ABSTRACT
Isolates from 475 cutaneous leishmaniasis (CL) patients from three endemic regions were studied by three typing techniques. The molecular analysis from lesion scrapings based on hsp70 PCR-restriction fragment length polymorphism (RFLP) showed that 78.1% (371/475) restriction patterns corresponded to Leishmania (Viannia) panamensis, 19% (90/475) to Leishmania (Viannia) guyanensis, and 3.0% (14/475) to Leishmania (Viannia) braziliensis. Promastigotes isolated by culture from lesions of 228 patients (48.0%, 228/475) were identified by multi-locus enzyme electrophoresis. Of them, 95.2% (217/228) were typified as L. (V.) panamensis, 1.3% (3/228) as L. (V.) guyanensis, 2.2% (5/228) as L. (V.) braziliensis, and 1.3% (3/228) as hybrids (L. [V.] braziliensis/L. [V.] panamensis). However, a partial sequencing analysis of the hsp70 gene from 77 selected samples showed 16.9% (13/77) typified as L. (V.) panamensis, 68.8% (53/77) as Leishmania (V.) sp., 1, 3.9% (3/77) as L. (V.) guyanensis, 1.3% (1/77) as L. (V.) braziliensis outlier, 2.6% (2/77) as Leishmania (Viannia) naiffi, 2.6% as (2/77) Leishmania (V.) sp., and 2 and 3.9% (3/77) hybrid isolates of L. (V.) braziliensis/L. (V.) guyanensis. These results confirm L. (V.) panamensis as the predominant species and cause of CL lesions in Panama and that L. (V.) guyanensis, L. (V.) braziliensis, and L. (V.) naiffi are circulating to a lower degree. Furthermore, the determination of parasite isolates belonging to atypical clusters and hybrid isolates suggests the circulation of genetic variants with important implications for the epidemiology and clinical follow-up of CL in Panama. No evidence of the existence of parasites of the Leishmania (Leishmania) mexicana complex in Panamanian territory was found in this study.
INTRODUCTION
Worldwide, more than 350 million people are considered at risk of contracting leishmaniasis, and some two million new cases occur yearly.1 Clinical features of this disease can vary significantly, reflecting the wide range of parasites in the genus Leishmania capable of infecting humans,2–5 the host immune response, and factors found in the saliva of the sand fly vector.6,7
Cutaneous leishmaniasis (CL) is the most common clinical form causing ulcers on exposed parts of the body and leaving scars in most cases. Moreover, some Leishmania species can disseminate from the cutaneous lesions to the nasopharyngeal mucosa and cause mucocutaneous leishmaniasis that can partially or totally destroy the mucous membranes of the nose, mouth, and throat cavities and surrounding tissues, causing serious disability.8
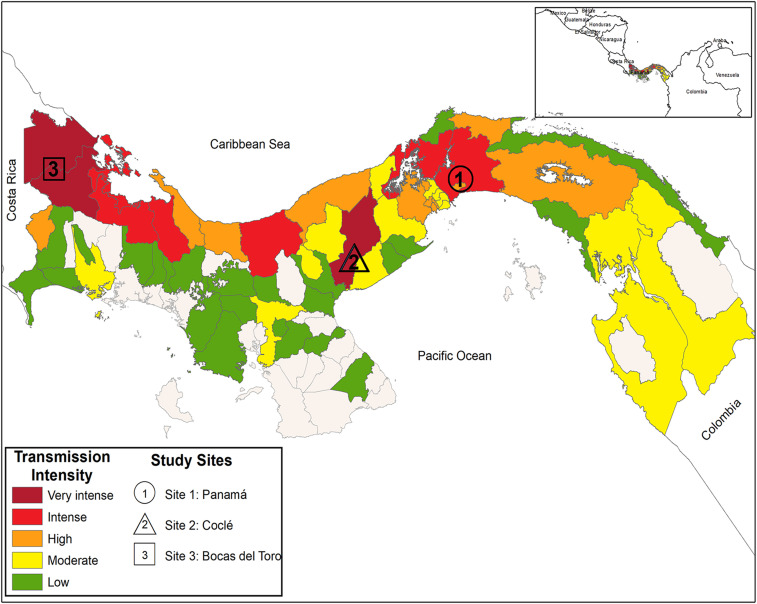
Cutaneous leishmaniasis is a worldwide neglected disease that is an emerging public health problem in Panama. There are 1,000–3,000 cases per year, but the health authorities consider that it could be 50% more.9 By 2011, the Pan American Health Organization (PAHO) reported that Panama was the country with the higher incidence of CL per 100,000 inhabitants in America.10 Panama, Bocas del Toro, and Cocle are the western provinces in Panama with the higher reported number of CL cases11 (Figure 1). Within these provinces, the transmission is concentrated in forested and rural areas, mainly among the marginalized population. In Panama, CL is a neglected disease, affecting the poorest, a situation similar to other endemic countries. This leads to an increase in morbidity, and inequity in a country that has already one of the highest levels of inequality in the region.12
Figure 1.
Map of Panama showing location of the study sites and the cutaneous leishmaniasis transmission intensity based on the median number of confirmed cases (2015–2017). Map was created using ArcGIS Desktop 10.6.1 software (ArcGIS Desktop© 10.6.1 software, 2018, Redlands, CA). Data were obtained from information reported by the Panamanian Ministry of Health-National Leishmaniasis Surveillance Services to the PAHO as published in PAHO, leishmaniasis, epidemiological report in the Americas, Washington, PAHO, 2019. Available at: www.paho.org/leishmaniasis. PAHO = Pan American Health Organization. This figure appears in color at www.ajtmh.org.
Leishmania (Viannia) panamensis, which has been considered a local variant of Leishmania (Viannia) guyanensis,13 is referred as the most widespread and main etiologic agent for most of the human cases in the country.14–16 Only a few sporadic human cases of CL caused by Leishmania (Viannia) braziliensis, Leishmania (Leishmania) amazonensis, and Leishmania (Leishmania) mexicana were reported decades ago using isoenzymic methods for species characterization.15,17–19 Other species such as Porcisia hertigi, originally identified as Leishmania hertigi,20 and Leishmania (Leishmania) aristidesi were detected in few wild mammals in 1965 and 1968, respectively.21–23 Also, Endotrypanum colombiensis, originally identified as Leishmania colombiensis,20 was isolated from four sand flies and a sloth between 1980 and 1986.24 More recently, Leishmania (Viannia) naiffi was identified in sand flies collected in Barro Colorado Island located in Gatún Lake Panama Canal and also in sand flies in a hyperendemic CL community in the district of Capira, province of Panama Oeste.25,26 Although the gold standard for diagnosis is still parasite visualization (smears/culture), identification of the species causing the infection is critical in this era of species-guided treatment recommendations from the WHO/PAHO.27 Likewise, characterization of the species can provide important complementary information about disease progression. In this regard, a significant association between the infecting Leishmania species and treatment outcome has been described.28–30 Furthermore, identification of the exact parasite species infecting humans and animal host, as well as sand fly vectors, also gains increasing importance for epidemiological surveillance and CL control.20,31–33
The Panama isthmus is the geographical link between Central and South America, providing a biologic corridor between Colombia and Costa Rica, that increases the risk for the entrance of pathogens, vectors, and reservoirs from Central and South America. Studies conducted in neighboring countries have reported human leishmaniasis caused by species not detected previously or of uncertain endemicity in Panama, as L. (L.) infantum, L. (V.) guyanensis, E. colombiensis, and L. (V.) braziliensis.24,34,35 However, for a better understanding of the identity and genetic characteristics of the causative agents of CL in Panama, thorough studies are necessary, especially in regions of high prevalence and using an adequate number of biological samples. To investigate the Leishmania species diversity causing human leishmaniasis in Panama, we typed parasites found in 475 human CL cases from different endemic areas, by hsp70 PCR-RFLP, multi-locus enzyme electrophoresis (MLEE), and hsp70 sequencing analysis. The results suggest that although L. (V.) panamensis is present in most of the evaluated cases, other Leishmania (Viannia) species and genetic variants causing localized CL are also circulating in Panama.
