Abstract
Ty1 mobile DNA element is the most abundant and mutagenic retrotransposon present in the genome of the budding yeast Saccharomyces cerevisiae. Protein regulator of Ty1 transposition 105 (Rtt105) associates with large subunit of RPA and facilitates its loading onto a single-stranded DNA at replication forks. Here, we dissect the role of RTT105 in the maintenance of genome stability under normal conditions and upon various replication stresses through multiple genetic analyses. RTT105 is essential for viability in cells experiencing replication problems and in cells lacking functional S-phase checkpoints and DNA repair pathways involving homologous recombination. Our genetic analyses also indicate that RTT105 is crucial when cohesion is affected and is required for the establishment of normal heterochromatic structures. Moreover, RTT105 plays a role in telomere maintenance as its function is important for the telomere elongation phenotype resulting from the Est1 tethering to telomeres. Genetic analyses indicate that rtt105Δ affects the growth of several rfa1 mutants but does not aggravate their telomere length defects. Analysis of the phenotypes of rtt105Δ cells expressing NLS-Rfa1 fusion protein reveals that RTT105 safeguards genome stability through its role in RPA nuclear import but also by directly affecting RPA function in genome stability maintenance during replication.
Keywords: Rtt105, replication, chromatin, cohesion, telomere
Introduction
The Ty1 element of the budding yeast Saccharomyces cerevisiae is the best-studied LTR-retrotransposon among the five species of retrotransposable elements present in budding yeast (Voytas and Boeke 1992; Curcio et al. 2015). The stability of Ty1-related sequences, which are the most repetitive components of the Saccharomyces cerevisiae genome is modulated by host factors. These factors influence retrotransposition either by promoting efficient Ty1 retromobility (retromobility host factors, RHF genes) or by maintaining transpositional dormancy (restrictors of Ty1 transposition, RTT genes) (Curcio et al. 2007; Nyswaner et al. 2008; Dakshinamurthy et al. 2010).
RTT genes are involved in different aspects of host–genome maintenance and the product of these genes function essentially during S phase of the cell cycle (Scholes et al. 2001). For example, RTT106 encodes a histone chaperone involved with CAF-1 in replication coupled chromatin assembly and in the integrity of advancing replication forks (Li et al. 2008; Clemente-Ruiz et al. 2011; Han et al. 2013). Rtt109 is a histone acetyl transferase that modifies lysine 56 of histone H3 (Driscoll et al. 2007). Rtt109 is required, with Rtt106, Rtt101, and Mms1 (Rtt108), during normal replication, and to modulate replisome function during replicative stress to promote cell survival in the presence of DNA damages (Han et al. 2013; Luciano et al. 2015). On the other hand, Rtt102 is a component of both SWI/SNF and RSC chromatin remodeling complexes involved in DNA replication stress response (Schubert et al. 2013) while Rtt103 associates with sites of DNA breaks and functions in the DNA damage response (Srividya et al. 2012). Rrm3 (Rtt104) travels with the fork and helps the replication fork traverse protein–DNA complexes (Azvolinsky et al. 2006). Elg1 (Rtt110) is a subunit of an alternative replication factor C complex important for DNA damage recovery during replication, involved in cohesion, and in telomere maintenance (Kanellis et al. 2003; Smolikov et al. 2004; Parnas et al. 2009). Rtt107 is implicated in DNA repair during S phase and recruits Smc5/6 to double-stranded breaks (DSB) (Leung et al. 2011).
A genome-wide analysis indicated that RTT105 exhibited genetic interaction with genes involved in genome maintenance (Collins et al. 2007). Immunoprecipitation and mass spectroscopy revealed that Rtt105 co-purifies with the three subunits of RPA and Kap95, the primary karyopherin responsible for RPA import (Li et al. 2018, 2019). The Rtt105/RPA interaction is required for the association of RPA with Kap95 and peaks in S phase (Li et al. 2018). Rtt105 was proposed to form an alternative adapter for RPA nuclear import, modulating RPA level in nucleus, and to assist RPA in adopting a more extended conformation to contact ssDNA (Li et al. 2018). Of note, Rtt105 is not present in the final RPA–ssDNA complex, revealing that the functions of Rtt105 are highly analogous to histone chaperones in regulating histone behaviors (Li et al. 2018). As a consequence, rtt105Δ cells die in the presence of hypomorph alleles of ORC2 or POL3, are sensitive to genotoxic agents such as HU, CPT, MMS, and bleomycin, and present a mild delay of S phase progression during DNA replication (Li et al. 2018). Importantly, while the binding level of RPA is strongly reduced in the absence of RTT105, no dramatic decrease of global DNA synthesis level under unperturbed replication forks is detected. These observations suggest that Rtt105 is more important for regulating RPA binding at perturbed replication forks where more and longer ssDNA intermediates are generated (Sogo et al. 2002). RPA is the main single-stranded DNA-binding protein involved in multiple processes including replication, transcription, recombination, checkpoints, telomere maintenance, elimination of G-rich DNA secondary structures, and DNA repair. To advance our understanding of the relationship between Rtt105 and RPA, it is crucial to determine the role of Rtt105 in the multiple functions of RPA and if these functions are directly affected through Rtt105 ability to chaperone RPA.
In this study, we further investigate the importance of RTT105 in replication and in various replication-coupled mechanisms in which RPA plays a critical role. We show that RTT105 is essential for the viability of cells when replisome progression or S-phase checkpoint is affected. Consistent with these results, homologous recombination (HR), but not nonhomologous end joining (NHEJ) is required to sustain the growth of rtt105Δ mutant. We further report multiple genetic interactions between RTT105, and genes involved in chromatin structure formation and in cohesion establishment. We also reveal that RTT105 is required for telomere elongation by telomerase likely at a step independent of telomerase recruitment but related to its function as RPA chaperon. Finally, our data show that RTT105 affects DNA metabolism and genome stability not only via its role in RPA nuclear import and indicate that RTT105 exerts a crucial role during replication.
Materials and methods
Strain construction
All strains used in this study are listed in Supplementary Table S1. To construct the pRS316-NLS-RFA1 plasmid allowing the expression of NLS-RFA1, a first PCR was used to amplify a fragment containing the promoter of RFA1 fused to an NLS (encoding PKKKRKV). A second PCR was performed to amplify the full RFA1 coding sequence. The primers were designed in order to generate overlapping sequences between the two PCR products. A third PCR combining the two previous PCR products as templates produced a fragment coding for NLS-RFA1 under the control of RFA1 promoter. This fragment was gel-purified, digested with BamHI-HF and HindIII-HF (BioLabs), and cloned into pRS316. The resulting pRS316-NLS-RFA1 vector was confirmed by DNA sequencing.
Fluorescence microscopy
All microscopy analyses were performed in liquid (SC synthetic media) using a Nikon Eclipse Ti microscope with a 100× objective. Images were collected using a Neo sCMOS camera (Andor). Exposure time was DIC: 100 ms; CFP: 500 ms. Images were analyzed using ImageJ on 2 D-maximum projections from 11-Z-stacks spaced 0.5µ each. Cells were prepared by growing at 30°C.
Telomere length and cell senescence analyses
Telomere length analysis and cell senescence assays were performed as described (Simon et al. 2021). Telomere lengths were determined with Quantity one software (Bio Rad) using a semi-log plot generated from the distance migrated on the same agarose gel by DNA fragments in the Eurogentec SmartLadder. Average lengths ± standard deviations are reported in Table 1. Liquid senescence assays were performed starting with the spore products of est1Δ/EST1 rtt105Δ/RTT105 diploid strain. The diploid strain has been propagated for at least 50 population doublings (PDs) on YPD plates to ensure homogeneous telomere length before sporulation. The senescence assay was performed as described by Simon et al. (2021).
Table 1.
Telomere length analysis of rtt105Δ, yku80Δ, and rtt105Δ yku80Δ mutants
| Mutant | Telomere length relative to WT (bp) |
|---|---|
| rtt 105Δ | −106 ± 14 |
| yku80Δ | −148 ± 13 |
| rtt105Δ yku80Δ | −167 ± 7 |
Values based on n = 7.
Analysis of CLB2-rfa1 protein level
Yeast cells were grown at 30°C in YPD to an OD600 = 0.8 and arrested in S-phase by adding 200 mM HU (Sigma) for 2 h. HU was removed to allow cells to progress synchronously through the cell cycle in the presence of 25 µg/ml nocodazole. Samples were taken at the indicated time point for FACS analyses to monitor the progression of the cell cycle and for protein extraction. Cells were then lysed by bead beating in the presence of 20% TCA. The pellets were recovered by centrifugation and incubated with 1× Laemmli buffer at 95°C for 5 min to recover proteins. Subsequently, proteins were separated on 10% poly-acrylamide gel (Life Technologies) followed by Western blotting with anti-Rfa1 antibody (Agrisera).
Protein chromatin-binding assay
Asynchronous cells were harvested and incubated in 3 ml of pre-spheroplasting buffer (100 mM PIPES (pH 9.6), 10 mM dithiothreitol (DTT) for 10 min at 30°C. After centrifugation, cells were resuspended in 2 ml of spheroplasting buffer (50 mM KH2PO4/K2HPO4 (pH 7.5), 0.6 M Sorbitol, 10 mM DTT, 0.5 mM PMSF) containing 10 µl of 10 mg/ml of Zymolase (AMSBIO) and incubated at 30°C for 25 min with gentle shaking. Spheroplasts were washed with 1 ml of cold Diffley buffer (20 mM PIPES (pH 6.8, 150 m KOAc, 2 mM MgOAc2, and 0.4 M Sorbitol) containing 1 mM PMSF and protease inhibitors, pelleted at 3000 rpm for 5 min at 4°C, resuspended in 600 µl of Diffley buffer containing 1% Triton X-100 and incubated on ice for 5 min with gentle mixing. Finally, lyzed nuclei were centrifugated at 13.000 rpm for 15 min at 4°C and the pellets corresponding to the chromatin-associated proteins fraction were resuspended in 1× Laemmli buffer.
Data availability
All data and method required to confirm the conclusions of this work are within the manuscript: Supplementary Table S1: Strains used in this study. Supplementary Figure S1: RTT105 is required for normal cell growth. Figure S2: RTT105 exhibits genetic interactions with S-phase checkpoint components. Supplementary Figure S3: RTT105 is important for cells affected in the replication-dependent nucleosome assembly process. Supplementary Figure S4: RTT105 is important for cells affected in the replication-dependent nucleosome assembly process but not in replication-independent nucleosome assembly. Supplementary Figure S5: RTT105 inactivation exacerbates the telomeric defects arising in the absence of YKU and EST1 genes. Supplementary Figure S6: RTT105 inactivation affects senescence and survivor formation in est1Δ cells. Supplementary Figure S7: Exogenous expression of rtt105Δ155-208 mutant failed to rescue the growth in rtt105Δ cells. Supplementary Figure S8: Cell cycle regulation of CLB2-rfa1. Supplementary Figure S9: Bringing Rfa1 into the nucleus rescues the viability of rfa1Δ and rfa1-D228Y rtt105Δ cells but not the growth defect and HU sensitivity in rtt105Δ cells. Supplementary Figure S10: Bringing Rfa1 into the nucleus rescues the growth of rtt105Δ cells affected in S-phase checkpoint, cohesion, or repair.
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13373021.
