Abstract
The current coronavirus disease (COVID-19) pandemic remains one of the most serious public health problems. Increasing evidence shows that infection by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) causes a very complex and multifaceted disease that requires detailed study. Nevertheless, experimental research on COVID-19 remains challenging due to the lack of appropriate animal models. Herein, we report novel humanized mice with Cre-dependent expression of hACE2, the main entry receptor of SARS-CoV-2. These mice carry hACE2 and GFP transgenes floxed by the STOP cassette, allowing them to be used as breeders for the creation of animals with tissue-specific coexpression of hACE2 and GFP. Moreover, inducible expression of hACE2 makes this line biosafe, whereas coexpression with GFP simplifies the detection of transgene-expressing cells. In our study, we tested our line by crossing with Ubi-Cre mice, characterized by tamoxifen-dependent ubiquitous activation of Cre recombinase. After tamoxifen administration, the copy number of the STOP cassette was decreased, and the offspring expressed hACE2 and GFP, confirming the efficiency of our system. We believe that our model can be a useful tool for studying COVID-19 pathogenesis because the selective expression of hACE2 can shed light on the roles of different tissues in SARS-CoV-2-associated complications. Obviously, it can also be used for preclinical trials of antiviral drugs and new vaccines.
Graphic abstract
Keywords: COVID-19, ACE2, Transgenes, SARS-CoV-2, Tamoxifen, Cre-recombinase
Introduction
Coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) remains one of the most critical challenges for the medical and scientific communities. Since the first cases of infected patients in Wuhan (Zhu et al., 2020), our understanding of COVID-19 pathogenesis has been repeatedly transformed by the results of clinical and molecular research. Most data were obtained from clinical observational studies, such as cohort and case–control studies. In silico (Shang et al. 2020; Meirson et al. 2020) and in vitro (Hoffmann et al. 2020; Suzuki 2020) research also clarified the mechanisms of interactions between the virus and human cells. Nevertheless, opportunities to perform experiments to reveal the details of COVID-19 pathogenesis are limited by the absence of available animal test systems.
Recently, SARS-CoV-2 was shown to effectively replicate in several species, including nonhuman primates, ferrets, dogs, and cats (Lakdawala et al. 2020). Macaques infected with the novel virus were used for testing an inactivated vaccine (Gao et al. 2020). A study in ferrets revealed unexpected details regarding the anti-SARS-CoV-2 immune response (Blanco-Melo et al. 2020). However, there are unfortunately no naturally SARS-CoV-2-sensitive species among the routinely used laboratory animals. Obviously, the most convenient, available, and popular laboratory test system requiring no special housing conditions is mice. As wild-type mice support transient coronavirus infection without clinical signs of the disease (Subbarao et al. 2004), the use of genetically modified strains expressing human angiotensin-converting enzyme 2 (hACE2) (McCray et al. 2007; Tseng et al. 2007; Menachery et al. 2016; Yang et al. 2007; Bao et al. 2020; Soldatov et al. 2020; Masemann et al. 2020), the entry receptor for SARS-CoV-2 (Lu et al. 2020), was proposed. Another proposed option was to deliver exogenous hACE2 to the alveolar epithelium via intranasal administration of replication-deficient adenovirus Ad5-hACE2 in wild-type mice (Sun et al. 2020; Hassan et al. 2020). Moreover, SARS-CoV-2 itself was genetically modified to effectively interact with murine ACE2 receptors, making any mouse suitable for COVID-19 studies without humanization (Dinnon et al. 2020; Gu et al. 2020).
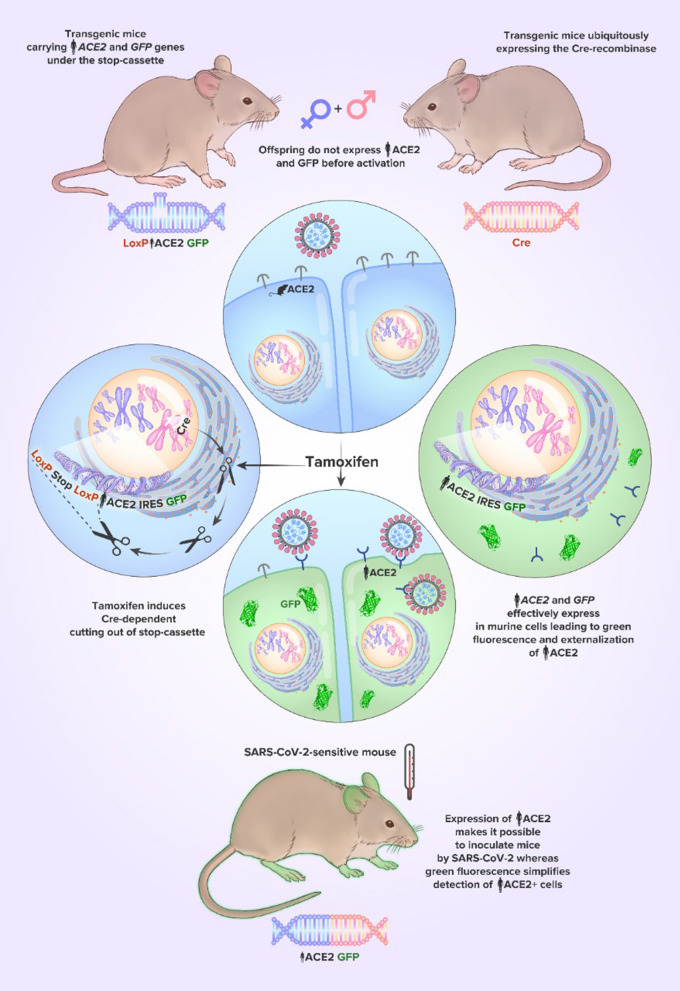
Here, we report a novel SARS-CoV-2-sensitive transgenic mouse strain, TgCAGLoxPStopACE2GFP (hereinafter referred to as hACE2(LoxP-Stop)), carrying hACE2 and green fluorescent protein (GFP) transgenes under the floxed STOP cassette. The basic hallmark of this strain is the possibility of temporal and spatial control of transgene activation, allowing adjustment of the hACE2 gene expression profile by breeding with different Cre strains. In our study, we crossed hACE2(LoxP-Stop) mice with Ubi-Cre/ERT2 mice, in which Cre/ERT2 recombinase is expressed ubiquitously but is able to excise the STOP cassette only after administration of tamoxifen (Cre/ERT2 needs tamoxifen to be translocated into the nucleus). Their offspring successfully expressed hACE2 and GFP after tamoxifen induction, proving that the system works.
