Summary
Dendritic cells (DCs) play an important role in linking innate and adaptive immunity. DCs can sense endogenous and exogenous antigens and present those antigens to T cells to induce an immune response or immune tolerance. During activation, alternative splicing (AS) in DCs is dramatically changed to induce cytokine secretion and upregulation of surface marker expression. PTBP1, an RNA‐binding protein, is essential in alternative splicing, but the function of PTBP1 in DCs is unknown. Here, we found that a specific deficiency of Ptbp1 in DCs could increase MHC II expression and perturb T‐cell homeostasis without affecting DC development. Functionally, Ptbp1 deletion in DCs could enhance antitumour immunity and asthma exacerbation. Mechanistically, we found that Pkm alternative splicing and a subset of Ifn response genes could be regulated by PTBP1. These findings revealed the function of PTBP1 in DCs and indicated that PTBP1 might be a novel therapeutic target for antitumour treatment.
Keywords: alternative splicing, asthma, dendritic cells, RNA‐binding protein, tumour
Ptbp1 deletion in DCs could enhance antitumour immunity and asthma exacerbation. Pkm alternative splicing and a subset of Ifn response genes could be regulated by PTBP1.
Abbreviations
- AS
alternative splicing
- DCs
dendritic cells
- DEGs
differentially expressed genes
- LPS
lipopolysaccharide
- RIP
RNA immunoprecipitation
INTRODUCTION
Dendritic cells (DCs) are efficient antigen‐presenting cells that can initiate adaptive immune responses. Indeed, DCs are the only kind of cells that can present antigens to resting T cells. 1 , 2 , 3 Thus, DCs are crucial for both adaptive immunity and immune tolerance in body homeostasis. Due to the essential role of DCs in linking innate and adaptive immunity, inappropriate activation of DCs can cause autoimmune diseases. 4 , 5 DCs can sense environmental antigens using pattern recognition receptors, such as Toll‐like receptors, which results in upregulation of the expression of MHC II, CD80 and CD86, and then migrate to lymphoid organs to activate T cells. 6 , 7 In the steady state, DCs can reduce the expression levels of surface makers and increase the production of regulatory T cells to maintain peripheral immune tolerance. The detailed mechanisms involving DCs in mediating immune tolerance have not been completely elucidated. 8 , 9 , 10
Alternative splicing of pre‐mRNA is of great importance for increasing gene expression diversity. 11 , 12 Splicing is precisely regulated by the binding of splicing factors to variable RNA exons or introns. 13 Thus, mutation of splicing factors can induce developmental abnormalities and diseases. 14 , 15 , 16 The temporal and spatial regulation of splicing is vital for the function and differentiation of immune cells. 17 , 18 DCs are important initiators of innate and adaptive immunity, and the transcriptional level in DCs changes dramatically to carry out an effective immune response. During activation, T cells undergo selective gene expression, which contributes to functional changes in their receptors, cytokines and ligands. 19 Moreover, the spliced genes in activated T cells have variable functions, especially those associated with cell cycle regulation. 20 Alternative splicing of CD45 induces the formation of homodimers on the cell surface, which leads to decreased signal transduction through TCRs. 21 By analysing the differentiation and activation of primary macrophages and THP‐1 cells using RNA‐Seq, Liu et al 22 demonstrated thousands of AS changes under stressful conditions. In tumour cells, Hu et al 23 found that A‐kinase anchor protein 8 could work as an RNA‐binding protein and regulate breast cancer metastasis. PTBP1, a member of a class of RNA‐binding proteins with pleiotropic functions, plays an important role in pre‐mRNA splicing. PTBP1 can interact with RNA via four RNA‐binding domains to regulate numerous alternative exons in mRNA transcripts. 24 PTBP1 can act in concert with c‐MYC to ensure the selection of B‐cell clones with the highest affinity for an antigen by promoting the proliferation of germinal centre B cells. 25 PTBP1 can also modulate MCL1 expression to regulate cellular apoptosis in response to antitubulin chemotherapeutics. 26 , 27 In drug‐resistant pancreatic ductal adenocarcinoma cells, PTBP1 expression is upregulated, and PTBP1 promotes PKM2 splicing. 28 PKM1 and PKM2 are two isoforms of the PKM pre‐mRNA produced by alternative splicing. PKM1 is inclusion of exon 9, and PKM2 is inclusion of exon 10. The main function of PKM2 is to promote glycolysis and tumorigenesis, while that of PKM1 is to promote oxidative phosphorylation. 29 , 30 More recent studies using conditional knockout mice have found that PKM2 deficiency accelerates tumour growth. 31 A new mouse model expressing only a specific pyruvate kinase isoform showed that the PKM isoform PKM1 confers a metabolic advantage and promotes KRASG12D‐induced or carcinogen‐initiated tumour growth. 32 Although the correlations between splicing factors and diseases have been reported, the mechanism of alternative splicing remains to be clarified.
In this study, we found that the RNA‐binding protein PTBP1 could regulate Pkm alternative splicing in DCs. Specific deletion of Ptbp1 in DCs could lead to enhanced MHC II expression and an increased population of activated T cells without impairing DC development. Furthermore, we found that PTBP1 deficiency in DCs could enhance DC function, which strengthened antitumour immunity. Additionally, in an OVA‐induced asthma model, PTBP1 deficiency increased the infiltration of immune cells compared with WT mice. These results show that PTBP1 functions as a regulator of DC function and that PTBP1 deficiency can disturb immunologic homeostasis.
MATERIALS AND METHODS
Mice
Ptbp1‐floxed mice were generated using the LoxP targeting system. Ptbp1‐floxed mice were crossed with CD11c‐Cre mice (The Jackson Laboratory) to produce DC‐specific deletion of Ptbp1. Mice were maintained in a specific pathogen‐free environment. For tumour models, B16‐F10 melanoma cells were cultured in DMEM supplemented with 10% FBS. Tumour cells were injected subcutaneously into eight‐week‐old WT and Ptbp1‐cKO mice (5 × 105 cells per mouse). The challenged mice were monitored for tumour size and the growth rate for 3 weeks. To establish an OVA‐induced asthma model, eight‐week‐old WT and Ptbp1‐cKO mice received intraperitoneal injections of OVA and Al(OH)3 three times over 3 weeks. In the fourth week, the mice were treated with inhaled 1% OVA (50 μL) three times.
Histology
Freshly acquired tissue samples from WT and Ptbp1‐cKO mice were fixed in 10% neutral buffered formalin. The tissue samples were embedded in paraffin and sectioned. The sections were stained with haematoxylin–eosin.
Intracellular cytokine staining
Mononuclear cells were acquired from the spleen and tumours of the indicated mice. These cells were cultured in PMA plus ionomycin in the presence of monensin. Then, intracellular cytokine staining was performed to identify T cells producing IFN‐γ by flow cytometric analysis.