MATERIALS AND METHODS
Study sites and samples.
The species of Leishmania present in cutaneous lesions from CL Panamanian patients were investigated. Study patients were part of a phase 3 clinical trial of a topical cream for the treatment of CL conducted in Panama, sponsored by the Surgeon General, Department of the Army, USA.36 The analyzed samples corresponded to 475 positive DNA lesion samples (kinetoplast DNA [KDNA] PCR/hsp70 PCR) and 228 promastigote-positive cultures from these patients. During the clinical trial, all lesion samples were initially evaluated by parasitological diagnostic procedures (smear/culture) and a PCR test that amplifies a specific sequence of Leishmania (Viannia) sp. KDNA37 (Supplemental Information 1). Lesion samples were collected between April 2013 and August 2015 from patients who attended one of the three study sites: Clinical Investigation Unit at the Gorgas Memorial Institute (ICGES) in Panama City, Panama Province (8°58′14″N, 79°32′1″W); Hospital Aquilino Tejeira in Penonome, Cocle Province (8°30′48″N, 80°21′3″W); and Health Center Materno Infantil Sandra Hernández in Changuinola (9°22′60″N, 82°31′60″W), Bocas del Toro Province (Figure 1).
Ethics.
As previously reported,36 the study and use of clinical samples were reviewed and approved by the National Review Board (Comité de Bioética de la Investigación, ICGES Panama City, Panama, assigned code PEI 50098, S-12-21), and by the Human Research Protections Office, U.S. Army Medical Research and Material Command. Written informed consent was obtained from all study participants and/or guardians before enrollment. Minors also provided assent to participate. All subjects gave permission to type and further use Leishmania parasites associated with their lesions.
Reference strains.
Leishmania (Viannia) reference strains used in this study as controls for hsp70-RFLP and phylogenetic analysis were as follows: L. (V.) panamensis (MHOM/PA/1998/WR2306), L. (V.) guyanensis (MHOM/BR/1975/M4147), L. (V.) braziliensis (MHOM/BR/1975/M2903), and Leishmania (Viannia) peruviana (MHOM/PE/2005/WR2771).
Molecular characterization methods.
DNA extraction.
DNA extraction of the skin scrapings in TE buffer was performed using the Qiagen QIAamp® DNA Blood Mini Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Leishmania DNA from culture-isolated parasites was purified using Wizard™ Genomic DNA Purification Kit (Promega, Madison, WI) according to the manufacturer’s instructions.
PCR-hsp70 detection of Leishmania genus.
PCR was performed with oligonucleotides F25 and R1310 that amplify a 1,286-bp product from the repeated gene heat shock protein 70 (hsp70) as previously described.38,39 Amplification reactions were performed in a final volume of 50 μL containing 25 μL of Go Taq Green Master Mix 2X (Promega), 0.6 μmol/L of each primer, 5 μL of DNA of clinical samples, and 1 ng of reference strains. Thermal cycling was performed in an Applied Biosystems® (Foster City, CA) 2,720 Thermal Cycler. When negative results or results with faint agarose gel band of amplification products were obtained, re-amplification was performed using the same conditions and primers mentioned earlier with 1–5 μL of amplified product to improve the sensitivity of the PCR or to have enough product for endonuclease digestion.
Hsp70-RFLP analysis.
Leishmania hsp70-RFLP characterization was performed as described elsewhere.38 In brief, an initial amplification was performed with the aforementioned hsp70 primers. The resulted amplicons were digested with Hae III and subsequently with BccI or RsaI endonucleases. Obtained restriction patterns were analyzed by electrophoresis and compared with reference strain patterns.
Multi-locus enzyme electrophoresis (MLEE).
Biochemical characterization by isoenzyme electrophoresis was performed in the Department of Leishmania Diagnosis at the Walter Reed Institute of Research, USA. In brief, parasite lysates were prepared from logarithmic-phase bulk cultures of isolated promastigotes, and their soluble enzymes were extracted. The isoenzymes were separated after electrophoresis on cellulose acetate as previously described.40,41 For each patient sample, the electrophoretic mobility banding pattern was compared with standard patterns of known Leishmania species (WHO reference strains). The following Leishmania enzymes were assessed: phosphogluconate dehydrogenase, mannose phosphate isomerase, glucose phosphate isomerase, and peptidase D. When necessary, additional enzymes (glucose-6-phosphate dehydrogenase, malic enzyme, aspartate aminotransferase, alanine aminotransferase, and phosphoglucomutase) were used to confirm the identifications made with initial enzymes. In addition, isolates that could not be identified with the reference strains were further analyzed using other enzymes, including acid phosphatase, fumarate hydratase, glutamic oxaloacetic transaminase, hexokinase, isocitrate dehydrogenase, lactate dehydrogenase, malate dehydrogenase, glutathione reductase 1 (GSR1), glutathione reductase 2 (GSR2), 6-phosphofructokinase, and pyruvate kinase.
Hsp70 sequencing and phylogenetic analysis.
The hsp70-amplified products were submitted to electrophoresis in 1.5% agarose gels in 1X TBE (89 mM Tris borate, 2 mM ethylenediaminetetraacetic acid [EDTA], pH 8.3). Product bands were excised from agarose gel and purified using the Qiaquick Gel Extraction Kit (Qiagen) following the manufacturer’s instructions. DNA sequencing of both strands was carried out using primers F25, R1310, 6F, and R61738,39; and BigDye Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems). Primers and deoxynucleotide triphosphates were removed using Xterminator Kit (Applied Biosystems). The clean sequencing reaction was run through an ABI 3130x sequencer (Foster City, CA). The chromatograms were edited by the assembling-to-reference tool of UGENE tool kit42 (Novosibirsk, Russia) using a trimming quality valor of 35. Sequences were then multiple-aligned using MAFFT software MAFFT (Multiple Alignment using Fast Fourier Transform, Philadelphia, PA) also included in the bioinformatic UGENE tool kit with a maximum number of iterative refinement of three and a gap penalty of 1.53. The generalised time-reversible model [GTR] (GTR + I + G) model was found as the best DNA evolution model by the program JModelTest 2 (Coruña, Spain).43 A phylogenetic tree reconstruction of Leishmania was implemented applying Bayesian inference with the Mr. Bayes v. 3.2 program (Bayesian inference of phylogeny, Rochester, NY). Ten Markov chains were proceeded for eight millions of generations, and trees were sampled for every 1,000 generations. Twenty-five percent of the sample tree were discarded, and the remaining trees were used to build up a consensus tree and calculation of posterior probabilities of clades. The results of Bayesian analyses were visualized using Figtree v. 1.4.2 (Edinburgh, Scotland). The hsp70 gene sequence Trypanozoma cruzi was used as root. Transformation of the leaves and schematic representation of the root were applied for visualization purposes.