Materials and methods
Design and manufacturing of the transgene DNA construct
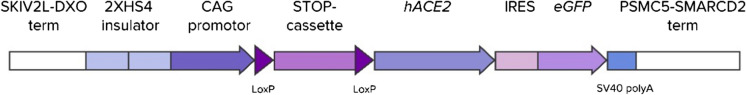
The DNA construct consisted of two basic parts: the pKB1 vector and the hACE2 ORF. pKB1, an ampicillin-resistant vector, was constructed for cloning genes intended for Cre-dependent expression. It contains insulators and terminators to protect the transgene from the position effect (Miyazaki et al. 1989; Chung et al. 1993; Deykin et al. 2019), the transcriptional unit under the control of the CAG promoter, and the STOP cassette (Lakso et al. 1992). The transcriptional unit consisted of multiple cloning sites for cloning of the transgene, the IRES element of the encephalomyocarditis virus in frame with the GFP gene and the SV40polyA signal. The pKB1 vector was constructed on the basis of the NIF vector (Zvartsev et al. 2019): we inserted a multiple cloning site (AgeI, EcoRV, MluI) between the STOP cassette and the IRES. Transgene expression in pKB1 is supposed to be activated only after excision of the STOP cassette by Cre recombinase.
Total RNA for hACE2 cloning was extracted from human kidney biopsy material. The tissue was homogenized with a Precellys 24 homogenizer, and RNA was extracted with the QIAGEN RNeasy Mini Kit. cDNA was synthesized with reverse transcriptase Superscript II (Invitrogen) and d(T)16 oligo. The hACE2 gene (2400 bp) was amplified with Q5 polymerase (NEB) using primers P15 and P16 (all primer sequences are presented in the Appendix Table 1). The amplified fragment was digested with restriction enzymes BshTI and MluI (Thermo Fisher Scientific) and inserted into the pKB1 vector digested with the same enzymes (Fig. 1).
Table 1.
Sequences of primers
| Code | Sequence 5′→3′ |
|---|---|
| P1 | AAAATTGTGTACCTTTAGCTTTTTA |
| P2 | CCTAGGAATGCTCGTCAAGA |
| P3 | CTTATGTGCACAAAGGTGAC |
| P4 | TGTGGCTGCAGAAAGTGA |
| P5 | GAGCGTTGGGCTTACCTCAC |
| P6 | GACTGCGGGTCGGCATGA |
| P7 | CCACCTCGATGACCTGCCCGG |
| P8 | CTGATGCCTCTGCTCCCCTTCC |
| P9 | GACTTCAAGACCGACCTGCG |
| P10 | TTGGGCATGATGGTGACACG |
| P11 | GCAGTACAGCCCCAAAATGG |
| P12 | CCTTTTCACCAGCAAGCT |
| P13 | GTTAGATCTGCTGCCACCGT |
| P14 | AGGTGGCAAGTGGTATTCCG |
| P15 | ATTAACCGGTATGTCAAGCTCTTCCTGGC |
| P16 | ATTAACGCGTCTAAAAGGAGGTCTGAACATCAT |
| P17 | GACGTCACCCGTTCTGTTG |
| P18 | AGGCAAATTTTGGTGTACGG |
| P19 | CTAGGCCACAGAATTGAAAGATCT |
| P20 | GTAGGTGGAAATTCTAGCATCATCC |
| P21 | CGACGTAAACGGCCACAAGT |
| P22 | GGCGGACTTGAAGAAGTCGT |
| P23 | GCGAGTCCATGTCACTCAGG |
| P24 | GTGTTGCCCTTTGGAGCTTG |
Fig. 1.
pKB1-hACE2 linear construct intended for transgenesis
The circular plasmid DNA was then digested with PvuI and gel purified; a 13.5 kbp linear fragment without the bacterial propagation elements was extracted with the QIAquick Gel Extraction Kit (Qiagen) and dissolved in TE buffer (Zvezdova et al. 2010) at a concentration of 1 ng/μl.
Animals, pronuclear microinjections, and embryo transfer
To generate primary transgenic animals, we used (CBAxC57BL/6)F1 zygotes obtained from 3-week-old CBA and C57BL/6 breeders (Stolbovaya breeding station, Russian Federation). The transgene DNA construct was introduced by pronuclear microinjections as described earlier (Zvezdova et al. 2010). Surviving zygotes were cultivated in M16 medium at 37 °C and 6% CO2 for 24 h and then transplanted into the oviducts of foster mothers at the 2-cell stage as previously described (Silaeva et al. 2018).
The B6. Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/1 J, also known as Ubi-Cre, mouse strain was purchased from the Jackson Laboratory (Goodwin et al. 2019). In this strain, chimeric Cre recombinase is expressed under the control of the UBC promoter. Chimeric Cre recombinase consists of a Cre recombinase domain and a domain representing a mutant form of the estrogen receptor, which lacks the ability to bind natural 17β-estradiol and binds only synthetic tamoxifen. Without tamoxifen, the protein is restricted to the cytoplasm, and only in the presence of tamoxifen translocates to the nucleus and performs recombination. Ubi-Cre mice can be maintained only in hemizygotes.
Animals were kept in the IGB RAN vivarium with artificial lighting (12 h/12 h mode) at a temperature of 21–23 °C and humidity of 38–50%; mice had free access to food and water.
