Abstract
Thyroid cancer (TC) is the most common type of endocrine malignancy in humans, and its relative incidence has increased continuously in recent years. However, the primary molecular mechanisms of thyroid tumorigenesis and progression remain unclear. Papillary TC (PTC) is the most common subtype of TC. Recent studies have reported that one of the tumorigenesis and progression mechanisms is driven by genetic alterations that regulate the TC cell signaling pathway. In the present study, RNA sequencing (RNA-seq) was performed on 79 paired PTC and adjacent normal thyroid tissues to further study the molecular mechanisms of TC. Reverse transcription-quantitative PCR was used to detect the expression levels of LEM domain containing 1 (LEMD1) in 47 paired PTC and adjacent normal thyroid tissue samples. Initial analysis revealed that LEMD1 expression was significantly upregulated in TC tissues compared with that in normal tissues. The results of the thyroid RNA-seq datasets from The Cancer Genome Atlas were consistent with the RNA-seq analysis results of the present study. High LEMD1 expression increased the risk of lymph node metastasis in patients with TC. The biological function of LEMD1 on cell proliferation, migration, invasion and apoptosis was investigated in vitro via small interfering RNA and overexpression vector. Gene set enrichment analysis indicated that high LEMD1 expression was associated with epithelial-mesenchymal transition (EMT) and the Wnt/β-catenin signaling pathway. Western blotting revealed that LEMD1 modulated the protein expression levels of E-cadherin, N-cadherin, vimentin, β-catenin and cleaved-caspase 3. In conclusion, the present results indicated that LEMD1 may drive TC cell tumorigenesis and progression by activating the Wnt/β-catenin signaling pathway and EMT.
Keywords: LEM domain containing 1, thyroid cancer, lymph node metastasis, epithelial-mesenchymal transition, Wnt/β-catenin
Introduction
Thyroid cancer (TC) originates from parafollicular or follicular thyroid cells, and it includes papillary TC (PTC), anaplastic TC, medullary TC, poorly differentiated TC and follicular TC (1). According to the Cancer Statistics report in 2019, 52,070 new cases were diagnosed and 2,170 deaths were recorded in the United States (2). A similar situation exists in China, where the incidence of TC was 1,700 per 100,000 people between 1981 and 2001 (3). PTC is the major histological type of TC that accounts for 80–85% of all TC cases, but this disease is highly curable (4,5). The prognosis of patients with PTC after total thyroidectomy with cervical node dissection is good, with a 10-year survival rate >90% (6). However, PTC still displays aggressive behaviors, such as metastasis and recurrence. Lymph node metastasis (LNM) occurs in 50% of patients with PTC, and the recurrence rate is up to 10% (7,8). Therefore, to avoid the loss of quality of life of patients, a new diagnostic or treatment method is essential for patients with TC.
Although numerous studies have been conducted on TC over the past decades (9,10), its underlying mechanisms remain unclear. With the development of RNA sequencing (RNA-seq), studies have revealed that the major mechanisms of genome alteration that underlie cancer pathogenesis are copy number gain/loss, chromosomal rearrangements and genetic mutations (11–13). Several vital genome alterations have been discovered (14,15). The mutations of B-type Raf kinase are the most well-known and important genome alterations that promote PTC tumorigenesis and progression by influencing the MAPK signaling pathway (16). Despite the numerous reported mechanisms of TC, the primary ones remain to be further elucidated (17,18).
LEM domain containing 1 (LEMD1) is a member of the cancer/testis gene family located on human chromosome 1q32.1, a member of the cancer/testis antigen (CTA) family. CTAs are commonly expressed in normal testis tissues and malignant tissues (19). Therefore, the cancer/testis gene LEMD1 may be a reasonable candidate target for the diagnosis and treatment of carcinoma. A previous study has reported that LEMD1 expression is widely upregulated in gastric cancer, prostate cancer, oral squamous cell carcinoma, anaplastic large cell lymphoma and colorectal cancer (20). In addition, the overexpression of LEMD1 promotes gastric cancer cell proliferation by activating the PI3K/AKT signaling pathway (20). Another study has suggested that LEMD1 promotes the tumorigenesis of anaplastic large-cell lymphoma, and that it is a target gene of microRNA-135b (21). Additionally, LEMD1 has a role in the maintenance of colorectal cancer stem-like cells (22).
To the best of our knowledge, the function of LEMD1 in PTC has not been investigated. Therefore, the present study aimed to investigate the function and the underlying mechanism of LEMD1 in the proliferation and migration of TC cells.
Materials and methods
Patients and tissue collection
Fresh tissue samples (47 paired PTC and non-tumor thyroid tissues) were provided by the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) between May 2018 and April 2019 as a local validated cohort. These samples were obtained from patients who were not undergoing chemotherapy nor radiotherapy. All of the samples were frozen in liquid nitrogen and stored at −80°C until processing. Tumor histological review was performed by two pathologists, and two senior pathologists confirmed the results. The patients were informed before the operation, and they provided written informed consent in accordance with the guidelines of the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University (approval no. 2012-57). The TC expression data and corresponding clinical information were obtained from The Cancer Genome Atlas (TCGA) portal (https://tcgadata.nci.nih.gov/tcga/). The TCGA gene expression data of 502 PTC samples and 58 normal samples were available.
RNA-seq
The next-generation sequencing data (79 paired PTC and non-tumor thyroid tissues) was from our unpublished study (Wang et al, unpublished data). Total RNA was extracted from 79 pairs of PTC and normal tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA concentration was measured using a DS-11 Spectrophotometer (DeNovix). After RNA quality was evaluated (Agilent 2100 Bioanalyzer; Agilent Technologies, Inc.; RNA integrity number >8.0), 1,000 ng of total RNA was used to construct cDNA libraries with the Ion Total RNA-Seq kit-v2 (Thermo Fisher Scientific, Inc.). Next-generation sequencing was conducted on Illumina Hiseq 2500 platform (125 bp paired end reads). Clean data (clean reads) were obtained by removing reads containing adapter, ploy-N and low quality reads from raw data. The clean data were then aligned to the human reference genome version (GRCh37) using the MapSplice program v2.1.6 (http://www.netlab.uky.edu/p/bioinfo/MapSplice/) (23).
Gene set enrichment analysis (GSEA)
GSEA v.4.0.3 software (http://www.broadinstitute.org/gsea) was used to analyze various potential biological gene sets that were associated with LEMD1 expression in TCGA dataset. The 502 PTC samples were divided into two groups based on the median LEMD1 expression (fragments per kilobase per million, 7.5610005855) in patients (high- and low-expression groups). The Kyoto Encyclopedia of Genes and Genomes subset of Canonical pathways (186 gene sets) was used for GSEA obtained from the Molecular Signature Database v7.1 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp).