The p distance among group of sequences was assessed using the software MegaX44 (Philadelphia, PA) set up to perform a bootstrap analysis of 1,000 replicates to estimate variation, including a Gamma correction of inter-site variation using four categories as parameter and also set up to delete gaps as missing treatment. Both transitions and transversions were included in the calculation of P-distances.
RESULTS
Hsp70-RFLP typing.
Four hundred seventy-five PCR products of hsp70 gene were digested with Hae III and subsequently with BccI or RsaI endonucleases. After comparing the digestion patterns with the ones observed in reference strains, 371 (78.1%) showed a restriction pattern coincident with L. (V.) panamensis, 90 (18.9%) with L. (V.) guyanensis, and 14 (3.0%) with L. (V.) braziliensis. Table 1 shows the hsp70-RFLP typing results obtained in each study site.
Table 1.
Characterization of Leishmania species by hsp70-RFLP analysis of skin scrapings from Panamanian cutaneous leishmaniasis patients
| Site of study | Typed samples | hsp70 PCR-RFLP* | ||
|---|---|---|---|---|
| BccI pattern | RsaI pattern | |||
| Leishmania panamensis | L. guyanensis | L. braziliensis | ||
| Panamá (site 1) | 266 | 210 (78.9%) | 46 (17.3%) | 10 (3.8%) |
| Coclé (site 2) | 161 | 115 (71.4%) | 42 (26.1%) | 4 (2.5%) |
| Bocas del Toro (site 3) | 48 | 46 (95.8%) | 2 (4.2%) | – |
| Total | 475 | 371 (78.1%) | 90 (18.9%) | 14 (3.0%) |
L. braziliensis = Leishmania braziliensis; L. guyanensis = Leishmania guyanensis.
After hsp 70 gene sequencing (1,245 bp), most samples typed as L. guyanensis or L. braziliensis regrouped into minor clusters near to reference strains.
Multi-locus enzyme electrophoresis (MLEE).
Two hundred twenty-eight positive cultures were typed by MLEE and compared with hsp70-RFLP typing results (Table 2). The isolates characterized as L. (V.) panamensis by MLEE (n = 202) showed a perfect agreement with hsp70-RFLP typing results. Three isolates characterized as L. (V.) guyanensis by hsp70-RFLP coincided with the MLEE typing result. However, 15 isolates considered L. (V.) guyanensis by hsp70-RFLP were characterized by MLEE as L. (V.) panamensis. Five isolates characterized by hsp70-RFLP as L. (V.) braziliensis were also confirmed by MLEE. In addition, three samples identified as L. (V.) braziliensis by hsp70-RFLP were assigned as L. (V.) braziliensis/L. (V.) panamensis hybrid isolates by MLEE.
Table 2.
Discrimination of Leishmania Viannia species from Panamanian cutaneous leishmaniasis patients by hsp70PRC-RFLP, MLEE, and sequencing
| Leishmania Viannia species | hsp70 PCR-RFLP (n = 228) | MLEE (n = 228) | Sequencing (n = 77) |
|---|---|---|---|
| L. panamensis | 202 | 202 | 13 |
| L. guyanensis | 15 | 15 (L. panamensis) | 15 + 38* (Leishmania sp. 1) |
| L. guyanensis | 3 | 3 | 3 |
| L. braziliensis | 5 | 5 | 1 (Leishmania braziliensis outlier) |
| 2 (Leishmania naiffi) | |||
| 2 (Leishmania sp. 2) | |||
| L. braziliensis | 3 | 3 (L. braziliensis/L. panamensis) | 3 (L. braziliensis/L. guyanensis) |
L. (V.) braziliensis = Leishmania (Viannia) braziliensis; L. (V.) guyanensis = Leishmania (Viannia) guyanensis; L. (V.) panamensis = Leishmania (Viannia) panamensis; MLEE = Multi-locus enzyme electrophoresis. Hsp70PCR-RFLP: PCR products were digested with Hae III and subsequently with BccI or RsaI endonucleases. Sequencing: hsp70 PCR-amplified product sequencing of both strands using primers F25, R1310, 6F and R617, followed by phylogenetic analysis. Results that do not match hsp70 RFLP analysis are shown in parentheses.
Besides 15 cultured samples, 38 DNA samples from skin scraping found as L. guyanensis by hsp70 PCR-RFLP were sequenced.
Hsp70 sequencing and phylogenetic analysis.
To confirm the hsp70-RFLP and MLEE typing results, we further sequenced 1,245 bp of the hsp70 gene from 39 isolated Leishmania parasites (positive cultures): 13 samples typed as L. (V.) panamensis by hsp70-RFLP and 26 samples typed as non-panamensis Leishmania by hsp70-RFLP (18 typed as L. [V.] guyanensis and eight as L. [V.] braziliensis). In addition, we sequenced the following reference strains: L. (V.) panamensis (WR 2306), L. (V.) guyanensis (M4147), L. (V.) braziliensis (M2903), and L. (V.) peruviana (WR 2771).