Tamoxifen treatment
Tamoxifen (Hexal) was administered by gavage at a dose of 1 mg/100 g once a day for 20 days (2% suspension formulations).
Genotyping
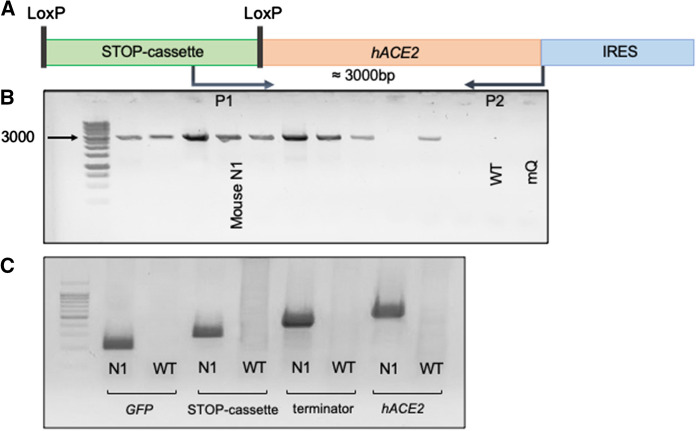
DNA samples were extracted from tail tissues samples. The first step of F0 genotyping was performed using the primer pair P1 and P2, specific to the 3′-region of the STOP cassette and the 5′-region of IRES, respectively (the 3000 bp PCR product band, Fig. 2). Advanced genotyping of F0 combined detection of the STOP cassette (P13, P14), the hACE2 ORF (P15, P16), eGFP (P21, P22), and the terminator (P23, P24).
Fig. 2.
F0 genotyping. a Primer positioning for primary genotyping; b F0 primary genotyping. Mouse N1 involved in further experiments is indicated; c Advanced genotyping of mouse N1. The presence of hACE2, GFP, the STOP cassette, and the terminator was confirmed
F1 genotyping was performed by detection of hACE2 and eGFP ORF presence as described above and Ubi-Cre presence according to the protocol recommended by the Jackson Laboratory using P17, P18, P19, and P20 primers. All Ubi-Cre probes had a 324 bp band as the internal positive control, and transgenic Ubi-Cre probes had a 475 bp band.
Copy number measurement
The transgene copy number was determined using logarithmic interpolation based on Ct for the transgene and reference genes with known copy numbers. We chose murine Hprt (1 copy per genome in males, 2 in females, primers P5 and P6), Hba1 (4 copies per genome, primers P7 and P8), and H3c7 (18 copies per genome, primers P9 and P10) genes as the reference for comparison. For qPCR, we used SYBR Green mix with HS Taq polymerase (Evrogen, Russia). We used primers P3 and P4 for hACE2 copy number measurement. In experiments assessing the Cre recombinase effectivity, we also measured the STOP cassette copy number with primers P13 and P14.
qPCR and gene expression measurement
Blood for evaluation of hACE2 expression was collected from the retro-orbital venous sinus under anesthesia induced by Zoletil (2 mg/100 g) + Xylazin (2 mg/100 g). One hundred microliters of blood samples were mixed with 1 ml of ammonium chloride-potassium RBC lysis buffer, incubated for 5 min at 25 °C and centrifuged for 5′ at 300 g at room temperature. The observed cell pellet was washed with PBS twice and used for downstream applications.
gDNA-free total RNA was extracted from blood cells with the RNeasy Plus Mini Kit (Qiagen). cDNA was synthesized with iScript™ Reverse Transcription Supermix (Bio-Rad), and real-time PCR was performed using SYBR GREEN mix (Evrogen) and primers P3, P4, P11, and P12. Real-time PCR with extracted RNA instead of cDNA was performed to prove the absence of genomic DNA contamination. The results were analyzed with CFX Manager Software (Bio-Rad).
Western blot
Peripheral blood mononuclear cells (PBMCs) of F1 mouse hACE2 (LoxP-Cre) were extracted as previously described (Silaeva et al. 2013). After RBC lysis, PBMCs were lysed in 200 μl of RIPA buffer (1 ml RIPA: 150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS (sodium dodecyl sulfate), 50 mM Tris, pH 8.0; 2 μl PIC, 20 μl PMSF). The protein concentration was measured by the Bradford assay, and a calibration curve was built using BSA serial dilutions. The samples (80 mg of protein each) were loaded into 8% PAAG. Electrophoresis was performed in SDS buffer at 120 V for 1.5 h and the proteins were then transferred to a nitrocellulose membrane (0.2 μm) at 250 mA for 1 h in transfer buffer with 20% MeOH. After the transfer, the membrane was incubated in 5% nonfat dried milk dissolved in TBS. The primary anti-hACE2 antibody PAB886Hu01 (Cloud-Clone Corp., USA-China) was diluted 1:1500 in 1% BSA in TBS buffer, and the membrane was stained for 18 h at 4 °C. The membrane was then washed in TBS buffer and incubated with secondary anti-rabbit antibodies (anti-rabbit IgG, HRP-linked antibody # 7074, Cell Signaling) diluted 1:1000 in 5% nonfat dried milk dissolved in TBS for 1 h. After washing in TBS buffer, chemiluminescent labels were activated with ECL and visualized in iBright.
FACS analysis
Extracted PBMCs were preincubated with Fc block (clone 2.4G2, BD Pharmingen, Franklin Lakes, NJ) (10 min, 4 °C), incubated with polyclonal anti-hACE2 antibodies (Cloud-clone Corp., Product No. PAB886Hu01) (40 min, 4 °C) and then stained with secondary fluorescently labeled antibodies (Invitrogen, Catalog # A-11010) (40 min, 4 °C). The analysis was performed on a BD FACSCanto II flow cytometer (BD Bioscience) using FACSDiva 6.0 software (BD Bioscience). Dead cells were excluded from the analysis based on the parameters of the forward and side scatter and staining with propidium iodide (BD Bioscience). At least 104 events per sample were collected to characterize the peripheral leukocyte populations. Data were processed using FlowJo 7.6 software (TreeStar Inc., Ashland, USA).
