Abstract
People with NR5A1 mutations experience testicular dysgenesis, ovotestes, or adrenal insufficiency, but we do not completely understand the origin of this phenotypic diversity. NR5A1 is expressed in gonadal soma precursor cells before expression of the sex-determining gene SRY. Many fish have two co-orthologs of NR5A1 that likely partitioned ancestral gene subfunctions between them. To explore ancestral roles of NR5A1, we knocked out nr5a1a and nr5a1b in zebrafish. Single-cell RNA-seq identified nr5a1a-expressing cells that co-expressed genes for steroid biosynthesis and the chemokine receptor Cxcl12a in 1-day postfertilization (dpf) embryos, as does the mammalian adrenal–gonadal (interrenal-gonadal) primordium. In 2dpf embryos, nr5a1a was expressed stronger in the interrenal-gonadal primordium than in the early hypothalamus but nr5a1b showed the reverse. Adult Leydig cells expressed both ohnologs and granulosa cells expressed nr5a1a stronger than nr5a1b. Mutants for nr5a1a lacked the interrenal, formed incompletely differentiated testes, had no Leydig cells, and grew far larger than normal fish. Mutants for nr5a1b formed a disorganized interrenal and their gonads completely disappeared. All homozygous mutant genotypes lacked secondary sex characteristics, including male breeding tubercles and female sex papillae, and had exceedingly low levels of estradiol, 11-ketotestosterone, and cortisol. RNA-seq showed that at 21dpf, some animals were developing as females and others were not, independent of nr5a1 genotype. By 35dpf, all mutant genotypes greatly under-expressed ovary-biased genes. Because adult nr5a1a mutants form gonads but lack an interrenal and conversely, adult nr5a1b mutants lack a gonad but have an interrenal, the adrenal, and gonadal functions of the ancestral nr5a1 gene partitioned between ohnologs after the teleost genome duplication, likely owing to reciprocal loss of ancestral tissue-specific regulatory elements. Identifying such elements could provide hints to otherwise unexplained cases of Differences in Sex Development.
Keywords: adreno-gonadal primordium, Differences in Sex Development, disorders of sex development, scRNA-seq, SF1, subfunctionalization, Genetics of Sex
Introduction
Sex determination (SD) involves several interacting cell types (germ cells, somatic cells), multiple organs (hypothalamus, pituitary, gonads, and adrenal), and impacts multiple traits, including disease susceptibility. We lack, however, full knowledge of the mechanisms by which genetic and environmental factors interact to establish an individual’s sex. In mammals, the Y-chromosome gene SRY generally initiates male development and without SRY, people usually become females (Sinclair et al. 1990). Less frequent outcomes, called Differences in Sex Development (DSD, or disorders of sex development), are a pediatric concern, occurring in one in 2000–5000 live births (Sax 2002; Baetens et al. 2019). DSDs can arise by mutations in sex determination pathway genes or by environmental factors (Marrocco et al. 2015; Baetens et al. 2019). Many cases of DSD involve gonadal dysgenesis, but the etiology of most cases is unknown (Rocha et al. 2011; Garcia-Acero et al. 2020).
The Undiagnosed Diseases Network case UDN365839 presented at the age of 16 years with primary hypogonadism, azoospermia, low serum testosterone (44 ng/dl), clinical obesity, and a 46, XX SRY-negative karyotype with a novel missense mutation in NR5A1 (p.R92W) (Bashamboo et al. 2016). NR5A1 [Nuclear Receptor Subfamily 5, Group A, Member 1, also called Steroidogenic Factor 1 (SF1) or AD4BP] is a nuclear receptor with no known ligand; mutations in NR5A1 can cause XX sex reversal (OMIM 617480) and XY sex reversal (OMIM 612965) (Achermann et al. 1999; Bashamboo et al. 2016), as well as adrenocortical insufficiency (OMIM 612964) (Guran et al. 2016) and premature ovarian or spermatogenic failure (OMIM 612964 and 613957). Null alleles of Nr5a1 in mouse result in the loss of the ventral medial hypothalamus and the adrenal–gonadal primordium (Ikeda et al. 1995; Val et al. 2003). We do not completely understand, however, what causes different NR5A1 mutations to result in different human phenotypes or the regulatory mechanisms that evoke NR5A1 expression in these organs.
Teleost fishes have two co-orthologs of NR5A1, called nr5a1a (ff1b) and nr5a1b (ff1d) (von Hofsten et al. 2001; Kuo et al. 2005), that originated in the teleost genome duplication event (TGD) (Amores et al. 1998; Postlethwait et al. 1999; Taylor et al. 2003; Jaillon et al. 2004). Here, we probed the question: What are the roles of the two zebrafish co-orthologs compared to those of the single-copy mammalian NR5A1 gene? Under one hypothesis (Force et al. 1999), the functions of Nr5a1 in the adrenal–gonadal primordium, adrenal, gonad, and hypothalamus were present in the last common ancestor of humans and zebrafish and partitioned between the two fish ohnologs after the TGD. Alternatively, some mammalian functions evolved after the divergence of zebrafish and human lineages and would be absent either from zebrafish or from human. To evaluate these possibilities, we studied single-cell transcriptomes in wild-type embryos and made mutations in both zebrafish nr5a1 duplicates, studying their gene expression patterns, phenotypes, transcriptomes, and hormone titers. The results showed that nr5a1a maintains the interrenal [the teleost adrenal cortex homologue that, along with chromaffin cells, represents the adrenal medulla and lies within the fish kidney (Chester Jones and Mosley 1980)]. Furthermore, without the wild-type allele of nr5a1b, the gonad disappeared from adult fish, leading to the loss of secondary sex characteristics. Both organs disappeared in double mutants. These results suggest that the ancestral functions of Nr5a1 included both adrenal and gonad, but that they partitioned between nr5a1a and nr5a1b in teleosts. Identification of conserved regulatory elements that specify interrenal vs. gonadal expression patterns in zebrafish NR5A1 ohnologs may help to identify sequences responsible for human DSDs of unknown origin.
Materials and methods
Mutagenesis
CRISPR/Cas9 mutagenesis generated deletions in zebrafish nr5a1a (ENSDARG00000103176) and nr5a1b (ENSDARG00000023362) (http://ensembl.org), using sites identified by ZiFiT Targeter (http://zifit.partners.org/ZiFiT/). Mutagenesis targeted region in nr5a1a exon-4 was: GTGCGTGCAGACCGGATGAG, and in nr5a1b exon-4: GTGCGGATAGGATGCGAGG, using gRNAs synthesized from DNA oligomer templates: aattaatacgactcactataGTGCGTGCAGACCGGATGAGgttttagagctagaaatagc for nr5a1a and aattaatacgactcactatagGTGCGGATAGGATGCGAGGgttttagagctagaaatagc nr5a1b (DT, Coralville, IA). MEGAscript T7 Transcription Kit transcribed gRNA and mMESSAGE mMACHINE T3 Transcription Kit (Thermo Fisher Scientific, Waltham, MA) synthesized Cas9 mRNA. Approximately, 2 nl of a solution containing 100 ng/µl Cas9 mRNA and 25 ng/µl of both nr5a1 gRNAs was co-microinjected into one-cell embryos of the AB strain. Genomic DNA from injected embryos at 24 hpf (hour postfertilization) provided template to amplify a 312-bp PCR fragment for nr5a1a gene (primers: F-ACACAAATGCATTATTCCTCTCTCT and R-CCTCCAGTTTGAAGCCGCTA); a 375-bp PCR fragment for nr5a1b (primers: F-TGGGGAAAAGAATTAACAGGGGT and R-GACGATGTTCGGATGGGTGT). Wild-type alleles have a BstCI recognition site for both nr5a1a and nr5a1b genes that are disrupted in nr5a1 mutant alleles. Sanger sequencing (GENEWIZ, Inc., NJ) verified mutations. We established stable lines for two noncomplementing alleles: deletion of 11 nucleotides designated nr5a1a(b1388) and a deletion of 8 nucleotides designated nr5a1b(b1389) (Figure 2C).
Figure 1.
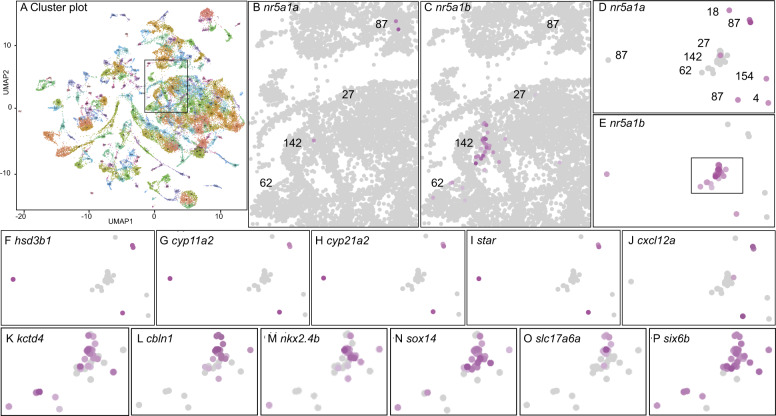
scRNA-seq identification of genes co-expressed with nr5a1a and nr5a1b. (A) In total, 220 clusters from scRNA-seq of whole animals at 24, 48, and 120 hpf (Farnsworth et al. 2020). The box indicates the portion enlarged in (B) and (C). (B) nr5a1a-expressing cells; color intensity is proportional to log expression level. (C) nr5a1b-expressing cells. (D) nr5a1a- and/or nr5a1b-expressing cells labeled only for nr5a1a-expression, with clusters numbered. (E) nr5a1a- and/or nr5a1b-expressing cells labeled only for nr5a1b-expression. The box indicates the portion enlarged in panels J–O. (F–I) Expression of the steroid biosynthesis genes hsd3b1, cyp11a2, cyp21a2, and star. (J) Cluster 87 cells labeled for the cytokine cxcl12a. (K–P) Expression of ventromedial hypothalamus genes kctd4, cbln1, nkx2.4b, sox14, slc17a6a, and six6b, respectively, in the boxed region of (E).
Histology and in situ hybridization
In situ hybridization was performed as described previously (Rodriguez-Mari et al. 2005) using the probes: nr5a1a (ENSDARG00000103176) using a 859-bp fragment including exon-2 to exon-6 (primers F-AAGTGTCCGGTTATCATTACGGCC and R-TGTCTGCAGATGTGATCCAGAAGC) (Yan et al. 2019); nr5a1b (ENSDARG00000023362) using a 821-bp fragment including partial exon 5 and 3′UTR (primers F-AACTTCTAGTGCTGGACTATGTTGCC and R-CATTTCTTAAGAGGCCACAGAGCGTA); amh (ENSDARG00000014357) (Yan et al. 2017); cyp11c1 (ENSDARG00000042014) (Wang and Orban 2007) using a 592-bp fragment located in exons 9–13 (primers F-CAGAGCCAACATCACTGAGCTGAT and R-GAAGGTGATTCTCGGTGGACACTC); cyp19a1a (ENSDARG00000041348) (Chiang et al. 2001b); cyp21a2 (ENSDARG00000037550) using a 875-bp fragment including partial exon 1 to exon 8 (F-TGGTCAGTGTTGTGCTATTGCTGT and R-TGAAGAAGTCAGTGTGCCACCTTC); ddx4, (vasa) (ENSDARG00000014373) (Yoon et al. 1997); nkx2.1 (ENSDARG00000019835) using a 612-bp fragment including partial exon (F-ACAGAACAATGTCGATGAGCCCTAAG andR-TTGAGTCGGAGTCAAGTGTATCATGC); nkx2.4a (ENSDARG00000075107) using a 1023-bp fragment including partial exon 1-2 (F-CCACGAGAACAGAGCTGATACAACAA and R-AGAAGTGTCTATCTGGCCCATACTGT); nkx2.4b (ENSDARG00000104107) using a 936-bp fragment including partial exon 1-2 (F-ATCGAGGAGACCTTCAAGAAGTTTGC and R-CACATATCTCCGTCCGTCAGTTGATT).
Histology used paraffin-embedded Bouin’s-fixed tissue sectioned at 10 µm and stained with hematoxylin and eosin (H&E) (Rodriguez-Mari et al. 2005). The gonadosomatic index was calculated as (gonad weight)(100)/(fish body weight). As a measure of gonad size, we cut cross-sections, counted the number of gonad-containing rows on a slide, identified the middle row, photographed each gonad from two sections in that row for four images, and used ImageJ to measure the area of each gonad twice using the freehand selection tool and calculated the area of the gonad with the measure function.
Transcriptomics
Fish homozygous for either nr5a1a(b1388), nr5a1b(b1389), double mutants or double wild-type siblings at 21 and 35 dpf were euthanized in Tricaine followed by isolating the gonad-containing trunk from just posterior of the pectoral fin to just anterior to the vent. Trunks from each fish were individually homogenized in 200 µl Trizol. Total RNA was extracted according to Amores et al. (2011) and enriched for mRNA using Dynabeads® Oligo(dt)25 (ThermoFisher). We constructed strand-specific, indexed, cDNA sequencing libraries (NEXTflex™ qRNA-seq kit, BIOO Scientific), quantified libraries by Qubit® fluorometer (Life Technologies), normalized libraries to 2.3 nM, multiplexed libraries, quality-checked libraries (Kapa Library Quantification Kit, Kapa Biosystems), and sequenced libraries in two lanes on an Illumina HiSeq 4000 using paired-end 100 base pair (bp) reads.
For single-cell RNA-seq, embryos from natural crosses were collected at 24, 48, and 120 hpf in two pools of 15 individuals per stage from the genotypes Tg(olig2: GFP)vu12 or Tg(elavl3: GCaMP6s). Embryos were dissociated and cell suspensions were prepared as described previously (Farnsworth et al. 2020). The University of Oregon Genomics and Cell Characterization core facility (https://gc3f.uoregon.edu/) separated cells and prepared libraries on a 10X Chromium platform using 10× v.2 chemistry targeting 10,000 cells. Fifteen cycles of PCR-amplified cDNA libraries that were sequenced on either an Illumina Hiseq (5/6 samples) or an Illumina Next-seq (1/6 samples). Reads were aligned to GRCz11_93 and bioinformatic analyses were done using CellRanger and Seurat as described previously (Farnsworth et al. 2020).
RNA-seq libraries were constructed from trunk tissue from eight individuals of 21-dpf wild-type fish, nr5a1a mutants, nr5a1b mutants, nr5a1ab double mutants, and eight individuals of 35-dpf wild-type fish, nr5a1a mutants, nr5a1b mutants, and nr5a1ab double mutants. In a preliminary principal component analysis, one 35-dpf wild-type individual was a substantial outlier and was subsequently ignored. The remaining 63 libraries produced 642,409,629 preprocessed reads, of which 473,982,972 reads aligned to zebrafish GRCz10 genome. Following correction for PCR duplicate reads, 413,607,158 reads remained, and 315,250,017 of these corrected reads aligned to protein-coding regions.
Bioinformatics
We used Dupligänger duplicate removal software (Sydes et al. 2019) to preprocess RNA-seq reads, to identify, and to remove BIOO inline unique molecular identifiers (UMIs), to remove read-through adapters (using cutadapt v1.18 (Martin 2011) with command line options: -n 3 -O 1 -m 30 -a AGATCGGAAGAGC-A AGATCGGAAGAGC –too-short-output –too-short-paired-output), and then to remove low-quality sequences at both the 5ʹ-ends and the 3ʹ-ends using Trimmomatic (v0.36) (Bolger et al. 2014), with command line options: LEADING : 10 TRAILING : 10 SLIDINGWINDOW : 5:10 MINLEN : 30. Dupligänger tracked the number of nucleotides removed from the 5ʹ-end and deleted reads <30 nt. STAR (version 2.7.0f, command line options: –outFilterMultimapNmax 1 –outSAMtype BAM Unsorted –alignIntronMax 1000000 –alignMatesGapMax 1000000) (Dobin et al. 2013) aligned processed PE reads to the zebrafish genome (GRCz11, Ensembl version 96) in a splice-aware manner. Dupligänger then deleted PCR duplicates from the sequence alignment file if (1) the read pair shares with another read its 5′ alignment starts for both R1 and R2 after correcting for 5′ trimming, and (2) the read pair shares the same R1 UMI and R2 UMI. Dupligänger forwarded de-duplicated sequence alignment files to HTSeq-count (Anders et al. 2015) (using command line options: -m intersection-strict –stranded=reverse) to obtain per-gene counts for protein-coding genes. DESeq2 (version 1.22.2) provided statistical differential expression analysis (Love et al. 2015). Reads are available at the Sequence Read Archive under project accession number PRJNA561212.