Results
The rtt105Δ mutant grows slowly at 25°C and is not sensitive to replication stress induced by the absence of PIF1 family helicases
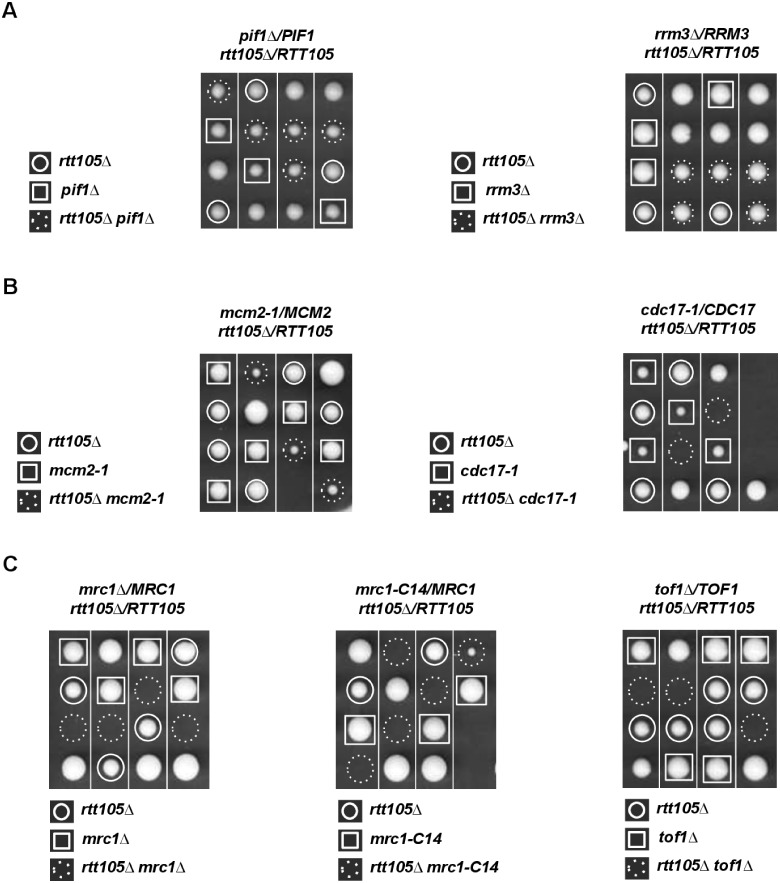
One allele of RTT105 was disrupted in a wild-type diploid yeast strain. After spore dissection, we noticed that rtt105Δ spores exhibited a slow-growth defect at 30°C (Supplementary Figure S1, upper panel, left). This slow growth phenotype was amplified at 25°C without affecting spore viability (Supplementary Figure S1, upper panel, right). We obtained similar results by deleting RTT105 directly in haploid cells (Supplementary Figure S1, lower panels). rtt105Δ cells were previously reported to be highly sensitive to DNA-damaging agents (Li et al. 2018). During replication, forks encounter natural impediments throughout the genome (Gadaleta and Noguchi 2017). We assessed the importance of RTT105 in the absence of Pif1 and Rrm3 that assist the replisome in the replication of difficult to replicate genomic regions. Both proteins perform overlapping and distinct roles in replication, repair, telomere length maintenance, and cohesion (reviewed in Muellner and Schmidt 2020). We found that RTT105 was dispensable for the growth of pif1Δ cells (Figure 1A, left) suggesting that Rtt105 and Pif1 work in the same pathway to counteract DNA damages arising from G4 DNA secondary structures (Maestroni et al. 2020). Surprisingly, we observed that RTT105 was also dispensable for rrm3Δ growth (Figure 1A, right). Indeed, in cells lacking Rrm3, chronic stalling of forks at protein–DNA barriers is associated with increased DNA damage and checkpoint activation (Ivessa et al. 2003; Azvolinsky et al. 2006; Schmidt and Kolodner 2006). These results indicate that RTT105 functions with specific type, and/or with a certain level of DNA damage.
Figure 1.
RTT105 inactivation results in cell lethality in different genetic contexts affecting replisome progression. (A) Members of the PIF1 DNA helicase family are not required for viability of yeast rtt105Δ cells. Tetrad dissection of the diploid strains pif1Δ/PIF1 rtt105Δ/RTT105 and rrm3Δ/RRM3 rtt105Δ/RTT105. In this and subsequent figures, the spores from a given tetrad are in vertical line in a YPD plate. Four representative tetrads are shown after 3 days at 30°C. (B) Genetic interaction of mcm2-1 and cdc17-1 with RTT105. The diploid strains mcm2-1/MCM2 rtt105Δ/RTT105 (left) and cdc17-1/CDC17 rtt105Δ/RTT105 (right) were sporulated and dissected. (C) RTT105 is required for viability in the absence of the replicative function of MRC1 or the Tof1-Csm3 complex. Left, tetrads from diploids heterozygous for mrc1Δ, and for rtt105Δ were dissected and analyzed as in (A). Center, tetrads from mrc1-C14, and rtt105Δ heterozygous diploids were dissected and analyzed. Right, tetrads from diploids heterozygous for tof1Δ, and for rtt105Δ were dissected and analyzed.
RTT105 exhibits synthetic genetic interactions with genes encoding replisome components
The rtt105Δ mutation is synthetic lethal when combined with mutations in either the origin recognition complex (ORC) or polymerase δ (Li et al. 2018). Having shown that the replisome component Rrm3 was not crucial for rtt105Δ cells, we investigated the importance of RTT105 for replication fork progression that depends on the replisome progression complex (RPC) (Gambus et al. 2006). The RPC consists of Mcm2–Mcm7 proteins, Mcm10, the go ichi ni san (GINS) complex, Cdc45, the trimeric complex of regulatory factors comprising Tof1, Csm3, and the checkpoint mediator Mrc1, Ctf4, Top1, and the histone chaperon FACT (Spt16 and Pob3). In agreement with the fact that RTT105 plays a role during replication (Li et al. 2018), we found that combining rtt105Δ with the thermosensitive mcm2-1 helicase mutant resulted in a marked slow growth phenotype (Figure 1B, left). We further found that rtt105Δ cdc17-1 double mutant was dead at 30°C (Figure 1B, right), indicating that RTT105 is important in cells experiencing lagging strand-induced replicative stress. Along the same line, cells lacking RTT105 were inviable in the absence of Mrc1 that promotes replisome progression at the leading strand (Yeeles et al. 2017) (Figure 1C, left). In addition to its role in replication, MRC1 is also required for checkpoint activation after DNA replicative stress. We found that the mrc1-C14 mutant that is compromised for its replication function but proficient for its checkpoint function (Naylor et al. 2009) was synthetically lethal or very slow growing with rtt105Δ (Figure 1C, middle). Finally, because Mrc1 together with Tof1/Csm3 also forms a complex required for fork protection (Calzada et al. 2005; Tourrière et al. 2005; Bando et al. 2009; Eickhoff et al. 2019), we analyzed the contribution of TOF1/CSM3 to rtt105Δ viability. Deleting TOF1 also caused synthetic lethality with rtt105Δ (Figure 1C, right) suggesting that the stability of the replication fork is strongly affected in absence of RTT105. Taken together, these genetic interactions show that RTT105 becomes crucial when replisome integrity is affected.
The S-phase checkpoint pathway is required for rtt105Δ viability
We next evaluated whether loss of RTT105 caused a synthetic interaction with mutations in the S-phase checkpoint. The S-phase checkpoint is divided into two branches: the DNA replication checkpoint (DRC), which is specific to S phase, and is mediated by MRC1, the RFC-CTF18 complex checkpoint mediator, and other fork components, and the DNA-damage checkpoint (DDC) which operates throughout the cell cycle and depends on the checkpoint mediator Rad9 (reviewed in Pardo et al. 2017). Both branches are activated by the sensor kinase Mec1 and converge on the effector kinase Rad53. We found that Mec1 and Rad53 are required for cell viability in the absence of RTT105 (Supplementary Figure S2, center).
Then, we evaluated the importance of the DRC in rtt105Δ by testing the ctf18Δ mutant, which is compromised for its DRC function. We found that ctf18Δ strongly affects rtt105Δ cell growth (Supplementary Figure S2, left). To assess the contribution of the DDC in rtt105Δ cells, we deleted RAD9 and found that DDC inactivation also strongly affects the viability of the rtt105Δ mutant (Supplementary Figure S2, right). These data indicate that in rtt105Δ cells both DRC and DDC are important to sustain their growth. These results support the notion that rtt105Δ cells exhibit replicative defects and associated DNA damages.
Homologous recombination is required for rtt105Δ viability in contrast to nonhomologous end joining
It has been recently shown that rtt105Δ cells exhibit a synthetic sick phenotype with mutations in key genes involved in HR and nonhomologous end joining suggesting that RTT105 have important role in these repair processes (Li et al. 2018).
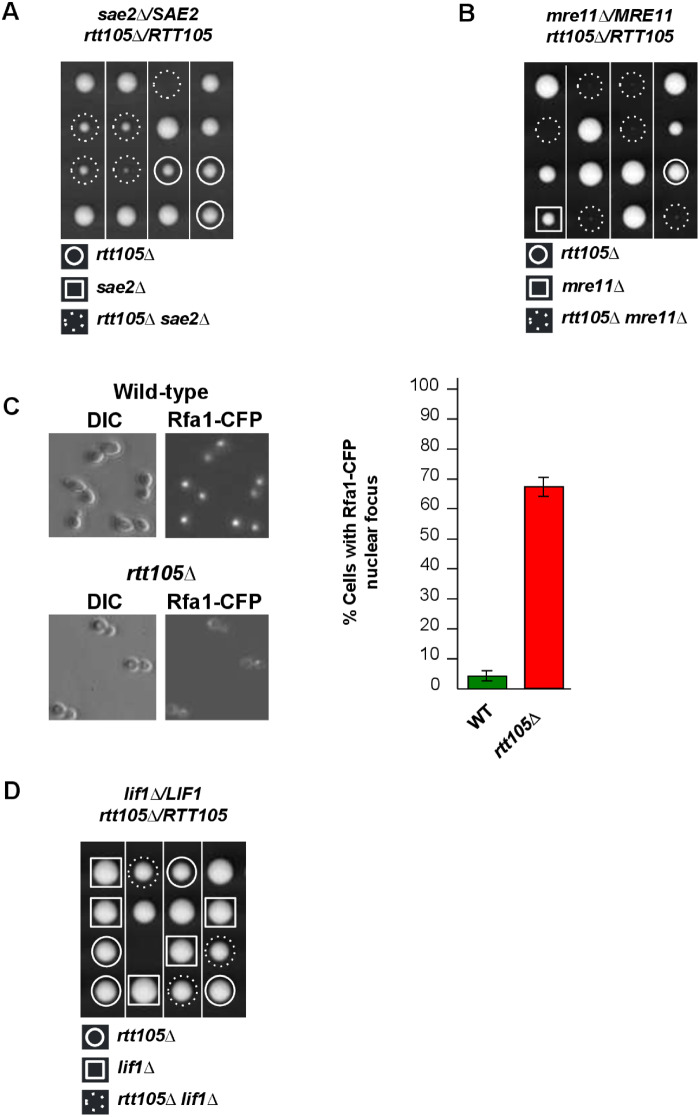
The MRX complex (Mre11-Rad50-Xrs2) and Sae2 function together to initiate end resection, an essential early step in homology-dependent repair of DSB (Longhese et al. 2010; Seeber et al. 2016; Gnügge and Symington 2017). Our results showed that rtt105Δ cells lacking either one of the MRX component or Sae2 were unable to grow or grew very poorly indicating that initial steps of HR are required to sustain the viability of rtt105Δ cells (Figure 2, A and B). Of note, this synthetic lethality could not be attributed to spore germination defects since mre11Δ rtt105Δ spores formed micro-colonies after 7 days at 30°C. To show that rtt105Δ-induced DNA damage was actually repaired by HR, we monitored nuclear localization of Rfa1-CFP that forms fluorescence foci representing DNA repair centres of multiple DSBs (Lisby et al. 2004). Despite the fact that deleting RTT105 gave a diffuse signal and reduced the level of RPA associated to the fork (Li et al. 2018, 2019) we found that rtt105Δ exhibited a very high frequency of spontaneous Rfa1-CFP foci compared to WT (67% vs 4%, respectively) (Figure 2C). We noted that some Rfa1-CFP foci appeared brighter (27%), likely reflecting abnormally long region of RPA-bound single-stranded DNA.