Statistical analysis
Statistical processing was not applied due to the limited number of animals (only one in each group).
Results
Generation of primary transgenic mice
A total of 1511 zygotes were microinjected and transplanted into 138 recipients. Twenty-four recipients gave birth to 48 pups (F0). Nine pups carried the transgene, and 4 of them gave F1 offspring.
Being aware of mosaic patterns of transgene insertion, we nevertheless evaluated the transgene copy number in the F0 generation. Seven of 9 transgenes had copy numbers, significantly exceeding 1—in the range between 4 and 27 copies per genome. This result indicates a high probability of multimers of the construct being inserted into the genome (Chen et al. 2001; Smirnov et al. 2020). Moreover, it is possible that in some mice, several independent insertions occurred. We expect that independently inserted sequences will be parted by chromosomal crossover in the next generations, whereas multimers will not be.
Breeding with Cre-mice and tamoxifen-induced activation
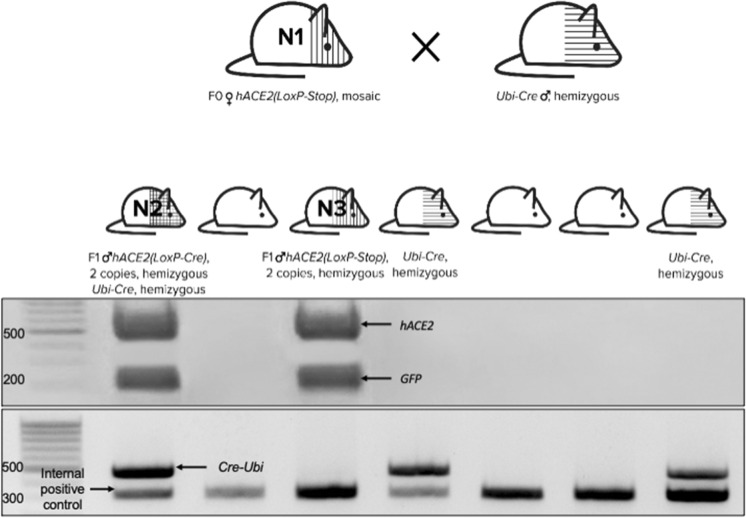
As an activator line, we used B6. Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/1 J, also known as Ubi-Cre, from the Jackson Laboratory (Goodwin et al., 2019). We crossed hACE2(LoxP-STOP) F0 transgenic female # 1190, further referred to as mouse N1, with 8.5 copies of the transgene per genome with the Ubi-Cre transgenic male #1720 and obtained a litter consisting of 7 pups. Two of them (# 2344, further referred to as mouse N2, and # 2346, further referred to as mouse N3) were hACE2 (LoxP-STOP)-positive, both with 2 copies of the transgene; 3 of them (mouse N2, # 2347, and # 2350) were Ubi-Cre-positive, and only mouse N2 was positive for both hACE2 (LoxP-STOP) and Ubi-Cre (further referred to as hACE2 (LoxP-Cre)) (Fig. 3). Then, we activated Cre recombinase in this mouse N2 with tamoxifen, and GFP fluorescence was observed in its skin via a UV lamp in just one week.
Fig. 3.
Scheme of transgenic mouse breeding and F1 genotyping. Seven pups characterized by hACE2 (≈500 bp band) and GFP (≈200 bp band) on the upper gel and by Cre-Ubi presence (2 bands in the transgenic mice, 1 band in wild-type mice) on the lower gel
Next, we measured the hACE2 (P3 and P4 primers) mRNA levels relative to the mHprt housekeeping gene (P11 and P12) in PBMCs by real-time PCR. We measured the hACE2 expression level in the mouse N2 hACE2(LoxP-Cre) after tamoxifen activation, in the mouse N3 hACE2(LoxP-STOP) with the transgene copy number, which was identical to that in mouse N2, and in a wild-type mouse (Fig. 4a; the hACE2 expression level after activation was assigned a value of 1). In mouse N2, hACE2 expression was much higher than that in the two other mice. hACE2 expression was not detected in wt mice. Transgenic mouse N3 without activation showed an expression level that was approximately 3% of that of the activated mouse N2. This probably indicated low-level leakage of the STOP cassette. Transgenic gDNA contamination was not the cause because the PCR product was not detectable without reverse transcription (data not shown). In mouse N2, we measured the copy numbers of hACE2 and the STOP cassette before and after activation. For this, we performed real-time PCR with the P3 and P4 primers for hACE2 and with the P13 and P14 primers for the STOP cassette using genomic DNA extracted before and after activation as the templates. We speculated that recombination would occur not only between adjacent LoxP sites but also between random sites in the case of multimeric insertion. However, we did not observe such recombination in our experiment. After tamoxifen activation, the STOP cassette copy number decreased by 30%, whereas the hACE2 copy number remained exactly the same (Fig. 4b). This experiment shows that tamoxifen induction works properly and effectively, and no transgene excision takes place.
Fig. 4.
