Abstract
Reactive oxygen species are toxic byproducts of aerobic respiration that are also important in mediating a diversity of cellular functions. Reactive oxygen species form an important component of plant defenses to inhibit microbial pathogens during pathogen–plant interactions. Tolerance to oxidative stress is likely to make a significant contribution to the viability and pathogenicity of plant pathogens, but the complex network of oxidative stress responses hinders identification of the genes contributing to this trait. Here, we employed a forward genetic approach to investigate the genetic architecture of oxidative stress tolerance in the fungal wheat pathogen Zymoseptoria tritici. We used quantitative trait locus (QTL) mapping of growth and melanization under axenic conditions in two cross-populations to identify genomic regions associated with tolerance to oxidative stress. We found that QTLs associated with growth under oxidative stress as well as inherent growth can affect oxidative stress tolerance, and we identified two uncharacterized genes in a major QTL associated with this trait. Our data suggest that melanization does not affect tolerance to oxidative stress, which differs from what was found for animal pathogens. This study provides a whole-genome perspective on the genetic basis of oxidative stress tolerance in a plant pathogen.
Keywords: quantitative trait locus mapping, oxidative stress, plant pathogen, fungi, Zymoseptoria tritici
Introduction
One of the biggest challenges in biology is to understand the genetic basis of phenotypic variation in quantitative traits, which are regulated by a complex network of genes (Mackay et al. 2009). Finding the causal variants affecting quantitative traits, known as quantitative trait loci (QTL), is an important goal for evolutionary biologists as well as plant and animal breeders (Slate 2004; Mackay et al. 2009). For decades, linkage mapping has been applied to identify QTLs by combining quantitative phenotypic data with genetic variants in cross-populations segregating different phenotypes (Slate 2004; Mackay et al. 2009).
Linkage mapping in fungal plant pathogens remains rare (Foulongne-Oriol 2012) though it can contribute to our understanding of plant–pathogen interactions and the development of improved methods of disease control (Croll and McDonald 2017; Genissel et al. 2017). Mapping in plant pathogens has been oriented mainly toward identifying genes that have large effects on virulence, especially avirulence effector genes that directly interact with plant resistance genes during gene-for-gene interactions (Linning et al. 2004; Gout et al. 2006; Fudal et al. 2007). Other genes encoding functions that affect pathogen virulence, such as manipulation of host immunity and tolerance of chemicals elicited by host defense (Dodds and Rathjen 2010), have been largely neglected. These genes may show additive effects that contribute to quantitative virulence, but can be difficult to detect due to their small individual effects in planta. An alternative way to identify these genes is to perform QTL mapping for a specific quantitative virulence trait in vitro, and then explore the functions of detected candidate QTL genes in planta.
Tolerance to reactive oxygen species (ROS) is an important trait that is likely to affect the virulence of plant pathogens. ROS are a group of highly reactive chemical species (e.g., H2O2, •, •OH) that can positively or negatively affect many cellular processes (Heller and Tudzynski 2011; Lambeth and Neish 2014). Historically, ROS were recognized as toxic byproducts of metabolic processes associated with an aerobic lifestyle that can oxidize macro-biomolecules and damage cellular structures, resulting in the disturbance of normal metabolism and ultimately cell death (Lambeth and Neish 2014). However, in recent decades, ROS were found to play positive roles in mediating a variety of cellular responses and interactions between different cells and organisms (Heller and Tudzynski 2011; Shapiguzov et al. 2012; Lambeth and Neish 2014).
For fungal plant pathogens, it is essential to maintain a beneficial ROS homeostasis. ROS are needed to regulate fungal development and fungal cell differentiation and to mediate the fungal response to environmental changes (Heller and Tudzynski 2011; Segal and Wilson 2018). Some necrotrophic pathogens are thought to benefit by inducing ROS to kill plant cells during infection (Williams et al. 2011). On the other hand, many plants produce ROS as part of their defense response to restrict the growth of pathogens during plant infection (Doehlemann and Hemetsberger 2013). Better knowledge of how plant pathogens maintain ROS homeostasis and tolerate oxidative stress imposed by their hosts during infection will improve our understanding of plant–pathogen interactions.
Several molecular mechanisms were reported to affect oxidative stress tolerance in fungi. Enzymes such as superoxide dismutases (SOD), catalases (CAT), and peroxidases, and small protein molecules such as thioredoxin and glutathione are known to reduce ROS (Thorpe et al. 2004; Heller and Tudzynski 2011; Papadakis and Workman 2015; Broxton and Culotta 2016). These ROS-related proteins were found to be largely regulated by the transcription factor Yap1 and MAPK pathways (Molina and Kahmann 2007; Segmüller et al. 2007; Walia and Calderone 2008; Lin et al. 2009; Ronen et al. 2013; Yu et al. 2017; Segal and Wilson 2018). Other mechanisms regulating oxidative stress responses were hypothesized when deletion of some of these genes did not result in observable effects on growth of the mutants under oxidative stress (Grant et al. 1997; Rolke et al. 2004; Heller and Tudzynski 2011; Gohari 2015; Segal and Wilson 2018). One of the major contributors to ROS production, the NADPH oxidase complex (NOX), was also reported to affect tolerance to oxidative stress in vitro (Chen et al. 2014; Lambeth and Neish 2014), but it is unclear how this would occur.
The pigment melanin was shown to protect animal pathogens from high doses of ROS during phagocytosis (Missall et al. 2004) and was suggested to protect pathogen spores from oxidative stress during plant infection and from abiotic stresses such as temperature extremes and UV radiation (Henson et al. 1999; Jacobson 2000). But it remains unclear whether melanin contributes to oxidative stress tolerance and its effect has not been analyzed quantitatively.
The response to oxidative stress is a quantitative trait that depends on a complex molecular network. Transcriptomic studies in fission yeast showed that hundreds of genes are induced by oxidative stress and that their expressions are affected by the ROS exposure time, dose, cellular location, and species (Chen et al. 2008). Investigating such a complex gene network based only on gene mutations is tedious and difficult. A different approach is to conduct QTL mapping to identify genes associated with tolerance to oxidative stress using a whole-genome perspective. Unfortunately, such genome-wide studies are rare in fungal species (Chen et al. 2003, 2008; Diezmann and Dietrich 2011), let alone in fungal plant pathogens, due to the difficulties in constructing suitable mapping populations, especially for fungal species that lack a sexual phase (Foulongne-Oriol 2012), as well as limitations in creating informative and reproducible quantitative assays for relevant phenotypes. We hypothesized that linkage mapping in plant fungal pathogens would provide more powerful insights into the evolution of this complex trait by enabling a whole-genome perspective. Combining genotypic and phenotypic variation in linkage mapping can enable identification of important genomic regions or genes that contribute a large fraction of the variation in oxidative stress tolerance. Mapping studies utilizing wild-type strains can provide insight into the evolution of this trait in natural populations. Utilizing more than one mapping population can enable a more comprehensive view of the genetic architecture of this trait.
The fungal wheat pathogen Zymoseptoria tritici, previously known as Mycosphaerella graminicola, is the most devastating crop pathogen in Europe (Fones and Gurr 2015). Zymoseptoria tritici causes septoria tritici leaf blotch which can reduce wheat yields by up to 50% (Eyal et al. 1987; Torriani et al. 2015). Zymoseptoria tritici has a long asymptomatic phase at the beginning of infection (Kema et al. 1996), sometimes called a biotrophic phase, lasting for 7- to 10-day post-inoculation (dpi). This phase is rapidly followed by the necrotrophic phase, in which leaf lesions develop and asexual fruiting bodies (pycnidia) become visible (Sanchez-Vallet et al. 2015; Kettles and Kanyuka 2016). Over the last 20 years, Z. tritici has developed into an important model fungal pathogen for genetic studies. Linkage mapping in this pathogen resulted in the identification of genes regulating virulence (Zhong et al. 2017; Meile et al. 2018; Stewart et al. 2018) and melanization (Lendenmann et al. 2014; Krishnan et al. 2018). Previous studies in Z. tritici illustrated the importance of ROS during plant infection (Shetty et al. 2003, 2007) and several homologs of genes encoding ROS scavengers in other fungi have been identified in Z. tritici (Mehrabi et al. 2006; Gohari 2015; Yang et al. 2015).
Here, we aimed to determine the genetic architecture of oxidative stress tolerance in Z. tritici and identify candidate genes affecting this trait. We performed quantitative trait locus (QTL) mapping of fungal growth and melanization in the presence or absence of hydrogen peroxide (H2O2) under axenic conditions in two segregating populations. Large QTLs with narrow confidence intervals were carefully analyzed to identify the most promising candidate genes associated with tolerance to oxidative stress.
