Abstract
Mutations in more than 150 genes are responsible for inherited hearing loss, with thousands of different, severe causal alleles that vary among populations. The Israeli Jewish population includes communities of diverse geographic origins, revealing a wide range of deafness-associated variants and enabling clinical characterization of the associated phenotypes. Our goal was to identify the genetic causes of inherited hearing loss in this population, and to determine relationships among genotype, phenotype, and ethnicity. Genomic DNA samples from informative relatives of 88 multiplex families, all of self-identified Jewish ancestry, with either non-syndromic or syndromic hearing loss, were sequenced for known and candidate deafness genes using the HEar-Seq gene panel. The genetic causes of hearing loss were identified for 60% of the families. One gene was encountered for the first time in human hearing loss: ATOH1 (Atonal), a basic helix-loop-helix transcription factor responsible for autosomal dominant progressive hearing loss in a five-generation family. Our results demonstrate that genomic sequencing with a gene panel dedicated to hearing loss is effective for genetic diagnoses in a diverse population. Comprehensive sequencing enables well-informed genetic counseling and clinical management by medical geneticists, otolaryngologists, audiologists, and speech therapists and can be integrated into newborn screening for deafness.
Keywords: Next-generation sequencing, Massively parallel sequencing, Diagnostics, Hearing, Deafness, gene panel, genomics
INTRODUCTION
Hearing loss is a leading cause of disability worldwide, with an estimated 466 million people suffering from a loss of greater than 40dB.1-3 Hearing loss can have dramatic effects on communication, levels of education, and psychosocial development; it is responsible for a subsequent decline in quality of life, particularly in an increasingly older population.4,5 Determining the causes of hearing loss is crucial for clinical management, genetic counseling, and potential prevention. More than 150 genes harbor variants causing non-syndromic hearing loss,6-8 and hundreds of genetic syndromes include hearing impairment.9 Virtually every population harbors deafness-causing alleles with worldwide distribution and others specific to the local population.10
The Jewish population of modern Israel is made up of communities that differ with respect to geographic origin, spoken language, and traditions. Ashkenazi Jews from Europe and North America, Sephardic Jews from North Africa (Morocco, Algeria, Libya, and Tunisia) and southern Europe (Italy, Greece, and Turkey), and Mizrahi Jews from the Middle East (Iran, Iraq, Syria, Yemen, and Egypt) all derive from the Jews who lived in the Middle East 4000 years ago, dispersing with the Babylonian exile in 586 BCE. 11,12 After the formation of the state of Israel in 1948, Jews from all these regions immigrated to the country. Today, roughly half of Jewish people live in Israel, yielding an Israeli Jewish population that is approximately 47% Ashkenazi, 30% Sephardi, and 23% Mizrahi.13,14 In a study conducted in Israel a few years after its founding, high rates of consanguineous marriage were observed, with the lowest rate (2.5%) among Ashkenazi Jews, and higher rates among non-Ashkenazi Jews, with the highest prevalence (29%) among Jews from Iraq.15 During the intervening years, inter-community marriages have become frequent, and consanguineous marriages are much less common.16
Centuries of endogamy within each of these communities led to high frequencies of recessive genetic traits, many due to community-specific founder mutations.13,17,18 The idea of that each Jewish ancestry has its own genetic blueprint is supported by studies revealing ancestry-specific polymorphisms and haplotypes. Among these are mutations for at least 40 diseases that are specific to individual Jewish communities.14 Population-specific mutations include some responsible for hearing loss, such as GJB2 c.167delT and TMC1 p.Ser647Phe, while other deafness-causing mutations, such as GJB2 c.35delG, are common in all Jewish ethnicities and elsewhere17,18,16.
GJB2 variants are the most prevalent cause of hereditary hearing loss worldwide and are responsible for ~30% of deafness in Jewish families.17,19,20 Hence in Israel, routine genetic testing has been for the two most common pathogenic variants, GJB2 c.35delG and GJB2 c.167delT. For hearing loss not explained by these alleles, high-throughput sequencing using hearing-loss-dedicated gene panels offers the opportunity to identify other disease-causing variants in hundreds of genes.21,22,18,23
The goal of this project was to identify the genetic causes of hearing loss in Israeli Jewish families with more than one affected relative (i.e. multiplex families) and to determine the number of genes responsible for hearing loss in the Israeli Jewish population as a whole. The long-term goal is to apply these results to development of guidelines for the molecular diagnosis of deafness in this population.
MATERIALS AND METHODS
Participants
Probands with hearing loss and their relatives were recruited from medical genetics clinics throughout Israel. All probands had a positive family history of hearing loss. Families were asked about their medical history, including family history of relevant symptoms, consanguinity, degree and symmetry of hearing loss, age of onset, use of hearing aids or cochlear implant, tinnitus, exposure to ototoxic drugs or noise, pathological conditions of the inner ear, and vestibular function. Hearing loss could be non-syndromic or syndromic, stable or progressive, and pre-lingual or post-lingual in onset. Probands or their parents gave written informed consent and provided a blood sample for DNA. GJB2 and GJB6 were evaluated by Sanger sequencing, and probands with hearing loss due to GJB2 were so advised and not sequenced with the gene panel.17 After these steps, 188 individuals from 88 multiplex families were evaluated with the HEar-Seq gene panels. Hearing controls from each Israeli Jewish ethnic group were identified from healthy, hearing individuals undergoing genetic screening at the Rabin and Sheba Medical Centers and from the National Laboratory for the Genetics of Israeli Populations (https://en-med.tau.ac.il/nlgip). An independent series of 105 individuals with hearing loss, treated at genetics clinics and not related to the 88 multiplex families, were included in order to estimate allele frequencies among Israeli Jewish deaf individuals.