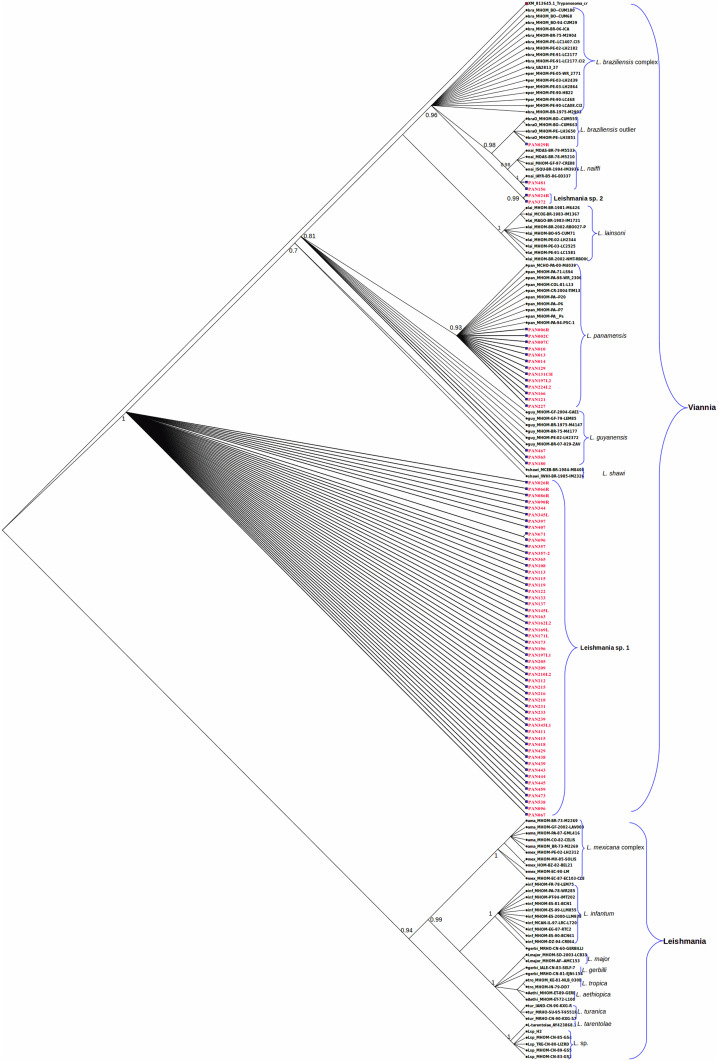
The sequences from the 13 samples that were typed as L. (V.) panamensis by hsp70-RFLP clearly clustered with reference sequences belonging to L. (V.) panamensis (Figure 2). Of the 18 samples that were typed as L. (V.) guyanensis by hsp70-RFLP, three grouped in the cluster of L. (V.) guyanensis reference strains. The remainder subset of 15 samples (here named as Leishmania [V.] sp. 1) clustered together in a distinct group, but close to L. (V.) guyanensis and Leishmania (Viannia) shawi reference strains (Figure 2). In addition, 38 lesion scraping samples identified as L. (V.) guyanensis by hsp70-RFLP were also sequenced. All of them clustered with the previously found Leishmania (V.) sp. 1 group.
Figure 2.
Hsp70-based Bayesian phylogenetic tree of Leishmania species obtained constructed under the GTR (Nst = 6) model with a gamma rate of four. The Bayesian consensus tree was searched by the program Mr. Bayes after 8,000,000 cycles of the Markov Chain Monte Carlo algorithm with a subsampling frequency of 1,000. Clade credibility values are shown as values at each clade node. Reference sequence codes appear in black color. All sequence codes obtained in this study are highlighted in red color. Reference sequences are abbreviated as follows: bra: Leishmania braziliensis; braO: Leishmania braziliensis outlier; guy: Leishmania guyanensis; lai: Leishmania lainsoni; nai: Leishmania naiffi; pan: Leishmania panamensis; per: Leishmania peruviana. This figure appears in color at www.ajtmh.org.
Sequencing results from the eight samples that were typed as L. (V.) braziliensis by hsp70-RFLP were as follows: one was grouped with L. (V.) braziliensis outlier reference strains, two clustered in the L. (V.) naiffi reference strains, and two samples were grouped together in a separate group (here named as Leishmania [V.] sp. 2), independent of the L. (V.) braziliensis/L. (V.) peruviana and L. (V.) braziliensis outlier reference sequences (Figure 2). Finally, the other three isolates identified as L. (V.) braziliensis by hsp70-RFLP and as hybrids by MLEE showed ambiguous nucleotides in their hsp70 sequences that suggest a possible event of genetic exchange between parental strains L. (V.) braziliensis and L. (V.) guyanensis. The sequences of these hybrid isolates were not included in the phylogenetic analysis presented in Figure 2. The different parasites found in the three regions, based on hsp70 sequence analysis, are presented in Table 3.
Table 3.
Characterization of Leishmania (Viannia) species by hsp70 gene sequencing from Panamanian cutaneous leishmaniasis patients
| Typed samples | hsp 70 gene sequencing (1,245 bp)* | |||||||
|---|---|---|---|---|---|---|---|---|
| Site of study | Leishmania (Viannia) panamensis | L. (V.) guyanensis | L. (V.) braziliensis outlier | Leishmania (Viannia) naiffi | L. (V.) braziliensis/guyanensis | L. (V.) sp. 1 | L. (V.) sp. 2 | |
| Panamá (site 1) | 51 | 13 | 2† | 1 | 1 | 2 | 31 | 1 |
| Coclé (site 2) | 23 | – | 1 | – | 1 | 1 | 20 | 1 |
| Bocas del Toro (site 3) | 2 | – | – | – | – | – | 2 | – |
| Total | 77 | 13 | 3 | 1 | 2 | 3 | 53 | 2 |
L. (V.) braziliensis = Leishmania (Viannia) braziliensis; L. (V.) guyanensis = Leishmania (Viannia) guyanensis.
Hsp70 PCR-amplified products sequencing of both strands using primers F25, R1310, 6F, and R617, followed by phylogenetic analysis.
One of these two cases was found to be imported from French Guyana.
Leishmania hsp70 sequences obtained in this work (N = 77) were deposited in GenBank-National Center for Biotechnology Information (accession numbers can be found in Supplementary Information 2). For the phylogenetic analysis, we also re-sequenced hsp70 gene of cultured reference strains, and in addition, we retrieved 92 Leishmania sp. hsp70 reference sequences from GenBank. The final alignment contained 168 sequences including 77 sequences obtained in this study. The Bayesian phylogenetic tree is shown in Figure 2.
DISCUSSION
In this study, we characterized the Leishmania parasites present in 475 CL Panamanian patients from different geographical areas. The findings confirm that L. (V.) panamensis is the species most often associated (78.1%) with CL cases from the evaluated endemic areas in Panama. However, we also described, based on hsp70 gene partial sequencing and MLEE, a sympatry of L. (V.) panamensis with L. (V.) braziliensis, L. (V.) naiffi, L. (V.) guyanensis, hybrid isolates, and two genetic variants (named here as Leishmania (V.) sp. 1 and Leishmania (V.) sp. 2) (Supplemental Information 2). No evidence of the existence of parasites of the L. (L.) mexicana complex was found.
The Bayesian phylogenetic analysis based on hsp70 gene revealed that 53 samples analyzed belonged to Leishmania (V.) sp. 1, a cluster close to L. (V.) guyanensis and L. (V.) shawi and appeared to be closely related to the last species (P = 0.033 and P = 0.028, respectively). On the other hand, two samples analyzed in this study grouped together in a new cluster denominated Leishmania (V.) sp. 2 near the group of L. (V.) braziliensis complex in the cladogram (Figure 2). It seems that these two samples belong to the L. (V.) braziliensis complex as suggested by the P distance between groups (P = 0.0012). Studies using other markers are ongoing to reveal the status of Leishmania (V.) sp. 1 and Leishmania (V.) sp. 2 groups in the L. (V.) guyanensis and L. (V.) braziliensis complexes, respectively.
The Leishmania (V.) sp. 1 genetic variant was detected in the three study sites, whereas samples of Leishmania (V.) sp. 2 were detected in sites 1 and 2 (Table 3). This finding, however, may be a sampling bias as the number of samples in site 3 was comparatively smaller. In general, it seems that the distribution of Leishmania (V.) sp. 1 and Leishmania (V.) sp. 2 is not restricted to particular endemic regions of Panama. However, the potential link between the eco-epidemiological characteristics of an endemic area and the frequency of CL cases induced by these genetic variants needs to be more adequately defined.
