Abstract
Background
The programmed cell death pathway necroptosis may synergize with the DNA damage response (DDR) in opposing tumor progression. While our basic mechanistic understanding of the necroptotic cell death advances rapidly, its prognostic implications have not been thoroughly examined in cancers.
Methods
We included 394 patients with stage I non-small-cell lung cancer (NSCLC) who underwent surgical tumor resection between 1 January 1997 and 31 December 2011 and measured expression levels of nine proteins involved in necroptosis and the DDR in primary samples from 394 patients using tissue microarray. Protein expression evaluated by using an H-score method was dichotomized by the median value. The overall survival as the endpoint was calculated from the time of diagnosis to the time of the last follow-up or death.
Results
We find that low-level expression of the necroptosis markers RIPK3 and PELI1 is associated with high risk of patient death. High-level expression of the key DDR factor p53 in combination with low-level expression of either RIPK3 or PELI1 increases the risk further. These gene expression effects appear to occur specifically in the squamous cell carcinoma (SCC) subtype of stage I NSCLC, while not observed in the non-SCC subtypes.
Conclusions
Low-level expression of such necroptosis factors as RIPK3 and PELI1 in combination with high-level expression of the DDR factor p53 can serve as a critical indicator in predicting survival of stage I NSCLC patients with the SCC subtype.
Keywords: DNA damage response (DDR), necroptosis, non-small-cell lung cancer (NSCLC), squamous cell carcinoma (SCC), survival
Introduction
“Hallmarks” of cancer has not only advanced our understanding of cancer biology but also provided a basis for biomarker development (1). Resistance to cell death, in particular, constitutes a crucial axis of the cancer hallmarks and indeed cancers develop diverse means to avoid “programmed” cell death (2). Acquisition of the ability of cancer cells to evade a form of the programmed cell death, apoptosis, which proceeds with a cascade of caspase activation, is well known as such a resistance to cancer cell death. Recent studies, however, have unveiled a qualitatively distinct, caspase-independent form of programmed cell death termed necroptosis, which would go awry in many cancers (3).
Genetic and biochemical studies have uncovered basic mechanisms by which necroptosis is regulated. We now know that necroptosis critically requires the formation of a microfilament-like protein complex called the necrosome. Phosphorylation in cis and trans between Receptor-Interacting Protein Kinases 1 and 3 (RIPK1 and RIPK3) is known to cause the necrosome to assemble. The necrosome in turn activates the pro-necroptotic kinase Mixed Lineage Kinase domain-Like protein (MLKL), leading to cell death with plasma membrane rupture, similarly to that of an unprogrammed mode of cell death called necrosis. The E3 ubiquitin ligase Pellino-1 (PELI1), on the other hand, dampens the necrosome activity by ubiquitinylating RIPK3 for proteasomal degradation (4,5). This suggests that an adequate level of key necrosome components crucially decide whether for the cell to engage the proper necroptotic cell death response.
While our basic mechanistic understanding of necroptosis has rapidly advanced, how it is related to survival of cancer patients remains less explored. Studies using preclinical models of select cancers such as leukemia and pancreatic cancer have elucidated the translational relevance of necroptosis (4,6,7). Notably, several cultured cell and preclinical models suggested an association of necroptosis with the DNA damage response (DDR), which is orchestrated by the “guardian of the genome” p53 (1,2,8,9). While radiation therapy and chemotherapy are presumed to mount the DDR, cancers can often develop a wide array of resistance to the DDR. The suggested association between necroptosis and the DDR thus necessitates further examination in cancer patients (10-13).
