Abstract
The eyespot patterns found on the wings of nymphalid butterflies are novel traits that originated first in hindwings and subsequently in forewings, suggesting that eyespot development might be dependent on Hox genes. Hindwings differ from forewings in the expression of Ultrabithorax (Ubx), but the function of this Hox gene in eyespot development as well as that of another Hox gene Antennapedia (Antp), expressed specifically in eyespots centers on both wings, are still unclear. We used CRISPR-Cas9 to target both genes in Bicyclus anynana butterflies. We show that Antp is essential for eyespot development on the forewings and for the differentiation of white centers and larger eyespots on hindwings, whereas Ubx is essential not only for the development of at least some hindwing eyespots but also for repressing the size of other eyespots. Additionally, Antp is essential for the development of silver scales in male wings. In summary, Antp and Ubx, in addition to their conserved roles in modifying serially homologous segments along the anterior–posterior axis of insects, have acquired a novel role in promoting the development of a new set of serial homologs, the eyespot patterns, in both forewings (Antp) and hindwings (Antp and Ubx) of B. anynana butterflies. We propose that the peculiar pattern of eyespot origins on hindwings first, followed by forewings, could be due to an initial co-option of Ubx into eyespot development followed by a later, partially redundant, co-option of Antp into the same network.
Keywords: Butterfly, Evodevo, Hox gene, Novel trait, CRISPR
Introduction
Hox genes are primarily known for their embryonic expression domains of broad stripes along the anterior–posterior axis of bilaterian animals and for giving body regions along this axis a unique identity (Lewis 1978; McIntyre et al. 2007; Mallo et al. 2010). In arthropod animals, these unique identities are often visualized by changes in the external appearance of serially homologous traits along the body, such as appendages. Hox genes have not been implicated in the origin of appendages, such as arthropod legs or insect wings, but rather in their modification or repression by tweaking the appendage’s gene regulatory networks (GRNs) (Akam 1995; Angelini and Kaufman 2005; Tomoyasu 2017). This is because the silencing of Hox genes changes an appendage’s identity, or leads to the development of additional appendages, rather than cause the loss of the appendage itself (Struhl 1982; Carroll et al. 1995; Tomoyasu et al. 2005; Ohde et al. 2013; Martin et al. 2016).
Aside from their well-known role as modifiers of arthropod appendages, Hox genes have also been implicated in the development of lineage-specific evolutionary novelties. Here, Hox genes have acquired novel expression domains, often post-embryonically, and appear to contribute to the development of novel traits. For example, a novel Sex combs reduced (Scr) expression domain in pupal legs is required to produce sex combs used for mating (Tanaka et al. 2011), or Scr expression in embryonic heads specifies salivary grands in Drosophila (Panzer et al. 1992). A novel expression domain of Ultrabithorax (Ubx) in the tibia of third thoracic larval legs leads to the development of the pollen basket in worker bees (Medved et al. 2014). The late pupal expression of Abd-B in Drosophila and Bombus abdominal epidermis leads, in both cases, to the origin of novel pigmentation patterns in the abdomen of these insects (Jeong et al. 2006; Tian et al. 2019). Abd-B also specifies posterior spiracle development in Drosophila during pupal development (Glassford et al. 2015). In these examples, and quite distinctly from their function as modifiers of serial homologs, when a Hox gene is disrupted, the novel trait is severely disrupted or lost.
Eyespots on the wings of nymphalid butterflies are an interesting example of both a novel trait and a serially repeated trait, but the molecular changes that led to the origin of eyespots are still unknown. Ancestral state reconstructions on a large phylogeny of ∼400 genera suggested that eyespots first originated in four to five wing sectors on the ventral side of the hindwings of an ancestral lineage of nymphalid butterflies before appearing on forewings or on dorsal sides of both wings (Oliver et al. 2014; Schachat et al. 2015). The origin of eyespots restricted to hindwings is intriguing and could suggest that hindwing-specific Hox genes might have been required for eyespot origins.
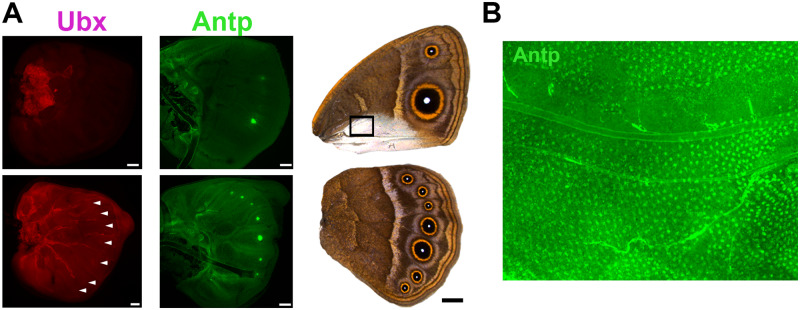
Butterflies have up to two known candidate Hox genes, Ubx and Antennapedia (Antp), expressed on their larval hindwings (Figure 1A, Weatherbee et al. 1999; Saenko et al. 2011). Ubx is expressed homogeneously across the whole hindwing, as observed in most other insects (Figure 1A, Prasad et al. 2016), whereas Antp has a novel and more specific expression pattern in the eyespot centers in both the fore- and hindwing in only a subset of butterflies with eyespots (Figure 1A, Saenko et al. 2011; Oliver et al. 2012; Shirai et al. 2012; Tong et al. 2014). Interestingly, in Bicyclus anynana, Ubx is additionally expressed at slightly elevated levels in the future eyespot centers in larval and pupal hindwings (Figure 1A, Tong et al. 2014), whereas Junonia coenia does not express Antp or elevate Ubx expression in their eyespots (Weatherbee et al. 1999; Oliver et al. 2012; Shirai et al. 2012; Tong et al. 2014). These observations suggest that Ubx homogeneous hindwing expression alone, but not Antp nor Ubx eyespot-specific expression, might have played some role in eyespot origins. However, Ubx cannot be responsible for the origin of eyespots on forewings because it is not expressed there in any of the species examined so far, unlike Antp.