Experimental procedures
Genomic analysis was carried out with the Hear-Seq gene panels, a custom design by the authors with version ranging from 178 to 372 genes 18,23,24 (Table S1). The BED file of Hear-Seq capture probe locations is freely available from the authors, and capture probes can be ordered directly from the manufacturer (Agilent Technologies, Santa Clara, CA, USA) with permission from the authors. Details of panel design and manufacture are included in Supporting Information.
Details of genomic analysis with the panels, of follow-up sequencing, and of the accompanying bioinformatics pipeline are also included in Supporting Information. Novel mutations of uncertain consequence in ATOH1 and MITF were evaluated by protein biochemistry and cell biology. Details of these methods are also included in Supporting Information.
RESULTS
Genetic diagnoses of hearing loss from gene panel sequencing
Genetic causes of hearing loss were identified for 60% (53/88) of the families evaluated by the HEar-Seq gene panels. These genetic diagnoses involved 57 different causal alleles in 27 different genes (Table S2, Figure S1). Most of the responsible alleles (32 of 57, or 56%) had not been previously reported from any population (Table 1). Of the novel variants, 50% were missense, 31% frameshifts, 9% nonsense, 9% copy number variants (CNVs), and 3% silent mutations that altered splicing. These diagnoses expanded the total number of genes known to be responsible for inherited loss in the Israeli Jewish population from seven17 to 32 (Table S3).
TABLE 1.
Novel variants discovered by the HEar-Seq gene panel in Israeli Jewish families with hearing loss
| Gene | Family | Inheritance* | Ethnicity** | Genomic coordinates (hg19) |
cDNA | Variant type | Effect | Allele freq controls of same ethnicity^ |
Allele freq in unrelated deaf^^ |
ClinVar ID |
ACMG classification (criteria)*** |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ATOH1 NM_005172.1 | HL263 | AD | M | chr4:94751102 | c.1030delC | Indel | His344fs17Ter | 0 | 0 | 813817 | P (PM2, PM4, PP1_S, PP3, PP4) |
| ATP2B2 NM_001001331.2 | DF328 | AD | A | chr3:10420936 | c.1033C>T | Nonsense | Gln345Ter | 0 | 0 | 1337668 | P (PM2, PM4, PP1_S, PP3, PP4) |
| CABP2 NM_001318496.1 | DF326 | AR | Mixed | chr11:67289435 | c.250G>A | Missense | Glu84Lys | 0 | 0 | 1337669 | P (PM2, PM3_S, PP1_M, PP3, PP4) |
| CEACAM16 NM_001039213.3 | DF301 | AR | M | chr19:45208901 Founder allele | c.703C>T | Missense | Arg235Cys | 0 | 0.008 | 236048 | P (PS4, PM2, PP1_S, PP3, PP4) |
| CLPP NM_006012.2 | DF313 | AR | S A |
chr19:6361758 chr19:6361914 |
c.173T>G (mat) c.233G>C (pat) |
Missense Missense |
Leu58Arg Arg78Pro |
0.004 0 |
0 0 |
500291 813818 |
1) LP (PM2, PM3_P, PP1_M, PP3, PP4) 2) LP (PM2, PM3_P, PP1_M, PP3, PP4) |
| COCH NM_004086.2 | HL1103 | AD | A | chr14:31355200 | c.1159C>T | Missense | Leu387Phe | 0 | 0 | 236036 | LP (PM2, PP1_M, PP3, PP4) |
| COL11A2 NM_080680.2 | HL1140 | AR | S M |
chr6:33152074 chr6:33138676 |
c.967insC c.3385G >A |
Indel Missense |
Thr323fs17Ter Gly1129Arg |
0 0 |
0 0 |
813820 813821 |
1) P (PVS1, PM2, PM3_P, PP1_M) 2) P (PM2, PM3_S, PP1_M, PP3, PP4) |
| EYA4 NM_172105.3 | HL21 | AD | Mixed | chr6:133783474 | c.441delC | Indel | Tyr148fs49Ter | 0 | 0 | 236032 | P (PVS1, PM2, PM4, PP1, PP3, PP4) |
| EYA4 NM_172105.3 | DF312 | AD | M | chr6:133844297 - 133844299 | c.1720_1722 delTACinsAAA | 2 bp changes in same codon | Tyr574Lys | 0 | 0 | 1164280 | LP (PM2, PM2, PP1, PP3, PP4) |
| GATA3 NM_002051.2 | HL738 | AD | A | chr10:8100707 | c.681ins35 | Indel | Glu228fs37Ter | 0 | 0 | 236031 | P (PVS1, PM2, PP1_M, PP3, PP4) |
| GATA3 NM_002051.2 | HL769 | AD | Mixed | chr10:8106009 | c.829G>A | Missense | Asp277Asn | 0 | 0 | 813823 | LP (PM2, PP1_S, PP3) |
| MITF NM_198159.2 | DF311 | AD | S | chr3:70001035 | c.935T>C | Missense | Leu312Pro | 0 | 0 | 547531 | P (PS4_P, PM2, PP1, PP3, PP4) |
| MITF NM_198159.2 | DF186 | AD | S | chr3:70014025 | c.1190delG | Indel | Gly397fs15Ter | 0 | 0 | 236050 | P (PVS1, PM2, PP1, PP3, PP4) |
| MITF NM_198159.2 | DF219 | AD | A | chr3:70005649 | c.981InsC | Indel | Leu327fs9Ter | 0 | 0 | 813825 | P (PVS1, PM2, PM4, PP1, PP3, PP4) |