Importantly, necroptotic factors appear to influence tumor progression differentially depending on individual tumor types (4,6,7). We focus our investigation on lung cancer, which is relatively less represented in preclinical necroptosis studies. Lung cancer is the leading cause of cancer-related deaths globally, constituting almost 25% of all cancer deaths (14). A major type of lung cancer, non-small-cell lung cancer (NSCLC) has a particularly grave prognosis albeit a slowly improving survival trend in recent years. Thus, development of prognostic biomarkers for the early-stage NSCLC is of interest in this field because the 5-year survival rate is only around 70% (15). In this vein, some factors such as those involved in DDR pathways have been explored in NSCLC patients as candidate prognostic markers (8,9). These factors, however, have not been clinically validated while biologically plausible. Importantly, the current adjuvant treatment does not seem to confer a survival advantage and treatment decisions are not guided by biomarkers (9). Squamous cell carcinoma (SCC) constitutes an aggressive subtype of NSCLC, distinguishing itself from non-SCCs not only by distinct histology and genomic landscapes but by unique clinical characteristics of age, smoking habit, and comorbid diseases (16,17). Given the significant differences between these subtypes, biomarkers specific to SCC are greatly needed.
To this end, we here investigated whether expression of necroptosis factors in combination with that of DDR factors has any association with survival of the patients with the stage I NSCLC. We present data about the relationship among expression levels of those proteins, NSCLC subtypes, and patient survival as follows and in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1027).
Methods
Patients and tissue specimens
Patients with pathological stage I NSCLC (pT1-2N0M0) who had been consecutively treated at the Inha University Hospital (Incheon, South Korea) were retrospectively included in the study (Figure S1). The patients who underwent surgical tumor resection between 1 January 1997 and 31 December 2011 were initially considered for inclusion. Cancers were staged according to the 7th edition Tumor-Node-Metastasis staging system (18). Data from each patient who had been followed up for at least 3 years after surgical resection and whose tumor had been completely resected (R0 resection) were included in the analysis. The patients were scheduled for follow-up every 3 months for 2 years and every 6 months thereafter. Data from the patients who had undergone limited resection (segmentectomy or wedge resection), received any anticancer treatment before surgical resection, or had a history of other cancers were excluded, resulting in the final sample size of 394 patients. All formalin-fixed, paraffin-embedded (FFPE) blocks were sent to a laboratory (SuperBioChips, Seoul, South Korea) for tissue microarray (TMA) construction and immunohistochemical staining. The study protocol was approved by the Institutional Review Board of the Inha University Hospital with waiving the informed consent requirement.
Clinical variables
Information about smoking habit, the Eastern Cooperative Oncology Group performance status, Charlson comorbidity index score (CCIS), tumor size, T stage, lymphatic or vascular invasion (from the pathological specimens), type of surgery, and use of adjuvant chemotherapy was collected (Table S1). To increase the quality of the information, radiological examination (e.g., chest computed tomography scan, positron-emission tomography), surgical pathology reports, and clinical information were reviewed independently for each patient.
Construction of tissue microarrays and immunohistochemistry
TMA construction by using individual FFPE samples was performed as previously described (8). Each tissue array block contained up to 50 specimens, which allowed a total of 394 specimens to be mounted on 20 blocks. Serial sections (4 µm) of the FFPE blocks were analyzed for protein expression with immunohistochemistry. A pathologist performed a test procedure by using a control human TMA panel (19) according to the manufacturer’s information and evaluated the quality of positive and negative controls in immunohistochemistry. Antibodies against RIPK3, MLKL, and PELI1 (F-7) were used due to their essential roles in the necrosome formation and modulation (3). In addition, antibodies against P53, γH2AX, ATM, Chk2pT68, BRCA1pS1423, and ERCC1 were used due to their well-defined roles in cellular responses to DNA damage (1,8,9). Information for the antibodies used in this study is summarized in Table S2. All immunohistochemical staining was performed with a BenchMark XT autostainer (Ventana Medical Systems, Tucson, AZ) and the i-View detection kit (Ventana Medical System) by following the standard operating procedure at SuperBioChips Laboratory, Seoul, South Korea.