Cell lines and cell culture
TPC-1 (RRID, CVCL_6298; human PTC cell line), BCPAP (RRID, CVCL_0153; human TC cell line), KTC-1 (RRID, CVCL_6300; human PTC cell line) and HTORI-3 (RRID, CVCL_4W02; normal human thyroid cell line) were used in the present study (24). KTC-1 and HTORI-3 cell lines were provided by The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. TPC-1 and BCPAP cell lines were obtained from Professor Mingzhao Xing (Johns Hopkins University School of Medicine, Baltimore, MA, USA). TPC-1, BCPAP and KTC-1 were cultured in RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin-streptomycin. HTORI-3 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS and 100 U/ml penicillin-streptomycin. The cells were cultured at a constant temperature of 37°C in a humidified incubator with 5% CO2.
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted from the 47 pairs of PTC and normal tissues or cells using TRIzol reagent in accordance with the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). The samples with A260/A280 of 1.8–2.0 were considered for further analysis. Subsequently, RT was performed using the ReverTra Ace qPCR RT kit (cat. no. FSQ-101; Toyobo Life Science) according to the manufacturer's protocol. qPCR was performed using Thunderbird SYBR qPCR Mix (cat. no. QPS-201; Toyobo Life Science) on Applied Biosystems 7500 (Thermo Fisher Scientific, Inc.). The thermocycling conditions of qPCR were as follows: 40 cycles of denaturation at 95°C for 15 sec, annealing at 60°C for 15 sec and extension at 72°C for 45 sec. The relative LEMD1 expression was normalized to GADPH. The 2−∆∆Cq method was used to calculate the quantification of mRNA expression (25). The primer sequences for PCR were as follows: LEMD1 forward, 5′-GCAAGAGCACCAAGCACCAG-3′ and reverse, 5′-TCAAGCCCACTGGGAAACCT-3′; GAPDH forward, 5′-GGTCGGAGTCAACGGATTTG-3′ and reverse, 5′-ATGAGCCCCAGCCTTCTCCAT-3′. All samples were run in triplicate.
Cell transfection
The negative control small interfering RNA (siRNA), the LEMD1-specific siRNA used for LEMD1-knockdown, the empty control vector [pEX-3 (PGCMV/MCS/Neo)] and the LEMD1 overexpression vector [pEX-3 (EcoRI/BamHI)-LEMD1] were purchased from Shanghai GenePharma Co., Ltd. The siRNA (20 µM/µl) was transfected into TC cell lines using Lipofectamine RNAiMAX (cat. no. 13778075; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocol for 8 h at 37°C. The vectors (1 µg/µl) were transfected into TC cell lines using Lipofectamine 3,000 (cat. no. L3000008; Thermo Fisher Scientific, Inc.) for 6 h at 37°C. The TC cells were seeded into 6-well plates and incubated at 37°C for 24 h before transfection (TPC-1, 4.0×104/well; BCPAP and KTC-1, 8.0×104/well). Subsequently, the transfected TC cells were incubated at 37°C for 48 h before being harvested for subsequent experimentation. The siRNA sequences were as follows: siRNA-LEMD1 sense, 5′-GCCCAAUACUACCUUCCACTT-3′ and antisense, 5′-GUGGAAGGUAGUAUUGGGCTT-3′; and the non-targeting negative control siRNA sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. The transfection efficiency was tested using RT-qPCR, as aforementioned. All experiments were performed in triplicate with three independent samples.
Cell Counting Kit-8 (CCK-8) proliferation assay
For the CCK-8 assay (cat. no. C0040; Beyotime Institute of Biotechnology), TPC-1 (1.25×103 cells/well), BCPAP-1 (1.25×103 cells/well) and KTC-1 (1.5×103 cells/well) cells transfected with siRNA or overexpression vector were plated into 96-well plates with the corresponding medium. After 24, 48, 72 and 96 h, the cellular viability of the transfected cells was tested by adding the CCK-8 solution (10 µl/well) and incubated for 3 h at 37°C. Subsequently, the absorbance was determined at a wavelength of 450 nm with SpectraMax M5 (Molecular Devices LLC).
Colony formation assay
The transfected cells were counted and seeded (1.25×103 cells/well for TPC-1 and BCPAP-1 cells, and 1.5×103 cells/well for KTC-1 cells) in 6-well plates. After the cells were incubated for 7–14 days at 37°C with 5% CO2, they were washed three times with PBS and fixed with 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) at room temperature for 30 min. Subsequently, all plates were washed with PBS and stained with 0.01% crystal violet staining solution at room temperature for 20 min.
Migration and invasion assays
The cell capacity of migration was detected using wound-healing and Transwell migration assays. Wound-healing assay was performed on 24-well plates. Cells (2×105) were plated into wells and cultured until 100% confluency was reached. A sterile 200-µl pipette tip was used to scratch wounds, and cells were incubated in serum-free RPMI 1640 medium at 37°C for 24 h. The results of the wound-healing assay were observed under a light microscope (DMi1; Leica Microsystems GmbH) using a fixed magnification of ×200. The migration rate was calculated as follows: (0 h wound area-24 h wound area)/0 h wound area × 100%. Transwell migration assay was conducted using 8-µm pore-size Transwell chambers (cat. no. 3422; Corning, Inc.) without Matrigel to evaluate the migratory ability. The transfected cells (3.5×104 cells/well for TPC-1, KTC-1 and BCPAP) were seeded in the upper chamber of each insert with 200 µl serum-free RPMI 1640 medium. Subsequently, the lower chambers were filled with 600 µl RPMI 1640 medium containing 10% FBS. After the cells were incubated for 24 h at 37°C, those that migrated were fixed with 4% paraformaldehyde and stained with 0.01% crystal violet staining solution both at room temperature for 15 min each.
Cell invasion was measured using 8-µm pore-size invasion chambers pre-coated with Matrigel® (cat. no. 354480; Corning, Inc.). The transfected tumor cell suspensions (4×104 cells/well for TPC1, BCPAP and KTC-1) in 200 µl serum-free RPMI-1640 medium were placed in the upper chamber, while the lower chamber was filled with 600 µl RPMI-1640 medium with 10% FBS. The same steps used for migration assays were used to fix and stain the cells on the bottom surface of the upper chamber. The results of the migration and invasion assays were observed under a light microscope (DMi1; Leica Microsystems GmbH) using a fixed magnification of ×200.