Figure 2.
RTT105 shows genetic interactions with components involved in DNA repair by homologous recombination. (A, B) sae2Δ/SAE2 rtt105Δ/RTT105 and mre11Δ/MRE11 rtt105Δ/RTT105 diploid strains were dissected and the resulting spores were incubated at 30°C for 3 days. (C) Left, Rfa1 foci are detected in rtt105Δ cells. Wild-type and rtt105Δ cells encoding Rfa1-CFP were analyzed with differential interference contrast (DIC) (left) and with florescence microscopy (right). Right, numbers indicate the percentage of cells that contained Rfa1-CFP foci. Rfa1 foci were analyzed in asynchronously growing cells. At least 200 cells were analyzed for each strain. (D) Genetic interaction of LIF1, with RTT105. lif1Δ/LIF1 rtt105Δ/RTT105 were dissected. Tetrads were grown at 30°C for 3 days.
We next investigated the requirement of the nonhomologous end-joining repair pathway for rtt105Δ viability. We found that RTT105 was dispensable for the growth of lif1Δ cells (Figure 2D). This result indicates that NHEJ per se is not important for rtt105Δ growth.
Collectively, these data strongly support the notion that the absence of RTT105 induces DSB and/or ssDNA gaps that form as a result of DNA replication perturbation and are repaired by HR but not by NHEJ (see Discussion).
Cohesion defects create a requirement for RTT105
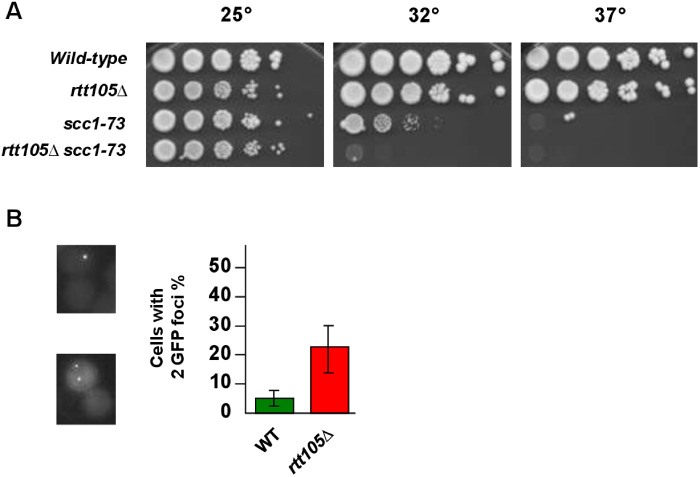
Considering that many replisome components are part of sister chromatid cohesion pathways (Xu et al. 2007) and that replication fork passage is intimately linked to the establishment of the sister chromatid cohesion, we further investigated the importance of RTT105 in cells experiencing cohesion defects. We focused on SCC1 (also known as MCD1), which encodes one of the essential subunits of the cohesin complex. We found that the ts allele scc1-73 which displays increased cohesion loss when shifted to the semi-restrictive temperature (32°C) (Michaelis et al. 1997) is lethal in the absence of RTT105 at this temperature (Figure 3A). We directly evaluated the cohesion defects in a rtt105Δ mutant using strains bearing Lac operator repeats integrated at a site near the centromere of chromosome III (Figure 3B). We found that rtt105Δ cells exhibited failure in cohesion (around 23%). Our results reveal that RTT105 is important to sustain the viability of cells with cohesion defects suggesting that RTT105 contributes to efficient sister chromatid cohesion.
Figure 3.
RTT105 is important for cells affected in sister chromatid cohesion. (A) Tenfold serial dilutions of exponentially growing cells with the indicated mutations were spotted onto YPD plates and incubated at 25°C, 32°C, or 37°C for 3 days. (B) Cohesion is affected in rtt105Δ cells. Sister chromatid cohesion was analyzed by monitoring the tagged centromere of chromosome III. Over 150 cells were counted for each experiment. The results represent the average of three independent experiments.
RTT105 genetically interacts with genes encoding histone chaperones and histone H3–H4 lysine mutants with defects in nucleosome assembly during replication
Cohesion and replication-coupled nucleosome assembly have been functionally linked (Zhang et al. 2017a, 2017b). H3K56ac is an important mark required for chromatin assembly (Chen and Tyler 2008; Li et al. 2008). This mark found in all newly synthesized histone H3 and deposited behind replication forks in S-phase is dependent on ASF1 and catalyzed by RTT109 (Driscoll et al. 2007; Han et al. 2007a, 2007b; Tsubota et al. 2007). To ascertain the importance of RTT105 in cells unable to acetylate H3K56, we analyzed the consequences of deleting either ASF1 or RTT109. We found that rtt105Δ cells showed significant growth defects when combined with asf1Δ and rtt109Δ (Supplementary Figure S3A). Consistent with this result, the substitution K56R in H3 also impaired the growth of rtt105Δ cells (Supplementary Figure S3B, compare green and red circles). We next analyzed the importance of other post-translational modifications on newly synthesized histones known to regulate the replication-coupled nucleosome assembly (Ai and Parthun 2004; Li et al. 2009). We observed that H3K9,14,18,23,27R and H4K5,8,12R mutations significantly reduced rtt105Δ growth (Supplementary Figure S4A). On the contrary, deletion of HIR1 or HIR2 which are involved in replication-independent nucleosome assembly (Green et al. 2005) caused no apparent effect on rtt105Δ growth (Supplementary Figure S4B). Taken together, these data support the idea that RTT105 is functionally linked to nucleosome assembly during replication.
RTT105 is required for heterochromatin silencing and genetically interacts with SIR complex
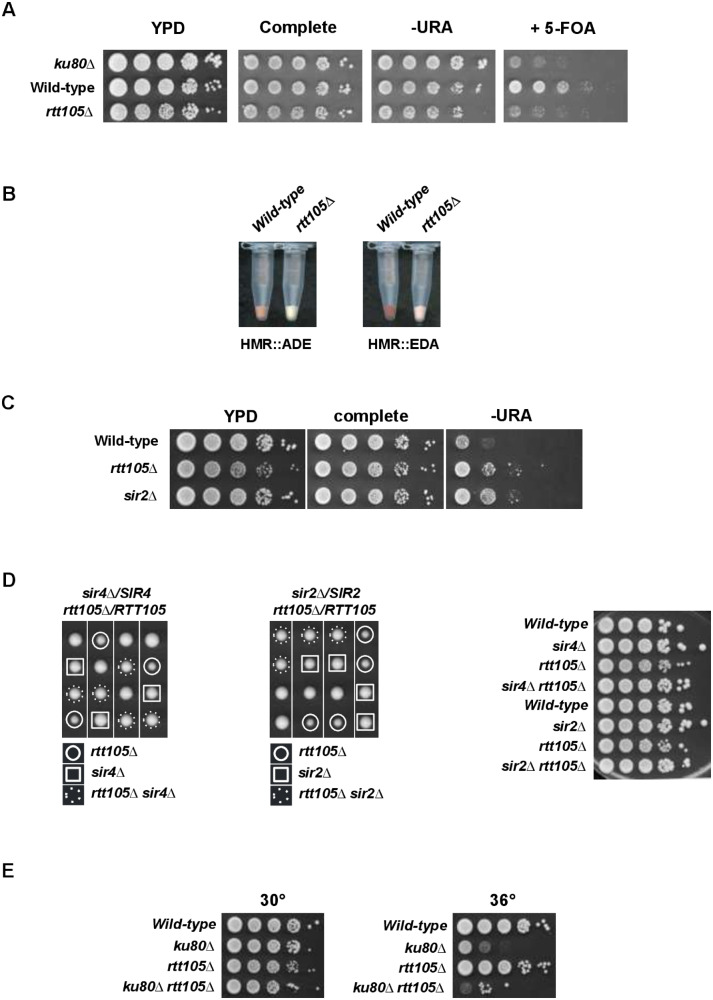
We tested if RTT105 is involved in gene silencing at the three heterochromatin-like loci in S. cerevisiae. We first ask whether RTT105 is required for telomeric position effect (TPE) using cells containing the reporter gene URA3 integrated at the left arm of telomere VII. We found that TPE was impaired in the rtt105Δ strain (Figure 4A). To determine whether silencing defects also occur at HMR in rtt105Δ cells, we used a color assay utilizing an ADE2 reporter in which the ADE2 expression is regulated by the HMR silencer (Sussel et al. 1993). As expected, we obtained pink colonies with the wild-type strain while rtt105Δ cells grew as white colonies indicating that silencing of ADE2 at HMR was lost (Figure 4B). We finally investigated if rtt105Δ cells show defects in rDNA silencing by using a reporter strain containing a URA3 gene inserted as a single copy at the rDNA locus (Chang and Winston 2011). When plated on minimal medium without uracil, WT cells showed slow growth whereas rtt105Δ cells (as well as sir2Δ cells used as control) were able to grow reflecting defects in rDNA silencing (Figure 4C).
Figure 4.
RTT105 is required for gene silencing and interacts genetically with SIR and YKU complexes. (A) Deletion of RTT105 reduces silencing of a telomere-proximal URA3 gene. Tenfold serial dilutions of exponentially growing cells were spotted onto YPD, complete, SD-URA, and 5-FOA plates and incubated at 30°C for 2 days. yku80Δ mutant was used as positive control. The absence of growth reveals a non-silenced state of the URA3 gene. (B) Deletion of RTT105 reduces ADE2 silencing at HMR. Strains containing the HMR::ADE2 allele or the HMR::2EDA allele (Sussel et al. 1993) were grown overnight in YPD liquid medium without additional adenine. A dark red/pink colony color indicates silencing of ADE2. A white/slightly pink colony color indicates a nonsilenced state of ADE2. 2EDA indicates that the ADE2 gene has been placed in a promotor-distal orientation with respect to the E-silencer (Sussel et al. 1993). (C) Deletion of RTT105 gene reduces silencing at rDNA. Tenfold serial dilutions of exponentially growing cells were spotted onto YPD, complete, and SD-URA plates and incubated at 30°C for 2 days. All the different strains carry an mURA3 reporter in a single copy within the rDNA (Chang and Winston 2011). Growth on medium without uracil (-URA) assesses the degree of reporter silencing. sir2Δ mutant was used as positive control. The absence of growth reveals a silenced state for the URA3 gene. (D) Deletion of SIR genes rescues rtt105Δ growth defect. Tetrad dissection of sir4Δ/SIR4 rtt105Δ/RTT105 (left) and sir2Δ/SIR2 rtt105Δ/RTT105 (center) diploid strains. Right, effects of sir2Δ and sir4Δ on viability of the rtt105Δ cells. Tenfold serial dilutions of exponentially growing cells were spotted onto YPD plates and incubated at 30°C for 3 days. (E) rtt105Δ yku80Δ mutant is not viable at 36°C. Tenfold serial dilutions of exponentially growing cells were spotted onto YPD plates and incubated at 30°C or 36°C for 3 days.
Subtelomeric regions, rDNA array, and the cryptic mating-type loci are all transcriptionally silenced by SIR proteins (reviewed in Gartenberg and Smith 2016). We assess the impact of heterochromatin disruption on growth of rtt105Δ cells by deleting SIR proteins. Strikingly, we uncover that deleting both SIR2 and SIR4 rescued the rtt105Δ growth defect.
Taken together, these results show that RTT105 is required for efficient silencing at the heterochromatic loci and suggest that the impaired localization of Sir proteins in the rtt105Δ contributes to its slow growth.
