Abstract
The long non-coding RNA (lncRNA) HOTAIR is an oncogene, that has been reported to be aberrantly expressed in multiple types of malignant tumor tissues. However, its expression and association with synovial sarcoma (SS) remains unclear. The present study aimed to elucidate the expression level of HOTAIR in SS tissues and also identify its role. Reverse transcription-quantitative PCR was used to detect the expression level of HOTAIR and microRNA (miR)-126 in 54 tissue samples from patients with SS, in 10 tissue samples from synovium tissues of normal patients, and in SW982 cells. The protein expression level was measured using western blot analysis and cellular immunofluorescence. Cellular proliferation, invasion and migration were assessed using MTT, Transwell and wound healing assays, respectively. HOTAIR was expressed at high levels in SS tissues. In contrast, miR-126 was expressed at low levels in SS tissues, and was negatively correlated with HOTAIR expression. HOTAIR knockdown in SW982 cells inhibited cellular proliferation in vitro, but also significantly increased the ratio of cells in the G1/G0 phase of the cell cycle, and decreased the ratio of cells in the G2/S phase. In addition, HOTAIR knockdown inhibited the invasion and migration of the SW982 cells, as observed in the Transwell and wound healing assays. Furthermore, HOTAIR knockdown increased miR-126 expression level and decreased the expression level of stromal cell-derived factor-1 (SDF-1) at the protein level. On the other hand, while miR-126-mimic decreased the protein expression level of SDF-1, miR-126-inhibitor increased its expression level in SW982 cells. Notably, HOTAIR knockdown or SDF-1 knockout significantly decreased the protein expression levels of CDK1, CDK2, cyclin D1, MMP-9, vimentin and N-cadherin, and significantly increased the protein expression levels of p21, p53 and E-cadherin in SW982 cells. HOTAIR was highly expressed in SS tissues, wherein it could promote the proliferation, invasion and migration of SS cells by increasing the expression of SDF-1 via miR-126 inhibition.
Keywords: synovial sarcomas, HOTAIR, microRNA126, stromal cell-derived factor-1, metastasis
Introduction
Synovial sarcoma (SS) is a malignant mesenchymal tumor that is characterized by partial epithelial differentiation, and it constitutes 8–10% of soft tissue sarcoma (1,2). Despite its low incidence, SS is not easy to diagnose, progresses rapidly, and has a low 5-year survival rate that ranges between 21–40% (1,2). While sensitive to a variety of chemotherapeutic drugs, distant metastasis is another malignant manifestation of SS. Patients with SS often exhibit distant metastasis at early stages, and the majority of metastatic sites are concentrated in the lymph nodes, lungs and the liver (3,4). In fact, the main cause of death in patients with SS is tumor recurrence and metastasis, wherein ~40% of patients with SS develop lung, liver and regional lymph node metastasis within 2 years (5,6). Therefore, elucidating the mechanisms underlying SS recurrence and metastasis would greatly improve the survival rate of patients.
Long non-coding RNAs (lncRNAs) are a type of RNA, that are >200 bp in length, which generally have no open reading frame and do not encode a protein. In addition, they are usually transcribed by RNA polymerase II and are composed of multiple spliced exons (7,8). Recently, various lncRNAs have been associated with the development of different types of cancer (7,9), and multiple studies have shown that lncRNA HOTAIR can promote the progression of neuroglioma (10,11). In this regard, the lncRNA HOTAIR has been shown to act as an oncogene, which is aberrantly expressed in multiple malignant tumor tissues, including colorectal cancer (12), pancreatic cancer (13), hepatocellular carcinoma (14), rhinitis (15) and cervical cancer (16). In addition, HOTAIR has been demonstrated to promote cancer metastasis (17). It has been shown that HOTAIR can regulate the biological characteristics of gastric cancer cells and glioma cells by binding to and inhibiting microRNA (miR)-126 expression (11,18). On the other hand, stromal cell-derived factor-1 (SDF-1), a target of miR-126 (19), has been shown to be highly expressed in SS tissues and to be associated with poor prognosis in patients with SS (20).
However, the expression of HOTAIR in SS tissues and its association with miR-126 remains unclear. In this regard, the hypothesis used in the present study was that the association between HOTAIR expression and the prognosis of patients with SS is associated with the regulation of SDF-1 via miR-126 inhibition.
