Abstract
As a highly diverse vertebrate class, bird species have adapted to various ecological systems. How this phenotypic diversity can be explained genetically is intensively debated and is likely grounded in differences in the genome content. Larger and more complex genomes could allow for greater genetic regulation that results in more phenotypic variety. Surprisingly, avian genomes are much smaller compared to other vertebrates but contain as many protein-coding genes as other vertebrates. This supports the notion that the phenotypic diversity is largely determined by selection on non-coding gene sequences. Transfer RNAs (tRNAs) represent a group of non-coding genes. However, the characteristics of tRNA genes across bird genomes have remained largely unexplored. Here, we exhaustively investigated the evolution and functional consequences of these crucial translational regulators within bird species and across vertebrates. Our dense sampling of 55 avian genomes representing each bird order revealed an average of 169 tRNA genes with at least 31% being actively used. Unlike other vertebrates, avian tRNA genes are reduced in number and complexity but are still in line with vertebrate wobble pairing strategies and mutation-driven codon usage. Our detailed phylogenetic analyses further uncovered that new tRNA genes can emerge through multiplication by transposable elements. Together, this study provides the first comprehensive avian and cross-vertebrate tRNA gene analyses and demonstrates that tRNA gene evolution is flexible albeit constrained within functional boundaries of general mechanisms in protein translation.
Keywords: tRNA annotation, tRNA gene usage, codon usage, transposons, vertebrate, comparative genomics
Significance
In accordance to the central dogma, organisms use a diverse set of transfer RNA (tRNA) genes to translate messenger RNAs (mRNA) into proteins. The evolution of tRNA diversity has mainly been studied on a kingdom level, comparing Bacteria, Archaea, and Eukarya. We present the first comprehensive overview of tRNA evolution across vertebrate classes. Surprisingly, we found that although the number of protein-coding genes is highly conserved across vertebrates, tRNA gene number and complexity is greatly reduced in bird genomes compared to other vertebrates. Despite this decrease millions of years ago, the pool of tRNA anticodons and mRNA codons is still balanced across bird species to ensure optimal translational efficiencies. Moreover, the repertoire of tRNA genes is still dynamically changing due to the activities of transposable elements that contribute to novel tRNA gene copies. This unexpected finding provides for the first time a link between genome evolvability and translation.
Introduction
With over 10,000 species, birds represent a large and diverse vertebrate class. Bird species experienced several bursts of diversification and adapted morphologically, ecologically, and behaviorally to a wide range of habitats (Tobias et al. 2020). Despite these enormous evolutionary expansions, avian genomes are small in size (about 1 Gb) with stable karyotypes and large syntenic regions, making them an exceptional study system for understanding vertebrate genome evolution (Koepfli et al. 2015). Recently, researchers have focused on the evolution of several types of non-coding elements of avian genomes (Kapusta and Suh 2017; Yusuf et al. 2020). However, transfer RNA (tRNA) genes as part of the non-coding gene family have received less attention.
tRNAs are indispensable molecules for delivering amino acids to the ribosomes during the translation of messenger RNA (mRNA) into proteins. Furthermore, recent studies have attributed additional vital functions of tRNAs in controlling gene expression and cellular stress responses (Raina and Ibba 2014; Kirchner and Ignatova 2015). tRNA genes are either clustered or scattered throughout the vertebrate genome (Coughlin et al. 2009; Tang et al. 2009; Bermudez-Santana et al. 2010; Kutter et al. 2011). After transcription by RNA polymerase III (Pol III), the typically 76–90 nucleotides (nt) long tRNA molecule folds into a secondary cloverleaf-like structure with several hairpin loops. One of these loops contains the anticodon, which is complementary to the codons in mRNAs. In general, tRNA genes can be classified into anticodon isoacceptor families based on the encoded 20 standard amino acid isotypes and selenocysteine. Not all tRNA genes encode for the 62 possible isoacceptors (Maraia and Arimbasseri 2017). For example, based on Pol III binding, mammals use about 46 different isoacceptors, depending on the species, and the additional tRNAGly(ACC) is only found in rodents (Kutter et al. 2011). The absence of certain isoacceptors is explained by wobble base pairing, in which the third anticodon position can deviate from the standard Watson–Crick base pairing, allowing for the translation of multiple synonymous codons by a single tRNA (Crick 1966). Furthermore, tRNA modifications at base 34 in the anticodon loop improve translational efficiencies (Grosjean et al. 2010; Novoa et al. 2012). Despite accounting for tRNA modification strategies, the gene copy number of particular tRNA isoacceptors can be highly variable (Marck and Grosjean 2002; Goodenbour and Pan 2006). Several studies have proposed that tRNA copy numbers are related to codon usage bias, which is explained by the use of synonymous codons at different frequencies in proteins (Hershberg and Petrov 2008). Balancing the pool of tRNA molecules with the demand of codons during protein synthesis can increase translational efficiency (Quax et al. 2015). Indeed, the number of tRNA isoacceptors is positively correlated with codon usage in bacteria and yeast (Ikemura 1985; Rocha 2004). This correlation appears to be weaker in eukaryotes (Kanaya et al. 2001; Reis et al. 2004) in which codon usage bias is driven by mutational biases, such as variation in GC content due to meiotic recombination (Hershberg and Petrov 2008; Pouyet et al. 2017). However, correlations based on tRNA gene copy numbers do not consider that the pool of tRNA isoacceptors is highly dynamic across cell types and developmental stages. When actual expression levels are taken into account, the abundance of tRNA isoacceptors correlates with the codon usage of expressed protein-coding genes in different tissues (Dittmar et al. 2006; Kutter et al. 2011; Schmitt et al. 2014; Rudolph et al. 2016).
Estimating the number of tRNA isoacceptors can be thwarted by the presence of transposable elements (TEs). As selfish and highly active genetic elements, TEs incorporate themselves into genomes by either a copy-paste or a cut-paste mechanism (Kazazian 2004) and thereby continuously give rise to new genomic loci. Specifically, short interspersed elements (SINEs) can create sequences that are difficult to discriminate from standard tRNA genes (Weiner 2002; Kramerov and Vassetzky 2005), which can hamper sequence analyses (Coughlin et al. 2009; Tang et al. 2009). Discriminating between standard and TE-associated tRNA genes is therefore important for explaining varying numbers of tRNA genes and the diversity of tRNA isoacceptors in vertebrate genome evolution.
In this study, we explored how the number and genomic distribution of tRNA genes across vertebrates contribute to genome evolution. Remarkably, bird genomes show an unprecedented reduction of tRNA gene number, yet tRNA gene usage is still in alignment with vertebrate codon usage to ensure optimal translational efficiency. Deeper inspection of the diversity and evolutionary dynamics of tRNA genes in 55 genomes from every bird order (Kraus and Wink 2015; Jarvis 2016; Ottenburghs et al. 2017) revealed striking differences and commonalities in tRNA gene evolution across narrow and wide evolutionary timescales that is in part driven by species-specific TE activity.
Results
The Quality of Bird Genome Assemblies Influences tRNA Gene Detection
We identified tRNA genes in the genomes of 55 bird species representing all bird orders across different evolutionary distances, which were selected based on the availability of high-quality genome assemblies (supplementary table S1–S4, Supplementary Material online). To predict tRNA genes, we used tRNAscan-SE 1.3 (Materials and Methods) that detects characteristic tRNA promoter sequences and structural features. To determine the reliability of tRNAscan-SE 1.3 in predicting tRNA genes, we compared our estimates with previously published data of three bird species (chicken, zebra finch, and turkey) (Chan and Lowe 2016). Significant relations (linear model, F=65–290, adjusted R2=0.75–0.93, P<0.01) supported that our estimates are consistent but also indicated variation due to differences in the quality of the genome assemblies (supplementary table S5, Supplementary Material online). We therefore inspected systematically whether differences in the quality of the genome assembly have an effect on identifying tRNA genes (supplementary fig. S1, Supplementary Material online). A significant relation between scaffold N50 length and number of tRNA genes (linear model, F=26.73, adjusted R2=0.32, P<0.01) suggested that better assembled genomes contain regions comprised of tRNA genes that are absent in lower quality genome assemblies. The absence of these tRNA genes may consequently lead to an underestimation in the number of tRNA genes in lower quality genome assemblies. We divided the genomes in our dataset into high- and low-quality avian groups separated by a scaffold N50 threshold of 10 kb (supplementary fig. S1A, Supplementary Material online). The high-quality avian group contained 34 genomes, which were assembled at the chromosome and scaffold level. We annotated a mean of 202 tRNA genes, ranging from 134 in house sparrow (Passer domesticus) to 377 in spot-billed duck (Anas zonorhyncha). In contrast, the 21 genomes in the low-quality avian group encompassed an average of 118 tRNA genes, ranging from 84 in American flamingo (Phoenicopterus ruber) to 183 in brown mesite (Mesitornis unicolor). The number of tRNA genes was significantly different in both groups (t-test, t=7.1, P<0.01, supplementary fig. S1A, Supplementary Material online), which underlined dependency on genome assembly quality.
