ABSTRACT
Fever is one of the most common reasons for pediatric consultation in Africa. Malaria incidence has now dropped considerably, yet etiologies of non-malarial febrile diseases are poorly documented. This pilot study aimed to 1) identify pathogens potentially associated with non-malarial fever in children younger than 10 years in the suburbs of Dakar and 2) describe the epidemiological characteristics of these patients. During the study period, all eligible children (< 10 years of age, body temperature ≥ 38°C, negative result for the malaria rapid diagnostic test, living in Guediawaye/Pikine for the previous four calendar months, not receiving any anti-infectious treatment since the onset of fever, and with parent’s consent to participate) presenting to the health post in Medina Gounass located in Guediawaye on Mondays and Fridays were included. In total, 106 children participated in the study, and PCR from nasopharyngeal swabs, hemoculture, C-reactive protein, blood cell counts, and quantitative buffy coat from blood samples and coproculture from stool samples were performed. In 70 (66%) children, at least one pathogen was isolated. Viruses were identified in 55 children, most commonly enteroviruses, rhinoviruses, and adenoviruses, and dengue virus was identified in three children. Only five children had bacterial infections, and 10 had bacterial and viral coinfections. Ninety-seven children (92%) received prescription for antibiotics. Many strains of bacteria were found to be resistant to several antibiotics. Despite limitations, this pilot study showed that pathogens potentially associated with non-malarial fever in children younger than 10 years near Dakar were predominantly viruses, most commonly upper respiratory infections, although bacteria accounted for a small proportion.
INTRODUCTION
In Africa, fever is one of the most frequent reasons for pediatric consultation and has been traditionally associated with malaria. As a result of improved malaria control in sub-Saharan African countries, the incidence of malaria has considerably decreased, leading to a reduction in the proportion of febrile illnesses attributable to malaria.1 However, the etiologies of non-malarial febrile diseases, including typhoid fever, pneumococcal bacteremia, influenza, yellow fever, dengue fever, or tick-borne fever (like borreliosis),2,3 are poorly documented because of the unavailability and/or high cost of diagnostic tests.
For several decades, the public health measures introduced to control malaria have contributed to the decline in prevalence.4 However, the trends in the prevalence of non-malarial febrile diseases across this same period are not well documented. In addition, the demographics of Africa are changing, with an increasing number of people moving to urban areas. By 2035, the proportion of the urban population in sub-Saharan Africa is projected to increase from 30% to more than 50%.5 As African countries become increasingly urbanized, the factors favoring non-malarial febrile diseases become more important, especially in overcrowded housing and slum settings in which there is often an inadequate supply of drinking water, malfunctioning sewage disposal systems, and lack of sanitation and removal of solid waste.6,7 In Senegal, there has been a significant reduction in the prevalence of malaria since 2008. Malaria parasite prevalence declined from 6% to 3% from 2008 to 2010 among children younger than 5 years.8 At the same time, recent studies have shown that the etiologies of non-malarial febrile diseases are poorly documented, especially in urban areas.8 Moreover, most of these infections tend to be systematically treated with antibiotics, which may facilitate antimicrobial resistance. Improved diagnostic testing would enable better understanding of non-malarial febrile diseases and improved management of patients.9
Objectives.
The objectives of this pilot study were 1) to identify pathogens potentially associated with non-malarial febrile diseases in children younger than 10 years living in peri-urban areas near Dakar, Senegal, and 2) to describe the epidemiological characteristics of these patients.
METHODS
Study design and location.
We conducted a cross-sectional study from September 2015 to March 2016 at the primary healthcare center in Medina Gounass, located in Guediawaye, in the peri-urban area of Dakar, Senegal. This period covered both the rainy season (September–October 2015) and the dry season (October 2015–March 2016). Guediawaye has close to 1,000,000 inhabitants, and the population density is 9,200 inhabitants/km2—a very high density population compared with the rest of Senegal. The population lives in an underserved peripheral zone, without drinking water with a very poor level of sanitation.
Inclusion criteria.
Children younger than 10 years, living in Guediawaye or Pikine for four successive calendar months, who presented to the primary health care in Medina Gounass on Mondays and Fridays with fever, who had not received any anti-infectious treatment since the onset of fever, who had a negative Plasmodium falciparum rapid diagnostic test (RDT) (Ref I13FRC25 Malaria Antigen P. falciparum HRP2 Card test, Gujarat, India), and whose mother or legal tutor provided informed consent for participation in the study were eligible for inclusion in the study. Because of logistical constraints, participants were recruited on Mondays and Fridays—the 2 days of the week with a higher number of consultations at the Medina Gounass healthcare center. During the data collection period, all children who met the eligibility criteria were consecutively included.