Materials and methods
Synovial sarcoma tissues and cell lines
As a prospective study, a total of 54 synovial sarcoma tissues from the limbs were collected from patients at the Fourth Hospital of Hebei Medical University (Shijiazhuang, China) between January 2011 and December 2017. The inclusion criteria were as follows: i) Synovial sarcoma identified by pathological diagnosis; ii) complete clinical and histological features, including sex, age, tumor size, histological grade, distant metastasis and the 8th Edition of American Joint Committee on Cancer (AJCC) (21) staging; iii) no radiotherapy, chemotherapy, immunotherapy or molecular targeting treatment prior to surgery; iv) regular follow-up or review following surgery and a clear cause or time of death; v) signed informed consent. The exclusion criteria were: i) Combined with other malignant tumors; ii) postoperative death due to non-tumor related causes; iii) combined with other severe diseases, such as chronic infection, organ disorders and cardiovascular and cerebrovascular diseases; iv) poor physical fitness, unable to tolerate surgery or other related examinations, poor postoperative mental state, and the prognosis affected by the basic conditions; v) postoperative follow-up is lost or the cause of death is unclear. In addition, 10 normal synovium tissues from the limbs were collected at the same hospital, over the same time period, from patients who underwent limb amputation with no history of joint disease. The tissues were snap-frozen in liquid nitrogen until required. Informed consent was obtained from all patients participating in the study or from their family members. The study was approved and monitored by the Ethics Committee of The Fourth Hospital of Hebei Medical University.
Wild-type (WT) SW982 (HTB-93; American Type Culture Collection) cell lines were cultured at 37°C and 5% CO2 in DMEM (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.). SDF-1 knockout (KO) SW982 cell lines were obtained from Synbio Technologies LLC., and were cultured under the same conditions as the WT-SW982. WT cells were used as the untreated control group.
Cell proliferation assay
A MTT Cell Proliferation and Cytotoxicity Assay kit (Beyotime Institute of Biotechnology) was used to assess cell proliferation according to the manufacturer's instructions. A total of 2×103 SW982 cells/well were initially seeded in a 96-well plate for the MTT assay. Following 24-h culture, the medium was removed, the cells were washed twice with PBS, and MTT reagent was added to incubate for ≥2 h at 37°C. The MTT solvent included in the assay kit was used to dissolve the purple formazan, and the absorbance was measured at 570 nm.
Cell transfection
Small interfering (si)-negative control (NC), si-HOTAIR-1, si-HOTAIR-2, miR-126-NC, miR-126-mimic and miR-126-inhibitor were designed and synthesized by Shenggong Bioengineering Co., Ltd. For siRNA or miRNA mimic/inhibitor transfection, 50 nmol/l siRNA, miRNA mimic or inhibitor was transfected into 2.5×106 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 6 h at 37°C according to the manufacturer's protocol. At 48 h post-transfection, the follow-up experiments were conducted. The sequences were as follows: si-NC: 5′-GAAACAUUAUGAAAAUCAAUU-3′; si-HOTAIR-1: 5′-AGACUAAGACGGAUAACGCGU-3′; si-HOTAIR-2: 5′-ACAAUAUCUACUUUGGAUCAC-3′; mimic-NC: 5′-AUCUCAACUUCGCGGCA-3′; miR-126-mimic: 5′-UCGUACCGUGAGUAAU-3′; inhibitor-NC: 5′-AUUGCUGGCUAAUAACG-3′; miR-126-inhibitor: 5′-AGCAUGGCACUCAUUA-3′.
Dual-luciferase reporter assay
First, WT-SDF-1 and mutant (MUT)-SDF-1 were integrated into pmirGLO plasmids (Promega Corporation) and co-transfected into cells (1×106 cells/well) with miR-126-NC, miR-126-mimic and miR-126-inhibitor (50 nmol/l) using Lipofectamine® 2000 for 6 h at 37°C. At 48 h post-transfection, the Dual-Lucy Assay kit (Beijing Solarbio Science & Technology Co., Ltd.) was used to detect the activity of luciferase according to the manufacturer's instructions. Briefly, cells were collected at 48 h post-transfection and lysed for 5 min on ice before centrifugation (12,000 × g for 1 min at room temperature) to collect the cell supernatant. Subsequently, 5X firefly or Renilla luciferase reaction solution were added to the cell lysate, and the enzyme activity was detected using a multifunctional microplate reader (Thermo Fisher Scientific, Inc.), and the enzyme activity was standardized to that of Renilla luciferase.
Reverse transcription-quantitative PCR (RT-qPCR)
The mRNA expression levels of HOTAIR and miR-126 were detected using the RT-qPCR Fluorescence quantitative-polymerase chain reaction kit (Hangzhou Bori Technology Co., Ltd.), as previously described (22). Total RNA was extracted using RNAiso Plus (Takara Bio, Inc.) and reverse transcribed into cDNA using the PrimeScript RT reagent kit with gDNA Eraser (Takara Bio, Inc.) at 37°C for 15 min and 85°C for 5 sec. U6 was used as a reference gene for miRNA (miR-126 and miR-429) measurements, whereas GAPDH was used as a reference gene for lncRNA HOTAIR. The following PCR primers were used: HOTAIR forward, 5′-GGCAAATGTCAGAGGGTT-3′ and reverse, 5′-GTGTAACAGGCAGGTGGA-3′; GAPDH forward, 5′-GGCTGTATTCCCCTCCATCG-3′ and reverse, 5′-CCAGTTGGTAACAATGCCATGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′; and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and miR-126 forward, 5′-ACACTCCAGCTGGGTCGTACCGTG-3′ and reverse, 5′-TGGTGTCGTGGAGTCG-3′. The 2−ΔΔCq method was used to calculate quantitative gene expression (23).