Apoptotic analysis
Apoptosis was measured using an Annexin-V-FITC/PI Apoptosis Detection kit (cat. no. 556547; BD Biosciences). The tumor cells were collected after transfection for 48 h. Subsequently, they were centrifuged at 1,000 × g for 3 min at 4°C, and the supernatant was removed. The cells were resuspended in PBS. These steps were repeated three times. Subsequently, 5 µl Annexin V-FITC and 5 µl PI were added to the solution to stain the cells for 15 min in the dark at room temperature in accordance with the manufacturer's protocol. The cells were analyzed using a BD FACScantoII flow cytometer (BD Biosciences) and FlowJo v10.0 software (FlowJo LLC). Apoptosis included early (Q3) and late apoptosis (Q2). Percentage of apoptosis=Q2 + Q3.
Protein extraction and western blot (WB) analysis
The cells were lysed in cell lysis buffer (cat. no. P0033; Beyotime Institute of Biotechnology) to extract the total cell protein. Lysate protein concentrations were measured using bicinchoninic acid assay. The protein samples (20 µg protein/lane) were separated via 10% SDS-PAGE and then were electro-transferred onto PVDF membranes. Subsequently, the membranes were blocked with 5% skimmed milk (cat. no. 232100; BD Biosciences) in TBS-Tween (TBST; 0.1% Tween-20) for 2 h at room temperature and then incubated with primary antibodies for 12 h at 4°C. Finally, after the membranes were rinsed three times in TBST, they were incubated with goat anti-mouse IgG/HRP (cat. no. SE131; 1:5,000; Beijing Solarbio Science & Technology Co., Ltd.) or goat anti-rabbit IgG/HRP secondary antibodies (cat. no. SE134; 1:5,000; Beijing Solarbio Science & Technology Co., Ltd.) for 2 h at room temperature. The target bands were detected using BeyoECL Plus (cat. no. P0018M; Beyotime Institute of Biotechnology) and analyzed using ImageJ v1.50b (National Institutes of Health). The primary antibodies were as follows: LEMD1 (1:500; cat. no. ab201206; Abcam), N-cadherin (1:1,000; cat. no. 22018-1-AP; ProteinTech Group, Inc.), E-cadherin (1:1,000; cat. no. 20874-1-AP; ProteinTech Group, Inc.), β-catenin (1:5,000; cat. no. 51067-2-AP; ProteinTech Group, Inc.), vimentin (1:8,000; cat. no. 60330-1-LG; ProteinTech Group, Inc.), Caspase 3/Cleaved caspase 3 (1:500; cat. no. 19677-1-AP; ProteinTech Group, Inc.) and β-actin (1:2,000; cat. no. 60008-1-Ig, ProteinTech Group, Inc.).
Statistical analysis
Statistical analyses were performed using SPSS software (v23.0; IBM Corp.). The χ2 test or Fisher's exact test were used to assess the association between clinicopathological characteristics and LEMD1 expression. The association between LEMD1 and LNM was analyzed using univariate and multivariate Cox regression analysis. Statistical significance was evaluated using Wilcoxon test, Mann-Whitney test, one-way ANOVA followed by Tukey's post-hoc test or Student's unpaired t-test. The diagnosis potential of LEMD1 was evaluated by receiver operating characteristic (ROC) curve using SPSS software. The optimal cut-off value of LEMD1 expression was determined by maximizing the Youden index value. Youden index=(sensitivity + specificity) −1. All experiments were repeated in triplicate at least three times. The data are presented as the mean ± SD. P<0.05 was considered to indicate a statistically significant difference.
Results
LEMD1 expression is upregulated in TC
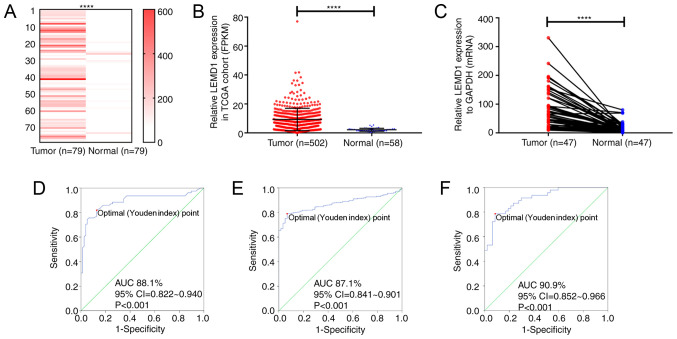
In the present study, RNA-seq was performed on 79 paired PTC and non-tumor thyroid tissues to detect the RNA expression, and the results revealed that LEMD1 expression in the PTC tissues was significantly upregulated compared with that in the paired non-cancer tissues (Fig. 1A; Table SI). The results from TCGA cohort were consistent with those from the RNA-seq dataset (Fig. 1B). Furthermore, LEMD1 expression in 47 paired PTC and non-tumor thyroid tissues was detected using RT-qPCR (Fig. 1C), revealing that LEMD1 expression was significantly upregulated in tumor compared with in normal tissues. The diagnostic potential of LEMD1 was evaluated using ROC curve analysis. In the RNA-seq cohort (sensitivity, 81.0; specificity, 87.3), TCGA dataset (sensitivity, 79.1; specificity, 93.1) and the local validated cohort (sensitivity, 78.7; specificity, 91.5), the ROC curve analyses indicated that LEMD1 expression may be a potential marker for TC from normal thyroid tissues (Fig. 1D-F). These results indicated that LEMD1 may be a potential gene for the diagnosis and treatment of TC.
Figure 1.
LEMD1 expression in TC tissues is upregulated compared with that in normal thyroid tissues. LEMD1 expression was significantly upregulated in the tumor tissues of (A) the RNA-seq cohort (Wilcoxon test), (B) TCGA dataset (Mann-Whitney test) and (C) the local validated cohort (Wilcoxon test). Receiver operating characteristic curve analysis of LEMD1 expression was used to identify TC tissues from normal thyroid tissues in (D) the RNA-seq cohort (sensitivity and specificity values were 81.0 and 87.3, respectively), (E) TCGA dataset (sensitivity and specificity values were 79.1 and 93.1, respectively) and (F) the local validated cohort (sensitivity and specificity values were 78.7 and 91.5, respectively). The optimal cut-off value of LEMD1 expression was determined by maximizing the Youden index value. Youden index=(sensitivity + specificity)-1. Data are shown as the mean ± SD of three independent experiments. ****P<0.0001. LEMD1, LEM domain containing 1; TC, thyroid cancer; TGCA, The Cancer Genome Atlas; FPKM, fragments per kilobase per million; AUC, area under curve.