We further tested whether the limitation in the quality of the genome assembly can be overcome by grouping tRNA genes into isoacceptor families. Our analysis revealed that isoacceptor families varied in the genomes of the high-quality avian group (range: from 41 in hoatzin (Opisthocomus hoazin) to 48 in North Island brown kiwi (Apteryx mantelli)) and low-quality avian group (range: from 35 in American flamingo (P. ruber) to 45 in brown mesite (M. unicolor)). This suggests that the number of tRNA isoacceptor families might have been underestimated in the low- compared to the high-quality avian genome assembly group (t-test, t=5.9, P <0.01, supplementary fig. S1B, Supplementary Material online) and varying numbers of detectable tRNA genes per genome cannot be fully compensated by grouping on the isoacceptor level.
Genome scaffolds are composed of contigs linked by “N” gaps and could miss potential tRNA genes on short contigs. We therefore repeated our analysis for contig N50 length of the sampled genomes. Our results did not show significant relations between contig N50 length and number of tRNA genes (linear model, F=3.5, adjusted R2=0.05, P=0.07, supplementary fig. S1C, Supplementary Material online) or isoacceptor families (linear model, F=2.93, adjusted R2=0.03, P=0.09, supplementary fig. S1D, Supplementary Material online). This demonstrates that most tRNA genes were annotated in the sampled genome assemblies and that inclusion of contig N50 length assemblies will not significantly improve our analysis. In summary, tRNA gene identification is robust in high-quality avian genome assemblies and trends can be supported by using low-quality avian genome assemblies.
Bird Genomes Contain Considerably Fewer tRNA Genes Than Other Vertebrates
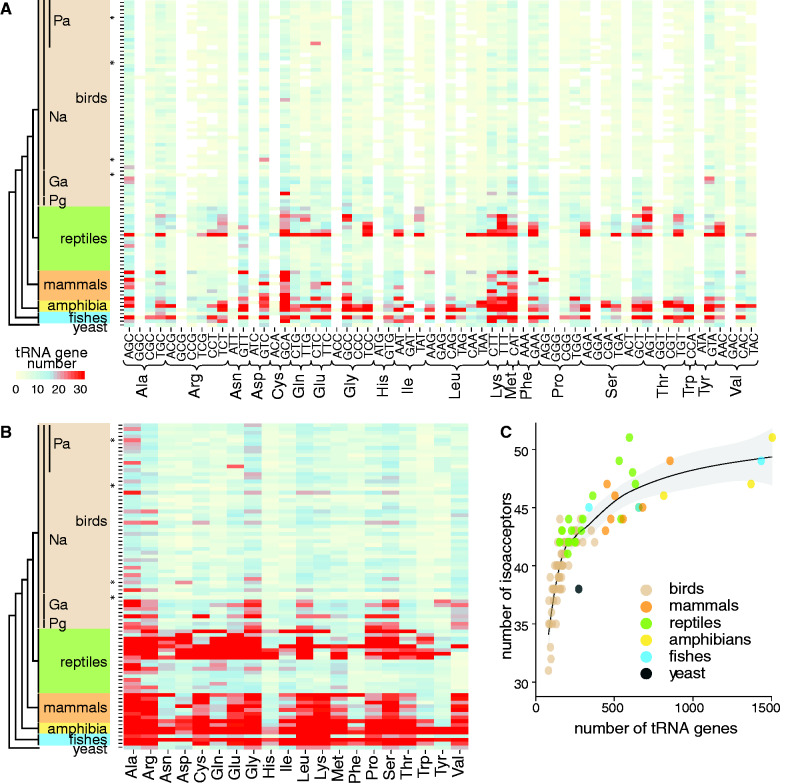
After exploring the number of tRNA genes in the low- and high-quality genomes of birds, we determined the number of tRNA genes in other vertebrate genome assemblies. On average, bird genomes house 169 tRNA genes (range: 84–377). Regardless of the genome assembly quality, the number of the tRNA genes in birds was significantly lower than in the genomes of reptiles (average=466; range: 155–1,610, divergence from birds 280 million years ago (MYA)), mammals (average=579; range: 445–855; 312 MYA), amphibians (average=1,229; range: 815–1,504; 352 MYA), and fishes (average=813; range: 343–1,438; 435 MYA) (high- and low-quality group versus vertebrate unpaired two-sample Wilcoxon test, W=3.5, P<0.01 and W=10.5, P<0.01, respectively). We noticed a difference in tRNA gene number in the reptile lineage, in which crocodile and turtle genomes contain more tRNA genes than lizard and snake genomes. The number of tRNA genes in crocodiles and turtle genomes was similar to mammals. In contrast, the tRNA gene number in lizard and snake genomes were comparable to the Galloanseres and Palaeognathae in birds or unicellular eukaryotes, such as yeast (275 tRNA genes; 1,105 MYA) (fig. 1A and B and supplementary table S1, Supplementary Material online). Remarkably, calculating the ratio of protein-coding and tRNA genes showed significantly higher values for bird genomes when compared to other vertebrate classes and yeast (Kruskal–Wallis test, χ2=34.82, P<0.01), suggesting that bird genomes have a more limited tRNA gene repertoire to translate mRNAs (fig. 1C). We did not detect any correlations between tRNA gene number and genome size (Spearman’s rank correlation, ρ=0.11, P=0.42), genomic GC content (Spearman’s rank correlation, ρ=0.15, P=0.28), weight (female, Spearman’s rank correlation, ρ=0.09, P=0.52; male, Spearman’s rank correlation, ρ=0.17, P=0.24), locomotion (flightless vs. flying, t-test, t=1.7, P=0.18), or geographic origin (Old World vs. New World, t-test, t=–1.4, P=0.19) (fig. 1D–H).
Fig. 1.
Varying number of tRNA genes in vertebrate genomes. (A) The phylogenetic tree (left) illustrates the evolutionary ancestry of Eupasseres (Eu), Passeriformes (Pa), Neoaves (Na), Galloanseres (Ga), Palaeognathae (Pg) within birds (brown), reptiles (green), mammals (orange), amphibians (yellow), fishes (blue), and yeast (grey). The horizontal bar plot shows the number of standard tRNA genes per genome (color-coded by vertebrate class) and TE-associated tRNAs families (black). Genomes marked by an asterisk indicate low-quality assemblies. Drawings of bird species investigated further in this study are inserted and scaled according to their height. (B) The density plot (top) shows the distribution (curve) and the median value (dot) of tRNA genes per vertebrate genome. (C) The boxplot displays the ratio of the number of protein-coding and tRNA genes per vertebrate genome. Plots correlating avian tRNA gene number to (D) genome size, (E) GC content, (F) weight (log-scale) (Dunning 2007) of females (red) and males (blue), (G) form of locomotion, and (H) geographic origin.
Bird tRNA Genes Represent Fewer Isoacceptor Families but All Isotype Classes
By grouping tRNA genes in bird genomes, we found on average 43 (range: 35–48) of the 61 possible tRNA isoacceptor families (fig. 2A). Irrespective of the genome assembly quality, significantly more tRNA isoacceptor families were found in the other vertebrate genomes (reptiles: 44–49, mammals: 43–49, amphibians: 46–51, and fishes: 45–49) (high- and low-quality group vs. vertebrate unpaired two-sample Wilcoxon test, W=119, P<0.01 and W=118.5, P<0.01, respectively). This suggests that genomes with a higher number of tRNA genes tend to harbor more tRNA isoacceptor families (fig. 2B and C).
Fig. 2.
Varying umber of tRNA isoacceptors and isotypes in vertebrate genomes. Heatmaps display the number of tRNA genes (yellow, low; blue, median; red, high, and white, genomic absence of genes for a tRNA isoacceptor family) grouped by (A) isoacceptors and by (B) isotypes for different vertebrate genomes ordered as in figure 1 and arranged into birds (brown), reptiles (green), mammals (orange), amphibians (yellow), fishes (blue), and yeast (grey). Order of species names as in figure 1A. Bird genomes marked by an asterisk (from top to bottom: zebra finch, red-legged seriema, brown mesite, and great crested grebe) deviate from anticodon-sparing pattern. (C) Plot displays the relationship between the number of tRNA genes (x-axis) and tRNA isoacceptors (y-axis) across vertebrate genomes and yeast (color-coded per vertebrate class). Regression (black line) and 95% confidence interval (grey) are shown.