Western blot analysis
Protein expression in SW982 cells was measured using western blot analysis as previously described (22), and GAPDH was used as the a loading control. Proteins were separated using 10% SDS-PAGE at 90 V for 1.5 h, and transferred on to PVDF membranes (EMD Millipore) at 300 mA for 1 h. The membranes were then blocked in 5% skimmed milk at room temperature for 1 h, then incubated with the primary antibody for 2 h at room temperature, followed by the secondary antibody for 1 h at room temperature. The antibodies used were as follows: Anti-CDK1 [EPR165] (1:1,000; cat. no. ab133327; Abcam), anti-Cdk2 [E304] (1:1,000; cat. no. ab32147; Abcam), anti-cyclin D1 (1:2,000; cat. no. ab226977, Abcam), anti-p21 [HUGO291] (1:1,500; cat. no. ab107099; Abcam), anti-p53 (DO-1) (1:500; cat. no. sc-126; Santa Cruz Biotechnology, Inc.), anti-MMP9 (1:1,000; cat. no. ab38898; Abcam), anti-vimentin [VI-10] (1:2,000; cat. no. ab20346; Abcam), anti-E-cadherin [4A2] (1:500; cat. no. ab231303; Abcam), anti-N-cadherin [EPR1791-4] (1:1,000; cat. no. ab76011; Abcam), SDF1/CXCL12 (D32F9) Rabbit mAb (1:1,500; cat. no. 3350; Cell Signaling Technology, Inc.), HRP Conjugate Goat Anti-Rabbit IgG (H+L) (1:3,000; cat. no. HS101-01) and HRP Conjugate Goat Anti-Mouse IgG (H+L) (1:3,000; cat. no. HS201-01) (both TransGen Biotech Co., Ltd.). The proteins were visualized with ECL solution (Beijing Xinjingke Biotechnologies Co., Ltd), followed by densitometry analysis using ImageJ v1.8.0 (National Institutes of Health). GAPDH was loaded as control.
Cell invasion assay
A 24 mm Transwell® with 3.0 µm Pore Polycarbonate Membrane Insert (Corning Inc.) was used to assess the invasive capacity of SW982 cells, as previously described (24). In brief, 25 µl Matrigel (diluted with 3X serum-free medium; Sigma-Aldrich; Merck KGaA) was added to the upper chamber of the Transwell plate (Corning, Inc.) for 30 min at 37°C. A total of 0.5×106 cells/ml were added into the Transwell upper chamber in serum-free DMEM, and media containing 20% FBS (Gibco; Thermo Fisher Scientific, Inc.) was added into the lower chamber. The plate was incubated at 37°C for 24 h. Subsequently, the Transwell insert was removed and washed twice with PBS, and 100% methanol was used for fixation for 30 min at room temperature, followed by drying. The membrane was stained with crystal violet for 20 min at room temperature, and the relative migration was determined by measuring the absorbance at 595 nm.
Wound healing assay
A total of 5×105 cells/well were seeded with 2 ml medium in a 6-well plate. A scratch perpendicular to the back of the horizontal line was created using a vertically positioned (not tilted) pipette tip against a ruler, and images were captured determine the distance the wound at the start of the assay using a light microscope (Olympus Corporation) (×40 magnification). Serum-free DMEM medium was added after washing three times with PBS to remove the scratched cells. Images were captured after incubation for 24 h at 37°C in a humidified incubator with 5% CO2. ImageJ v1.8.0 was used for quantification.
Cell cycle and apoptosis assay
The cell cycle phases were detected using the Cell Cycle and Apoptosis Analysis kit (cat. no. 40301ES50; Shanghai Yeasen Biotechnology Co., Ltd.) using the MACSQuant® Analyzer 10 Flow cytometer (Miltenyi Biotec GmbH), according to the manufacturer's instructions. Data were analyzed by FlowJo X 10.0.7 software (Beckman Coulter, Inc.).
Immunofluorescence assay
A total of 3×104 cells/well were seeded onto a Lab-Tek Chambered cover glass (cat. no. LOT1228622; Thermo Fisher Scientific, Inc.). After incubation for 24 h, the medium was removed, and the cells were washed twice with PBS. The cells were incubated with 0.3% Triton X-100 in PBS-Tween-20 (PBS-T) for 30 min at room temperature, washed twice with PBS-T and incubated with the SDF-1 antibody (1:50; cat. no. ab155090; Abcam) for 30 min at room temperature. After washing three times with PBS-T, the cells were incubated with the Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488) secondary antibody (1:500; cat. no. ab150077; Abcam) for 1 h at room temperature. The nuclei were finally stained with 5 µg/ml DAPI for 5 min at room temperature. A Leica TCS SP5 microscope with the LAS AF Lite v4.0 image browser software (DMI3000; Leica Microsystems GmbH) was used for fluorescence detection and analysis.