LEMD1 expression is associated with clinicopathological characteristics in patients with TC
The association between LEMD1 expression and the clinicopathological features of patients with TC was analyzed in TCGA dataset and the local validated cohort. According to LEMD1 expression, the RNA-seq results of LEMD1 expression from TCGA dataset were divided into two groups, namely the high- (n=251) and low-expression (n=251) groups. The results revealed that LEMD1 expression was associated with histological type (P<0.001), tumor size (P=0.039), LNM (P<0.001) and disease stage (P=0.001) (Table I). However, no significant association was observed between LEMD1 expression and sex, age and distant metastasis. In the local validated cohort, LEMD1 expression was associated with LNM (P=0.006) and disease stage (P=0.002) (Table II). All analyses supported that LEMD1 may act as a cancer-promoting gene in TC.
Table I.
Association between LEMD1 expression and clinicopathological features in The Cancer Genome Atlas cohort (n=502).
| High LEMD1 | Low LEMD1 | |||
|---|---|---|---|---|
| Clinicopathological features | expression (n=251) | expression (n=251) | χ2 | P-value |
| Histological type | 30.288 | <0.001a | ||
| Classical | 206 | 150 | ||
| Other types | 45 | 101 | ||
| Age, years | <0.001 | >0.999a | ||
| Mean ± SD | 47.55±16.03 | 47.13±15.66 | ||
| ≤45 | 113 | 113 | ||
| >45 | 138 | 138 | ||
| Sex | 0.010 | 0.920a | ||
| Male | 67 | 68 | ||
| Female | 184 | 183 | ||
| Tumor size, mm | 4.277 | 0.039a | ||
| ≥20 | 189 | 168 | ||
| <20 | 62 | 83 | ||
| Lymph node metastasis | 35.179 | <0.001a | ||
| Yes | 144 | 78 | ||
| No | 107 | 173 | ||
| Distant metastasis | NA | 0.504b | ||
| Yes | 3 | 6 | ||
| No | 248 | 245 | ||
| AJCC disease stage | 10.992 | 0.001a | ||
| I+II | 150 | 185 | ||
| III+IV | 101 | 66 |
χ2 test.
Fisher's exact test. LEMD1, LEM domain containing 1; AJCC, American Joint Committee on Cancer; NA, not applicable.
Table II.
Association between LEMD1 expression and clinicopathological features in the local validated cohort (n=47).
| High LEMD1 | Low LEMD1 | |||
|---|---|---|---|---|
| Clinicopathological features | expression (n=23) | expression (n=24) | χ2 | P-value |
| Histological type | NA | NA | ||
| Classical | 23 | 24 | ||
| Age, years | 0.180 | 0.671a | ||
| Mean ± SD | 47.56±14.43 | 48.23±14.86 | ||
| ≤45 | 15 | 14 | ||
| >45 | 11 | 10 | ||
| Sex | 0.295 | 0.587a | ||
| Male | 6 | 8 | ||
| Female | 17 | 16 | ||
| Tumor size, mm | 0.236 | 0.627a | ||
| ≥20 | 15 | 14 | ||
| <20 | 8 | 10 | ||
| Lymph node metastasis | 7.671 | 0.006a | ||
| Yes | 16 | 7 | ||
| No | 7 | 17 | ||
| Distant metastasis | NA | NA | ||
| No | 23 | 24 | ||
| AJCC disease stage | NA | 0.002b | ||
| I+II | 15 | 24 | ||
| III+IV | 8 | 0 |
χ2 test.
Fisher's exact test. LEMD1, LEM domain containing 1; AJCC, American Joint Committee on Cancer; NA, not applicable.
High LEMD1 expression aggravates the risk of LNM in TC
The association between LNM and LEMD1 expression was evaluated using logistic regression analysis. The results of the univariate logistic regression analysis are shown in Table III. The significant variables associated with LNM were LEMD1 expression odds ratio (OR), 2.985; 95% CI, 2.070–4.305, P<0.001), histological type (OR, 2.649; 95% CI, 1.747–4.017; P<0.001), age (OR, 0.653; 95% CI, 0.458–0.931; P=0.019), sex (OR, 0.63; 95% CI, 0.424–0.937; P=0.023) and tumor size (OR, 2.283; 95% CI, 1.514–3.441; P<0.001) (Table III). Meanwhile, multivariate logistic regression analysis was performed using the parameters with significant variables in univariate logistic regression. The results indicated that LEMD1 expression (OR, 2.581; 95% CI, 1.752–3.804; P<0.001), histological type (OR, 2.128; 95% CI, 1.365–3.318; P=0.001), age (OR, 0.647; 95% CI, 0.441–0.950; P=0.026), sex (OR, 0.627; 95% CI, 0.410–0.960; P=0.032) and tumor size (OR, 2.206; 95% CI, 1.429–3.406; P<0.001) were independent high-risk factors of LNM (Table IV).
Table III.
Univariate logistic regression analysis for the risk of lymph node metastasis.
| Factor | Odds ratio | 95% CI | P-value |
|---|---|---|---|
| LEMD1 expression (high vs. low) | 2.985 | 2.070–4.305 | <0.001a |
| Histological type (PTC vs. other TC subtypes) | 2.649 | 1.747–4.017 | <0.001a |
| Age (≤45 vs. >45 years) | 0.653 | 0.458–0.931 | 0.019 a |
| Sex (male vs. female) | 0.630 | 0.424–0.937 | 0.023a |
| AJCC disease stage (III+IV vs. I+II) | 1.009 | 2.468–3.803 | 0.989 |
| Tumor size (≥20 vs. <20 mm) | 2.283 | 1.514–3.441 | <0.001a |
P<0.05. LEMD1, LEM domain containing 1; AJCC, American Joint Committee on Cancer.
Table IV.
Multivariate logistic regression analysis for the risk of lymph node metastasis.
| Factor | Odds ratio | 95% CI | P-value |
|---|---|---|---|
| LEMD1 expression (high vs. low) | 2.581 | 1.752–3.804 | <0.001a |
| Histological type (PTC vs. other TC subtypes) | 2.128 | 1.365–3.318 | 0.001a |
| Age (≤45 vs. >45 years) | 0.647 | 0.441–0.950 | 0.026a |
| Sex (male vs. female) | 0.627 | 0.410–0.960 | 0.032a |
| Tumor size (≥20 vs. <20 mm) | 2.206 | 1.429–3.406 | <0.001a |
P<0.05. LEMD1, LEM domain containing 1; AJCC, American Joint Committee on Cancer.