Endocrinology
Sex steroids were extracted from zebrafish homogenates according to the flowchart in (Newman et al., 2008). Individual flash-frozen fish were macerated using scissors and a knife on a cold cutting board, transferred to cold glass test tubes, and weighed. Water and HPLC-grade methanol were added to the tissue and this mixture was homogenized and sonicated. Tubes were shaken on a multi-tube vortexer (Glas-Col Large Capacity Mixer, speed set at 500 rpm; Glas-Col, Terre Haute, IN, USA) for 1 h at room temperature and then stored overnight at 4 °C. Before use, tubes were shaken again and then centrifuged at 1500 g for 15 min at 4 °C. In brief, 1 ml of supernatant was combined with 10 ml water and then extracted using solid-phase extraction (SPE columns: Agilent Bond Elut-C18 OH, 500 mg 3 ml, cat # 12102046). Eluates were dried in a Savant SpeedVac Concentrator (model SDP121P; Thermo Fisher Scientific, Waltham, MA, USA) at 35°C and then stored at −80°C. One day prior to assay, samples were resuspended in 500-µl assay buffer (X065 buffer; Arbor Assays, Ann Arbor, MI, USA) and shaken at 500 rpm for 1 h at room temperature. After storage at 4°C overnight, samples were shaken and then assayed. Both male and female extracts were diluted 1:4 for cortisol. Female extracts were diluted 1:2 for both E2 and 11-KT. Male extracts were assayed undiluted for E2 and diluted 1000× for 11-KT. All extracts were run undiluted for both E2 and 11-KT. Commercially available EIAs were used to measure cortisol (Cortisol ELISA Kit #K003, Arbor Assays, Ann Arbor, MI), estradiol (E2; Estradiol ELISA Kit #K030, Arbor Assays, Ann Arbor, MI), and 11-ketotestosterone (11-KT; 11-keto Testosterone ELISA Kit #582751, Cayman Chemical, Ann Arbor, MI), according to the manufacturer’s instructions. These kits were validated for zebrafish homogenate extract using tests of parallelism and standard addition (Hunt et al. 2017). Any sample that exceeded 10% coefficient of variation between duplicates or was outside the range of the standard curve was reanalyzed. Intra- and inter-assay variation for cortisol was 2.2% and 5.2%, respectively. Intra- and inter-assay variation for E2 was 2.6% and 3.1%, respectively. Intra- and inter-assay variation for 11-KT was 2.5% and 13%, respectively.
Data availability
The Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) contains RNA-seq reads under accession number PRJNA561212. Supplementary Tables list differentially expressed genes. The scRNA-seq data are publicly available at https://cells.ucsc.edu/?ds=zebrafish-dev. Additional data and code relevant to the Atlas can be accessed at: https://www.adammillerlab.com/. Work was performed under the University of Oregon Institutional Animal Care and Use Committee (IACUC) protocol #14-08 R. Mutant strains are available on request. Data should be cited according to the citation of this article.
Supplementary material is available at figshare: https://doi.org/10.25386/genetics.13236983.
Results
Searching for the adrenal–gonadal primordium
In mammals, NR5A1-expressing cells give rise to the adrenal and to the gonadal soma before the expression of the sex-determining gene SRY (Ikeda et al. 1994; Hatano et al. 1996; Sekido and Lovell-Badge 2008). To identify the earliest nr5a1-expressing cells in the zebrafish embryo and the adrenal (interrenal)-gonadal primordium, we performed single-cell RNA-seq (scRNA-seq) on whole bodies of wild-type animals at 1, 2, and 5 dpf (days post-fertilization) (Farnsworth et al. 2020). Analyses identified 220 clusters of transcriptionally related cells in the zebrafish scRNA-seq atlas (“Atlas,” Figure 1A), but only four clusters had multiple cells expressing nr5a1a (ENSDARG00000103176, ff1b), or nr5a1b (ENSDARG00000023362, ff1d), or both (Figure 1, B and C). We bioinformatically isolated cells expressing nr5a1 ohnologs (Figure 1, D and E) and identified genes differentially expressed (DE) compared to all other cells in the Atlas.
In nr5a1a-expressing cells, 43 genes were differentially over-expressed (Padj < 0.05), the top six of which were related to steroid biosynthesis (hsd3b1, cyp11a2, cyp21a2, star, fdx1b, and interestingly, nr5a1b), and all were expressed in Cluster 87 (Figure 1, F–I and Supplementary Table S1). The expression of CYP21A2 (cyp21a2) is restricted to the human adrenal cortex and the fish interrenal (Fagerberg et al. 2014; Eachus et al. 2017), and FDX1 is expressed almost exclusively in the human adrenal (Fagerberg et al. 2014). Human orthologs of the other top six genes are expressed in both adrenal and gonad (Fagerberg et al. 2014). This result suggests the hypothesis that nr5a1a-expressing cells may mark the zebrafish adrenal (interrenal)–gonadal precursor. Cells in Cluster 87 also significantly differentially expressed six1b (Supplementary Table S1), whose mammalian ortholog marks the adrenal–gonadal primordium (Kobayashi et al. 2007; Fujimoto et al. 2013), and they strongly expressed cxcl12a, the ligand that guides migrating primordial germ cells to the gonadal soma in the celomic epithelium (Boldajipour et al. 2011) (Figure 1J) although cxcl12a was not DE with respect to other cells in the atlas because it also helps guide the migration of vascular, hematopoietic, and neural cells (Aiuti et al. 1997; Tachibana et al. 1998; Zou et al. 1998; Peled et al. 1999; Knaut et al. 2005; Lieberam et al. 2005; Siekmann et al. 2009; Walters et al. 2010). Cluster 87 cells also differentially expressed prox1b (Supplementary Table S1), a proposed coregulator of nr5a1a (Liu et al. 2003). These scRNA-seq results are predicted by the hypothesis that Cluster 87 contains the adrenal (interrenal)–gonadal primordium and its derivatives.
In nr5a1b-expressing cells, 630 genes were differentially coexpressed by scRNA-seq (Padj < 0.05) (Supplementary Table S2). The 11 most DE genes from the 32 nr5a1b-expressing cells in Clusters 27 [1dpf (days postfertilization)], 62 (1, 2, and 5 dpf), and 142 (1 and 2 dpf) included ventromedial hypothalamus (VMH) genes in zebrafish and/or mammals [kctd4, cbln1, sox1a, nkx2.4b, sox14, nkx2.1, slc17a6a, sox1b, six6a, six6b, lhx5 (Karlstrom et al. 2003; Segal et al. 2005; Kurrasch et al. 2007; Toro et al. 2009; Appelbaum et al. 2010; Larder et al. 2011; Machluf et al. 2011; Armant et al. 2013; Manoli and Driever 2014; Sun et al. 2015; Chen et al. 2017)] (Figure 1, K–P and Supplementary Table S2). In mouse, Nr5a1 is the most specific marker for the VMH (Segal et al. 2005), but the role of Nr5a1 in the VMH is not yet understood (Budefeld et al. 2012). The rx3 gene that was shown to interact with nr5a1b in zebrafish hindbrain development was strongly DE in nr5a1b-expressing cells (Muthu et al. 2016), and Six6 expression in the VMH is necessary for normal fertility in mouse (Xie et al. 2015). These results show that the expression of nr5a1a and nr5a1b already differs at 1 dpf in the hypothalamus. Genes co-expressed with nr5a1 ohnologs provide a resource to further investigate the roles of Nr5a1 in hypothalamus development.
Mutagenesis to identify functions of nr5a1 ohnologs
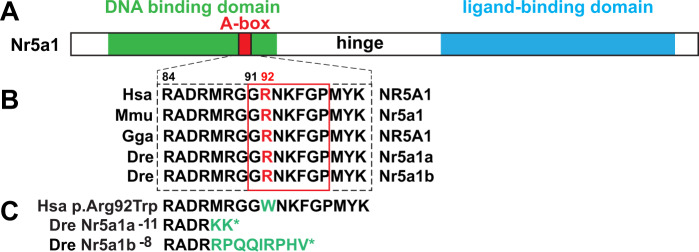
Having identified single-cell transcriptomes in nr5a1-expressing cells, we wanted to learn the phenotypes of mutants lacking these gene functions. CRISPR/Cas9 mutagenesis induced premature stop codon alleles in nr5a1a and nr5a1b. Mutation targets were in the conserved A-box within the DNA-binding domain (Lin and Achermann 2008) near the site of the R92W mutation that results in variable testis development in 46, XX humans (Bashamboo et al. 2016; Miyado et al. 2016; Werner et al. 2017). The amino acid sequence of the human A-box and its surroundings is precisely conserved in both zebrafish ohnologs (Figure 2, A and B). Supplementary Figure S1 shows CRISPR target sites and sequence traces for an 11-base pair (bp) deletion in nr5a1a (the nr5a1a−11) allele, and an 8 bp deletion in nr5a1b (the nr5a1b−8 allele). Both deletions should produce an 87-residue polypeptide with two out-of-frame residues in nr5a1a and nine in nr5a1b (Figure 2C). Predicted proteins disrupt the DNA-binding domain and eliminate the ligand-binding domain and hence should be null activity alleles.
Figure 2.
CRISPR/Cas9-induced nr5a1a and nr5a1b mutants. (A) Consequences of induced mutations on Nr5a1 proteins (DNA-binding domain, green; Abox, red; ligand-binding domain, blue). (B) Amino acid sequences surrounding the zebrafish CRISPR target sites for orthologs in human (Hsa), mouse (Mmu), chicken (Gga), and zebrafish (Dre). The human mutations R84H and G91S are dominant and cause 46, XY sex reversal; R92W is a dominant mutation resulting in variable testis development in 46, XX individuals; and R92Q is a recessive that causes adrenal insufficiency, 46, XX sex reversal, and 46, XY sex reversal (Lin and Achermann 2008; Bashamboo et al. 2016; Miyado et al. 2016; Werner et al. 2017). (C) The zebrafish nr5a1a−11 and nr5a1b−8 deletion alleles are predicted to result in polypeptides with short out-of-frame sequences (green) that are truncated owing to a premature stop codon (*) before or within the A-box.
Embryonic expression patterns of nr5a1a and nr5a1b
Gene expression in wild types
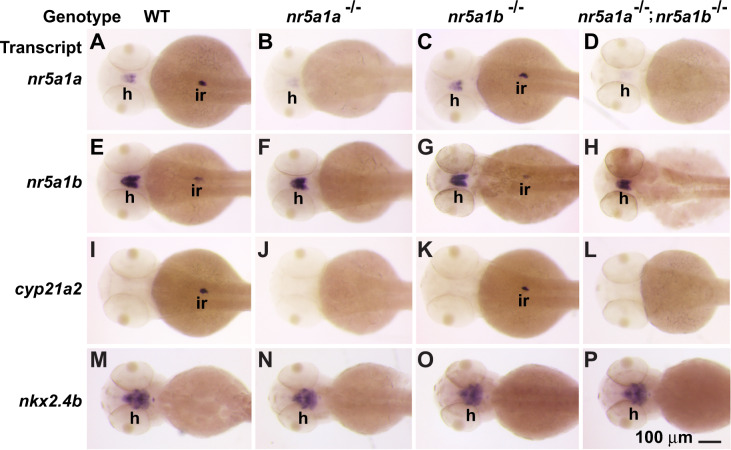
To verify scRNA-seq results, we performed in situ hybridization experiments in wild types. Results showed that at 48 hpf (hours postfertilization), nr5a1a and nr5a1b were both expressed in the hypothalamus and in the left-biased interrenal (Figure 3, A and E), confirming previously published results (von Hofsten et al. 2001; Chai et al. 2003; Hsu et al. 2003; Kuo et al. 2005; Kurrasch et al. 2007; Chai and Chan 2000; von Hofsten et al. 2002; Liu and Guo 2006; Wang et al. 2007; Chiu et al. 2012; Nakamoto et al. 2012). The mammalian adrenal enzyme Cyp21a2 converts progesterone to an aldosterone precursor (Miller and Auchus 2011), although teleosts lack aldosterone (Jiang et al. 1998; Bridgham et al. 2006). Expression of cyp21a2 in zebrafish (Weger et al. 2018) confirmed that the abdominal expression domain of nr5a1 ohnologs is the interrenal (Figure 3I), consistent with the results of scRNA-seq (Figure 1E). To identify nr5a1-expressing cell types in the brain, we studied the hypothalamus marker genes nkx2.4b (see also Figure 1M for scRNA-seq), nkx2.4a, and nkx2.1 (Rohr et al. 2001; Kurrasch et al. 2007, 2009). These nkx2 paralogs were expressed in domains that encompassed those of nr5a1 ohnologs (Figure 3M and Supplementary Figure S2), and hence confirm the expression of nr5a1a and nr5a1b in the hypothalamus. In 48 hpf wild-type embryos, nr5a1a expression was more intense in the interrenal than in the hypothalamus, but the reverse was true for nr5a1b (Figure 3, A and E).
Figure 3.
Gene expression patterns in embryos at 48 hpf. (A, E, I, M) Wild-type embryos. (B, F, J, N) nr5a1a−11 mutant embryos. (C, G, K, O) nr5a1b−8 mutant embryos. (D, H, L, P) nr5a1a−11; nr5a1b−8 double mutant embryos. In situ hybridization for nr5a1a (A, B, C, D), nr5a1b (E, F, G, H), cyp21a2 (I, J, K, L), nkx2.4b (M, N, O, P). interrenal development depends on nr5a1a, but hypothalamus expression of nkx2-family marker genes is not strongly affected in either single mutant or double mutants. Based on three clutches, each containing 20–25 wild types, 42–48 heterozygotes, 19–24 nr5a1a mutants, 18–27 nr5a1b mutants, and 8–12 double mutant embryos. h, hypothalamus; ir, interrenal. Scale bar in P represents 100 μm.
Gene expression in nr5a1a mutants
Expression of nr5a1a transcript virtually disappeared from the interrenal and was far lower in the hypothalamus in nr5a1a mutants compared to wild-type siblings hybridized in the same experiment (Figure 3B). This result suggests that nr5a1a function is required to maintain nr5a1a-expressing cells or that the message is unstable. Transcripts from nr5a1b also disappeared from the interrenal in nr5a1a mutants but were merely reduced in the hypothalamus (Figure 3F). Greatly decreased expression of the interrenal marker cyp21a2 confirmed defective interrenal development in nr5a1a mutants (Figure 3J). In contrast, nr5a1a mutants showed no obvious expression changes for hypothalamus markers (nkx2.4b, Figure 3N; nkx2.4a, nkx2.1, Supplementary Figure S1). We conclude that nr5a1a is necessary for normal development of the embryonic interrenal but seems less important for hypothalamus gene expression patterns, confirming morpholino studies (Hsu et al. 2003).
Gene expression in nr5a1b mutants
Expression patterns of nr5a1a, nr5a1b, and nkx2.4b in the hypothalamus of 48hpf nr5a1b mutants were similar to those in wild types (Figure 3, A, C, E, G, M, and O), providing no evidence that nr5a1b strongly affects the development of the hypothalamus despite its expression in the hypothalamus as early as 1 dpf (Figure 1, C, E, and L). In the interrenal, nr5a1b mutants had only slightly reduced expression of nr5a1a, nr5a1b, or cyp21a2 (Figure 3, A, C, E, G, I, and K). We conclude that nr5a1b appears to be less important for interrenal development than nr5a1a.
Gene expression in double mutants
In brief, 48 hpf embryos lacking both nr5a1a and nr5a1b activity (Figure 3, D, H, L, and P) lacked the expression of genes marking interrenal development but had little effect on the expression of nr5a1a, nr5a1b, or nkx2.4b in the hypothalamus (Figure 3, B, F, J, and N). We conclude that at 48 hpf, nr5a1a is important for interrenal development and that neither nr5a1a nor nr5a1b is required for apparently normal expression of examined hypothalamus marker genes (Supplementary Figure S2).
nr5a1 Ohnologs in juvenile gonad development
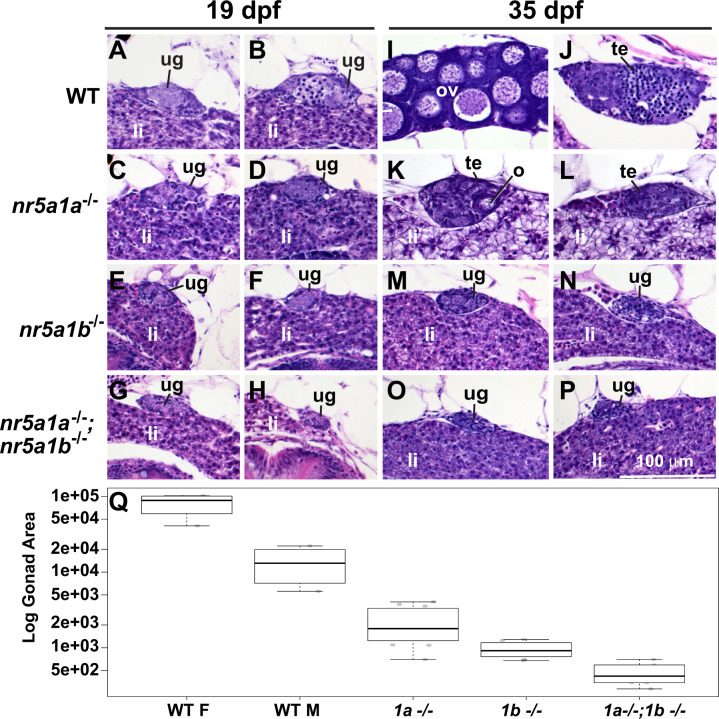
To learn the roles of nr5a1 ohnologs in juveniles, we made histological sections of wild types and mutants (Figure 4). Gonads in AB strain wild-type juveniles at 19 dpf are undifferentiated, but they are beginning to transition from the juvenile ovary stage to differentiated gonads (Takahashi 1977; Uchida et al. 2002; Rodriguez-Mari et al. 2005; Orban et al. 2009) (Figure was 4, A and B). Mutant gonads at 19 dpf lacking nr5a1a activity (Figure 4, C and D) were somewhat smaller than wild-type gonads and were undifferentiated. Gonads in nr5a1b mutants were even smaller (Figure 4, E and F), and gonads in double mutants were smaller still (Figure 4, G and H). These results show that both zebrafish nr5a1 ohnologs are required for juvenile gonads to reach normal size.