Data collection.
Following recruitment into the study, a questionnaire was completed by one of the parents. This covered the following: 1) clinical and sociodemographic data; 2) the child’s medical history, including the use of antibiotics; and 3) socioeconomic status of the family (household income, level of education of the parents, indicators related to living conditions: urbanization, density, removal of solid waste, and waste water, sanitation, drinking water, electricity, and the presence of animals in the home (domestic animals and livestock).
Laboratory evaluations.
For each child, a venous blood sample was collected on inclusion into the study in two blood culture flasks (aerobic and anaerobic BacTAlert equipment, bioMérieux, Durham, NC) using a tube fitted with a fin (2.5 mL of venous blood for each flask) plus three tubes: one with EDTA and two without anticoagulant (dry tubes). In addition, one nasopharyngeal swab was collected.
Additional sampling was performed when other symptoms, in addition to fever, were reported, on indication of the attending physician: 1) a pus swab in the case of abscesses or rash with suppuration, 2) a stool sample in the case of diarrhea, and 3) a urinary tract swab for suspicion of urinary infection (micturition, urinary frequency, and polyuria).
The blood sample was analyzed for cell blood count to identify anemia (hemoglobin [Hb] level < 12 g/dL) and hyperleukocytosis (leukocytes > 10 giga per liter [G/L]) with an Automation XN 1000 Sysmex® (Sysmex Corporation, Kobe, Japan), and for the C-reactive protein (CRP) level to identify inflammation (CRP level ≥ 10 mg/L) with an Automation c4000 Abbott Architect® (Abbott Diagnostics, North Chicago, IL). Quantitative buffy coat (QBC) tests (QBC™ Malaria Test, Drucker Diagnostics, Philipsburg, PA) were performed to detect Plasmodium parasite and bacteria of genus Borrelia (morphological aspects). All laboratory analyses were conducted at the Institut Pasteur of Dakar.
Bacteriological analysis.
Fluid from positive blood culture flasks, pus, and nasopharyngeal swabs were inoculated on adequate culture media to isolate bacteria on the basis of microscopic examination after Gram staining. Different culture media (chocolate agar with IsoVitaleX™ BD® [Dickinson and Company, Spark, MD], Columbia CAN agar with 5% sheep blood BD®, Columbia agar [Biomérieux, Durham, NC] with 5% sheep blood BD, and Bromocresol purple lactose agar BD®, Becton Dickinson GmbH, Heidelberg, Germany) were inoculated and incubated in aerobic or anaerobic environments (5–10% CO2) for 24–48 hours depending on the bacteria sought. Stool samples were inoculated on Hektoen enteric agar BD® (Hektoen Institute for Medical Research, Chicago, IL) and Rambach™ (Sigma-Aldrich, Saint-Louis, MO) agar chromogenic media and on Muller Kauffmann broth for detection of bacterial pathogens associated with diarrhea.
Bacteria were identified based on morphological characteristics (cocci or bacilli), cultural (appearance of colonies and type of hemolysis), biochemical (using API® Biomerieux galleries, Biomerieux, Durham, NC), or antigenic (agglutination latex Strepslide II Fumouze Diagnostics® for grouping beta-hemolytic streptococci and Pastorex® Biorad, Biorad, Marnes-la-Coquette, France for Staphylococcus aureus) properties. All bacterial isolates underwent antibiotic susceptibility testing by using the disk diffusion method according to the Antibiogram Committee of the French Society for Microbiology 2016 guidelines and the European Committee on Antimicrobial Susceptibility Testing guidelines. Antibiotic disks were obtained from Bio-Rad laboratories (Marnes-la-Coquette, France).
Viruses detection.
For detection of arboviruses, real-time PCR (RT-PCR) assays for dengue virus (DENV),10 yellow fever virus (YFV),11 chikungunya virus (CHIKV),12 Rift Valley fever virus (RVFV),13 and Zika virus (ZIKV)14 were performed on sera samples after RNA extraction using the QIAamp viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. In brief, the detection was performed using ABI7500 with the QuantiTect Probe RT-PCR Master Mix (Qiagen), with the following temperature profiles for all RT-qPCR assays: RT at 50°C for 10 minutes, activation at 95°C for 15 minutes, and 45 cycles of two-step PCR—at 95°C for 15 seconds and 60°C for 1 minute. In addition, the presence of DENV, YFV, RVFV, CHIKV, and ZIKV IgM in samples was assessed by a capture ELISA Immunoglobulin M antibody-capture enzyme-linked immunosorbent assay [MAC-ELISA]), following a published protocol.15
RNA extracts from nasopharyngeal swabs were subjected to detection of respiratory viruses using a two-step real-time multiplex RT-PCR using Anyplex RV16 (Seegene, Seoul, South Korea) following the published protocol16 performed on the Bio-Rad CFX-96 thermocycler (Bio-Rad Laboratories). In brief, we used multiplex PCR targeting 16 respiratory virus distributed across two panels (A and B): Adenoviruses, Influenza (A and B), Parainfluenza virus (1, 2, 3, and 4), and Rhinoviruses (A, B, and C) on panel A; and Respiratory syncytial viruses (A and B), Bocavirus, Metapneumovirus, Enterovirus, and Coronavirus (229E, NL63, and OC43) on panel B.