ELISA
Cytokine levels in BALF were determined using ELISA kits (Elabscience Biotechnology Co., Ltd), as previously described. 33
Flow cytometry
The primary antibodies used for flow cytometry were as follows: anti‐F4/80 (clone BM8, BioLegend), anti‐CD11c (clone N418, BioLegend), anti‐MHC II (clone 10‐3.6, BioLegend), anti‐PDCA1 (clone 927, BioLegend), anti‐Gr1 (clone RB6‐8C5, BioLegend), anti‐CD4 (clone GK1.5, BioLegend), anti‐CD8a (clone 53‐6.7, BioLegend), anti‐IFN‐γ (clone XMG1.2, BioLegend), anti‐CD44 (clone IM7, BioLegend), anti‐CD80 (clone 16‐10A1, BioLegend), anti‐CD62L (clone MEL‐14, BioLegend), anti‐CD11b (clone M1/70, BioLegend), anti‐CD86 (clone GL‐1, BioLegend), anti‐I‐A/I‐E (clone M5/114.15.2, BioLegend), DC Marker (clone 33D1, BioLegend), anti‐Siglec‐H (clone 551, BioLegend), anti‐CD3ε (clone 145‐2C11, BioLegend), anti‐CD19 (clone 6D5, BioLegend), anti‐Ly‐6G (clone 1A8, BioLegend) and anti‐CD335 (clone 29A1.4, BioLegend). Mononuclear cells were isolated from the spleen and draining lymph nodes. A total of 1 × 106 cells were incubated with 1·5 mg mL−1 antibody for 30 min at 4°C. For intracellular staining, cells were fixed and permeabilized using fixation/permeabilization buffer (eBioscience) for 30 min at 4°C. The cells were then washed and stained for 30 min at 4°C with antibodies diluted with permeabilization buffer. The cell suspensions were analysed on an LSRII flow cytometer (BD Biosciences), and the data were analysed using FlowJo.
Western blotting
CD11c+ dendritic cells were purified from spleens and lysed in Laemmli sample buffer (cat 161‐0737, Bio‐Rad). Then, the lysates were subjected to gel electrophoresis (SDS‐PAGE) and electrotransferred to a polyvinylidene difluoride membrane (GE Healthcare). The transferred membranes were blocked with 5% fat‐free skim milk for 2 h at room temperature. The membranes were then incubated with a primary antibody at 4°C overnight. After washing three times with PBST (PBS with 0·001% Tween‐20), horseradish peroxidase‐conjugated secondary antibodies were added and incubated at room temperature for 1 h. After washing three times with PBST, the signals were detected using an ECL kit (cat WP20005, Invitrogen).
PTBP1 RIP
RIP is an antibody‐based technique used to map in vivo RNA–protein interactions. The experiments were conducted by using the Magna RIP™ RNA‐Binding Protein Immunoprecipitation Kit (Sigma). Briefly, 30 × 106 DC2.4 cells were lysed in RIP lysis buffer (cat cs203176) with 0·5 μL protease inhibitor cocktail and 0·25 μL RNase inhibitor. Magnetic beads were prepared for immunoprecipitation and purification of RNA for qPCR.
Isolation of Spleen cDCs
Spleens from WT or Ptbp1‐cKO mice were digested with 0·5 mg/mL Liberase (cat 423610, Roche) and 100 U/mL DNaseⅠ (cat D8071, Solarbio) in RPMI‐1640 medium at 37°C for 45 min. The red blood cells were removed with red blood cell lysis buffer. The mononuclear cells were counted and incubated with anti‐CD11c and anti‐MHC II antibodies at 4°C for 30 min. After the incubation, the cells were washed with PBS. Then, CD11c and MHC II double‐positive cells were sorted on a new FACSAria III (BD).
RNA extraction and quantitative real‐time PCR (qRT‐PCR)
Total RNA was extracted from DCs with TRIzol™ (cat 15596026, Invitrogen) and reverse‐transcribed using TransScript II One‐Step gDNA Removal and cDNA Synthesis SuperMix (cat AH311‐03, TransGen Biotech). qRT‐PCR was performed using a SYBR Green Master Mix Kit (cat A25742, Thermo Fisher Scientific), and the assay was conducted with the QuantStudio® 5 Real‐Time PCR System (Life Technologies). The primers used for this study are shown in Table 1.
Table 1.
Gene‐specific primers used for quantitative PCR
| Gene | Primer |
|---|---|
| Ifnα‐F | 5′‐TGACCTCAAAGCCTGTGTGATG‐3′ |
| Ifnα‐R | 5′‐AAGTATTTCCTCACAGCCAGCAG‐3′ |
| Ifnβ‐F | 5′‐AGCTCCAAGAAAGGACGAACAT‐3′ |
| Ifnβ‐R | 5′‐GCCCTGTAGGTGAGGTTGATCT‐3′ |
| Irf7‐F | 5′‐CAGCGAGTGCTGTTTGGAGAC‐3′ |
| Irf7‐R | 5′‐AAGTTCGTACACCTTATGCGG‐3′ |
| Irf1‐F | 5′‐CCCACAGAAGAGCATAGCAC‐3′ |
| Irf1‐R | 5′‐AGCAGTTCTTTGGGAATAGG‐3′ |
| Actin‐F | 5′‐CGTGAAAAGATGACCCAGATCA‐3′ |
| Actin‐R | 5′‐CACAGCCTGGATGGCTACGT‐3′ |
| Ifi202b‐F | 5′‐CCGGGAAACACCATTGCTTT‐3′ |
| Ifi202b‐R | 5′‐GGCTCTTCACCTCAGACACG‐3′ |
| Slfn8‐F | 5′CCCAGATTTCTCACGCCAGT‐3′ |
| Slfn8‐R | 5′‐GGATGCTGCATGGAGGGTTA‐3′ |
| Oas2‐F | 5′‐TTTACCCCCAAAGTACGCCC‐3′ |
| Oas2‐R | 5′‐ATGCAGAGCTGCCGGTATTT‐3′ |
| Oas3‐F | 5′‐GAACAGCAAGGTGGCCTTTG‐3′ |
| Oas3‐R | 5′‐CCTCAGGAGTCCTTGTGCAG‐3′ |
| Il10‐F | 5′‐CCAGAGCCACATGCTCCTAGA‐3′ |
| Il10‐R | 5′‐GGTCCTTTGTTTGAAAGAAAGTCTTC‐3′ |
| Il6‐F | 5′‐TGAACAACGATGATGCACTTGC‐3′ |
| Il6‐R | 5′‐GCTATGGTACTCCAGAAGACC‐3′ |
| Tnf‐F | 5′‐CATCTTCTCAAAATTCGAGTGACAA‐3′ |
| Tnf‐R | 5′‐CCAGCTGCTCCTCCACTTG‐3′ |
| Il23a‐F | 5′‐CGTATCCAGTGTGAAGATGGTT‐3′ |
| Il23a‐R | 5′‐CTATCAGGGAGTAGAGCAGGCT‐3′ |
| Il12a‐F | 5′‐ACTAGAGAGACTTCTTCCACAACAA‐3′ |
| Il12a‐R | 5′‐CACAGGGTCATCATCAAAGAC‐3′ |
| Il12β‐F | 5′‐GGAGACACCAGCAAAACGAT‐3′ |
| Il12β‐R | 5′‐TCCAGATTCAGACTCCAGGG‐3′ |
| Il1β‐F | 5′‐AAGCCTCGTGCTGTCGGACC‐3′ |
| Il1β‐R | 5′‐TGAGGCCCAAGGCCACAGGT‐3′ |
| MusD‐F | 5′‐GATTGGTGGAAGTTTAGCTAGCAT‐3′ |
| MusD‐R | 5′‐TAGCATTCTCATAAGCCAATTGCAT‐3′ |
| IAP Pol‐F | 5′‐CTTGCCCTTAAAGGTCTAAAAGCA‐3′ |
| IAP Pol‐R | 5′‐GCGGTATAAGGTACAATTAAAAGATATGG‐3′ |
