Orchestra of cytokines regulates phenotypic and functional plasticity of non‐pathogenic and pathogenic Th17 cells in human health and diseases.
Keywords: Cytokine modulation, Human Th17, inflammation, T‐cell plasticity
Summary
Upon activation, naïve CD4+ T helper (Th) cells differentiate into distinct Th effector cell lineages depending on the local cytokine environment. However, these polarized Th cells can also adapt their function and phenotype depending on the changing cytokine environment, demonstrating functional plasticity. Here, Th17 cells, which play a critical role in host protection from extracellular pathogens and in autoimmune disorders, are of particular interest. While being able to shift phenotype within their lineage, Th17 cells can also acquire characteristics of Th1, Th2, T follicular helper (Tfh) or regulatory T cells. Th17 cell identity is determined by a spectrum of extracellular signals, including cytokines, which are critical orchestrators of cellular immune responses. Cytokine induces changes in epigenetic, transcriptional, translational and metabolomic parameters. How these signals are integrated to determine Th17 plasticity is not well defined, yet this is a crucial point of investigation as it represents a potential target to treat autoimmune and inflammatory diseases. The goal of this review was to discuss how cytokines regulate intracellular networks, focusing on the regulation of lineage‐specific transcription factors, chromatin remodelling and metabolism, to control human Th17 cell plasticity. We discuss the importance of Th17 plasticity in autoimmunity and cancer and present current strategies and challenges in targeting pathogenic Th17 cells with cytokine‐based approaches, considering human genetic variants associated with altered Th17 differentiation. Finally, we discuss how modulating Th17 plasticity rather than targeting the Th17 lineage as a whole might preserve its essential immune function while purging its adverse effects.
Abbreviations
- APC
antigen‐presenting cell
- AS
ankylosing spondylitis
- CAPS
cryopyrin‐associated periodic syndrome
- CMC
chronic mucocutaneous candidiasis
- CyTOF
cytometry by time of flight
- FFA
free fatty acid
- GM‐CSF
granulocyte–macrophage colony‐stimulating factor
- IFN‐γ
interferon‐γ
- IBD
inflammatory bowel disease
- IL
interleukin
- MALT
mucosa‐associated lymphoid tissue
- MS
multiple sclerosis
- PBMC
peripheral blood mononuclear cell
- PsA
psoriatic arthritis
- RA
rheumatoid arthritis
- RORγt
retinoid‐related orphan receptor γt
- Sc
single cell
- SLE
systemic lupus erythematosus
- seq
sequencing
- STAT
signal transducer and activator of transcription
- SNP
single nucleotide polymorphism
- TCM
central memory T cell
- TCR
T‐cell receptor
- TF
transcription factor
- TGF‐β
transforming growth factor‐β, Th, T helper
- TNF‐α
tumour necrosis factor‐α
- TRM
tissue‐resident memory T cell
- Treg
regulatory T cell
INTRODUCTION
T helper (Th) CD4+ T cells are major players of the adaptive immune system, orchestrating the immunity against pathogens. 1 Following their maturation in the thymus, naïve Th cells circulate through secondary lymphoid organs, including the spleen, lymph nodes and the mucosa‐associated lymphoid tissue (MALT) until they recognize antigen presented on the surface of antigen‐presenting cells (APCs). Following activation, naïve Th cells differentiate into different effector lineages to carry out specific immunomodulatory roles. Th lineage fate decision is controlled by (a) the identity and activation states of APCs, (b) the magnitude of T‐cell receptor (TCR) signalling and 3) environmental signals. 2 , 3 Here, the local cytokine milieu is critical in determining T‐cell differentiation into immunoregulatory cells (T regulatory (Treg)) or pro‐inflammatory Th lineages (e.g. Th1, Th2, Th17). 3 These different Th lineages, in turn, produce specific cytokines that define their effector function. TGF‐β (transforming growth factor‐β) and interleukin‐2 (IL‐2) drive differentiation of naïve T cells into suppressive Tregs secreting IL‐10 to maintain immune homeostasis. IL‐12 drives differentiation of (IFN)‐γ‐producing Th1 that is critical for clearance of intracellular pathogens. IL‐4 promotes differentiation into IL‐4, IL‐5 and IL‐13 producing Th2 cells, necessary for targeting parasite infections. 4 , 5 IL‐12, IL‐6, IL‐23 and IL‐21 polarize IL‐21‐producing Tfh cells that provide B‐cell help for optimal humoral immunity. 6 IL‐17‐producing Th17 cells differentiate from naïve Th cells in the presence of cytokines such as TGF‐β and IL‐6 7 , 8 and contribute to host mucosal immunity against bacterial and fungal pathogens. 3 Immunodeficient patients suffering from Job`s syndrome, incapable of generating Th17 cells, show recurrent bacterial, fungal and chronic viral infections. 9 However, since their discovery in 2005, Th17 cells have also been implicated in the development of autoimmunity and cancer pathogenesis. 10 , 11 , 12 This apparent functional heterogeneity has sparked interest to better understand Th17 biology in order to manipulate it for therapeutic purposes. A defining feature of Th17 cells is their phenotypic flexibility, here defined as plasticity. While it was initially believed that Th cells remained committed to their acquired lineage, 5 , 13 differentiated effector Th cells can acquire functional features of other Th lineage characteristics.
Most of our knowledge of Th17 biology is based on mouse studies. Deciphering the molecular mechanism and signalling pathways in human Th17 subsets has not been as straightforward. Key similarities and differences in the differentiation programmes of mouse and human Th subsets from naïve Th precursors are discussed elsewhere. 14 In this review, we will explore the contribution of cytokines and their downstream effects (e.g. epigenetic modifications, transcription factor (TF) programmes and metabolic activity) in determining the plasticity and effector function of human Th17 cells. We highlight current therapeutic strategies targeting Th17 cells in the clinic to modulate Th17 plasticity for the treatment of inflammatory diseases and cancer.
HETEROGENEITY AND FUNCTION OF MATURE HUMAN TH17 SUBSETS
While there are few Th17 cells in the circulation, they are abundant in mucosal tissues. 15 Here, they orchestrate the balance between tolerance and inflammation, mucosal repair and healing. 16 Based on the microbiota, pathogens, other environmental factors or genetic predisposition, Th17 cells can acquire phenotypes and effector cytokine profiles that can either be detrimental or re‐establish immune homeostasis. 3 Single nucleotide polymorphisms (SNPs) in genes associated with the Th17 pathway have been linked to autoimmune and inflammatory diseases, 17 , 18 but also to immunodeficiency. 19 Accordingly, IL‐17 levels are increased in inflammatory lesions of patients suffering from multiple sclerosis (MS), psoriasis, rheumatoid arthritis (RA), inflammatory bowel diseases (IBDs) and Sjögren´s syndrome [31–34]; in contrast, impaired Th17 responses are associated with chronic mucocutaneous candidiasis (CMC) and bacterial infections in the lung and skin. 20
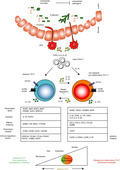
While all human in vivo Th17 subsets express CCR6 and the surface marker CD161, they can be distinguished by their differential expression of the chemokine receptors CCR4 and CXCR3: classical Th17 cells (CCR4+CXCR3−) express high levels of IL‐17 and low levels of IFN‐γ; non‐classical Th17.1 cells (CCR4−CXCR3+, also called Th17.1 or Th1/Th17) produce low levels of IL‐17 and large amounts of IFN‐γ; and double‐positive (CCR4+CXCR3+) and double‐negative (CCR4‐CXCR3‐) cells have been identified as Th17 cells in a precursor or transitional state. 21 , 22 , 23 , 24 Such heterogeneity can either be generated during initial priming from naïve CD4+ T cells, or it can be the result of plastic events affecting Th effector/memory cells (Figure 1).
Figure 1.
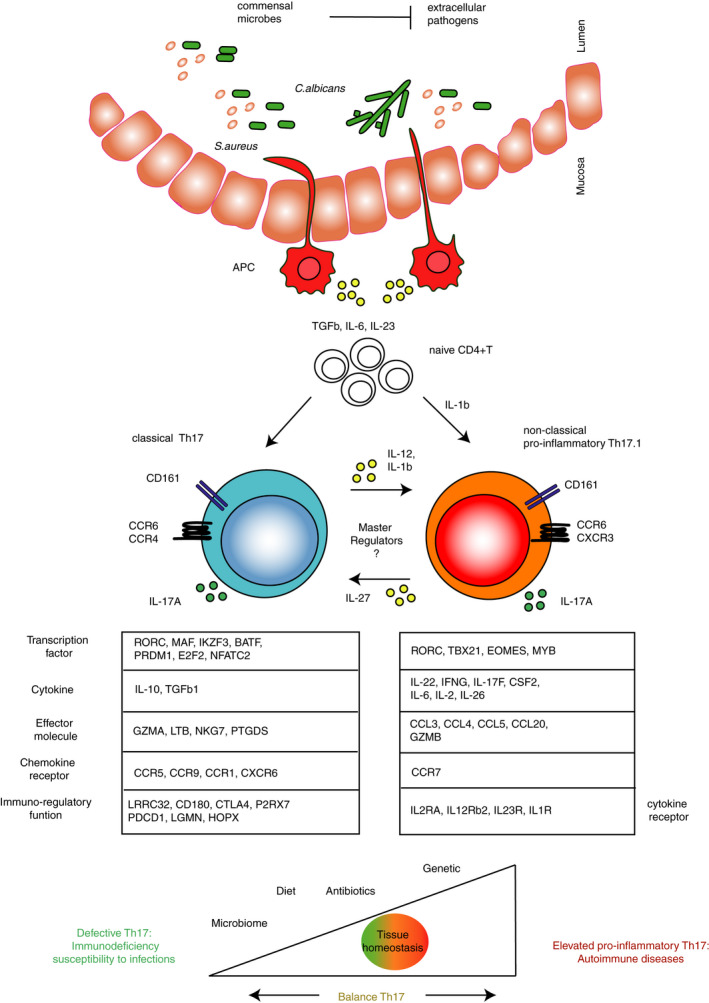
Heterogeneity, function and plasticity of human Th17 subsets. All ex vivo human Th17 cells express CCR6 and CD161 and produce IL‐17A required for pathogen clearance; however, classical Th17 cells have a differential transcriptional profile compared with non‐classical Th17.1 cells (genes listed below the two subsets). Classical Th17 cells can be isolated ex vivo (through CD161, CCR6 and CCR4) from blood and tissue of healthy individuals or generated in vitro in response to Staphylococcus aureus (S.aureus). They express the transcription factors (TFs) MAF, AHR and IKZF3 implicated in the regulation of immunoregulatory genes (e.g. CTLA4, LRRC32 and others), cytokines (e.g. IL‐10, TGF‐β1) and effector molecules (e.g. GMZA,LTB, NKG7, PTGDS). Non‐classical pro‐inflammatory Th17.1 is an heterogenous population characterized by IFN‐γ production. Ex vivo Th17.1 cells express CXCR3 and coexpress IL‐17 and IFN‐γ, and they can be generated in vitro from Candida albicans (C. albicans), although without expressing CXCR3. Pro‐inflammatory Th17.1 cells have a transcriptional profile similar to murine pathogenic Th17 cells, coexpressing RORγt and T‐bet, IL‐23R, IL‐1R and IL‐12Rβ2. Expression of EOMES is a ‘marker’ of acquired Th1‐like phenotype. They express pro‐inflammatory cytokine (e.g. CSF2, IL‐22, IL‐6, IL‐17F,IL‐2 and IL‐26) and effector molecules (e.g. GZMB and CCL20). Th17 cells can be generated directly after pathogen sensing by antigen‐presenting cells (APCs) in the mucosa, leading to TGF‐β, IL‐6, IL‐23 and IL‐1β production, or acquire a pro‐inflammatory phenotype resulting from plastic events (e.g. in the presence of pro‐inflammatory cytokines such as IL‐12 and IL‐1β). In vitro human Th17.1 plasticity towards IL‐10‐secreting phenotype is achieved in the presence of IL‐27. So far, specific master regulators directing the Th17 cell phenotypical switch from immunoregulatory to pro‐inflammatory and vice versa have not yet been identified. Overall, microbiota composition, the nature of invading pathogens, other environmental factors (e.g. diet and antibiotics) or genetic variants shape Th17 cell phenotype and function to maintain immune homeostasis. However, compromised Th17 cell function can lead to immunodeficiency; in contrast, elevated pro‐inflammatory Th17 cells can contribute to autoimmune diseases.