| MYO6 NM_004999.3 | DF305 | AD | A | chr6:76538307 | c.238C>T | Nonsense | Arg80Ter | 0 | 0 | 178957 | P (PVS1, PM2, PP1, PP3, PP4) |
| MYO6 NM_004999.3 | HL1133 | AD | S | chr6:76568683 | c.1452insT | Nonsense | Asn485Ter | 0 | 0 | 1337671 | P (PVS1, PM2, PM4, PP1_M, PP3, PP4) |
| MYO6 NM_004999.3 | HL1274 | AD | A | chr6:76568710 | c.1473del3insC | Indel | skip exon 14, Glu461fs13Ter | 0 | 0 | 236034 | P (PVS1, PM2, PM4, PP1, PP3) |
| MYO6 NM_004999.3 | HL158 | AD | A | chr6:76624636 | c.3765delC | Indel | Cys1256fs28Ter | 0 | 0 | 813826 | P (PVS1, PM2, PM4, PP1, PP3, PP4) |
| MYO15A NM_016239.3 | HL72 | AR | M | chr17:18054799 | c.7751_8224del 3446ins23 | CNV | Gln2583fs19Ter | 0 | 0.003 | 236039 | P (PVS1, PM2, PM4, PP1, PP3, PP4) |
| MYO15A NM_016239.3 | DF327 DF317 |
AR | A | chr17:18069748 Founder allele |
c.9861C>T | Silent | Gly3287Gly | 0 | 0.003 | 228276/45777 | P (PM2_P, PM3_S, PP1_S, PP3) |
| PCDH15 NM_001142769.1 | HL1134 | AR | M | chr10:56287598 | c.146T>C | Missense | Val49Ala | 0 | 0.003 | 450626 | LP (PM2, PM3_P, PP1, PP3, PP4) |
| SLC26A4 NM_000441.1 | HL1132 HL1327 |
AR | A | chr7:107312627 | c.349C>T | Missense | Leu117Phe | 0.007 | 0.013 | 43555 | P (PS4, PM1, PM3, PM3_P, PP1_S, PP3, PP4) |
| SOX10 NM_006941.3 | HL971 | AD | A | chr22: 38379660 | c.125_132del8 | Indel | Leu42fs21Ter | 0 | 813829 | P (PVS1, PM2, PM4, PP3, PP4) | |
| STRC NM_153700.2 | HL927 | AR | Mixed | chr15:43,892,353 -43,910,998 (min) | del entire gene | CNV | - | 0.003 | 1164296 | P (PVS1, PM1, PM3, PM4, PP1, PP3, PP4) | |
| TECTA NM_005422.2 | DF303 | AR | A | chr11:120979969 | c.248C>T | Missense | Thr83Met | 0.003 | 813831 | LP (PM2, PM3_P, PP1_M, PP3, PP4) | |
| TECTA NM_005422.2 | HL277 | AD | S | chr11:121000866 | c.2887G>A | Missense | Ala963Thr | 0 | 0 | 236059 | LP (PM2, PP1_M, PP3, PP4) |
| TECTA NM_005422.2 | DF183 | AD | A | chr11:121058558 | c.6017A>G | Missense | Asp2006Gly | 0 | 0 | 236033 | LP (PM2, PM5, PP1, PP3, PP4) |
| TJP2 NM_001170414.2 | DF180 | AD | S | chr9:71704982 -71840362 dup | dup exons 1-6 | CNV | - | 0 | 0 | 236035 | P (PS4_P, PM1, PM2, PM4, PP1_S, PP3, PP4) |
| TMC1 NM_138691.2 | DF193 | AR | Mixed | chr9:75263573 | c.15insA | Indel | Val6fs12Ter | 0 | 0.003 | 236041 | P (PVS1, PM2, PM4, PP1, PP3, PP4) |
| TMC1 NM_138691.2 | HL1159 | AR | A | chr9:75369733 | c.674C>T | Missense | Pro225Leu | 0 | 0 | 424807 | P (PM2, PM3_S, PP1_M, PP3, PP4) |
Recessive, R; Dominant, D
A, Ashkenazi; M, Mizrahi; S, Sephardi
ACMG classification and criteria legend: P, pathogenic; LP, likely pathogenic; PVS, very strong pathogenicity evidence; PS, strong pathogenicity evidence; PM, moderate pathogenicity evidence; PP, supporting pathogenicity evidence; _S, strong; _M, moderate; _P, supporting
Number of ethnicity-matched hearing controls ranges from 163-500 individuals (326-1000 chromosomes)
Number of unrelated deaf is 105 individuals (210 chromosomes)
Oza AM, DiStefano MT, Hemphill SE et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat. 2018;39(11):1593-1613.
Richards S, Aziz N, Bale S et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424.
ATOH1, a new gene for human hearing loss
Panel sequencing revealed involvement of a new gene for human hearing loss. ATOH1 (Atonal) encodes a basic helix-loop-helix transcription factor that is essential for neuronal development in the cerebellum.25 Heterozygosity for any of several different variants of Atoh1 leads to hearing deficits in mice, some with syndromic features.26,27 ATOH1 was included on the panel because mutations in the mouse ortholog lead to hearing loss. In Iraqi Jewish kindred HL263, ATOH c.1030delC co-segregated over five generations with progressive non-syndromic hearing loss, with onset at birth or early childhood (Figure 1A,B). Based on whole exome sequencing, no other potentially damaging variant in any gene co-segregated with hearing loss in this family. ATOH c.1030delC causes a frameshift that alters the last ten residues of the normally 354-amino acid protein and adds six residues to its length before a stop.