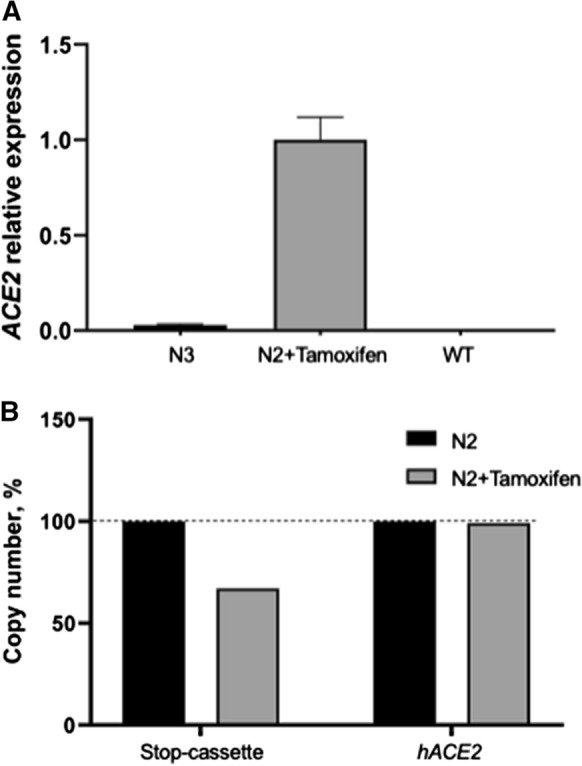
Effect of tamoxifen administration on transgene activation. a hACE2 expression was found in only hACE2(LoxP-Cre) mice (N2) after tamoxifen activation. B) The Stop-cassette copy number in hACE2(LoxP-Cre) mice (N2) after tamoxifen activation was decreased by 30%, whereas the hACE2 copy number did not change
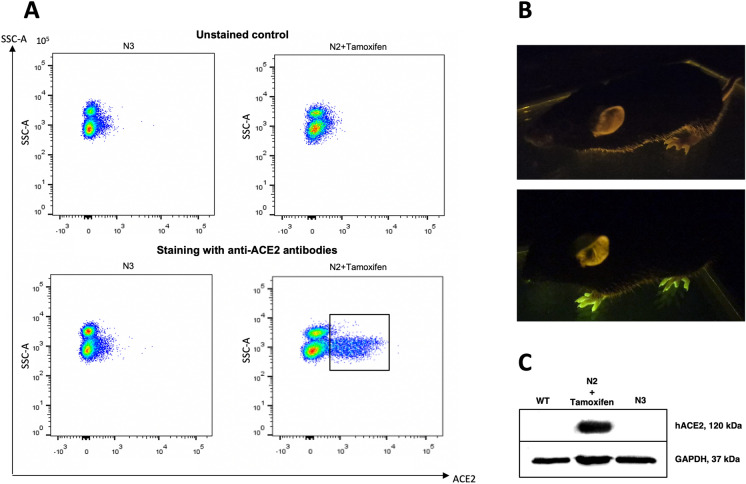
To explore transgenic hACE2 at the protein level, we performed Western blot analysis with polyclonal anti-hACE2 rabbit antibodies (Cloud-Clone Corp., USA-China). We detected a band of approximately 120 kDa corresponding to the glycosylated hACE2 form only in the blood of the activated mouse N2. This band was not detected in the wild-type mice or in the nonactivated transgenic mouse N3 (Fig. 5). Glycosylation is an important process for membrane proteins such as ACE2. The weight of the detected band affirmed proper posttranslational modifications because the unglycosylated protein has a molecular weight of approximately 85–90 kDa (Warner 2004).
Fig. 5.
Detection of hACE2 and GFP. a hACE2 presence on the surface of PBMCs. a Activated transgenic mouse N2 hACE2(LoxP-Cre) (right lower panel) compared to the nonactivated transgenic mouse N3 hACE2(LoxP-Stop) (left lower panel). Unstained controls are represented in the upper panel. b The N2 mouse skin in the UV chamber before (upper) and after (lower) tamoxifen induction. c Western blot analysis of hACE2 expression in PBMCs. Glycosylated 120 kDa hACE2 in the upper panel and 37 kDa GAPDH in the lower panel. A WT mouse is shown on the left, an activated transgenic mouse is shown in the middle, a nonactivated transgenic mouse is shown on the right
Finally, we measured hACE2 expression by flow cytometry with the same primary antibodies. We observed that 16% of blood mononuclei in mouse N2 were stained positively for hACE2, whereas no hACE2 + cells were detected in the nonactivated mouse N3 or in the wt mouse. As hACE2 was not detected in mouse N3 (Fig. 5) by Western blot or flow cytometry, we can conclude that there was no STOP cassette leakage at the protein level. The difference between mRNA and protein levels can be explained by the SV40polyA sequence in the 3′ part of the STOP cassette. Even if low-rate transcription occurs from the CAG promoter, translation of this transcript terminates at SV40polyA before the ribosome reaches the hACE2 ORF. The fact that STOP cassette leakage was observed at only the mRNA level and not at the protein level supports our claim that our animals satisfy the requirement of tamoxifen-induced expression. As we stained the cells without fixation and permeabilization, we presume that the antibodies reacted with only hACE2 translocated to the cell surface. This is important because only translocated hACE2 can serve as an entry point for the coronavirus, and there are doubts regarding whether transgenic hACE2 can be properly processed in different types of cells (Fig. 5).
Discussion
In December 2019, Wuhan, the capital of Hubei Province in China, became the center of the outbreak of pneumonia with an unknown etiology. By January 7, 2020, Chinese scientists identified the causative infectious agent as a novel strain of coronavirus and named it SARS-CoV-2. SARS-CoV-2 causes COVID-19, a very complex disease with a wide spectrum of symptoms. At first, SARS-CoV-2 invades the epithelium of the upper and lower airways, but in the case of a severe course, it reaches the gas exchange units of the lungs and infects alveolar type II cells (Mason 2020).
Many reports show that immune cells (Mortaz et al. 2020; Liu et al. 2020; Ruan et al. 2020; Pontelli et al. 2020), vascular endothelium (Libby et al. 2020, Wichmann et al. 2020; Al-Ani et al. 2020; Varga et al. 2020; Ackermann et al. 2020; Monteil et al. 2020) and the nervous system (Iadecola et al. 2020; Song et al. 2020, preprint) can also be distinguished as important targets of SARS-CoV-2. Moreover, the list of potential targets for COVID-19 is growing. There are reports of the involvement of other organs with high expression of ACE2, such as the intestines, kidneys, and heart (Zaim et al. 2020). As ACE2 is highly expressed in tissues of the male reproductive system, there are concerns that SARS-CoV-2 can cause infertility in men (Sun 2020; Dutta et al. 2020). This suggestion is partially confirmed by cases of orchitis following the outbreak of SARS-CoV in 2002 (Xu et al. 2006) and sex hormone imbalance in male patients with COVID-19 (Ma et al. 2020). Therefore, COVID-19 has a very complex and multifaceted pathogenesis. The revelation of its pathways is an essential way in the search for effective approaches to therapy.