Evaluation of immunohistochemistry
TMA samples containing more than 500 malignant cells were considered for evaluation. Scanned images of the stained slides were viewed using the Aperio ImageScope program (version 11.2.0.782; Aperio Technologies, Vista, CA, USA) at the 20× objective magnification. Automated digital image analysis was performed using the Genie Classifier and Nuclear v9 algorithm (Aperio Technologies). The percentage of stained cells and the intensity of staining were estimated in the cytoplasm for MLKL and PELI1 and in the nucleus for RIPK3, p53, γH2AX, ATM, Chk2, BRCA1, and ERCC1 by two pathologists (HJJ and LK). The evaluators were blinded to each patient’s results for the clinical and pathological variables and the survival status.
Protein expression was evaluated by using an H-score method. The percentage of tumor cells with positive cytoplasmic or nuclear staining on each TMA core was assigned a score of either 0 (0% stained tumor cells), 0.1 (1–9%), 0.5 (10–49%), or 1 (≥50%). The staining intensity was assigned a score of either 0 (weakest intensity), 1, 2, or 3 (strongest intensity). Each H-score was obtained by multiplying the proportion by the staining intensity. The median values of all mean H-scores were used as the cut-off values for the classification of expression of the nine proteins as either “low” or “high” (Figure S2).
Survival measurements
The overall survival as the endpoint of this study was calculated from the time of diagnosis to the time of the last follow-up or death due to any cause. Dates of death were obtained principally from review of the medical records. To increase the quality of the information regarding the survival status of patients lost to follow-up, missing information was either obtained by contacting the patients or their relatives by phone or mail (13 patients) or obtained from the Ministry of Public Administration and Security, South Korea (29 patients).
Statistical analysis
Distributions of the clinical variables and levels of protein expression between SCC and non-SCC groups were tested using either chi-square test or Mann-Whitney U test as appropriate as possible. Cox model was performed to estimate HR and its 95% CI for each clinical variable and protein. Proteins that were significant at 5% level in the univariate Cox model were included in the multivariate Cox model. Performance of the model was evaluated using Harrell’s c-index. Statistical significance was accepted when P<0.05. To evaluate significance of each protein, P values were corrected using False Discovery Rate (FDR) for multiple comparisons (20). Proteins with FDR-corrected P value (q value) <0.05 were considered significant. Statistical analyses were performed using a statistical software package (SPSS version 19.0, SPSS, Chicago, IL).
Results
Characteristics of patients
Clinical characteristics of 394 patients with the stage I NSCLC (197 with SCC, 175 with adenocarcinoma, and 22 with other histology) are presented in Table 1. The median age of the patients was 66 years. 160 patients (40.6%) have since died. The median survival time was 9.8 years. Each patient was classified into either the SCC or the non-SCC group. The proportions of never smokers and women were greater in the non-SCC group (82% and 81% of the entire cohort, respectively, chi-square P<0.001) than in the SCC group. The non-SCC group also had greater proportions of patients who were younger, had smaller tumors, had lower T stages, and/or received lobectomy. Associations between survival, age, size of tumor, and T stage were statistically significant (Table S2). The Gender and Charlson comorbidity index score (CCIS), smoking habit, histology results, lymphatic or blood vessel invasion, type of surgery, and use of adjuvant chemotherapy were not statistically significant prognostic factors, however.
Table 1. Baseline characteristics of the patients with stage I NSCLC according to histology.