Materials and methods
Phenotyping for oxidative stress tolerance in vitro
Two crosses among four Swiss strains of Z. tritici (1E4 × 1A5 and 3D1 × 3D7) generated 249 and 257 offspring, respectively, as reported in previous studies (Lendenmann et al. 2014, 2015, 2016). The four parents and their 506 progeny were divided into 20 sets of strains to phenotype for oxidative stress tolerance as follows. Spores were recovered from glycerol stocks kept at −80°C and grown in YPD (yeast potato dextrose) media for 5 days at 18°C on a shaker at 120 rpm. The spore suspensions were filtered through a double layer of sterilized cheesecloth and then centrifuged at 3273 g for 15 min. The spore pellets were diluted to a concentration of 200 spores per ml, and 200 µl of the spore solution was added to each PDA (Difco potato dextrose agar) Petri plate to obtain around 40 colonies per plate. A hydrogen peroxide solution was used to create the oxidative stress environment. For each liter of autoclaved and cooled (<60°C) PDA media, 113 µl of 30% H2O2 (Sigma–Aldrich Chemio GmbH) was added to produce a final concentration of 1.0 mM H2O2. Unamended PDA was used as the control environment. Each strain was inoculated onto three replicate Petri plates for each environment. All plates were incubated at 18°C in the dark. Digital images of each plate were acquired at 8 and 12 dpi as described by Lendenmann et al. (2014).
To measure colony area (cm2) and the degree of melanization, all images were processed in MatLab R2017b (The MathWorks, Inc., Natick, Massachusetts, United States) using a purpose-developed script (Supplementary File S1) based on instructions given in the Image Segmentation Tutorial by Image Analyst (https://ch.mathworks.com/matlabcentral/fileexchange/25157-image-segmentation-tutorial). Colony radii were calculated as radii = . To measure melanization of each colony, gray scale values were obtained from RGB color values by using the rgb2gray function in MatLab. Higher gray values indicate lower melanization on the 0–255 gray scale, where 0 is completely black and 255 is completely white. The average colony radius and average gray value for each isolate under each condition was calculated for each plate, and the mean value across the three replicates was used as the phenotype input for growth and melanization in the QTL mapping. We also calculated the growth rate and melanization rate between 8 and 12 dpi, as the colony growth between these time points was found to be linear in a previous study (Lendenmann et al. 2015). As most strains showed different growth patterns under stressed and non-stressed conditions, the relative values of growth or melanization, representing the percentage of growth reduction or increase in melanization due to oxidative stress, were calculated for each strain to estimate differences in sensitivity to oxidative stress among offspring. All phenotypes used as inputs for QTL mapping were calculated as shown below, and all statistical analyses were performed in R (R Core Team 2018).
Growth = Mean colony radius
Melanization = Mean colony gray value
Growth_Rate = (Radius12dpi − Radius8dpi)/4
Melanization_Rate = (Gray12dpi − Gray8dpi)/4
Value (relative) = Value (in oxidative stress environment)/Value (in control environment)
To estimate the phenotypic variation resulting from the genetic variation, broad-sense heritability was calculated as (Bloom et al. 2013), where is the genetic variance and is the residual variance. The calculation of these two variance components was performed using mixed models with strain, replicate and set as random effects in the sommer package (Covarrubias-Pazaran 2016) in R. To detect transgressive segregation, we performed Dunnett’s test to identify any segregants with mean values either significantly higher or lower than the parents (P < 0.001) (Johansen-Morris and Latta 2006). The Dunnett’s tests were performed by using replicates in the multcomp package (Hothorn et al. 2008) in R.
Genotype data and QTL mapping
All QTL analyses were performed using the R/qtl package (Broman et al. 2003) in R, following the instructions in “A Guide to QTL Mapping with R/qtl” (Broman and Sen 2009). Genes located within the 95% Bayes credibility intervals were identified according to the genome annotations of the reference parental strains (Plissonneau et al. 2018).
SNP data used for QTL mapping were obtained from RADseq data generated previously (Lendenmann et al. 2014), but using the finished genome sequence of one of the parental strains (Plissonneau et al. 2018) as the reference genome for each cross (1A5 for the 1E4 × 1A5 cross and 3D7 for the 3D1 × 3D7 cross). The SNP markers were generated and filtered as described in previous studies (Zhong et al. 2017; Meile et al. 2018). This provided 35,030 SNP markers in the 1E4 × 1A5 cross and 57,513 SNP markers in the 3D1 × 3D7 cross, leading to average marker distances of 0.31 and 0.10 cM (1145 and 658 bp) respectively. Single-QTL genome scans using standard interval mapping were performed via the “scanone” function to provide an overview of the genetic architecture of each trait. The significance of each QTL was assessed using 1000 permutation tests across the entire genome.
To detect additional QTLs and explore possible interactions among QTLs, we applied multiple-QTL models. We selected markers with a minimum distance of 0.0001 cM using the function “pickMarkerSubset” in the r/qtl package, leading to 8699 markers for the 1E4 × 1A5 cross and 9593 markers for the 3D1 × 3D7 cross. The penalized LOD score was calculated using “scantwo” with 1000 permutations across the whole genome with “method = ‘hk’”. Then the penalized LOD score criterion with significance level of 5% was estimated via the function “calc.penalties” from the permutation results. The best fit QTL model was then selected by using both forward and backward stepwise algorithms with the function “stepwiseqtl.”
Identification of candidate genes associated with oxidative stress tolerance
The confidence intervals of all QTLs were compared taking into consideration the QTL estimated effects. QTLs with overlapping confidence intervals (>2/3 of their length) and with the same estimated effects among similar traits were considered to be the same QTL. Unique QTLs which had no overlaps in confidence intervals were identified among the same QTLs.
In this study, we considered the fraction of growth retained by a fungal strain when grown under oxidative stress as a measure of its tolerance to oxidative stress. Hence, all factors that affect the growth under oxidative stress and the growth difference between the stressed and the control conditions are considered as affecting the tolerance, which means all QTLs detected in growth under oxidative stress and relative growth were recognized as QTLs related to oxidative stress tolerance. We classified QTLs that were found only in the presence of H2O2 as oxidative stress-specific QTLs. We classified all QTLs related to growth under the control conditions as general growth-related QTLs. Only QTLs related to oxidative stress tolerance were considered in the search for candidate genes. Genes were sought in the unique QTLs having the narrowest confidence intervals.
Gene models (Plissonneau et al. 2018) within the largest QTLs with the narrowest confidence intervals were confirmed by using previously published in planta expression data (Palma-Guerrero et al. 2017) at 7 dpi and in vitro expression data (Francisco et al. 2019) obtained for the four parental strains. The protein sequence of all candidate genes were BLASTed to the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm their functional domains and to search for homology to functionally characterized genes. Variation in DNA and protein sequences between the two parental strains for each cross was identified by performing sequence alignments (CLC Sequence Viewer 7.6.1, QIAGEN Aarhus A/S). Genes with at least one sequence variant between the two parental strains in either the protein-encoding sequence or the 5′- or 3′-UTR regions were considered to be candidates to explain the observed marker-trait associations. Candidates encoding domains involved in ROS generation and elimination, cellular growth, and transcription regulation of ROS- and growth-related genes were considered to be top candidates.
In planta expression of candidate genes
To infer the possible in planta function of candidate genes, we used the in planta expression data obtained in a previous study using the 3D7 parent (Palma-Guerrero et al. 2016), which consisted of five time points, covering the complete life cycle of the fungus in wheat leaves. A detailed analysis of the RNAseq data was given in Palma-Guerrero et al. 2016. We compared the expression of the top candidate genes to genes known to be involved in the oxidative stress response in fungi to infer their in planta functions in oxidative stress response.
Data availability
The genotype data, marker information, and phenotype data of the two cross-populations are available at figshare DOI: https://doi.org/10.25386/genetics.11862129.
Results
Tolerance to oxidative stress is affected by both oxidative stress response and inherent growth rate
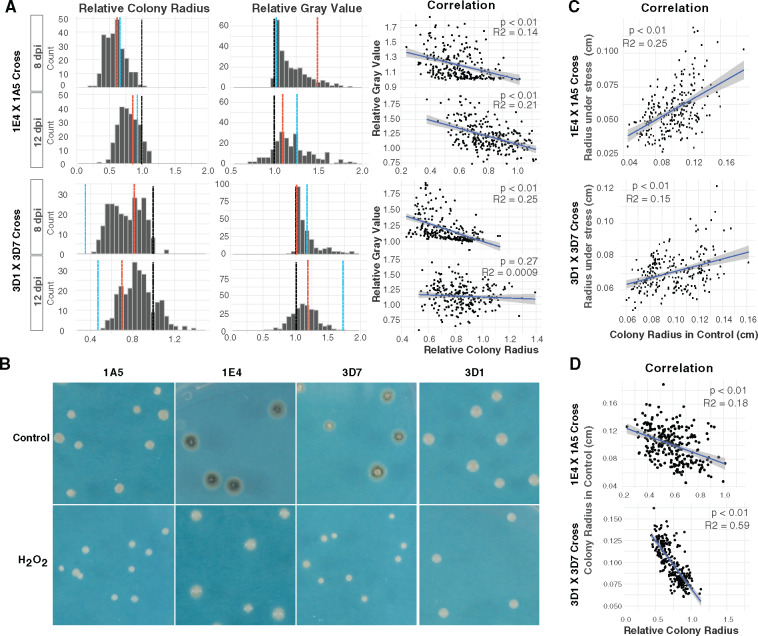
Two segregating F1 populations of Z. tritici were scored for growth and melanization in the presence and absence of hydrogen peroxide at 8 and 12 dpi. All traits exhibited high broad-sense heritability (0.98–0.56, Supplementary Table S1), indicating that the phenotypic variation among offspring was mainly due to genetic variation. Transgressive segregation was found in the 1A5 × 1E4 cross, with a total of 94 offspring from this cross exhibiting growth inhibition significantly (P < 0.001) lower than 1E4 (0.62 ± 0.04) or higher than 1A5 (0.68 ± 0.01) (Figure 1, A and B). Only eight offspring from the 3D1 × 3D7 cross had significantly (P < 0.001) higher growth inhibition than 3D1 (0.81 ± 0.03), and no offspring had significantly lower growth inhibition than 3D7 (0.52 ± 0.02) (Figure 1, A and B).