Disruption of RTT05 affects telomere maintenance
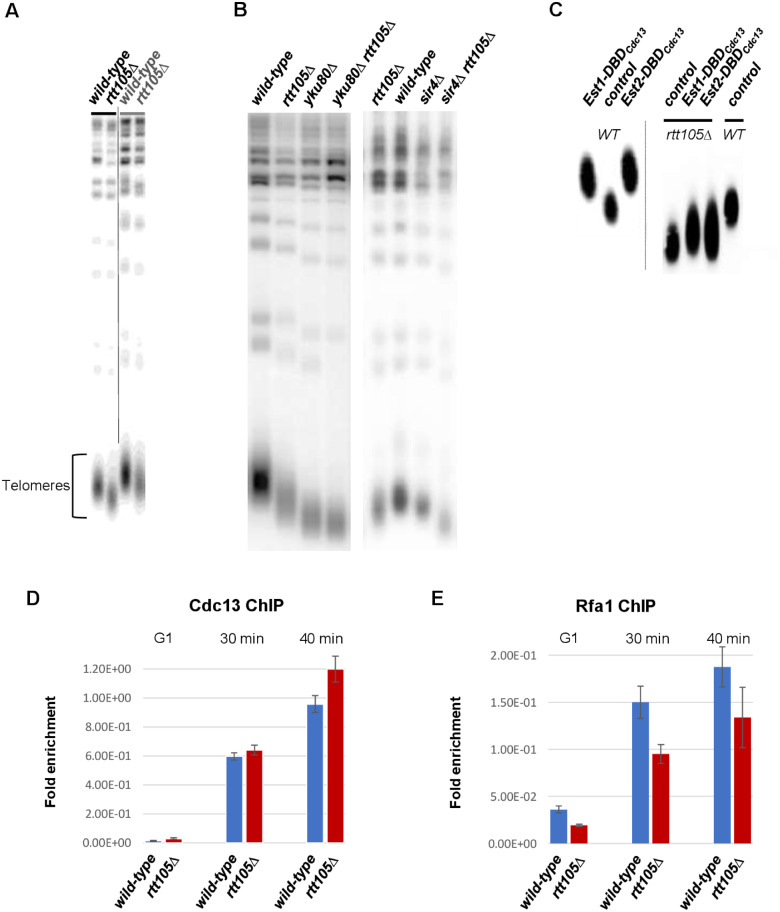
We analyzed the ts phenotype of the yku80Δ which reflects telomere structure defects in the rtt105Δ (Gravel et al. 1998; Polotnianka et al. 1998). At 30°C, a permissive temperature for ku80Δ mutant, the double mutant rtt105Δ ku80Δ grew normally while at 36°C (the restrictive temperature) the rtt105Δ deletion abolished the ability of ku80Δ mutant to form colonies (Figure 4E) suggesting that the telomere structure defect of the ku80Δ mutant was aggravated by inactivation of RTT105. Interestingly, we often but not systematically noted the emergence of colonies in the double mutant that could reflect possible activation of telomerase-independent maintenance of telomeres in rtt105Δ yku80Δ cells. Since overexpression of telomerase was reported to suppress yku80Δ ts phenotype (Nugent et al. 1998; Teo and Jackson 2001), we investigated if rtt105Δ affected telomere length. As shown in Figure 5A, loss of RTT105 resulted in shortening of telomeres revealing that RTT105 positively regulates telomere length. In S. cerevisiae, recruitment of telomerase to telomeres was reported to be mediated by two pathways involving yKu-TLC1/Sir4 and Est1/Cdc13 interactions (Hass and Zappulla 2015; Chen et al. 2018). In addition to its role in recruiting telomerase, YKU that associates with TLC1 regulates TLC1 nuclear retention and also telomerase subunit Est1 accumulation at telomeres (Stellwagen 2003; Fisher et al. 2004; Gallardo et al. 2008; Lemon et al. 2019). We examined the genetic interaction between RTT105, YKU80, and SIR4 genes related to telomere length. As previously described (Longhese et al. 2000; Hass and Zappulla 2015; Chen et al. 2018), we found that sir4Δ slightly decreases telomere size, and that yku80Δ cells have short but stable telomere (Figure 5B). We discovered that deleting RTT105 diminished telomere length of sir4Δ mutant (Figure 5B, right) and slightly reduced telomere shortening of yku80Δ cells (Figure 5B, left and Table 1). These results suggest that RTT105 acts in a pathway different than the yKu-TLC1/Sir4 recruitment pathway. We also evaluated the telomere size of yku80Δ rtt105Δ emerging colonies arising at 36°C in the spot assays (refer to Figure 4E, right). When these yku80Δ rtt105Δ emerging colonies were grown in liquid cultures, their growing colonies exhibited amplification of the tandemly repeated Y' short and Y' long subtelomeric elements (Type I survivors) with the disappearance of X-only telomeres. These results suggested that deleting RTT105 in yku cells abolished telomerase activity at nonpermissive temperature leading to the appearance of Type I survivors (Supplementary Figure S5A) (Lundblad and Blackburn, 1993; Fellerhoff et al. 2000). We next inspected if telomere lengthening occurred in rtt105Δ cells when telomerase recruitment was bypassed by a fusion between Est1 or Est2 and the DNA-binding domain of Cdc13 (Evans and Lundblad 1999). We transformed wild-type and rtt105Δ cells with a plasmid expressing the hybrid proteins (Est1-DBDCdc13 or Est2-DBDCdc13). In wild-type cells, expression of the Est1-DBDCdc13 or Est2-DBDCdc13 protein caused elongated telomeres as previously reported (Evans and Lundblad 1999). In contrast, in all rtt105Δ clones (n = 5) that we analyzed, artificial tethering of Est1 or Est2 led only to a modest lengthening of telomere size (Figure 5C) suggesting that RTT105 is required for telomerase action when telomerase is artificially tethered to telomeres by the Est1-DBDCdc13 or Est2-DBDCdc13 fusion proteins.
Figure 5.
RTT105 is required for telomere maintenance. (A) Telomere length of rtt105Δ cells measured by southern blotting. In this and subsequent figures, genomic DNA from each cell culture was digested by XhoI, separated on a 0.8% agarose gel, and hybridized to a poly(GT) telomere specific probe. Two different genetic backgrounds were used. Left (black), W303 background. Right (grey), LPY917 backgound. (B) Telomere length analysis of rtt105Δ in yku80Δ, and sir4Δ cells. (C) Tethering of Est1 and Est2 in rtt105Δ cells. Telomere length of wt and rtt105Δ strains transformed with either pVL1120 which directs the expression of Est1-DBDCDC13 or pVL1107 which directs the expression of Est2-DBDCDC13 (Evans and Lundblad 1999). Controls: wild-type and rtt105Δ strains were transformed with the empty plasmid. (D, E) Cdc13 binding at telomere is not reduced in rtt105Δ cells while binding of RPA is compromised. Cells were synchronized in G1-phase of the cell cycle using alpha-factor and subsequently released in YPD. ChIP experiments were performed at t = 0 min (G1), t = 30 min (S), and t = 40 min (late S) using an antibody either against the MYC-Tag of Cdc13-Myc (D) or against Rfa1 (Agrisera) (E). The immunoprecipitated DNA was quantified with real time PCR using primers amplifying the left arm of telomere XV (Tel-XV-L) (Bianchi and Shore 2007). Experiments were performed in triplicate.
We next monitored the binding of Cdc13 and RPA at telomeres by performing ChIP experiments in WT and rtt105Δ cells. Telomere-ChIP experiments revealed that RTT105 deletion did not decrease Cdc13 binding to telomeres (Figure 5D) strongly favoring the hypothesis that Rtt105 acts at step independent of telomerase recruitment by the Cdc13 pathway. We also found that the absence of Rtt105 caused a decrease in the amount of RPA associated with telomeres (Figure 5E). Taken together, these data indicate that the role of RTT105 in telomere maintenance likely relies on RPA and is independent of the canonical telomerase recruitment pathways.
Afterward, we analyzed the impact of RTT105 inactivation on replicative senescence by analyzing the senescence profiles of est1Δ cells in the absence of RTT105. As expected, the est1Δ single mutant showed a decrease in growth over generations. Analysis of rtt105Δ est1Δ spore colonies revealed that growth defect of the double mutant was more severe than the one of the single est1Δ mutant (Supplementary Figure S5B). As expected, we did not notice signs of senescence in rtt105Δ cells that can grow indefinitely. In agreement with these observations, we found that recombined telomeres appeared more quickly in rtt105Δ est1Δ double mutant compared to est1Δ single mutant (Supplementary Figure S5C). Our results show that the deletion of RTT105 accelerates replicative senescence without altering the rate of telomere shortening. To confirm these results, we performed liquid senescence assays. Wild-type, rtt105Δ, est1Δ, and rtt105Δ est1Δ spores arising from rtt105Δ/RTT105 est1Δ/EST1 diploid strain with an EST1-expressing plasmid were isolated after micromanipulation on YPD plate and then propagated in liquid cultures for around 120 PDs via serial dilution every 24 h (Aguilera et al. 2020). As observed on YPD plates, we did not notice any sign of senescence in rtt105Δ cells (Figures 6 and Supplementary Figure S6A, green lines). As expected, proliferation of est1Δ mutant declined progressively until the cells reached crisis after about 70 PDs before formation of the survivors (black lines). Deleting RTT105 in est1Δ cells increased the rate of senescence as indicated by the early appearance of the crisis that appeared after about 50 PDs (blue lines). Interestingly, we noticed that the double rtt105Δ est1Δ mutant stayed for prolonged time in crisis before appearance of survivors, which reflect defects in survivor formation (Figures 6 and Supplementary Figure S6A, blue lines). These results highlight the role of RTT105 in telomere replication that is particularly manifested in the absence of telomerase activity (Simon et al. 2016).
Figure 6.
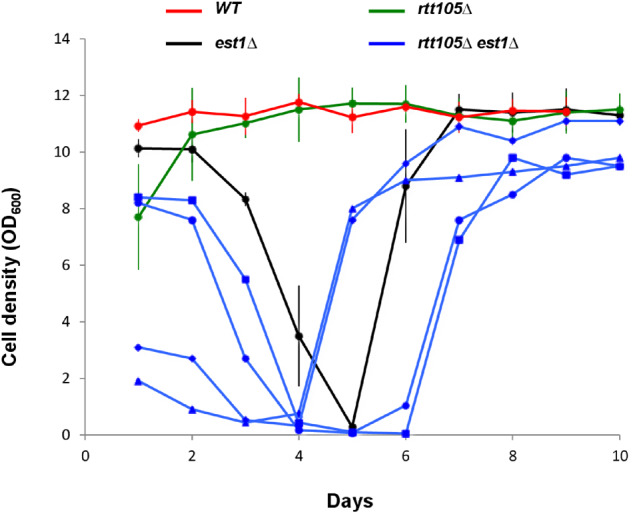
RTT105 inactivation affects senescence and survivor formation in est1Δ. Mean senescence profiles of the WT, rtt105Δ, and est1Δ clones, and senescence profiles of the rtt105Δ est1Δ clones analyzed in the course of this study. At least three clones were analyzed in each case. Each clone was isolated by sporulation of a heterozygous diploid strain and subsequently propagated in liquid culture through daily serial dilutions. OD600 was measured every day to estimate the cell density reached in 24 h.
Finally, to compare the telomere structure in rtt105Δ est1Δ and est1Δ telomeres during replicative senescence, telomere length was analyzed at different time points of the senescence kinetics by southern blot. As expected, at the later time points the liquid culture of est1Δ cells gave rise to long TG1-3 tracts, heterogeneous in length, corresponding to type II survivors (Lundblad and Blackburn, 1993). Using several independently isolated clones, we found that similarly to est1Δ clones, rtt105Δ est1Δ clones produced type II survivors (Supplementary Figure S6B).
Taken together these results unveils the importance of RTT105 in telomere length maintenance according to a scenario in which RTT105 favors the restart of stalled replication forks at eroded telomeres in cells lacking telomerase activity (Simon et al. 2016).
