E. coli cytoplasmic membranes based on Rumbley et al. 199740, with modifications as described in Hards et al. 201841. E. coli cytoplasmic membranes were prepared from strain GO105/pJRhisA in which cytochrome bo3 is overexpressed. E. coli was aerobically grown to mid-log phase at 37 °C in LB medium supplemented with 500 μM CuSO4 and 100 μg ml−1 carbenicillin. Cells were harvested by centrifugation at 10,000 × g for 10 min and the pellets were washed and repelleted twice with buffer A (20 mM (3-N-morpholino-propanesulfonic acid (MOPS), 30 mM Na2SO4, pH 7.4). Cells were then resuspended in buffer A containing a mini protease inhibitor tablet (cOmplete) per 50 mL, 0.1 mM phenylmethylsulfonyl fluoride, 0.1 mg/ml pancreatic DNase, and lysed by two passages through a French Press at 20,000 psi. Any debris and unbroken cells were removed by centrifugation at 10,000 × g for 30 min. The supernatant was then ultracentrifuged (200,000 × g, 45 min, 4 °C) and the membrane pellet resuspended in buffer B (20 mM MOPS, 30 mM Na2SO4, 25% w/w sucrose, pH 7.4). The suspension was applied to the top of a 30% w/w to 55% w/w sucrose gradient and ultracentrifugation (130,000 × g, 16 h, 4 °C) with no deceleration or breaking to separate inner membrane from outer membrane. The inner membrane fraction was removed from the sucrose gradient and washed 3 times with buffer A by ultracentrifugation (200,000 × g, 45 min, 4 °C). Inner membranes were then resuspended in buffer A and either used immediately for purification or stored in aliquots at − 80 °C until use. To extract cytochrome bo3, inner membrane fractions were diluted to 5 mg/mL protein content with solubilization buffer (20 mM Tris HCl, pH 8.0, 5 mM MgSO4, 10% glycerol, 300 mM NaCl, 1% DDM, 10 mM imidazole) and incubated at 30 °C for 30 min with gentle inversion every 5 min. The unsolubilized material was removed by ultracentrifugation (200,000 × g, 45 min, 4 °C), and the supernatant was applied to a Nickel-Sepharose High Performance (GE Healthcare) column that was previously washed with water and equilibrated with IMAC buffer (50 mM Tris/HCl, pH 8.0, 5 mM MgSO4, 10% glycerol, 0.01% DDM, 300 mM NaCl) containing 10 mM imidazole. To remove contaminating proteins, the resin was washed with IMAC buffer containing 30 mM imidazole and 150 mM NaCl, and cytochrome bo3 was eluted with IMAC buffer containing with 200 mM imidazole, 150 mM NaCl, and 20% glycerol. The red cytochrome bo3 containing fractions were pooled and concentrated to 6.57 mg mL−1 using an Amicon Ultra centrifugal filter devices with100,000 Da molecular weight cutoff.
Oxygen consumption activity assay
Oxygen consumption by purified cytochrome bd and cytochrome bo3 was measured using a Clark-type electrode as previously described in Lu et al.51, with modifications as in Goojani et al.31. Briefly, the electrode was fully aerated (212 μM O2 at 37 °C) and calibrated with sodium hydrosulfite. The purified enzymes (final conc: 2 nM for cytochrome bd from E. coli, 10 nM for cytochrome bd from G. thermodenitricifans, 2.8 nM for cytochrome bd from C. glutamicum, 5 nM for cytochrome bo3) were pre-incubated for three minutes with CL (and with inhibitors, if applicable) in a pre-warmed (37 °C) buffer containing 50 mM 3-N-morpholino-propanesulfonic acid (MOPS), 100 mM NaCl and 0.025% DDM, pH 7.5. Ubiquinone-1 (Sigma) and menaquinone-1 (Santa Cruz Biotechnology) were dissolved in absolute ethanol (20 mM stock) and the reducing agent dithiothreitol (1 M stock) in 50 mM HEPES (4- (2-hydroxyethyl) piperazine-1-ethanesulfonic acid), pH 7.75. Quinone stock and DTT stock were mixed in 1:1 volume ratio and incubated for 3 min (ubiquinone-1/DTT) or 6 min (menaquinone-1/DTT) at 37 °C. The oxygen consumption reaction was initiated by adding the quinone/DTT mixture (final concentration 200 μM quinone and 10 mM DTT) to the assay mixture, respiration was measured for 3 min. The enzymatic activity was calculated from the slope in the period 30 s—60 s after starting the reaction (linear approximation).