LEMD1 promotes the proliferation of TC cell lines
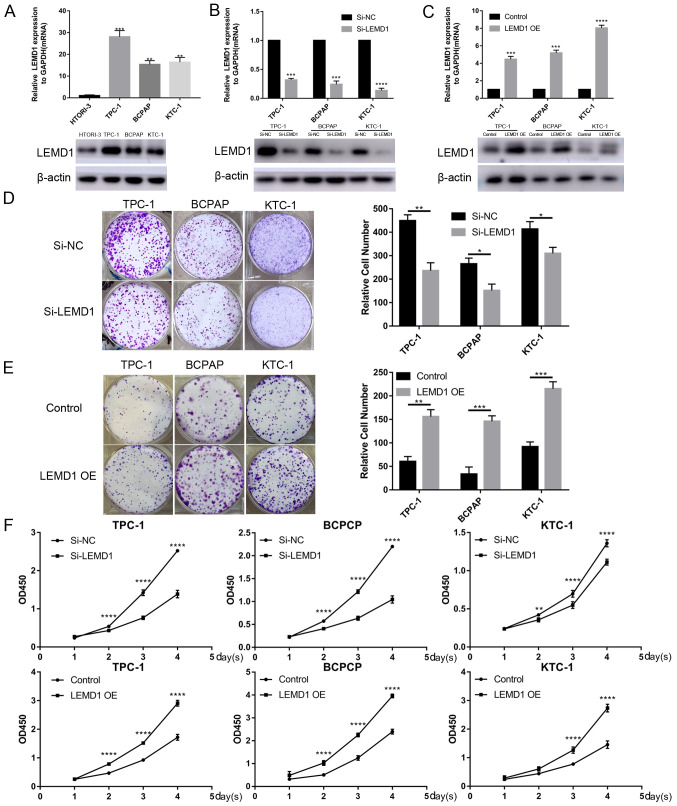
Given that LEMD1 expression was generally upregulated in TC, LEMD1 may serve an important role in TC tumorigenesis and progression. Thus, LEMD1 expression in TC cell lines and a normal human thyroid cell line (HTORI-3) was assessed at the mRNA and protein levels. LEMD1 expression was significantly upregulated in TPC-1, BCPAP and KTC-1 cells compared with in HTORI-3 cells (Fig. 2A). Subsequently, si-LEMD1 or LEMD1 overexpression vector were chosen to knock down or overexpress, respectively, LEMD1 expression in the TC cell lines, and the transfection efficiency was evaluated via RT-qPCR and WB (Fig. 2B and C). Subsequently, CCK-8 and colony formation assays were performed. The results revealed that LEMD1-knockdown significantly inhibited cell proliferation and colony formation, while LEMD1 overexpression promoted the proliferation and colony formation of TPC-1, BCPAP and KTC-1 cell lines (Fig. 2D-F).
Figure 2.
TC cell transfection with LEMD1 siRNA or overexpression vector reveals that LEMD1 promotes TC cell proliferation. (A) Relative mRNA and protein LEMD1 expression in HTORI-3 (normal human thyroid cell line) and three TC cell lines. (B) LEMD1 expression was detected via RT-qPCR and WB in TPC-1, BCPAP and KTC-1 cell lines transfected with siRNA. (C) Transfection efficiency of LEMD1 overexpression vector in TC cell lines detected via RT-qPCR and WB. Results of colony formation assays for TC cells transfected with (D) Si-LEMD1 or (E) LEMD1 OE vector. (F) Cell proliferation was measured using the Cell Counting Kit-8 assay in TC cells transfected with Si-LEMD1 or LEMD1 OE vector. The data are shown as the mean ± SD of three independent experiments. Statistical significance was evaluated using one-way ANOVA followed by Tukey's post-hoc test for comparisons among multiple groups or Student's unpaired t-test for comparisons between two groups. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 vs. HTORI-3, Si-NC or control. LEMD1, LEM domain containing 1; TC, thyroid cancer; siRNA, small interfering RNA; RT-qPCR, reverse transcription quantitative PCR; OE, overexpression; WB, western blot; NC, negative control; OD, optical density.
LEMD1 enhances the migratory and invasive capacities of TC cells in vitro
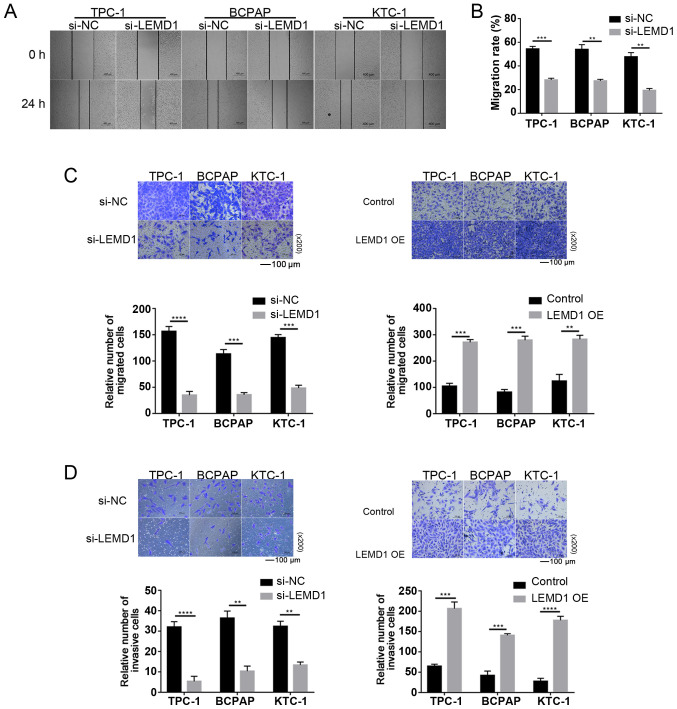
Considering the significant association between LNM and LEMD1 expression, the function of LEMD1 in the migratory and invasive capacities of TC cell lines was assessed. As shown in Fig. 3A-C, LEMD1-knockdown significantly decreased the migratory ability of TC cells compared with that of the control cells. The invasive ability was similarly inhibited by LEMD1-knockdown in TC cell lines (Fig. 3D). With the overexpression of LEMD1, the experiments exhibited the opposite results (Fig. 3C and D), with significantly increased migratory and invasive capacities. The current results indicated that LEMD1 may serve an important role in the migratory and invasive capacities of TC cell lines.
Figure 3.
