Abstract
Understanding the basic interactions between engineered nanoparticles (ENPs) and biological systems is essential for evaluating ENPs’ safety and developing better nanomedicine. Profound interactions between ENPs and biomolecules such as proteins are inevitable to occur when ENPs are administered or exposed to biological systems, for example, through intravenous injection, oral, or respiration. As a key component of these interactions, protein corona (PC) is immediately formed surrounding the outlayer of ENPs. PC formation is crucial because it gives ENPs a new biological identity by altering not only the physiochemical properties, but also the biobehaviors of ENPs. In the past two decades, most investigations about PC formation were carried out with in vitro systems which could not represent the true events occurring within in vivo systems. Most recently, studies of in vivo PC formation were reported, and it was found that the protein compositions and structures were very different from those formed in vitro. Herein, we provide an in-time review of the recent investigations of this in vivo PC formation of ENPs. In this review, commonly used characterization methods and compositions of in vivo PC are summarized firstly. Next, we highlight the impacts of the in vivo PC formation on absorption, blood circulation, biodistribution, metabolism, and toxicity of administered ENPs. We also introduce the applications of modulating in vivo PC formation in nanomedicine. We further discuss the challenges and future perspectives.
Keywords: protein corona, engineered nanoparticles, nano-bio interactions, nanomedicine, biobehaviors, nanotoxicology
Introduction
Engineered nanoparticles (ENPs) with unique physical and chemical properties have been widely developed in the fields of energy (Pomerantseva et al., 2019; Wang et al., 2020), electronics (Kamyshny and Magdassi, 2019), materials (Wang et al., 2019), biomedicine (Lee et al., 2012; Pelaz et al., 2017; Wong et al., 2020), food and agriculture (Peters et al., 2016; Lowry et al., 2019), and so on. More and more ENP-based biomedical products were put into the markets or translated to clinic. Despite the rapid developments in these fields, there remain a number of challenges in nanotechnology development. The key issues are the health risk and safety concern of ENPs, especially in the field of nanomedicine (Susan et al., 2018; Ahmad et al., 2019). Numerous studies have reported the ENP-associated toxicity to zebra fish (Lakshmi et al., 2019; Bakshi, 2020), macroalgae (Sfriso et al., 2019), animals (Jia et al., 2017; Teng et al., 2019), and even human beings (Guo et al., 2020). To avoid these ENP-caused toxicities and promote the development of nanomedicine, it is imperative to thoroughly understand the basic interactions between ENPs and the physiological environment, biomolecules, tissues, and organs. Therefore, investigations about ENPs-physiological systems interactions stay at the forefront of nanomedicine and nanotoxicology research.
Once entering into the physiological environment, including blood, interstitial fluid, and intracellular environment, ENPs are expected to interact with biomolecules such as lipids, metabolites, sugars, and especially proteins by van der Waals forces, electrostatic, hydrogen bonding, and hydrophilic/hydrophobic interactions because of the high surface energy and unique surface chemistry of ENPs, and after that biomolecular corona are formed (Nel et al., 2009; Monopoli et al., 2012; Shemetov et al., 2012; Hadjidemetriou and Kostarelos, 2017). Until now, the protein corona (PC) is the most important class of biomolecular corona formed and has mostly been studied. Before the PC was proposed in 2007, ENPs had been considered to interact with living cells directly, and the physiochemical properties of the pristine ENPs were considered to determine their biological effects (Cedervall et al., 2007). However, this understanding is far from the truth. After exposed to a complete cell culture medium in vitro or blood in vivo, tens of thousands of proteins immediately reached onto the outlayer of ENPs and PC were formed in a dynamic process (Yu et al., 2020). According to the relative affinity, PC was divided into hard and soft types (Docter et al., 2015; Kokkinopoulou et al., 2017). Hard corona is made by a protein fraction strongly bound to the surface, while soft corona is formed by loosely bound proteins, maybe via protein–protein interaction. PC significantly alters the identity of ENPs, including physicochemical properties and the bioidentity of ENPs (Caracciolo G. et al., 2016; Liu et al., 2020; Szekeres et al., 2020). First, PC formation alters the size, zeta potential, morphology, and aggregation states of ENPs. Second, PC formation alters the interactions between ENPs and physiological systems, and the subsequent biobehaviors of ENPs (Chinen et al., 2017; Nayak et al., 2019). For example, adsorption of opsonin proteins enhanced the cellular uptake of ENPs by phagocytes, and subsequently accelerate the clearance (Vu et al., 2019). In addition, special protein can be recruited by ENPs with special properties which give ENPs specificity capacity (Wang et al., 2017). Therefore, investigation of interaction mechanism between ENPs and protein molecules and its biological effects is critical to understand the interactions between ENPs and living organisms, which is of great significance for evaluating the safety of ENPs and the application of ENPs in biomedical field.
Until now, most of studies are investigated on the PC formation on ENPs and their biological effects in vitro. However, there are significant differences between in vitro and in vivo PC formation, not only quantities of adsorbed protein, but also the composition and structures (Amici et al., 2017; Wang M. et al., 2018). Furthermore, the subsequent impacts on bio-nano interactions are also vary in vitro and in vivo (Raoufi et al., 2018). Thus, in this review, we focus on the research of in vivo PC formation and their impacts on the biobehaviors of administered ENPs. To investigate the in vivo PC formation, characterization techniques are different from the in vitro PC analytical approaches, which were discussed in the first part. Next, we summarize the influences of in vivo PC formation on absorption, blood circulation, biodistribution, metabolism, and toxicity of administered or exposed ENPs. Moreover, in vivo PC formation can be modulated by the physicochemical properties of ENPs. We then further discuss the applications of in vivo PC formation for targeted delivery and personalized medicine. Finally, the major research gaps, challenges, and future perspectives of in vivo PC formation are discussed.
Characterization of in vivo PC Formation
Characterization of in vivo PC formation and protein-bound ENPs is a key step to understand the formation mechanism and the function of protein composition (Brun and Sicard-Roselli, 2014; Kokkinopoulou et al., 2017). Analytical methods of PC protein composition and structure can be categorized into in situ and ex situ characterizations (Sakulkhu et al., 2014; Carril et al., 2017). Ex situ techniques separate protein-bound ENPs from the physiological environment and then cleave the bound proteins for further characterization. On the contrary, in situ technique directly provide relevant information about PC formation when ENPs disperse into physiological environment.
For ex situ characterization, separation of PC-ENP complexes from in vivo physiology environment is one of the main obstacles in characterizing the in vivo PC formation. Magnetic separation method was employed to separate protein-bound magnetic ENPs, which can avoid disrupting loosely bound protein during centrifugation. Sakulkhu et al. (2014) employed magnetic separation to obtain PC-ENP complexes from rat sera, and then PC compositions were analyzed by nano-LC-MS/MS. They found that ENPs with positive charge adsorbed 32 types of proteins, while neutral and negative charged ENPs adsorbed 55 and 51 types of proteins, respectively. Low molecular weight (<30 kDa) proteins are the most amount of proteins for all ENPs adsorbed in vivo, while 90–120 kDa proteins were the least. The top 5 bound proteins were listed in Table 1. Hemoglobin subunit, such as alpha-1/2, beta-1, and beta-2 are all in the top 5 bound proteins. Size-exclusion chromatography (SEC) combined with membrane ultrafiltration were usually employed to separate non-magnetic protein-bound ENPs. For example, PC-AuNP complexes recovered from blood can be separated from unbound proteins by SEC, followed by membrane ultrafiltration (García-Álvarez et al., 2018). They found that the total number of identified proteins were 298 and 246 for AuNRs with diameters of 40 and 70 nm, respectively. AuNSs with diameters of 40 nm and 70 nm adsorbed 406 and 215 types of proteins, respectively. Serum albumin and alpha-2-macroglobulin were the top 2 bound proteins for all AuNPs (Table 1). Proteins with MW < 80 kDa contributed to 75–80% of the PC. Besides, PC-liposome complexes can also be separated form plasma by SEC. The types of unique proteins identified in vivo formed coronas of bare-, PEG-, and monoclonal antibody targeted-liposomes NPs were 453, 478, and 511, respectively (Hadjidemetriou et al., 2015). Apolipoproteins were the most abundant classes of protein in vivo PC of all types of liposomes NPs. Separation of protein-bound ENPs from plasma may inevitably interfere with the composition of the PC, resulting in the loss of weak-binding proteins, which is inaccurate for the subsequent analysis. It is also difficult to separate PC-ENP complexes from tissues/organs other than blood.
TABLE 1.
Top-five adsorbed proteins of some ENPs with different properties and the separation methods.