Figure 1.
Variation in growth and in melanization under oxidative stress. (A) The distribution of the relative growth (left panels) and relative melanization (middle panels) traits in the 1E4 × 1A5 cross and the 3D1 × 3D7 cross at 8- and 12-day post-inoculation (dpi). Relative colony radius (x axis) less than 1 indicates that oxidative stress inhibited the growth, and vice versa. Relative melanization value (x axis) less than 1 indicates more melanization under oxidative stress. The black dashed line stands for 1, indicating that the value under oxidative condition equals the value under the control condition. The blue and red dashed lines represent the two parental values: the blue dashed line stands for either 1A5 or 3D7 and the red dashed line stands for either 1E4 or 3D1, depending on the cross-population. The linear correlations between relative growth and relative melanization are shown in the right panel. (B) Phenotypes of the parental strains under control conditions and under oxidative stress. (C) Significant correlations between growth in the control condition and growth under oxidative stress at 8 dpi. (D) Significant correlation between relative growth and growth under control conditions at 8 dpi.
Growth of nearly all strains was inhibited (i.e., relative growth was <1) by 1 mM H2O2 at 8 dpi in both crosses. But the degree of inhibition varied among progeny strains (Figure 1A, left panel), as also found in their parental strains (Figure 1B), suggesting different capacities to tolerate oxidative stress. At 8 dpi, only one offspring from the 1E4 × 1A5 cross and five offspring from the 3D1 × 3D7 cross did not show growth inhibition under oxidative stress. At 12 dpi, 25 offspring from the 1E4 × 1A5 cross and 47 offspring from the 3D1 × 3D7 cross showed an increase in growth under oxidative stress compared to the control (i.e., relative growth was >1), suggesting a varying capacity among the offspring to negate the effects of oxidative stress. Weak but significant correlations between growth under control and oxidative stress conditions were found in both segregating populations (Figure 1C), suggesting inherent differences in growth rates among strains irrespective of their environment. Growth in the control environment and growth inhibition under oxidative stress were negatively correlated in both segregating populations (Figure 1D) at both time points, indicating that the strains that grew the fastest in the control environment were the most affected by oxidative stress. The 3D1 × 3D7 cross had the strongest correlation (R2 = 0.59). To determine whether differences among strains in relative growth result from differences in growth under the control condition, we compared phenotypes of the eight strains with the highest relative growth value with phenotypes of the eight strains with the lowest relative growth value from the 3D1 × 3D7 cross (Supplementary Figure S1). Larger differences in colony growth were found in the control environment compared to the oxidative stress environment (Supplementary Figure S1), suggesting that the difference in relative growth largely reflects inherent growth differences under non-stressful conditions. Taking together, these results suggest that variation in growth under oxidative stress is affected by both inherent fungal growth rate and the oxidative stress response.
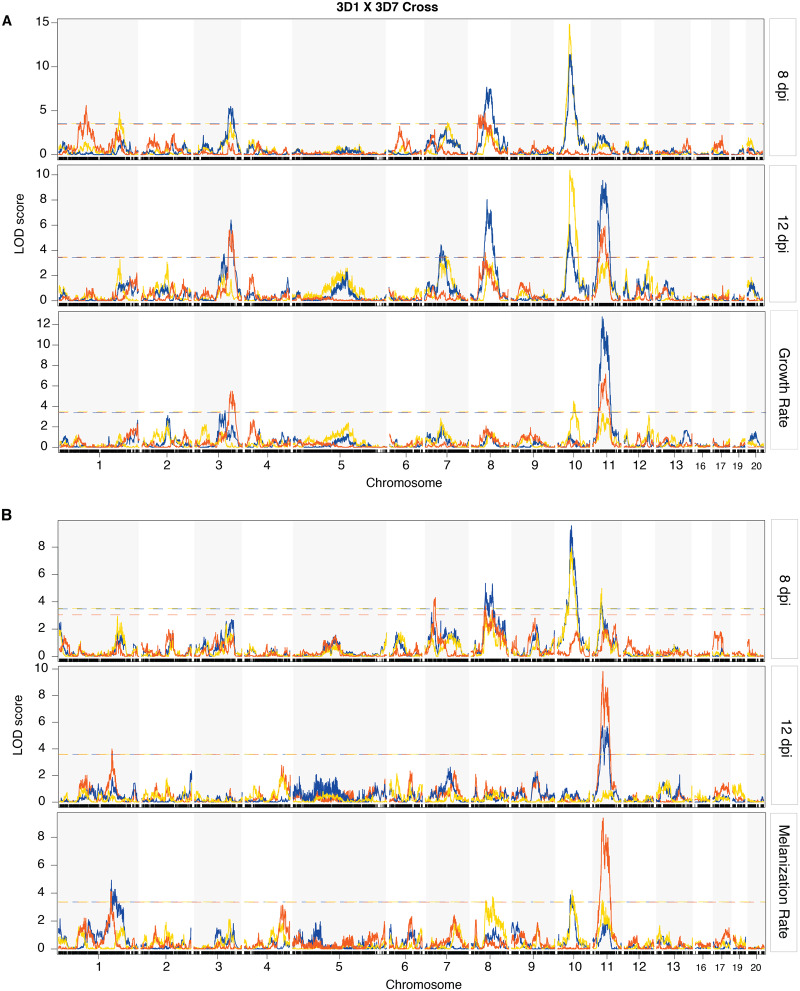
The combined effect of the oxidative stress response and inherent fungal growth rate was validated by the QTL scans. Simple interval scans provided an overview of the genetic architecture of growth under oxidative stress and under control conditions (Figures 2A and 3A), identifying QTLs that were specific for oxidative stress as well as QTLs associated with growth in the absence of H2O2 (Supplementary Table S2). Multiple QTL scans confirmed these findings and separated several QTLs that were linked (Supplementary Table S4). The chromosome 1, 8, and 12 QTLs in the 1A5 × 1E4 cross (Figure 2A and Table 1) in addition to the chromosome 1 and 7 QTLs in the 3D1 × 3D7 cross (Figure 3A and Table 1) were characterized as QTLs associated only with oxidative stress. The chromosome 2 and 3 QTLs for the 1A5 × 1E4 cross (Figure 2A and Table 1) and the chromosome 8 and 11 QTLs for the 3D1 × 3D7 cross (Figure 3A and Table 1) were categorized as inherent growth-related QTLs that were also associated with growth under oxidative stress.
Figure 2.
Genetic architecture of growth- and melanization-related traits under simple interval mapping in the 1E4 × 1A5 cross. (A) Interval mapping of growth-related traits. (B) Interval mapping of melanization-related traits. Chromosomes 1–13 are the core chromosomes of Zymoseptoria tritici, and they are present in both crosses, while chromosomes 14–21 are accessory chromosomes that may disappear in either cross. The blue line shows the mapping results for colony radius or melanization under control conditions, the orange line shows the mapping results for colony radius or melanization under oxidative stress, and the yellow line shows the mapping results for relative traits. The vertical axis shows the log10 likelihood ratio (LOD score), and the horizontal axis indicates the chromosome number. The horizontal dashed lines in the figure indicate the LOD threshold estimated from 1000 permutations of the genome-wide scan.
Figure 3.
Genetic architecture of growth- and melanization-related traits under simple interval mapping in the 3D1 × 3D7 cross. (A) Interval mapping of growth-related traits. (B) Interval mapping of melanization-related traits. Chromosomes 1–13 are the core chromosomes of Zymoseptoria tritici, and they are present in both crosses, while chromosomes 14–21 are accessory chromosomes that may disappear in either cross. The blue line presents the mapping results for colony radius or melanization under control conditions, the orange line presents the mapping results for colony radius or melanization under oxidative stress and the yellow line presents the mapping results for relative traits. The vertical axis shows the log10 likelihood ratio (LOD score), and the horizontal axis indicates the chromosome number. The horizontal dashed lines in the figure indicate the LOD threshold estimated from 1000 permutations of the genome-wide scan.
Table 1.