Despite the varying number of tRNA isoacceptor families across low- and high-quality avian genome assemblies, all mRNA codons can be decoded during protein translation due to wobble pairing. Preferences for certain tRNA isoacceptor families were in line with the general wobble pairing strategies in eukaryotic genomes (fig. 2), such as G34 anticodon sparing where the enzyme hetADAT catalyzes the conversion of adenine-34 to inosine-34 in specific isoacceptors. This conversion enables position 34 to wobble with adenine, cytosine, and uridine. Because of the selection for tRNA isoacceptors modified by hetADAT, members from the same isoacceptor family encoding a G at position 34 are expected to be absent or occur at lower frequencies (Grosjean et al. 2010; Novoa et al. 2012). However, we observed exceptions from the general wobble pairing strategies. First, the red-legged seriema (Cariama cristata) genome contained more tRNA genes encoding for isoacceptor family tRNASer(GGA) than for tRNASer(AGA). Second, an equal number of tRNA genes corresponded to isoacceptor families tRNAArg(GCG) and tRNAArg(ACG) in great crested grebe (Podiceps cristatus). Third, the brown mesite (M. unicolor) genome housed the same number of tRNA genes for isoacceptor families tRNAHis(ATG) and tRNAHis(GTG). Finally, the zebra finch (Taeniopygia guttata) genome encompassed the same number of tRNA genes for isoacceptor families tRNACys(ACA) and tRNACys(GCA). Apart from zebra finch, the genomes of these exceptions belonged to the low-quality group. Further improvements in bird genome assemblies might reveal the presence of more tRNA isoacceptor families, which could be closer to the number found in other vertebrates.
tRNA genes identified in the bird genomes represented all 20 standard amino acid isotype classes. Each isotype class is encoded by 1–37 tRNA genes in birds (fig. 2B), whereas this range was much broader in the other vertebrates (reptiles: 2–187, mammals: 3–207, amphibians: 13–225, fishes: 4–151 tRNA genes). Some bird lineages showed an overrepresentation of gene copies in certain tRNA isotype classes, defined as the top 90% percentile in an isotype class. Repeating this analysis by excluding low-quality genomes did not change the resulting patterns. We observed an overrepresentation of gene copies for particular isotype classes in the flightless cormorant (Phalacrocorax harrisi), downy woodpecker (Picoides pubescens), and in the two early-branching avian lineages Palaeognathae and Galloanseres irrespective of the filtering parameters used.
In summary, in comparison to other vertebrate genomes, bird genomes are composed of fewer tRNA genes and have less diverse isoacceptor families but harbor conserved isotype classes. This pointed toward a reduction in tRNA gene and isoacceptor number and complexity but maintained functional constraint on tRNA isotype classes in early avian evolution (figs. 1 and 2).
Microsynteny Reveals Highly Conserved tRNA Clusters in Bird Genomes
Previous reports described that multiple copies of tRNA genes are arranged in distinct clusters to spatially organize mammalian genomes (Dixon et al. 2012; Raab et al. 2012). We therefore investigated the arrangements of tRNA genes across chromosome models of high-quality avian (chicken, turkey, guineafowl, zebra finch, collared flycatcher, great tit, and house sparrow), reptilian (anole lizard), and mammalian (mouse and human) genome assemblies. For each genome, we defined a cluster of tRNA genes when the neighboring tRNA gene is within a 1, 5, or 10 kb window (Materials and Methods). Several chromosomes within the genome of an inspected species showed that the distribution of tRNA genes was significantly different from a random distribution (Fisher Exact Test, P value, supplementary table S6, Supplementary Material online), which confirmed frequent clustering of tRNA genes. In some cases, the significance was dependent on the window size, which can be explained by the density of tRNA clusters (i.e. the distance between neighboring tRNA genes). For example, the distribution of tRNA genes on chromosome 7 in the chicken genome was significantly different from a random distribution for window sizes of 5 kb and 10 kb but not for a window size of 1 kb. This means that tRNA genes in the chicken genome are frequently clustered within a distance of 1 kb and 5 kb. Within the bird clades, we detected frequent clustering of tRNA genes in the sampled Galloanseres compared to Passeriformes that diverged about 100 MYA (Jarvis et al. 2014) (fig. 3A and B and supplementary fig. S2, Supplementary Material online). Clustering was irrespective of window size. Galloanseres genomes had a wider range of tRNA clusters (range: 2–22 tRNA genes per cluster), whereas Passeriformes tRNA genes occurred in narrower clusters (range: 2–12 tRNA genes per cluster). Generally, we observed that many tRNA genes occur in pairs. Smaller size clusters were composed of a homogenous set of tRNA genes (i.e. tRNA genes belonging to the same isoacceptor families) and larger size clusters were more heterogenous (i.e. tRNA genes belonging to different isoacceptor families) (fig. 3A and B and supplementary fig. S2, Supplementary Material online). In addition, we uncovered tRNA gene clusters on chicken chromosomes 1, 2, 7, 15, and 18 that were shared by the sampled chromosome-level representatives of Galloanseres (i.e. turkey and guineafowl) and Passeriformes (i.e. great tit, starling, collared flycatcher, house sparrow, and zebra finch). Figure 3C and D depicts an example of such a tRNA cluster on chromosome 15 of the chicken genome, residing in between the neighboring protein-coding genes BRI3BP and UBC.
Fig. 3.
tRNA gene clustering and conservation in bird genomes. Histograms for (A) chicken and (B) zebra finch genome represent the number of tRNA genes (N) per cluster (x-axis) versus tRNA cluster frequency (y-axis) that can be either heterogenous (diverse tRNA isoacceptor families, black bar) or homogenous (same tRNA isoacceptor family, grey bar) in composition. (C) UCSC and NCBI genome browser tracks depict a representative example of a H3K4me3-bound tRNA gene cluster (highlighted in dark-grey) located between the BRI3BP and UBC genes (black) on chromosome 15 in the chicken genome. An unannotated gene (grey) is located in between both genes. Exons are shown as black boxes and transcriptional start sites with arrows. H3K4me3 binding is shown as red enrichment below input DNA (as baseline). The syntenic regions and phylogenetic relationship in eight bird species is displayed. The vertebrate conservation track shows degree of base pair conservation (77 species), sequence constraint (blue) and divergence (red). (D) Magnification of the tRNA gene cluster in (C). tRNA isotypes and isoacceptors for each tRNA gene are annotated. Altered tRNA gene annotation in flycatcher (yellow) and tRNA gene cluster duplication in zebra finch are emphasized. (E) MAFFT pairwise alignments between the syntenic region on chicken chromosome 15 and zebra finch. Conserved tRNA cluster is shaded in grey and the 10 kb flanking regions are shown. The x- and y-axis show base pair alignments in kb. (F) Horizontal bar chart represents percentages of chicken-anchored tRNA genes that are conserved in bird species for which UCSC syntenic blocks (pairwise BlastZ alignments) were available. (G) Evolutionary tree shows the degree of conservation of tRNA genes (in percentage) using the 23 Sauropsida whole‐genome alignment referenced to the collared flycatcher genome. Approximate lineage divergence (unscaled) in million years (MY) is indicated.
To confirm the cross-species syntenic relationship of these genomic regions, we aligned this tRNA cluster plus the 10 kb flanking regions of all tested bird species against chicken chromosome 15 by using MAFFT (fig. 3E and supplementary fig. S3, Supplementary Material online). Our results showed a high degree of conservation within this microsyntenic region. To assess the overall evolutionary divergence of tRNA genes across bird genomes, we used UCSC pairwise synteny nets available for chicken-anchored genome comparisons (Chiaromonte et al. 2002). We identified that 51% of all tRNA genes (n=301) identified in the chicken genome were unique when compared to turkey (n=355, 37 MYA), which is its closest relative within the Galloanseres. Between 22% and 48% of all tRNA genes remained conserved between chicken and other Neoaves (80 MYA) (fig. 3F and supplementary table S7, Supplementary Material online). This variation in tRNA gene conservation can be explained by the number of tRNA genes identified in their respective genomes, for example Galloanseres genomes contain on average 301 (range: 211–376) tRNA genes, whereas Neoaves genomes house on average 150 (range: 83–262) tRNA genes. Furthermore, there are differences in genome evolution dynamics, for example, the genomes of passerines (order Passeriformes) evolve about 50% faster than the genome of other bird species (Zhang et al. 2014). It has also been reported that intrachromosomal rearrangements are most prevalent in the zebra finch genome (Kapusta and Suh 2017). In addition to the local and pairwise comparisons, we used the 23 Sauropsida whole-genome alignments (Green et al. 2014; Craig et al. 2018) to estimate the evolutionary ancestry of tRNA genes. Similar to the pairwise comparisons, we observed that the majority of tRNA genes (57.5%) were species-unique, 40.3% were Neoaves-specific (80 MYA), and 5.5% were deeply rooted in the Sauropsida (352 MYA) (fig. 3G).