| Characteristics | Categories | Total, n (%) | SCC, n (%) | Non-SCC, n (%) | Statistics* | P value |
|---|---|---|---|---|---|---|
| Age, years | Median (IQR) | 66 (60–72) | 67 (61–73) | 65 (57–72) | −2.390 | 0.017a |
| Gender | Men | 291 (73.9) | 177 (60.8) | 114 (39.2) | 52.173 | <0.001 |
| Women | 103 (26.1) | 20 (19.4) | 83 (80.6) | |||
| Smoking habit | Never | 99 (28.0) | 18 (18.2) | 81 (81.8) | 47.714 | <0.001 |
| Ever | 254 (72.0) | 150 (59.1) | 104 (40.9) | |||
| ECOG performance status | 0–1 | 326 (97.0) | 161 (49.4) | 165 (50.6) | 1.459 | 0.338 |
| 2 or more | 10 (3.0) | 3 (30.0) | 7 (70.0) | |||
| CCIS | 0 | 108 (35.8) | 50 (46.3) | 58 (53.7) | 2.604 | 0.272 |
| 1 | 162 (53.6) | 82 (50.6) | 80 (49.4) | |||
| 2 or more | 32 (10.6) | 20 (62.5) | 12 (37.5) | |||
| Tumor size, cm | Median (IQR) | 3 (2.3–4.2) | 3.5 (2.5–4.5) | 2.9 (2.2–4.0) | −2.784 | 0.005a |
| T stage | 1 | 147 (37.3) | 60 (40.8) | 87 (59.2) | 7.911 | 0.007 |
| 2 | 247 (62.7) | 137 (55.5) | 110 (44.5) | |||
| Lymphatic invasion | No | 274 (75.7) | 137 (50.0) | 137 (50.0) | 0.310 | 0.625 |
| Yes | 88 (24.3) | 47 (53.4) | 41 (46.6) | |||
| Vascular invasion | No | 267 (73.8) | 133 (49.8) | 134 (50.2) | 0.420 | 0.551 |
| Yes | 95 (26.2) | 51 (53.7) | 44 (46.3) | |||
| Type of surgery | Lobectomy | 324 (82.2) | 152 (46.9) | 172 (53.1) | 11.022 | 0.004 |
| Bilobectomy | 33 (8.4) | 17 (51.5) | 16 (48.5) | |||
| Pneumonectomy | 37 (9.4) | 28 (75.7) | 9 (24.3) | |||
| Adjuvant chemotherapy | No | 346 (88.0) | 174 (50.3) | 172 (49.7) | 0.030 | 0.878 |
| Yes | 47 (12.0) | 23 (48.9) | 24 (51.1) | |||
| MST | Years (95% CIs) | 9.8 (7.9–11.8) | 10.1 (7.4–12.8) | 9.1 (NA) | 0.180 | 0.671b |
| Death | No | 234 (59.4) | 118 (50.4) | 116 (49.6) | 0.042 | 0.918 |
| Yes | 160 (40.6) | 79 (49.4) | 81 (50.6) |
P values obtained by chi-square test, except: a, Mann-Whitney U test; b, log-rank test. *, the z-value was indicated for age and tumor size, and the chi-square value was indicated for other variables. SCC, squamous cell carcinoma; IQR, interquartile range; ECOG, Eastern Cooperative Oncology Group; CCIS, Charlson comorbidity index score; MST, median survival time; NA, not applicable.
Expression of proteins involved in necroptosis or the DDR
Expression levels of proteins (low versus high) were compared between the SCC and the non-SCC groups using tissue microarray (Table 2). High-level expression of RIPK3, p53, γH2AX, ATM, Chk2, BRCA1, and ERCC1 was more common in the SCC group than in the non-SCC group. By contrast, low-level expression of MLKL and PELI1 was more common in the SCC group than in the non-SCC group.
Table 2. Expression of proteins involved in necroptosis and DNA damage response according to histology.