Figure 1.
ATOH1 c.1030delC and age-related hearing loss. A, Family HL263 with progressive sensorineural hearing loss in five generations. Filled symbols represent individuals with hearing loss. V represents the variant allele and N the normal allele. The number under each individual is the birth year. B, Average hearing thresholds of family members of various ages heterozygous for the mutation. C, HEK293 cells transfected with expression constructs for ATOH1WT or ATOH1c.1030delC. Anti-ATOH1 labeling in green indicates nuclear localization of both wild type and mutant ATOH1 (arrows). D, left column: Cochlear explant from an Atoh1−/− mouse established at E14 and transfected with an ATOH1c.1030delC expression construct. Transfected cells (red) are present in a region that is consistent with the location of the organ of Corti in a wildtype cochlea (shaded region, OC), as well as in several patches that are located outside of the organ of Corti (arrows). Counter-staining for the hair cell marker Myo7a indicates that many transfected cells have developed as hair cells. Right column: High magnification images of the boxed region demonstrating expression of Myo7a (green) and ATOH1 (blue) in a patch of transfected cells. E, Western blot of ATOH1 protein extracted from HEK293T cells transfected with wild type or mutant ATOH1, after 1-5 hours treatment with 1mM cycloheximide. F, Quantification of the results of part C. Statistical test was repeated measures ANOVA with post-hoc Holm-sidak correction for multiple comparisons.
To examine the effects of this mutation, expression constructs for wild type and mutant ATOH1 were transfected into HEK293 cells. Antibody localization indicated nuclear translocation of both wild-type and mutant ATOH1 (Figure 1C). Next, to determine the effects of the mutation on protein function, cochlear explants were established at E14 or E15 from Atoh1−/− animals and transfected with either the ATOH1WT or ATOH1c.1030delC expression construct. (Atoh1−/− cochleae were used to ensure no potential effects from the endogenous gene.) Results indicated induction of Myo7A+ hair cells in response to expression of either construct (Figure 1D). However, western analysis revealed a significantly slower rate of degradation for mutant ATOH1 protein compared to wild-type ATOH1 protein (P=0.032, multiple t-test with Holm-Sidak correction, two biological replicates) (Figure 1E,F).
Complex relationships of genotypes to phenotypes
For genes responsible for syndromic hearing loss, different variants in the same gene revealed new relationships of genotypes to phenotypes. Three families with mutations in MITF illustrate these complexities (Figure 2). Damaging variants of MITF can cause autosomal dominant Waardenburg type 2A and Tietz albinism/deafness syndromes, both of which are highly heterogeneous clinically (Figure 2A,B). In family DF311, three relatives heterozygous for MITF c.935T>C, p.(Leu312Pro) had severe to profound sensorineural hearing loss with congenital albinism. (Hearing loss of DF311.02 is due to a mutation in CDH23.) Leu312 is a completely conserved residue in the middle of the MITF basic helix-loop-helix (bHLH) domain. A proline at this position would likely break the helix and preclude proper DNA binding and possibly preclude dimerization as well. In family DF219, three relatives heterozygous for MITF c.981insC, p.(Leu327fs9Ter) demonstrated the same hearing loss and albinism. A frameshift at residue 327 would lead to truncation in the middle of the bHLH domain and loss of normal protein function. In contrast, in family DF186, three relatives heterozygous for MITF c.1190delG, p.(Gly397fs15Ter) also demonstrated congenital sensorineural hearing loss but no pigmentation signs other than hair whitening of the mother in her twenties. Truncation due to frameshift at residue 397 is distal to the bHLH domain, so its consequences to protein function were unknown. A transactivation assay of the protein encoded by MITF c.1190delG, p.(Gly397fs15Ter) indicated that the transcriptional potential of the mutant protein is greatly impaired compared to that of wild-type MITF (Figure 2C, 2D).
Figure 2.
MITF1 variants associated with hearing loss and Waardenburg Syndrome Type 2A / Tietz Syndrome in three families. A, Pedigrees of families DF311, DF186, and DF219, indicating variation in syndromic features. B, Hearing thresholds by age, reflecting severe to profound hearing loss in all affected individuals. C, Transactivation assay using a tyrosinase promoter and luciferase reporter revealing that the transcriptional potential of protein encoded by MITF c.1190delG is greatly impaired compared to the wild-type protein. Statistical test was one-tailed student’s t-test. D, Western blot analysis indicates similar levels of wild type and mutant MITF protein.