Herein, we describe novel transgenic mice with Cre-dependent coexpression of hACE2 and GFP. This is the first transgenic strain allowing spatial and temporal control of hACE2 expression. Previous lines constitutively expressed the hACE2 transgene under the control of nonspecific promoters K18 (McCray et al. 2007) and CAG (Tseng et al. 2007) and a specific murine Ace2 promoter (Yang et al. 2007). Although existing strains can serve for preclinical trials of anti-SARS-CoV-2 agents and research on some details of virus invasion, they are not appropriate to reveal the contribution of different tissues to the pathogenesis of SARS-CoV-2 infection. The present hACE2(LoxP-Stop) mice were designed mostly to study the role of different tissues in the development of COVID-19 symptoms and complications. For instance, specific activation of hACE2 in endothelial, immune, or alveolar cells can help to reveal the primary causative tissue, the involvement of which leads to acute respiratory distress syndrome in COVID-19. Another option, for example, is to study the fertility of SARS-CoV-2-infected male mice with testicular-specific activation of hACE2. In this way, fertility will depend only on testicular but not on systemic and behavioral changes caused by COVID-19.
hACE2(LoxP-Stop) mice must be crossed with Cre- expression strains to initiate the transcription of transgenes. In brief, Cre strains are available transgenic animals expressing Cre recombinase, an enzyme that catalyzes site-specific recombination between two LoxP sites and, as a result, deletion of the LoxP-limited sequence in the proper orientation. Thus, if Cre recombinase is expressed, it excises the STOP cassette, resulting in the expression of hACE2 and GFP. In our study, we examined the F1 generation obtained after crossing the hACE2(LoxP-Stop) F0 mouse with the Ubi-Cre mouse. The human ubiquitin C (UBC) promoter that drives Cre expression in this strain provides a relatively consistent expression level across different cell types. One of the offspring carried both hACE2GFP and Cre alleles. In this mouse, the expression of hACE2 and GFP was then successfully triggered by tamoxifen administration, at least in blood mononuclei and skin.
Choosing Ubi-Cre/ERT2 as the first strain to excise the floxed STOP cassette, we were guided by considerations of biosafety because tamoxifen-dependent activation is a two-level defense against SARS-CoV-2 infection, simplifying work with animals. Before activation, the mice were epidemiologically safe for laboratory staff, and workers were safe for the mice. Thus, it minimizes all the risks related to undesired infection of the mice by SARS-CoV-2 before placing them in specific conditions of a virological laboratory. However, other Cre activators can be chosen according to different research aims in further studies.
In this study, after activation, we detected STOP cassette excision, hACE2 mRNA presence, and hACE2 protein assembly. Western blot analysis found hACE2 with a molecular weight of approximately 120 kDa, indicating proper protein glycosylation. Using FACS, we confirmed that hACE2 is located on the cell surface, which means that transgenic protein successfully underwent surface translocation and can interact with SARS-CoV-2. Interestingly, our dosage scheme of tamoxifen administration led to relatively modest activation of the transgene, but it can be increased in further experiments if required (Donocoff et al. 2020). Thus, tamoxifen-dependent activation is an additional option to adjust transgene expression and, therefore, SARS-CoV-2 sensitivity.
Developing our strategy for the generation of mice with Cre-dependent activation of hACE2, we were guided by several points. First, we decided to coexpress hACE2 with GFP because we think it simplifies visualization procedures. GFP fluorescence confirms transgene activation, allowing visualization of hACE2-expressing tissues without the use of complicated and expensive immunostaining protocols. In our study, we demonstrated that activation of hACE2/GFP expression can be confirmed just by fluorescence in transgenic mouse skin. Notably, some authors notice that floxed transgenes can be expressed at a small level even before Cre-dependent activation (Van Hove et al., 2019), so coexpression with GFP also allows immediate detection of this leak. In our study, we did not perform the analysis of hACE2 expression before tamoxifen administration because we tried to minimize all the invasive procedures with valuable N2 transgenic mice. Nevertheless, we did not detect any green fluorescence of the skin of the nonactivated transgene in the UV chamber.
Finally, recognizing that our strain is a hybrid of CBA and C57Bl/6 J, we see that it must be returned to an inbred background for more representative research results in further studies. The most appropriate background to return to is C57Bl/6 J because almost all of the activator Cre lines are contained on this background.
Conclusions
SARS-CoV-2-sensitive murine models are an important component of studies on COVID-19. In our laboratory, a novel mouse strain that conditionally coexpresses hACE2 and GFP was obtained. We believe that our strain, hACE2(LoxP-Stop), can serve as a useful tool for studying the roles of different tissues in COVID-19 pathogenesis. Moreover, Cre-dependent expression of hACE2 makes hACE2(LoxP-Stop) mice convenient for housing because before activation, they cannot serve as a reservoir for SARS-CoV-2 and do not require specific biosafety conditions. Finally, coexpression with GFP simplifies the visualization of hACE2-expressing tissues. We are ready to transfer hACE2(LoxP-STOP) mice to any research group of interest. We believe that our hACE2(LoxP-STOP) strain has a number of unique properties that can be useful in preclinical trials and especially useful for studying COVID-19 pathogenesis.