Figure 1.
Expression pattern of Ubx and Antp in wings. (A) Expression pattern of Ubx and Antp on the larval wings and corresponding adult male wings. Ubx is expressed homogeneously across the hindwings and more intensely in the eyespot centers. Antp is expressed in all eyespot centers. (B) Expression of Antp in a section of a Wt male pupal forewing corresponding to the rectangle in (A). Antp is expressed intensely in silver scale building cells (arrowheads) during the pupal stage (image courtesy of Xiaoling Tong).
To date, neither Ubx nor Antp has been directly targeted for loss-of-function experiments in any butterfly species, and the role of these genes in eyespot development remains unclear. Here, we investigate the functions of Antp and Ubx in eyespot development in B. anynana with CRISPR-Cas9 to create mosaic mutants (crispants). We show that Antp is required for eyespots to form on the forewings, whereas Ubx has both a repressive and activating role on eyespot formation on the hindwings, depending on the wing sector. By integrating these results with previous work detailing the origin of eyespots across wings and wing surfaces of nymphalid butterflies, we propose that Hox genes were probably implicated in the origin of these novel traits in hindwings (first) and forewings (second) during butterfly diversification.
Materials and methods
Butterfly husbandry
Bicyclus anynana, originally collected in Malawi, have been reared in the lab since 1988. The caterpillars were fed on young corn plans and adults on mashed banana. Bicyclus anynana were reared at 27°C and 60% humidity in a 12:12 light:dark cycle.
sgRNA design
sgRNA target sequences were selected based on their GC content (around 60%) and the number of mismatch sequences relative to other sequences in the genome (>3 sites). In addition, we selected target sequences that started with a guanidine for subsequent in vitro transcription by T7 RNA polymerase.
sgRNA production
The template for in vitro transcription of sgRNA was made with a PCR method described in Matsuoka and Monteiro (2018) and Banerjee and Monteiro (2018; containing a video protocol). The forward primer contains a T7 RNA polymerase binding site and a sgRNA target site (GAAATTAATACGACTCACTATAGNN19GTTTTAGAGCTAGAAATAGC). The reverse primer contains the remainder of sgRNA sequence (AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC). PCR was performed with Q5 High-Fidelity DNA Polymerase (NEB) in 100 µl reaction volumes. After checking with gel electrophoresis, the PCR product was purified with the Gene JET PCR Purification Kit (Thermo Fisher). In vitro transcription was performed with T7 RNA polymerase (NEB), using 500 ng of purified PCR product as a template for overnight. After DNase I treatment to remove the template DNA, the RNA was precipitated with ethanol. The RNA was then suspended in RNase-free water and stored at −80°C.
Cas9 mRNA production
The plasmid pT3TS-nCas9n (Addgene) was linearized with XbaI (NEB) and purified by phenol/chloroform purification and ethanol precipitation. In vitro transcription of mRNA was performed using the mMESSAGEmMACHINE T3 Kit (Ambion). One microgram of linearized plasmid was used as a template, and a poly(A) tail was added to the synthesized mRNA by using the Poly(A) Tailing Kit (Thermo Fisher). The A-tailed RNA was purified by lithium chloride precipitation and then dissolved to RNase-free water and stored at −80°C.
Microinjection
Eggs were laid on corn leaves for 30 min. Within 2–3 h after egg laying, sgRNA and Cas9 mRNA were co-injected into embryos. At that stage, the embryo is a syncytium and cells membranes will only appear around 4–5 h after egg laying (Holzem et al. 2019). The concentrations of sgRNA and Cas9 are listed in Table 1. Food dye was added to the injection solution for better visualization. The injections were performed while the eggs were submerged in PBS. The injected eggs were incubated at 27°C in PBS, transferred onto moist cotton on the next day, and further incubated at 27°C. The hatched caterpillars were moved to corn leaves and reared at 27°C with a 12:12 h light:dark cycle and 60% relative humidity.
Table 1.
Summary of injections performed. Injections with a yellow guide serve as controls for hatching rate and frequency of crispant phenotypes
| Target | sgRNA final concentration (µg/µl) |
Cas9 mRNA final concentration (µg/µl) |
No. of injected embryos | No. of hatched larvae | No. of adults | No. of adults showing phenotype |
|---|---|---|---|---|---|---|
| Antp #1 | 0.5 | 1 | 176 | 61 | 18 | 1 |
| Antp #1 | 0.1 | 0.2 | 111 | 47 | 28 | 1 |
| Antp #1 | 0.1 | 0.2 | 146 | 59 | 10 | 1 |
| Antp #2 | 0.1 | 0.2 | 161 | 38 | 7 | 3 |
| Antp #1 | 0.1 | 0.2 | 138 | 40 | 12 | 2 |
| Ubx #1, 2 | 0.4 each | 0.8 | 151 | 59 | 21 | 1 |
| Ubx #1, 2 | 0.3 each | 0.6 | 142 | 17 | 5 | 0 |
| Ubx #1, 2 | 0.25 each | 0.5 | 182 | 46 | 22 | 1 |
| Ubx #1 | 0.5 | 0.5 | 137 | 63 | 17 | 4 |
| yellow | 0.5 | 0.5 | 53 | 35 | 13 | 8 |
Table 2.