| Proteins | Level | SCC, n (%) | Non-SCC, n (%) | Chi-square value | P value |
|---|---|---|---|---|---|
| RIPK3 | Low | 89 (44.3) | 112 (55.7) | 5.401 | 0.026 |
| High | 107 (56.0) | 84 (44.0) | |||
| MLKL | Low | 115 (58.7) | 81 (41.3) | 12.178 | 0.001 |
| High | 80 (41.0) | 115 (59.0) | |||
| PELI1 | Low | 116 (59.5) | 79 (40.5) | 14.385 | <0.001 |
| High | 79 (40.3) | 117 (59.7) | |||
| p53 | Low | 83 (42.3) | 113 (57.7) | 8.902 | 0.003 |
| High | 112 (56.9) | 83 (42.1) | |||
| BRCA1 | Low | 102 (45.7) | 121 (54.3) | 3.730 | 0.067 |
| High | 95 (55.6) | 76 (44.4) | |||
| H2AX | Low | 96 (43.4) | 125 (56.6) | 8.667 | 0.004 |
| High | 101 (58.4) | 72(41.6) | |||
| ATM | Low | 103 (46.4) | 119 (53.6) | 2.642 | 0.127 |
| High | 94 (54.7) | 78 (45.3) | |||
| Chk2 | Low | 101 (40.4) | 149 (59.6) | 26.004 | <0.001 |
| High | 96 (67.1) | 47 (32.9) | |||
| ERCC1 | Low | 95 (41.3) | 135 (58.7) | 15.860 | <0.001 |
| High | 100 (61.7) | 62 (38.3) |
Expression of the proteins was classified as low and high by using a criterion of median value of all mean H-scores. P values obtained by chi-square test. SCC, squamous cell carcinoma.
Association of necroptosis and DDR proteins with patient survival
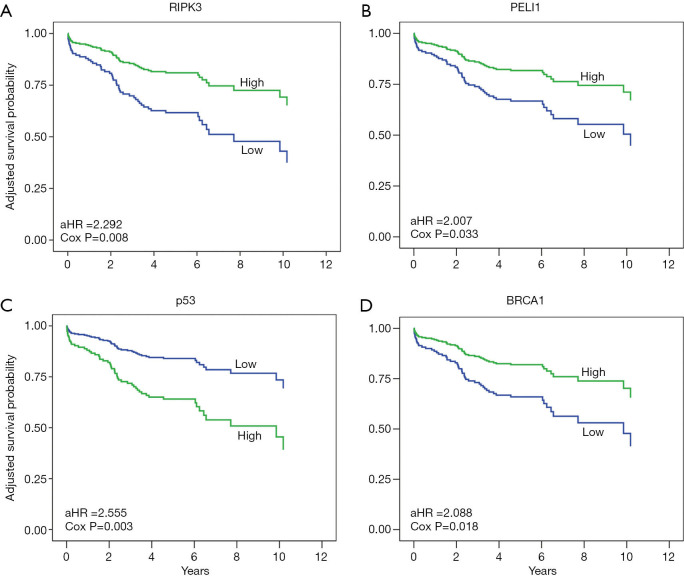
Analysis of the entire cohort revealed that expression of neither necroptosis nor DDR proteins was significant for the survival of the patients at the 5% confidence level (Table S3). However, histology results indicated that low-level expression of RIPK3, PELI1, or BRCA1 in conjunction with high-level expression of p53 was significantly detected in the univariate Cox proportional hazards model (Cox model) for the SCC-group patients (P=0.011, 0.019, 0.029, and 0.016, respectively). However, the effect of such protein expression was not apparent in the patients of the non-SCC group. In the SCC group, the prognostic effect of expression of these four proteins were statistically significant after adjusting clinical or pathological variables [adjusted hazard ratio (aHR) (95% CI)], P value, and P value corrected by FDR (q value) were 2.292 (1.242–4.228), 0.008, and 0.016, respectively, for low RIPK3; 2.007 (1.058–3.806), 0.033, and 0.033 for low PELI1; 2.555 (1.382–4.721), 0.003, and 0.012 for high p53; and 2.088 (1.133–3.850), 0.018, and 0.024 for low BRCA1 (Figure 1, Table 3). The estimated Harrell’s c-index was 0.73 (0.69–0.77), indicating that the final model was accurate enough.
Figure 1.
Survival of patients with stage I SCC according to the expression of (A) RIPK3, (B) PELI1, (C) p53 and (D) BRCA1 in adjusted models. aHR: hazard ratio after adjusting age, gender, smoking habit, Eastern Cooperative Oncology Group performance status, Charlson comorbidity index score, tumor size, T stage, lymphatic or vascular invasion, type of surgery, and adjuvant chemotherapy.