On the other hand, one sample characterized by hsp70-RFLP as L. (V.) braziliensis and by hsp70 sequencing as L. (V.) braziliensis “outlier” was collected in site 1 of the study. Epidemiological data suggest that this patient came from the region of Panama Oeste Province (near Panama City), confirming that currently L. (V.) braziliensis is infecting humans in Panamanian territory. In a previous 1990 study, L. (V.) braziliensis was reported near the border with Colombia, but the real origin of this sample was not totally defined.45 The L. (V.) braziliensis “outlier” cluster was originally named because in 2012, some isolates were identified as L. (V.) braziliensis by MLEE, but using molecular analyses, they clustered separately from the main L. (V.) braziliensis–L. (V.) peruviana clade.46 Also, it is known that parasites belonging to L. (V.) braziliensis “outlier” group can produce mucocutaneous lesions.39 The genetic diversity of L. (V.) braziliensis has been reported in isolates even from very close geographical areas.47 Thus, determining the degree of genetic variability presented by this species in Panamanian endemic regions will improve our knowledge about the epidemiology of CL in this country.
Another important typing result from this study were two samples identified initially by hsp70-RFLP as L. (V.) braziliensis, which after hsp70 sequencing were finally confirmed as L. (V.) naiffi. In this sense, it has been reported that the Rsa I enzyme used in our hsp70-RFLP approach is unable to distinguish between L. (V.) braziliensis and L. (V.) naiffi.38 However, the nucleotide sequence of the 1,245-bp hsp70 amplified product performed during the present study ruled out the possibility of misidentification of this species. It is also important to mention that a recent study using hsp70-RFLP describes that L. (V.) naiffi isolates can present polymorphisms that must be taken into consideration.48 The epidemiological data revealed that one of these two patients was infected in Darién Province near the Colombian border, whereas the other came from Cocle Province (Table 3). It should be emphasized that L. (V.) naiffi has been previously detected in sand flies in Barro Colorado Island located in Gatún Lake Panama Canal and in a hyperendemic CL community in the district of Capira, province of Panama Oeste.25,26 Cutaneous leishmaniasis cases caused by L. (V.) naiffi evolves with a benign clinical course, and there is currently no association observed between L. (V.) naiffi and mucosal leishmaniasis.49 However, therapeutic failure of leishmaniasis caused by L. (V.) naiffi has been reported.49 This species had not been reported as a CL etiological agent previously in Panama; nevertheless, it has been frequently described in Brazil, French Guyana, Surinam, and Ecuador.50–54
We also identified by hsp70-RFLP and sequencing three cases of CL caused by L. (V.) guyanensis in Panamanian patients. According to the epidemiological data, one of these patients acquired the infection in French Guyana where the infection with L. (V.) guyanensis is naturally endemic. This is the first report of an imported case of CL caused by L. (V.) guyanensis in Panama. The other two patients acquired the infection in Panama and Cocle regions (Table 3). In South America, L. (V.) guyanensis has been reported as responsible of CL in Colombia, Brazil, French Guyana, Surinam, Ecuador, Peru, and Venezuela52,55,56 transmitted by Nyssomyia umbratilis and Nyssomyia anduzei.54 The first of these vector species has not been found yet in Panama, but the second one was reported since 1972.57 Nevertheless, it is possible that other phlebomine sand fly species, from the about 76 species described in Panama,58 are able to transmit L. (V.) guyanensis. Early reports mention that L. (V.) guyanensis is eliminated with difficulty by pentavalent antimonials, with patients frequently needing many courses of treatment. By contrast, L. (V.) panamensis is in most cases susceptible to these drugs.59
Finally, three isolates were typified as hybrids (L. [V.] braziliensis/L. [V.] panamensis) after analysis by MLEE. Two of these patients came from site 1 and one from site 2 (Table 3), and were originally characterized by hsp70-RFLP as L. (V.) braziliensis. However, sequencing analysis of an hsp70 region showed a large number of ambiguities that were consistent with the hybrid condition of these isolates. Nevertheless, the sequences of analysis suggest that the most likely parental isolates were L. (V.) braziliensis and L. (V.) guyanensis. Although genetic exchange in Leishmania parasites is considered to be an uncommon event in South America,60 CL caused by hybrid Leishmania species has been reported in some countries of this region.61–65 Moreover, hybrid Leishmania species has been reported infecting sand fly vectors.66 Interestingly, these Leishmania hybrids are reported for the first time in Panama; however, we are unaware of the epidemiological and clinical consequences associated with them. In this sense, it is important to consider that Leishmania hybrids can have a strong selective advantage, capable of enhancing its virulence, modifying its tissue tropism, and conferring them drug resistance.66 Furthermore, hybrids can infect and be transmitted by new vectors, which could change the geographical distribution of the disease.66
Leishmania (V.) braziliensis, L. (V.) guyanensis, and L. (V.) panamensis are responsible for causing different clinical manifestations as localized CL (the most benign form), disseminated leishmaniasis, and mucosal leishmaniasis.67 Although L. (V.) guyanensis and L. (V.) panamensis are able to invade mucosal tissues,14,45,68 their involvement in mucosal disease is less destructive, differing from the chronic and severe forms characteristically caused by L. (V.) braziliensis.69 In Panama, mucosal involvement has been observed in 4.2% of positive patients from where L. (V.) panamensis has been demonstrated.22 However, based on the findings presented in this study, it is important to highlight that L. (V.) braziliensis and L. (V.) braziliensis outlier should be considered as potential etiological agents in mucosal leishmaniasis cases reported in Panama.
Relapses, therapeutic failure, and treatment resistance have been reported in human leishmaniasis caused by species of Leishmania subgenus Viannia.69–74 In this sense, the biology, clinical relevance, and response to treatment of the non–L. (V.) panamensis parasites (including genetic variants “Leishmania [V.] sp. 1, Leishmania [V.] sp. 2” and hybrids isolates) described in this study are uncertain, and therefore need further evaluation.
Although the hsp70 methodology used in this study has been recommended for typing Leishmania spp. from regions where many circulating species are endemic,75 its discriminatory power is not sufficient to clearly determine the phylogenetic position of some Leishmania species/genetic variants.48,76 Probably, the analyzed region of the hsp70 gene (1,245 pb) presents a low polymorphism to readily distinguish between closely related Leishmania species. In this regard, complementary studies using more powerful molecular analysis tools, like multi-locus sequence typing and complete genome sequencing,22–24,77,78 are necessary to clearly define the phylogenetic position of the Leishmania isolates (here referred as Leishmania (V.) sp. 1, Leishmania (V.) sp. 2) and hybrids.
The genetic variability of the Leishmania (Viannia) parasites has been already reported in South America.47,77,79–81 The presence and frequency of these species/species variants are linked to eco-biological conditions not clearly defined, or only partially in some studies.47 Nevertheless, like other pathogens, the genetic diversity could be linked to particular eco-epidemiological patterns, virulence, pathogenicity, and drug responses. In conclusion, the results here presented not only improve our knowledge about the genetic diversity of the parasites that cause localized CL in Panama but also highlight the need to perform additional studies with others molecular methodologies that confirm and extend these findings.