Statistical analysis
The data was analyzed using SPSS v20.0 software (IBM, Corp.). Pairwise comparisons between groups were performed using either the unpaired Student's t-test, the χ2 test or Fisher's exact test, while multi-group statistical analysis was performed using one-way ANOVA followed by the Tukey's post hoc test. Pearson's coefficient was used for correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
HOTAIR is highly expressed in SS tissues
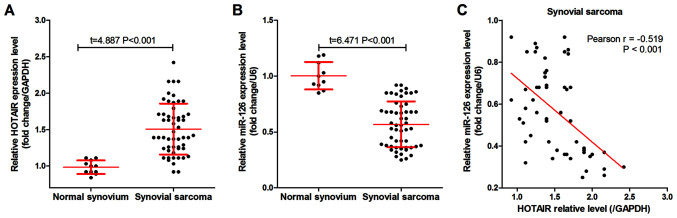
As shown in Fig. 1A, HOTAIR was highly expressed in the 54 SS tissues compared with that in the 10 normal synovium tissues. By contrast, miR-126 was expressed at low levels in the SS tissues (Fig. 1B) compared with that in the normal synovium tissues, and was negatively correlated with HOTAIR expression level in the SS tissues (Fig. 1C). In addition, patients with SS were divided into groups based on the median HOTAIR or miR-126 expression. Analysis using either χ2 or Fisher's exact tests found that both HOTAIR and miR-126 expression levels were significantly associated with histological grade, distant metastasis and AJCC staging in the patients with SS (Table I).
Figure 1.
HOTAIR and miR-126 expression level in SS and normal synovium tissues. Reverse transcription-quantitative PCR was used to measure (A) HOTAIR and (B) miR-126 expression level in SS (n=54) and normal synovium tissue (n=10). (C) Correlation between HOTAIR and miR-126 expression level in SS tissues. SS, synovial sarcoma; miR, microRNA.
Table 1.
Clinicopathological variables and HOTAIR/miR-126 expression level.
| HOTAIR | miR-126 | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Number | Low, n | High, n | P-value | Low, n | High, n | P-value |
| Sex | 0.846 | 0.409 | |||||
| Female | 23 | 12 | 11 | 13 | 10 | ||
| Male | 31 | 17 | 14 | 14 | 17 | ||
| Age, years | 0.816 | 0.413 | |||||
| ≥30 | 25 | 13 | 12 | 14 | 11 | ||
| <30 | 29 | 16 | 13 | 13 | 16 | ||
| Tumor size, cm | 0.918 | 0.580 | |||||
| <5 | 32 | 17 | 15 | 15 | 17 | ||
| ≥5 | 22 | 12 | 10 | 12 | 10 | ||
| Histological grade | 0.033 | 0.021 | |||||
| I | 12 | 3 | 9 | 10 | 2 | ||
| II | 20 | 8 | 12 | 9 | 11 | ||
| III | 22 | 15 | 7 | 8 | 14 | ||
| Distant metastasis | 0.001 | 0.001 | |||||
| No | 34 | 24 | 10 | 11 | 23 | ||
| Yes | 20 | 5 | 15 | 16 | 4 | ||
| AJCC staging | 0.015 | 0.003 | |||||
| I/II | 25 | 9 | 16 | 7 | 18 | ||
| III/IV | 29 | 20 | 9 | 20 | 9 | ||
miR, microRNA; AJCC, American Joint Committee on Cancer (8th edition).
HOTAIR knockdown inhibits cellular proliferation and blocks the cells in the G1 phase
A MTT assay was used to measure the proliferation of the SW982 cells in vitro following knockdown of HOTAIR, using si-HOTAIR-1 and si-HOTAIR-2 (Fig. 2A). The results showed that HOTAIR knockdown significantly reduced the cell proliferation of the SW982 cells (Fig. 2B). In addition, HOTAIR knockdown significantly increased the ratio of the SW982 cells in the G1/G0 phase of the cell cycle (Fig. 2C). In addition, HOTAIR knockdown decreased the protein expression level of CDK1, CDK2 and cyclin D1, while it increased the expression level of p21 and p53 (Fig. 2D).
Figure 2.
Effects of HOTAIR expression on the proliferation, cell cycle and cell cycle-related protein expression of the SS cells in vitro. (A) HOTAIR expression level in the SW982 cells transfected with si-NC or 2 siRNAs. (B) The proliferation of the SW982 cells was measured using MTT assays. (C) The cell cycle of SW982 was detected using flow cytometry. (D) Western blot analysis was used to measure CDK1, CDK2, cyclin D1, p21, and p53 protein expression levels in the SW982 cells. *P<0.05, **P<0.01, ***P<0.001 vs. si-NC. si, short inhibiting; NC, negative control; OD, optical density.