Figure 4.
Histology of gonads in 19 and 35 dpf wild-type and nr5a1-mutant fish. (A–H) 19 dpf. (I–P) 35 dpf. In 19-dpf juveniles, gonads in all (8/8 fish) wild-type fish (WT) were undifferentiated (e.g., A, B). Gonads in all nr5a1a−11 mutants (6/6 fish) (e.g., C, D), all nr5a1b−8 mutants (6/6 fish) (e.g., E, F) and in all (4/4 fish) nr5a1a−11; nr5a1b−8 double mutants (e.g., G, H) were undifferentiated like wild-type gonads but all mutant gonads were smaller than wild-type sibling gonads, especially in nr5a1b−8 mutants and nr5a1a−11; nr5a1b−8 double mutants (e.g., E–H). At 35-dpf (I–P), wild-type fish contained gonads that were clearly either ovaries (4/4 fish) (e.g., I) or testis (4/4 fish) (e.g., J). In 35-dpf nr5a1a−11 mutants, all fish examined had smaller testis (13/13 fish) (e.g., K, L, Q); In 35-dpf nr5a1b−8 mutants (8/8 fish), gonads were still undifferentiated (e.g., M, N, Q); In 35-dpf nr5a1a−11; nr5a1b−8 double mutants, gonads were still undifferentiated and were even smaller than earlier (4/4 fish) (e.g., O, P, Q). (Q), Log cross-sectional area of the gonad in arbitrary units for 35-dpf wild-type females (4/4 fish) and males (4/4 fish); for 35-dpf nr5a1a−11 mutants (13/13 fish); 35-dpf nr5a1b−8 mutants (8/8 fish), and 35-dpf nr5a1a−11; nr5a1b−8 double mutants (4/4 fish). Gonad sizes were significantly smaller in nr5a1 mutants compared to wild-type siblings. Outer bars, minimum and maximum; thick bar, median; lower box edge, first quartile; upper box edge third quartile. li, liver; ov, ovary; te, testis; ug, undifferentiated gonad. Statistical significance: different letters (a–e) signify differences at P < 0.05. Scale bar in P is 100 µm for all panels.
Gonads in postmetamorphic wild types at 35 dpf had differentiated into either ovaries with stage I oocytes (Figure was 4I) or testes with tubules containing developing spermatogonia (Figure 4J). In contrast, gonads in 35dpf nr5a1a mutants remained small, and although they formed immature oocytes like juvenile wild types, these oocytes did not mature; nr5a1a mutant gonads appeared to be on a delayed pathway to become a small testis (Figure 4, K, I, L, and Q). At 35 dpf, nr5a1b mutant gonads remained undifferentiated and were no larger than those in 19 dpf wild types (Figure 4, M and N). Gonads in 35 dpf double-mutant fish were smaller than in either single mutant (Figure 4, O–Q). We conclude that Nr5a1 action is necessary for ovary development and gonad growth. Furthermore, each nr5a1 ohnolog has independent functions in gonad development because the double-mutant phenotype is more severe than either single-mutant phenotype, and because nr5a1a is required to form ovaries but not testes, whereas nr5a1b is required for gonad differentiation in either sex-specific direction.
nr5a1 Ohnologs and secondary sex characteristics
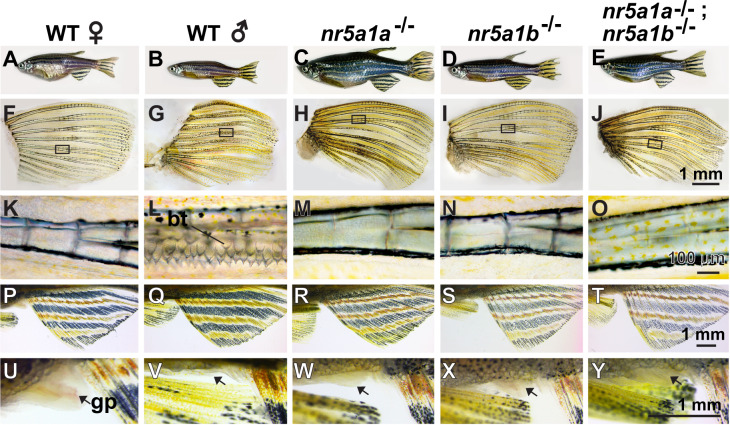
To evaluate the roles of nr5a1 ohnologs in adults, we examined secondary sex characteristics at 8 mpf (months postfertilization). All three mutant genotypes had body shapes more similar to wild-type males than wild-type females, but coloration was not strongly masculine (Figure was 5, A–J). Wild-type males, but not females, had breeding tubercles (Figure 5, F, G, K, and L) (McMillan et al. 2015), but mutants for either or both nr5a1 ohnologs lacked breeding tubercles (Figure was 5, H–J and M–O). Females have a longer genital papilla than males do (Figure 5, U and V) (McMillan et al. 2015), but mutants for either or both nr5a1 ohnologs had genital papillae of intermediate size (Figure 5, W–Y). We conclude that secondary sex characteristics were absent or ambiguous in nr5a1 mutants.
Figure 5.
Secondary sex characteristics. (A–E) 8-mpf adult zebrafish. (A) Wild-type female. (B) Wild-type male. (C) nr5a1a−11 mutant. (D) nr5a1b−8 mutant. (E) nr5a1a−11; nr5a1b−8 double mutant. Among mutants, body shape was male-like but body color was not. (F–O) Pectoral fin of 8-mpf fish. (F) wild-type female, (G) wild-type male, (H) nr5a1a−11 mutant, (I) nr5a1b−8 mutant, and (J) nr5a1a−11; nr5a1b−8 double mutant. (K–O) Boxed region of pectoral fin in higher magnification, showing breeding tubercles (bt) in (I) wild-type male but not in (K) wild-type female or in any nr5a1 mutants (M–O). (P–T) Anal fins dissected from (P) wild-type female, (Q) wild-type male, (R) nr5a1a−11 mutant, (S) nr5a1b−8 mutant, and (T) nr5a1a−11; nr5a1b−8 double mutant. (U–Y) Genital papilla (gp, arrows) located anterior to the anal fin. (U) Long genital papilla in wild-type female compared to (V) wild-type male and (W–Y) nr5a1 mutants. Based on five fish of each genotype, a total of 25 fish. bt, breeding tubercles; gp, genital papilla. Scale bar in J for F–J: 1 mm; scale bar in O for K–O: 100 μm; scale bar in T for P–T; scale bar in Y for U–Y: 1 mm.
nr5a1 Ohnologs and body size
By 11 mpf, nr5a1 mutants were enormous compared to co-housed wild-type siblings. At 8 mpf, nr5a1a mutants were three times the mass of wild-type males and 35% longer (Figure 5A vs. C, Supplementary Figure S3). The nr5a1b mutants were somewhat larger than normal males, as they were double mutants. Body cavities of nr5a1b mutants were filled with fat cells and had abnormal gonads or no gonad at all. It is possible that when free cholesterol is not used for making steroid hormones, the excess becomes esterified and stored in lipid droplets (Hatano et al. 2016). The lack of the stress hormone cortisol might also contribute to more efficient calorie usage and hence more growth.
nr5a1 Ohnologs and adult gonadogenesis
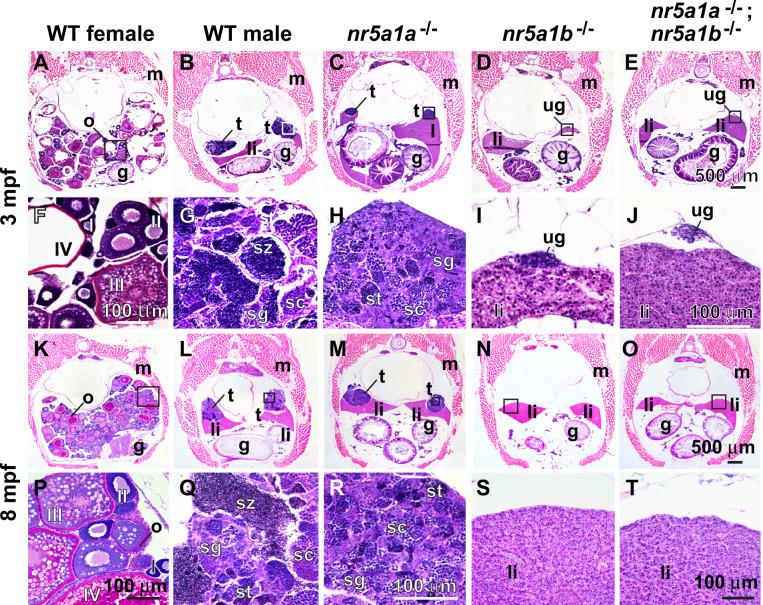
Given the ambiguous sex phenotypes of nr5a1 mutants, we wondered about adult gonads. Adult gonads of 3-mpf nr5a1a mutants contained testes tubules with a few spermatocytes, but they lacked mature spermatozoa already possessed by wild-type male siblings (Figure 6, B, C, G, and H). At 8 mpf, nr5a1a mutant gonads had still not developed any mature spermatozoa (Figure was 6, L, M, Q, and R). We conclude that nr5a1a is necessary for the maintenance of oocytes, for female development, and for sperm maturation.
Figure 6.
Histology of gonads in adult zebrafish. (A–J) 3-mpf adult zebrafish. (A, F) wild-type female, (B, G) wild-type male. (C, H) nr5a1a−11 mutant (n.8). (D, I) nr5a1b−8 mutant (n.5). (E, J) nr5a1a−11; nr5a1b−8 double mutant (n.4). (K–T) 8-mpf adult zebrafish. (K, P) Wild-type female, (L, Q) wild-type male, (M, R); nr5a1a−11 mutant (n.8). (N, S); nr5a1b−8 mutant (n.8). (O, T) nr5a1a−11; nr5a1b−8 double mutant (n.4). (A–E) Low magnification at 3 mpf, (F–J), high magnification of boxed region in AE at 3 mpf (K–O), low magnification at 8 mpf (P–T), high magnification of boxed region at (K–O) at 8 mpf. Crosssections of 3mpf (A, F) and 8-mpf wild-type female siblings (K, P) show maturing (stage-I and -II) and vitellogenic (stage-III and -IV) follicles. Wild-type male siblings at 3mpf (B, G) and 8 mpf (L, Q) had formed all spermatogenic stages. Mutant nr5a1a males at 3 mpf at low (C), and high magnification (H) show that older animals had developed more immature sperm and spermatocytes, compared to wild-type siblings (B, C, G, H). I, II, III, IV: ovarian follicle stages 1–4; g, gut; li, liver; o, ovary; s, Sertoli cells; sc, spermatocytes; sg, spermatogonia; st, spermatids; sz, spermatozoa; t, testis. Scale bar in (E) for (A–E): 500 μm; scale bar in (J) for (F–J): 100 mm; scale bar in (O) for (K–O): 500 mm; scale bar in (R) for (P–R): 100 μm; scale bar in (T) for (S) and (T): 100 μm.
The gonadal phenotype of nr5a1b mutants was more severe than that of nr5a1a mutants. At 3 mpf, gonads in nr5a1b mutants continued to be small and undifferentiated (Figure 6, D and I) like they were at 35 dpf. By 8 mpf, nr5a1b mutants, remarkably, had no gonad at all (Figure 6, N and S). The gonadal phenotype of double mutants was similar to that of nr5a1b mutants (Figure 6, E, J, O, and T). We conclude that nr5a1b function is required for gonad differentiation, growth, and maintenance.
To test the fertility of nr5a1a mutants, we set up single pair crosses between mutants and wild-type females. All six single-pair control crosses of 4.5 mpf wild-type male siblings mated to AB strain wild-type females produced eggs with an average of 150 ± 114 (SD) viable offspring among 173 ± 121 deposited eggs with 83%±11 (SD) eggs, developing per clutch. In contrast, 14 crosses of 4.5 mpf nr5a1a mutants produced no eggs in any cross. Similarly, crosses of 11-mpf wild-type siblings produced an average of 150 ± 35 (SD) viable offspring among 155 ± 36 deposited eggs, with 97%±2 (SD) of the eggs developing; in contrast, 10 crosses of 11-mpf nr5a1a mutants to AB females yielded no eggs. These experiments are consistent with the results of histology, which showed that nr5a1a mutants lacked mature sperm (Figure 6), and furthermore show that nr5a1a mutants did not stimulate wild-type females to lay eggs, consistent with their lack of male-breeding tubercles and other masculine traits (Figure 5).
Gene expression in nr5a1 mutant adult gonads
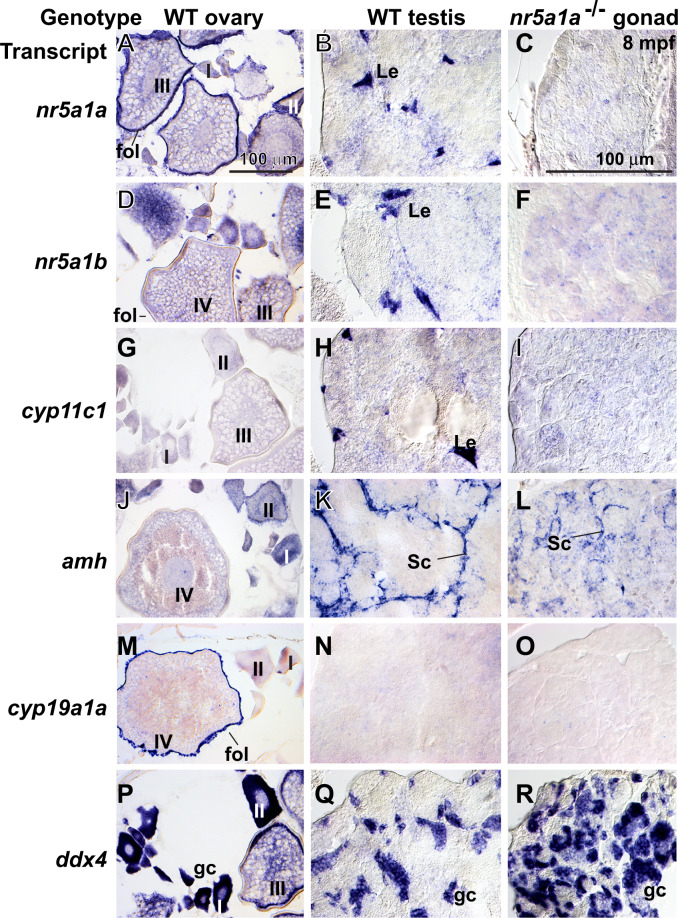
To help interpret adult mutant phenotypes, we studied gene expression patterns. In 8-mpf wild-type adults, granulosa and theca cells in ovaries and Leydig cells in testes expressed nr5a1a (Figure 7, A and B) and young oocytes and Leydig cells expressed nr5a1b (Figure 7, D and E) (Kuo et al. 2005; Quek and Chan 2009; Yan et al. 2019). Testes in nr5a1a mutants did not express nr5a1a or nr5a1b or the Leydig cell marker cyp11c1 (Wang and Orban 2007) (Figure , G–I). We conclude that each nr5a1 ohnolog is required for Leydig cell development. In wild types, Sertoli cells express amh (Rodriguez-Mari et al. 2005) (von Hofsten et al. 2005a; Schulz et al. 2007; Yan et al. 2017, 2019) (Figure K). In nr5a1a mutant gonads, amh expression appeared in fewer, less organized cells than in wild types (Figure 7, K and L), consistent with the loss of normal Leydig cells. The granulosa cell marker cyp19a1a is not expressed in normal testes (Chiang et al. 2001a,b) and was not expressed in nr5a1a mutant gonads (Figure 7, M–O). Expression of the germ cell marker ddx4 (vasa) (Yoon et al. 1997) showed that nr5a1a mutant testis tubules contained a substantial number of disorganized germ cells. The nr5a1b mutants had no gonads at 8 mpf, precluding investigation of gonadal gene expression. These in situ hybridization experiments show that nr5a1a is necessary for the development of Leydig cells and secondarily for the organization of amh-expressing Sertoli cells.
Figure 7.
Gene expression patterns in adult gonads at 8 mpf. (A, D, G, J, M, P) Wild-type ovaries. (B, E, H, K, N, Q) Wild-type testis. (C, F, I, L, O, R) nr5a1a−11 mutant testis. In situ hybridization using probes for (A, B, C) nr5a1a, (D, E, F) nr5a1b, (G, H, I) cyp11c1, (J, K, L) amh, (M, N, O) cyp19a1a, and (P, Q, R) ddx4. Scale bar in A (for all wild-type ovaries) and scale bar in C (for all testis panels) represents 100 μm. I, II, III, IV: ovarian follicle stages 1–4; fol, follicle cell; gc, germ cell; Le, Leydig cell; Sc, Sertoli cell.