Case definition.
Body temperature was considered the axillary temperature plus 0.5°C, in accordance with the WHO recommendations, and fever was defined as a body temperature greater than or equal to 38°C. For the purposes of this study, WHO references were used for influenza or flu syndrome17—defined as the presence of fever associated with sore throat or cough. Global acute malnutrition was defined as a weight-for-height Z-score of −2 or lower. Bacterial infection was determined based on the detection of at least one bacterium in the blood sample or other biological samples, and viral infection was determined based on the detection of at least one virus in one or more biological samples. Arboviral infection was determined by a positive PCR test for one of the five arboviruses tested. Coinfection was determined based on the detection of virus and bacteria in one or more biological samples.
Ethical considerations.
A written consent form was signed by the parent or legal guardian for each child before any procedure related to the study was conducted. The study protocol was approved by the National Ethics Committee of the Senegalese Ministry of Health.
Statistical analysis.
Descriptive statistics were used for the general characteristics of the children, as well as their clinical presentation, diagnosis, and treatments prescribed. Age was treated as a binary variable, with a cutoff at 2 years, derived from the median age in the study population: 23 months.
Characteristics of children with and without bacterial infection and with and without viral infection were compared using the chi-squared test or Fisher’s exact test for categorical variables and Student’s T-test for continuous variables. Univariate analyses identified factors associated with viral and bacterial infections. For these analyses, participants with coinfection were removed. All variables potentially associated with infection with a P-value < 0.25 were included in a backward logistic regression analysis. A multinomial model (with virus, bacteria, and no pathogens identified) was not able to be performed because of sample size constraints. All tests were two-tailed, and significance was considered as P < 0.05. All statistical analyses were performed using R software 3.3.3.
RESULTS
Clinical characteristics.
During the study period, 106 children were enrolled in the study, 60% of whom were female. Age ranged from 3 to 116 months, with an average of 32 (±26 SD) months. Fifty-five children (52%) were younger than 24 months.
The median time to consultation after the onset of fever was 2 (range: 0–8) days. In addition to fever, symptoms reported included influenza-like illness (70%), rhinorrhea (64%), cough (62%), and vomiting (33%). Among the 88 children younger than 5 years, 23 (32%) had global acute malnutrition. Further characteristics of the children included in the study are shown in Table 1.
Table 1.
General characteristics of children included in the study (n = 106)
| Characteristic | n (%) |
|---|---|
| Median age (months) [IQ1, IQ3] | 31.6 [10.7–48.8] |
| Male | 42 (40) |
| Global acute malnutrition* | 23/73 (32) |
| Median time between onset of fever and medical consultation (days) [IQ1, IQ3] | 2 [1–2] |
| Median body temperature (°C) [IQ1, IQ3] | 38.6 [38.3–39.1] |
| Clinical signs | |
| Flu syndrome (two missing) | 73/104 (70) |
| Rhinorrhea | 68 (64) |
| Cough | 66 (62) |
| Vomiting | 35 (33) |
| Abdominal pain | 27 (25) |
| Diarrhea | 27 (25) |
| Headache | 24 (23) |
| Sore throat | 15 (14) |
| Antibiotic prescription | 97 (92) |
| Laboratory values | |
| Inflammation† | 67 (63.2) |
| Anemia‡ | 67/96 (70) |
In the population of children younger than 5 years (n = 88).
C-reactive protein level ≥ 10 mg/L.
Hemoglobin level < 12 g/dL.
Samples taken.
In this study, a total of 102 nasopharyngeal swabs, three stool samples, and 106 blood samples were collected from the 106 children. Results of PCR from nasopharyngeal swabs, hemoculture, CRP, blood cell counts, and QBC tests were available for 106, 102, 98, 104, and 104 children, respectively. Coproculture was performed on the three stool samples collected. For arbovirus detection, PCR tests were performed for 104 blood samples, and ELISA was performed on the three samples that were positive by PCR for DENV.
Biological data.