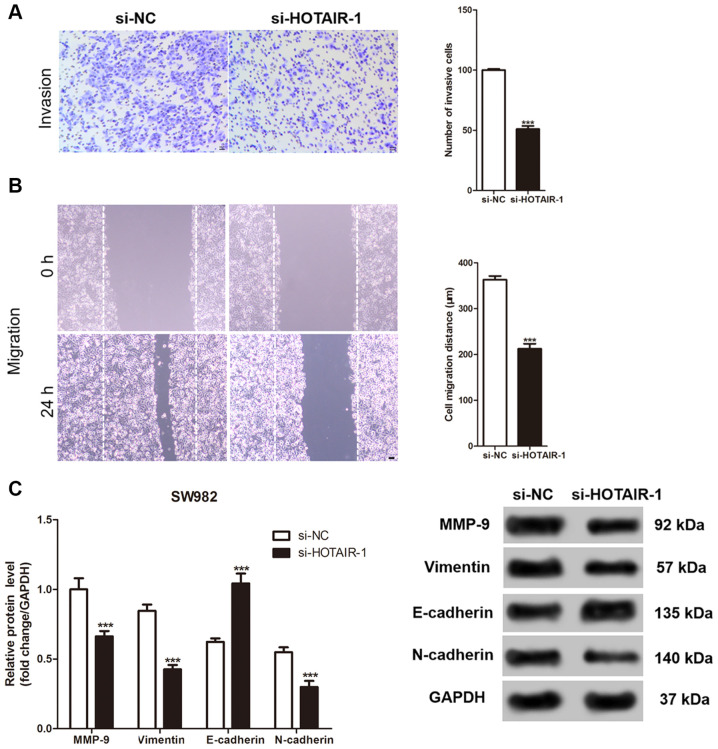
HOTAIR knockdown inhibits invasion and migration of the SS cells
The Transwell and wound healing assays were used to evaluate the invasive and migration abilities of the SS cells following knockdown of HOTAIR, respectively. As shown in Fig. 3A and B, the number of invasive SS cells was significantly reduced after HOTAIR knockdown. In addition, the migration distance by the SS cells was also significantly reduced. Similarly, the expression level of the proteins involved in cellular invasion and migration were measured. While HOTAIR knockdown significantly decreased the protein expression level of MMP-9, vimentin and N-cadherin, it increased the expression level of E-cadherin in the SW982 cells compared with that in the control cells (Fig. 3C).
Figure 3.
Effect of HOTAIR expression on migration and invasion of the SS cells in vitro. (A) Matrigel and (B) cell-scratch assays were used to evaluate the invasion and migration of the SW982 cells, respectively. (C) MMP-9, vimentin, E-cadherin and N-cadherin protein expression level in the SW982 cells transfected with si-NC and si-HOTAIR-1. Scale bar, 50 µm; ***P<0.001 vs. si-NC. si, short inhibiting; NC, negative control.
HOTAIR promotes SDF-1 expression level by inhibiting miR-126
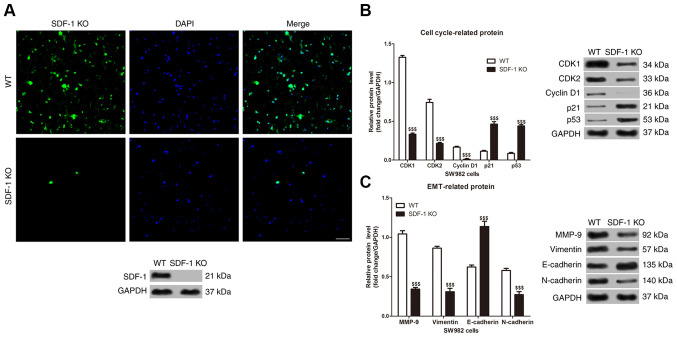
HOTAIR knockdown significantly increased miR-126 expression level in the SW982 cells (Fig. 4A). Notably, sequence analysis showed that HOTAIR and SDF-1 contained complementary sequences to miR-126 (Fig. 4B). In accordance with the sequence analysis, a luciferase gene reporter assay indicated that that the miR-126-mimics significantly reduced, and the miR-126-inhibitor significantly increased the luciferase activity in SW982 cells transfected with WT-SDF-1, whereas the miR-126-mimics and miR-126-inhibitor did not affect the luciferase activity in SW982 cells transfected with MUT-SDF-1 (Fig. 4C and D). Both HOTAIR knockdown and miR-126-mimics significantly decreased SDF-1 protein expression level, whereas treatment with miR-126 inhibitor significantly increased the protein expression level in the SW982 cells (Fig. 4E). Furthermore, immunofluorescence and western blot analysis showed that the expression of SDF-1 protein in SDF-1 KO SW982 cells was significantly lower than that of wild-type SW982 cells (Fig. 5A). In addition, SDF-1 knockdown suppressed the protein expression level of CDK1, CDK2, cyclin D1, MMP-9, vimentin and N-cadherin, while it induced the expression level of p21, p53 and E-cadherin in the SW982 cells (Fig. 5B and C).