Development of the interrenal in nr5a1 mutants
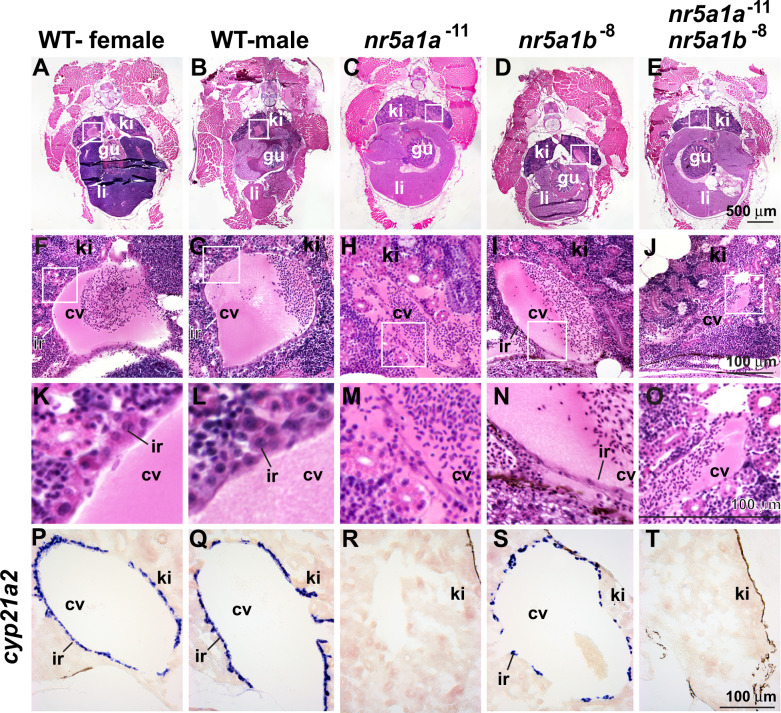
Because in situ hybridization experiments (Figure 3) showed that nr5a1 paralogs are expressed in the embryonic interrenal, we investigated this organ in adults. interrenal cells express nr5a1 paralogs, reside within the head kidney, and occupy a cell layer surrounding the posterior cardinal veins in male and female zebrafish (Figure 8, A, B, F, G, K, and L) (Chester Jones and Mosley 1980; Hsu et al. 2003; Menke et al. 2011; Eachus et al. 2017). The interrenal produces corticosteroids using the interrenal-specific gene cyp21a2 (Eachus et al. 2017) (Figure 11, P and Q). In nr5a1a adult mutants, however, histology failed to show any interrenal cells around the cardinal vein (Figure 8, C, H, and M) confirmed by in situ hybridization experiments that showed no expression of cyp21a2 (Figure 8R), mimicking 2-dpf nr5a1a mutant embryos (Figure is 3, B, F, and J). In contrast, nr5a1b mutants had interrenal cells visible in histological sections and cyp21a2-expressing cells surrounding the cardinal vein although they were fewer and more scattered than in wild types (Figure 8, D, I, N, and S). Double mutants, like nr5a1a mutants, also appeared to lack an interrenal, (Figure 8, E, J, O, and T). We conclude that nr5a1a action is necessary for the development of the interrenal and that nr5a1b function is required for fully normal interrenal development.
Figure 8.
Nr5a1 activity is required for normal interrenal morphology in adult zebrafish. (A–O) Cross-sections of 8-mpf adult zebrafish histology and (P–T) in situ hybridization for cyp21a2. (A, F, K, P) Wild-type females. (B, G, L, Q) Wild-type males. (C, H, M, R) nr5a1a−11 mutants. (D, I, N, S) nr5a1b−8 mutants. (E, J, O, T) nr5a1a−11; nr5a1b−8 double mutants. Histological sections of anterior trunk: (A–E) at low magnification; white box expanded in (F–J) medium magnification; and white box expanded in (K–O) high magnification. (P–T) In situ hybridization of cyp21a2 to interrenal cells. cv, cardinal vein; gu, gut; ir, interrenal; li, liver; ki, kidney. Scale bar in (E) for (A–E); scale bar in J for F–J; scale bar in (O) for (K–O); scale bar in (T) for (P–T). All scale bars: 100 μm. Based on five fish of each genotype for morphology (25 fish), and four fish of each genotype for gene expression (20 fish).
Endocrinology of nr5a1 mutants
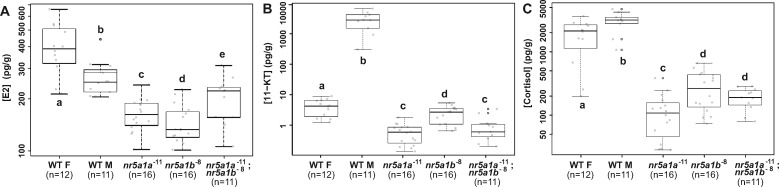
The analysis of secondary sexual characteristics, gonad development, interrenal morphologies, and gene expression all suggested profoundly aberrant steroid biosynthesis in nr5a1 mutants. To determine which hormone levels are regulated by which nr5a1 ohnolog, we assayed estrogen (E2), 11-keto testosterone [11-KT, the primary fish androgen (Borg 1994)], and cortisol [the primary corticosteroid in actinopterygian fishes (Sangalang et al. 1971; Idler and Truscott 1972; Hanson and Fleming 1979; Barton et al. 1998)]. Tests assayed entire bodies of individual wild-type and mutant adults at 4.5 and 12 mpf. Assays showed that estrogen levels in 4.5-mpf wild-type females were far greater than those in wild-type males, but that estrogen levels in single and double nr5a1 mutants were substantially lower than in either wild-type sex (Figure 9A). Levels of 11-KT were several hundred times lower in wild-type females than in wild-type males, and interestingly, were still lower in both single-mutant genotypes and in double mutants (Figure 9B). Cortisol concentrations were about 70% higher in wild-type males than in wild-type females and, on average, cortisol levels in single and double nr5a1 mutants were only about 7% of those in wild-type males, with nr5a1a mutants being more strongly affected than nr5a1b mutants (Figure 9C). Consistent with in situ hybridization and histology data, nr5a1a mutants had less than half the concentration of cortisol as found in nr5a1b mutants (Figure 9C). The results for 12-mpf fish were similar to those for 4.5-mpf fish (Supplementary Figure S4). We conclude that both zebrafish co-orthologs of NR5A1 are critical for normal levels of estrogen and 11-keto testosterone, and owing to the low levels of both sex steroids, mutant fish are neither masculinized nor feminized. Cortisol assays showed that both nr5a1 co-orthologs are important to maintain cortisol levels, but that nr5a1a is more important than nr5a1b in mature adult zebrafish (Figure 9C).
Figure 9.
Loss of function of nr5a1 ohnologs disrupts whole-body concentrations of sex steroids and cortisol at 4.5 mpf (log scale). (A) Estradiol (E2, pg/g wet weight). (B) 11-Keto testosterone (11-KT) (pg/g wet weight). (C) Cortisol (pg/g wet weight). WT F (wild-type females, n.12); WT M (wild-type males, n.11); nr5a1a−11 (n.11); nr51b−8 (n.16); nr5a1a−11; nr5a1b−8 double mutants (n.11). Different letters (a–e) signify statistically different groups at P < 0.05.
Transcriptomic analysis of nr5a1 mutants
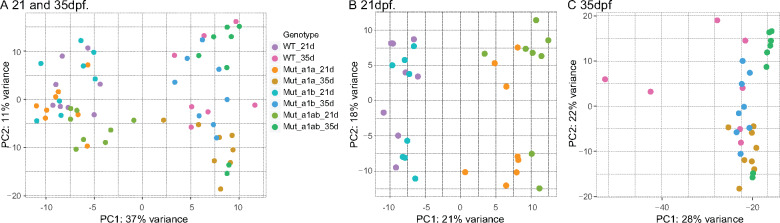
To understand how nr5a1 ohnologs regulate the expression of other genes, we made RNA-seq libraries from fish trunks, which included interrenal, gonad, liver, and other viscera. We made 64 sequencing libraries, encompassing four homozygous genotypes (wild types, nr5a1a mutants, nr5a1b mutants, nr5a1ab double mutants), two ages (21 and 35 dpf), and eight individuals for each time point and genotype although one 35-dpf wild-type sample failed. Two-way similarity clustering by regularized log-transformed Euclidean distances and principal component analysis separated all 21-dpf animals from all 35-dpf samples in the PC1 axis, which explained 37% of the variance (Figure 10A). This result is consistent with the histological analyses (Figure 4), showing substantial changes in gonads during this period. In the PC2 axis, all nr5a1a mutants at 35 dpf and all double mutants at 21 dpf were in the lower half of the plot (Figure 10A), suggesting that interrenal functions were driving PC2.
Figure 10.
Principal component analysis of RNA-seq data for juvenile and young adult wild-type and nr5a1 mutants. Each dot represents a different individual fish. (A) Analysis of all samples together. (B) Analysis of 21-dpf samples. (C) Analysis of 35-dpf samples. WT, wild type; 1a, nr5a1a mutants; 1b, nr5a1b mutants; 1ab, nr5a1a; nr5a1b double mutants; 21d, 21 dpf; 35d, 35 dpf.
Transcriptomics of 21-dpf zebrafish juveniles
At 21 dpf, principal component analysis separated the 32 samples into approximately four quadrants (Figure 10B).
The PC1 axis separated individuals into two groups: Group21L on the left in Figure 10B, containing wild types plus nr5a1b mutants, vs Group21R on the right, containing nr5a1a mutants plus double mutants (Figure 10B). Genes differentially over-expressed in Group21L versus Group21R included the “female” genes zp2.2 (zona pellucida-2.2, 7216-fold up) and vtg5 (vitellogenin, 21-fold up); the steroid biosynthesis genes cyp17a1 (13-fold up), hsd3b1 (12-fold up); the interrenal gene cyp21a2 (6-fold up); and nr5a1a (2.5-fold up). We conclude that: (1) Group21L individuals had more advanced ovary development than Group21R; (2) Group21L juveniles were expressing interrenal steroid biosynthesis genes but Group21R juveniles had not formed a functioning interrenal; (3) the nr5a1a phenotype drives the double-mutant phenotype because these two genotypes associated together in Group21R in PC1; and (4) at 21 dpf, the gonadal phenotype of nr5a1b in the context of the entire trunk is not distinct enough to distinguish its transcriptome from wild types.
The PC2 axis at 21 dpf also separated samples into two groups, each of which had some representatives from all four genotypes (Figure 10B), suggesting that some factors other than genotype was driving the PC2 axis. The six most over-expressed genes in Group21B (bottom) compared to Group21T (top) were the uncharacterized genes si:ch211-250e5.16, zgc:175135, zgc:171977, CU467646.2, zgc:173544, plus zp2.2, which were over-expressed from 9,005 to 11,957-fold. Because wild-type adult ovaries over-expressed this gene set vs wild-type adult testes from 938 to 3,159-fold (Yan et al. 2019), we conclude that Group21B animals had initiated development as females but Group21T juveniles had not. This finding reflects a similar situation in a study of amh mutants and wild types in which transcriptomes from similar 21-dpf trunks separated samples into developing female vs not-female groups, both of which contained both amh mutants and wild types (Yan et al. 2019). We conclude that in an AB genetic background, some 21-dpf individuals have already embarked on a female developmental pathway without respect to the action of either nr5a1a (Figure 0B) or amh (Yan et al. 2019).
nr5a1a mutants compared to wild-type siblings at 21 dpf had 1071 DE genes (Padj < 0.1, Supplementary Table S3). Regarding genes over-expressed in nr5a1a mutants, the top five genes all encode uncharacterized proteins (zgc:175135, si:ch73-160p18.3, si:dkeyp-46h3.8, zgc:171750, si:dkey-229d11.3; from 48,846- to 94,500-fold up) and were also greatly over-expressed in adult wild-type ovaries versus adult wild-type testes (1,045- to 3,159-fold) (Yan et al. 2019), suggesting that some ovary functions are inhibited by the normal allele of nr5a1a at 21 dpf. The first over-expressed known gene was fosb (23-fold up), and it was paradoxically under-expressed (63-fold) in adult wild-type ovaries versus testes (Yan et al. 2019), whereas fosab, fosl1a, and jdp2b were also among the first 33 most over-expressed genes. FOS family genes help regulate the levels of mRNA for both Star and Cyp19a1 in mammalian ovaries and hence the rate of estrogen production (Beshay et al. 2007; Manna et al. 2009; Patel et al. 2009). Other over-expressed genes included sox21a, which is up-regulated 7.9-fold by the oocyte maturation-inducing steroid MIS (Klangnurak and Tokumoto 2017), and metabolism-related genes including the fatty acid elongase gene elovl7b, glucokinase gck, and the potential diabetes-related hepatic gluconeogenesis regulators foxq1a, and foxq1b (Cui et al. 2016), all among the 25 most over-expressed genes. This result suggests metabolic compensation for altered cortisol and sex steroid metabolism in nr5a1a mutants.
Regarding under-expressed genes in 21dpf nr5a1a mutants, results identified the ovarian follicle tight junction gene cldnd (15,049-fold down) and the retinoic acid signaling gene retsatl (6,208-fold down) (Sreenivasan et al. 2008), consistent with disrupted ovary organization and retinoic acid control of meiosis. Additional nr5a1a under-expressed genes in the top 11 included star (54-fold down), whose promotor is enhanced by Nr5a1 in mouse (Yang et al. 2010), the steroidogenic enzyme cyp17a1 (20-fold down), and the estrogen-responsive liver-expressed yolk protein gene vtg5 (13-fold down). The interrenal gene cyp21a2 trended downward (6-fold) in nr5a1a mutants (Pval = 0.00013). We conclude that at 21 dpf, Nr5a1a is necessary for inhibiting some ovarian developmental steps and for the production of steroid hormones, likely from both the gonad and the interrenal, supporting data from histology and in situ hybridization studies (Figures 4 and 8).
Gene Ontology clustering for 21dpf nr5a1a mutants compared to wild-type siblings identified metabolic processes as the top three clusters (glycerol biosynthetic process, cellular glucose homoeostasis, and gluconeogenesis). These clusters included pck1 and pck2, which encode phosphoenolpyruvate carboxykinase enzymes that help control gluconeogenesis. The fourth cluster was “response to glucocorticoid.” These findings suggest that disturbed interrenal development in nr5a1a mutants greatly altered metabolism throughout the trunk. Continuing alteration of metabolism likely led to the enormous body size and fat cell accumulation in nr5a1a mutants at 4.5 and 11 mpf (Supplementary Figure S3).
nr5a1b Mutants vs. wild-type siblings at 21 dpf had 78 DE genes (Supplementary Table S4). Regarding over-expressed genes, the second most differentially expressed gene (CABZ01059627.1) was also over-expressed in adult wild-type ovaries versus testes (1,713-fold). The oocyte gene ca15b (Wang et al. 2013) was massively over-expressed [234,946,505-fold up in nr5a1b vs wild types and 1,337-fold up in wild-type adult ovaries vs. wild-type testes (Yan et al. 2019)]). The fifth most over-expressed gene encodes an egg coat protein (zp2.2, 9,474,925-fold up). As in nr5a1a mutants, fosb was high on the list of over-expressed genes in nr5a1b mutants (10-fold). Regarding under-expressed genes in 21-dpf nr5a1b mutants, the most under-expressed was the ovary gene cldnd, mimicking nr5a1a mutants, with cyp17a1 in seventh place. This mix of ovary genes both over- and under-expressed in nr5a1b mutants vs wild types suggests that gonads had initiated oocyte development in nr5a1b mutants at 21,dpf, but that regulation of oogenesis was greatly disturbed. Gene Ontology clustering identified no significant DE clusters, perhaps because of the low number of DE genes.
nr5a1a; nr5a1b Double mutants vs wild types at 21 dpf had 2,218 DE genes (Supplementary Table S5). Regarding over-expressed genes in double mutants, 6 of the top 22 were also among the 22 most over-expressed genes comparing nr5a1a mutants to wild types (si:ch73-160p18.3, si:dkeyp-46h3.8, si:ch211-125e6.5, gck, sox21a, zgc:153642) and seven comparing nr5a1b mutants to wild types (zgc:171446, CABZ01059627.1, ca15b, zgc:175135, si:ch73-160p18.3, si:dkeyp-46h3.8, nr0b2b). Several of these genes were greatly over-expressed in wild-type ovaries versus wild-type testes (Yan et al. 2019), suggesting that at 21 dpf, double mutants had begun to embark on a female pathway. Under-expressed genes in double mutants included some female genes, like the yolk protein gene vtg5 (35-fold down), consistent with low levels of estrogen. Some “male” genes were also under-expressed, including gsdf (21-fold down), the male sex determinant in some fish (Myosho et al. 2012; Rondeau et al. 2013), suggesting disruption of the male developmental pathway. Of the 22 most under-expressed genes in double mutants, 12 (nr1d2a, si:dkey-242g16.2, cdkn1d, muc5.2, vtg5, zgc:153932, si:ch211-89o9.4, cyp17a1, klf9, cyp2k6, fkbp5, star, cldnd) were also among the 22 most under-expressed genes in nr5a1a mutants, but only one (cyp17a1) was also among the 22 most under-expressed in nr5a1b mutants, suggesting that nr5a1a has a stronger effect in double mutants. Gene Ontology clustering for 21-dpf double mutants identified the same four metabolic GO clusters as for nr5a1a mutants.