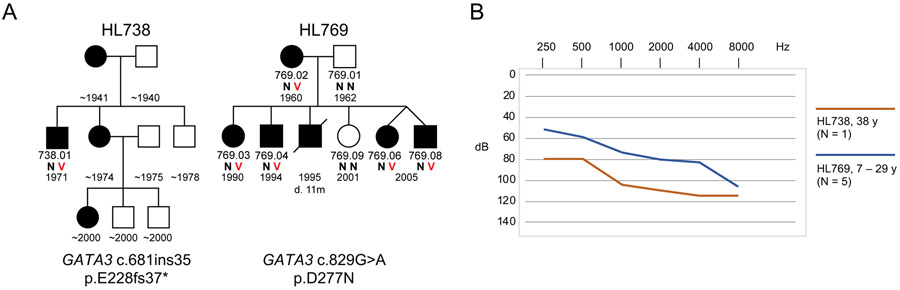
GATA3 variants present an analogous story. GATA3 is responsible for an autosomal dominant syndrome including hypoparathyroidism, sensorineural deafness, and renal dysplasia (HDR), for which different alleles are associated with a wide spectrum of phenotypes.28 Two families in our series reflect this heterogeneity (Figure 3). In family HL738, the proband (DF738.01), heterozygous for GATA3 c.681ins35, p.(Glu228fs37Ter), and his mother both demonstrated congenital severe-to-profound hearing loss and kidney dysplasia. (Medical records for the sister and niece of the proband were not available.) In contrast, in family HL769, all relatives heterozygous for GATA3 c.829G>A, p.(Asp277Asn), demonstrated severe-to-profound hearing loss, but to date no renal or parathyroid problems.
Figure 3.
GATA3 variants associated with nonsyndromic and syndromic hearing loss in two families. A, Pedigrees of families HL738 and HL769. B, Hearing thresholds by age, reflecting severe to profound hearing loss in all affected individuals.
CLPP is associated with Perrault syndrome, characterized by sensorineural hearing loss (SNHL) and infertility in both females and males.29 In family DF313, affected siblings were both compound heterozygous for novel variants CLPP c.173T>G, p.(Leu58Arg) and c.233G>C, p.(Arg78Pro) (Figure S1). These siblings are young, age 3y, and thus far have profound hearing loss and auditory neuropathy but no other symptoms. It is not clear whether they are pre-symptomatic for infertility or if this genotype leads to nonsyndromic hearing loss.
SOX10 is responsible for autosomal dominant Waardenburg syndrome types 2E30 and 4C,31 and for Kallmann syndrome.32 The adult proband of family HL971 and his mother, both with congenital profound hearing loss, are heterozygous for novel variant SOX10 c.125_132del8, p.(Leu42fs21Ter) (Figure S1). Neither the proband nor his mother initially reported any symptoms other than deafness. Upon detecting this SOX1 variant and asking the proband about his sense of smell, we discovered that both he and his deaf mother have anosmia, characteristic of Kallmann syndrome.
USH2A is responsible for Usher syndrome type 2A but signs of retinitis pigmentosa (RP) depend on genotype and on age. In family HL149, the proband, age 3y is compound heterozygous for USH2A c.3368A>G, p.(Tyr1123Cys)33 and USH2A c.240_241insGTAC, a known pathogenic variant associated with SHL34 (Table S2). His mother, in her early thirties, is homozygous for USH2A c.240_241insGTAC, with sloping mild-to-severe hearing loss and mild RP. The proband also has a sloping mild-to moderate hearing loss, but as yet no signs of RP.
TECTA can be responsible for autosomal dominant or autosomal recessive hearing loss. Among families with dominant hearing loss, missense mutations in the TECTA zonadhesion domain (amino acid residues 260-1694) are associated with high-frequency hearing loss, while missense mutations in the TECTA zona pellicuda domain (residues 1795-2059) are associated with mid-frequency hearing impairment35-37. This correspondence obtains for family HL277 (p.Ala963Thr) and family DF193 (p.Asp2006Gly) (Figure S1).
Contributions of founder alleles
Founder alleles of ancestral Jewish communities continue to contribute to hearing loss in Israeli Jewish families. Table S3 lists the 75 variants in 32 genes of the families in the study, distributed among different communities. Some variants were private to one family, whereas others were more common but limited to one community, reflecting a founder effect (Table S4). Among founder mutations, the principal contribution to hearing loss was of course from GJB2 alleles, but founder mutations in other genes also contributed. For example, CEACAM16 c.703C>T, p.(Arg235Cys) was responsible for recessive hearing loss in family DF301, of Jewish Iranian ancestry (Figure S1). This allele was subsequently identified in children with hearing loss from other families of Jewish Iranian ancestry evaluated in clinics. Another example is OTOF c.5193-1G>A, responsible for recessive hearing loss with auditory neuropathy in family HL1015, of Syrian Jewish ancestry (Figure S1). This allele was heterozygous in three of 184 hearing controls of Syrian Jewish ancestry (allele frequency 0.008), but was absent from hearing controls of all other Jewish ethnicities and absent from gnomAD. It is likely a founder mutation among Syrian Jews.
A quite common founder allele in the Ashkenazi community is SLC26A4 c.349C>T, p.(Leu117Phe), with allele frequency 0.005 in the Ashkenazi Jewish population compared to 0.0002 in other gnomAD populations. No homozygotes for this variant have been observed among hearing individuals of any ancestry. This missense occurs in a completely conserved residue in the first transmembrane domain of SLC26A4, but its consequence has been uncertain. Families HL1132 and HL1327 were informative for this variant (Figure S1). Of the five children with hearing loss in these families, four are homozygous for SLC26A4 p.(Leu117Phe). These deaf children are the first homozygotes reported for this variant. HL1327.09, who has profound hearing loss but is heterozygous for SLC26A4 p.(Leu117Phe), remains unexplained. She has no other mutation in SLC26A4. She may be a phenocopy for inherited hearing loss in this family. Homozygosity for SLC26A4 p.(Leu117Phe) has also been observed in four other Ashkenazi families during screening for childhood hearing loss by genetics clinics in Israel. We speculate that SLC26A4 c.349C>T, p.(Leu117Phe), or a non-coding regulatory mutation of SLC26A4 in tight linkage disequilibrium with it, is pathogenic for nonsyndromic hearing loss.38-40,41
Figure 4 presents genotype-phenotype-ancestry associations for all variants encountered in the study.