Sixty-seven subjects (41%) had anemia, including one with severe anemia (Hb ≤ 6 g/dL). C-reactive protein was more than 10 mg/L in 67 children (63.2%), among whom 17 had levels between 100 and 425 mg/L.
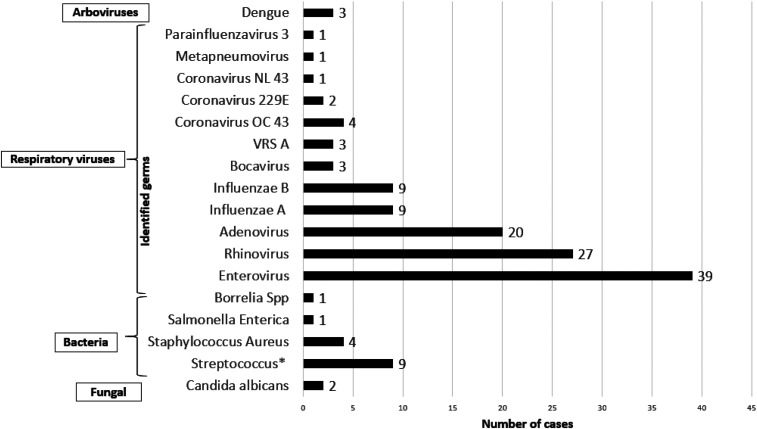
Pathogens identified (Figure 1).
Figure 1.
Distribution of pathogens isolated.
At least one pathogen was isolated in 70 children (66%) by PCR, hemoculture, culture, or QBC test. Viruses were identified in 65 (61% [51–71%]) children. Among these, respiratory viruses identified from nasopharyngeal samples accounted for the highest number of identified pathogens, with enteroviruses, rhinoviruses, and adenoviruses in 39 (38%), 27 (26%), and 20 (20%), respectively, identified in children. Among the 102 subjects with nasopharyngeal swabs collected, 29 children had two or three viruses. Influenza viruses (A and B) were frequently identified along with adenovirus, rhinovirus, or enterovirus. Three children had a positive PCR test for DENV serotype 1, but all three samples were negative by ELISA.
Bacteria were identified in 15 (14.2%; 95% CI [8.1–22.2]) children. Four subjects (3.7%; n = 106) had a bacterium isolated from the blood: S. aureus (n = 2), Salmonella enterica (n = 1), and one strain of Borrelia spp. (n = 1) identified by the QBC test. Seven poolable strains of beta-hemolytic streptococci were identified in the nasopharynx: group A streptococci (or Streptococcus agalactiae) (n = 2), group C streptococci (n = 2), group F streptococci (n = 2), and group G streptococci (n = 1). Four strains of bacteria were isolated from pus and boil samples: group A streptococci (n = 2) and S. aureus (n = 2). Candida albicans were found in nasopharyngeal swabs from two children and associated with parainfluenza 2 and group A streptococci, respectively, and high levels of leucocytes (19 G/L) in both children.
Coinfection.
Detection of both bacterial and viral pathogens was observed in 10 children. Pathogens identified in the cases of coinfection were S. aureus, different groups of beta-hemolytic streptococci (A, C, F, and G), S. enterica, and C. albicans with viruses (Influenza A, Influenza B, Enterovirus, Rhinovirus, Coronavirus, and parainfluenza virus 2) (Table 2).
Table 2.
Pathogens identified in children with bacterial and viral coinfection
| Children, n | Bacteria | Viruses | |||
|---|---|---|---|---|---|
| Blood | Nasopharynx | Pus | Nasopharynx | ||
| 1 | – | Group F streptococci | – | Influenza B | Enterovirus |
| 2 | – | – | S. aureus | Enterovirus | – |
| 3 | – | – | Group A streptococci | Enterovirus | – |
| 4 | – | Group A streptococci | Influenza A | Rhinovirus | |
| 5 | – | – | S. aureus | Enterovirus | Coronavirus 229E |
| 6 | Salmonella enterica | Group F streptococci | – | Influenza A | Enterovirus |
| 7 | – | C. albicans | Group A streptococci | Enterovirus | – |
| 8 | S. aureus | – | – | Coronavirus NL 43 | – |
| 9 | – | C. albicans | – | Parainfluenza virus 2 | – |
| 10 | – | Group C streptococci | – | Enterovirus | Coronavirus 229E |
C. albicans = Candida albicans; S. aureus = Staphylococcus aureus.
Treatments at healthcare center and antimicrobial resistance.
Ninety-seven children (92%) were prescribed at least one antibiotic following the consultation. In terms of antimicrobial resistance, all isolated staphylococci strains (n = 4) showed resistance to penicillin G. The isolated strains of Streptococcus (n = 9) showed resistance to erythromycin (five strains), sulfamethoxazole–trimethoprim (three strains), and chloramphenicol (five strains).