Summary of crispant phenotypes
| Targeted gene | Individual | Phenotype |
|---|---|---|
| Antp | Figure 2B | Loss of Cu1 eyespot on the ventral side of forewing |
| Figure 2E | Complete or partial loss of white eyespot center on the ventral side of hindwings | |
| Figure 2F |
- Loss of Cu1 eyespot on the ventral side of forewing - Transformation of silver scales into brown scales on the dorsal side of forewing |
|
| Supplementary Figure S2A | - Loss of white (and gold) scales of eyespots on the ventral side of hindwing | |
| Supplementary Figure S2B |
- Partial loss of white eyespot center (Cu1 eyespot) on the ventral side of forewing - Transformation of sliver scales into brown scales on the ventral side of forewing - Loss of white and gold scales of eyespot on the ventral side of hindwing |
|
| Supplementary Figure S2C |
- Partial loss of Cu1 eyespots on the ventral side of forewings - Loss of white eyespot center on the dorsal side of hindwing |
|
| Supplementary Figure S2D and Figure 2K lower |
- Loss of M1 eyespot on the dorsal side of the forewing - Loss of white eyespot center on the dorsal side of the hindwing - Loss of white eyespot centers on the ventral side of hindwing |
|
|
Supplementary Figure S2E and Figure 2K upper |
- Partial loss of Cu1 eyespot on the ventral side of forewing - Loss of white eyespot centers on the ventral side of the hindwing - Loss of Cu1 eyespot on the dorsal side of the forewing |
|
| Ubx | Figure 3A |
- Enlargement of Cu1 eyespot on the ventral side of the hindwing - Disappearance of M3 eyespot on the ventral side of the hindwing - Ectopic silver scales were generated on the ventral side of the hindwing |
| Figure 3H |
- Loss of Rs eyespot on the ventral side of hindwing - Enlargement of M1 eyespot on the ventral side of hindwing - Ectopic eyespots were generated in M1, M2, and Cu1 wing sectors on the dorsal side of the hindwing resembling the ventral side of the forewing |
|
| Supplementary Figure S3A |
- Enlargement of M1 eyespot on the ventral side of hindwing *This individual has abnormal gold color spreading from Cu1 eyespot on the ventral side of forewing probably due to damage during wing development |
|
| Supplementary Figure S3B | - Ectopic eyespot was generated in M1 wing sector on the ventral side of hindwing resembling the larger size of the M1 eyespot on the ventral forewing | |
| Supplementary Figure S3C |
- Decrease in the M3 eyespot size on the ventral side of hindwing - Enlargement of Cu1 eyespot on the ventral side of hindwing |
|
| Supplementary Figure S3D |
- Ectopic eyespots were generated in a double M1 wing sector on the ventral and dorsal hindwing *This individual is a spontaneous mutant having an extra vein below M1 wing sector, resulting in additional eyespots visible on both sides of the wing. The M1 eyespot on the dorsal surface resembles in size more the ventral forewing M1 eyespot than the dorsal forewing M1 eyespot |
Detection of indel mutations
Genomic DNA was extracted with the SDS and Proteinase K method from a pool of five injected embryos that did not hatch. About 250 bp of sequence spanning the target sequence was amplified with PCRBIO Taq Mix Red (PCRBIOSYSTEMS), and PCR conditions were optimized until there was no smear, primer dimers or extra bands. Primers for those analyses are listed in Table 3. The PCR products were purified with the Gene JET PCR Purification Kit (Thermo Fisher). Two hundred nanograms of PCR product was denatured and re-annealed in 10× NEB2 buffer. One microliter of T7 endonuclease I (NEB) was added to the sample, while 1 µl of MQ water was added to a negative control. Immediately after the incubation for 15 min at 37°C, all the reactions were analyzed on a 3% agarose gel. Amplicons that showed positive cleavage from the T7 endonuclease I assay were subcloned into the pGEM-Teasy Vector (Promega) through TA cloning. For each target, we picked eight colonies and extracted the plasmid with a traditional alkali-SDS method and performed a PEG precipitation. Sequence analysis was performed with the BIGDYE terminator kit and a 3730xl DNA Analyzer (Thermo Fisher).
Table 3.
Primer list
| Primer name | Sequence (5′ → 3′) (underline: PAM sequence) |
|---|---|
| sgRNA target sequence for Antp #1 | GGGTAAGGCATGCCAGGGGCGGG |
| sgRNA target sequence for Antp #2 | GCGACCAGCAGCTCAGGCCCGGG |
| sgRNA target sequence for Ubx #1 | GGCTGCCACGGAGGCGTCGTAGG |
| sgRNA target sequence for Ubx #2 | GGCGTGCACCAGGGCGGTGGCGG |
| Genotype for Antp Fw | AGCTTGCACGAGGGGTACA |
| Genotype for Antp Rv | GAGTACCTGCGACGGAAGC |
| Genotype for Ubx Fw | CACCGTATCCGTTCTGCTG |
| Genotype for Ubx Rv | TCGGGTGACGTTTAATAGGC |
Immunohistochemistry for wing tissues
Larval wing tissues were dissected in PBS buffer under the microscope. The samples were fixed in 4% formaldehyde/Fix buffer (0.1 M PIPES pH 6.9, 1 mM EGTA pH 6.9, 1.0% Triton x-100, 2 mM MgSO4) for 30 min on ice. The samples were washed with 0.02% PBSTx (PBS + Triton x-100) for three times in every 10 min and then the samples were kept in 5% BSA/PBSTx for 1 h at room temperature as a blocking reaction. The samples were replaced into the 5% BSA/PBSTx with primary antibody and incubated at 4°C for overnight. We used a mouse monoclonal anti-Antp 4C3 (at 1:200; Developmental Studies Hybridoma Bank) and a rabbit anti-J. coenia Ubx antibody (at 1:500; a gift from L. Shashidhara). The wings were washed with PBSTx for three times in every 10 min. Then, replace the PBSTx to the 5% BSA/PBSTx as a blocking reaction for 1 h at room temperature, and then replace it into the 5% BSA/PBSTx with appropriate secondary antibody (1:200), and incubated at 4°C for 2 h. The wings were washed for three times in every 10 min, and the wings were mounted in ProLong Gold mounting media. The images were taken under Olympus FV3000.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13203650.