Supplemental tables and figures
ACKNOWLEDGMENTS
We thank all study participants and the local field and clinical staff in the Clinical Investigation Unit at the Gorgas Memorial Institute (ICGES); Hospital Aquilino Tejeira in Penonomé, Coclé Province; and Health Center Materno Infantil Sandra Hernández in Changuinola, Bocas del Toro Province. We acknowledge the National Research System (SNI-SENACYT-PANAMA) for supporting J. E. C., Y. M., J. A. S., J. M. P., N. S., and A. S. We also thank Alberto Cumbrera for the help with the design of the maps.
Note: Supplemental information and table appear at www.ajtmh.org.
REFERENCES
- 1.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M; WHO Leishmaniasis Control Team , 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7: e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey MS, Lockwood DNJ, 2007. Cutaneous leishmaniasis. Clin Dermatol 2: 203–211. [DOI] [PubMed] [Google Scholar]
- 3.Leon LL, Machado GM, Barral A, de Carvalho-Paes LE, Grimaldi Júnior G, 1992. Antigenic differences among Leishmania amazonensis isolates and their relationship with distinct clinical forms of the disease. Mem Inst Oswaldo Cruz 87: 229–234. [DOI] [PubMed] [Google Scholar]
- 4.Passos VM, Fernandes O, Lacerda PA, Volpini AC, Gontijo CM, Degrave W, Romanha AJ, 1999. Leishmania (Viannia) braziliensis is the predominant species infecting patients with American cutaneous leishmaniasis in the State of Minas Gerais, Southeast Brazil. Acta Trop 72: 251–258. [DOI] [PubMed] [Google Scholar]
- 5.Berman J, 2006. Visceral leishmaniasis in the New World and Africa. Indian J Med Res 123: 289–294. [PubMed] [Google Scholar]
- 6.Vanloubbeeck Y, Jones DE, 2004. The immunology of Leishmania infection and the implications for vaccine development. Ann N Y Acad Sci 1026: 267–272. [DOI] [PubMed] [Google Scholar]
- 7.de Moura TR, et al. 2007. Enhanced Leishmania braziliensis infection following pre-exposure to sandfly saliva. PLoS Negl Trop Dis 1: e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David CV, Craft N, 2009. Cutaneous and mucocutaneous leishmaniasis. Dermatol Ther 22: 491–502. [DOI] [PubMed] [Google Scholar]
- 9.Organización Panamericana de la Salud/Organización Mundial de la Salud , 2015. Guía para el abordaje Integral de la Leishmaniasis en Panamá, 66. [Google Scholar]
- 10.Organización Panamericana de la Salud, 2013. LEISHMANIASIS Informe Epidemiológico de Las Américas. Washington, DC: Organización Panamericana de la Salud. Available at: http://www.paho.org/hq/index.php?option=com_topics&view=readall&cid=6722&Itemid=&lang=es. Accessed September 28, 2020.
- 11.Yamada K, Valderrama A, Gottdenker N, Cerezo L, Minakawa N, Saldaña A, Calzada JE, Chaves LF, 2016. Macroecological patterns of American cutaneous leishmaniasis transmission across the health areas of Panamá (1980–2012). Parasite Epidemiol Control 1: 42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hotez PJ, Woc-Colburn L, Bottazzi ME, 2014. Neglected tropical diseases in Central America and Panama: review of their prevalence, populations at risk and impact on regional development. Int J Parasitol 44: 597–603. [DOI] [PubMed] [Google Scholar]
- 13.Van der Auwera G, Dujardin JC, 2015. Species typing in dermal leishmaniasis. Clin Microbiol Rev 28: 265–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen HA, Johnson CM, de Vásquez AM, 1984. Cutaneous leishmaniasis in Panama. A brief review. Rev Med Panama 9: 182–187. [PubMed] [Google Scholar]
- 15.Petersen J, Johnson CM, de Vásquez AM, Sáenz R, 1987. Cutaneous leishmaniasis caused by Leishmania mexicana amazonensis in Panama. Rev Med Panama 112: 158–164. [PubMed] [Google Scholar]
- 16.Miranda A, Carrasco R, Paz H, Pascale JM, Samudio F, Saldaña A, Santamaría G, Mendoza Y, Calzada JE, 2009. Molecular epidemiology of American tegumentary leishmaniasis in Panama. Am J Trop Med Hyg 81: 565–571. [DOI] [PubMed] [Google Scholar]
- 17.Kreutzer RD, Souraty N, Semko ME, 1987. Biochemical identities and differences among Leishmania species and subspecies. Am J Trop Med Hyg 36: 22–32. [DOI] [PubMed] [Google Scholar]
- 18.de Vasquez AM, Saenz RE, Petersen JL, Christensen HA, Johnson CM, 1990. Leishmania mexicana complex: human infections in the Republic of Panama. Am J Trop Med Hyg 43: 619–622. [DOI] [PubMed] [Google Scholar]
- 19.Christensen HA, de Vasquez AM, Petersen JL, 1999. Short report epidemiologic studies on cutaneous leishmaniasis in eastern Panama. Am J Trop Med Hyg 60: 54–57. [DOI] [PubMed] [Google Scholar]
- 20.Espinosa OA, Serrano MG, Camargo EP, Teixeira MMG, Shaw JJ, 2018. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology 145: 430–442. [DOI] [PubMed] [Google Scholar]
- 21.Herrer A, 1971. Leishmania hertigi sp. n., from the tropical porcupine, Coendou rothschildi Thomas. J Parasitol 57: 626–629. [PubMed] [Google Scholar]
- 22.Herrer A, Telford SR, Christensen HA, 1971. Enzootic cutaneous leishmaniasis in eastern Panama. I. Investigation of the infection among forest mammals. Ann Trop Med Parasitol 65: 349–358. [DOI] [PubMed] [Google Scholar]
- 23.Christensen HA, Fairchild GB, Herrer A, Johnson CM, Young DG, de Vásquez AM, 1983. The ecology of cutaneous leishmaniasis in the Republic of Panama. J Med Entomol 20: 463–484. [DOI] [PubMed] [Google Scholar]
- 24.Kreutzer RD, Corredor A, Grimaldi G, Grogl M, Rowton ED, Young DG, Morales A, McMahon-Pratt D, Guzman H, Tesh RB, 1991. Characterization of Leishmania colombiensis sp. n (Kinetoplastida: Trypanosomatidae), a new parasite infecting humans, animals, and phlebotomine sand flies in Colombia and Panama. Am J Trop Med Hyg 44: 662–675. [DOI] [PubMed] [Google Scholar]
- 25.Azpurua J, De La Cruz D, Valderama A, Windsor D, 2010. Lutzomyia sand fly diversity and rates of infection by Wolbachia and an exotic Leishmania species on Barro Colorado Island, Panama. PLoS Negl Trop Dis 4: e627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigg CA, Calzada JE, Saldaña A, Perea M, Chaves LF, Valderrama A, 2019. Leishmania spp. infection rate and feeding patterns of sand flies (Diptera: Psychodidae) from a hyperendemic cutaneous leishmaniasis community in Panamá. Am J Trop Med Hyg 100: 798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Organización Panamericana de la Salud, 2019. LEISHMANIASIS Informe Epidemiológico de Las Américas. Washington, DC: Organización Panamericana de la Salud. Available at: http://www.paho.org/hq/index.php?option=com_topics&view=readall&cid=6722&Itemid=&lang=es. Accessed September 28, 2020.