Table 3. Proteins involved in necroptosis and DNA damage response and survival of patients with stage I SCC: Multivariate Cox model.
| Proteins | Level | Wald | aHR (95% CI) | P value | Q value* |
|---|---|---|---|---|---|
| RIPK3 | Low vs. high | 7.045 | 2.292 (1.242–4.228) | 0.008 | 0.016 |
| PELI1 | Low vs. high | 4.546 | 2.007 (1.058–3.806) | 0.033 | 0.033 |
| p53 | High vs. low | 8.961 | 2.555 (1.382–4.721) | 0.003 | 0.012 |
| BRCA1 | Low vs. high | 5.565 | 2.088 (1.133–3.850) | 0.018 | 0.024 |
Expression of the proteins was classified as low and high by using a criterion of median value of all mean H-scores. *, FDR corrected P values after adjustment for multiple comparisons. aHR, hazard ratio after adjusting for age, gender, smoking habit, Eastern Cooperative Oncology Group performance status, Charlson comorbidity Index score, tumor size, T stage, lymphatic or vascular invasion, type of surgery, and adjuvant chemotherapy.
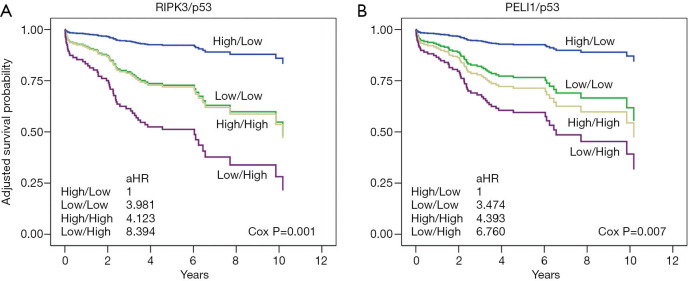
Combined effects of necroptosis and DDR protein expression on patient survival
Combined effects of a protein pair were analyzed to determine whether any combination of proteins involved in necroptosis and the DDR pathway had a synergy on patient survival. We noted that the combination of RIPK3 with p53 potentiated the multiplicative effects in the patients with SCC (P value and q value, 0.001 and 0.005) (Figure 2A, Table S4). Patients with low RIPK3 and high p53 expression had markedly worse survival than patients with high RIPK3 and low p53 expression [aHR (95% CI): 8.394 (2.856–24.677)] (Figure 2A, Table S4). Similarly, the combination of PELI1 with p53 potentiated the multiplicative effects in patients with SCC [P value and q value, and aHR (95% CI), 0.007, 0.018, and 6.760 (2.173–21.024) for low PELI1 and high p53] (Figure 2B, Table S4). The combined effect of RIPK3 with BRCA1 was associated with survival of the patients with SCC, but its significance was not observed (Table S4).
Figure 2.
Interaction effects of (A) RIPK3 or (B) PELI1 and p53 on the survival of SCC patients in adjusted models. aHR: hazard ratio after adjusting age, gender, smoking habit, Eastern Cooperative Oncology Group performance status, Charlson comorbidity index score, tumor size, T stage, lymphatic or vascular invasion, type of surgery, and adjuvant chemotherapy.
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of the Inha University Hospital (2016-12-011) and the informed consent requirement was waived.
Discussion
In summary, our analysis has revealed that expression of the proteins involved in necroptosis, such as RIPK3, is a robust indicator of poor prognosis for patients with the stage I SCC, but not for patients with non-SCC. We note that the effect of low-level expression of necroptotic factors such as RIPK3 on the increased death risk becomes clearer when the same patient displays high-level p53 expression.