Summary of the unique QTLs associated with oxidative stress tolerance in Zymoseptoria tritici
| Cross | Dpi | Trait | Chr | Highest LOD | Var. (%) | QTL Effect | Confidence Interval (bp) | No. Genes | Contr. | Melan. | Possible Candidate |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1E4 × 1A5 | 8, 12 | Radius, Relative _radius | 1 | 6 | 6 | + | 1,850,952–2,398,478 | 214 | N | N | Ferric reductases, Calcineurin-like phosphoesterase |
| 8, 12 | Radius | 2 | 4 | 4 | + | 1,156,179–2,578,978 | 492 | Y | Y | ||
| 8, 12 | Radius | 3 | 5 | 6 | − | 356,096–1,345,213 | 264 | Y | Y | Cu/Zn SOD, Fe/Mn SOD, Cyclin | |
| 12 | Relative_radius | 3 | 3 | 6 | − | 1,710,431–1,854,263 | 37 | N | Y | ||
| 8 | Relative_radius | 8 | 5 | 8 | − | 281,510–346,108 | 17 | Y | Y | Glucose-6-phosphate 1-dehydrogenase (zwf1) | |
| 8, 12 | Radius, Relative _radius | 8 | 20 | 24 | + | 532,118–559,333 | 17 | N | Y | Glutathione synthetase, Sak1, Zinc fingers, Swi5 | |
| 12 | Radius | 12 | 4 | 4 | + | 562,413–1,301,393 | 234 | N | N | Tyrosine protein phosphatase (yvh1), Tyrosine-protein phosphatase SIW14 | |
| 3D1 × 3D7 | 8 | Radius | 1 | 6 | 9 | − | 1,634,586–2,068,459 | 124 | N | N | Catalase-peroxidase (ZtCpx2) |
| 8, 12 | Radius, Relative _radius | 1 | 6 | 6 | − | 5,190,476–5,431,363 | 78 | N | N | Glutamate-cysteine ligase | |
| 12 | Relative_radius | 2 | 6 | 5 | − | 795,640–2,118,190 | 437 | Y | N | ||
| 8 | Relative_radius | 3 | 7 | 9 | + | 2,982,889–3,365,105 | 135 | Y | N | Catalase (ZtCat1) | |
| 12 | Relative_radius | 3 | 5 | 5 | − | 2,192,700–2,712,400 | 192 | Y | N | ||
| 12 | Relative_radius | 5 | 5 | 4 | + | 1,216,748–1,777,899 | 211 | N | N | ||
| 8 | Radius | 7 | 3 | 5 | + | 218,051–1,190,215 | 281 | N | Y | ||
| 8, 12 | Relative_radius | 7 | 6 | 6 | − | 1,379,714–1,900,515 | 184 | N | N | ||
| 8, 12 | Radius | 8 | 5 | 7 | − | 834,823–1,121,753 | 114 | Y | Y | Catalase-peroxidase (ZtCpx1) | |
| 8 | Relative_radius | 10 | 16 | 20 | − | 749,521–758,289 | 5 | Y | Y | Ferric reductase (NOX) | |
| 12 | Radius, Relative _radius | 11 | 8 | 12 | − | 463,800–741,646 | 92 | Y | Y | Tyrosinase, Nuclear transport protein (Msn5) | |
| 12 | Relative_radius | 12 | 5 | 4 | + | 1,142,717–1,211,904 | 27 | N | N | Tyrosine protein phosphatase (yvh1), Tyrosine protein phosphatase SIW14 |
Dpi, days post inoculation; Chr, chromosome; Var., percentage of variance explained by the QTL; QTL effect, the minus indicates that the 1A5 or 3D7 allele results in a lower value of the phenotype, and vice versa; No. gene, number of genes in the confidence interval; Contr., the confidence interval overlaps with QTLs found in the control condition; Melan., the confidence interval overlaps with QTLs found in the melanization under stressed condition; Y, yes; N, no; Radius, radius of colonies under stressed conditions.
Not all of the QTLs associated with growth under control conditions were significantly associated with growth under oxidative stress (Figures 2A and 3A and Supplementary Table S4). For example, the simple interval mapping detected a chromosome 8 QTL with large effects (LOD >8) under both ROS stress and control conditions (Figure 2A). But the multiple QTL model separated multiple peaks on the chromosome 8 QTL under control conditions, with the result that none of the chromosome 8 QTLs in the control environment had overlapping confidence intervals with the chromosome 8 QTL found under ROS stress (Figure 4A and Supplementary Table S4). This result suggests that the chromosome 8 QTLs with large effects on growth under control conditions do not contribute to growth under oxidative stress to a similar extent. The same pattern was found for inherent growth-related QTLs on chromosomes 5, 6, and 7 in the 1A5 × 1E4 cross and on chromosome 10 in the 3D1 × 3D7 cross (Supplementary Table S4).
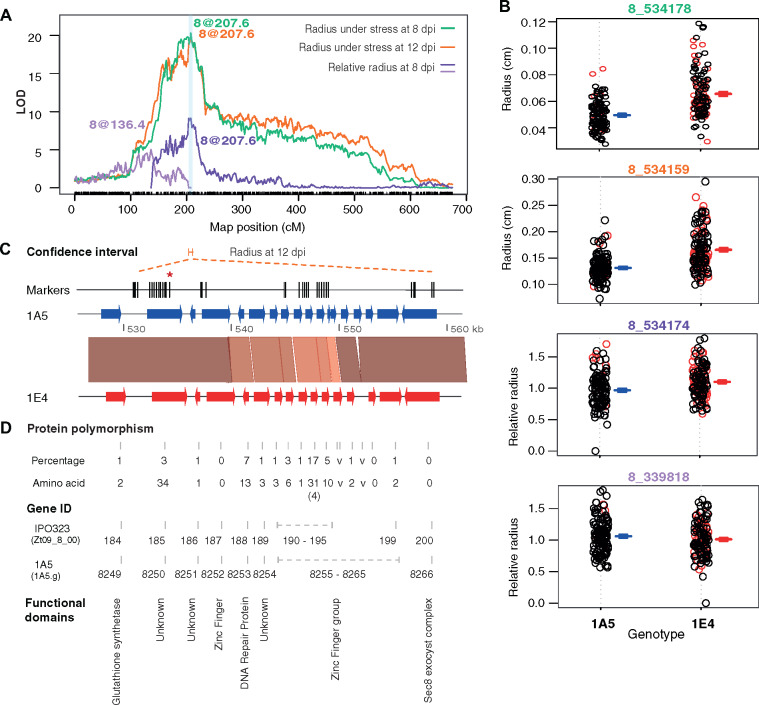
Figure 4.
The chromosome 8 QTLs for growth under oxidative stress in the 1E4 × 1A5 cross. (A) LOD plot of the chromosome 8 QTLs that are identified by multiple QTL mapping for growth under oxidative stress at 8 dpi (green) and 12 dpi (orange) and in relative growth at 8 dpi (purple). The y axis indicates the LOD score, and the x axis indicates the genetic distance (centiMorgans) along the chromosome. Three detected QTLs have the highest LOD at 207.6 cM, and one detected QTL has its highest LOD at 136.4 cM. The light blue vertical line indicates the confidence interval of the chromosome 8 QTL identified from the growth under oxidative stress at 12 dpi, which is the narrowest confidence interval among these chromosome 8 QTLs. (B) Allele effects corresponding to the QTLs shown in (A). Numbers on the top of each plot (e.g., 8_534178) are markers with the highest LOD in each QTL. For example, marker 8_534178 indicates that this marker is at 534,178 bp on chromosome 8. Each closed circle represents a different offspring. Red circle indicates that the genotype at the locus is missing and was inferred based on a single imputation. (C) SNP markers and synteny of the confidence intervals marked in light blue in (A). Markers are shown as vertical lines. The red asterisk indicates the location of the highest LOD. Synteny plot shows the DNA polymorphisms in this genomic region between the two parents. The thick arrows represent genes. The red segments indicate regions with sequence identity greater than 90%. The darker the red color, the lower the degree of polymorphism in the genomic region. (D) Protein polymorphisms, gene IDs, and functional domains of the 17 genes within the confidence interval. Percentage indicates the percentage of the polymorphic amino acids in each protein, and the amino acid indicates the number of polymorphic amino acids (not including indels) between the two parental strains. The number in brackets indicates the number of amino acids missing (indels) in either parent. Polymorphism less than 1% appears as 1%, and polymorphism larger than 50% appears as “v” in the figure. Only the different parts of the gene ID were used in the figure, and the full gene IDs should be, for example, Zt09_8_00184 in strain IPO323 and 1A5.g8249 in strain 1A5. Genes 1A5.g8260-8264 were missing in the annotation of IPO323 genome.