| Line1‐F | 5′‐TTTGGGACACAATGAAAGCA‐3′ |
| Line1‐R | 5′‐CTGCCGTCTACTCCTCTTGG‐3′ |
| MuERV‐L‐F | 5′‐TTTCTCAAGGCCCACCAATAGT‐3′ |
| MuERV‐L‐R | 5′‐GACACCTTTTTTAACTATGCGAGCT‐3′ |
| Mda5‐F | 5′‐CTACGCACTTTCCCAGTGGAT‐3′ |
| Mda5‐R | 5′‐TGTTCAGTCTGAGTCATGGGC‐3′ |
Library construction and processing of RNA‐Seq data
CD11c and MHC II double‐positive DCs from the spleen of CD11c Cre+/− Ptbp1 fl/fl cKO and CD11c Cre−/− Ptbp1 fl/fl control mice were sorted by FACS. Total RNA was extracted using TRIzol (cat 15596026, Invitrogen). Library construction and data processing were performed by Novogene (Beijing, China), and the libraries were then sequenced with an Illumina HiSeq platform that generated 150‐bp paired‐end reads. Quality control of raw sequence data was performed with FastQC (v0.11.8; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and trimming of raw sequence data was carried out with Trim Galore (v0.3.6; http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with default parameters. Then, the clean reads were aligned to the mouse genome GRCm38 using HISAT2, 34 and the annotation GTF file GENCODE.vM17.gtf was downloaded from https://www.gencodegenes.org/. After that, StringTie was used to quantify gene and transcript isoform expression based on the aligned reads. 34
Identification of differentially expressed genes (DEGs) and gene functional annotation analysis
A gene count matrix was used as the input data to identify DEGs between WT and cKO mice with the Bioconductor package DESeq2 (v 1.24.0). 35 The DESeq2 results were considered for only genes expressed as a minimum level of 1 FPKM in any of the samples. p‐values were adjusted using the Benjamini and Hochberg method, and the threshold was set at padj < 0·05. Then, significant DEGs were used to perform GO enrichment analysis with the Bioconductor package clusterProfiler (v3.12.0) and DAVID (https://david.ncifcrf.gov/).
Identification of differential alternative splicing
Alternative splicing events were performed by rMATS (v4.0.2) with default parameters. 36 The spliced events with an FDR < 0·05 and inclusion level difference greater than 0·2 were identified as significant differentially alternatively spliced events.
Statistical analysis
Statistical significance was calculated with a 2‐tailed Student’s t test. Differences with a p value < 0·05 were considered statistically significant.
RESULTS
Ptbp1 deletion enhances MHC II expression in DCs
To assess the functions of PTBP1 in DCs, we generated Ptbp1‐cKO mice by crossing Ptbp1 flox/flox mice with CD11c‐Cre mice (Figure S1A). The protein expression of PTBP1 was lost in the cKO mice (Figure S1B). We then examined the role of PTBP1 in the development of other lineages. We found that the frequencies of DCs, neutrophils and macrophages in the bone marrow showed no differences between WT and cKO mice (Figure 1A). The frequencies of conventional DCs and plasmacytoid DCs (pDCs) in the spleen were also comparable between WT and cKO mice (Figure 1B,C). DCs have been recently classified into cDC1 and myeloid DCs (cDC2), 37 we examined the spleen and found no obvious differences between WT and cKO mice (Figure 1D and Figure S1C). We then detected surface marker expression and found that MHC II expression increased with Ptbp1 deletion in DCs (Figure 1E). Other surface markers, such as CD80 and CD86, exhibited little change between WT and cKO mice (Figure 1F,G). Thus, these data indicated that PTBP1 was dispensable for DC development but did change MHC II expression.
Figure 1.
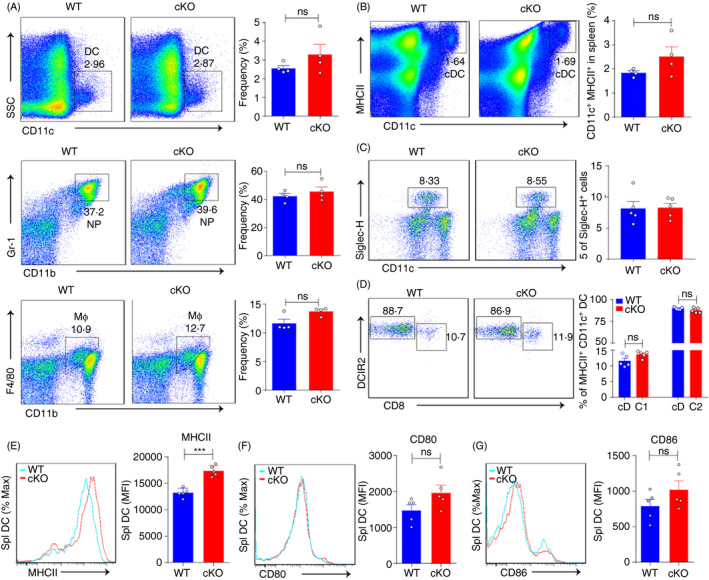
Ptbp1 deletion enhances MHC II expression in DCs. (A) Flow cytometric analysis of the frequencies of DCs, neutrophils and macrophages in the bone marrow of WT and cKO mice. (B‐D) Flow cytometric analysis of the frequencies of conventional DCs, plasmacytoid DCs (pDCs), cDC1 and cDC2 in the spleen of WT and cKO mice. (E‐G) Flow cytometric analysis of MHC II, CD80 and CD86 expression in DCs from the spleen of WT and cKO mice. MFI, mean fluorescence intensity. Data are representative of at least three independent experiments. All data are shown as the mean ± SEM; * indicates p < 0·05, ** indicates p < 0·01, n.s., not significant. Statistical significance was evaluated by Student’s t tests.
Ptbp1 deletion in DCs perturbs T‐cell homeostasis
As important antigen‐presenting cells, DCs play an important role in regulating T‐cell function. Thus, we analysed T‐cell homeostasis after Ptbp1 deletion in DCs. Ptbp1 deletion did not impair the development of T cells in the thymus, as WT and cKO mice showed no differences in CD8+ or CD4+ T cells or in the double‐positive or double‐negative population (Figure 2A). However, we found that the size and total cell number of the spleen and lymph nodes were increased in cKO mice (Figure 2B,C). Additionally, the populations of activated and memory‐like CD4+ and CD8+ T cells were increased in cKO mice, and the total numbers of CD4+ and CD8+ T cells in the spleen were increased in cKO mice (Figure 2D,E and Figure S2A). Consistent with T‐cell homeostasis results, we found minor inflammatory responses in the liver and lungs (Figure S2B). In conclusion, these results demonstrated that DC‐specific Ptbp1 deletion could disturb T‐cell homeostasis.