Transcriptomics of 35-dpf zebrafish young adults
Principal component analysis of 35-dpf fish (Figure 10C) showed that in the PC1 axis, three of the seven wild types were further to the left than any other fish; the other four wild types nested with single-mutant individuals in PC1; and six of the eight double mutants clustered at the far right. This result suggests that individuals sorted along PC1 according to the level of gonad maturation and interrenal function. The broad dispersion of wild types along PC1 suggests that different wild-type individuals may have varied greatly in gonadal maturation. Along PC2, all nr5a1a mutants were in the lower half of the plot.
nr5a1a mutants at 35 dpf had 1,404 DE genes vs wild-type siblings (Supplementary Table S6). Regarding over-expressed genes, human orthologs of several highly over-expressed genes (ly6m6, adm2b, nxph2b; 8- to 15-fold) have their strongest expression in the human ovary (Fagerberg et al. 2014). In contrast, other genes over-expressed in mutants, including the steroid-breakdown gene cyp2k6 (11-fold) (Wang-Buhler et al. 2005), and several unstudied genes (si:ch211-219a15.4, si:ch211-161h7.5, si:ch211-114l13.3; 6- to 8-fold up) were under-expressed in wild-type ovaries versus wild-type testes (7- to 11-fold down) (Yan et al. 2019). Regarding under-expressed genes in 35 dpf nr5a1a mutants, the top gene was zgc:175135, which was, in contrast, the most over-expressed gene in this genotype at 21 dpf. Because zgc:175135 is over-expressed (1,081-fold) in normal ovaries versus testes (Yan et al. 2019), this finding likely reflects the presence of immature oocytes at 21 dpf that were dying or de-differentiating at 35 dpf. The fourth most under-expressed gene in 35dpf nr5a1a mutant trunks was ccna1, which in mouse is necessary exclusively for meiosis in male germ cells (Liu et al. 1998), suggesting a decrease in male meiosis in 35 dpf nr5a1a mutants. Several oocyte genes were also under-expressed (e.g., bucl2, buc, and wee2 (10- to 132-fold), and many zona pellucida genes (e.g., zp3.2, 66-fold). Many steroid biosynthesis genes were also under-expressed in 35-dpf nr5a1a mutants (cyp11a1, cyp17a1, star, hsd3b1, cyp11c1, cyp11a2, cyp17a2, 14- to 95-fold down) including the interrenal gene cyp21a2 (7-fold down). Gene Ontology clustering for 35dpf nr5a1a mutants vs wild types identified 35 functional clusters with under-expression of corticosteroid genes dominating the top three categories. The fourth cluster, “long-chain fatty-acyl-CoA biosynthetic process,” contained five genes encoding enzymes that elongate very long fatty acids (Elovl), possibly associated with the accumulation of fat in older nr5a1a mutants. We conclude that gene expression patterns in nr5a1a mutants showed a chaotic mix of processes involved in shutting down the gonad and interrenal between 21 and 35 dpf in nr5a1a mutants and altering energy metabolism.
nr5a1b Mutants at 35 dpf compared to wild types had 260 DE genes. Regarding over-expressed genes, the four strongest (rfesd, mylpfb, oacyl, si:ch211-161h7.5; 4- to 8-fold up) are also over-expressed in testes relative to ovary [7- to 14-fold up (Yan et al. 2019)], suggesting that oocytes were disappearing in nr5a1b mutants as gonads became testes. Regarding under-expressed genes, fold changes for the lowest 10 for nr5a1b mutants versus wild types were massive [log2fold of -26 to -49 for cldnd, zar1l (zygote arrest 1 like), and many uncharacterized genes; Supplementary Table S7]; these genes were all over-expressed in wild-type ovary versus testis by an average of 1737-fold (Yan et al. 2019). Additional under-expressed genes in nr5a1b mutants included other oocyte-expressed genes (ddx4, bmp15, dazl, nanog, and several zona pellucida genes, from 37- to 584-fold down), in contrast to under-expressed genes in nr5a1a mutants, which included many steroid biosynthesis genes. Gene Ontology clustering identified as top clusters several oocyte functions [“binding of sperm to zona pellucida,” “positive regulation of acrosome reaction,” and “egg coat formation”]. This result is consistent with histology, which suggested that a main role of nr5a1b is to support oocyte development rather than interrenal steroid biosynthesis.
Direct comparisons of nr5a1a mutants to nr5a1b mutants identified over-expressed ovary genes (e.g. cldnd and zar1l, log2FoldChange = 30.3 and 20.1, respectively), under-expressed steroidogenesis genes (e.g., vtg5, cyp17a1, star, fold change 12- to 58-fold), and under-expressed lipid metabolism genes (klf9, lpin1, fold change of 7- to 13-fold), verifying the role of nr5a1a in interrenal and lipid biology and nr5a1b in oocyte development.
nr5a1a; nr5a1b Double mutants at 35 dpf showed 2,219 DE genes with respect to wild-type siblings (Supplementary Table S9). Regarding over-expressed genes, the top gene (si:ch211-253p18.2) was the third most over-expressed in 35 dpf nr5a1b mutants and the third gene (si:ch211-125e6.5) was the most over-expressed in 35-dpf nr5a1a mutants. Some other genes in the top 10 (sid1, oacyl, rfesd) were also in the top 10 over-expressed genes in nr5a1a or nr5a1b vs wild types, showing that double mutants combined the effects of each single mutant. Regarding under-expressed genes, some were massively changed in double mutants at 35 dpf, including the germ cell gene ca15b (log2fold change, −37.5), in contrast to 21 dpf, when ca15b was massively over-expressed comparing double mutants to wild types (log2fold change, +28.0). The zona pellucida gene zp2.2 (log2fold change, −32.8) was also massively under-expressed in 35-dpf double mutants. Other under-expressed genes included the male meiosis gene ccna1, several endocrine genes (cyp17a1, cyp11a1), and the estrogen-responsive gene vtg5. These results suggest that double mutants had more oocytes than wild types at 21 dpf but the situation reversed by 35 dfp. One of the most consistently under-expressed genes in nr5a1a mutants and double mutants at both 21 and 35 dpf was cyp2k1 (in the 93rd to 99th percentile); its apparent human ortholog CYP2W1 is highly expressed in normal and cancerous adrenal glands (Ronchi et al. 2014). This finding is expected by the lack of the interrenal in nr5a1a-containing mutants. Gene Ontology clustering identified the top three categories as “C2 steroid hormone metabolic process,” “glucocorticoid metabolic process,” and “gluconeogenesis,” similar to nr5a1a single mutants. The fourth category, “binding of sperm to zona pellucida,” mimicked nr5a1b results, reflecting loss of both genes.
Remarkably large bodies of nr5a1a mutants (Figure 6) might be due to the mis-expression of downstream growth regulating genes. The pituitary hormone somatotropin (growth hormone, GH) regulates growth in many fish species (Canosa et al. 2007). GH binds to GH receptors (GHRs) in target cells and can stimulate the liver to produce insulin-like growth factors, which can promote growth (Picha et al. 2008; Fuentes et al. 2013). Although the large size of nr5a1 mutants does not appear until later, the trunk transcriptomes of 21 and 35 dpf nr5a1a mutants, surprisingly, under-expressed the igf-related genes igf2a, igfals, igfbp7, and igf2bp2a by 1.3- to 2.5-fold down with respect to wild types (Supplementary Tables S3 and S6). Understanding the mechanisms that lead to the continuing growth of nr5a1a mutants will require further analysis.
Discussion
Nr5a1 and the adreno-gonadal primordium
In mammals, the gonadal soma and the adrenal cortex arise from the adreno-gonadal primordium as a thickening of the celomic epithelium that expresses Nr5a1 before the activation of Sry (Ikeda et al. 1994; Hatano et al. 1996). The rostral portion of the adreno-gonadal primordium forms the anlage of the adrenal cortex (Morohashi 1997). In mouse, scRNA-seq of Nr5a1-expressing cells showed that they become Sertoli and granulosa cells using a common, non-sex-specific transcriptomic program before they acquire sex-specific identity (Stevant et al. 2019). An adreno-gonadal primordium has not been demonstrated in teleosts. Our zebrafish scRNA-seq experiments showed that nr5a1a-expressing cells are already specifically expressing gonadal and adrenal cortex genes (hsd3b1, cyp11a2, cyp21a2, nr5a1b, star, fdx1b) at 1 dpf, 3 weeks before the acquisition of sex identity. Far fewer nr5a1b-expressing cells were in this cluster as verified by in situ hybridization experiments on 2-dpf embryos (Chai and Chan 2000; von Hofsten et al. 2001; Chai et al. 2003; Hsu et al. 2003; von Hofsten et al. 2005b; Quek and Chan 2009). The mammalian adreno-gonadal primordium expresses Nr5a1 in the context of Wt1, Emx2, Six1, Six4, Gata4, Cbx2, Cited2, Odd1, and Lhx9 (Kreidberg et al. 1993; Birk et al. 2000; Schnabel et al. 2003; Katoh-Fukui et al. 2005; Buaas et al. 2009; Fujimoto et al. 2013; Hu et al. 2013). Among these genes, only six1b was DE (Supplementary Table S1) in nr5a1a- or in nr5a1b-expressing cells in zebrafish embryos, providing little support for a teleost adreno-gonadal primordium because other genes, like hsd3b1 and cyp11a2, are expressed in both organs, so at 5 dpf, nr5a1a-expressing cells might be precursors of the interrenal but not the gonad. Arguing against that interpretation is the finding that some Cluster 87 nr5a1-expressing cells in zebrafish embryos were co-expressing cxcl12a, encoding the ligand secreted by the genital ridge that guides migrating primordial germ cells expressing the receptor cxcr4b to the gonadal soma in the celomic epithelium (Boldajipour et al. 2011). In the Atlas, cxcr4b is expressed in 1-dpf germ cells (Cluster 219) as expected (Farnsworth et al. 2020). Future cell-lineage experiments are necessary learn whether the gonad and interrenal share a common origin in zebrafish and the earliest time at which the somatic component of the gonad arises, which is at least by 10 dpf because gata4 and nr5a1a are co-expressed in the somatic gonad at this time (Leerberg et al. 2017).
Nr5a1 and the hypothalamus
For nr5a1a-expressing cells, the most strongly DE genes in scRNA-seq were related to steroid biogenesis, but for nr5a1b-exressing cells, the strongest DE genes were related to the ventro-medial hypothalamus (VMH); in situ hybridization experiments verified this result. In mouse, Nr5a1 is expressed both in the adreno-gonadal primordium and in the VMH, and hence these zebrafish experiments show first, that these two roles of Nr5a1 already existed in the last common ancestor of zebrafish and mammals, and second, that after the teleost genome duplication, the two roles partitioned between the two ohnologs, with regulatory elements controlling one expression domain going to nr5a1a and the other to nr5a1b as predicted by the process of subfunctionalization (Force et al. 1999).
nr5a1a mutants and transcriptional adaptation
Some mutations that result in nonsense-mediated transcript decay exhibit genetic compensation by transcriptional adaptation, the upregulation of related genes independent of feedback loops involving proteins (El-Brolosy et al. 2019). Our nr5a1a mutant allele generated a premature stop codon seven codons prior to the stop codon formed by our nr5a1b allele (Figure 2C). In situ hybridization experiments showed reduced quantities of nr5a1a transcript in nr5a1a mutant embryos at 48 hpf (Figure 3B), suggesting transcript instability and thus a potential for transcriptional adaptation that might ameliorate the mutant phenotype owing to the upregulation of a paralog (El-Brolosy et al. 2019). RNA-seq analysis confirmed the reduction of nr5a1a mutant transcript (3.9- and 6.3-fold at 35 dpf in nr5a1a mutants and double mutants, respectively), but showed that transcripts for paralogs that might perform transcriptional adaptation (nr5a1b, nr5a2, nr6a1b, and nr6a1a) were not differentially expressed in any mutant combination. This observation makes transcriptional adaptation unlikely for our nr5a1a−11 allele. The quantity of nr5a1b message was not differentially expressed in any mutant genotype relative to wild-type siblings in RNA-seq experiments, as expected if it was not subject to transcriptional adaptation; in addition, none of the paralogs mentioned above were differentially expressed. We conclude that the mutant alleles we studied for both genes are unlikely to have been partially rescued by transcriptional adaptation and thus, owing to the early stops, represent null activity mutations.
Contrasting phenotypes of nr5a1a and nr5a1b mutants
Newborn mice totally lacking Nr5a1 have no adrenal glands or gonads but do have a uterus, a vagina and oviducts regardless of genetic sex (Luo et al. 1994; Sadovsky et al. 1995; Shinoda et al. 1995). In addition, the VMH of mouse Nr5a1 mutants is smaller than normal and disorganized and the pituitary lacks cells that secrete gonadotrophins. If ancestral functions of Nr5a1 partitioned between the two zebrafish co-orthologs of Nr5a1a, we would expect some of these phenotypes to be associated with nr5a1a mutants and others to accompany nr5a1b mutations. Results indeed showed that nr5a1a mutants lost the adrenal and nr5a1b mutants lacked gonads. Furthermore, unlike mouse Nr5a1 mutants, zebrafish nr5a1b mutants appeared to lack sex-specific traits, including male sex tubercles and pigmentation and female-specific sex papillae. Coupled with exceedingly low sex steroid titers, nr5a1b and double mutant fish seemed to lack both primary and secondary sex characteristics.
Most homozygous null Nr5a1 mutant mice die shortly after birth owing to adrenal deficiency (Luo et al. 1994; Sadovsky et al. 1995; Shinoda et al. 1995), but tissue-specific knockouts survive longer. Nr5a1 knockout in Leydig cells led to hypoplastic testes and sterility and knockout in granulosa cells decreased the number of ovarian follicles and resulted in failed ovulation (Jeyasuria et al. 2004; Anamthathmakula et al. 2019). The knockout of Nr5a1 specifically in the VMH led to increased anxiety-like behaviors and susceptibility to high-fat diet-induced obesity (Kim et al. 2009, 2011). The phenotypes of zebrafish nr5a1 mutants were a mixture of these phenotypes, with nr5a1a mutants, showing enormous growth, accumulation of abdominal fat cells, aberrant gonad differentiation, and loss of the interrenal, whereas nr5a1b mutants lost their gonads completely and developed an abnormal interrenal. Zebrafish double mutants combined these phenotypes and mimicked the situation in complete knockout mice. These phenotypes are as predicted by the duplication, degeneration, and complementation hypothesis (Force et al. 1999).
Nr5a1 and growth control
In mammals, NR5A1-expressing cells in the VMH help regulate food intake and body weight (King 2006). The VMH-specific knockout of Nr5a1 in mice leads to late-onset obesity, especially when fed a high fat diet (Kim et al. 2011). VMH-specific Nr5a1 knockout mice were not longer than littermate controls, but they were more adipose. Similarly, nr5a1a mutant zebrafish were large, some nearly three times the mass of their wild-type siblings; in contrast to NR5A1 mouse mutants, however, zebrafish nr5a1a mutants also grew longer, perhaps related to indeterminant growth in fish. The mechanism for weight gain in Nr5a1-mutant mice likely involves leptin, which is expressed in adipocytes in mammals and directly activates leptin receptors on Nr5a1-expressing VMH cells: mice lacking leptin receptors on these cells gained body weight and were sensitive to diet-induced obesity (Dhillon et al. 2006; Bingham et al. 2008). These observations raise the hypothesis that without Nr5a1a activity, cells in the zebrafish VMH do not express leptin receptor and thus cannot detect leptin or satiety, and hence continue to acquire energy from food and grow to the enormous observed sizes. Evidence to evaluate that hypothesis, however, is mixed. Zebrafish and other teleosts generally have one gene encoding the leptin receptor (lepr) and two genes encoding leptin (lepa, lepb) (Gorissen et al. 2009; Liu et al. 2010). Arguing against the hypothesis is the finding that zebrafish bearing a premature stop codon in lepr were the same size as controls (Michel et al. 2016). Similarly, medaka lepr mutants were longer and heavier than wild-type controls up to 7 weeks posthatching, but by 9 weeks, the two genotypes were the same size (Chisada et al. 2014). On the other hand, zebrafish lepa mutants were about 10% longer and 15% heavier than wild-type siblings at 6 mpf (Audira et al. 2018), as expected from the hypothesis. Mechanisms leading to the massive increase in mutant size may be related to the observed substantial mis-regulation of long-chain fatty acid genes and gluconeogenesis genes in nr5a1a mutant zebrafish, reflecting the finding that in mouse, nearly all genes in the glycolytic pathway are regulated by Nr5a1 (Baba et al. 2014). A further conundrum is that our scRNA-seq and in situ hybridization experiments showed that nr5a1b is expressed more broadly than nr5a1a in the embryonic hypothalamus but nr5a1b mutants were of near normal size. We hypothesize that in adults, nr5a1a is expressed in hypothalamic cell types that help regulate growth but nr5a1b is expressed in hypothalamic cells with different functions. Future work is essential to understand the mechanism that leads to greatly increased body size in zebrafish nr5a1 mutants.
Nr5a1 and sex steroids
Nr5a1 targets steroidogenesis genes in mammals (Clemens et al. 1994; Sugawara et al. 1996; Parker et al. 2002). Accordingly, in zebrafish adults, nr5a1a and nr5a1b mutants had greatly reduced titers of both estrogen and 11-ketotestosterone. As a consequence, these mutants lacked secondary sex characteristics mediated by these hormones, including the androgen-induced characters of male coloration and sex tubercles on male pectoral fins (McMillan et al. 2015), and the estrogen-induced trait of extended urogenital papillae in females (Brion et al. 2004), as if these mutants had no somatic sex. Similarly, expression of the liver-expressed, estrogen-induced (Brion et al. 2004; Hao et al. 2013) yolk protein gene vtg5 (Meng et al. 2010) was greatly reduced in 21 and 35 dpf nr5a1a-bearing mutants, almost 100-fold down in double mutants, consistent with low estrogen titers. Vitellogenin genes were not differentially expressed in nr5a1b mutants despite their small gonads at 21 and 35 dpf and low estrogen titer at 4.5 months (Figure 9A), and hence it is possible that the small gonads of nr5a1b mutants were producing estrogen in juveniles but not in young adults.