Results
Antp crispants lose forewing eyespots and produce smaller hindwing eyespots with no white center
Antp crispant butterflies had distinct phenotypes on fore- and hindwings. On the forewings, Antp crispants lost both the anterior (M1) and posterior (Cu1) eyespots on the ventral surfaces (Figure 2, B and F and Supplementary Figure S2D). Loss of eyespots was also observed on dorsal surfaces of forewings (Figure 2K and Supplementary Figure S2, C and D). Furthermore, the silver scales observed at the posterior end of the male ventral forewings were transformed to brown background scales, which are observed at this location only in females (Figure 2, F and I and Supplementary Figure S2B). In male B. anynana, the expression of Antp in the early pupal stage was associated with the development of silvery scales on the forewings (Figures 1B and 2C), which was a previously undocumented expression pattern for this gene. On the hindwings of Antp crispants, the white eyespot centers were missing in every eyespot (on both ventral and dorsal surfaces) (Figure, 2E, H, and K and Supplementary Figure S2, A, B, D, and E), and the overall size of the eyespots was reduced (Figure 2H compared to Figure 2G), but the eyespots were never lost (Figure 2H). Eyespots on the dorsal surface of hindwings often lack a distinct black and gold ring so we assume Antp merely removed the white center in these eyespots (Figure 2K and Supplementary Figure S2, C and D). These results indicate that Antp is essential for eyespot development and silver scale development in forewings, whereas in hindwings, Antp seems to be only required for the differentiation of the white central scales in eyespots and for increasing the size of the eyespots.
Figure 2.
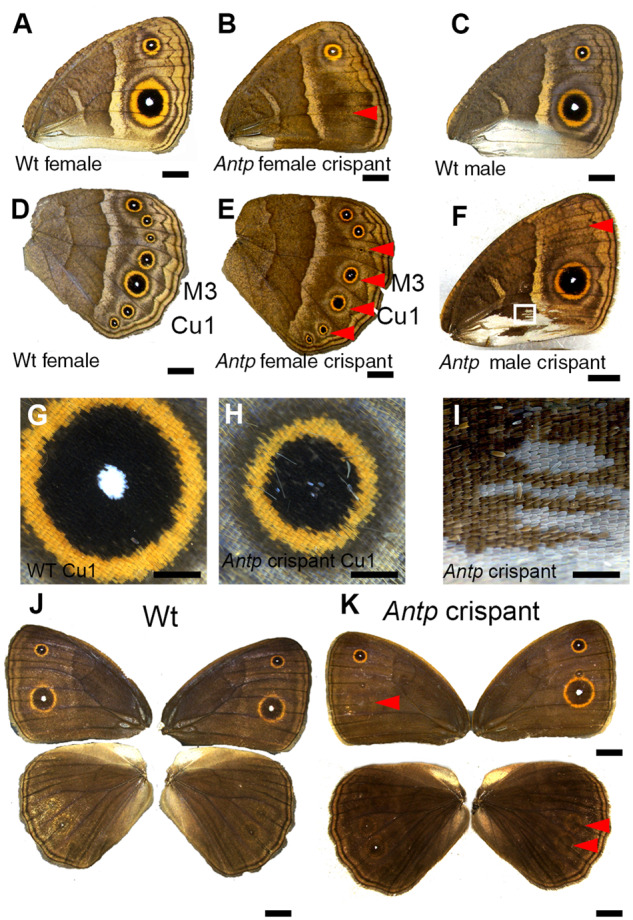
Antp crispant phenotypes on adult wings. (A) Ventral side of a Wt female forewing. (B) Ventral side of Antp female crispant forewing. Red arrowhead indicates the position where an eyespot is missing. (C) Ventral side of a Wt male forewing. (D) Ventral side of a Wt female hindwing. (E) Ventral side of Antp crispant male hindwing. Antp crispants partially or completely lost white eyespot centers, but eyespots were never missing. (F) Male-specific silver scales on the posterior part of the forewing were partially transformed into brown background scales (close up in I). (G) Highly magnified pictures of eyespots from D. (H) Highly magnified pictures of eyespots from E. The white eyespot center was completely or partially lost, and the eyespots were reduced in size. (J, K) Dorsal side of Wt and Antp crispant fore- and hindwings. Eyespots were lost on the dorsal side for both fore- and hindwings (red arrowheads). Crispant phenotypes are summarized in Table 2. Scale bars in A–F, J, and K: 5 mm; scale bars in G–I: 1 mm.