Figure 2.
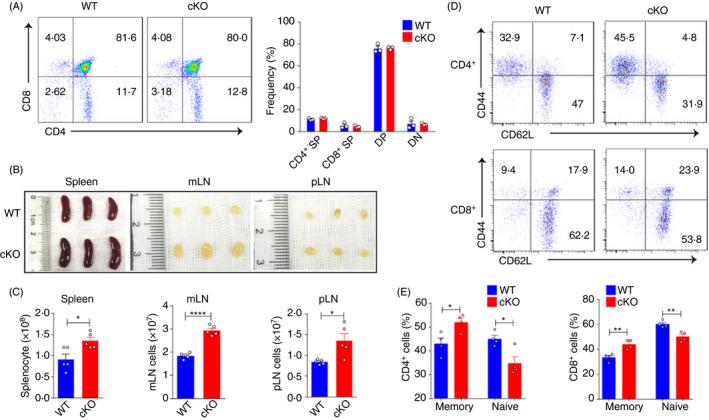
Ptbp1 deletion in DCs perturbs T‐cell homeostasis. (A) FACS analysis of thymocytes in WT and cKO mice, with the percentages of CD4 and CD8 single‐positive, double‐positive and double‐negative populations shown. (B‐C) Representative images and total cell numbers of the spleen, peripheral lymph nodes and mesenteric lymph nodes of WT and cKO mice. (D‐E) Flow cytometric analysis of the frequencies of naïve and memory‐like CD4+ T and CD8+ T cells in WT and cKO mice. Data are representative of at least three independent experiments. All data are shown as the mean ± SEM; * indicates p < 0·05, ** indicates p < 0·01, n.s., not significant. Statistical significance was evaluated by Student’s t tests.
PTBP1 deficiency can enhance antitumour immunity
DCs have the ability to present antigens to T cells and are necessary in antitumour immunity. In the tumour microenvironment, DCs increase the expression of costimulatory molecules and secretion of soluble factors. 39 , 40 , 41 Simultaneously, DCs can sense and present tumour antigens on MHC molecules to activate T cells. However, DCs are often found to be tolerogenic or dysfunctional in the tumour microenvironment, which is mainly due to immune suppression. 41
Although the expression of PTBP1 is increased in some tumours, 43 , 44 , 45 its function in DCs remains unknown. Here, we used a model of mice challenged with B16F10 melanoma cells. PTBP1 deficiency in DCs suppressed tumour growth and increased the survival rate compared with WT mice (Figure 3A–C). Consistently, the frequencies of IFN‐γ‐producing tumour‐infiltrating CD4+ and CD8+ T cells were also increased in cKO mice (Figure 3D,E and Figure S3A). Collectively, these results demonstrated that targeting PTBP1 in DCs could be an efficient approach to enhance antitumour immunity.
Figure 3.
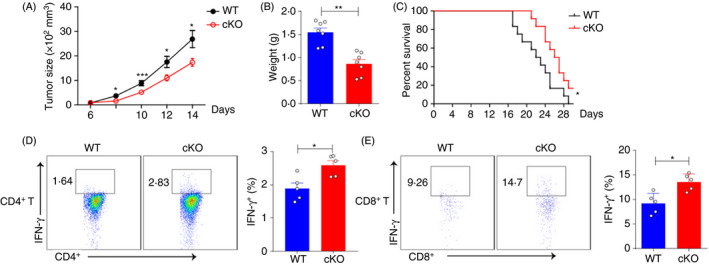
Ptbp1 deletion in DCs promotes antitumour immunity. (A‐B) Tumour growth and tumour weight of WT and cKO mice challenged subcutaneously with B16 melanoma cells. (C) Survival curves of WT and cKO mice challenged subcutaneously with B16 melanoma cells. (D‐E) FACS analysis of the percentages of IFN‐γ‐producing CD4+ T and CD8+ T cells in tumours from WT and cKO mice challenged subcutaneously with B16 melanoma cells. Data are representative of at least three independent experiments. All data are shown as the mean ± SEM; * indicates p < 0·05, ** indicates p < 0·01, n.s., not significant. Statistical significance was evaluated by Student’s t tests.
PTBP1 deficiency enhances the asthma‐induced immune response
Alternative RNA splicing plays important roles during the activation and maturation of immune cells, and abnormal alternative splicing can induce autoimmune disorders. 46 , 47 PTBP1, an important RNA‐binding protein, can regulate RNA splicing and stability. 44 , 48 , 49 The functions of PTBP1 in DCs in asthma remain unknown. Here, we challenged mice with OVA and Al(OH)3 by intraperitoneal injection for 3 weeks, followed by aerosol inhalation for another week (Figure 4A). After establishing OVA‐induced asthma, eosinophils, lymphocytes and neutrophils were recruited into the lungs and could induce lung injury. Ptbp1 deletion enhanced the recruitment of immune cells, especially that of eosinophils (Figure 4B,C). Furthermore, increased protein levels of IL‐4 and IL‐13 in OVA‐treated cKO mice were observed compared with WT mice (Figure 4D). Thus, Ptbp1 deficiency could enhance the recruitment of immune cells, indicating that Ptbp1 deletion could enhance the immune response.
Figure 4.
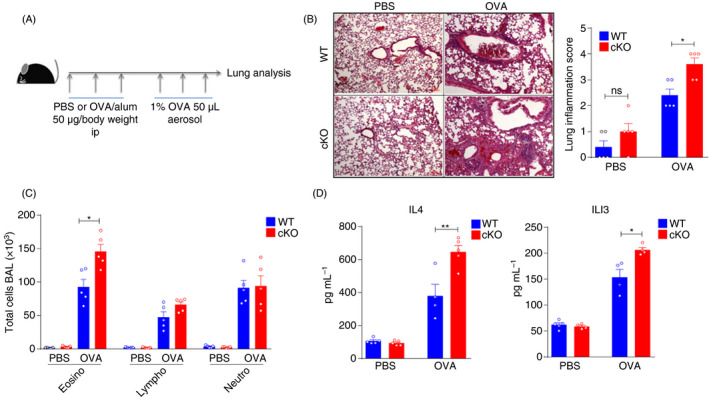
PTBP1 deficiency enhances the asthma‐induced immune response. (A) The procedure for the OVA‐induced asthma model. WT and cKO mice were intraperitoneally injected with OVA and Al(OH)3 for 3 weeks, followed by inhaled OVA treatment for 1 week. (B) Haematoxylin–eosin staining and inflammation scores of tissue samples from the lungs of WT and cKO mice after asthma induction with OVA. (C) The cell numbers of eosinophils, neutrophils and lymphocytes in the lungs of WT and cKO asthma model mice induced with OVA. (D) Protein levels of IL‐4 and IL‐13 in BALF. Data are representative of at least three independent experiments. All data are shown as the mean ± SEM; * indicates p < 0·05, ** indicates p < 0·01, n.s., not significant. Statistical significance was evaluated by Student’s t tests.