Studies using cancer cell lines and animal models have shown that RIPK3 expression has diverse effects in tumor progression (4,6,7). Our data show that the SCC patients with low RIPK3 expression have an approximately 2.3-times increased risk of death relative to those with high RIPK3 expression. This observation indicates that as found in patients with esophageal SCC, RIPK3 inhibits tumor progression in the patients with SCC (10). Expression of another necroptosis factor PELI1 is also known to have diverse effects on survival in patients with diffuse large B cell lymphoma or melanoma (21,22). We found that the SCC patients with low expression of PELI1 had an approximately twice increased death risk when compared with those with high expression. Diversities of tumor types or tumor microenvironments may account for differences in the roles for RIPK3 and PELI1 in tumor progression.
Our data also reveal a synergistic effect of expression of either RIPK3 or PELI1 with that of p53. Patients with low-RIPK3 and high-p53 expression showed an approximately 8.4-times increased death risk relative to those with high-RIPK3 and low-p53 expression. Similarly, the combination of low PELI1 expression and high p53 expression increased the risk of death up to 6.7 times when compared with that of high-PELI1 and low-p53 expressions. Notably, a large proportion of patients (approximately 45%) had either low-RIPK3 and high-p53 expression or high-RIPK3 and low-p53 expression. This result suggests that the use of these biomarker combinations could help better classify the patients with SCC for their prognosis. The result that the combination of low RIPK3 and high p53 expression can promote tumor progression was expected because high expression of p53 indicates an abnormal function. Either RIPK3 or PELI1 may somehow interact with p53 in the patients with SCC. Indeed, it was reported that in the human fimbrial epithelium RIPK3 function depends on the status of p53 and PELI1 can regulate p53 function by promoting cytoplasmic localization of MDMX (12,22). Given these, the potential interaction between either RIPK3 or PELI1 and p53 thus merits further investigation using cell lines and preclinical models of lung cancer.
Our findings should be interpreted with caution. Of note, we here adopted a retrospective study design. Therefore, we did not evaluate recurrence free survival as alternative endpoint because of the lack of stringent strategy for follow-up. The results were not validated using an independent cohort. However, we think the sample size was large enough to evaluate the effects of protein expression. Also, our present study was designed to maintain the homogeneity of the study population. We used stringent criteria for both inclusion and exclusion of patients’ data to minimize selection biases. Information on clinical or pathological variables as potential confounders was comprehensively collected and extensively considered during the analysis. We did not provide a biological basis that might directly supports the differential prognostic effects between the two histological groups. However, our findings may be supported by previous mechanistic studies with cancer cell lines or animal models (4-7,10-13,22,23) as well as the differences in genomic architectures and clinical profiles between these groups (16,17). All in all, our findings demonstrate a clear translational relevance of the decreased expression levels of select necroptosis factors in predicting a poor prognosis of the SCC-subtype NSCLC.
Conclusions
This is the first study to demonstrate prognostic implications of necroptosis proteins and their association with expression of DDR proteins in patients with stage I NSCLC. Our results indicate that RIPK3 and PELI1 or their individual interaction with p53 can help classify patients with SCC into groups of either low or high death risk, providing a novel insight for clinicians into which patients would require thorough follow-ups.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank SuperBioChips (Seoul, South Korea) for the service of tissue microarray (TMA) construction and immunohistochemical staining of FFPE blocks.
Funding: This work was supported by a grant (NRF-2017R1E1A1A01074863) from the National Research Foundation of Korea (NRF).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of the Inha University Hospital (2016-12-011) and the informed consent requirement was waived.