In conclusion, our results showed high conservation in the distribution of tRNA genes across the avian genomes and revealed much stronger cross-species synteny in comparison to mammals (Kutter et al. 2011).
A Large Fraction of Avian tRNA Genes Are Expressed
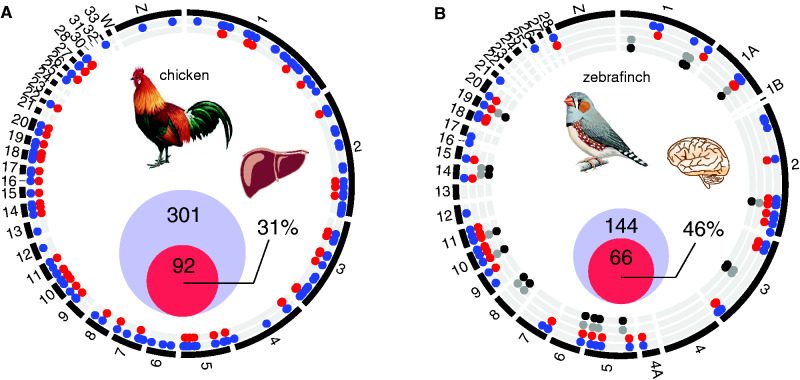
tRNA genes exist in multiple copies and are identical in sequence when belonging to the same isoacceptor family. As a consequence, sequencing reads will map to multiple tRNA gene locations. RNA-based methods are therefore inadequate to determine individual tRNA gene transcription. However, the 5' and 3' flanking regions of tRNA genes contain unique sequence information. We therefore generated and analyzed sequencing data obtained from chromatin immunoprecipitation (ChIP-seq) of factors binding to these unique genomic regions to demarcate tRNA gene usage (Materials and Methods). Nuclear-encoded Pol III-transcribed tRNA genes reside in active chromatin, which is frequently marked by histone 3 lysine 4 trimethylation (H3K4me3) (Barski et al. 2010; Moqtaderi et al. 2010; Oler et al. 2010; Kutter et al. 2011; Canella et al. 2012; Schmitt et al. 2014). We first mapped occupancy of H3K4me3 to tRNA genes in mammalian genomes. Our analysis showed that 20% (91/464) and 39% (170/435) of all annotated tRNA genes are marked by H3K4me3 in human and mouse livers, respectively (supplementary fig. S4, table S8, Supplementary Material online). By mapping Pol III binding to the genome, a higher number of actively used tRNA genes can be detected in livers of human (63%, 292/464) and mouse (72%, 311/435). Significant correlations between our estimates and previous studies (Kutter et al. 2011; Rudolph et al. 2016) showed that H3K4me3-enrichment is a reliable proxy for tRNA gene activity (human, linear model, adjusted R2=0.30, P<0.01; mouse, linear model, adjusted R2=0.72, P<0.01).
We next quantified tRNA gene usage in bird genomes by mapping H3K4me3 ChIP-seq data to the chicken and zebra finch genome (supplementary table S8, Supplementary Material online). We found that 31% (92/301) and 46% (66/144) of all tRNA genes were occupied by H3K4me3 in chicken liver and zebra finch brain, respectively (fig. 4). We permutated 100 genomic regions to rule out the possibility of a random association between H3K4me3 binding and tRNA gene locations. Of these permutated genomic regions, a mean of 0.23% (SD=0.25) for chicken and 0.08% (SD=0.17) for zebra finch overlapped with H3K4me3 signals, which is significantly lower than the observed 31% for chicken and 46% for zebra finch. Since all tRNA isoacceptor families and isotype classes were bound by H3K4me3, we reasoned that we have obtained a reliable estimate of actively used tRNA genes (supplementary fig. S5, Supplementary Material online). tRNA genes unbound by H3K4me3 could be either active in cell types not profiled in this study or reside in heterochromatic regions.
Fig. 4.
tRNA gene usage in bird genomes. Circular representation of the (A) chicken and (B) zebra finch genome. Each chromosome is proportionally scaled to its length. Tracks inserted in the circle represent the genomic location of annotated (blue) and H3K4me3-marked genes in chicken liver and zebra finch brain (red). In zebra finch, we annotated TE-associated tRNA genes (grey), which were marked by H3K4me3 (black). Two-way Venn diagrams show the proportional diameter of annotated (blue) and H3K4me3-marked tRNA genes (red). The percentage of H3K4me3-marked tRNA genes is depicted. Chromosome W in zebra finch has not been assembled.
Lineage-Specific tRNA Isoacceptors Contain Features of Different SINE-Derived TEs
Before filtering low-quality tRNA gene candidates, we noticed that several species exhibited an overrepresentation of certain tRNA isoacceptor families (supplementary fig. S6, Supplementary Material online). All Eupasseres (i.e. Passeriformes excluding rifleman, Acanthisitta chloris) showed an excess of tRNAIle(AAT) genes. In addition to the Eupasseres tRNAIle(AAT) genes (range: 20–175), we noted that golden-collared manakin (Manacus vitellinus) contained over 230 tRNAGlu(CTC) genes. An overrepresentation of 577 tRNAIle(AAT) genes was also apparent in Dalmatian pelican (Pelecanus crispus). In the genome of bar-tailed trogon (Apaloderma vittatum) a high number of tRNA genes were detectable, which belong to isoacceptor families tRNAAla(GGC) (n=676), tRNAIle(GAT) (n=1,147), tRNAIle(AAT) (n=67), tRNALeu(AAG) (n=27), tRNALeu(GAG) (n=34), tRNAPhe(AAA) (n=23), tRNAPhe(GAA) (n=53), tRNASer(GCT) (n=38), tRNAThr(GGT) (n=89) tRNAVal(GAC) (n=1,352), and tRNAVal(AAC) (n=1,365). Close inspection of the proximate flanking regions of these overrepresented tRNA genes revealed the presence of full-length SINE sequences directly positioned around the tRNA gene. We extended our analysis to all annotated tRNA genes and screened for overlaps with SINE sequences. This has led to the identification of novel lineage-specific PeleSINE1 in Dalmatian pelican and ApalSINE1 in bar-tailed trogon, which explained the overrepresentation of all eleven tRNA isoacceptors. In addition, we also detected the previously reported clade-specific TguSINE1 in Eupasseres and lineage-specific ManaSINE1 in manakin (Warren et al. 2010; Suh et al. 2016; Kapusta and Suh 2017) (supplementary fig. S6D, Supplementary Material online). tRNAIle was the most frequently occurring isotype class but the tRNAIle isoacceptor families were differently used across lineages, in that tRNAIle(GAT) was present in bar-tailed trogon whereas tRNAIle(AAT) was found in Eupasseres and Dalmatian pelican.