Low levels of sex steroids in nr5a1 mutants can be explained by downregulation of steroidogenesis genes mimicking mouse Nr5a1 mutants [Star (Caron et al. 1997), Cyp11a1 (Clemens et al. 1994), Cyp17a1 (Park et al. 2010), Cyp19a1 (Lynch et al. 1993), Hsd3b1 (Buaas et al. 2012), and Hsd3b2 (Martin et al. 2005)]. The only zebrafish orthologs of these mouse genes that were not DE in nr5a1a mutants and double mutants were hsd3b2 and cyp19a1a, both of which had exceptionally low base-mean counts that prevented adequate statistical tests. The cyp19a1a gene was also not mentioned to be induced by estrogen treatments in zebrafish larvae or adult males (Brion et al. 2004; Hao et al. 2013). The depression of steroidogenic gene expression in nr5a1a mutants compared to wild types was much stronger at 35 dpf than at 21 dpf, demonstrating the increasing failure of mutants during this critical period for sex determination. In nr5a1b mutants, only cyp11a1, hsd3b1, and cyp17a1 among sex steroid genes were DE, and again the effect was stronger at 35 dpf than at 21 dpf.
Nr5a1 and cortisol
The stress hormone cortisol is secreted from the adrenal cortex, which is lacking from nr5a1a mutants; correspondingly, wild types had 25-times more cortisol than their nr5a1a siblings. The cortisol-synthesizing enzyme gene cyp21a2 was not DE at 21 dpf in any of the three mutant genotypes but was downregulated about sevenfold at 35 dpf in nr5a1a mutants and double mutants, which eventually had no interrenal; it was not DE in nr5a1b mutants, which developed a small interrenal. Zebrafish 5-dpf larvae treated with cortisol upregulate a number of genes, including fkbp5, tsc22d3, socs3a, and cyp2k6 (Hartig et al. 2016); as expected for cortisol-stimulated genes, these genes were all under-expressed in nr5a1a mutants (4- to 99-fold), in accordance with low cortisol levels in these mutants. These cortisol-sensitive genes, as well as cyp21a2, were not mis-expressed in nr5a1b mutants, which has a small interrenal.
Gonad development in nr5a1a mutants
Oocyte development has already begun in 19-dpf zebrafish AB-strain gonads because by this time, they are already expressing zona pellucida genes (zp2 and zp3 paralogs) (Liu et al. 2006). In nr5a1a mutant genotypes, zp3 genes were not DE in mutants vs wild types at 21 dpf, as expected if nr5a1a mutants began to develop oocytes like wild types do. In nr5a1a mutants, however, many zp genes were under-expressed at 35 dp (20- to 698-fold), showing that they failed to maintain oocyte development. Although 35dpf nr5a1a mutants under-expressed many zp3 genes (zp3a.1, zp3b, zp3c, zp3d.1, zp3d.2, zp3.2), they did not mis-express the seven zp2 genes that are strongly over-expressed in adult wild-type ovaries vs. testes [zp2.1, zp2.2, zp2.3, zp2.5, zp2.6, zp2l1, zp2l2 (Yan et al. 2019)] or the zp-gene regulator figla (Qin et al. 2018). Although figla regulates both zp2 and zp3 gene families and estrogen regulates neither (Liu et al. 2006), the mechanism that leads to their differential regulation in nr5a1 mutants remains to be discovered. Like zp genes, many meiosis genes (fanci, sycp1, dazl, mns1, eme1, wee2) were not mis-expressed in 21dp nr5a1 mutants but were under-expressed in 35-dpf mutants, as were the oocyte genes buc (Marlow and Mullins 2008; Bontems et al. 2009) and bmp15 (Clelland et al. 2006; Dranow et al. 2016). These results give the general picture that gene expression in nr5a1a mutant gonads is not disrupted severely enough to be distinguished from that in wild types at 21 dpf but by 35 dpf, many more mutant fish than wild types were transitioning to male phenotypes.
Loss of nr5a1a gene activity affected some gonadal cell types more than others. In situ hybridization experiments showed that in adult nr5a1a mutants, Leydig cells were missing, which was confirmed by transcriptomic studies, showing that the Leydig cell marker cyp11c1 (Wang and Orban 2007) was downregulated 16- to 19-fold by 35 dpf in nr5a1a mutants. In contrast, in situ hybridization studies showed that amh-expressing Sertoli cells were merely fewer and less-well organized in nr5a1a adult mutants than in wild types, confirmed by RNA-seq (2.3- to 4.4-fold). We conclude that nr5a1a is necessary for Leydig cell development and that effects on amh-expressing Sertoli cells are likely to be secondary.
Gonad development in nr5a1b mutants
Histology showed that nr5a1b activity is essential for gonads to move beyond the juvenile ovary stage. This conclusion is supported by RNA-seq, which showed that the top 10 genes over-expressed in 21 dpf nr5a1b mutants were also upregulated about a thousand-fold or more in wild-type ovaries vs testes (Yan et al. 2019), as expected if nr5a1b mutants had started making oocytes before wild types do; by 35 dpf, however, most of these genes were among the 20 most downregulated in nr5a1b mutants, as expected if early oocytes had disappeared. By 3 mpf, gonads in nr5a1b mutants were not much larger than at 35 dpf, and by 8 mpf, gonads were not detected at all, similar to the “vanishing testis syndrome” of some human NR5A1 alleles (Philibert et al. 2007). We conclude that nr5a1b action is required for the proliferation and maintenance of the gonad, whereas nr5a1a is required for the maintenance of just oocytes. In adult wild types, nr5a1b is expressed in Leydig cells, but we do not yet know whether gonad loss in nr5a1b mutants is due to cell-autonomous action of nr5a1b in the gonad or to a non-cell-autonomous action owing to its role in the hypothalamus. Construction of genetic mosaics would answer that question.
Sex determination and zebrafish nr5a1 ohnologs
Results showed that all nr5a1a mutants developed a testis, and hence we conclude that nr5a1a is required for ovary development in AB strain zebrafish. A male-biased sex ratio also occurs after mutation of a number of other gonadal genes, including homologs of genes in the female branch of the mammalian sex-determination pathway [e.g., wnt4a (Kossack et al. 2019), nr0b1 (Chen et al. 2016), foxl2a; foxl2b double mutants; cyp19a1a (Dranow et al. 2016; Lau et al. 2016; Yin et al. 2017)]. In contrast, mutations in zebrafish orthologs of mammalian male-pathway genes provide female-biased sex ratios [ar, encoding the androgen receptor (Crowder et al. 2018), dmrt1 (Webster et al. 2017), amh (Lin et al. 2017; Yan et al. 2019)]. Mutants for genes necessary for uncommitted germ cell survival or proliferation also tend to develop as males [nanos3 (Dranow et al. 2013), fgf24 (Leerberg et al. 2017), piwil1 (Houwing et al. 2007), piwil2 (Houwing et al. 2008), ddx4 (Hartung et al. 2014), prmt5 (Zhu et al. 2019)]. Male development, sometimes with fertility, can come from mutations in genes that are important for early oocyte survival, [e.g. bmp15 (Dranow et al. 2016), figla (Qin et al. 2018), and 15 different Fanconi anemia genes (Rodriguez-Mari et al. 2010; Rodriguez-Mari et al. 2011; Botthof et al. 2017; Ramanagoudr-Bhojappa et al. 2018)]. This observation led to the hypothesis that a large number of germ cells is important for female sex determination in AB strain zebrafish because a proportion of them will form oocytes that enter meiosis and emit a signal to the gonadal soma that maintains female development (Slanchev et al. 2005; Siegfried and Nusslein-Volhard 2008; Rodriguez-Mari et al. 2010). In line with this hypothesis, the expression of germ-cell marker genes [e.g. ddx4, piwil1, piwil2, tdrd1, (Yoon et al. 1997; Houwing et al. 2007, 2008; Huang et al. 2011)], including oocyte-specific genes (bmp15, buc, dazl, wee2), were not differentially expressed at 21 dpf, but had drastically reduced expression by 35 dpf in nr5a1a and nr5a1b mutants (9- and 26-fold, respectively), consistent with germ cell loss, especially oocyte loss, over these 2 weeks as confirmed by histology. Principal component analysis also separated 21-dpf individuals into fish embarking on a female developmental pathway vs. those not yet developing as females independent of the action of nr5a1a, as was found earlier for amh (Yan et al. 2019). Oocytes in transitioning gonads die by Tp53-mediated apoptosis (Rodriguez-Mari et al. 2010; Rodriguez-Mari and Postlethwait 2011; Sun et al. 2013; Miao et al. 2017). In accordance with this finding, several apoptosis-related genes (tp53, casp7, casp6l1) were upregulated significantly, although to a small degree (1.2- to 1.6-fold). Taken together, these histological and gene expression studies support the hypothesis that zebrafish nr5a1 mutants begin to develop testes owing to death of oocytes.
Although nr5a1a mutants developed testes with developing spermatocytes, they failed to develop male secondary sex characteristics. Gonads in nr5a1b mutants failed to increase in size from 21 dpf and eventually disappeared and hence these animals and double mutants appeared to have no sex at all: they had no gonads and no secondary sex characteristics.
Subfunctionalization and Nr5a1
The last common ancestor of mammals and zebrafish had a single Nr5a1 gene, but the teleost genome duplication produced two ohnologs, nr5a1a and nr5a1b, which survive in zebrafish. The duplication, degeneration, complementation hypothesis (Force et al. 1999) predicts that one mechanism for the survival of both gene copies after a gene duplication event is the reciprocal partitioning of essential ancestral gene subfunctions between the two duplicates, which can evolve further in diverging lineages (Force et al. 1999; Postlethwait et al. 2004; Glasauer and Neuhauss 2014; Freeling et al. 2015; Braasch et al. 2018). Mutations in the single-copy NR5A1 gene in human cause a large range of gonadal phenotypes, including primary ovarian insufficiency and lack of ovary formation in 46, XX individuals and partial testicular dysgenesis in 46, XY karyotypes leading to being raised as a girl (Achermann et al. 1999; Bashamboo et al. 2016; Fabbri-Scallet et al. 2020), but only infrequently leads to adrenal dysfunction (only 10 of 175 patients tested) (Fabbri-Scallet et al. 2020). Nr5a1 mutations in mouse give gonadal phenotypes similar to NR5A1 mutations in humans, but have a stronger adrenal phenotype, with death shortly after birth (Luo et al. 1994; Shinoda et al. 1995; Val et al. 2003). The early death of nr5a1 mouse mutants is likely because they lack the adrenal hormone aldosterone, which is essential for salt and water balance in mammals. Fish, however, achieve salt and water balance without aldosterone, and hence the lack of the adrenal homolog in fish is not lethal. Like mammals, Nile tilapia has a single copy of nr5a1 whose relationship to zebrafish nr5a1 ohnologs has not previously been identified; our analysis shows that the tilapia gene is an ortholog of nr5a1a in the genomic context adgrd2 nr5a1a nr6a1a mir-181b-2 and that tilapia lost nr5a1b and nr6a1b in the ancestral context nr6a1b nr5a1b psmb7 nek6 lhx2a dennd1a after the cichlid lineage diverged from the medaka lineage. Nile tilapia individuals developing from zygotes injected with CRISPR/Cas9 reagents targeting nr5a1a showed a high rate of mutation associated with gonadal dysgenesis and few steroidogenic cells coupled with feminization of XY fish and sex reversal of XX fish (Xie et al. 2016), similar to the zebrafish nr5a1a mutants. Given the essential nature of nr5a1b in zebrafish, it is difficult to understand how tilapia manages to develop gonads without the function of nr5a1b.
The reciprocal loss of the interrenal and the gonad in zebrafish nr5a1a and nr5a1b mutants, respectively, likely reflects differential roles of nr5a1a and nr5a1b in the specification of key cell types in the adrenal–gonadal (interrenal-gonadal) primordium at or before the bipotential gonad stage (Kreidberg et al. 1993; Birk et al. 2000; Schnabel et al. 2003; Katoh-Fukui et al. 2005; Buaas et al. 2009; Fujimoto et al. 2013; Hu et al. 2013). The milder phenotypes—oocyte loss leading to the development of testes that are abnormal owing to aberrant Leydig cell development in nr5a1a mutants and the reciprocal reduction and disorganization of the interrenal in nr5a1b mutants—likely come from the later roles of Nr5a1 in the regulation of steroid biosynthesis, which was greatly reduced in both genotypes. These differences are associated with differential expression patterns of the two nr5a1 ohnologs that may represent the subfunctionalization events that preserved both gene copies. Resolving these issues will require future single-cell transcriptomics experiments and functional analyses of promotors.
Acknowledgments
This article is dedicated to the 70th anniversary of the first publication of our coauthor Ruth BreMiller (née R.A. Allen), who performed and analyzed histology for this study (White and Allen 1951). The authors thank Tim Mason, director of the University of Oregon Aquatic Animal Care Services, for excellent zebrafish husbandry, and Doug Turnbull and Maggie Weitzman of the University of Oregon Genomics and Cell Characterization Core Facility for help with transcriptomics.
Funding
Genomics-related research supported by R01OD011116 (J.H.P.). Gonadal development research supported by R01GM085318 (J.H.P.). scRNA-seq work supported by R24OD026591 (J.H.P. and A.C.M.). UDN-related research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Numbers U01HG007709 (Baylor College of Medicine), U01HG007530 (UDN Coordinating Center), U54NS093793 (Model Organism Screening Center, J.H.P. and M.W.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of interest
None declared.