Most knowledge about human memory Th17 cells is based on in vitro studies using circulating memory T cells (TCM17) rather than long‐lived tissue‐resident memory Th17 (TRM17) cells. TRM17 cells provide immediate response to bacterial and fungal reinfections at mucosal sites but might equally contribute to tissue damage and amplification of autoimmune diseases.25, 26, 27, 28 Whether different memory Th17 subsets have different potential for plasticity remains to be investigated.
Classical immunoregulatory Th17 cells
Immunoregulatory Th17 are present in blood and tissue of healthy individuals 29 and can be generated in vitro from naïve CD4+ T cells in response to Staphylococcus aureus (S.aureus)‐pulsed monocytes 30 (Figure 1). They express IL‐17 and immunomodulatory genes normally associated with Tregs (e.g. CTLA‐4, LRRC32). Classical Th17 cells are characterized by expression of the transcription factors (TFs) MAF, AHR and IKZF3 (encoding for Aiolos), which have been implicated in IL‐10 gene regulation in human Th cells.29, 31, 32 IL‐10‐producing Th17 cells contribute to tissue homeostasis as loss‐of‐function mutations in the IL‐10 or IL‐10R genes lead to infantile onset of IBD in humans. 33 In MS patients, Th17 cells isolated from blood had reduced expression of IL‐10 as compared to healthy controls. Conversely, higher expression of IL‐10 in Th17 cells positively correlated with clinically stable disease. 31 It is currently not well understood, in which environmental cues direct the differentiation of immunoregulatory Th17 cells in vivo.
Non‐classical pro‐inflammatory Th17.1
Ex vivo non‐classical human Th17.1 cells that express both IL‐17 and IFN‐γ have been found at sites of inflammation. 34 Given their transcriptional similarity to murine pathogenic Th17 cells, 35 human pro‐inflammatory Th17.1 cells are believed to contribute to the pathogenesis of human autoimmune diseases.31, 36 Pro‐inflammatory Th17 cells show double‐positive Th1/Th17 features as they coexpress the TF retinoid‐related orphan receptor γt (RORγt) and T‐bet, as well as IL‐12Rβ2 and IL‐23R.21, 22, 37 They are characterized by coexpression of pro‐inflammatory cytokines such as granulocyte–macrophage colony‐stimulating factor (GM‐CSF), IL‐26, CCL20 and IL‐22. 38 Expression of these cytokines depends on the secretion of IL‐6, TGF‐β, IL‐1β, IL‐12 and IL‐23 by APCs during differentiation or during reactivation of already‐differentiated classical Th17 cells.3, 7, 8, 30 Cells with similar cytokine production profile can be generated in vitro from human naïve CD4+ T cells using Candida albicans (C. albicans)‐pulsed monocytes, however without expressing CXCR3. 30 Alternatively, in vitro polarization with Mycobacterium tuberculosis induces a population of Th1* (to be read as ‘star’) which coexpress T‐bet and RORγt, CXCR3 and CCR6 and produce IFN‐γ (but not IL‐17). Furthermore, patients with RORC loss‐of‐function mutations lack Th1*, 39 suggesting that they could derive from Th17 cells through plastic events in a IL‐12‐, TNF‐α‐ and/or IL‐1β‐dependent manner. 40 It is believed that phenotypic instability predisposes Th17 cells to acquire a pro‐inflammatory phenotype in chronically inflamed tissues.
In addition to Th17‐to‐Th1 cell plasticity, Th17 cells can acquire characteristics of other Th or Treg subsets.
Th17 cell acquisition of other Th cell identify
Th17‐to‐Th2 plasticity
Circulating CD4+ memory T cells producing both IL‐4 and IL‐17, as well as CRTh2 and CCR6, GATA3 and RORγt, have been identified in patients with allergic asthma.41, 42 However, it is still unclear whether Th17/Th2 cells derive from Th17 or Th2, and whether IL‐4 (driving Th17 to Th2 cells) or IL‐1β, IL‐6, IL‐21 (driving Th2 to Th17 cells) or additional cytokines are needed. An in‐depth understanding of the origin and the role of these double cytokine producers in the human lung could be vital for targeting respiratory diseases, such as neutrophilic asthma.
Th17‐to‐Tfh plasticity
A fraction of circulating memory human Tfh cells expresses CXCR5 and CCR6, BCL6 and RORγt, and produces IL‐21, IL‐22 and IL‐17 (hence termed Tfh17).6, 43 It remains to be explored whether Tfh17 cells originate from Tfh or Th17 cells. The Th17‐to‐Tfh plasticity could have implications for Th17‐mediated autoantibody generation.
Th17‐to‐Treg plasticity
Evidence from fate mapping experiments in mice identified Th17‐to‐T regulatory cell transdifferentiation events. 44 While this phenomenon has yet to be reported in humans, studies on skin samples from psoriasis patients show IL‐17 production in Treg‐like cells. 45 Moreover, the analysis of synovial tissue from active RA patients revealed the presence of IL‐17+ Foxp3+ T cells. 46 For details on the role and origin of IL‐17‐producing Treg cells in human diseases, see reference. 47 A deeper understanding of mechanisms underlying the Th17‐Treg transdifferentiation in human conditions might be critical to restore tolerance to self in autoimmune diseases.
CURRENT METHODS TO STUDY HUMAN TH17 CELL HETEROGENEITY AND PLASTICITY
The study of human Th17 heterogeneity and plasticity requires different sets of tools (Figure 2). While heterogeneity can be resolved by deep phenotyping of Th17 populations in a cross‐sectional manner, plasticity requires the longitudinal study of a defined Th17 cell population. Due to the heterogeneity of circulating effector/memory Th17 cells, the study of human Th17 plasticity ex vivo is complicated and warrants controlled in vitro systems. 3
Figure 2.
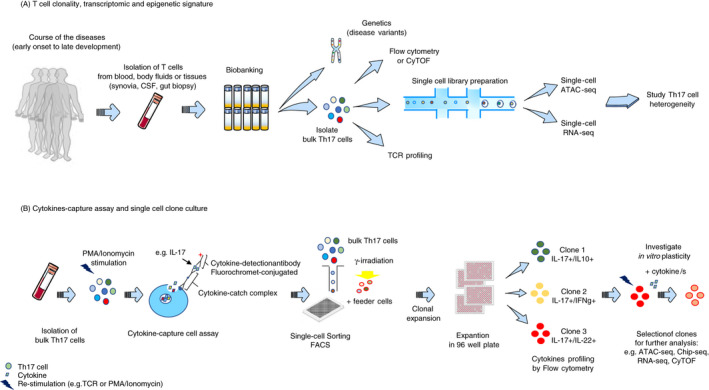
Current methods to study human Th17 cell heterogeneity and plasticity. A) To resolve the heterogeneity and dynamic of specific Th17 populations in a disease setting, it is relevant to isolate T cells from blood, other body fluids or tissues (e.g. synovia, cerebrospinal fluid (CSF), gut biopsy) at several time‐points (e.g. before and after treatment, at multiple disease stages). Collected cells can be biobanked for traceability and used at later time‐points. Genetic analysis can be performed to identify risk variants associated with the disease and altered Th17 cell phenotype and function, and, in parallel, perform TCRVβ sequencing to investigate clonality. Flow cytometry (FACS) or mass cytometry (CyTOF) allows to immunophenotype bulk Th17 cell populations to dissect their complexity. Th17 cell heterogeneity can be resolved by ex vivo epigenetic and transcriptional deep phenotyping at single‐cell level combining, for example, ATAC‐seq and RNA‐seq. Isolation of cytokine‐specific Th17 cells by cytokine capture assay allows in vitro generation of homogeneous Th17 clones. Isolation of bulk Th17 cells from, for example, blood and their stimulation with PMA/ionomycin allow single‐cell sorting, by FACS, of specific cytokine‐expressing Th17 cells and culturing with feeder cells. Following clonal expansion, each clone is transferred and expanded in a 96‐well plate. Th17 clones can be immunophenotyped before and after restimulation, which allows clone selection for further investigation. Plasticity of a defined Th17 clone can be investigated after reactivation and culturing with cytokines using CyTOF, RNA‐seq, ATAC‐seq and Chip‐seq.
Methods to study Th17 heterogeneity
Single‐cell RNA‐sequencing (scRNA‐seq) has proven a powerful technology to study heterogeneity and pathogenicity of mouse Th17 cells, 48 and it holds promise for resolving functional and cellular heterogeneity in human tissues as well. 49 Generating ‘transcriptomic’ and ‘epigenomic’ signatures on single‐cell (sc) level using, for example, scRNA‐seq, scChIP‐seq from chromatin modifications or scATAC‐seq will allow to identify phenotypically distinct Th17 cell subsets50, 51 at a given location and time‐point.
A powerful tool to study human Th17 heterogeneity at the protein level is the cytokine capture assay. 24 Isolation and immunophenotyping of cytokine‐specific Th17 cells using flow cytometry or mass cytometry (CyTOF) 52 allows to link function to phenotype in Th17 cells at a single‐cell level. 24 Further, single‐cell sorting of cytokine‐secreting cells can be used to generate single‐cell clone cultures of Th17 cells (Figure 2).
Methods to study Th17 plasticity
The generation of human Th17 cell clones provides the opportunity to study the plasticity of a homogenous Th17 cell population. 29 Single‐cell clones can be expanded in vitro using feeder cells (e.g. immortalized peripheral blood mononuclear cells, PBMCs) and can be immunophenotyped before and after reactivation by, for example, flow cytometry, proteomics, RNA‐seq and ATAC‐seq. This method was used to address the plasticity of IL‐10+ and IL‐10− Th17 clones when cultured in the presence of IL‐1β and IL‐27, respectively. 29
High‐throughput sequencing technologies now allow examination of antigen receptor repertoires by resolving the T‐cell receptor β variable region (TCRVβ) at single‐nucleotide level. 53 This method relies on two basic principles of T‐cell biology: (a) every T‐cell clone has a unique TCR sequence, and (b) clonal expansion following cognate antigen recognition. In healthy individuals, the TCR repertoire is polyclonal with little overrepresentation of T cells deriving from a single CD4+ T‐cell clone. 54 In the context of infections, cancer or immunological disorders, 55 T cells that have been activated and underwent clonal expansion will be overrepresented. Hence, assessing Th17 cell clonality coupled with transcriptional and epigenetic sequencing during the course of a disease or therapy will allow the study of plasticity in T cells in vivo (Figure 2).
In the following sections, we will discuss the downstream effects of cytokines involved in human Th17 plasticity, with particular emphasis to Th17‐to‐Th1 plasticity.
In this review, human memory Th17 cells are defined as CD4+ RA45CD̄ CCR6+CCR4+CXCR3̄ memory cells unless reported otherwise. Single‐cell memory Th17 clones were derived from these cells.
CYTOKINE‐MEDIATED SIGNALLING IN TH17 CELLS AND TARGETING FOR AUTOIMMUNITY
Th17 cells and their developmental plasticity are regulated by distinct combinations of cytokines and environmental cues. Here, we discuss the effects of two distinct classes: (1) cytokines that promote heterogeneity and induce plasticity towards pro‐inflammatory Th17 cells; and (2) effector cytokines released by Th17 cells in driving autoimmunity and inflammation. For both categories, we reference to completed or ongoing clinical trials that have tried to ameliorate autoimmune and inflammatory disease by cytokine targeting (Table 1).
Table 1.