Acknowledgements
Authors are thankful to Alexandra Proskura (Institute for Urology and Reproductive Health, Sechenov University, Russia) for human kidney samples, to dr. Dimitry Mazurov (Institute of Gene Biology, Russian academy of sciences, Russia) for providing antibodies, to Ekaterina Varlamova (Institute of Gene Biology, Russian academy of sciences, Russia) for laboratory support, to researchers of the State Research Center of Virology and Biotechnology VECTOR for critical discussion. This work was supported by the grant 075-15-2019-1661 from the Ministry of Science and Higher Education of the Russian Federation (all animal experiments) and Institute of functional genomics of the Lomonosov Moscow State University state funding (in part related to the preparation of hACE2 cDNA). Co-workers of the Laboratory of Genome Editing for veterinary and biomedicine were supported by the Belgorod Scientific and Educational Center "Innovative Solutions in the Agro-Industrial Complex". The study was performed using the equipment of IGB RAS facilities supported by the Ministry of Science and Higher Education of the Russian Federation.
Appendix
See Table 1
Author contributions
AD: main idea, design of the experiments, embryo transfer, obtaining the results, writing the article. DK: animal surgery, animal care, tamoxifen administration, pronuclear microinjections, tissue sampling. AB: molecular biology, DNA transgenic construcrion development, writing the article. EK: animal surgery, animal care, animal breeding, animal handling, obstetrics procedures, veterinary, blood collection. MK: molecular biology, DNA transgenic construcrion development. AK: FASC, writing the article. YuS: FASC. PS: cDNA cloning. LI: bioinformatics, mathematical proceeding. SV: main idea, design of the experiments, graphical design, writing the article.
Declarations
Conflict of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Ethical approval
Human biomaterials were obtained in the process of medical intervention under the supervision of the Local Ethics Committee of Sechenov (Medical) University. All animal experiments were carried out in accordance with the rules of humane treatment and were approved and controlled by the IGB RAS Bioethics Commission (the conclusion of the Bioethical Commission # 06-01/02-1 dated 02/03/2020). The animal experiments did not pose any biological hazards and did not require a biosafety facility.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
AlexandraV. Bruter, DianaS. Korshunova and MarinaV. Kubekina have contributed equally to this work
Contributor Information
Alexandra V. Bruter, Email: bruter@genebiology.ru
Diana S. Korshunova, Email: korshunova@genebiology.ru
Marina V. Kubekina, Email: kubekina@genebiology.ru
Petr V. Sergiev, Email: petya@belozersky.msu.ru
Anastasiia A. Kalinina, Email: aakalinina89@gmail.com
Leonid A. Ilchuk, Email: lechuk12@gmail.com
Yuliya Yu. Silaeva, Email: silaeva@genebiology.ru
Eugenii N. Korshunov, Email: pharmsoldatov@gmail.com
Vladislav O. Soldatov, Email: korshunov@genebiology.ru
Alexey V. Deykin, Email: deikin@igb.ac.ru
References
- Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A, Li WW, Li VW, Mentzer SJ, Jonigk D. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432.PMID:32437596;PMCID:PMC7412750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ani F, Chehade S, Lazo-Langner A. Thrombosis risk associated with COVID-19 infection. A Scoping Rev Thromb Res. 2020;192:152–160. doi: 10.1016/j.thromres.2020.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Deng W, Huang B, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Yant SR, He CY, Meuse L, Shen S, Linear KMA, DN, As concatemerize in vivo and result in sustained transgene expression in mouse liver. MolTher. 2001;3:403–410. doi: 10.1006/mthe.2001.0278. [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G. A 5' element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74(3):505–514. doi: 10.1016/0092-8674(93)80052-G. [DOI] [PubMed] [Google Scholar]
- Deykin A, Tikhonov M, Kalmykov V, Korobko I, Georgiev P, Maksimenko O. Transcription termination sequences support the expression of transgene product secreted with milk. Transgenic Res. 2019;28(3–4):401–410. doi: 10.1007/s11248-019-00122-9. [DOI] [PubMed] [Google Scholar]
- Dinnon KH, Leist SR, Schäfer A, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donocoff RS, Teteloshvili N, Chung H, Shoulson R, Creusot RJ. Optimization of tamoxifen-induced Cre activity and its effect on immune cell populations. Sci Rep. 2020;10(1):15244. doi: 10.1038/s41598-020-72179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Sengupta P (2020) SARS-CoV-2 and Male Infertility: Possible Multifaceted Pathology. Reprod Sci 1–4. doi:10.1007/s43032-020-00261-z [DOI] [PMC free article] [PubMed]
- Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, Li Y, Zhu L, Wang N, Lv Z, Gao H, Ge X, Kan B, Hu Y, Liu J, Cai F, Jiang D, Yin Y, Qin C, Li J, Gong X, Lou X, Shi W, Wu D, Zhang H, Zhu L, Deng W, Li Y, Lu J, Li C, Wang X, Yin W, Zhang Y, Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin LO, Splinter E, Davis TL, Urban R, He H, Braun RE, Chesler EJ, Kumar V, van Min M, Ndukum J, Philip VM, Reinholdt LG, Svenson K, White JK, Sasner M, Lutz C, Murray SA. Large-scale discovery of mouse transgenic integration sites reveals frequent structural variation and insertional mutagenesis. Genome Res. 2019;29(3):494–505. doi: 10.1101/gr.233866.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AO, et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadecola C, Anrather J, Kamel H. Effects of COVID-19 on the nervous system. Cell. 2020;183(1):16–27.e1. doi: 10.1016/j.cell.2020.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala SS, Menachery VD. The search for a COVID-19 animal model. Science. 2020;368(6494):942–943. doi: 10.1126/science.abc6141. [DOI] [PubMed] [Google Scholar]