Figure 4.
Distribution of hearing loss variants in Ashkenazi, Mizrahi, Sephardi, and mixed Jewish communities. Numbers in parentheses are the number of probands of each ethnicity. Numbers of cases are listed next to each gene.
DISCUSSION
Genetic diagnosis will play an increasing role in treatment of both congenital and later onset hearing loss. Success of cochlear implant may depend on the genetic cause of the hearing loss.42 The clinical application of gene therapy for some forms of hearing loss may prove feasible,43,44 but its application depends on correct genetic diagnosis. Gene panel-based sequencing increased the yield of genetic diagnoses from 23%17 to 60% of familial hearing loss in the Israeli Jewish population. The analysis revealed 57 different pathogenic variants in 27 genes, with most variants not previously reported, and increased the number of genes known to cause hearing loss in the Jewish population from seven to 32.
Yield from the HEar-Seq gene panel compares favorably with whole exome sequencing (WES).45 Costs of WES have decreased in recent years, but far fewer patients can be sequenced simultaneously with high coverage by WES than with a gene panel. Gene panels have proven effective for genetic diagnosis of a tremendous variety of conditions, ranging from inherited predisposition to cancer46 to inherited eye disorders.47 Gene panel sequencing also minimizes the frequency of incidental findings,48 which can introduce legal, ethical, and social dilemmas. Nevertheless, for families not solved by panel sequencing, WES is the next step in searching for a genetic diagnosis.
The discovery that mutation of ATOH1 can cause human hearing loss adds to understanding the role of this transcription factor in mammalian hearing. ATOH1 is crucial for the development and differentiation of inner-ear hair cells49 and is first expressed in the nascent organ of Corti. Loss of Atoh1 in mice causes hearing impairment, cerebellar and cochlear malformations, and death,25 while conditional deletion of Atoh1 leads to lack of differentiated inner ear hair cells,50 and the naturally occurring mutation Atoh1 p.Met200Ile causes hearing loss, progressive cerebellar atrophy, and trembling.26 The ATOH1 mutation of family HL263 yields a protein with an abnormal C-terminus associated with an abnormally slow degradation rate. This is consistent with previous observations that Atoh1 protein stability is regulated by its interaction with the E3 ubiquitin ligase Huwe1 at phosphorylation sites S328 and S339 (human S331 and S342).51,52 In this context, a mouse with mutation at the phosphorylation site Atoh1 S193 was shown to have late-onset deafness.27 In wild-type mice, expression of Atoh1 ceases by the end of the first postnatal week. Induction of Atoh1 at the neonatal stage causes formation of immature ectopic hair cells, with randomized stereocilia orientation and reduced basolateral measured currents.53,54 Atoh1 is also known to positively auto-regulate its own expression.55 We showed that the human ATOH1 mutation increases the stability of the protein through decreased degradation. We hypothesize that persistent untimely expression of ATOH1 may generate immature ectopic hair cells that interfere with development of normal hair cells.56,57
Early diagnosis enables caregivers to learn whether children are likely to develop symptoms other than hearing loss and so plan for specialized education and treatment. Among our families, pathogenic variants in CLPP, SOX10, and USH2A, which cause syndromic hearing loss, were detected in children prior to onset of additional symptoms. Similarly, a missense mutation in GATA3 was present in a family with relatives ages 15-60y with apparently non-syndromic hearing loss. Ages at onset of the syndromic features of GATA3 are variable, with mean age of onset during childhood or early teenage years, but possible onset of the full spectrum of phenotypes as late as age 50.58 It is still possible, therefore, that the affected relatives of this family will develop syndromic features. It is also possible that mild mutations in GATA3 could cause non-syndromic hearing loss.
In summary, the diversity of the Israeli Jewish population is reflected in the diversity of mutations that have been revealed to be responsible for hearing loss. These variants include mutations private to one family, founder mutations of ancestral communities, and mutations appearing in families of all ancestries. Our results are informative for genetic counselors, medical geneticists, audiologists and otolaryngologists caring for families with inherited hearing loss. Genetic diagnosis can be integrated with history, physical examination, and audiometry to guide management of patients with hearing loss. The results can also assist in developing guidelines for genetic screening of newborns with possible hearing loss, in Israel and elsewhere.
Supplementary Material
Table S1. Genes on HEar-Seq panels
Table S2. Variants identified by HEar-Seq in Israeli Jewish families
Table S3. All variants leading to hearing loss in Israeli Jewish Families in the study
Table S4. Founder mutations in Israeli Jewish families
Figure S1. Pedigrees, audiograms and genotypes of families with hearing loss. N = wild type; V = variant.
ACKNOWLEDGEMENTS
The authors wish to thank all the families for their participation in this study. This research was supported by grants from the National Institutes of Health/National Institute of Deafness and Communication Disorders (NIDCD) R01DC011835 (K.B.A., M.C.K., M.N.K.), the Intramural Program at the NIDCD, DC000059 (M.W.K.), the Israel Precision Medicine Program or the Israel Science Foundation, ISF3499-19 (K.B.A.), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine (K.B.A.), the Hedrich Charitable Trust (K.B.A.), and travel grants from the University of Washington Virginia Bloedel Hearing Research Institute (K.B.A., R.C.).