Cytokine‐mediated signalling in Th17 cells and targeting for autoimmunity
| Targeted cytokine | Potential drug (company) | Indication/status | Identifier |
|---|---|---|---|
| Cytokine driving Th17 plasticity | |||
| IL−12p40 and IL−23p40 | Ustekinumab (Janssen) | Psoriasis/ Phase IIIA | NCT01009086 |
| NCT01077362 | |||
| Psoriatic arthritis / Phase IIIA | NCT01009086 | ||
| NCT01077362 | |||
| Ankylosing spondylitis/ Phase III | NCT02438787 | ||
| Ulcerative colitis/ Phase IIIA | NCT02407236 | ||
| Crohn´s disease/ Phase IIIA | NCT02407236 | ||
| SLE/ Phase III | NCT03517722 | ||
| Primary Sjögren's syndrome/Phase I | NCT04093531 | ||
| IL−23 p19 | Tildrakizumab | Plaque psoriasis/ Phase IIIA | NCT01729754 |
| (Merck/Sun Pharma) | Psoriatic arthritis/ Phase III | NCT04314544 | |
| Ankylosing spondylitis/ Phase II‐III | NCT03552276 | ||
| Guselkumab (Janssen) | Plaque psoriasis/ Phase IIIA | NCT02207244 | |
| Psoriatic arthritis/ Phase III | NCT03158285 | ||
| Crohn´s disease/ Phase III | NCT04397263 | ||
| Ulcerative colitis/ Phase II‐III | NCT04033445 | ||
| Lupus nephritis/ Phase II | NCT04376827 | ||
| Brazikumab (MEDI2070/AMG139) (AstraZeneca) | Crohn´s disease/ Phase II | NCT01714726 | |
| Psoriasis/ Phase I | NCT01094093 | ||
| Risankizumab (BI655066) (Boehringer Ingelheim, AbbVie) | Plaque psoriasis/ Phase IIIA | NCT02684357 | |
| NCT02672852 | |||
| Psoriatic arthritis/ Phase III | NCT03671148 | ||
| Ankylosing spondylitis/ Phase II | NCT02047110 | ||
| Ulcerative colitis/ Phase II‐III | NCT03398148 | ||
| Mirikizumab (LY3074828) (Lilly) | Asthma/ Phase II | NCT02443298 | |
| Crohn´s disease/ Phase III | NCT03105128 | ||
| Psoriasis/ Phase III | NCT03556202 | ||
| Crohn´s disease/ Phase III | NCT03926130 | ||
| Ulcerative colitis/ Phase III | NCT03518086 | ||
| IL−1b | Canakinumab (Novartis) | SJIA / Phase IIIA | NCT00886769 |
| Gouty arthritis/ Phase III | NCT01362608 | ||
| Type 1 diabetes/ Phase II | NCT00947427 | ||
| IL−1R | Anakinra | Rheumatoid arthritis/ Phase IIIA | 153 |
| (Swedish Orphan Biovitrum) | Asthma/ Phase I‐II | NCT03513471 | |
| SJIA / Phase I‐II | NCT00339157 | ||
| Rilonacept | Juvenile idiopathic arthritis/ Phase II | NCT00534495 | |
| Th17.1‐derived effector cytokines | |||
| IL−17A |
Secukinumab (Novartis) |
Psoriatic arthritis/ Phase IIIA | NCT02404350 |
| Ankylosing spondylitis/ Phase IIIA | NCT01358175 | ||
| Psoriasis / Phase IIIA | NCT01365455 | ||
| NCT01358578 | |||
| Rheumatoid arthritis/ Phase III | NCT01350804 | ||
| Ixekizumab (Lilly) | Ankylosing spondylitis/ Phase IIIA | NCT02757352 | |
| Psoriasis/ Phase IIIA | NCT01474512, NCT01597245 | ||
| Psoriatic arthritis /Phase IIIA | NCT01695239 | ||
| CNTO6785 (Janssen) | Rheumatoid arthritis/ Phase II | NCT01909427 | |
| COPD/ Phase II | NCT01966549 | ||
| NCT01828086 | |||
| CJM112 (Novartis) | Psoriasis/ Phase I‐II | NCT03299686 | |
| Asthma/ Phase II | |||
| IL−17A and IL−17F | Bimekizumab (UCB) | Rheumatoid arthritis/ Phase II | NCT02430909 |
| Ankylosing spondylitis/ Phase III | NCT03928743 | ||
| Psoriasis/ Phase III | NCT03598790 | ||
| Psoriatic arthritis/ Phase III | NCT04009499 | ||
| IL−17RA | Brodalumab (AstraZeneca) | Plaque psoriasis/ Phase IIIA | NCT04305327 |
| Psoriatic arthritis/ Phase III | NCT02024646 | ||
| IFN‐g | Fontolizumab | Crohn´s disease/ Phase II | NCT00072943 |
| (PDL BioPharma) | Rheumatoid arthritis/ Phase IIT | NCT00281294 | |
| AMG811 | Psoriasis/ Phase I | NCT01510951 | |
| (Amgen) | SLE/ Phase I | NCT02291588 | |
| GM‐CSF | Otilimab (GSK3196165) (GlaxoSmithKline) | Rheumatoid arthritis/ Phase III | NCT04333147 |
| Namilumab (Takeda) | Rheumatoid arthritis/ Phase II | NCT02379091 | |
| Psoriasis/ Phase II | NCT02129777 | ||
| Otilimab | Multiple sclerosis/ Phase I‐II | NCT01517282 | |
| MOR103 (MorphoSys AG) | Rheumatoid arthritis/ Phase I‐II | NCT01023256 | |
| Asthma/ Phase II | NCT01603277 | ||
| Lenzilumab (KB003) (Humanigen) | Rheumatoid arthritis/ Phase I | NCT01357759 | |
| GM‐CSF‐R | Mavrilimumab (CAM−3001) (Cambridge Antibody Technology, MedImmune) | Rheumatoid arthritis/ Phase IIT | NCT01712399 |
| NCT01050998 | |||
| NCT01706926 | |||
| IL−22 | Fezakinumab (Pfizer) | Atopic dermatitis/ Phase II | NCT01941537 |
| Psoriasis/ Phase I | NCT00563524 | ||
| TNF‐a | Adalimumab (AbbVie) or infliximab (Johnson& Johnson) | Rheumatoid arthritis/ Phase IIIA | NCT00195702 |
| Juvenile idiopathic arthritis/ Phase IIIA | NCT00048542 | ||
| Psoriatic arthritis/ Phase IIIA | NCT00235885 | ||
| Ankylosing spondylitis/ Phase IIIA | NCT00195819 | ||
| Crohn's disease/ Phase IIIA | NCT00207662 | ||
| Ulcerative colitis/ Phase IIIA | NCT00385736 | ||
| Plaque psoriasis/ Phase IIIA | NCT01680159 | ||
| IBD/ Phase IIIA | NCT00805766 | ||
| Certolizumab pegol (UCB) | Psoriatic arthritis/ Phase IIIA | NCT01087788 | |
| Plaque psoriasis/ Phase IIIA | NCT02326298 | ||
| Ankylosing spondylitis/ Phase IIIA | NCT02505542 | ||
| Rheumatoid arthritis/ Phase IIIA | NCT00160693 | ||
| Crohn's disease/ Phase IIIA | NCT00160524 | ||
| Axial spondyloarthritis/ Phase IIIA | NCT01087762 | ||
| Golimumab (Janssen) | Ulcerative colitis/ Phase II | NCT01090154 | |
| Rheumatoid arthritis/ Phase IIIA | NCT00299546 | ||
| Psoriatic arthritis/ Phase IIIA | NCT00265096 | ||
| Ankylosing spondylitis/ Phase IIIA | NCT01871649 | ||
| Ulcerative colitis/ Phase IIIA | NCT00487539 | ||
| Ozoralizumab (Ablynx) | Juvenile arthritis/ Phase III | NCT02277444 | |
| Rheumatoid arthritis/ Phase III | NCT04077567 | ||
| TNFR | Etanercept (Amgen) | Rheumatoid arthritis/ Phase IIIA | NCT02378506 |
| Juvenile idiopathic arthritis/ Phase IIIA | NCT03780959 | ||
| Psoriatic arthritis/ Phase IIIA | NCT00317499 | ||
| Ankylosing spondylitis/ Phase IIIA | NCT01258738 | ||
| Plaque psoriasis/ Phase IIIA | NCT01241591 | ||
Clinical trialA, approved by FDA; Clinical trialT, terminated; IL‐12 p40, p40 subunit of interleukin‐12; IL‐23 p19, p19 subunit of interleukin‐23; IL‐23 p40, p40 subunit of interleukin‐23; IL‐17A, interleukin‐17A; IL‐17F, interleukin‐17F; IL‐17RA, IL‐17 receptor A; IFN‐γ, interferon γ; GM‐CSF, granulocyte–macrophage colony‐stimulating factor; IL‐22, interleukin‐22; TNF‐α, tumour necrosis factor‐α; TNFR, tumour necrosis factor receptor; COPD, chronic obstructive pulmonary disease; SLE, systemic lupus erythematosus; SJIA, systemic juvenile idiopathic arthritis.
Cytokines driving Th17 plasticity
Members of the IL‐12 family (e.g. IL‐12, IL‐23 and IL‐27) are pleiotropic mediators,56, 57 which are produced by myeloid cells and play a role in shaping human memory Th17 cell plasticity. All IL‐12 family cytokines consist of an α‐ and a β‐cytokine subunit, which can be shared among family members. While IL‐12 and IL‐23 share the p40 β‐subunit, they differ in their α‐subunit (IL‐12p35 and IL‐23p19, respectively). 58 Interestingly, IL‐12 and IL‐23 receptors also share a common β‐subunit (IL‐12Rβ1), while having a specific α‐subunit (IL‐12Rβ2 and IL‐23R), respectively. Upon receptor binding, both IL‐12 and IL‐23 activate both STAT3 and STAT4 signalling pathways, with IL‐12 predominantly signalling through STAT4 and IL‐23 signalling through STAT3. 56 IL‐12 induces IFN‐γ production in T and NK cells and is increased at sites of chronic inflammation. 59 IL‐12 can contribute to the conversion of Th17 to non‐canonical Th1 cells as stimulation of both human Th17 and Th17.1 cells with IL‐12 downregulates RORγt and IL‐17, while upregulating expression of IFN‐γ and T‐bet.22, 60
IL‐23 is dispensable for initial Th17 differentiation but plays a key role in the stabilization and expansion of pathogenic Th17 cells. 12 IL‐23‐dependent STAT3 signalling contributes to lineage stabilization, while STAT4 activation promotes IFN‐γ expression and plasticity towards a pro‐inflammatory phenotype. 61 This mechanism requires upregulation of IL‐23R by pre‐Th17 cells. 61 In human Th17 cells, IL‐23 synergizes with TGF‐β and IL‐1β to induce upregulation of IL‐23R during differentiation. 8 The importance of the IL‐23/IL‐23R signalling axis is highlighted by the finding that multiple gene polymorphisms in the IL‐23R gene correlate with an increase in intestinal Th17.1 cells and an increased risk to develop IBD, psoriasis, RA, psoriatic arthritis (PsA) and ankylosing spondylitis (AS). 17 However, mechanistic studies elucidating how these SNPs contribute to disease pathogenesis are still missing. Clinical trials using monoclonal antibodies either targeting the IL‐12p40 subunit (shared by IL‐12 and IL‐23) or targeting IL‐23p19 yielded good results in the treatment of several autoimmune diseases, including Crohn´s disease, psoriasis and psoriatic arthritis (Table 1).