PTBP1 regulates Ifn response genes in DCs
To further clarify the function of PTBP1 in DCs, we examined gene induction by LPS. PTBP1 deficiency impaired Ifnα expression while promoting the induction of several immunostimulatory cytokines after treatment with the TLR4 ligand LPS (Figure 5A). The expression of the surface markers CD80 and CD86 was increased in PTBP1‐deficient DCs compared with WT DCs after LPS treatment (Figure 5B). To clarify the genes regulated by PTBP1 in vivo, we sorted CD11c and MHC II double‐positive DCs from the spleen for RNA sequencing. Before further analysis, t‐distributed stochastic neighbour embedding analysis was performed on expressed genes in DCs (FPKM > 1). Samples from WT or cKO mice clustered closely together (Figure 5C), indicating that the samples from each strain were very closely related. This analysis also showed that the transcriptomes of WT and cKO mice were distinct (Figure 5C). To further analyse the differences in the transcriptome profiles of WT and cKO DCs, we analysed RNA‐Seq data using DESeq2, a Bioconductor package for detecting differentially expressed genes in replicated count data. Comparison of WT and cKO DCs revealed 288 genes with significantly increased mRNA expression (FDR < 0·05, fold change > 2; Figure 5D). Then, gene ontology (GO) analysis was performed based on these differentially expressed genes, and we identified several associated enriched GO terms, which provided insight into the biological processes changed after Ptbp1 deletion. Among these significantly enriched GO terms, 'Response to interferon‐beta' and 'Cellular response to interferon‐beta' were upregulated in cKO DCs (Figure 5E). Gene set enrichment analysis (GSEA) results also showed that 'Interferon_alpha_response' and 'Interferon_gamma_response' were significantly enriched in cKO DCs (Figure 5F). To verify these results, we performed real‐time PCR analysis of DCs from WT and cKO mice, and the interferon response‐related genes Ifi202b, Oas2, Oas3 and Irf1 exhibited significantly upregulated expression in the cKO DCs (Figure 5G).
Figure 5.
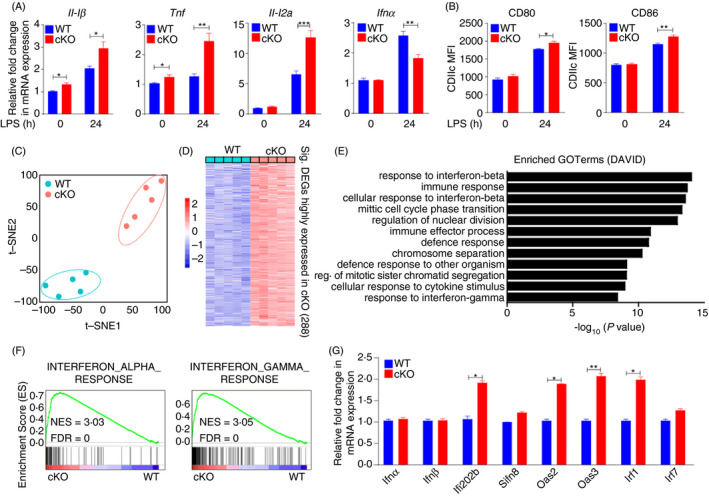
PTBP1 regulates Ifn response genes in DCs. (A) qRT‐PCR analysis of the genes of LPS (500 ng/mL)‐treated DCs (24 h) from the spleen of WT or cKO mice. (B) Flow cytometric analysis of CD80 and CD86 expression in LPS (500 ng/mL)‐treated DCs (24 h) from the spleen of WT or cKO mice. (C) T‐distributed stochastic neighbour embedding analysis of the genes expressed in DCs with comparison between WT and cKO mice (FPKM > 1). (D) Analysis of increased expression in transcriptome profiles among cKO cells (FDR < 0·05, fold change > 2). (E) Gene ontology (GO) analysis of upregulated genes enriched in cKO mice. (F) Significant enrichment of 'Interferon_gamma_response' and 'Interferon_alpha_response' in KO cells determined by gene set enrichment analysis (GSEA). (G) qRT‐PCR analysis of the interferon response‐related genes Ifi202b, Oas2, Oas3 and Irf1 in DCs from WT and cKO mice. Data are representative of at least three independent experiments. All data are shown as the mean ± SEM; * indicates p < 0·05, ** indicates p < 0·01, n.s., not significant. Statistical significance was evaluated by Student’s t tests.
PTBP1 regulates Pkm alternative splicing in DCs
The differentially expressed genes between WT and cKO mice could help us clarify the molecular mechanisms of PTBP1 in DCs (Figure 6A). However, considering that PTBP1 is an RNA‐binding protein, we also analysed the differences in inclusion levels between WT and cKO mice by rMATS, 36 a computational tool used to detect differential alternative splicing events from RNA‐Seq data. Ultimately, we found 240 significant AS events that were skipped more and 360 significant AS events that were more included with an inclusion level difference greater than 20% in cKO compared with WT DCs, indicating enhanced AS changes due to Ptbp1 deletion (Figure 6B). Next, we compared the genes involved in these significantly different alternative splicing events with the significantly differentially expressed genes and found that 33 genes had both different AS events and different mRNA expression levels (Figure 6C). GO enrichment analysis of these 33 overlapping genes revealed immune regulation‐associated terms (Figure 6D). Additionally, we also found 59 genes involved in the interferon alpha response and 105 genes involved in the interferon gamma response that were not only differentially expressed between WT and cKO DCs (fold change > 1·2, FDR < 0·05) (Figure 6E, left) but also belonged to different AS events (inclusion level difference > 10%, FDR < 0·05) (Figure 6E, middle). Taken together, our data suggested that the DC‐specific deficiency in PTBP1 could alter the immune response functions of DCs by regulating mRNA expression and AS events in these cells.
Figure 6.