Factors associated with viral or bacterial infection.
In both univariate and multivariate analyses, none of the variables were found to be associated with bacterial infection (Table 3) or with viral infections (Table 4).
Table 3.
Factors associated with bacterial infection
| Variables | n | /N | OR (95% CI) | P-value | |
|---|---|---|---|---|---|
| Age (years) | < 2 | 2 | /21 | 1.75 (0.18; 23.49) | 0.65 |
| ≥ 2 | 3 | /19 | |||
| Gender | Male | 1 | /15 | 2.49 (0.22; 134.07) | 0.63 |
| Female | 4 | /26 | |||
| C-reactive protein (mg/L) | < 10 | 4 | /39 | 1.71 (0.46; 7.94) | 0.56 |
| ≥ 10 | 11 | /67 | |||
| Anemia (hemoglobin g/dL) | < 12 | 3 | /22 | 1.05 (0.07; 10.63) | 0.99 |
| ≥ 12 | 2 | /14 | |||
| Leucocytes (G/L) | ≤ 10 | 2 | /19 | 1.33 (0.13; 17.75) | 0.99 |
| > 10 | 3 | /22 | |||
| Flu | No | 2 | /15 | 0.85 (0.08; 11.43) | 0.99 |
| Yes | 3 | /26 | |||
Table 4.
Factors associated with viral infection (n = 91)
| Variable | n/ | N | OR (95% CI) | P-value | |
|---|---|---|---|---|---|
| C-reactive protein (mg/L) | < 10 | 25/ | 39 | 0.83 (0.33; 2.01) | 0.68 |
| ≥ 6 | 40/ | 67 | |||
| Anemia (hemoglobin g/dL) | < 12 | 36/ | 55 | 0.62 (0.21; 1.78) | 0.33 |
| ≥ 12 | 14/ | 26 | |||
| Flu | Yes | 39/ | 62 | 1.56 (0.57; 4.32) | 0.35 |
| No | 14/ | 27 | |||
| Age (years) | < 2 | 30/ | 49 | 0.98 (0.39; 2.53) | 0.99 |
| ≥ 2 | 25/ | 41 | |||
| Gender | Male | 24/ | 38 | 0.82 (0.32; 2.10) | 0.67 |
| Female | 31/ | 53 | |||
DISCUSSION
To our knowledge, this pilot study is the first to be conducted in Senegal to identify pathogens potentially associated with non-malarial febrile disease in children. In this study, we found that flu-like syndrome and acute respiratory infection were the primary reasons for consultation of children with fever, accounting for 60–70% of consultations, respectively. This was followed by digestive tract symptoms with 25% reporting abdominal pain and diarrhea and 33% reporting vomiting (n = 104 children). Our results were similar to those reported in Tanzania in 2014 in a study of 1,005 children younger than 10 years, 62.2% had an acute respiratory infection and only 10.3% (n = 1,005) had gastroenteritis.18 The results were also similar to those of a study of outpatients in a Tanzanian hospital, in which 64% (339/530) of children younger than 12 years had respiratory tract complaints and among 428 children younger than 5 years, 27.3% had digestive tract symptoms.19
Overall, at least one pathogen was isolated in 70 children (66%) by PCR, hemoculture, culture, or QBC test. This level of successful pathogen identification was likely the result of the proximity of the study site to the Institut Pasteur de Dakar and was comparable with other studies, with the exception of one study conducted in 2013 in northeastern Tanzania in which the cause of infection was identified in only 7.5% (65/867) of patients aged 2–59 months.20
Among pathogens identified, the most common were respiratory viruses (61%; 95% CI [51–71]), followed by bacteria (14.2%; 95% CI [8.1–22.2]), and bacterial and viral coinfections (10%). These results were consistent with previously reported data. In Tanzania, Mahende et al.20 and D’Acremont et al.18 reported that more than half of respiratory infections in children younger than 5 years had a viral origin. Furthermore, Crump et al.21 found bacteria in only 9.8% of all pediatric admissions with fever in Tanzania.
In our study, we identified 38% and 27% of children with enterovirus and rhinovirus, respectively. In a large cohort of children conducted from 2007 to 2010 in Kenya, which followed up children until 3 years of age, a significant share of causes of respiratory infections was found to be viral.22,23 In Senegal (2012–2014), Fall et al.24 also observed that among children younger than 5 years, rhinovirus and enterovirus infections accounted for 33.7% and 20.4%, respectively.