Ubx crispants led to the homeotic transformation of hindwings into forewings by both the activation and repression of eyespots
Ubx crispants showed the predicted homeotic phenotype, e.g. of hindwings acquiring a forewing identity (Figure 3A). In particular, hindwing M3 and Rs eyespots disappeared (Figure 3, A–C and H–J), and hindwing Cu1 eyespots became as large as the corresponding Cu1 eyespots on the forewing (Figure 3, D, E, and H–J). On the other hand, some Ubx crispants generated ectopic eyespots on the dorsal side of the hindwing (Figure 3L and Supplementary Figure S3, B and D). Dorsal eyespots are normally observed on the forewing, but the ectopic M1 eyespot was larger than the homologous eyespot on the forewing (Figure 3K and Supplementary Figure S3B) and an additional (M2) eyespot was visible with no corresponding forewing dorsal eyespot (Figure 3L and Supplementary Figure S3D). This crispant individual appears to have modified its dorsal hindwing into the ventral forewing instead (Figure 3H, right wing), where two eyespots are present in the M1 and M2 sectors. In addition, male-specific silver scales, which are normally present on the posterior edge of the ventral forewing, developed ectopically at a homologous posterior position in ventral hindwings of crispant males (Figure 3F compared to Figure 3G). These results indicate that Ubx has both repressive as well as activating effects on eyespots, depending on their position on the wing, e.g. Ubx has a repressive role on M1, M2, and Cu1 eyespot size for both dorsal and ventral surfaces, and an activating, essential role on Rs and M3 eyespot development. In addition, Ubx is a repressor of silver scale development on posterior ventral hindwings.
Figure 3.
Ubx crispant phenotypes on adult wings. (A) The hindwing on the left is highly mutated, whereas the wings on the right are not affected and represent the Wt wing pattern. (B) The M3 eyespot from the Ubx crispant is missing, resembling the forewing, while it is present on the other wing (C). White dotted lines indicate the position of veins bordering the M3 sector. (D) The Cu1 hindwing eyespot became enlarged, resembling the forewing Cu1 eyespot, relative to the non-mutated Cu1 eyespot on the wing shown in (E). (F, G) Enlargements of the black dotted areas in A. (F) Ectopic silver scales were generated on the posterior hindwing of the Ubx crispant instead of brown scales normally found at this position in non-mutated wings (G). (H) The hindwing on the right is mutated. (J) The Rs eyespot from the Ubx crispant is missing, but the M1 eyespot became enlarged compared to the other wing (I). (K) The hindwing on the left is highly mutated, in which ectopic eyespots were observed in M1, M2, and Cu1 wing sectors. (L) The size of the dorsal hindwing ectopic eyespots is slightly bigger than the homologous ones on the dorsal side of the forewing. (M) Eyespots are not observed in the other wing. Crispant phenotypes are summarized in Table 2. Scale bars in A, H, and K: 5 mm; scale bars in B–G, I, J, L, and M: 1 mm.
Discussion
Here, we show that the Hox genes Antp and Ubx have evolved novel expression domains and functions in B. anynana beyond those connected to anterior–posterior axis patterning of embryos that give segments and appendages along this axis their unique identities. Our results implicate these genes in the development of silver scales, and in the development of a morphological innovation within the Lepidoptera, eyespots, pointing to their likely involvement in eyespot origins.
Antp is essential for eyespot development on forewings only
Antp had been hypothesized to function in eyespot development in fore- and hindwings because of its expression pattern in the eyespot centers of both larval (Figure 1A, Saenko et al. 2011; Oliver et al. 2012; Shirai et al. 2012) and pupal wings (Tong et al. 2014). Here, we provide the first functional evidence supporting this hypothesis: Antp is an essential gene for eyespot development in forewings. In hindwings, however, Antp is only required for the differentiation of the white eyespot centers and for an increase in eyespot size. The different functions of Antp in fore- and hindwing eyespots may be related to the isolated expression of Antp in forewing eyespots, and the co-expression of Antp and Ubx in hindwing eyespots. On the forewings, Antp is likely to be required for both focal establishment (in the larval stage) and the production of the morphogenetic signal in early pupal stages that differentiates the color rings. It has been suggested that the white scales at the eyespot centers may need high levels, the black scales moderate levels, and golden scales lower levels of a morphogenetic signal to differentiate into their respective colors (French and Brakefield 1995; Brakefield et al. 1996). On the hindwings, however, Ubx might be able to partially substitute for these roles of Antp. When Antp activity is removed, eyespot foci are still able to differentiate but might not be able to generate enough signal to differentiate the central white scales, nor to reach the same number of cells away from the center, leading to overall smaller eyespots.
Ubx acts both as a repressor and as an essential gene for eyespot development
We showed that hindwing M1, M2, and Cu1 eyespots became larger in Ubx crispants (Figure 3, D, J, and L and Supplementary Figure S3, A–C), while Rs and M3 eyespots disappeared (Figure 3, B and J), suggesting opposite and location-specific effects of Ubx on eyespot development. The enlargement of Cu1 eyespots is consistent with a previously proposed repressor function for Ubx on both J. coenia and B. anynana Cu1 eyespots (Weatherbee et al. 1999; Tong et al. 2014). These prior experiments made use of a spontaneous mutant line of J. coenia that developed patches of forewing color patterns on the hindwing and also lacked Ubx expression in clones of hindwing cells (the nature of the mutation still remains to be characterized). When clones of transformed cells included Cu1 eyespots, these eyespots were transformed into larger eyespots bearing the size and colors of forewing eyespots (Weatherbee et al. 1999). Furthermore, in B. anynana, the overexpression of Ubx caused a reduction of Cu1 eyespot size in both fore- and hindwings, again suggesting a repressor function of Ubx on Cu1 eyespot size regulation (Tong et al. 2014). However, the disappearance of Rs and M3 hindwing eyespots in Ubx crispants clearly supports a novel, previously undocumented, eyespot-promoting function for this gene. While the disappearance of these two eyespots is expected of a “homeotic” mutation, in this case, the homeosis represents cells in the region of these two eyespots (Rs and M3) becoming Hox free, as the corresponding homologous regions of the forewing do not have any other known Hox gene expression at these positions. This result supports, thus, an eyespot promoting function for Ubx at these positions on the hindwing.