Literature cited
- Achermann JC, Ito M, Ito M, Hindmarsh PC, Jameson JL. 1999. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat Genet. 22:125–126. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Webb IJ, Bleul C, Springer T, Gutierrez-Ramos JC. 1997. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J Exp Med. 185:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics. 188:799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science. 282:1711–1714. [DOI] [PubMed] [Google Scholar]
- Anamthathmakula P, Miryala CSJ, Moreci RS, Kyathanahalli C, Hassan SS et al. 2019. Steroidogenic factor 1 (Nr5a1) is required for sertoli cell survival post sex determination. Sci Rep. 9:4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum L, Wang G, Yokogawa T, Skariah GM, Smith SJ et al. 2010. Circadian and homeostatic regulation of structural synaptic plasticity in hypocretin neurons. Neuron. 68:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armant O, Marz M, Schmidt R, Ferg M, Diotel N et al. 2013. Genome-wide, whole mount in situ analysis of transcriptional regulators in zebrafish embryos. Dev Biol. 380:351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audira G, Sarasamma S, Chen JR, Juniardi S, Sampurna BP et al. 2018. Zebrafish mutants carrying leptin a (lepa) gene deficiency display obesity, anxiety, less aggression and fear, and circadian rhythm and color preference dysregulation. Int J Mol Sci. 19:4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Otake H, Sato T, Miyabayashi K, Shishido Y, et al. 2014. Glycolytic genes are targets of the nuclear receptor Ad4BP/SF-1. Nat Commun. 5:3634. [DOI] [PubMed] [Google Scholar]
- Baetens D, Verdin H, De Baere E, Cools M. 2019. Update on the genetics of differences of sex development (DSD). Best Pract Res Clin Endocrinol Metab. 33:101271. [DOI] [PubMed] [Google Scholar]
- Barton BA, Rahn AB, Feist G, Bollig H, Schreck CB. 1998. Physiological stress responses of the freshwater chondrostean paddlefish (Polyodon spathula) to acute physical disturbances. Comp Biochem Physiol. 120:355–363. [Google Scholar]
- Bashamboo A, Donohoue PA, Vilain E, Rojo S, Calvel P, Members of UDN et al. 2016. A recurrent p.Arg92Trp variant in steroidogenic factor-1 (NR5A1) can act as a molecular switch in human sex development. Hum Mol Genet. 25:5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshay VE, Havelock JC, Sirianni R, Ye P, Suzuki T et al. 2007. The mechanism for protein kinase C inhibition of androgen production and 17alpha-hydroxylase expression in a theca cell tumor model. J Clin Endocrinol Metab. 92:4802–4809. [DOI] [PubMed] [Google Scholar]
- Bingham NC, Anderson KK, Reuter AL, Stallings NR, Parker KL. 2008. Selective loss of leptin receptors in the ventromedial hypothalamic nucleus results in increased adiposity and a metabolic syndrome. Endocrinology. 149:2138–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birk OS, Casiano DE, Wassif CA, Cogliati T, Zhao L et al. 2000. The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature. 403:909–913. [DOI] [PubMed] [Google Scholar]
- Boldajipour B, Doitsidou M, Tarbashevich K, Laguri C, Yu SR et al. 2011. Cxcl12 evolution–subfunctionalization of a ligand through altered interaction with the chemokine receptor. Development. 138:2909–2914. [DOI] [PubMed] [Google Scholar]
- Bolger A, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T et al. 2009. Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol. 19:414–422. [DOI] [PubMed] [Google Scholar]
- Borg B. 1994. Mini review—androgens in teleost fishes. Comp Biochem Physiol. 109:219–245. [Google Scholar]
- Botthof JG, Bielczyk-Maczyńska E, Ferreira L, Cvejic A. 2017. Loss of the homologous recombination gene rad51 leads to Fanconi anemia-like symptoms in zebrafish. Proc Natl Acad Sci USA. 114:E4452–E4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Bobe J, Guiguen Y, Postlethwait JH. 2018. Reply to: ‘Subfunctionalization versus neofunctionalization after whole-genome duplication’. Nat Genet. 50:910–911. [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. 2006. Evolution of hormone-receptor complexity by molecular exploitation. Science. 312:97–101. [DOI] [PubMed] [Google Scholar]
- Brion F, Tyler CR, Palazzi X, Laillet B, Porcher JM et al. 2004. Impacts of 17beta-estradiol, including environmentally relevant concentrations, on reproduction after exposure during embryo-larval-, juvenile- and adult-life stages in zebrafish (Danio rerio). Aquat Toxicol. 68:193–217. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Gardiner JR, Clayton S, Val P, Swain A. 2012. In vivo evidence for the crucial role of SF1 in steroid-producing cells of the testis, ovary and adrenal gland. Development. 139:4561–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas FW, Val P, Swain A. 2009. The transcription co-factor CITED2 functions during sex determination and early gonad development. Hum Mol Genet. 18:2989–3001. [DOI] [PubMed] [Google Scholar]
- Budefeld T, Tobet SA, Majdic G. 2012. Steroidogenic factor 1 and the central nervous system. J Neuroendocrinol. 24:225–235. [DOI] [PubMed] [Google Scholar]
- Canosa LF, Chang JP, Peter RE. 2007. Neuroendocrine control of growth hormone in fish. Gen Comp Endocrinol. 151:1–26. [DOI] [PubMed] [Google Scholar]
- Caron KM, Ikeda Y, Soo SC, Stocco DM, Parker KL et al. 1997. Characterization of the promoter region of the mouse gene encoding the steroidogenic acute regulatory protein. Mol Endocrinol. 11:138–147. [DOI] [PubMed] [Google Scholar]
- Chai C, Chan WK. 2000. Developmental expression of a novel Ftz-F1 homologue, ff1b (NR5A4), in the zebrafish Danio rerio. Mech Dev. 91:421–426. [DOI] [PubMed] [Google Scholar]
- Chai C, Liu YW, Chan WK. 2003. Ff1b is required for the development of steroidogenic component of the zebrafish interrenal organ. Dev Biol. 260:226–244. [DOI] [PubMed] [Google Scholar]
- Chen R, Wu X, Jiang L, Zhang Y. 2017. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 18:3227–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang H, Wang F, Zhang W, Peng G. 2016. nr0b1 (DAX1) mutation in zebrafish causes female-to-male sex reversal through abnormal gonadal proliferation and differentiation. Mol Cell Endocrinol. 433:105–116. [DOI] [PubMed] [Google Scholar]
- Chester Jones I, Mosley W. 1980. The interrenal gland in Pisces. In: Jones IC, Henderson IW, editors. General, Comparative and Clinical Endocrinology of the Adrenal Cortex. London, New York: Academic Press. p. 395–523. [Google Scholar]
- Chiang EF, Yan YL, Guiguen Y, Postlethwait J, Chung B. 2001a. Two Cyp19 (P450 aromatase) genes on duplicated zebrafish chromosomes are expressed in ovary or brain. Mol Biol Evol. 18:542–550. [DOI] [PubMed] [Google Scholar]
- Chiang EF, Yan YL, Tong SK, Hsiao PH, Guiguen Y et al. 2001b. Characterization of duplicated zebrafish cyp19 genes. J Exp Zool. 290:709–714. [DOI] [PubMed] [Google Scholar]
- Chisada S, Kurokawa T, Murashita K, Ronnestad I, Taniguchi Y et al. 2014. Leptin receptor-deficient (knockout) medaka, Oryzias latipes, show chronical up-regulated levels of orexigenic neuropeptides, elevated food intake and stage specific effects on growth and fat allocation. Gen Comp Endocrinol. 195:9–20. [PubMed] [Google Scholar]
- Chiu CH, Chou CW, Takada S, Liu YW. 2012. Development and fibronectin signaling requirements of the zebrafish interrenal vessel. PLoS One. 7:e43040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clelland E, Kohli G, Campbell RK, Sharma S, Shimasaki S et al. 2006. Bone morphogenetic protein-15 in the zebrafish ovary: complementary deoxyribonucleic acid cloning, genomic organization, tissue distribution, and role in oocyte maturation. Endocrinology. 147:201–209. [DOI] [PubMed] [Google Scholar]
- Clemens JW, Lala DS, Parker KL, Richards JS. 1994. Steroidogenic factor-1 binding and transcriptional activity of the cholesterol side-chain cleavage promoter in rat granulosa cells. Endocrinology. 134:1499–1508. [DOI] [PubMed] [Google Scholar]
- Crowder CM, Lassiter CS, Gorelick DA. 2018. Nuclear androgen receptor regulates testes organization and oocyte maturation in zebrafish. Endocrinology. 159:980–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Qiao A, Jiao T, Zhang H, Xue Y et al. 2016. The hepatic FOXQ1 transcription factor regulates glucose metabolism in mice. Diabetologia. 59:2229–2239. [DOI] [PubMed] [Google Scholar]
- Dhillon H, Zigman JM, Ye C, Lee CE, McGovern RA et al. 2006. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron. 49:191–203. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranow DB, Hu K, Bird AM, Lawry ST, Adams MT et al. 2016. Bmp15 is an oocyte-produced signal required for maintenance of the adult female sexual phenotype in zebrafish. PLoS Genet. 12:e1006323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranow DB, Tucker RP, Draper BW. 2013. Germ cells are required to maintain a stable sexual phenotype in adult zebrafish. Dev Biol. 376:43–50. [DOI] [PubMed] [Google Scholar]
- Eachus H, Zaucker A, Oakes JA, Griffin A, Weger M et al. 2017. Genetic disruption of 21-hydroxylase in zebrafish causes interrenal hyperplasia. Endocrinology. 158:4165–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S et al. 2019. Genetic compensation triggered by mutant mRNA degradation. Nature. 568:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri‐Scallet H, Sousa LM, Maciel‐Guerra AT, Guerra‐Júnior G, Mello MP. 2020. Mutation update for the NR5A1 gene involved in DSD and infertility. Hum Mutat. 41:58–68. [DOI] [PubMed] [Google Scholar]
- Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth DR, Saunders LM, Miller AC. 2020. A single-cell transcriptome atlas for zebrafish development. Dev Biol. 459:100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL et al. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 151:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Scanlon MJ, Fowler JE. 2015. Fractionation and subfunctionalization following genome duplications: mechanisms that drive gene content and their consequences. Curr Opin Genet Dev. 35:110–118. [DOI] [PubMed] [Google Scholar]
- Fuentes EN, Valdes JA, Molina A, Bjornsson BT. 2013. Regulation of skeletal muscle growth in fish by the growth hormone–insulin-like growth factor system. Gen Comp Endocrinol. 192:136–148. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y, Tanaka SS, Yamaguchi YL, Kobayashi H, Kuroki S et al. 2013. Homeoproteins Six1 and Six4 regulate male sex determination and mouse gonadal development. Dev Cell. 26:416–430. [DOI] [PubMed] [Google Scholar]
- Garcia-Acero M, Moreno-Nino O, Suarez-Obando F, Molina M, Manotas MC et al. 2020. Disorders of sex development: genetic characterization of a patient cohort. Mol Med Rep. 21:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasauer SM, Neuhauss SC. 2014. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics. 289:1045–1060. [DOI] [PubMed] [Google Scholar]
- Gorissen M, Bernier NJ, Nabuurs SB, Flik G, Huising MO. 2009. Two divergent leptin paralogues in zebrafish (Danio rerio) that originate early in teleostean evolution. J Endocrinol. 201:329–339. [DOI] [PubMed] [Google Scholar]
- Guran T, Buonocore F, Saka N, Ozbek MN, Aycan Z et al. 2016. Rare causes of primary adrenal insufficiency: genetic and clinical characterization of a large nationwide cohort. J Clin Endocrinol Metab. 101:284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RC, Fleming WR. 1979. Serum cortisol levels of juvenile bowfin, Amia calva: effects of hypophysectomy, hormone replacement and environmental salinity. Comp Biochem Physiol. 63:499–502. [Google Scholar]
- Hao R, Bondesson M, Singh AV, Riu A, McCollum CW et al. 2013. Identification of estrogen target genes during zebrafish embryonic development through transcriptomic analysis. PLoS One. 8:e79020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig EI, Zhu S, King BL, Coffman JA. 2016. Cortisol-treated zebrafish embryos develop into pro-inflammatory adults with aberrant immune gene regulation. Biol Open. 5:1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung O, Forbes MM, Marlow FL. 2014. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol Reprod Dev. 81:946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano M, Migita T, Ohishi T, Shima Y, Ogawa Y et al. 2016. SF-1 deficiency causes lipid accumulation in Leydig cells via suppression of STAR and CYP11A1. Endocrine. 54:484–496. [DOI] [PubMed] [Google Scholar]
- Hatano O, Takakusu A, Nomura M, Morohashi K. 1996. Identical origin of adrenal cortex and gonad revealed by expression profiles of Ad4BP/SF-1. Genes Cells. 1:663–671. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 27:2702–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 129:69–82. [DOI] [PubMed] [Google Scholar]
- Hsu HJ, Lin G, Chung BC. 2003. Parallel early development of zebrafish interrenal glands and pronephros: differential control by wt1 and ff1b. Development. 130:2107–2116. [DOI] [PubMed] [Google Scholar]
- Hu YC, Okumura LM, Page DC. 2013. Gata4 is required for formation of the genital ridge in mice. PLoS Genet. 9:e1003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Houwing S, Kaaij LJ, Meppelink A, Redl S et al. 2011. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J. 30:3298–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt KE, Lysiak NS, Robbins J, Moore MJ, Seton RE et al. 2017. Multiple steroid and thyroid hormones detected in baleen from eight whale species. Conserv Physiol. 5:cox061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idler DR, Truscott B. 1972. Corticosteroids in fish. In: Idler DR, editor. Steroids in Nonmammalian Vertebrates. New York: Academic Press. p. 126–252. [Google Scholar]
- Ikeda Y, Luo X, Abbud R, Nilson JH, Parker KL. 1995. The nuclear receptor steroidogenic factor 1 is essential for the formation of the ventromedial hypothalamic nucleus. Mol Endocrinol. 9:478–486. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Shen WH, Ingraham HA, Parker KL. 1994. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol Endocrinol. 8:654–662. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 431:946–957. [DOI] [PubMed] [Google Scholar]
- Jeyasuria P, Ikeda Y, Jamin SP, Zhao L, De Rooij DG et al. 2004. Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol Endocrinol. 18:1610–1619. [DOI] [PubMed] [Google Scholar]
- Jiang JQ, Young G, Kobayashi T, Nagahama Y. 1998. Eel (Anguilla japonica) testis 11beta-hydroxylase gene is expressed in interrenal tissue and its product lacks aldosterone synthesizing activity. Mol Cell Endocrinol. 146:207–211. [DOI] [PubMed] [Google Scholar]
- Karlstrom RO, Tyurina OV, Kawakami A, Nishioka N, Talbot WS et al. 2003. Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development. 130:1549–1564. [DOI] [PubMed] [Google Scholar]
- Katoh-Fukui Y, Owaki A, Toyama Y, Kusaka M, Shinohara Y et al. 2005. Mouse Polycomb M33 is required for splenic vascular and adrenal gland formation through regulating Ad4BP/SF1 expression. Blood. 106:1612–1620. [DOI] [PubMed] [Google Scholar]
- Kim KW, Zhao L, Donato J Jr, Kohno D, Xu Y et al. 2011. Steroidogenic factor 1 directs programs regulating diet-induced thermogenesis and leptin action in the ventral medial hypothalamic nucleus. Proc Natl Acad Sci USA. 108:10673–10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Zhao L, Parker KL. 2009. Central nervous system-specific knockout of steroidogenic factor 1. Mol Cell Endocrinol. 300:132–136. [DOI] [PubMed] [Google Scholar]
- King BM. 2006. The rise, fall, and resurrection of the ventromedial hypothalamus in the regulation of feeding behavior and body weight. Physiol Behav. 87:221–244. [DOI] [PubMed] [Google Scholar]
- Klangnurak W, Tokumoto T. 2017. Fine selection of up-regulated genes during ovulation by in vivo induction of oocyte maturation and ovulation in zebrafish. Zool Lett. 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut H, Blader P, Strahle U, Schier AF. 2005. Assembly of trigeminal sensory ganglia by chemokine signaling. Neuron. 47:653–666. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kawakami K, Asashima M, Nishinakamura R. 2007. Six1 and Six4 are essential for Gdnf expression in the metanephric mesenchyme and ureteric bud formation, while Six1 deficiency alone causes mesonephric-tubule defects. Mech Dev. 124:290–303. [DOI] [PubMed] [Google Scholar]
- Kossack ME, High SK, Hopton RE, Yan YL, Postlethwait JH et al. 2019. Female sex development and reproductive duct formation depend on Wnt4a in zebrafish. Genetics. 211:219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J et al. 1993. WT-1 is required for early kidney development. Cell. 74:679–691. [DOI] [PubMed] [Google Scholar]
- Kuo MW, Postlethwait J, Lee WC, Lou SW, Chan WK et al. 2005. Gene duplication, gene loss and evolution of expression domains in the vertebrate nuclear receptor NR5A (Ftz-F1) family. Biochem J. 389:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrasch DM, Cheung CC, Lee FY, Tran PV, Hata K et al. 2007. The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning. J Neurosci. 27:13624–13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrasch DM, Nevin LM, Wong JS, Baier H, Ingraham HA. 2009. Neuroendocrine transcriptional programs adapt dynamically to the supply and demand for neuropeptides as revealed in NSF mutant zebrafish. Neural Dev. 4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder R, Clark DD, Miller NL, Mellon PL. 2011. Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6. J Neurosci. 31:426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau ES, Zhang Z, Qin M, Ge W. 2016. Knockout of zebrafish ovarian aromatase gene (cyp19a1a) by TALEN and CRISPR/Cas9 leads to all-male offspring due to failed ovarian differentiation. Sci Rep. 6:37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leerberg DM, Sano K, Draper BW. 2017. Fibroblast growth factor signaling is required for early somatic gonad development in zebrafish. PLoS Genet. 13:e1006993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberam I, Agalliu D, Nagasawa T, Ericson J, Jessell TM. 2005. A Cxcl12-CXCR4 chemokine signaling pathway defines the initial trajectory of mammalian motor axons. Neuron. 47:667–679. [DOI] [PubMed] [Google Scholar]
- Lin L, Achermann JC. 2008. Steroidogenic factor-1 (SF-1, Ad4BP, NR5A1) and disorders of testis development. Sex Dev. 2:200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Mei J, Li Z, Zhang X, Zhou L et al. 2017. Distinct and cooperative roles of amh and dmrt1 in self-renewal and differentiation of male germ cells in zebrafish. Genetics. 207:1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Matzuk MM, Sung WK, Guo Q, Wang P et al. 1998. Cyclin A1 is required for meiosis in the male mouse. Nat Genet. 20:377–380. [DOI] [PubMed] [Google Scholar]
- Liu Q, Chen Y, Copeland D, Ball H, Duff RJ et al. 2010. Expression of leptin receptor gene in developing and adult zebrafish. Gen Comp Endocrinol. 166:346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang H, Gong Z. 2006. Tandem-repeated zebrafish zp3 genes possess oocyte-specific promoters and are insensitive to estrogen induction. Biol Reprod. 74:1016–1025. [DOI] [PubMed] [Google Scholar]