Interestingly, in all cases, the tRNA gene sequence was positioned 5' in the TE. Similar to tRNAs, active SINEs also contain Pol III promoter sequences and are therefore Pol III transcribed (Kramerov and Vassetzky 2011). In the absence of Pol III ChIP-seq data, we profiled H3K4me3 binding to inspect active usage of the TE-associated tRNA genes and found that 71% (20/28) were H3K4me3 occupied in zebra finch brain (fig. 4B). This suggests that transcriptionally active TEs containing full-length tRNA genes can contribute to shaping the pool of available tRNA genes in the genome.
tRNA Structure Predictions Reveal Evolutionary History of TEs
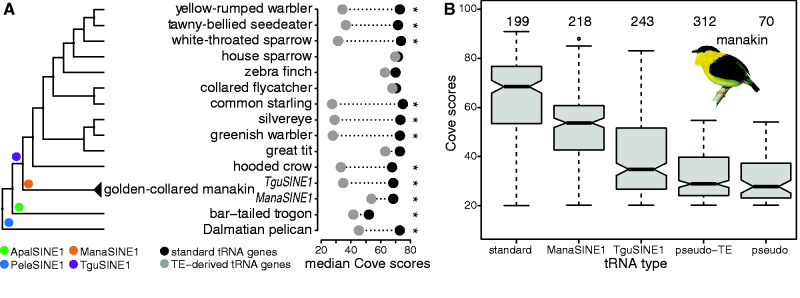
The probability of the predicted gene to function as a bona fide tRNA is calculated by the Cove program as part of tRNAscan-SE. The Cove score is assigned to each tRNA gene candidate based on the ability of a given sequence to form tRNA stem-loop structures and the presence of Pol III promoter and terminator sequences. Active tRNA genes have therefore high Cove scores. In contrast, inactive tRNA genes might have accumulated mutations and decay into pseudogenes, which leads to lower Cove scores. Similarly, TE-associated tRNA genes that have been silenced by epigenetic control mechanisms will gradually accumulate mutations after the TE insertion event, which also results in lower Cove scores. We took advantage of this feature to unravel the evolutionary history of full-length TE-associated tRNA genes by interrogating bird genomes with detectable TE-associated tRNA genes (14 of 55) (figs. 4B and 5A). In these bird genomes, TE-associated tRNA genes had significantly lower Cove scores (median 36.5) than standard (median 71.7) tRNA genes (Kruskal–Wallis Test, supplementary table S9, Supplementary Material online). However, the difference was not significant in zebra finch, collared flycatcher, house sparrow, and great tit (fig. 5A) for which we assume that the genome assembly quality might have impacted the calculation of Cove scores for TE-associated tRNA genes. However, there was no significant correlation between measures of genome assembly quality and the calculation of Cove scores across all avian genome assemblies (scaffold N50, Spearman’s rank correlation, ρ=0.06, P=0.6; contig N50, Spearman’s rank correlation, ρ=0.02, P=0.9).
Fig. 5.
Evolution of TE-associated tRNAs. (A) Right: phylogenetic tree displaying bird genomes with detectable TE-derived tRNA genes. Presence of branch-specific TEs is highlighted at the root of the clades (circles in different colors) and the name of the SINEs giving rise to TE-derived tRNA genes (legend bottom left). Left: median Cove scores of standard (black circle) and TE-derived (grey circle) tRNA genes per species are shown. Asterisks highlights significantly lower median Cove scores of TE-associated tRNA genes compared to standard tRNA genes. (B) Boxplots show decrease of Cove scores from standard tRNA genes to pseudogenes in the golden-collared manakin genome. ManaSINE1-associated tRNA genes have a higher Cove score than TguSINE1-associated tRNA genes because they appeared later in the phylogenetic history and thus accumulated fewer mutations. Numbers above each box entail the total gene number interrogated per tRNA type.
A remarkable example is the presence of two tRNA-containing TE families (TguSINE1 and ManaSINE1) in the golden-collared manakin genome (fig. 5A and B). These TE-associated tRNA genes have been active at different times during Passeriformes evolution (Suh et al. 2016, 2017). TguSINE1 was active about 30 MYA, while the activity of ManaSINE1 is more recent (about 5 MYA). Based on these temporal activity patterns, we hypothesized that the Cove scores for TguSINE1-derived tRNAs are lower compared to ManaSINE1-derived tRNAs since the evolutionarily older TguSINE1-derived tRNAs have had more time to accumulate mutations. Importantly, since we compared the Cove scores of the TE-associated tRNA genes within the same genome, we circumvented any potential biases in genome assembly quality. In line with our hypothesis, we calculated significantly different Cove scores between the Mana- and TguSINE1-derived tRNAs (post-hoc Tukey test, P<0.01, fig. 5B, supplementary table S10, Supplementary Material online). Our measurements confirmed that TguSINE1 was active before ManaSINE1 (Suh et al. 2016, 2017) in the manakin genome and then accumulated more mutations, which resulted in lower Cove scores. Similar trends were confirmed when determining Cove scores in the other bird genomes with TE-associated tRNA genes (supplementary table S10, Supplementary Material online). Importantly, our analysis and these notable examples illustrate that tRNA genes can be used to study the evolutionary history and biology of TEs.
Codon Usage in Birds Is Determined by Mutational Biases
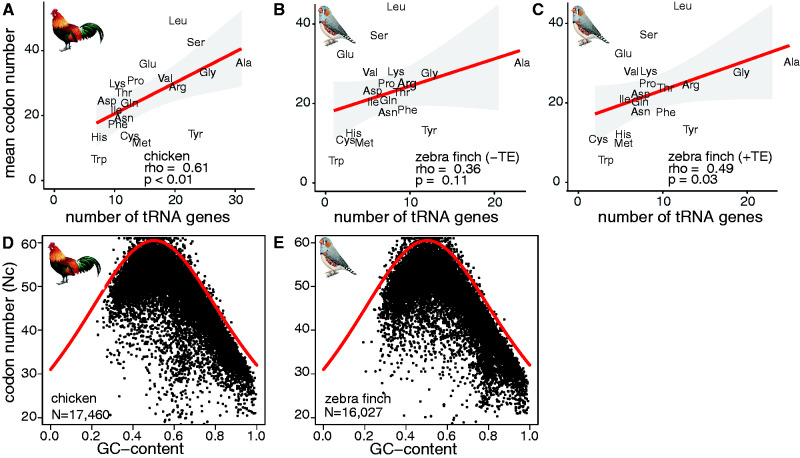
To infer whether codon usage in birds is driven by translational selection or mutational biases, we tested the correlations between codon usage and number of tRNA genes in four species (chicken, turkey, zebra finch, and collared flycatcher) for which protein-coding gene annotations were available. We determined significant correlations between the number of tRNA genes per isotype and the number of amino acids in codons of protein-coding genes in chicken (Spearman’s rank correlation coefficients, ρ=0.61, P<0.01) (fig. 6A) and turkey (Spearman’s rank correlation coefficients, ρ=0.72, P<0.01) (supplementary fig. S7A, Supplementary Material online). The correlations were not significant in zebra finch (Spearman’s rank correlation coefficients, ρ=0.36, P=0.11) (fig. 6B) and collared flycatcher (Spearman’s rank correlation coefficients, ρ=0.35, P=0.13) (supplementary fig. S7C, Supplementary Material online). Notably, the correlation improved when accounting for TE-derived tRNA genes for zebra finch (Spearman’s rank correlation coefficients, ρ=0.49, P=0.03) (fig. 6C) but not for collared flycatcher (Spearman’s rank correlation coefficients, ρ=0.34, P=0.14) (supplementary fig. S7C, Supplementary Material online). In general, the correlations were similar when analyzing either all annotated genes or only genes that have been mapped to chromosomes (supplementary table S11, Supplementary Material online).
Fig. 6.
Codon usage correlates with tRNA gene number and transcriptomic GC content in birds. Amino acid usage in codons of protein-coding genes (y-axis) are plotted against the number of tRNA genes per isotype (abbreviated by three-letter code) (x-axis) for (A) chicken, (B) zebra finch without (–TE), and (C) with (+TE) TE-associated tRNA genes. Regression (red line), 95% confidence interval (grey area), Spearman’s rank correlation coefficients (rho) and P values (P) are shown. For (D) chicken and (E) zebra finch, the effective number of codons (Nc) per protein-coding gene (N, y-axis) (black points) is plotted against the proportional GC content at the third codon position (x-axis). The red curve indicates the expected relationship if mutational biases affect codon usage.
Although significant for chicken and turkey, the correlations suggested that codon usage bias is not solely driven by translational selection. Next, we argued that if mutational biases are also influencing codon usage, a strong relationship between codon usage and GC content would be expected (Wright 1990; Smith et al. 2013). We therefore tested for GC content and effective codon numbers across all protein-coding genes. If a relationship exists, protein-coding genes follow a parabolic distribution. We first tested this trend for all annotated protein-coding genes in human and mouse (supplementary fig. S8A and B, table S12, Supplementary Material online) and found that the interrogated protein-coding genes were located on or close to the GC curve and thus are under GC content constraints. Our results corroborated previous observations for a limited number of mammalian protein-coding genes (Wright 1990). In contrast, yeast does not adhere to this relationship (supplementary fig. S8C, table S12, Supplementary Material online) as indicated before (Wright 1990). Similar to mammals and reptiles (supplementary fig. S8, table S12, Supplementary Material online), we found that protein-coding genes of four sampled bird species followed the distribution along the GC curve (fig. 6D and E, supplementary fig. S8E and F, table S12, Supplementary Material online). We concluded that similar to other vertebrates, but in contrast to yeast, codon usage bias in birds is driven by a combination of translational selection and mutational biases acting on GC content.