Genetic interactions between RTT105 and RPA mutants
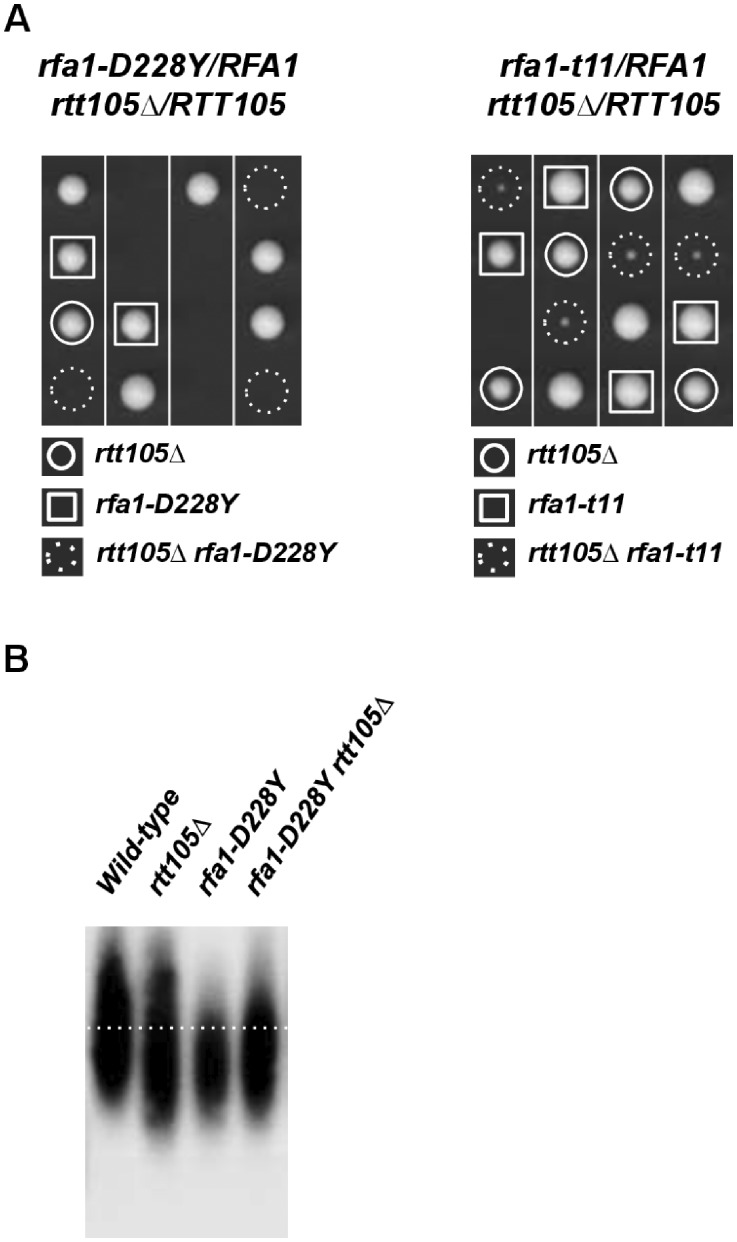
Because Rtt105 functions as an “RPA chaperone” (Li et al. 2018, 2019), we conducted epistasis analysis between rtt105Δ and rfa1 mutants. We first analyzed the genetic interaction between rtt105Δ and the rfa1-D228Y mutation reported to decrease the affinity of Rfa1 complex to ssDNA (Smith and Rothstein 1999; Audry et al. 2015). We found that most rfa1-D228Y rtt105Δ cells did not form visible colonies after 3 days at 30°C or formed microcolonies that grew extremely slowly (Figure 7A, left). This result suggests that decreasing the level of RPA is toxic for the cell when RPA ssDNA binding activity is compromised. We also noticed that replication-proficient but recombination-defective rfa1-t11 (K45E) mutant (Vanoli et al. 2010) was very sick in the absence of RTT105 (Figure 7A, right) confirming the necessity of recombinational process for normal growth in rtt105Δ cells. Since rfa1-D228Y allele was previously reported to shorten telomeres (Smith and Rothstein 2000; Luciano et al. 2012), we examined telomeres of rfa1-D228Y rtt105Δ cells. No synergistic reduction in telomere length occurred in the rfa1-D228Y rtt105Δ double mutant (Figure 7B) suggesting that the reduction in telomere size observed in rtt105Δ cells is related to RPA functions.
Figure 7.
rtt105Δ displays genetic interactions with rfa1 alleles. (A) Genetic interaction of RFA1 with RTT105. rfa1-D228Y/RFA1 rtt105Δ/RTT105 (left), and rfa1-t11/RFA1 rtt105Δ/RTT105 (right) were dissected and incubated 30°C for 3 days. (B) Deleting RTT105 does not increase the telomere length defect of rfa1-D228Y cells. Teloblots were performed as in Figure 5. The dashed line indicates wild-type telomere position.
Reducing the level of RPA in S-phase phenocopies rtt105Δ
The question that arises from all the above results is whether these effects are indeed related to RPA functions. To answer this question, we analyzed if the rtt105Δ155-208 mutant whose association to Rfa1 is compromised (Li et al. 2018) mimics the rtt105Δ mutation. After sporulating rtt105Δ/RTT105 diploid strains containing a plasmid either expressing RTT105 or the rtt105Δ155-208 mutant, we observed that cells carrying the rtt105Δ155-208 had similar phenotypes as rtt105Δ cells (Table 2, Supplementary Figure S7). This result suggests that the observed phenotypes described in the absence of RTT105 are related to a lack of interaction between Rtt105 and Rfa1.
Table 2.
Genetic dependence on rtt105Δ155-208 viability. rtt105Δ/RTT105 strains heterozygous for the indicated deletions expressing rtt105Δ155-208 or RTT105 from a centromeric plasmid were sporulated, dissected, and the genotype of the variable spores were determined
| Mutant | Growth defect | Growth defect | Growth defect |
|---|---|---|---|
| with rtt105Δ + | with rtt105Δ + | with rtt105Δ + | |
| rtt105Δ155-208 | RTT105 | Empty vector | |
| orc5-1 | Synthetic lethal | − | Synthetic lethal |
| mcm2-1 | +++ | − | +++ |
| cdc17-1 | Synthetic lethal | − | Synthetic lethal |
| mrc1Δ | Synthetic lethal | − | Synthetic lethal |
| tof1Δ | Synthetic lethal | − | Synthetic lethal |
| ctf18Δ | Synthetic lethal | − | Synthetic lethal |
| rad9Δ | +++ | − | Synthetic lethal |
| rad53-K227A | +++ | − | Synthetic lethal |
| rad52Δ | Synthetic lethal | − | Synthetic lethal |
| mre11Δ | Synthetic lethal | − | Synthetic lethal |
| sae2Δ | +++ | − | +++ |
| lif1Δ | − | − | − |
| scc1-73* | Synthetic lethal | − | Synthetic lethal |
| asf1Δ | +++ | − | +++ |
| sir4Δ | − | − | − |
A “−” represents no effect on growth over the individual single mutants. A “+” represents a synthetic effect on growth. More “+” indicate a more dramatic synthetic effect in comparison with other strain tested. All the analyses were conducted at 30°C excepted for those marked by a “*”, which were conducted at 32°C.
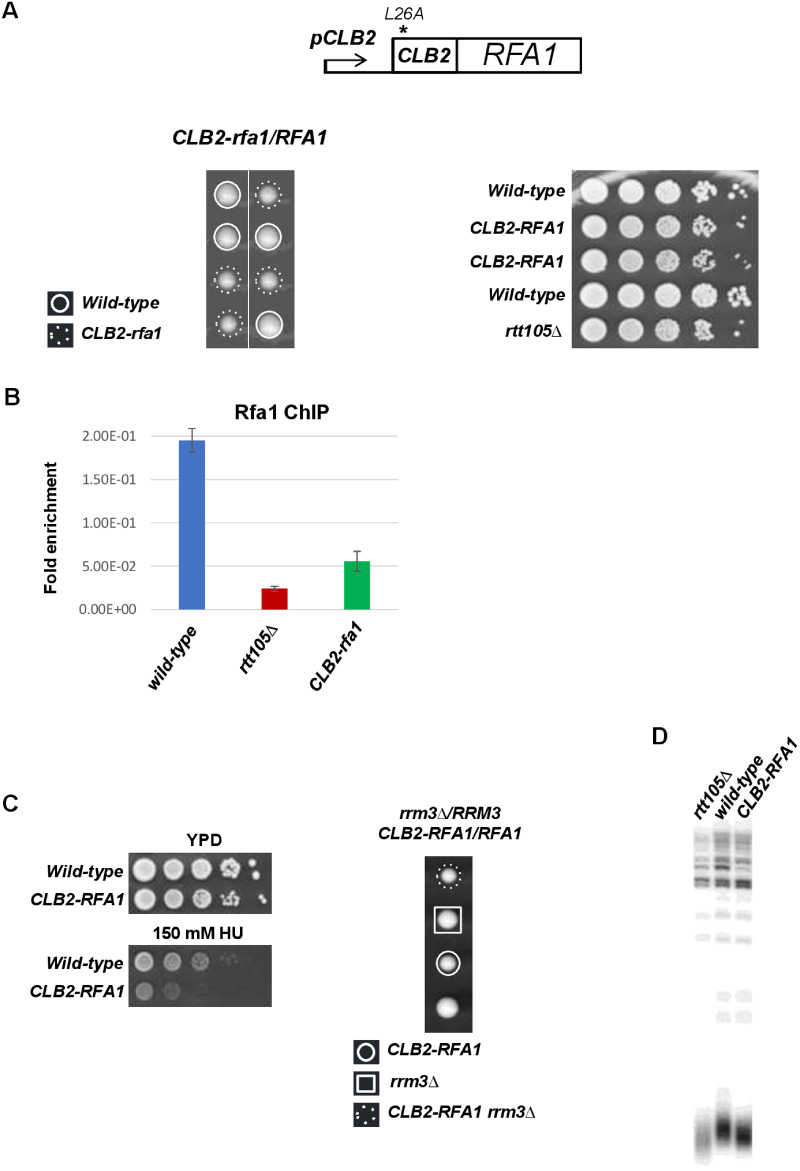
To confirm these observations, we examined if reducing the RPA levels specifically during S-phase gave rise to similar phenotype as those obtained in rtt105Δ cells. We took advantage of results indicating that restricting the expression of genes in G2/M phase can lead to potentially hypomorphic alleles in S phase (as a consequence of their reduced expression in S-phase). We used a diploid strain to swap the promoter of one allele of RFA1 to create CLB2-rfa1 allele in which RFA1 is under the control of the mitotic Clb2 promoter and fused to 5ʹ region of CLB2 encoding Clb2 degron (Hombauer et al. 2011). We found that CLB2-rfa1 cells were viable and exhibited modest growth defect (Figure 8A). These results indicate that the amounts of CLB2-rfa1 persists in S-phase as shown in Supplementary Figure S8, and that N-terminal CLB2 fusion per se was not sufficient to compromise vital S-phase function of RFA1. We performed chromatin immunoprecipitation (ChIP) to measure the level of RPA binding at the early origin ARS607 after replication fork stalling in CLB2-rfa1 cells. ChIP-qPCR revealed that the association of Rfa1 subunit with replicating DNA (analyzed after HU treatment) was significantly reduced in CLB2-rfa1 cells as shown for rtt105Δ (Li et al. 2018) but slightly higher than in rtt105Δ background (Figure 8B). We next examined the sensitivity of CLB2-rfa1 cells in cells experiencing replicative stress. We found that similarly to rtt105Δ, CLB2-rfa1 cells were sensitive to chronic exposure to HU but not to replicative damages arising in cells lacking RRM3 helicase (Figure 8C). We further conducted an extensive genetic analysis with CLB2-rfa1 (Table 3). We found that orc5-1, cdc17-1, mrc1Δ, tof1Δ, ctf18Δ, rad53-K227A, rad52Δ, mre11Δ, sae2Δ, scc1-73, asf1Δ, and rtt109Δ mutations all strongly affected the viability or growth of CLB2-rfa1 cells in contrast to lif1Δ, and sir4Δ that did not (Table 3). Having shown that the deletion of RTT105 reduced telomere length, we analyzed the average size of telomeres in CLB2-rfa1 cells and found that CLB2-rfa1 caused a shortening of telomere length (Figure 8D). Consistent with our ChIP-qPCR experiment showing more Rfa1 signal in CLB2-rfa1 than in rtt105Δ mutant, we observed a smaller telomeres size reduction in CLB2-rfa1 compared to rtt105Δ (Figure 8B). Altogether, these results show that CLB2-rfa1 and rtt105Δ cells exhibit similar phenotypes and suggest that the observed phenotypes described in the absence of RTT105 are due to lack of RPA binding at fork.