Other inherent growth-related QTLs were detected in the relative growth-related traits. The genome scan for relative growth in the 3D1 × 3D7 cross at 8 dpi detected a chromosome 10 QTL with large effect (Figure 3A and Supplementary Tables S2 and S4). This QTL was also detected for growth under control conditions at 8 dpi, but was not detected for growth under ROS stress (Figures 3A and 5A; Supplementary Tables S2 and S4). Analyzing the allele effect of these QTLs showed that the 3D7 allele is associated with higher growth under the control condition and lower relative growth than the 3D1 allele (reflecting higher growth inhibition) (Figure 5B). But the effects of these two alleles at this locus are not significantly different in growth under the stress (Figure 5B). This result is in agreement with our observation that strain 3D1 is less sensitive to oxidative stress than strain 3D7 (Figure 1B). Another general growth-related QTL which was also detected in the relative growth is the chromosome 8 QTL in the 1E4 × 1A5 cross (Figure 4A). This QTL for relative growth overlapped with the QTL for growth under the control condition (Table 1 and Supplementary Table S4), but showed an opposite QTL effect from the other chromosome 8 QTL which is recognized as specific for oxidative stress response (Figure 4B). These results suggest that different alleles in general growth-related QTLs could affect the oxidative stress tolerance of Z. tritici.
Figure 5.
The chromosome 10 QTL for relative growth at 8 dpi in the 3D1 × 3D7 cross. (A) LOD curves of the chromosome 10 QTL identified by multiple QTL mapping in relative growth at 8 dpi (blue) and growth in control condition at 8 dpi (orange). The highest LOD for each QTL is at 112.5 and 123.9 cM, respectively. (B) Allelic effects for QTLs in (A) at the marker with the highest LOD. The effect of marker at 112.5 cM (752,685 bp) on chromosome 10 in growth under oxidative stress at 8 dpi is also shown, though no significant QTL was detected for this trait on chromosome 10, suggesting that the mean of strains with different alleles are not significantly different. Each closed circle represents an individual. (C) Markers and synteny plot of the confidence interval marked in (A). The asterisk indicates the peak marker. In the synteny plot of the confidence interval region between the two parental strains, the darker the red color of the segments, the lower the degree of polymorphism in the genomic region. (D) Polymorphisms in protein and UTR regions, gene IDs, and the functional domains of candidate genes in (C). Numbers outside brackets indicate the number of substitutions. Numbers in brackets indicate either the percentage of polymorphic amino acids in the entire protein or the number of nucleotides missing in one of the parental strains. Only the different parts of the gene IDs were used in the figure, and the full gene IDs should be, for example, Zt09_10_00216 in strain IPO323 and 1A5.g9786 in strain 3D7. (E) Expression values in reads per kilobase of transcript per million mapped reads (RPKM) of the five genes in yeast sucrose broth medium (YSB, control), minimal medium (starvation), and in planta infection at 7 dpi. The expression data in minimal medium and YSB (Francisco et al. 2019) and the in planta infection data (Palma-Guerrero et al. 2017) were collected from three replicates in previous studies. The asterisk indicates the significant difference (P < 0.05) in expression between the two parental strains.
Significant QTL interactions, indicating epistasic effects, were only detected in growth under the control conditions (Supplementary Table S4). All unique QTLs (i.e., lacking overlapping confidence intervals) associated with oxidative stress tolerance were summarized in Table 1, showing the narrowest confidence intervals. Detailed information about results from genome scan by interval mapping is presented in Supplementary Table S2, and the results from genome scan by multiple QTL mapping is presented in Supplementary Table S4.
Higher melanization does not consistently result in higher growth under oxidative stress in Z. tritici
Melanization decreased significantly (relative gray value >1) under oxidative stress at 8 dpi in both segregating populations (Figure 1A, middle panel), though four offspring from the 3D1 × 3D7 cross showed increased melanization under oxidative stress. At 12 dpi, 36 progeny from the 1E4 × 1A5 cross and 62 progeny from the 3D1 × 3D7 cross became more melanized under oxidative stress (relative gray value <1), indicating that oxidative stress induced more melanization for those strains at a later stage of growth. To assess how melanin production correlates with colony growth under oxidative stress, linear regressions were conducted between relative growth and relative melanization for both time points. A significant negative correlation (P < 0.01) was found between relative gray value and relative growth in both crosses at 8 dpi (Figure 1A, right panel), indicating that a higher reduction in melanization is associated with a higher inhibition in growth under oxidative stress. However, significant correlations between the absolute growth and absolute melanization were also found at 8 dpi for both crosses under both oxidative stress and control conditions (Supplementary Figure S2), indicating that melanization is associated with growth independent of oxidative stress. In both crosses under oxidative stress at 12 dpi, the strains with stress-induced melanization (relative melanization <1) did not consistently show a better growth (relative growth >1) than under the control condition. On the contrary, most of those strains showed decreased growth under oxidative stress (relative growth <1) (Figure 1A, right panel). In addition, strains with an increased growth under oxidative stress (relative growth >1) did not consistently show an increase in melanization (relative melanization <1) (Figure 1A, right panel). Overall, these results suggest that the degree of melanization is not associated with tolerance to oxidative stress.
The genome scans for melanization also indicated a correlation between growth and melanization, as QTLs for melanization largely overlapped with QTLs for growth (Figures 2 and 3). The melanization QTLs on chromosomes 2, 8, and 12 in the 1A5 × 1E4 cross and the melanization QTLs on chromosomes 8 and 11 in the 3D1 × 3D7 cross overlapped with the QTLs found for growth under both stress and control conditions (Supplementary Tables S2–S5). As found for the growth-related traits, some QTLs for melanization under control conditions overlapped with QTLs for relative melanization, for example, the chromosome 10 QTL in the 3D1 × 3D7 cross (Figure 3B, yellow line). The multiple QTL model also detected several significant QTLs associated with melanization only under oxidative stress (Supplementary Table S5), including the QTLs on chromosomes 6 and 9 in the 3D1 × 3D7 cross and the QTL on chromosome 5 in the 1A5 × 1E4 cross. These QTLs suggest that melanization is also affected by oxidative stress. Results from simple interval mapping and from multiple QTL mapping are presented in Supplementary Tables S3 and S5, respectively.
New genes related to oxidative stress tolerance were identified in the QTLs
Only QTLs associated with tolerance to oxidative stress (Table 1) were explored for possible candidate genes. Using the genome annotation created by Plissonneau et al. (2018), functional domains of the top candidate genes were confirmed by protein BLAST in the NCBI database. Many genes related to ROS elimination (e.g., SODs and CAT) and fungal growth regulation (e.g., putative homologs to zwf1, yvh1, and Msn5) were found in the QTLs (Table 1).
We made a more detailed analysis of the QTLs with LOD >10 and with narrow confidence intervals containing fewer than 20 genes. The oxidative stress-specific chromosome 8 QTL (LOD = 15.8) in the 1A5 × 1E4 cross contains 17 genes (Figure 4, C and D). The peak markers of this QTL were located in gene 1A5.g8250 (Zt09_8_00185) (Figure 4, A and C), which has an unknown functional domain (Figure 4D). This gene is next to a glutathione synthetase (1A5.g8249), which may regulate glutathione production during oxidative stress. Many genes in this confidence interval have no homology to characterized genes (Figure 4D). A high number of DNA polymorphisms between the two parental strains, which could affect the expression of genes, protein structures, or protein maturation processes, were also found in this confidence interval (Figure 4C).
The chromosome 10 QTL (LOD = 15) detected for relative growth at 8 dpi in the 3D1 × 3D7 cross contained only five genes (Figure 5, C and D). Protein BLAST analysis of these genes showed that none of them were homologous to genes characterized in other fungi. The gene 3D7.g9787 contains a NOX_Duox_like_FAD_NADP domain (E value = 2.36e-43) and a ferric reductase domain belonging to the cytochrome b superfamily (E value = 4.83e-25). NOX domains are known to be involved in regulating fungal growth and cell development in fungi (Egan et al. 2007; Segmüller et al. 2008; Ryder et al. 2013; Samalova et al. 2014). Protein BLAST analyses between this NOX and characterized NOXs such as NOXa in Z. tritici (Choi et al. 2016) and NOXB (CAP12517) in Botrytis cinerea (Segmüller et al. 2008) showed low similarities (E value = 5e-07, Coverage = 30%, Identity = 23%). The functions of the MFS transporter and the acetyltransferase, which are respectively related to protein/carbohydrate transport and transcriptional elongation, could also affect fungal growth. Many polymorphisms found in this QTL affect both DNA and protein sequences between the two parental strains (Figure 5, C and D). Significant differences in expression between the two parental strains were found for all five genes under different growth conditions (Palma-Guerrero et al. 2017; Francisco et al. 2019) (Figure 5E), suggesting that polymorphisms in the promoter regions of these genes could also affect the associated phenotype.
The in planta expression patterns of the top candidate genes match ROS burst timing during plant–pathogen interactions
To infer the function of our top candidate genes in response to oxidative stress in planta, we compared their expression patterns with the expression patterns of genes that are homologs to characterized ROS scavengers in other fungi. These homologs include the CAT-peroxidases, glutathione peroxidase, CAT, and SODs in Z. tritici. We also included homologs of the two major regulators of oxidative stress response, ZtYap1 (Yang et al. 2015) and MgHog1 (Mehrabi et al. 2006), and the characterized homolog NOXa in Z. tritici (Choi et al. 2016). A previous study reported bursts of hydrogen peroxide during the latent phase (around 5 dpi) and the start of the necrotrophic phase (around 13 dpi) for a compatible infection (Shetty et al. 2003). We assumed that top candidate genes from oxidative stress-specific QTLs were highly expressed during ROS bursts and that they have expression patterns matching the expression patterns of ROS scavengers.