IL‐27 is a heterodimeric cytokine comprised of the p28 and Epstein–Barr virus‐induced gene 3 (EBI3) subunits. 62 IL‐27 mediates its effects by signalling through IL‐27Rα and the gp130 receptor. 63 IL‐27 specifically activates STAT1, STAT3 and to a lesser extent STAT4. 64 While IL‐27 seems to have a pro‐inflammatory effect by promoting a Th1‐like phenotype, it is now also recognized as an anti‐inflammatory agent, as it induces IL‐10 in human T cells. 65 In addition, IL‐27‐induced downregulation of IL‐22 and upregulation of IL‐10 in both human memory CD4+ T cells and memory IL‐10+ Th17 clones in vitro 29, 66 suggests a role for IL‐27 in shaping Th17 plasticity. To date, no clinical trial investigates IL‐27 as a target for autoimmune disease.
IL‐1β is an inflammasome‐related cytokine produced by myeloid cells. IL‐1β synergizes with IL‐6 and IL‐23 to induce differentiation of pro‐inflammatory Th17 cells. It activates the p38 mitogen‐activated protein kinase (MAPK) and the AKT‐mTOR pathways, thus promoting the expansion and effector functions of Th17 cells.67, 68 Moreover, IL‐1β‐dependent signalling induces IRF4, which in turn promotes pro‐inflammatory Th17 differentiation while inhibiting Foxp3+ expression. 69 In fact, IL‐1β modulates human memory Th17 plasticity in vitro and in vivo by downregulating IL‐10 production in classical Th17 cells. 70 Targeting IL‐1 signalling, both by blocking its receptor (anakinra) and by specifically neutralizing IL‐1β, has proven to be effective in several autoimmune diseases (Table 1). Indeed, patients suffering from Schnitzler syndrome or cryopyrin‐associated periodic syndrome (CAPS) have decreased numbers of circulating IL‐10‐producing Th17 cells, likely due to high IL‐1β levels.30, 71 The authors showed that defective IL‐10 production by the Th17 cells can be restored in vivo by IL‐1β antibody‐blocking therapy. 71 However, whether the effect of IL‐1β antibody‐blocking therapy on Th17 plasticity drives clinical efficacy is still a matter of debate.
IL‐2 is a pleiotropic cytokine that is necessary for T‐cell proliferation but influences Th cell differentiation by modulating cytokine receptor expression. 72 While IL‐2 supports Th1 differentiation through STAT5‐dependent upregulation of IL‐12Rβ2 and T‐bet, it negatively impacts Th17 cell differentiation by downregulating subunits of the IL‐6R, IL‐6Rα and gp130. In vitro, mouse Th17 cells differentiated in the presence of IL‐2 maintain the expression of IL‐12β2, possibly allowing the acquisition of IFN‐γ production if stimulated by IL‐12. 73 In human memory T cells, IL‐2 promotes expression of GM‐CSF in a STAT5‐dependent manner 38 contributing to Th17 plasticity towards pro‐inflammatory phenotype. Furthermore, IL‐2 may play a role in human memory Th17 cell expansion. 74 It remains to be investigated whether it drives memory Th17 cells towards a pro‐inflammatory phenotype in human inflamed tissues. In contrast, immunoregulatory low dose of IL‐2, which favours Treg over Th17 differentiation, is under investigation in Crohn´s diseases (NCT02424396), relapsing–remitting MS (NCT02424396), SLE (NCT02424396) and type I diabetes (NCT02265809).
IL‐21 is produced by pre‐Th17 cells and promotes development and expansion of mature mouse and human Th17 cells in an autocrine manner by inducing IL‐23R expression in a STAT3‐dependent manner.75, 76 Increased serum levels of IL‐21 in several autoinflammatory diseases, including RA and SLE, suggest a pro‐inflammatory role of IL‐21. In fact, treatment with monoclonal antibodies against IL‐21 significantly improved inflammation in patients with active RA (NNC01140006/ Phase II (Novo Nordisk A/S)). However, its role in Th17 plasticity is not clear.
Roles of signature cytokines produced by pro‐inflammatory Th17 cells
The IL‐17 family comprises six members: IL‐17A, IL‐17B, IL‐17C, IL‐17D, IL‐17E and IL‐17F, which initiate an inflammatory response by inducing the release of mediators from IL‐17R‐expressing, non‐haematopoietic cells (e.g. cytokines, such as IL‐6, IL‐1, GM‐CSF and TNF‐α; chemokines such as CXCL1, CXCL2, CXCL5, CCL2, CCL7, CCL20 and IL‐8; antimicrobial peptides (AMPs); and matrix metalloproteinases (MMPs)). 77 In the gut, IL‐17 contributes to the maintenance of intestinal barrier integrity. 78 Given the range of biological effects of IL‐17, which is produced by both pro‐inflammatory and immunoregulatory Th17 cells, clinical trials with IL‐17‐ or IL‐17R‐targeting antibodies both worsened or improved disease symptoms, depending on the type of inflammatory disease. While anti‐IL‐17 treatment was efficacious in psoriasis, psoriatic arthritis, ankylosing spondylitis and lichen planus,79, 80, 81, 82 it had a moderate or even detrimental effect in patients with rheumatoid arthritis and Crohn´s disease, respectively. This highlights the complexity of targeting the entire Th17 lineage in autoimmune diseases.
TNF‐α is a pro‐inflammatory cytokine produced by immune and epithelial cells in response to infection and tissue injury, 83 and is one of the major effector cytokines secreted by pathogenic Th17 cells. It initiates production and release of IL‐1, IL‐6, IL‐8 and GM‐CSF. expression and is detected at the sites of inflammation in RA, Crohn´s disease and psoriasis.84, 85, 86 Monoclonal antibody‐based anti‐TNF therapy has emerged as standard of care for many autoimmune diseases. Interestingly, anti‐TNF treatment of human CD4+ T cells in vitro favours differentiation of IL‐17/IL‐10 double‐positive Th17 cells and prevented accumulation of non‐classical Th1 cells. 34
IL‐22 is member of the IL‐10 family, binding to the IL‐22R1 and IL‐10R2 to propagate downstream signals through STAT3 signalling. 87 IL‐22 maintains mucosal barrier integrity, cell survival, proliferation and synthesis of antimicrobial peptides by targeting mostly non‐haematopoietic cells expressing IL‐22R in the skin, intestine, liver, lung and kidney.88, 89 IL‐22 is induced by the IL‐23/IL‐23R axis, which is hyperactive in autoimmune diseases. Serum levels of IL‐22 correlate with disease activity in Crohn´s disease 90 ; additionally, patients with Crohn´s disease carrying an IL‐23R risk allele have increased levels of serum IL‐22. 91 Furthermore, IL‐22 serum levels correlate with disease severity in psoriasis, possibly due to the effects of IL‐22 on hyperplasia and abnormal differentiation of keratinocytes. 92 Thus, IL‐22 acts in concert with IL‐17 at the site of inflammation 93 and multiple clinical trials currently investigate the efficacy of targeting IL‐22 in autoimmune diseases such as Crohn´s disease, SLE, rheumatoid arthritis and psoriasis, yet critical role of IL‐22 in epithelial reaeration and wound healing remains a concern.
GM‐CSF was originally identified as a haematopoietic growth factor and immune modulator produced by T and B cells, monocytes, macrophages, endothelial cells and fibroblasts. 94 GM‐CSF can polarize inflammatory M1‐like macrophages to produce pro‐inflammatory cytokines and induce mixed Th1/Th17 responses.95, 96, 97 It has been linked to autoimmune tissue damage and encephalomyelitis in mice and humans.38, 98, 99 Mouse pathogenic Th17 cells can produce GM‐CSF,98, 99 and human memory IL‐10–Th17 clones express significantly more CSF2 (encoding for GM‐CSF) compared with IL‐10+Th17 cells. 29 However, data from in vitro cultures of human naïve T cells suggest that GM‐CSF is either expressed alone in a ‘GM‐CSF‐only’ (ThGM‐CSF) subset or coexpressed with IFN‐γ, associating with Th1 rather than Th17 cells. ThGM‐CSF can be isolated ex vivo as CCR10+CCR4+CXCR3–CCR6– cells and might derive from plastic Th1 or pro‐inflammatory Th17 cells.38, 100 Overall, GM‐CSF is an attractive target for treatment of autoimmune diseases.101, 102, 103 However, there are some concerns about possible treatment‐related side‐effects; thus, better patient stratification is warranted to improve treatment results.
IFN‐γ is a classical Th1 cytokine 104 and is expressed by pro‐inflammatory Th17 cells.30, 59 Clinical trials targeting IFN‐γ have demonstrated efficacy, however, to a lesser extent than blocking IL‐12p40‐dependent upstream signalling, thus terminated (Table 1).
In the following two sections, we will discuss downstream effects of cytokine on changes in Th17 metabolism and epigenetics during T‐cell plasticity.
Other microenvironmental factors as a source of Th17 cell plasticity
Recent evidence suggests that microenvironmental factors other than cytokines (e.g. oxygen, vitamins, pollutants, sodium chloride (NaCl), potassium, diet‐derived metabolites and commensal microbiota) shape T‐cell identity and plasticity. 105 For instance, high NaCl concentrations have an important function for Th17 differentiation and plasticity. NaCl boosts IL‐17 production in in vitro‐differentiated mouse CD4+ T cells and in human memory CD4+ T cells where it also enhances Th2 and suppresses Th1 cell responses.106, 107 In human memory Th17 cells, NaCl, in the presence of TGF‐β, promotes their immunoregulatory function through the NFAT5‐SGK1‐FoxP3 pathway. However, in pro‐inflammatory conditions and the absence of TGF‐β, NaCl boosts pro‐inflammatory cytokine production in both human and mouse Th17 cells and promotes pathogenicity in an EAE model. 108 Also, oxygen concentrations appear to impact on Th17 cell phenotype, as mouse Th17 cells cultured in hypoxic conditions (1% O2) produced significantly more IL‐10 than Th17 cells cultured at 21% O2. 109 Thus, NaCl and O2 are two examples of cytokine‐independent factors, which modulate Th17 phenotype or plasticity in a context‐dependent manner. More examples are discussed elsewhere. 105
METABOLIC SHIFTS BETWEEN PRO‐ AND ANTI‐INFLAMMATORY TH17 CELLS
A major downstream effect of cytokine signalling is a change in cellular metabolism. 110 However, the specific metabolic needs of different Th subsets are less well understood. PI3 K‐AKT‐mTOR signalling pathways and the hypoxia‐induced factor 1α (Hif1α) are important to determine T‐cell differentiation into Th or Treg lineages by shifting cellular metabolism from oxidative phosphorylation to the glycolysis‐based Warburg effect. 110 Intriguingly, multiple cytokines, such as IL‐2, IL‐1β, IL‐21 and TGF‐β, involved in Th17 differentiation and plasticity promote PI3 K‐AKT‐mTORC1 activation. 111 In mouse settings, the PI3 K‐AKT‐mTORC1‐S6 K1/2 axis promotes Th17 cell differentiation through the inhibition of Gfi1 (a negative regulator of Th17 cells) and the nuclear translocation of RORγ. 112 How this pathway alters pro‐inflammatory phenotypes of Th17 cells in human autoimmune diseases is yet to be explored.
Several mouse studies have highlighted different metabolic requirements for Th17 cells, other effectors T cells and regulatory T cells.113, 114 In mouse settings, anti‐inflammatory Th17 cells have a metabolic profile similar to quiescent or memory cells (mainly using oxidative phosphorylation), while pro‐inflammatory Th17 cells are highly glycolytic and glutaminolytic effector cells with increased mTORC1 activation.115, 116, 117. Here, glutaminase, a key enzyme of glutamine metabolism, promotes mouse Th17 cell differentiation, while restraining Th1 cells. These events are accompanied by altered chromatin accessibility and gene expression and suggest possible implications for Th17‐Th1 plasticity. 118 In addition, calcium signalling‐dependent mitochondrial regulation and oxidative phosphorylation also play crucial roles in the differentiation of pathogenic Th17 cells in mice. 119
Tissue‐specific metabolic adaptations might also contribute to plasticity of different memory Th17 cell subsets. Circulating mouse TCM relies on oxidative phosphorylation of endogenous free fatty acids (FFA), while skin‐resident TRM metabolizes exogenous FFA to support their long‐term survival. 120 Whether this has (in)direct implications on the ability of mouse and human memory Th17 cells to acquire new phenotypes remains a matter of investigation.