Discussion
This study provides the first comprehensive overview of tRNA diversity and evolution in vertebrates and deeply interrogates recently annotated genomes of major bird lineages. Overall bird genomes encode fewer tRNA genes and have fewer isoacceptor families than other vertebrates (figs. 1 and 2). This reduction could be a by-product of an evolutionary trend toward smaller genomes in birds through deletions of non-coding DNA. Alternatively, the lower numbers of tRNA genes could be a result of the avian genome structure. Based on their length, avian chromosomes can be divided into macro- (>50 Mb), intermediate- (20–40 Mb), and micro-chromosomes (<20 Mb) (International Chicken Genome Sequencing Consortium 2004). Micro-chromosomes, which tend to be absent in mammals, are difficult to sequence and to assemble but might house more tRNA genes (Warren et al. 2017; Peona et al. 2018). Our analyses could be partly impeded by varying quality of genome assemblies in that lower quality genome assemblies contained on average fewer tRNA genes. We accounted for this bias in our analysis. Although this did not markedly influence our results, it does emphasize the urge for improving already existing genome assemblies for non-model organisms. This may lead to a more detailed picture of tRNA gene evolution across the tree of life since genomic regions with long stretches of repetitive content could possibly harbor more tRNA genes (Korlach et al. 2017; Warren et al. 2017; Weissensteiner et al. 2017).
The high-quality chicken genome was used as a benchmark (Warren et al. 2017). We annotated 301 tRNA genes in the chicken genome, of which about half remain in chicken-specific genomic locations. At least 92 (31%) were actively used in liver. This number is probably a conservative estimate because the ChIP-seq approach in our study investigated only H3K4me3-binding sites. However, the tendency of not using all annotated tRNA genes is similar to mammals. A ChIP-seq experiment specifically targeting tRNA genes, for example by profiling Pol III binding, will likely uncover a higher number. However, the antigen-binding sites of the currently available Pol III antibodies required for a ChIP-seq experiment do not recognize Pol III subunits outside the mammalian lineage due to differences in the epitope sequences (data not shown).
Our comparison of tRNA genes and complexity in birds with other vertebrate classes conform to the overall eukaryotic patterns, such as similar wobble pairing strategies as well as codon usage bias driven by translational selection and mutational biases. The heteromeric deaminases complex (ADAT) catalyzes the conversion of adenine-34 to inosine-34 in the tRNA anticodon loop and enables this position to wobble with adenine, cytosine, and uridine (Gerber and Keller 2001). Mammals possess three ADAT genes, whereas birds have one or two (supplementary table S13, Supplementary Material online). The emergence of enzymes such as ADATs probably allowed for the efficient use of tRNA isoacceptors to improve translation efficiencies.
Our genome-wide survey of tRNA genes uncovered several lineage-specific TEs. Since they were bound by H3K4me3, we presume that they are transcriptionally active. The detected TEs are associated with particular tRNA isoacceptors. For example, all Eupasseres genomes contained a SINE element derived from tRNAIle(AAT) (Suh et al. 2017). Similarly, Dalmatian pelican and bar-tailed trogon genomes had a SINE element derived from tRNAIle(AAT) and tRNAIle(GAT), respectively. The golden-collared manakin genome housed tRNAs derived from the Eupasseres-specific TguSINE1 and the suboscine-specific ManaSINE1 (Suh et al. 2017, 2016). Different TE activity patterns were found in our analyses, in which we showed that Cove scores for TguSINE1-associated tRNAs were significantly lower compared to ManaSINE1-associated tRNAs (fig. 5B). The reason could be that the evolutionarily older TguSINE1-associated tRNAs have had more time to accumulate mutations after their genomic insertion.
Based on these patterns, we propose a model for TE-tRNA coevolution in certain lineages. This model consists of three phases. First, a TE recruits a copy of a tRNA gene for its own mobilization and increases its copy number in the genome. Second, the TE is silenced by epigenetic control mechanisms. Third, some TE-associated tRNA genes decay into pseudogenes, while others remain transcriptionally active and become coopted for their original tRNA function. The first phase of this model is clearly supported by our results. Lineages with a tRNA-associated TE showed a dramatic increase in the number of intact tRNA genes for particular tRNA isoacceptors. For instance, the bar-tailed trogon genome accommodates 760 tRNAAla and 2,750 tRNAVal candidate genes due to the activity of ApalSINE1. The second phase of the model, which encompasses the silencing of the TE by epigenetic control mechanisms, requires further investigation. TE silencing could be mediated by several possible mechanisms, such as heterochromatization by histones, changes in DNA methylation patterns, or activation of small RNAs (such as piwi-interacting RNAs) (Ernst et al. 2017; Kapusta and Suh 2017). How birds deal with TE-associated tRNA genes that could still retrotranspose remains to be determined. The third phase of the TE-tRNA coevolution model concerns the fate of TE-associated tRNA genes. Our results indicate that the majority of these genes become inactive and slowly decay into pseudogenes by accumulating mutations over time. However, some TE-derived tRNA genes might escape silencing and remain transcriptionally active. ChIP-seq data from chicken and zebra finch indicated that several TE-derived tRNA genes are actively transcribed. It remains to be determined whether this activity is due to functioning as tRNA gene or simply as active TE. The former would suggest domestication of particular TE-derived tRNA genes which could drive the evolution of tRNA gene number and complexity.
Materials and Methods
Identification of tRNA Genes
We used the program tRNAscan-SE 1.3 to identify tRNA genes in assembled vertebrate genomes available at the National Centre for Biotechnology Information (NCBI) including 8 mammals, 84 birds, 17 reptiles, 3 amphibians, 3 fish, and 1 fungus (supplementary table S1, Supplementary Material online). From the 84 initially analyzed bird genomes, we selected at least one genome per bird order. If multiple genomes per bird order were available, we opted for the highest quality genome assembly (based on scaffold N50 and contig N50 values) for further analyses, resulting in a final data set of 55 bird genomes. To pinpoint candidate tRNA genes in genomic sequences, tRNAscan-SE 1.3 uses two algorithms (tRNAscan 1.4 and EufindtRNA) that detect intragenic RNA Pol III promoters and terminators and consider both primary sequence and secondary structure information evaluated by a covariance model. This approach identifies 99% of all tRNA genes in a DNA sequence with less than one false positive per 15 Gigabases (Lowe and Eddy 1997). The covariance model assigns a Cove score to each tRNA gene, which indicates the quality of a predicted tRNA gene. Candidate genes that score low are considered pseudogenes. We applied a Cove score threshold value of 50 to remove low-quality tRNA gene predictions (Kutter et al. 2011). To infer the reliability of tRNAscan-SE 1.3, we compared our results with previously published data from the Genomic tRNA Database (Chan and Lowe 2016).
Before filtering low-quality tRNA genes, several species exhibited overrepresentations of particular tRNA genes. Close inspection revealed that these genes belong to specific TEs (hereafter referred to as TE-associated tRNA gene candidates), which we removed using the following approach. First, tRNA genes and flanking regions were aligned with MAFFT (Katoh et al. 2019). The resulting alignments were manually curated in MEGA6 (Tamura et al. 2013) and BLASTN (Altschul et al. 1990) and aligned against the TE consensus sequence to determine sequence similarity. TE consensus sequences were based on previously published data (TguSINE1 from zebra finch (Warren et al. 2010) and ManaSINE1 from manakin (Suh et al. 2016)) except for ApalSINE1 from bar-tailed trogon and PeleSINE1 from Dalmatian pelican (supplementary table S2, Supplementary Material online). ApalSINE1 and PeleSINE1 were identified through manually curated consensus sequences from multicopy tRNA gene alignments including flanking sequences, similar to previous in-depth TE annotations of birds (International Chicken Genome Sequencing Consortium 2004; Warren et al. 2010; Suh et al. 2018). We applied a lenient (60%) and a stringent (90%) similarity threshold to remove putative TE-associated tRNA genes. To test the reliability of this filtering, we applied the same approach to closely related species that do not contain TE-associated tRNA genes (Kapusta and Suh 2017) (supplementary table S3, Supplementary Material online).
tRNA Gene Cluster Analysis
The distribution of tRNA genes was computed for the chromosome-assembled genomes of chicken, turkey, guineafowl, zebra finch, collared flycatcher, great tit, and house sparrow (supplementary table S4, Supplementary Material online). We only considered tRNA genes mapping to a chromosome, that is, not located on unplaced contigs or scaffolds. We assigned tRNA genes to the same cluster if the distance between them was less than the genome-wide median distance between consecutive tRNA genes. For each cluster, we determined its size and content. If all tRNA genes of a cluster code for the same amino acid, the content was considered homogeneous. If clusters contained tRNA genes that code for different amino acids, the content was considered heterogeneous.