LEMD1 enhances cell migration and invasion in TC cell lines. (A) Wound healing assays were used to detect cell migration in Si-LEMD1-transfected TC cells (scale bar, 400 µm). (B) Relative quantification of migration rate in wound healing assays. Migration rate=(0 h wound area-24 h wound area)/0 h wound area × 100%. Transwell (C) migration and (D) invasion assays were used to detect cell migration and invasion in TC cells transfected with Si-LEMD1 or LEMD1 OE vector (scale bar, 100 µm; magnification, ×200). The data are shown as the mean ± SD of three independent experiments. Student's unpaired t-test was used for statistical analyses. **P<0.01, ***P<0.001 and ****P<0.0001 vs. Si-NC or control. LEMD1, LEM domain containing 1; TC, thyroid cancer; siRNA, small interfering RNA; OE, overexpression; NC, negative control.
LEMD1-knockdown induces apoptosis of TC cells in vitro
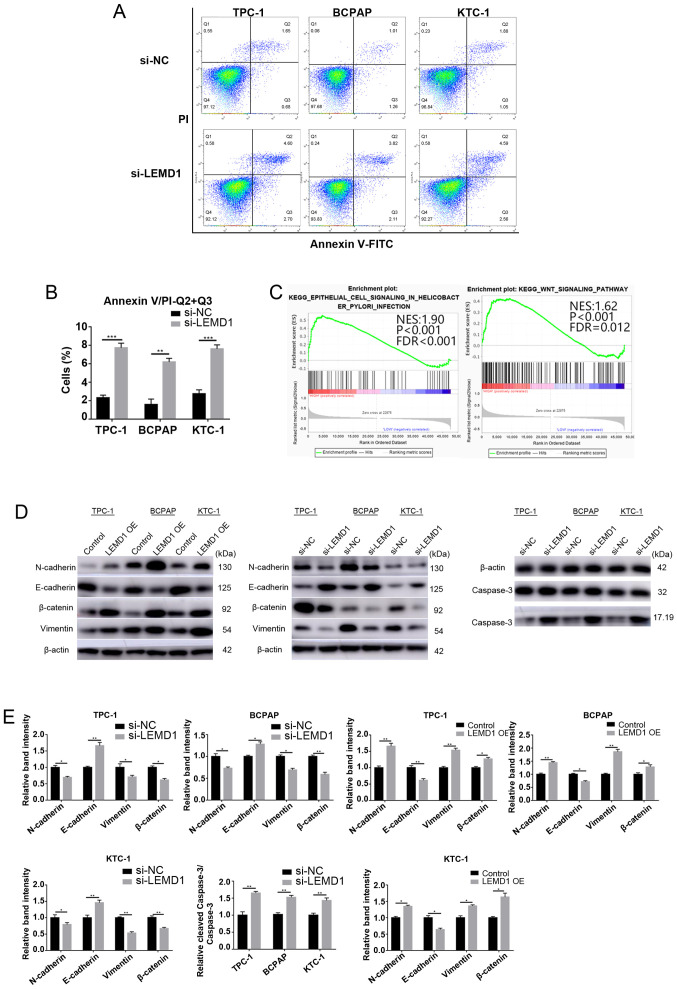
The effect of LEMD1 on the apoptosis of TC cell lines was analyzed via flow cytometry and WB analysis (caspase 3 and cleaved-caspase 3) to further explore the function of this gene. LEMD1-knockdown led to a significant increase in the apoptosis of TPC-1, BCPAP and KTC-1 cells (Fig. 4A, D and E). The results indicated that the apoptosis suppressed by LEMD1 may be responsible for TC progression.
Figure 4.
LEMD1-knockdown induces apoptosis of thyroid cancer cells in vitro, and LEMD1 regulates EMT and the Wnt/β-catenin signaling pathway. (A) Flow cytometry analysis of the apoptotic rates of the TPC-1, BCPAP and KTC-1 cells transfected with Si-NC or Si-LEMD1. (B) Quantitative results of the apoptotic cell percentages. Apoptosis included early (Q3) and late apoptosis (Q2). Percentage of apoptosis=Q2 + Q3. (C) Gene set enrichment analysis plots revealed that high LEMD1 expression was positively associated with EMT and the Wnt/β-catenin signaling pathway. (D) Protein expression levels of vimentin, N-cadherin, E-cadherin, β-catenin, caspase 3 and cleaved-caspase 3 were evaluated using western blotting, with β-actin serving as a loading control. (E) Relative quantitative analysis of protein expression. All experiments were repeated in triplicate at least three times. The data are presented as the mean ± SD. Student's unpaired t-test was used for statistical analyses. *P<0.05, **P<0.01 and ***P<0.001 vs. Si-NC or control. LEMD1, LEM domain containing 1; OE, overexpression; NC, negative control; EMT, epithelial-mesenchymal transition; NES, normalized enrichment score; FDR, false discovery rate.
LEMD1 promotes TC progression by regulating EMT in vitro
EMT is an indispensable process for cancer cell metastasis (26). As shown in Tables I and II, LEMD1 expression was significantly associated with LNM. GSEA was performed on the basis of TCGA dataset to explore the potential mechanism by which LEMD1 promoted TC metastasis. The results revealed that the upregulation of LEMD1 expression was closely associated with EMT and the Wnt/β-catenin signaling pathway (Fig. 4C). The essential specific markers of EMT (E-cadherin, N-cadherin and vimentin) were detected in TC cells with LEMD1-knockdown or overexpression compared with the respective control cells using WB. The cells with LEMD1-knockdown expressed significantly lower expression levels of N-cadherin and vimentin, and significantly higher expression levels of E-cadherin compared with the respective negative control cells (Fig. 4D and E). With the overexpression of LEMD1, the WB results exhibited opposite results (Fig. 4D and E), with significantly higher expression levels of N-cadherin and vimentin, and significantly lower expression levels of E-cadherin. These findings demonstrated that LEMD1 regulated EMT in TC cell lines.
Effects of LEMD1 on activation of the Wnt/β-catenin signaling pathway
The Wnt/β-catenin signaling pathway is one of the classical pathways that drive EMT in malignant tumors (27). The GSEA results indicated that LEMD1 expression was associated with the Wnt/β-catenin signaling pathway. Several pathway-specific markers that regulate EMT were assessed using WB to further study the molecular mechanisms of LEMD1 in TC. The results revealed that LEMD1-knockdown significantly decreased β-catenin expression (Fig. 4D and E). On the other hand, the overexpression of LEMD1 significantly upregulated β-catenin expression (Fig. 4D and E). Overall, the results demonstrated that LEMD1 was able to activate the Wnt/β-catenin signaling pathway and EMT in TC cell lines in vitro.