All of our top candidate genes were expressed in planta in strain 3D7 (Supplementary Figure S3). The gene 1A5.g8250 from the oxidative stress-specific QTL on chromosome 8 in the 1A5 × 1E4 cross reached the highest expression at 14 dpi, together with the glutathione synthetase and three SODs (Supplementary Figure S3B), matching the timing of the second ROS burst reported previously (Shetty et al. 2003). The ferric reductase (3D7.g9787) from the inherent growth-related QTL on chromosome 10 was highly expressed at 7 dpi, together with genes such as CAT 1 (ZtCat1), ZtCpx1 and Yap1 (Supplementary Figure S3B), matching the timing of the first ROS burst reported previously (Shetty et al. 2003). The gene 3D7.g9786 (without a functional domain), the MFS, and the acetyltransferase showed the highest expression at the early necrotrophic phase (11 dpi), together with CAT-peroxidase 2 and glutathione peroxidase (Supplementary Figure S3B).
Discussion
Most cellular plant pathogens are exposed to a ROS burst as part of the plant’s defense response. Thus, tolerance to oxidative stress is important for plant pathogens, and the degree of tolerance may be associated with pathogenicity and viability during host infection (Heller and Tudzynski 2011; Segal and Wilson 2018). For the fungal pathogen Z. tritici, a previous study found that infiltration of hydrogen peroxide decreased disease symptoms and delayed fungal sporulation, while infiltration of CAT into infected leaves increased disease symptoms and accelerated disease development (Shetty et al. 2007). This result suggested that tolerance to oxidative stress could affect virulence of Z. tritici. Several genes affecting hydrogen peroxide tolerance of Z. tritici have already been identified, including CAT-peroxidases (ZtCpx1, ZtCpx2), CAT (ZtCat1 and ZtCat2), and oxidative stress regulators (ZtYap1 and MgHog1) (Mehrabi et al. 2006; Gohari 2015; Yang et al. 2015). Interestingly, ZtYap1 was found to be essential for in vitro tolerance to oxidative stress but not for in planta virulence (Yang et al. 2015), suggesting that other ROS management systems may exist in Z. tritici. Zymoseptoria tritici is known to produce melanin during its in vitro vegetative growth (Lendenmann et al. 2014; Krishnan et al. 2018), but the role of melanin in protecting Z. tritici from oxidative stress was not previously explored. In this study, we used QTL mapping of growth and melanization in two segregating populations of Z. tritici to investigate the genetic basis of tolerance to oxidative stress.
Reproducibility of results from independent QTL analyses
We reproduced in this study many of the QTL peaks for growth and melanization identified in previous studies (Lendenmann et al. 2014, 2015, 2016). Previous QTL mapping investigations used PDA media with or without fungicides and growth at either 15°C or 22°C. Our new study used the same fungal strains and the same culture media, but with growth at 18°C in the presence or absence of H2O2.
Three of the QTLs for growth in the control 18°C environment in this study overlapped with the QTLs found in the earlier studies, suggesting that they are the same QTLs. These QTLs include the chromosome 8 QTL in the 1E4 × 1A5 cross and the QTLs on chromosomes 10 and 11 in the 3D1 × 3D7 cross. All of these QTLs were found for inherent growth at 15°C and 22°C (Lendenmann et al. 2016) and for fungicide sensitivity (Lendenmann et al. 2015). Re-mapping of the thermal sensitivity trait of Lendenmann et al. 2016 using our new genetic map resulted in the same confidence interval for the chromosome 10 QTL as was found for the oxidative stress sensitivity trait in this study. The melanization QTLs on chromosome 2 in the 1E4 × 1A5 cross and on chromosome 11 in the 3D1 × 3D7 cross overlapped with the melanization QTLs identified previously (Lendenmann et al. 2014). This reproducibility of QTLs between independent studies conducted under similar experimental conditions illustrates the robustness and accuracy of our phenotyping methods. The overlap of QTLs for growth-related traits under different environmental conditions suggests that these QTLs contain genes that generally regulate growth under many different conditions.
Evidence for antioxidants affecting tolerance to oxidative stress
The growth of fungal pathogens in plants may be restricted due to ROS bursts produced during the plant defense response (Doehlemann and Hemetsberger 2013). Pathogen strains that can maintain growth under oxidative stress may be more virulent than less tolerant strains. Many studies found that antioxidants such as CAT, peroxidase, and SOD were more expressed under oxidative stress in other fungal systems (Chen et al. 2003, 2008; Fountain et al. 2016). In this study, we found genes encoding these antioxidants within our QTLs (Table 1), suggesting they may play a role in the oxidative stress response of Z. tritici. Genes involved in glutathione production such as glutamate-cysteine ligase (3D7.g1830) and glutathione synthetase (1A5.g8249), which encode the two enzymes known to be involved in the biosynthesis of glutathione (Lu 2013), were also found in our QTLs (Table 1). Glutathione was shown to play an essential role in maintaining the cellular redox environment (Aquilano et al. 2014; Corso and Acco 2018) during fungal cell development (Samalova et al. 2014) and the oxidative stress response in fungi (Grant et al. 1997; Gutiérrez-Escobedo et al. 2013; Bakti et al. 2017). Though the functions of these two genes will need to be confirmed in future molecular studies, the discovery of these genes in our QTLs suggests that glutathione plays an important role in oxidative stress tolerance in Z. tritici.
A detailed analysis of one of the oxidative stress-specific QTLs in the 1E4 × 1A5 cross identified an uncharacterized gene (1A5.g8250) that may also affect glutathione production (Figure 4). Peak markers of this QTL from three traits (e.g., growth under oxidative stress at 8 and 12 dpi, and relative growth at 8 dpi) were located in this gene, which was found next to the homolog of glutathione synthetase in the genome (Figure 4C). Their nearby locations in the genome (Figure 4C) and the similarity in the in planta expression patterns (Supplementary Figure S3) suggests a possibility of cooperative functions for these two genes.
Previous studies had reported a weak ROS burst during the latent phase (5–7 dpi) and a strong burst during the necrotrophic phase (from around 13 dpi) during a compatible infection in Z. tritici (Shetty et al. 2003). But no study has reported on genes in Z. tritici that may be involved in ROS tolerance during these two bursts. In our study, we found a correlation between the timing of the first ROS burst and the peak of expression (7 dpi) of ZtCat1, ZtCpx1, and NOX (3D7.g9787), and between the second ROS burst and the peak of expression (14 dpi) of glutathione synthetase, the top candidate gene 1A5.g8250 and two SODs (Supplementary Figure S3). These patterns suggest the corresponding proteins may contribute to tolerance to the exogenous ROS produced by plant defenses. Interestingly, the two SODs found in the general growth-related QTLs were highly expressed during the late necrotrophic phase when the rapid formation of asexual fruiting bodies takes place, suggesting that they may be associated with elimination of endogenous ROS produced by respiration during intensive fungal growth. To better understand the roles played by these candidate genes in tolerance to oxidative stress, future studies should consider the effect of different doses of ROS on Z. tritici growth, and conduct additional ROS measurements in planta.
The effect of general growth-related genes on tolerance to oxidative stress
Several of the growth QTLs found under non-stressful conditions were also found for growth under oxidative stress (Figure 2, A and B and Table 1). Possible explanations for these QTLs include that they contain genes with pleiotropic effects in managing cell response to different environmental conditions or that they generally affect growth in all environments. Exploring the possible candidate genes for these QTLs matched the two possible explanations mentioned above, as homologs to a general stress regulator Msn5 and two SODs were found in their confidence intervals (Table 1). Msn5 was found associated with multiple environmental stress response in fission yeast (Chen et al. 2003, 2008), and the two SODs are likely related to the elimination of endogenous ROS during growth (as discussed above).
Some QTLs with large effects on growth under non-stressful condition, such as the chromosome 8 QTLs in the 1E4 × 1A5 cross and the chromosome 10 QTL in the 3D1 × 3D7 cross, were not found when growth occurred under oxidative stress (Figures 2A and 3A). These QTLs were detected for relative growth, suggesting that different alleles at these loci affect the growth difference between stressed and non-stressful conditions. This likely reflects changes in fungal metabolism and development that occur under oxidative stress (Chen et al. 2003; Fountain et al. 2016; Segal and Wilson 2018). These changes could explain why the strains growing the fastest in the control environment were the most sensitive to oxidative stress in the 3D1 × 3D7 cross at 8 dpi (Figure 1D), where the difference in relative growth largely reflected growth differences under non-stressful conditions (Supplementary Figure S2).