Supplementary Information
Acknowledgement
A.H.A is indebted to Royal Embassy of Saudi Arabia – Cultural Bureau in Netherlands and King Abdulaziz University for their support. The authors wish to thank Henk Hakvoort (VU Amsterdam) for technical assistance.
Author contributions
A.H.A. and A.G.H. performed experiments; A.H.A., H.L., A.G.H. and H.G.G. designed experiments and/or analyzed data; D.G.G.M. J.S. and D.B. supervised and coordinated experiments; A.H.A. D.G.G.M and D.B. wrote the manuscript with contributions from all co-authors, D.B. and D.G.G.M. supervised the overall research.
Funding
This work was partly supported by a Grant-in-Aid for Scientific Research (C) (16K07299 to J.S.) from the Japan Society for the Promotion of Science.
Data availability
The original data describing rates measured in this study are compiled in a supplementary file (Suppl. Table 1). Original time courses generated during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Amer H. Asseri and Albert Godoy-Hernandez.
Contributor Information
Duncan G. G. McMillan, Email: D.G.G.McMillan@tudelft.nl
Dirk Bald, Email: d.bald@vu.nl.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-87354-0.
References
- 1.Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Arias-Cartin R, et al. Cardiolipin binding in bacterial respiratory complexes: structural and functional implications. Biochim Biophys Acta. 2012;1817:1937–1949. doi: 10.1016/j.bbabio.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Laganowsky A, et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musatov A, Sedlak E. Role of cardiolipin in stability of integral membrane proteins. Biochimie. 2017;142:102–111. doi: 10.1016/j.biochi.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Díaz-Quintana A, et al. Wheel and deal in the mitochondrial inner membranes: the tale of cytochrome c and Cardiolipin. Oxid. Med. Cell Longev. 2020;2020:6813405. doi: 10.1155/2020/6813405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy TL, Webber MM. Solubilization, purification, and characterization of succinate dehydrogenase from membranes of Mycobacterium phlei. J. Bacteriol. 1986;167:1–6. doi: 10.1128/JB.167.1.1-6.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAuley KE, et al. Structural details of an interaction between cardiolipin and an integral membrane protein. Proc. Natl. Acad. Sci. USA. 1999;96:14706–14711. doi: 10.1073/pnas.96.26.14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esfahani M, et al. Lipid-protein interactions in membranes: interaction of phospholipids with respiratory enzymes of Escherichia coli membrane. J. Biol. Chem. 1977;252:3194–3198. doi: 10.1016/S0021-9258(17)40370-X. [DOI] [PubMed] [Google Scholar]
- 9.Kita K, et al. Terminal oxidases of Escherichia coli aerobic respiratory chain. I. Purification and properties of cytochrome b562-o complex from cells in the early exponential phase of aerobic growth. J Biol Chem. 1984;259:3368–3374. doi: 10.1016/S0021-9258(17)43304-7. [DOI] [PubMed] [Google Scholar]
- 10.Dancey GF, Shapiro BM. Specific phospholipid requirement for activity of the purified respiratory chain NADH dehydrogenase of Escherichia coli. Biochim Biophys. Acta. 1977;487:368–377. doi: 10.1016/0005-2760(77)90013-3. [DOI] [PubMed] [Google Scholar]
- 11.Arias-Cartin R, et al. Cardiolipin-based respiratory complex activation in bacteria. Proc. Natl. Acad. Sci. USA. 2011;108:7781–7786. doi: 10.1073/pnas.1010427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jormakka M, et al. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 13.Yankovskaya V, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 14.Borisov VB, et al. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 2011;1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giuffrè A, et al. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim Biophys Acta. 2014;1837:1178–1187. doi: 10.1016/j.bbabio.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 16.D’mello R, et al. The cytochrome bdquinol oxidase in Escherichia colihas an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology. 1996;142:755–763. doi: 10.1099/00221287-142-4-755. [DOI] [PubMed] [Google Scholar]
- 17.Borisov VB, et al. Redox control of fast ligand dissociation from Escherichia coli cytochrome bd. Biochem. Biophys. Res. Commun. 2007;355:97–102. doi: 10.1016/j.bbrc.2007.01.118. [DOI] [PubMed] [Google Scholar]