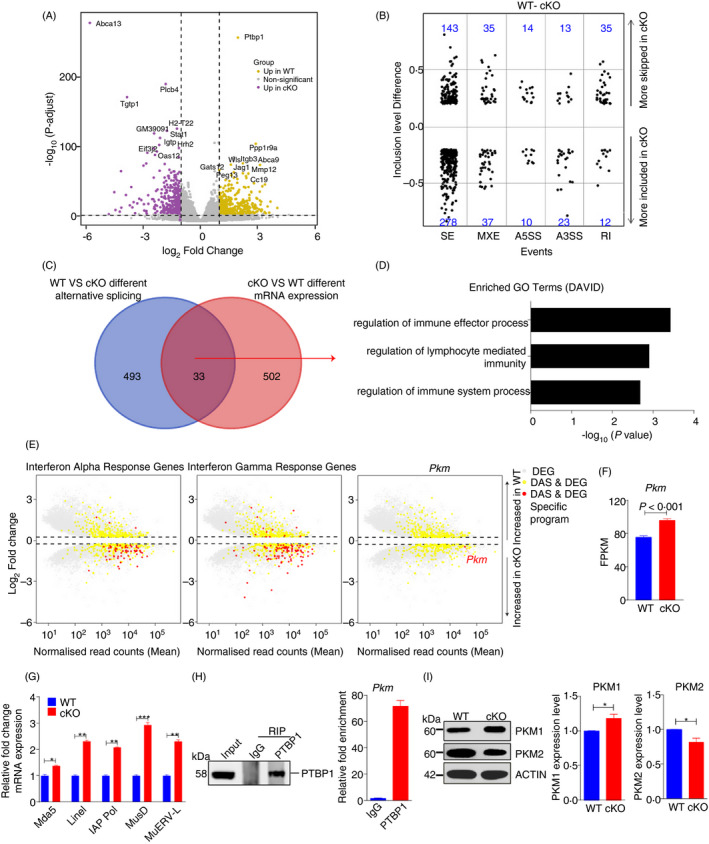
PTBP1 regulates Pkm alternative splicing in DCs. (A) Volcano plot comparing changes in mRNA expression between WT and cKO mice (n = 5, respectively). (B) Changes in AS between WT and cKO mice for skipped exons (SE), mutually exclusive exons (MXE), alternative 5′ and 3′ splice sites (A5SS and A3SS, respectively) and retained introns (RI). (C) The number of genes with both different AS results and different mRNA expression results. (D) Overlapping genes enriched in immune functions of DCs. (E) Different AS (DAS) and differentially expressed genes (DEG) between WT and cKO DCs. Grey dots represent DEGs, yellow dots represent DEGs with different AS, and red dots represent specific programmes belonging to DAS and DEGs. (F) The FPKM of Pkm between WT and cKO mice. (G) Relative mRNA expression of the indicated endogenous retroviruses in WT and cKO in DCs of spleen. (H) RIP of PTBP1 in DC2.4 cells and enrichment of Pkm. (I) The protein levels of PKM1 and PKM2 in DCs from WT and cKO mice.
Next, we focused on exploring the possible regulators of the immune response functions described above. Previous studies have shown that Pkm2 deletion in mice can increase systemic inflammation and promote metabolic distress. 49 In endothelial cells, loss of pyruvate kinase M2 leads to endogenous retrovirus expression and induction of interferon‐stimulated gene (ISG) expression. 51 , 52 These findings led us to hypothesize that the loss of PTBP1 in DCs may induce the downregulation of PKM2 expression and eventually alter immune response functions. To test this hypothesis, we first analysed the expression level and AS of Pkm and found that Pkm showed different mRNA expression levels and different AS events between WT and cKO mice (Figure 6E, right). The quantitative data also revealed that Pkm expression was significantly increased in PTBP1‐deficient DCs (Figure 6F). As PKM2 deficiency could reduce DNA methylation and increase the expression of endogenous retroviral elements, 50 we found Ptbp1 deletion increased the expression of endogenous retroviral elements in DCs (Figure 6G). Then, we performed RIP‐PCR with the DC2.4 cell line and found that PTBP1 could bind to the pre‐mRNA of Pkm (Figure 6H). Furthermore, we found that the protein level of PKM2 was downregulated by Ptbp1 deletion in DCs (Figure 6I), which is consistent with prior studies. 50 , 51 , 52 Taken together, these data indicated that PTBP1 deficiency in DCs enhanced their Ifn response ability via Pkm alternative splicing.
DISCUSSION
The mechanism by which PTBP1 regulates the development and function of DCs is unknown. In this study, we reported the function of PTBP1 in DCs. PTBP1 deficiency in DCs upregulated MHC II expression without influencing DC development. However, the frequencies of activated CD4+ and CD8+ T cells were increased. In B16F10 melanoma cell‐challenged mice, Ptbp1‐specific deletion in DCs resulted in a remarkable reduction in tumour growth, and the populations of T cells expressing IFN‐γ were increased in tumours. With the asthma model, we found that Ptbp1‐deleted mice exhibited increased immune cell recruitment to the lungs. These data indicated that PTBP1 deficiency in DCs could promote DC immune function. By sorting DCs from spleens and performing RNA‐Seq, we found that Ifn response genes showed an increase in expression in cKO mice. The alternative splicing of Pkm changed with Ptbp1 deletion. The decreased of PKM2 resulted in expression of endogenous retroviral elements and enhanced immune response. Furthermore, these data suggested that PTBP1 might be a novel therapeutic target for tumour treatment.
DCs are specialized cells that present endogenous and exogenous antigens to T cells. The ability of DCs to present exogenous antigens on MHC I is critical for the ability of the immune system to defend against intracellular viruses. While presentation of endogenous antigens on MHC II is related to autophagy, ATG5 deficiency can impair this pathway. 53 , 54 In the steady state, DCs also maintain immune tolerance. It has been proposed that DCs may directly present or cross‐present self‐Ags for negative selection of thymocytes. 54 In addition, peripheral DCs may recirculate into the thymus and induce clonal deletion of T cells and Treg proliferation. 55 Nevertheless, complete deletion of DCs does not influence the peripheral T‐cell repertoire, indicating that DCs do not play a dominant role in peripheral immune tolerance. 8 In our experiments, we found that Ptbp1 deletion could perturb T‐cell homeostasis without affecting T‐cell development. The deletion of Ptbp1 enhanced the function of DCs, which was consistent with the antitumour activity of DCs. We also found that in the OVA‐induced asthma model, Ptbp1 deletion in DCs exacerbated damage to the lungs. Therefore, PTBP1 could be a novel target for autoimmune disease therapy.
We observed a significant phenotype in cKO mice, which was perturbed T‐cell homeostasis. PTBP1 deficiency in DCs did not impair T‐cell development in the thymus, but the populations of activated and memory T cells were increased. PTBP1‐mediated alternative splicing can regulate the pro‐tumorigenic effects of senescent cells, and inhibiting PTBP1 can reduce the risk of tumorigenesis. 56 In colon cancer patients, PTBP1 is highly expressed and can accelerate cancer cell growth. Inhibition of PTBP1 can activate caspase‐3 and PARP1 to induce apoptosis. 57 Pancreatic ductal adenocarcinoma (PDAC) is an aggressive disease, and some drug‐resistant subpopulations can develop after chronic exposure to gemcitabine. The mechanism underlying this development of resistance is that highly expressed PTBP1 can regulate the splicing of PKM to increase the level of the PKM2 variant. Knockdown of PTBP1 expression abolishes drug resistance and promotes splicing of the PKM1 variant. 28 PTBP1 is also highly expressed in blood outgrowth endothelial cells (BOECs) from patients. PTBP1‐mediated alternative splicing of the PKM gene can increase PKM2 expression, which leads to abnormal growth and enhanced glycolysis in pulmonary artery endothelial cells. 58 PTBP1 also plays an essential role during CD4+ T‐cell activation by regulating the expression of CD40L. 59 During B‐cell selection in germinal centres, the expression of PTBP1 is upregulated, which is necessary for B‐cell receptor (BCR)‐mediated antibody production. 25 , 61 Nonetheless, other than Pkm, it is likely that PTBP1 may regulate other genes via splicing to enhance DC function, which merit further investigations in the near future. The last but not the least, given that CD11c is also expressed in cell types other than DCs, we cannot rule out the possibility that these other cells also contribute to the phenotypes observed in Ptbp1 fl/fl: CD11c‐Cre mice. 61
In conclusion, we demonstrated the function of PTBP1 in DCs involved in regulating T‐cell homeostasis. We also found that targeting PTBP1 in DCs promoted antitumour immunity. These findings provide additional evidence for the role of PTBP1 in regulating the immune response.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Supporting information
Figure S1. Genotyping of CD11c Cre/Ptbp1 f/f mice. (A) PCR analysis of PTBP1 and CD11c‐Cre to test the genotypes of WT and cKO mice. (B) Immunoblotting for PTBP1 in DCs from WT and cKO mice. (C) Gating strategy for pDCs, cDC1 and cDC2 in the spleen.