Footnotes
CONFLICT OF INTEREST
The authors declare they have no conflict of interest.
ETHICS APPROVAL
The Ethics Committee of Tel Aviv University, the Helsinki Committee of the Israel Ministry of Health, and the Human Subjects Division of the University of Washington approved the study.
DATA AVAILABILITY STATEMENT
Novel variants are available at ClinVar (www.ncbi.nlm.nih.gov/clinvar/).
REFERENCES
- 1.Cunningham LL & Tucci DL. Hearing loss in adults. N Engl J Med. 2017;377(25):2465–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD 2015 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1545–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Deafness and Hearing Loss. Updated March 2020. Accessed May 2, 2020. http://www.who.int/news-room/fact-sheets/detail/deafness-and-hearing-loss
- 4.Shukla A, Harper M, Pedersen E et al. Hearing loss, loneliness, and social isolation: A systematic review. Otolaryngol Head Neck Surg. 2020;194599820910377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deal JA, Betz J, Yaffe K et al. Hearing impairment and incident dementia and cognitive decline in older adults: The Health ABC Study. J Gerontol A Biol Sci Med Sci. 2017;72(5):703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.an Camp G & Smith RJH. Hereditary Hearing Loss Homepage. Updated April 2020. Accessed May 2, 2020. http://hereditaryhearingloss.org [Google Scholar]
- 7.Deafness Variation Database. Accessed May 2, 2020. http://deafnessvariationdatabase.org/
- 8.Azaiez H, Booth KT, Ephraim SS et al. Genomic landscape and mutational signatures of deafness-associated genes. Am J Hum Genet. 2018;103(4):484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koffler T, Ushakov K & Avraham KB. Genetics of hearing loss: Syndromic. Otolaryngol Clin North Am. 2015;48(6):1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shearer AE, Eppsteiner RW, Booth KT et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet. 2014;95(4):445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostrer H & Skorecki K. The population genetics of the Jewish people. Hum Genet. 2013;132(2):119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer MF, Redd AJ, Wood ET et al. Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci U S A. 2000;97(12):6769–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlotogora J Genetics and genomic medicine in Israel. Mol Genet Genomic Med. 2014;2(2):85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostrer H A genetic profile of contemporary Jewish populations. Nat Rev Genet. 2001;2(11):891–898. [DOI] [PubMed] [Google Scholar]
- 15.Goldschmidt E, Ronen A & Ronen I. Changing marriage systems in the Jewish communities of Israel. Ann Hum Genet. 1960;24(191–204. [DOI] [PubMed] [Google Scholar]
- 16.Cohen T, Vardi-Saliternik R & Friedlander Y. Consanguinity, intracommunity and intercommunity marriages in a population sample of Israeli Jews. Ann Hum Biol. 2004;31(1):38–48. [DOI] [PubMed] [Google Scholar]
- 17.Brownstein Z & Avraham KB. Deafness genes in Israel: implications for diagnostics in the clinic. Pediatr Res. 2009;66(2):128–134. [DOI] [PubMed] [Google Scholar]
- 18.Brownstein Z, Friedman LM, Shahin H et al. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 2011;12(9):R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobe T, Vreugde S, Shahin H et al. The prevalence and expression of inherited connexin 26 mutations associated with nonsyndromic hearing loss in the Israeli population. Hum Genet. 2000;106(1):50–57. [DOI] [PubMed] [Google Scholar]
- 20.Estivill X, Fortina P, Surrey S et al. Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet. 1998;351(9100):394–398. [DOI] [PubMed] [Google Scholar]
- 21.Shearer AE & Smith RJ. Massively parallel sequencing for genetic diagnosis of hearing loss: The new standard of care. Otolaryngol Head Neck Surg. 2015;153(2):175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shearer AE, DeLuca AP, Hildebrand MS et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci USA. 2010;107(49):21104–21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brownstein Z, Abu-Rayyan A, Karfunkel-Doron D et al. Novel myosin mutations for hereditary hearing loss revealed by targeted genomic capture and massively parallel sequencing. Eur J Hum Genet. 2014;22(6):768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danial-Farran N, Brownstein Z, Gulsuner S et al. Genetics of hearing loss in the Arab population of Northern Israel. Eur J Hum Genet. 2018;26(12):1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben-Arie N, Bellen HJ, Armstrong DL et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390(6656):169–172. [DOI] [PubMed] [Google Scholar]
- 26.Sheykholeslami K, Thimmappa V, Nava C et al. A new mutation of the Atoh1 gene in mice with normal life span allows analysis of inner ear and cerebellar phenotype in aging. PLoS One. 2013;8(11):e79791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie WR, Jen HI, Seymour ML et al. An Atoh1-S193A phospho-mutant allele causes hearing deficits and motor impairment. J Neurosci. 2017;37(36):8583–8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muroya K, Hasegawa T, Ito Y et al. GATA3 abnormalities and the phenotypic spectrum of HDR syndrome. J Med Genet. 2001;38(6):374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenkinson EM, Rehman AU, Walsh T et al. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am J Hum Genet. 2013;92(4):605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bondurand N, Dastot-Le Moal F, Stanchina L et al. Deletions at the SOX10 gene locus cause Waardenburg syndrome types 2 and 4. Am J Hum Genet. 2007;81(6):1169–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pingault V, Bondurand N, Kuhlbrodt K et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18(2):171–173. [DOI] [PubMed] [Google Scholar]