Bacteria were identified in four children (3.7%) by hemoculture. These results were in accordance with data reported in Tanzania, in which 3.2% of children had bacteria isolated by blood culture.20 Hildenwall et al.19 also identified a low percentage (1.3% of 1,028 pediatric outpatients) of bacteria by blood culture. These low percentages of bacterial infections do not necessarily reflect the true burden of bacterial infections as they do not account for the presence of fragile or intracellular or non-detectable bacteria and non-bacteremic bacteria. It could also reflect the sensitivity of hemoculture (87.2%) to detect bacteria.
We found three cases of DENV serotype 1. Across the same period, Burkina Faso and Ivory Coast reported DENV epidemics.25 The detection of cases of DENV infection in our study might be linked to the geographical proximity of those countries. Furthermore, in northern Tanzania, CHIKV was present in 7.9% of febrile pediatric admissions.21 In our study, for all three cases, ELISA tests were negative, suggesting that all three were in the acute phase of infection.
In our study, 97 children (92%) received a prescription for antibiotics, although bacteria could only be identified in 15 (14%). We found resistance among strains of staphylococci to penicillin G, whereas Streptococcus strains (n = 9) showed resistance to erythromycin (five strains), sulfamethoxazole–trimethoprim (three strains), and chloramphenicol (five strains). We could not draw conclusions as to the overuse of antibiotics, but from a public health perspective, it nonetheless suggests that many received a prescription for an antibiotic that might not necessarily be needed. The inappropriate prescription of antibiotics is now understood to be one factor which facilitates antimicrobial resistance.20 This is particularly problematic in areas with limited access to a broad range of antimicrobial treatments. It is therefore increasingly important to monitor resistance to antibiotics or at least to those routinely used in the population.26 This finding was consistent with previous reports of antibiotic prescription. In the same area of our study (Guediawaye), a community survey conducted in 2016 by Padget et al.27 found antibiotic prescription among children younger than 2 years to be approximately 92%. In a study on community-acquired infectious diarrhea in children younger than 5 years in Senegal in 2007–2008, 50% of the antibiotic prescriptions given to children younger than 5 years were not needed.28 In 2016 in France, among pediatric patients with upper respiratory tract infections, antibiotic prescription was given to 59% of patients, of which 76% were deemed inappropriate.29 In 2013 in Burkina Faso, among children younger than 5 years with diarrhea, the development of resistance of Salmonella and Shigella strains to ciprofloxacin, sulfamethoxazole–trimethoprim, and amoxicillin was shown.30 In 2007, mortality from resistant bacteria in children in Tanzania was shown to begin to be significant.31
Our results highlight the need to improve the routine diagnosis of bacterial and viral infections in children with non-malarial fever. This also needs to be accompanied by continuing education of health professionals on effective and suitable antibiotic prescription practices.32 This will allow for improved antibiotic prescription algorithms, including first-line antibiotics, which are currently inadequate and inappropriate, as demonstrated by Ndir and et al.33 in a major pediatric hospital in Senegal in 2016.
In addition, we identified 2% of fungal infections, exclusively in the nasopharynx. But the remaining question relates to the link between fungal pathogens and fever, especially in our case in which bacteria and viruses were also identified. In this respect, Candida species could be considered as a facilitator of bacterial or viral infection. Among 870 pediatric and adult febrile admissions to two hospitals in northern Tanzania between September 2007 and August 2008, fungal pathogens accounted for 2.9% of those found in the blood.20
Limitations of the study.
Overall, we attempted to identify pathogens present in biological samples of children with non-malarial febrile illness. However, apart from pathogens identified by hemoculture, we cannot assume that the pathogens identified were necessarily responsible for fever in these children. Furthermore, many serious bacterial illnesses (e.g., leptospirosis, rickettsiosis, and meningitis) are not bacteremic, and the methods used would not have been able to identify them. The same applies to the use of PCR, in that PCR may detect fragments of a pathogen, rather than the pathogen that was the cause of fever. Similarly, the collection of only one nasopharyngeal swab may have been insufficient to detect and confirm the presence of microorganisms, especially respiratory viruses as the window of PCR positivity is often very narrow. Moreover, intracellular germs were also not investigated. Additional tests such as the procalcitonin test for bacterial infection could better clarify the diagnosis. As a pilot study, the sample size was small largely because of financial constraints. As a result, it did not have sufficient statistical power to identify factors associated with the identification of bacteria and/or viruses. It was conducted over a period of 6 months, which did not allow for seasonality. For this reason, it would be useful to extend a similar study beyond 1 year and include more children.