| ENPs | Properties | Administration route and sampling time | Separation method | Top-five adsorbed proteins (Relative protein abundance) | References |
| SPIONs | 90 nm, positive charged | 10 min after intravenous injection | magnetic separation | Hemoglobin subunit beta-2 (16.76%) Hemoglobin subunit alpha-1/2 (16.50%) Hemoglobin subunit beta-1 (13.04%) Apolipoprotein E (9.99%) Fibrinogen alpha chain (7.90) | Sakulkhu et al., 2014 |
| 95 nm, neutral | 10 min after intravenous injection | magnetic separation | Fibrinogen beta chain (9.05%) Hemoglobin subunit beta-2 (8.86%) Hemoglobin subunit alpha-1/2 (8.44%) Fibrinogen gamma chain (7.30%) Hemoglobin subunit beta-1 (6.94%) | ||
| 91 nm, negative charged | 10 min after intravenous injection | magnetic separation | Hemoglobin subunit beta-2 (10.23%) Hemoglobin subunit alpha-1/2 (9.33%) Hemoglobin subunit beta-1 (8.96%) Apolipoprotein A-II (7.15%) Apolipoprotein E (5.72%) | ||
| AuNRs | 40 nm, PEG-COOH coated | 10 min after intravenous injection | SEC and membrane ultrafiltration | Serum albumin (5.16%) Alpha-2-macroglobulin (4.30%) Fibrinogen beta chain (2.29%) Apolipoprotein A-I (2.27%) Complement factor H (2.12%) | García-Álvarez et al., 2018 |
| 70 nm, PEG-COOH coated | 10 min after intravenous injection | SEC and membrane ultrafiltration | Serum albumin (7.52%) Alpha-2-macroglobulin (6.13%) Serine protease inhibitor A3K (4.83%) Apolipoprotein A-I (3.24%) Fibrinogen beta chain (2.51%) | ||
| AuNSs | 40 nm, PEG-COOH coated | 10 min after intravenous injection | SEC and membrane ultrafiltration | Serum albumin (3.71%) Alpha-2-macroglobulin (3.70%) Serine protease inhibitor A3K (2.44%) Fibrinogen beta chain (2.37%) Apolipoprotein E (2.31%) | |
| 70 nm, PEG-COOH coated | 10 min after intravenous injection | SEC and membrane ultrafiltration | Serum albumin (8.19%) Alpha-2-macroglobulin (7.80%) Serine protease inhibitor A3K (6.01%) Fibrinogen beta chain (3.99%) Alpha-1B-glycoprotein (3.11%) | ||
| AmBisome< | 100 nm | 10 min after intravenous injection | SEC and membrane ultrafiltration | Serum albumin (4.07%) Fibrinogen beta chain (2.28%) Apolipoprotein C-III (2.14%) Actin, cytoplasmic 2 (2.11%) Fibrinogen gamma chain (1.81%) | Amici et al., 2017 |
| polystyrene NPs | 80.77 nm, PEG coated | 10 min after intravenous injection | SEC and membrane ultrafiltration | ApoE protein (9.83%) Apolipoprotein C-IV (7.56%) Apolipoprotein A-IV (7.29%) Clusterin (5.31%) Hemoglobin subunit beta-2 (4.08%) | Zhang et al., 2018 |
| 89.5 nm, modified with LT7 | 10 min after intravenous injection | SEC and membrane ultrafiltration | ApoE protein (7.54%) Apolipoprotein A-IV (7.54%) Clusterin (6.62%) Albumin 1 (3.89%) Apolipoprotein C-IV (2.59%) | ||
| 83.61 nm Modified with DT7 | 10 min after intravenous injection | SEC and membrane ultrafiltration | Apolipoprotein A-IV (7.14%) Clusterin (6.57%) ApoE protein (6.32%) Apolipoprotein C-IV (2.52%) Albumin 1 (2.34%) | ||
| 92.47 nm, modified with Tf | 10 min after intravenous injection | SEC and membrane ultrafiltration | Hemoglobin subunit beta-2 (8.65%) Albumin 1 (7.69%) Clusterin (5.22%) Apolipoprotein A-IV (5.20%) ApoE protein (4.61%) | ||
| Liposome | 127.77 nm | 10 min after intravenous injection | SEC and membrane ultrafiltration | Apolipoprotein C-III (4.93%) Apolipoprotein E (3.54%) Hemoglobin subunit beta-1 (3.34%) Beta-globin (3.17%) Alpha-2-macroglobulin (2.96%) | Hadjidemetriou et al., 2015 |
| 119.53 nm, PEG coated, | 10 min after intravenous injection | SEC and membrane ultrafiltration | Apolipoprotein C-III (4.53%) Apolipoprotein E (3.46%) Hemoglobin subunit beta-1 (2.89%) Alpha-globin 1 (2.24%) Alpha-2-macroglobulin (2.15%) | ||
| 121.73 nm, monoclonal antibody (IgG) targeted | 10 min after intravenous injection | SEC and membrane ultrafiltration | Apolipoprotein E (2.79%) Apolipoprotein C-III (2.66%) Alpha-2-macroglobulin (2.49%) Hemoglobin subunit beta-1 (2.44%) Apolipoprotein C-IV (1.58%) | ||
| PEGylated liposomal doxorubicin | 115 nm | 10 min after intravenous injection | SEC and membrane ultrafiltration | Alpha-2-macroglobulin (8.02%) Apolipoprotein C-III (6.37%) Hemoglobin subunit beta-1 (5.79%) Apolipoprotein E (PE = 1 SV = 2) (5.57%) Beta-globin, Hbbt1 (A8DUK2) (4.48%) | Hadjidemetriou et al., 2016 |
| 115 nm | 1 h after intravenous injection | SEC and membrane ultrafiltration | Apolipoprotein E (PE = 2 SV = 1) (8.19%) Alpha-2-macroglobulin (7.66%) Apolipoprotein C-III (4.86%) Serum albumin (4.41%) Apolipoprotein E (PE = 1 SV = 2) (3.87%) | ||
| 115 nm | 3 h after intravenous injection | SEC and membrane ultrafiltration | Hemoglobin subunit beta-1 (8.58%) Apolipoprotein E (PE = 2 SV = 1) (7.30%) Apolipoprotein C-III (6.65%) Alpha-2-macroglobulin (6.42%) Beta-globin, Hbbt1 (A8DUK2) (6.02%) |
After separation, the characterization methods for protein-bound ENPs and cleaved proteins were similar to that used in in vitro PC analysis. For example, size of PC-ENP complexes can be characterized by transmission electron microscopy (TEM) (Mahmoudi et al., 2011) and atomic force microscopy (AFM) (Guan et al., 2015). Polyacrylamide gel electrophoresis (PAGE) and liquid chromatography tandem mass spectrometry (LC-MS/MS) (Walkey et al., 2014; Saha et al., 2016; Pinals et al., 2020) are commonly used for the identification and quantification of individual proteins in the PC after the separation of adsorbed proteins from the surface of ENPs. Isothermal titration calorimetry (ITC) (Fleischer and Payne, 2014) and SEC (Shakiba et al., 2018) can be used to evaluate the strength and adsorption kinetics of the interaction between ENPs and proteins. Furthermore, conformational changes of proteins was determined by circular dichroism (CD) spectroscopy (Yan et al., 2013; Fleischer and Payne, 2014) and fourier transform infrared spectroscopy (FTIR) (Wang et al., 2012), nuclear magnetic resonance (NRM) (Brancolini et al., 2012), and enzyme activity determination (Gagner et al., 2011). These traditional techniques are used to determine the structural and physicochemical parameters of PC.
Unlike ex situ technique, in situ technique may be more suitable for characterization of PC in biofluids. For example, the current understanding of the biological identity that ENPs may acquire in a given biological milieu is mostly inferred from hard corona. However, because classical approach based on ENPs separation from the biological medium fails to detect the composition of soft corona and illustrate their biological relevance. In recent, in situ techniques were employed to character soft corona. For example, a new method using cryoTEM and synchrotron-radiation CD was developed to analyze the weakly bound proteins and reveal molecular basis of soft corona in situ (Sanchez-Guzman et al., 2020). Soft corona proteins were altered by ENPs based on shifting the equilibrium toward the unfolded states at physiological temperature. Besides, in situ click-chemistry reaction was also used to identify soft corona and hard corona on the surface of silica and polystyrene NPs (Mohammad-Beigi et al., 2020). They found that soft corona and hard corona were distinguished by different binding strength but not special proteins.
In addition, the hydrodynamic radius of PC-ENP complexes in biofluid can be analyzed by 19F diffusion-ordered NMR (Carril et al., 2017), fluorescence correlation spectroscopy (Shang and Nienhaus, 2017) and high-speed dark-field microscopy (Lin et al., 2019). Molecular motifs can be detected by flow cytometry combined with microfluidics in biological milieu (Lo Giudice et al., 2016). Protein adsorption behavior, including affinity and adsorption orientation can be analyzed by in situ fluorescence resonance energy transfer (FRET) (Qu et al., 2020). Time-evolution of PC formation can be detected by microfluidics combined with confocal laser scanning microscopy (Weiss et al., 2019).
Despite these technologies are established to analyze the ENP-bound PC formation, several limitations remain. First, most of them don’t take the protein conformational changes into account. In fact, the formation of PC-ENP complexes can affect the higher structures of protein molecules. For example, secondary structures of bovine serum albumin (BSA) molecules were broken by Au NPs and Ag NPs after adsorbing on NPs (Treuel et al., 2010). Carbon nanotubes induced significant changes in the secondary structure of bovine fibrinogen and Ig, with a decrease in the α-helical content and an increase in the β-sheet (Ge et al., 2011). Therefore, the in vivo conformational changes of PC should be considered in the analysis process. Second, the analysis of PC formation is still lacking a proper modeling of the interaction kinetics. This is a challenging task because a large number of molecules are involved and dynamic exchanges occur in a short period of time. Only scarce studies are carried out in this regard. For example, the kinetics of the PC formed on Silica NPs were studied by combing experiments with simulations and theory in a ternary solution made of HSA, Tf, and Fib. Theoretical model correctly reproduced competitive protein replacement and observed a memory effect in the final corona composition, which was proved by independent experiments (Vilanova et al., 2016). Besides, Duan et al. (2020) found a descriptor based on fluorescence change from fluorescamine labeling on a protein, which helped build machine learning models to predict the composition of PC, even in heterogeneous nanomaterials. More appropriate models need to be investigated to predict PC behaviors and their influence on nano-bio interactions in complex environment in vivo.
In our opinion, obtaining an accurate PC information in vivo remains a challenge due to the rapid and dynamic, complex and often weak interactions between ENPs and proteins. Development of appropriate in situ analytical approaches that can address these issues are highly demanded.
Impacts of in vivo PC on Biobehaviors of Administered ENPs
Absorption
Absorption is the process in which ENPs enters the blood from the site of administration. Intravenous injection (IV) seems to be the most recurrent application route for most ENPs and this process is not related to the absorption. However, PC formation should be taken into account in non-IV routes of administration such as oral and inhale. After non-IV routes of administration, ENPs must translocate through the mucus layer before reaching the surface of the epithelium, and then transepithelial absorption may occur (García-Díaz et al., 2017). So far, most of reports focused on the study of PC upon plasma or blood contact, whereas studies involving the biomolecular corona in non-IV administration, especially studies in vivo are still scarce.
After oral administration, ENPs encounter gastrointestinal (GI) fluids with multiple components including zwitterionic phospholipids, bile salts, carbohydrates, and mucin. These GI fluids can adsorb on the surface of ENPs, and therefore, impact their properties and may impede their passage through the intestinal mucosa. For example, after oral administration of PEG-GNPs (PGNPs), intestinal mucin adsorbed on their surface and induced aggregation to form nanoclustering (Figure 1A; Yang et al., 2018). Compared with the monodispersive PGNPs, mucin adsorbed nanoclustering entered into epithelial cell through endocytosis, and transported through retrograde pathway [PM-Golgi/ER-PM (plasma membrane)]. Therefore, transcellular capability of PGNP was inhibited by mucin, while cell uptake by epithelial cell was enhanced. Covered ENPs with native proteins could prevent the adsorption of mucus. Wang A. et al. (2018) found that pre-coating of liposomes with a corona of BSA could improve their mucus-penetration and intestinal absorption ability.
FIGURE 1.