To test whether tRNA genes cluster on chromosomes, we divided each chromosome into windows of 1, 5 and 10 kb size. Next, we counted the number of tRNA genes in each window. The resulting distribution was compared to a random distribution by means of a Fisher Exact test. This analysis was only performed on chromosomes that contain at least 10 tRNA genes. For comparison, we repeated this analysis on the genomes of anole lizard, mouse and human. Given the higher stability of avian karyotypes relative to mammals (Ellegren 2010), we expected to find several evolutionary conserved tRNA clusters.
Mapping of Orthologous tRNA Genes
First, we compared the genomic regions of tRNA clusters on chicken chromosomes 1, 2, 7, 15 and 18 plus the 10 kb flanking regions with the chromosome-level assemblies of Galloanseres (turkey and guineafowl) and Passeriformes (great tit, starling, collared flycatcher, house sparrow, and zebra finch) by using MAFFT alignments (FFT-NS-2 method, version 7) and the adjust direction function to ensure proper orientation of all sequences (Katoh et al. 2019). Second, we assessed the overall evolutionary divergence of tRNA genes across bird genomes. We used UCSC’s BlastZ-based pairwise genome alignments (Chiaromonte et al. 2002) to identify syntenic tRNA genes between chicken and bird species (turkey, collared flycatcher, house sparrow, and zebra finch) for which previously calculated nets were available (supplementary table S7, Supplementary Material online). Genomic regions of chicken tRNA genes (without flanking regions) were queried. We only considered top-level blocks without gaps in the genomic regions of tRNA genes irrespective whether the entire block was reversed in the genome. tRNA genes within the conserved block had to be of the same isotype. Finally, we investigated the conservation of tRNA genes on a larger evolutionary scale by assessing precalculated conserved elements (CEs) in the collared flycatcher genome as part of a 23 Sauropsida whole genome alignment (Craig et al. 2018). We used the intersect-function in bedtools (version 2.29.2) with default settings to query the locations of collared flycatcher tRNA genes with the genomic coordinates of these CEs. This allowed us to infer whether tRNA genes were either species-unique or present in the lineages of Sauropsida (352 MYA), Archosaura (237 MYA), Aves (98 MYA), Neoaves (80 MYA), or Passerea (65 MYA).
Tissue Preparation
Chicken tissue samples were provided from the Poultry Production Unit at the BBSRC Institute of Animal Health, Compton, UK. Healthy livers from chicken (two females, two years old) were perfused with 1× phosphate-buffered saline (PBS) and cut in small pieces. Liver tissues were crosslinked in 1% formaldehyde (v/v) and neutralized by adding 250 mM glycine. Afterwards, cells were homogenized by tissue douncing and washed twice with 1× PBS.
Chromatin Immunoprecipitation Followed by High-Throughput Sequencing (ChIP-Seq) Library Preparation
Cells were lysed and sonicated to an average size of 250 bp Misonix 240 Sonicator 3000. ChIP-seq assays were performed as previously described (Kutter et al. 2011) using an antibody against tri-methylation of lysine 4 on histone 3 (H3K4me3, Millipore 05-1339). The immunoprecipitated cells were end-repaired, A-tailed, ligated to the sequencing adapters, amplified by 18 cycles of PCR, and size-selected (200–300 bp). DNA fragments were 45 bp single-end read sequenced on an Illumina Genome Analyser IIx according to manufacturer’s instructions (detailed under ArrayExpress submission E-MTAB-8106).
Short-Read Alignment and Peak Calling
To estimate the percentage of expressed tRNA genes, we analyzed H3K4me3 ChIP-seq generated from chicken livers for this study (E-MTAB-8106) and publicly available zebra finch brain tissues (GSE91399) (Kelly et al. 2018). Fastq files were aligned to their respective reference genomes (galGal5 and taeGut2) using HISAT2 (version 2.1.0) with default settings (Kim et al. 2015). Peaks were called with MACS2 (Feng et al. 2012) using an effective genome size of 1.2e9 (option –g) and filtering of binning intervals by a q-value of 0.01. tRNA genes plus their 100 bp flanking regions overlapping with an H3K4me3-enriched region were considered as actively expressed. To benchmark this approach, we repeated these analyses on published human and mouse liver data (E-MTAB-2633) (Villar et al. 2015) and compared these results with a previous study that specifically targeted tRNA genes by characterizing Pol III binding sites (E-MTAB-958) (Kutter et al. 2011).
To generate a background model for stochastic H3K4me3 binding events, we permutated 100 sets of 144 regions (75 bp each) from the chicken and zebra finch genome without filtering for any annotated genic regions. These sets plus their 100 bp flanking regions were overlapped with H3K4me3 peaks and binding events were scored.
Evolution of TEs
Because most TE-associated tRNA gene predictions are of low-quality (Cove score below the threshold of 50), analyses were performed on the unfiltered data sets. For each genome, tRNA gene candidates were divided into four categories: standard tRNA genes, TE-associated tRNA genes, pseudogenes and TE-associated pseudogenes. Differences in the Cove scores between these categories were tested with a non-parametric Kruskal-Wallis Test followed by a Tukey post-hoc test for multiple comparisons.
Codon Usage
We calculated codon usage per gene in four species (chicken, turkey, zebra finch, and collared flycatcher) for which gene annotations were available on the Ensembl BioMart database (release 93) (Zerbino et al. 2018). When multiple transcripts of one gene were present, we calculated the codon usage for the longest transcript. The codon usage was then correlated with the number of tRNA genes on an amino acid level. Analyses were performed on a data set containing all genes and a data set containing only genes that have been mapped to chromosomes.
Next, we calculated the effective number of codons per gene (Nc) following the formula derived by Wright (1990). Nc is a measure that quantifies the departure of a gene from random usage of synonymous codons. Its value ranges from 20 to 61, for example, an extremely biased gene uses only 20 codons (i.e. one per amino acid), whereas an unbiased gene uses all 61 codons equally. Because selection leads to a reduction in randomness of a sequence, Nc provides a reliable way to test for selection on codon usage. The relationship between Nc and GC-content at the third codon position (GC3) with no selection can be approximated by the following formula (Wright 1990; Reis et al. 2004), where x corresponds to GC3:
To test the validity of this approach, we compared our results with the original analysis of Wright (1990) on human and yeast. In addition, we assessed this relationship in anole lizard and mouse.
Data Visualization
Computational analyses were performed using Perl version 5.16.3 (www.perl.org), Python version 3.6.0 (www.python.org), R version 3.5.0 (www. r-project.org), and R packages: ggplot2, cowplot, plotly, gplots, Rmisc, ggtree, jpeg, ggpubr, grDevices, grid, OmicCircos, heatmaply, RColorBrewer, pBrackets, and vcd.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr. Venugopal Nair and Dr. Sarah Leigh Brown for help in obtaining the chicken tissue samples. Duncan Odom’s group and the Cambridge Institute-Genomics facility for sequencing the H3K4me3 ChIP-seq libraries and providing the fastq files. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project SNIC 2017/7-108. Leif Andersson (Uppsala University), Diem Nguyen (Uppsala University) and Duncan Odom (DKFZ Heidelberg) for critical feedback on the manuscript. Bird drawings were used with permission of “Handbook of Birds of the World” (del Hoyo et al. 2018). This work was supported by the Knut & Alice Wallenberg foundation (KAW 2016.0174), Ruth & Richard Julin foundation (2017-00358 and 2018-00328, CK), SFO-SciLifeLab fellow research program (SFO_2016-003, CK), Chinese Government Scholarship (2016-KG-01, KG, CK), Swedish Research Council Vetenskapsrådet (2016-05139, AS and 2019-05165, CK), and Swedish Research Council Formas (2017-01597, AS).
Author Contributions
J.O., K.G., A.S., and C.K. conceived experiments. J.O., K.G., and C.K. performed experiments. J.O., A.S., and C.K. analyzed the data. J.O., K.G., A.S., and C.K. wrote the paper.