- 28.Berman JD, 1997. Human leishmaniasis: clinical, diagnostic, and chemotherapeutic developments in the last 10 years. Clin Infect Dis 24: 684–703. [DOI] [PubMed] [Google Scholar]
- 29.Arevalo J, et al. 2007. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J Infect Dis 195: 1846–1851. [DOI] [PubMed] [Google Scholar]
- 30.Croft SL, Yardley V, Kendrick H, 2002. Drug sensitivity of Leishmania species: some unresolved problems, 2002. Trans R Soc Trop Med Hyg 96 (Suppl 1): S127–S129. [DOI] [PubMed] [Google Scholar]
- 31.Montalvo AM, Fraga J, Monzote L, Montano I, De Doncker S, Dujardin JC, Van der Auwera G, 2010. Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology 137: 1159–1168. [DOI] [PubMed] [Google Scholar]
- 32.Quaresma PF, et al. 2018. Distinct genetic profiles of Leishmania (Viannia) braziliensis associate with clinical variations in cutaneous-leishmaniasis patients from an endemic area in Brazil. Parasitology 145: 1161–1169. [DOI] [PubMed] [Google Scholar]
- 33.Rêgo FD, Cristina A, Mariano V, Antônia A, Pereira S, Quaresma PF, Pascoal-xavier MA, Shaw JJ, Maria C, Gontijo F, 2018. Genetic variant strains of Leishmania (Viannia) braziliensis exhibit distinct biological behaviors. Parasitol Res 117: 3157–3168. [DOI] [PubMed] [Google Scholar]
- 34.Corredor A, Kreutzer RD, Tesh RB, Boshell J, Palau MT, Caceres E, Duque S, Pelaez D, Rodriguez G, Nichols S, 1990. Distribution and etiology of leishmaniasis in Colombia. Am J Trop Med Hyg 42: 206–214. [DOI] [PubMed] [Google Scholar]
- 35.Programa de Estudio y Control de Enfermedades tropicales (PECET), 2010. Manual Diagnóstico y Control de la Leishmaniasis en Centroamérica. Medellín, Colombia: Universidad de Antioquía, 209.
- 36.Sosa N, et al. 2019. Topical paromomycin for New World cutaneous leishmaniasis. PLoS Negl Trop Dis 13: e0007253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vergel C, Walker J, Saravia NG, 2005. Amplification of human DNA by primers targeted to Leishmania kinetoplast DNA and post-genome considerations in the detection of parasites by a polymerase chain reaction. Am J Trop Med Hyg 72: 423–429. [PubMed] [Google Scholar]
- 38.Montalvo AM, Fraga J, Maes I, Dujardin JC, Van der Auwera G, 2012. Three new sensitive and specific heat-shock protein 70 PCRs for global Leishmania species identification. Eur J Clin Microbiol Infect Dis 231: 1453–1461. [DOI] [PubMed] [Google Scholar]
- 39.Van der Auwera G, Maes I, De Doncker S, Ravel C, Cnops L, Van Esbroeck M, Van Gompel A, Clerinx J, Dujardin JC, 2013. Heat-shock protein 70 gene sequencing for Leishmania species typing in European tropical infectious disease clinics. Euro Surveill 18: 20543. [DOI] [PubMed] [Google Scholar]
- 40.Kreutzer RD, Christensen HA, 1980. Characterization of Leishmania spp. by isozyme electrophoresis. Am J Trop Med Hyg 29: 199–208. [DOI] [PubMed] [Google Scholar]
- 41.Kreutzer RD, Semko ME, Hendricks LD, Wright N, 1983. Identification of Leishmania spp. by multiple isozyme analysis. Am J Trop Med Hyg 32: 703–715. [DOI] [PubMed] [Google Scholar]
- 42.Okonechnikov K, et al. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28: 1166–1167. [DOI] [PubMed] [Google Scholar]
- 43.Darriba D, Taboada GL, Doallo R, Posada D, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Li M, Knyaz C, Tamura K, 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santrich C, Segura I, Arias AL, Saravia NG, 1990. Mucosal disease caused by Leishmania braziliensis guyanensis. Am J Trop Med Hyg 42: 51–55. [DOI] [PubMed] [Google Scholar]
- 46.Odiwuor S, Veland N, Maes I, Arévalo J, Dujardin JC, Van der Auwera G, 2012. Evolution of the Leishmania braziliensis species complex from amplified fragment length polymorphisms, and clinical implications. Infect Genet Evol 12: 1994–2002. [DOI] [PubMed] [Google Scholar]
- 47.Figueiredo de Sá BSL, Rezende AM, de Melo Neto OP, de Brito MEF, Brandão Filho SP, 2019. Identification of divergent Leishmania (Viannia) braziliensis ecotypes derived from a geographically restricted area through whole genome analysis. PLoS Negl Trop Dis 13: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espada CR, Ortiz PA, Shaw JJ, Barral AMP, Costa JML, Uliana SRB, Coelho AC, 2018. Identification of Leishmania (Viannia) species and clinical isolates of Leishmania (Leishmania) amazonensis from Brazil using PCR-RFLP of the heat-shock protein 70 gene reveals some unexpected observations. Diagn Microbiol Infect Dis 91: 312–318. [DOI] [PubMed] [Google Scholar]
- 49.Fagundes-Silva GA, Sierra Romero GA, Cupolillo E, Gadelha Yamashita EP, Gomes-Silva A, De Oliveira Guerra JA, Da-Cruz AM, 2015. Leishmania (Viannia) naiffi: rare enough to be neglected? Mem Inst Oswaldo Cruz 110: 797–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naiff RD, Freitas RA, Naiff MF, Arias JR, Barrett TV, Momen H, Grimaldi Júnior G, 1989. Epidemiological and nosological aspects of Leishmania naiffi Lainson and Shaw, 1989. Mem Inst Oswaldo Cruz 86: 317–321. [DOI] [PubMed] [Google Scholar]
- 51.Pratlong F, Deniau M, Darie H, Eichenlaub S, Pröll S, Garrabe E, le Guyadec T, Dedet JP, 2002. Human cutaneous leishmaniasis caused by Leishmania naiffi is wide-spread in South America. Ann Trop Med Parasitol 96: 781–785. [DOI] [PubMed] [Google Scholar]
- 52.Lainson R, 2010. The Neotropical Leishmania species: a brief historical review of their discovery, ecology and taxonomy. Rev Pan-Amazônica Saúde 1: 13–32. [Google Scholar]
- 53.van Thiel PPAM, van Gool T, Kager PA, Bart A, 2010. First cases of cutaneous leishmaniasis caused by Leishmania (Viannia) naiffi infection in Surinam. Am J Trop Med Hyg 82: 588–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato H, Calvopiña M, Criollo H, Hashiguchi Y, 2013. First human cases of Leishmania (Viannia) naiffi infection in Ecuador and identification of its suspected vector species. Acta Trop 128: 710–713. [DOI] [PubMed] [Google Scholar]
- 55.Silveira FT, Lainson R, Corbett CEP, 2004. Clinical and immunopathological spectrum of American cutaneous leishmaniasis with special reference to the disease in Amazonian Brazil: a review. Mem Inst Oswaldo Cruz 99: 239–251. [DOI] [PubMed] [Google Scholar]