Footnotes
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1027
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1027
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1027). The authors have no conflicts of interest to declare.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 2.Green DR, Evan GI. A matter of life and death. Cancer Cell 2002;1:19-30. 10.1016/S1535-6108(02)00024-7 [DOI] [PubMed] [Google Scholar]
- 3.Linkermann A, Green DR. Necroptosis. N Engl J Med 2014;370:455-65. 10.1056/NEJMra1310050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeon YK, Kim CK, Hwang KR, et al. Pellino-1 promotes lung carcinogenesis via the stabilization of Slug and Snail through K63-mediated polyubiquitination. Cell Death Differ 2017;24:469-80. 10.1038/cdd.2016.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi SW, Park HH, Kim S, et al. PELI1 Selectively Targets Kinase-Active RIP3 for Ubiquitylation-Dependent Proteasomal Degradation. Mol Cell 2018;70:920-35.e7. 10.1016/j.molcel.2018.05.016 [DOI] [PubMed] [Google Scholar]
- 6.Hockendorf U, Yabal M, Herold T, et al. RIPK3 Restricts Myeloid Leukemogenesis by Promoting Cell Death and Differentiation of Leukemia Initiating Cells. Cancer Cell 2016;30:75-91. 10.1016/j.ccell.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 7.Seifert L, Werba G, Tiwari S, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016;532:245-9. 10.1038/nature17403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi CM, Yang SC, Jo HJ, et al. Proteins involved in DNA damage response pathways and survival of stage I non-small-cell lung cancer patients. Ann Oncol 2012;23:2088-93. 10.1093/annonc/mdr606 [DOI] [PubMed] [Google Scholar]
- 9.Friboulet L, Olaussen KA, Pignon JP, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med 2013;368:1101-10. 10.1056/NEJMoa1214271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, Zhai L, Ma S, et al. Down-regulation of RIP3 potentiates cisplatin chemoresistance by triggering HSP90-ERK pathway mediated DNA repair in esophageal squamous cell carcinoma. Cancer Lett 2018;418:97-108. 10.1016/j.canlet.2018.01.022 [DOI] [PubMed] [Google Scholar]
- 11.Wang HH, Wu ZQ, Qian D, et al. Ablative Hypofractionated Radiation Therapy Enhances Non-Small Cell Lung Cancer Cell Killing via Preferential Stimulation of Necroptosis In Vitro and In Vivo. Int J Radiat Oncol Biol Phys 2018;101:49-62. 10.1016/j.ijrobp.2018.01.036 [DOI] [PubMed] [Google Scholar]
- 12.Wu NY, Huang HS, Chao TH, et al. Progesterone Prevents High-Grade Serous Ovarian Cancer by Inducing Necroptosis of p53-Defective Fallopian Tube Epithelial Cells. Cell Rep 2017;18:2557-65. 10.1016/j.celrep.2017.02.049 [DOI] [PubMed] [Google Scholar]
- 13.Koo GB, Morgan MJ, Lee DG, et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res 2015;25:707-25. 10.1038/cr.2015.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 15.Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. 10.1016/j.jtho.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 16.Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. 10.1038/ng.3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Socinski MA, Obasaju C, Gandara D, et al. Clinicopathologic Features of Advanced Squamous NSCLC. J Thorac Oncol 2016;11:1411-22. 10.1016/j.jtho.2016.05.024 [DOI] [PubMed] [Google Scholar]
- 18.Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2:706-14. 10.1097/JTO.0b013e31812f3c1a [DOI] [PubMed] [Google Scholar]
- 19.Lee HS, Kim WH, Kang GH. Hepatocyte expressions in hepatocellular carcinomas, gastrointestinal neoplasms, and non-neoplastic gastrointestinal mucosa: its role as a diagnostic marker. J Korean Med Sci 2003;18:842-8. 10.3346/jkms.2003.18.6.842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal statistical society: series B (Methodological) 1995;57:289-300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 21.Park HY, Go H, Song HR, et al. Pellino 1 promotes lymphomagenesis by deregulating BCL6 polyubiquitination. J Clin Invest 2014;124:4976-88. 10.1172/JCI75667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Tavana O, Sun SC, et al. Peli1 modulates the subcellular localization and activity of Mdmx. Cancer Res 2018;78:2897-910. 10.1158/0008-5472.CAN-17-3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber K, Roelandt R, Bruggeman I, et al. Nuclear RIPK3 and MLKL contribute to cytosolic necrosome formation and necroptosis. Communications Biology 2018;1:6. 10.1038/s42003-017-0007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as