Ubx is negatively regulating eyespot size in the M1, M2, and Cu1 wing sectors of both dorsal and ventral surfaces, but it does it in slightly different ways. Ubx seems to have a simple homeotic effect on the ventral side, reverting hindwing eyespots into the size of the corresponding forewing eyespots; however, on the dorsal side, Ubx may be interacting with a dorsal selector, apterous A (apA) (or any of its regulators), promoting its expression on the dorsal surface of the wing. Previously we showed that apA represses eyespot development on the dorsal side of hindwings, as apA crispants led to the development of seven ectopic eyespots on this surface with the same size as the ventral eyespots (Prakash and Monteiro, 2018). Ubx and ap are not regulating each other in the Drosophila wing (Weatherbee et al. 1998), but we speculate that Ubx is repressing the size of M1, M2, and Cu1 eyespots and also positively regulating apA expression in eyespots on the dorsal surface, where disruption of Ubx function causes a decrease in apA levels, resulting in a slight de-repression of eyespot size on the dorsal side, making these eyespots resemble the size of ventral eyespots, and in this case, the size of ventral forewing eyespots. Removal of Ubx causes, thus, the transformation of the dorsal hindwing into the identity of the ventral forewing.
While we have no clear insight for why Ubx acts in such different ways toward eyespots in different sectors of the wing, we speculate that these different modes of action might be accomplished in three possible ways: (1) by the Ubx protein having a direct activating role on genes from the eyespot GRN in the Rs and M3 sectors; (2) by the Ubx protein having distinct types of downstream targets that indirectly affect the eyespot GRN: different sector-specific selector genes that are present in only some sectors of the wing (e.g. engrailed, invected, spalt, optomotor blind, or their downstream targets) (Carroll et al. 1994; Keys et al. 1999; Monteiro 2015; Özsu and Monteiro 2017; Banerjee and Monteiro 2020) and which, in turn, interact with the eyespot GRN by activating it or repressing it; or (3) by the Ubx protein using these sector-specific selectors as cofactors to either activate or repress the eyespot GRN in different ways in the different sectors (Mann et al. 2009). In support of the direct or indirect activating role of Ubx on genes of the eyespot GRN we previously observed that ectopic expression of Ubx in patches of cells in the early pupal forewing led to ectopic expression of both Distal-less (Dll) and Spalt (Sal) (Tong et al. 2014), two essential proteins for eyespot development (Connahs et al. 2019; Zhang and Reed 2016). In summary, we propose that Ubx functions as both an activator and a repressor of eyespots in species such as Bicyclus and Junonia, via its interactions with the eyespot GRN directly or via sector-specific selector genes that modulate its mode of action.
Antp promotes, while Ubx represses silver scale development
Silver scales develop only in males on the ventral posterior side of forewings and on the dorsal anterior side of hindwings, closely associated with scent glands that synthesize and release male sex pheromones (Figure 1C) (Dion et al. 2016). In Antp crispants, patches of silver scales on the forewing were changed to brown scales (Figure 2, F and I and Supplementary Figure S2B), whereas in Ubx crispants, ectopic silver scales were generated at the posterior end of the hindwing, which are normally covered by brown scales (Figure 3F). These results suggest that Antp promotes, whereas Ubx represses silver scale development. Our lab recently showed that the male isoform of the sex determination pathway gene, doublesex (dsx), is also required for silver scale development in B. anynana males on both fore- and hindwings (Prakash and Monteiro 2020). In addition, apA regulates silver scale development in a surface-specific manner in males: it represses silver scale development from dorsal forewings and promotes silver scale development on dorsal hindwings (Prakash and Monteiro 2018). We speculate that the male isoform of dsx activates Antp expression at the posterior end of the forewing to produce silver scales. Ubx might repress the expression of Antp, dsx, or any of their downstream targets, at the posterior end of the hindwing to prevent the generation of silver scales at the homologous hindwing region.