Figure 4.
HOTAIR regulates SDF-1 expression level via miR-126 in the SS cells in vitro. (A) Effect of HOTAIR expression level on miR-126 expression level in the SW892 cells in vitro. ***P<0.001 vs. si-NC group. (B) The complementary sequences in HOTAIR and miR-126 with SDF-1 were identified, then a WT-SDF-1 3′-UTR or MUT-SDF-1 3′-UTR luciferase reporter vector with the miR-126 binding sites was constructed. (C) miR-126 expression level in the SW982 cells. ###P<0.001 vs. mimic-NC; &&&P<0.001 vs. inhibitor-NC. (D) Luciferase activity was detected in the SW982 cells with transfected with miR-126-NC, -mimics and -inhibitor. ###P<0.001 vs. mimic-NC (WT); &&&P<0.001 vs. inhibitor-NC (WT). (E) SDF-1 protein expression level in the SW982 cells with different treatment. $$$P<0.001 vs. inhibitor-NC group. WT, wild-type; MUT, mutant; miR, microRNA; UTR, untranslated region; NC, negative control.
Figure 5.
Effects of SDF-1 knockout on cell cycle- and EMT-related protein expression level in synovial sarcoma cells. (A) Measurement of SDF-1 protein expression level using cellular immunofluorescence and western blot in SW892 cells. Scale bar, 100 µm. Western blot analysis was used to detect the expression level of (B) cell cycle- and (C) ETM-related proteins in the SW892 cells. $$$P<0.001. KO, knockout; WT, wild-type; EMT, epithelial-mesenchymal transition.
Discussion
It has been shown in our previous study that SDF-1 was highly expressed in SS tissues, which was also associated with poor prognosis in patients with SS (20). Notably, SDF-1 was a target gene of miR-126. Indeed, van Solingen et al (25) showed that miR-126 increased the number of endothelial cells by inhibiting SDF-1 expression in the endothelium. Similarly, Zhang et al (19) demonstrated that miR-126 could inhibit the recruitment of mesenchymal stem cells and inflammatory monocytes into the tumor tissues by inhibiting SDF-1 expression level, thereby inhibiting the metastasis of breast cancer cells. Notably, HOTAIR was shown to promote the metastasis of cancer cells by inhibiting miR-126 expression level in different malignant tumor cells, such as breast, gastric and liver cancer cells (17). In the present study, it was shown that HOTAIR mRNA was highly expressed and was negatively correlated with miR-126 expression level in SS tissues, but was also significantly associated with histological grade, distant metastasis and AJCC staging in patients with SS. These results suggested that HOTAIR was associated with the development of SS.
The knockdown of HOTAIR in the SS cells inhibited proliferation, and the invasion and migration abilities of the SS cells in vitro, which is consistent with previous studies reporting HOTAIR as an oncogene in various malignancies. For example, Gupta et al (17) showed that HOTAIR could promote cancer metastasis through epigenetic reprograming of the cellular chromatin. As a non-coding RNA, HOTAIR may only act indirectly to regulate cellular functions through miRNA and mRNA regulation. In this regard, the results from the present study showed that HOTAIR knockdown could increase miR-126 expression level, which is consistent with previous studies reporting miR-126 to be a target of HOTAIR (11,18).
miRNAs are non-coding, single-stranded RNAs, that are 20–25 nucleotides in length, which can regulate cellular proliferation, differentiation, apoptosis, metabolism, invasion and migration through the post-transcriptional regulation of target genes (26,27). Multiple miRNAs have been recognized as biomarkers for the diagnosis of multiple diseases and have been demonstrated to serve important roles in the development of cancer, acute myocardial infarction and rheumatoid arthritis (28). Previous studies have shown (29,30) that the development of breast cancer was associated with the abnormal expression of miR-1271 and miR-192-5p. miR-126, a tumor suppressor, was shown to be abnormally expressed in numerous tumors, where it regulated multiple biological processes in tumors, such as proliferation, apoptosis, invasion, and migration (31). Feng et al (32) demonstrated that miR-126 could inhibit the proliferation, invasion and migration of gastric cancer cells by inducing cell-cycle arrest at the G1/G0 phase. Hamada et al (33), on the other hand, showed that miR-126 could play a tumor suppressor role in pancreatic tumor cells through the regulation of ADAM9. Taken together, these findings indicated that HOTAIR could regulate the proliferation, invasion and migration of SS cells via miR-126 regulation.
There have been two major interaction mechanisms reported between lncRNA and miRNA. The first mechanism suggested that lncRNA could act as ‘molecular sponges’ to block the post-transcriptional inhibitory effects of miRNAs on downstream target genes. These molecular sponge lncRNAs are called competitive endogenous RNA (34). The other mechanism suggested that lncRNAs and miRNAs could increase the stability of target genes by competing with miRNAs to bind target genes (34). In the present study, it was shown that HOTAIR knockdown increased miR-126 expression level, and that miR-126-mimics inhibited the protein expression level of SDF-1 in the SS cells. This suggested that the high expression level of HOTAIR may act as an oncogene in the SS cells by promoting SDF-1 expression level by regulating miR-126. Furthermore, it was also shown that SDF-1 knockout and HOTAIR knockdown resulted in similar molecular signatures in the SW982 cells, whereby they significantly reduced CDK1, CDK2, cyclin D1, MMP-9, vimentin and N-cadherin protein expression levels, while inducing p21, p53 and E-cadherin expression level.