Knowledge about metabolic requirements of human Th17 cells is more limited, especially in the context of diseases. A recent study showed that activin‐A signalling and CD73‐dependent molecular pathways restrain pro‐inflammatory human Th17 cells in MS patients counteracting HIF1α activity, critical for glycolysis. These events result in increased anti‐inflammatory genes through AhR, STAT3 and c‐Maf and subsequent IL‐10 secretion (in naïve CD4+ T cells polarized towards Th17 cell lineage). 121 In the coming years, it will be vital to discover other metabolic factors that shift the balance between human anti‐ and pro‐inflammatory Th17 subsets, determine plastic events in health and diseases and explore the role of cytokine signalling driving metabolic changes.
EPIGENETIC AND TRANSCRIPTIONAL REGULATION
The transcriptional regulation of Th17 effector functions involves pioneer TFs, which promote chromatin remodelling and thus allow secondary transcription factors to access regulatory genetic elements.
Mechanistic studies suggest that the expression of pro‐ or anti‐inflammatory cytokines in Th17 cells is mutually exclusive. Using reactivated human memory Th17 clones in vitro, IL‐10 expression correlated with downregulation of IL‐17 and upregulation of the TF c‐Maf. 29 While overexpressing c‐Maf boosted IL‐10 production, silencing of c‐Maf led to a reduction in IL‐10. Conversely, downregulation of c‐Maf increased expression of pro‐inflammatory cytokines such as IFN‐γ, IL‐22 and GM‐CSF. 29 In another study, IL‐10 expression was upregulated after TCR engagement and inversely correlated with IL‐17 expression through downmodulation of RORγt, a mechanism dependent on IL‐2/STAT5 signalling. 30 In line with this, pharmacological inhibition seems to reduce pro‐inflammatory cytokine expression (e.g. IL‐22) in Th17 cells (defined as IL‐17A‐expressing cells generated from mononuclear cells from the synovial fluid (SFMC) of patients with AS and PsA cultured in Th17‐promoting conditions 6 days before RORγt inhibitor treatment). 122 These findings suggest a role for RORγt in the transcriptional regulation of pro‐ and anti‐inflammatory cytokines. It remains to be explored whether RORγt directly regulates IL‐10 expression in human Th17 cells, as seen in mouse. 123 RORγt inhibitors are currently investigated as therapeutic approach for inflammatory autoimmune disease such as psoriasis. 124
Human memory Th17 clones cultured in vitro with IL‐12 produced IFN‐γ, an observation that was not accompanied by permissive epigenetic modifications at the IFNG locus. 125 However, converting in vitro‐generated Th17 cells into these ‘non‐classical’ Th1 cells led to an increase in DNA methylation at the IL‐17A and RORC2 gene loci, while both TBX21 (encoding for T‐bet) and IFNG genes were demethylated. 126 These non‐classical Th1 cells could not be reverted into Th17 as the TF Eomes prevented further chromatin remodelling, a mechanism that required IL‐2/IL‐12 signalling. 127
These findings suggest that cytokines can induce epigenetic changes and stabilize the expression profiles depending on the permissiveness of the epigenetic landscape in the target cell or the differentiation state of the cells.29, 127 In fact, a global mapping of H3K4me3 (permissive) and H3K27me3 (repressive) histone marks in different CD4+ T‐cell lineages revealed the presence of a mixed, ‘poised’ state in the promoter of lineage‐specific transcription factors, which support the hypothesis of epigenetic plasticity. For instance, in mouse Th17 cells, the Foxp3 promoter is not epigenetically repressed possibly allowing for Th17‐to‐Treg cell plasticity. 128
A recent study showed that systemic inhibition of histone demethylases (corresponding to a genome‐wide increase in histone methylation, likely correlated with transcriptional repression) inhibits Th17 differentiation, by downregulation of RORC, IL‐17 and IFNG expression. Histone demethylase inhibitors had a similar effect in vitro on pro‐inflammatory cytokine (e.g. IL‐17, IL‐22 and TNF‐α) expression in Th cells enriched from ankylosing spondylitis patients. 129 However, the reduction in pro‐inflammatory cytokines was accompanied by a metabolic switch towards an anergic cell state, suggesting that both might impact on effector function in T lymphocytes. 129 Cytokines themselves can induce epigenetic changes, for example, by activating DNA methyltransferases (DNMTs) downstream of the cytokine signalling network. 130 Whether this plays a role in human Th17 plasticity is currently unknown. It is of great interest to understand whether those epigenetic regulators can be specifically targeted to modulate pro‐inflammatory Th17 cells. Similarly, it remains to be understood which other key TFs govern the function of pro‐ versus anti‐inflammatory Th17 cells.
PARADOXICAL ROLE OF TH17 SUBSETS IN TUMOUR IMMUNITY
In addition to their well‐appreciated role in autoimmunity, the pluripotent nature of Th17 cells has motivated extensive investigation into their biological roles and functions in cancer. Th17 cells are prevalent in many different types of cancer, including melanoma, pancreatic cancer, prostate cancer, and colon and gastric cancer. 131 This suggests that the microenvironment within tumours promotes the recruitment and/or expansion of Th17 cells. Recent studies suggest that tumours secrete Th17‐attracting chemokines, including CCL17, CCL20, MCP‐1 and RANTES, 132 and support Th17 development through secretion of cytokines, including IL‐1β, TGF‐β and IL‐6. 131 Other cytokine‐independent factors such as hypoxic tumour microenvironment and tumour‐derived glutamine‐dependent metabolites also promote development of Th17 cells.118, 133 Tumour‐infiltrating Th17 cells can exert both pro‐ or antitumour effects depending on the types of cancer, 131 suggesting functional heterogeneity within tumour‐associated Th17 cells. The dual nature of Th17 cells in both promoting tumour growth and mounting an antitumour response has been discussed extensively elsewhere.134, 135 Here, we summarize Th17 phenotypes that are associated with different cancer types (Table 2). On the one hand, Th17 cells were found to directly promote tumour growth in cancers such as hepatocellular carcinoma, and colorectal and pancreatic cancers by inducing vascularization and immunosuppressive activities through secretion of effector cytokines, including IL‐17A and IL‐10.132, 134, 136, 137 On the other hand, in some tumour types such as ovarian, prostrate and colorectal cancers, Th17 cells can induce antitumor immune responses through recruitment of cytotoxic effector T cells and production of effector cytokines, including IFN‐γ.138, 139 These functions are associated with increased chances of survival of cancer patients. Notably, a recent study highlighted that the frequency of Th17 in colorectal tumour cells themselves was not predictive of the number or proportion of Th17 cells in the tumour tissue. The authors demonstrated that although the increased frequency of intra‐epithelial Th17 cells is associated with a pronounced cytotoxic T‐cell infiltration and prolonged survival in colorectal cancer, the stromal Th17 infiltrates were not.140, 141 Hence, the functional plasticity of Th17 cells within the same tumour types depending on their location and the cytokine milieu at those sites may allow a pro‐ or antitumorigenic effect.
Table 2.
Overview of pleiotropic function of Th17 cells in different tumours
| Type of cancer | Outcomes | Th17‐derived cytokine profiles | Effect of Th17 cells on tumour | References |
|---|---|---|---|---|
| Ovarian | Protective | IL−17, IFN‐γ, GM‐CSF, IL−2 | Increased intratumoral Th17 cell numbers were associated with improved survival rates. | 138, 154, 155 |
| Colorectal | Protective | TNF‐α, IL−21, IL−22, GM‐CSF, IFN‐γ and IL−8 | Positive contribution of tumour‐infiltrating Th17 cells in mediating tumour immunity through the recruitment of cytotoxic CD8+ T cells. | 132, 140, 156, 157 |
| Gastric | Adverse | IL−17, IL−21 | Th17 was shown to promote tumour growth via secretion through upregulation of VEGF prostaglandins. | 158, 159, 160 |
| Non‐Hodgkin lymphoma | Protective | IL−17, IFN‐γ (?) | Lymphoma limits Th17 generation and maintenance to mediate tolerance. | 146, 161 |
| Hodgkin lymphoma (HL) | N/A | IL−17, IL−1, GITR | Compared with Epstein–Barr virus (EBV) + HL patients, EBV‐ HL patients upregulated IL−23 that displayed a pro‐inflammatory Th17 phenotype. | 162 |
| Multiple myeloma | Adverse | IL−17, IFN‐γ | Higher numbers of Th17 cells were associated with tumour growth and impeded host tumour immunity | 144, 163 |
| Acute myeloid leukaemia | Adverse | IL−17, IL−10 | Increased frequencies of Th17 cells were associated with poor prognosis. Th17 promotes tumour growth via secretion of IL−17, while Th7‐producing IL−10 suppresses immune activity. | 143, 164 |
N/A, not applicable; IL‐17, interleukin‐17; IFN‐γ, interferon γ; GM‐CSF, granulocyte–macrophage colony‐stimulating factor; IL‐2, interleukin‐2; IL‐22, interleukin‐22; IL‐23, interleukin‐23; TNF‐α, tumour necrosis factor‐α; IL‐10, interleukin‐10.
Th17 cells and their associated cytokines have also been implicated in the pathogenesis of haematological cancers, including acute myeloid leukaemia (AML), lymphoma and multiple myeloma (MM). Here, an increased frequency of Th17 cells in AML patients is associated with tumour burden.142, 143 Furthermore, Th17 cell frequency was reduced in patients who achieved complete remission following chemotherapy. Th17 cell numbers and associated cytokine levels (e.g. IL‐17, IL‐21, IL‐22 and IL‐23) are also increased in the blood and bone marrow of myeloma patients. 144 Elevated inflammatory cytokine levels within bone marrow niches of MM patients support development of Th17 cells, which in turn sustains myeloma growth via secretion of cytokines, possibly by secretion of IL‐17 and IL‐21. 145 In contrast to AML and MM, lymphoma B cells were shown to inhibit differentiation of Th17 cells in favour of promoting immunosuppressive Treg cells to promote disease progression. 146 Whether Th17 cells transdifferentiate into Treg cells is not clear. These studies overwhelmingly suggest that Th17 plasticity and functional heterogeneity are regulated by tumour‐specific microenvironment.
Efforts to develop therapies that modulate the tumour microenvironment in order to promote antitumour T‐cell functions, while inhibiting their tumour‐promoting potential, are undergoing. Current checkpoint blockade therapies have been shown to boost Th17‐mediated antitumour responses by enhancing their production of IFN‐γ, IL‐17 and TNF‐α.147, 148 Furthermore, in mouse models of adoptive T‐cell therapy, Th17 cells grown in vitro under pro‐inflammatory conditions exhibited potent antitumour activity upon adoptive transfer in vivo.149, 150 The systemic administration of the Th‐supporting cytokine IL‐23 or autocrine secretion of IL‐23 by CAR‐T cells mediates antitumour immunity in solid tumour models.151, 152 Future work characterizing functional heterogeneity Th17 cells in different cancer settings will be required to define the therapeutic potential of targeting Th17 plasticity to mount effective anticancer response.
CONCLUDING REMARKS
Understanding mechanisms behind the context‐dependent generation of anti‐inflammatory and pro‐inflammatory Th17 cells is at the core of future treatments for autoimmunity. In this review, we highlighted the interconnected relationship between cytokines, intracellular signalling and metabolic pathways, epigenetic regulators and transcription factors, which together orchestrate the ever‐changing human Th17 cell identity. A thorough characterization of the epigenetic status of Th17 cells from autoimmune patients and healthy controls, from both inflamed and non‐inflamed tissue, would provide a better understanding of how cytokine‐mediated changes are linked to disease pathogenesis, which is critical for designing immunomodulatory therapies for chronic inflammatory diseases.
Funding Information
AstraZeneca R&D postdoc program (SC), Fondation Arc Pour La Recherche Sur Le Cancer (S.M.), Ligue Nationale Contre Le Cancer (S.M.), Cancéropôle Nord‐Ouest (S.M.).