No urinary tract infections were identified probably because of the age of most of the children who were not able to complain of common urinary tract infection symptoms: dysuria or burning sensation. Moreover, it was difficult to collect urine systematically for young children younger than 1 year. In our study, no child was younger than 3 months. This is likely explained by the organization of the health system. Usually, neonates (0–2 months) are not usually taken to primary health centers, like the one used for this study, for medical consultation, rather they are usually seen at maternity wards or specialized neonate centers. Furthermore, most of the reported diarrheal cases were anamnestic diarrhea (the mothers being unable to clearly define the symptoms of diarrhea). This may explain why stool collection was low.
CONCLUSION
In our study, pathogens potentially associated with non-malarial fever in children younger than 10 years in peri-urban areas of Dakar were predominantly viruses, and the most common type of infections was upper respiratory infection. We observed that bacterial species associated with a non-malarial febrile disease accounted for a small proportion. It is therefore imperative to develop RDTs and to improve diagnostic algorithms for bacterial and viral infections as well as the education of health professionals on effective prescription of antibiotics. As we could not draw definitive conclusions as to the overuse of antibiotics, further studies are needed to further investigate antibiotic prescription practices and antimicrobial resistance.
ACKNOWLEDGMENTS
We would like to thank all the workers in the epidemiology unit of the IPD (Institut Pasteur de Dakar) and the staff of the Medina Gounass Health Center and the director of the Guediawaye health district. We would also like to thank all those who have contributed, either directly or indirectly, to the completion of this work.
REFERENCES
- 1.d’Acremont V, Lengeler C, Genton B, 2019. Reduction in the proportion of fevers associated with Plasmodium falciparum parasitaemia in Africa: a systematic review. Malar J 9: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joshi R, Colford JM, Jr., Reingold AL, Kalantri S, 2008. Nonmalarial acute undifferentiated fever in a rural hospital in central India: diagnostic uncertainty and overtreatment with antimalarial agents. Am J Trop Med Hyg 78: 393–399. [PubMed] [Google Scholar]
- 3.White LJ, Newton PN, Maude RJ, Pan-Ngum W, Fried JR, Mayxay M, Maude RR, Day NP, 2012. Defining disease heterogeneity to guide the empirical treatment of febrile illness in resource poor settings. PLoS One 7: e44545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gething PW, Battle KE, Bhatt S, Smith DL, Eisele TP, Cibulskis RE, Simon I, 2014. Hay declining malaria in Africa: improving the measurement of progress. Malar J 13: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parnell S, Walawege R, 2011. Sub-Saharan African urbanisation and global environmental change. Glob Env Change 21: S12–S20. [Google Scholar]
- 6.Patel RB, Burke TF, 2009. Urbanization, an emerging humanitarian disaster. N Engl J Med 361: 741–743. [DOI] [PubMed] [Google Scholar]
- 7.Alirol E, Getaz L, Stoll B, Chappuis F, Loutan L, 2011. Urbanisation and infectious diseases in a globalised world. Lancet Infect Dis 11: 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thwing J, et al. 2017. Declines in malaria burden and all-cause child mortality following increases in control Interventions in Senegal, 2005–2010. Am J Trop Med Hyg 97 (Suppl 3): 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breurec S, et al. 2016. Aaetiology and epidemiology of diarrhea in hospitalized children from low income country: a matched case-control study in Central African Republic. PLoS Negl Trop Dis 10: e0004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner D, de With K, Huzly D, Hufert F, Weidmann M, Breisinger S, Eppinger S, Kern WV, Bauer TM, 2004. Nosocomial acquisition of dengue. Emerg Infect Dis 10: 1872–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weidmann M, Faye O, Faye O, Kranaster R, Marx A, Nunes MRT, Vasconcelos PF, Hufert FT, Sall AA, 2010. Improved LNA probe-based assay for the detection of African and South American yellow fever virus strains. J Clin Virol 48: 187–192. [DOI] [PubMed] [Google Scholar]
- 12.Shu PY, Yang CF, Su CL, Chen CY, Chang SF, Tsai KH, Cheng CH, Huang J, 2008. Two imported chikungunya cases, Taiwan. Emerg Infect Dis 14: 1325–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weidmann M, Sanchez-Seco MP, Sall AA, Ly PO, Thiongane Y, Lô MM, Schley H, Hufert FT, 2008. Rapid detection of important human pathogenic Phleboviruses. J Clin Virol 41: 138–142. [DOI] [PubMed] [Google Scholar]