Figure 8.
Phenotypes of CLB2-rfa1 cells. (A) Top, schematic representation of the CLB2-rfa1 construct used in this study. Down, RTT105 is required for normal cell growth. Left, tetrad dissection of the diploid strain CLB2-rfa1/RFA1. Right, 10-fold serial dilutions of exponentially growing haploid cells were spotted. (B) Reduced binding of Rfa1 at fork in CLB2-rfa1 cells. Asynchronous cells were blocked in S-phase with 200 mM HU. ChIP experiments were performed in triplicate using an antibody against Rfa1 (Agrisera) and the resulting DNA was quantified with real time PCR using primers amplifying ARS607 (CGTGCGGCAGTATAAGTTCA and GCAGGATCGACCTGACTCTT). (C) CLB2-rfa1 mutant viability is affected by HU but not by RRM3 inactivation. Left, 10-fold serial dilutions of exponentially growing cells were spotted onto YPD plate or 150 mM HU plate. Right, tetrad dissection of the diploid strain CLB2-rfa1/RFA1 rrm3Δ/RRM3. Plates were incubated at 30°C for 3 days. (D) Effect of CLB2-rfa1 on telomere length measured by Southern blotting.
Table 3.
Genetic dependence on CLB2-rfa1 viability. CLB2-rfa1/RFA1 strains heterozygous for the indicated deletions were sporulated, dissected, and the genotype of the variable spores were determined
| Mutant | Growth defect |
|---|---|
| with CLB2-rfa1 | |
| orc5-1 | Synthetic lethal |
| cdc17-1 | Synthetic lethal |
| mrc1Δ | Synthetic lethal |
| tof1Δ | Synthetic lethal |
| ctf18Δ | Synthetic lethal |
| rad53-K227A | +++ |
| rad52Δ | Synthetic lethal |
| mre11Δ | Synthetic lethal |
| sae2Δ | +++ |
| lif1Δ | − |
| scc1-73* | Synthetic lethal |
| asf1Δ | Synthetic lethal |
| sir4Δ | − |
A “−" represents no effect on growth over the individual single mutants. A “+” represents a synthetic effect on growth. More “+” indicates a more dramatic synthetic effect in comparison with other strain tested. All the analyses were conducted at 30°C excepted for those marked by a “*,” which were conducted at 32°C.
Targeting Rfa1 into the nucleus only partially rescues rtt105Δ phenotypes
To distinguish whether the defects in DNA metabolism that we reported in the absence of RTT105 were the consequence of the RPA nuclear import defect or resulted from a loss of function of Rtt105 in genome stability maintenance, we expressed a fusion protein constituted by a nuclear localization signal (NLS, PKKKRKV) fused to the N-terminal part of the full-length Rfa1 sequence. We observed that Rfa1 levels in the nucleus and Rfa1-bound to chromatin were both increased in rtt105Δ cells expressing NLS-Rfa1 indicating that NLS fused to Rfa1 promoted its import (Supplementary Figure S9A). Moreover, NLS-Rfa1 rescued the growth of rfa1Δ cells and the lethality of rfa1-D228Y rtt105Δ mutant showing that the fusion protein is functional (Supplementary Figure S9B).
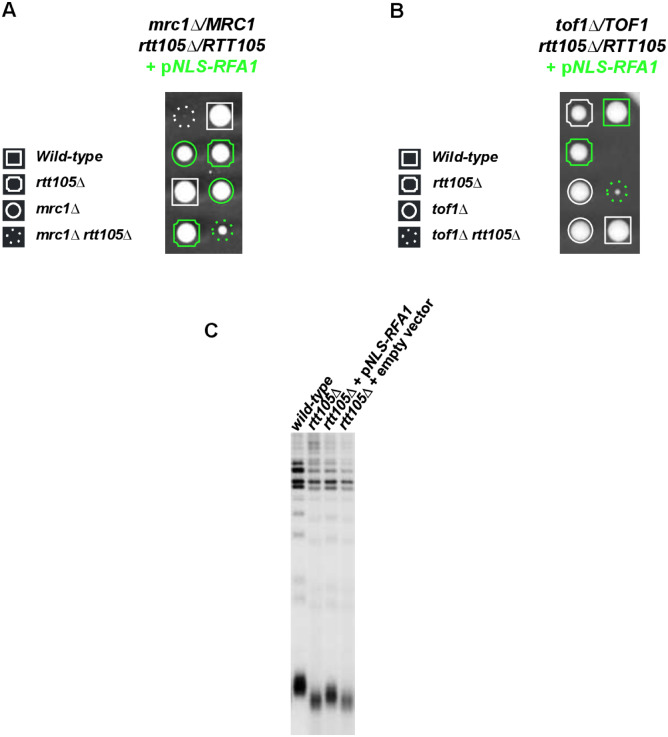
We further found that expressing NLS-Rfa1 failed to rescue the growth defect at 25°C and sensitivity to HU of rtt105Δ cells (Supplementary Figure S9C), suggesting that the impact on replication due to the absence of Rtt105 is not fully related to its role in importing RPA to the nucleus and to its role in mediating RPA binding to ssDNA. We further asked whether NLS-Rfa1 suppressed the genome stability defects displayed in rtt105Δ cells. We found that in the absence of RTT105, NLS-Rfa1 efficiently restored the viability of ctf18Δ, scc1-73 and rad52Δ mutants, respectively required for activation of the replication checkpoint, cohesion and repair (Supplementary Figure S10). Likewise, we explored the growth of rtt105Δ NLS-RFA1 cells devoid of MRC1 or TOF1, two genes important to prevent chromosome fragility through their multiple functions during replication (Tourrière et al. 2005; Pardo et al. 2017; Puddu et al. 2017; Yeeles et al. 2017; Gellon et al. 2019). mrc1Δ rtt105Δ and tof1Δ rtt105 expressing NLS-Rfa1 were able to form colonies after 3 days at 30°C (Figure 9, A and B) indicating that NLS-Rfa1 could restore viability in mrc1Δ rtt105Δ and tof1Δ rtt105Δ mutants. However, mrc1Δ rtt105Δ and tof1Δ rtt105Δ cells expressing NLS-Rfa1 exhibited severe growth defects (Figure 9, A and B) revealing that NLS-Rfa1 failed to rescue growth in the absence of RTT105 and suggesting that Rtt105 functions in replication independently of its known RPA chaperone function. This result was consistent with the fact that NLS-Rfa1 does not rescue the HU sensitivity in rtt105Δ cells (Supplementary Figure S9C, bottom). Finally, we also evaluated the telomere size of rtt105Δ cells expressing NLS-Rfa1 and found that NLS-Rfa1 exogenous expression suppressed only partially telomere length defect in rtt105Δ (Figure 9C). Altogether these results show that bringing Rfa1 to the nucleus and on chromatin only partly rescued some of the phenotypes of rtt105Δ cells pointing out that the pleiotropic phenotypes arising in rtt105Δ cells are not exclusively due to the function of Rtt105 in chaperoning RPA during DNA metabolism. Collectively these results strongly suggest that in addition to its role in RPA nuclear import and in the regulation of RPA binding to DNA replication forks Rtt105 exerts a role in the maintenance of genome stability during S phase either by directly affecting RPA function and/or through an unknown function independent of RPA.
Figure 9.
Bringing Rfa1 into the nucleus does not rescue the growth of rtt105Δ cells in absence of MRC1 and TOF1. (A) Genetic interaction of mrc1Δ with RTT105. The diploid strains mrc1Δ/MRC1 rtt105Δ/RTT105 expressing the fusion protein NLS-Rfa1 was sporulated and dissected. (B) Genetic interaction of tof1Δ with RTT105. The diploid strain tof1Δ/TOF1 rtt105Δ/RTT105, expressing the fusion protein NLS-Rfa1 was sporulated, and dissected. The green color indicates spores expressing the NLS-Rfa1 fusion protein. Plates were incubated at 30°C for 3 days. (C) Effect of NLS-Rfa1 expression in rtt105Δ cells on telomere length measured by Southern blotting.
Discussion
Our extensive genetic analysis reveals that RTT105 is important for replication and multiple vital co-replicational events by facilitating RPA nuclear localization and by supporting RPA function during replication.
We show that the weakening of coupling between MCM helicase and DNA polymerases is lethal in the absence of RTT105. In addition, we show neither Pif1 nor Rrm3, whose function is to assist fork progression across pausing sites, is essential for the growth of rtt105Δ cells despite the fact that these cells are sensitive to exogenous DNA-damaging agents. One explanation could be the number of damages arising in pif1Δ and rrm3Δ cells is less important than damages created by exogenous DNA damaging agents. The fact that RRM3 deletion affects cell viability in the absence of genes involved in replication, checkpoint, or repair (Torres et al. 2004; Schmidt and Kolodner, 2006) rather suggests that RTT105 function is required for cells undergoing specific replicative damages. Another possibility could be that replicative damages induced by either rtt105Δ or rrm3Δ are similar and therefore processed in the same way.
Our study reveals that both branches of S-Phase checkpoint, DRC, and DNA damage checkpoint are required for rtt105Δ viability. These interactions strengthen the notion that RTT105 carries out an important function during DNA replication. Along the same line, Mre11 and Sae2 that initiate resection at stalled forks are critical in cells lacking RTT105 (Mimitou and Symington 2008; Tittel-Elmer et al. 2009; Bentsen et al. 2013; Delamarre et al. 2020), confirming the importance of recombination in cells lacking RTT105 (Li et al. 2018). The fact that yku80Δ rtt105Δ cells generated after the sporulation of the diploid yku80Δ/YKU80 rtt105Δ/RTT105 exhibited growth defects (Li et al. 2018) could have suggested that NHEJ was required for the growth of rtt105Δ cells. Nevertheless, our discovery that lif1Δ rtt105Δ grew normally at 30°C indicates that NHEJ is not required for rtt105Δ fitness. Considering that NHEJ predominantly operates in G1 while Rtt105 acts in S-phase (Chiruvella et al. 2013), it is not so surprising that NHEJ is not required to sustain the growth of rtt105Δ cells. We also report in this work that deleting RTT105 aggravates cohesion defect of scc1-73 cells consistent with the fact that a number of replication proteins play important roles in sister chromatid cohesion. However, because cohesion establishment factors localize to replication forks to promote fork restart (Lengronne et al. 2006; Gambus et al. 2009; Terret et al. 2009; Frattini et al. 2017) it is likely that repair of broken replication forks which arise in the absence of RTT105 requires an intact cohesion (Klein et al. 1999; Sjögren and Nasmyth 2001).