- Liu YW, Gao W, Teh HL, Tan JH, Chan WK. 2003. Prox1 is a novel coregulator of Ff1b and is involved in the embryonic development of the zebra fish interrenal primordium. Mol Cell Biol. 23:7243–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YW, Guo L. 2006. Endothelium is required for the promotion of interrenal morphogenetic movement during early zebrafish development. Dev Biol. 297:44–58. [DOI] [PubMed] [Google Scholar]
- Love MI, Anders S, Kim V, Huber W. 2015. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Res. 4:1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Ikeda Y, Parker KL. 1994. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 77:481–490. [DOI] [PubMed] [Google Scholar]
- Lynch JP, Lala DS, Peluso JJ, Luo W, Parker KL et al. 1993. Steroidogenic factor 1, an orphan nuclear receptor, regulates the expression of the rat aromatase gene in gonadal tissues. Mol Endocrinol. 7:776–786. [DOI] [PubMed] [Google Scholar]
- Machluf Y, Gutnick A, Levkowitz G. 2011. Development of the zebrafish hypothalamus. Ann N Y Acad Sci. 1220:93–105. [DOI] [PubMed] [Google Scholar]
- Manna PR, Dyson MT, Stocco DM. 2009. Role of basic leucine zipper proteins in transcriptional regulation of the steroidogenic acute regulatory protein gene. Mol Cell Endocrinol. 302:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli M, Driever W. 2014. nkx2.1 and nkx2.4 genes function partially redundant during development of the zebrafish hypothalamus, preoptic region, and pallidum. Front Neuroanat. 8:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC. 2008. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol. 321:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco G, Grammatico P, Vallasciani S, Gulia C, Zangari A et al. 2015. Environmental, parental and gestational factors that influence the occurrence of hypospadias in male patients. J Pediatr Urol. 11:12–19. [DOI] [PubMed] [Google Scholar]
- Martin LJ, Taniguchi H, Robert NM, Simard J, Tremblay JJ et al. 2005. GATA factors and the nuclear receptors, steroidogenic factor 1/liver receptor homolog 1, are key mutual partners in the regulation of the human 3beta-hydroxysteroid dehydrogenase type 2 promoter. Mol Endocrinol. 19:2358–2370. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–10. [Google Scholar]
- McMillan SC, Geraudie J, Akimenko MA. 2015. Pectoral fin breeding tubercle clusters: a method to determine zebrafish sex. Zebrafish. 12:121–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Bartholomew C, Craft JA. 2010. Differential expression of vitellogenin and oestrogen receptor genes in the liver of zebrafish, Danio rerio. Anal Bioanal Chem. 396:625–630. [DOI] [PubMed] [Google Scholar]
- Menke AL, Spitsbergen JM, Wolterbeek AP, Woutersen RA. 2011. Normal anatomy and histology of the adult zebrafish. Toxicol Pathol. 39:759–775. [DOI] [PubMed] [Google Scholar]
- Miao L, Yuan Y, Cheng F, Fang J, Zhou F et al. 2017. Translation repression by maternal RNA binding protein Zar1 is essential for early oogenesis in zebrafish. Development. 144:128–138. [DOI] [PubMed] [Google Scholar]
- Michel M, Page-McCaw PS, Chen W, Cone RD. 2016. Leptin signaling regulates glucose homeostasis, but not adipostasis, in the zebrafish. Proc Natl Acad Sci USA. 113:3084–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL, Auchus RJ. 2011. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev. 32:81–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyado M, Inui M, Igarashi M, Katoh-Fukui Y, Takasawa K et al. 2016. The p.R92W variant of NR5A1/Nr5a1 induces testicular development of 46,XX gonads in humans, but not in mice: phenotypic comparison of human patients and mutation-induced mice. Biol Sex Differ. 7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K. 1997. The ontogenesis of the steroidogenic tissues. Genes Cells. 2:95–106. [DOI] [PubMed] [Google Scholar]
- Muthu V, Eachus H, Ellis P, Brown S, Placzek M. 2016. Rx3 and Shh direct anisotropic growth and specification in the zebrafish tuberal/anterior hypothalamus. Development. 143:2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y et al. 2012. Tracing the emergence of a novel sex-determining gene in medaka. Oryzias luzonensis. Genetics. 191:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto M, Fukasawa M, Tanaka S, Shimamori K, Suzuki A et al. 2012. Expression of 3beta-hydroxysteroid dehydrogenase (hsd3b), star and ad4bp/sf-1 during gonadal development in medaka (Oryzias latipes). Gen Comp Endocrinol. 176:222–230. [DOI] [PubMed] [Google Scholar]
- Newman A, ChinEH, SchmidtKL, BondL, Wynne-Edwards KE, . et al. 2008. Analysis of steroids in songbird plasma and brain by coupling solid phase extraction to radioimmunoassay. Gen. Comp. Endocrinol. 155:503–510. [DOI] [PubMed] [Google Scholar]
- Orban L, Sreenivasan R, Olsson PE. 2009. Long and winding roads: testis differentiation in zebrafish. Mol Cell Endocrinol. 312:35–41. [DOI] [PubMed] [Google Scholar]
- Park M, Shin E, Won M, Kim JH, Go H et al. 2010. FOXL2 interacts with steroidogenic factor-1 (SF-1) and represses SF-1-induced CYP17 transcription in granulosa cells. Mol Endocrinol. 24:1024–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker KL, Rice DA, Lala DS, Ikeda Y, Luo X et al. 2002. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog Horm Res. 57:19–36. [DOI] [PubMed] [Google Scholar]
- Patel SS, Beshay VE, Escobar JC, Suzuki T, Carr BR. 2009. Molecular mechanism for repression of 17alpha-hydroxylase expression and androstenedione production in granulosa cells. J Clin Endocrinol Metab. 94:5163–5168. [DOI] [PubMed] [Google Scholar]
- Peled A, Petit I, Kollet O, Magid M, Ponomaryov T et al. 1999. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 283:845–848. [DOI] [PubMed] [Google Scholar]
- Philibert P, Zenaty D, Lin L, Soskin S, Audran F et al. 2007. Mutational analysis of steroidogenic factor 1 (NR5a1) in 24 boys with bilateral anorchia: a French collaborative study. Hum Reprod. 22:3255–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picha ME, Turano MJ, Beckman BR, Borski RJ. 2008. Endocrine biomarkers of growth and applications to aquaculture: a minireview of growth hormone, insulin-like growth factor (IGF)-I, and IGF-binding proteins as potential growth indicators in fish. N Am J Aquacult. 70:196–211. [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. 2004. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 20:481–490. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Force A, Yan YL. 1999. The zebrafish genome. Methods Cell Biol. 60:149–163. [PubMed] [Google Scholar]
- Qin M, Zhang Z, Song W, Wong QW, Chen W et al. 2018. Roles of figla/figla in juvenile ovary development and follicle formation during zebrafish gonadogenesis. Endocrinology. 159:3699–3722. [DOI] [PubMed] [Google Scholar]
- Quek SI, Chan WK. 2009. Transcriptional activation of zebrafish cyp11a1 promoter is dependent on the nuclear receptor Ff1b. J Mol Endocrinol. 43:121–130. [DOI] [PubMed] [Google Scholar]
- Ramanagoudr-Bhojappa R, Carrington B, Ramaswami M, Bishop K, Robbins GM et al. 2018. Multiplexed CRISPR/Cas9-mediated knockout of 19 Fanconi anemia pathway genes in zebrafish revealed their roles in growth, sexual development and fertility. PLoS Genet. 14:e1007821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha VB, Guerra-Junior G, Marques-de-Faria AP, de Mello MP, Maciel-Guerra AT. 2011. Complete gonadal dysgenesis in clinical practice: the 46,XY karyotype accounts for more than one third of cases. Fertil Steril. 96:1431–1434. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mari A, Canestro C, Bremiller RA, Nguyen-Johnson A, Asakawa K et al. 2010. Sex reversal in zebrafish fancl mutants is caused by Tp53-mediated germ cell apoptosis. PLoS Genet. 6:e1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mari A, Postlethwait JH. 2011. The role of Fanconi anemia/BRCA genes in zebrafish sex determination. Methods Cell Biol. 105:461–490. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mari A, Wilson C, Titus TA, Canestro C, BreMiller RA et al. 2011. Roles of brca2 (fancd1) in oocyte nuclear architecture, gametogenesis, gonad tumors, and genome stability in zebrafish. PLoS Genet. 7:e1001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mari A, Yan YL, Bremiller RA, Wilson C, Canestro C et al. 2005. Characterization and expression pattern of zebrafish Anti-Mullerian hormone (Amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr Patterns. 5:655–667. [DOI] [PubMed] [Google Scholar]
- Rohr KB, Barth KA, Varga ZM, Wilson SW. 2001. The nodal pathway acts upstream of hedgehog signaling to specify ventral telencephalic identity. Neuron. 29:341–351. [DOI] [PubMed] [Google Scholar]
- Ronchi CL, Sbiera S, Volante M, Steinhauer S, Scott-Wild V et al. 2014. CYP2W1 is highly expressed in adrenal glands and is positively associated with the response to mitotane in adrenocortical carcinoma. PLoS One. 9:e105855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondeau EB, Messmer AM, Sanderson DS, Jantzen SG, von Schalburg KR et al. 2013. Genomics of sablefish (Anoplopoma fimbria): expressed genes, mitochondrial phylogeny, linkage map and identification of a putative sex gene. BMC Genomics. 14:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA et al. 1995. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci USA. 92:10939–10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangalang GB, Weisbart M, Idler DR. 1971. Steroids of a chondrostean: corticosteroids and testosterone in the plasma of the American Atlantic sturgeon, Acipenser oxyrhynchus Mitchill. J Endocrinol. 50:413–421. [DOI] [PubMed] [Google Scholar]
- Sax L. 2002. How common is intersex? a response to Anne Fausto-Sterling. J Sex Res. 39:174–178. [DOI] [PubMed] [Google Scholar]
- Schnabel CA, Selleri L, Cleary ML. 2003. Pbx1 is essential for adrenal development and urogenital differentiation. Genesis. 37:123–130. [DOI] [PubMed] [Google Scholar]
- Schulz RW, Bogerd J, Male R, Ball J, Fenske M et al. 2007. Estrogen-induced alterations in amh and dmrt1 expression signal for disruption in male sexual development in the zebrafish. Environ Sci Technol. 41:6305–6310. [DOI] [PubMed] [Google Scholar]
- Segal JP, Stallings NR, Lee CE, Zhao L, Socci N et al. 2005. Use of laser-capture microdissection for the identification of marker genes for the ventromedial hypothalamic nucleus. J Neurosci. 25:4181–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. 2008. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 453:930–934. [DOI] [PubMed] [Google Scholar]
- Shinoda K, Lei H, Yoshii H, Nomura M, Nagano M et al. 1995. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev Dyn. 204:22–29. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Nusslein-Volhard C. 2008. Germ line control of female sex determination in zebrafish. Dev Biol. 324:277–287. [DOI] [PubMed] [Google Scholar]
- Siekmann AF, Standley C, Fogarty KE, Wolfe SA, Lawson ND. 2009. Chemokine signaling guides regional patterning of the first embryonic artery. Genes Dev. 23:2272–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL et al. 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 346:240–244. [DOI] [PubMed] [Google Scholar]
- Slanchev K, Stebler J, de la Cueva-Mendez G, Raz E. 2005. Development without germ cells: the role of the germ line in zebrafish sex differentiation. Proc Natl Acad Sci USA. 102:4074–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan R, Cai M, Bartfai R, Wang X, Christoffels A et al. 2008. Transcriptomic analyses reveal novel genes with sexually dimorphic expression in the zebrafish gonad and brain. PLoS One. 3:e1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevant I, Kuhne F, Greenfield A, Chaboissier MC, Dermitzakis ET et al. 2019. Dissecting cell lineage specification and sex fate determination in gonadal somatic cells using single-cell transcriptomics. Cell Rep. 26:3272–3283.e3273. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Holt JA, Kiriakidou M, Strauss JF 3rd, 1996. Steroidogenic factor 1-dependent promoter activity of the human steroidogenic acute regulatory protein (StAR) gene. Biochemistry. 35:9052–9059. [DOI] [PubMed] [Google Scholar]
- Sun D, Zhang Y, Wang C, Hua X, Zhang XA et al. 2013. Sox9-related signaling controls zebrafish juvenile ovary-testis transformation. Cell Death Dis. 4:e930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Chen F, Peng G. 2015. Conserved noncoding sequences regulate lhx5 expression in the zebrafish forebrain. PLoS One. 10:e0132525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydes J, Batzel P, Titus TA, Postlethwait JH. 2019. Dupligänger: a reference genome-based, UMI-cognizant, 5′-trimming-aware PCR duplicate removal pipeline. (Version 0.97). In: Zenodo CR, editor, doi: 10.5281/zenodo.3988328.
- Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, et al. 1998. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 393:591–594. [DOI] [PubMed] [Google Scholar]
- Takahashi H. 1977. Juvenile hermaphroditism in the zebrafish, Brachydanio rerio. Bull Fac Fish Hokkaido Univ. 28:57–65. [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. 2003. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 13:382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro S, Wegner J, Muller M, Westerfield M, Varga ZM. 2009. Identification of differentially expressed genes in the zebrafish hypothalamic-pituitary axis. Gene Expr Patterns. 9:200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida D, Yamashita M, Kitano T, Iguchi T. 2002. Oocyte apoptosis during the transition from ovary-like tissue to testes during sex differentiation of juvenile zebrafish. J Exp Biol. 205:711–718. [DOI] [PubMed] [Google Scholar]
- Val P, Lefrancois-Martinez AM, Veyssiere G, Martinez A. 2003. SF-1 a key player in the development and differentiation of steroidogenic tissues. Nucl Recept. 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hofsten J, Jones I, Karlsson J, Olsson PE. 2001. Developmental expression patterns of FTZ-F1 homologues in zebrafish (Danio rerio). Gen Comp Endocrinol. 121:146–155. [DOI] [PubMed] [Google Scholar]
- von Hofsten J, Karlsson J, Jones I, Olsson PE. 2002. Expression and regulation of fushi tarazu factor-1 and steroidogenic genes during reproduction in Arctic char (Salvelinus alpinus). Biol Reprod. 67:1297–1304. [DOI] [PubMed] [Google Scholar]
- von Hofsten J, Larsson A, Olsson PE. 2005a. Novel steroidogenic factor-1 homolog (ff1d) is coexpressed with anti-Mullerian hormone (AMH) in zebrafish. Dev Dyn. 233:595–604. [DOI] [PubMed] [Google Scholar]
- von Hofsten J, Modig C, Larsson A, Karlsson J, Olsson PE. 2005b. Determination of the expression pattern of the dual promoter of zebrafish fushi tarazu factor-1a following microinjections into zebrafish one cell stage embryos. Gen Comp Endocrinol. 142:222–226. [DOI] [PubMed] [Google Scholar]
- Walters KB, Green JM, Surfus JC, Yoo SK, Huttenlocher A. 2010. Live imaging of neutrophil motility in a zebrafish model of WHIM syndrome. Blood. 116:2803–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DS, Kobayashi T, Zhou LY, Paul-Prasanth B, Ijiri S et al. 2007. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol Endocrinol. 21:712–725. [DOI] [PubMed] [Google Scholar]
- Wang H, Teng Y, Xie Y, Wang B, Leng Y et al. 2013. Characterization of the carbonic anhydrases 15b expressed in PGCs during early zebrafish development. Theriogenology. 79:443–452. [DOI] [PubMed] [Google Scholar]
- Wang XG, Orban L. 2007. Anti-Mullerian hormone and 11 beta-hydroxylase show reciprocal expression to that of aromatase in the transforming gonad of zebrafish males. Dev Dyn. 236:1329–1338. [DOI] [PubMed] [Google Scholar]
- Wang-Buhler JL, Lee SJ, Chung WG, Stevens JF, Tseng HP et al. 2005. CYP2K6 from zebrafish (Danio rerio): cloning, mapping, developmental/tissue expression, and aflatoxin B1 activation by baculovirus expressed enzyme. Comp Biochem Physiol C Toxicol Pharmacol. 140:207–219. [DOI] [PubMed] [Google Scholar]
- Webster KA, Schach U, Ordaz A, Steinfeld JS, Draper BW et al. 2017. Dmrt1 is necessary for male sexual development in zebrafish. Dev Biol. 422:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger M, Diotel N, Weger BD, Beil T, Zaucker A et al. 2018. Expression and activity profiling of the steroidogenic enzymes of glucocorticoid biosynthesis and the fdx1 co-factors in zebrafish. J Neuroendocrinol. 30:e12586. [DOI] [PubMed] [Google Scholar]
- Werner R, Monig I, Lunstedt R, Wunsch L, Thorns C et al. 2017. New NR5A1 mutations and phenotypic variations of gonadal dysgenesis. PLoS One. 12:e0176720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RT, Allen RA. 1951. An improved clinical microscope for sectioning frozen tissue. Stain Technol. 26:137–138. [PubMed] [Google Scholar]
- Xie H, Hoffmann HM, Meadows JD, Mayo SL, Trang C et al. 2015. Homeodomain proteins SIX3 and SIX6 regulate gonadotrope-specific genes during pituitary development. Mol Endocrinol. 29:842–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie QP, He X, Sui YN, Chen LL, Sun LN et al. 2016. Haploinsufficiency of SF-1 causes female to male sex reversal in Nile tilapia. Oreochromis niloticus. Endocrinology. 157:2500–2514. [DOI] [PubMed] [Google Scholar]
- Yan YL, Batzel P, Titus T, Sydes J, Desvignes T et al. 2019. A hormone that lost its receptor: anti-Mullerian hormone (AMH) in zebrafish gonad development and sex determination. Genetics. 213:529–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YL, Desvignes T, Bremiller R, Wilson C, Dillon D et al. 2017. Gonadal soma controls ovarian follicle proliferation through Gsdf in zebrafish. Dev Dyn. 246:925–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Gutierrez NM, Wang L, Ellsworth BS, Wang CM. 2010. Synergistic activation of the Mc2r promoter by FOXL2 and NR5A1 in mice. Biol Reprod. 83:842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Tang H, Liu Y, Chen Y, Li G et al. 2017. Targeted disruption of aromatase reveals dual functions of cyp19a1a during sex differentiation in zebrafish. Endocrinology. 158:3030–3041. [DOI] [PubMed] [Google Scholar]
- Yoon C, Kawakami K, Hopkins N. 1997. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 124:3157–3165. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang D, Liu X, Yu G, Cai X et al. 2019. Zebrafish prmt5 arginine methyltransferase is essential for germ cell development. Development. 146:dev179572. [DOI] [PubMed] [Google Scholar]
- Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. 1998. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 393:595–599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) contains RNA-seq reads under accession number PRJNA561212. Supplementary Tables list differentially expressed genes. The scRNA-seq data are publicly available at https://cells.ucsc.edu/?ds=zebrafish-dev. Additional data and code relevant to the Atlas can be accessed at: https://www.adammillerlab.com/. Work was performed under the University of Oregon Institutional Animal Care and Use Committee (IACUC) protocol #14-08 R. Mutant strains are available on request. Data should be cited according to the citation of this article.
Supplementary material is available at figshare: https://doi.org/10.25386/genetics.13236983.