Protein corona impacts in vivo biobehaviors of ENPs. (A) Schematic diagram of intestinal mucus altering the endocytosis and transcytosis characteristics of PGNPs. Adapted with permission from Yang et al. (2018). (B) PC affects circulation time. Coagulation and complement proteins that were adsorbed initially on the surface of SMPN-1 were released during the circulation and accumulation of lipoproteins and acute phase reactants happened over 48 h on the surface. In contrast, short-circulating SMPN-9 had high abundance of complement proteins and acute phase reactants at 48 h. Adapted with permission from Abbina et al. (2020). (C) PC influenced the biodistribution of G-rich and poly-T SNAs. Compared to poly-T SNAs, G-rich SNAs adsorbed more amounts of apolipoprotein B100, complement factor H, and complement C3, and less human serum albumin, which induced more accumulation in liver and spleen. *p < 0.05. Adapted with permission from Chinen et al. (2017). (D) The major part of MaPSi was cleared fast from liver just a few minutes after injection, whereas DPEG-MaPSi decreased the removal rate and prevented the fast decline of theT2* relaxation rates. Adapted with permission from Nissinen et al. (2016).
The respiratory tract lining fluid is the first biological matter that ENPs encounter after inhale route of administration. This layer is mainly composed of pulmonary surfactants which is a complex mixture of phospholipids, proteins, and cholesterol. The adsorption of these components on inhaled ENPs likely modulates the bioavailability. One study found that polyethylene glycol-coated AuNPs with different outermost regions (Au@PEG-X NPs) adsorb similar types of bronchoalveolar lavage fluid (BALF) proteins, but different composition of PC (Yin et al., 2019). PC composition such as surfactant protein D (SP-D), carbonyl reductase (NADPH), albumin and surfactant protein A (SP-A), have strong correlations with intrapulmonary cellular uptake. For example, adsorption of SP-D may promote the cell uptake by neutrophils in the lavaged lung. Adsorption of NADPH may promote the cell uptake by total macrophage and M2 macrophage in the lavaged lung, but have the opposite effect for macrophage in BALF. At the same time, albumin in the PC shows strong negative correlations with the association of Au@PEG-X NPs to dendritic cells and endothelial cell in the lavaged lung, as well as neutrophils in BALF. SP-A in the PC shows strong positive correlation with the association of Au@PEG-X NPs to dendritic cells in BALF. However, another study reported that SP-A had no effect on cell uptake of PLGA/PLA@PEG NPs by adherent cells in BALF (Ruge et al., 2016).
These studies indicate that the absorbed mucus components can affect epithelial uptake of the ENPs and alter kinetics of the ENPs’ transportation. Thus, it is critical to understand whether the process of PC formation during ENP absorption would subsequently modulates the behavior and fate of ENPs in vivo, such as blood circulation, biodistribution and metabolism. In many cases, the in vivo performance of ENPs does not meet expectations although passed in vitro examinations because of overlooking this issue. Therefore, the transport through the mucus layer may be the first factor to consider for design of ENPs in non-IV administered nanomedicine.
Blood Circulation
The ENPs enter blood following absorption. The formation of PC might change the interactions between ENPs and tissues/cells (Walkey et al., 2014). Thus, PC will consequently impact the circulation time of ENPs in the body. Compositions of PC such as complement protein, fibrinogen and immune globulin, which called opsonin, may enhance the eliminated of ENPs through the reticuloendothelial system (RES) (Vu et al., 2019). If the amount of these proteins in PC is high, it will accelerate the identification and phagocytosis of ENPs by macrophages, resulting in quick clearing from the body. On the contrary, if the PC is rich in another type of protein such as albumin and apolipoprotein, the blood circulation time of NPs will be prolonged in vivo (Bertrand et al., 2017).
PEG and other polymer modifications on ENPs are widely used to reduce protein adsorption, complement activation, and therefore prolong ENP circulation time in blood. For example, Zhou et al. (2018) presented a hierarchical PEG modified ENPs which significantly enhance ENPs circulation time compared to ENPs with signal PEG layer. The fluctuating PEG segments reduce protein binding affinity with ENPs, rather than the total amount of adsorbed proteins, which could prolong circulation time. Except for PEG, soft single molecule polymer nanomaterials (SMPNs) which were prepared with macroinitiator, hyperbranched polyglycerol (HPG), was also presented to enhance circulation time in blood (Figure 1B; Abbina et al., 2020). PC composition of ENPs at the biointerface is highly dynamic and remodeled while in circulation. SMPNs with longer circulation are able to clear some of the opsonins that adsorbed on the surface of ENPs initially, which then evade the immune system and can prolong circulation time in blood.
In addition, biomimetic liposomes and leukosomes (which was obtained from leukocyte membrane), were also used to prolong blood circulation time (Corbo et al., 2017). The amount and number of protein types adsorbed on liposomes were about 45 and 42% more than that of leukosomes respectively 1 h after injections. Although compositions of PC, such as immunoglobulin gamma (IgG), coagulation factors, and complement proteins have been identified on both liposomes and leukosomes, blood circulation time of leukosomes was longer than that of liposomes. One reason was the different abundance of these proteins in the PCs of leukosomes and liposomes. Another reason was that IgG and other proteins were adsorbed on leukosomes through a manner of receptor-to-ligand binding, which reduced the interactions with macrophages and avoid opsonization process. Generally, construction of NPs for medicine applications should consider reducing or controlling adsorption of total proteins, especially opsonins, or pre-coating NPs with selected proteins, so as to prolong the blood circulation time, which is a key factor to improve the performance of NPs in vivo.
Biodistribution
Biodistribution is the process in which protein-absorbed ENPs circulating from blood to tissue fluids or intracellular fluids. Generally, based on the process of opsonization, the main biodistribution organs of ENPs are liver and spleen. For example, guaninerich (G-rich) oligonucleotides and poly-thymine (poly-T) oligonucleotides were used to modify AuNPs, and two spherical nucleic acids (SNAs) were obtained, named G-rich SNAs and poly-T SNAs. It was found that G-rich SNAs adsorbed more proteins than poly-T SNAs both in amounts and in types, including complement proteins, which caused more NPs accumulate in the liver and spleen (Figure 1C; Chinen et al., 2017). Eight hour after injection, more than 85% of the injected dose of G-rich SNAs was found in the liver and spleen, compared to approximately 55% of the injected dose of poly-T SNAs. Moreover, in IgM sensitized mice, anti-PEG IgMs coated ENPs adsorbed complement proteins, such as C1q-A, C1q-B, and C1q-C, which involved in classical complement activation pathway, and the amounts were 1.3-fold more than those in control IgM naive mice. This difference caused the amount of PEGylated NPs accumulated in liver in IgM sensitized mice to increase nearly 3 folds compared with that in control mice (Grenier et al., 2018).
On the contrary, conjugation with blood proteins, such as albumin and apolipoprotein E, remarkably reduces recognition of ENPs by mononuclear phagocytic system. One study found that liver retention of polyelectrolyte-multilayer-coated AuNPs with a diameter of 15 nm was reduced by about 23 and 43% after NPs were conjugated with apolipoprotein E and albumin (Schäffler et al., 2014). Furthermore, albumin conjugation significantly increases translocation into the brain and lung. However, another study found that despite pre-incubated with mouse serum before administration, CTAB-AuNRs mainly accumulated in liver (Cai et al., 2016). Some of the CTAB-AuNRs escaped the phagocytosis mainly due to the adsorption of mouse serum albumin in process of pre-incubation, but the particles were found in the hepatocytes. Interestingly, hollow mesoporous silica nanoparticle (HMSNs) loaded with perfluoro-15-crown-5-ether (PFCE) adsorbed more proteins than the bare particles, especially the amount of apolipoproteins A-1 and A-2. These proteins only induced ENPs to accumulate in liver 24 h after injection, but in other RES organs like spleen or lung (Pochert et al., 2017). In summary, the biodistribution of ENPs determines the organ/tissue specific therapeutic or toxic effects, and the PC formation plays an important role. Undesirable biodistribution of NPs in disease treatment applications will reduce their therapeutic effect and even induce toxicity. Therefore, controlling PC formation in nanomedicine can provide more precise guidelines for improving the specificity and efficiency of NPs for targeted biodistribution.
Metabolism
Engineered nanoparticles structural change and eventual excretion in either original form or metabolites through different pathways is the process of degradation or metabolism, also known as biotransformations. PC is known to affect the biotransformation of ENPs in vivo. Until now, most of studies revealed that the PC impacts ENPs’ metabolic and degradation rate through affecting blood circulation and biodistribution. For example, the MRI traceable superparamagnetic mesoporous silicon nanoparticles (MaPSi) adsorbed proteins that were mainly associated with liver activity, immune response, coagulation, and wound response. Those were distinguished from that of dual PEGylation (DPEG) modified MaPSi (DPEG-MaPSi) (which were enriched with glycoproteins CD59, CD44, CD47, CD93, and CD36). Such difference caused DPEG-MaPSi to have significantly longer circulation half-life (241 min) than that of MaPSi (1 min) (Figure 1D; Nissinen et al., 2016). Moreover, the composition of PC on the surface of NPs@PEG was rich in albumin, which is different with that on NPs@Glc (enrich fibrinogen). It induced NPs@PEG core to have a faster degradation rate than NPs@Glc, in both liver and spleen (Stepien et al., 2018). Reducing protein binding affinity can also increase circulation half-life of ENPs. Compared with PLGA-PEG NPs (100% methoxy-PLGA20K-PEG5K), maleimide-functionalized PLGA-PEG NPs can increase circulation half-life from 3.48 to 22.2 h by reducing protein binding affinity (Zhou et al., 2018). Until now, the impact of PC on ENPs biotransformations is mainly focus on in vitro experiments. Many issues in vivo remain needed to be unraveled. Since PC is endogenous and dynamical, and is the outermost layer of NPs, it is involved in the early process of biodegradation of ENPs. Thus, their degradation products may affect the fate of ENPs in vivo. Furthermore, various types of protein compositions may have different effects on the biodegradation rate of ENPs in vivo. Therefore, optimization of the products and rate of ENPs’ biodegradation though controlling PC formation requires more attentions in future clinical application.