AUTHOR CONTRIBUTIONS
S.C., S.P. and S.M. conducted the literature search and wrote the paper. U.G., S.P. and S.M. reviewed the paper.
CONFLICT OF INTEREST
S.C. is a fellow of the AstraZeneca R&D postdoc programme. S.C., S.P. and U.G. are employees of AstraZeneca and may own stock or stock options.
Senior author: Suman Mitra
Contributor Information
Silvia Preite, Email: silvia.preite@astrazeneca.com.
Suman Mitra, Email: suman.mitra@inserm.fr.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- 1. Raphael I, Joern RR, Forsthuber TG. Memory CD4(+) T cells in immunity and autoimmune diseases. Cells 2020;9(3):531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhaumik S, Basu R. Cellular and molecular dynamics of Th17 differentiation and its developmental plasticity in the intestinal immune response. Front Immunol. 2017;8:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sallusto F. Heterogeneity of human CD4(+) T cells against microbes. Annu Rev Immunol. 2016;34:317–34. [DOI] [PubMed] [Google Scholar]
- 4. Mosmann TR, Coffman RL. Heterogeneity of cytokine secretion patterns and functions of helper T cells. Adv Immunol. 1989;46:111–47. [DOI] [PubMed] [Google Scholar]
- 5. Romagnani S. Human TH1 and TH2 subsets: doubt no more. Immunol Today. 1991;12(8):256–7. [DOI] [PubMed] [Google Scholar]
- 6. Ma CS, Wong N, Rao G, Avery DT, Torpy J, Hambridge T, et al. Monogenic mutations differentially affect the quantity and quality of T follicular helper cells in patients with human primary immunodeficiencies. J Allergy Clin Immunol. 2015;136(4):993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)‐17 cells requires transforming growth factor‐beta and induction of the nuclear receptor RORgammat. Nat Immunol. 2008;9(6):641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Volpe E, Servant N, Zollinger R, Bogiatzi SI, Hupe P, Barillot E, et al. A critical function for transforming growth factor‐beta, interleukin 23 and proinflammatory cytokines in driving and modulating human T(H)‐17 responses. Nat Immunol. 2008;9(6):650–7. [DOI] [PubMed] [Google Scholar]
- 9. Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper‐IgE syndrome. Nature 2008;452(7188):773–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, et al. Interleukin‐23 rather than interleukin‐12 is the critical cytokine for autoimmune inflammation of the brain. Nature 2003;421:744–8. [DOI] [PubMed] [Google Scholar]
- 11. Dong C. Diversification of T‐helper‐cell lineages: finding the family root of IL‐17‐producing cells. Nat Rev Immunol. 2006;6:329–33. [DOI] [PubMed] [Google Scholar]
- 12. Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, et al. IL‐23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–73. [DOI] [PubMed] [Google Scholar]
- 14. Schmitt N, Ueno H. Regulation of human helper T cell subset differentiation by cytokines. Curr Opin Immunol. 2015;34:130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol. 2013;8:477–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brockmann L, Giannou AD, Gagliani N, Huber S. Regulation of TH17 Cells and Associated Cytokines in Wound Healing, Tissue Regeneration, and Carcinogenesis. Int J Mol Sci. 2017;18:1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel DD, Kuchroo VK. Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity 2015;43:1040–51. [DOI] [PubMed] [Google Scholar]
- 18. Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, et al. Stability of the regulatory T cell lineage in vivo. Science 2010;329:1667–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J, Casanova JL, Puel A. Mucocutaneous IL‐17 immunity in mice and humans: host defense vs. excessive inflammation. Mucosal Immunol. 2018;11:581–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanna S, Etzioni A. New host defense mechanisms against Candida species clarify the basis of clinical phenotypes. J Allergy Clin Immunol. 2011;127:1433–7. [DOI] [PubMed] [Google Scholar]
- 21. Acosta‐Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, et al. Surface phenotype and antigenic specificity of human interleukin 17‐producing T helper memory cells. Nat Immunol. 2007;8:639–46. [DOI] [PubMed] [Google Scholar]
- 22. Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maggi L, Santarlasci V, Capone M, Peired A, Frosali F, Crome SQ, et al. CD161 is a marker of all human IL‐17‐producing T‐cell subsets and is induced by RORC. Eur J Immunol. 2010;40:2174–81. [DOI] [PubMed] [Google Scholar]
- 24. Cosmi L, De Palma R, Santarlasci V, Maggi L, Capone M, Frosali F, et al. Human interleukin 17‐producing cells originate from a CD161+CD4+ T cell precursor. J Exp Med. 2008;205:1903–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amezcua Vesely MC, Pallis P, Bielecki P, Low JS, Zhao J, Harman CCD, et al. Effector Th17 cells give rise to long‐lived TRM cells that are essential for an immediate response against bacterial infection. Cell 2019;178:1176–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bishu S, El Zaatari M, Hayashi A, Hou G, Bowers N, Kinnucan J, et al. CD4+ Tissue‐resident memory T cells expand and are a major source of mucosal tumour necrosis factor alpha in active Crohn's disease. J Crohns Colitis. 2019;13:905–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krebs CF, Reimers D, Zhao Y, Paust HJ, Bartsch P, Nunez S, et al. Pathogen‐induced tissue‐resident memory TH17 (TRM17) cells amplify autoimmune kidney disease. Sci Immunol. 2020;5:eaba4163. [DOI] [PubMed] [Google Scholar]
- 28. Ueno K, Urai M, Sadamoto S, Shinozaki M, Takatsuka S, Abe M, et al. A dendritic cell‐based systemic vaccine induces long‐lived lung‐resident memory Th17 cells and ameliorates pulmonary mycosis. Mucosal Immunol. 2019;12(1):265–76. [DOI] [PubMed] [Google Scholar]
- 29. Aschenbrenner D, Foglierini M, Jarrossay D, Hu D, Weiner HL, Kuchroo VK, et al. An immunoregulatory and tissue‐residency program modulated by c‐MAF in human TH17 cells. Nat Immunol. 2018;19(10):1126–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, et al. Pathogen‐induced human TH17 cells produce IFN‐gamma or IL‐10 and are regulated by IL‐1beta. Nature 2012;484(7395):514–8. [DOI] [PubMed] [Google Scholar]
- 31. Hu D, Notarbartolo S, Croonenborghs T, Patel B, Cialic R, Yang TH, et al. Transcriptional signature of human pro‐inflammatory TH17 cells identifies reduced IL10 gene expression in multiple sclerosis. Nat Commun. 2017;8(1):1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gandhi R, Kumar D, Burns EJ, Nadeau M, Dake B, Laroni A, et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell‐like and Foxp3(+) regulatory T cells. Nat Immunol. 2010;11(9):846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glocker EO, Kotlarz D, Klein C, Shah N, Grimbacher B. IL‐10 and IL‐10 receptor defects in humans. Ann N Y Acad Sci. 2011;1246:102–7. [DOI] [PubMed] [Google Scholar]
- 34. Evans HG, Roostalu U, Walter GJ, Gullick NJ, Frederiksen KS, Roberts CA, et al. TNF‐alpha blockade induces IL‐10 expression in human CD4+ T cells. Nat Commun. 2014;5:3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13(10):991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin‐Orozco N, Chung Y, Chang SH, Wang YH, Dong C. Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur J Immunol. 2009;39(1):216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maggi L, Santarlasci V, Capone M, Rossi MC, Querci V, Mazzoni A, et al. Distinctive features of classic and nonclassic (Th17 derived) human Th1 cells. Eur J Immunol. 2012;42(12):3180–8. [DOI] [PubMed] [Google Scholar]
- 38. Noster R, Riedel R, Mashreghi MF, Radbruch H, Harms L, Haftmann C, et al. IL‐17 and GM‐CSF expression are antagonistically regulated by human T helper cells. Sci Transl Med. 2014;6(241):241ra80. [DOI] [PubMed] [Google Scholar]
- 39. Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, et al. IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi‐allelic RORC mutations. Science 2015;349(6248):606–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sallusto F, Cassotta A, Hoces D, Foglierini M, Lanzavecchia A. Do Memory CD4 T cells keep their cell‐type programming: plasticity versus fate commitment? T‐cell heterogeneity, plasticity, and selection in humans. Cold Spring Harb Perspect Biol. 2018;10(3):a029421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cosmi L, Liotta F, Maggi E, Romagnani S, Annunziato F. Th17 cells: new players in asthma pathogenesis. Allergy 2011;66(8):989–98. [DOI] [PubMed] [Google Scholar]
- 42. Wang YH, Voo KS, Liu B, Chen CY, Uygungil B, Spoede W, et al. A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL‐17 cytokine and promote the exacerbation of chronic allergic asthma. J Exp Med. 2010;207(11):2479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 2011;34(1):108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 2015;523(7559):221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Voo KS, Wang YH, Santori FR, Boggiano C, Wang YH, Arima K, et al. Identification of IL‐17‐producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci USA. 2009;106(12):4793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh‐hora M, Kodama T, et al. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat Med. 2014;20(1):62–8. [DOI] [PubMed] [Google Scholar]
- 47. Jung MK, Kwak JE, Shin EC. IL‐17A‐Producing Foxp3(+) Regulatory T Cells and Human Diseases. Immune Netw. 2017;17(5):276–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, et al. Single‐cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 2015;163(6):1400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stubbington MJT, Rozenblatt‐Rosen O, Regev A, Teichmann SA. Single‐cell transcriptomics to explore the immune system in health and disease. Science 2017;358(6359):58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Han A, Glanville J, Hansmann L, Davis MM. Linking T‐cell receptor sequence to functional phenotype at the single‐cell level. Nat Biotechnol. 2014;32(7):684–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim SM, Bhonsle L, Besgen P, Nickel J, Backes A, Held K, et al. Analysis of the paired TCR alpha‐ and beta‐chains of single human T cells. PLoS One 2012;7(5):e37338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Palit S, Heuser C, de Almeida GP, Theis FJ, Zielinski CE. Meeting the challenges of high‐dimensional single‐cell data analysis in immunology. Front Immunol. 2019;10:1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Becattini S, Latorre D, Mele F, Foglierini M, De Gregorio C, Cassotta A, et al. T cell immunity. Functional heterogeneity of human memory CD4(+) T cell clones primed by pathogens or vaccines. Science 2015;347(6220):400–6. [DOI] [PubMed] [Google Scholar]
- 54. Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, et al. Comprehensive assessment of T‐cell receptor beta‐chain diversity in alphabeta T cells. Blood 2009;114(19):4099–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Attaf M, Huseby E, Sewell AK. alphabeta T cell receptors as predictors of health and disease. Cell Mol Immunol. 2015;12(4):391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tait Wojno ED, Hunter CA, Stumhofer JS. The immunobiology of the interleukin‐12 family: room for discovery. Immunity 2019;50(4):851–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kobayashi M, Fitz L, Ryan M, Hewick RM, Clark SC, Chan S, et al. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J Exp Med. 1989;170(3):827–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, et al. Novel p19 protein engages IL‐12p40 to form a cytokine, IL‐23, with biological activities similar as well as distinct from IL‐12. Immunity 2000;13(5):715–25. [DOI] [PubMed] [Google Scholar]