- 14.Faye O, Faye O, Diallo D, Diallo D, Weidmann M, Sall AA, 2013. Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol J 10: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ba F, Loucoubar C, Faye O, Fall G, Mbaye RNPN, Sembene M, Diallo M, Balde AT, Sall AA, Faye O, 2018. Retrospective analysis of febrile patients reveals unnoticed epidemic of zika fever in Dielmo, Senegal, 2000. Clin Microbiol Infect Dis 3: 1–9. [Google Scholar]
- 16.Niang MN, Diop NS, Fall A, Kiori DE, Sarr FD, Sy S, Goudiaby D, Barry MA, Fall M, Dia N, 2017. Respiratory viruses in patients with influenza-like illness in Senegal: focus on human respiratory adenoviruses. PLoS One 12: e0174287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO , 2013. Global Epidemiological Surveillance Standards for Influenza. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 18.D’Acremont V, Kilowoko M, Kyungu E, Philipina S, Sangu W, Kahama-Maro J, Lengeler C, Cherpillod P, Kaiser L, Genton B, 2014. Beyond malaria--causes of fever in outpatient Tanzanian children. N Engl J Med 370: 809–817. [DOI] [PubMed] [Google Scholar]
- 19.Hildenwall H, Amos B, Mtove G, Muro F, Cederlund K, Reyburn H, 2016. Causes of non-malarial febrile illness in outpatients in Tanzania. Trop Med Int Health 21: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahende C, Ngasala B, Lusingu J, Butichi A, Lushino P, Lemnge M, Premji Z, 2014. Aetiology of acute febrile episodes in children attending Korogwe district hospital in north-eastern Tanzania. PLoS One 9: e104197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crump JA, et al. 2013. Aaetiology of severe non-malaria febrile illness in northern Tanzania: a prospective cohort study. PLoS Negl Trop Dis 7: e2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feikin DR, et al. 2012. Aaetiology and Incidence of viral and bacterial acute respiratory illness among older children and adults in rural western Kenya, 2007–2010. PLoS One 7: e43656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feikin DR, et al. 2013. Viral and bacterial causes of severe acute respiratory illness among children aged less than 5 years in a high malaria prevalence area of western Kenya, 2007–2010. Pediatr Infect Dis J 32: e14–e19. [DOI] [PubMed] [Google Scholar]
- 24.Fall A, et al. 2016. Enteroviruses and rhinoviruses: molecular epidemiology of the most influenza-like illness associated viruses in Senegal. Am J Trop Med Hyg 95: 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim JK, et al. 2019. Clinical and epidemiologic characteristics associated with dengue during and outside the 2016 outbreak identified in health facility-based surveillance in Ouagadougou, Burkina Faso. PLoS Negl Trop Dis 13: e0007882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padget M, Guillemot D, Delarocque-Astagneau E, 2016. Measuring antibiotic consumption in low-income countries: a systematic review and integrative approach. Int J Antimicrob Agents 48: 27–32. [DOI] [PubMed] [Google Scholar]
- 27.Padget M, Tamarelle J, Herindrainy P, Ndir A, Diene Sarr F, Richard V, Piola P, Guillemot D, Delarocque-Astagneau E; BIRDY Study Group , 2017. A community survey of antibiotic consumption among children in Madagascar and Senegal: the importance of healthcare access and care quality. J Antimicrob Chemother 72: 564–573. [DOI] [PubMed] [Google Scholar]
- 28.Sire JM, Garin B, Chartier L, Fall NK, Tall A, Seck A, Weill FX, Breurec S, Vray M, 2013. Community-acquired infectious diarrhoea in children under 5 years of age in Dakar, Senegal. Paediatr Int Child Health 33: 139–144. [DOI] [PubMed] [Google Scholar]
- 29.Marc C, Vrignaud B, Levieux K, Robine A, Gras-Le Guen C, Launay E, 2016. Inappropriate prescription of antibiotics in pediatric practice: analysis of the prescriptions in primary care. J Child Health Care 20: 530–536. [DOI] [PubMed] [Google Scholar]
- 30.Bonkoungou IJ, Haukka K, Österblad M, Hakanen AJ, Traoré AS, Barro N, Siitonen A, 2013. Bacterial and viral aetiology of childhood diarrhea in Ouagadougou, Burkina Faso. BMC Pediatr 13: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blomberg B, et al. 2007. Antimicrobial resistance predicts death in Tanzanian children with bloodstream infections: a prospective cohort study. BMC Infect Dis 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blomberg B, 2008. Antimicrobial resistance in developing countries. Tidsskr Nor Laegeforen 128: 2462–2466. [PubMed] [Google Scholar]
- 33.Ndir A, Diop A, Faye PM, Cissé MF, Ndoye B, Astagneau P, 2016. Epidemiology and burden of bloodstream infections caused by extended-spectrum beta-lactamase producing Enterobacteriaceae in a pediatric hospital in Senegal. PLoS One 11: e0143729. [DOI] [PMC free article] [PubMed] [Google Scholar]