Toxicity to Immune Systems and Others
The formation of PC impacts not only on the pharmacokinetics of ENPs but also ENPs’ toxicity including immunotoxicity. In general, PC formation changes the nanoparticle-cell interactions and may reduce cytotoxicity of ENPs through inhibiting their agglomeration and increasing the stability. The PC layer may also shelter ENP’s surface, thus reduce cytotoxicity caused by surface chemistry or metal ion release. Complement system is involved in the specific and non-specific immune mechanisms of the body (Merle et al., 2015a, b). As expected, complement proteins were shown to be involved in identifying and inducing the elimination process of ENPs. Thus, immunotoxicity is one of the main toxicity of ENPs induced by PC in body. For example, PEG-coated iron oxide NPs (IONP-PEG) can trigger complement activation and induces an inflammatory response (increased in proinflammatory cytokines such as IL-1β, IL-6, and TNF-α) (Rivera et al., 2019). Magnetic nanoparticle (MNP)-infiltrated bone regeneration scaffolds could also adsorb inflammatory related protein such as alpha-2-HS-glycoprotein, haptoglobin, and complement components, which can activate the immune system (Figure 2A; Zhu et al., 2019). In addition, Toy et al. (2019) found that imidazole-aceticacid (IAA) modification polyethyleneimine (bPEI) NPs adsorbed less fibrinogen than bPEI NPs, and thus have reduced immune activation and expression of chemokine expression. Moreover, blood hemolysis, liver toxicity, and anaphylactic responses were also reduced. However, for IAA-modified chitosan NPs, their toxicity was decreased mainly through the balance of TLR and complement activation.
FIGURE 2.
A few examples of PC-induced toxicity. (A) In vivo PC formation influenced immune-modulating osteogenesis by magnetic nanoparticle (MNP)-infiltrated bone regeneration scaffolds. Adapted with permission from Zhu et al. (2019). (B) SiNP-100 specifically recruited TGF-β1 to their corona and subsequently induced lung fibrosis (N-SiNPs, amination-SiNPs; P-SiNPs, polyetherimide-SiNPs). Adapted with permission from Wang et al. (2017).
Besides immunotoxicity, other types toxicity can also be regulated by PC in vivo. For example, one study explored SiNPs with a diameter of 100 nm and special chemistry such as hydration (H-SiNPs), dextran (D-SiNPs), and gelatin (G-SiNPs), can induce lung fibrosis by specifically recruit and enrich transforming growth factor β1 (TGF-β1) into their PC in lung (Figure 2B; Wang et al., 2017). Besides, SiNP-100 not only kept biological activities of TGF-β1 in binding cell receptors and triggering lung fibrosis, but also slowed degradation rate and prolonged activation of the TGF-β/Smad2 pathway which directly promotes lung fibrosis. In addition, amorphous silica NPs with a diameters of 70 nm (nSP70) induced acute lethality and abnormal activation of coagulation cascade, which attributed to the special affinity of silica NPs for coagulation factor XII in blood (Yoshida et al., 2015). In another case, the amino modified nSP70 showed lower toxicity compared to nSP70 due to fewer coagulation factor XII adsorbed on the surface. Therefore, toxic effects have emerged to be one of the main problems that need to be solved in nanomedicine (Dhawan et al., 2018). Since PC is the outermost surface that interacts with cells and tissues, it affects the toxicity of ENPs as well as their functions. On one hand, PC can decrease the toxicity of ENPs, which may improve their performance. On the other hand, PC may also shield their targeting capability and induce immunotoxicity. Therefore, precise understanding and taking the advantages of PC provide opportunities as well as challenges in the design of medical-used ENPs.
Modulation and Applications of in vivo PC
Modulation of in vivo PC
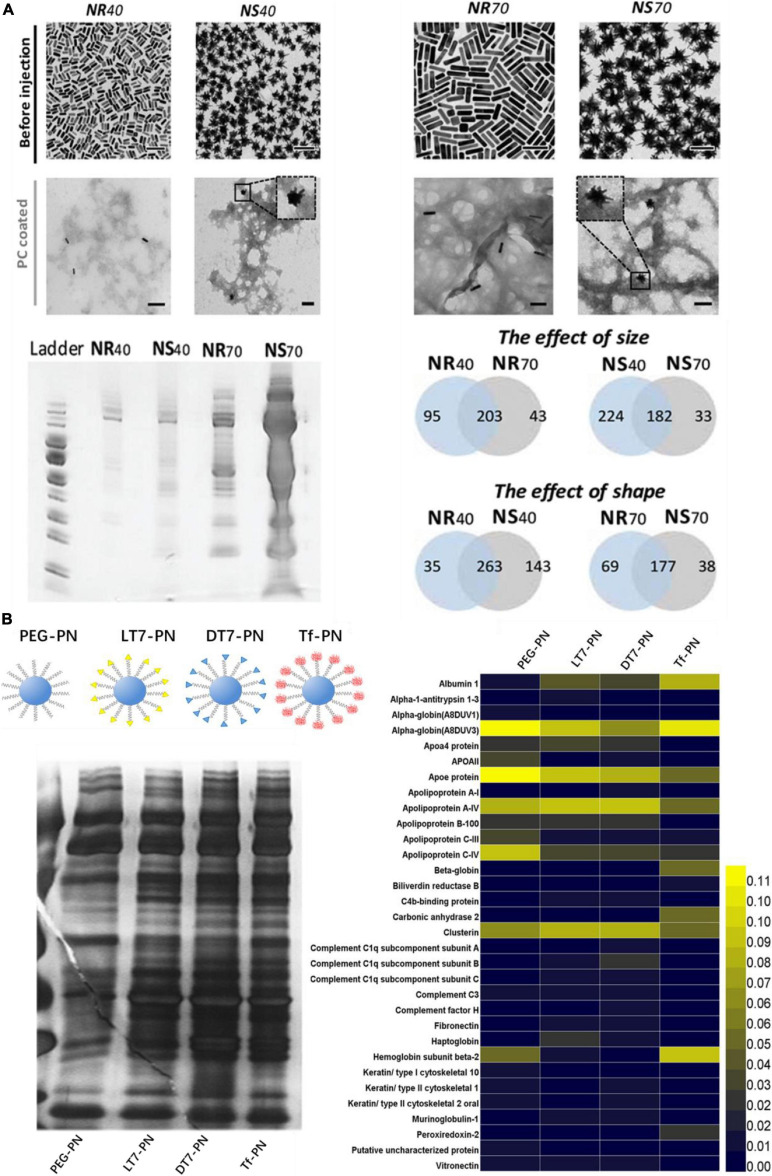
Since PC plays an essential role in regulating bio-nano interactions in vivo, modulation or control the PC formation is an urgent need to address. As a dynamic process, the PC formation is not only highly dependent on the properties of ENPs, but also on the biological environment. Physicochemical characteristics of ENPs such as size, shape, composition and surface chemistry modulate PC composition (Yu et al., 2020). ENPs size determines their surface curvature and also affects the surface area. Size is an important factor that affect not only the number but also the types of proteins adsorbed on the surface of ENPs in vivo. One study presented the effect of size (40 and 70 nm) on the formation of AuNSs in vivo (García-Álvarez et al., 2018). The amount of proteins adsorbed on AuNSs with a diameter of 70 nm were about 9 folds more than that of AuNSs with a diameter of 40 nm. On the contrary, protein types that adsorb on AuNSs with a diameter of 40 nm was about 88.8% higher than that of AuNSs with a diameter of 70 nm (Figure 3A). The similar effect of size on PC also presented on AuNRs with the same size and surface chemistry. Besides, due to surface curvature effect, unlike flat surfaces, proteins adsorb on the surface of ENPs alter their conformations to fit the curved surface, and this result in different protein binding affinities. It has been found that 35 nm AuNPs covered with artificial virus NPs (AVNs), which generated by integrating lipids, showed a much more prominent soft corona formation than that of 80 nm AuNPs with the same coating (Xu et al., 2016).
FIGURE 3.
Physicochemical properties of ENPs affect the in vivo PC formation. (A) TEM images of AuNPs with different size and shape and they adsorbed protein amounts and types in vivo. Adapted with permission from García-Álvarez et al. (2018). (B) Four types of polystyrene NPs modified with PEG and Tf receptor (TfR)-targeting ligands (LT7, DT7, and Tf) and their corresponding in vivo PC formation. Adapted with permission from Zhang et al. (2018).
The morphology or shape has a great impact on the structure and composition of PC in vivo. One study showed that amount of protein adsorbed on AuNSs with a diameter of 40 and 70 nm were 2 and 9 folds more than that of AuNRs with the same size, respectively (Figure 3A). It may attribute to the higher surface area for NSs Furthermore, with a diameter of 40 nm, the number of protein adsorbed on the surface of AuNSs was about 36% more than that of AuNRs (García-Álvarez et al., 2018). On the contrary, the number of proteins adsorbed on the surface of AuNRs with a diameter of 70 nm was about 14% more than that of AuNSs with the same size.
Due to the various properties, surface chemistry is another crucial factor influencing the formation and evolution of PC on ENPs. Surface charge is an important factor for regulating PC in vivo. It has been found that the types of protein adsorbed on neutral SPIONs and negatively charged SPIONs were about 72 and 59% more than that of positively charged SPIONs in vivo, respectively (Sakulkhu et al., 2014). Apolipoprotein A-II was found in the PC of neutral and negatively charged SPIONs, but not in the PC of positively charged SPIONs. The PCs formed on the surface of positively charged NPs contained considerably higher amounts of low molecular weight (<30 kDa) proteins, which is different from neutral SPIONs (high amount of protein with molecular weight <30 kDa and 50∼70 kDa). In addition, ligand density can also regulate PC in vivo. Bertrand et al. (2017) synthesized PEG–PLGA NPs with different PEG densities, from10 to 50 PEG chains per 100 nm2, and they found that PEG–PLGA NPs with higher PEG densities appeared to have lower relative abundance of apolipoprotein E. Zhang et al. (2018) synthesized four types of polystyrene NPs modified with PEG and Tf receptor (TfR)-targeting ligands (LT7, DT7, and Tf), and they found that different amounts and types of proteins adsorbed on the NPs, in which PEG-modified polystyrene NPs have the lease PC adsorbed (Figure 3B). Moreover, complement proteins were significantly abundant in PC of DT7 modified NPs.
Besides the properties of ENPs, environmental factors are also important for regulating PC. Generally, temperature, pH, fluidics, and concentration of protein are the main environmental factors for regulating PC in vitro. However, in vivo, PC is strongly affected by personal features in blood, such as a patient’s specific disease. Therefore, different compositions of PC can also be found in different individuals, even incubated with the same NPs. This gives rise to a concept of personalized PC (PPC). Colapicchioni et al. investigated the composition of PC on AmBisome-like liposomes in breast, gastric, and pancreatic cancer patients (Caracciolo V. et al., 2016). They found liposomes adsorb more protein in pancreatic cancer patients than that in breast and gastric patients. Especially for proteins with molecular weight ∼37 kDa, which are associated with immunoglobulin alpha (IgA) and IgG.