- 59. Cosmi L, Cimaz R, Maggi L, Santarlasci V, Capone M, Borriello F, et al. Evidence of the transient nature of the Th17 phenotype of CD4+CD161+ T cells in the synovial fluid of patients with juvenile idiopathic arthritis. Arthritis Rheum. 2011;63(8):2504–15. [DOI] [PubMed] [Google Scholar]
- 60. Nistala K, Adams S, Cambrook H, Ursu S, Olivito B, de Jager W, et al. Th17 plasticity in human autoimmune arthritis is driven by the inflammatory environment. Proc Natl Acad Sci USA. 2010;107(33):14751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, et al. Induction of pathogenic TH17 cells by inducible salt‐sensing kinase SGK1. Nature 2013;496(7446):513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pflanz S, Timans JC, Cheung J, Rosales R, Kanzler H, Gilbert J, et al. IL‐27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4+ T cells. Immunity 2002;16(6):779–90. [DOI] [PubMed] [Google Scholar]
- 63. Pflanz S, Hibbert L, Mattson J, Rosales R, Vaisberg E, Bazan JF, et al. WSX‐1 and glycoprotein 130 constitute a signal‐transducing receptor for IL‐27. J Immunol. 2004;172(4):2225–31. [DOI] [PubMed] [Google Scholar]
- 64. Hibbert L, Pflanz S, De Waal MR, Kastelein RA. IL‐27 and IFN‐alpha signal via Stat1 and Stat3 and induce T‐Bet and IL‐12Rbeta2 in naive T cells. J Interferon Cytokine Res. 2003;23(9):513–22. [DOI] [PubMed] [Google Scholar]
- 65. Yoshida H, Hunter CA. The immunobiology of interleukin‐27. Annu Rev Immunol. 2015;33:417–43. [DOI] [PubMed] [Google Scholar]
- 66. Wang H, Li Z, Yang B, Yu S, Wu C. IL‐27 suppresses the production of IL‐22 in human CD4(+) T cells by inducing the expression of SOCS1. Immunol Lett. 2013;152(2):96–103. [DOI] [PubMed] [Google Scholar]
- 67. Gulen MF, Kang Z, Bulek K, Youzhong W, Kim TW, Chen Y, et al. The receptor SIGIRR suppresses Th17 cell proliferation via inhibition of the interleukin‐1 receptor pathway and mTOR kinase activation. Immunity 2010;32(1):54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Muranski P, Restifo NP. Essentials of Th17 cell commitment and plasticity. Blood 2013;121(13):2402–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, et al. The development of inflammatory T(H)‐17 cells requires interferon‐regulatory factor 4. Nat Immunol. 2007;8(9):958–66. [DOI] [PubMed] [Google Scholar]
- 70. Gattorno M, Piccini A, Lasiglie D, Tassi S, Brisca G, Carta S, et al. The pattern of response to anti‐interleukin‐1 treatment distinguishes two subsets of patients with systemic‐onset juvenile idiopathic arthritis. Arthritis Rheum. 2008;58(5):1505–15. [DOI] [PubMed] [Google Scholar]
- 71. Noster R, de Koning HD, Maier E, Prelog M, Lainka E, Zielinski CE. Dysregulation of proinflammatory versus anti‐inflammatory human TH17 cell functionalities in the autoinflammatory Schnitzler syndrome. J Allergy Clin Immunol. 2016;138(4):1161–9. [DOI] [PubMed] [Google Scholar]
- 72. Mitra S, Leonard WJ. Biology of IL‐2 and its therapeutic modulation: Mechanisms and strategies. J Leukoc Biol. 2018;103(4):643–55. [DOI] [PubMed] [Google Scholar]
- 73. Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL‐2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12(6):551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Amadi‐Obi A, Yu CR, Liu X, Mahdi RM, Clarke GL, Nussenblatt RB, et al. TH17 cells contribute to uveitis and scleritis and are expanded by IL‐2 and inhibited by IL‐27/STAT1. Nat Med. 2007;13(6):711–8. [DOI] [PubMed] [Google Scholar]
- 75. Yang L, Anderson DE, Baecher‐Allan C, Hastings WD, Bettelli E, Oukka M, et al. IL‐21 and TGF‐beta are required for differentiation of human T(H)17 cells. Nature 2008;454(7202):350–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, et al. Essential autocrine regulation by IL‐21 in the generation of inflammatory T cells. Nature 2007;448(7152):480–3. [DOI] [PubMed] [Google Scholar]
- 77. Monin L, Gaffen SL. Interleukin 17 family cytokines: signaling mechanisms, biological activities, and therapeutic implications. Cold Spring Harbor Perspectives in Biology 2018;10(4):a028522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chang SH, Dong C. Signaling of interleukin‐17 family cytokines in immunity and inflammation. Cell Signal. 2011;23(7):1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Solimani F, Pollmann R, Schmidt T, Schmidt A, Zheng X, Savai R, et al. Therapeutic targeting of Th17/Tc17 cells leads to clinical improvement of lichen planus. Front Immunol. 2019;10:1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Braun J, Baraliakos X, Deodhar A, Baeten D, Sieper J, Emery P, et al. Effect of secukinumab on clinical and radiographic outcomes in ankylosing spondylitis: 2‐year results from the randomised phase III MEASURE 1 study. Ann Rheum Dis. 2017;76(6):1070–7. [DOI] [PubMed] [Google Scholar]
- 81. Langley RG, Elewski BE, Lebwohl M, Reich K, Griffiths CE, Papp K, et al. Secukinumab in plaque psoriasis–results of two phase 3 trials. N Engl J Med. 2014;371(4):326–38. [DOI] [PubMed] [Google Scholar]
- 82. Mease P, van der Heijde D, Landewe R, Mpofu S, Rahman P, Tahir H, et al. Secukinumab improves active psoriatic arthritis symptoms and inhibits radiographic progression: primary results from the randomised, double‐blind, phase III FUTURE 5 study. Ann Rheum Dis. 2018;77(6):890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Papadakis KA, Targan SR. Tumor necrosis factor: biology and therapeutic inhibitors. Gastroenterology 2000;119(4):1148–57. [DOI] [PubMed] [Google Scholar]
- 84. Breese EJ, Michie CA, Nicholls SW, Murch SH, Williams CB, Domizio P, et al. Tumor necrosis factor alpha‐producing cells in the intestinal mucosa of children with inflammatory bowel disease. Gastroenterology 1994;106(6):1455–66. [DOI] [PubMed] [Google Scholar]
- 85. Di Giovine FS, Nuki G, Duff GW. Tumour necrosis factor in synovial exudates. Ann Rheum Dis. 1988;47(9):768–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kristensen M, Chu CQ, Eedy DJ, Feldmann M, Brennan FM, Breathnach SM. Localization of tumour necrosis factor‐alpha (TNF‐alpha) and its receptors in normal and psoriatic skin: epidermal cells express the 55‐kD but not the 75‐kD TNF receptor. Clin Exp Immunol. 1993;94(2):354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dumoutier L, Louahed J, Renauld JC. Cloning and characterization of IL‐10‐related T cell‐derived inducible factor (IL‐TIF), a novel cytokine structurally related to IL‐10 and inducible by IL‐9. J Immunol. 2000;164(4):1814–9. [DOI] [PubMed] [Google Scholar]
- 88. Andoh A, Zhang Z, Inatomi O, Fujino S, Deguchi Y, Araki Y, et al. Interleukin‐22, a member of the IL‐10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology 2005;129(3):969–84. [DOI] [PubMed] [Google Scholar]
- 89. Brand S, Dambacher J, Beigel F, Zitzmann K, Heeg MH, Weiss TS, et al. IL‐22‐mediated liver cell regeneration is abrogated by SOCS‐1/3 overexpression in vitro. Am J Physiol Gastrointest Liver Physiol. 2007;292(4):G1019–G1028. [DOI] [PubMed] [Google Scholar]
- 90. Schmechel S, Konrad A, Diegelmann J, Glas J, Wetzke M, Paschos E, et al. Linking genetic susceptibility to Crohn's disease with Th17 cell function: IL‐22 serum levels are increased in Crohn's disease and correlate with disease activity and IL23R genotype status. Inflamm Bowel Dis. 2008;14(2):204–12. [DOI] [PubMed] [Google Scholar]
- 91. Sarin R, Wu X, Abraham C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T‐cell functional responses. Proc Natl Acad Sci USA. 2011;108(23):9560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Boniface K, Bernard FX, Garcia M, Gurney AL, Lecron JC, Morel F. IL‐22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol. 2005;174(6):3695–702. [DOI] [PubMed] [Google Scholar]
- 93. Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi‐Joannopoulos K, Collins M, et al. Interleukin (IL)‐22 and IL‐17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203(10):2271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Demetri GD, Antman KH. Granulocyte‐macrophage colony‐stimulating factor (GM‐CSF): preclinical and clinical investigations. Semin Oncol. 1992;19(4):362–85. [PubMed] [Google Scholar]
- 95. Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte‐macrophage colony‐stimulating factor (CSF) and macrophage CSF‐dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol. 2007;178(8):5245–52. [DOI] [PubMed] [Google Scholar]
- 96. Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1‐TH17 responses. Nat Immunol. 2011;12(3):231–8. [DOI] [PubMed] [Google Scholar]
- 97. Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, et al. Human IL‐23‐producing type 1 macrophages promote but IL‐10‐producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci USA. 2004;101(13):4560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, et al. RORgammat drives production of the cytokine GM‐CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12(6):560–7. [DOI] [PubMed] [Google Scholar]
- 99. El‐Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, et al. The encephalitogenicity of T(H)17 cells is dependent on IL‐1‐ and IL‐23‐induced production of the cytokine GM‐CSF. Nat Immunol. 2011;12(6):568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sheng W, Yang F, Zhou Y, Yang H, Low PY, Kemeny DM, et al. STAT5 programs a distinct subset of GM‐CSF‐producing T helper cells that is essential for autoimmune neuroinflammation. Cell Res. 2014;24(12):1387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Behrens F, Tak PP, Ostergaard M, Stoilov R, Wiland P, Huizinga TW, et al. MOR103, a human monoclonal antibody to granulocyte‐macrophage colony‐stimulating factor, in the treatment of patients with moderate rheumatoid arthritis: results of a phase Ib/IIa randomised, double‐blind, placebo‐controlled, dose‐escalation trial. Ann Rheum Dis. 2015;74(6):1058–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Constantinescu CS, Asher A, Fryze W, Kozubski W, Wagner F, Aram J, et al. Randomized phase 1b trial of MOR103, a human antibody to GM‐CSF, in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. 2015;2(4):e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Papp KA, Gooderham M, Jenkins R, Vender R, Szepietowski JC, Wagner T, et al. Granulocyte‐macrophage colony‐stimulating factor (GM‐CSF) as a therapeutic target in psoriasis: randomized, controlled investigation using namilumab, a specific human anti‐GM‐CSF monoclonal antibody. Br J Dermatol. 2019;180(6):1352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rengarajan J, Szabo SJ, Glimcher LH. Transcriptional regulation of Th1/Th2 polarization. Immunol Today. 2000;21(10):479–83. [DOI] [PubMed] [Google Scholar]
- 105. Bonelli M, Shih HY, Hirahara K, Singelton K, Laurence A, Poholek A, et al. Helper T cell plasticity: impact of extrinsic and intrinsic signals on transcriptomes and epigenomes. Curr Top Microbiol Immunol. 2014;381:279–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, et al. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 2013;496(7446):518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Matthias J, Maul J, Noster R, Meinl H, Chao YY, Gerstenberg H, et al. Sodium chloride is an ionic checkpoint for human TH2 cells and shapes the atopic skin microenvironment. Sci Transl Med. 2019;11(480):eaau0683. [DOI] [PubMed] [Google Scholar]
- 108. Matthias J, Heink S, Picard F, Zeitrag J, Kolz A, Chao YY, et al. Salt generates antiinflammatory Th17 cells but amplifies pathogenicity in proinflammatory cytokine microenvironments. J Clin Invest. 2020;130(9):4587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Volchenkov R, Nygaard V, Sener Z, Skalhegg BS. Th17 polarization under hypoxia results in increased IL‐10 production in a pathogen‐independent manner. Front Immunol. 2017;8:698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Geltink RIK, Kyle RL, Pearce EL. Unraveling the complex interplay between T cell metabolism and function. Annu Rev Immunol. 2018;36:461–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell 2017;170(4):605–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, et al. PI3K‐Akt‐mTORC1‐S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORgamma. Cell Rep. 2012;1(4):360–73. [DOI] [PubMed] [Google Scholar]