Figure S2. Total cell numbers of CD4+ and CD8+ T cells in the spleen. (A) Total cell numbers of CD4+ and CD8+ T cells in the spleen of WT and cKO mice. (B) Haematoxylin–eosin staining of tissue samples from the liver and lungs of WT and cKO mice. Data are representative of at least three independent experiments.
Figure S3. Gating strategy for immune cells in tumours. (A) Gating strategy for IFN‐γ‐producing CD4+ T and CD8+ T cells in tumours.
ACKNOWLEDGEMENTS
We thank X. Feng for helping with our project. S.L and Z.Y designed and supervised the study; G.G and X.C carried out the experiment and performed the data analysis; G.G, X.C, P.N, L.Y, W.J, L.J, Z.Y and S.L wrote the manuscript; and S.L and Z.Y revised the manuscript. This research was supported by the National Key Research and Development Program of China (2019YFA0508603, 2017YFA0103102 and 2016YFA0102300); the CAMS Innovation Fund for Medical Sciences (CIFMS) (2016‐I2M‐3‐002, 2019‐I2M‐1‐006, 2016‐I2M‐1‐018 and 2017‐I2M‐1‐015); and the National Natural Science Foundation of China (91754114, 31671441, 81870089, 81890990 and 81700105).
Guangfeng Geng and Changlu Xu contributed equally to this study.
Contributor Information
Yushan Zhu, Email: zhuys@nankai.edu.cn.
Lihong Shi, Email: shilihongxys@ihcams.ac.cn.
DATA AVAILABILITY STATEMENT
The RNA‐Seq data reported in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE153401. Other data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gallegos AM, Bevan MJ. Central tolerance to tissue‐specific antigens mediated by direct and indirect antigen presentation. J Exp Med. 2004;200:1039–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thery C, Amigorena S. The cell biology of antigen presentation in dendritic cells. Curr Opin Immunol. 2001;13:45–51. [DOI] [PubMed] [Google Scholar]
- 4. Sozzani S, Del Prete A, Bosisio D. Dendritic cell recruitment and activation in autoimmunity. J Autoimmun. 2017;85:126–40. [DOI] [PubMed] [Google Scholar]
- 5. Fu Y, Zhan X, Wang Y, Jiang X, Liu M, Yang Y, et al. NLRC3 expression in dendritic cells attenuates CD4(+) T cell response and autoimmunity. EMBO J. 2019;38:e101397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bak SP, Barnkob MS, Bai A, Higham EM, Wittrup KD, Chen J. Differential requirement for CD70 and CD80/CD86 in dendritic cell‐mediated activation of tumor‐tolerized CD8 T cells. J Immunol. 2012;189:1708–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hochrein H, O'Keeffe M. Dendritic cell subsets and toll‐like receptors. Handb Exp Pharmacol. 2008;183:153–79. [DOI] [PubMed] [Google Scholar]
- 8. Birnberg T, Bar‐On L, Sapoznikov A, Caton ML, Cervantes‐Barragan L, Makia D, et al. Lack of conventional dendritic cells is compatible with normal development and T cell homeostasis, but causes myeloid proliferative syndrome. Immunity. 2008;29:986–97. [DOI] [PubMed] [Google Scholar]
- 9. Proietto AI, van Dommelen S, Wu L. The impact of circulating dendritic cells on the development and differentiation of thymocytes. Immunol Cell Biol. 2009;87:39–45. [DOI] [PubMed] [Google Scholar]
- 10. Zou T, Caton AJ, Koretzky GA, Kambayashi T. Dendritic cells induce regulatory T cell proliferation through antigen‐dependent and ‐independent interactions. J Immunol. 2010;185:2790–9. [DOI] [PubMed] [Google Scholar]
- 11. Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, et al. Single‐cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013;498:236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frumkin I, Yofe I, Bar‐Ziv R, Gurvich Y, Lu YY, Voichek Y, et al. Evolution of intron splicing towards optimized gene expression is based on various Cis‐ and Trans‐molecular mechanisms. PLoS Biol. 2019;17:e3000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dehm SM. mRNA splicing variants: exploiting modularity to outwit cancer therapy. Can Res. 2013;73:5309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee SC, Abdel‐Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35:2413–27. [DOI] [PubMed] [Google Scholar]
- 17. Schaub A, Glasmacher E. Splicing in immune cells‐mechanistic insights and emerging topics. Int Immunol. 2017;29:173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chauhan K, Kalam H, Dutt R, Kumar D. RNA splicing: a new paradigm in host‐pathogen interactions. J Mol Biol. 2019;431:1565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. Global analysis of alternative splicing during T‐cell activation. RNA. 2007;13:563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katsuyama T, Li H, Comte D, Tsokos GC, Moulton VR. Splicing factor SRSF1 controls T cell hyperactivity and systemic autoimmunity. J Clin Investig. 2019;129:5411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu H, Lorenzini PA, Zhang F, Xu S, Wong MSM, Zheng J, et al. Alternative splicing analysis in human monocytes and macrophages reveals MBNL1 as major regulator. Nucleic Acids Res. 2018;46:6069–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu X, Harvey SE, Zheng R, Lyu J, Grzeskowiak CL, Powell E, et al. The RNA‐binding protein AKAP8 suppresses tumor metastasis by antagonizing EMT‐associated alternative splicing. Nat Commun. 2020;11:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson F, Smith CW. A splicing repressor domain in polypyrimidine tract‐binding protein. J Biol Chem. 2006;281:800–6. [DOI] [PubMed] [Google Scholar]
- 25. Monzon‐Casanova E, Screen M, Diaz‐Munoz MD, Coulson RMR, Bell SE, Lamers G, et al. The RNA‐binding protein PTBP1 is necessary for B cell selection in germinal centers. Nat Immunol. 2018;19:267–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cui J, Placzek WJ. PTBP1 modulation of MCL1 expression regulates cellular apoptosis induced by antitubulin chemotherapeutics. Cell Death Differ. 2016;23:1681–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cui J, Placzek WJ. PTBP1 enhances miR‐101‐guided AGO2 targeting to MCL1 and promotes miR‐101‐induced apoptosis. Cell Death Dis. 2018;9:552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Calabretta S, Bielli P, Passacantilli I, Pilozzi E, Fendrich V, Capurso G, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016;35:2031–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. [DOI] [PubMed] [Google Scholar]
- 30. David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c‐Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, et al. PKM2 isoform‐specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morita M, Sato T, Nomura M, Sakamoto Y, Inoue Y, Tanaka R, et al. PKM1 confers metabolic advantages and promotes cell‐autonomous tumor cell growth. Cancer Cell. 2018;33:355–67 e7. [DOI] [PubMed] [Google Scholar]
- 33. Park SJ, Im DS. Blockage of sphingosine‐1‐phosphate receptor 2 attenuates allergic asthma in mice. Br J Pharmacol. 2019;176:938–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gilbert C, Svejstrup JQ. RNA immunoprecipitation for determining RNA‐protein associations in vivo. Current Protocols in Molecular Biology. 2006(p. Unit 27.4, Chapter 27). [DOI] [PubMed] [Google Scholar]