The modulation of in vivo PC is a comprehensive task, as ENPs are complex systems by possessing parameters from several dimensions including size, shape, composition, surface chemistry, etc. Among these properties, surface chemistry is heavily investigated as the ENP’s biological properties are largely surface-driven. Even a single surface chemistry change can alter the surface charge, hydrophobicity, hydrogen bonding, π bonding, or topography structure of an ENP. Because a construct of ENP involves multiple aspects of parameters, it’s extremely challenging to obtain a thorough relationships between ENPs’ physiochemical properties and PC formation. Thus, some observations achieved from one type of ENP may not extend to another ENP without a complete control of these parameters. Besides, the complex biofluids including protein density, types, and structures can subtly differ from one to another in different studies. Furthermore, as these nanomedicine research in small animals are ultimately used in humans, it is also critical to understand the physiological differences between different species regarding the in vivo PC formation.
Applications of in vivo PC
Adsorption of protein on the surface of ENPs and formation of PC-ENP complex in vivo seem inevitable. Thus, we can obtain specific biofunctionalized PC-ENP complex with unique advantages in biomedical applications. At present, the study of PC has gradually evolved from identification to application. There have been applied researches in the field of biomedicine such as drug carriers, targeting and personalized therapy based on PC.
Engineered nanoparticles used for targeted delivery of cancer drugs in vivo have been extensively investigated (Chou et al., 2011; Yu et al., 2012). There are two main targeting strategies for ENPs’ delivery to tumor: passive targeting and active targeting. However, most investigations remain at the laboratory stage. One possible reason is that PC rapidly formed on the surface of the carrier, thereby impeding their targeting (Salvati et al., 2013; Su et al., 2016, 2018). For passive targeting, ENPs target tumors by enhanced permeability and retention (EPR) effect. However, proteins such as immunoglobulins, fibrinogen, and complement can facilitate the clearance of ENPs by MPS. Thus, decrease of the adsorption of these types of protein on EPS can be used to enhance their passive targeting. For example, Takeuchi et al. (2017) prepared molecularly imprinted nanogels (MIP-NGs) with the capability of albumin recognition, which allows the particles to be immediately cloaked by albumin corona after injection, and thus reduces phagocytosis. As a result, most of MIP-NGs accumulated in tumor tissue, and almost no retention in liver tissue. Besides, PEG-conjugated lipid nanoparticle (LNP) adsorbed apolipoproteins in blood and successfully delivered small interfering RNA (siRNA) to LDLR-expressed HepG2 tumors and, with the accumulation ratio of tumor to all organs up by about 30% (Chen et al., 2019). Tesarova et al. (2020) found that PAS [proline (P), alanine (A), and serine (S)] modified ferritin (FRT)-based nanocarriers (PAS-10-FRTElli) reduced PC formation and did not activate complement C3 in mice. They were used to deliver cytostatic alkaloid ellipticine (Elli) in vivo. Compared with Elli alone or FRTElli, the amount in Elli internalization after treatment with PAS-10-FRTElli in tumor increased by nearly 3 folds. On the contrary, the amount in Elli internalization after treatment with PAS-10-FRTElli in liver decreased by 58% compared with that of non-encapsulated Elli. Compared to unmodified FRTElli, the amount in Elli internalization in spleen after treatment with PAS-10-FRTElli decreased by about 55%. Besides, DTX encapsulated in maleimide-modified ENPs were also used to target breast tumor site/cells (Li et al., 2018). Compared with unmodified ENPs, maleimide-modified ENPs adsorbed higher amount of low-immunogenic albumin, which protected ENPs from phagocytosis and prevented accelerated blood clearance. Then, maleimide-modified NPs carrier specially accumulated in tumor by albumin receptor-mediated active targeting or passive targeting in tumor tissue, and released DTX in tumor cell, followed by enhancing antitumor activity. Therefore, for drugs delivered by passive targeting, in vivo PC formation should be considered. Decreasing the composition of immunoglobulins, fibrinogen, and complement will prolong the blood circulation time and improve the targeting efficiency. Some surface modifications to modulate PC formation are needed in nanocarrier construction.
For active targeting, ENPs are modified with affinity ligands for specific recognition by the targeted cells via receptor mediated pathways. PC can also impact active targeting by their various compositions. For example, through controlling surface PC, Zhang et al. (2015) delivered antisense oligonucleotide (ASO) using the retinol-conjugated polyetheramine NPs system for the treatment of liver fibrosis (Figures 4A–D). This nanocarrier can selectively recruit retinoic alcohol binding protein 4 in the protein environment, which targets hepatic stellate cells (HSC). Then the ASO was released in HSC specifically and therefore effectively inhibits the expression of type I collagen, and thus reduces the liver fibrosis in mouse models. In addition, after modified with retinol, poly(beta-amino ester) polymers combined with terminal oligopeptides (OM-PBAE) increased the adsorption of apolipoproteins in the corona, thus enhanced active targeting to the liver, where receptors for these proteins are located (Fornaguera et al., 2019). 2-mercapto ethanol modified AuNSs can reduce the amount of adsorbed PC, and thus enhanced the active targeting ability of 2Rb17c nanobody to epidermal growth factor 2 receptor expressing tumor cells (D’Hollander et al., 2017).
FIGURE 4.
Sample applications of in vivo PC formation in nanomedicine. (A) Schematic diagram showing the fate of ASO-loaded retinol-conjugated polyetherimine (RAP) NPs after intravenous administered. (B) ASO (naked or delivered by RAP NPs) distribution in different organs. (C,D) Time-dependent ASO (naked or delivered by RAP NPs) concentrations in (C) the blood and (D) the liver. Adapted with permission from Zhang et al. (2015). (E) Schematic illustration of nanomedicine development strategy based on systematic analysis of patient-personalized PCs. Adapted with permission from Ren et al. (2019).
Since PC formation can highly impact the adsorption, biodistribution, and activity of nanocarriers, engineered PC formation and controlled PC compositions have raised much attention and huge interests recently. One of most common strategies is surface chemistry modulation. The above research works using this approach successfully modulated PC formation and improved the passive or active targeting efficiency of nanocarriers. Besides, PC pre-coating before nanocarriers were administered has emerged as a novel and facile strategy to engineer PC formation in vivo. For example, pre-coating of ApoE reduced the adsorption of albumin and immunoglobulin E on graphene/gold NPs, which enhanced the blood circulation time and tumor targeting (Lu et al., 2019).
Personalized therapy based on in vivo PC formation is another application. As ENP-plasma protein interactions are strongly influenced by the composition of PC, such interactions are highly dependent on the source of plasma. Therefore, it is speculated that different NP-protein complex exist between individuals, especially patients with specific diseases. Thus, understanding of PPC not only benefits the nanotherapy treatment efficacy but also aid in disease diagnosis. For example, Ren et al. (2019) found that Gd@C82(OH)22 NPs adsorbed more C1q in lung cancer patient than that in healthy human. In lung cancer patient, binding affinity between C1q and Gd@C82(OH)22 is significantly high, and secondary protein structure of C1q was abnormal, which subsequently influences the biological functions of C1q such as immune response. Thus, C1q can be as a specific biomarker for cancer diagnosis and cancer immune therapy (Figure 4E). Furthermore, the amount and composition of PC adsorbed on poly(ethylene glycol) (PEG)-coated mesoporous silica with a diameter of 100 nm (PEG-MS-100) were different from plasma samples of 23 healthy donors (Ju et al., 2020). Such changes of PC on PEGylated doxorubicin-encapsulated liposomes, especially for immunoglobulins (IGKV2-29, IGHV4-34, IGHG1) and complement proteins (C4B, C3), can impact particle-immune cell interactions. Thus, based on the diversity of plasm proteins in patients with different disease or in healthy person, a new class of size- and shape-tunable personalized protein NPs (PNP) was designed and made from patient-derived proteins (Lazarovits et al., 2019). The PNPs are advantageous because they are biodegradable, biocompatible, modifiable and non-toxic in vivo. They can be further combined with unique molecular fingerprints from different human patients, providing a huge potential in personalized nanomedicine.
Conclusion and Perspectives
After administered or exposed to living organisms, ENPs’ biobehaviors are highly related to their activity and safety. Besides a synthetic identity, PC endows ENPs a new biological identity, and subsequently alters the biobehaviors of ENPs. PC-related biological effects are dependent on their compositions and structures. Therefore, PC characterizations are critical. However, obtaining the accurate PC information remains a challenge, due to rapid and dynamic exchanges of proteins, limit of sample processing time, and weak interactions in some cases. Accurate characterization of in vivo PC formation is more difficult compared to in vitro characterization due to their complex environment. Until now, pioneer works on this aspect were focused on PC formation in blood. As compositions of PC significantly change over time in different cells, tissues, and organs, the spatial-temporal specific PC information still cannot be obtained. As the blood PC formation is dynamic, the findings of relationships between PC and biological effects of ENPs with static approaches may not be accurate. Moreover, PC compositions were identified from a pool of ENPs, which reveal the average level of protein adsorption. In fact, unlike small molecules, each individual ENP may vary slightly on physiochemical properties and the in vivo PC formation on each ENP is likely to be different, which poses another challenge of understanding the mechanisms.
In vivo PC formation is known to determine the in vivo behaviors of ENPs including absorption, blood circulation, biodistribution, metabolism, and toxicity, which are important pharmacokinetic parameters for ENPs used for nanomedicine development. So far, only a few studies have obtained the relationships between PC compositions and in vivo behaviors to predict the ENPs’ biological effects. Furthermore, PC is not only one layer of adsorbed proteins but can be two or more layers. The outmost layer of proteins directly influences the ENPs’ biobehaviors. Therefore, mostly reported relationship studies obtained from analysis of the total PC might not be accurate. Future research should pay attention to distinguish the inner adsorbed proteins and outer adsorbed proteins.
Engineered nanoparticles can be functionalized by specific targeting moieties to target disease sites in nanomedicine, however, the in vivo PC formation can completely or partially shield the targeting ability of ENPs. Therefore, modulating in vivo PC formation to reduce the negative influences of PC is important. In vivo PC formation can be modulated by the size, morphology, charge, and surface chemistry of ENPs. Besides, recruiting specific proteins to realize targeting delivery of ENPs has become a new concept. Additionally, in vivo PC formation can also be affected by different types of blood as the plasma proteome is different between individuals. In this case, a systematic investigation of disease-related in vivo PC formation is beneficial for diagnosis and therapeutics, as well as speeding up the clinical translation of ENPs. In the future, we can envision disease-specific and in vivo PC-based diagnostic and therapeutic ENP agents.