- 56.Montalvo AM, Fraga J, Montano I, Monzote L, Van der Auwera G, Marín M, Muskus C, 2016. Molecular identification of Leishmania spp. clinical isolates from Colombia based on hsp70 gene. Biomedica 36: 37–44. [DOI] [PubMed] [Google Scholar]
- 57.Christensen HA, 1972. Check list of the phlebotomine sand flies (Díptera: Psychodidae) of Panama including two species not previously reported. Mosq News 32: 88–89. [Google Scholar]
- 58.Dutari LC, Loaiza JR, 2014. American cutaneous leishmaniasis in Panama: a historical review of entomological studies on anthropophilic Lutzomyia sand fly species. Parasit Vectors 7: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lainson R, Shaw J, 1987. Evolution, classification and geographical distribution. Peter W, Killick-Kendrick R, eds. The Leishmaniasis in Biology and Medicine. London, United Kingdom: Academic Press, 1–120. [Google Scholar]
- 60.Akhoundi M, et al. 2017. Leishmania infections: molecular targets and diagnosis. Mol Aspects Med 57: 1–29. [DOI] [PubMed] [Google Scholar]
- 61.Belli AA, Miles MA, Kelly JM, 1994. A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology 109: 435–442. [DOI] [PubMed] [Google Scholar]
- 62.Delgado O, Cupolillo E, Bonfante-Garrido R, Silva S, Belfort E, Grimaldi Júnior G, Momen H, 1997. Cutaneous leishmaniasis in Venezuela caused by infection with a new hybrid between Leishmania (Viannia) braziliensis and L. (V.) guyanensis. Mem Inst Oswaldo Cruz 92: 581–582. [DOI] [PubMed] [Google Scholar]
- 63.Kato H, Cáceres AG, Hashiguchi Y, 2016. First evidence of a hybrid of Leishmania (Viannia) braziliensis/L. (V.) peruviana DNA detected from the phlebotomine sand fly Lutzomyia tejadai in Peru. PLoS Negl Trop Dis 10: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nolder D, Roncal N, Davies CR, Llanos-Cuentas A, Miles MA, 2007. Multiple hybrid genotypes of Leishmania (Viannia) in a focus of mucocutaneous leishmaniasis. Am J Trop Med Hyg 76: 573–578. [PubMed] [Google Scholar]
- 65.Dujardin JC, Bañuls AL, Llanos-Cuentas A, Alvarez E, DeDoncker S, Jacquet D, Le Ray D, Arevalo J, Tibayrenc M, 1995. Putative Leishmania hybrids in the eastern Andean valley of Huanuco, Peru. Acta Trop 59: 293–307. [DOI] [PubMed] [Google Scholar]
- 66.Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM, Sacks DL, 2009. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324: 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torres-Guerrero E, Quintanilla-Cedillo MR, Ruiz-Esmenjaud J, Arenas R, 2017. Leishmaniasis: a review. F1000Res 6: 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osorio LE, Castillo CM, Ochoa MT, 1998. Mucosal leishmaniasis due to Leishmania (Viannia) panamensis in Colombia: clinical characteristics. Am J Trop Med Hyg 59: 49–52. [DOI] [PubMed] [Google Scholar]
- 69.Marsden PD, 1986. Mucosal leishmaniasis (“espundia” Escomel, 1911). Trans R Soc Trop Med Hyg 80: 859–876. [DOI] [PubMed] [Google Scholar]
- 70.Souza AS, Giudice A, Pereira JM, Guimarães LH, de Jesus AR, de Moura TR, Wilson ME, Carvalho EM, Almeida RP, 2010. Resistance of Leishmania (Viannia) braziliensis to nitric oxide: correlation with antimony therapy and TNF-alpha production. BMC Infect Dis 10: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fernández O, Diaz-Toro Y, Valderrama L, Ovalle C, Valderrama M, Castillo H, Perez M, Saravia NG, 2012. Novel approach to in vitro drug susceptibility assessment of clinical strains of Leishmania spp. J Clin Microbiol 50: 2207–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peláez RG, Muskus CE, Cuervo P, Marín-Villa M, 2012. Differential expression of proteins in Leishmania (Viannia) panamensis associated with mechanisms of resistance to meglumine antimoniate. Biomedica 32: 418–429. [DOI] [PubMed] [Google Scholar]
- 73.Fernández OL, Diaz-Toro Y, Ovalle C, Valderrama L, Muvdi S, Rodríguez I, Gomez MA, Saravia NG, 2014. Miltefosine and antimonial drug susceptibility of Leishmania Viannia species and populations in regions of high transmission in Colombia. PLoS Negl Trop Dis 8: e2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Couto DV, Medeiros MZ, Barbosa AB, Faria C, Vicari S, 2014. American tegumentary leishmaniasis - a case of therapeutic challenge. An Bras Dermatol 89: 974–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montalvo AM, Fraga J, Tirado D, Blandón G, Alba A, Van der Auwera G, Vélez ID, Muskus C, 2017. Detection and identification of Leishmania spp.: application of two hsp70-based PCR-RFLP protocols to clinical samples from the New World. Parasitol Res 116: 1843–1848. [DOI] [PubMed] [Google Scholar]
- 76.Van der Auwera G, et al. 2016. Comparison of Leishmania typing results obtained from 16 European clinical laboratories in 2014. Euro Surveill 21: 30418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herrera G, Hernández C, Ayala MS, Flórez C, Teherán AA, Ramírez JD, 2017. Evaluation of a multilocus sequence typing (MLST) scheme for Leishmania (Viannia) braziliensis and Leishmania (Viannia) panamensis in Colombia. Parasit Vectors 10: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Restrepo CM, Llanes A, De La Guardia C, Lleonart R, 2015. Genome-wide discovery and development of polymorphic microsatellites from Leishmania panamensis parasites circulating in central Panama. Parasit Vectors 8: 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patino LH, Muñoz M, Muskus C, Méndez C, Ramírez JD, 2020. Intraspecific genomic divergence and minor structural variations in Leishmania (Viannia) panamensis. Genes (Basel) 11: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuhls K, Cupolillo E, Silva SO, Schweynoch C, Boité MC, Mello MN, Mauricio I, Miles M, Wirth T, Schönian G, 2013. Population structure and evidence for both clonality and recombination among Brazilian strains of the subgenus Leishmania (Viannia). PLoS Negl Trop Dis 7: e2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cupolillo E, Brahim LR, Toaldo CB, de Oliveira-Neto MP, de Brito MEF, Falqueto A, de Farias Naiff M, Grimaldi G, 2003. Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. J Clin Microbiol 41: 3126–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.