Taken together, our genetic analyses suggest that the absence of RTT105 affects replication and leads to the emergence of DSBs, which are subsequently repaired by HR.
rtt105Δ mutation exhibits synthetic defects with mutations involved in the regulation of replication-coupled nucleosome assembly. On the contrary, deletion of the HIR complex, which is involved in replication-independent nucleosome assembly, did not induce a growth defect in rtt105Δ cells. These data highlight the importance of RTT105 in a chromatin assembly process linked to replication, and ruled out a role for RTT105 in replication-independent chromatin assembly. We further show that RTT105 is required for efficient transcriptional silencing at the three heterochromatic regions that are transcriptionally silenced by the SIR proteins. Cells exhibiting defects in replication, in nucleosome assembly, and in sister chromatid cohesion have defective transcriptional silencing (Zhang et al. 2000; Sharp et al. 2001; Suter et al. 2004; Huang et al. 2007; Burgess et al. 2012). Because these processes are affected in rtt105Δ cells, it is possible that RTT105 deletion affects silencing through its impact on these mechanisms. Curiously, we observed that deleting SIR2 or SIR4 improved the growth of rtt105Δ cells, which pinpoint Sir proteins contribution to rtt105Δ mutant fitness reduction. Deleting Sir2 and to a lesser extend Sir4 were reported to suppress cdc6-4 ts lethality (Pappas et al. 2004), and to target sensitive origins on chromosome III and VI (Crampton et al. 2008). Recently, it was shown that Sir2 inactivation rescued MCM loading at most euchromatic regions in the context of a cdc6-4 mutant (Hoggard et al. 2018). These results therefore reinforce the notion that Rtt105 targets DNA replication.
We next showed that deleting RTT105 reduces telomere length. Telomerase has two main recruitment pathways assisting and providing enzyme access to telomere. An essential pathway requires interaction of Est1 with Cdc13 (Evans and Lundblad 1999). The other pathway requires yKu80 and its binding to Sir4 (Peterson et al. 2001; Stellwagen 2003; Fisher et al. 2004; Hass and Zappulla 2015; Chen et al. 2018). Our genetic analysis between RTT105 and SIR4 suggests that RTT105 promotes telomere lengthening independently of TLC1-Ku-Sir4 pathway. We further found that the telomere overelongation phenotype conferred by the artificial tethering of Est1 and Est2 via Est1-DBDCdc13 or Est2-DBDCdc13 fusion proteins was partially suppressed by rtt105Δ. Since deletion of RTT105 does not impair Cdc13 binding at telomeres, this result suggests that the telomere shortening observed in the absence of RTT105 is due to a decrease in telomerase activity independent of its recruitment. We also observed that in est1Δ cells absence of RTT105 accelerates senescence without affecting the kinetics of telomere shortening suggesting that RTT105 has a particular role in telomeres replication that are known to be prone to replication stress (Maestroni et al. 2017). We found that similarly to est1Δ cells, est1Δ rtt105Δ cells produced type II survivors but with a delayed kinetics suggesting that the absence of RTT105 affects the appearance of survivors.
RPA protects and stabilizes ssDNA generated during DNA metabolism. Our genetic epistasis analysis between rtt105Δ mutant and rfa1-t11 mutant revealed strong negative genetic interactions (respect to growth). Because rfa1-t11 mutant is replication proficient but defective in recombination repair (Lee et al. 1998; Umezu et al. 1998; Kantake et al. 2003) we assume that the sickness of the rfa1-t11 rtt105Δ double mutant is due to the inability of rfa1-t11 mutant to repair replicative damages provoked by the absence of Rtt105. Consistent with this hypothesis, we have shown that rtt105Δ displays also synthetic lethality phenotype with mre11Δ, which is itself functionally epistatic with rfa1-t11 (for survival of replication fork stress or DSB recovery) (Seeber et al. 2016). Genetic epistasis analysis between rtt105Δ and rfa1-D228Y also pinpoints strong negative genetic interaction (respect to growth). Since RPA level bound to ssDNA and replication forks is reduced in rfa1-D228Y cells (Audry et al. 2015; Ruff et al. 2016), the synthetic lethality/sickness between rfa1-D228Y and rtt105Δ could be at least in part a consequence of the lower affinity of RPA for ssDNA. We also report the absence of a synergistic reduction in telomere length when the rfa1-D228Y mutant allele is combined with null mutation of RTT105. This result suggests that the negative effect on telomere length associated to RTT105 deletion is related to RPA function at telomeres (Schramke et al. 2004; Luciano et al. 2012).
Genetic analyses that we have conducted with rtt105Δ155-208 (Li et al. 2018) and CLB2-rfa1 mutants in which both exhibit reduced level of RPA on ssDNA reveal that these two mutants phenocopy the rtt105Δ mutant. Because both alleles show genetic interactions similar to those of rtt105Δ, we assume that the pleiotropic effects observed in rtt105Δ cells are mostly related to the role of Rtt105 in chaperoning RPA a notion reinforced by recent observation that RTT105 and RPA both play a role in removing G4 structures (Maestroni et al. 2020). The fact that rtt105Δ155-208 phenocopies rtt105Δ but cannot rescue nuclear localization defect of Rfa1 (Li et al. 2018), could suggest that the role of Rtt105 in RPA nuclear import is the cause of the observed phenotypes. However, since Rtt105–Rfa1 interaction occurs predominantly in the nucleus and is also required to promote the binding of RPA to ssDNA (Li et al. 2018) one cannot distinguish whether the role of Rtt105 is related to defect in the nuclear localization of RPA or to a more direct effect on genome stability.
We observed that NLS-Rfa1 rescues rtt105Δ combined to either scc1-73, or rad52Δ suggesting that Rtt105 contributes to cohesion, and repair by chaperoning RPA. We propose that the lethality of rad52Δ rtt105Δ cells is a consequence of replicative damages leading to the emergence of DSBs, which cannot be repaired by HR in the absence of Rad52. Therefore, we consider that Rtt105 is not directly involved in Rad52-dependent HR repair pathways. NLS-Rfa1 also rescues the viability of rfa1-D228Y rtt105Δ double mutant, however the double mutant exhibits a slow growth. Given that rfa1-D228Y mutant is not functional for break-induced replication, which involves long ssDNA intermediates, but is largely functional for both intra-homologue gene conversion and single strand annealing (Ruff et al. 2016), we favor the idea that rfa1-D228Y mutant goes to the nucleus and that Rtt105 helps rfa1-D228Y cells to perform its essential function. Finally, we show that NLS-Rfa1 does not rescue the sensitivity to HU of rtt105Δ cells and only poorly rescues the growth of mutants affected in factors protecting stalled forks (mrc1Δ, tof1Δ, ctf18Δ). The fact that Ctf18 and Mrc1 act in separate pathways to maintain stability of repeat sequences (Gellon et al. 2011; Stokes et al. 2020) could explain the difference in RPA requirement observed between ctf18Δ rtt105Δ and mrc1Δ rtt105Δ cells. On their side, Mrc1 and Tof1 are both crucial for preventing fork breakage in the presence of secondary structures and are equally important for preventing instability at long repeat sequences (Gellon et al. 2019). It is tempting to speculate that as Mrc1 and Tof1, Rtt105 could have a role in fork stabilization when long ssDNA intermediates are generated and that this role is not related to RPA. This could explain the extreme instability that we have recently observed in presence of G4 structures in rtt105Δ cells (Maestroni et al. 2020).
In summary, our studies reveal that Rtt105 guards genome stability through multiple mechanisms. We have shown that in the absence of RTT105 yeast cells require the two branches of the S phase checkpoint and HR to survive, indicating the presence of replicative defects. Furthermore, our detailed genetic analyses demonstrate that RTT105 is important for several vital mechanisms intimately connected to replication fork progression, as sister chromatid cohesion and replication-dependent nucleosome assembly. We also point out the importance of RTT105 in heterochromatin silencing and in telomere-length maintenance. This work reveals novel roles for RTT105 during DNA metabolism and show that the pleiotropic effects of loss of RTT105 are not only related to Rtt105's role in chaperoning Rfa1. Rtt105 may exert a crucial role in the maintenance of genome stability during replication by directly affecting RPA function and/or through an unknown function, independent of RPA.
Acknowledgments
Thanks to Marie-Noëlle Simon for critically reading the manuscript. We thank Victoria Lundblad for providing plasmids pVL1107 and pVL1120 and Richard Kolodner for providing plasmid pRDK1598. We acknowledge Zhiguo Zhang for ZGY1077 and ZGY1069 strains, David Shore for YLS409 and YLS410 strains, Fred Winston for L1139 and L1140 strains, Alain Verreault for HMY140 strain, Sue Biggins for SBY885 strain, and Carl Mann for anti-Clb2. Thanks to Pierre-Marie Dehé for helpful discussions and much more, to Nagham Ghaddar, and to Michel-Hervé Moimême for permanent support. V.G. laboratory is supported by the Ligue Nationale Contre le Cancer (Equipe labellisée).
Conflicts of interest
The authors declare that they have no conflict of interest.
Literature cited
- Aguilera P, Whalen J, Minguet C, Churikov D, Freudenreich C, et al. 2020. The nuclear pore complex prevents sister chromatid recombination during replicative senescence. Nat Commun. 11:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai X, Parthun MR. 2004. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol Cell. 14:195–205. [DOI] [PubMed] [Google Scholar]
- Audry J, Maestroni L, Delagoutte E, Gauthier T, Nakamura TM, et al. 2015. RPA prevents G-rich structure formation at lagging-strand telomeres to allow maintenance of chromosome ends. EMBO J. 34:1942–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA. 2006. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 20:3104–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando M, Katou Y, Komata M, Tanaka H, Itoh T, et al. 2009. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem. 284:34355–34365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsen IB, Nielsen I, Lisby M, Nielsen HB, Gupta SS, et al. 2013. MRX protects fork integrity at protein-DNA barriers, and its absence causes checkpoint activation dependent on chromatin context. Nucleic Acids Res. 41:3173–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Shore D. 2007. Early replication of short telomeres in budding yeast. Cell. 128:1051–1062. [DOI] [PubMed] [Google Scholar]
- Burgess RJ, Zhou H, Han J, Li Q, Zhang Z. 2012. The SCFDia2 ubiquitin E3 ligase ubiquitylates Sir4 and functions in transcriptional silencing. (A. L. Kirchmaier, Ed). PLoS Genet. 8:e1002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19:1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, Winston F. 2011. Spt10 and Spt21 are required for transcriptional silencing in Saccharomyces cerevisiae. Eukaryot Cell. 10:118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Tyler J. 2008. Chromatin reassembly signals the end of DNA repair. Cell Cycle. 7:3792–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xue J, Churikov D, Hass EP, Shi S, et al. 2018. Structural insights into yeast telomerase recruitment to telomeres. Cell. 172:331–343.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE. 2013. Repair of double-strand breaks by end joining. Perspect Biol. 5:a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Ruiz M, González-Prieto R, Prado F. 2011. Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. (J. E. Haber, Ed). PLoS Genet. 7:e1002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, et al. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 446:806–810. [DOI] [PubMed] [Google Scholar]
- Crampton A, Chang F, Pappas DL, Frisch RL, Weinreich M. 2008. An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol Cell. 30:156–166. [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Kenny AE, Moore S, Garfinkel DJ, Weintraub M, et al. 2007. S-phase checkpoint pathways stimulate the mobility of the retrovirus-like transposon Ty1. Mol Cell Biol. 27:8874–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Lutz S, Lesage P. 2015. The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microb Spectr. 3:1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakshinamurthy A, Nyswaner KM, Farabaugh PJ, Garfinkel DJ. 2010. BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae. Genetics. 185:1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delamarre A, Barthe A, de la Roche Saint André C, Luciano P, Forey R, et al. 2020. MRX increases chromatin accessibility at stalled replication forks to promote nascent DNA resection and cohesin loading. Mol Cell. 77:395–410. [DOI] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 315:649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff P, Kose HB, Martino F, Petojevic T, Abid Ali F, et al. 2019. Molecular basis for ATP-hydrolysis-driven DNA translocation by the CMG helicase of the eukaryotic replisome. Cell Rep. 28:2673–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. 1999. Est1 and Cdc13 as comediators of telomerase access. Science. 286:117–120. [DOI] [PubMed] [Google Scholar]
- Fellerhoff B, Eckardt-Schupp F, Friedl AA. 2000. Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics. 154:1039–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Taggart AKP, Zakian VA. 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol. 11:1198–1205. [DOI] [PubMed] [Google Scholar]
- Frattini C, Villa-Hernandez S, Pellicano G, Jossen R, Katou Y, et al. 2017. Cohesin ubiquitylation and mobilization facilitate stalled replication fork dynamics. Mol Cell. 68:758–772. [DOI] [PubMed] [Google Scholar]
- Gadaleta M-C, Noguchi E. 2017. Regulation of DNA replication through natural impediments in the eukaryotic genome. Genes. 8:98. [Google Scholar]
- Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P. 2008. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 27:748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, et al. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 8:358–366. [DOI] [PubMed] [Google Scholar]