Cyclin network systems, including CDKs and CDKIs, play important roles in the regulation of the G1/S phase of the cell cycle (35,36). p21 is a negative regulator of the cell cycle and is a member of the CDKI family. p21 can induce cell cycle arrest and block cell proliferation by binding to cyclins, CDKs or cyclin-CDKs complexes (37,38). In addition, the p21 gene has a specific binding site to the p53 protein and is considered one of the most important target genes of p53; when the cell is subjected to various stressors (such as chemical drugs or radiation), the WT p53 protein acts on the p21 gene and rapidly induces its expression (39). The p21 protein, encoded by the p21 gene, is a cell cycle inhibitor that is currently considered to have the broadest kinase inhibitor activity (39). Indeed, p21 can bind to and inhibit a wide range of cyclin-CDK complexes, such as cyclinD1-CDK4, cyclinE-CDK2, and cyclinA-CDK2, but also has a weak inhibitory activity against cyclinB-related complexes. p21 inhibits the activity of cyclinD1-CDK4 and cyclinE-CDK2, and hence, can block Rb protein phosphorylation and the release of the E2F protein, thereby inducing cell cycle arrest in the G1 phase (40). On the other hand, MMP-9, vimentin, E-cadherin and N-cadherin are key regulators of epithelial-mesenchymal transition, and hence, can regulate the metastatic ability of cancer cells (41,42).
In summary, the results from the present study showed that HOTAIR could be highly expressed in SS tissues, where it can promote the proliferation, invasion and migration of the SS cells by promoting SDF-1 protein expression through the regulation of miR-126 expression. Despite these important results, the number of SS tissues, as well as their freezing time and conditions may affect the expression level of HOTAIR and miR-126. Therefore, future studies, using larger sample sizes and more cell lines are required to further support the findings.
Acknowledgements
Not applicable.
Funding Statement
This research was supported by the fund of Youth Science and Technology Project of Hebei Provincial Department of Health (grant no. 20170692).
Funding
This research was supported by the fund of Youth Science and Technology Project of Hebei Provincial Department of Health (grant no. 20170692).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QF conceived and designed the current study and contributed to writing the manuscript. QF and DW performed the experiments. QF and DW confirm the authenticity of all the raw data. DW, PG, ZZ and JF analyzed and interpreted the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved and monitored by the Ethics Committee of The Fourth Hospital of Hebei Medical University (Hebei, China). Informed consent was provided by all the patients participating in the study or from their family members.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Palmerini E, Staals EL, Alberghini M, Zanella L, Ferrari C, Benassi MS, Picci P, Mercuri M, Bacci G, Ferrari S. Synovial sarcoma: Retrospective analysis of 250 patients treated at a single institution. Cancer. 2009;115:2988–2998. doi: 10.1002/cncr.24370. [DOI] [PubMed] [Google Scholar]
- 2.Bergh P, Meis-Kindblom JM, Gherlinzoni F, Berlin O, Bacchini P, Bertoni F, Gunterberg B, Kindblom LG. Synovial sarcoma: Identification of low and high risk groups. Cancer. 1999;85:2596–2607. doi: 10.1002/(SICI)1097-0142(19990615)85:12<2596::AID-CNCR16>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 3.Baccari-Ezzine S, Chelbi E, Bouzaidi K. Intracardiac metastasis of primary synovial sarcoma of the lung. Asian Cardiovasc Thorac Ann. 2013;21:623–623. doi: 10.1177/0218492312461006. [DOI] [PubMed] [Google Scholar]
- 4.Amankwah EK, Conley AP, Reed DR. Epidemiology and therapies for metastatic sarcoma. Clin Epidemiol. 2013;5:147–162. doi: 10.2147/CLEP.S28390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wushou A, Miao XC. Tumor size predicts prognosis of head and neck synovial cell sarcoma. Oncol Lett. 2015;9:381–386. doi: 10.3892/ol.2014.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakri A, Shinagare AB, Krajewski KM, Howard SA, Jagannathan JP, Hornick JL, Ramaiya NH. Synovial sarcoma: Imaging features of common and uncommon primary sites, metastatic patterns, and treatment response. AJR Am J Roentgenol. 2012;199:208–215. doi: 10.2214/AJR.11.8039. [DOI] [PubMed] [Google Scholar]