Author Contributions
XB, QM, and GS designed this work of review. XB and JW performed the literature search of the databases. XB and GS wrote the manuscript. QM revised the manuscript. All authors have read and approved to publish the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (22076085) and Shandong Provincial Natural Science Foundation of China (ZR2020QB138).
References
- Abbina S., Takeuchi L. E., Anilkumar P., Yu K., Rogalski J. C., Shenoi R. A., et al. (2020). Blood circulation of soft nanomaterials is governed by dynamic remodeling of protein opsonins at nano-biointerface. Nat. Commun. 11:3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F., Wang X., Li W. (2019). Toxico-Metabolomics of Engineered Nanomaterials: progress and Challenges. Adv. Funct. Mater. 29:201904268 [Google Scholar]
- Amici A., Caracciolo G., Digiacomo L., Gambini V., Marchini C., Tilio M., et al. (2017). In vivo protein corona patterns of lipid nanoparticles. RSC Adv. 7:1137–1145 [Google Scholar]
- Bakshi M. S. (2020). Impact of nanomaterials on ecosystems: mechanistic aspects in vivo. Environ. Res. 182:109099. 10.1016/j.envres.2019.109099 [DOI] [PubMed] [Google Scholar]
- Bertrand N., Grenier P., Mahmoudi M., Lima E. M., Appel E. A., Dormont F., et al. (2017). Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nat. Commun. 8:777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancolini G., Kokh D. B., Calzolai L., Wade R. C., Corni S. (2012). Docking of Ubiquitin to Gold Nanoparticles. ACS Nano. 6 9863–9878. 10.1021/nn303444b [DOI] [PubMed] [Google Scholar]
- Brun E., Sicard-Roselli C. (2014). Could nanoparticle corona characterization help for biological consequence prediction? Cancer Nanotechnol. 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Ma Y., Wu Z., Ding Y., Zhang P., He X., et al. (2016). Protein corona influences liver accumulation and hepatotoxicity of gold nanorods. NanoImpact 3-4 40–46. 10.1016/j.impact.2016.09.005 [DOI] [Google Scholar]
- Caracciolo V., Caracciolo G., Digiacomo L., Tilio M., et al. (2016). Personalized liposome-protein corona in the blood of breast, gastric and pancreatic cancer patients. Int. J. Biochem. Cell Biol. 75 180–187. 10.1016/j.biocel.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Caracciolo G., Farokhzad O. C., Mahmoudi M. (2016). Biological Identity of Nanoparticles In Vivo: clinical Implications of the Protein Corona. Trends. Biotechnol. 35 257–264. 10.1016/j.tibtech.2016.08.011 [DOI] [PubMed] [Google Scholar]
- Carril M., Padro D., Del Pino P., Carrillo-Carrion C., Gallego M., Parak W. J. (2017). In situ detection of the protein corona in complex environments. Nat. Commun. 8:1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedervall T., Lynch I., Lindman S., Berggård T., Thulin E., Nilsson H., et al. (2007). Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. 104 2050–2055. 10.1073/pnas.0608582104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Parayath N., Ganesh S., Wang W., Amiji M. M. (2019). Role of Apolipoprotein- and Vitronectin-Enriched Protein Corona on Lipid Nanoparticles for In Vivo Targeted Delivery and Transfection of Oligonucleotides in Murine Tumor Models. Nanoscale 11 18806–18824. 10.1039/c9nr05788a [DOI] [PubMed] [Google Scholar]
- Chinen A. B., Guan C. M., Ko C. H., Mirkin C. A. (2017). The Impact of Protein Corona Formation on the Macrophage Cellular Uptake and Biodistribution of Spherical Nucleic Acids. Small 13:1603847. 10.1002/smll.201603847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou L. Y. T., Ming K., Chan W. C. W. (2011). Strategies for the intracellular delivery of nanoparticles. Chem. Soc. Rev. 40 233–245. [DOI] [PubMed] [Google Scholar]
- Corbo C., Molinaro R., Taraballi F., Toledano Furman N. E., Hartman K. A., Sherman M. B., et al. (2017). Unveiling the in Vivo Protein Corona of Circulating Leukocyte-like Carriers. ACS Nano. 11 3262–3273. 10.1021/acsnano.7b00376 [DOI] [PubMed] [Google Scholar]
- Dhawan A., Shanker R., Singh S., Kumar A. (2018). Nanobiotechnology: Human Health and the Environment. United Kingdom: Taylor & Francis group; 1–34. [Google Scholar]
- D’Hollander A., Jans H., Velde G. V., Verstraete C., Massa S., Devoogdt N., et al. (2017). Limiting the protein corona: a successful strategy for invivo active targeting of anti-HER2 nanobody-functionalized nanostars. Biomaterials 123 15–23. 10.1016/j.biomaterials.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Docter D., Westmeier D., Markiewicz M., Stolte S., Knauer S. K., Stauber R. H. (2015). The nanoparticle biomolecule corona: lessons learned - challenge accepted? Chem. Soc. Rev. 46 6094–6121. 10.1039/c5cs00217f [DOI] [PubMed] [Google Scholar]
- Duan Y. K., Coreas R., Liu Y., Bitounis D., Zhang Z. Y., Parviz D., et al. (2020). Prediction of protein corona on nanomaterials by machine learning using novel descriptors. NanoImpact 12:100207. 10.1016/j.impact.2020.100207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer C. C., Payne C. K. (2014). Secondary Structure of Corona Proteins Determines the Cell Surface Receptors Used by Nanoparticles. J. Phys. Chem. B 118 14017–14026. 10.1021/jp502624n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornaguera C., -Rebollo M. G., Lázaro M. A., Cascante A., Rubio N., Blanco J., et al. (2019). In Vivo Retargeting of Poly(beta aminoester) (OM-PBAE) Nanoparticles is Influenced by Protein Corona. Adv. Healthcare Mater. 8:1900849. 10.1002/adhm.201900849 [DOI] [PubMed] [Google Scholar]
- Gagner J. E., Lopez M. D., Dordick J. S., Siegel R. W. (2011). Effect of gold nanoparticle morphology on adsorbed protein structure and function. Biomaterials 32 7241–7252. 10.1016/j.biomaterials.2011.05.091 [DOI] [PubMed] [Google Scholar]
- García-Álvarez R., Hadjidemetriou M., Sánchez-Iglesias A., Liz-Marzán L. M., Kostarelos K. (2018). In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape. Nanoscale 10 1256–1264. 10.1039/c7nr08322j [DOI] [PubMed] [Google Scholar]
- García-Díaz M., Birch D., Wan F., Nielsen H. M. R. (2017). The role of mucus as an invisible cloak to transepithelial drug delivery by nanoparticles. Adv. Drug Deliv. Rev. 124 107–124. 10.1016/j.addr.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Ge C. C., Du J. F., Zhao L. N., Wang L. M., Liu Y., Li D. H., et al. (2011). Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 108 16968–16973. 10.1073/pnas.1105270108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier P., Viana I. M. O., Lima E. M., Bertrand N. (2018). Anti-polyethylene glycol antibodies alter the protein corona deposited on nanoparticles and the physiological pathways regulating their fate in vivo. J. Contr. Release 287 121–131. 10.1016/j.jconrel.2018.08.022 [DOI] [PubMed] [Google Scholar]
- Guan G., Zhang S., Liu S., Cai Y., Low M., Teng C. P., et al. (2015). Protein Induces Layer-by-Layer Exfoliation of Transition Metal Dichalcogenides. J. Am. Chem. Soc. 137 6152–6155. 10.1021/jacs.5b02780 [DOI] [PubMed] [Google Scholar]
- Guo C., Liu Y., Li Y. (2020). Adverse effects of amorphous silica nanoparticles: focus on human cardiovascular health. J. Hazard. Mater. 406 124626–124626. 10.1016/j.jhazmat.2020.124626 [DOI] [PubMed] [Google Scholar]
- Hadjidemetriou M., Al-Ahmady Z., Mazza M., Collins R. F., Dawson K., Kostarelos K. (2015). In Vivo Biomolecule Corona around Blood-Circulating, Clinically Used and Antibody-Targeted Lipid Bilayer Nanoscale Vesicles. ACS Nano. 9 8142–8156. 10.1021/acsnano.5b03300 [DOI] [PubMed] [Google Scholar]
- Hadjidemetriou M., Kostarelos K. (2017). Nanomedicine: evolution of the nanoparticle corona. Nat. Nanotechnol. 12 288–290. 10.1038/nnano.2017.61 [DOI] [PubMed] [Google Scholar]
- Hadjidemetriou M., Zahraa A. A., Kostarelos K. (2016). Time-evolution of in vivo protein corona onto blood-circulating PEGylated liposomal doxorubicin (DOXIL) nanoparticles. Nanoscale 8 6948–6957. 10.1039/c5nr09158f [DOI] [PubMed] [Google Scholar]
- Jia J., Li F., Zhou H., Bai Y., Liu S., Jiang Y., et al. (2017). Oral Exposure to Silver Nanoparticles or Silver Ions May Aggravate Fatty Liver Disease in Overweight Mice. Environ. Sci. Technol. 51 9334. 10.1021/acs.est.7b02752 [DOI] [PubMed] [Google Scholar]
- Ju Y., Kelly H. G., Dagley L. F., Reynaldi A., Schlub T. E., Spall S. K., et al. (2020). Person-Specific Biomolecular Coronas Modulate Nanoparticle Interactions with Immune Cells in Human Blood. ACS Nano. 14 15723–15737. 10.1021/acsnano.0c06679 [DOI] [PubMed] [Google Scholar]
- Kamyshny A., Magdassi S. (2019). Conductive nanomaterials for 2D and 3D printed flexible electronics. Chem. Soc. Rev. 48 1712–1740. 10.1039/c8cs00738a [DOI] [PubMed] [Google Scholar]
- Kokkinopoulou M., Simon J., Landfester K., Mailander V., Lieberwirth I. (2017). Visualization of the protein corona: towards a biomolecular understanding of nanoparticle-cell-interactions. Nanoscale 9:25. [DOI] [PubMed] [Google Scholar]