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, et al. 2009. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 28:2992–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR, Smith JS. 2016. The nuts and bolts of transcriptionally silent chromatin in Saccharomyces cerevisiae. Genetics. 203:1563–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon L, Kaushal S, Cebrián J, Lahiri M, Mirkin SM, et al. 2019. Mrc1 and Tof1 prevent fragility and instability at long CAG repeats by their fork stabilizing function. Nucleic Acids Res. 47:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon L, Razidlo DF, Gleeson O, Verra L, Schulz D, et al. 2011. New functions of Ctf18-RFC in preserving genome stability outside its role in sister chromatid cohesion. PLoS Genet. 7:e1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnügge R, Symington LS. 2017. Keeping it real: MRX-Sae2 clipping of natural substrates. Genes Dev. 31:2311–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Larrivée M, Labrecque P, Wellinger RJ. 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 280:741–744. [DOI] [PubMed] [Google Scholar]
- Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, et al. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 15:2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhang H, Zhang H, Wang Z, Zhou H, et al. 2013. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell. 155:817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu R-M, et al. 2007a. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 315:653–655. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu R-M, Zhang Z. 2007b. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 282:28587–28596. [DOI] [PubMed] [Google Scholar]
- Hass EP, Zappulla DC. 2015. The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. Elife. 4:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard TA, Chang F, Perry KR, Subramanian S, Kenworthy J, et al. 2018. Yeast heterochromatin regulators Sir2 and Sir3 act directly at euchromatic DNA replication origins. (G. P. Copenhaver, Ed.). PLoS Genet. 14:e1007418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H, Srivatsan A, Putnam S, Kolodner RD. 2011. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 334:1713–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhou H, Tarara J, Zhang Z. 2007. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 26:2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, et al. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 12:1525–1536. [DOI] [PubMed] [Google Scholar]
- Kanellis P, Agyei R, Durocher D. 2003. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol. 13:1583–1595. [DOI] [PubMed] [Google Scholar]
- Kantake N, Sugiyama T, Kolodner RD, Kowalczykowski SC. 2003. The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein. J Biol Chem. 278:23410–23417. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, et al. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 98:91–103. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, et al. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 94:399–409. [DOI] [PubMed] [Google Scholar]
- Lemon LD, Morris DK, Bertuch AA. 2019. Loss of Ku's DNA end binding activity affects telomere length via destabilizing telomere-bound Est1 rather than altering TLC1 homeostasis. Sci Rep. 9:10607–10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, et al. 2006. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 23:787–799. [DOI] [PubMed] [Google Scholar]
- Leung GP, Lee L, Schmidt TI, Shirahige K, Kobor MS. 2011. Rtt107 is required for recruitment of the SMC5/6 complex to DNA double strand breaks. J Biol Chem. 286:26250–26257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Fazly AM, Zhou H, Huang S, Zhang Z, et al. 2009. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. (C. E. Pearson, Ed). PLoS Genet. 5:e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, et al. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 134:244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Dong Z, Yang S, Feng J, Li Q. 2019. Chaperoning RPA during DNA metabolism. Curr Genet. 65:857–864. [DOI] [PubMed] [Google Scholar]
- Li S, Xu Z, Xu J, Zuo L, Yu C, et al. 2018. Rtt105 functions as a chaperone for replication protein A to preserve genome stability. EMBO J. 37:e99154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 118:699–713. [DOI] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Manfrini N, Clerici M. 2010. Mechanisms and regulation of DNA end resection. EMBO J. 29:2864–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Paciotti V, Neecke H, Lucchini G. 2000. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics. 155:1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano P, Coulon S, Faure V, Corda Y, Bos J, et al. 2012. RPA facilitates telomerase activity at chromosome ends in budding and fission yeasts. EMBO J. 31:2034–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano P, Dehé P-M, Audebert S, Géli V, Corda Y. 2015. Replisome function during replicative stress is modulated by histone H3 lysine 56 acetylation through Ctf4. Genetics. 199:1047–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. 1993. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell. 73:347–360. [DOI] [PubMed] [Google Scholar]
- Maestroni L, Audry J, Luciano P, Coulon S, Géli V, et al. 2020. RPA and Pif1 cooperate to remove G-rich structures at both leading and lagging strand. Cell Stress. 4:48–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestroni L, Audry J, Matmati S, Arcangioli B, Géli V, et al. 2017. Eroded telomeres are rearranged in quiescent fission yeast cells through duplications of subtelomeric sequences. Nat Commun. 8:1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 91:35–45. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 455:770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellner J, Schmidt K. 2020. Yeast genome maintenance by the multifunctional PIF1 DNA helicase family. Genes. 11:224.doi:10.3390/genes11020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor ML, Li J-M, Osborn AJ, Elledge SJ. 2009. Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. Proc Natl Acad Sci USA. 106:12765–12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, et al. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 8:657–660. [DOI] [PubMed] [Google Scholar]
- Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. 2008. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics. 178:197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas DL, Frisch R, Weinreich M. 2004. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 18:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Crabbé L, Pasero P. 2017. Signaling pathways of replication stress in yeast. FEMS Yeast Res. 17:fow101. [DOI] [PubMed] [Google Scholar]
- Parnas O, Zipin-Roitman A, Mazor Y, Liefshitz B, Ben-Aroya S, et al. 2009. The ELG1 clamp loader plays a role in sister chromatid cohesion. (C. Fairhead, Ed). PLoS One. 4:e5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, et al. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 27:64–67. [DOI] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ. 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 8:831–834. [DOI] [PubMed] [Google Scholar]
- Puddu F, SalgueroI, HerzogM, GeislerN J, Costanzo V, . et al. 2017. Chromatin determinants impart camptothecin sensitivity. EMBO Rep. 18:1000–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff P, Donnianni RA, Glancy E, Oh J, Symington LS. 2016. RPA stabilization of single-stranded DNA is critical for break-induced replication. Cell Rep. 17:3359–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KH, Kolodner RD. 2006. Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc Natl Acad Sci USA. 103:18196–18201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DT, Banerjee M, Bowen B, Curcio MJ. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 159:1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, et al. 2004. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat Genet. 36:46–54. [DOI] [PubMed] [Google Scholar]
- Schubert HL, Wittmeyer J, Kasten MM, Hinata K, Rawling DC, et al. 2013. Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler. Proc Natl Acad Sci USA. 110:3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A, Hegnauer AM, Hustedt N, Deshpande I, Poli J, et al. 2016. RPA mediates recruitment of MRX to forks and double-strand breaks to hold sister chromatids together. Mol Cell. 64:951–966. [DOI] [PubMed] [Google Scholar]
- Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 11:463–473. [DOI] [PubMed] [Google Scholar]
- Simon M-N, Churikov D, Géli V. 2016. Replication stress as a source of telomere recombination during replicative senescence in Saccharomyces cerevisiae. FEMS Yeast Res. 16:fow085. [DOI] [PubMed] [Google Scholar]
- Simon M-N, Churikov D, Géli V. 2021. Analysis of recombination at yeast telomeres. Methods Mol Biol. 2153:395–402. [DOI] [PubMed] [Google Scholar]
- Sjögren C, Nasmyth K. 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 11:991–995. [DOI] [PubMed] [Google Scholar]
- Smith J, Rothstein R. 1999. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics. 151:447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Zou H, Rothstein R. 2000. Characterization of genetic interactions with RFA1: the role of RPA in DNA replication and telomere maintenance. Biochimie. 82:71–78. [DOI] [PubMed] [Google Scholar]
- Smolikov S, Mazor Y, Krauskopf A. 2004. ELG1, a regulator of genome stability, has a role in telomere length regulation and in silencing. Proc Natl Acad Sci USA. 101:1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 297:599–602. [DOI] [PubMed] [Google Scholar]
- Srividya I, Tirupataiah S, Mishra K. 2012. Yeast transcription termination factor Rtt103 functions in DNA damage response. (A. J. Lustig, Ed). PLoS One. 7:e31288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen AE. 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17:2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes K, Winczura A, Song B, Piccoli GD, Grabarczyk DB. 2020. Ctf18-RFC and DNA pol e form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication. Nucleic Acids Res. 48:8128–8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Vannier D, Shore D. 1993. Epigenetic switching of transcriptional states: cis- and trans-acting factors affecting establishment of silencing at the HMR locus in Saccharomyces cerevisiae. Mol Cell Biol. 13:3919–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Tong A, Chang M, Yu L, Brown GW, et al. 2004. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics. 167:579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo SH, Jackson SP. 2001. Telomerase subunit overexpression suppresses telomere-specific checkpoint activation in the yeast yku80 mutant. EMBO Rep. 2:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terret M-E, Sherwood R, Rahman S, Qin J, Jallepalli PV. 2009. Cohesin acetylation speeds the replication fork. Nature. 462:231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M, Alabert C, Pasero P, Cobb JA. 2009. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J. 28:1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JZ, Schnakenberg SL, Zakian VA. 2004. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol Cell Biol. 24:3198–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Versini G, Cordón-Preciado V, Alabert C, Pasero P. 2005. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 19:699–706. [DOI] [PubMed] [Google Scholar]
- Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, et al. 2007. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 25:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Sugawara N, Chen C, Haber JE, Kolodner RD. 1998. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 148:989–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. 2010. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. (J. E. Haber, Ed). PLoS Genet. 6:e1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF, Boeke JD. 1992. Yeast retrotransposon revealed. Nature. 358:717–717. [DOI] [PubMed] [Google Scholar]
- Xu H, Boone C, Brown GW. 2007. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics. 176:1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JTP, Janska A, Early A, Diffley JFX. 2017. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell. 65:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shi D, Li X, Ding L, Tang J, et al. 2017a. Rtt101-Mms1-Mms22 coordinates replication-coupled sister chromatid cohesion and nucleosome assembly. EMBO Rep. 18:1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yeung CHL, Wu L, Yuen KWY. 2017b. E3 ubiquitin ligase Bre1 couples sister chromatid cohesion establishment to DNA replication in Saccharomyces cerevisiae. Elife. 6:1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shibahara K, Stillman B. 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 408:221–225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and method required to confirm the conclusions of this work are within the manuscript: Supplementary Table S1: Strains used in this study. Supplementary Figure S1: RTT105 is required for normal cell growth. Figure S2: RTT105 exhibits genetic interactions with S-phase checkpoint components. Supplementary Figure S3: RTT105 is important for cells affected in the replication-dependent nucleosome assembly process. Supplementary Figure S4: RTT105 is important for cells affected in the replication-dependent nucleosome assembly process but not in replication-independent nucleosome assembly. Supplementary Figure S5: RTT105 inactivation exacerbates the telomeric defects arising in the absence of YKU and EST1 genes. Supplementary Figure S6: RTT105 inactivation affects senescence and survivor formation in est1Δ cells. Supplementary Figure S7: Exogenous expression of rtt105Δ155-208 mutant failed to rescue the growth in rtt105Δ cells. Supplementary Figure S8: Cell cycle regulation of CLB2-rfa1. Supplementary Figure S9: Bringing Rfa1 into the nucleus rescues the viability of rfa1Δ and rfa1-D228Y rtt105Δ cells but not the growth defect and HU sensitivity in rtt105Δ cells. Supplementary Figure S10: Bringing Rfa1 into the nucleus rescues the growth of rtt105Δ cells affected in S-phase checkpoint, cohesion, or repair.
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13373021.