- 113. Wei J, Raynor J, Nguyen TL, Chi H. Nutrient and metabolic sensing in T cell responses. Front Immunol. 2017;8:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. de Oliveira BV, Dos Santos FA, Degasperi GR. Deciphering targets of Th17 cells fate: From metabolism to nuclear receptors. Scand J Immunol. 2019;90(4):e12793. [DOI] [PubMed] [Google Scholar]
- 115. Omenetti S, Bussi C, Metidji A, Iseppon A, Lee S, Tolaini M, et al. The intestine harbors functionally distinct homeostatic tissue‐resident and inflammatory Th17 cells. Immunity 2019;51(1):77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Karmaus PWF, Chen X, Lim SA, Herrada AA, Nguyen TM, Xu B, et al. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 2019;565(7737):101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 2014;40(5):692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase‐dependent metabolism. Cell 2018;175(7):1780–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kaufmann U, Kahlfuss S, Yang J, Ivanova E, Koralov SB, Feske S. Calcium signaling controls pathogenic Th17 cell‐mediated inflammation by regulating mitochondrial function. Cell Metab. 2019;29(5):1104–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Pan Y, Kupper TS. Metabolic reprogramming and longevity of tissue‐resident memory T cells. Front Immunol. 2018;9:1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Morianos I, Trochoutsou AI, Papadopoulou G, Semitekolou M, Banos A, Konstantopoulos D, et al. Activin‐A limits Th17 pathogenicity and autoimmune neuroinflammation via CD39 and CD73 ectonucleotidases and Hif1‐alpha‐dependent pathways. Proc Natl Acad Sci USA. 2020;117(22):12269–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. de Wit J, Al‐Mossawi MH, Huhn MH, Arancibia‐Carcamo CV, Doig K, Kendrick B, et al. RORgammat inhibitors suppress T(H)17 responses in inflammatory arthritis and inflammatory bowel disease. J Allergy Clin Immunol. 2016;137(3):960–3. [DOI] [PubMed] [Google Scholar]
- 123. Sun M, He C, Chen L, Yang W, Wu W, Chen F, et al. RORgammat represses IL‐10 production in Th17 cells to maintain their pathogenicity in inducing intestinal inflammation. J Immunol. 2019;202(1):79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Capone A, Volpe E. Transcriptional regulators of T helper 17 cell differentiation in health and autoimmune diseases. Front Immunol. 2020;11:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Cohen CJ, Crome SQ, MacDonald KG, Dai EL, Mager DL, Levings MK. Human Th1 and Th17 cells exhibit epigenetic stability at signature cytokine and transcription factor loci. J Immunol. 2011;187(11):5615–26. [DOI] [PubMed] [Google Scholar]
- 126. Mazzoni A, Santarlasci V, Maggi L, Capone M, Rossi MC, Querci V, et al. Demethylation of the RORC2 and IL17A in human CD4+ T lymphocytes defines Th17 origin of nonclassic Th1 cells. J Immunol. 2015;194(7):3116–26. [DOI] [PubMed] [Google Scholar]
- 127. Mazzoni A, Maggi L, Siracusa F, Ramazzotti M, Rossi MC, Santarlasci V, et al. Eomes controls the development of Th17‐derived (non‐classic) Th1 cells during chronic inflammation. Eur J Immunol. 2019;49(1):79–95. [DOI] [PubMed] [Google Scholar]
- 128. Wei G, Wei L, Zhu J, Zang C, Hu‐Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009;30(1):155–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cribbs AP, Terlecki‐Zaniewicz S, Philpott M, Baardman J, Ahern D, Lindow M, et al. Histone H3K27me3 demethylases regulate human Th17 cell development and effector functions by impacting on metabolism. Proc Natl Acad Sci USA. 2020;117(11):6056–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Dees C, Potter S, Zhang Y, Bergmann C, Zhou X, Luber M, et al. TGF‐beta‐induced epigenetic deregulation of SOCS3 facilitates STAT3 signaling to promote fibrosis. J Clin Invest. 2020;130(5):2347–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Guery L, Hugues S. Th17 Cell Plasticity and Functions in Cancer Immunity. Biomed Res Int. 2015;2015:314620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Su X, Ye J, Hsueh EC, Zhang Y, Hoft DF, Peng G. Tumor microenvironments direct the recruitment and expansion of human Th17 cells. J Immunol. 2010;184(3):1630–41. [DOI] [PubMed] [Google Scholar]
- 133. Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, et al. Control of T(H)17/T(reg) balance by hypoxia‐inducible factor 1. Cell 2011;146(5):772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Bailey SR, Nelson MH, Himes RA, Li Z, Mehrotra S, Paulos CM. Th17 cells in cancer: the ultimate identity crisis. Front Immunol. 2014;5:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ye J, Livergood RS, Peng G. The role and regulation of human Th17 cells in tumor immunity. Am J Pathol. 2013;182(1):10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chang SH, Mirabolfathinejad SG, Katta H, Cumpian AM, Gong L, Caetano MS, et al. T helper 17 cells play a critical pathogenic role in lung cancer. Proc Natl Acad Sci USA. 2014;111(15):5664–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Kryczek I, Wei S, Szeliga W, Vatan L, Zou W. Endogenous IL‐17 contributes to reduced tumor growth and metastasis. Blood 2009;114(2):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kryczek I, Banerjee M, Cheng P, Vatan L, Szeliga W, Wei S, et al. Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood 2009;114(6):1141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71(4):1263–71. [DOI] [PubMed] [Google Scholar]
- 140. Amicarella F, Muraro MG, Hirt C, Cremonesi E, Padovan E, Mele V, et al. Dual role of tumour‐infiltrating T helper 17 cells in human colorectal cancer. Gut 2017;66(4):692–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer 2020;20(11):662–80. [DOI] [PubMed] [Google Scholar]
- 142. Han Y, Ye A, Bi L, Wu J, Yu K, Zhang S. Th17 cells and interleukin‐17 increase with poor prognosis in patients with acute myeloid leukemia. Cancer Sci. 2014;105(8):933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wu C, Wang S, Wang F, Chen Q, Peng S, Zhang Y, et al. Increased frequencies of T helper type 17 cells in the peripheral blood of patients with acute myeloid leukaemia. Clin Exp Immunol. 2009;158(2):199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Prabhala RH, Pelluru D, Fulciniti M, Prabhala HK, Nanjappa P, Song W, et al. Elevated IL‐17 produced by TH17 cells promotes myeloma cell growth and inhibits immune function in multiple myeloma. Blood 2010;115(26):5385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Brenne AT, Ro TB, Waage A, Sundan A, Borset M, Hjorth‐Hansen H. Interleukin‐21 is a growth and survival factor for human myeloma cells. Blood 2002;99(10):3756–62. [DOI] [PubMed] [Google Scholar]
- 146. Yang ZZ, Novak AJ, Ziesmer SC, Witzig TE, Ansell SM. Malignant B cells skew the balance of regulatory T cells and TH17 cells in B‐cell non‐Hodgkin's lymphoma. Cancer Res. 2009;69(13):5522–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Dulos J, Carven GJ, van Boxtel SJ, Evers S, Driessen‐Engels LJ, Hobo W, et al. PD‐1 blockade augments Th1 and Th17 and suppresses Th2 responses in peripheral blood from patients with prostate and advanced melanoma cancer. J Immunother. 2012;35(2):169–78. [DOI] [PubMed] [Google Scholar]
- 148. Krieg C, Nowicka M, Guglietta S, Schindler S, Hartmann FJ, Weber LM, et al. High‐dimensional single‐cell analysis predicts response to anti‐PD‐1 immunotherapy. Nat Med. 2018;24(2):144–53. [DOI] [PubMed] [Google Scholar]
- 149. Martin‐Orozco N, Muranski P, Chung Y, Yang XO, Yamazaki T, Lu S, et al. T helper 17 cells promote cytotoxic T cell activation in tumor immunity. Immunity 2009;31(5):787–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Muranski P, Boni A, Antony PA, Cassard L, Irvine KR, Kaiser A, et al. Tumor‐specific Th17‐polarized cells eradicate large established melanoma. Blood 2008;112(2):362–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Kaiga T, Sato M, Kaneda H, Iwakura Y, Takayama T, Tahara H. Systemic administration of IL‐23 induces potent antitumor immunity primarily mediated through Th1‐type response in association with the endogenously expressed IL‐12. J Immunol. 2007;178(12):7571–80. [DOI] [PubMed] [Google Scholar]
- 152. Ma X, Shou P, Smith C, Chen Y, Du H, Sun C, et al. Interleukin‐23 engineering improves CAR T cell function in solid tumors. Nat Biotechnol. 2020;38(4):448–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Bresnihan B, Newmark R, Robbins S, Genant HK. Effects of anakinra monotherapy on joint damage in patients with rheumatoid arthritis. Extension of a 24‐week randomized, placebo‐controlled trial. J Rheumatol. 2004;31(6):1103–11. [PubMed] [Google Scholar]
- 154. Droeser RA, Guth U, Eppenberger‐Castori S, Stadlmann S, Hirt C, Terracciano L, et al. High IL‐17‐positive tumor immune cell infiltration is indicative for chemosensitivity of ovarian carcinoma. J Cancer Res Clin Oncol. 2013;139(8):1295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Miyahara Y, Odunsi K, Chen W, Peng G, Matsuzaki J, Wang RF. Generation and regulation of human CD4+ IL‐17‐producing T cells in ovarian cancer. Proc Natl Acad Sci USA. 2008;105(40):15505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Lin Y, Xu J, Su H, Zhong W, Yuan Y, Yu Z, et al. Interleukin‐17 is a favorable prognostic marker for colorectal cancer. Clin Transl Oncol. 2015;17(1):50–6. [DOI] [PubMed] [Google Scholar]
- 157. Tseng JY, Yang CY, Liang SC, Liu RS, Yang SH, Lin JK, et al. Interleukin‐17A modulates circulating tumor cells in tumor draining vein of colorectal cancers and affects metastases. Clin Cancer Res. 2014;20(11):2885–97. [DOI] [PubMed] [Google Scholar]
- 158. Iida T, Iwahashi M, Katsuda M, Ishida K, Nakamori M, Nakamura M, et al. Tumor‐infiltrating CD4+ Th17 cells produce IL‐17 in tumor microenvironment and promote tumor progression in human gastric cancer. Oncol Rep. 2011;25(5):1271–7. [DOI] [PubMed] [Google Scholar]
- 159. Li Q, Li Q, Chen J, Liu Y, Zhao X, Tan B, et al. Prevalence of Th17 and Treg cells in gastric cancer patients and its correlation with clinical parameters. Oncol Rep. 2013;30(3):1215–22. [DOI] [PubMed] [Google Scholar]
- 160. Su Z, Sun Y, Zhu H, Liu Y, Lin X, Shen H, et al. Th17 cell expansion in gastric cancer may contribute to cancer development and metastasis. Immunol Res. 2014;58(1):118–24. [DOI] [PubMed] [Google Scholar]
- 161. Alvarez E, Moga E, Barquinero J, Sierra J, Briones J. Dendritic and tumor cell fusions transduced with adenovirus encoding CD40L eradicate B‐cell lymphoma and induce a Th17‐type response. Gene Ther. 2010;17(4):469–77. [DOI] [PubMed] [Google Scholar]
- 162. Duffield AS, Ascierto ML, Anders RA, Taube JM, Meeker AK, Chen S, et al. Th17 immune microenvironment in Epstein‐Barr virus‐negative Hodgkin lymphoma: implications for immunotherapy. Blood Adv. 2017;1(17):1324–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Dhodapkar KM, Barbuto S, Matthews P, Kukreja A, Mazumder A, Vesole D, et al. Dendritic cells mediate the induction of polyfunctional human IL17‐producing cells (Th17‐1 cells) enriched in the bone marrow of patients with myeloma. Blood 2008;112(7):2878–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Musuraca G, De Matteis S, Napolitano R, Papayannidis C, Guadagnuolo V, Fabbri F, et al. IL‐17/IL‐10 double‐producing T cells: new link between infections, immunosuppression and acute myeloid leukemia. J Transl Med. 2015;13:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