- 18.Mason MG, et al. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat. Chem. Biol. 2009;5:94–96. doi: 10.1038/nchembio.135. [DOI] [PubMed] [Google Scholar]
- 19.Wall, et al. arc-dependent thermal regulation and extragenic suppression of the Escherichia coli cytochrome d operon. J. Bacteriol. 1992;174:6554–6562. doi: 10.1128/JB.174.20.6554-6562.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borisov VB, et al. Cytochrome bd oxidase from Escherichia coli displays high catalase activity: an additional defense against oxidative stress. FEBS Lett. 2013;587:2214–2218. doi: 10.1016/j.febslet.2013.05.047. [DOI] [PubMed] [Google Scholar]
- 21.Al-Attar S, et al. Cytochrome bd displays significant quinol peroxide activity. Sci. Rep. 2016;6:27631. doi: 10.1038/srep27631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korshunov S, et al. The cytochrome bd oxidase of Escherichia coli prevents respiratory inhibition by endogenous and exogenous hydrogen sulfide. Mol. Microbiol. 2016;101:62–77. doi: 10.1111/mmi.13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forte E, et al. The terminal oxidase cytochrome bd promotes sulfide-resistant bacterial respiration and growth. Sci. Rep. 2016;6:23788. doi: 10.1038/srep23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shepherd M, et al. The cytochrome bd I respiratory oxidase augments survival of multidrug-resistant Escherichia coli during infection. Sci. Rep. 2016;6:35285. doi: 10.1038/srep35285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller MJ, Gennis RB. The purification and characterization of the cytochrome d terminal oxidase complex of the Escherichia coli aerobic respiratory chain. J. Biol. Chem. 1983;258:9159–9165. doi: 10.1016/S0021-9258(17)44645-X. [DOI] [PubMed] [Google Scholar]
- 26.Kita K, et al. Terminal oxidases of Escherichia coli aerobic respiratory chain. II. Purification and properties of cytochrome b558-d complex from cells grown with limited oxygen and evidence of branched electron-carrying systems. J. Biol. Chem. 1984;259:3375–3381. doi: 10.1016/S0021-9258(17)43305-9. [DOI] [PubMed] [Google Scholar]
- 27.Jünemann S, Wrigglesworth JM. Antimycin inhibition of the cytochrome bd complex from Azotobacter vinelandii indicates the presence of a branched electron transfer pathway for the oxidation of ubiquinol. FEBS Lett. 1994;345:198–202. doi: 10.1016/0014-5793(94)00372-6. [DOI] [PubMed] [Google Scholar]
- 28.Kusumoto K, et al. Menaquinol oxidase activity and primary structure of cytochrome bd from the amino-acid fermenting bacterium Corynebacterium glutamicum. Arch. Microbiol. 2000;173:390–397. doi: 10.1007/s002030000161. [DOI] [PubMed] [Google Scholar]
- 29.Sakamoto J, et al. Cytochrome bd-type quinol oxidase in a mutant of Bacillus stearothermophilus deficient in caa3-type cytochrome c oxidase. FEMS Microbiol. 1996;143:151–158. doi: 10.1111/j.1574-6968.1996.tb08474.x. [DOI] [PubMed] [Google Scholar]
- 30.Safarian S, et al. Structure of a bd oxidase indicates similar mechanisms for membrane integrated oxygenreductases. Science. 2016;352:583–586. doi: 10.1126/science.aaf2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoeser J, et al. Subunit CydX of Escherchia coli cytochrome bd ubiquinol oxidase is essential for assembly and stability of the di-heme active site. FEBS Lett. 2014;588:1537–1541. doi: 10.1016/j.febslet.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 32.Goojani HG, et al. The carboxy-terminal insert in the Q- loop is needed for functionality of Escherichia coli cytochrome bd-I. Biochim. Biophys. Acta. 2020;1861:148175. doi: 10.1016/j.bbabio.2020.148175. [DOI] [PubMed] [Google Scholar]
- 33.Meunier B, et al. New inhibitors of the quinol oxidation sites of bacterial cytochromes bo and bd. Biochemistry. 1995;34:1076–1083. doi: 10.1021/bi00003a044. [DOI] [PubMed] [Google Scholar]
- 34.Mogi T, et al. Probing the ubiquinol-binding site in cytochrome bd by site-directed mutagenesis. Biochemistry. 2006;45:7924–7930. doi: 10.1021/bi060192w. [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto, N. Sone, Biochemical and molecular features of terminal oxidases, in: D. Zannoni (Ed.), Respiration in Archaea and Bacteria, Vol.1: Diversity of Prokaryotic Electron Transport Carriers, Series: Advances in Photosynthesis & Respiration, Vol. 15, Kluwer Academic Publishers, The Netherlands, 2004, pp. 87–113 (2004).