Discussion
The incidence of TC has increased rapidly in recent years. A previous study has predicted that TC would replace colorectal cancer as the fourth leading type of cancer by 2030 (28). Although almost all patients with PTC have a favorable prognosis, LNM remains a pressing issue for some patients (29). Thus, understanding the molecular pathogenesis underlying PTC LNM and distant metastasis is essential for the development of improved diagnostic and therapeutic strategies to treat patients with PTC. Increasing studies have revealed that LEMD1 serves a vital role in the tumorigenesis and progression of several types of cancer (20–22), but the function of LEMD1 in PTC is poorly understood. The evolution of RNA-seq technologies provided an effective method to further elucidate the underlying PTC molecular mechanisms. RNA-seq was previously conducted on 79 paired PTC and non-tumor thyroid tissues, and LEMD1 expression in PTC tissues was significantly upregulated compared with that in normal tissues. This phenomenon was also observed in TCGA dataset. RT-qPCR and WB analysis revealed that LEMD1 was highly expressed in TC cell lines and tissues. All these data indicated that LEMD1 may play a critical role in TC tumorigenesis and progression.
EMT can result in the loss of cell adhesion and promote the invasion and metastasis of cancer (30). Upregulation of mesenchymal markers (N-cadherin and vimentin) and downregulation of epithelial markers (E-cadherin) have been reported in different types of tumor during EMT (27,31). According to a study by Sasahira et al (32), LEMD1 downregulation may promote the invasive and metastatic ability of oral squamous cell carcinoma. The GSEA results of the present study revealed that high LEMD1 expression was positively associated with the EMT signaling pathway in PTC tissues (TCGA cohort). However, to the best of our knowledge, the association between LEMD1 and EMT in TC has not been studied.
LEMD1 acts as a cancer-promoting gene in several types of tumor, such as gastric cancer, prostate cancer, oral squamous cell carcinoma, anaplastic large-cell lymphoma and colorectal cancer (33,34). Given that human leucocyte antigen is not expressed in testis, LEMD1 (cancer/testis antigen) is an ideal target for TC treatment (35). However, although several studies have reported that LEMD1 expression is significantly associated with tumor cell invasive ability and endothelial transmigration in multiple types of tumor, the metastatic mechanisms of LEMD1 in TC have not been investigated (20,22,32–34). In the present study, siRNA was used to knock down LEMD1 expression in TC cell lines. Gain of function assays were also performed using overexpression vectors. The results revealed that LEMD1 promoted TC cell proliferation in vitro. In addition, the migratory and invasive capacities of TC cell lines were increased following LEMD1 overexpression in vitro. LEMD1-knockdown led to a significant increase in the protein expression levels of E-cadherin and cleaved-caspase 3, and a significant decrease in N-cadherin, vimentin and β-catenin protein expression, while LEMD1 overexpression resulted in the opposite results. All these results indicated that LEMD1 promoted TC cell migration and invasion by regulating EMT and the Wnt/β-catenin signaling pathway.
However, there are some limitations in the present study. The Wnt signaling pathway is complex and complicated. According to previous studies, the protein expression levels of WNT1 and WNT3A, the traditionally potent activators of Wnt/β-catenin signaling, did not change after transfection with siRNA or overexpression vector (36,37). The proteins that activate β-catenin-dependent signaling require further research. Furthermore, since fresh TC tissues from patients were hard to obtain, immunohistochemical staining of the tissue sections could not be performed. Additionally, a co-immunoprecipitation experiment should be performed to evaluate the association between LEMD1 and β-catenin. Furthermore, animal experiments should be performed to verify the biological function of LEMD1 in vivo. The molecular mechanisms of LEMD1 affecting the metastasis of TC require further research in future studies.
In conclusion, the present study revealed that LEMD1 expression was positively associated with LNM in TCGA cohort and the local validated cohort. LEMD1-knockdown inhibited TC cell proliferation and migration, and induced apoptosis by suppressing EMT and the Wnt/β-catenin signaling pathway. On the other hand, with the overexpression of LEMD1, opposite results were obtained. Therefore, the current results suggested that LEMD1 may be a potential biomarker for the diagnosis and treatment of TC.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Wenchao Cai (Emergency Department, The First Affiliated Hospital of Wenzhou Medical University) for the instrument support.
Funding Statement
The present study was financially supported by grants from the Wenzhou Science and Technology Bureau (grant no. Y20180460) and the Natural Science Foundation of Zhejiang Province (grant no. LY17H160053).
Funding
The present study was financially supported by grants from the Wenzhou Science and Technology Bureau (grant no. Y20180460) and the Natural Science Foundation of Zhejiang Province (grant no. LY17H160053).
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due to restrictions on data sharing imposed by the funding body, but are available from the corresponding author on reasonable request.