- Lakshmi K., Kadirvelu K., Mohan P. S. (2019). Reclaimable La: ZnO/PAN nanofiber catalyst for photodegradation of methyl paraoxon and its toxicological evaluation utilizing early life stages of zebra fish (Danio rerio). Chem. Engin. J. 357 724–736. 10.1016/j.cej.2018.09.201 [DOI] [Google Scholar]
- Lazarovits J., Chen Y. Y., Song F., Ngo W., Tavares A. J., Zhang Y., et al. (2019). Synthesis of Patient-Specific Nanomaterials. Nano. Lett. 19 116–123. [DOI] [PubMed] [Google Scholar]
- Lee D. E., Koo H., Sun I.-C., Ryu J. H., Kim K., Kwon I. C. (2012). Multifunctional nanoparticles for multimodal imaging and theragnosis. Chem. Soc. Rev. 41 2656–2672. 10.1039/c2cs15261d [DOI] [PubMed] [Google Scholar]
- Li Z., Li D., Li Q., Luo C., Li J., Kou L., et al. (2018). In situ low-immunogenic albumin-conjugating-corona guiding nanoparticles for tumor-targeting chemotherapy. Biomater. Sci. 6 2681–2693. 10.1039/c8bm00692j [DOI] [PubMed] [Google Scholar]
- Lin X., Pan Q., He Y. (2019). In situ detection of protein corona on single particle by rotational diffusivity. Nanoscale 11 18367–18374. 10.1039/c9nr06072c [DOI] [PubMed] [Google Scholar]
- Liu S., Wang Z., Jiang X., Gan J., Tian X., Xing Z., et al. (2020). Denatured corona proteins mediate the intracellular bioactivities of nanoparticles via the unfolded protein response. Biomaterials 265:120452. 10.1016/j.biomaterials.2020.120452 [DOI] [PubMed] [Google Scholar]
- Lo Giudice M. C., Herda L. M., Polo E., Dawson K. A. (2016). In situ characterization of nanoparticle biomolecular interactions in complex biological media by flow cytometry. Nat. Commun. 7:13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry G. V., Avellan A., Gilbertson L. M. (2019). Opportunities and challenges for nanotechnology in the agri-tech revolution. Nat. Nanotechnol. 14:517. 10.1038/s41565-019-0461-7 [DOI] [PubMed] [Google Scholar]
- Lu X., Xu P. P., Ding H. M., Yu Y. S., Huo D., Ma Y. Q. (2019). Tailoring the component of protein corona via simple chemistry. Nat. Commun. 10:4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi M., Shokrgozar M. A., Sardari S., Moghadam M. K., Vali H., Laurent S., et al. (2011). Irreversible changes in protein conformation due to interaction with superparamagnetic iron oxide nanoparticles. Nanoscale 3 1127–1138. [DOI] [PubMed] [Google Scholar]
- Merle N. S., Elizabeth C. S., Veronique F. B., Roumenina L. T. (2015a). Complement System Part I – Molecular Mechanisms of Activation and Regulation. Front. Immunol. 6:262. 10.3389/fimmu.2015.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle N. S., Remi N., Lise H. M., Veronique F. B., Roumenina L. T. (2015b). Complement System Part II: role in Immunity. Front. Immunol. 6:257. 10.3389/fimmu.2015.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad-Beigi H., Hayashi Y., Zeuthen C. M., Eskandari H., Scavenius C., Juul-Madsen K., et al. (2020). Mapping and identification of soft corona proteins at nanoparticles and their impact on cellular association. Nat. Commun. 11:4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monopoli M. P., Åberg C., Salvati A., Dawson K. A. (2012). Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 7 779–786. 10.1038/nnano.2012.207 [DOI] [PubMed] [Google Scholar]
- Nayak P. S., Borah S. M., Gogoi H., Asthana S., Bhatnagar R., Jha A. N., et al. (2019). Lactoferrin adsorption onto silver nanoparticle interface: implications of corona on protein conformation, nanoparticle cytotoxicity and the formulation adjuvanticity. Chem. Engin. J. 361 470–484. 10.1016/j.cej.2018.12.084 [DOI] [Google Scholar]
- Nel A. E., Mdler L., Velegol D., Xia T., Thompson M. (2009). Understanding Biophysicochemical Interactions at the Nano-Bio Interface. Nat. Mater. 8 543–557. 10.1038/nmat2442 [DOI] [PubMed] [Google Scholar]
- Nissinen T., Nkki S., Laakso H., Kuiauskas D., Lehto V. P. (2016). Tailored Dual PEGylation of Inorganic Porous Nanocarriers for Extremely Long Blood Circulation in Vivo. ACS Appl. Mater. Interf. 8 32723–32731. 10.1021/acsami.6b12481 [DOI] [PubMed] [Google Scholar]
- Pelaz B., Alexiou C., Alvarez-Puebla R. A., Alves F., Andrews A. M., Ashraf S., et al. (2017). Diverse Applications of Nanomedicine. ACS Nano. 11 2313–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R. J. B., Bouwmeester H., Gottardo S., Amenta V., Arena M., Brandhoff P., et al. (2016). Nanomaterials for products and application in agriculture, feed and food. Trends Food Sci. Technol. 54 155–164. 10.1016/j.tifs.2016.06.008 [DOI] [Google Scholar]
- Pinals R. L., Yang D., Rosenberg D. J., Chaudhary T., Crothers A. R., Iavarone A. T., et al. (2020). Quantitative Protein Corona Composition and Dynamics on Carbon Nanotubes in Biological Environments. Angewandte Chem. 59 2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochert A., Vernikouskaya I., Pascher F., Rasche V., Lindén M. (2017). Cargo-influences on the biodistribution of hollow mesoporous silica nanoparticles as studied by quantitative 19F-magnetic resonance imaging. J. Coll. Interf. Sci. 488 1–9. 10.1016/j.jcis.2016.10.085 [DOI] [PubMed] [Google Scholar]
- Pomerantseva E., Bonaccorso F., Feng X., Cui Y., Gogotsi Y. (2019). Energy storage: the future enabled by nanomaterials. Science 366:6468. [DOI] [PubMed] [Google Scholar]
- Qu S., Sun F., Qiao Z., Li J., Shang L. (2020). In Situ Investigation on the Protein Corona Formation of Quantum Dots by Using Fluorescence Resonance Energy Transfer. Small 16:1907633. 10.1002/smll.201907633 [DOI] [PubMed] [Google Scholar]
- Raoufi M., Hajipour M. J., Kamali Shahri S. M., Schoen I., Linn U., Mahmoudi M. (2018). Probing fibronectin conformation on a protein corona layer around nanoparticles. Nanoscale 10 1228–1233. 10.1039/c7nr06970g [DOI] [PubMed] [Google Scholar]
- Ren J., Cai R., Wang J., Daniyal M., Baimanov D., Liu Y., et al. (2019). Precision Nanomedicine Development Based on Specific Opsonization of Human Cancer Patient-Personalized Protein Coronas. Nano. Lett. 19 4692–4701. 10.1021/acs.nanolett.9b01774 [DOI] [PubMed] [Google Scholar]
- Rivera V. E., Rodrguez A. S., Ramrez M. U., Lozano O., Ruiz A. D. V. (2019). Plasma protein adsorption on Fe 3 O 4 -PEG nanoparticles activates the complement system and induces an inflammatory response. Int. J. Nanomed. 14:2055. 10.2147/ijn.s192214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruge C. A., Hillaireau H., Grabowski N., Beck-Broichsitter M., Cañadas O., Tsapis N., et al. (2016). Pulmonary Surfactant Protein A-Mediated Enrichment of Surface-Decorated Polymeric Nanoparticles in Alveolar Macrophages. Mole. Pharm. 13 4168–4178. 10.1021/acs.molpharmaceut.6b00773 [DOI] [PubMed] [Google Scholar]
- Saha K., Rahimi M., Yazdani M., Kim S. T., Moyano D. F., Hou S., et al. (2016). Regulation of Macrophage Recognition through the Interplay of Nanoparticle Surface Functionality and Protein Corona. ACS Nano. 10 4421–4430. 10.1021/acsnano.6b00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakulkhu U., Maurizi L., Mahmoudi M., Motazacker M., Vries M., Gramoun A., et al. (2014). Ex situ evaluation of the composition of protein corona of intravenously injected superparamagnetic nanoparticles in rats. Nanoscale 6 11439–11450. 10.1039/c4nr02793k [DOI] [PubMed] [Google Scholar]
- Salvati A., Pitek A. S., Monopoli M. P., Prapainop K., Bombelli F. B., Hristov D. R., et al. (2013). Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 8 137–143. 10.1038/nnano.2012.237 [DOI] [PubMed] [Google Scholar]
- Sanchez-Guzman D., Giraudon–Colas G., Marichal L., Boulard Y., Wien F., Degrouard J., et al. (2020). In Situ Analysis of Weakly Bound Proteins Reveals Molecular Basis of Soft Corona Formation. ACS Nano. 14 9073–9088. 10.1021/acsnano.0c04165 [DOI] [PubMed] [Google Scholar]
- Schäffler M., Sousa F., Wenk A., Sitia L., Hirn S., Schleh C., et al. (2014). Blood protein coating of gold nanoparticles as potential tool for organ targeting. Biomaterials 35 3455–3466. [DOI] [PubMed] [Google Scholar]
- Sfriso A. A., Mistri M., Munari C., Moro I., Wahsha M., Sfriso A., et al. (2019). Hazardous effects of silver nanoparticles for primary producers in transitional water systems: the case of the seaweed Ulva rigida C. Agardh. Environ. Int. 131 104942. 10.1016/j.envint.2019.104942 [DOI] [PubMed] [Google Scholar]
- Shakiba S., Hakimian A., Barco L. R., Louie S. M. (2018). Dynamic Intermolecular Interactions Control Adsorption from Mixtures of Natural Organic Matter and Protein onto Titanium Dioxide Nanoparticles. Environ. Sci. Technol. 52 14158–14168. 10.1021/acs.est.8b04014 [DOI] [PubMed] [Google Scholar]
- Shang L., Nienhaus G. U. (2017). In Situ Characterization of Protein Adsorption onto Nanoparticles by Fluorescence Correlation Spectroscopy. Account. Chem. Res. 50 387–395. 10.1021/acs.accounts.6b00579 [DOI] [PubMed] [Google Scholar]
- Shemetov A. A., Nabiev I., Sukhanova A. (2012). Molecular Interaction of Proteins and Peptides with Nanoparticles. ACS Nano. 6 4585–4602. 10.1021/nn300415x [DOI] [PubMed] [Google Scholar]