A correlation between growth rate under non-stressful condition and sensitivity to stress has also been reported in bacteria and plants (Shipley and Keddy 1988; Berney et al. 2006). Our study provided a relatively small genomic region that may explain this phenomenon in fungi. The chromosome 10 QTL in the 3D1 × 3D7 cross was strongly associated with sensitivity to oxidative stress. Interestingly, a previous study found the same QTL associated with temperature sensitivity (Lendenmann et al. 2016). We hypothesized that this QTL contains genes that can make significant contributions to fungal growth, but are sensitive to several stresses. The expression of two (3D7.g9786 and gene 3D7.g9787) of the five genes in this QTL decreased significantly under starvation stress compared to the control (YSB) condition (Figure 5E). Gene 3D7.g9787 showed the highest polymorphisms in both protein sequence and the 5′-UTR region among the five candidate genes (Figure 5, C and D), and its expression was significantly different between the two parental strains (Figure 5E). These polymorphisms in DNA sequence and expression level between the two parents could result in different phenotypic effects (Figure 5B). Gene 3D7.g9787 has a NOX domain, which has been shown to affect fungal cell differentiation and hyphal tip growth (Egan et al. 2007; Segmüller et al. 2008; Ryder et al. 2013; Samalova et al. 2014). These functions are highly associated with the colony growth measured in this study. For these reasons, we consider 3D7.g9787 to be our top candidate gene to explain this QTL. Future molecular studies of the function of this gene in vitro and in planta will provide more insight into its effects on growth and stress sensitivity in Z. tritici.
The effects of melanization on tolerance to oxidative stress
Though melanin was suggested to act as a physiological redox buffer that helps protect fungal cells against ROS (Jacobson 2000; Missall et al. 2004), to our knowledge no empirical evidence has been presented to support this assumption in fungi. In our study, we did not find evidence that higher melanization (relative melanization <1) enhances fungal tolerance to oxidative stress. Strains with higher growth were not consistently observed to exhibit higher melanization under oxidative stress compared to the control condition (Figure 1A). Correlations were found between absolute melanization and absolute growth in both environments (Supplementary Figure S2), which could simply reflect a correlation between relative melanization and relative growth (Figure 1A). Our correlation data and QTL mapping results suggest a strong association between melanization and growth in Z. tritici (Figures 2B and 3B; Supplementary Tables S3 and S5), which confirms previous findings (Lendenmann et al. 2014; Krishnan et al. 2018). This link between melanization and growth under oxidative stress was also reported in a study on Aspergillus flavus, in which the expression of many secondary metabolites were found correlated with fungal biomass (Fountain et al. 2016).
However, this tight link between melanization and growth suggests that melanization may contribute to the oxidative stress tolerance by reducing the degree of growth inhibition induced by the stress. A study that identified the Zmr1 gene regulating melanin production in Z. tritici showed that mutants with less melanization had higher growth inhibition under fungicide stress than the wild-type (Krishnan et al. 2018), suggesting the positive effect of melanization in fungicide tolerance. This positive effect could be easily explained by the strong association between growth and melanization under stressful conditions, considering the possible pleiotropic effect of genes regulating growth and melanization (Lendenmann et al. 2014; Krishnan et al. 2018).
Conclusion
In this study, we identified genomic regions associated with variation in oxidative stress tolerance using QTL mapping in two crosses of Z. tritici. We found that genomic regions specific for oxidative stress response and associated with fungal growth regulation are related to the tolerance to oxidative stress. Additionally, our data did not suggest that melanization could assist the fungal growth under oxidative stress. Based on our findings, we hypothesize that genes related to antioxidants and fungal cell growth could both contribute to the variation in oxidative stress tolerance.
Acknowledgments
We thank Tiziana Valeria Vonlanthen, Bethan Turnbull, Susanne Dora, Sarah Furler, Alexandra Waltenspühl, and Jasmin Wiedmer for helping to conduct experiments. Thanks to Dr Xin Ma and Dr Carolina Francisco for providing the analyzed RNAseq expression data. We are grateful to Dr Antoine Branca, Dr Andrea Sánchez Vallet, and Prof. Daniel Croll for critical reading of the manuscript.
Funding
This research was supported by the Swiss National Science Foundation (31003 A_155955 granted to B.A.M.).
Conflicts of interest
None declared.
Literature Cited
- Aquilano K, Baldelli S, Ciriolo MR. 2014. Glutathione: new roles in redox signaling for an old antioxidant. Front Pharmacol. 5:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakti F, Király A, Orosz E, Miskei M, Emri T, et al. 2017. Study on the glutathione metabolism of the filamentous fungus Aspergillus nidulans. Acta Microbiol Immunol Hung. 64:255–272. [DOI] [PubMed] [Google Scholar]
- Berney M, Weilenmann HU, Ihssen J, Bassin C, Egli T. 2006. Specific growth rate determines the sensitivity of Escherichia coli to thermal, UVA, and solar disinfection. Appl Environ Microbiol. 72:2586–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Ehrenreich IM, Loo WT, Lite TLV, Kruglyak L. 2013. Finding the sources of missing heritability in a yeast cross. Nature. 494:234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Sen S. 2009. A Guide to QTL Mapping with R/QTL. New York: Springer. [Google Scholar]
- Broman KW, Wu H, Sen Ś, Churchill GA. 2003. R/QTL: QTL mapping in experimental crosses. Bioinformatics. 19:889–890. [DOI] [PubMed] [Google Scholar]
- Broxton CN, Culotta VC. 2016. SOD enzymes and microbial pathogens: surviving the oxidative storm of infection. PLoS Pathog. 12:e1005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Toone WM, Mata J, Lyne R, Burns G, et al. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 14:214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wilkinson CR, Watt S, Penkett CJ, Toone WM, et al. 2008. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol Biol Cell. 19:308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-H, Yang SL, Chung K-R. 2014. Resistance to oxidative stress via regulating siderophore-mediated iron acquisition by the citrus fungal pathogen Alternaria alternata. Microbiology. 160:970–979. [DOI] [PubMed] [Google Scholar]
- Choi Y-E, Lee C, Goodwin SB. 2016. Generation of reactive oxygen species via NOXa is important for development and pathogenicity of Mycosphaerella graminicola. Mycobiology. 44:38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corso CR, Acco A. 2018. Glutathione system in animal model of solid tumors: from regulation to therapeutic target. Crit Rev Oncol Hematol. 128:43–57. [DOI] [PubMed] [Google Scholar]
- Covarrubias-Pazaran G. 2016. Genome-assisted prediction of quantitative traits using the R package sommer. PLoS One 11:e0156744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, McDonald BA. 2017. The genetic basis of local adaptation for pathogenic fungi in agricultural ecosystems. Mol Ecol. 26:2027–2040. [DOI] [PubMed] [Google Scholar]
- Diezmann S, Dietrich FS. 2011. Oxidative stress survival in a clinical Saccharomyces cerevisiae isolate is influenced by a major quantitative trait nucleotide. Genetics. 188:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 11:539–548. [DOI] [PubMed] [Google Scholar]
- Doehlemann G, Hemetsberger C. 2013. Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 198:1001–1016. [DOI] [PubMed] [Google Scholar]
- Egan MJ, Wang Z-Y, Jones MA, Smirnoff N, Talbot NJ. 2007. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc Natl Acad Sci U S A. 104:11772–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal Z, Scharen AL, Prescott JM, van Ginkel M. 1987. The Septaria Diseases of Wheat: Concepts and Methods of Disease Management. Mexico, D.F.: CIMMYT. [Google Scholar]
- Foulongne-Oriol M. 2012. Genetic linkage mapping in fungi: current state, applications, and future trends. Appl Microbiol Biotechnol. 95:891–904. [DOI] [PubMed] [Google Scholar]
- Fountain JC, Bajaj P, Nayak SN, Yang L, Pandey MK, et al. 2016. Responses of Aspergillus flavus to oxidative stress are related to fungal development regulator, antioxidant enzyme, and secondary metabolite biosynthetic gene expression. Front Microbiol. 7:2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco CS, Ma X, Zwyssig MM, McDonald BA, Palma-Guerrero J. 2019. Morphological changes in response to environmental stresses in the fungal plant pathogen Zymoseptoria tritici. Sci Rep. 9:9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudal I, Ross S, Gout L, Blaise F, Kuhn ML, et al. 2007. Heterochromatin-like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map-based cloning of AvrLm6. Mol Plant Microbe Interact. 20:459–470. [DOI] [PubMed] [Google Scholar]
- Fones H, Gurr S. 2015. The impact of Septoria tritici Blotch disease on wheat: an EU perspective. Fungal Genet Biol. 79:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout L, Fudal I, Kuhn ML, Blaise F, Eckert M, et al. 2006. Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol. 60:67–80. [DOI] [PubMed] [Google Scholar]
- Genissel A, Confais J, Lebrun MH, Gout L. 2017. Association genetics in plant pathogens: minding the gap between the natural variation and the molecular function. Front Plant Sci. 8:1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohari AM. 2015. Functional analysis of CAT-peroxidase encoding genes in the fungal wheat pathogen Zymoseptoria tritici [Doctoral dissertation, Wageningen University] https://edepot.wur.nl/364257.