- 7.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, et al. An atlas of human long non-coding RNAs with accurate 5′ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From bio-markers to therapeutic targets. Int J Cancer. 2017;140:1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 10.Rao AKDM, Rajkumar T, Mani S. Perspectives of long non-coding RNAs in cancer. Mol Biol Rep. 2017;44:203–218. doi: 10.1007/s11033-017-4103-6. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Cui S, Wan T, Li X, Tian W, Zhang R, Luo L, Shi Y. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote glioma progression by sponging miR-126-5p. J Cell Physiol. 2018;233:6822–6831. doi: 10.1002/jcp.26432. [DOI] [PubMed] [Google Scholar]
- 12.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 13.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long Non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 15.Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumor Biol. 2016 Nov 30; doi: 10.1007/s13277-016-5448-5. (Epub ahead of print). doi: 10.1007/s13277-016-5448-5. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, Li C, Chong M, Ibrahim T, Mercatali L, et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol. 2013;15:284–294. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Q, Guo P, Wang J, Zhang X, Yang HC, Feng JG. High expression of SDF-1 and VEGF is associated with poor prognosis in patients with synovial sarcomas. Exp Ther Med. 2018;15:2597–2603. doi: 10.3892/etm.2018.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka K, Ozaki T. New TNM classification (AJCC eighth edition) of bone and soft tissue sarcomas: JCOG Bone and Soft Tissue Tumor Study Group. Jpn J Clin Oncol. 2019;49:103–107. doi: 10.1093/jjco/hyy157. [DOI] [PubMed] [Google Scholar]
- 22.Tao J, Zhang J, Ling Y, McCall CE, Liu TF. Mitochondrial Sirtuin 4 resolves immune tolerance in monocytes by rebalancing glycolysis and glucose oxidation homeostasis. Front Immunol. 2018;9:419. doi: 10.3389/fimmu.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Xie Z, Chen W, Chen Y, Wang X, Gao W, Liu Y. miR-768-3p is involved in the proliferation, invasion and migration of non-small cell lung carcinomas. Int J Oncol. 2017;51:1574–1582. doi: 10.3892/ijo.2017.4133. [DOI] [PubMed] [Google Scholar]
- 25.van Solingen C, de Boer HC, Bijkerk R, Monge M, van Oeveren-Rietdijk AM, Seghers L, de Vries MR, van der Veer EP, Quax PH, Rabelink TJ, van Zonneveld AJ. MicroRNA-126 modulates endothelial SDF-1 expression and mobilization of Sca-1(+)/Lin(−) progenitor cells in ischaemia. Cardiovasc Res. 2011;92:449–455. doi: 10.1093/cvr/cvr227. [DOI] [PubMed] [Google Scholar]
- 26.Bartel DP. MicroRNA: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2009;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2016;231:25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Xu L, Jiang L. miR-1271 promotes non-small-cell lung cancer cell proliferation and invasion via targeting HOXA5. Biochem Biophys Res Commun. 2015;458:714–719. doi: 10.1016/j.bbrc.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 30.Ye M, Zhang J, Zhang J, Miao Q, Yao L, Zhang J. Curcumin promotes apoptosis by activating the p53-miR-192-5p/215-XIAP pathway in non-small cell lung cancer. Cancer Lett. 2015;357:196–205. doi: 10.1016/j.canlet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 31.Meister J, Schmidt MH. miR-126 and miR-126*: New players in cancer. ScientificWorldJournal. 2014;10:2090–2100. doi: 10.1100/tsw.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, Masamune A, Kikuta K, Kume K, Shimosegawa T. miR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10:3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- 34.Erratum: The lncRNA XIST exhibits oncogenic properties via regulation of miR-449a and Bcl-2 in human non-small cell lung cancer. Acta Pharmacol Sin. 2017;38:443. doi: 10.1038/aps.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim S, Kaldis P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz GK, Shah MA. Targeting the cell cycle: A new approach to cancer therapy. J Clin Oncol. 2005;23:9408–9421. doi: 10.1200/JCO.2005.18.341. [DOI] [PubMed] [Google Scholar]
- 37.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 38.Kim EM, Jung CH, Kim J, Hwang SG, Park JK, Um HD. The p53/p21 complex regulates cancer cell invasion and apoptosis by targeting Bcl-2 family proteins. Cancer Res. 2017;77:3092–3100. doi: 10.1158/0008-5472.CAN-16-2098. [DOI] [PubMed] [Google Scholar]
- 39.Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Dutto I, Tillhon M, Cazzalini O, Stivala LA, Prosperi E. Biology of the cell cycle inhibitor p21(CDKN1A): Molecular mechanisms and relevance in chemical toxicology. Arch Toxicol. 2015;89:155–178. doi: 10.1007/s00204-014-1430-4. [DOI] [PubMed] [Google Scholar]
- 41.Zavadil J, Haley J, Kalluri R, Muthuswamy SK, Thompson E. Epithelial-mesenchymal transition. Cancer Res. 2008;68:9574–9577. doi: 10.1158/0008-5472.CAN-08-2316. [DOI] [PubMed] [Google Scholar]
- 42.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.