- 35. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript‐level expression analysis of RNA‐seq experiments with HISAT. StringTie and Ballgown. Nat Protocols. 2016;11:1650–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Homma R, Yasui K. Cellular receptors for animal viruses. Tanpakushitsu kakusan koso Protein, nucleic acid, enzyme. 1968;13:543–53. [PubMed] [Google Scholar]
- 37. Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA‐Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farren MR, Carlson LM, Lee KP. Tumor‐mediated inhibition of dendritic cell differentiation is mediated by down regulation of protein kinase C beta II expression. Immunol Res. 2010;46:165–76. [DOI] [PubMed] [Google Scholar]
- 40. Aymeric L, Apetoh L, Ghiringhelli F, Tesniere A, Martins I, Kroemer G, et al. Tumor cell death and ATP release prime dendritic cells and efficient anticancer immunity. Can Res. 2010;70:855–8. [DOI] [PubMed] [Google Scholar]
- 41. Imai K, Minamiya Y, Koyota S, Ito M, Saito H, Sato Y, et al. Inhibition of dendritic cell migration by transforming growth factor‐beta1 increases tumor‐draining lymph node metastasis. J Exp Clin Cancer Res: CR. 2012;31:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. 2020;20:7–24. [DOI] [PubMed] [Google Scholar]
- 43. Wang X, Li Y, Fan Y, Yu X, Mao X, Jin F. PTBP1 promotes the growth of breast cancer cells through the PTEN/Akt pathway and autophagy. J Cell Physiol. 2018;233:8930–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cho CY, Chung SY, Lin S, Huang JS, Chen YL, Jiang SS, et al. PTBP1‐mediated regulation of AXL mRNA stability plays a role in lung tumorigenesis. Sci Rep. 2019;9:16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bielli P, Panzeri V, Lattanzio R, Mutascio S, Pieraccioli M, Volpe E, et al. The splicing factor PTBP1 promotes expression of oncogenic splice variants and predicts poor prognosis in patients with non‐muscle‐invasive bladder cancer. Clinical Cancer Res: Off J Am Assoc Cancer Res. 2018;24:5422–32. [DOI] [PubMed] [Google Scholar]
- 46. Agrebi N, Ben‐Mustapha I, Matoussi N, Dhouib N, Ben‐Ali M, Mekki N, et al. Rare splicing defects of FAS underly severe recessive autoimmune lymphoproliferative syndrome. Clin Immunol. 2017;183:17–23. [DOI] [PubMed] [Google Scholar]
- 47. Evsyukova I, Somarelli JA, Gregory SG, Garcia‐Blanco MA. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biol. 2010;7:462–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sayed ME, Yuan L, Robin JD, Tedone E, Batten K, Dahlson N, et al. NOVA1 directs PTBP1 to hTERT pre‐mRNA and promotes telomerase activity in cancer cells. Oncogene. 2019;38:2937–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang L, Yang Z, Trottier J, Barbier O, Wang L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology. 2017;65:604–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dayton TL, Gocheva V, Miller KM, Israelsen WJ, Bhutkar A, Clish CB, et al. Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev. 2016;30:1020–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stone OA, El‐Brolosy M, Wilhelm K, Liu X, Romao AM, Grillo E, et al. Loss of pyruvate kinase M2 limits growth and triggers innate immune signaling in endothelial cells. Nat Commun. 2018;9:4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Radoshevich L, Dussurget O. Cytosolic Innate Immune Sensing and Signaling upon Infection. Front Microbiol. 2016;7:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 2005;307:593–6. [DOI] [PubMed] [Google Scholar]
- 54. Oh DS, Lee HK. Autophagy protein ATG5 regulates CD36 expression and anti‐tumor MHC class II antigen presentation in dendritic cells. Autophagy. 2019;15:2091–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lopes N, Charaix J, Cedile O, Serge A, Irla M. Lymphotoxin alpha fine‐tunes T cell clonal deletion by regulating thymic entry of antigen‐presenting cells. Nat Commun. 2018;9:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Proietto AI, van Dommelen S, Zhou P, Rizzitelli A, D'Amico A, Steptoe RJ, et al. Dendritic cells in the thymus contribute to T‐regulatory cell induction. Proc Natl Acad Sci USA. 2008;105:19869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Georgilis A, Klotz S, Hanley CJ, Herranz N, Weirich B, Morancho B, et al. PTBP1‐mediated alternative splicing regulates the inflammatory secretome and the pro‐tumorigenic effects of senescent cells. Cancer Cell. 2018;34:85–102 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li X, Han F, Liu W, Shi X. PTBP1 promotes tumorigenesis by regulating apoptosis and cell cycle in colon cancer. Bull Cancer. 2018;105:1193–201. [DOI] [PubMed] [Google Scholar]
- 59. Caruso P, Dunmore BJ, Schlosser K, Schoors S, Dos Santos C, Perez‐Iratxeta C, et al. Identification of microRNA‐124 as a major regulator of enhanced endothelial cell glycolysis in pulmonary arterial hypertension via PTBP1 (polypyrimidine tract binding protein) and pyruvate kinase M2. Circulation. 2017;136:2451–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. La Porta J, Matus‐Nicodemos R, Valentin‐Acevedo A, Covey LR. The RNA‐binding protein, polypyrimidine tract‐binding protein 1 (PTBP1) is a key regulator of CD4 T cell activation. PLoS One. 2016;11:e0158708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sasanuma H, Ozawa M, Yoshida N. RNA‐binding protein Ptbp1 is essential for BCR‐mediated antibody production. Int Immunol 2019;31:157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Throsby M, Herbelin A, Pleau JM, Dardenne M. CD11c+ eosinophils in the murine thymus: developmental regulation and recruitment upon MHC class I‐restricted thymocyte deletion. J Immunol. 2000;165:1965–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Genotyping of CD11c Cre/Ptbp1 f/f mice. (A) PCR analysis of PTBP1 and CD11c‐Cre to test the genotypes of WT and cKO mice. (B) Immunoblotting for PTBP1 in DCs from WT and cKO mice. (C) Gating strategy for pDCs, cDC1 and cDC2 in the spleen.
Figure S2. Total cell numbers of CD4+ and CD8+ T cells in the spleen. (A) Total cell numbers of CD4+ and CD8+ T cells in the spleen of WT and cKO mice. (B) Haematoxylin–eosin staining of tissue samples from the liver and lungs of WT and cKO mice. Data are representative of at least three independent experiments.
Figure S3. Gating strategy for immune cells in tumours. (A) Gating strategy for IFN‐γ‐producing CD4+ T and CD8+ T cells in tumours.
Data Availability Statement
The RNA‐Seq data reported in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE153401. Other data that support the findings of this study are available from the corresponding author upon reasonable request.