- Grant CM, MacIver FH, Dawes IW. 1997. Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to an accumulation of the dipeptide gamma-glutamylcysteine. Mol Biol Cell. 8:1699–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Escobedo G, Orta-Zavalza E, Castaño I, Alejandro DLP. 2013. Role of glutathione in the oxidative stress response in the fungal pathogen Candida glabrata. Curr Genet. 59:91–106. [DOI] [PubMed] [Google Scholar]
- Heller J, Tudzynski P. 2011. Reactive oxygen species in phytopathogenic fungi: signaling, development and disease. Annu Rev Phytopathol. 49:369–390. [DOI] [PubMed] [Google Scholar]
- Henson JM, Butler MJ, Day AW. 1999. The dark side of the mycelium: melanins of phytopathogenic fungi. Annu Rev Phytopathol. 37:447–471. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom J. 50:346–363. [DOI] [PubMed] [Google Scholar]
- Jacobson ES. 2000. Pathogenic roles for fungal melanins. Clin Microbiol Rev. 13:708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Morris AD, Latta RG. 2006. Fitness consequences of hybridization between ecotypes of Avena barbata: hybrid breakdown, hybrid vigor, and transgressive segregation. Evolution. 60:1585–1595. [PubMed] [Google Scholar]
- Kema GHJ, Yu DZ, Rijkenberg HJ, Shaw MW, Baayen RP. 1996. Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology. 86:777–786. [Google Scholar]
- Kettles GJ, Kanyuka K. 2016. Dissecting the molecular interactions between wheat and the fungal pathogen Zymoseptoria tritici. Front Plant Sci. 7:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan P, Meile L, Plissonneau C, Ma X, Hartmann FE, et al. 2018. Transposable element insertions shape gene regulation and melanin production in a fungal pathogen of wheat. BMC Biol. 16:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth JD, Neish AS. 2014. Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. Annu Rev Pathol Mech Dis. 9:119–145. [DOI] [PubMed] [Google Scholar]
- Lendenmann MH, Croll D, Stewart EL, McDonald BA. 2014. Quantitative trait locus mapping of melanization in the plant pathogenic fungus Zymoseptoria tritici. G3 (Bethesda). 4:2519–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendenmann MH, Croll D, McDonald BA. 2015. QTL mapping of fungicide sensitivity reveals novel genes and pleiotropy with melanization in the pathogen Zymoseptoria tritici. Fungal Genet Biol. 80:53–67. [DOI] [PubMed] [Google Scholar]
- Lendenmann MH, Croll D, Palma-Guerrero J, Stewart EL, McDonald BA. 2016. QTL mapping of temperature sensitivity reveals candidate genes for thermal adaptation and growth morphology in the plant pathogenic fungus Zymoseptoria tritici. Heredity. 116:384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-H, Yang SL, Chung K-R. 2009. The YAP1 homolog–mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol Plant Microbe Interact. 22:942–952. [DOI] [PubMed] [Google Scholar]
- Linning R, Lin D, Lee N, Abdennadher M, Gaudet D, et al. 2004. Marker-based cloning of the region containing the UhAvr1 avirulence gene from the basidiomycete barley pathogen Ustilago hordei. Genetics. 166:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC. 2013. Glutathione synthesis. Biochim Biophys Acta. 1830:3143–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. 2009. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 10:565–577. [DOI] [PubMed] [Google Scholar]
- Mehrabi R, Zwiers LH, de Waard MA, Kema GH. 2006. MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol Plant Microbe Interact. 19:1262–1269. [DOI] [PubMed] [Google Scholar]
- Meile L, Croll D, Brunner PC, Plissonneau C, Hartmann FE, et al. 2018. A fungal avirulence factor encoded in a highly plastic genomic region triggers partial resistance to septoria tritici blotch. New Phytol. 219:1048–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missall TA, Lodge JK, McEwen JE. 2004. Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Eukaryot Cell. 3:835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina L, Kahmann R. 2007. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19:2293–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Guerrero J, Torriani SF, Zala M, Carter D, Courbot M, et al. 2016. Comparative transcriptomic analyses of Zymoseptoria tritici strains show complex lifestyle transitions and intraspecific variability in transcription profiles. Mol Plant Pathol. 17:845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Guerrero J, Ma X, Torriani SF, Zala M, Francisco CS, et al. 2017. Comparative transcriptome analyses in Zymoseptoria tritici reveal significant differences in gene expression among strains during plant infection. Mol Plant Microbe Interact. 30:231–244. [DOI] [PubMed] [Google Scholar]
- Papadakis MA, Workman CT. 2015. Oxidative stress response pathways: fission yeast as archetype. Crit Rev Microbiol. 41:520–535. [DOI] [PubMed] [Google Scholar]
- Plissonneau C, Hartmann FE, Croll D. 2018. Pangenome analyses of the wheat pathogen Zymoseptoria tritici reveal the structural basis of a highly plastic eukaryotic genome. BMC Biol. 16:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/. [Google Scholar]
- Rolke Y, Liu S, Quidde T, Williamson B, Schouten A, et al. 2004. Functional analysis of H2O2-generating systems in Botrytis cinerea: the major Cu-Zn-superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensable. Mol Plant Pathol. 5:17–27. [DOI] [PubMed] [Google Scholar]
- Ronen M, Shalaby S, Horwitz BA. 2013. Role of the transcription factor ChAP1 in cytoplasmic redox homeostasis: imaging with a genetically encoded sensor in the maize pathogen Cochliobolus heterostrophus. Mol Plant Pathol. 14:786–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder LS, Dagdas YF, Mentlak TA, Kershaw MJ, Thornton CR, et al. 2013. NADPH oxidases regulate septin-mediated cytoskeletal remodeling during plant infection by the rice blast fungus. Proc Natl Acad Sci U S A. 110:3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samalova M, Meyer AJ, Gurr SJ, Fricker MD. 2014. Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol. 201:556–573. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vallet A, McDonald MC, Solomon PS, McDonald BA. 2015. Is Zymoseptoria tritici a hemibiotroph? Fungal Genet Biol. 79:29–32. [DOI] [PubMed] [Google Scholar]
- Segal LM, Wilson RA. 2018. Reactive oxygen species metabolism and plant-fungal interactions. Fungal Genet Biol. 110:1–9. [DOI] [PubMed] [Google Scholar]
- Segmüller N, Ellendorf U, Tudzynski B, Tudzynski P. 2007. BcSAK1, a stress-activated mitogen-activated protein kinase, is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryot Cell. 6:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segmüller N, Kokkelink L, Giesbert S, Odinius D, van Kan J, et al. 2008. NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol Plant Microbe Interact. 21:808–819. [DOI] [PubMed] [Google Scholar]
- Shapiguzov A, Vainonen JP, Wrzaczek M, Kangasjärvi J. 2012. ROS-talk – how the apoplast, the chloroplast, and the nucleus get the message through. Front Plant Sci. 3:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty NP, Kristensen BK, Newman M-A, Møller K, Gregersen PL, et al. 2003. Association of hydrogen peroxide with restriction of Septoria tritici in resistant wheat. Physiol Mol Plant Pathol. 62:333–346. [Google Scholar]
- Shetty NP, Mehrabi R, Lütken H, Haldrup A, Kema GH, et al. 2007. Role of hydrogen peroxide during the interaction between the hemibiotrophic fungal pathogen Septoria tritici and wheat. New Phytol. 174:637–647. [DOI] [PubMed] [Google Scholar]
- Shipley B, Keddy P. 1988. The relationship between relative growth rate and sensitivity to nutrient stress in twenty-eight species of emergent macrophytes. J Ecol. 76:1101–1110. [Google Scholar]
- Slate J. 2004. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol Ecol. 14:363–379. [DOI] [PubMed] [Google Scholar]
- Stewart EL, Croll D, Lendenmann MH, Sanchez-Vallet A, Hartmann FE, et al. 2018. Quantitative trait locus mapping reveals complex genetic architecture of quantitative virulence in the wheat pathogen Zymoseptoria tritici. Mol Plant Pathol. 19:201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci U S A. 101:6564–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani SFF, Melichar JP, Mills C, Pain N, Sierotzki H, et al. 2015. Zymoseptoria tritici: a major threat to wheat production, integrated approaches to control. Fungal Genet Biol. 79:8–12. [DOI] [PubMed] [Google Scholar]
- Walia A, Calderone R. 2008. The SSK2 MAPKKK of Candida albicans is required for oxidant adaptation in vitro. FEMS Yeast Res. 8:287–299. [DOI] [PubMed] [Google Scholar]
- Williams B, Kabbage M, Kim HJ, Britt R, Dickman MB. 2011. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7:e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Li W, Derbyshire M, Larsen MR, Rudd JJ, et al. 2015. Unraveling incompatibility between wheat and the fungal pathogen Zymoseptoria tritici through apoplastic proteomics. BMC Genomics. 16:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P-L, Wang CL, Chen PY, Lee MH. 2017. YAP1 homologue-mediated redox sensing is crucial for a successful infection by Monilinia fructicola. Mol Plant Pathol. 18:783–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Marcel TC, Hartmann FE, Ma X, Plissonneau C, et al. 2017. A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytol. 214:619–631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genotype data, marker information, and phenotype data of the two cross-populations are available at figshare DOI: https://doi.org/10.25386/genetics.11862129.