Possible functions of Antp and Ubx in eyespot evolution
Antp and Ubx phenotypes give us additional insights about the evolution of eyespot number and location on the wings of nymphalid butterflies. Here we propose that as forewings and hindwings differ in the expression of a key Hox gene, Ubx, which was shown here to be required to activate eyespot deployment (in Rs and M3 sectors), and previously able to ectopically activate Dll and sal on the forewing (Tong et al. 2014), this gene might have been essential for the origin of eyespots, which were initially restricted to the hindwings (Figure 4B) (Oliver et al. 2014). Recent work has identified a reaction–diffusion mechanism involving Dll in promoting the differentiation of eyespot centers in larval wings (Connahs et al. 2019). If novel binding sites for Ubx (or for a Ubx cofactor or target) evolved in the regulatory regions of any of the genes involved in this reaction–diffusion mechanism, and if this led to the stabilization of Dll expression in eyespot centers, this might have aided the origin of eyespots in hindwings only. Eyespots appear to have subsequently evolved on forewings multiple times independently in different lineages (Schachat et al. 2015). We propose that Antp was required for eyespots to eventually originate on the forewings of butterflies from the subfamilies Satyrinae and Biblidinae (Figure 4C). The independent co-option of Antp to the eyespot GRN in these lineages may have allowed eyespots to be activated, for the first time on the forewings, as this Hox gene might have substituted for the activating role of Ubx on this novel wing surface, and also led to an increase in the overall size of hindwing eyespots. The recruitment of Antp to the eyespot GRN might also have led to the origin of the white centers (Figure 4C), at least in the lineage leading to B. anynana. Genes other than Antp might have allowed eyespots to emerge in forewings in other lineages of nymphalids that do not express Antp in eyespots, such as Junonia. Furthermore, negative regulators of the eyespot GRN that are expressed exclusively on dorsal wing surfaces, such as apA, might have further limited the origin of eyespots to the ventral surfaces of wings (Prakash and Monteiro 2018). Finally, eyespots appear to have originated on the dorsal surfaces via the repression of apA (via a yet unidentified mechanism), as observed in the region of the anterior (M1) and posterior (Cu1) eyespot centers in B. anynana (Prakash and Monteiro 2018). Variation in eyespot size and number between forewings and hindwings has likely further evolved via novel interactions between the Hox genes and the sector-specific selector genes that still need to be identified (Figure 4D). Further comparative investigations will be needed to test whether the currently known role of Ubx as a repressor of Cu1 eyespot size in two divergent lineages of nymphalids (Bicyclus and Junonia) represents the ancestral function of this gene, or whether this is a more derived function that evolved separately in each lineage. It is worth noting that close relatives of each of these species can vary dramatically in the relative size of their Cu1 forewing and hindwing eyespots, arguing for two separate origins for this function in Ubx.
Figure 4.
Possible functions of Ubx and Antp in eyespot origins. Ancestral state reconstructions suggested that eyespots first originated in four to five wing sectors on the ventral side of the hindwings (Oliver et al. 2014). Eyespots later appeared on the ventral sides of the forewing (and later on the dorsal sides of both wings) (Oliver et al. 2014). (A) Common ancestral nymphalid butterflies did not have eyespot on their wings. (B) We propose that Ubx, shown here to be essential for the activation of some hindwing eyespots, was instrumental in the origin of eyespots restricted to hindwings. (C) Once Antp was co-opted to the eyespot GRN, its functional similarity to Ubx permitted eyespots to develop on the forewings (in lineages of butterfly that express this gene in eyespot centers) and also to become larger (in hindwings) as well as acquire a white center. (D) Size and number of eyespot are further evolved by probably through novel interactions between the Hox genes and the sector-specific selector genes.
In conclusion, we report novel functions for the Hox genes Antp and Ubx that implicate these genes in the development of nymphalid eyespots. Our data also shed some light on the mechanisms that led to the evolution of differences in eyespot number, size, and morphology between fore- and hindwings. Our work also implicates Antp in the development of white eyespot centers and silver scales in B. anynana. Finally, we propose a novel hypothesis for eyespot origins: that Ubx was essential in restricting the origin of eyespots to hindwings, and that the recruitment of Antp to the eyespot GRN led to redundancy of function (with Ubx) and to the appearance of eyespots on forewings, at least in the satyrid lineage of butterflies. This hypothesis will need to be tested with double knockouts of both Ubx and Antp in B. anynana, something still difficult to do. Future comparative work should examine how the function and deployment of these Hox genes has evolved at a finer scale across a butterfly phylogeny to test the eyespot origin hypothesis further.
Acknowledgments
We thank Xiaoling Tong for providing a high-resolution image of a pupal wing stained with Antp, and Thomas Werner and Anupama Prakash for comments on the manuscript.
Funding
Funding was received from the Ministry of Education, Singapore (grant MOE2015-T2-2-159) and the National Research Foundation, Singapore (award NRF-NRFI05-2019-0006).
Conflicts of interest
We have no conflict of interest.
Literature cited
- Akam M. 1995. Hox genes and the evolution of diverse body plans. Philos Trans R Soc Lond B Biol Sci. 349:313–319. [DOI] [PubMed] [Google Scholar]
- Angelini DR, Kaufman TC.. 2005. Comparative developmental genetics and the evolution of arthropod body plans. Annu Rev Genet. 39:95–119. [DOI] [PubMed] [Google Scholar]
- Banerjee TD, Monteiro A.. 2018. CRISPR-Cas9 mediated genome editing in Bicyclus anynana butterflies. Methods Protoc. 15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee TD, Monteiro A.. 2020. Molecular mechanism underlying venation patterning in butterflies. bioRxiv. 10.1101/2020.01.02.892760. [DOI]
- Brakefield PM, Gates J, Keys D, Kesbeke F, Wijngaarden PJ, et al. 1996. Development, plasticity and evolution of butterfly eyespot patterns. Nature. 384:236–242. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Gates J, Keys DN, Paddock SW, Panganiban GE, et al. 1994. Pattern formation and eyespot determination in butterfly wings. Science. 265:109–114. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Weatherbee SD, Langeland JA.. 1995. Homeotic genes and the regulation and evolution of insect wing number. Nature. 375:58–61. [DOI] [PubMed] [Google Scholar]