Authors' contributions
MX, BL and DZ wrote the manuscript and performed the main experiments. JW and XWZ collected and analyzed the raw data. WH and CL performed the experiments and revised the article. XHZ and JQ designed the study. XHZ and JQ confirm the authenticity of the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China; approval no. 2012-57). Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lloyd RV, Osamura RY, Klöppel G, Rosai J, editors. WHO Classification of Tumours of Endocrine Organs. 4th Edition. Vol. 10. IARC Press; Lyon: 2017. [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Qian B, He M, Dong S, Wang J, Chen K. Incidence and mortality of thyroid cancers in Tianjin from 1981 to 2001. Chin J Endocrinol Metab. 2005;21:432–434. [Google Scholar]
- 4.Sosa JA, Udelsman R. Papillary thyroid cancer. Surg Oncol Clin N Am. 2006;15:585–601. doi: 10.1016/j.soc.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Hinson AM, Massoll NA, Jolly LA, Stack BC, Jr, Bodenner DL, Franco AT. Structural alterations in tumor-draining lymph nodes before papillary thyroid carcinoma metastasis. Head Neck. 2017;39:1639–1646. doi: 10.1002/hed.24807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albores-Saavedra J, Henson DE, Glazer E, Schwartz AM. Changing patterns in the incidence and survival of thyroid cancer with follicular phenotype-papillary, follicular, and anaplastic: A morphological and epidemiological study. Endocr Pathol. 2007;18:1–7. doi: 10.1007/s12022-007-0002-z. [DOI] [PubMed] [Google Scholar]
- 7.LiVolsi VA. Papillary thyroid carcinoma: An update. Mod Pathol. 2011;24(Suppl 2):S1–S9. doi: 10.1038/modpathol.2010.129. [DOI] [PubMed] [Google Scholar]
- 8.Hong IK, Eun YG, Chung DH, Kwon KH, Kim DY. Association of the oncostatin m receptor gene polymorphisms with papillary thyroid cancer in the korean population. Clin Exp Otorhinolaryngol. 2011;4:193–198. doi: 10.3342/ceo.2011.4.4.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho AS, Luu M, Barrios L, Chen I, Melany M, Ali N, Patio C, Chen Y, Bose S, Fan X, et al. Incidence and mortality risk spectrum across aggressive variants of papillary thyroid carcinoma. JAMA Oncol. 2020;6:706–713. doi: 10.1001/jamaoncol.2019.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choden S, Keelawat S, Jung CK, Bychkov A. VE1 immunohistochemistry improves the limit of genotyping for detecting BRAFV600E mutation in papillary thyroid cancer. Cancers (Basel) 2020;12:596. doi: 10.3390/cancers12030596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 12.You JS, Jones PA. Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Hou P, Ji M, Guan H, Studeman K, Jensen K, Vasko V, El-Naggar AK, Xing M. Highly prevalent genetic alterations in receptor tyrosine kinases and phosphatidylinositol 3-kinase/akt and mitogen-activated protein kinase pathways in anaplastic and follicular thyroid cancers. J Clin Endocrinol Metab. 2008;93:3106–3116. doi: 10.1210/jc.2008-0273. [DOI] [PubMed] [Google Scholar]
- 15.Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer. 2005;12:245–262. doi: 10.1677/erc.1.0978. [DOI] [PubMed] [Google Scholar]
- 16.Xie H, Wei B, Shen H, Gao Y, Wang L, Liu H. BRAF mutation in papillary thyroid carcinoma (PTC) and its association with clinicopathological features and systemic inflammation response index (SIRI) Am J Transl Res. 2018;10:2726–2736. [PMC free article] [PubMed] [Google Scholar]
- 17.Salvatore G, Giannini R, Faviana P, Caleo A, Migliaccio I, Fagin JA, Nikiforov YE, Troncone G, Palombini L, Basolo F, Santoro M. Analysis of BRAF point mutation and RET/PTC rearrangement refines the fine-needle aspiration diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab. 2004;89:5175–5180. doi: 10.1210/jc.2003-032221. [DOI] [PubMed] [Google Scholar]
- 18.Lian EY, Maritan SM, Cockburn JG, Kasaian K, Crupi MJ, Hurlbut D, Jones SJ, Wiseman SM, Mulligan LM. Differential roles of RET isoforms in medullary and papillary thyroid carcinomas. Endocr Relat Cancer. 2017;24:53–69. doi: 10.1530/ERC-16-0393. [DOI] [PubMed] [Google Scholar]
- 19.Chang Y, Wang X, Xu Y, Yang L, Qian Q, Ju S, Chen Y, Chen S, Qin N, Ma Z, et al. Comprehensive characterization of cancer-testis genes in testicular germ cell tumor. Cancer Med. 2019;8:3511–3519. doi: 10.1002/cam4.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q, Ge Y, Chen X, Wang L, Xia Y, Xu Z, Li Z, Wang W, Yang L, Zhang D, Xu Z. LEM domain containing 1 promotes proliferation via activating the PI3K/Akt signaling pathway in gastric cancer. J Cell Biochem. 2019;120:15190–15201. doi: 10.1002/jcb.28783. [DOI] [PubMed] [Google Scholar]
- 21.Matsuyama H, Suzuki HI, Nishimori H, Noguchi M, Yao T, Komatsu N, Mano H, Sugimoto K, Miyazono K. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood. 2011;118:6881–6892. doi: 10.1182/blood-2011-05-354654. [DOI] [PubMed] [Google Scholar]
- 22.Takeda R, Hirohashi Y, Shen M, Wang L, Ogawa T, Murai A, Yamamoto E, Kubo T, Nakatsugawa M, Kanaseki T, et al. Identification and functional analysis of variants of a cancer/testis antigen LEMD1 in colorectal cancer stem-like cells. Biochem Biophys Res Commun. 2017;485:651–657. doi: 10.1016/j.bbrc.2017.02.081. [DOI] [PubMed] [Google Scholar]
- 23.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Staveren WC, Solis DW, Delys L, Duprez L, Andry G, Franc B, Thomas G, Libert F, Dumont JE, Detours V, Maenhaut C. Human thyroid tumor cell lines derived from different tumor types present a common dedifferentiated phenotype. Cancer Res. 2007;67:8113–8120. doi: 10.1158/0008-5472.CAN-06-4026. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Georgakopoulos-Soares I, Chartoumpekis DV, Kyriazopoulou V, Zaravinos A. EMT factors and metabolic pathways in cancer. Front Oncol. 2020;10:499. doi: 10.3389/fonc.2020.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 29.Lundgren CI, Hall P, Dickman PW, Zedenius J. Clinically significant prognostic factors for differentiated thyroid carcinoma: A population-based, nested case-control study. Cancer. 2006;106:524–531. doi: 10.1002/cncr.21653. [DOI] [PubMed] [Google Scholar]
- 30.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang Y, Massague J. Epithelial-mesenchymal transitions: Twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Sasahira T, Kurihara M, Nakashima C, Kirita T, Kuniyasu H. LEM domain containing 1 promotes oral squamous cell carcinoma invasion and endothelial transmigration. Br J Cancer. 2016;115:52–58. doi: 10.1038/bjc.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuki D, Lin YM, Fujii Y, Nakamura Y, Furukawa Y. Isolation of LEM domain-containing 1, a novel testis-specific gene expressed in colorectal cancers. Oncol Rep. 2004;12:275–280. [PubMed] [Google Scholar]
- 34.Ghafouri-Fard S, Ousati Ashtiani Z, Sabah Golian B, Hasheminasab SM, Modarressi MH. Expression of two testis-specific genes, SPATA19 and LEMD1, in prostate cancer. Arch Med Res. 2010;41:195–200. doi: 10.1016/j.arcmed.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Hirohashi Y, Torigoe T, Tsukahara T, Kanaseki T, Kochin V, Sato N. Immune responses to human cancer stem-like cells/cancer-initiating cells. Cancer Sci. 2016;107:12–17. doi: 10.1111/cas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Najdi R, Proffitt K, Sprowl S, Kaur S, Yu J, Covey TM, Virshup DM, Waterman ML. A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation. 2012;84:203–213. doi: 10.1016/j.diff.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to restrictions on data sharing imposed by the funding body, but are available from the corresponding author on reasonable request.