Data Availability
ArrayExpress Accession
H3K4me3 ChIP-seq (chicken): E-MTAB-8106
H3K4me3 ChIP-seq (zebra finch): GSE91399
H3K4me3 ChIP-seq (mammals): E-MTAB-2633
Pol III ChIP-seq (mammals): E-MTAB-958
Code Accessibility
github.com/JenteOttie/Avian_tRNAs
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Barski A, et al. 2010. Pol II and its associated epigenetic marks are present at Pol III–transcribed noncoding RNA genes. Nat Struct Mol Biol 17(5):629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez-Santana C, et al. 2010. Genomic organization of eukaryotic tRNAs. BMC Genomics 11(1):270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canella D, et al. ; The CycliX Consortium. 2012. A multiplicity of factors contributes to selective RNA polymerase III occupancy of a subset of RNA polymerase III genes in mouse liver. Genome Res 22(4):666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM.. 2016. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44(D1):D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaromonte F, Yap VB, Miller W.. 2002. Scoring pairwise genomic sequence alignments. Biocomputing 7:115–126. [DOI] [PubMed] [Google Scholar]
- Coughlin DJ, Babak T, Nihranz C, Hughes TR, Engelke DR.. 2009. Prediction and verification of mouse tRNA gene families. RNA Biol 6(2):195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig RJ, Suh A, Wang M, Ellegren H.. 2018. Natural selection beyond genes: identification and analyses of evolutionarily conserved elements in the genome of the collared flycatcher (Ficedula albicollis). Mol Ecol 27(2):476–492. [DOI] [PubMed] [Google Scholar]
- Crick F. 1966. Codon-anticodon pairing: the wobble hypothesis. J Mol Biol 19(2):548–555. [DOI] [PubMed] [Google Scholar]
- del Hoyo J, Elliott A, Sargatal J, Christie D, de Juana E.. 2018. Handbook of birds of the world. Barcelona: Lynx Edicions. [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T.. 2006. Tissue-specific differences in human transfer RNA expression. PLoS Genet 2(12):e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, et al. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485(7398):376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning J. 2007. CRC handbook of avian body masses. 2nd ed. Boca Raton, (FL: ): CRC Press. [Google Scholar]
- Ellegren H. 2010. Evolutionary stasis: the stable chromosomes of birds. Trends Ecol Evol 25(5):283–291. [DOI] [PubMed] [Google Scholar]
- Ernst C, Odom DT, Kutter C.. 2017. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat Commun 8(1):1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS.. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7(9):1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Keller W.. 2001. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci 26(6):376–384. [DOI] [PubMed] [Google Scholar]
- Goodenbour JM, Pan T.. 2006. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res 34(21):6137–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among Archosaurs. Science 346(6215):1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, de Crécy-Lagard V, Marck C.. 2010. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett 584(2):252–264. [DOI] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA.. 2008. Selection on codon bias. Annu Rev Genet 42(1):287–299. [DOI] [PubMed] [Google Scholar]
- Ikemura T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. [DOI] [PubMed] [Google Scholar]
- Jarvis ED. 2016. Perspectives from the avian phylogenomics project: questions that can be answered with sequencing all genomes of a vertebrate class. Annu Rev Anim Biosci 4(1):45–59. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215):1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T.. 2001. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol 53(4–5):290–298. [DOI] [PubMed] [Google Scholar]
- Kapusta A, Suh A.. 2017. Evolution of bird genomes – a transposon’s-eye view. Ann NY Acad Sci 1389(1):164–185. [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD.. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20(4):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H. 2004. Mobile elements: drivers of genome evolution. Science 303(5664):1626–1632. [DOI] [PubMed] [Google Scholar]
- Kelly TK, Ahmadiantehrani S, Blattler A, London SE.. 2018. Epigenetic regulation of transcriptional plasticity associated with developmental song learning. Proc R Soc B 285(1878):20180160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S, Ignatova Z.. 2015. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 16(2):98–112. [DOI] [PubMed] [Google Scholar]
- Koepfli K-P, Paten B, O’Brien SJ, O’Brien SJ; the Genome 10K Community of Scientists. 2015. The genome 10K project: a way forward. Annu Rev Anim Biosci 3(1):57–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlach J, et al. 2017. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. Gigascience 6(10):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS.. 2005. Short retroposons in eukaryotic genomes. Int Rev Cytol 247:165–221. [DOI] [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS.. 2011. Origin and evolution of SINEs in eukaryotic genomes. Heredity (Edinb) 107(6):487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus RHS, Wink M.. 2015. Avian genomics: fledging into the wild! J Ornithol 156(4):851–865. [Google Scholar]
- Kutter C, et al. 2011. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat Genet 43(10):948–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R, Arimbasseri A.. 2017. Factors that shape eukaryotic tRNAomes: processing, modification and anticodon–codon use. Biomolecules 7(4):26. [Google Scholar]
- Marck C, Grosjean H.. 2002. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8(10):1189–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, et al. 2010. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17(5):635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L.. 2012. A role for tRNA modifications in genome structure and codon usage. Cell 149(1):202–213. [DOI] [PubMed] [Google Scholar]
- Oler AJ, et al. 2010. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17(5):620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenburghs J, et al. 2017. Avian introgression in the genomic era. Avian Res 8(1):30. [Google Scholar]
- Peona V, Weissensteiner M, Suh A.. 2018. How complete are ‘complete’ genome assemblies? An avian perspective. Mol Ecol Resour 18(6):1188–1195. [DOI] [PubMed] [Google Scholar]
- Pouyet F, Mouchiroud D, Duret L, Sémon M.. 2017. Recombination, meiotic expression and human codon usage. Elife 6:e27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TEF, Claassens NJ, Söll D, van der Oost J.. 2015. Codon bias as a means to fine-tune gene expression. Mol Cell 59(2):149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab J, et al. 2012. Human tRNA genes function as chromatin insulators. Embo J 31(2):330–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina M, Ibba M.. 2014. tRNAs as regulators of biological processes. Front Genet 5:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis MD, Savva R, Wernisch L.. 2004. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res 32(17):5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC. 2004. Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res 14(11):2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KLM, et al. 2016. Codon-driven translational efficiency is stable across diverse mammalian cell states. PLoS Genet 12(5):e1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt BM, et al. 2014. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA-tRNA interface. Genome Res 24(11):1797–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, et al. 2013. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet 45(4):415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A, et al. 2016. Ancient horizontal transfers of retrotransposons between birds and ancestors of human pathogenic nematodes. Nat Commun 7:11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A, et al. 2017. De-novo emergence of SINE retroposons during the early evolution of passerine birds. Mob DNA 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A, Smeds L, Ellegren H.. 2018. Abundant recent activity of retrovirus-like retrotransposons within and among flycatcher species implies a rich source of structural variation in songbird genomes. Mol Ecol 27(1):99–111. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang DT, Glazov EA, McWilliam SM, Barris WC, Dalrymple BP.. 2009. Analysis of the complement and molecular evolution of tRNA genes in cow. BMC Genomics 10(1):188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias JA, Ottenburghs J, Pigot AL.. 2020. Avian diversity: speciation, macroevolution, and ecological function. Annu Rev Ecol Evol Syst 51(1):533–560. [Google Scholar]
- Villar D, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160(3):554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. 2010. The genome of a songbird. Nature 464(7289):757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. 2017. A new chicken genome assembly provides insight into avian genome structure. G3 7:109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM. 2002. SINEs and LINEs: the art of biting the hand that feeds you. Curr Opin Cell Biol 14(3):343–350. [DOI] [PubMed] [Google Scholar]
- Weissensteiner MH, et al. 2017. Combination of short-read, long-read, and optical mapping assemblies reveals large-scale tandem repeat arrays with population genetic implications. Genome Res 27(5):697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. 1990. The ‘effective number of codons’ used in a gene. Gene 87(1):23–29. [DOI] [PubMed] [Google Scholar]
- Yusuf L, et al. 2020. Noncoding regions underpin avian bill shape diversification at macroevolutionary scales. Genome Res 30(4):553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, et al. 2018. Ensembl 2018. Nucleic Acids Res 46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, et al. ; Avian Genome Consortium. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346(6215):1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ArrayExpress Accession
H3K4me3 ChIP-seq (chicken): E-MTAB-8106
H3K4me3 ChIP-seq (zebra finch): GSE91399
H3K4me3 ChIP-seq (mammals): E-MTAB-2633
Pol III ChIP-seq (mammals): E-MTAB-958
Code Accessibility
github.com/JenteOttie/Avian_tRNAs