- 32.Vaaralahti K, Tommiska J, Tillmann V et al. De novo SOX10 nonsense mutation in a patient with Kallmann syndrome and hearing loss. Pediatr Res. 2014;76(1):115–116. [DOI] [PubMed] [Google Scholar]
- 33.Behar DM, Davidov B, Brownstein Z et al. The many faces of sensorineural hearing loss: one founder and two novel mutations affecting one family of mixed Jewish ancestry. Genet Test Mol Biomarkers. 2014;18(2):123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auslender N, Bandah D, Rizel L et al. Four USH2A founder mutations underlie the majority of Usher syndrome type 2 cases among non-Ashkenazi Jews. Genet Test. 2008;12(2):289–294. [DOI] [PubMed] [Google Scholar]
- 35.Sagong B, Park R, Kim YH et al. Two novel missense mutations in the TECTA gene in Korean families with autosomal dominant nonsyndromic hearing loss. Ann Clin Lab Sci. 2010;40(4):380–385. [PubMed] [Google Scholar]
- 36.Yamamoto N, Mutai H, Namba K et al. Prevalence of TECTA mutation in patients with mid-frequency sensorineural hearing loss. Orphanet J Rare Dis. 2017;12(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim AR, Chang MY, Koo JW, Oh SH & Choi BY. Novel TECTA mutations identified in stable sensorineural hearing loss and their clinical implications. Audiol Neurootol. 2015;20(1):17–25. [DOI] [PubMed] [Google Scholar]
- 38.Taylor JP, Metcalfe RA, Watson PF, Weetman AP & Trembath RC. Mutations of the PDS gene, encoding pendrin, are associated with protein mislocalization and loss of iodide efflux: implications for thyroid dysfunction in Pendred syndrome. J Clin Endocrinol Metab. 2002;87(4):1778–1784. [DOI] [PubMed] [Google Scholar]
- 39.Reardon W, OMahoney CF, Trembath R, Jan H & Phelps PD. Enlarged vestibular aqueduct: a radiological marker of pendred syndrome, and mutation of the PDS gene. QJM. 2000;93(2):99–104. [DOI] [PubMed] [Google Scholar]
- 40.Albert S, Blons H, Jonard L et al. SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. Eur J Hum Genet. 2006;14(6):773–779. [DOI] [PubMed] [Google Scholar]
- 41.Costa MD, Pereira JB, Pala M et al. A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages. Nat Commun. 2013;4(2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyagawa M, Nishio SY & Usami S. A comprehensive study on the etiology of patients receiving cochlear implantation with special emphasis on genetic epidemiology. Otol Neurotol. 2016;37(2):e126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schilder AGM, Blackshaw H, Lenarz T et al. Biological therapies of the inner ear: What otologists need to consider. Otol Neurotol. 2018;39(2):135–137. [DOI] [PubMed] [Google Scholar]
- 44.Taiber S & Avraham KB. Genetic therapies for hearing loss: Accomplishments and remaining challenges. Neurosci Lett. 2019;713(134527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis MA, Nolan LS, Cadge BA et al. Whole exome sequencing in adult-onset hearing loss reveals a high load of predicted pathogenic variants in known deafness-associated genes and identifies new candidate genes. BMC Med Genomics. 2018;11(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh T, Lee MK, Casadei S et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107(28):12629–12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Consugar MB, Navarro-Gomez D, Place EM et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet Med. 2015;17(4):253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green RC, Berg JS, Grody WW et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen P, Johnson JE, Zoghbi HY & Segil N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129(10):2495–2505. [DOI] [PubMed] [Google Scholar]
- 50.Pan N, Jahan I, Kersigo J et al. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear Res. 2011;275(1-2):66–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forget A, Bihannic L, Cigna SM et al. Shh signaling protects Atoh1 from degradation mediated by the E3 ubiquitin ligase Huwe1 in neural precursors. Dev Cell. 2014;29(6):649–661. [DOI] [PubMed] [Google Scholar]
- 52.Cheng YF, Tong M & Edge AS. Destabilization of Atoh1 by E3 ubiquitin ligase Huwe1 and casein kinase 1 is essential for normal sensory hair cell development. J Biol Chem. 2016;291(40):21096–21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelly MC, Chang Q, Pan A, Lin X & Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci. 2012;32(19):6699–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z, Dearman JA, Cox BC et al. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J Neurosci. 2012;32(19):6600–6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helms AW, Abney AL, Ben-Arie N, Zoghbi HY & Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127(6):1185–1196. [DOI] [PubMed] [Google Scholar]
- 56.Zhong C, Fu Y, Pan W, Yu J & Wang J. Atoh1 and other related key regulators in the development of auditory sensory epithelium in the mammalian inner ear: function and interplay. Dev Biol. 2019;446(2):133–141. [DOI] [PubMed] [Google Scholar]
- 57.Wu DK & Kelley MW. Molecular mechanisms of inner ear development. Cold Spring Harb Perspect Biol. 2012;4(8):a008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lemos MC & Thakker RV. Hypoparathyroidism, deafness and renal dysplasia (HDR) syndrome: 20 years after the identification of the first GATA3 mutations. Hum Mutat. 2020; [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes on HEar-Seq panels
Table S2. Variants identified by HEar-Seq in Israeli Jewish families
Table S3. All variants leading to hearing loss in Israeli Jewish Families in the study
Table S4. Founder mutations in Israeli Jewish families
Figure S1. Pedigrees, audiograms and genotypes of families with hearing loss. N = wild type; V = variant.
Data Availability Statement
Novel variants are available at ClinVar (www.ncbi.nlm.nih.gov/clinvar/).