- Connahs H, Tlili S, van Creij J, Loo TYJ, Banerjee TD, et al. 2019. Activation of butterfly eyespots by Distal-less is consistent with a reaction-diffusion process. Development. 146:dev169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion E, Monteiro A, Yew JY.. 2016. Phenotypic plasticity in sex pheromone production in Bicyclus anynana butterflies. Sci Rep. 6:39002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French V, Brakefield PM.. 1995. Eyespot development on butterfly wings: the focal signal. Dev Biol. 168:112–123. [DOI] [PubMed] [Google Scholar]
- Glassford WJ, Johnson WC, Dall NR, Jacquelyn SS, Liu Y, et al. 2015. Co-option of an ancestral Hox-regulated network underlies a recently evolved morphological novelty. Dev Cell. 34:520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzem M, Braak N, Brattström O, McGregor AP, Breuker CJ.. 2019. Wnt gene expression during early embryogenesis in the nymphalid butterfly Bicyclus anynana. Front Ecol Evol. 7:468. [Google Scholar]
- Jeong S, Rokas A, Carroll SB.. 2006. Regulation of body pigmentation by the abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 125:1387–1399. [DOI] [PubMed] [Google Scholar]
- Keys DN, Lewis DL, Selegue JE, Pearson BJ, Goodrich LV, et al. 1999. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science. 283:532–534. [DOI] [PubMed] [Google Scholar]
- Lewis EB. 1978. A gene complex controlling segmentation in Drosophila. Nature. 276:565–570. [DOI] [PubMed] [Google Scholar]
- Mallo M, Wellik DM, Deschamps J.. 2010. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 344:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R.. 2009. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 88:63–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Serano JM, Jarvis E, Bruce HS, Wang J, et al. 2016. CRISPR/Cas9 mutagenesis reveals versatile roles of Hox genes in Crustacean limb specification and evolution. Curr Biol. 26:14–26. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Monteiro A.. 2018. Melanin pathway genes regulate color and morphology of butterfly wing scales. Cell Rep. 24:56–65. [DOI] [PubMed] [Google Scholar]
- McIntyre DC, Rakshit S, Yallowitz AR, Loken L, Jeannotte L, et al. 2007. Hox patterning of the vertebrate rib cage. Development. 134:2981–2989. [DOI] [PubMed] [Google Scholar]
- Medved V, Huang ZY, Popadic A.. 2014. Ubx promotes corbicular development in Apis mellifera. Biol Lett. 10:20131021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro A. 2015. Origin, development, and evolution of butterfly eyespots. Annu Rev Entomol. 60:253–271. [DOI] [PubMed] [Google Scholar]
- Ohde T, Yaginuma T, Niimi T.. 2013. Insect morphological diversification through the modification of wing serial homologs. Science. 340:495–498. [DOI] [PubMed] [Google Scholar]
- Oliver JC, Tong XL, Gall LF, Piel WH, Monteiro A.. 2012. A single origin for nymphalid butterfly eyespots followed by widespread loss of associated gene expression. PLoS Genet. 8:e1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JC, Beaulieu JM, Gall LF, Piel WH, Monteiro A.. 2014. Nymphalid eyespot serial homologues originate as a few individualized modules. Proc R Soc B. 281:20133262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özsu N, Monteiro A.. 2017. Wound healing, calcium signaling, and other novel pathways are associated with the formation of butterfly eyespots. BMC Genomics. 18:788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer S, Weigel D, Beckendorf SK.. 1992. Organogenesis in Drosophila melanogaster: embryonic salivary gland determination is controlled by homeotic and dorsoventral patterning genes. Development. 114:49–57. [DOI] [PubMed] [Google Scholar]
- Prakash A, Monteiro A.. 2018. apterous A specifies dorsal wing patterns and sexual traits in butterflies. Proc R Soc B. 285:20172685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, Monteiro A.. 2020. Doublesex mediates the development of sex-specific pheromone organs in Bicyclus butterflies via multiple mechanisms. Mol Biol Evol. 37:1694–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad N, Tarikere S, Khanale D, Habib F, Shashidhara LS.. 2016. A comparative genomic analysis of targets of Hox protein Ultrabithorax amongst distant insect species. Sci Rep. 6:27885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenko SV, Marialva MS, Beldade P.. 2011. Involvement of the conserved Hox gene Antennapedia in the development and evolution of a novel trait. Evodevo. 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachat SR, Oliver JC, Monteiro A.. 2015. Nymphalid eyespots are co-opted to novel wing locations following a similar pattern in independent lineages. BMC Evol Biol. 15:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai LT, Saenko SV, Keller RA, Jerónimo MA, Brakefield PM, et al. 2012. Evolutionary history of the recruitment of conserved developmental genes in association to the formation and diversification of a novel trait. BMC Evol Biol. 12:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. 1982. Genes controlling segmental specification in the Drosophila thorax. Proc Natl Acad Sci U S A. 79:7380–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Barmina O, Sanders LE, Arbeitman MN, Kopp A.. 2011. Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 9:e1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Rahman SR, Ezray BD, Franzini L, Strange JP, et al. 2019. A homeotic shift late in development drives mimetic color variation in a bumble bee. Proc Natl Acad Sci U S A. 116:11857–11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu Y, Wheeler SR, Denell RE.. 2005. Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature. 433:643–647. [DOI] [PubMed] [Google Scholar]
- Tomoyasu Y. 2017. Ultrabithorax and the evolution of insect forewing/hindwing differentiation. Curr Opin Insect Sci. 19:8–15. [DOI] [PubMed] [Google Scholar]
- Tong X, Hrycaj S, Podlaha O, Popadic A, Monteiro A.. 2014. Over-expression of Ultrabithorax alters embryonic body plan and wing patterns in the butterfly Bicyclus anynana. Dev Biol. 394:357–366. [DOI] [PubMed] [Google Scholar]
- Weatherbee SD, Halder G, Kim J, Hudson A, Carroll S.. 1998. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 12:1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee SD, Nijhout HF, Grunert LW, Halder G, Galant R, et al. 1999. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr Biol. 9:109–115. [DOI] [PubMed] [Google Scholar]
- Zhang L, Reed RD.. 2016. Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat Commun. 7:11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13203650.