- 36.Arutyunyan AM, et al. Optical and magneto-optical activity of cytochrome bd from Geobacillus thermodenitrificans. Biochim Biophys. Acta. 2012;1817:2087–2094. doi: 10.1016/j.bbabio.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Degli Esposti M, et al. Molecular evolution of cytochrome bd oxidases across proteobacterial genomes. Genome Biol. Evol. 2015;7:801–820. doi: 10.1093/gbe/evv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakamoto J, et al. Gene structure and quinol oxidase activity of a cytochrome bd-type oxidase from Bacillus stearothermophilus. Biochim. Biophys. Acta. 1999;1411:147–158. doi: 10.1016/S0005-2728(99)00012-2. [DOI] [PubMed] [Google Scholar]
- 39.Abramson J, et al. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat. Struct. Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- 40.Rumbley JN, et al. One-step purification of histidine-tagged cytochrome bo3 from Escherichia coli and demonstration that associated quinone is not required for the structural integrity of the oxidase. Biochim. Biophys. Acta. 1997;1340:131–142. doi: 10.1016/S0167-4838(97)00036-8. [DOI] [PubMed] [Google Scholar]
- 41.Hards K, et al. Bactericidal mode of action of bedaquiline. J. Antimicrob. Chemother. 2015;70:2028–2037. doi: 10.1093/jac/dkv054. [DOI] [PubMed] [Google Scholar]
- 42.Safarian S, et al. Active site rearrangement and structural divergence in prokaryotic oxidases. Science. 2019;366:100–104. doi: 10.1126/science.aay0967. [DOI] [PubMed] [Google Scholar]
- 43.Pfeiffer K, et al. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 44.Duncan AL, et al. Cardiolipin binds selectively but transiently to conserved lysine residues in the rotor of metazoan ATP synthases. Proc. Natl. Acad. Sci. USA. 2016;113:8687–8692. doi: 10.1073/pnas.1608396113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theßeling A, et al. Homologous bd oxidases share the same architecture but differ in mechanism. Nat. Commun. 2019;10:5138. doi: 10.1038/s41467-019-13122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood JM. Perspective: challenges and opportunities for the study of cardiolipin, a key player in bacterial cell structure and function. Curr. Genet. 2018 doi: 10.1007/s00294-018-0811-2. [DOI] [PubMed] [Google Scholar]
- 47.Gold VA, et al. The action of cardiolipin on the bacterial translocon. Proc. Natl. Acad. Sci. USA. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lenn T, et al. Clustering and dynamics of cytochrome bd-I complexes in the Escherichia coli plasma membrane in vivo. Mol. Microbiol. 2008;70:1397–1407. doi: 10.1111/j.1365-2958.2008.06486.x. [DOI] [PubMed] [Google Scholar]
- 49.Li XW, et al. Synthesis and biological activities of the respiratory chain inhibitor aurachin D and new ring versus chain analogues. Beilstein J. Org. Chem. 2013;9:1551–1558. doi: 10.3762/bjoc.9.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabashima Y, et al. Correlation between proton translocation and growth: genetic analysis of the respiratory chain of Corynebacterium glutamicum. J. Biochem. 2009;146:845–855. doi: 10.1093/jb/mvp140. [DOI] [PubMed] [Google Scholar]
- 51.Lu P, et al. The anti-mycobacterial activity of the cytochrome bcc inhibitor Q203 can be enhanced by small-molecule inhibition of cytochrome bd. Sci. Rep. 2018;8:2625. doi: 10.1038/s41598-018-20989-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.https://biorender.com/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data describing rates measured in this study are compiled in a supplementary file (Suppl. Table 1). Original time courses generated during the current study are available from the corresponding author on reasonable request.