- Lakso M, Sauer B, Mosinger B, Jr, et al. Targeted oncogene activation by site-specific recombination in transgenic mice. ProcNatlAcadSci USA. 1992;89(14):6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby P, Lüscher T. COVID-19 is, in the end, an endothelial disease. Eur Heart J. 2020;41(32):3038–3044. doi: 10.1093/eurheartj/ehaa623.PMID:32882706;PMCID:PMC7470753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Xie W, Li D, Shi L, Mao Y, Xiong Y, et al (2020) Effect of SARS-CoV-2 infection upon male gonadal function: a single center-based study. medRxiv. 10.1101/2020.03.21.20037267
- Masemann D, Ludwig S, Boergeling Y. Advances in transgenic mouse models to study infections by human pathogenic viruses. Int J MolSci. 2020;21:9289. doi: 10.3390/ijms21239289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RJ. Pathogenesis of COVID-19 from a cell biology perspective. EurRespir J. 2020;55(4):2000607. doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray PB, Jr, Pewe L, Wohlford-Lenane C, et al. J Virol. 2007;81(2):813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirson T, Bomze D, Markel G. Structural basis of SARS-CoV-2 spike protein induced by ACE2. Bioinformatics. 2020;20:744. doi: 10.1093/bioinformatics/btaa744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery VD, et al. SARS-like WIV1-CoV poised for human emergence. ProcNatlAcadSci. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki J, Takaki S, Araki K, Tashiro F, Tominaga A, Takatsu K, Yamamura K. Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene. 1989;79(2):269–277. doi: 10.1016/0378-1119(89)90209-6. [DOI] [PubMed] [Google Scholar]
- Monteil VKH, Prado P, Hagelkrüys A, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortaz E, Tabarsi P, Varahram M, Folkerts G, Adcock IM. The immune response and immunopathology of COVID-19. Front Immunol. 2020;11:2037. doi: 10.3389/fimmu.2020.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontelli MC, et al. Infection of human lymphomononuclear cells by SARS-CoV-2. Biorxiv. 2020 doi: 10.1101/2020.07.28.225912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Yan K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intens Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silaeva YY, Kalinina AA, Vagida MS, et al. Decrease in pool of T lymphocytes with surface phenotypes of effector and central memory cells under Influence of TCR transgenic β-chain expression. Biochem Moscow. 2013;78:549–559. doi: 10.1134/S0006297913050143. [DOI] [PubMed] [Google Scholar]
- Silaeva YY, Kirikovich YK, Skuratovskaya LN, et al. Optimal number of embryos for transplantation in obtaining genetic-modified mice and goats. Russ J Dev Biol. 2018;49:356–361. doi: 10.1134/S106236041806005X. [DOI] [Google Scholar]
- Smirnov A, Fishman V, Yunusova A, et al. DNA barcoding reveals that injected transgenes are predominantly processed by homologous recombination in mouse zygote. Nucleic Acids Res. 2020;48(2):719–735. doi: 10.1093/nar/gkz1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov VO, Kubekina MV, Silaeva YuYu, Bruter AV, Deykin AV. On the way from SARS-CoV-sensitive mice to murine COVID-19 model. Res Results Pharmacol. 2020;6(2):1–7. doi: 10.3897/rrpharmacology.6.53633. [DOI] [Google Scholar]
- Song E, Zhang C, Israelow B, Lu P, Weizman O-E, Liu F, Dai Y, Szigeti-Buck K, Yasumoto Y, Wang G, et al. Neuroinvasive potential of SARS-CoV-2 revealed in a human brain organoid model. BioRxiv. 2020 doi: 10.1101/2020.06.25.169946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K, McAuliffe J, Vogel L, et al. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78(7):3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. The hypothesis that SARS-CoV-2 affects male reproductive ability by regulating autophagy. Med Hypotheses. 2020;143:110083. doi: 10.1016/j.mehy.2020.110083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182:734–743. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T. Generation of human bronchial organoids for SARS-CoV-2 research. BioRxiv. 2020 doi: 10.1101/2020.05.25.115600. [DOI] [Google Scholar]
- Tseng CT, et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove H, Antunes ARP, De Vlaminck K, Scheyltjens I, Van Ginderachter JA, Movahedi K. Identifying the variables that drive tamoxifen-independent CreERT2 recombination: implications for microglial fate mapping and gene deletions. Eur J Immunol. 2020;50(3):459–463. doi: 10.1002/eji.201948162. [DOI] [PubMed] [Google Scholar]
- Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka F, Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5.PMID:32325026;PMCID:PMC7172722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner FJ, Smith AI, Hooper NM, Turner AJ. Angiotensin-converting enzyme-2: a molecular and cellular perspective. Cell Mol Life Sci. 2004;61(21):2704–2713. doi: 10.1007/s00018-004-4240-7.PMID:15549171;PMCID:PMC7079784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann D, Sperhake JP, Lütgehetmann M, et al. autopsy findings and venous thromboembolism in patients with COVID-19: A prospective cohort study. Ann Intern Med. 2020;173(4):268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Qi L, Chi X, Yang J, Wei X, Gong E, Peh S, Gu J. Orchitis: a complication of severe acute respiratory syndrome (SARS) BiolReprod. 2006;74(2):410–416. doi: 10.1095/biolreprod.105.044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XH, et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp Med. 2007;57:450–459. [PubMed] [Google Scholar]
- Zaim S, Chong JH, Sankaranarayanan V, Harky A. COVID-19 and multiorgan response. CurrProblCardiol. 2020;45(8):100618. doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2019;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvartsev RV, Korshunova DS, Gorshkova EA, Nosenko MA, Korneev KV, Maksimenko OG, Korobko IV, Kuprash DV, Drutskaya MS, Nedospasov SA, Deikin AV. Neonatal lethality and inflammatory phenotype of the new transgenic mice with overexpression of human interleukin-6 in myeloid cells. Dokl Biochem Biophys. 2019;483(1):344–347. doi: 10.1134/S1607672918060157. [DOI] [PubMed] [Google Scholar]
- Zvezdova ES, Silaeva YY, Vagida MS, et al. Generation of transgenic animals expressing the α and β chains of the autoreactive T-cell receptor. MolBiol. 2010;44:277–286. doi: 10.1134/S0026893310020135. [DOI] [PubMed] [Google Scholar]