- Stepien G., Moros M., Pérez-Hernández M., Monge M., Gutiérrez L., Fratila R. M., et al. (2018). Effect of Surface Chemistry and Associated Protein Corona on the Long-Term Biodegradation of Iron Oxide Nanoparticles In Vivo. ACS Appl. Mater. Interf. 10 4548–4560. 10.1021/acsami.7b18648 [DOI] [PubMed] [Google Scholar]
- Su G., Jiang H., Xu B., Yu Y., Chen X. (2018). Effects of Protein Corona on Active and Passive Targeting of Cyclic RGD Peptide-Functionalized PEGylation Nanoparticles. Mole. Pharm. 15 5019–5030. 10.1021/acs.molpharmaceut.8b00612 [DOI] [PubMed] [Google Scholar]
- Su G., Zhou X., Zhou H., Li Y., Zhang X., Liu Y., et al. (2016). Size-Dependent Facilitation of Cancer Cell Targeting by Proteins Adsorbed on Nanoparticles. ACS Appl. Mater. Interf. 8 30037–30047. 10.1021/acsami.6b10967 [DOI] [PubMed] [Google Scholar]
- Susan H., De M. M. B. C., Metselaar J. M., Gert S. (2018). Current Trends and Challenges in the Clinical Translation of Nanoparticulate Nanomedicines: pathways for Translational Development and Commercialization. Front. Pharmacol. 9:790. 10.3389/fphar.2018.00790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres G. P., Stephan W., Guttmann P., Spedalieri C., Drescher D., Živanovića V., et al. (2020). Relating the composition and interface interactions in the hard corona of gold nanoparticles to the induced response mechanisms in living cells. Nanoscale 12 17450–17461. 10.1039/d0nr03581e [DOI] [PubMed] [Google Scholar]
- Takeuchi P. D. T., Kitayama Y., Sasao R., Yamada T., Toh K., Matsumoto Y., et al. (2017). Molecularly Imprinted Nanogels Acquire Stealth In Situ by Cloaking Themselves with Native Dysopsonic Proteins. Angewandte Chem. 129 7194–7198. 10.1002/ange.201700647 [DOI] [PubMed] [Google Scholar]
- Teng C., Jia J., Wang Z., Sharma V. K., Yan B. (2019). Size-dependent maternal-fetal transfer and fetal developmental toxicity of ZnO nanoparticles after oral exposures in pregnant mice. Ecotoxicol. Environ. Safety 182:109439. 10.1016/j.ecoenv.2019.109439 [DOI] [PubMed] [Google Scholar]
- Tesarova B., Dostalova S., Smidova V., Goliasova Z., Skubalova Z., Michalkova H., et al. (2020). Surface-PASylation of ferritin to form stealth nanovehicles enhances in vivo therapeutic performance of encapsulated ellipticine. Appl. Mater. Today 18:100501. 10.1016/j.apmt.2019.100501 [DOI] [Google Scholar]
- Toy R., Pradhan P., Ramesh V., Di Paolo N. C., Lash B., Liu J., et al. (2019). Modification of primary amines to higher order amines reduces in vivo hematological and immunotoxicity of cationic nanocarriers through TLR4 and complement pathways. Biomaterials 225:119512. 10.1016/j.biomaterials.2019.119512 [DOI] [PubMed] [Google Scholar]
- Treuel L., Malissek M., Gebauer J. S., Zellner R. (2010). The influence of surface composition of nanoparticles on their interactions with serum albumin. Chemphyschem 11 3093–3099. 10.1002/cphc.201000174 [DOI] [PubMed] [Google Scholar]
- Vilanova O., Mittag J. J., Kelly P. M., Milani S., Dawson K. A., Rädler J. O., et al. (2016). Understanding the Kinetics of Protein–Nanoparticle Corona Formation. ACS Nano. 10 10842–10850. 10.1021/acsnano.6b04858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu V. P., Gifford G. B., Chen F., Benasutti H., Wang G., Groman E. V., et al. (2019). Immunoglobulin deposition on biomolecule corona determines complement opsonization efficiency of preclinical and clinical nanoparticles. Nat. Nanotechnol. 14 260–268. 10.1038/s41565-018-0344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkey C. D., Olsen J. B., Song F., Liu R., Guo H., Olsen D. W. H., et al. (2014). Protein Corona Fingerprinting Predicts the Cellular Interaction of Gold and Silver Nanoparticles. ACS Nano. 8 2439–2455. 10.1021/nn406018q [DOI] [PubMed] [Google Scholar]
- Wang A., Yang T., Fan W., Yang Y., Zhu Q., Guo S., et al. (2018). Protein Corona Liposomes Achieve Efficient Oral Insulin Delivery by Overcoming Mucus and Epithelial Barriers. Adv. Healthcare Mater. 8:1801123. 10.1002/adhm.201801123 [DOI] [PubMed] [Google Scholar]
- Wang M., Gustafsson O. J. R., Pilkington E. H., Kkinen A., Ke P. C. (2018). Nanoparticle-Proteome In Vitro and In Vivo. J. Mater. Chem. B 6 6026–6041. [DOI] [PubMed] [Google Scholar]
- Wang H., Fu J., Wang C., Wang J., Yang A., Li C., et al. (2020). A binder-free high silicon content flexible anode for Li-ion batteries. Energ. Environ. Sci. 13 848–858. 10.1039/c9ee02615k [DOI] [Google Scholar]
- Wang Q.-W., Zhang H.-B., Liu J., Zhao S., Xie X., Liu L., et al. (2019). Multifunctional and Water-Resistant MXene-Decorated Polyester Textiles with Outstanding Electromagnetic Interference Shielding and Joule Heating Performances. Adv. Funct. Mater. 29:1806819. 10.1002/adfm.201806819 [DOI] [Google Scholar]
- Wang T., Bai J., Jiang X., Nienhaus G. U. (2012). Cellular Uptake of Nanoparticles by Membrane Penetration: a Study Combining Confocal Microscopy with FTIR Spectroelectrochemistry. ACS Nano 6 1251–1259. 10.1021/nn203892h [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang C., Liu S., He W., Wang L., Gan J., et al. (2017). Specifically Formed Corona on Silica Nanoparticles Enhances Transforming Growth Factor β1 Activity in Triggering Lung Fibrosis. ACS Nano 11 1659–1672. 10.1021/acsnano.6b07461 [DOI] [PubMed] [Google Scholar]
- Weiss A. C. G., Krüger K., Besford Q. A., Schlenk M., Kempe K., Förster S., et al. (2019). In Situ Characterization of Protein Corona Formation on Silica Microparticles Using Confocal Laser Scanning Microscopy Combined with Microfluidics. ACS Appl. Materi. Interf. 11 2459–2469. 10.1021/acsami.8b14307 [DOI] [PubMed] [Google Scholar]
- Wong X. Y., Sena-Torralba A., lvarez-Diduk R., Muthoosamy K., Merkoçi A. (2020). Nanomaterials for Nanotheranostics: tuning Their Properties According to Disease Needs. ACS Nano. 14 2585–2627. 10.1021/acsnano.9b08133 [DOI] [PubMed] [Google Scholar]
- Xu F., Reiser M., Yu X., Gummuluru S., Wetzler L., Reinhard B. M. (2016). Lipid-Mediated Targeting with Membrane-Wrapped Nanoparticles in the Presence of Corona Formation. ACS Nano 10 1189–1200. 10.1021/acsnano.5b06501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Gause K. T., Kamphuis M. M. J., Ang C.-S., O’brien-Simpson N. M., Lenzo J. C., et al. (2013). Differential Roles of the Protein Corona in the Cellular Uptake of Nanoporous Polymer Particles by Monocyte and Macrophage Cell Lines. ACS Nano 7 10960–10970. 10.1021/nn404481f [DOI] [PubMed] [Google Scholar]
- Yang D., Liu D., Qin M., Chen B., Song S., Dai W., et al. (2018). Intestinal Mucin Induces More Endocytosis but Less Transcytosis of Nanoparticles across Enterocytes by Triggering Nanoclustering and Strengthening the Retrograde Pathway. ACS Appl. Mater. Interf. 10 11443–11456. 10.1021/acsami.7b19153 [DOI] [PubMed] [Google Scholar]
- Yin B., Chan C. K. W., Liu S., Hong H., Wong S. H. D., Lee L. K. C., et al. (2019). Intrapulmonary Cellular-Level Distribution of Inhaled Nanoparticles with Defined Functional Groups and Its Correlations with Protein Corona and Inflammatory Response. ACS Nano 13 14048–14069. 10.1021/acsnano.9b06424 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Yoshioka Y., Morishita Y., Aoyama M., Tochigi S., Hirai T., et al. (2015). Protein corona changes mediated by surface modification of amorphous silica nanoparticles suppress acute toxicity and activation of intrinsic coagulation cascade in mice. Nanotechnology 26:245101. 10.1088/0957-4484/26/24/245101 [DOI] [PubMed] [Google Scholar]
- Yu M. K., Park J., Jon S. (2012). Targeting Strategies for Multifunctional Nanoparticles in Cancer Imaging and Therapy. Theranostics 2 3–44. 10.7150/thno.3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Zhao L., Guo C., Yan B., Su G. (2020). Regulating Protein Corona Formation and Dynamic Protein Exchange by Controlling Nanoparticle Hydrophobicity. Front. Bioengin. Biotechnol. 8:210. 10.3389/fbioe.2020.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wu T., Yu W., Ruan S., He Q., Gao H. (2018). Ligand Size and Conformation Affect the Behavior of Nanoparticles Coated with in Vitro and in Vivo Protein Corona. ACS Appl. Mater. Interf. 10 9094–9103. 10.1021/acsami.7b16096 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang C., Zha Y., Hu W., Gao Z., Zang Y., et al. (2015). Corona-Directed Nucleic Acid Delivery into Hepatic Stellate Cells for Liver Fibrosis Therapy. Acs Nano 9 2405–2419. 10.1021/nn505166x [DOI] [PubMed] [Google Scholar]
- Zhou H., Fan Z., Li P. Y., Deng J., Arhontoulis D. C., Li C. Y., et al. (2018). Dense and Dynamic Polyethylene Glycol Shells Cloak Nanoparticles from Uptake by Liver Endothelial Cells for Long Blood Circulation. ACS Nano 12 10130–10141. 10.1021/acsnano.8b04947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Jiang P., Luo B., Lan F., He J., Wu Y. (2019). Dynamic protein corona influences immune-modulating osteogenesis in magnetic nanoparticle (MNP)-infiltrated bone regeneration scaffoldsin vivo. Nanoscale 11 6817–6827. 10.1039/c8nr08614a [DOI] [PubMed] [Google Scholar]